Abstract
Objectives Evidence reveals that microRNAs (miRNAs) are abnormally expressed in lung adenocarcinoma (LUAD) tissue and are crucial in LUAD occurrence. Therefore, this study aims to find the miRNA which could regulate LUAD and to further explore its regulatory mechanism, thus providing a potential molecular target for LUAD. Methods miR-9-5p and ID4 expression in LUAD cells were measured by real-time quantitative PCR and western blot. Cell functional assays were conducted to detect the biological functions of LUAD cells. A dual-luciferase reporter assay was utilized to validate the binding relationship between miR-9-5p and ID4. Results miR-9-5p was highly expressed whereas ID4 was lowly expressed in LUAD. miR-9-5p facilitated LUAD cell progression by targeting ID4. Conclusion miR-9-5p promotes LUAD cell progression by modulating ID4 and may become a potential target for LUAD.
Keywords: miR-9-5p, ID4, lung adenocarcinoma, migration, proliferation, invasion
Introduction
Lung cancer has become a major cause resulting in high mortality in world.1,2 Lung adenocarcinoma (LUAD) approximately makes up 50% of total lung cancer cases, which has overtaken lung squamous cell carcinoma (LACC).3,4 Despite the latest advancements in diagnosis, the prognosis of lung cancer patients is still poor.5,6 Hence, finding new therapeutic approaches for lung cancer patients is in urgent need.
MicroRNAs (miRNAs) can hinder the target gene level through binding to the 3′-untranslated region (UTR) and inducing translational suppression. 7 Up to now, a lot of miRNAs have been confirmed to be implicated with cell proliferation, differentiation, apoptosis, migration, cell cycle, and other biological processes. 8 Besides, by mediating target gene expression, loads of miRNAs are uncovered to play an inhibitory or promoting role in the malignant progression of human carcinoma. 9 For instance, miR-26a modulates ovarian cancer cell behaviors by targeting TCF12. 10 miR-125 suppresses colorectal cancer progression through TAZ. 11 Besides, research suggests that miRNA is important in the development of LUAD. For example, miR-218 modulates epithelial-mesenchymal transition and PI3K/Akt pathway by targeting BMI-1 to suppress LUAD progression. 12 miR-142-3p suppresses LUAD cell progression through targeting NR2F6. 13 Collectively, these findings fully clarify the significance of miRNAs in LUAD and that miRNA-dependent treatment can be one of the therapeutic approaches for LUAD.
miR-9-5p is proven to be able to mediate the development of cancers, by regulating different mRNAs14–16 Currently, there have been few studies on miR-9-5p and LUAD except that one study displayed an increase of miR-9-5p in LUAD. 17 Given this, mechanism by which miR-9-5p regulates LUAD is yet to be confirmed and validated. We attempted to validate the regulatory function of miR-9-5p in the disease as well as explored related downstream regulatory mechanisms.
Materials and Methods
Bioinformatics Approach
LUAD miRNA and corresponding clinical data (normal: n = 58, tumor: n = 522) were acquired from the TCGA-LUAD dataset. miRDB, TargetScan, and miRTarBase were used in the prediction of downstream targets of the target miRNA. Meanwhile, DE_mRNAs were screened with the “DESeq2R” package, and normal samples were taken as control (|logFC|>1.5, Padj<.05). The screened results were then intersected with predicted mRNAs for identification of the mRNA of interest.
Cell Incubation
Human bronchial epithelial cell line BEAS-2B (BNCC338205), and human LUAD cell lines including A549 (BNCC337696), Calu-3 (BNCC338514), and NCI-H1299 (BNCC100268) were all provided by BeNa Culture Collection (Beijing, China). Cells were all incubated in DMEM (Invitrogen) with 10% fetal bovine serum (FBS) under standard conditions. Cells at the fourth passage were used for follow-up experiments.
Cell Transfection
miR-9-5p-mimic, miR-9-5p-inhibitor, and corresponding NCs (NC-mimic, NC-inhibitor) were obtained from GenePharma (China). The over-expression vector of ID4 (oe-ID4) was synthesized by GenePharma (China). Using Lipofectamine® 3000 reagent kit (Invitrogen, USA) to transfect above plasmids or oligonucleotide fragments into LUAD cells. Later, real-time quantitative PCR (qRT-PCR) or western blot was performed to verify transfection efficiency.
