Abstract
Type 2 diabetes is a multifactorial disease with multiple underlying aetiologies. To address this heterogeneity, investigators of a previous study clustered people with diabetes according to five diabetes subtypes. The aim of the current study is to investigate the etiology of these clusters by comparing their molecular signatures. In three independent cohorts, in total 15,940 individuals were clustered based on five clinical characteristics. In a subset, genetic (N = 12,828), metabolomic (N = 2,945), lipidomic (N = 2,593), and proteomic (N = 1,170) data were obtained in plasma. For each data type, each cluster was compared with the other four clusters as the reference. The insulin-resistant cluster showed the most distinct molecular signature, with higher branched-chain amino acid, diacylglycerol, and triacylglycerol levels and aberrant protein levels in plasma were enriched for proteins in the intracellular PI3K/Akt pathway. The obese cluster showed higher levels of cytokines. The mild diabetes cluster with high HDL showed the most beneficial molecular profile with effects opposite of those seen in the insulin-resistant cluster. This study shows that clustering people with type 2 diabetes can identify underlying molecular mechanisms related to pancreatic islets, liver, and adipose tissue metabolism. This provides novel biological insights into the diverse aetiological processes that would not be evident when type 2 diabetes is viewed as a homogeneous disease.
Introduction
Type 2 diabetes is a multifactorial disease with multiple underlying aetiologies (1,2). In an attempt to address this heterogeneity, investigators of a recent study stratified people with any form of diabetes into five clusters based on six clinical variables: age, GAD antibodies, BMI, HbA1c, insulin resistance (HOMA2 of insulin resistance), and β-cell function estimates (HOMA2 of β-cell function) (3). Based on this work, we clustered and cross validated individuals into five clusters in three large cohorts based on age, BMI, random or fasting C-peptide, HbA1c, and HDL, largely reproducing the All New Diabetics in Scania (ANDIS) clusters and using more readily measured clinical variables (4).
The results reported in the original and subsequent articles showed that people in different clusters had different risks for a number of diabetes-related outcomes (3,5–7). The autoimmunity and insulin-deficient cluster was defined by a high HbA1c at diagnosis, had ketoacidosis and retinopathy (7) more often, and progressed more rapidly onto insulin compared to the other clusters (3). The insulin-resistant cluster showed a higher frequency of nonalcoholic fatty liver disease, and people in this group were at increased risk of developing chronic kidney disease (3). The differences in progression and characteristics of the different clusters suggest that these groups represent different underlying aetiologies. For example, differences in genotype frequency across clusters based on candidate loci were observed and this was further illustrated in a follow-up study where it was shown that individuals in different clusters have differences in portioned polygenic risk scores for diabetes-related outcomes (3,8).
A systematic deconvolution of the different etiological processes underlying the clusters is currently lacking. To address this, we investigate each cluster’s molecular signature using metabolomics, lipidomics, proteomics, and genomics to better understand the underlying aetiological processes representative of patients with diabetes in that cluster.
Research Design and Methods
Cohort Descriptions
Data from 15,940 individuals from three cohorts, the Hoorn Diabetes Care System (DCS) (Netherlands), Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS) (Scotland), and ANDIS (Sweden), were used in this cross-sectional study. Inclusion criteria for IMI-RHAPSODY were age of diagnosis ≥35 years, clinical data available within 2 years after diagnosis, GAD−, no missing data in one of the five clinical variables used for clustering, and the presence of genome-wide association study (GWAS) data. Individuals were clustered using K-means clustering based on five clinical characteristics: age at sampling, BMI, HbA1c, HDL, and C-peptide. Of note, C-peptide was included in the clustering as a proxy of insulin resistance, while HDL has previously been recognized as a risk factor for time to insulin requirement. Details on the cohorts and clustering have previously been published (4). Briefly, DCS is an open prospective cohort that started in 1998 comprising >14,000 individuals with type 2 diabetes from the northwest part of the Netherlands (9). The Ethical Review Committee of the VU University Medical Center, Amsterdam, approved the study. People visit DCS annually as part of routine care. GoDARTS is a study comprising individuals with diabetes from the Tayside region of Scotland (N = 391,274; January 1996) who were added to the Diabetes Audit and Research in Tayside Scotland (DARTS) register (10). The GoDARTS study was approved by the Tayside Medical Ethics Committee. Longitudinal retrospective and prospective anonymized data were collected, including data on prescribing, biochemistry, and clinical data. In ANDIS, people were recruited with incident diabetes within Scania County, Sweden, from January 2008 to November 2016.
Molecular Measures
An overview of the sample selection procedure is given in Supplementary Fig. 1A. Individuals were selected based on the shortest time between diagnosis date and sampling date without taking into account cluster assignment. Analysis of small charged molecule analytes (metabolomics, ultrahigh-performance liquid chromatography–tandem mass spectrometry [UHLPC-MS/MS]) was performed in the largest set (N = 2,945), followed by lipidomics (N = 2,593 [Lipotype lipidomics platform; Lipotype, Dresden, Germany]) and proteomics (N = 1,170 [somascan Platform; SomaLogic]). Of note, the smaller sets were selected from the larger set based on the samples being collected closest to the time of diagnosis, so in the smallest set of 1,170, GWAS, metabolomics, lipidomics, and proteomics were available (Supplementary Fig. 1A). Molecular measures were taken close to diagnosis (Supplementary Table 2). Quality control (QC) was performed in a similar way for metabolomics, lipidomics, and proteomics. A participant’s data were excluded if their profile was a strong outlier based on principal components analysis and the data of the individual measurements was clearly distinct from the other samples.
Genetic Data
In DCS, genetic data were generated with the Illumina HumanCoreExome array. In GoDARTS genetic data were generated with the Affymetrix Genome-Wide Human SNP Array 6.0 and the Illumina HumanOmniExpress array. In ANDIS, genotyping was performed with InfiniumCoreExome-24 v1-1 BeadChip arrays (Illumina, San Diego, CA) at Lund University Diabetes Centre. Samples were excluded for ambiguous gender, call rate <95%, and any duplicate or related individuals (pi_hat ≥ 0.2). Single nucleotide polymorphisms (SNPs) were excluded for monomorphic SNPs, SNPs with a minor allele frequency (MAF) <0.05, and SNPs with missingness rate >0.05. Differences in diabetes-related genetic risk were based on 403 relatively independent diabetes-associated SNPs identified in a recent large GWAS meta-analysis (11). Genetic data were imputed using the Michigan Imputation Server against the reference panel Human Reference Consortium R1.1 with default settings, i.e., phasing with Eagle v2.3 and population of European descent (12). SNPs with minor allele frequency <5% were discarded from the analyses, leaving 394 SNPs across the three studies.
