Abstract
Evolutionary radiations have intrigued biologists for more than a century, yet our understanding of the drivers of radiating diversification is still limited. We investigate the roles of environmental and species-intrinsic factors in driving the rapid radiation of Saussurea (Asteraceae) by deploying a number of palaeoenvironment-, diversity- and trait-dependent models, as well as ecological distribution data. We show that three main clades of Saussurea began to diversify in the Miocene almost simultaneously, with increasing diversification rates (DRs) negatively dependent on palaeotemperature but not dependent on species diversity. Our trait-dependent models detect some adaptive morphological innovations associated with DR shifts, while indicating additional unobserved traits are also likely driving diversification. Accounting for ecological niche data, we further reveal that accelerations in DRs are correlated with niche breadth and the size of species' range. Our results point out a macroevolutionary scenario where both adaptive morphological evolution and ecological opportunities provided by palaeoenvironmental fluctuations triggered an exceptionally radiating diversification. Our study highlights the importance of integrating phylogenomic, morphological, ecological and model-based approaches to illustrate evolutionary dynamics of lineages in biodiversity hotspots.
Keywords: rapid radiation, Saussurea, diversification rates, ecological opportunities, morphological innovations, the Qinghai-Tibet Plateau
1. Introduction
The diversification pattern of species-rich rapid radiations reflects the evolutionary dynamics of biodiversity hotspots [1]. Understanding how these radiating lineages formed in response to historical processes can advance our knowledge of adaptive evolution and enhance our ability to predict threats to biodiversity posed by global warming [2]. Mountainous regions represent just one-eighth of terrestrial land surface but are home to one-third of all species and exceptional species-rich radiations [3]. Particularly enigmatic is the Qinghai-Tibet Plateau (QTP), characterized by a complex geographical history and encompassing areas with remarkably high biodiversity [4,5]. This region includes the Himalaya and Hengduan Mountains which are listed as two of the 36 global biodiversity hotspots [4]. While a plethora of studies have suggested that diversification of plants on the QTP have evolved in association with the process of plateau uplifting (reviewed by Wen et al. [6]), the underlying mechanisms responsible for high species diversity in a short period of geologic time, and the interactions with geography, climate, ecology and biotic factors remain seldom examined.
Evolutionary radiations are often correlated with environmental forces, such as abrupt changes in climate or tectonic events that drive speciation and extinction rates, and/or species-intrinsic factors such as species interactions and key innovations [7,8]. Previous studies of plant diversification mostly provide only a temporal framework associating rapid radiations within the span of geologic history [4]. Employing model-based approaches that allow diversification rates (DRs) to continuously vary along with dependence on palaeoenvironmental variables is essential to determine how DRs are affected by environmental changes [9,10]. In addition to abiotic factors, diversification shifts are often correlated with the evolution of certain functional traits, such as higher rates of diversification in geophytic monocots [11], diversification promoted by polyploidization within Allium [12], and changes of pollinators, fruit types and elevation in Andean bellflowers [13]. Furthermore, the inclusion of ecological niche data is crucial due to the interplay between historical processes and intrinsic species interactions [7,14].
Here, we explore the drivers of a rapid radiation by examining the roles of abiotic (environmental) and biotic (species-intrinsic) factors in accelerating DRs of the species-rich genus Saussurea DC (Asteraceae). Saussurea is one of the most diverse genera in Asteraceae, serving as an ideal study system for investigating the evolutionary patterns of rapid radiations. The genus comprises approximately 400 species that are distributed in Asia, Europe and North America, with the highest diversity in the QTP [15–17]. Uncertainty in the number of species has largely been attributed to the complex taxonomy of related QTP taxa, indicative of a recent radiation. Saussurea includes four morphology-based subgenera (i.e. Amphilaena, Eriocoryne, Saussurea and Theodorea), exhibiting extraordinary morphological diversity. For example, the ‘snow lotuses’ or ‘greenhouse plants’, S. subg. Amphilaena, are characterized by the capitula hidden in semi-transparent leafy bracts [15,17]. Saussurea is present in virtually all habitats, including steppes, moist forests, cold and dry alpine meadows, and scree slopes above 5000 m, demonstrating a highly adaptive capability [15]. Previous studies suggested that adaptive morphological traits were the result of convergent adaptation to diverse environments in the QTP [16], yet the contributions of these traits to the high level of diversity in Saussurea are still elusive. While biogeographic analyses inferred that Saussurea originated during the early-middle Miocene in the Hengduan Mountains [18], limited information about macroevolutionary patterns related to historical climate and geologic processes are available due to the lack of modelling DRs.
A robust phylogenetic framework is the basis for large-scale analyses of macroevolutionary patterns [19], yet previous studies mainly relied on fragment DNA markers (e.g. [9,11,12]) which provide insufficient phylogenetic signal and yield parallel relationships for phylogenies of rapid radiations. In this study, we reconstructed a robust time-calibrated phylogeny of Saussurea using complete plastomes to explore the role of abiotic and biotic factors in this exceptional evolutionary radiation. If evolutionary dynamics are driven primarily by abrupt abiotic perturbations, we expect DR shifts to follow major climate or geological changes and show a strong dependence on palaeoenvironmental variables. Conversely, if biotic factors are the dominant drivers of evolution, we expect diversification shifts to be correlated with the evolution of functional traits and/or the increase of species diversity. In a joint-effect scenario, both ecological opportunities provided by climatic fluctuations and species-intrinsic factors can be responsible for the rapid radiation. To test these hypotheses, we deployed a series of palaeoenvironment-, diversity- and trait-dependent models, as well as ecological distribution data (electronic supplementary material, table S1), particularly with regard to the following questions. (1) When did the radiation of Saussurea start? (2) Was the radiating process associated with QTP tectonics, palaeoclimate fluctuation or species diversity? (3) Is there evidence that adaptive trait innovations accelerated the DR? And (4) how do ecological factors play a role in the rapid radiation of Saussurea?
2. Methods
(a) . Plastome sampling, sequencing and assembly
We sequenced plastomes for 65 species and downloaded an additional 161 plastomes from GenBank (accessed 29 November 2019); collectively these species include 199 taxa of Saussurea and 27 outgroups. Collection details of the specimens are provided in electronic supplementary material, table S2. We combined treatment evidence for all Saussurea names from The Plant List (http://www.theplantlist.org/), Flora of Pan-Himalaya [17] and Flora of China [15], in the priority order: Flora of China > Flora of Pan-Himalaya > The Plant List. Since the goal was species-level estimates of DRs, subspecific records were combined into a single species.
