Abstract
In addition to facilitating the nuclear export of incompletely spliced viral mRNAs, equine infectious anemia virus (EIAV) Rev regulates alternative splicing of the third exon of the tat/rev mRNA. In the presence of Rev, this exon of the bicistronic RNA is skipped in a fraction of the spliced mRNAs. In this report, the cis-acting requirements for exon 3 usage were correlated with sequences necessary for Rev binding and transport of incompletely spliced RNA. The presence of a purine-rich exon splicing enhancer (ESE) was required for exon 3 recognition, and the addition of Rev inhibited exon 3 splicing. Glutathione-S-transferase (GST)-Rev bound to probes containing the ESE, and mutation of GAA repeats to GCA within the ESE inhibited both exon 3 recognition in RNA splicing experiments and GST-Rev binding in vitro. These results suggest that Rev regulates alternative splicing by binding at or near the ESE to block SR protein-ESE interactions. A 57-nucleotide sequence containing the ESE was sufficient to mediate Rev-dependent nuclear export of incompletely spliced RNAs. Rev export activity was significantly inhibited by mutation of the ESE or by trans-complementation with SF2/ASF. These results indicate that the ESE functions as a Rev-responsive element and demonstrate that EIAV Rev mediates exon 3 exclusion through protein-RNA interactions required for efficient export of incompletely spliced viral RNAs.
Retroviruses utilize a variety of mechanisms to differentially express numerous proteins from relatively small genomes which possess a single transcriptional start site. These mechanisms include, but are not limited to, the use of polyprotein precursors, ribosomal frameshifting (19), alternative start codons (6), bicistronic mRNAs (6), and alternative splicing (29). Alternative splicing allows the production of multiple viral mRNAs from a single RNA precursor. The simplest retroviruses, such as murine leukemia virus, express only two mRNAs, an unspliced RNA which serves both as mRNA for the Gag and Pol proteins and as progeny viral RNA, and a singly spliced mRNA which encodes the env gene. In contrast, complex retroviruses, such as human immunodeficiency virus type 1 (HIV-1), produce at least 20 different mRNAs, including several multiply spliced RNAs that encode small regulatory proteins (29).
In all retroviruses, alternative splicing requires the presence of suboptimal splice sites, allowing expression of several mRNAs from a single pre-RNA. Regulation of suboptimal splice sites is a complex process mediated in part by cis-acting RNA sequences that can either enhance or repress recognition of a splice site by the splicing machinery. Exon-splicing enhancers (ESEs) and silencers (ESSs) have been described for many virus and cellular RNAs (2, 23). ESEs typically are purine-rich sequences embedded within alternatively spliced exons. The purine-rich sequences mediate exon recognition through interactions with members of the SR protein family of splicing factors. SR proteins are both essential splicing factors and regulators of alternative splicing (reviewed in reference 16). Binding of SR proteins to an ESE recruits essential splicing factors to suboptimal splice sites near ESE sequences, resulting in exon inclusion of alternatively spliced exons.
