Abstract
Retinoblastoma protein (Rb) plays important roles in cell cycle progression and cellular differentiation. It may also participate in M phase events, although heretofore only circumstantial evidence has suggested such involvement. Here we show that Rb interacts, through an IxCxE motif and specifically during G2/M phase, with hsHec1p, a protein essential for proper chromosome segregation. The interaction between Rb and hsHec1p was reconstituted in a yeast strain in which human hsHEC1 rescues the null mutation of scHEC1. Expression of Rb reduced chromosome segregation errors fivefold in yeast cells sustained by a temperature-sensitive (ts) hshec1-113 allele and enhanced the ability of wild-type hsHec1p to suppress lethality caused by a ts smc1 mutation. The interaction between Hec1p and Smc1p was important for the specific DNA-binding activity of Smc1p. Expression of Rb restored part of the inactivated function of hshec1-113p and thereby increased the DNA-binding activity of Smc1p. Rb thus increased the fidelity of chromosome segregation mediated by hsHec1p in a heterologous yeast system.
Genetic instability is one of the most important hallmarks of cancer. It occurs at two different levels. On one level, increased mutation rates result from defective repair of damaged DNA or replication errors, which leads to missense, nonsense, or other small but functionally important mutations in several types of cancer. On another level, improper segregation of whole chromosomes or pieces of chromosomes during mitosis leads to aneuploidy or translocations, traits commonly observed in cancers (35). Chromosome segregation is controlled by a large group of proteins that together coordinate M phase progression (43, 44, 48, 58). Loss of function of key proteins important for the structure and dynamics of mitotic chromosomes would be expected to lead to cell death and thus to prevent passage of mutations of such fundamental proteins to daughter cells. Loss of function of proteins that play subtler regulatory roles in mitosis, however, may not be immediately lethal but instead may lead to high frequencies of chromosome abnormalities and to neoplasia.
Associations of oncoproteins or tumor suppressors with the process of chromosome segregation provide possible links between carcinogenesis and chromosomal instability. Recent studies suggest that both p53 and retinoblastoma protein (Rb) play important roles in the prevention of aneuploidy in human and rodent cells (12, 28, 34, 56). When treated with microtubule-destabilizing agents, cells lacking functional Rb or p53 do not finish mitosis properly but nonetheless enter a new cell cycle, leading to hyperploidy (28, 34). p53 has been found to be associated with centrosome duplication activity (15) and mitotic or postmitotic checkpoint control (18, 34), loss of these functions would result in aberrant mitosis and contribute to the observed increase in ploidy. Similarly, the propensity of Rb-deficient cells to become hyperploid is most likely due to the loss of a novel function of Rb in M phase of the cell cycle, although supportive evidence remains scarce.
Study of the function of Rb has been centered on the progression of G1 phase (17, 23, 52). However, accumulating evidence has suggested potential functional roles for Rb during other phases of the cell cycle (27, 31, 46), especially during M phase. First, the functional, hypophosphorylated form of Rb is present at this phase of the cell cycle (8, 38). Second, hypophosphorylated Rb is associated with at least three cellular proteins that have crucial functions in M phase progression (50). One example is the human H-nuc2 (also called hCDC27) protein (9), a subunit of the anaphase-promoting complex that controls the onset of sister chromatid separation and metaphase-anaphase transition by degradation of specific substrates (30, 32). Another Rb-associated protein, protein phosphatase 1α catalytic subunit (13), is important for kinetochore function, chromosome segregation, and M phase progression, as demonstrated by the abnormal phenotype resulting from the mutational inactivation of its yeast homolog (2, 3, 49, 51). Lastly, mitosin (also called CENP-F), a kinetochore protein (60), also interacts specifically with Rb during M phase.
However, the mechanisms by which Rb plays a role in chromosome segregation and M phase progression remain elusive. The current approach of counting total chromosome numbers by karyotyping or detecting a specific chromosome by fluorescent in situ hybridization can only display the status of chromosome instability, which may not necessarily be a direct consequence of a certain gene defect in mammalian cells. To determine whether the loss of a gene function is responsible for improper chromosome segregation, a method for monitoring the dynamic transmission of a specific chromosome marker is required. Any attempt to select for mammalian cells carrying an integrated exogenous chromosome marker, however, bears the risk of immortalizing a primary cell line or making the genetic content of a tumor cell line even more unpredictable. Studies of chromosome segregation in mammalian cells are therefore complicated. On the other hand, methods for monitoring the dynamic transmission of chromosomes in yeast cells are feasible, and the genetic manipulation of a given gene in yeast can be accomplished without affecting the rest of the gene population and chromosome structures. Moreover, the basic machinery for chromosome segregation is conserved between mammals and yeast (42).
In this study, a yeast assay system for investigating the role of Rb in chromosome segregation was established, based on the study of hsHec1p. hsHec1p, isolated from a screen for proteins interacting with Rb (10, 13), is a coiled-coil protein crucial for proper mitosis (10, 11, 59). Inactivation of hsHec1p leads to disruption of M phase progression (10). The homolog of hsHec1p in Saccharomyces cerevisiae, scHec1p (also called Ndc80 or Tid3), has a similar essential function (57, 59), and hsHEC1 is able to rescue the lethality caused by the null mutation of scHEC1 (59). Yeast cells carrying a mutant allele of human or yeast HEC1 segregate their chromosomes aberrantly (57, 59). At the nonpermissive temperature, significant mitotic delay, unequal nuclear division, and decreased viability were observed in yeast cells carrying hshec1-113, a temperature-sensitive mutant allele of human HEC1 (59). Increased frequencies of chromosome segregation errors were also detected in the hshec1-113 mutant at permissive temperatures. Hec1p has been found to interact physically or genetically with a number of proteins important for G2/M progression and chromosome segregation, including SMC (structural maintenance of chromosomes) proteins and yeast centromere protein Ctf19p (10, 26, 59). A potential role for Hec1p in modulating chromosome segregation in part through interactions with SMC proteins has been suggested (59). There is no protein with sequence similarity to Rb in the entire S. cerevisiae genome. Without interference from endogenous Rb, yeast strains in which the null mutation of scHEC1 has been complemented by hsHEC1 (59) therefore provide useful tools to address the consequence of the interaction between Rb and hsHec1p for chromosome segregation.
The biological significance of the interaction between Rb and hsHec1p is demonstrated here by reconstitution of these proteins in a heterologous in vivo yeast system. Expression of Rb resulted in a decrease in the rate of chromosome segregation errors in cells carrying a mutant form of hsHec1p and an increase in the survival rate of smc1 mutant cells with defects in chromosome segregation. These results suggest that Rb plays a positive regulatory role in chromosome segregation.
MATERIALS AND METHODS
Strains and plasmids.
