Summary
Autophagy is a conserved biological process that maintains cell homeostasis by targeting macromolecules for lysosome-mediated degradation. The levels of autophagy are relatively lower under normal conditions than under stress conditions (e.g., starvation), as autophagy is usually stimulated after multiple stresses. However, many autophagy-related regulators are still expressed under normal conditions. Although these regulators have been studied deeply in autophagy regulation, the nonautophagic roles of these regulators under normal conditions remain incompletely understood. Here, we found that autophagy-related 5 (ATG5), which is a key regulator of autophagy, regulates c-Myc protein degradation under normal conditions through the ubiquitin-proteasome pathway. We also found that ATG5 binds c-Myc and recruits the E3 ubiquitin-protein ligase FBW7 to promote c-Myc degradation. Moreover, ATG5-mediated degradation of c-Myc limits cell growth under normal conditions and is essential for embryonic stem cell differentiation. Therefore, this study reveals a nonautophagic role of ATG5 in regulating of c-Myc protein degradation.
Subject areas: Cell biology, Functional aspects of cell biology
Graphical abstract
Highlights
-
•
ATG5 differentially regulates cell growth between normal and starvation conditions
-
•
ATG5 recruits FBW7 to regulate c-Myc protein degradation under normal conditions
-
•
ATG5-mediated degradation of c-Myc limits cell growth under normal conditions
-
•
ATG5 negatively regulates the protein level of c-Myc during ESC differentiation
Cell biology; Functional aspects of cell biology
Introduction
Autophagy is an evolutionarily conserved biological process by which proteins, other macromolecules, or even cell organelles are degraded through the lysosome-mediated degradation pathway (Klionsky, 2007; Yang and Klionsky, 2010a). Autophagy plays important roles in maintaining cell survival, especially when cells encounter multiple stress conditions, such as nutrient deprivation or pathogen infection. Cells can degrade distinct macromolecules by activating autophagy to produce essential crude components for cell synthesis and survival to overcome the starvation condition (Fimia et al., 2013; Johansen and Lamark, 2011; Mehrpour et al., 2010). Autophagy is also employed by host cells to defend against infection by viruses or other types of pathogens by directly targeting intracellular pathogens for degradation (Deretic et al., 2013; Pradel et al., 2020). In addition, dysregulation of autophagy has been reported to be involved in multiple physiological and pathological processes, such as aging, cancer, and neurodegenerative disorders, and autophagy-related therapies are increasingly being developed (Mizushima and Levine, 2020).
Activation of autophagy can be divided into multiple steps, including autophagosome formation and elongation, autophagosome-lysosome fusion, and mature autolysosome formation, and several membrane-related components and their rearrangements are involved (Mehrpour et al., 2010; Yang and Klionsky, 2010b). The molecular mechanisms of autophagy have been widely studied, and several autophagy-related genes (ATGs) that play important roles in the regulation of autophagy during distinct steps have been identified. Among them, the roles of ATG5 during the activation of autophagy have been extensively studied. ATG5 plays critical roles in regulating the maturation of autophagosomes. In mammals, the ubiquitin E1-like enzyme ATG7 (autophagy related 7) collaborates with the E2-like enzyme ATG10 (autophagy related 10) to synergistically catalyze the covalent conjugation of ATG12 (autophagy related 12) to ATG5. The conjugated ATG5-ATG12 complex further targets autophagosome vesicles, recruits other autophagy-related proteins, and eventually regulates the maturation of autophagosomes (Mizushima et al., 1998; Shintani et al., 1999; Tanida et al., 1999).
c-Myc is a well-known oncoprotein that strongly regulates cell growth, differentiation, genome instability, apoptosis, etc. (Evan et al., 1994). c-Myc functions as a transcription factor usually through heterodimerization with Max (MYC associated factor X), and several cancer-related genes are regulated by the c-Myc-Max complex (Meyer and Penn, 2008). The protein level of c-Myc is tightly controlled in human cells, and the ubiquitin-proteasome pathway has been reported to be involved in the regulation of c-Myc protein degradation (Yeh et al., 2018). Several E3 ubiquitin ligases and deubiquitinating enzymes have been identified, such as FBW7, SKP2 (S-phase-kinase-associated protein 2), CHIP (STIP1 homology and U-box-containing protein 1), TRIM32 (tripartite motif containing 32), and USP28 (ubiquitin-specific peptidase 28) (Farrell and Sears, 2014). Among these proteins, FBW7 is the best-studied E3 ubiquitin ligase regulating c-Myc protein degradation. FBW7 belongs to the SCF (Skp1-Cullin-F-box) E3 ubiquitin ligase complex and can bind, ubiquitinate, and destabilize the c-Myc protein (Welcker et al., 2004; Yada et al., 2004).
As mentioned earlier, autophagy is usually activated under numerous stress conditions. However, many autophagy regulators, such as ATG5, are still highly expressed under normal culture conditions. In addition, the biological roles of these autophagy-related regulators under normal conditions are still largely unclear. In this study, we found that the autophagy regulator ATG5 negatively regulates the protein stability of c-Myc, likely by recruiting the E3 ubiquitin ligase FBW7. The regulation of c-Myc stability by ATG5 plays critical roles in the regulation of cell growth under normal culture conditions and in embryonic stem cell differentiation.
Results
ATG5 showed distinct effects on cell growth between normal culture conditions and serum starvation (autophagy-activated) conditions
To determine the biological roles of ATG5 under normal conditions, we used serum starvation to induce autophagy (Chen et al., 2014; Liu et al., 2012) for comparison. LC3 (microtubule-associated protein 1 light chain 3) is a well-known autophagy marker and is widely used in studies of autophagy. When autophagy is activated, LC3 usually exhibits a typical punctate distribution in human cells, and lipidation of LC3 occurs to generate the low-molecular-weight protein LC3-II. Therefore, the punctate distribution and the ratio of LC3-II to LC3-I are considered markers for assessing autophagy activity. We first compared autophagy activity between these two conditions by evaluating the autophagy marker LC3, and autophagy activity was indeed elevated under serum starvation conditions, as evidenced by the increased LC3-II to LC3-I ratio (Figure S1A), and more LC3 puncta (Figure S1B) were observed in cells under serum starvation conditions. Moreover, we also evaluated the difference in autophagic flux between normal culture conditions and serum starvation conditions by using the reporter plasmid LC3-DsRed2-GFP (Wang et al., 2019a). As GFP is sensitive to the acidic environment of autophagosomes while DsRed2 is not, fewer GFP puncta than DsRed2 puncta were detected under serum starvation conditions, suggesting that autophagic flux was elevated after serum starvation (Figure S1C).
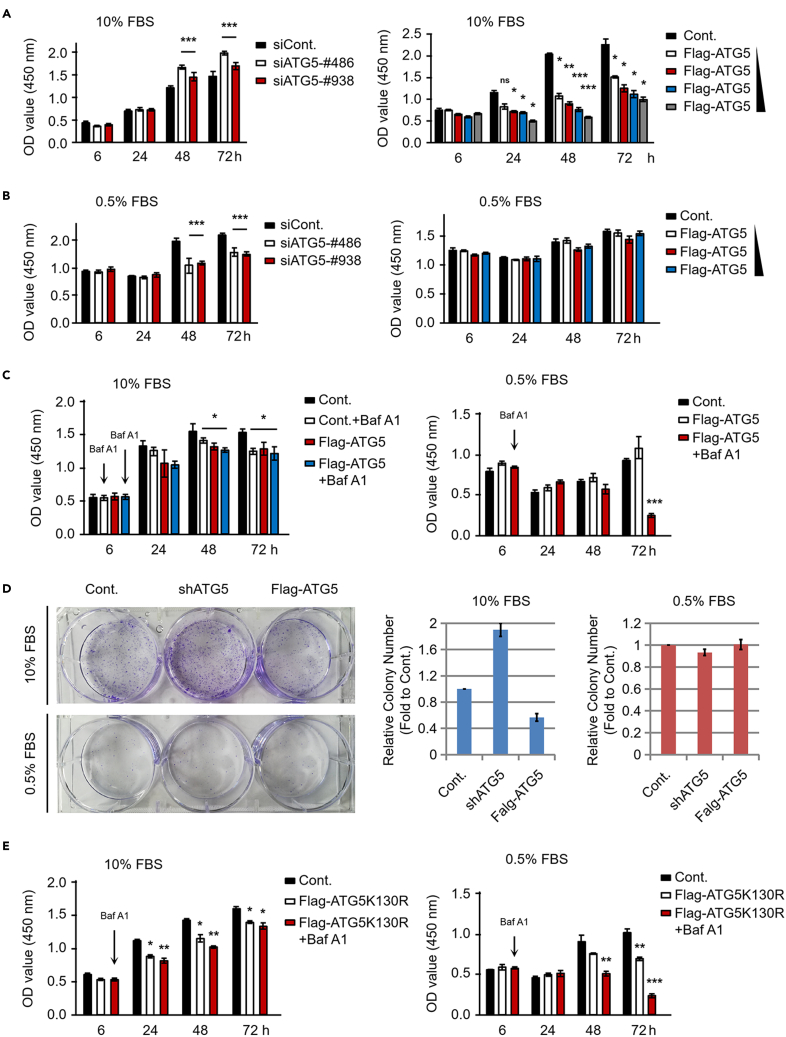
We next compared the effects of ATG5 on cell growth under both normal culture conditions and serum starvation conditions. Intriguingly, opposite effects were observed between these two conditions. First, our cell counting kit 8 (CCK-8) assays showed that ATG5 negatively regulates the growth of HeLa cells under normal culture conditions, as the cell growth was enhanced in ATG5-depleted cells (Figure 1A, left) and decreased in ATG5-overexpressing cells (Figure 1A, right). Unlike under normal culture conditions, ATG5 is likely required for cell growth under serum starvation conditions, as loss of ATG5 significantly inhibited cell growth (Figure 1B, left). However, unexpectedly, no obvious effect on cell growth was observed when ATG5 was overexpressed under serum starvation conditions (Figure 1B, right). We speculated that the effect of ATG5 on cell growth under serum starvation conditions was likely mediated through its regulation of autophagy and that the autophagy activity induced by serum starvation may be sufficient for the maintenance of cell growth. Further elevation of autophagy by ATG5 overexpression may not be beneficial for cell growth, as excessive autophagy is also related to cell death (Deng et al., 2020). Our results also partially supported this speculation, as blocking autophagy with the autophagy inhibitor bafilomycin A1 (Baf A1) significantly impaired the growth of ATG5-overexpressing HeLa cells under serum starvation conditions (Figure 1C, right), suggesting that autophagy activity is essential for cell growth under serum starvation conditions. Moreover, loss of ATG5 also resulted in an impaired cell growth under serum starvation conditions, and treatment with Baf A1 further decreased the cell growth of ATG5-depleted cells (Figure S2), further supporting the aforementioned idea. In contrast, we found no obvious effect of Baf A1 on ATG5-regulated cell growth under normal culture conditions (Figure 1C, left), suggesting that autophagy-independent mechanisms are likely involved in ATG5-regulated cell growth under normal culture conditions. The knockdown and overexpression efficiencies were also determined (Figure S3). It is worth mentioning that the effects of ATG5 on autophagy were more marked under serum starvation conditions than under normal culture conditions, as the LC3-II to LC3-I ratio was more severely affected under serum starvation conditions especially upon ATG5 depletion (Figure S3), further supporting the hypothesis that autophagy was activated under serum starvation conditions. It was worth mentioning that the major form of endogenous ATG5 is the ATG5-ATG12-conjugated form, and it was hard to detect the free ATG5 (nonconjugated with ATG12) (Figure S3). Our observation is consistent to a previous report suggesting that the endogenous-free ATG5 is almost completely degraded by the proteasomes (Wible et al., 2019). Moreover, we further performed colony formation assays. Similar to the results of the CCK-8 assays, depletion of ATG5 increased the colony-forming ability of HeLa cells under normal culture conditions, whereas ATG5 overexpression decreased the colony-forming ability (Figure 1D, left upper, and middle histogram). However, colony formation was impaired when cells were cultured under serum starvation conditions, as only a small number of colonies were formed (Figure 1D, left lower, and right histogram). Therefore, it was difficult to compare the effects of ATG5 on the colony-forming ability under serum starvation conditions. Together, our results indicate that ATG5 plays distinct roles between normal culture conditions and serum-starvation-induced autophagy activation conditions. ATG5 negatively regulates cell growth under normal culture conditions but is required for cell growth under serum-starvation-induced autophagy activation conditions. To further support the hypothesis that ATG5 regulates cell growth in normal culture conditions in an autophagy-independent manner (Figure 1C, left), the ATG5K130R mutant (lose the ability to form the ATG5-ATG12 conjugate) was used to examine its effect on cell growth. We found that similar to the effect of wild-type ATG5, an impaired cell growth was observed in ATG5K130R-overexpressing cells under normal culture conditions, and Baf A1 did not rescue the effect (Figure 1E, left). However, under serum starvation conditions, in consistent to the effect of wild-type ATG5, overexpression of ATG5K130R impaired the cell growth of HeLa cells (Figure 1E, right). We speculated that the ATG5K130R mutant may competitively inhibit the function of endogenous ATG5 in controlling autophagy in serum starvation conditions. Moreover, treatment with Baf A1 in ATG5K130R-overexpressing cells also impaired the cell growth, further supporting the essential roles of autophagy in maintaining cell growth under serum starvation conditions (Figure 1E, right).
