Abstract
The yeast poly(A) binding protein Pab1p mediates the interactions between the 5′ cap structure and the 3′ poly(A) tail of mRNA, whose structures synergistically activate translation in vivo and in vitro. We found that deletion of the PAT1 (YCR077c) gene suppresses a PAB1 gene deletion and that Pat1p is required for the normal initiation of translation. A fraction of Pat1p cosediments with free 40S ribosomal subunits on sucrose gradients. The PAT1 gene is not essential for viability, although disruption of the gene severely impairs translation initiation in vivo, resulting in the accumulation of 80S ribosomes and in a large decrease in the amounts of heavier polysomes. Pat1p contributes to the efficiency of translation in a yeast cell-free system. However, the synergy between the cap structure and the poly(A) tail is maintained in vitro in the absence of Pat1p. Analysis of translation initiation intermediates on gradients indicates that Pat1p acts at a step before or during the recruitment of the 40S ribosomal subunit by the mRNA, a step which may be independent of that involving Pab1p. We conclude that Pat1p is a new factor involved in protein synthesis and that Pat1p might be required for promoting the formation or the stabilization of the preinitiation translation complexes.
Translational control of gene expression operates most frequently during the initiation phase of protein synthesis. The recruitment of the 43S preinitiation complex (40S ribosomal subunit, initiator methionyl-tRNA, and initiation factors) by the capped 5′ end of mRNA and the scanning of the 5′ untranslated region until the initiator codon is found are the main rate-limiting steps (for a review, see reference 28). Studies of the yeast Saccharomyces cerevisiae implicate 3′ poly(A) tails in the joining of the 40S ribosomal subunits to the 5′ end of mRNA (19, 42). The two mRNA ends are brought together by specific protein-protein interactions. The multicomponent eukaryotic initiation factor 4F (eIF4F) initiation complex binds to the cap through the eIF4E subunit, and the eIF4G subunit acts as a bridge both between eIF4F and the 40S ribosomal subunit and between the 5′ and 3′ ends of mRNA through specific interactions with the Pab1p, which is associated with the poly(A) tails (16, 47). Thus, a capped and polyadenylated RNA can be made circular in the presence of an eIF4E-eIF4G-Pab1p complex (52). This is consistent with the model in which mRNA forms a closed loop to facilitate translation initiation (19). The interactions between the two mRNA ends result in a synergistic enhancement of protein synthesis in vivo and in vitro (10, 46, 48). Moreover, this synergy is essential for translation in vitro when mRNAs compete each other for ribosome binding and when neither the cap structure nor the poly(A) tail alone is able to promote efficient protein synthesis (34, 35). Thus poly(A)-associated Pab1p is necessary for the stimulation of translation initiation and for the recruitment of the 40S ribosomal subunit by the mRNA.
Pab1p also has an essential function in mRNA turnover. In yeast, translation-dependent decay of most mRNAs is initiated by 3′ deadenylation, followed by 5′ decapping and exonucleolytic digestion in the 5′ to 3′ direction (26). Pab1p is involved in controlling poly(A) tail degradation and in protecting of mRNAs from decapping (7). Pab1p also contributes to nuclear mRNA 3′-end processing by controlling the length of the poly(A) tails synthesized (1, 29), in association with the Pab1p-dependent poly(A) nuclease, PAN (5). Pab1p is always found associated with the poly(A) tails during these various processes. However, recent results show that Pab1p is able to prevent mRNA decay independently of the poly(A) tail (8), which may function to locate Pab1p and to tether it to its site of activity.
The properties of genes, mutations, or deletions that suppress the lethality of a PAB1 deletion support the model in which the essential role of Pab1p is the stimulation of translation initiation. These suppressors can be grouped into two main classes based on their role in the control of protein synthesis, but both are consistent with the translational machinery being deficient in the absence of Pab1p. One class of suppressors inhibits 5′-end decapping, making mRNAs more stable (4, 14). These mutations may modify the equilibrium between protein synthesis and mRNA turnover: the increase in mRNA levels counteracts the lower translation rate due to the absence of Pab1p. The second class of suppressors is genes directly involved in translation. They mostly involve the 60S ribosomal subunit by affecting its production (40, 41, 53). The increased concentration of free 40S subunits is assumed to compensate for the defect in the Pab1p function of joining the 43S preinitiation complex to mRNA. pab1Δ suppression by the deletion of the PBP1 gene is an exception, and it can be attributed to nuclear effects on the regulation of polyadenylation, even though a fraction of Pab1p is found associated with polysomes (24). In this report we describe PAT1 as a new gene, the deletion of which can bypass the PAB1 gene function. Pat1p is involved in translation initiation, but it is not associated with the production of the 60S ribosomal subunit. We discuss how deletion of the PAT1 gene can suppress a PAB1 gene deletion.
MATERIALS AND METHODS
Yeast methods.
The parent strain was W303-1B (ura3-1 trp1-1 ade2-1 leu2-3, 112 his3-11, 15). The PAT1 gene was disrupted by HIS3, and the PAB1 gene was disrupted by HIS3 or CAN1-100. Standard genetic techniques were used (44). Strains were grown at 30°C in supplemented minimal medium. To select bypass suppressors of the PAB1 gene deletion, the pab1Δ strain containing a plasmid carrying PAB1 and URA3 was irradiated with UV light to yield about 1% cell survival and then grown in the presence of 5-fluoro-orotic acid (5FOA). 5FOA-resistant Ura− colonies were screened for a recessive pab1Δ suppression phenotype (3).
RNA isolation, poly(A) and cap selection, and analysis of mRNA poly(A) tail lengths.
Total yeast RNA was isolated and mRNAs were analyzed on Northern blots by standard methods (43). For polysomal RNA, polysomal extracts were prepared as described below. Polysomes were pelleted by centrifugation through a 20% sucrose pad at 100,000 × g for 30 min. RNA was purified from the pellet by phenol extraction. To measure the amounts of poly(A)- and cap-containing RNA, 1 ml of yeast culture was labeled for 5 min with 5 μCi of [8-3H]adenine at a concentration of 24 Ci/mmol (Amersham). Poly(A)+ RNA was purified by batch chromatography on oligo(dT) cellulose (43). Cap+ RNA was isolated by immunoprecipitation with anti-cap antibodies as described by Muhlrad et al. (31). Poly(A) tail length was analyzed by RNA 3′-end labeling with [32P]pCp (Amersham) and T4 RNA ligase, RNase digestion, and poly(A) fractionation on a denaturing polyacrylamide gel as previously described (2).
Protein procedures, antibodies, and polysome analysis.
Gel electrophoresis, protein transfers to membranes (Protean membrane; Schleicher and Schuell), and binding conditions for antibodies were as described by Harlow and Lane (13). Proteins were detected by enhanced chemiluminescence with the Super Signal substrate (Pierce). Specific polyclonal antibodies were obtained against total Pab1p, Ssm1p, and Rna15p and against amino acids 230 to 790 of Pat1p produced in Escherichia coli. The recombinant Pat1 fragment (60 kDa) was found in inclusion bodies and purified as described previously for insoluble proteins (13). Production and purification of antibodies were as previously described (1, 2). Anti-Gar1p and anti-Nam1p antibodies were generous gifts from M. Ferrer and G. Dujardin, respectively. Anti-cap antibodies were produced from 7-methylguanosine 5′ monophosphate (7m-GMP) cross-linked to bovine serum albumin, as described by Meredith and Erlanger (27), and purified by chromatography on 7m-GMP agarose.
Nuclear and cytoplasmic fractions were isolated by lysis of spheroblasts, followed by separation on Ficoll gradients, as described by Hurt et al. (17). Polysome extracts were prepared as previously described from cells grown to an optical density at 600 nm (OD600) of between 0.6 and 0.8 and clarified by centrifugation at 20,000 × g for 20 min. Extracts were fractionated through 5 to 50% sucrose gradients by centrifugation at 39,000 rpm, for 3 h, at 4°C in a Beckman SW41 rotor and analyzed by monitoring the A260 (33). Cycloheximide was omitted from the culture medium before cell harvesting, and the extraction buffer and sucrose gradients were as indicated in the text below.
Preparation of cell-free translation extracts.