S. cerevisiae haploid and diploid strains carrying the hshec1-113 mutant have been described previously (59). A new yeast strain, 4bWHL273 (matx ade2 lys2 ura3 trp1 smc1-2::LEU2), is one of the meiotic segregates of the diploid strain from the mating between 3bAS273 (a gift from D. Koshland) and YPH1015 (a gift from P. Hieter). The full-length 2.8-kb RB cDNA, or the cDNA for the H209 mutant RB derivative (Cys706 changed to Phe), was inserted in two sets of plasmids, p415GAL1 (41) and pESC::TRP1 (Stratagene), by use of BamHI and SalI. The resultant plasmids were used to transform the above-mentioned strains. By a procedure described previously (21), cells were cultured in 2% raffinose overnight at 25°C before Rb expression was induced in medium containing additional 2% galactose for the indicated number of hours. The YEp195-GC15C plasmid was generated by inserting the GAL1 promoter, hsHEC1 cDNA (59), and CYC1 terminator (41) into the YEplac195 vector (16) for the expression of hsHec1p. The YEp195-GEKC plasmid was generated by site-directed mutagenesis for the expression of the hshec1-EK mutant (Glu234 changed to Lys). To express myc-tagged Smc1p, the full-length SMC1 was generated by PCR, sequenced, and fused with the c-myc tag in the pESC plasmid.
Immunoprecipitation and immunoblotting.
The preparation of yeast cell lysates, immunoprecipitation, and immunoblotting have been described previously (59). Human hsHec1p, S. cerevisiae Smc1p, and human Rb were precipitated or immunoblotted with anti-hsHec1p monoclonal antibody (MAb) 9G3, mouse anti-Smc1p antiserum (59), and anti-Rb MAb 11D7, respectively.
Human bladder carcinoma T24 cells were cultured and synchronized at different stages of the cell cycle as described previously (8, 10). Cells were lysed and immunoprecipitated by procedures described previously (8, 10).
For immunoprecipitation and immunoblotting of human SMC1 (hSMC1) from T24 cells, mouse anti-hSMC1 antiserum was obtained from mice immunized with glutathione S-transferase (GST) protein fused with the peptide region of hSMC1 isolated from a yeast two-hybrid screen (10, 11, 59).
Colony sectoring assays.
Colony sectoring assays were used to measure the frequencies of chromosome missegregation, as described previously (33, 59). Five single pink colonies of each diploid strain that contains a homozygous ade2-101 ochre color mutation and a dispensable chromosome fragment carrying a copy of SUP11 were picked and cultured to log phase in histidine-free supplemented minimal medium at 25°C for 3 days. Cells were diluted and incubated at 30°C for 4 h (one generation) in fresh medium supplied with histidine and containing 2% galactose and 2% raffinose to induce the expression of Rb or the H209 mutant. An aliquot of culture was then removed and plated on medium containing 6 mg of adenine per liter. The plates were incubated at 30°C for 6 days and at 4°C overnight before observation. The remaining cultures were used for detecting the expression of Rb or for examining the interaction between Rb and hsHec1p as described above.
Immunoaffinity purification.
Yeast cell lysate was prepared as described previously (59). Smc1p was partially purified from this lysate with mouse anti-Smc1p polyclonal antibodies by immunoaffinity chromatography, according to modification of a procedure described previously (25, 29). Antibodies were incubated with 50 μl of protein A-Sepharose beads for 2 h at 4°C and washed twice with 1 ml of Tris-buffered saline (50 mM Tris [pH 8.0], 125 mM NaCl). A 1.5-ml portion of cell lysate (about 50 mg of total protein) was added to the antibody–protein A-Sepharose beads and incubated for another 1 h at 4°C. The mixture was then loaded on a minicolumn and washed sequentially with 4 ml of XBE2 buffer (20 mM potassium HEPES [pH 7.7], 0.1 M KCl, 10% glycerol, 2 mM MgCl2, 5 mM EGTA), 0.5 ml of XBE2 with 0.4 M KCl, and 0.5 ml of XBE2. For elution, 150 μl of XBE2 containing a 4-μg/μl concentration of a GST fusion with the C-terminal region of Smc1p (59) was used. Fifty microliters of elution buffer was first allowed to flow in, and then the other 100 μl was loaded. After incubation at 4°C for 4 h, the elution buffer was allowed to flow through and collected. The elution product was incubated with 100 μl of glutathione-Sepharose that was prewashed with XBE2 for 1 h at 4°C three times to completely remove the GST fusion protein.
For multiple samples in the same experiment, equal numbers of yeast cells that contained comparable amounts of total proteins were lysed. The cell lysates were added to the antibody–protein A-Sepharose beads for immunoaffinity purification as described above. The eluted products from each sample were calibrated with comparable protein concentrations for use in gel shift assays. To minimize the effect of any quantitative variations, the amount of Smc1p in each purified product was adjusted according to the immunoblotting results.
Gel mobility shift assay.
Approximately 2 μl of the purified product described above was incubated with the 230-bp M13 replicative-form (RF) DNA fragment in 20 μl of XBE2 buffer with 0.5 mg of bovine serum albumin per ml. The 230-bp M13 RF DNA fragment was digested from the same region of M13 genomic DNA described previously (1), although HindIII was used instead of EcoRI. The DNA fragment was end labeled with [α-32P]dCTP by the fill-in reaction with Klenow enzymes. DNA fragments labeled with 10,000 to 20,000 cpm were used as substrates. For competition, unlabeled 230-bp M13 fragment, a 220-bp pUC19 fragment digested with AvaII from a pUC19-derived vector (1), and a 240-bp CEN3 fragment (bp, 113925 to 114168) generated by PCR amplification from yeast genomic DNA with the primers described previously (40) were used. The DNA-protein reaction mixtures were loaded on a 5.5% acrylamide gel and run at 4°C in a buffer containing 20 mM HEPES [pH 7.5] and 0.1 mM EDTA. The results were quantified using a densitometer and ImageQuant v1.1 (Molecular Dynamics).
RESULTS
hsHec1p specifically interacts with Rb through the IxCxE motif.
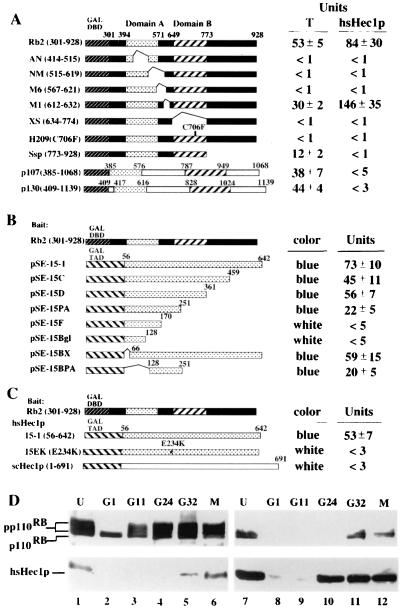
hsHec1p was originally identified using Rb as the bait in a yeast two-hybrid screen (10, 13). In order to determine the specific region of Rb required for binding hsHec1p, a deletion set that had previously been used to delineate the binding domain for protein phosphatase 1α was employed (13). Amino acids 301 to 928 of the Rb protein and several carboxy-terminal deletion mutants, as well as the H209 point mutant with residue 706 changed from cysteine to phenylalanine, were fused with the yeast Gal4 DNA-binding domain. Full-length hsHec1p protein was fused with the Gal4 transactivation domain. The results showed that Rb uses the same T-antigen-binding domain to interact with hsHec1p, and the H209 point mutation abolished this binding (Fig. 1A). hsHec1p sequences required for binding Rb were also determined in a reciprocal manner, using a series of hsHec1p deletion mutants. These mutants showed that the central region of hsHec1p, from amino acids 128 to 251, binds to Rb (Fig. 1B). Two Rb-related proteins, p107 and p130 (14, 20, 36, 39), did not interact with hsHec1p (Fig. 1A).