Figure 1.
ATG5 shows opposite effects on cell growth under normal culture conditions and serum starvation conditions
(A) ATG5 was depleted or overexpressed in HeLa cells with two ATG5-specific siRNAs (left) or a Flag-tagged ATG5 construct (right), respectively. HeLa cells transfected with a control siRNA (left) or Flag control construct (right) were used as the corresponding controls. Cells were further cultured under normal culture conditions for the indicated durations, and the relative cell growth ability was examined by a cell counting kit 8 (CCK-8) assay.
(B) HeLa cells transfected as described in A were cultured under serum starvation conditions for the indicated durations. The relative cell growth ability was examined as described in (A).
(C) HeLa cells were transfected with a Flag control or a Flag-tagged ATG5 construct and cultured under normal conditions (left) or serum starvation conditions (right) for 6 h. Cells were then treated with or without 1 μM Baf A1 as indicated and were further cultured for different durations as indicated. The relative cell growth ability was examined by CCK-8 assays as described in (A).
(D) HeLa cells stably transfected with an ATG5-specific shRNA or a Flag-tagged ATG5 construct were generated, and a colony formation assay was performed with cells cultured under normal conditions (left upper) or serum starvation conditions (left lower) for 7 days. The colonies were photographed (left), and the relative quantification of the number of colonies is shown (middle and right).
(E) HeLa cells were transfected with a Flag control or a Flag-tagged ATG5K130R construct and cultured under normal conditions (left) or serum starvation conditions (right) for 6 h. Cells were then treated with or without 1 μM Baf A1 as indicated and were further cultured for different durations as indicated. The relative cell growth ability was examined by CCK-8 assays as described in (A). Statistics: all experiments were repeated more than 3 times. The bars and error bars indicate the mean ± s.d. values; n = 3 independent replicates. Two-tailed unpaired Student's t test was performed. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns: nonsignificant.
In addition, we examined the effects of ATG5 on cell migration under both normal and serum starvation conditions. Similar effects of ATG5 that negatively regulates cell migration under both normal culture conditions and serum starvation conditions were observed (Figure S4). The effects of ATG5 on cell migration seemed to be autophagy independent, as the autophagy inhibitor Baf A1 showed no obvious effects on the decrease in cell migration induced by Flag-ATG5 overexpression (Figure S4). It is worth mentioning that treatment with only serum starvation also enhanced cell migration activity (Figure S4), which is consistent with a previous report (Tong et al., 2019).
ATG5 negatively regulates the protein level and transcriptional activity of c-Myc under normal culture conditions
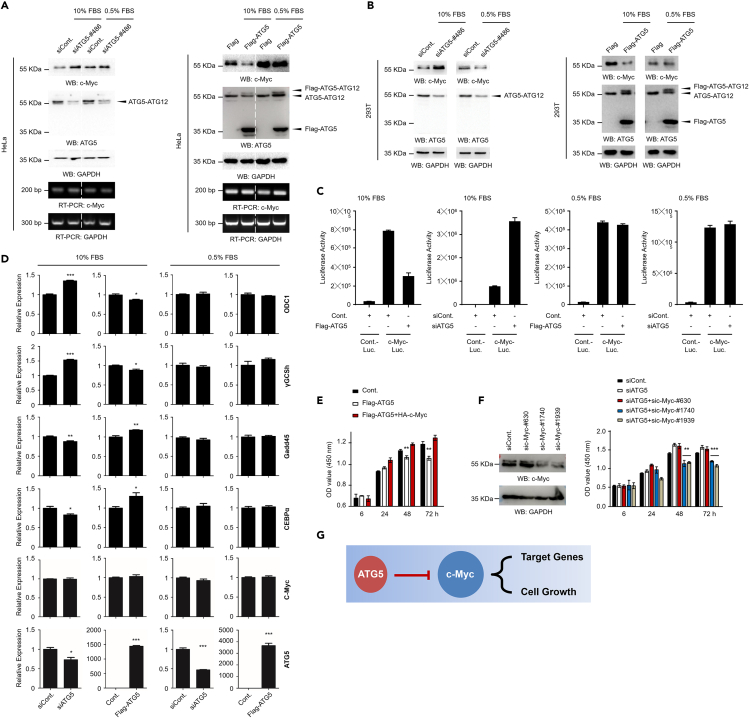
To understand the molecular mechanisms by which ATG5 negatively regulates cell growth under normal culture conditions, we evaluated the effects of ATG5 on multiple key regulators of cell growth, including c-Myc, Ras, and p53 (data not shown). Of great interest, we found that the oncoprotein c-Myc was markedly affected by ATG5. The protein level of c-Myc was increased in ATG5-depleted HeLa cells but was decreased in ATG5-overexpressing HeLa cells, especially under normal culture conditions, whereas under serum starvation conditions, the c-Myc protein level was not changed as obvious as in normal conditions after altering the ATG5 protein level (Figure 2A, upper). This negative regulation of c-Myc by ATG5 was consistent with the effects of ATG5 on cell growth under normal culture conditions (Figure 1A). It is worth mentioning that ATG5-mediated regulation of c-Myc is likely achieved through a posttranscriptional pathway, as the mRNA levels of c-Myc were not obviously affected by ATG5 under either normal culture or serum starvation conditions (Figure 2A, lower). We also evaluated the negative regulation of c-Myc by ATG5 in 293T cells, and results similar to those in HeLa cells were observed (Figure 2B). Moreover, the ATG12 protein level was also examined upon alteration of ATG5 expression. Our data showed that, similar as ATG5, the major form of ATG12 in human cells is also the ATG5-conjugated form (ATG5-ATG12), and only very weak free ATG12 (nonconjugated with ATG5) was detected (Figure S5). More interestingly, we found that the protein level of the free form of ATG12 was negatively changed in parallel with the alteration of ATG5 protein level, as when ATG5 was overexpressed, the protein level of free ATG12 was decreased, whereas the protein level of free ATG12 was increased when ATG5 was depleted (Figure S5). This observation is reasonable, as ATG12 and ATG5 usually form the ATG5-ATG12 conjugate; if the protein level of one protein was altered, the protein level of another protein could be reversely changed. For instance, if ATG5 was overexpressed in human cells, more ATG12 could be needed to conjugate with ATG5. Therefore, a decreased free ATG12 protein level could be detected (Figure S5, left). Reversely, if ATG5 was depleted, more free ATG12 could be released from the ATG5-ATG12 conjugate. Thus, an increased free ATG12 could be detected (Figure S5, right).
Figure 2.
ATG5 negatively regulates the protein level of c-Myc, especially under normal culture conditions
(A) HeLa cells were transfected with an ATG5-specific siRNA or a Flag-tagged ATG5 construct and were further cultured under normal conditions or serum starvation conditions for 48 h. Western blot analyses or RT-PCR analyses were then performed.
(B) 293T cells were transfected with an ATG5-specific siRNA or a Flag-tagged ATG5 construct and were further cultured under normal conditions or serum starvation conditions for 48 h. Western blot analyses were then performed.
(C) Flag-tagged ATG5 or ATG5-specific siRNA-transfected 293T cells were further transfected with a control luciferase reporter construct or a c-Myc-targeted luciferase reporter construct as indicated. Cells were then cultured under normal conditions or serum starvation conditions for 48 h. Luciferase activity was evaluated.
(D) 293T cells were transfected with an ATG5-specific siRNA or a Flag-tagged ATG5 construct and were further cultured under normal conditions or serum starvation conditions for 48 h as indicated. Cells were then lysed and subjected to quantitative RT-PCR analyses.