Extracts were prepared essentially as described by Tuite and Plesset (49). Spheroblasts were prepared from yeast cultures (OD600 ≈ 1.5). After a 10-min period of recovery in rich culture medium supplemented with 1 M sorbitol, spheroblasts were resuspended in a volume of buffer A (30 mM HEPES, 100 mM KOAc, 2 mM MgOAc, and 2 mM dithiothreitol) one times the cell weight. A volume of glass beads equal to that of the cell suspension was added, and spheroblasts were lysed by eight cycles of shaking by hand for 15 s and 1 min of cooling on ice. The lysate was clarified by two centrifugations, one at 30,000 × g for 15 min and one at 100,000 × g for 33 min. The supernatant was chromatographed on a G-25 Sephadex column (2 by 25 cm for a 4-ml sample) in buffer A containing 20% glycerol and 1 mM phenylmethylsulfonyl fluoride. Fractions with OD260s of ≥80 were pooled, and aliquots were frozen at −70°C.
mRNA transcription in vitro.
Luciferase (LUC) mRNAs were prepared from the plasmid pT7-Luc minus 3′UTR-A50, kindly provided by D. R. Gallie (10), by using the T7 RNA production system (Promega). Radioactive MFA2 mRNAs were prepared, as described by Tarun, Jr., and Sachs (46), from the plasmid pAS225, a generous gift from A. B. Sachs, in the presence of 35S-UTP (1,000 Ci/mmol; Amersham). RNAs were purified over Micro Biospin30 columns (Bio-Rad) and analyzed by gel electrophoresis.
In vitro translation.
Translation assays were performed essentially as described by Tarun, Jr., and Sachs (46). Treatment by micrococcal nuclease (Boehringer) was optimized for each extract. For 35S-labeled translation, 15-μl reaction mixtures containing 3 μM methionine were incubated with 500 ng of yeast polysomal RNA and 10 μCi of [35S]methionine (3,000 Ci/mmol; Amersham). For luciferase synthesis, 15-μl reaction mixtures containing 40 μM methionine were supplemented with 50 ng of LUC mRNA. Luminescence was measured with the LUC assay reagent (Promega) on a Lumat LB 9501 luminometer. For translation of poly(U), 15-μl reaction mixtures containing 20 μM phenylalanine and 12 mM MgOAc were incubated with 2 μg of poly(U) and 2 μCi of [3H]phenylalanine (140 Ci/mmol; Amersham). For immunoneutralization, purified antibodies in buffer A were incubated with extracts for 20 min at 4°C prior to the start of the translation reaction. For reconstitution of translation extract activity, purified recombinant Pat1 peptide was concentrated by electrophoresis on a gel and transferred to a membrane (about 20 μg of protein per cm2). The membrane was blocked to avoid nonspecific adsorption, washed, and incubated with purified Pat1p antibodies in phosphate-buffered saline (0.5 ml for a 2-cm2 membrane). A blot with E. coli proteins was used as control. Treated antibodies were dialyzed against buffer A and tested for immunoneutralization as described above.
Translation initiation assays and sucrose gradient analysis.
As previously described (12, 46), 30-μl volumes of translation extract, with 1 mM cycloheximide and with or without antibodies or other inhibitors (1 mM guanylyl-imido diphosphate [GMPPNP], 5 μM edeine, or 50 mM EDTA), were incubated with 35S-labeled MFA2 mRNA (105 cpm ≈ 2 ng) for 15 min at 25°C. Reaction mixtures were diluted with 100 μl of cold buffer A with 0.075% glutaraldehyde and fractionated after 5 min on linear 10 to 30% sucrose gradients by centrifugation at 40,000 rpm for 3 h at 4°C in an SW41 Beckman rotor. Gradients were analyzed by measuring the A260. Radioactivity in each fraction was measured by direct counting in scintillation fluid.
RESULTS
Identification of a pat1 mutant as a new suppressor of a PAB1 gene deletion.
To identify new genes that functionally interact with the PAB1 gene, we screened yeast genomic mutations for those that suppressed a PAB1 deletion. Cells bearing a genomic deletion of PAB1 but carrying the PAB1 gene on a plasmid also carrying the URA3 gene were mutagenized by UV light treatment. The plasmid carrying the URA3 and PAB1 genes was then eliminated in the presence of 5FOA, and viable strains were isolated. Those showing both the suppression of the PAB1 deletion phenotype and temperature sensitivity were selected for further characterization. We verified that the alterations were due to mutations at a single genetic locus by checking that both phenotypes cosegregated in backcrosses of the mutants with the wild-type strain. The wild-type genes corresponding to the mutated genes were cloned by screening for yeast genomic DNA fragments that restored the normal growth phenotype. The thermosensitivity of two mutant strains was rescued by the product of the YCR077c gene, which also reversed the suppression of lethality of the pab1Δ mutation. The YCR077c gene product was previously identified in a two-hybrid screen as interacting with topoisomerase II, and it was named Pat1 (protein associated with topoisomerase II) (51). The PAT1 gene maps on chromosome III (GenBank accession number X59720). It codes for a 797-amino-acid protein whose sequence is unusually rich in proline and glutamine residues. A BLAST search for homologous proteins showed extensive similarity to an open reading frame encoding a putative protein of 744 amino acids in Schizosaccharomyces pombe (GenBank accession number AL021839). The primary sequences of the two deduced proteins shared 47% identity and 55% similarity. There is a short region of potential coiled-coil structure between residues 717 and 732 in the C terminus of the predicted Pat1p (88% probability) (23) which matches the coiled-coil regions of several proteins.
Deletion of the PAT1 gene also suppressed the lethality of the pab1Δ mutation. PAT1 is not essential for cell viability. However, the deletion had a phenotype by itself in the genetic background used. It was cold and heat sensitive and it also had much slower growth at 30°C (doubling time, 6 h) than the wild-type strain (doubling time, 1.8 h). However, spontaneous suppressors of the growth phenotype were frequent. The double mutant pat1Δ/pab1Δ strain had the same phenotype but with a slower growth rate (doubling time, 7.5 h at a permissive temperature). All experiments reported below were done with pat1Δ and pat1Δ/pab1Δ strains at 30°C.
Pat1p is involved in translation initiation.
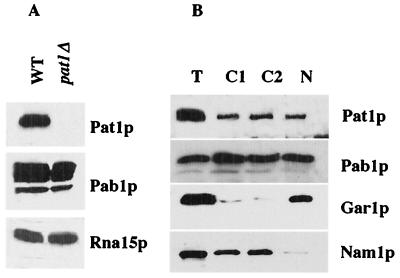
Purified Pat1p antibodies recognized a single protein in a whole-protein extract of the wild-type cells, a protein which was not detected in cells of the pat1Δ strain by Western blotting (Fig. 1A). The apparent molecular mass of this protein was 97 kDa, whereas the predicted mass is 88 kDa. This aberrant migration on sodium dodecyl sulfate-polyacrylamide gels has previously been described and attributed in part to the presence of numerous proline residues (38). Rna15p and Pab1p were used as controls, and the amounts detected were identical for both strains, indicating that there was no visible protein degradation in the pat1Δ strain. In particular, Pab1p was stable in the absence of the Pat1p.
FIG. 1.
Subcellular localization of Pat1p. (A) Western blotting analysis of Pat1p, Pab1p, and Rna15p in 25 μg of proteins extracted from wild-type cells (WT) and from cells deficient in Pat1p (pat1Δ); (B) Western blotting analysis of Pat1p and Pab1p in a total extract (T), two cytoplasmic fractions (C1 and C2), and purified nucleus (N) prepared from wild-type cells. Equivalent percentages of fractions were analyzed on the gel. Nucleolar Gar1p and mitochondrial Nam1p were used as fractionation controls.