FIG. 1.
Specific interaction between Rb and hsHec1p. (A) hsHec1p and T antigen bind to similar regions of the Rb protein. The Gal4 DNA-binding domain (DBD) (amino acids 1 to 147; stippled box) was fused to various Rb mutants, p107 (amino acids 385 to 1068), or p130 (amino acids 409 to 1139). The simian virus 40 T-antigen-binding domains A and B are shown as shaded and hatched boxes, respectively. hsHec1p or T antigen (13) was expressed as a Gal4 transactivation domain fusion protein and used to test for interaction with Rb fusion proteins in yeast two-hybrid assays. Transformants were grown in liquid cultures and used for o-nitrophenyl-β-d-galactopyranoside quantitation of β-galactosidase activity as described previously (13). (B) Various hsHec1p mutants were fused with the Gal4 transactivation domain (TAD) (hatched box). Rb (amino acids 301 to 928) was expressed as the fusion with the Gal4 DNA-binding domain used for panel A. (C) Rb was expressed as the same fusion used for panel B. Wild-type hsHec1p (15-1), an hsHec1p mutant with amino acid 234 changed from E to K (15EK), and scHec1p were fused with the Gal4 transactivation domain. (D) Cell cycle-dependent interaction between Rb and hsHec1p. T24 cells were density arrested at G1 (lanes 2 and 8) and then released for reentry into the cell cycle. At different time points after release as indicated above the lanes, 5 × 106 cells were collected, lysed, and immunoprecipitated with anti-Rb MAb 11D7 (lanes 1 to 6) or with anti-hsHec1p MAb 9G3 (lanes 7 to 12). The immune complexes were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting with MAb 11D7 (upper panel) or with 9G3 (lower panel). G11 represents 11 h after release and corresponds to G1, G24 marks 24 h after release and corresponds to S, and G32 marks 32 h after release and corresponds to G2. M phase lysates (lanes 6 and 12) were obtained from cells treated with nocodazole (0.4 μg/ml). Lanes 1 and 7, unsynchronized cells.
The region of hsHec1p that binds Rb was not conserved in yeast scHec1p. Thus, it is likely that the interaction between Hec1p and Rb is not conserved in yeast. To test this notion, a yeast two-hybrid assay was performed using the above-described construct, with the Rb sequence fused with the Gal4 DNA-binding domain and a plasmid for the expression of yeast scHec1p sequence fused with the Gal4 transactivation domain. As predicted, yeast scHec1p failed to bind Rb (Fig. 1C).
An examination of the hsHec1p sequence showed that it contains an IxCxE motif, which has been implicated as the specific Rb-binding site in many proteins (reviewed in reference 5). This motif is not found in yeast scHec1p, suggesting that the inability of Rb to bind to scHec1p may be due to the lack of the IxCxE sequence. To verify this possibility, a point mutant with residue 234 changed from glutamic acid to lysine in this motif was tested in a yeast two-hybrid assay. This mutation abolished the ability of hsHec1p to bind to Rb (Fig. 1C). However, hshec1-EK, with this mutation, was able to rescue the yeast schec1 null mutant (data not shown) and generate the strain WHL101EK. This indicated that hsHec1p proteins, with or without an Rb-binding site, are able to perform their essential cellular function in yeast.
Rb and hsHec1p interact at G2/M phase in mammalian cells.
The interaction between Rb and hsHec1p was also examined by coimmunoprecipitation following cell cycle progression. As shown in Fig. 1D, hsHec1p binds to Rb specifically at G2/M phase in human bladder carcinoma T24 cells, which were synchronized as described previously (8). Similar to most of other Rb-associated proteins, hsHec1p binds specifically to the hypophosphorylated form of Rb that reappears during M phase.
The specific interaction between Rb and hsHec1p is reconstituted in yeast.
Wild-type Rb and the H209 mutant Rb were expressed under control of the GAL1 promoter through LEU2-selectable plasmids (p415GAL1) in the same yeast strain that carries an hshec1-113 mutant allele (59). As a negative control, wild-type Rb was also expressed in a yeast strain carrying the hshec1-113EK allele, which encodes a hshec1-113p without Rb-binding activity. The hshec1-113EK cells demonstrated no apparent difference in the temperature-sensitive (ts) phenotype compared with the hshec1-113 cells (59).
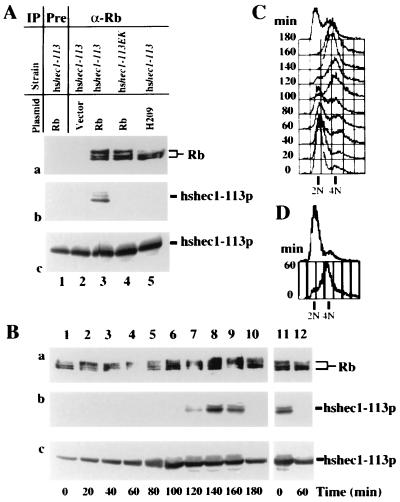
Wild-type Rb coimmunoprecipitated with hshec1-113p but not with hshec1-113EK. The H209 mutant of Rb failed to form a complex with hshec1-113p (Fig. 2A, panel b). Consistent with a previous report (21), both hypophosphorylated and hyperphosphorylated forms of Rb were detected in these unsynchronized cells, but the H209 mutant was deficient in hyperphosphorylated forms. The abundance of hshec1-113p protein did not vary significantly when either Rb or the H209 mutant was expressed (Fig. 2A, panel c). These results suggested that the specific interaction between hsHec1p and Rb could be reconstituted in yeast cells.
FIG. 2.
Reconstitution of the interaction between Rb and hsHec1p in yeast. (A) Specific interaction between hsHec1p and Rb. Yeast cells were diluted to an optical density at 600 nm of 0.75 in fresh medium with 2% galactose and 2% raffinose and then cultured at 30°C for 4 h. Aliquots of cell lysate were immunoprecipitated (IP) with nonspecific IgG (lane 1) or with anti-Rb (α-Rb) MAb 11D7 (lanes 2 to 5) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The same blot was probed with MAb 11D7 for Rb (a) or MAb 9G3 for hshec1-113p (b). Panel c shows the endogenous level of hshec1-113p in the cells used in panels a and b. For each lane in panel c, aliquots of the same lysates used in panel a were immunoprecipitated and immunoblotted with 9G3. The yeast strains and plasmids used to express Rb are indicated for each lane. (B) The yeast cells carrying the hshec1-113 allele and an Rb expression vector were treated for 5 h with 0.1 M hydroxyurea or 20 μg of nocodazole per ml in medium containing 2% galactose and 2% raffinose. At different time points after release (indicated under each lane [lanes 1 to 10, release from hydroxyurea; lanes 11 and 12, release from nocodazole]), cells were collected, lysed, and immunoprecipitated with α-Rb MAb 11D7. The immunoprecipitates were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting with 11D7 for Rb (a) or with 9G3 for hshec1-113p (b). Aliquots of the same lysates were immunoprecipitated and immunoblotted with 9G3 (c). hshec1-113p was co-precipitated by Rb specifically at 120 to 160 min after release from hydroxyurea treatment, corresponding to G2/M phase, or metaphase arrest by nocodazole (time zero, lane 11). (C and D) The DNA content of the same cells used for panel B was analyzed by fluorescence-activated cell sorting as described previously (59).
