Summary
We present a protocol for measuring the activity of the mechanistic target of rapamycin (mTOR) pathway in ex vivo isolated mouse primary hepatocytes. It can be used as a tool for genetic, pharmacological, metabolomic, and signal transduction procedures. We discuss critical aspects for improving yield, viability, and modulation of the mTOR pathway. This protocol can be adapted to other signaling cascades and is compatible with multiple readouts.
For complete details on the use and execution of this protocol, please refer to Ortega-Molina et al. (2021).
Subject areas: Cell culture, Cell isolation, Cell-based Assays, Metabolism, Metabolomics, Signal Transduction
Graphical abstract
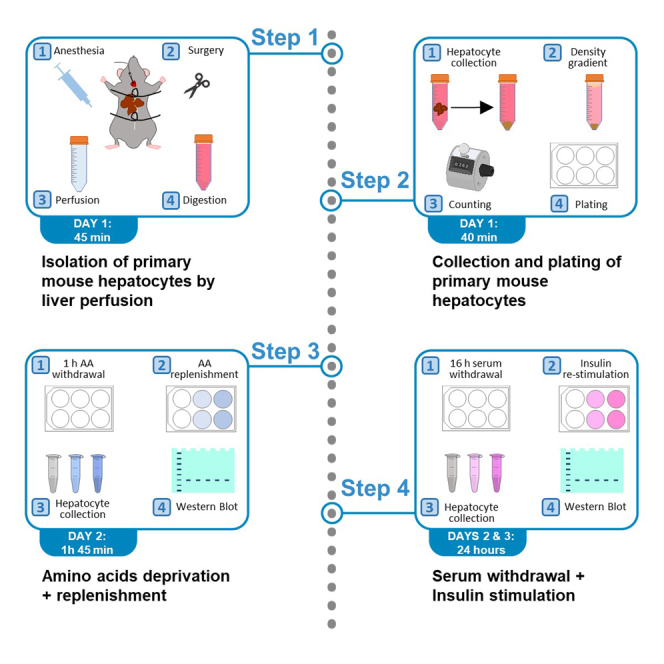
Highlights
-
•
Describes the purification and culture of primary hepatocytes from mice
-
•
Describes the assessment of mTORC1 activity manipulating nutrient and growth factor signals
-
•
Protocol adaptable to other signaling cascades and compatible with multiple readouts
We present a protocol for measuring the activity of the mechanistic target of rapamycin (mTOR) pathway in ex vivo isolated mouse primary hepatocytes. It can be used as a tool for genetic, pharmacological, metabolomic, and signal transduction procedures. We discuss critical aspects for improving yield, viability, and modulation of the mTOR pathway. This protocol can be adapted to other signaling cascades and is compatible with multiple readouts.
Before you begin
Tissue culture room
Timing: 3 days
-
1.Prepare the solutions for perfusion and digestion of the liver and the hepatocyte medium.
-
a.Perfusion Solution: Mix 500 mL of Hank Balanced Salt Solution 1× (HBSS) without Ca2+ without Mg2+ without Phenol red with 5 mL 1M HEPES and 5 mL of 0.1M EGTA.
-
b.Digestion Solution: Mix 500 mL of Williams E Medium with 5 mL 1M HEPES. Each 50 mL of this solution must be supplemented with 40 mg of Collagenase Type I.Optional: Aliquot Perfusion Solution and Digestion Solution in 50 mL centrifuge tubes. Each tube contains the volume necessary for the perfusion of two mouse livers.Note: Prepare envelopes containing 40 mg of Collagenase Type I. Each envelope contains the Collagenase necessary to digest two mouse livers. Immediately before the first step of isolation of primary mouse hepatocytes add the content of one envelope to one 50 mL tube of Digestion Solution and mix well.
 CRITICAL: The Collagenase must be added to the Digestion Solution immediately before the start of the isolation procedure. It must be completely dissolved and warmed to obtain the best digestion efficiency.
CRITICAL: The Collagenase must be added to the Digestion Solution immediately before the start of the isolation procedure. It must be completely dissolved and warmed to obtain the best digestion efficiency. -
c.Rinsing Solution: Mix 500 mL of sterile PBS with 5 mL 5,000 U/mL Penicillin-Streptomycin and 500 μL 50 mg/mL Gentamycin.
-
d.Gradient Solution: Mix 22.5 mL Percoll with 2.5 mL HBSS 10× per mouse.
 CRITICAL: Gradient Solution must be prepared immediately before the density-gradient separation.
CRITICAL: Gradient Solution must be prepared immediately before the density-gradient separation. -
e.Hepatocyte Attachment Medium: Mix 200 mL DMEM high glucose, 200 mL F12 Nutrient mix Ham Medium, 100 mg 0.02% BSA fatty acid free, 255 mg NaHCO3, 25 mL 100 mM Sodium Pyruvate, 9 mL 1M HEPES, 5 mL 200 mM L-Glutamine, 5 mL 5,000 U/mL Penicillin-Streptomycin, 500 μL 50 mg/mL Gentamycin and 50 mL fetal bovine serum (FBS). Filter the medium through a 0.22 μm filter.
-
a.
Store all the solutions at 4°C.
-
2.Prepare cell culture plates for primary hepatocytes.
-
a.Mix 1 mL of rat tail Collagen Type I with 49 mL of Rinsing Solution and homogenize by repeated pipetting up and down.
-
b.Add 2 mL of the mix in each well of a 6 multi-well plate. Ensure that the bottom of the well is completely covered.Note: For the assessment of mTOR activity, 6 multi-well plates yield sufficient whole protein lysates for western blotting (around 250 μg), but 10 cm, 12-well and 24-well plates are also suitable for hepatocyte plating.
-
c.Incubate the plates at 37°C during 3 h under sterile conditions inside the cell culture incubator.
-
d.Sterilize the plates inside the cell culture hood by UV light exposure for 30 min.
-
e.Transfer the plates to a 4°C fridge. Close and cover each plate with parafilm.Note: Collagen must be aspirated from cell plates before the plating of the primary hepatocytes in step 17.
-
a.
-
3.Prepare solutions for amino acid withdrawal and for FBS withdrawal.
-
a.Dialyzed FBS (dFBS):
-
i.Take a SnakeSkin Dialysis Tube and cut the required length of membrane from the packaging tube.
-
ii.Close the tube in one end using the dialysis clips or by making a tight knot.Note: Ensure that the tube is properly closed before pouring the FBS inside.
-
iii.Pour 100 mL of FBS inside the tube and close the tube in the open end.
-
iv.Place the tube in a beaker with 5L of PBS 1× and put it in a magnetic mixer. Transfer to a 4°C room.Note: Ensure that the external surface of the whole tube is in contact with the PBS.
-
v.Set a slow spin in the magnetic mixer and leave 16 h.
-
vi.The following day take out the tube carefully. Discard the PBS 1× and fill the beaker with 5L of fresh cold PBS 1×.
-
vii.Return the beaker in the magnetic mixer at 4°C.
-
viii.Set a slow spin in the magnetic mixer and leave 16 h.
-
ix.The following day take out the tube carefully. Open it by one side and pour the content into a clean beaker.
-
x.Filter the dFBS through a 0.22 μm filter inside a cell culture hood.
-
xi.Aliquot the dFBS in 15 mL conical tubes to avoid to multiple freeze-thaw cycles and freeze at −20°C.
-
i.
-
b.Pre-starvation Medium: Dissolve 3.85 g DMEM/F12 without Amino acids Medium in 450 mL of ddH2O. Supplement the medium with 255 mg NaHCO3, 9 mL 1M HEPES and 5 mL 2.5M Glucose. Filter the medium through 0.22 μm filter.
-
c.Amino acid Starvation Medium: Mix 90 mL of Pre-starvation Medium with 10 mL of dFBS. Supplement the medium with 100 μL of 100 μM insulin.Note: DMEM/F12 without Amino acids, supplemented with dFBS, is better suited to perform amino acid starvation on primary hepatocytes, as compared to other media (see below; DAY 2 Amino acid deprivation). Moreover, the strength of acute activation of mTORC1 further improves when medium is supplemented with insulin.Note: Specific metabolite withdrawal procedures can be performed to assess the effect on mTORC1 activity. For example, single amino acid drop-out experiments can be performed following the same procedure but incorporating all other amino acids (except for the one to be deprived of) in the Amino acid Starvation Medium.