The subcellular location of Pat1p was investigated by cell organelle fractionation on a Ficoll-sucrose gradient (17). The nuclear fraction (Fig. 1B, lane N) was separated from cytoplasmic components, which were partially separated into two main fractions: one fraction was enriched in soluble proteins, and the other was enriched in mitochondria and microsomes (Fig. 1B, lanes C1 and C2). Fractionation was checked by testing for the presence of organellar markers. Gar1p a nucleolar RNP protein, was found mostly in the nuclear fraction, and the mitochondrial Nam1p was found in both cytosolic fractions. We estimated cross-contamination to be less than 10% for the nucleus-cytoplasm fractionation by semiquantitative Western blotting (data not shown). As expected, Pab1p was found both in the nucleus and the cytoplasm (39). Pat1p was found in the same cell fractions. Comparison with the Gar1p and Nam1p markers indicated that Pat1p's presence in both the nucleus and the cytoplasm was not due to contamination, even though the fractionation was not totally quantitative. Proteins were not recovered in stoichiometric ratios, particularly in the cytoplasmic fractions, and it was impossible to determine the primary location of Pat1p in the cells from these experiments. However, Pat1p has been previously described as a cytosolic protein, partly associated with membranes, and it has not been detected in the nucleus (38). This contradiction may be due to differences in the fractionation procedure. Even though the method used here has been shown to separate the nuclei from cytosolic contaminants, we cannot exclude the possibility that the nuclear fraction was partly contaminated by endoplasmic reticulum proteins. Moreover, antibodies used in the earlier experiments were raised against only an oligopeptide from the N terminus of Pat1p and therefore may give a signal too weak for the protein to be identified in the nucleus by Western blotting.
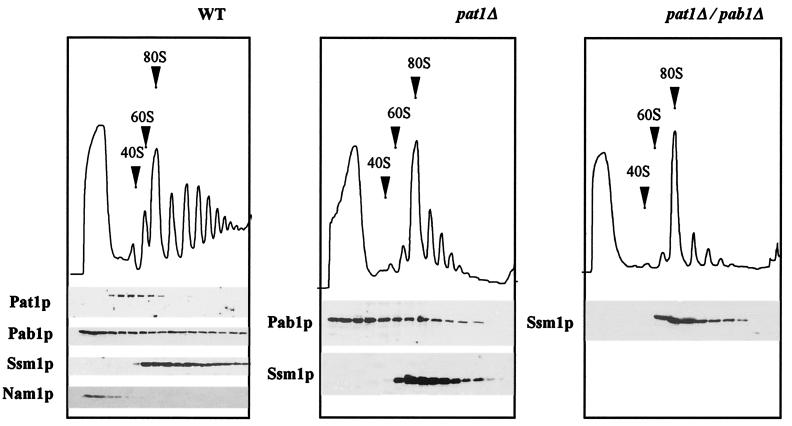
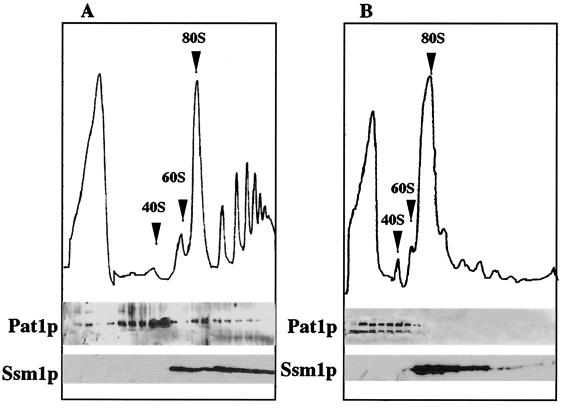
Poly(A) tail-associated Pab1p is present in the cytoplasm on mRNAs translated by ribosomes. We determined whether Pat1p, which seemed functionally linked to Pab1p, was also present in the polysomal fraction. The polysome content of wild-type cells was analyzed by centrifugation on sucrose gradients (Fig. 2). Proteins were probed by Western blotting. As previously shown, Pab1p was found throughout the gradient as both a soluble and a polysome-associated protein (24). Ssm1p, a 60S ribosomal subunit protein (33), was detected in fractions containing the free 60S subunit and in the polysomes. Nam1p, a mitochondrial contaminant, was present essentially in the upper fractions with the soluble proteins. Pat1p was also present in the gradient, but its distribution was very different from that of Pab1p. It migrated almost exclusively at the level of the free 40S subunit. This association was destroyed by EDTA treatment, which dissociated ribosomal subunits from mRNA and translation initiation factors (data not shown). When large quantities of the polysomal fraction were analyzed (Fig. 3A), Pat1p accumulated mainly in both the 40S subunit-containing fractions and the adjacent heavier fractions corresponding to the 43 to 48S region of the gradient. It was also found in complexes lighter than the 40S subunit and detected in small amounts in monosomes and polysomes. In the experiment whose results are shown in Fig. 3B, cycloheximide was omitted during polysome extraction and from the sucrose gradient. Under these conditions elongation was completed in vitro without reinitiation of new cycles of synthesis. Then polysomes ran off from mRNA, and 43 to 48S and 80S particles accumulated. These 80S ribosomes were inactive for translation, and they were not associated with mRNA (30, 37). Pat1p was detected in the soluble proteins and in the small complex-containing fractions until the 43 to 48S region, and it was absent from the heavier fractions. The second smaller band (molecular mass, about 60 kDa), found in these fractions, corresponded to a Pat1p fragment. It seemed to be a degradation product that was also observed in protein extracts denatured under mild conditions (data not shown). These results indicate that the presence of Pat1p in polysomes depended on active translation. It could not be due to the association of this protein with high-molecular-weight complexes that copurified with polysomes and that were unrelated to the translation complexes. This is consistent with Pat1p being associated, at least in part, with preinitiation complexes. Pat1p was also detected in polysomes, given that preinitiation complexes were present in polysomes on mRNAs bearing several other 80S ribosomes.
FIG. 2.
Pat1p fractionates with the 40S ribosomal subunit on a gradient. Deletion of the gene results in a decrease of heavier polysomes. Polysomes extracted from wild-type cells (WT) and from cells deficient in Pat1p (pat1Δ) and in both Pat1p and Pab1p (pat1Δ/pab1Δ) were separated on sucrose gradients. Sedimentation proceeded from left to right. 40S and 60S subunits and 80S monoribosomes are indicated by arrows on OD260 profiles. Fractions were tested for Pat1p and Pab1p by Western blotting. The 60S ribosomal protein Ssm1p and nonpolysomal Nam1p were used as controls.
FIG. 3.
Sedimentation of Pat1p in gradient varies with the conditions under which polysomes are dissociated. Polysomes were extracted and analyzed as described in the legend to Fig. 2. Cycloheximide was either added to the culture medium and to the extraction buffer and gradient (A) or omitted from them (B).
We first investigated whether protein synthesis was impaired in pat1Δ and pat1Δ/pab1Δ strains, by following the incorporation of 14C-amino acids into cells during logarithmic growth. Protein synthesis in both mutant strains was only about 30% of that in wild-type cells (data not shown). This corresponds to the lower growth rates of these mutants (see above). We analyzed the polysome profiles of the pat1Δ and pat1Δ/pab1Δ strains (Fig. 2). They were different from that of the wild-type strain but similar to each other, with fewer large polysomes and many more monosomes. This pattern is characteristic of blocked translation initiation. The reduced rate of initiation without a change in the rate of elongation leads to a decrease in polysome size and an increase in the amount of free 80S ribosomes, as already shown for mutant strains involved in translation initiation (50, 53). On the other hand, a wild-type profile of polysomes was found in slow-growing strains; the pap1-1 strain, mutated in poly(A) polymerase, had a doubling time of 4.5 h (36), and the rpb1-1 strain, mutated in RNA polymerase II, had a doubling time of 4.8 h (data not shown). Thus, a deficit in Pat1p alone inhibited translation initiation even though Pab1p was present on ribosome-bound mRNAs, although the paucity of large polysomes was more marked in the pat1Δ/pab1Δ double mutant strain than in the pat1Δ single mutant. pat1Δ appears to be a novel bypass suppressor of the PAB1 deletion involved in translation initiation but with properties different from those of previously described suppressors. Pat1p is not a ribosomal protein, and it does not seem to be involved in the assembly of ribosomal subunits or in the stability of ribosomes. The pat1Δ strain does not show the reduced level of 60S ribosomal subunit characteristic of the spb1-spb7 and sos1Δ suppressors of pab1Δ (40, 53). The ratio between 18S and 23S rRNAs is not modified in the mutant pat1Δ strain (data not shown).
Structure of mRNAs is not globally modified in the pat1Δ strain.