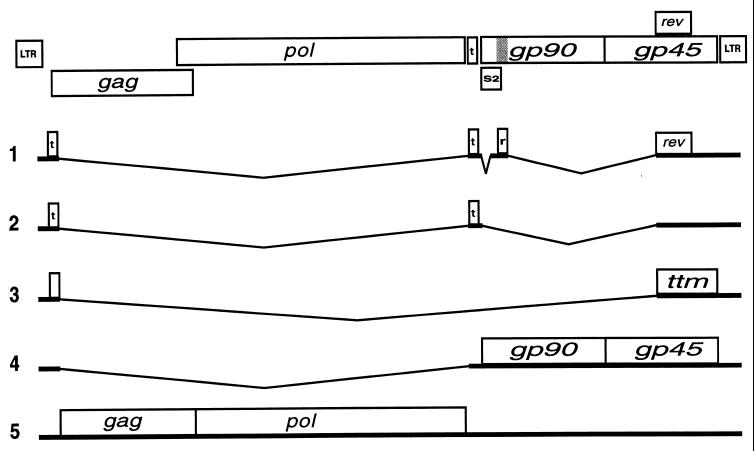
In addition to the above mechanisms, complex retroviruses utilize Rev-like pathways to differentially regulate expression of incompletely spliced RNAs encoding virion structural and enzymatic proteins and progeny RNA molecules (reviewed in reference 9). The prototype member of this family, HIV-1 Rev, binds to the viral pre-mRNA at a specific sequence called the Rev-responsive element (RRE) (8, 35), multimerizes (24, 34), and facilitates export of incompletely spliced RNAs from the nucleus via a CRM1-mediated pathway (13, 14). Equine infectious anemia virus (EIAV) Rev is functionally homologous to HIV-1 Rev (15) but is less well characterized. EIAV Rev is a 165-amino-acid protein translated from exons 3 and 4 of a multiply spliced, four-exon, bicistronic mRNA which also encodes the trans-activating protein Tat (Fig. 1) (6). In addition to promoting nuclear export of incompletely spliced RNA, EIAV Rev also regulates inclusion of exon 3 of the multiply spliced RNA. In the presence of Rev, a multiply spliced mRNA lacking exon 3 is produced (22). Rev variants which are nuclear export signal (NES) defective have been shown to mediate alternative splicing (4, 18); however, it is not known if the alternative splicing function is required for nuclear export activity. Exon 3 is flanked by a suboptimal splice acceptor and contains a purine-rich, ESE-like motif which has been shown to interact with the SR protein SF2/ASF (17). Gontarek and Derse have proposed that EIAV Rev-mediated skipping of exon 3 is a consequence of Rev-RNA interactions which directly or indirectly inhibit SF2/ASF (17). We previously mapped an RRE of Rev to a 534-nucleotide (nt) region containing exon 3 (4), suggesting the possibility that Rev mediates exon 3 skipping by binding at or near the purine-rich sequence to disrupt SR protein interactions necessary for exon 3 recognition.
FIG. 1.
Organization and splicing patterns of EIAV. Schematic of EIAV genome with open reading frames (ORFs). The tat ORFs are indicated with a t, and the first exon of rev is marked with an r (location in genome indicated by the shaded region). Splicing patterns and genes expressed are indicated. The ttm ORF encodes a truncated transmembrane protein of unknown function (3). LTR, long terminal repeat.
Here, we further delineate the role of Rev in exon 3 alternative splicing. Our results indicate that the purine-rich sequence in exon 3 is required for the utilization of the exon 3 splice acceptor, confirming the presence of an ESE within exon 3. RNA gel mobility shift assays and nuclear export assays demonstrate that Rev binds to the ESE and that this binding facilitates RNA export. Together, these results indicate that the exon 3 ESE is an RRE of EIAV. Therefore, Rev mediates exon 3 alternative splicing by binding the viral pre-mRNA at the ESE/RRE and interfering with SR protein-ESE interactions.
MATERIALS AND METHODS
PCR and plasmid construction.
All plasmid constructs were confirmed by sequence analysis (Iowa State University DNA Synthesis and Sequencing Facility). DNA templates for splicing substrates were amplified from p33k, a subclone of the p26 EIAV proviral clone described previously (5). Unless otherwise indicated, PCRs were performed as directed by the manufacturer (Perkin Elmer, Foster City, Calif.) using 1 μM primers. Standard PCRs consisted of 25 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 50°C, and 1 min of extension at 72°C, followed by an additional cycle with a prolonged, 5-min extension. All DNA templates for splicing substrates used a common 5′ primer, CTGAAGGCAATCCAACAAGG, and individual 3′ primers to generate the substrates shown in Fig. 2A. The 3′ primers used and the region of EIAV amplified were: CTCTCTATGATAAGCTTC, EIAV nt 5233 to 5793; CCAGTAGTTCCTGCTAAGCA, nt 5233 to 5573; TTTCCACCAGTCATTTCTTC, nt 5233 to 5535; and CAGGTTCATTTCTTGGTCT, nt 5233 to 5490. All nucleotide numbering is based on that of Kawakami et al. (20). After PCR, fragments were TA cloned into the pGEM-T Easy vector as directed by the manufacturer (Promega, Madison, Wis.).