(E) HeLa cells transfected with a Flag control construct, a Flag-tagged ATG5 construct, or a Flag-tagged ATG5 construct together with an HA-tagged c-Myc construct were cultured under normal conditions for the indicated durations. The relative cell growth ability was examined by CCK-8 assays.
(F) HeLa cells were transfected with 3 individual c-Myc-specific siRNAs for 48 h. Western blot analyses were performed to determine the knockdown efficiency of the c-Myc siRNAs (left). HeLa cells transfected with a control siRNA, an ATG5-specific siRNA, or an ATG5-specific siRNA together with a c-Myc-specific siRNA as indicated were cultured under normal conditions for the indicated durations. The relative cell growth ability was examined by CCK-8 assays (right).
(G) Predicted model. Under normal culture conditions, ATG5 negatively regulates c-Myc and further affects the expression of c-Myc downstream target genes and cell growth. Statistics: all experiments were repeated more than 3 times. The bars and error bars indicate the mean ± s.d. values; n = 3 independent replicates. Two-tailed unpaired Student's t test was performed. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
The identity of c-Myc as a well-known transcription factor, together with the aforementioned observation that ATG5 negatively regulates the c-Myc protein level under normal culture conditions, prompted us to examine whether ATG5 regulates the transcription factor activity of c-Myc. We first performed luciferase reporter assays by using a c-Myc-specific luciferase reporter construct containing multiple binding sites specifically recognized by c-Myc (#11544ES03, YEASEN, China) and a control luciferase reporter construct (#11401ES60, YEASEN, China). Consistent with the effects of ATG5 on c-Myc protein levels, ATG5 also negatively regulated the transcription factor activity of c-Myc, especially under normal culture conditions. The activity of the c-Myc-targeted luciferase reporter was downregulated in ATG5-overexpressing cells but upregulated in ATG5-depleted cells (Figure 2C, left two histograms). However, no obvious changes in the activity of the c-Myc-targeted luciferase reporter were observed under serum starvation conditions after changing the expression of ATG5 (Figure 2C, right two histograms). We next examined whether the reported downstream targets, including genes positively regulated by c-Myc (ODC1 and γGCSh) and genes negatively regulated by c-Myc (Gadd45 and C/EBPα) (Dang, 1999), are affected by ATG5. Consistently, we found that all the evaluated c-Myc downstream targets were inversely affected by ATG5 under normal culture conditions (Figure 2D, left), whereas no obvious changes were observed under serum starvation conditions (Figure 2D, right). Therefore, these data suggested that ATG5 negatively regulates the protein level and transcription factor activity of c-Myc. Considering the findings that ATG5 negatively regulates cell growth (Figures 1A–1D) and the protein level and transcription factor activity of c-Myc (Figures 2A–2D) under normal culture conditions, we next determined whether the negative regulation of cell growth by ATG5 is achieved through the regulation of c-Myc. Our results indicated that the growth impairment induced by ATG5 overexpression under normal culture conditions was at least partially rescued by further overexpression of c-Myc (Figure 2E) and that the loss of ATG5-induced enhancement of cell growth was partially abolished by further depletion of c-Myc (Figure 2F). Taken together, our results indicate that the negative regulation of cell growth by ATG5 is likely achieved through its negative regulation of the oncoprotein c-Myc (Figure 2G).
ATG5 positively regulates c-Myc protein degradation under normal culture conditions
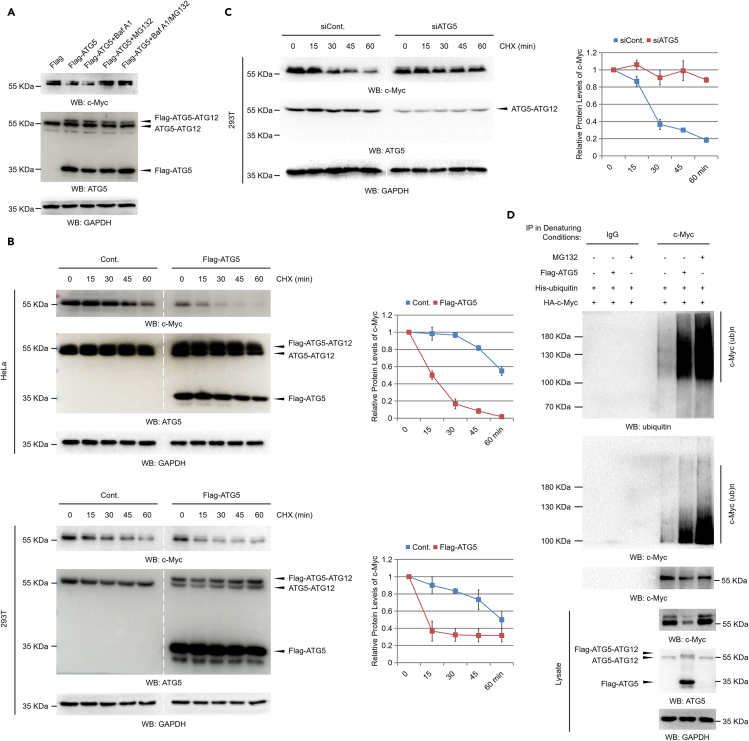
As the negative regulation of the c-Myc protein level by ATG5 is likely not achieved through the regulation of the c-Myc mRNA level (Figure 2A, lower) and to determine how ATG5 negatively regulates the protein level of c-Myc under normal culture conditions, we next evaluated whether protein degradation is involved by using multiple protein degradation inhibitors. Our data revealed that the decrease in the c-Myc protein level induced by ATG5 overexpression was completely abolished by the 26S proteasome inhibitor MG132, bortezomib, and carfilzomib, whereas the autophagy-lysosome pathway inhibitor Baf A1 showed no obvious effect on the ATG5-regulated c-Myc protein level (Figures 3A and S6). These data suggested that the negative regulation of the c-Myc protein level by ATG5 under normal culture conditions is likely achieved through the 26S proteasome-mediated protein degradation pathway, further partially supporting the idea that ATG5 likely regulates cell growth in an autophagy-independent manner. To further validate this hypothesis, we next examined the effect of ATG5 on the c-Myc protein degradation rate by chase assays. We found that in both HeLa and 293T cells, overexpression of ATG5 increased the protein degradation rate of c-Myc under normal culture conditions (Figure 3B). Moreover, in contrast to ATG5 overexpression, loss of ATG5 expression inhibited c-Myc protein degradation (Figure 3C). It was worth mentioning that, as shown in Figure S3, the major form of endogenous ATG5 is the ATG5-ATG12-conjugated form; this is consistent to a previous report suggesting that the endogenous-free ATG5 (nonconjugated with ATG12) is almost completely degraded by the proteasomes (Wible et al., 2019). In addition, we examined the effect of ATG5 on c-Myc ubiquitination, and consistent with the effect on c-Myc protein degradation, our data revealed that ATG5 promoted the ubiquitination of c-Myc under normal culture conditions (Figure 3D). Taken together, these results indicate that ATG5 negatively regulates the c-Myc protein level likely through the ubiquitination-proteasome pathway under normal culture conditions.
Figure 3.
ATG5 regulates c-Myc protein degradation through the ubiquitination-proteasome pathway
(A) 293T cells were transfected with a Flag control or a Flag-tagged ATG5 construct for 48 h. Cells were then treated with or without 1 μM Baf A1 (bafilomycin A1), 25 μM MG132, or both as indicated for another 10 h. Cell extracts were prepared, and western blot analyses were performed.
(B) HeLa and 293T cells transfected with a Flag control construct or a Flag-tagged ATG5 construct were treated with 50 μg/mL CHX (cycloheximide) for the indicated durations (chase assays). Cells were then lysed and subjected to western blot analyses. Relative protein levels of c-Myc were analyzed with ImageJ software (right).
(C) 293T cells transfected with a control siRNA or an ATG5-specific siRNA were treated with 50 μg/mL CHX for the indicated durations (chase assays). Cells were then lysed and subjected to western blot analyses. Relative protein levels of c-Myc were analyzed with ImageJ software (right).
(D) An HA-tagged c-Myc and an His-tagged ubiquitin construct were transfected with or without a Flag-tagged ATG5 construct into 293T cells as indicated for 48 h. Cells were then treated with or without 25 μM MG132 for another 10 h. Cells were lysed under denaturing conditions, and the c-Myc protein was immunoprecipitated (IP) with c-Myc-specific antibodies. IP with immunoglobulin G (IgG) antibodies was performed as a negative control. The precipitated proteins were subjected to western blot analyses with the indicated antibodies. Statistics: all experiments were repeated more than 3 times. The bars and error bars indicate the mean ± s.d. values; n = 3 independent replicates.