To explore whether M phase-specific binding exists in yeast cells expressing Rb, the interaction was examined during cell cycle progression. Cells from the strain carrying the hshec1-113 allele were induced to express Rb and then synchronized in early S phase by treatment with hydroxyurea. After release from treatment, an equal aliquot of cells was taken out every 20 min (Fig. 2B; lanes 1 to 10). hshec1-113p was coimmunoprecipitated by anti-Rb MAb in cells that entered M phase, according to DNA content analysis (Fig. 2C and D), and morphology was observed under the microscope. Similarly, hshec1-113p and Rb were coimmunoprecipited in cells synchronized at metaphase with nocodazole (Fig. 2B, lanes 11 and 12) but not in cells released from nocodazole treatment for 1 h. These results indicated that the M phase-specific interaction between Rb and hsHec1p can also be reconstituted in yeast cells.
Rb specifically enhances the fidelity of chromosome segregation.
The reconstitution of the specific interaction between Rb and hsHec1p in yeast cells provided an in vivo system for investigation of the consequence of this interaction. If hsHec1p plays a crucial role in maintaining the fidelity of chromosome segregation as described previously (59), we surmised that Rb modulates hsHec1p and enhances this activity. To test this hypothesis, we examined the rate of chromosome missegregation by using the colony sectoring assay (33) after induction of Rb expression in the hshec1-113 diploid strain. This strain was chosen because of its higher rate of chromosome segregation errors during mitosis (59). The total numbers of pink colonies (representing 1:1 segregation of a single dispensable chromosome fragment carried by this yeast strain), half-pink, half-red sectored colonies (representing 1:0 segregation), and half-white, half-red sectored colonies (representing 2:0 segregation) were counted. The rates of chromosome loss and nondisjunction in the first division were determined by the frequencies of half-pink, half-red colonies and half-white, half-red colonies, respectively. As shown in Table 1, Rb expression decreased the frequency of chromosome segregation errors due to chromosome loss or nondisjunction by approximately fivefold. In contrast, expression of the H209 mutant of Rb or the vector alone had no effect. As another control, we examined the influence of Rb expression on the strain carrying the hshec-113EK mutant; it had no significant effect.
TABLE 1.
Rb reduces chromosome segregation errors
| Relevant genotype | Plasmid | No. of coloniesa | Missegregation rate (%)
|
|
|---|---|---|---|---|
| 1:0 eventsb | 2:0 eventsc | |||
| hsHEC1 | 6,096 | 0.02 | 0.01 | |
| hshec1-113 | Vector | 6,687 | 1.47 | 0.56 |
| hshec1-113 | Rb | 12,641 | 0.31 | 0.10 |
| hshec1-113 | H209 | 4,299 | 1.54 | 0.60 |
| hshec1-113EK | Vector | 5,392 | 1.48 | 0.46 |
| hshec1-113EK | Rb | 6,554 | 1.51 | 0.50 |
Total number of pink colonies.
Number of half-red, half-pink colonies divided by total number of pink colonies.
Number of half-red, half-white colonies divided by total number of pink colonies.
The observed difference in the frequencies of chromosome missegregation is not due to the variable cell growth or cell cycle status, because expression of Rb or H209 has no significant effects on these processes in yeast cells carrying either wild-type HEC1 alleles (21) or the mutant hshec1-113 allele (data not shown). Therefore, the fidelity of chromosome segregation is enhanced specifically by interaction between Rb and hsHec1p.
Rb enhances the ability of hsHec1p to suppress lethality caused by an smc1 mutation.
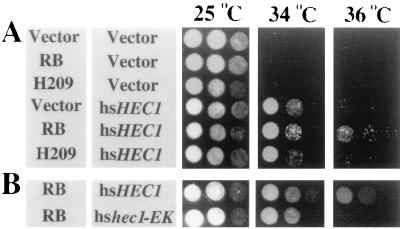
hsHec1p plays an essential role in chromosome segregation in part through interacting with SMC1 protein, which, in a complex with SMC3, is involved in sister chromatid cohesion (24, 37, 54). The mutated hec1p fails to interact with Smc1p physically in the hshec1-113 mutant cells at the nonpermissive temperature (59). Overexpression of Hec1p suppresses the lethal phenotype of the smc1-2 mutant strain (1-1bAS172) at 37°C (55, 59). If Rb enhances the activity of mutated hshec1p in the maintenance of proper chromosome segregation, it is likely that Rb also enhances the activity of wild-type hsHec1p in suppression of defective chromosome segregation due to the smc1 mutation. To test this hypothesis, we employed the yeast strain 2bAS273, which also carries the smc1-2 mutant allele and has a lethal phenotype at temperatures above 33°C (D. Koshland, personal communication). The isogenic strain 4bWHL273, carrying the same smc1-2 allele, was generated from 2bAS273 and transformed by plasmids expressing hsHec1p and Rb under control of the GAL1 promoter. Cells overexpressing hsHec1p grew at 34°C, a nonpermissive temperature for this smc1 mutant strain, while cells not overexpressing hsHec1p failed to grow, whether Rb was expressed or not (Fig. 3A). Interestingly, if the temperature was raised further to 35 to 36°C, cells overexpressing hsHec1p also failed to grow. Cells overexpressing both hsHec1p and Rb, however, continued to grow, whereas cells expressing the H209 mutant hardly survived at this higher temperature. Overexpression of hshec1-EK suppressed the smc1-2 mutant at 34°C, but coexpression of Rb and hshec1-EK did not suppress it if the temperature was raised further (Fig. 3B). These results suggested that the specific interaction between Rb and wild-type hsHec1p results in the enhancement of the fidelity of chromosome segregation, probably mediated by the interaction between Hec1p and Smc1p.
FIG. 3.
Rb suppresses the ts phenotype of the smc1-2 mutant through hsHec1p. (A) 4bWHL273 (smc1-2) cells were double transformed by hsHEC1 in a GAL1-inducible and URA3-selectable vector (a YEplac195-based vector), by RB or the H209 RB mutant cDNA in a GAL1-inducible and TRP1-selectable vector (pESC::TRP1), or by the vectors alone. (B) 4bWHL273 cells were double transformed by hsHEC1 or hshec1-EK in the GAL1-inducible and URA3-selectable vector and by Rb in the GAL1-inducible and TRP1-selectable vector. Different dilutions of log-phase cells grown at 25°C were inoculated on three plates with 2% galactose in the same manner and incubated at 25, 34, or 36°C.
Specific binding of Smc1p to highly structured DNA.
SMC1 protein has been suggested to associate preferentially with highly structured DNA regions of chromatin, such as AT-rich DNA, bent DNA, and scaffold-associated regions (1, 22, 24, 29), and to mediate intermolecular cross-linking in sister chromatid cohesion (24). An in vitro binding assay for investigation of SMC1 DNA-binding activity has been established by using a 230-bp M13 RF DNA fragment (bp 6001 to 6231), which has a very high potential to form secondary structures, e.g., stem-loops, and therefore mimics highly structured DNA regions (1). The carboxyl-terminal region of SMC1 protein had been shown to mediate this specific DNA-binding activity (1).