We first verified that the alteration in translation in cells deficient in Pat1p was not due to the absence of mRNAs. The amounts of mRNA in the wild-type, pat1Δ, and pat1Δ/pab1Δ cells in exponential growth were compared by Northern blotting. Equivalent proportions of stable PGK1 and ACT1 mRNAs (half-lives of about 30 and 20 min, respectively) and unstable URA3 mRNA (half-life of 3 min) were found in total RNA by phosphorimaging (data not shown).
The defect in translation initiation in the absence of Pat1p could be due either to a faulty translation initiation factor or to an abnormal structure of translatable mRNAs. We first analyzed the poly(A) tail lengths of total cellular mRNA by 3′-end labeling, degradation of the mRNA body, and poly(A) separation by electrophoresis on polyacrylamide gels. The same broad size distribution was observed in both wild-type and pat1Δ strains, indicating that the mRNA polyadenylation/deadenylation ratio was not substantially modified by this mutation (data not shown). However pat1Δ/pab1Δ cells showed long poly(A) tails with an abnormal length distribution similar to that in a temperature-sensitive pab1 mutant at the restrictive temperature and that in strains lacking PAB1 in the presence of spb1-spb7 and pbp1Δ suppressors (24, 40). Thus the poly(A) shortening is a Pab1p-dependent reaction, which is not rescued in suppressive conditions in the absence of Pat1p. These results confirm that the accumulation of long poly(A) tails is not related to the lethality associated with PAB1 deletion.
Steady-state accumulation of mRNAs in the cytoplasm and their polyadenylation were normal in the pat1Δ strain, suggesting that the equilibrium between synthesis and degradation and the equilibrium between adenylation and deadenylation of mRNAs were maintained in this mutant. We determined the capping and polyadenylation status of new transcripts. Cells in logarithmic growth were labeled with [3H]adenine for 5 min, and total RNAs were purified. We measured first the extent of mRNA polyadenylation by binding poly(A)+ RNAs to oligo(dT) cellulose and second the extent of capping by immunoprecipitation of cap+ mRNAs with an excess of antibodies directed against the 5′ cap structure. The proportions of capped (26 to 28%) and polyadenylated (20 to 22%) labeled RNAs were similar in all strains (average of four independent determinations). Moreover, 90% of poly(A)+ RNAs, isolated from each strain on oligo(dT) cellulose, were immunoprecipitated with anti-cap antibodies and were therefore capped. 70% of cap+ RNAs, selected by specific anti-cap antibodies, were retained on oligo(dT) cellulose as polyadenylated RNA (data not shown). We found no extensive differences in the structure of mRNA between the pat1Δ strain and a wild-type strain.
Total cellular RNAs and RNAs present in polysomes, including mRNAs in ongoing translation, were purified from the three strains. They were used as the substrates in a yeast RNA-dependent cell-free translation system purified from a wild-type strain (see below); no significant difference was found between the rates of protein synthesis (data not shown). These results strongly suggest that cells deficient in Pat1p are defective for translation initiation and that the phenotype is not due to the absence of translatable mRNAs.
Pat1p is involved in efficiency of translation in vitro.
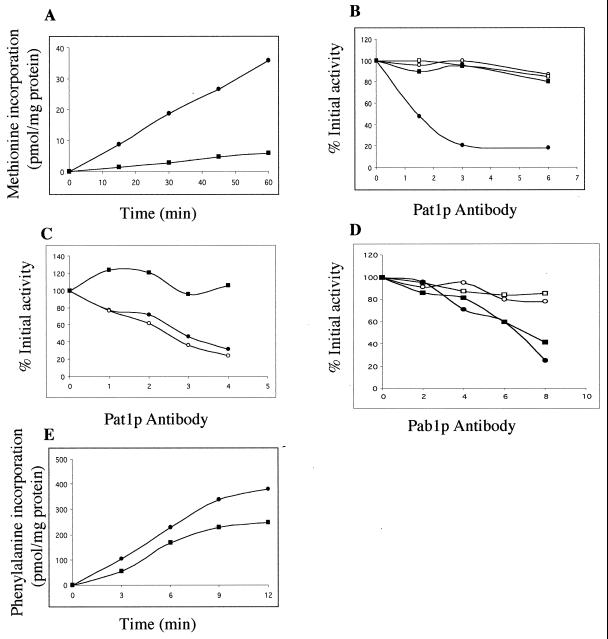
We studied translation in a yeast cell-free system deficient in Pat1p. An active cell-free translation system was obtained using a wild-type yeast strain. It actively translated exogenously added mRNAs once endogenous RNA had been removed by nuclease treatment. Unlike the results previously reported (18), background translation was less than 10% of total protein synthesis in our reference strain and translational activity could be directly measured by [35S]methionine incorporation into acid-insoluble materials. In a first set of experiments, exogenous yeast mRNAs extracted from the polysomal fraction of wild-type cells were used as a template; methionine incorporation was linear over a 60-min period in the optimal conditions described in the Materials and Methods section (Fig. 4A). It was much more difficult to obtain an active cell-free translation system from the pat1Δ strain. Spheroblasts were unable to recover metabolically when prepared from the strain deficient in Pat1p. A similar problem has been already described for a pab1 temperature-sensitive mutant, and it possibly reflected the defects in translation machinery in these two mutants (40). We tested the rate of protein synthesis in spheroblasts, immediately after preparation, by [35S]methionine pulse labeling. Translational activity recovered within 15 min in the wild-type spheroblasts and was maintained for 3 to 4 h. In contrast the activity in pat1Δ spheroblasts was stable for only 20 to 30 min and then decreased rapidly (data not shown), so translation extracts were prepared by spheroblast lysis after a short 10-min period of recovery following zymolyase treatment. This method reproducibly yielded active cell-free translation from the pat1Δ strain: [35S]methionine incorporation was linear for more than 60 min and activity was 15% of that of a similarly prepared wild-type extract (Fig. 4A). This is consistent with the differences in the protein synthesis rates in vivo; intact cells deficient in Pat1p synthesized three to four fewer polypeptides than wild-type cells did (see above). We were unable to obtain active translation extracts from the pat1Δ/pab1Δ cells, which did not recover protein synthesis activity in spheroblasts, indicating that the lethality is enhanced in the absence of Pab1p.
FIG. 4.
Pat1p antibodies inhibit cell-free translation of exogenous yeast mRNAs in extracts of wild-type cells. (A) Translation of exogenous yeast polysomal mRNAs. Nuclease-treated extracts prepared from wild-type cells (●) and from cells deficient in Pat1p (■) were programmed with yeast polysomal RNA. [35S]methionine incorporation into acid-insoluble materials was measured as a function of time. (B) Pat1p antibodies inhibit translation of exogenous yeast polysomal mRNAs in the wild-type extract. Nuclease-treated extracts were incubated with the indicated amounts of antibody (in micrograms), and the reaction was started by addition of yeast polysomal RNA. [35S]methionine incorporation was determined after 30 min of incubation at 28°C. Data are expressed as percentages of activity in the absence of antibody and are averages of the results of five independent experiments. Pat1p antibodies were added in wild-type (●) and pat1Δ (■) extracts; corresponding amounts of preimmune serum were added in wild-type (○) and pat1Δ (□) extracts. (C) Pat1p antibodies, depleted by contact with recombinant Pat1 peptide, do not inhibit translation of exogenous yeast mRNA in extract of wild-type cells. Pat1p antibodies were incubated with Pat1 peptide blotted onto a membrane as described in the text. Untreated (●) and depleted (■) antibodies and antibodies mock depleted on a control membrane (○) were added in wild-type extracts. The effects on translation were measured as described for panel B. Data are averages from two independent experiments. (D) Pab1p antibodies inhibit translation of exogenous yeast polysomal mRNAs in both wild-type extracts and extracts deficient in Pat1p. The effects of Pab1p antibodies on translation were measured as described for panel B. Pab1p antibodies were added in wild-type (●) and pat1Δ (■) extracts, and the corresponding preimmune serum in wild-type (○) and pat1Δ (□) extracts. (E) Translation of poly(U). Nuclease-treated extracts prepared from wild-type cells (●) and from cells deficient in Pat1p (■) were programmed with poly(U). [3H]phenylalanine incorporation into acid-insoluble materials was measured as a function of time.