ATG5 interacts with c-Myc under normal culture conditions
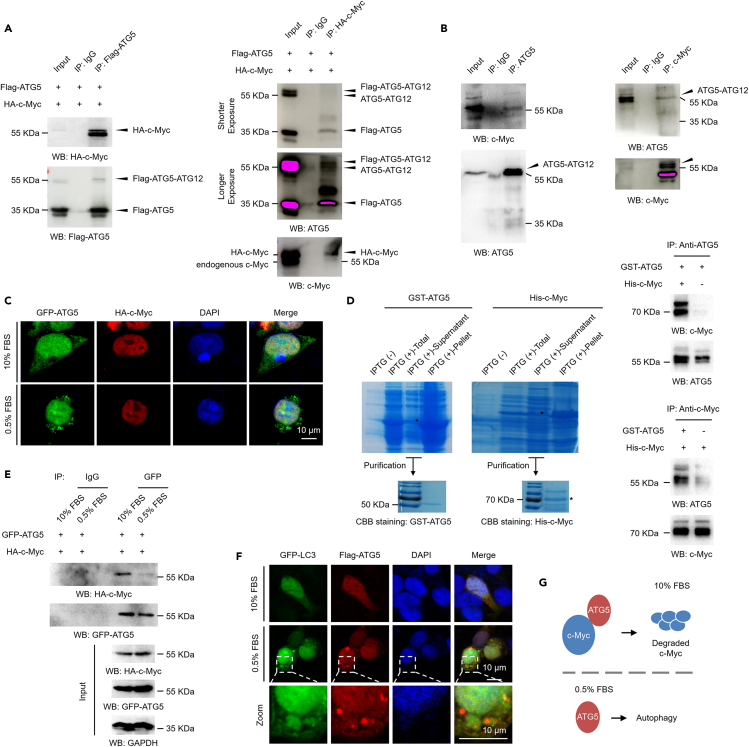
To further determine the molecular mechanisms by which ATG5 regulates c-Myc degradation, we next evaluated whether ATG5 interacts with c-Myc under normal culture conditions. We first transfected 293T cells with a GFP-tagged ATG5 construct and an HA-tagged c-Myc construct, and reverse coimmunoprecipitation (co-IP) assays were performed with antibodies against Flag and HA. Our data suggested that exogenously expressed GFP-tagged ATG5 indeed interacts with HA-tagged c-Myc in vivo (Figure 4A). We further examined the interaction between endogenous ATG5 and c-Myc by reverse co-IP assays with antibodies against ATG5 and c-Myc. Consistent with the above results, ATG5 also interacted with c-Myc endogenously (Figure 4B). It was worth mentioning that c-Myc likely interacts with both the ATG12-conjugated ATG5 and free ATG5, as both forms can be precipitated by c-Myc protein (Figure 4A, right and Figure 4B, right), suggesting that ATG5 interacts with c-Myc independently on its conjugation form. To further substantiate the in vivo interaction between ATG5 and c-Myc, immunofluorescence (IF) analysis was performed to determine the colocalization of these two proteins. Although the ATG5 protein is distributed throughout the cell, while c-Myc is a nuclear protein, ATG5 and c-Myc colocalization signals were observed in the cell nucleus under normal culture conditions (Figure 4C, upper). However, when cells were cultured under serum starvation conditions, the distribution of ATG5 changed remarkably with a typical punctate distribution in the cytoplasm (Figure 4C, lower). We next purified GST-tagged ATG5 and His-tagged c-Myc proteins expressed in Escherichia coli (Figure 4D, left) and performed in vitro binding assays to determine whether ATG5 can directly interact with c-Myc in vitro. Our results indicated that ATG5 can indeed interact with c-Myc directly in vitro (Figure 4D, right). Therefore, these data confirmed that ATG5 directly interacts with c-Myc both in vitro and in vivo. We next tested the response dynamics of the ATG5-c-Myc interaction upon serum starvation. Very interestingly, our results revealed that the interaction between ATG5 and c-Myc was abolished after serum starvation (Figure 4E). We suspected that dissociation of the ATG5 protein from the c-Myc protein after serum starvation may contribute to autophagy, and our IF assay at least partially supported this speculation. We found that the distribution of ATG5 puncta increased after serum starvation, and these ATG5 puncta were distributed near the autophagosome marker LC3 (Figure 4F). Taken together, our results suggest that ATG5 interacts with c-Myc under normal culture conditions and further affects c-Myc protein degradation, whereas under serum starvation conditions, the interaction between ATG5 and c-Myc is disrupted, and the released ATG5 may primarily contribute to autophagy (Figure 4G).
Figure 4.
ATG5 interacts with c-Myc under normal culture conditions
(A) 293T cells were transfected with a Flag-tagged ATG5 and an HA-tagged c-Myc constructs for 48 h. Cells were lysed and subjected to coimmunoprecipitation (co-IP) assays with antibodies against Flag (left) or HA (right) as indicated.
(B) Co-IP assays were performed in 293T cells with antibodies against ATG5 (left) or c-Myc (right) to determine the interaction between endogenous ATG5 and c-Myc proteins.
(C) 293T cells were transfected with a GFP-tagged ATG5 and an HA-tagged c-Myc construct for 48 h under normal or serum starvation conditions. Immunofluorescence (IF) analysis was then performed to evaluate the localization of ATG5 (green) and c-Myc (red). The blue signal indicates the nucleolus (stained with DAPI).
(D) GST-tagged ATG5 and His-tagged c-Myc were expressed in E coli and purified (left two panels). The two proteins were co-incubated, and IP assays with antibodies against ATG5 or c-Myc (right two panels) were performed as indicated to evaluate the direct binding between ATG5 and c-Myc in vitro.
(E) 293T cells co-transfected with a GFP-tagged ATG5 and an HA-tagged c-Myc were further cultured under normal conditions or serum starvation conditions as indicated. Cell extracts were prepared, and co-IP assays with antibodies against GFP were performed. Co-IP with IgG antibodies was performed as a negative control.
(F) 293T cells transfected with a flag-tagged ATG5 and a GFP-tagged LC3 were cultured under normal conditions or serum starvation conditions as indicated. IF analysis was performed to evaluate the localization of ATG5 (red) and LC3 (green). DAPI staining was used to indicate the nucleolus (blue).
(G) Molecular model. ATG5 interacts with c-Myc and promotes c-Myc protein degradation under normal culture conditions. However, when cells are subjected to serum starvation to activate autophagy, ATG5 may be released from c-Myc and mainly contribute to autophagy.
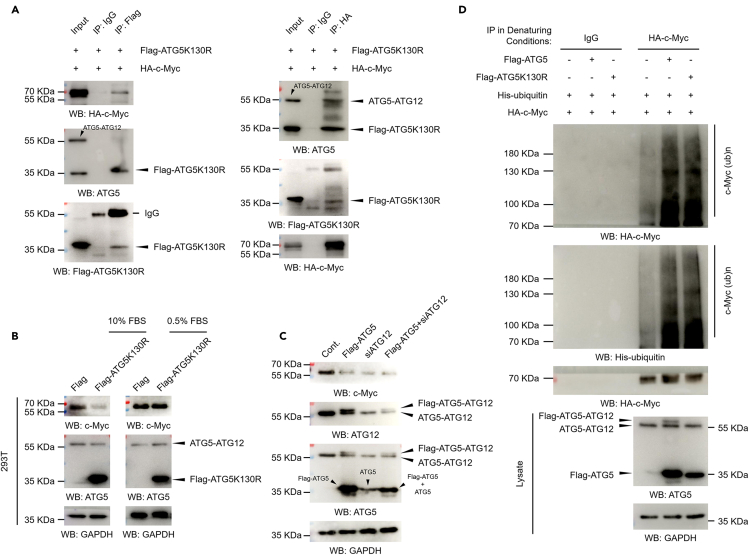
To further support the idea that ATG5 regulates c-Myc protein degradation in an autophagy-independent manner, the ATG12-conjugation-deficient mutant ATG5K130R was used. First, reverse co-IP analyses were performed in HA-c-Myc and Flag-ATG5K130R-transfected 293T cells. Consistent to our hypothesis, we found that c-Myc also interacts with ATG5K130R mutant (Figure 5A). In accordance with the ATG5K30R-c-Myc interaction, and similar to the effect of wild-type ATG5, ATG5K130R mutant also negatively regulates c-Myc protein level under normal culture conditions (Figure 5B). Furthermore, we knocked down the expression of ATG12 after ATG5 overexpression to determine whether the observed negative regulation of c-Myc by ATG5 in normal culture conditions is dependent on the existence of ATG12. However, our data indicated that loss of ATG12 does not affect the effect of ATG5 on c-Myc protein level (Figure 5C). It was worth mentioning that, when ATG12 was knocked down, an amount of free ATG5 appeared, and the protein level of c-Myc also decreased (Figure 5C, line “siATG12”). We speculated that the decrease of c-Myc after ATG12 depletion might be due to the increased free ATG5. Moreover, as wild-type ATG5, ATG5K130R mutant also promotes the ubiquitination of c-Myc under normal culture conditions (Figure 5D). Thus, these data suggested that ATG5 regulates c-Myc protein level likely independently on its conjugation status, supporting an autophagy-independent manner.
Figure 5.
ATG5 negatively regulates c-Myc independent of its conjugation with ATG12
(A) 293T cells were transfected with a Flag-tagged ATG5K130R and an HA-tagged c-Myc construct for 48 h. Cells were lysed and subjected to co-immunoprecipitation (co-IP) assays with antibodies against Flag (left) or HA (right) as indicated.
(B) 293T cells transfected with a Flag control and a Flag-tagged ATG5K130R construct were further cultured under normal conditions or serum starvation conditions for 48 h. Western blot analyses were then performed.
(C) 293T cells were transfected with a Flag-tagged ATG5 construct or an ATG12-specific siRNA or together for 48 h. Western blot analyses were then performed.
(D) An HA-tagged c-Myc and an His-tagged ubiquitin construct were transfected with a Flag-tagged ATG5 or ATG5K130R mutant construct into 293T cells as indicated for 48 h. Cells were lysed under denaturing conditions, and the c-Myc protein was immunoprecipitated (IP) with HA-tag-specific antibodies. IP with IgG antibodies was performed as a negative control. The precipitated proteins were subjected to western blot analyses with the indicated antibodies.