Total genomic DNA was extracted from silica-gel dried leaves with a modified hexadecyltrimethylammonium bromide (CTAB) method [20]. Purified DNA was fragmented and short-insert (500 bp) libraries were constructed per the manufacturer's instructions (Illumina). Libraries were quantified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and sequenced on an Illumina HiSeq 4000 platform at Novogene Co., Ltd. in Kunming, Yunnan, China. Raw reads were directly assembled with the organellar assembler NovoPlasty v.2.7.2 [21], using a seed-and-extend algorithm employing the plastome sequence of Saussurea japonica (Genbank accession: MH926107.1), with all parameters kept at default settings. Sequences of assembled plastome were initially annotated using plastid genome annotator [22] and then manually checked in Geneious v.9.0.5 [23].
(b) . Estimates of divergence times
Whole-plastome sequences were used to estimate divergence times, as our prior study suggested that including noncoding regions maximizes the power to resolve relationships of Saussurea [16]. Plastome sequences containing one inverted repeat region were aligned using MAFFT v.7.22 [24]. Poorly aligned regions were removed with trimAL v.1.2 [25] using the command ‘-automated1’. Age estimates were obtained using Markov chain Monte Carlo (MCMC) analysis in BEAST v.1.10.4 [26]. Two reliable fossil calibrations were assigned: (1) the crown age of Carduus–Cirsium group was set to a minimum age of 14 million years ago (Ma) based on the Middle Miocene achenes of Cirsium [27,28]; (2) the split between Centaurea and Carthamus was calibrated with a minimum age of 5 Ma, based on records of pollen and achenes of Centaurea dating to the Early Pliocene [29]. Both fossil calibrations were defined with lognormal distributions [30], placing the 95% height posterior density (HPD) for the node ages at 14.19–19.18 Ma and 5.19–10.18 Ma. Additionally, the crown age of Cardueae was set to 39.2 Ma as a secondary calibration with a normal distribution (95% HPD: 35.91–43.49 Ma) based on the estimation by Barres et al. [28]. A GTR + I + Γ nucleotide substitution model, uncorrelated relaxed lognormal clock and a birth–death (BD) model for the tree prior were applied [26]. The MCMC analysis was run for 500 million generations, sampling every 10 000 generations, and the first 10 000 samples were discarded as burn-in. Convergence of the MCMC runs were checked using Tracer v.1.6 [31]. A maximum clade credibility tree was then reconstructed in Treeannotator v.1.8.4 [32], with annotated median age and 95% HPD.
(c) . Estimates of diversification rate
BAMM v.2.5.0 [33] was used to explore the diversification dynamics of Saussurea. Prior values were selected using the ‘setBAMMpriors’ function using BAMMtools v.2.1.7 [34] implemented in R [35]. Due to the controversial species number in Saussurea, the incomplete taxon sampling was set to 0.5 for all subsequent analyses. The MCMC was run for 500 million generations and sampled every 50 000 generations. Post-run analyses were performed in BAMMtools, with an initial 10% discarded as burn-in, and the remaining data assessed for convergence and ESS values greater than 200. Rates-through-time plots were generated using ‘PlotRateThroughTime’ function for the entire genus as well as three phylogenetic clades (see the Results). In addition, we used TESS v.2.1 [36] as a complement to BAMM rates-through-time to detect abrupt changes in speciation and extinction rates by applying the R scripts of Condamine et al. [8]. Speciation rates of Saussurea were obtained using the ‘getTipRates’ function. Considering recent criticism relating to the statistical methods for lineage-specific diversification models like BAMM [37] (but also see [38]), we employed the semiparametric species-level lineage DR to calculate speciation rates [39]. We fit our BAMM tip rates and DR results into a linear model using phylogenetic generalized least squares (PGLS) under a Brownian motion model in APE v. 5.5 [40] to validate the correlation between these two methods. Moreover, phylogenetic analysis of variance (ANOVA) was performed to determine whether differences of species DRs among four traditional subgenera were significant using Phytools v. 0.7-80 [41].
(d) . Palaeoenvironment-dependent analyses
Seven palaeoenvironment-dependent models were tested in RPANDA v. 1.9 [10]: BD model with constant λ (speciation rate) and μ (extinction rate) (i); BD model with λ dependent on time (ii) and environment (iii) exponentially, and constant μ; BD model with constant λ, and μ dependent to time (iv) and environment (v) exponentially; and BD model with λ and μ dependent to time (vi) and environment (vii) exponentially. We used the equations: λ(E) = λ0 × eαE and μ(E) = μ0 × eβE for modelling exponential dependence, in which λ0 and μ0 are the speciation and extinction rates for a given environmental variable. The values of α and β are the rates of change in speciation and extinction according to the environment, with positive values indicating a positive effect of the environment on speciation or extinction [10]. We used global climate change over the last 12 million years (Myr) estimated from relative proportions of different oxygen isotopes [42] as palaeoenvironmental variables and randomly sampled 500 trees from the BEAST posterior distribution (outgroups removed) to accommodate phylogenetic and dating uncertainties. The R package PSPLINE v. 1.0 [43] was used to visualize the variation of speciation rates with palaeoenvironmental variables. In addition, we tested the effect of diversity dependence on the diversification history of Saussurea using the method of Etienne et al. [44] implemented in the R package DDD v. 2.7. Six different models were applied, including a constant speciation and extinction rates model (CSE) and five diversity dependence models: speciation linearly dependent on diversity without extinction (DDL), speciation linearly dependent on diversity with extinction (DDL + E), speciation exponentially dependent on diversity with extinction (DDX + E), speciation independent of diversity and extinction linearly dependent on diversity (DD + EL), and speciation independent of diversity and extinction exponentially dependent on diversity (DD + EX).
(e) . Trait-dependent analyses
Eight morphological characters were selected and coded based on descriptions in eFloras (http://www.efloras.org/), herbarium specimens and taxonomic literature, or manually checked directly using online herbarium specimens from the Chinese Virtual Herbarium (http://www.cvh.ac.cn/), JSTOR (https://plants.jstor.org/) and field collections (electronic supplementary material, table S3). These characters included four binary traits: stemless (0) versus cauliferous (1), stem glabrous (0) versus densely hairy (1), the absence (0) versus presence (1) of leafy bracts, and capitula solitary (0) versus numerous (1); and four multi-state traits: leaf margin entire (1) versus pinnately lobed (2) versus both types (3), leaves glabrous (1) versus sparsely hairy (2) versus densely hairy (3), phyllary in less than 5 (1) versus 5 (2) versus 6 (3) versus greater than 6 (4) rows, and phyllary glabrous (1) versus sparsely hairy (2) versus densely hairy (3) versus appendaged (4).