Metabolomics
Fifteen small charged molecules were measured in plasma with targeted UHLPC-MS/MS (Steno Diabetes Center) (13). In DCS, 1,267 individuals were included for metabolomics measurements. All passed QC, and data for 1,230 individuals overlapped with the cluster data. In GoDARTS, 898 individuals were included in the analysis; 1 failed QC, and among the 897 remaining individuals, data for 894 overlapped with the cluster data. In ANDIS, 896 individuals were included in the analysis; 4 failed QC, and of the 892 remaining samples, 821 overlapped with the cluster data.
Lipidomics
A total of 614 plasma lipids common to the three cohorts were determined with use of an Q Exactive mass spectrometer (Thermo Fisher Scientific) equipped with a TriVersa NanoMate ion source (Advion Biosciences) on the Lipotype lipidomics platform (14). Samples were divided into analytical batches of 84 samples each. Lipid identification was performed on unprocessed mass spectra files with LipotypeXplorer (15). Only lipid identifications with a signal-to-noise ratio >5, and a signal intensity fivefold higher than in corresponding blank samples, were considered for further data analysis. Batch correction was applied using eight reference samples per 96-well. Amounts were also corrected for analytical drift if the P value of the slope was <0.05 with an R2 >0.75 and the relative drift was >5%. In DCS, 900 individuals were included for lipidomics measurements; all passed QC, and data for 877 overlapped with the cluster data. In GoDARTS, 898 individuals were included in the analysis; 1 failed QC, and of the 897 remaining samples, 894 overlapped with one of the clusters. In ANDIS, 896 individuals were included in the analysis; 5 failed QC, and of the 891 remaining samples, 820 overlapped with one of the clusters. Lipid nomenclature is used as previously described, and SwissLipids database identifiers are provided (16) (Supplementary Table 1). After QC 162 lipid species were used in this study. The median coefficient of subspecies variation of the 162 lipids used as accessed by reference samples was 9.49% across all three cohorts.
Protein Measurements
Protein levels (1,195 proteins) in plasma were measured on the SomaLogic somascan platform (Boulder, CO) in 600 individuals each, for both DCS and GoDARTS. Individuals were removed if they were strong outliers based on a principal components analysis. In DCS, 600 individuals were included for proteomics measurements, 11 failed QC, and data for 573 overlapped with data for one of the clusters. In GoDARTS, 600 individuals were included in the analysis; 1 failed QC, and of the 599 remaining samples, 597 overlapped with one of the clusters.
Statistical Analysis
Molecular data were log transformed and z-scaled before analysis on a federated node system. Data for each of the cohorts were stored on a local node using Opal, an open-source data warehouse (Open Source Software for BioBanks [OBiBa]). A central node responsible for federated node access, user administration, and software deployment was set up at SIB Swiss Institute of Bioinformatics. Clinical and molecular data were harmonized according to the CDISC Study Data Tabulation Model (www.cdisc.org).
To identify molecular measures specific for a cluster, a generalized linear model was used to test each of the molecular measures in each cluster, where cluster i was compared against reference group j, where j was a combined group of the other clusters. Effect sizes represent change per log SD of the tested molecular measure. Genetic data were not transformed and represent change in allele frequency. For example, cluster 1 was compared with clusters 2–5 combined and cluster 2 was compared with clusters 1 and 3–5. Main results presented are based on an unadjusted model (log and z-scaled). Next, as an exploratory sensitivity analysis, models were adjusted for the extreme characteristic of a cluster to investigate whether the observed effect was independent of the extreme characteristic. This was only done for those clusters with extreme characteristics. Models were run on each of the cohorts separately and meta-analyzed with the R package meta (17). Meta-analyzed P values were adjusted with the Benjamini-Hochberg procedure, and a false discovery rate (FDR)-adjusted P value <0.05 was considered significant.
Partitioned polygenic risk scores (pPRS) were obtained from Udler et al. (18). In each individual cohort, dosages of SNPs were multiplied with the scores for each cluster, which resulted in a risk score per individual for each of the five clusters: β-cell (30 SNPs), proinsulin (7 SNPs), obesity (5 SNPs), lipodystrophy (20 SNPs), and liver (5 SNPs). Differences in pPRS were tested with a linear model for one cluster with the other clusters as the reference group. Next, results from the three cohorts were meta-analyzed with use of the metagen function from the meta package. P values were Bonferroni adjusted and considered significant at Padjusted < 0.05.
Pathway enrichment on the proteomics was performed based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with the use of the R package STRINGdb (1.24.0). The entire SomaLogic set (1,195 proteins) was used as the background set. P values of enriched pathways were adjusted using the Benjamini-Hochberg procedure, and an FDR-adjusted P value <0.05 was considered significant.
Effect sizes of proteins associated with estimated glomerular filtration rate (eGFR) and incident cardiovascular disease (CVD) were obtained from Yang et al. (2020) (19). Up- and downregulated proteins in each of the clusters (PFDR < 0.05) were selected. Effect sizes in the current study were compared to Yang et al. obtained 1) correlation coefficient of protein levels and eGFR and 2) hazard ratios (HRs) from the Cox proportional hazards models for CVD in individuals without chronic kidney disease (19).
Analyses were performed with R statistics (version 3.6.2). Figures were produced with the R packages ggplot2 (v3.3.0) and omicCircos (v1.22.0).
Data and Resource Availability
The data sets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author upon reasonable request.
Results
In this cross-sectional study, 15,940 individuals from three cohorts were included as previously described (4). We reproduced the original ANDIS severe insulin-deficient (SIDD), severe insulin resistance (SIRD), and mild obesity-related diabetes (MOD) clusters and refined the mild age-related diabetes (MARD) cluster into two: a subset with high HDL (MDH) and one without any particular defining features (MD). The characteristics of the clusters and those of the individuals used for molecular characterization (genetic data, metabolites, lipids, and proteins) are given in Supplementary Table 2 and Supplementary Table 3.