Real-Time Quantitative PCR
Total RNA was isolated using Trizol solution (TaKaRa) and then came with reverse transcription of mRNA and miRNA into cDNA by m-MLV reverse transcriptase kits (TaKaRa) and SuperScript II kits (Invitrogen), respectively. qRT-PCR was completed on the Applied Biosystems 7500 with TaKaRa SYBR Premix E × Taq kit. U6 and GAPDH were internal references for miR-9-5p and ID4, respectively. Primers were exhibited in Table 1.
Table 1.
Primer Sequences.
| Gene | Sequences | |
|---|---|---|
| miR-9-5p | Forward: 5′-CGAGCTCTGTGTGTGTGTGTGTGTG-3′ | |
| Reverse: 5′-TTCCGCGGCCGCTATGGCCGAGGAA-3′ | ||
| U6 | Forward: 5′-CTCGCTTCGGCAGCACA-3′ | |
| Reverse: 5′-AACGCTTCACGAATTTGCGT-3′ | ||
| ID4 | Forward: 5′-CGATGAAGGCGGTGAGCC-3′ | |
| Reverse: 5′-CCAGGCTGTGGATCTTCGT-3′ | ||
| GAPDH | Forward: 5′-GACCTGACCTGCCGTCTA-3′ | |
| Reverse: 5′-AGGAGTGGGTGTCGCTGT-3′ | ||
Western Blot
Briefly, after cell lysis with RIPA + a protease inhibitor, centrifugation of the lysate was performed at 4°C for 10 min at 12 000 r/min. The obtained protein samples were quantitated with a BCA kit (Thermo Fisher Scientific, MA, USA), boiled at 95°C, and sequentially separated by SDS-PAGE, which were loaded to nitrocellulose membranes. Then, membranes were silked in 5% skimmed milk. Membranes were reacted with primary antibodies (ID4 [ab220166, 1:1000, Abcam, Cambridge, UK] and GAPDH [ab181602, 1:10 000, Abcam, Cambridge, UK]) overnight. Later, they were cultured with horseradish peroxidase (HRP)-conjugated secondary antibody goat anti-rabbit IgG (ab6721, 1:3000, Abcam, Cambridge, UK) for 120 min. At last, an enhanced chemiluminescence detection Kit (Solarbio, Beijing, China) was used for protein band visualization.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT)
Cells (2 × 104 cells/mL) were inoculated on 96-well plates. 10 μL MTT (5 mg/mL) was added at specific time points (0, 24, 48, and 72 h) after cell culture. Cells were maintained at 37°C for 4 h. The optical density (OD) value at 490 nm was assayed with a microplate reader.
Colony Formation Assay
Transfected cells (2 × 102 cells/mL) were placed on 6-well plates. The medium was replaced every 4 d and discarded after 12 d. Cells were then treated with 4% paraformaldehyde for 15 min and 0.1% crystal violet for 10 min. Finally, crystal violet was rinsed off with PBS, and cells were photographed.
Wound Healing Assay
Calu-3 cells were digested after transfection and 2 ml of cells (4 × 105 cells/mL) were used. With 90% cell confluence, we made a scratch with the tip of a sterile 200 μL pipette. Floating cells were rinsed with PBS. Remained cells were cultured in an FBS-free medium under standard conditions. An inverted microscope was used to capture photos of the scratch area at 0 and 48 h. Cell migration rate was assessed using the Image J software.
Transwell Assay
Calu-3 cells were digested after transfection and 200 μL of cells in FBS-free medium were then inoculated into the upper chamber of Transwell laid with a matrix. In the lower chamber, 650 μL medium plus 10% FBS was added. 24 h after continuous cell culture, cells in the upper chamber were swabbed, after which cells invaded were treated with paraformaldehyde (4%) for 15 min and stained with crystal violet (0.1%) for 10 min. Finally, 5 random visual fields were observed under a microscope to count invading cells.
1.10. Dual-Luciferase Reporter Assay
Using pmirGLO vectors (Promega, WI, USA) to generate ID4-wt and ID4-mut vectors. ID4-wt/ID4-mut and miR-9-5p mimic/NC mimic were transfected into cells. Then, the Dual-Luciferase Reporter Assay kit was used for activity assessment.