Shifts in DRs of binary traits were investigated using the hidden state speciation and extinction (HiSSE) model, which allows for testing hypotheses related to both the effects of the observed traits as well as incorporating unmeasured factors [45]. As described by Beaulieu & O'Meara [45], 25 models were tested in the R package HISSE v.1.9.10: a full HiSSE model allowing all states to vary independently; four binary state speciation and extinction-like models that excluded hidden states or constrained specific parameters of λ, μ and transition rates (q); four null models of character-independent diversification; and 16 models assuming hidden states with a variety of constrained values for λ, μ and q (electronic supplementary material, table S4). The best-fitting model was selected based on likelihood-ratio tests under a chi-square distribution and corrected Akaike's information criterion (AICc) [46]. Speciation, extinction and net DRs were calculated by model-averaging the marginal ancestral state reconstruction. We used the nonparametric FiSSE model (Fast, intuitive SSE model) [47] as a complementary analyses for measuring the robustness of our HiSSE results by calculating the speciation rates for the binary traits and determining if statistical differences between the two states for each character existed. For multi-state traits, multi-state speciation and extinction (MuSSE) analyses were performed in DIVERSITREE v.0.9.10 [48] by fitting four distinct models with subsequent ANOVA testing: a null model with fully constrained variables; a full model allowing all variables to change independently; a model constraining each μ to be equal (free λ) and a model constraining the λ values for each state to be equal (free μ). Further estimates for the parameters of λ, μ and net DRs (λ − μ) for each state were obtained in a Bayesian approach by incorporating a MCMC analysis with an exponential prior with 5000 generations.
(f) . Ecological distribution data and association with diversification rates
We used the ‘occ_search’ function of RGBIF v.1.3.0 [49] to retrieve latitude and longitude coordinates for Saussurea species from global biodiversity information facility (GBIF) on October 28, 2020 (GBIF references are presented in the electronic supplementary material, data S1). We extracted data records that were georeferenced and excluded any coordinates with zero and/or integer latitude and longitude. Duplicate collections and records with coordinate system issues were deleted manually to improve data reliability. We removed geographic outliers defined by boxplots of species occurrences in R. Range size of each species was estimated by applying a 5 km buffer around each locality using the ‘gBuffer’ function of RGEOS v.0.5-5 (https://CRAN.R-project.org/package=rgeos) following the descriptions of Testo et al. [50]. Range size data were log-transformed before analysis to overcome skewed distributions [50]. We extracted the values of 19 bioclimatic variables (from 1970 to 2000) from WorldClim (http://worldclim.org) using RASTER v.2.6-6 (https://CRAN.R-project.org/package=raster) and calculated a mean value for each species. Highly correlated variables were identified with a Pearson's correlation coefficient greater than 0.75, and subsequently removed. The remaining eight bioclimatic variables were annual temperature (BIO1), mean diurnal range (BIO2), isothermality (BIO3), max temperature of warmest month (BIO5), annual precipitation (BIO12), precipitation seasonality (BIO15), precipitation of warmest quarter (BIO18) and precipitation of coldest quarter (BIO19). The main variation of bioclimatic variables represented by climate lability was estimated by extracting the first two principal components (PC1 and PC2) from a principal component analysis in R. To calculate the ecological niche breadth, we first estimated environmental niche using generalized linear modelling in the R package ENMTOOLS v.1.0.2 [51], and then measured the spatial heterogeneity of the distribution of suitability scores using Levins's B metrics [52] (‘raster.breadth’ function).
To demonstrate whether ecological factors drove rapid diversification of Saussurea, a series of quantitative state speciation and extinction (QuaSSE) [48] and ES-sim [53] analyses were performed. Five models of QuaSSE with increasing complexity were constructed in DIVERSITREE to fit the changes in speciation rates with climate lability (PCs of bioclimatic variables), niche breadth and species range size. For the ES-sim analyses, in addition to the default inverse equal splits statistic [53], the DR statistic was also used as a reliable estimator to investigate correlations between speciation rate and continuous ecological factors using the R-scripts retrieved from Sun et al. [9].
3. Results
(a) . Divergence time and diversification rate
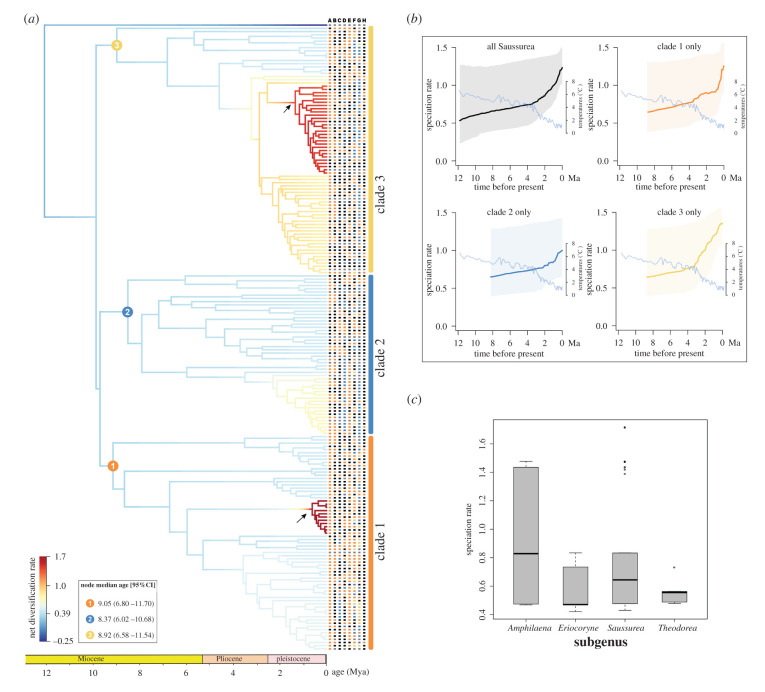
Our tree topology shows that the four traditional morphology-based subgenera of Saussurea are paraphyletic, indicating adaptive traits have occurred multiple times. Three phylogenetic clades were resolved (clades 1, 2 and 3; electronic supplementary material, figure S1). Estimated divergence times show a median stem age of ca 11.79 Ma (95% HPD, 8.38–15.35 Ma) for Saussurea. The three clades began to diversify in parallel during the Miocene (ca 9.05 Ma, ca 8.37 Ma and ca 8.92 Ma, respectively; electronic supplementary material, figures S1 and S2). Net DRs estimated by BAMM revealed two shifts in diversification within Saussurea having high posterior probability (figure 1a; electronic supplementary material, figure S3). Rates-through-time plots showed that DRs of the three clades accelerated during the Pleistocene (figure 1a,b). TESS results indicated that the speciation shifts had high posterior probabilities during the Pleistocene (ca 2–4 Ma), consistent with the BAMM results (electronic supplementary material, figure S4). The PGLS showed a significant positive correlation between BAMM tip rates and DR results (correlation coefficient: 0.313, p < 0.001**). Phylogenetic ANOVA of BAMM tip rates showed that speciation rates of S. subg. Amphilaena (0.945 events Myr–1 per lineage) was highest among four morphological-based subgenera (F = 6.698, p = 0.002**; figure 1d; electronic supplementary material, table S4). However, the DR statistic revealed no significant difference (F = 3.285, p = 0.120; electronic supplementary material, figure S5 and table S5) between the four morphological-based subgenera.