SIDD
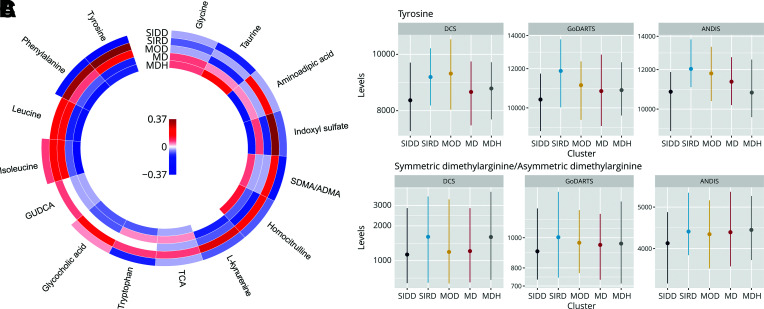
For SIDD, no differences were observed in allele frequency of known type 2 diabetes loci compared with the other clusters (Supplementary Table 4) or in the pPRS (Supplementary Fig. 2). Two metabolites, tyrosine (Fig. 1A and Fig. 1B) and asymmetric/symmetric dimethylarginine (Fig. 1C and Supplementary Table 5), were significantly lower in SIDD versus all other clusters. The effect sizes attenuated slightly after adjustment for the primary variable HbA1c that defined the SIDD cluster (Supplementary Fig. 3B and Supplementary Table 5). Of the lipids, eight were downregulated and one upregulated. Out of the eight downregulated lipids, three belonged to the sphingomyelin class, four belonged to the phosphatidylcholine (PC) class, and one was a cholesterol ester (Fig. 2A and Supplementary Table 6). The sole upregulated lipid was a cholesterol ester (CE 20:2;0). Seven of nine lipids remained significant after adjustment (Supplementary Fig. 3C and Supplementary Table 6). Finally, eight proteins were differentially expressed, with four up- and four downregulated (Supplementary Fig. 4A–D), where the effect sizes remained similar after adjustment (Supplementary Fig. 3D and Supplementary Table 7).
Figure 1.
Metabolite levels in the five clusters. A: Change in metabolites levels in each of the clusters versus all others. Colors represent effect size in log SD; red, upregulation, and blue, downregulation. GUDCA, glycoursodeoxycholic acid; SDMA/ADMA, symmetric dimethylarginine/asymmetric dimethylarginine; TCA, taurocholic acid. B: Levels of tyrosine in DCS, GoDARTS, and ANDIS. SIDD and SIRD PFDR < 0.05. C: Levels of (a)symmetric dimethylarginine. SIDD and SIRD PFDR ≤ 0.05. Dots represent the median, and the vertical line represents the interquartile range.
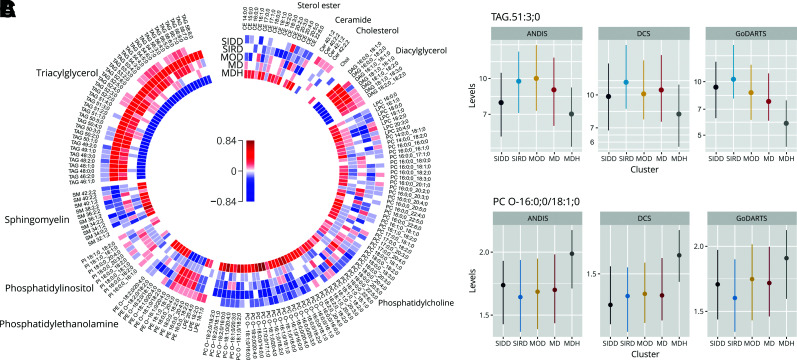
Figure 2.
Lipid levels in the five clusters. A: Change in lipid levels in each of the clusters versus all others. Colors represent effect size in log SD: red, upregulation, and blue, downregulation. B: Levels of TAG 51:3;0 in DCS, GoDARTS, and ANDIS. SIRD, MOD, and MDH PFDR ≤ 0.05. C: Levels of PC O-16:0;0/18:1;0. SIRD, MOD, and MDH PFDR ≤ 0.05. Dots represent the median, and the vertical line represents the interquartile range.
SIRD
The SIRD cluster was characterized by a strong and distinct molecular signature of insulin resistance. The pPRS for β-cell function and proinsulin (18) were decreased in the SIRD cluster relative to other clusters (β-cell, β 1.41 [95% CI −2.21 to −0.62]; proinsulin, −0.28 [−0.41 to −0.15]) (Supplementary Fig. 2), indicating genetically higher β-cell function in the SIRD group. Five diabetes-associated SNPs all showed a lower risk allele frequency. The top SNP (rs3802177-A) of SIRD mapped to the protective allele in SLC30A8 (Supplementary Table 4 and Table 1). In a sensitivity analysis, only the SLC30A8 variant remained significant after adjustment for C-peptide (Supplementary Fig. 3A, Supplementary Table 4, and Table 1). The SIRD cluster showed eight upregulated metabolites, including four amino acids, i.e., tyrosine, leucine, isoleucine, and phenylalanine (Fig. 1A and Supplementary Fig. 5A and B). Two were metabolites of the amino acid l-tryptophan, i.e., l-kynurenine and indoxyl sulfate. Adjustment for C-peptide attenuated the effect (Supplementary Fig. 3B and Supplementary Table 5).
Eighty-nine lipids were changed in SIRD, with 45 (50.6%) upregulated and 44 downregulated (49.4%) (Fig. 2A). Of the 45 upregulated lipids, 43 were in the diacylglycerol (DAG) and triacylglycerol (TAG) class, with TAG 51:3;0 as the strongest associating lipid (Fig. 2B and Supplementary Table 6), while the remaining two upregulated lipids were PCs containing the n-3 fatty acid docosahexaenoic acid (22:6;0, PC 18:0;0_22:6;0, PC 16:0;0_22:6;0). Of the 44 downregulated lipids, the most represented were the PC class (27 lipids [61.4%]), especially with the ether PCs (38.6%), with PC O-16:0;0/18:1;0 being the strongest downregulated lipid (Fig. 2A and C and Supplementary Table 6). Also, most ether phosphatidylethanolamines (four lipids [9.1%]) and sphingomyelin species (seven lipids [15.9%]) were downregulated. The changes in lipids seemed to be dependent on the high C-peptide levels, with effect sizes of DAGs and TAGs close to zero after adjustment for the latter (Supplementary Fig. 3C and Supplementary Table 6).