Statistical Analysis
Statistical analysis and visualization of experimental data from 3 independent replicates were completed using GraphPad Prism 7 software (CA). All data met normal distribution and were represented by mean ± SD. The t-test was conducted to compare data differences between the 2 groups. Analysis of variance was conducted to compare data differences among multiple groups. Kruskal-Wallis test was used to analyze the correlation between miR-9-5p/ID4 and clinicopathological characteristics of LUAD patients. P < .05 indicated statistically significant.
Results
miR-9-5p is at a High Level in LUAD
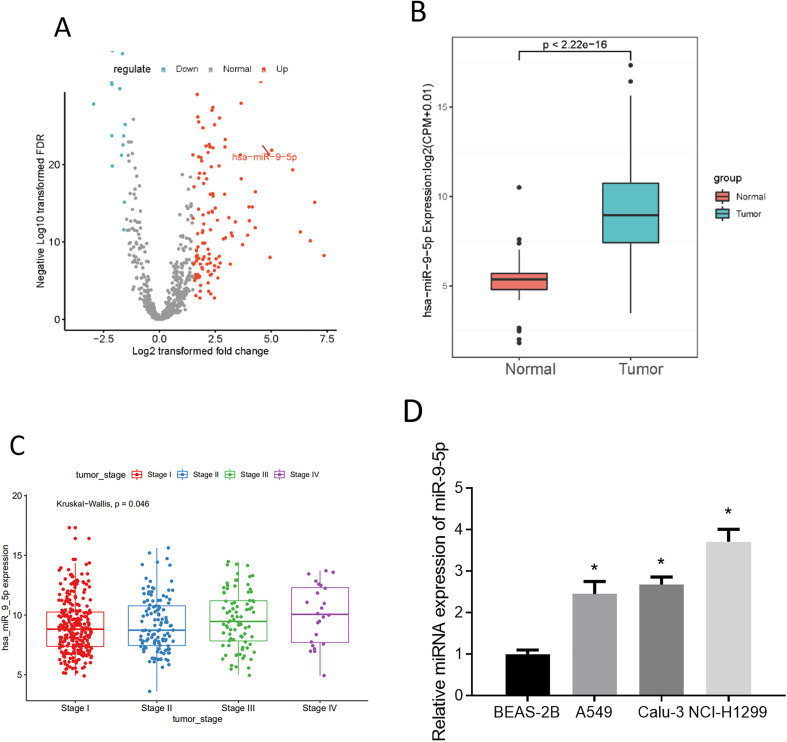
After data acquirements from the bioinformatics database, differential gene expression analysis performed by the “edgeR” package suggested that 164 DE_miRNAs were acquired (Figure 1A). There are few reports about miR-9-5p in LUAD. In addition, compared with normal samples, miR-9-5p in tumor samples was markedly increased (Figure 1B), and the miR-9-5p level was significantly different in different stages (Figure 1C). Hence, miR-9-5p was taken as a research target. Additionally, we assayed miR-9-5p levels in control cells BEAS-2B, human nonmetastatic LUAD cell lines, and human metastatic LUAD cell line NCI-H1299 through qRT-PCR. The result demonstrated miR-9-5p expression was increased in LUAD cells (Figure 1D). Due to the fact that tumor cell behaviors are more often observed in early tumor stages, the Calu-3 cell line which had a relatively high miR-9-5p expression of the 2 nonmetastatic cell lines was chosen for our further study.
Figure 1.
miR-9-5p is at a high level in lung adenocarcinoma (LUAD)
(A) Volcano plot of 164 DE_miRNAs; (B) miR-9-5p level in TCGA-LUAD (red: normal group, and blue: tumor group); (C) relationship between miR-9-5p and LUAD staging; (D) miR-9-5p level in BEAS-2B, A549, Calu-3, and NCI-H1299; * P < .05.