Figure 1.
Diversification dynamics of Saussurea inferred from BAMM analysis. (a) Two shifts in DRs (represented by arrows). The timing for the origins of the three major clades of the genus is provided. The character states of eight morphological traits are plotted on the tree, as follows. A: stemless (black) versus stemmed (orange), B: stem glabrous (black) versus densely hairy (orange), C: leaf margin entire (black) versus pinnately lobed (orange) versus both types (blue), D: leaves glabrous (black) versus sparsely hairy (orange) versus densely hairy (blue), E: capitula solitary (black) versus numerous (orange), F: phyllary in less than 5 (black) versus 5 (orange) versus 6 (blue) versus greater than 6 (yellow) rows, G: phyllary glabrous (black) versus sparsely hairy (orange) versus densely hairy (blue) versus appendaged (yellow) and H: the absence (black) versus presence (orange) of leafy bracts. Unknown character states are coloured in grey. (b) Rates-through-time plots of all Saussurea species and three main clades separately, with trends in global climate change over 12 million years depicted. (c) BAMM tip rates of four morphology-based subgenera of Saussurea. (Online version in colour.)
(b) . Palaeoenvironment-dependent diversification
Of the seven RPANDA models, a model with palaeotemperature-dependent speciation fit the data best (electronic supplementary material, table S6). The best-fit model indicated that past temperature and speciation rates were negatively exponentially dependent (α = −0.093, AICc = 654.971), while the extinction rate remained constant, suggesting extinction was likely unaffected by temperature fluctuations. This best-fit model demonstrated a diversification regime in which speciation rates had opposite responses to changes of temperature over time and increased toward the present (electronic supplementary material, figure S6), consistent with the results from the BAMM rates-through-time analysis (figure 1b). However, the constant BD model cannot be rejected because the difference in AICc between the best-fit model and the constant BD model is small (ΔAICc = 0.672; electronic supplementary material, table S6). The diversity dependence analysis showed that the constant speciation and extinction rates model (CSE) obtained the best support compared to five diversity dependence models (AICc = 639.556; electronic supplementary material, table S7), indicating that accumulating diversity did not influence DRs of Saussurea.
(c) . Trait-dependent diversification
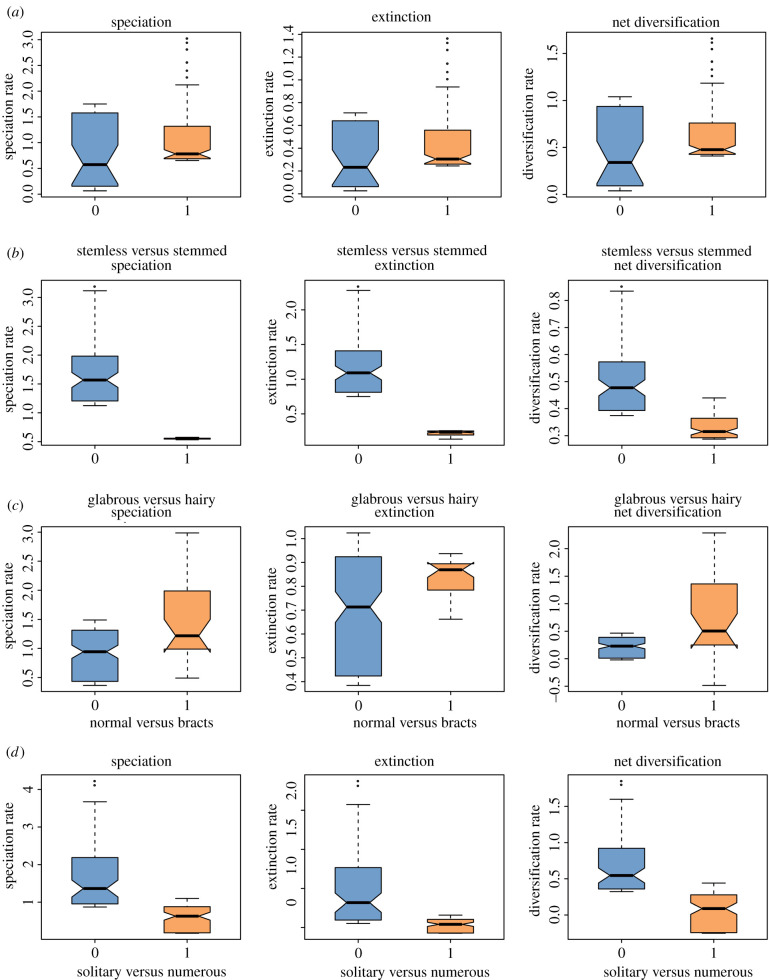
The best model for the binary traits was the full HiSSE model with unique speciation, extinction and transition rates between the two observed character states and hidden states (AICc: 741.520, 816.623, 739.764 and 714.435, respectively; electronic supplementary material, table S8). The calculated mean speciation, extinction and net DR values suggest that species with stems (λ = 1.089, μ = 0.382), glabrous stem (λ = 1.723, μ = 1.210), leafy bracts (λ = 1.395, μ = 0.875) and solitary capitula (λ = 1.692, μ = 0.566) have higher mean rates of speciation, extinction and net diversification (figure 2; electronic supplementary material, table S9). These traits are generally observed in the subgenus Amphilaena (snow lotus), congruent with the highest speciation rates among the four polyphyletic subgenera detected by BAMM tip rates. While the full HiSSE model showed observed differences in DRs between states, the analyses also indicated unobserved traits are driving the diversification. The FiSSE results supported the patterns of speciation rates revealed by HiSSE, but the only significant difference between states was solitary capitula (λ = 1.083) and numerous capitula (λ = 0.783) (p = 0.024; electronic supplementary material, table S9).
Figure 2.
Binary trait-dependent diversification of Saussurea inferred from HiSSE analysis. Speciation, extinction and net DRs are calculated by the model-averaged marginal ancestral state reconstruction for four binary traits: (a) stemless (0) versus stemmed (1), (b) stem glabrous (0) versus densely hairy (1), (c) the absence (0) versus presence (1) of leafy bracts, and (d) capitula solitary (0) versus numerous (1). (Online version in colour.)