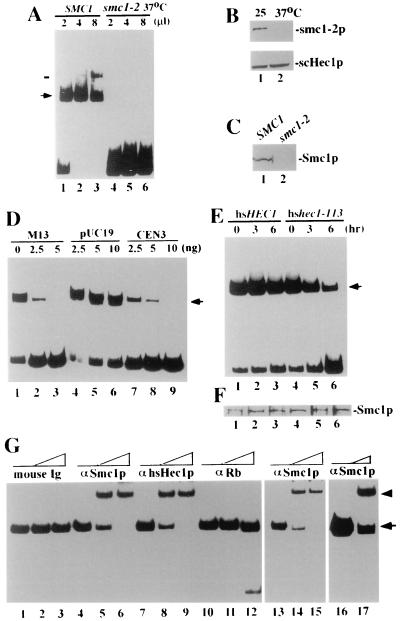
To partially purify the Smc1p protein complex from yeast cell lysates, anti-Smc1p polyclonal antibodies (59) and a single-step immunoaffinity approach (25, 29) were employed. The affinity-bound proteins were eluted by the use of a highly concentrated GST fusion protein that had been used as the antigen to raise the antibodies (59). Excessive GST fusion protein was subsequently removed with glutathione-Sepharose. The affinity-purified fraction (APF) was incubated with the 230-bp M13 RF DNA fragment. Specific DNA-binding activity for the APF was detected by gel mobility shift assays (Fig. 4A, lanes 1 to 3). The abundance of the specific DNA-protein complex increased when more APF was added. Meanwhile, the mobility of the DNA-protein complex decreased and formed a more slowly migrating band (Fig. 4A, lane 3, bar). This stoichiometric effect is consistent with previous observations for the DNA-binding activity of another SMC-containing complex, the 13S condensin in Xenopus (29). This DNA-protein complex is not likely to be contaminated by the eluting antigen, which, encompassing the C-terminal DNA-binding region of Smc1p (1), formed a faster-migrating complex with the same DNA substrate (data not shown).
FIG. 4.
DNA-binding activity of Smc1p and Hec1p complexes. (A) DNA-binding activity of Smc1p purified from equal numbers of wild-type cells (lanes 1 to 3) and smc1-2 ts mutant cells (lanes 4 to 6) with a 230-bp M13 RF DNA fragment. The amount of concentrated protein in each lane is shown, and the position of the DNA-protein complex is indicated (arrow). Note that a slower-mobility complex (bar) appeared when more protein was added. (B) Immunoblotting by mouse anti-Smc1p polyclonal antibodies (upper panel) or by anti-scHec1p polyclonal antibodies (lower panel) of lysates from smc1-2 ts mutant cells cultured at 25°C (lane 1) or 37°C (lane 2) for 6 h. (C) Immunoblotting of Smc1p purified from wild-type (lane 1) or smc1-2 mutant (lane 2) cells cultured at 37°C for 6 h. (D) Competition of DNA-binding activity by unlabeled DNA fragments. The amount of competitor DNA added in each reaction is indicated above each lane. (E) DNA-binding activity of Smc1p purified from wild-type hsHEC1 cells (lanes 1 to 3) or from the hshec1-113 mutant cells (lanes 4 to 6) with the 230-bp M13 fragment. Cells were cultured at 25°C until log-phase growth and then shifted to 37°C for 0, 3, and 6 h before harvest. (F) Comparable amounts of Smc1p in each of the APFs were measured by immunoblotting with anti-Smc1p antibodies and were used for panel E. (G) Antibody supershift assay. Anti-Smc1p (αSmc1p) antibodies and anti-hsHec1p MAb 9G3 supershifted the DNA-protein complex formed by APF from hshec1-113 cells expressing Rb (lanes 1 to 12), but mouse IgG or anti-Rb MAb 11D7 did not. Anti-Smc1p also supershifted the DNA-binding complex formed by APF from hshec1-113 cells not expressing Rb (lanes 13 to 15) and by APF from the wild-type hsHEC1 cells (lanes 16 and 17). Lanes 1, 4, 7, 10, 13, and 16, no antibodies; lanes 2 and 3, 0.5 and 1 μg of mouse IgG, respectively, lanes 5 and 6, 0.5 and 1 μg anti-Smc1p antibody, respectively; lanes 8 and 9, 0.5 and 1 μg of 9G3, respectively; lanes 11 and 12, 0.5 and 1 μg of 11D7, respectively; lanes 14 and 15, 0.5 and 1 μg of anti-Smc1p antibody, respectively; lane 17, 0.5 μg of anti-Smc1p antibody. The original shift is indicated by an arrow, and the antibody supershift is indicated by an arrowhead.
In order to determine the specificity of this DNA-binding activity, we also tested the APF from the smc1-2 mutant cells cultured at 37°C, with smc1p inactivated (55). Our observation suggested that this mutated protein is unstable and barely detectable in these mutant cells cultured for 6 h at 37°C (Fig. 4B). No Smc1p-containing complex was obtained from these cells using the same purification procedure (Fig. 4C), and therefore, no DNA-binding activity was detected (Fig. 4A, lanes 4 to 6).
To determine whether this Smc1p-associated activity is specific to the highly structured DNA, the DNA-binding activity was competed by unlabeled DNA fragments containing the scaffold-associated region of S. cerevisiae CEN3. This centromere region was suggested to be a preferential binding site of SMC proteins and was able to compete with the M13 fragment in the in vitro DNA-binding assay of recombinant SMC1 (1). As shown in Fig. 4D, the Smc1p-associated DNA-binding activity that we detected in the yeast cells can also be competed by unlabeled CEN3 DNA and M13 DNA fragment but not by the region on pUC19 DNA with the least potential to form the secondary structures (1).
We also used a similar procedure to partially purify myc-tagged Smc1p from yeast cells overexpressing myc-Smc1p by use of anti-c-myc MAb and elution with the corresponding peptide. myc-Smc1p has the same DNA-binding activity (data not shown).
Hec1p modulates specific DNA-binding activity of Smc1p.
To test whether a deficiency in Hec1p activity affects the function of Smc1p, we examined the activity of Smc1p in the hshec1-113 mutant yeast cells and compared it with that in cells expressing wild-type hsHec1p. Cells expressing wild-type hsHec1p or mutant hshec1-113p were cultured at the permissive temperature (25°C) and then shifted to 37°C for different periods of time. Equal numbers of cells were harvested and lysed. The resultant cell lysates from different samples contained comparable amounts of total proteins and were subjected to affinity purification of Smc1p. The DNA-binding activity of Smc1p in the wild-type cells did not change significantly after the cells were shifted to 37°C. In the hshec1-113 mutant cells, however, this Smc1p activity dramatically decreased, and only less than 20% remained after 6 h at 37°C (Fig. 4E). The amount of Smc1p expressed in the hshec1-113 cells was comparable to that in the wild-type cells (Fig. 4F), suggesting that the functional defect resulted specifically because of the mutated hec1p. The mobilities of the DNA-binding complexes from the wild-type cells and the mutant hshec1-113 cells were very similar; only if gel electrophoresis was prolonged more than usual could they be distinguished (data not shown). It is therefore likely that Hec1p is present in the DNA-binding complex. As shown in Fig. 4G, anti-Hec1p antibodies and anti-Smc1p antibodies, but not anti-Rb antibodies or mouse immunoglobulin G (IgG), were able to supershift the DNA-binding complex. These results suggest that Hec1p is present in the complex with Smc1p to mediate the DNA-binding activity. Similar results were observed with APF from yeast cells expressing the wild-type Hec1p or cells not expressing Rb (Fig. 4G, lanes 13 to 17).