FIG. 2.
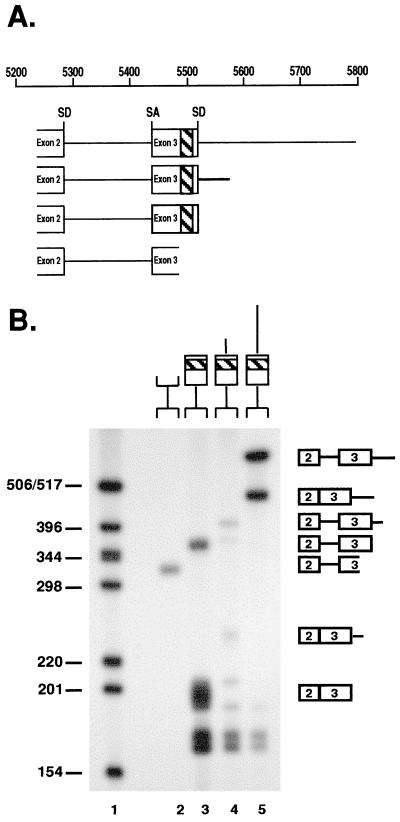
Exon 3 splicing requires the purine-rich sequence. (A) Diagram of RNA substrates used for in vitro splicing, showing the locations of exons 2 and 3. All substrates contain the exon 2 splice donor and exon 3 splice acceptor. The approximate location of the purine-rich sequence is highlighted. (B) After incubation for 2 h in HeLa cell splicing extracts, RNA products were electrophoresed through 4% polyacrylamide gels and visualized by autoradiography. The locations of spliced and unspliced products are shown. The fastest-migrating products in lanes 3 to 5 are intron products resulting from splicing. Sizes are shown at the left (in nucleotides).
The expression plasmid pRevWT was described previously as pcH21 (4). pDM138, pERRE-All (EIAV nt 5280 to 7534), and pERRE-1 (nt 5280 to 5834) have also been described previously (4). To construct pERRE-1A, primers containing a ClaI restriction site were used to amplify EIAV nt 5281 to 5795. The ERRE-1A 5′ primer was GGATCGATTTGATATATGGGATTATT, and the 3′ primer was GGATCGATCTCTCTATGATAAGCTTC (ClaI sites are underlined). The minRRE sequence (EIAV nt 5485 to 5540) was synthesized as complementary oligonucleotides with ClaI extensions on the 5′ and 3′ ends. The oligonucleotides were heated at 95°C for 5 min and annealed by slow cooling. The fragment was phosphorylated and then ligated with pDM138. The pGST-Rev expression vector contains a cDNA cloned in frame into the BamHI site of the glutathione-S-transferase (GST) fusion vector pGEX-3X (Amersham Pharmacia Biotech, Piscataway, N.J.).
ESE mutants were constructed by PCR-Ligation-PCR mutagenesis according to the methods described by Ali et al. (1) using internal primers designed with the specified mutations shown in Fig. 4A. The two regions were amplified with Vent DNA polymerase (New England Biolabs, Beverly, Mass.). The 3′ fragment was phosphorylated and ligated with the 5′ fragment, and 2 μl of the ligation reaction mixture was PCR amplified with the outer primers described above to amplify EIAV nt 5233 to 5793. Amplicons were cloned into pGEM-T. To construct the mutant pDM138 constructs, the pERRE-1A primers described above were used to PCR amplify the respective mutants from the pGEM-T background and clone them into the ClaI restriction site of pDM138.