ATG5 recruits the E3 ubiquitin-protein ligase FBW7 to regulate c-Myc protein degradation under normal culture conditions
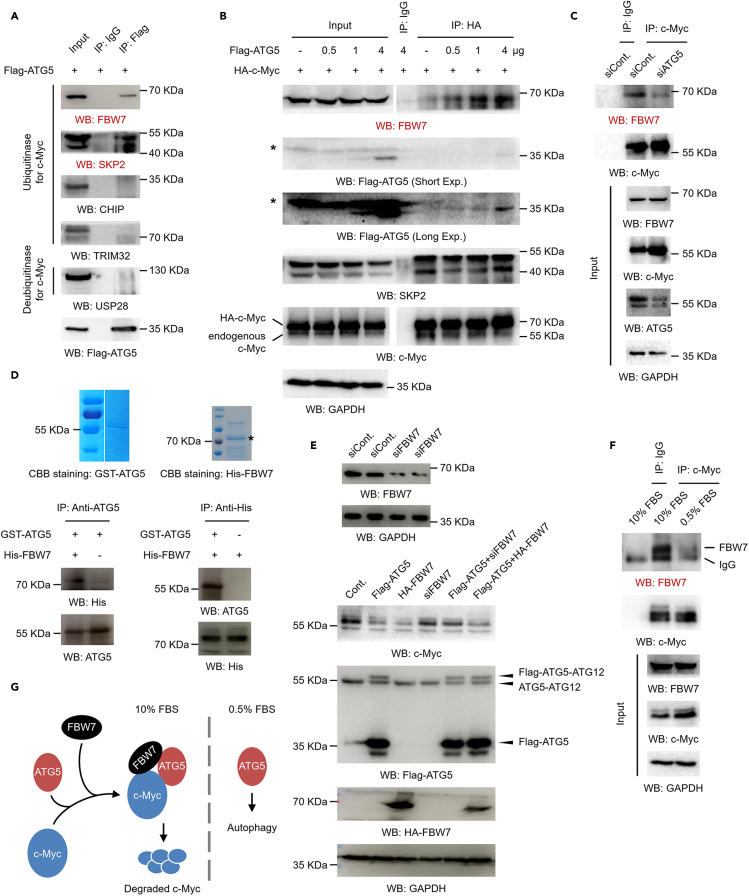
To further determine the detailed molecular mechanism by which ATG5 regulates c-Myc protein degradation under normal culture conditions, we examined whether ATG5 associates with the reported E3 ubiquitin-protein ligases (including FBW7, SKP2, CHIP, and TRIM32) (Farrell and Sears, 2014; Paul et al., 2013; Schwamborn et al., 2009; von der Lehr et al., 2003; Yada et al., 2004) and deubiquitinases (including USP28) (Farrell and Sears, 2014; Popov et al., 2007) of c-Myc in human cells. Our data indicated that two E3 ubiquitin-protein ligases, FBW7 and SKP2, were associated with ATG5 in 293T cells (Figure 6A). As ATG5 negatively regulates c-Myc protein levels, we therefore suspected that ATG5 promotes the interaction between c-Myc and FBW7 or SKP2. Partially as expected, only the binding of FBW7 to c-Myc was enhanced by ATG5 overexpression, whereas no obvious changes in the SKP2-c-Myc interaction were observed upon ATG5 overexpression (Figure 6B). Moreover, the opposite result was obtained after ATG5 depletion: loss of ATG5 abolished the interaction between FBW7 and c-Myc, suggesting that ATG5 is likely essential for this interaction (Figure 6C). Therefore, these data suggested that ATG5 may specifically recruit FBW7 to c-Myc under normal culture conditions and is essential for the FBW7-c-Myc interaction. In addition, we purified GST-tagged ATG5 and His-tagged FBW7 proteins expressed in E coli (Figure 6D, upper) and performed in vitro binding assays to determine the direct interaction between ATG5 and FBW7. Our data showed that ATG5 directly interacted with FBW7 in vitro (Figure 6D, lower). Together with the findings that ATG5 directly interacted with both c-Myc (Figures 4A–4D) and FBW7 (Figures 6A and 6D) and that ATG5 promoted and was essential for the interaction between FBW7 and c-Myc (Figures 6B and 6C), our results support the possibility that ATG5 may act as an adaptor protein linking FBW7 and c-Myc. Next, to determine whether FBW7 is required for the negative regulation of c-Myc protein levels by ATG5, a rescue experiment was performed with a FBW7-specific siRNA. The knockdown efficiency of siFBW7 was confirmed (Figure 6E, upper), and very interestingly, our data revealed that loss of FBW7 indeed abolished the decrease in the c-Myc protein level induced by ATG5 overexpression under normal culture conditions (Figure 6E, lower). As ATG5 promoted and was required for the interaction between c-Myc and FBW7 (Figures 6B and 6C), and the interaction between ATG5 and c-Myc was abolished when cells are cultured under serum starvation conditions (Figure 4E), we next investigated the dynamics of the c-Myc-FBW7 interaction in response to serum starvation. Our data indicated that the interaction of c-Myc and FBW7 was also impaired after serum starvation (Figure 6F). Taken together, our results demonstrate that ATG5 directly interacts with both c-Myc and its E3 ubiquitin-protein ligase FBW7 and further promotes the interaction between c-Myc and FBW7, likely contributing to c-Myc protein degradation via this mechanism (Figure 6G).
Figure 6.
ATG5 recruits the E3 protein-ubiquitin ligase FBW7 to bind c-Myc
(A) 293T cells were transfected with a Flag-tagged ATG5 for 48 h. Cells were lysed, and co-IP assays were performed with antibodies against Flag. IgG antibodies were used as a negative control.
(B) 293T cells were transfected with an HA-tagged c-Myc construct together with increasing amounts of a Flag-tagged ATG5 construct for 48 h. Cells were lysed, and co-IP assays were performed with antibodies against HA. IgG antibodies were used as a negative control.
(C) 293T cells were transfected with an ATG5-specific siRNA or a control siRNA for 48 h. Cells were lysed, and co-IP assays were performed with antibodies against c-Myc. IgG antibodies were used as a negative control.
(D) GST-tagged ATG5 and His-tagged FBW7 were expressed in E coli and purified (upper). The two proteins were co-incubated, and IP assays with antibodies against ATG5 or His (lower) were performed as indicated to evaluate the direct binding between ATG5 and FBW7 in vitro.
(E) A FBW7-specific siRNA was transfected into 293T cells for 48 h. Cells were then lysed, and the RNAi efficiency was determined by western blot analysis (upper). 293T cells were transfected with a Flag-tagged ATG5 construct, a HA-tagged FBW7 construct, FBW7-specific siRNA, or distinct combinations as indicated for 48 h. Cell extracts were prepared, and western blot analyses were performed (lower).
(F) 293T cells were cultured under normal conditions or serum starvation conditions as indicated. Cells were harvested, and co-IP assays were performed with antibodies against c-Myc. IgG antibodies were used as a negative control.
(G) Molecular model. Under normal culture conditions, ATG5 recruits FBW7 to c-Myc and further promotes c-Myc protein degradation. However, under serum starvation conditions, ATG5 may mainly contribute to the regulation of autophagy.
ATG5 negatively regulates the protein level of c-Myc during embryonic stem cell differentiation and is essential for ESC differentiation
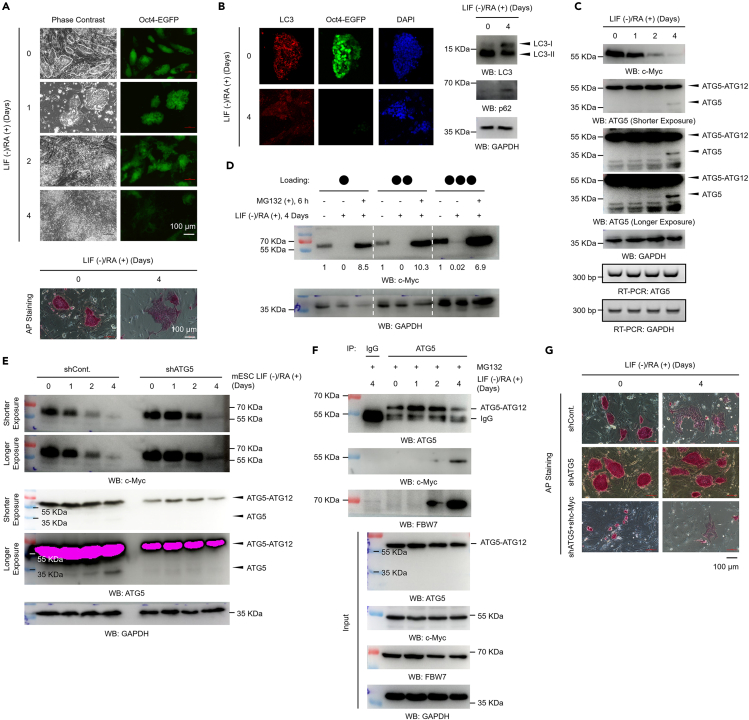
To verify the observed negative regulation of c-Myc protein levels by ATG5 under relatively physiological conditions, an ESC differentiation system was employed, as autophagy activity has been reported to show a significant decrease during ESC differentiation (Liu et al., 2017; Salemi et al., 2012). We established an ESC differentiation system by using LIF (leukemia inhibitory factor) depletion together with RA (retinoic acid) incubation in the Oct4-EGFP mouse ES (Oct4-EGFP mES) cell line. The Oct4-EGFP mES cell line was established by isolation of mES cells from Oct4-EGFP transgenic mice, which express enhanced green fluorescent protein (EGFP) under the control of the Oct4 promoter (Szabo et al., 2002) (https://www.jax.org/strain/004654). Therefore, the expression of EGFP can reflect the level of Oct4. First, we confirmed the differentiation efficiency of our ESC differentiation system by examining the expression of Oct4-regulated EGFP, the colony phenotype, and the alkaline phosphatase (AP) staining intensity. Our data showed that the ESC differentiation system was successfully established, as an obvious differentiated phenotype and a decrease in the AP staining intensity were observed after LIF depletion and RA incubation. Moreover, the expression of EGFP was decreased after LIF depletion and RA incubation, suggesting that the protein expression of Oct4 was downregulated (Figure 7A). Next, we compared the autophagy activity between ESCs and differentiated ESCs. Consistent with previous reports (Liu et al., 2017; Salemi et al., 2012), our data showed that autophagy activity was indeed impaired after ESC differentiation, as decreased numbers of LC3 puncta and decreased LC3-II to LC3-I ratios were observed in differentiated ESCs (Figure 7B). In addition, the protein level of p62 (sequestosome 1), whose degradation is a marker for autophagic flux, was elevated after ESC differentiation, further supporting the impairment of autophagic flux in differentiated ESCs (Figure 7B). We next examined the dynamics of the ATG5 and c-Myc protein levels during ESC differentiation. We found that the protein level of c-Myc decreased strikingly during ESC differentiation, which is consistent with the well-recognized understanding that c-Myc is a stem cell marker (Figure 7C, upper). However, the level of the ATG5-ATG12 conjugate showed no obvious change during ESC differentiation (Figure 7C, upper). More interestingly, a band corresponding to free ATG5 was detected after ESC differentiation (Figure 7C, upper), and we suspected that this free ATG5 may be generated by cleavage of ATG5-ATG12 conjugates, as autophagy was impaired after ESC differentiation. The RT-PCR results also support this hypothesis, as no obvious changes on ATG5 mRNA level were observed (Figure 7C, lower). To determine whether protein degradation mechanisms mediate the decrease in the c-Myc protein level during ESC differentiation, the proteasome inhibitor MG132 was used. Our data revealed that MG132 completely blocked the decrease in the c-Myc protein level after ESC differentiation, suggesting that the 26S proteasome-mediated protein degradation pathway indeed contributes to the decrease in the c-Myc protein level during ESC differentiation (Figure 7D). As our results indicated that ATG5 regulates c-Myc protein degradation through the ubiquitination-proteasome pathway (Figures 2, 3, 4, 5, and 6), we next examined whether ATG5 participates in the regulation of c-Myc protein degradation during ESC differentiation. Our data suggested that ATG5 is indeed required for the regulation of c-Myc protein degradation during ESC differentiation, as depletion of ATG5 inhibited the decrease in the c-Myc protein level during ESC differentiation (Figure 7E). It is worth mentioning that the lower band below the band corresponding to the ATG5-ATG12 conjugate that appeared after ESC differentiation indeed corresponded to free ATG5, as transfection of ATG5-specific shRNA also resulted in depletion of the lower band (Figure 7E). Furthermore, we next investigated the dynamics of the interaction among ATG5, c-Myc, and FBW7 during ESC differentiation. We found that in parallel with the degradation of c-Myc during ESC differentiation, the interaction among ATG5, c-Myc, and FBW7 was markedly enhanced (Figure 7F). It is worth mentioning that the lower band corresponding to free ATG5 disappeared in ESCs treated with MG132, suggesting that the cleavage of free ATG5 from ATG5-ATG12 conjugates may depend on proteasome activity (Figure 7F, “Input” panel). However, more studies are needed to determine the detailed underlying mechanisms. In addition, we sought to determine whether the observed regulation of c-Myc protein degradation by ATG5 contributes to ESC differentiation. Our results revealed that depletion of ATG5 strikingly abolished ESC differentiation, and although the growth of ATG5-depleted cells with further loss of c-Myc was greatly impaired, the surviving cells showed a normal differentiation phenotype (Figure 7G), suggesting that the regulation of ESC differentiation by ATG5 is at least partially achieved through c-Myc. This hypothesis is reasonable, as c-Myc is a well-known stem cell marker and is essential for the maintenance of ESCs. Taken together, these data indicate that the negative regulation of c-Myc by ATG5 is essential for ESC differentiation.