miR-9-5p Fosters LUAD Cell Progression
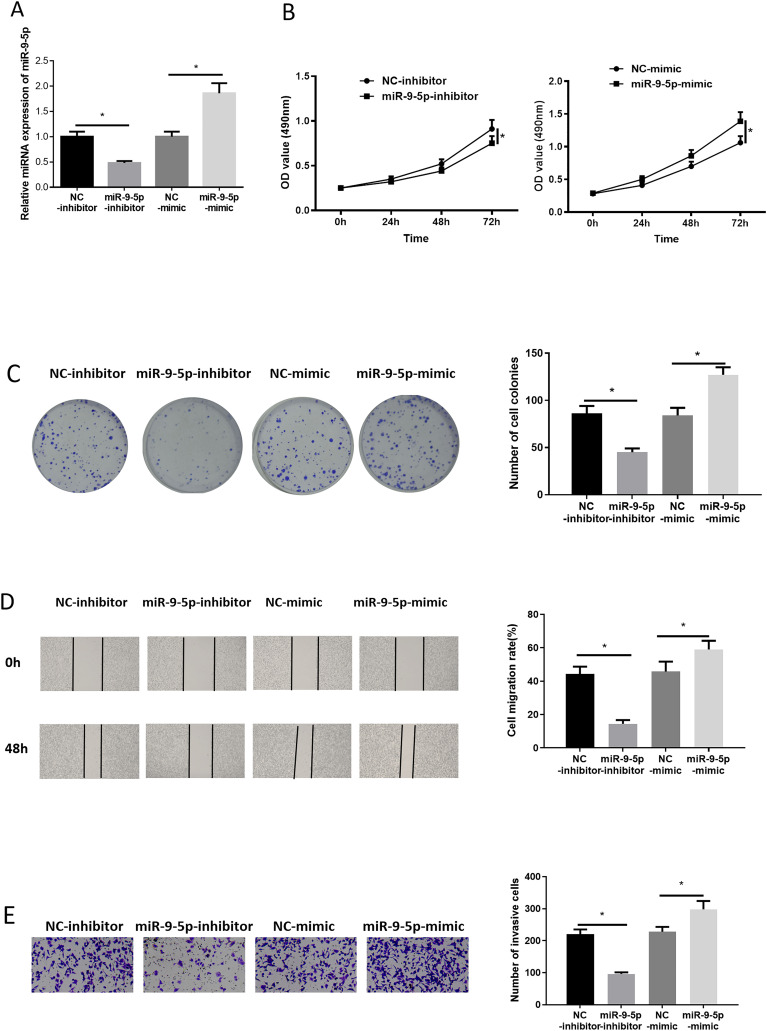
We firstly transfected NC-inhibitor/miR-9-5p-inhibitor, and NC-mimic/miR-9-5p-mimic into LUAD cells, respectively. Then, transfection efficiency was measured by applying qRT-PCR. Results unveiled that miR-9-5p in LUAD cells was down-regulated after miR-9-5p-inhibitor treatment, while it was up-regulated after miR-9-5p-mimic treatment (Figure 2A). MTT and colony formation assays unveiled that suppressing/over-expressing miR-9-5p reduced/promoted cell proliferative activity and colony formation ability (Figure 2B and C). Similarly, we found in wound healing and Transwell assays that suppressing/over-expressing miR-9-5p decreased/facilitated cell migratory and invasive abilities (Figure 2D and E). Collectively, forced miR-9-5p expression facilitated LUAD cell progression.
Figure 2.
miR-9-5p promotes lung adenocarcinoma (LUAD) cell growth
(A) miR-9-5p level in LUAD cells after transfection; (B, C) effect of repressing/over-expressing miR-9-5p on proliferative activity and colony formation ability of LUAD cells, respectively; (D, E) impacts of suppressed/over-expressed miR-9-5p on migratory and invasive potentials of LUAD cells; * P < .05.