In the MuSSE analyses, AMOVAs preferred models constraining μ to be equal and allowing λ to vary, compared with either null models and full models (AICc: 951.950, 895.464, 986.781 and 1016.269, respectively; electronic supplementary material, table S10). The reconstructions of the probability density of net DRs (λ − μ) showed that some traits (i.e. leaf margin and phyllary type) have an overlap in net DRs among examined character states (electronic supplementary material, figure S7). Species with glabrous leaves have higher net DRs than sparsely or densely hairy species, consistent with higher mean rates for glabrous stem in the HiSSE analysis. For the phyllary character, the glabrous state also showed higher net DRs than sparsely or densely hairy states despite some overlapping, and the rates of phyllary with six rows are higher than the remaining character states (electronic supplementary material, figure S7).
(d) . Ecological drivers of diversification
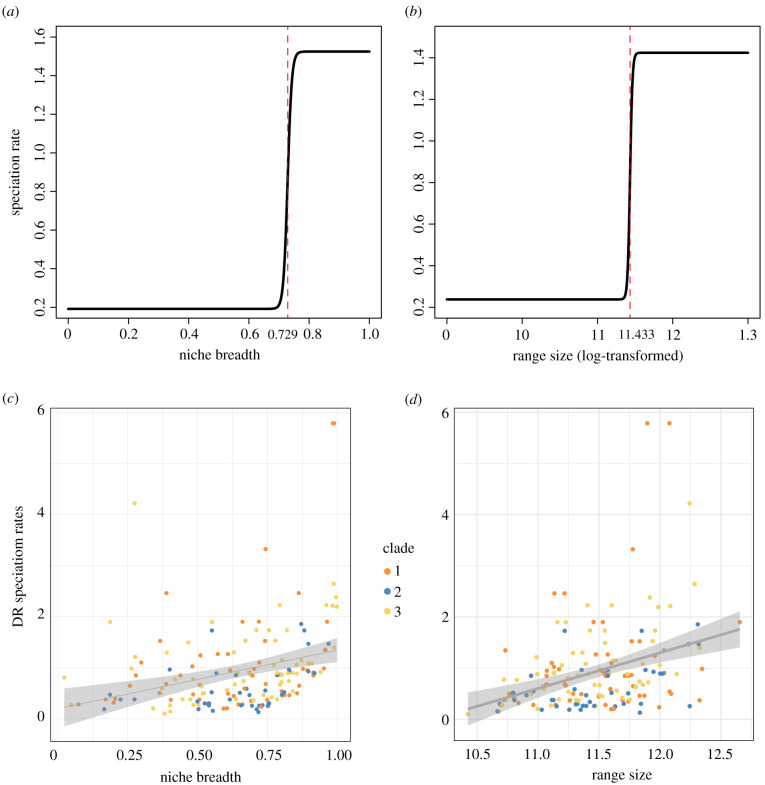
We correlated climate lability (PCs of bioclimatic variables), niche breadth and size of species range with speciation rates of Saussurea (electronic supplementary material, table S11). A total of 5352 distribution records of Saussurea passed the filtering criteria and were retained for ecological analyses. The first two PCs of bioclimatic variables explained 75.7% of the total climate variation in Saussurea (electronic supplementary material, figure S8a). Among the eight retained bioclimatic variables, the precipitation of warmest quarter (BIO18) had the largest contribution to the first two PCs, followed by annual precipitation (BIO12) and mean diurnal range (BIO2) (electronic supplementary material, figure S8b). Under the QuaSSE analyses, PC1 of the climate variables showed a significant positive linear (l.m = 0.330, AICc = 1240.548, p-value = 0.005**) relationship with speciation rate, while PC2 of the climate variables favoured a constant model (AICc = 1183.524, p-value = 0.953) (electronic supplementary material, table S12). Both niche breadth (AICc = 529.532, p-value less than 0.000**) and species range size (AICc = 700.671, p-value less than 0.000**) showed a significant positive sigmoidal (with drift) relationships with speciation rate (electronic supplementary material, table S12). The best sigmoidal models showed that the speciation rates of Saussurea maintained a low stable state until the niche breadth and distribution range reached 0.729 and 11.433 (log-transformed), respectively (midpoints; figure 3a,b). Under the ES-sim tests, both the DR statistic and the default inverse equal splits statistic revealed the same correlation pattern, in which both niche breadth (ρ = 0.363 and 0.387, p-value = 0.027* and 0.019*) and range size (ρ = 0.399 and 0.411, p-value = 0.018* and 0.011*) showed significant positive relationships with speciation rates (figure 3c,d), while the correlation between speciation rates and climate lability (climate PC1: ρ = 0.170 and 0.188, p-value = 0.359 and 0.335; climate PC2: ρ = 0.098 and 0.095, p-value = 0.649 and 0.635) was not significant (electronic supplementary material, table S13).
Figure 3.
Speciation rates of Saussurea correlated with ecological factors based on the QuaSSE best-fitted model and ES-sim tests. Both (a) niche breadth and (b) species range size (log-transformed) show positive sigmoidal curves in QuaSSE analysis with the midpoints (represented by the red dashed line) of 0.729 and 11.433 on the x-axis, respectively. EM-sim tests show significant positive relationships between DR speciation rates and (c) niche breadth and (d) species range size. Species from three clades are in different colours. (Online version in colour.)
4. Discussion
In this study, we took an integrative approach to address the role that environmental conditions and species-intrinsic interactions played in the evolutionary radiation of Saussurea. Specifically, we sought to determine when the radiation of Saussurea began and when major shifts occurred. Our dated phylogeny and BAMM analyses suggest an origin in the Miocene with increased diversification during the Pleistocene. These results were supported with the TESS analyses that showed a higher posterior probability for shifts in speciation during the Pleistocene. The diversification of Saussurea was negatively dependent on the palaeoclimate according to RPANDA, and was not dependent on species diversity. Moreover, several character states are directly associated with increases in diversification or speciation including leafy bracts, solitary capitula and glabrous stems. Unobserved traits appear to also impact diversification according to the HiSSE analyses. In addition to specific morphological traits, niche breadth and range size positively impact speciation rates according to QuaSSE. Overall, our results reveal a Miocene diversification pattern in which increased speciation rates are related to global climate changes, clade-specific traits and ecological factors, suggesting both adaptive morphological evolution and ecological opportunities provided by palaeoenvironmental fluctuations triggered an exceptionally radiating diversification of Saussurea species. Of note, while we included a global sampling fraction to account for effects due to incomplete sampling, our species sampling of Saussurea remains limited, introducing possible biases, which are inherent in any large-scale biodiversity analyses [11]. Nonetheless, our study highlights the importance of combining biotic and abiotic factors rather than a single factor in explaining the macroevolutionary pattern of a radiating lineage.