To determine whether pat1Δ extract lacked a soluble factor, we tested translation by adding an equal mixture of wild-type and mutant extracts under conditions in which protein synthesis responded linearly to the mRNA concentrations. Translational activities were strictly additive (data not shown). Thus, factors present in the wild-type extract did not rescue translation by the mutant. These findings suggest that the Pat1p is present either in limiting amounts or associated with a stable complex that is not reconstituted in vitro. It is also possible that other limiting proteins were absent from or unstable in the pat1Δ extract. The translational activity of the wild-type extract was not inhibited, indicating that the defect in protein synthesis in pat1Δ extracts was not due to enhanced degradation of added mRNA.
We performed immunoneutralization experiments to confirm the involvement of Pat1p in translation in vitro. Various amounts of purified Pat1p antibodies were added to an RNA-dependent wild-type cell-free system and the extracts were programmed with exogenous yeast polysomal mRNA. Translation was strongly inhibited, whereas no inhibition was observed after the addition of nonspecific antibodies purified from the preimmune serum. Anti-Pat1p antibodies had no effect on pat1Δ extracts treated under the same conditions (Fig. 4B). Until now, it has not been possible to express the whole Pat1p in E. coli and thus rescue a mutant extract by the use of recombinant protein. Moreover, the Pat1p fragment, expressed in bacteria and used as antigen, was insoluble in the in vitro translation buffer, regardless of the purification conditions tested. It was impossible to test a direct competition between the recombinant Pat1 peptide and the endogenous Pat1 protein towards Pat1p antibodies in a translation extract. We therefore performed the competition in two steps. Purified recombinant Pat1 peptide was first blotted on membrane and incubated with purified Pat1p antibodies as described in Materials and Methods. Then the original and treated antibodies were tested for Pat1p immunoneutralization in a wild-type extract. Antibodies incubated with the Pat1 peptide fastened on a membrane did not inhibit translation, whereas the antibodies remained active after incubation with a control membrane (Fig. 4C). These results strongly indicated that Pat1p is required for efficient in vitro protein synthesis.
We undertook immunoneutralization with Pab1p antibodies in extracts containing or lacking Pat1p (Fig. 4D). As previously described (46), the translation activity of a wild-type extract was progressively abolished by the immunoneutralization of Pab1p. Pab1p neutralization also inhibited protein synthesis in the absence of Pat1p and the extent of inhibition was similar in both extracts at equivalent antibody concentrations. These results indicated that Pab1p might be involved in the stimulation of translation even in the absence of Pat1p. This finding was not expected for several reasons, as follows. (i) PAT1 gene deletion overcomes the Pab1p requirement for viability, which almost certainly concerns Pab1p function in translation. (ii) The translation initiation is inhibited in the pat1Δ strain even in the presence of Pab1p in vivo. (iii) The protein synthesis rate is very low in a Pat1p-deficient cell-free system containing a normal level of Pab1p.
We compared translation of poly(U) in both wild-type and mutant extracts under conditions whereby phenylalanine incorporation in polypeptides was independent of initiation factors (22) (Fig. 4E). Activity in a pat1Δ extract was about 60% of that in a wild-type extract. The translation rate was relatively much higher on the homopolymer than on exogenous yeast mRNA, which requires a complete cycle of translation (Fig. 4A). This suggests that the elongation phase was less defective than the initiation phase in the absence of Pat1p.
Pat1p is required for cap- and poly(A)-dependent translation in vitro.
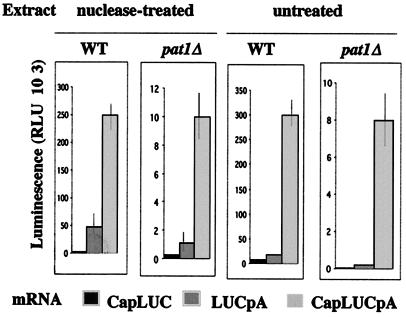
Protein synthesis in a yeast wild-type cell extract reflects the effects of the cap structure and poly(A) tails on translational efficiency and the functional synergy between the capped and polyadenylated ends of mRNA found in vivo (18, 46). This allowed us to ask whether Pat1p was required for cap- or poly(A)-dependent translation in vitro. We used an mRNA encoding the firefly luciferase protein, which was assayed enzymatically to measure translation rates. Three forms of this mRNA were used: a 5′ cap structure (CapLUC mRNA), a 3′ 50-nt-long poly(A) tail (LUCpA mRNA) or a structure containing both (CapLUCpA mRNA). Translation was first compared under noncompetitive conditions, after endogenous mRNAs had been degraded by nuclease treatment (Fig. 5). The results obtained with the wild-type extract agreed with previously published findings. The capped mRNA was the least efficient substrate. The presence of a poly(A) tail increased translation about 10-fold. The cap structure increased the translation of the poly(A) mRNA four- to sixfold. The same relative values were obtained with the pat1Δ extract, except that the translation of LUCpA mRNA was relatively lower, only five- to sevenfold higher than that of CapLUC mRNA. However, this difference was small compared to the experimental variability and does not seem significant. As previously shown with a wild-type extract (34, 35), the synergy between the cap structure and the poly(A) tail was maintained and it promoted efficient translation in the presence of endogenous mRNA. In contrast, protein synthesis was not activated by poly(A) tails alone under these competitive conditions (Fig. 5). Analogous results were obtained in the absence of Pat1p. The protein synthesis rate of CapLUCpA mRNA was comparable under competitive and noncompetitive conditions, and the translation efficiency of heterologous luciferase mRNAs was not modified by competition with homologous yeast mRNAs in pat1Δ extracts.
FIG. 5.
The cap structure and the poly(A) tail synergistically activate cell-free translation in extracts deficient in Pat1p. Extracts from wild-type cells (WT) and from cells deficient in Pat1p (pat1Δ) were assayed for their ability to translate LUC mRNA carrying a cap structure (CapLUC) or a poly(A) tail (LUCpA) or both (CapLUCpA) under the same conditions, in which protein synthesis responded linearly to the mRNA concentrations. Luciferase production was determined by luminescence. Activities were measured under noncompetitive conditions in nuclease-treated extracts and under competitive conditions in the presence of endogenous RNA in untreated extracts. Five repeat experiments were performed. Error bars represent variations (about 15 to 20%) between individual experiments. RLU, relative light units.
The synthesis rate in the mutant extract was about 30-fold lower than that in the wild-type extract, regardless of the structure of the LUC mRNA or the conditions of synthesis used. This difference was much higher than that observed when yeast polysomal mRNA was used as substrate (the synthesis rate was only eightfold lower) (Fig. 4A). This difference was not due to differences in LUC mRNA stability. 35S-labeled MFA2 mRNAs were not degraded by incubation in either extract (as shown by results of centrifugation of initiation complexes in sucrose gradients [see Fig. 7 and 8]). LUC mRNAs were functional in the mutant extract and were translated into active luciferase, without loss of activity, in a mixture of both extracts (data not shown). The translation rate of CapLUCpA mRNA was not inhibited by competition with endogenous mRNAs (Fig. 5); thus, it is unlikely that the difference was due to an inadequate structure of LUC mRNA. The difference in the translation rates might reflect intrinsic variations existing in the translatability of the mRNAs, which may be amplified by the poorer translation efficiency of the pat1Δ extract.
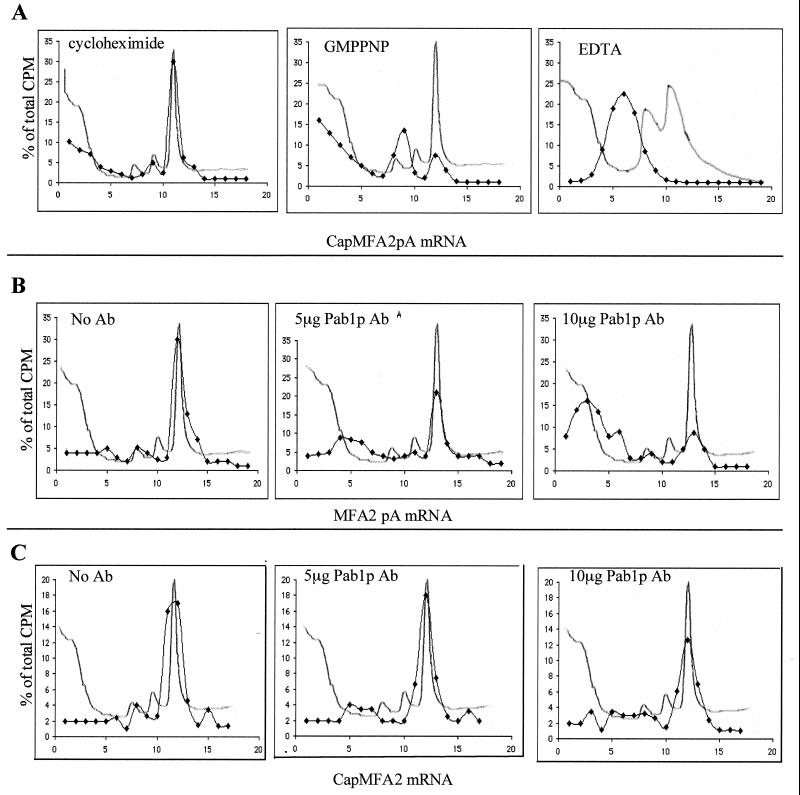
FIG. 7.