-
d.Amino acid cocktail:
-
i.Prepare a 50× Essential Amino acid mix stock:50× AA mix
Amino acid Amount Final concentration (stock) Final concentration (when added to amino acid starvation medium) C (L-Cystine-2HCl) 325 mg 3,250 mg/L 207.52 μM F (L-Phenylalanine) 75 mg 750 mg/L 90.81 μM H (L-Histidine) 75 mg 750 mg/L 71.56 μM I (L-Isoleucine) 250 mg 2,500 mg/L 381.19 μM K (L-Lysine-HCl) 200 mg 2,000 mg/L 218.99 μM L (L-Leucine) 250 mg 2,500 mg/L 381.19 μM M (L-Methionine) 75 mg 750 mg/L 100.53 μM Q (L-Glutamine) 1,500 mg 15,000 mg/L 2052.83 μM R (L-Arginine) 1,000 mg 10,000 mg/L 1148.11 μM T (L-Threonine) 100 mg 1,000 mg/L 167.89 μM V (L-Valine) 100 mg 1,000 mg/L 170.72 μM W (L-Tryptophan) 25 mg 250 mg/L 24.48 μM Y (L-Tyrosine-2Na-2H2O) 145 mg 1,450 mg/L 160.05 μM ddH2O 100 mL N/A N/A Total 100 mL 50× 1× Note: aliquot and store the mix at −20°C. -
ii.Prepare a 100× Non-Essential Amino acid mix stock:100× AA mix
Amino acid Amount Final concentration (stock) Final concentration (when added to amino acid starvation medium) D (L-Aspartic Acid) 200 mg 2,000 mg/L 150.26 μM E (L-Glutamic Acid) 200 mg 2,000 mg/L 135.93 μM G (Glycine) 100 mg 1,000 mg/L 133.21 μM N (L-Asparagine) 500 mg 5,000 mg/L 378.44 μM P (L-Proline) 200 mg 2,000 mg/L 173.72 μM S (L-Serine) 300 mg 3,000 mg/L 285.47 μM ddH2O 100 mL N/A N/A Total 100 mL 100× 1× Note: aliquot and store the mix at −20°C. -
iii.Mix 1000 μL of 100× Non-Essential Amino acids and 2000 μL of 50× Essential Amino acids in 7 mL of Amino acid Starvation Medium to have a 10× amino acid solution.
-
i.
-
e.FBS withdrawal Medium: Mix 200 mL DMEM high glucose, 200 mL F12 Nutrient mix Ham Medium, 100 mg 0.02% BSA fatty acid free, 255 mg NaHCO3, 25 mL 100 mM Sodium Pyruvate, 9 mL 1M HEPES, 5 mL 200 mM L-Glutamine, 5 mL 5,000 U/mL Penicillin-Streptomycin and 500 μL 50 mg/mL Gentamycin. Filter the medium with 0.22 μm filter.Note: The composition of the FBS withdrawal Medium is the same as in the Hepatocyte Attachment Medium but without FBS.
-
a.
-
4.
Prepare Protein Lysis Buffer:
Protein Lysis Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| HEPES-KOH 1M pH 7.4 | 50 mM | 25 mL |
| NaCl | 40 mM | 1.175 g |
| EDTA 0.5 M | 2 mM | 2 mL |
| Sodium orthovanadate (NaVO4) 150 mM ∗ | 1,5 mM | 500 μL per 50 mL |
| Sodium Fluoride (NaF) 0.5M | 50 mM | 50 mL |
| Sodium Pyrophosphate | 10 mM | 1.325 g |
| β-Glycerophosphate disodium salt hydrate 1M ∗ | 10 mM | 500 μL per 50 mL |
| Triton X-100 | 1% | 5 mL |
| Sodium deoxycholate | 1% | 5 g |
| Complete Mini Protease Inhibitor Cocktail ∗ | N/A | 2 tablets per 50 mL |
| Sodium dodecyl sulfate (SDS) 10% | 0.1 % | 5 mL |
| ddH2O | N/A | 408.5 mL |
| Total | N/A | 500 mL |
Note: store at −20°C.
Note: Reagents marked with ∗ in the Protein Lysis Buffer Table are to be added immediately before using the Protein Lysis Buffer.
Animal facility
Timing: 10 min
-
5.Mouse housing:
-
a.All animal procedures carried out were performed according to protocols approved by the CNIO-ISCIII Ethics Committee for Research and Animal Welfare (CEIyBA) and the Autonomous Community of Madrid (CAM). Protocol number PROEX15/18.
-
b.Mice were housed under specific pathogen-free conditions at 22°C on a regular 12 h light and 12 h dark cycle. 7 to 10-week-old mice were used in this protocol.
-
a.
Note: Optimal age of mice for isolating primary hepatocytes is 7–10 weeks old. In younger mice the veins are smaller and more fragile, and veins in older mice have increased deposition of visceral fat, making more difficult the surgery process and the cannulation. Cannulation gauge and perfusion speed may need to be optimized for younger and older mice.
-
6.
Preparation of anesthesia: Mix 500 μL of 100 mg/mL Ketamine, 250 μL of 20 mg/mL Xylazine and 4.25 mL Saline Solution.
-
7.
Sterilization of the perfusion line: Wash the perfusion tube of the pump with 70% ethanol. Maintain the 70% ethanol solution in the tube for 16 h.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-p70 S6 Kinase (Thr389) (108D2) Rabbit mAb (dilution 1:500) | Cell Signaling Technology | Cat#9234; RRID: AB_2269803 |
| P70 S6 Kinase (49D7) Rabbit mAb (dilution 1:500) | Cell Signaling Technology | Cat#2708; RRID: AB_390722 |
| Phospho-S6 Ribosomal Protein (Ser235/236) Antibody (dilution 1:1000) | Cell Signaling Technology | Cat#2211; RRID: AB_331679 |
| S6 Ribosomal protein (5G10) Rabbit mAb (dilution 1:1000) | Cell Signaling Technology | Cat#2217; RRID: AB_331355 |
| Phospho-4E-BP1 (Thr37/46) (236B4) Rabbit mAb (dilution 1:500) | Cell Signaling Technology | Cat#2855 |
| 4E-BP1 (53H11) Rabbit mAb (dilution 1:1000) | Cell Signaling Technology | Cat#9644; RRID: AB_2097841 |
| Phospho-Akt (Ser473) (D9E) XP Rabbit mAb (dilution 1:1000) | Cell Signaling Technology | Cat#4060; RRID: AB_2315049 |
| Akt (pan) (C67E7) Rabbit mAb (dilution 1:1000) | Cell Signaling Technology | Cat#4691; RRID: AB_915783 |
| Vinculin (dilution 1:5000) | Sigma-Aldrich | Cat#V9131; RRID: AB_477629 |
| Chemicals, peptides, and recombinant proteins | ||
| Collagen, Type I solution from rat tail | Sigma-Aldrich | Cat#C3867 |
| PBS – Dulbecco’s Phosphate Buffered Saline | Sigma-Aldrich | Cat#D8537 |
| Penicillin-Streptomycin (5,000 U/mL) | Thermo Fisher | Cat#15070063 |
| Gentamycin solution 50 mg/mL | Sigma-Aldrich | Cat#G1397 |
| HBSS (1×), no calcium, no magnesium, no phenol red | Thermo Fisher Scientific | Cat#14175053 |
| HEPES Buffer 1M | Lonza | Cat#BE17-737E |
| Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid | Sigma-Aldrich | Cat#E3889 |
| William’s E Medium, no glutamine | Thermo Fisher Scientific | Cat#22551022 |
| Collagenase, Type I | Worthington-biochem | Cat#LS004196 |
| Percoll | Sigma-Aldrich | Cat#GE17-0891-01 |
| HBSS (10×), no calcium, no magnesium, no phenol red | Thermo Fisher Scientific | Cat#14185052 |
| DMEM, high glucose | Thermo Fisher Scientific | Cat#41965039 |
| Ham’s F-12 Nutrient Mix | Thermo Fisher Scientific | Cat#21765029 |