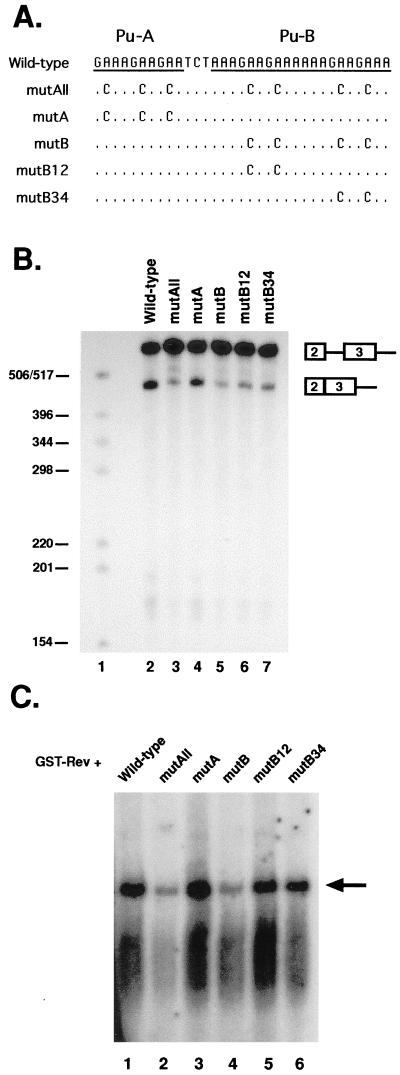
FIG. 4.
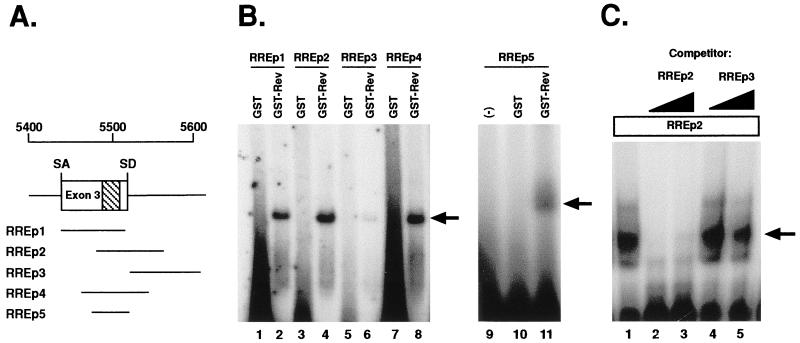
GST-Rev binds the ESE in exon 3. (A) Locations of the RNA probes, relative to exon 3, used in RNA gel mobility shift assays. The purine-rich ESE sequence is highlighted. SA, splice acceptor; SD, splice donor. (B) After incubation with GST or GST-Rev, radiolabeled RNAs were electrophoresed through 8% native polyacrylamide gels. The locations of GST-Rev-RNA complexes are indicated with arrows. (C) Competition assays were performed with either 0.5 or 1 μg of the indicated excess of unlabeled competitor RNAs. Competitors were mixed with GST-Rev 10 min prior to the addition of radiolabeled probe.
pSF2/ASF was generated from the PET9c-SF2 plasmid, obtained from Adrian Krainer, Cold Spring Harbor Laboratory (21). The cDNA region corresponding to SF2/ASF was cloned as two fragments, BglII-KpnI (−100 to +242; numbering based on +1 at the initiation AUG) and KpnI-BamHI (+242 to a BamHI site downstream of the UAA terminator). These were inserted into the eukaryotic expression vector pCMV5 (provided by Mark Stinski, University of Iowa) which had been cleaved with BglII and BamHI.
Synthesis of RNA substrates.
The plasmids containing the splicing substrates were digested with SpeI (Gibco-BRL, Rockville, Md.) to create linearized templates for transcription of RNA splicing substrates. In vitro run-off RNA transcripts labeled with [32P]UTP (Amersham Pharmacia Biotech) were generated as previously described (2). DNA templates for RNA binding analysis were amplified by PCR from p33k using 5′ primers containing a T7 promoter site (a diagram of the substrates is shown in Fig. 3A). The primers used for the substrates were: 5′ primer, TAATACGACTCACTATAGGGAGGAACAGCATGGCAGAATCG, and 3′ primer, TTTCCACCAGTCATTTCTTC (RREp1, nt 5443 to 5535); 5′ primer, TAATACGACTCACTATAGGGAGGTGAAAGAAGAATCTAAAG, and 3′ primer, CCACCAAAGTATTCCTCC (RREp2, nt 5489 to 5589); 5′ primer, TAATACGACTCACTATAGGGAGGTGACTGGTGGAAAATAGG, and 3′ primer, CCCTATATAATGTTGCTG (RREp3, nt 5523 to 5622); 5′ primer, TAATACGACTCACTATAGGGAGGCGGAGGAAGCAAGAGACC, and 3′ primer, CCTGCTAAGCATAACAGA (RREp4, nt 5458 to 5565). The T7 promoter is underlined in the 5′ primers. Amplified DNA was phenol-chloroform extracted, ethanol precipitated, and resuspended in RNase-free distilled water. The RREp5 DNA fragment was synthesized as two complementary oligonucleotides containing the T7 promoter attached to EIAV nt 5485 to 5540. Complementary DNA fragments were combined at equal molar amounts, heated at 95°C for 5 min, and then slowly cooled to anneal.