ID4 is a Direct Target of miR-9-5p
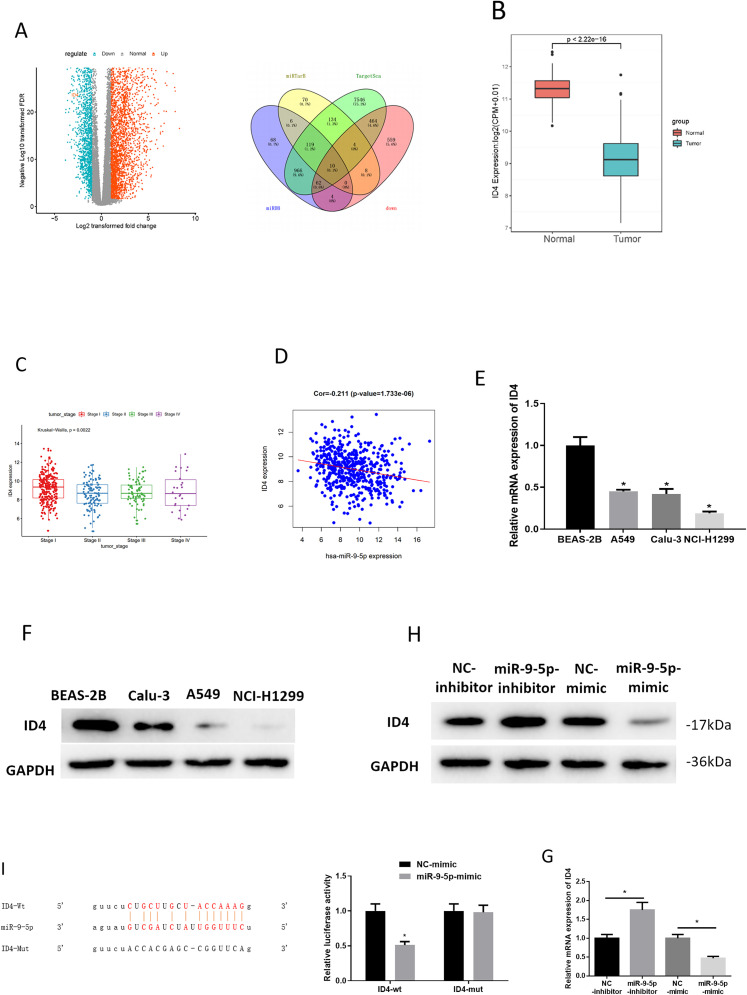
We predicted downstream targets of miR-9-5p by employing TargetScan, miRDB, and miRTarBase databases. Meanwhile, differential analysis of mRNAs in the TCGA-LUAD dataset was conducted (Figure 3A), and then the down-regulated DE_mRNAs in LUAD were intersected with the predicted mRNAs. As a result, 10 mRNAs (TGFBR2, AMOTL1, PTX3, MYOCD, SLC19A3, CD34, STARD13, ID4, CBX7, and COLEC12) could serve as the regulatory targets of miR-9-5p (Figure 3A). Among these 10 mRNAs, ID4 was found to be poorly expressed in LUAD (Figure 3B). ID4 expression was significantly different in different stages (Figure 3C). In addition, Pearson correlation analysis noted that ID4 was inversely associated with miR-9-5p in LUAD tissue (Figure 3D). Research showed that ID4 could inhibit pancreatic cancer cell proliferation, migration, and invasion. 18 Therefore, it was hypothesized that miR-9-5p regulates ID4 in LUAD cells. To confirm the hypothesis, we assessed the ID4 level in control cells and cancer cell lines through qRT-PCR and western blot. ID4 level was poor in all LUAD cells (Figure 3E and F). After that, ID4 level was measured upon miR-9-5p silencing/overexpression in LUAD cells, which uncover that silencing/over-expressing miR-9-5p up-regulated/down-regulated ID4 (Figure 3G and H). These results validated the inverse correlation of miR-9-5p and ID4. Subsequently, dual-luciferase reporter assay unveiled that luciferase activity of ID4-wt was suppressed by over-expression of miR-9-5p whereas that of ID4-mut was not affected (Figure 3I), which suggested that miR-9-5p could target ID4. Taken together, ID4 was directly targeted by miR-9-5p.
Figure 3.
ID4 is targeted directly by miR-9-5p
(A) Volcano plot of 1111 DE_mRNAs in TCGA-LUAD, and downregulated DE_mRNAs and predicted target mRNAs were intersected; (B) ID4 expression in TCGA-LUAD (red: normal group, and blue: tumor group); (C) relationship between ID4 expression and lung adenocarcinoma (LUAD) staging; (D) correlation of miR-9-5p and ID4 in tumor tissue; (E, F) ID4 mRNA and protein levels in BEAS-2B, A549, Calu-3, and NCI-H1299; (G, H) ID4 mRNA and protein expression when inhibiting/over-expressing miR-9-5p in LUAD cells; (I) dual-luciferase reporter assay verified relationship of miR-9-5p; * P < .05.