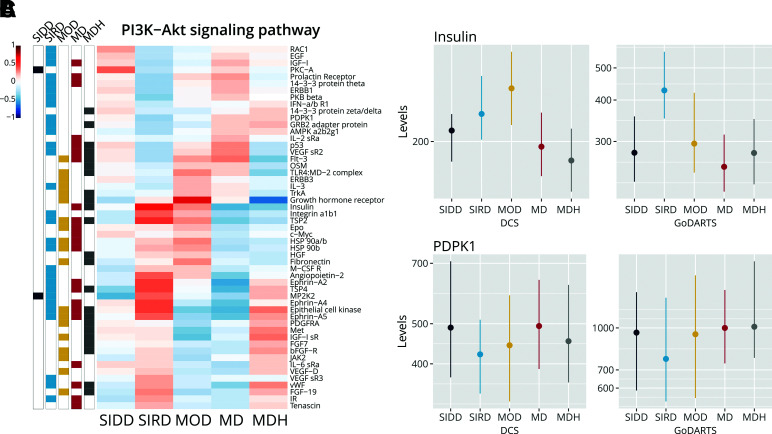
Of the 1,195 plasma proteins investigated, 367 proteins were differentially expressed, with 158 proteins downregulated and 209 upregulated. Several top proteins were upregulated independent of C-peptide levels, including two metalloproteinases, matrix metalloproteinase-7 (MMP-7) and MMP-12, and MIC-1 (Supplementary Table 7). Metalloproteinases are associated with multiple physiological processes but also with atherosclerosis and diabetes-related nephropathy (20,21). MIC-1 (GDF-15) is known to be associated with insulin resistance (22). The identified proteins showed a strong enrichment in pathways, including cytokine–cytokine receptor interaction (50 proteins, PFDR = 8.69 · 10−56), chemokine signaling pathway (26 proteins, PFDR = 1.81 · 10−34), Axon guidance (26 proteins, PFDR = 3.55 · 10−34) and PI3K-Akt signaling pathway (29 proteins, PFDR = 1.05 · 10−29). There was a significant reduction in 3-phosphoinositide–dependent protein kinase 1 (PDPK1) (Fig. 3A and C), which, when activated by insulin, activates Akt/PKB and increases glucose uptake via GLUT4 (23). Plasma Akt itself was also decreased in SIRD (Fig. 3A). Insulin tended to be higher in SIRD, although this was not significant (Fig. 3B), while the insulin receptor was significantly upregulated (Fig. 3A). In the downstream signaling cascade of the PI3K-Akt pathway, PDPK1 (Fig. 3C), RAC1, AMPK, HSP90, 14-3-3, and p53 were differentially expressed (Supplementary Fig. 5C–I). Of note, the proteins associated with SIRD were only modestly driven by C-peptide levels (Supplementary Fig. 3D).
Figure 3.
Proteins in the PI3K/Akt pathway in the five clusters. A: Effect sizes of proteins in the PI3K/Akt pathway (PFDR = 1.05 · 10−29) with upregulation (red) in the cluster vs. all others and downregulation (blue). Bars on the left indicate whether proteins are statistically significant in a specific cluster. Dots represent the median, and the vertical line represents the interquartile range. B: Levels of insulin in DCS, GoDARTS, and ANDIS. MDH PFDR ≤ 0.05. C: Levels of PDPK1. Dots represent the median, and the vertical line represents the interquartile range. SIRD PFDR ≤ 0.05.
Next, we overlapped identified proteins with those previously associated with eGFR and incident CVD (19). Proteins upregulated in SIRD were previously associated with lower eGFR levels, including cystatin C (ρ = −0.74, P = 1.12 · 10−163), tumor necrosis factor receptor superfamily member 1A (TNF sR-I) (ρ = −0.65, P = 2.51 · 10−114), and neuroblastoma suppressor of tumorigenicity 1 (DAN) (ρ = −0.64, P = 2.29 · 10−109) (Supplementary Fig. 7A). Conversely, proteins positively associated with eGFR were downregulated including epidermal growth factor receptor (ERBB1) (ρ = 0.44, P = 1.96 · 10−46) and α-2-antiplasmin (ρ = 0.41, P = 1.42 · 10−38). For incident CVD, angiopoietin-2 (HR 1.66, P = 2.20 · 10−16) and MMP-12 (HR 1.65, P = 2.20 · 10−16) were upregulated risk factors in SIRD, while ERBB1 (HR 0.59, P = 2.20 · 10−16) was protective for CVD and downregulated in SIRD (Supplementary Fig. 7B).
MOD
In MOD, the pPRS for obesity was significantly higher (β 0.51 [95% CI 0.34–0.68]) (Supplementary Fig. 2) compared with other clusters. Individual diabetes-associated risk alleles associated with high BMI were also more frequent in MOD, i.e., FTO (rs1421085-C) and the MC4R locus (rs523288-T) (Supplementary Table 4 and Table 1). Of note, both loci are also in the pPRS, although different SNPs in linkage disequilibrium. Naturally, adjustment for BMI attenuated the effect size for both SNPs (Supplementary Fig. 3A and Supplementary Table 4).
Isoleucine was the sole metabolite that was differentially upregulated in MOD (Fig. 1A, Supplementary Fig. 4A, and Supplementary Table 5), and this difference was completely eliminated after adjustment for BMI. The lipid profile of the MOD cluster was largely similar to the SIRD cluster (Fig. 2A and Supplementary Table 6); i.e., in MOD, acyl phosphatidylethanolamine species were upregulated, but not the ether phosphatidylethanolamines. Cholesterol esters and PC species containing the n-3 fatty acids eicosapentaenoic acid (20:5;0) and docosahexaenoic acid (22:6;0) were downregulated, while these were upregulated or not significantly changed in the SIRD cluster. However, cholesterol esters and PC species containing 20:3;0 fatty acids are upregulated in MOD while downregulated or not significantly changed in the SIRD cluster. In total, 61 lipids were affected, of which 40 were upregulated. Among these, the DAGs (15%) and TAGs (57.5%) were strongly enriched. Of the 21 downregulated lipids, the majority were PCs (61.9%). The effect size for DAG and TAG changes were strongly reduced after adjustment for BMI (Supplementary Fig. 3C and Supplementary Table 6). Interestingly, the largest effect size was seen in the TAGs with the lowest number of acyl chain carbons and double bonds (Supplementary Fig. 6A and B), while the TAGs with more acyl chain carbons and double bonds were not significantly altered in MOD. In a previous study, saturated or monounsaturated TAGs were associated with increased diabetes risk, including TAG 46:1, TAG 48:0, and TAG 48:1, which were also significantly upregulated in the MOD cluster (24).