FIG. 3.
Rev inhibits exon 3 splicing. GST-Rev or GST was added at the indicated concentrations to the splicing reaction mixtures. The locations of the splicing products are indicated on the right. Sizes are shown at the left (in nucleotides).
Expression and purification of GST-Rev.
Escherichia coli BL21 transformed with the pGST-Rev expression vector was grown overnight at 1/10 of the final culture volume in NZY broth containing ampicillin (0.1 mg/ml). The next day, cells were brought up to the final volume, grown for an additional 3 h, and then induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 5 h. After induction, cells were washed three times and resuspended in 50 mM Tris (pH 8.0)–50 mM NaCl (TN buffer). Cells were lysed by sonication, and the supernatant was clarified by centrifugation at 10,000 × g. GST-Rev was purified by binding to glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech) overnight and washed three times with TN buffer. The fusion protein was eluted with 15 mM reduced glutathione in 50 mM Tris (pH 8.0), concentrated with a 30-kDa cut-off filter concentrator (Millipore, Bedford, Mass.), and dialyzed against TN buffer. Protein expression was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with convalescent anti-EIAV or anti-GST antiserum (Amersham Pharmacia Biotech), which detected expression of the GST-Rev fusion protein and several minor bands, including GST alone (data not shown). In some cases, the fusion protein was digested with 4 U of Factor Xa protease (Amersham Pharmacia Biotech) per bead bed volume while bound to glutathione-Sepharose beads. Excess GST and GST-Rev fusion protein were removed with the glutathione-Sepharose beads, and the supernatant, containing cut Rev protein, was concentrated and dialyzed as described above.
In vitro splicing and gel electrophoresis.
Splicing reactions were carried out as previously described (2). In brief, approximately 8 fmol of EIAV RNA substrates was incubated for 2 h at 30°C with 60% (vol/vol) nuclear extract in Dignam's buffer D (12) containing 20 mM creatine phosphate, 3 mM MgCl2, 0.8 mM ATP, and 2.6% (wt/vol) polyvinyl alcohol. In some experiments, EIAV Rev protein was diluted in buffer D (12) and added to the splicing reaction mixtures at the indicated concentrations. RNAs were analyzed on 4% polyacrylamide gels containing 7 M urea.
RNA binding assays and gel electrophoresis.
RNA-protein interactions were determined in 1× RNA binding buffer, containing 10 mM HEPES-KOH (pH 7.5), 100 mM KCl, 1 mM MgCl2, 0.5 mM EDTA, 1 mM dithiothreitol, 50 μg of E. coli tRNA per μl, and 10% glycerol. RNA was in vitro transcribed in the presence of [32P]UTP as described above. From 100 ng to 2 mg of GST or GST-Rev fusion protein was incubated with approximately 106 cpm of RNA probe on ice for 15 min. The reactions were loaded directly onto an 8% native 100 mM Tris–glycine–polyacrylamide gel (37.5:1 acrylamide-bisacrylamide cross-linking ratio) which had been prerun for 1 h. The samples were electrophoresed for an additional 3 h. The gel was fixed in 20% ethanol–10% acetic acid for 15 min, dried, and exposed to X-ray film with an intensifying screen.
CAT assays.