(A) Separation of translation initiation intermediates by centrifugation through sucrose gradients. Translation extracts from wild-type strains containing the indicated inhibitors were programmed with 35S-labeled CapMFA2pA mRNA. Formation of initiation complexes was analyzed on 10 to 30% sucrose gradients. Sedimentation proceeded from left to right. OD260 profiles were monitored to determine positions of 40S, 60S, and 80S particles (line without symbol). Radioactivity was determined in each fraction by direct counting, and data are expressed as the percentage of total radioactivity in the gradient (⧫). Labeled RNA cosedimented with the 80S initiation complex in the presence of cycloheximide, the 48S preinitiation complex in the presence of GMPPNP, and free mRNP in the presence of EDTA. (B and C) Pab1p antibodies inhibit 40S ribosomal subunit binding to MFA2pA mRNA but not to CapMFA2 mRNA. Translation extracts from wild-type cells containing the indicated amounts of Pab1p antibody were programmed with MFA2pA mRNA (B) or CapMFA2 mRNA (C) in the presence of cycloheximide and analyzed on sucrose gradients as described for panel A. Ab, antibody.
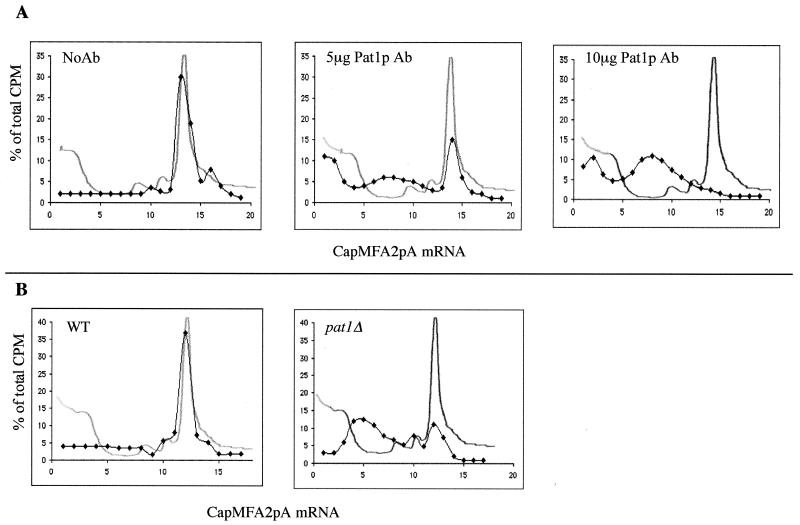
FIG. 8.
(A) Pat1p antibodies inhibit 40S ribosomal subunit binding to CapMFA2pA mRNA. Translation extracts from wild-type cells containing the indicated amounts of Pat1p antibodies were programmed with CapMFA2pA mRNA in the presence of cycloheximide. Initiation complexes were analyzed on sucrose gradients as described in the legend to Fig. 7A. (B) Translation extracts deficient in Pat1p are unable to form stable initiation complexes. Extracts from wild-type cells (WT) and cells deficient in Pat1p (pat1Δ) were programmed with amounts of CapMFA2pA mRNA 10 times those used in the experiments for which the results are shown in panel A. Formation of initiation complexes was analyzed as described in the legend to Fig. 7A. Ab, antibody.
Exogenous poly(A) is a specific inhibitor of translation initiation of poly(A)+ RNAs but not of poly(A)− RNAs, and it is supposed to act by limiting the availability of Pab1 protein (32). Similarly the cap analog m7GpppG prevents the translation of capped mRNAs by inhibiting the binding of eIF4E initiation factor to 5′ ends, but it has no inhibitory effect on the translation of uncapped mRNAs (46). We tested CapLUCpA mRNA translation in the presence of these two inhibitors. Inhibition was similar in RNA-dependent translation extracts from the wild-type and pat1Δ strains (data not shown). This confirmed that translation initiation in extracts deficient for Pat1p involves recognition of both the cap structure and the poly(A) tails. Thus, the inefficiency of translation is not due to a bypassing of the stimulatory functions of the 5′ and 3′ ends of the mRNA.
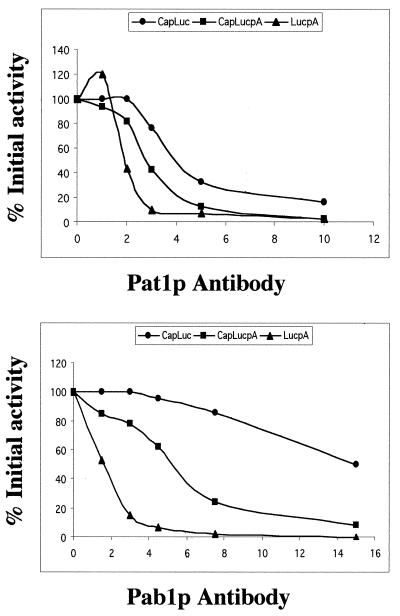
We studied the effects of various concentrations of Pat1p and Pab1p antibodies on the translation of the three forms of LUC mRNA (Fig. 6). As previously shown (46), translation of LUCpA mRNA was abolished by low concentrations of Pab1p antibodies. That of CapLUC mRNA was only sensitive to high antibody concentrations. Inhibition was intermediate with CapLUCpA mRNA. Experiments with Pat1p antibodies gave different results. Neutralization of Pat1p inhibited protein synthesis from all LUC mRNA species. Translation of CapLUC and CapLUCpA mRNAs was similarly affected. Low concentrations of antibodies weakly but reproducibly stimulated LUCpA mRNA translation; as the antibody concentration increased, however, the translation activity fell more steeply. Nonspecific antibodies used as a control had no effect on protein synthesis (data not shown).
FIG. 6.
Pat1p antibodies inhibit cap- and poly(A) tail-dependent translations. Nuclease-treated extracts from wild-type cells were incubated with the indicated amounts (in micrograms) of Pab1p or Pat1p antibody. Translation was initiated by addition of LUC mRNA with the indicated structure. Luminescence was measured after 30 min of incubation at 28°C. Data are expressed as percentages of activity in the absence of antibody. These 100% values fall within the ranges indicated by the error bars shown in Fig. 5.
We conclude that the loss of Pat1p function strongly diminishes the efficiency of protein synthesis. This effect is global, and both cap- and poly(A)-dependent translations are affected. The synergy involving the recognition of the 5′ and 3′ ends of mRNA is not abolished in the absence of Pat1p and continues to require Pab1p since Pab1p antibodies inhibited protein synthesis in extracts deficient in Pat1p (Fig. 4D).
Pat1p is required for 40S ribosomal subunit joining to capped and/or polyadenylated mRNA.