miR-9-5p Promotes LUAD Cell Progression by Targeting ID4
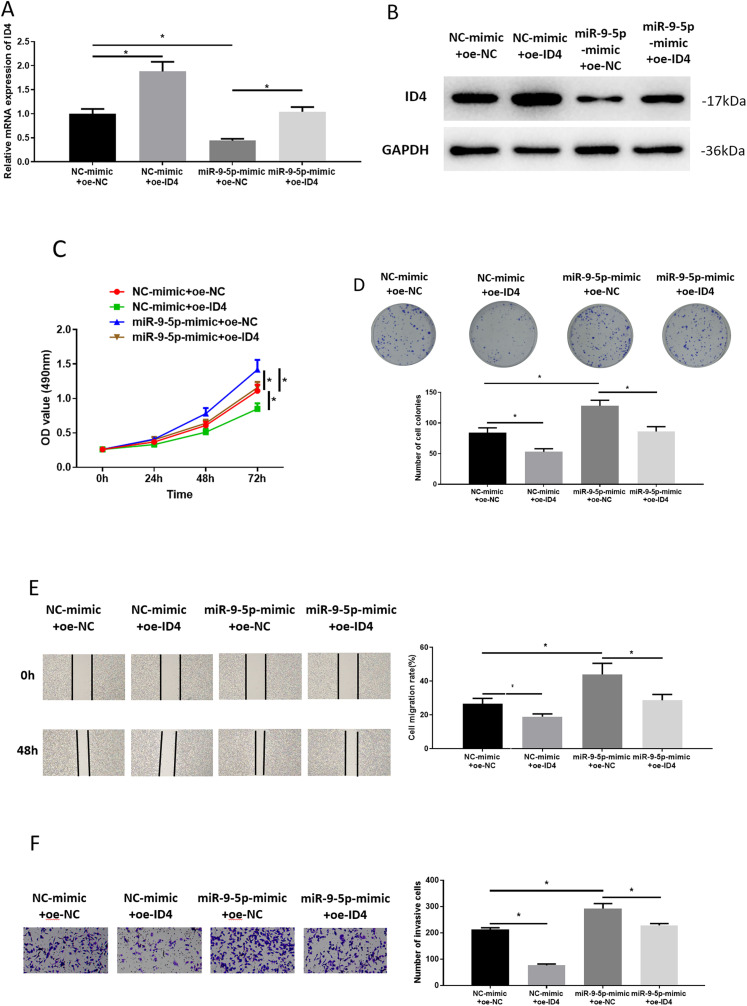
We proved in the section “ID4 is a Direct Target of miR-9-5p” that ID4 was a direct target of miR-9-5p, and then we observed whether miR-9-5p could promote LUAD cell behaviors by targeting ID4. After transfection, the ID4 level in these groups was assayed through qRT-PCR and western blot. ID4 expression in the NC-mimic + oe-ID4 group was markedly higher than that in the control group. ID4 expression was significantly lower after miR-9-5p overexpression than that in the control group. ID4 expression in the miR-9-5p-mimic + oe-ID4 group was significantly higher than that in the miR-9-5p-mimic + oe-NC group (Figure 4A and B). From the MTT assay and colony formation assay, we noted that ID4 hindered proliferation and colony formation of LUAD cells. Simultaneously increased miR-9-5p and ID4 expression could reverse promoting impacts of miR-9-5p on proliferation and colony formation of LUAD cells (Figure 4C and D). In wound healing and Transwell assays, we also observed that ID4 reduced cell migratory and invasive abilities, which could be rescued by miR-9-5p overexpression (Figure 4E and F). Hence, miR-9-5p could promote LUAD cell progression by suppressing ID4.
Figure 4.
miR-9-5p promotes lung adenocarcinoma (LUAD) cell progression via targeting ID4
(A, B) Detection of ID4 mRNA and protein in transfection groups; (C to F) the proliferative activity, colony formation, migratory, and invasive potentials of LUAD cells, respectively; * P < .05.