Transient transfections and chloramphenicol acetyltransferase (CAT) assays were performed with human embryonic kidney 293 cells and canine fetal thymus (Cf2th) cells. Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and penicillin-streptomycin. CAT assays with 293 cells were performed as previously described (4). Briefly, 1 μg of either pcDNA3 (Invitrogen, Carlsbad, Calif.) or pRevWT was transfected by calcium phosphate coprecipitation with 0.2 μg of pDM138 reporter plasmid, 0.2 μg of pCH110, and 0.6 μg of pUC19. Two days posttransfection, cells were harvested, resuspended in 0.3 ml of 0.25 M Tris (pH 7.5), lysed by freeze/thawing, and assayed for β-galactosidase activity to normalize CAT assays for transfection efficiency. Normalized lysates were assayed for CAT activity with 3 μl of [14C]chloramphenicol and 1 mM acetyl coenzyme A. Acetylated products were separated by thin-layer chromatography, and the percent acetylation was quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Experiments were performed in triplicate, and the results summarize a minimum of six independent transfections.
Cf2th cells were used for in vivo competition assays. Cells were transfected with 0.5 ng of pRevWT, 0.3 μg of pERRE-1 reporter plasmid, and 10-fold increasing concentrations (0 to 100 ng) of pSF2/ASF with 4 μl of TransIT-LT reagent (Mirus Corporation, Madison, Wis.). Each reaction mixture also included 0.2 μg each of pCH110 and pUC19 to equalize the total amount of DNA per reaction. Two days posttransfections, cells were harvested, and lysates were assayed for β-galactosidase activity as above. Normalized lysates were assayed for CAT activity with a commercially available CAT enzyme-linked immunosorbent assay (ELISA) kit (Roche Molecular Biochemicals).
RESULTS
Purine-rich sequence required for exon 3 recognition.
Exon 3 of the bicistronic, four-exon EIAV mRNA contains a purine-rich sequence which resembles an ESE. Previous reports showed that the SR protein SF2/ASF cross-links to the ESE-like sequence in vitro and suggested that the ESE-like sequence may enhance exon 3 inclusion during pre-mRNA splicing (17). To further investigate the cis-acting requirements for exon 3 inclusion, we constructed a series of DNA templates to generate radiolabeled RNA substrates for in vitro splicing. All substrates contained the exon 2 splice donor, the intervening intron, exon 3, and downstream sequences. Nested 3′ deletions were made to identify splicing enhancer sequences present within or downstream of exon 3 (Fig. 2A). Splicing of radiolabeled substrates was assayed in vitro with HeLa cell nuclear extracts, which include all SR proteins. All constructs containing the purine-rich sequence were spliced (Fig. 2B, lanes 3 to 5), whereas no splicing was observed for the substrate lacking the purine-rich sequence (Fig. 2B, lane 2). This is consistent with the hypothesis that the purine sequence functions as an ESE and is required for exon 3 inclusion in the multiply spliced four-exon transcript. Taken together with previous work (17), this suggests that exon 3 recognition requires SF2/ASF interactions at the ESE. The addition of as little as 100 ng of Rev to a splicing reaction mixture containing the largest splicing substrate inhibited exon 3 recognition (Fig. 3). This confirms earlier, in vivo observations of Rev-mediated changes of alternative splicing (17) and indicates that Rev is the only viral protein necessary for exon 3 skipping.
Rev binds the ESE.