| Bovine Serum Albumin essentially fatty acid free | Sigma-Aldrich | Cat#A6003 |
| Sodium bicarbonate (NaHCO3) | Merck | Cat#1063291000 |
| Sodium pyruvate solution 100 mM | Sigma-Aldrich | Cat#S8636 |
| L-Glutamine solution 200 mM | Sigma-Aldrich | Cat#G7513 |
| Fetal Bovine Serum (FBS) | Hyclone | Cat#SV30160.03 |
| Dulbecco’s MEM (DMEM) F-12 without Amino acids, Glucose, L-Glutamine, Sodium Bicarbonate, HEPES, Sodium | USBiological Life Sciences | Cat#D9807-10 |
| D-(+)-Glucose solution | Sigma-Aldrich | Cat#G8769 |
| C (L-Cystine-2HCl) | Sigma-Aldrich | Cat#C6727 |
| F (L-Phenylalanine) | Sigma-Aldrich | Cat#P5482 |
| H (L-Histidine) | Sigma-Aldrich | Cat#H5659 |
| I (L-Isoleucine) | Sigma-Aldrich | Cat#I7403 |
| K (L-Lysine-HCl) | Sigma-Aldrich | Cat#L8662 |
| L (L-Leucine) | Sigma-Aldrich | Cat#L8912 |
| M (L-Methionine) | Sigma-Aldrich | Cat#M5308 |
| Q (L-Glutamine) | Sigma-Aldrich | Cat#G3126 |
| R (L-Arginine) | Sigma-Aldrich | Cat#A8094 |
| T (L-Threonine) | Sigma-Aldrich | Cat#T8441 |
| V (L-Valine) | Sigma-Aldrich | Cat#V0513 |
| W (L-Tryptophan) | Sigma-Aldrich | Cat#T8941 |
| Y (L-Tyrosine-2Na-2H2O) | Sigma-Aldrich | Cat#T8566 |
| D (L-Aspartic Acid) | Sigma-Aldrich | Cat#A8949 |
| E (L-Glutamic Acid) | Sigma-Aldrich | Cat#G1251 |
| G (Glycine) | Sigma-Aldrich | Cat#G7126 |
| N (L-Asparagine) | Sigma-Aldrich | Cat#A0884 |
| P (L-Proline) | Sigma-Aldrich | Cat#P0380 |
| S (L-Serine) | Sigma-Aldrich | Cat#S4500 |
| HEPES | Sigma-Aldrich | Cat#H3375 |
| Sodium chloride | Sigma-Aldrich | Cat#S9625 |
| EDTA 0.5 M, pH 8 | Thermo Fisher Scientific | Cat#15575020 |
| Sodium orthovanadate (NaVO4) | Sigma-Aldrich | Cat#S6508 |
| Sodium Fluoride (NaF) | Sigma-Aldrich | Cat#S7920 |
| Sodium Pyrophosphate tetra-basic decahydrate | Sigma-Aldrich | Cat#S6422 |
| β-Glycerophosphate disodium salt hydrate | Sigma-Aldrich | Cat#G9422 |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| Sodium deoxycholate | Sigma-Aldrich | Cat#D6750 |
| Complete Mini Protease Inhibitor Cocktail | Roche | Cat #11836153001 |
| Sodium dodecyl sulfate (SDS) | Merck | Cat#151-21-3 |
| 70% Ethanol | N/A | N/A |
| Ketamine | Imalgene 1000 | N/A |
| Xylazine | Rompur | N/A |
| Physiological saline solution | B Braun | Cat #607189.2 |
| Trypan Blue solution | Sigma-Aldrich | Cat#T8154 |
| Insulin human | Sigma-Aldrich | Cat#I2643 |
| SnakeSkin Dialysis Tubing, 3.5K MWCO, 35 mm dry I.D., 35 feet | Thermo Fisher Scientific | Cat #88244 |
| Critical commercial assays | ||
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23225 |
| Primary mouse hepatocytes | This paper | N/A |
| Experimental models: Organisms/strains | ||
| C57BL/6J (7–10 weeks) | Jackson Laboratory | Cat#JAX:000664; RRID: IMSR_JAX:000664 |
| Software and algorithms | ||
| ImageJ software | ImageJ | https://imagej.nih.gov/ij/ |
| Other | ||
| 6-well Clear Multiwell Plate | Life Sciences | Cat#353046 |
| 500 mL Vacuum Filter/Storage Bottle System, 0.22 μm Pore 33.2 cm2 PES Membrane, Sterile, 12/Case | Life Sciences | Cat#431097 |
| Corning Polypropylene Centrifuge Tubes, Sterile (50 mL) | Fisher Scientific | Cat#10509891 |
| Masterflex C/L Analog Variable-Speed Pump with Dual-Channel Pump Head for Microbore Tubing Pump, 10–60 rpm; 12 VDC | Cole-Parmer | Cat#EW-77120-62 |
| Water bath Thermo Haake | Thermo Fisher Scientific | Cat#003–2859 |
| Sterile needles 23G ×1” – Nr. 16 0.6 mm × 25 mm | BD Microlance 3 | Cat#300800 |
| Sterile surgical non-absorbable sutures 2/0 USP 90 cm | LorcaMarín | Cat#55184-50U |
| Intravascular Catheter BD Vialon 22GA 0.9 × 25 mm | Clinimark SL | Cat#381223 |
| ROCKER 3D digital (Shaker) | IKA | Cat#0004001000 |
| Fine Scissors – Sharp-Blunt | Fine Science Tools | Cat#14028-10 |
| Dumont #5 – Fine Forceps | Fine Science Tools | Cat#11254-20 |
| Corning 100 μm Cell Strainer, Yellow, Sterile, Individually Packaged, 50/Case | Life Sciences | Cat#431752 |
| TC20 Automated cell counter | Bio-Rad | Cat#145-0101 |
| Cell Lifter with J-Hook Blade | VWR | Cat#76036-004 |
| Eppendorf 5810 Centrifuge | Merck | Cat#EP5810000320 |
| Eppendorf 5424R Refrigerated Centrifuge | Marshall Scientific | Cat#EPP-5424R |
| ELMI RM-2L Intelli-mixer Large | ELMI | Cat#ELMI RM-2L |
| Parafilm | Merck | Cat#P7793 |
Step-by-step method details
Day 1: Isolation of primary mouse hepatocytes by liver perfusion
Timing: 45 min (per mouse)
This major step allows to obtain digested livers from mice. Following anesthesia, two surgical knots are performed in the abdominal and the thoracic cavities of the mouse; the heart is cannulated through the left atrium and the liver is perfused and digested in situ.
-
1.
Preheating: Warm the Perfusion Solution and the Digestion Solution in the water bath at 37°C. The water bath should be inside a laminar flow hood to assure sterile conditions of the solutions. Keep both solutions inside the bath during the whole isolation procedure.
CRITICAL: Collagenase must be added to the Digestion Solution immediately before the start of the isolation procedure. It must be completely dissolved and warmed before dissection to obtain the best digestion efficiency. We recommend to add the Collagenase before anesthetizing the mouse See troubleshooting 1.
-
2.
Anesthesia: Anesthetize the mouse according to your institution’s approved animal protocol; for example, through intraperitoneal injection of Ketamine/Xylazine (100 μL/ 10 g animal weight). Mice lose consciousness within 15 min after the injection.
Note: Pre-warm the anesthesia before injecting the mouse to avoid thermal shock.
Note: Confirm that the mouse is completely anesthetized before continuing with the step 3. Loss of the righting and palpebral reflexes, relaxation in the muscular tone, disappearance of pedal reflex and a decrease in the rate and depth of respiration are common readouts of successful anesthesia. If the mouse still maintains reflex 15 min after the injection, we recommend to additionally inject 50 μL of Ketamine/Xylazine.
-
3.