We compared the formation of initiation complexes in extracts containing or lacking Pat1p. RNA-dependent translation extracts were programmed with the 35S-labeled MFA2 mRNA synthesized with either a cap structure (CapMFA2 mRNA), a 100-adenine-long poly(A) tail (MFA2pA mRNA) or both (CapMFA2pA mRNA). Extracts were added in excess so that all the transcripts, whatever their structure, were recruited by ribosomes to form initiation complexes. Ribosome-mRNA association was analyzed by centrifugation on sucrose gradients to separate 40S and 60S ribosomal subunits and 80S ribosomes. RNA migration on gradients was directly assayed by counting radioactivity in each fraction. The data are plotted as the percentage of total radioactivity present in each gradient to facilitate comparisons. As previously shown (12, 46), various initiation intermediates can be isolated in the presence of the following inhibitors (Fig. 7A): (i) cycloheximide, which inhibits peptide chain elongation, leading to the accumulation of the true RNA-associated 80S initiation complexes ready to engage in the elongation phase (all the experiments for which the results are shown below were performed in the presence of cycloheximide); (ii) GMPPNP, a nonhydrolyzable GTP analog that blocks 60S subunit binding, resulting in the accumulation of 48S intermediates (labeled mRNA comigrated essentially with the 40S subunit; the second heaviest peak observed corresponded to two 40S molecules bound to a single mRNA); and (iii) EDTA (mRNPs are released in the presence of EDTA, which dissociates ribosomes). Use of these inhibitors allows one to distinguish between free nontranslating mRNPs, 48S preinitiation complexes, and full 80S initiation complexes.
The formation of initiation complexes was analyzed in wild-type translation extracts in the presence of increasing amounts of Pab1p antibodies. The sedimentation of MFA2pA mRNA progressively shifted from 80S to the free mRNP position (Fig. 7B). The migration of CapMFA2 mRNA was not significantly modified and the 80S initiation complexes were stable under the same conditions (Fig. 7C). Preimmune serum had no effect on mRNA sedimentation (data not shown). As previously shown (46), immunoneutralization of Pab1p blocked the binding of the 40S ribosomal subunit to polyadenylated mRNP and not to capped mRNP.
We studied the effect of Pat1p immunoneutralization on the formation of initiation complexes using CapMFA2pA mRNAs (Fig. 8A). mRNAs sedimented as a broad peak overlapping 40S and mRNP positions that progressively shifted to the free mRNA position with increasing concentrations of Pat1p antibodies. Nonspecific antibodies purified from preimmune serum did not affect RNA migration (data not shown). Identical results were obtained in the presence of CapMFA2 or MFA2pA mRNA under the same conditions (data not shown). We tested the formation of the initiation complexes in the pat1Δ extract. Experimental conditions were modified to increase the competition between mRNAs for the formation of initiation complexes. The mRNA/translation extract ratio was 10-fold higher than that used in the experiments for which results are reported in Fig. 7 and 8A. Total CapMFA2pA mRNA was recruited to form 80S initiation complexes in wild-type extract under these conditions. In contrast, extract from pat1Δ cells was unable to form large amounts of 80S initiation complexes and about 80% of the labeled mRNA was found in lighter fractions and sedimented essentially as mRNP (Fig. 8B).
These results are consistent with a function of Pat1p in translation initiation seen in vivo. In addition, they seem to indicate that Pat1p acts at an early step in the process. 48S particles did not accumulate in large amounts in the absence of the protein, as would be expected if 60S subunit binding were blocked. Pat1p might be required to stabilize the association between the 40S ribosomal subunit and the mRNP. Most mRNA is found as free mRNP, and few stable 80S complexes were isolated from the pat1Δ extract, although a portion of the mRNA comigrated with the 40S subunit. Moreover, the pat1Δ strain grew so slowly that there might be defects in the extract for other reasons. Both Pab1p and Pat1p play a part in the formation of translation initiation complexes in vitro. However, Pab1p is involved mainly in poly(A) tail-dependent translation, whereas Pat1p would be required whatever the mRNA structure.
DISCUSSION
We selected mutations suppressing the lethality associated with a PAB1 gene deletion. We found among them mutations and a deletion in PAT1 gene. The deletion of this gene, even when not associated with the PAB1 deletion, presents a phenotype of slow growth and of translation impairment in vivo as well as in vitro in extracts from the pat1Δ strain. The same impairment can be seen in wild-type extracts treated by Pat1p antibodies; this shows that the translation defect seen in the pat1Δ mutant is not a secondary effect due to a more general metabolic defect.
Different results indicate that Pat1p is involved in translation initiation, as follows. (i) The polysome profile from a pat1Δ strain was characteristic of an inhibition of the translation initiation, with a reduction of the size of polysomes and an accumulation of 80S ribosomes (Fig. 2). (ii) In an in vitro translation extract deficient in Pat1p the translation of poly(U), which is independent of initiation factors (22), was not inhibited compared to the strong inhibition seen for yeast mRNAs which require initiation factors (Fig. 4A and E). (iii) An analysis of initiation complexes on sucrose gradients showed a lack of complete 80S initiation complexes, a strong reduction of 48S intermediary complexes, and an accumulation of free mRNAs lighter than the 40S ribosomal subunit (Fig. 8). All these observations can be interpreted to indicate a defect in the fixation of the 40S subunits on the mRNAs.
The in vitro association of the 40S subunits to the 5′ end of mRNAs has been thoroughly described. It depends on the 5′ cap structure, the 3′ poly(A) tail, and the proteins to which they are associated. The poly(A)-binding protein, Pab1, is thus involved in the joining of the 40S subunit to polyadenylated mRNAs and not to mRNAs which are only capped (46). This is not the case for Pat1p, since its neutralization affects to the same extent the formation of the initiation complexes on capped MFA2, polyadenylated MFA2, and both capped and polyadenylated MFA2 mRNAs (Fig. 8). These results are consistent with those obtained with the different luciferase transcripts (Fig. 6). It seems that, in contrast to that of Pab1p, the function of Pat1p in in vitro translation is not directly related to the 5′ and 3′ end interactions stabilizing the 48S preinitiation complex. This conclusion is further strengthened by the translation behavior of Pat1p-deficient extracts, as follows. (i) The extracts showed the synergistic enhancement of translation due to the interactions between the 5′ cap structure and the polyadenylated 3′ end (Fig. 5). (ii) The translation level of capped and polyadenylated LUC mRNAs was maintained in the presence of endogenous mRNAs (Fig. 5) (the efficiency of translation depends on 5′ to 3′ interactions in these conditions of competition) (34, 35). (iii) Pab1p antibodies inhibited translation, showing that the poly(A)-bound Pab1p is at least partly involved in translation stimulation independently from Pat1p.
In lysates prepared with cells pretreated with cycloheximide, Pat1p sedimented mainly at the level of the 40S subunit and of the 43 to 48S complexes but was also present in smaller quantities in the next lighter fractions as well as in monosomes and polysomes (Fig. 3A). In absence of cycloheximide, when ribosome runoff occurred and the translation reinitiation rate was very low (37), Pat1p was only seen at the level of the 43 to 48S complexes and in the lighter fractions (Fig. 3B). It was notably absent from the 80S ribosomes known to be inactive and not associated with mRNAs under these conditions. This strongly suggests that Pat1p is physically associated with polysomes in active translation. Moreover, the amounts of Pat1p dropped with the polysome size, suggesting a stoichiometry of 1, like the initiation complex. A significant amount of Pat1p, increasing in runoff conditions, was seen in fractions lighter than the 40S subunit, in which the protein was likely engaged in protein complexes. Pat1p might be associated with the 43 to 48S complex in a dynamic process depending on active translation. This would be consistent with the very low amounts of 80S and 43 to 48S complexes which have been isolated on gradients from extracts of the pat1Δ strain (Fig. 8B). In this strain, the affected step would occur before or during the fixation of the 40S subunit to mRNA. Pat1p might be required for stabilizing the translation preinitiation complex.
After the completion of this work, it was shown that the PAT1 gene has been isolated under the name of MRT1, a gene whose mutants increase mRNA stability and suppress a PAB1 deletion. A study of the turnover of some mRNAs in conditions allowing the characterization of degradation intermediates led to the conclusion that Pat1p (Mrt1p) stimulates decapping in the main pathway of mRNA degradation, where deadenylation precedes decapping and the exonucleolytic degradation of the mRNA body in the 5′ to 3′ direction (14). Pat1p (Mrt1p) has been found in a multiprotein complex, where it is associated with the Dcp1 decapping enzyme and with several Sm-like proteins named Lsm (W. He, S. Tharum, A. Mayes, D. Dunkley, P. Lennetz, J. Beggs, and R. Parker, Abstr. 4th Annu. Meet. RNA Soc., abstr. RNA 99, 1999). Among them, Lsm1p (also known as Spb8p) is also associated with decapping regulation and the deletion of this gene suppresses a PAB1 deletion (4).