Gontarek and Derse have suggested that binding of EIAV Rev to a region of the viral pre-mRNA near the ESE results in either direct or indirect inhibition of SR protein function (17). In previous work, we identified an RRE region spanning exon 3 (nt 5280 to 5834) (4). This favors a mechanism by which Rev-RRE interactions disrupt SF2/ASF binding at the ESE. To examine whether Rev binds at or near the ESE, we generated a series of RNA probes and tested for Rev-RNA interactions by RNA gel mobility shift assays with a bacterially expressed and purified GST-Rev fusion protein. The locations of the RNA probes relative to exon 3 and the ESE are shown in Fig. 4A. GST-Rev bound to exon 3 probes RREp1 and RREp2, both of which contain the ESE; however, no binding was observed with the GST negative control (Fig. 4B, lanes 1 to 4). Minor binding was observed with RREp3 (Fig. 4B, lane 6), which has sequences immediately downstream of the ESE (nt 5523 to 5622), but lacks the purine-rich region. The binding site was further delineated to a 57-nt region of viral RNA by using two smaller ESE-containing probes, RREp4 and RREp5. GST-Rev interacted with both probes (Fig. 4B, lanes 8 and 11), further suggesting that Rev binds at or near the ESE. To confirm the specificity of binding, gel shift assays were performed with RREp2 in the presence of excess unlabeled competitor RREp2 or RREp3 (Fig. 4C). Excess unlabeled RREp2 inhibited GST-Rev binding (Fig. 4C, lanes 2 and 3), whereas no inhibition was observed with RREp3 (Fig. 4C, lanes 4 and 5), demonstrating that the binding of GST-Rev to the ESE-containing RREp2 is specific. No slower-migrating bands, indicative of Rev multimerization, were observed in any of the RNA binding analyses, although multimerization was readily observed with HIV-1 Rev when used as a positive control (data not shown). The minor bands in Fig. 4C represent GST-Rev and degraded by-products. Overall, these results demonstrate that Rev specifically interacts with the viral RNA at or near the ESE.
Mutagenesis of the ESE reduces exon 3 splicing and Rev binding.
The finding that GST-Rev bound to a 57-nt region containing the ESE suggested that a Rev-RNA interaction was directly competing with SF2/ASF for binding at the ESE. If so, there should be similar sequence requirements for exon 3 recognition and Rev binding. The ESE contains two purine-rich sequences (designated A and B), which include seven GAA repeats (Fig. 5A). GAA repeats have been shown to be important for SF2/ASF recognition of ESE sequences in other systems (reviewed in reference 16). Therefore, we constructed five ESE mutations in the largest splicing construct and the RREp4 RNA probe fragment which contained various GAA to GCA mutations (Fig. 5A). The mutant templates were tested for in vitro splicing and GST-Rev binding. Mutation of all GAA motifs (mutAll) or the B-purine stretch (mutB) resulted in a decrease in both exon 3 in vitro splicing (Fig. 5B, lanes 3 and 5) and GST-Rev binding (Fig. 5C, lanes 2 and 4). Mutation of the GAA repeats in only the B purine stretch resulted in a more modest reduction in both in vitro splicing and GST-Rev binding (Fig. 5B, lanes 6 and 7, and 5C, lanes 5 and 6). Mutation of GAA repeats in the A region (mutA) did not appear to significantly affect either exon 3 splicing or GST-Rev binding in vitro (Fig. 5B, lane 4, and 5C, lane 3), suggesting that the B purine stretch alone contains cis-acting sequences necessary for exon 3 recognition and GST-Rev binding. The finding that each mutation had comparable effects in both assays suggests similar requirements in the ESE for both exon 3 recognition and Rev binding, further supporting a model of Rev inhibition of splicing through direct competition with SR proteins for binding at the ESE.
FIG. 5.
In vitro splicing and RNA binding of ESE mutants. (A) Sequence of two purine stretches (designated A and B) in exon 3. GAA repeats were mutated to GCA in the largest splicing construct (Fig. 2A) and RNA probe RREp4 (Fig. 4A). (B) In vitro splicing analysis of mutant ESE constructs. The locations of splicing products are indicated. Sizes are shown at the left (in nucleotides). (C) RNA gel mobility shift assays detecting GST-Rev binding to the mutant probes. The arrow points to the location of shifted RNAs.
ESE can function as an RRE to mediate RNA nuclear export.