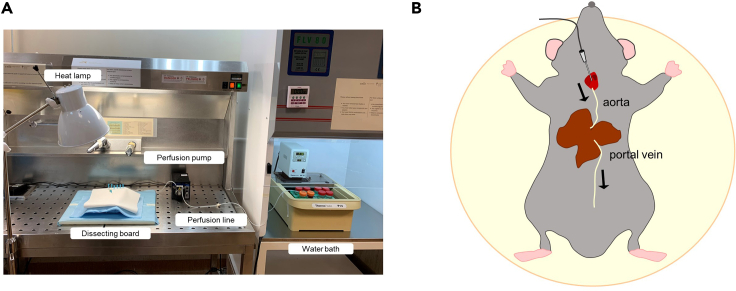
Preparation of the perfusion pump: Extensively wash the perfusion line with Rinsing Solution to eliminate 70% ethanol. Place one end of the perfusion line inside the Perfusion Solution located inside the water bath. Fill the perfusion line with Perfusion Solution before opening the mouse peritoneal cavity. Connect the outlet end of the perfusion line to the catheter (Figure 1A).
CRITICAL: Bubbles in the pumping tube can compromise digestion efficiency and viability of the isolated hepatocytes. See troubleshooting 2 for instructions on how to avoid the presence of bubbles.
-
4.
Positioning: Immobilize the anesthetized mouse by using needles to attach the upper and lower limbs on a dissecting board. Spray the mouse with 70% ethanol to ease dissection (Methods video S1 – part 1).
Figure 1.
Isolation of primary mouse hepatocytes by liver perfusion
(A) Working area for isolation of primary hepatocytes from mouse. Most relevant materials are indicated.
(B) Cartoon showing the direction of the non-recirculating circuit from aorta artery to portal vein.
Note: If primary hepatocytes from more than one mouse are going to be isolated on a day, anesthetize the second mouse at this step. The 15 min of anesthesia of the second mouse will fit with the 15 minutes of dissection, surgery, cannulation, perfusion, digestion and collection of the liver from the first mouse.
-
5.
Dissection: Using surgical scissors and forceps, open the abdominal cavity and carefully lean the small intestine to expose inferior vena cava and portal vein. With a syringe, add approximately 2 mL of Rising Solution into the abdominal cavity of the mouse to prevent contaminations (Methods video S1 – part 2).
Note: All the surgical material must be sterilized with 70% ethanol or by dry heating before use.
-
6.Surgery: For stable liver perfusion, perform two surgical knots in the abdominal and the thoracic cavities (Methods video S1 – part 3):
-
a.Abdominal knot: Take the needle and thread (Sterile surgical non-absorbable sutures) and pass the needle under the inferior cava vein and the left kidney (located at researcher’s right side). Pass the needle above the kidney located at your left and tie a loose knot.
-
b.Thoracic knot: Open the thoracic cavity vertically cutting the ribs on both sides. Heart must not be damaged, so slow and gentle scissor movements are needed. Remove the ribs to facilitate posterior cannulation. Pass the needle under the visible aorta artery above the liver and tie a loose knot.
-
a.
Note: Ensure the thoracic knot is over the end of the catheter. Otherwise, the circuit would be blocked.
-
7.Cannulation: Make an incision in the left atrium of the heart and cannulate.
-
a.Switch on the pump to begin a very slow perfusion (1 mL/min) of the mouse liver with Perfusion Solution. Secure the catheter with two needles.
-
b.Once the mouse is cannulated, tight the thoracic knot to secure the catheter above the liver; this allows the catheter to remain stable during posterior perfusion and digestion.
-
c.Then, tight the abdominal knot below the liver and cut inferior vena cava below the knot. Wait until the portal vein becomes distended, and then cut the portal vein (Methods video S2). At the end of this step a non-recirculating circuit from aorta artery to portal vein has been created (Figure 1B).
-
a.
CRITICAL: Proper cannulation of the aorta is crucial for a successful perfusion. Verify that no bubbles are introduced to avoid an incomplete digestion. See troubleshooting 2.
Note: Cannulation of the inferior cava vein instead of the left atrium for the isolation of primary mouse hepatocytes has been described in other protocols (Charni-Natan and Goldstein, 2020; Shen et al., 2021).
-
8.
Liver perfusion: Perfuse approximately 25 mL of Perfusion Solution at 5 mL/min of speed (Methods video S3 – part 1).
Note: If the surgery, cannulation and initial perfusion were performed correctly, the liver should turn pale. The Perfusion Solution should leave the circuit through the portal vein.
Optional: Add Rinsing Solution to the abdominal cavity of the mouse regularly to prevent contamination.
Optional: A soft massage of the liver while perfusing, favors posterior liver digestion.
-
9.
Liver digestion: Switch on the heat lamp facing the mouse liver. Switch the end of the perfusion line from the Perfusion Solution to the Digestion Solution and perfuse the liver with 25 mL of this solution at 3.5 mL/min (Methods video S3 – part 2).
Note: When switching the perfusion line from the Perfusion Solution to the Digestion Solution, briefly turn off the perfusion pump to avoid bubbles.
Note: The optimal pump speed depends on the diameter of the tube. In this protocol, pump speed was optimized according to the perfusion pump used (Masterflex C/L Analog Variable-Speed Pump with Dual-Channel Pump Head for Microbore Tubing Pump, 10–60 rpm; 12 VDC).
-
10.
Liver collection: Once the perfusion with the Digestion Solution has finished, collect the mouse liver carefully. Remove gallbladder and the leftovers of tissue from diaphragm. Transfer the liver to 25 mL of Hepatocyte Attachment Medium in a 50 mL centrifuge tube in ice (Methods video S3 – part 3).
Note: Carefully remove the liver avoiding puncturing/cutting the small intestine to prevent bacterial contamination.
CRITICAL: After digestion, liver texture should be soft and easily disaggregated by shaking the 50 mL centrifuge tube. If the liver has a dense-viscous texture, the perfusion failed and the yield for cell culture will be very low. See troubleshooting 3.
-
11.
Liver disaggregation: To favor release of hepatocytes, place the 50 mL tube containing the liver in a shaker (ROCKER 3D digital) for 5 min on ice.
Note: After finishing liver perfusion of one mouse, and before starting the procedure with another mouse, wash the perfusion line of the pump with Rinsing Solution to eliminate the rest of Collagenase from Digestion Solution.
Day 1: Collection and plating of primary mouse hepatocytes
Timing: 35–40 min (plus 6 h for incubation)
This major step allows to culture primary hepatocytes from the digested liver obtained in the previous step. Hepatocytes are filtered and separated by a density gradient; cells are counted and plated at a suitable confluence to perform the signaling experiment in the following step.
Note: All the steps of this section must be performed in a sterile cell culture hood (Methods video S4).
-
12.
Hepatocyte filtration: Filter the mouse liver through a 100 μm cell strainer transferring the sample to a clean 50 mL centrifuge tube. Favor liver disaggregation by gently smashing the liver with the plunger of a 10 mL syringe on top of the cell strainer. Use 10 mL extra of Hepatocyte Attachment Medium to clean the initial centrifuge tube and recover the possible remaining cells.
-
13.
Hepatocyte collection: Centrifuge the filtered hepatocytes at 50 ×g for 5 min at 20ºC–25ºC (Figure 2A). Aspirate the supernatant and resuspend the cell pellets with 25 mL of Hepatocyte Attachment Medium.
Note: Prepare the Gradient Solution while cells are centrifuging.
-
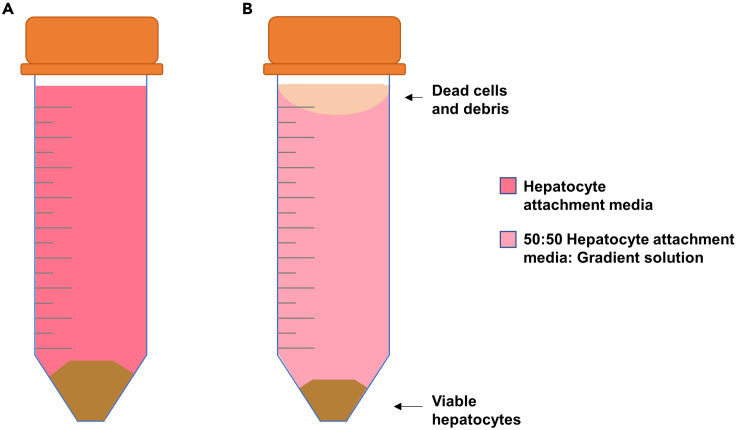
14.
Density-gradient separation: Add 25 mL of Gradient Solution slowly to the resuspended cells without mixing. Homogenize the solution by flipping the centrifuge tube 10 times. Centrifuge at 50 × g for 10 min at 20ºC–25ºC with deceleration. At the end of the centrifugation, viable hepatocytes will be pelleted and dead cells will remain in the upper fraction of the supernatant (Figure 2B).