Discussion
Studies that have been done over the recent few years confirm the close correlation between the part of miRNAs and malignant tumors, including the regulation of miRNAs on tumor growth and metastasis.19,20 As a member of numerous miRNAs, miR-9-5p was confirmed as a modulatory factor of neural progenitor cells. 21 Deeper exploration into miR-9-5p uncovered that miR-9-5p modulates other cancers, such as colorectal cancer, cervical cancer, NSCLC, carcinoma of paranasal sinuses, and so on, in promotion of tumor proliferation, invasion, and metastasis.14,22–24 Here, we validated the up-regulation of miR-9-5p in LUAD. Additionally, the influence of miR-9-5p on the biological functions of LUAD cells was assayed by cell functional assays. It was suggested that miR-9-5p could enhance the proliferative activity, colony formation ability, migratory and invasive properties of LUAD cells. Collectively, miR-9-5p is able to facilitate LUAD cell progression.
ID4 can act as a dominant inverse modulatory factor of a basic HLH transcription factor. 25 Up to now, research has uncovered that ID4 can play an important regulatory role in cancers. For example, ID4 can facilitate HCC cell proliferation, 26 suppress prostate cancer cell proliferation, and raise cell apoptosis. 27 However, no research has reported the correlation between ID4 and LUAD. Herein, after confirming the oncogenic role of miR-9-5p, we explored targets of miR-9-5p and identified ID4. Meanwhile, rescue experiments unveiled the reverse effect of ID4 on miR-9-5p in exacerbating LUAD progression. Given the literature which displayed that miR-9-5p in NSCLC suppresses TGFBR2 to induce cell proliferation, invasion, and metastasis, 22 we reasoned that promoting effect of miR-9-5p on LUAD cell behaviors was partially realized by suppressing ID4 expression.
In conclusion, miR-9-5p expression was at a high level in LUAD and could promote LUAD cell progression by targeting ID4, which provides a theoretical basis for the possibility that miR-9-5p/ID4 axis is a possible target for LUAD.
Supplemental Material
Supplemental material, sj-docx-1-tct-10.1177_15330338211048592 for miR-9-5p Promotes Lung Adenocarcinoma Cell Proliferation, Migration and Invasion by Targeting ID4 by Kai Zhu, Jinlan Lin, Shengjia Chen and Qian Xu in Technology in Cancer Research & Treatment
Footnotes
Availability of Data and Materials: The data used to support the findings of this study are included within the article. The data and materials in the current study are available from the corresponding author on reasonable request.
Authors’ Contributions: KZ contributed to the study design. JL conducted the literature search. SC acquired the data. QX wrote the article. KX performed data analysis and drafted. QX revised the article. All the authors gave the final approval of the version to be submitted.
Consent for Publication: All authors consent to submit the manuscript for publication.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship and/or publication of this article.
ORCID iD: Qian Xu https://orcid.org/0000-0001-5068-4103
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115-132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Harlow BE, Flythe MD, Aiken GE. Effect of biochanin A on corn grain (Zea mays) fermentation by bovine rumen amylolytic bacteria. J Appl Microbiol. 2017;122(4):870-880. doi: 10.1111/jam.13397 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5-29. doi: 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87-108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 5.Li-Ming X, Zhao LJ, Simone CB, et al. Receipt of thoracic radiation therapy and radiotherapy dose are correlated with outcomes in a retrospective study of three hundred and six patients with extensive stage small-cell lung cancer. Radiother Oncol. 2017;125(2):331-337. doi: 10.1016/j.radonc.2017.10.005 [DOI] [PubMed] [Google Scholar]
- 6.Bernhardt D, Adeberg S, Bozorgmehr F, et al. Outcome and prognostic factors in single brain metastases from small-cell lung cancer. Strahlenther Onkol. 2018;194(2):98-106. doi: 10.1007/s00066-017-1228-4 [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857-866. doi: 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281-297. doi: 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999-3004. doi: 10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao S, Bian T, Su M, Liu Y, Zhang Y. Mir26a inhibits ovarian cancer cell proliferation, migration and invasion by targeting TCF12. Oncol Rep. 2020;43(1):368-374. doi: 10.3892/or.2019.7417 [DOI] [PubMed] [Google Scholar]