Of the 1,195 proteins, 261 were differentially expressed in MOD with the majority downregulated (158 proteins [60.5%]) (Supplementary Table 7). After adjustment for BMI, several remained significant, although their effect sizes were attenuated, including NCAM-120, DKK3, and CRDL1 (Supplementary Fig. 3D and Supplementary Table 7). DKK3 has been associated with increased adipogenesis in fat cells (25). CRDL1 has been shown to be predictive of β-cell function (26). The role of NCAM-120 is largely unclear. The strongest enrichment was found for cytokine–cytokine receptor interaction, with 38 proteins (42.7%, PFDR = 2.08 · 10−43) overlapping (Supplementary Fig. 8). The strongest upregulated proteins in this pathway were leptin (Supplementary Fig. 4B), growth hormone receptor, and interleukin-1 receptor antagonist protein, while interleukin-1 receptor type 1 (IL-1 sRi) was downregulated. Adjustment for BMI influenced the effect size of several proteins, including leptin, FABP, and CRP (Supplementary Fig. 3D and Supplementary Table 7). Finally, upregulated proteins identified in MOD were generally positively associated with eGFR and protective for CVD, including the growth hormone receptor (HR 0.62, P = 2.20 · 10−16) (Supplementary Fig. 7).
MDH
The MDH cluster showed a higher pPRS relative to the other clusters for β-cell function (β 0.61 [95% CI 0.33–0.38]) (Supplementary Fig. 2). Among the diabetes-associated SNPs, a lower risk allele frequency was observed for a SNP near LPL (rs10096633-T) (Supplementary Table 4 and Supplementary Table 1). With respect to metabolite, lipid, and peptide levels the MDH cluster showed opposite effects compared with the SIRD and MOD cluster. The amino acids that were upregulated in SIRD were generally downregulated in MDH (Fig. 1A and Supplementary Table 5). Only the difference in isoleucine level was significant and phenylalanine borderline insignificant. In addition, taurine was significantly upregulated in MDH. After adjustment for HDL the effect sizes strongly attenuated (Supplementary Fig. 3B and Supplementary Table 5).
Of the 162 lipids, 135 lipids were affected in MDH, with 52 downregulated and 83 upregulated (Supplementary Table 6). Opposite SIRD and MOD, DAGs (13.5%), TAGs (73.1%), and acyl phosphatidylethanolamines (9.6%) were downregulated in MDH, while PCs (65.1%) were upregulated, especially the ether PCs (PC O-, 25.6%) (Supplementary Table 6). The TAGs with a smaller number of acyl chain carbons and double bonds showed the lowest protein levels versus the other clusters, while the differences attenuated with increasing number of acyl chain carbons and double bonds (Supplementary Fig. 6A and B). In addition, upregulation was seen for cholesterol esters (13.3%), sphingomyelins (10.8%), and all ether phosphatidylethanolamines (9.6%), which point in the opposite direction in the SIRD cluster (Supplementary Table 6). Adjustment for HDL strongly decreased the effect size for DAGs and TAGs (Supplementary Fig. 3C and Supplementary Table 6).
Of the 1,195 proteins, 270 were differentially expressed in the MDH cluster (119 down- and 151 upregulated). The effect size of the proteins changed very modestly after adjustment for HDL (Supplementary Fig. 3D). The peptide profile of the MDH cluster was opposite that of MOD (Supplementary Fig. 9 [r = −0.82]). As such, among the top proteins similar proteins were identified, such as CRDL1, that remained significant after adjustment for HDL. The pathway enrichment resembled that of SIRD and MOD, with enrichment for cytokine–cytokine receptor interaction (31 proteins, PFDR = 7.04 · 10−32), pathways in cancer (22 proteins, PFDR = 2.35 · 10−24), and the PI3K-Akt signaling pathway (22 proteins, PFDR = 5.56 · 10−23). In the PI3K-Akt signaling pathway, growth hormone receptor was downregulated, as well as insulin (Fig. 3B and Supplementary Table 7). Effect sizes were generally not solely driven by increased HDL levels (Supplementary Fig. 3D and Supplementary Table 7). MDH-associated proteins in relation to eGFR showed a pattern similar to that of SIRD (Supplementary Fig. 7A and B), with proteins associated with lower eGFR being upregulated as well as proteins associated with higher risk for CVD, the latter including Follistatin-related protein 3 (HR 1.55, P = 2.20 · 10−16) and HCC-1 (HR 1.54, P = 2.20 · 10−16).
MD
The MD cluster was generally less well-defined, with only one significant SNP and no significant pPRSs, lipids, or metabolites. There was a higher risk allele frequency (C allele) in MD—opposite that of MDH—compared with the other clusters near the LPL gene (rs10096633-T) (Supplementary Table 4). In contrast to the few signals for lipids or metabolites, 354 proteins were differentially expressed in the MD cluster, with the majority downregulated (209 proteins [59.0%]). Enrichment was found for Axon guidance (20 proteins, PFDR = 1.12 · 10−30), cytokine–cytokine receptor interaction (25 proteins, PFDR = 3.48 · 10−25), and PI3K-Akt signaling pathway (21 proteins, PFDR = 4.28 · 10−23). While similar pathways were found to be enriched in comparison with the SIRD cluster, the effect sizes were correlated but reversed (r = −0.88) (Supplementary Fig. 9). In line with this, insulin and its receptor were significantly downregulated in MD. Finally, in MD upregulated proteins were generally associated with better eGFR levels and lower risk for CVD (Supplementary Fig. 7A and B).
Discussion
Based on five clinical variables, people with type 2 diabetes from three large European cohorts were assigned to five separate clusters. The molecular phenotyping of the clusters revealed that, in addition to differences in clinical characteristics, there were also profound differences in underlying molecular profiles, which related to pancreatic islet biology (in SIDD), liver (in SIRD), and adipose tissue metabolism (in MOD and MDH).