Rb, through hsHec1p, enhances the DNA-binding activity of Smc1p.
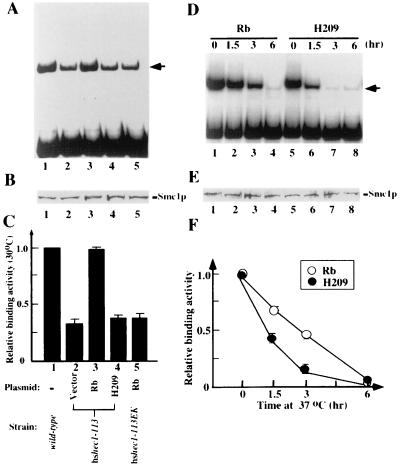
If Rb enhances the fidelity of chromosome segregation, which may be mediated by the DNA-binding activity of Smc1p through hsHec1p, this Smc1p activity should increase in the cells expressing Rb. To test this hypothesis, we examined the DNA-binding activity of Smc1p in the same Rb-reconstituted yeast strains that had been tested for frequencies of chromosome missegregation. As in the colony sectoring assay, the cells were cultured at 30°C for 8 h while either Rb or the H209 mutant was induced. In the hshec1-113 cells carrying only the empty vector, the DNA-binding activity of Smc1p decreased dramatically, to 30 to 40% of the wild-type level (Fig. 5A and C). These results are consistent with the abnormally high frequencies of chromosome missegregation in the same cells at the permissive temperature (Table 1). In cells expressing wild-type Rb, however, Smc1p DNA-binding activity was restored nearly to normal levels. Expression of the H209 mutant had no such effect, nor did wild-type Rb expression alone affect cells carrying the hshec1-113EK allele, which encodes a protein that cannot bind to Rb.
FIG. 5.
Rb enhances the DNA-binding activity of Smc1p through hsHec1p. (A) DNA-binding ability of Smc1p purified from equal numbers of cells expressing various forms of Rb and hsHec1p. Expression of Rb and the H209 mutant was induced by addition of 2% galactose to the medium, and cells were cultured at 30°C in this medium for 8 h before harvest. (B) Immunoblot showing that comparable amounts of Smc1p were detected in each of the affinity-purified products used for panel A. (C) Histogram showing relative binding activity of Smc1p in each lane of panel A. (D) DNA-binding ability of Smc1p purified from hshec1-113 cells expressing Rb (lanes 1 to 4) or the H209 mutant (lanes 5 to 8). Cells were cultured at 25°C in medium containing 2% galactose for 8 h to induce the expression of wild-type Rb or the H209 mutant and then shifted to 37°C for 0, 1.5, 3, or 6 h before harvest. (E) Immunoblot showing comparable amounts of Smc1p in each of the APFs used for panel D. (F) Effect of Rb on the DNA-binding activity of Smc1p in hshec1-113 cells. The relative DNA-binding activity of Smc1p indicates the ratio between the quantified density result of each lane in panel D and that of lane 1 for Rb or lane 5 for H209. Bars represent standard errors from three separate experiments.
To further corroborate this finding, the dynamic effect of Rb on the activity of hsHec1p was examined. hshec1-113 cells were cultured at 25°C in medium containing 2% galactose to induce the expression of Rb or the H209 mutant and then shifted to 37°C for different periods of time. As in the previous experiment (Fig. 4E), the DNA-binding activity of Smc1p began to decrease after cells were shifted to 37°C (Fig. 5D and F). This decrease of Smc1p activity, however, was significantly retarded during the first 3 h at 37°C in the cells expressing Rb compared with the cells expressing H209 mutant (Fig. 5F). By 6 h, Smc1p activity in both strains was very low. This result suggested that Rb can restore much of the activity impaired by mutation of hshec1p but cannot by itself complement the complete loss of hshec1p. Interestingly, Rb was not found in the Smc1p DNA-binding complex, since anti-Rb antibodies were not able to supershift the complex formed by the APF from hshec1-113 cells expressing wild-type Rb (Fig. 4G). Rb thus appears to function like a chaperone, consistent with a previous proposal (6).
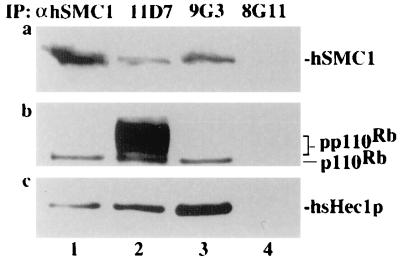
The above results suggest that a potential role of Rb in the modulation of SMC1 through hsHec1p exists and that both Hec1p and SMC1 proteins are functionally conserved from yeast to humans (24, 54, 59). It is therefore likely that Rb, Hec1p, and SMC1 may form a single complex in mammalian cells. To test this notion, hsHec1p and human SMC1 protein were coimmunoprecipitated with each other in human T24 cells (Fig. 6), consistent with our previous observation showing that the Hec1p-SMC1 interaction is conserved (59). Interestingly, the hypophosphorylated form of Rb was also coimmunoprecipitated. As a control, anti-GST MAb 8G11 did not coimmunoprecipitate any of these proteins (Fig. 6). These results suggest that Rb is present in a complex with Hec1p and SMC1 and support a potential role for Rb in modulating the activity of SMC1.
FIG. 6.
Rb, hsHec1p, and hSMC1 protein form a complex in human cells. Asynchronous fast-growing human T24 cells (6 × 106) were lysed and immunoprecipitated (IP) by mouse anti-hSMC1 (αhSMC1) antiserum (lane 1), anti-Rb MAb 11D7 (lane 2), anti-hsHec1p MAb 9G3 (lane 3), and anti-GST MAb 8G11 (lane 4). The immunocomplexes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting with anti-hSMC1 antiserum to detect human SMC1 (a), with 11D7 to detect Rb (b), and with 9G3 to detect hsHec1p (c).
DISCUSSION
In this study, we have employed a yeast system to address the function of Rb in chromosome segregation. Expression of Rb reduced chromosome segregation errors in cells carrying a mutant form of hsHec1p and enhanced the survival rate of smc1 mutant cells with defects in chromosome segregation. Complexes of Hec1p and Smc1p play essential roles in chromosome segregation. Rb appears to chaperone Hec1p and indirectly to enhance the DNA-binding activity of Smc1p. These results reveal a novel biological activity of Rb intimately linked to its role in carcinogenesis and cancer progression.