CRITICAL: Double-check that the deceleration step is set in the centrifuge. Otherwise, the gradient will be lost and the hepatocyte pellet will be contaminated with dead cells.
Note: This step is an improvement over other protocols, in which viable hepatocytes are not separated from dead cells through a density gradient. Percoll-based separation results in higher viability of the isolated hepatocytes.
-
15.Hepatocyte clean-up: Centrifuge the hepatocytes to remove dead cells.
-
a.Aspirate the supernatant very carefully to remove dead cells and resuspend the pellet using 20 mL of Hepatocyte Attachment Medium. Centrifuge the cells at 50 × g for 5 min at 20ºC–25ºC.
-
b.Aspirate supernatant and resuspend the pellet with Hepatocyte Attachment Medium. In this step the volume for resuspending the hepatocytes depends on the pellet size.
-
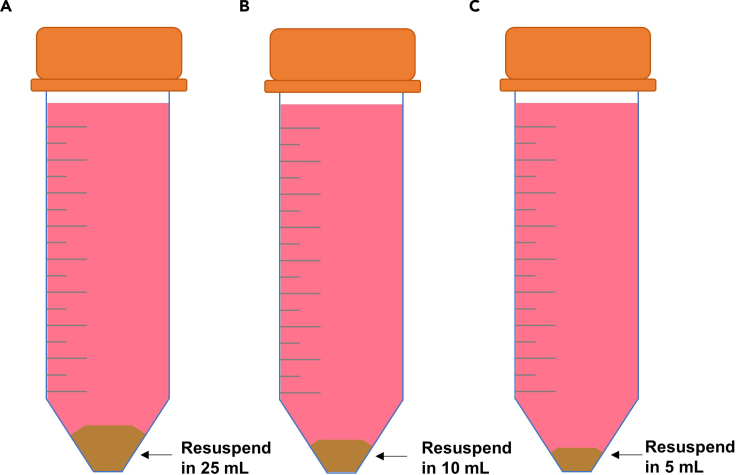
a.
Note: A successful isolation will require 25 mL of Hepatocyte Attachment Medium for resuspending the hepatocyte pellet (Figure 3A). If the yield of isolation is lower, volume can be decreased to 10 mL (Figure 3B) or 5 mL (Figure 3C) of Hepatocyte Attachment Medium.
Figure 2.
Hepatocyte collection and density-gradient separation
(A) Filtered hepatocytes collected from liver are centrifuged. The cartoon shows the pellet with total cells from liver.
(B) The pellet in (A) is resuspended in 50:50 Hepatocyte Attachment Medium: Gradient Solution and centrifuged. The cartoon shows the pellet containing viable hepatocytes; the supernatant is composed by dead cells and debris.
Figure 3.
Hepatocyte clean up
The volume of Hepatocyte Attachment Medium for resuspending hepatocytes depends on the size of the pellet obtained in step 15.
(A) A successful isolation requires a resuspension volume of 25 mL of Hepatocyte Attachment Medium.
(B) If the yield of isolation is lower, volume can be decreased to 10 mL.
(C) If the isolation was limited, the volume of resuspension should not exceed 5 mL.
-
16.
Assess hepatocyte viability: Gently homogenize the hepatocyte suspension by pipetting up and down with a 1000 μL pipette. Stain the hepatocytes with 10 μL of cell sample with 10 μL of Trypan Blue staining solution. Count both dead (dark blue) and live (translucent) cells using a TC20 Automated cell counter to calculate the survival rate. Alternatively, a classical hemocytometer could be used. Hepatocyte viability should be around 85%.
Note: Repeat the staining and cell counting twice or three times on each sample and average the numbers obtained. Variability in sampling can lead you to inaccurate quantification of the hepatocyte concentration and to differences in confluence upon plating.
-
17.
Hepatocyte plating: Plate 300,000 live hepatocytes per well in the collagen-coated 6 multi-well plates with 2 mL of Hepatocyte Attachment Medium. Replace the medium after 6 h and culture hepatocytes for another 16 h with Hepatocyte Attachment Medium before the signaling experiment.
CRITICAL: Confluence of hepatocytes must not be higher than 300,000 cells per well, because mTORC1 activity is affected by confluence (Figures 4A and 4B). See troubleshooting 4.
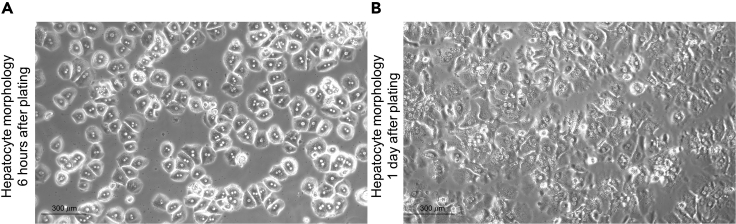
Figure 4.
Hepatocyte plating
Liver cells should be plated at a confluence of 300,000 hepatocytes per 3.5 cm well for signaling experiments.
(A) Within 6 h after hepatocyte plating, cells attach to the plate and acquire a round shape (Scale bar 300 μm).
(B) 16–20 h after hepatocyte plating, cells have acquired their hexagonal shape (Scale bar 300 μm).
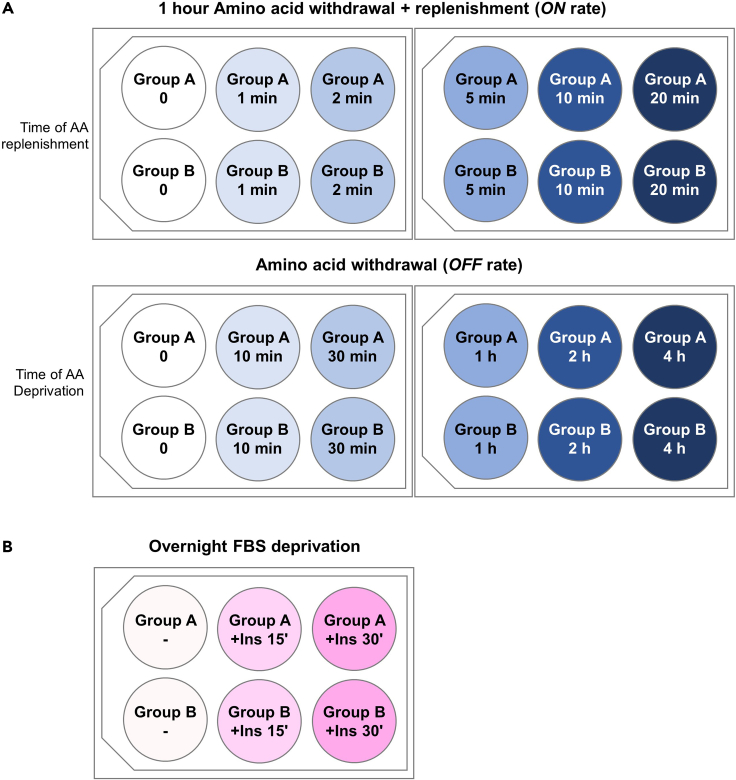
Note: If mTORC1 signaling will be determined in different conditions (e.g., mice of different genotypes, under different treatments, etc., we strongly recommend to plate all groups in the same plate. If so, upon processing the plate, different conditions would have simultaneous, equal handling (Figures 5A and 5B).
Figure 5.
Scheme of hepatocyte plating for signaling experiments
Groups A and B are shown as an example to distribute different experimental groups and to achieve equal handling.
(A) Amino acid deprivation of mouse primary hepatocytes. Top: mTOR ON rate. Down: mTOR OFF rate.
(B) Serum withdrawal + insulin stimulation of mouse primary hepatocytes.
Optional: Add one extra well in the plate as positive control for the signaling experiments. These hepatocytes will be cultured in complete Hepatocyte Attachment Medium during the whole experiment.
Note: Before changing the medium 6 hours after the plating, we recommend to wash each well with Rinsing Solution to prevent potential contaminations.
Day 2: Amino acid deprivation + replenishment of primary mouse hepatocytes
Timing: 1 h 45 min for ON rate; 4 h for OFF rate
This major step allows for a time-lapse assessment of nutrient-dependent activation of the mTOR signaling pathway through amino acid deprivation followed by replenishment.