Our experiments do not bear directly on mRNA stability, but an in vivo global study on the length of the poly(A) tails and on the amounts of capped or polyadenylated mRNAs in our pat1Δ (mrt1Δ) strain has not shown differences able to explain the modification of the polysome profile of this strain compared to that of the wild-type strain. Moreover, the mRNAs extracted from these two strains present the same efficiency of translation in yeast extracts able to respond to translation stimulation by capping and polyadenylation. Our translation experiments have been done using homologous as well as heterologous exogenous mRNAs. All these data argue against the possibility that the observed translation defects are a direct consequence of a structural difference in the translated mRNAs. Thus, the pleiotropic phenotype of pat1 (mrt1) mutants could just be a new example of the strong connection between translation and mRNA turnover (6, 20). The following are notable: (i) drugs and mutations inhibiting translation elongation also inhibit mRNA degradation (20). (ii) the stabilizing or destabilizing effect of specific sequences present in mRNA appears only when translation has begun (15, 21). (iii) some of the proteins involved in mRNA turnover, like Xrn1p and Upf1p, are associated with polysomes (25) and (iv) mRNA degradation intermediates are found in polysomes, indicating that the degradation can occur during mRNA translation (25).
Pat1p (Mrt1p) could be one of the numerous proteins necessary for the equilibrium between mRNA translation and degradation. It could both stabilize the initiation complex and localize the decapping and, maybe, the exonucleolitic enzymes to their sites of action. Pab1p also demonstrates these two properties, being both necessary to recruit the 40S subunit to mRNAs and involved in deadenylation and decapping (7, 46). The exact mechanism by which a deletion of PAT1 (MRT1) (and maybe also LSM1 [SPB8]) suppresses a PAB1 deletion remains to be found. It could involve an inhibition of the mRNA turnover that allows the transcripts to reach a level counteracting the stabilization defect due to the lack of Pab1p. It could also involve modifications of the translation initiation complexes, reducing the role of Pab1p in this process. Both mechanisms could in fact act synergistically to bypass the necessity of the Pab1p.
Pat1p was first characterized by its interaction in a two-hybrid screen with topoisomerase II involved in DNA metabolism (51). A direct relationship of this interaction with translation is not very likely. It is probable that Pat1p (Mrt1p) belongs to the group of yeast proteins which have pleiotropic functions like the exonuclease Xrn1 implicated in mRNA degradation and meiosis (45) or the Dcp2p (also known as Psu1p) involved in decapping and transcription (9, 11). There is a growing evidence, now that the yeast genome is under sharp scrutiny, that a significant number of its proteins display multifunctional properties.
ACKNOWLEDGMENTS
We thank A. B. Sachs, D. R. Gallie, and A. G. Hinnebush for their gifts of plasmids and M. Ferrer and G. Dujardin for their gifts of antibodies.
This work was supported by the Centre National de la Recherche Scientifique and the Association pour la Recherche sur le Cancer (grant 9922).
REFERENCES
- 1.Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amrani N, Minet M, Wyers F, Dufour M E, Aggerbeck L P, Lacroute F. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol Cell Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bach M L, Lacroute F, Botstein D. Evidence for transcriptional regulation of orotidine-5′-phosphate decarboxylase in yeast by hybridization of mRNA to the yeast structural gene cloned in Escherichia coli. Proc Natl Acad Sci USA. 1979;76:386–390. doi: 10.1073/pnas.76.1.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeck R, Lapeyre B, Brown C E, Sachs A B. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown C E, Sachs A B. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 8.Coller J M, Gray N K, Wickens M P. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 11.Gaudon C, Chambon P, Losson R. Role of the essential yeast protein PSU1 in p6 transcriptional enhancement by the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1999;18:2229–2240. doi: 10.1093/emboj/18.8.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray N K, Hentze M W. Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and cALAS mRNAs. EMBO J. 1994;13:3882–3891. doi: 10.1002/j.1460-2075.1994.tb06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlow E, Lane D. Antibodies, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 14.Hatfield L, Beelman C A, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennigan A N, Jacobson A. Functional mapping of the translation-dependent instability element of yeast MATα1 mRNA. Mol Cell Biol. 1996;16:3833–3843. doi: 10.1128/mcb.16.7.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hentze M W. eIF4G: a multipurpose ribosome adapter. Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 17.Hurt E C, McDowall A, Schimmang T. Nucleolar and nuclear envelop proteins of the yeast Saccharomyces cerevisiae. Eur J Cell Biol. 1988;46:554–563. [PubMed] [Google Scholar]
- 18.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobson A. Poly(A) metabolism and translation: the closed-loop model. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 20.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–740. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 21.LaGrandeur T, Parker R. The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context of the translational start codon. RNA. 1999;5:420–433. doi: 10.1017/s1355838299981748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leibowitz M J, Barbone F P, Georgopoulos D. In vitro protein synthesis. Methods Enzymol. 1991;194:536–545. doi: 10.1016/0076-6879(91)94040-j. [DOI] [PubMed] [Google Scholar]
- 23.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 24.Mangus D A, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)− binding protein, regulates polyadenylation. Mol Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangus D A, Jacobson A. Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods. 1999;17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy J E G. Posttranscriptional control of gene expression in yeast. Microbiol Mol Biol Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meredith R D, Erlanger B F. Isolation and characterization of rabbit anti-m7G-5′-P antibodies of high apparent affinity. Nucleic Acids Res. 1979;6:2179–2191. doi: 10.1093/nar/6.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merrick M C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 29.Minvielle-Sebastia L, Preker P J, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessager RNA 3′-end formation. Proc Natl Acad Sci USA. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moldave K. Eukaryotic protein synthesis. Annu Rev Biochem. 1985;54:1109–1149. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- 31.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the MFA2 gene leads to decapping followed by 5′-3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 32.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petitjean A, Bonneaud N, Lacroute F. The duplicated Saccharomyces cerevisiae gene SSM1 encodes the eucaryotic homolog of the eubacterial and archaebacterial L1 ribosomal protein. Mol Cell Biol. 1995;15:5071–5081. doi: 10.1128/mcb.15.9.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preiss T, Hentze M W. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 35.Preiss T, Muckenthaler M, Hentze M W. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proweller A, Butler S. Efficient translation of poly(A)-deficient mRNAs in Saccharomyces cerevisiae. Genes Dev. 1994;8:2629–2640. doi: 10.1101/gad.8.21.2629. [DOI] [PubMed] [Google Scholar]
- 37.Ramirez M, Wek R C, Hinnebusch A G. Ribosome association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3027–3036. doi: 10.1128/mcb.11.6.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez-Cousino N, Lill R, Neupert W, Court D A. Identification and initial characterization of the cytosolic protein Ycr77p. Yeast. 1995;11:581–585. doi: 10.1002/yea.320110608. [DOI] [PubMed] [Google Scholar]
- 39.Sachs A B, Bond M W, Kornberg R D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 40.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A)-shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 41.Sachs A B, Davis R W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247:1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- 42.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 45.Solinger J A, Pascolini D, Heyer W D. Active-site mutations in the Xrn1p exoribonuclease of Saccharomyces cerevisiae reveal a specific role in meiosis. Mol Cell Biol. 1999;19:5930–5942. doi: 10.1128/mcb.19.9.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarun S Z, Jr, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 47.Tarun S Z, Sachs B A. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 48.Tarun S Z, Wells S E, Deardoff J A, Sachs A B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tuite M F, Plesset J. mRNA-dependent yeast cell-free translation systems: theory and practice. Yeast. 1986;2:35–52. doi: 10.1002/yea.320020103. [DOI] [PubMed] [Google Scholar]
- 50.Vornlocher H P, Hanachi P, Ribeiro S, Hershey J W. A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiae TIF32 and TIF31 genes. J Biol Chem. 1999;274:16802–16812. doi: 10.1074/jbc.274.24.16802. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Watt P M, Louis E J, Borts R H, Hickson I D. Pat1: a topoisomerase II-associated protein required for faithful chromosome transmission in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:4791–4797. doi: 10.1093/nar/24.23.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells S E, Hillner P E, Vale R D, Sachs A B. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 53.Zhong T, Arndt K T. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]