- 11.Yang M, Tang X, Wang Z, et al. miR-125 inhibits colorectal cancer proliferation and invasion by targeting TAZ. Biosci Rep. 2019;39(12):BSR20190193. 10.1042/BSR20190193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu L, Sun HB, Xu ZN, et al. MicroRNA-218 regulates the epithelial-to-mesenchymal transition and the PI3K/Akt signaling pathway to suppress lung adenocarcinoma progression by directly targeting BMI-1. Eur Rev Med Pharmacol Sci. 2019;23(18):7978-7988. doi: 10.26355/eurrev_201909_19014 [DOI] [PubMed] [Google Scholar]
- 13.Jin C, Xiao L, Zhou Z, et al. MiR-142-3p suppresses the proliferation, migration and invasion through inhibition of NR2F6 in lung adenocarcinoma. Hum Cell. 2019;32(4):437-446. doi: 10.1007/s13577-019-00258-0 [DOI] [PubMed] [Google Scholar]
- 14.Wang M, Gao Q, Chen Y, et al. PAK4, A target of miR-9-5p, promotes cell proliferation and inhibits apoptosis in colorectal cancer. Cell Mol Biol Lett. 2019;24:58. doi: 10.1186/s11658-019-0182-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Liu T, Liu Y, Wei C, et al. LncRNA HULC promotes the progression of gastric cancer by regulating miR-9-5p/MYH9 axis. Biomed Pharmacother. 2020;121:109607. doi: 10.1016/j.biopha.2019.109607 [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Chen JY, Zhong Y, Xie L, Li JS. lncRNA MEG3 inhibits the growth of hepatocellular carcinoma cells by sponging miR-9-5p to upregulate SOX11. Braz J Med Biol Res. 2019;52(10):e8631. doi: 10.1590/1414-431X20198631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeybek A, Oz N, Kalemci S, et al. Diagnostic value of MiR-125b as a potential biomarker for stage I lung adenocarcinoma. Curr Mol Med. 2019;19(3):216-227. doi: 10.2174/1566524019666190314113800 [DOI] [PubMed] [Google Scholar]
- 18.Cheng D, Fan J, Ma Y, et al. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci. 2019;9:28. doi: 10.1186/s13578-019-0290-2 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Han TS, Hur K, Xu Z, et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut. 2015;64(2):203-214. doi: 10.1136/gutjnl-2013-306640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou M, Magi-Galluzzi C. Prostatic adenocarcinoma, prostatic intraepithelial neoplasia, and intraductal carcinoma. Surg Pathol Clin. 2008;1(1):43-75. doi: 10.1016/j.path.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 21.Hang C, Yan HS, Gong C, et al. MicroRNA-9 inhibits gastric cancer cell proliferation and migration by targeting neuropilin-1. Exp Ther Med. 2019;18(4):2524-2530. doi: 10.3892/etm.2019.7841 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Li G, Wu F, Yang H, Deng X, Yuan Y. MiR-9-5p promotes cell growth and metastasis in non-small cell lung cancer through the repression of TGFBR2. Biomed Pharmacother. 2017;96:1170-1178. doi: 10.1016/j.biopha.2017.11.105 [DOI] [PubMed] [Google Scholar]
- 23.Wei YQ, Jiao XL, Zhang SL, et al. MiR-9-5p could promote angiogenesis and radiosensitivity in cervical cancer by targeting SOCS5. Eur Rev Med Pharmacol Sci. 2019;23(17):7314-7326. doi: 10.26355/eurrev_201909_18837 [DOI] [PubMed] [Google Scholar]
- 24.Kovarikova J, Baranova I, Laco J, et al. Deregulation of selected microRNAs in sinonasal squamous cell carcinoma: searching for potential prognostic biomarkers. Folia Biol (Praha). 2019;65(3):142-151. [DOI] [PubMed] [Google Scholar]
- 25.Riechmann V, van Cruchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 1994;22(5):749-755. doi: 10.1093/nar/22.5.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Zhang LX, Liu XQ, et al. Id4 promotes cell proliferation in hepatocellular carcinoma. Chin J Cancer. 2017;36(1):19. doi: 10.1186/s40880-017-0186-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carey JP, Asirvatham AJ, Galm O, Ghogomu TA, Chaudhary J. Inhibitor of differentiation 4 (Id4) is a potential tumor suppressor in prostate cancer. BMC Cancer. 2009;9:173. doi: 10.1186/1471-2407-9-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tct-10.1177_15330338211048592 for miR-9-5p Promotes Lung Adenocarcinoma Cell Proliferation, Migration and Invasion by Targeting ID4 by Kai Zhu, Jinlan Lin, Shengjia Chen and Qian Xu in Technology in Cancer Research & Treatment