The lack of an Rb homolog in yeast allowed us to address Rb function using yeast machinery as a powerful assay tool, without interference from endogenous Rb. Mechanisms similar to those governing Rb phosphorylation in mammalian cells have been demonstrated in yeast (21). However, no significant differences in cell morphology, growth rate, cell cycle progression, or mating pheromone response were observed in yeast cells expressing human wild-type or mutant Rb. These results suggest that Rb does not exert a function when yeast lacks specific cellular mediators of the antiproliferation and differentiation functions of Rb during G1 phase. Alternatively, potential mediators of such functions in yeast are unable to interact with Rb; such is the case with yeast Hec1p, which has no Rb-binding motif. In either case, the lack of both Rb and mediators of Rb function in yeast made it possible to exploit the yeast cell as an assay system for chromosome segregation and to reconstitute the interaction between hsHec1p and Rb in this system. This assay system ensures that the observed phenomena are direct and specific consequences of Rb expression and are specifically mediated by hsHec1p.
Expression of Rb decreased the frequency of chromosome segregation errors fivefold but was insufficient alone to rescue the yeast cells completely from aberrant mitosis. The fivefold enhancement is likely reminiscent of the physiological effect from a high-level regulator on the basic machinery for chromosome segregation. This improvement in the fidelity of chromosome segregation, however, would be quite significant in higher organisms, considering the millions of cells undergoing mitosis or meiosis daily. In the case of a lack of functional Rb, chromosome segregation errors in mammalian cells are expected to occur at a frequency similar to that for the wild-type yeast. Apparently, a higher fidelity of chromosome segregation is required for higher organisms to avoid errors in the more complicated chromosome segregation.
The biochemical mechanisms by which Rb modulates the activity of Hec1p and by which Hec1p modulates the activity of Smc1p remain to be elucidated. Our studies of DNA-binding activity of Smc1p from cells with different genetic backgrounds have provided some important clues leading to the understanding of these biochemical mechanisms. The DNA-binding activity of SMC1 is suggested to serve as a biochemical basis for its function in the chromatin assembly essential for sister chromatid cohesion and chromosome segregation (24). The modulation of this activity will undoubtedly affect the biological function of SMC1 in chromosome segregation, although other functions of Smc1p may also be influenced. It has been suggested that SMC1 forms complexes with various proteins, most of which, however, have not been revealed (24, 54). Our results indicate that Hec1p is present in the DNA-binding complex of Smc1p and also suggest that Hec1p is important for the biochemical activity of this complex. Consistently, the interaction between Hec1p and Smc1p is critical for proper chromosome segregation (59). Although purified recombinant protein containing the C-terminal region of SMC1 was shown to have the DNA-binding activity (1), it is likely that SMC1 requires other cofactors, such as Hec1p, to enhance its activity for a more stable binding of structured DNA. Rb appears to enhance the DNA-binding activity of Smc1p through Hec1p. This positive regulatory effect of Rb has also been observed in a number of transcription factors, such as MyoD (47), the glucocorticoid receptor (53), C/EBPβ (6), NF-IL6 (7), and c-jun (45). Our results showing that Rb is not a component of the DNA-binding complex formed by Smc1p and Hec1p suggest that Rb may serve as a chaperone for Hec1p, presumably by stabilizing its active conformation. Taken together, the results presented here suggest a potential role of Rb in regulation of SMC1 through hsHec1p.
The functional analysis of Rb in the heterologous yeast system is further supported by the in vivo interaction between Rb, Hec1p, and SMC1 in mammalian cells. The presence of Rb in a complex with Hec1p and Smc1p suggests the relevance of the novel Rb function revealed by the yeast study to mammalian cells where Rb exists. The complex formed between Rb and SMC1 indicates a biological role of Rb in the SMC1 activity, although Rb is not present in the DNA-binding complex of SMC1. Rb thus appears to modulate the activity of SMC1 before SMC1 binds to chromatin DNA. Unlike the activities of Hec1p and SMC1, this M phase activity of Rb does not appear to be required by either yeast or mammalian cells for their basic machinery of chromosome segregation or for cell survival. Nevertheless, such an activity of Rb in regulating SMC1 could be important for higher fidelity of chromosome segregation and higher integrity of mitotic chromosome structures in mammalian cells.
Whether loss of Rb function leads to a decrease in the fidelity of chromosome segregation in mammalian cells remains to be explored. Nonetheless, such an activity for Rb may explain in part the abnormal process of mitosis observed in Rb-deficient fibroblasts and the chromosome abnormalities observed in Rb-deficient tumor cells (12, 28). It may also provide some clues for explaining observations that the majority of human retinoblastomas losing the wild-type allele and reduplicating the mutant allele early in the course of carcingenesis result from nondisjunction and misproportioning of sister chromatids (4, 19). Interestingly, yeast or human cells lacking functional Hec1p complete mitosis with unseparated or unequally separated chromosomes and enter a new cell cycle, leading to hyperploidy and aneuploidy (10, 57, 59). This is similar to the phenomenon observed in Rb-deficient fibroblasts treated with nocodazole (12, 28). Since neither loss of Rb function nor loss of Hec1p function appears to affect the mitotic checkpoint control (28, 59), microtubule-destabilizing agents probably challenge the chromosome segregation process in Rb-deficient cells and thereby induce a high frequency of aberrant mitosis. These results indirectly support the role of Rb in chromosome segregation.
Taken together, the results of this study, using a heterologous yeast system, provide a useful assay and more direct evidence for revealing the mechanistic process of a novel function of Rb in chromosome segregation. The study thereby contributes to explaining a new critical role of Rb in human carcinogenesis.
ACKNOWLEDGMENTS
The first two authors contributed equally to this report.
We thank O. Cohen-Fix, R. D. Gietz, D. Koshland, P. Hieter, C. Holm, A. M. Hoyt, C. Mann, A. Murray, and A. Strunnikov for yeast strains, antibodies, plasmid vectors, and assistance in yeast genetic analysis, and we thank T. Boyer for his critical reading of the manuscript.
This work was supported by NIH grants (EY05758 and CA58318), and L.Z. was supported by a predoctoral training grant from the U.S. Army (DAMD17-99-1-9402).