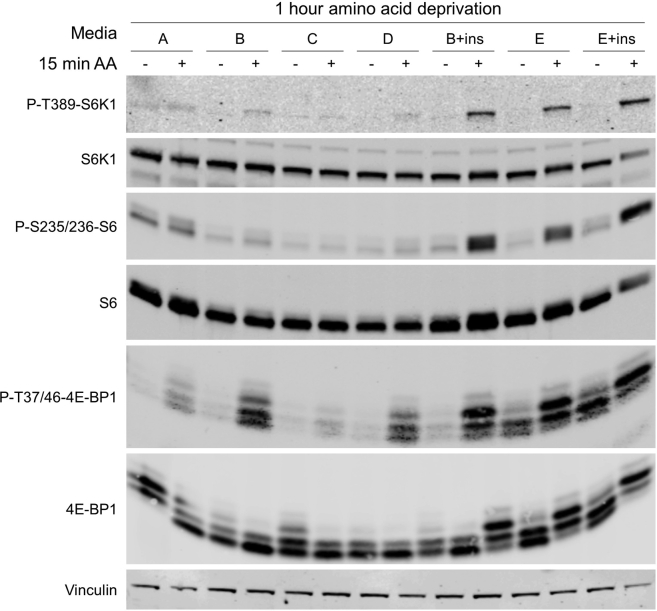
Note: We have previously performed nutrient signaling experiments on other primary cultures of mouse cells: Mouse Embryonic Fibroblasts (de la Calle Arregui et al., 2021) (Ortega-Molina et al., 2021) and naïve B lymphocytes (Ortega-Molina et al., 2019), using standard RPMI without amino acids, supplemented with dFBS. However, this medium resulted suboptimal for the assessment of mTORC1 activity in primary hepatocytes from adult mice: poor acute activation of mTORC1 was observed with this medium and with several iterations aimed to more closely resemble Hepatocyte Attachment Medium (Table 1 and Figure 6, See troubleshooting 5). In contrast, a wide dynamic range of activation/inhibition suitable for the assessment of nutrient-dependent activation of mTORC1 was obtained with Amino acid Starvation Medium (CELL MEDIUM E with insulin in Table 1).
Table 1.
Composition of cell culture media tested to optimize amino acid starvation on primary mouse hepatocytes
| Name | Medium | Supplements |
|---|---|---|
| CELL MEDIUM A | RPMI R8999-04A | Glucose, Penicillin/Streptamycin, Gentamycin, dFBS |
| CELL MEDIUM B | RPMI R8999-04A | Glucose, NaHCO3, BSA, Sodium Pyruvate, HEPES, Penicillin/Streptamycin, Gentamycin, dFBS |
| CELL MEDIUM C | RPMI R8999-04A | Glucose, HEPES, Penicillin/Streptamycin, Gentamycin, dFBS |
| CELL MEDIUM D | RPMI R8999-04A | Glucose, NaHCO3, Sodium Pyruvate, HEPES, Penicillin/Streptamycin, Gentamycin, dFBS |
| CELL MEDIUM B with insulin | RPMI R8999-04A | Glucose, NaHCO3, BSA, Sodium Pyruvate, HEPES, Insulin, Penicillin/Streptamycin, Gentamycin, dFBS |
| CELL MEDIUM E | DMEM/F12 D9807-10 | Glucose, NaHCO3, HEPES, dFBS |
| CELL MEDIUM E with insulin (Amino acid Starvation Medium) | DMEM/F12 D9807-10 | Glucose, NaHCO3, HEPES, Insulin, dFBS |
Figure 6.
Assessment of mTORC1 activity in the cell culture media tested and sub-optimal to perform amino acid starvation on primary mouse hepatocytes
DMEM/F12 is best suited to perform amino acid starvation on primary hepatocytes when supplemented with insulin (CELL MEDIUM E with Ins, Amino acid Starvation Medium).
-
18.
Hepatocyte washing: The next day, rinse cultured hepatocytes three times with Pre-starvation Medium. Aspirate thoroughly the medium in each wash to remove any leftover of the Hepatocyte Attachment Medium (containing non-dialyzed FBS and amino acids).
-
19.
Amino acid starvation: Add to each well 2 mL of Amino acid Starvation Medium. Return the plates to the incubator for 1 h.
Note: During this incubation prepare the Amino acid cocktail.
-
20.
Amino acid replenishment: 1 h after incubation of hepatocytes in the Amino acid Starvation Medium, add 200 μL of Amino acids cocktail, or 200 μL of Amino acid Starvation Medium (without amino acids) to control, non-replenished hepatocytes. In our experiments, we perform re-stimulation during 1, 2, 5, 10 and 20 min (Figure 5A top).
Note: Timing of plate processing is critical. Start the amino acid re-stimulation with the longest timepoint. In our experiment the longest replenishment is 20 min. 10 min after adding back amino acids in the “20 min” wells, we add back amino acids to the “10 min” wells. 5 min after this condition we add back amino acids to the “5 min” wells. 3 min after this condition we add back amino acids to the “2 min” wells. Finally, 1 min after this condition we add back amino acids to the “1 min” wells.
Optional: In case you have included an additional well for high mTORC1 activity, replace the medium 16 hours after plating, adding to each well 2 mL of Hepatocyte Attachment Medium and incubate for 1 hour.
-
21.Hepatocyte collection: Once the timepoints are finished, place the cell plates on ice.
-
a.Wash twice with cold PBS. Aspirate thoroughly the PBS in each wash for minimal PBS leftover volume.
-
b.Add 150 μL of cold, freshly prepared Protein Lysis Buffer to each well of the plate.
-
c.Scrape hepatocytes with cell lifters.
-
d.Collect the hepatocytes in 1.5 mL centrifuge tubes.
-
a.
Note: At this point the cells could be stored at −80°C, or proceed to protein extraction.
-
22.Protein extraction:
-
a.Incubate the samples at 4°C during 10 min with rotation in the ELMI Intelli Mixer.
-
b.Centrifuge at 4°C during 10 min at 21,130 g.
-
c.Transfer supernatant to a new 1.5 mL centrifuge tube.
-
a.
-
23.
Protein quantification: Quantify protein concentration with Pierce BCA method or an equivalent method.
Note: Protein concentration from isolated primary hepatocytes should be approximately 1.5 μg/μL.
-
24.
Western Blot: Load 8 μg of protein in each lane in a 16% polyacrylamide gel and proceed to run and transfer a standard Western Blot. Probe for levels of Phospho-Thr389-p70 S6 Kinase (1:500), total S6 Kinase (1:500), Phospho-Ser235/236-S6 (1:1000), total S6 (1:1000), Phospho-Thr37/46-4E-BP1 (1:500) and total 4E-BP1 (1:1000) See troubleshooting 5 (Figure 7A).