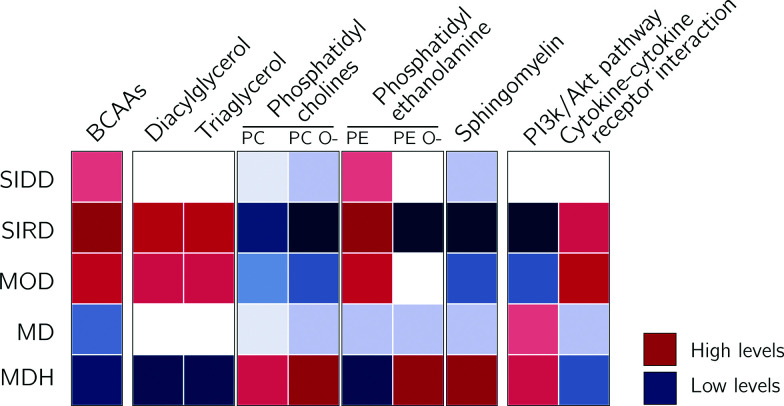
The SIRD cluster was characterized by a molecular profile that fits with insulin resistance, i.e., upregulation of DAGs, branched-chain amino acids (BCAAs), and insulin and downregulation of PI3K-Akt pathway–related proteins and PCs. The MOD cluster showed overlap with the SIRD cluster, but with a more pronounced molecular profile of obesity. Individuals in the MDH cluster showed the opposite effect of SIRD and MOD, with, relative to the other clusters, low levels of TAG, DAG, and BCAAs but higher levels of ether PCs and phosphatidylethanolamines, sphingomyelins, and cholesterol esters. The results were in part, but not fully, driven by the identifying characteristic of the cluster, except for SIDD, which showed consistent results after adjustment for HbA1c. For example, effect sizes of TAGs and DAGs in SIRD and MDH were influenced by adjustment for C-peptide and HDL, respectively. The lower frequency of diabetes-associated risk alleles could be explained by the fact that most diabetes SNPs are associated with reduced insulin secretion. People in the SIRD cluster have diabetes not because of lower insulin secretion but, rather, because of high insulin resistance (and consequent greater β-cell function).
The SIDD cluster was characterized by greater insulin sensitivity and lower β-cell function than the other clusters based on the clinical characteristics. SIDD was characterized by low tyrosine levels and (a)symmetric dimethylargine, CE 16:1;0, PC O-34;1, and PC O-34;2 compared with the other clusters; higher levels of these metabolites and lipids have been associated with higher type 2 diabetes risk (27–30). Higher CE 16:1;0 has also been associated with higher fasting plasma glucose and 2-h postloading glucose (31). Moreover, in SIDD, CRP was downregulated, and this is in line with a previous report that CRP levels are generally higher in those with insulin resistance and not low secretion (32).
The SIRD—and to some extent the MOD—cluster showed opposing metabolite, lipid, and protein profiles compared with the MDH cluster (Fig. 4). The SIRD cluster was characterized by a molecular signature compatible with insulin resistance inside cells. In SIRD, the frequency of protective alleles was higher for HOMA2 of β-cell function–associated variants. Evidence was found for downregulation of insulin-mediated glucose uptake across the different omics levels, where, for example, higher levels of BCAA and DAG/TAG were observed. BCAAs have been shown to be risk factors for developing incident type 2 diabetes in observational studies; their causal role has also been suggested (33). Both BCAAs and DAG inhibit insulin receptor substrate 1 (IRS1) (34). DAGs activate PKC isoforms, which inhibit PI3K activation by phosphorylating the inhibitory serine 307 of IRS1 instead of tyrosine (34,35). BCAAs target the intramuscular mammalian target of rapamycin/ribosomal protein S6 kinase β-1 (mTOR/p70S6K) signaling pathway, as shown in in vitro and rodent in vivo studies, which also inhibits the PI3K/Akt pathway via IRS1 and IRS2, depending on the cell type (34). Inhibition of PI3K/Akt reduces the GLUT4 translocation. In SIRD, multiple proteins were downregulated in PI3K/Akt and the GLUT4 translocation pathway, including Akt, PDPK1, and RAC1, while insulin was strongly upregulated (36,37). Furthermore, upregulation was seen in three ephrin family members (ephrin A2, A2, A5). Inhibition of the ephrin receptors has been shown to enhance glucose-stimulated insulin secretion in mice (38). Although these results might suggest changes in the insulin or glucose responsiveness of relevant metabolic tissues (e.g., muscle, liver, or adipose), proteins were measured in plasma in the current study and, as such, are unlikely to reflect changes in intracellular signaling. Future studies will be needed to determine the tissue(s) of origin of these biomarkers and the mechanisms through which they are released. For example, tissue-specific knockout of proteins identified in plasma in cell lines or model organisms might provide insight into both the role and tissue of origin. The higher BMI in individuals in the MOD cluster was in line with the higher allele frequency of variants associated with a higher BMI, i.e., variants near FTO and MC4R. Interestingly, variants near TM6SF2 were also associated with this cluster. TM6SF2 is known to be associated with nonalcoholic steatohepatitis (39). The metabolic and lipid profile of MOD resembled that of SIRD. An interesting observation was that the number of acyl chain carbons and double bonds was associated with the effect size in some clusters, in particular MOD and MDH. In MOD lipids with a higher number of acyl chain carbons and double bonds the effect size was much lower compared with those with lower numbers. These findings are in line with those of a previous publication, which showed that TAGs with a lower number of acyl chain carbons and double bonds were elevated in T2D case versus control subjects (24). In addition, lipids that were associated with increased diabetes risk were generally saturated or monounsaturated fatty acids (24). MOD was further characterized by upregulation of leptin, growth hormone receptor, and multiple interleukins and IL-1Ra. People with a high BMI have high levels of leptin, which may be a marker of leptin resistance (40). IL-1Ra is negatively correlated with QUICKI, where higher levels associate with higher insulin resistance (32).
Figure 4.
Schematic overview of the results in the current study. BCAAs, DAGs, TAGs, and phosphatidylethanolamine (PE) were upregulated in SIRD and to a lesser extend MOD, while being downregulated in MDH. PE O-, sphingomyelins, and proteins associated with the PI3K/Akt pathway were downregulated in SIRD. In MOD, proteins were found upregulated that have been associated with cytokine-cytokine interaction.
The MDH cluster was the cluster with the most beneficial profile and had a molecular signature of insulin sensitivity. This cluster had high HDL levels, low BCAA levels, low DAGs, and high levels of ether PCs relative to the other clusters (Fig. 4). Regarding the peptide level, the effects were opposite those of the MOD cluster. The MDH cluster displayed high levels of anti-inflammatory fatty acids, which have been associated with improved insulin sensitivity in animal studies (41–43).