Our results showed that rapid diversification of Saussurea occurred in parallel during the Miocene, a period with extensive climatic fluctuations and tectonic movement on the QTP. Recent large-scale studies of species diversification have provided convincing evidence for a Miocene diversification in plant lineages [2] as well as amphibians and reptiles [54]. A hypothesis for the rich biodiversity found in mountainous regions like the QTP is uplift-driven diversification—that orogenic activities created diverse habitats favouring rapid in situ speciation of resident lineages [5]. Extensive plateau uplift in the Miocene further intensified summer monsoons, which increased the precipitation for erosion through river incision, leading to greater topographic relief. These events would have promoted the differentiation of microhabitats associated with elevational gradients and slope aspects, increasing the availability of habitats for radiating species [2].
The rates of species diversification were revealed to be negatively correlated with palaeotemperature and accelerate sharply in the Pleistocene toward the present. While a constant rates model cannot be rejected from RPANDA analysis, it is evident that some clades show shifts in DRs in the Miocene and Pleistocene from the BAMM and TESS results. The lack of ability to reject a constant rate model may be due to the heterogeneous geography of the QTP which could be obscuring larger evolutionary patterns within the Asteraceae [55]. However, the negative correlation between palaeotemperature and DRs in Saussurea does not seem surprising given the high species diversity found in high elevations. Nevertheless, this insight is progressive, as it represents one of the few attempts to explicitly quantify the relationship between lineage diversification and a palaeoenvironmental variable. Geological evidence suggests that after 15 Ma, global cooling and the further rise of QTP progressively led to more open, herb-rich vegetation as the modern high plateau formed with its cool, dry climate [56]. Thus, diversification among Saussurea clades could have been driven by increased ecological niches as suitable cold habitats became available. In addition, the diversification of Saussurea is not limited by the accumulation of species diversity, implying that the genus is possibly still in the evolutionary process of speciation.
Trait-dependent analyses demonstrated that species exhibiting cauliferous plant, glabrous stem, leafy bracts and solitary capitula have higher speciation rates. These traits are usually observed in the ‘snow lotus’, which are characterized by attractive leafy bracts, the symbols of snow mountains [15,17]. Despite having significant taxonomic characteristics, a snow lotus is a non-monophyletic group, demonstrating that these adaptive traits have multiple origins and arose by convergent evolution. In fact, specialized leafy bracts, the so-called ‘glasshouse’ morphology, are prevalent among many alpine species, such as in members of Lamiaceae, Asteraceae and Polygonaceae [57]. Leafy bracts reportedly protect pollen grains from damage by UV-B radiation and rain, promote pollen germination by maintaining warmth and facilitate the development of fertilized ovules during seed development [57]. Additionally, the subgenus Amphilaena show the highest speciation rates according to the tip rates analyses in BAMM. The species in this subgenus largely possess the traits of glabrous stems, leafy bracts and leaf margin entire, whereas these traits are less prevalent in other groups. Although trait-dependent analyses showed several adaptive traits driving an increase in speciation rate, unobserved traits also appear to be facilitating rapid diversification, such as two rows of pappus and small achenes that are common across the whole genus promoting the dispersal power of seeds to increase reproductive capacity [17]. This highlights the important roles of morphological diversity in the evolutionary history of Saussurea. Morphological diversity is an essential but often neglected aspect of biodiversity [58]. Our work provides a valuable guide for conservation efforts in the protection of morphological diversity of organisms, especially in the context of exacerbated biodiversity loss due to global warming.
Furthermore, our results suggest a positive relationship between speciation rate and niche breadth as well as species range, supporting the hypothesis of ecological opportunity-driven radiation. Among the few studies that have tested a niche breadth–diversification relationship, a clear consensus has not been reached. One argument for low niche breadth lineages having greater DRs is that they are more likely to suffer from resource limitations and more susceptible to range fragmentation, and thus resulting in allopatric speciation occurring more frequently [59]. An alternative view is that species with high niche breadth typically have larger range sizes and are therefore more likely to have their ranges fragmented by ecological or geographical barriers over evolutionary time, promoting allopatric speciation [60]. Combining our results, we argue that wider ecological niches can help species diverging in the QTP cope with climatic fluctuations, occupy microhabitats and promote morphological divergence. Hence, we conclude that the current mega-diversity of Saussurea is the result of interactions between global palaeoclimate, species morphological innovations and ecological opportunities.
Supplementary Material
Acknowledgements
We are grateful to all the collectors of Saussurea morphological and distribution data. We thank Ting-Shen Han for help in the visualization of figure 3. We thank Jun-tong Chen and Kai Xue for providing the photos of Saussurea species. We also thank the members of the alpine research group of KIB, Jianwen Zhang, Zhuo Zhou, Hongliang Chen, Lishen Qian, Lu Sun and Yongzeng Zhang for helping with sample collection, and Yazhou Zhang for helping with species identification.
Contributor Information
Tao Deng, Email: dengtao@mail.kib.ac.cn.
Hengchang Wang, Email: hcwang@wbgcas.cn.
Hang Sun, Email: sunhang@mail.kib.ac.cn.
Data accessibility
All newly sequenced plastomes were deposited in the National Center for Biotechnology Information (NCBI) database with accession numbers provided in electronic supplementary material, table S1. R scripts used in this study are available on GitHub (https://github.com/ZhangXu-CAS/Saussurea-diversification.git).
The data are provided in the electronic supplementary material [61].
Authors' contributions
X.Z.: formal analysis, investigation, methodology, resources, software, visualization, writing-original draft, writing-review and editing; J.B.L.: formal analysis, methodology, software, validation, writing-review and editing; Y.S.: investigation, validation, writing-review and editing; H.Z.: investigation and resources; N.L.: investigation and resources; T.K.: investigation, resources; X.H.: investigation and resources; T.D.: data curation, funding acquisition, investigation, resources and supervision; H.W.: conceptualization, data curation, funding acquisition, investigation, supervision, writing-review and editing; H.S.: conceptualization, data curation, funding acquisition, project administration, resources, supervision, writing-review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This study was supported by the Second Tibetan Plateau Scientific Expedition and Research program (grant no. 2019QZKK0502), the Strategic Priority Research Program of Chinese Academy of Sciences (grant no. XDA20050203), the Key Projects of the Joint Fund of the National Natural Science Foundation of China (grant no. U1802232), the Youth Innovation Promotion Association of Chinese Academy of Sciences (grant no. 2019382) and the Young Academic and Technical Leader Raising Foundation of Yunnan Province (grant no. 2019HB039).