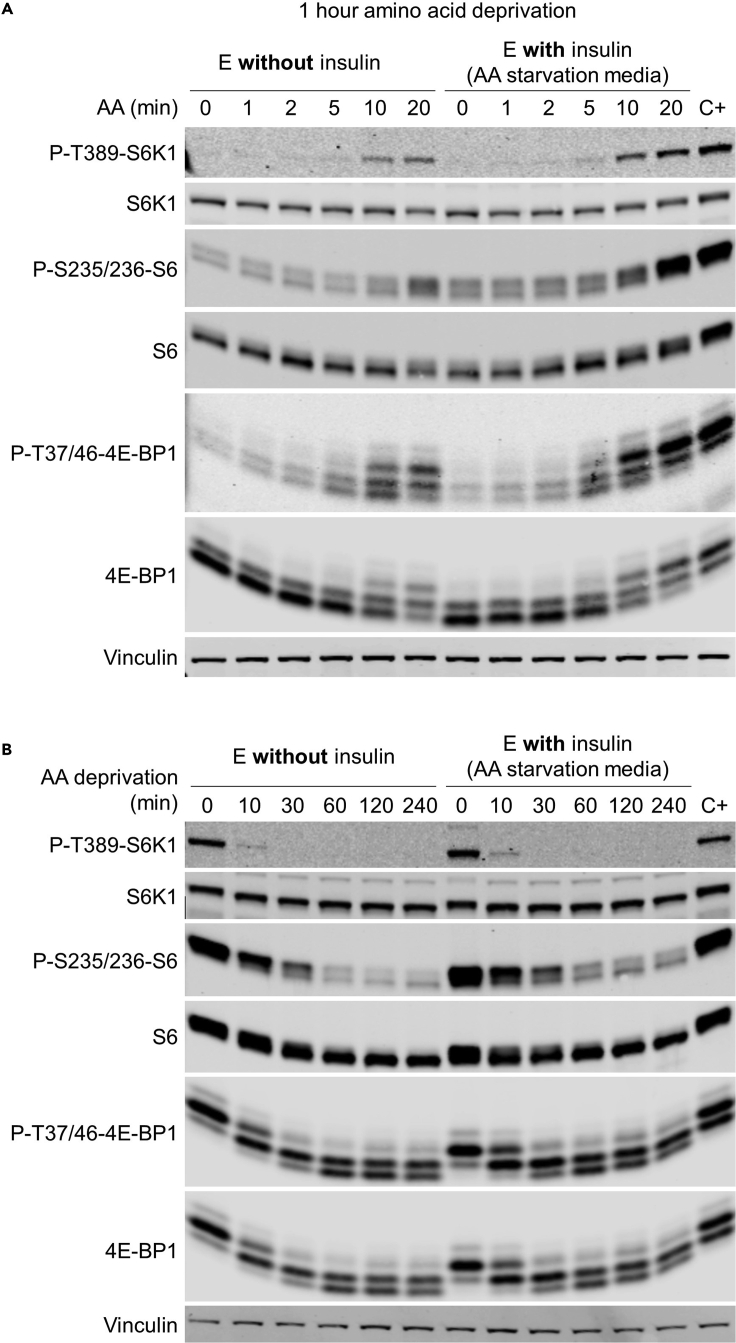
Optional: As an alternative to the assessment of the ON rate of amino acid-dependent mTOR activation, it is possible to assess the OFF rate. In such case, the experiment is performed identically until the step 19. Primary hepatocytes are deprived of all amino acids for 0 min, 10 min, 30 min, 1h, 2h and 4h (Figure 5A down, Figure 7B).
Figure 7.
Assessment of mTORC1 activity by amino acid deprivation/stimulation in DMEM/F12 medium without or with insulin
(A) mTORC1 ON rate.
(B) mTORC1 OFF rate.
Days 2 and 3: Serum withdrawal + insulin stimulation of primary mouse hepatocytes
Timing: 24 h
This major step allows to assess growth factor-dependent activation of the mTOR signaling pathway through withdrawal of FBS followed by a 10-min stimulation with insulin.
∗ Continue from step 17.
-
25.
Change medium: 16–24 h after plating, replace the medium (Hepatocyte Attachment Medium).
-
26.
Hepatocyte washing: 4 h after changing the medium, rinse cultured hepatocytes three times with FBS withdrawal Medium. Thoroughly aspirate the medium in each wash to remove any leftover of FBS from the Hepatocyte Attachment Medium.
-
27.
Serum withdrawal: Add 2 mL of FBS withdrawal Medium to each well. Return the plates to the incubator and leave for 16 h.
-
28.
Insulin re-stimulation: After 16 h, add 2 μL of 100 μM insulin in the corresponding wells during 15 or 30 min (Figure 5B). 10% FBS can be used instead of insulin for re-stimulation.
Optional: In case you have included an additional well for high mTORC1 activity, replace the medium 20 and 4 hours after plating, adding to each well 2 mL of Hepatocyte Attachment Medium (containing FBS).
-
29.Hepatocyte collection: After 15 or 30 min, place the cell plate on ice.
-
a.Wash twice with cold PBS. Aspirate thoroughly the PBS in each wash for minimal PBS leftover volume.
-
b.Add 150 μL of Protein Lysis Buffer to each well of the plate.
-
c.Collect the hepatocytes with cell lifters in 1.5 mL centrifuge tubes.
-
a.
Note: At this point the cells could be stored at −80°C, or proceed to protein extraction.
-
30.Protein extraction:
-
a.Incubate the samples at 4°C during 10 min with rotation in the ELMI Intelli Mixer.
-
b.Centrifuge at 4°C during 10 min at 21,130 g.
-
c.Transfer supernatant to a new 1.5 mL centrifuge tube.
-
a.
-
31.
Protein quantification: Quantify protein concentration with Pierce BCA method.
-
32.
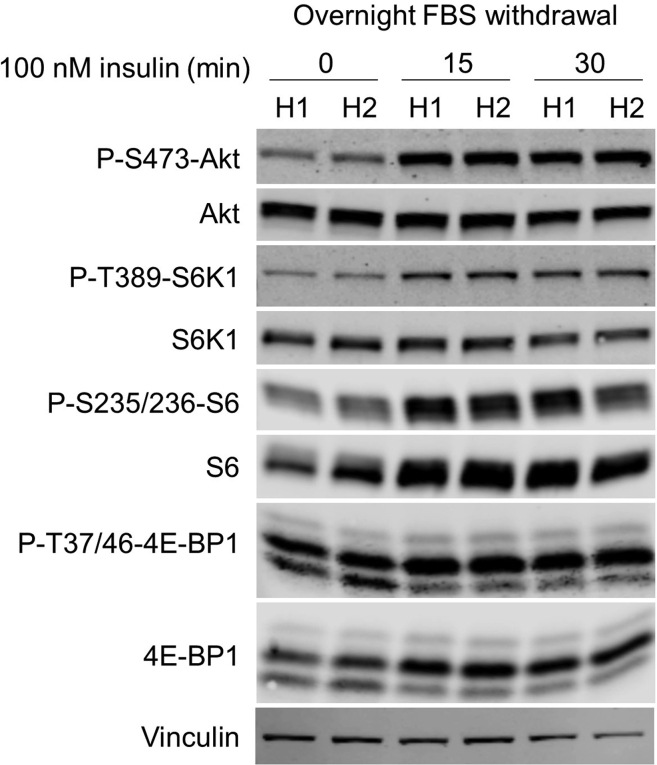
Western Blot: Load 8 μg of protein in each lane in a 16% polyacrylamide gel and proceed to run and transfer a standard Western Blot. Probe for levels of Phospho-Thr389-p70 S6 Kinase (1:500), total S6 Kinase (1:500), Phospho-Ser235/236-S6 (1:1000), total S6 (1:1000), Phospho-Thr37/46-4E-BP1 (1:500), total 4E-BP1 (1:1000), Phospho-Ser473-Akt (1:1000) and total Akt (1:1000) (Figure 8).
Figure 8.
Assessment of mTORC1 activity by serum withdrawal + insulin stimulation
Two different samples of hepatocytes (H1 & H2) are shown for each condition.
The isolated primary hepatocytes can be alternatively used for other molecular purposes independently of the assessment of mTOR activity. This protocol can be applied to assess the activity of additional signaling pathways, by exposing the primary hepatocytes to different experimental manipulations (chemical, physical and pharmacological). We have previously used this in vitro system to measure ex vivo cellular respiration (de la Calle Arregui et al., 2021), as well as to evaluate specific proteins through immunofluorescence, gene expression through RT-qPCR or whole transcriptome profiling, and metabolomic analysis.