Figure 7.
The observed ATG5-c-Myc regulatory axis is involved in the regulation of mouse embryonic stem cell (ESC) differentiation
(A) An Oct4-EGFP mouse ES (Oct4-EGFP-mES) cell line was used to establish the ESC differentiation system. Briefly, ESCs were cultured for the indicated durations in differentiation medium from which LIF (leukemia inhibitory factor) was removed and to which RA (retinoic acid, 1 μM) was added. The differentiation efficiency was determined by analyzing the morphologies of the colonies (upper left), the expression of Oct4-regulated EGFP (upper right), and the staining of AP (alkaline phosphatase) (lower).
(B) Oct4-EGFP-mESCs were differentiated for 4 days as described in (A). IF analyses (left) were then performed to determine the levels of LC3 (red) and GFP-Oct4 (green). DAPI staining indicates the nucleolus (blue). Western blot analyses were also performed with antibodies as indicated (right).
(C) Oct4-EGFP-mESCs were differentiated as described in (A). Western blot and RT-PCR analyses were performed with antibodies as indicated.
(D) Oct4-EGFP-mESCs were differentiated for 4 days as described in (A). Cells were then treated with 25 μM MG132 for 6 h. Western blot analyses were performed with antibodies as indicated.
(E) Oct4-EGFP-mESCs were stably transfected with an ATG5-specific shRNA or a control shRNA. Cells were then differentiated as described in A by removing LIF and adding RA. Western blot analyses were performed with antibodies as indicated.
(F) Oct4-EGFP-mESCs were differentiated as described in (A). Cells were then treated with 25 μM MG132 for 6 h. Co-IP assays with antibodies against ATG5 were performed. IgG antibodies were used as a negative control.
(G) Oct4-EGFP-mESCs were stably transfected with an ATG5-specific shRNA, an ATG5-specific shRNA combined with a c-Myc-specific shRNA, or a control shRNA as indicated. Cells were then differentiated for 4 days as described in A by removing LIF and adding RA. AP staining assays were performed to determine the differentiation efficiency.
Discussion
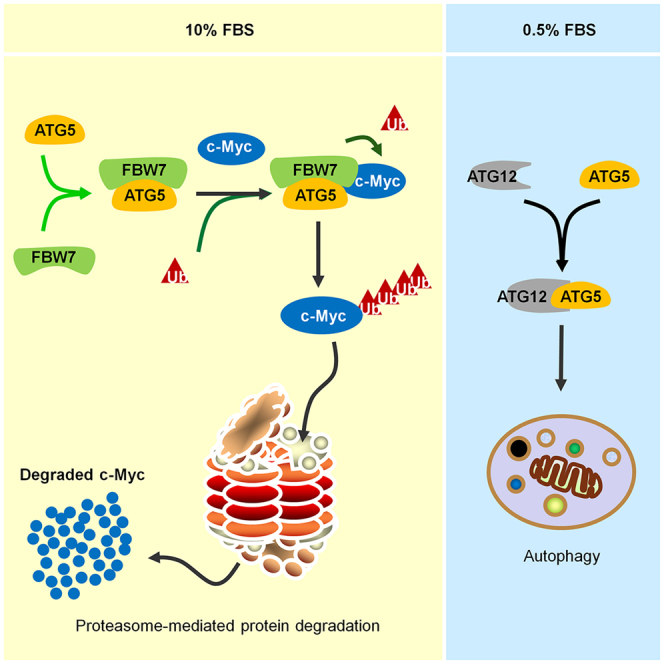
Autophagy is usually activated under distinct stress conditions, such as starvation and infection, and is maintained at a relatively low level under normal conditions (Kroemer et al., 2010; Ravanan et al., 2017). However, the protein levels of some autophagy-related regulators are still highly under normal conditions (e.g., the protein level of ATG5 was comparable between normal culture conditions and serum starvation conditions; Figures 2A and 2B, S3). The biological functions of these autophagy-related proteins under normal conditions are largely unclear. In this study, we demonstrated that the autophagy-related regulator ATG5 negatively regulates the protein level of c-Myc through the ubiquitination-proteasome pathway under normal culture conditions in an autophagy-independent manner (Figures 2, 3, 4, 5, and 6). Moreover, the observed ATG5-c-Myc regulatory axis further participates in the regulation of cell growth, especially under normal culture conditions (Figures 1 and 2) and in ESC differentiation (Figure 7).
ATG5 is a key component of the ATG12 ubiquitin-like conjugation system and plays essential roles in the regulation of autophagy. ATG5 is covalently conjugated by ATG12 on the acceptor lysine residue via an isopeptide bond, forming the ATG5-ATG12 conjugate. This conjugation is catalyzed by the E1-like enzyme ATG7 and the E2-like enzyme ATG10 in an ATP-dependent manner (Mizushima, 2020; Mizushima and Levine, 2020). In this study, we found that in both HeLa and 293T cells, ATG5 is present mainly as ATG12-conjugated ATG5 (the ATG5-ATG12 conjugate) and that exogenously overexpressed Flag-tagged ATG5 was present as both free ATG5 and ATG5-ATG12 conjugates (Figures 2A and 2B, 3A–3C, and S3). Intriguingly, our data indicated that both ATG12-conjugated and free ATG5 can likely associate with c-Myc, as endogenous immunoprecipitation (IP) assays clearly indicated that c-Myc associates with ATG5-ATG12 conjugates in vivo (Figure 4B) and IP assays with exogenously overexpressed proteins showed that HA-tagged c-Myc associates with both free ATG5 (Flag-tagged free ATG5) and ATG12-conjugated ATG5 in vivo (Figure 4A, right). Moreover, the ATG5K130R mutant also interacts with c-Myc (Figure 5A). Therefore, these data suggested that the conjugation of ATG12 likely does not affect the interaction between c-Myc and ATG5.
The negative regulation of c-Myc by ATG5 likely occurs specifically under normal culture conditions, as when cells were cultured under serum starvation conditions, the interaction between ATG5 and c-Myc was disrupted (Figure 4E), and the ATG5-c-Myc regulatory axis was therefore abolished (Figures 2A–2D). Moreover, our data indicated that ATG5 released from c-Myc after serum starvation may further participate in the regulation of autophagy, as the dispersed distribution of ATG5 under normal culture conditions changed to a punctate distribution, and the observed ATG5 puncta were localized near puncta of the autophagosome marker LC3 (Figure 4F). However, the underlying mechanisms by which the observed functional shift of ATG5 between normal culture conditions and serum starvation conditions is triggered are still unclear, and further studies are needed.
Autophagy activity has been reported to be decreased during ESC differentiation (Liu et al., 2017; Salemi et al., 2012). It was reasonable that autophagy activity was enhanced in cells with the “stem” status, as these cells robustly proliferate, and ESCs require more crude components for cell synthesis. However, the roles of the autophagy-related regulators released when autophagy activity is decreased after differentiation are largely unknown. Of great interest, our data indicate that in differentiated ESCs, the autophagy-related regulator ATG5 is likely essential for the decrease in c-Myc during ESC differentiation (Figure 7E). We found that the decrease in c-Myc was likely achieved mainly through the regulation of c-Myc protein degradation, as when proteasome activity was blocked, the decrease in c-Myc was completely abolished (Figure 7D). Moreover, the interaction between ATG5 and c-Myc was markedly enhanced after ESC differentiation (Figure 7F), and the negative regulatory ATG5-c-Myc axis was likely required for ESC differentiation (Figure 7G). Thus, our results revealed a novel mechanism by which ATG5 regulates ESC differentiation by negatively regulating c-Myc in an autophagy-independent manner.
In conclusion, in this study, we identified a novel nonautophagic role of ATG5 in the regulation of c-Myc protein levels through the ubiquitination-proteasome pathway, which further plays critical roles in the regulation of cell growth, especially under normal culture conditions, and in ESC differentiation.