REFERENCES
- 1.Akhmedov A T, Frei C, Tsai-Pflugfelder M, Kemper M B, Gasser S M, Jessberger R. Structural maintenance of chromosome's protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998;273:24088–24094. doi: 10.1074/jbc.273.37.24088. [DOI] [PubMed] [Google Scholar]
- 2.Baker S H, Frederick D L, Bloecher A, Tatchell K. Alanine-scanning mutagenesis of protein phosphatase type 1 in the yeast Saccharomyces cerevisiae. Genetics. 1997;145:615–626. doi: 10.1093/genetics/145.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloecher A, Tatchell K. Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Genes Dev. 1999;13:517–522. doi: 10.1101/gad.13.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavenee W K, Dryja T P, Phillips R A, Benedict W F, Godbout R, Gallie B L, Murphree A L, Strong L C, White R L. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen P-L, Riley D J, Lee W-H. The retinoblastoma protein as a fundamental mediator of growth and differentiation signals. Crit Rev Eukaryot Gene Express. 1995;5:79–95. [PubMed] [Google Scholar]
- 6.Chen P-L, Riley D J, Lee W-H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2745–2752. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 7.Chen P-L, Riley D J, Chen-Kiang S, Lee W-H. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen P-L, Scully P, Shew J-Y, Wang J Y-J, Lee W-H. Phosphorylation of the retinoblastoma gene product is modulated during cell cycle and cellular differentiation. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 9.Chen P-L, Ueng Y-C, Durfee T, Chen K-C, Yang-Feng T, Lee W-H. Identification of a human homologue of yeast nuc2 which interacts with the retinoblastoma protein in a specific manner. Cell Growth Differ. 1995;6:199–210. [PubMed] [Google Scholar]
- 10.Chen Y, Riley D J, Chen P-L, Lee W-H. HEC, a novel nuclear protein rich in leucine haptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Sharp Z D, Lee W-H. HEC binds to the seventh regulatory subunit of the 26 S proteasome and modulates the proteolysis of mitotic cyclins. J Biol Chem. 1997;272:24081–24087. doi: 10.1074/jbc.272.38.24081. [DOI] [PubMed] [Google Scholar]
- 12.Di Leonardo A S, Khan S H, Linke S P, Greco V, Seidita G, Wahl G M. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 13.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 14.Ewen M E, Xing Y, Lawrence J B, Livingston D M. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 15.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude G F. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 16.Gietz R D, Sugino A. New yeast-E. coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restrictive sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 17.Goodrich D W, Wang N P, Qian Y-W, Lee E Y-H P, Lee W-H. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell. 1991;67:293–302. doi: 10.1016/0092-8674(91)90181-w. [DOI] [PubMed] [Google Scholar]
- 18.Gualberto A, Aldape K, Kozakiewicz K, Tlsty T D. An oncogenic form of p53 confers a dominant, gain-of-function phenotype that disrupts spindle checkpoint control. Proc Natl Acad Sci USA. 1998;95:5166–5171. doi: 10.1073/pnas.95.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagstrom S A, Dryja T P. Mitotic recombination map of 13cen-13q14 derived from an investigation of loss of heterozygosity in retinoblastomas. Proc Natl Acad Sci USA. 1999;96:2952–2957. doi: 10.1073/pnas.96.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannon G J, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 21.Hatakeyama M, Brill J A, Fink G R, Weinberg R A. Collaboration of G1 cyclins in the functional inactivation of the retinoblastoma protein. Genes Dev. 1994;8:1759–1771. doi: 10.1101/gad.8.15.1759. [DOI] [PubMed] [Google Scholar]
- 22.Heck M M S. Condensins, cohesins, and chromosome architecture: how to make and break a mitotic chromosome. Cell. 1997;91:5–8. doi: 10.1016/s0092-8674(01)80002-7. [DOI] [PubMed] [Google Scholar]
- 23.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 24.Hirano T. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- 25.Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 26.Hyland K M, Kingsbury J, Koshland D, Hieter P. Ctf19p: a novel kinetochore protein in Saccharomyces serevisiae and potential link between the kinetochore and mitotic spindle. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karantza V, Maroo A, Fay D, Sedivy J M. Overproduction of Rb protein after the G1/S boundary causes G2 arrest. Mol Cell Biol. 1993;13:6640–6652. doi: 10.1128/mcb.13.11.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan S H, Wahl G M. p53 and pRb prevent rereplication in response to microtubule inhibitors by mediating a reversible G1 arrest. Cancer Res. 1998;58:396–401. [PubMed] [Google Scholar]
- 29.Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 30.King R W, Deshaies R J, Peters J, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 31.Knudsen E S, Buckmaster C, Chen T-T, Feramisco J R, Wang J Y-J. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 1998;12:2278–2292. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koepp D M, Harper J W, Elledge S J. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 33.Koshland D, Hieter P. Visual assay for chromosome ploidy. Methods Enzymol. 1987;155:351–372. doi: 10.1016/0076-6879(87)55024-8. [DOI] [PubMed] [Google Scholar]
- 34.Lanni J S, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lengauer C, Kinzler K W, Vogelstein B. Genetic instability in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Graham C, Lacy S, Duncan M V, Whyte P. The adenovirus E1A-associated 130-kD protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A and E. Genes Dev. 1993;7:2366–2377. doi: 10.1101/gad.7.12a.2366. [DOI] [PubMed] [Google Scholar]
- 37.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludlow J W, Glendening C L, Livingston D M, DeCaprio J A. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol Cell Biol. 1993;13:367–372. doi: 10.1128/mcb.13.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayol X, Grana X, Baldi A, Sang N, Hu Q, Giordano A. Cloning of a new member of the retinoblastoma gene family (pRB2) which binds to the E1A transforming domain. Oncogene. 1993;8:2561–2566. [PubMed] [Google Scholar]
- 40.Meluh P B, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray A W, Szostak J W. Chromosome segregation in mitosis and meiosis. Annu Rev Cell Biol. 1985;1:289–315. doi: 10.1146/annurev.cb.01.110185.001445. [DOI] [PubMed] [Google Scholar]
- 43.Murray A W. The genetics of cell cycle checkpoints. Curr Opin Genet Dev. 1995;5:5–11. doi: 10.1016/s0959-437x(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 44.Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1995;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 45.Nead M A, Baglia L A, Antinore M J, Ludlow J W, McCance D J. Rb binds c-Jun and activates transcription. EMBO J. 1998;17:2342–2352. doi: 10.1093/emboj/17.8.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niculescu A B, III, Chen X, Smeets M, Hengst L, Prives C, Reed S I. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreplication. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novitch B G, Mulligan G J, Jacks T, Lassar A B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nurse P. Ordering S phase and M phase in the cell cycle. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 49.Ohkura H, Kinoshita N, Miyatani S, Toda T, Yanagida M. The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell. 1989;57:997–1007. doi: 10.1016/0092-8674(89)90338-3. [DOI] [PubMed] [Google Scholar]
- 50.Riley D J, Lee E Y-H P, Lee W-H. The retinoblastoma protein: more than a tumor suppressor. Annu Rev Cell Biol. 1994;10:1–29. doi: 10.1146/annurev.cb.10.110194.000245. [DOI] [PubMed] [Google Scholar]
- 51.Sassoon I, Severin F F, Andrews P D, Taba M-R, Kaplan K B, Ashford A J, Stark M J R, Sorger P K, Hyman A A. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 53.Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 54.Strunnikov A V. SMC proteins and chromosome structure. Trends Cell Biol. 1998;8:454–459. doi: 10.1016/s0962-8924(98)01370-1. [DOI] [PubMed] [Google Scholar]
- 55.Strunnikov A V, Larionov V L, Koshland D. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas J T, Laimins L A. Human papillomavirus oncoproteins E6 and E7 independently abrogate the mitotic spindle checkpoint. J Virol. 1998;72:1131–1137. doi: 10.1128/jvi.72.2.1131-1137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wigge P A, Jensen O N, Holmes S, Soues S, Mann M, Kilmartin J V. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yanagida M. Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol. 1998;8:144–149. doi: 10.1016/s0962-8924(98)01236-7. [DOI] [PubMed] [Google Scholar]
- 59.Zheng L, Chen Y, Lee W-H. Hec1p, an evolutionarily conserved coiled-coil protein, modulates chromosome segregation through interaction with SMC proteins. Mol Cell Biol. 1999;19:5417–5428. doi: 10.1128/mcb.19.8.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu X, Mancini M A, Chang K-H, Liu C-Y, Chen C-F, Shan B, Jones D, Yang-Feng T L, Lee W-H. Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progression. Mol Cell Biol. 1995;15:5017–5029. doi: 10.1128/mcb.15.9.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]