References
- 1.Linder HP, Verboom GA. 2015. The evolution of regional species richness: the history of the Southern African Flora. Annu. Rev. Ecol. Evol. Syst 46, 393-412. ( 10.1146/annurev-ecolsys-112414-054322) [DOI] [Google Scholar]
- 2.Ding WN, Ree RH, Spicer RA, Xing YW. 2020. Ancient orogenic and monsoon-driven assembly of the world's richest temperate alpine flora. Science 369, 578-581. ( 10.1126/science.abb4484) [DOI] [PubMed] [Google Scholar]
- 3.Antonelli A. 2015. Biodiversity: multiple origins of mountain life. Nature 524, 300-301. ( 10.1038/nature14645) [DOI] [PubMed] [Google Scholar]
- 4.Favre A, Packert M, Pauls SU, Jahnig SC, Uhl D, Michalak I, Muellner-Riehl AN. 2015. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. Camb. Phil. Soc. 90, 236-253. ( 10.1111/brv.12107) [DOI] [PubMed] [Google Scholar]
- 5.Xing Y, Ree RH. 2017. Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl Acad. Sci. USA 114, E3444-E3451. ( 10.1073/pnas.1616063114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen J, Zhang JQ, Nie ZL, Zhong Y, Sun H. 2014. Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front. Genet. 5, 4. ( 10.3389/fgene.2014.00004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muellner-Riehl AN, Schnitzler J, Kissling WD, Mosbrugger V, Rijsdijk KF, Seijmonsbergen AC, Versteegh H, Favre A. 2019. Origins of global mountain plant biodiversity: testing the ‘mountain-geobiodiversity hypothesis’. J. Biogeogr. 46, 2826-2838. ( 10.1111/jbi.13715) [DOI] [Google Scholar]
- 8.Condamine FL, Rolland J, Hohna S, Sperling FAH, Sanmartin I. 2018. Testing the role of the Red Queen and Court Jester as drivers of the macroevolution of apollo butterflies. Syst. Biol. 67, 940-964. ( 10.1093/sysbio/syy009) [DOI] [PubMed] [Google Scholar]
- 9.Sun M, Folk RA, Gitzendanner MA, Soltis PS, Chen Z, Soltis DE, Guralnick RP. 2020. Recent accelerated diversification in rosids occurred outside the tropics. Nat. Commun. 11, 3333. ( 10.1038/s41467-020-17116-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condamine FL, Rolland J, Morlon H. 2013. Macroevolutionary perspectives to environmental change. Ecol. Lett. 16(Suppl 1), 72-85. ( 10.1111/ele.12062) [DOI] [PubMed] [Google Scholar]
- 11.Howard CC, Landis JB, Beaulieu JM, Cellinese N. 2020. Geophytism in monocots leads to higher rates of diversification. New Phytol. 225, 1023-1032. ( 10.1111/nph.16155) [DOI] [PubMed] [Google Scholar]
- 12.Han TS, Zheng QJ, Onstein RE, Rojas-Andres BM, Hauenschild F, Muellner-Riehl AN, Xing YW. 2020. Polyploidy promotes species diversification of Allium through ecological shifts. New Phytol. 225, 571-583. ( 10.1111/nph.16098) [DOI] [PubMed] [Google Scholar]
- 13.Lagomarsino LP, Condamine FL, Antonelli A, Mulch A, Davis CC. 2016. The abiotic and biotic drivers of rapid diversification in Andean bellflowers (Campanulaceae). New Phytol. 210, 1430-1442. ( 10.1111/nph.13920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folk RA, Stubbs RL, Mort ME, Cellinese N, Allen JM, Soltis PS, Soltis DE, Guralnick RP. 2019. Rates of niche and phenotype evolution lag behind diversification in a temperate radiation. Proc. Natl Acad. Sci. USA 116, 10 874-10 882. ( 10.1073/pnas.1817999116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Z, Raab-Straube EV. 2011. Cardueae. In Flora of China (eds Wu ZY, Raven PH, Hong DY), pp. 42-194. Beijing, People's Republic of China: Science Press. [Google Scholar]
- 16.Zhang X, et al. 2019. Plastome phylogenomics of Saussurea (Asteraceae: Cardueae). BMC Plant Biol. 19, 290. ( 10.1186/s12870-019-1896-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen YS. 2015. Asteraceae II Saussurea. In Flora of Pan-Himalaya (eds Hong D-Y, Sun H, Watson M, Wen J, Zhang X-C). Beijing, People's Republic of China: Science Press. [Google Scholar]
- 18.Xu LS, Herrando-Moraira S, Susanna A, Galbany-Casals M, Chen YS. 2019. Phylogeny, origin and dispersal of Saussurea (Asteraceae) based on chloroplast genome data. Mol. Phylogenet. Evol. 141, 106613. ( 10.1016/j.ympev.2019.106613) [DOI] [PubMed] [Google Scholar]
- 19.Koenen EJM, et al. 2020. Large-scale genomic sequence data resolve the deepest divergences in the legume phylogeny and support a near-simultaneous evolutionary origin of all six subfamilies. New Phytol. 225, 1355-1369. ( 10.1111/nph.16290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol. Ecol. Resour. 14, 1024-1031. ( 10.1111/1755-0998.12251) [DOI] [PubMed] [Google Scholar]
- 21.Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18. ( 10.1093/nar/gkw955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 50. ( 10.1186/s13007-019-0435-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearse M, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647-1649. ( 10.1093/bioinformatics/bts199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772-780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972-1973. ( 10.1093/bioinformatics/btp348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016. ( 10.1093/ve/vey016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mai DH. 1995. Tertiäre vegetationsgeschichte Europas: methoden und ergebnisse. Stuttgart, Germany: Gustav Fischer Verlag. [Google Scholar]
- 28.Barres L, Sanmartin I, Anderson CL, Susanna A, Buerki S, Galbany-Casals M, Vilatersana R. 2013. Reconstructing the evolution and biogeographic history of tribe Cardueae (Compositae). Am. J. Bot. 100, 867-882. ( 10.3732/ajb.1200058) [DOI] [PubMed] [Google Scholar]
- 29.Popescu SM. 2002. Repetitive changes in Early Pliocene vegetation revealed by high-resolution pollen analysis: revised cyclostratigraphy of southwestern Romania. Rev. Palaeobot. Palyno. 120, 181-202. ( 10.1016/S0034-6667(01)00142-7) [DOI] [Google Scholar]
- 30.Parham JF, et al. 2012. Best practices for justifying fossil calibrations. Syst. Biol. 61, 346-359. ( 10.1093/sysbio/syr107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901-904. ( 10.1093/sysbio/syy032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rambaut A, Drummond A. 2010. Treeannotator version 1.6. 1. Edinburgh, UK: University of Edinburgh. See http://beast.bio.ed.ac.uk. [Google Scholar]