Limitations of the study
In this study, we found that, under normal culture conditions, ATG5 regulates the protein degradation of c-Myc through the proteasome-mediated pathway by recruiting the E3 ubiquitin-ligase FBW7. When cells were cultured under serum starvation conditions, the c-Myc degradation machinery composed by ATG5-FBW7 wase disrupted, and the released ATG5 may mainly participate in the regulation of autophagy. However, how the extracellular nutrition signals are transduced to the ATG5-FBW7-c-Myc regulatory axis to allow cells to sense the extracellular nutrition changes is still unclear. Thus, more studies are needed to elucidate the signaling pathways that connect the extracellular nutrition signals to the ATG5-FBW7-c-Myc regulatory axis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ATG5 (rabbit) | Abcam | Cat #: ab108327; RRID: AB_2650499 |
| ATG5 (mouse) | Proteintech | Cat #: 66744-1-lg; RRID: AB_2882092 |
| c-Myc | Abcam | Cat #: ab32072; RRID: AB_731658 |
| FBW7 | Abcam | Cat #: ab109617; RRID: AB_2687519 |
| HA (rabbit) | CST | Cat #: 3724; RRID: AB_1549585 |
| HA (mouse) | Abmart | Cat #: M20003; RRID: AB_2864345 |
| Flag (rabbit) | CST | Cat #: 14793; RRID: AB_2572291 |
| Flag (mouse) | Abmart | Cat #: M20008; RRID: AB_2713960 |
| LC3B | CST | Cat #: 2775; RRID: AB_915950 |
| p62 | CST | Cat #: 5114; RRID: AB_10624872 |
| USP28 | Proteintech | Cat #: 17707-1-AP; RRID: AB_2272676 |
| SKP2 | Proteintech | Cat #: 15010-1-AP; RRID: AB_2187647 |
| TRIM32 | Proteintech | Cat #: 10326-1-AP; RRID: AB_2208005 |
| CHIP | Proteintech | Cat #: 55430-1-AP; RRID: AB_10949225 |
| GST | ABclonal | Cat #: AE006; RRID: AB_2771923 |
| His | Proteintech | Cat #: 66005-1-lg; RRID: AB_11232599 |
| IgG (rabbit) | Beyotime | Cat #: A7028; no RRID was obtained |
| IgG (mouse) | Beyotime | Cat #: A7007; no RRID was obtained |
| Alexa Fluor 594-conjugated secondary antibodies | ZSGB-BIO | Cat #: ZF-0516; no RRID was obtained |
| Alexa Fluor 594-conjugated secondary antibodies | ZSGB-BIO | Cat #: ZF-0513; no RRID was obtained |
| HRP-conjugated secondary antibodies | ZSGB-BIO | Cat #: ZB-2301; no RRID was obtained |
| HRP-conjugated secondary antibodies | ZSGB-BIO | Cat #: ZB-2305; no RRID was obtained |
| Bacterial and virus strains | ||
| DH5ɑ | TianGen | CB101 |
| BL21(DE3) | TianGen | CB105 |
| ATG5 shRNA (mouse) Lentiviral Particles | Santa Cruz | sc-41446-V |
| c-Myc shRNA (mouse) Lentiviral Particles | Santa Cruz | sc-29227-V |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | HyClone | SH30243.01 |
| Lipo8000 | Beyotime | C0533 |
| Polybrene | Beyotime | C0351 |
| CCK-8 | Beyotime | C0038 |
| RIPA buffer | Beyotime | P0013B |
| PMSF | Beyotime | ST506 |
| TRIzol reagent | Thermo Fisher | 15596018 |
| GelRed | Vazyme | GR501 |
| ECL | CWBio | CW0049M |
| FBS | Every Green | 11011-8611 |
| Penicillin and streptomycin | Beyotime | C0222 |
| L-GlutaMAX | Gibco | 35050079 |
| NEAA | Gibco | 11140050 |
| β-mercaptoethanol | Solarbio | M8210 |
| Sodium pyruvate | Gibco | 11360070 |
| LIF | Beyotime | P6339 |
| Retinoic acid (RA) | MCE | HY-14649 |
| Experimental models: Cell lines | ||
| HEK-293T cells | Chinese National Collection of Authenticated Cell Lines | GNHu17 |
| HeLa cells | Chinese National Collection of Authenticated Cell Lines | TCHu187 |
| OG2 mouse embryonic stem cells (ESCs) | The Jackson Laboratory | 004654 |
| Recombinant DNA | ||
| GST-ATG5 plasmid | This paper | N/A |
| His-c-Myc plasmid | This paper | N/A |
| His-FBW7 plasmid | This paper | N/A |
| Software and algorithms | ||
| ImageJ | Open Source | https://imagej.nih.gov/ij |
| Prism | GraphPad | https://www.graphpad.com |
| Other | ||
| Protein A/G agarose beads | Santa Cruz Biotechnology | sc-2003 |
| Glutathione Sepharose 4B Beads | GE Healthcare | 71024800-EG |
| Luciferase Reporter Gene Assay Kit | Yeasen | 11401ES60 |
| Alkaline phosphatase detection kit | Merck | SCR004 |
| BCA Protein Assay Kit | CWBio | CW0014S |
| Reverse transcription kit | Takara | RR036A |
| ChamQTM SYBR Color qPCR Master Mix | Vazyme | 411-02/03 |
| PVDF membranes | Merck | ISEQ00010 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Su Chen (chensubio@163.com, or chenlab@henu.edu.cn, or chensu@xjtu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject
Cell culture and transfection
HEK293T human embryonic kidney cells and HeLa human cervical carcinoma cells were cultured at 37°C in DMEM supplemented with 10% fetal bovine serum (FBS) (normal culture conditions) or 0.5% FBS (serum starvation conditions) together with 1% penicillin and streptomycin in a 5% CO2 incubator.
OG2 mouse embryonic stem cells (ESCs) were cultured at 37°C in DMEM supplemented with 15% FBS, 1% penicillin and streptomycin, 1% L-GlutaMAX, 1% NEAA (nonessential amino acids), 1% 10 mM β-mercaptoethanol, 1% sodium pyruvate, and 1000 U/ml LIF (Leukemia Inhibitory Factor) in a 5% CO2 incubator. Embryonic stem cells were induced to differentiate with 1 μM retinoic acid (RA).
Transfection of cells with constructs or siRNAs was performed by using Lipo8000 transfection reagent according to the manufacturer's standard protocol. Infection of cells with lentivirus was performed by using 5 μg/ml polybrene according to the manufacturer's standard protocol.
Methods details
Plasmid construction
The cDNAs of human FBW7, c-Myc and ATG5 were amplified by RT-PCR from total RNA extracted from 293T cells. FBW7 cDNA was inserted into the pCMV-HA vector. ATG5 cDNA was inserted into the pET-42b (+) vector containing a GST tag. FBW7 and c-Myc cDNAs were inserted into the pET-42b (+) vector containing a His tag. The sequences of the primers are listed in Table S1.
Knockdown of gene expression
shRNA against ATG5 and siRNAs against ATG5, c-Myc and FBW7 were designed by and purchased from GenePharma Company (Shanghai, China). The sequences of the siRNAs and shRNAs are listed in Table S2.
Cell growth assay
Cell growth assays were performed with a CCK-8 Kit. Briefly, 3×103 cells/well were plated into 96-well plates and cultured at 37°C. Then, 10 μL of CCK-8 reagent was added to the medium, and the cells were incubated for 1 h at 37°C. The absorbance (OD value) was measured at 450 nm using a Multiskan MK3 microplate reader (Thermo Fisher Scientific, USA). Three independent CCK-8 assays were performed with three technical replicates.
Colony formation assay
A total of 1×103 cells/well were seeded into six-well plates and incubated for 5 days. The cells were washed twice with PBS and stained with crystal violet staining solution at room temperature for 5 min. The number of colonies (>50 cells) was counted under a light microscope at 40× magnification.
Cell migration assay
A total of 1×106 cells were seeded into the upper chambers of Transwell plates (8-μm pore size, 6.5 mm diameter) (#03318032, Corning Life Sciences, USA) and cultured overnight under normal conditions or serum starvation conditions. Medium supplemented with 10% FBS was placed into the lower chambers. After further incubation for 24 h, the invaded cells were fixed with 100% methanol for 10 min at room temperature, stained with 0.5% crystal violet for 30 min at room temperature, and washed with water to remove excess dye. Stained cells were visualized by using an optical microscope.
Antibodies and western blot analysis
Antibodies against ATG5, c-Myc, and FBW7 were purchased from Abcam (USA). Antibodies against the HA tag, the Flag tag, LC3B, and p62 were purchased from Cell Signaling Technology (USA). Antibodies against the HA tag (mouse) and Flag tag (mouse) were purchased from Abmart. Antibodies against ATG5 (mouse), USP28, SKP2, TRIM32, CHIP, and the His tag were purchased from Proteintech (China).
Cells were lysed in RIPA buffer containing 1 mM PMSF. The protein concentration in the supernatant was measured using a BCA Protein Assay Kit. Samples were loaded into 10% or 15% SDS-PAGE gels to resolve proteins. Different amounts of total protein were loaded in each experiment to facilitate the detection of different target proteins. After electrophoresis, the proteins were transferred to PVDF membranes and hybridized with primary antibodies at a dilution of 1:1000. HRP-conjugated secondary antibodies were used at a dilution of 1:5000. An ECL detection system was used to detect the signals on the membranes.
RT-PCR assay
Cells were lysed for isolation of total RNA using TRIzol reagent according to the manufacturer's instructions. Reverse transcription was performed using a reverse transcription kit. Briefly, total RNA (1 μg) was reverse transcribed into cDNA in a volume of 20 μL. Each 25 μL PCR mixture contained 1 μL of cDNA. For semiquantitative analysis, the PCR products were loaded onto a 2% agarose gel, stained with GelRed, and imaged. The sequences of the primers used for PCR assays are listed in Table S3.
Quantitative RT-PCR assay
Cells were lysed for isolation of total RNA using TRIzol reagent according to the manufacturer's instructions. Reverse transcription was performed using a reverse transcription kit. Briefly, total RNA (1 μg) was reverse transcribed into cDNA in a volume of 20 μL. qPCR was subsequently performed in triplicate using the ChamQTM SYBR Color qPCR Master mix on a QuantStudio™ 5 Real-Time PCR system (Thermo). Each qPCR reaction was performed in a final volume of 20 μl, containing 10 μl ChamQ SYBR Color qPCR Master mix (2×), 0.5 μl forward and reverse primers (10 μM), 8 μl DEPC treated water and 1 μl cDNA. The following thermocycling conditions were used for the qPCR: 95°C for 30 sec, 40 cycles of 95°C for 10 sec and 60°C for 30 sec.
Luciferase reporter assay
Luciferase activity was measured using a Cytation 5 microplate reader (BioTek, USA). All luciferase emission measurements were performed using a Luciferase Reporter Gene Assay Kit. Cells were washed with 150 μL of phosphate-buffered saline (PBS) per well in 96-well plates. Cells were then lysed using 30 μL of lysis buffer per well in 96-well plates. The necessary amount of lysate required for the experiment was transferred to white 96-well plates for luminescence measurement.