In the study by Ahlqvist et al. (3), the SIRD cluster was associated with poorer renal function. In the current study we compare the identified proteins with proteins previously associated with eGFR levels and CVD risk (19). We show that proteins identified in the current study upregulated in the SIRD and MDH cluster are generally associated with lower eGFR levels and higher risk for CVD and, conversely, those downregulated in these two clusters are associated with higher eGFR levels and lower CVD risk. An explanation may be that individuals in the SIRD and MDH cluster are generally older compared with those in the other three clusters. These results also further confirm the added value of adding HDL to the clustering, as the MOD and MD cluster were much more alike than MD and MDH. The proteins upregulated in the MD and MOD cluster were associated with higher eGFR levels and lower CVD risk.
The strengths of the current study include the large number of individuals, the use of multiple cohorts, and the use of multiple molecular layers to characterize the clusters. A limitation is that the identified markers are measured in plasma and, as such, they cannot be directly linked to specific metabolic tissues. Second, while we adjusted models for the characteristic of that cluster to identify markers that were not simply proxies of the clinical features that defined the cluster, we cannot estimate whether we were able to fully adjust for that characteristic. Third, in the current study we compared the levels of molecular measures between individuals with type 2 diabetes and not relative to those of healthy control subjects. We therefore cannot infer which cluster would be closest to the general population. Fourth, we use a validated quantitative method to measure metabolites that have previously been linked to diabetes, but the limitation of this targeted method is that other metabolites are not measured. As such, we may have missed metabolites with differential levels across clusters. Finally, the cohorts used are mainly comprised of people of European descent and these results may not be generalizable to other populations.
Conclusion
In the current study, clusters were identified in three cohorts, based on five different clinical characteristics. The underlying molecular signatures of each cluster were markedly different (Fig. 4), suggesting different underlying etiopathological processes. As expected, the identified molecular signatures reflected the underlying phenotype to some extent but often remained associated after adjustment. Our study provides important new granularity on the likely molecular processes involved in diabetes pathology in each of the diabetes subgroups.
Article Information
Acknowledgments. The authors acknowledge the support of the Health Informatics Centre, University of Dundee, for managing and supplying the anonymized data.
Funding. This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement no. 115881 (IMI-RHAPSODY). This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation program and EFPIA. This work is supported by the Swiss State Secretariat for Education, Research and Innovation (SERI) under contract number 16.0097-2. E.R.P. was supported by a Wellcome Trust investigator award (102820/Z/13/Z). G.A.R. was supported by a Wellcome Trust Senior Investigator Award (WT098424AIA) and Investigator Award (212625/Z/18/Z), MRC program grants (MR/R022259/1, MR/J0003042/1, MR/L020149/1), and Diabetes UK project grants (BDA/11/0004210, BDA/15/0005275, BDA 16/0005485). This DCS study was in part supported by a grant from the Foundation for the National Institutes of Health through the Accelerating Medicines Partnership (no. HART17AMP).
The opinions expressed and arguments employed herein do not necessarily reflect the official views of these funding bodies.
Duality of Interest. K.S. is CEO of Lipotype. K.S. and C.K. are shareholders of Lipotype. M.J.G. is an employee of Lipotype. G.A.R. has received grant funding and consultancy fees from Sun Pharmaceuticals and Les Laboratoires Servier. M.K.H. is an employee of Janssen Research & Development, LLC. A.F. and I.P. are employees of Eli Lilly Regional Operations. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. R.C.S., L.A.D., J.W.J.B., L.M.’t.H., and E.R.P. designed the study and drafted the manuscript. R.C.S., L.A.D., H.F., G.A.B., and M.Å. performed the analyses. I.D., D.K., and M.I. set up a federated node system for data analysis. R.C.S., L.A.D., H.F., M.J.G., E.A., A.A., D.M.A., M.K., F.M., T.S., A.W., C.L.Q., and M.I. were involved in the data preprocessing and QC. G.N.G., A.F., M.K.H., D.M.A., I.P., T.J.P., V.L., L.G., B.T., P.W.F., and G.A.R. contributed to the data acquisition and project logistics. M.J.G., C.K., and K.S. generated the Lipotype data. A.A., T.S. A.W., P.R., and C.L.Q. generated the metabolomics data. All authors contributed to data interpretation. All authors critically revised the manuscript and approved the final version. R.C.S. and L.A.D. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
R.C.S., L.A.D., L.M.’t.H., and E.R.P. contributed equally.
This article contains supplementary material online at https://doi.org/10.2337/figshare.15113193.
References
- 1. McCarthy MI. Painting a new picture of personalised medicine for diabetes. Diabetologia 2017;60:793–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pearson ER. Type 2 diabetes: a multifaceted disease. Diabetologia 2019;62:1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahlqvist E, Storm P, Käräjämäki A, et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 2018;6:361–369 [DOI] [PubMed] [Google Scholar]
- 4. Slieker RC, Donnelly LA, Fitipaldi H, et al. Replication and cross-validation of T2D subtypes based on clinical variables: an IMI-RHAPSODY study. medRxiv 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dennis JM, Shields BM, Henley WE, Jones AG, Hattersley ATH. Disease progression and treatment response in data-driven subgroups of type 2 diabetes compared with models based on simple clinical features: an analysis using clinical trial data. Lancet Diabetes Endocrinol 2019;7:442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zaharia OP, Strassburger K, Strom A, et al.; German Diabetes Study Group . Risk of diabetes-associated diseases in subgroups of patients with recent-onset diabetes: a 5-year follow-up study. Lancet Diabetes Endocrinol 2019;7:684–694 [DOI] [PubMed] [Google Scholar]
- 7. Safai N, Ali A, Rossing P, Ridderstråle M. Stratification of type 2 diabetes based on routine clinical markers. Diabetes Res Clin Pract 2018;141:275–283 [DOI] [PubMed] [Google Scholar]
- 8. Aly DM, Dwivedi OP, Prasad RB, et al. Aetiological differences between novel subtypes of diabetes derived from genetic associations. 30 September 2020 [preprint]. medRxiv:2020.09.29.20203935 [Google Scholar]