- 33.Rabosky DL. 2014. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE 9, e89543. ( 10.1371/journal.pone.0089543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabosky DL, Grundler M, Anderson C, Title P, Shi JJ, Brown JW, Huang H, Larson JG. 2014. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5, 701-707. ( 10.1111/2041-210x.12199) [DOI] [Google Scholar]
- 35.Team RC. 2018. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 36.Hohna S, May MR, Moore BR. 2016. TESS: an R package for efficiently simulating phylogenetic trees and performing Bayesian inference of lineage diversification rates. Bioinformatics 32, 789-791. ( 10.1093/bioinformatics/btv651) [DOI] [PubMed] [Google Scholar]
- 37.Moore BR, Hohna S, May MR, Rannala B, Huelsenbeck JP. 2016. Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proc. Natl Acad. Sci. USA 113, 9569-9574. ( 10.1073/pnas.1518659113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabosky DL, Mitchell JS, Chang J. 2017. Is BAMM flawed? Theoretical and practical concerns in the analysis of multi-rate diversification models. Syst. Biol. 66, 477-498. ( 10.1093/sysbio/syx037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. 2012. The global diversity of birds in space and time. Nature 491, 444-448. ( 10.1038/nature11631) [DOI] [PubMed] [Google Scholar]
- 40.Paradis E, Schliep K. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526-528. ( 10.1093/bioinformatics/bty633) [DOI] [PubMed] [Google Scholar]
- 41.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217-223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 42.Zachos JC, Dickens GR, Zeebe RE. 2008. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 451, 279-283. ( 10.1038/nature06588) [DOI] [PubMed] [Google Scholar]
- 43.Ben J, Roberto GG. 2008. PSPLINE: Stata module providing a penalized spline scatterplot smoother based on linear mixed model technology. See https://boris.unibe.ch/69410. [Google Scholar]
- 44.Etienne RS, Haegeman B, Stadler T, Aze T, Pearson PN, Purvis A, Phillimore AB. 2012. Diversity-dependence brings molecular phylogenies closer to agreement with the fossil record. Proc. R. Soc. B 279, 1300-1309. ( 10.1098/rspb.2011.1439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beaulieu JM, O'Meara BC. 2016. Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Syst. Biol. 65, 583-601. ( 10.1093/sysbio/syw022) [DOI] [PubMed] [Google Scholar]
- 46.Akaike H. 1974. A new look at the statistical model identification. IEEE Trans. Autom. Control 19, 716-723. ( 10.1109/tac.1974.1100705) [DOI] [Google Scholar]
- 47.Rabosky DL, Goldberg EE. 2017. FiSSE: a simple nonparametric test for the effects of a binary character on lineage diversification rates. Evolution 71, 1432-1442. ( 10.1111/evo.13227) [DOI] [PubMed] [Google Scholar]
- 48.FitzJohn RG. 2012. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3, 1084-1092. ( 10.1111/j.2041-210X.2012.00234.x) [DOI] [Google Scholar]
- 49.Chamberlain SA, Boettiger C. 2017. R Python, and Ruby clients for GBIF species occurrence data. PeerJ Preprints 5, e3304v3301. ( 10.7287/peerj.preprints.3304v1) [DOI] [Google Scholar]
- 50.Testo WL, Sessa E, Barrington DS. 2019. The rise of the Andes promoted rapid diversification in Neotropical Phlegmariurus (Lycopodiaceae). New Phytol. 222, 604-613. ( 10.1111/nph.15544) [DOI] [PubMed] [Google Scholar]
- 51.Warren DL, Glor RE, Turelli M. 2010. ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33, 607-611. ( 10.1111/j.1600-0587.2009.06142.x) [DOI] [Google Scholar]
- 52.Levins R. 1968. Evolution in changing environments. Princeton, NJ: Princeton University Press. [Google Scholar]
- 53.Harvey MG, Rabosky DL. 2018. Continuous traits and speciation rates: alternatives to state-dependent diversification models. Methods Ecol. Evol. 9, 984-993. ( 10.1111/2041-210x.12949) [DOI] [Google Scholar]
- 54.Xu W, et al. 2020. Herpetological phylogeographic analyses support a Miocene focal point of Himalayan uplift and biological diversification. Natl Sci. Rev. 8, nwaa263. ( 10.1093/nsr/nwaa263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandel JR, Dikow RB, Siniscalchi CM, Thapa R, Watson LE, Funk VA. 2019. A fully resolved backbone phylogeny reveals numerous dispersals and explosive diversifications throughout the history of Asteraceae. Proc. Natl Acad. Sci. USA 116, 14 083-14 088. ( 10.1073/PNAS.1903871116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spicer RA, Farnsworth A, Su T. 2020. Cenozoic topography, monsoons and biodiversity conservation within the Tibetan Region: an evolving story. Plant Divers. 42, 229-254. ( 10.1016/j.pld.2020.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun H, Niu Y, Chen YS, Song B, Liu CQ, Peng DL, Chen JG, Yang Y. 2014. Survival and reproduction of plant species in the Qinghai-Tibet Plateau. J. Syst. Evol. 52, 378-396. ( 10.1111/jse.12092) [DOI] [Google Scholar]
- 58.Chartier M, von Balthazar M, Sontag S, Lofstrand S, Palme T, Jabbour F, Sauquet H, Schonenberger J. 2021. Global patterns and a latitudinal gradient of flower disparity: perspectives from the angiosperm order Ericales. New Phytol. 230, 821-831. ( 10.1111/nph.17195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vrba ES. 1987. Ecology in relation to speciation rates: some case histories of Miocene-Recent mammal clades. Evol. Ecol. 1, 283-300. ( 10.1007/bf02071554) [DOI] [Google Scholar]
- 60.Slatyer RA, Hirst M, Sexton JP. 2013. Niche breadth predicts geographical range size: a general ecological pattern. Ecol. Lett. 16, 1104-1114. ( 10.1111/ele.12140) [DOI] [PubMed] [Google Scholar]
- 61.Zhang X, et al. 2021. Macroevolutionary pattern of Saussurea (Asteraceae) provides insights into the drivers of radiating diversification. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Zhang X, et al. 2021. Macroevolutionary pattern of Saussurea (Asteraceae) provides insights into the drivers of radiating diversification. Figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All newly sequenced plastomes were deposited in the National Center for Biotechnology Information (NCBI) database with accession numbers provided in electronic supplementary material, table S1. R scripts used in this study are available on GitHub (https://github.com/ZhangXu-CAS/Saussurea-diversification.git).
The data are provided in the electronic supplementary material [61].