Expected outcomes
A typical hepatocyte yield from a young C57BL/6 mouse is 25 × 106 cells with a viability of 85%. Yield of hepatocyte isolation in genetically-engineered mice may be lower, particularly if livers have excess fibrous tissue and/or hepatic damage. Hepatocytes adhere to the surface of the well 6 h after plating, looking sparsely plated, at the optimal confluence for performing signaling experiments (Figures 4A and 4B). Mouse primary hepatocytes do not survive in cell culture for more than 3 days, and trypsinization/replacing is not feasible. Hence, signaling experiments should be performed one or two days after cell plating.
mTOR signaling will be evaluated by the phosphorylation status of main targets: Phospho-Thr389-p70 S6 Kinase, Phospho-Ser235/236-S6 proteins and Phospho-Thr37/46-4E-BP1. Total protein levels will be used to normalize phosphorylation levels. In the amino acid deprivation protocol of mouse primary hepatocytes (ON rate), time 0, without re-stimulation, mTOR pathway should be off, revealed by a very faint band for Phospho-Thr389-p70-S6 Kinase and Phospho-Ser235/236-S6 (Figure 7A). One, 2, 5, 10 and 20 min after amino acid replenishment, a gradual increase in the signals of Phospho-Thr389-p70-S6 Kinase and Phospho-Ser235/236-S6 should be observed, reaching the maximum levels in the 20-min timepoint, together with a slight upwards shift. By 10 or 20 min after stimulation, mTORC1 activity should be similar to that of the positive control (C+) of hepatocytes cultured with Hepatocyte Attachment Medium (Figure 7A). During the same 1-to-20-min time lapse, Phospho-Thr37/46-4E-BP1 levels should also increase progressively. In addition to an increase in overall Phospho-Thr37/46-4E-BP1 bands intensity, the ratio of the upper (phosphorylated)-to-lower (unphosphorylated) bands should also gradually increase. This change in upper-to-lower ratio should also be appreciable in the membrane incubated with the antibody raised against total 4E-BP1 (Figure 7A) Seetroubleshooting 4 & 5.
In the amino acid deprivation protocol of mouse primary hepatocytes (OFF rate), similar results will be observed, with a gradual decrease in the signals of Phospho-Thr389-p70-S6 Kinase and Phospho-Ser235/236-S6, reaching the minimum levels already at 60-min timepoint (Figure 7B). During the same 0-to-240-min time lapse, Phospho-Thr37/46-4E-BP1 levels should also decrease progressively (Figure 7B).
In the FBS withdrawal of mouse primary hepatocytes (time - without re-stimulation) mTOR pathway should be off. 15 and 30 min of insulin stimulation will switch on the mTOR pathway, which should be observed as described for amino acid-replenishment (Figure 8).
Quantification and statistical analysis
Western Blot bands can be quantified using ImageJ software. The intensities of the phosphorylation levels of p70 S6 Kinase and Akt should be made relative to the total p70 S6 Kinase and total Akt, respectively, or to a loading control, such as β-actin or Vinculin. We discourage quantifying total S6 levels, and we always use another loading control for Phospho-Ser235/236-S6. For the quantification of phosphorylation levels of 4E-BP1, the upper-band-to-lower-band ratio can be quantified.
Limitations
If hepatocyte yield is low or cellular density is too low or too high, downstream signaling experiments may be compromised. Similarly, if the viability of the cells is not the suitable, signaling may not be reliable.
Troubleshooting
Problem 1
The cannulation is successful but liver is not properly digested.
Potential solution
Check that Collagenase in the Digestion Solution is completely dissolved. Temperature is a key factor in this process, confirm that the Digestion Solution is at 37°C during the whole perfusion procedure (Figure 1A). We recommend adding the Collagenase before anesthetizing the mouse to allow complete solution.
Problem 2
Bubbles appear in the tube of the perfusion pump before liver perfusion.
Potential solution
To remove bubbles from the tube, halt the perfusion pump before the bubbles reach the catheter connected to the mouse heart. Gently pull the pump tube back from the catheter, switch on the pump and allow Perfusion Solution to drain until all bubbles are eliminated. Before reconnecting the tube to the catheter, fill the catheter with perfusion buffer. It is critical to eliminate all bubbles to successfully perfuse the liver and to guarantee the viability of the resulting isolated hepatocytes.
Problem 3
Final liver texture after digestion is not soft and tissue is not easily disaggregated.
Potential solution
Before starting the digestion step, verify the perfusion flow ensuring that the Perfusion Solution is leaving the circuit through the portal vein.
Problem 4
High variability in mTOR signaling is observed between replicate samples.
Potential solution
Maintaining identical hepatocyte confluence across the wells is critical when performing these signaling experiments. Hepatocytes should not exceed 300,000 per 3.5 cm well (Figures 4A and 4B). We recommend to repeat the counting of dead and viable cells with Trypan Blue twice or three times per sample. In addition, simultaneous processing of all wells belonging to the same condition (e.g., time 1 min) is important for minimizing the potential of producing technical artifacts, particularly with the early time points. Replenishment of amino acids should be performed as fast as possible, as cells are removed from the incubators and transiently exposed to atmospheric concentration of CO2 and to room/hood temperature.
Problem 5
mTOR signaling in hepatocytes does not reach high levels of activation upon stimulation.
Potential solution
The levels of activation of mTOR pathway after 20 min of amino acid replenishment should be similar to those of the positive control hepatocytes (cultured with Hepatocyte Attachment Medium). (Figures 7A and 7B). We have seen that supplementation of the Amino acid Starvation Medium with insulin is important for robust mTOR activation after amino acid replenishment without undermining nutrient-dependent control on mTORC1 activity.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alejo Efeyan; aefeyan@cnio.es
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank CNIO Animal Facility for excellent technical support. Research was supported by the RETOS Projects Program of the Spanish Ministry of Science, Innovation and Universities, the Spanish State Research Agency (AEI/10.13039/ 501100011033) co-funded by the European Regional Development Fund (SAF2015-67538-R and PID2019-104012RB-I00), the EU-H2020 Program (ERC-2014-STG-638891), an Excellence Network Grant from MICIU/AEI (SAF2016-81975-REDT), a Ramon y Cajal Award from MICIU/AEI (RYC-2013-13546), a Spanish Association Against Cancer Research Scientific Foundation laboratory grant (LABAE16001EFEY/AECC), Beca de Investigación en Oncología Olivia Roddom, a FERO Grant for Research in Oncology (to A.E.), EFSD/Lilly European Diabetes Research Programme, MICIU (PID2019-104399RB-I00), Fundación AECC PROYE19047SABI, and Comunidad de Madrid IMMUNOTHERCAN-CM B2017/BMD-3733 (to G.S.). A.E. and G.S. are EMBO Young Investigators. A.B.P.-G., M.C., C.d.l.C.A., and L.d.P.-R. are recipients of Ayudas de contratos predoctorales para la formación de doctores from MICIU/AEI (BES-2017-081381, BES-2017-079711, BES-2015-073776, and PRE-2019-090891).
Author contributions
A.B.P.-G. performed and optimized the experiments with support from M.C., C.d.l.C.A., and L.d.P.-R. A.B.P.-G. wrote the manuscript and M.C., G.S., and A.E. edited the manuscript. A.E. and G.S. conceived and supervised the work.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100918.
Contributor Information
Ana Belén Plata-Gómez, Email: abplata@cnio.es.
Guadalupe Sabio, Email: guadalupe.sabio@cnic.es.
Alejo Efeyan, Email: aefeyan@cnio.es.
Data and code availability
This study did not generate any data set or code.
References
- Charni-Natan M., Goldstein I. Protocol for primary mouse hepatocyte isolation. STAR Protoc. 2020;1:100086. doi: 10.1016/j.xpro.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Calle Arregui C., Plata-Gómez A.B., Deleyto-Seldas N., García F., Abril-Garrido J., Rodriguez E., Nemazanyy I., Tribouillard L., Martino A., Caleiras E. Limited survival and impaired hepatic fasting metabolism in mice with constitutive Rag GTPase signaling. Nat. Commun. 2021;12:3660. doi: 10.1038/s41467-021-23857-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A., Deleyto-Seldas N., Carreras J., Sanz A., Lebrero-Fernández C., Menéndez C., Vandenberg A., Fernández-Ruiz B., Marín-Arraiza L., de la Calle Arregui C., Plata-Gómez A.B. Oncogenic Rag GTPase signalling enhances B cell activation and drives follicular lymphoma sensitive to pharmacological inhibition of mTOR. Nat. Metab. 2019;1:775–789. doi: 10.1038/s42255-019-0098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A., Lebrero-Fernández C., Sanz A., Deleyto-Seldas N., Plata-Gómez A.B., Menéndez C., Graña-Castro O., Caleiras E., Efeyan A. Inhibition of Rag GTPase Signaling in Mice Suppresses B Cell Responses and Lymphomagenesis with Minimal Detrimental trade-offs. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Liu W., Zuo J., Han J., Chao Z. Protocol for visualizing newly synthesized proteins in primary mouse hepatocytes. STAR Protoc. 2021;2:100616. doi: 10.1016/j.xpro.2021.100616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any data set or code.