- 9. van der Heijden AA, Rauh SP, Dekker JM, et al. The Hoorn Diabetes Care System (DCS) cohort. A prospective cohort of persons with type 2 diabetes treated in primary care in the Netherlands. BMJ Open 2017;7:e015599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hébert HL, Shepherd B, Milburn K, et al. Cohort profile: Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS). Int J Epidemiol 2018;47:380–381j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet 2016;48:1284–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahonen L, Jäntti S, Suvitaival T, et al. Targeted clinical metabolite profiling platform for the stratification of diabetic patients. Metabolites 2019;9:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Surma MA, Herzog R, Vasilj A, et al. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur J Lipid Sci Technol 2015;117:1540–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herzog R, Schwudke D, Schuhmann K, et al. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol 2011;12:R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aimo L, Liechti R, Hyka-Nouspikel N, et al. The SwissLipids knowledgebase for lipid biology. Bioinformatics 2015;31:2860–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schwarzer G. meta: an R package for meta-analysis. R News 2007;7:40–45 [Google Scholar]
- 18. Udler MS, Kim J, von Grotthuss M, et al. Clustering of type 2 diabetes genetic loci by multi-trait associations identifies disease mechanisms and subtypes: a soft clustering analysis. 2018;15:e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang J, Brody EN, Murthy AC, et al. Impact of kidney function on the blood proteome and on protein cardiovascular risk biomarkers in patients with stable coronary heart disease. J Am Heart Assoc 2020;9:e016463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu W, Wei R, Wang L, Lu J, Liu H, Zhang W. Correlations of MMP-1, MMP-3, and MMP-12 with the degree of atherosclerosis, plaque stability and cardiovascular and cerebrovascular events. Exp Ther Med 2018;15:1994–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McLennan SV, Kelly DJ, Schache M, et al. Advanced glycation end products decrease mesangial cell MMP-7: a role in matrix accumulation in diabetic nephropathy? Kidney Int 2007;72:481–488 [DOI] [PubMed] [Google Scholar]
- 22. Carstensen M, Herder C, Brunner EJ, et al. Macrophage inhibitory cytokine-1 is increased in individuals before type 2 diabetes diagnosis but is not an independent predictor of type 2 diabetes: the Whitehall II study. Eur J Endocrinol 2010;162:913–917 [DOI] [PubMed] [Google Scholar]
- 23. Beg M, Abdullah N, Thowfeik FS, Altorki NK, McGraw TE. Distinct Akt phosphorylation states are required for insulin regulated Glut4 and Glut1-mediated glucose uptake. eLife 2017;6:e26896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rhee EP, Cheng S, Larson MG, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest 2011;121:1402–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang X, Fu C, Liu W, et al. Chemerin-induced angiogenesis and adipogenesis in 3 T3-L1 preadipocytes is mediated by lncRNA Meg3 through regulating Dickkopf-3 by sponging miR-217. Toxicol Appl Pharmacol 2019;385:114815. [DOI] [PubMed] [Google Scholar]
- 26. Belongie KJ, Ferrannini E, Johnson K, Andrade-Gordon P, Hansen MK, Petrie JR. Identification of novel biomarkers to monitor β-cell function and enable early detection of type 2 diabetes risk. PLoS One 2017;12:e0182932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fiodorenko-Dumas Z, Dumas I, Mastej K, Adamiec R. Physical activity - related changes in ADMA and vWF levels in patients with type 2 diabetes: a preliminary study. Adv Clin Exp Med 2017;26:601–608 [DOI] [PubMed] [Google Scholar]
- 28. Yu E, Ruiz-Canela M, Razquin C, et al. Changes in arginine are inversely associated with type 2 diabetes: a case-cohort study in the PREDIMED trial. Diabetes Obes Metab 2019;21:397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruiz-Canela M, Guasch-Ferré M, Toledo E, et al. Plasma branched chain/aromatic amino acids, enriched Mediterranean diet and risk of type 2 diabetes: case-cohort study within the PREDIMED Trial. 2018;61:1560–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng Y, Ceglarek U, Huang T, et al. Weight-loss diets and 2-y changes in circulating amino acids in 2 randomized intervention trials. Am J Clin Nutr 2016;103:505–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meikle PJ, Wong G, Barlow CK, et al. Plasma lipid profiling shows similar associations with prediabetes and type 2 diabetes. PLoS One 2013;8:e74341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saltevo J, Laakso M, Jokelainen J, Keinänen-Kiukaanniemi S, Kumpusalo E, Vanhala M. Levels of adiponectin, C-reactive protein and interleukin-1 receptor antagonist are associated with insulin sensitivity: a population-based study. Diabetes Metab Res Rev 2008;24:378–383 [DOI] [PubMed] [Google Scholar]
- 33. Lotta LA, Scott RA, Sharp SJ, et al. Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. PLoS Med 2016;13:e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gancheva S, Jelenik T, Álvarez-Hernández E, Roden M. Interorgan metabolic crosstalk in human insulin resistance. Physiol Rev 2018;98:1371–1415 [DOI] [PubMed] [Google Scholar]
- 35. Li Y, Soos TJ, Li X, et al. Protein kinase C Theta inhibits insulin signaling by phosphorylating IRS1 at Ser(1101). J Biol Chem 2004;279:45304–45307 [DOI] [PubMed] [Google Scholar]
- 36. Sylow L, Nielsen IL, Kleinert M, et al. Rac1 governs exercise-stimulated glucose uptake in skeletal muscle through regulation of GLUT4 translocation in mice. J Physiol 2016;594:4997–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ueda S, Kitazawa S, Ishida K, et al. Crucial role of the small GTPase Rac1 in insulin-stimulated translocation of glucose transporter 4 to the mouse skeletal muscle sarcolemma. FASEB J 2010;24:2254–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jain R, Jain D, Liu Q, et al. Pharmacological inhibition of Eph receptors enhances glucose-stimulated insulin secretion from mouse and human pancreatic islets. Diabetologia 2013;56:1350–1355 [DOI] [PubMed] [Google Scholar]
- 39. Koo BK, Joo SK, Kim D, et al. Additive effects of PNPLA3 and TM6SF2 on the histological severity of non-alcoholic fatty liver disease. J Gastroenterol Hepatol 2018;33:1277–1285 [DOI] [PubMed] [Google Scholar]
- 40. Gruzdeva O, Borodkina D, Uchasova E, Dyleva Y, Barbarash O. Leptin resistance: underlying mechanisms and diagnosis. Diabetes Metab Syndr Obes 2019;12:191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lalia AZ, Lanza IR. Insulin-sensitizing effects of omega-3 fatty acids: lost in translation? Nutrients 2016;8:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oh DY, Talukdar S, Bae EJ, et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 2010;142:687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Storlien LH, Kraegen EW, Chisholm DJ, Ford GL, Bruce DG, Pascoe WS. Fish oil prevents insulin resistance induced by high-fat feeding in rats. Science 1987;237:885–888 [DOI] [PubMed] [Google Scholar]