Coimmunoprecipitation (co-IP) assays
co-IP assays were performed as previously described (Jing et al., 2020; Wang et al., 2019b). Briefly, cells were transfected with the indicated constructs using Lipo8000. After 48 h, cells were harvested, washed 3 times with ice-cold PBS, and resuspended in 800 μL of RIPA buffer containing 1 mM PMSF. Cell lysates were subjected to immunoprecipitation at 4°C overnight with the indicated antibodies or with normal IgG as a negative control. Protein A/G agarose beads were subsequently added and incubated for another 3 h. The solution was centrifuged to harvest the agarose beads after they were washed 5 times with fresh ice-cold lysis buffer. The precipitated proteins were released by boiling in loading buffer and resolved via SDS-PAGE. Western blot analyses were performed using the related antibodies.
In vitro protein binding assays
In vitro protein binding assays were performed as we previously described (Jing et al., 2020). Briefly, GST-tagged ATG5, His-tagged c-Myc, and His-tagged FBW7 proteins were expressed in E. coli and were purified, and the purified proteins were verified with Coomassie brilliant blue (CBB) staining assays. The purified His-tagged c-Myc or FBW7 proteins were individually mixed with GST-tagged ATG5 or control GST proteins for 4 h at 4°C. Antibodies against ATG5, c-Myc, or the His-tag were then added to the mixtures and incubated overnight. Protein A/G agarose beads were subsequently added and incubated for another 3 h. The solutions were centrifuged to harvest the agarose beads after they were washed 5 times with fresh ice-cooled lysis buffer. The precipitated proteins were released by boiling in loading buffer and resolved via SDS-PAGE. Western blot analyses were performed using the related antibodies.
Immunofluorescence (IF) analysis
IF analyses were performed as we previously described (Jing et al., 2020; Wang et al., 2019b). Briefly, cells grown on culture slides were fixed with 4% paraformaldehyde and were then permeabilized with 0.2% Triton X-100, both for 10 min at room temperature. Cells were then washed and incubated with 3% BSA for 1 h at room temperature. Specific antibodies were used to detect the target proteins by incubation with cells at 4°C overnight. The cells were then washed with PBS 3 times, and Alexa Fluor 594-conjugated secondary antibodieswere used to detect the primary antibodies. After a wash step, the cells were examined with a Leica DMRE confocal laser scanning microscope (Leica, Germany).
AP staining assays
AP staining assays were performed according to methods described in one of our previous reports with an alkaline phosphatase detection kit (Chen et al., 2017). Briefly, mouse embryonic stem cells (ESCs) or differentiated ESCs were fixed for 1 min. After washing, the cells were stained with alkaline phosphatase solution and imaged with an optical microscope.
Quantification and statistical analysis
All statistical analyses were analyzed with the Graphpad Prism 8.0 software by using the method of unpaired Student's t-test to define the statistical significance of the differences between groups. All experiments were repeated, n represents the number of independent replicates. Bars and error bars in the figures represent mean ± s.d. value. p value < 0.05 was considered to be statistically significant, and p value were shown on the figures as asterisks or ns as following: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ns, no significance.
Acknowledgments
We thank Dr Zhiji Chang from Tsinghua University for kindly providing the Flag-tagged ATG5 construct, Dr Wuhan Xiao from the Institute of Hydrobiology, Chinese Academy of Sciences, for kindly providing the HA-tagged c-Myc construct, and Dr Zhuohua Zhang from Central South University for kindly providing the LC3-DsRed2-GFP reporter construct. This work was supported by the National Natural Science Foundation of China (Grant No.: 81972650 and 81773009), the Supporting Fund for Teachers' Research of Jining Medical University (Grant No.: JYFC2019KJ002 and JYFC2018KJ010), the Research Fund for Lin He's Academician Workstation of New Medicine and Clinical Translation in Jining Medical University (Grant No.: JYHL2018MS13 and JYHL2018MS14), the Shandong Provincial Natural Science Foundation (Grant No.: ZR2019PH039 and ZR2020MH212), and the Jining Key Science and Technology Project (2019SMNS005).
Author contributions
SC designed the study; SL, LZ, GZ, and SZ performed the experiments; GS, XH, WD, YX, NX, BZ, and JD prepared the materials; YW, WC, and SC analyzed the data; SC and SL wrote the manuscript.
Declaration of interests
The authors declare that they have no conflicts of interest.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103296.
Contributor Information
Yequan Wang, Email: wangyequan1103@163.com.
Wen Cui, Email: cuiwenmdd@163.com.
Su Chen, Email: chensubio@163.com, chenlab@henu.edu.cn, chensu@xjtu.edu.cn.
Supplemental information
Data and code availability
-
•
All data produced in this study are included in the published article and its supplementary information, or are available from the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Chen R., Zou Y., Mao D., Sun D., Gao G., Shi J., Liu X., Zhu C., Yang M., Ye W. The general amino acid control pathway regulates mTOR and autophagy during serum/glutamine starvation. J. Cell Biol. 2014;206:173–182. doi: 10.1083/jcb.201403009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Jing Y., Kang X., Yang L., Wang D.L., Zhang W., Zhang L., Chen P., Chang J.F., Yang X.M. Histone H2B monoubiquitination is a critical epigenetic switch for the regulation of autophagy. Nucleic Acids Res. 2017;45:1144–1158. doi: 10.1093/nar/gkw1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S., Liu J., Wu X., Lu W. Golgi apparatus: a potential therapeutic target for autophagy-associated neurological diseases. Front Cell Dev Biol. 2020;8:564975. doi: 10.3389/fcell.2020.564975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Saitoh T., Akira S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G., Harrington E., Fanidi A., Land H., Amati B., Bennett M. Integrated control of cell proliferation and cell death by the c-myc oncogene. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994;345:269–275. doi: 10.1098/rstb.1994.0105. [DOI] [PubMed] [Google Scholar]
- Farrell A.S., Sears R.C. MYC degradation. Cold Spring Harb Perspect. Med. 2014;4:a014365. doi: 10.1101/cshperspect.a014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia G.M., Kroemer G., Piacentini M. Molecular mechanisms of selective autophagy. Cell Death Differ. 2013;20:1–2. doi: 10.1038/cdd.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y.Y., Cai F.F., Zhang L., Han J., Yang L., Tang F., Li Y.B., Chang J.F., Sun F., Yang X.M. Epigenetic regulation of the Warburg effect by H2B monoubiquitination. Cell Death Differ. 2020;27:1660–1676. doi: 10.1038/s41418-019-0450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Kroemer G., Marino G., Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Liu K., Gu H., Wang W., Gong J., Zhu Y., Zhao Q., Cao J., Han C., Gao F. High autophagic flux guards ESC identity through coordinating autophagy machinery gene program by FOXO1. Cell Death Differ. 2017;24:1672–1680. doi: 10.1038/cdd.2017.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.Y., Chen C.L., Yang T.T., Huang W.C., Hsieh C.Y., Shen W.J., Tsai T.T., Shieh C.C., Lin C.F. Albumin prevents reactive oxygen species-induced mitochondrial damage, autophagy, and apoptosis during serum starvation. Apoptosis. 2012;17:1156–1169. doi: 10.1007/s10495-012-0758-6. [DOI] [PubMed] [Google Scholar]
- Mehrpour M., Esclatine A., Beau I., Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- Meyer N., Penn L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020;63:1–10. doi: 10.1016/j.ceb.2019.12.001. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B. Autophagy in human diseases. N. Engl. J. Med. 2020;383:1564–1576. doi: 10.1056/NEJMra2022774. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Paul I., Ahmed S.F., Bhowmik A., Deb S., Ghosh M.K. The ubiquitin ligase CHIP regulates c-Myc stability and transcriptional activity. Oncogene. 2013;32:1284–1295. doi: 10.1038/onc.2012.144. [DOI] [PubMed] [Google Scholar]
- Popov N., Wanzel M., Madiredjo M., Zhang D., Beijersbergen R., Bernards R., Moll R., Elledge S.J., Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- Pradel B., Robert-Hebmann V., Espert L. Regulation of innate immune responses by autophagy: a goldmine for viruses. Front Immunol. 2020;11:578038. doi: 10.3389/fimmu.2020.578038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanan P., Srikumar I.F., Talwar P. Autophagy: the spotlight for cellular stress responses. Life Sci. 2017;188:53–67. doi: 10.1016/j.lfs.2017.08.029. [DOI] [PubMed] [Google Scholar]
- Salemi S., Yousefi S., Constantinescu M.A., Fey M.F., Simon H.U. Autophagy is required for self-renewal and differentiation of adult human stem cells. Cell Res. 2012;22:432–435. doi: 10.1038/cr.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn J.C., Berezikov E., Knoblich J.A. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Mizushima N., Ogawa Y., Matsuura A., Noda T., Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.E., Hubner K., Scholer H., Mann J.R. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 2002;115:157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., Kominami E. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Yin H., Hossain M.A., Wang Y., Wu F., Dong X., Gao S., Zhan K., He W. Starvation-induced autophagy promotes the invasion and migration of human bladder cancer cells via TGF-beta1/Smad3-mediated epithelial-mesenchymal transition activation. J. Cell Biochem. 2019;120:5118–5127. doi: 10.1002/jcb.27788. [DOI] [PubMed] [Google Scholar]
- von der Lehr N., Johansson S., Wu S., Bahram F., Castell A., Cetinkaya C., Hydbring P., Weidung I., Nakayama K., Nakayama K.I. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol. Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- Wang R., Tan J., Chen T., Han H., Tian R., Tan Y., Wu Y., Cui J., Chen F., Li J. ATP13A2 facilitates HDAC6 recruitment to lysosome to promote autophagosome-lysosome fusion. J. Cell Biol. 2019;218:267–284. doi: 10.1083/jcb.201804165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yang L., Zhang X., Cui W., Liu Y., Sun Q.R., He Q., Zhao S., Zhang G.A., Wang Y. Epigenetic regulation of ferroptosis by H2B monoubiquitination and p53. EMBO Rep. 2019;20:e47563. doi: 10.15252/embr.201847563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M., Orian A., Jin J., Grim J.E., Harper J.W., Eisenman R.N., Clurman B.E. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wible D.J., Chao H.P., Tang D.G., Bratton S.B. ATG5 cancer mutations and alternative mRNA splicing reveal a conjugation switch that regulates ATG12-ATG5-ATG16L1 complex assembly and autophagy. Cell Discov. 2019;5:42. doi: 10.1038/s41421-019-0110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., Ishida N., Okumura F., Nakayama K., Nakayama K.I. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C.H., Bellon M., Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol. Cancer. 2018;17:115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data produced in this study are included in the published article and its supplementary information, or are available from the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.