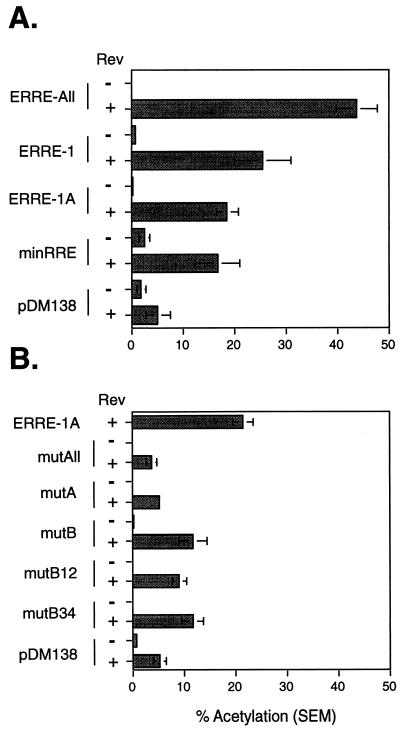
In other complex retroviruses, Rev functions to regulate the export of incompletely spliced RNAs via interaction with the viral pre-mRNA at a specific sequence called the RRE. We had previously used the HIV-1-derived pDM138 reporter system to preliminarily map the RRE of EIAV to a region which overlapped exon 3 (ERRE-1) (4). However, this fragment possessed only 52% of the activity of a reporter containing a much larger fragment of EIAV (ERRE-All), suggesting that sequences further downstream enhanced Rev-mediated export. The RNA binding data given above suggested that the functional sequence within ERRE-1 was the ESE sequence. To test this, we constructed a pDM138 reporter plasmid, minRRE, which contains only 57 nt of EIAV (nt 5485 to 5540, RREp5), spanning the ESE and the remainder of exon 3. Transient-transfection assays in 293 cells demonstrated that the minRRE reporter produced levels of CAT activity comparable to those by ERRE-1 (Fig. 6A), but only 35% of the activity with ERRE-All. This indicates that minRRE contains the functional RRE in ERRE-1 but additional elements outside ERRE-1 may be required for full export activity.
FIG. 6.
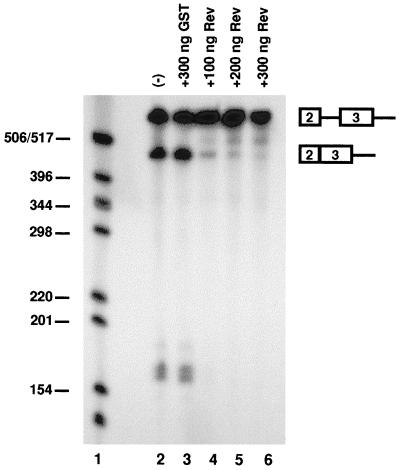
EIAV ESE can function as an RRE. (A) pDM138-derived reporter vectors containing various regions of the EIAV genome were used in transient transfections with and without Rev, and CAT assays were performed in 293 cells as described in Materials and Methods. The results are presented as the percent acetylation. Experiments were performed in triplicate, and the results represent the mean of at least nine independent transfections. Error bars denote the standard error of the mean. (B) The ESE mutations indicated in Fig. 5A were also introduced into the ERRE-1A reporter vector and assayed for CAT activity in the presence and absence of Rev as described for panel A.
To confirm that the ESE is the RRE within minRRE, we introduced the GAA-to-GCA mutations used for in vitro splicing and RNA binding assays (Fig. 5A) into a reporter vector containing the same sequences present in the largest splicing substrate (Fig. 2A). This vector, ERRE-1A, is 41 nt shorter than ERRE-1 but exhibited similar levels of activity (Fig. 6A). In all cases, mutation of the GAA repeats in the ESE significantly reduced Rev-dependent nuclear export activity (P < 0.01) (Fig. 6B). The greatest reduction in activity was seen in constructs containing mutations of all seven GAA repeats (mutAll) or the three repeats in the A purine stretch (mutA). The reduction in activity in mutA indicates that this region, while not necessary for GST-Rev binding, is required for RNA nuclear export. Mutations in the B region (mutB, B12, and B34) also significantly reduced activity. Therefore, we conclude that the B purine stretch functions in GST-Rev binding, exon 3 inclusion, and Rev-dependent nuclear export. Together, these results indicate that the ESE sequence acts as an RRE and that Rev mediates alternative splicing by binding at or near the ESE to disrupt SF2/ASF interactions.
SR proteins inhibit Rev-dependent nuclear export.
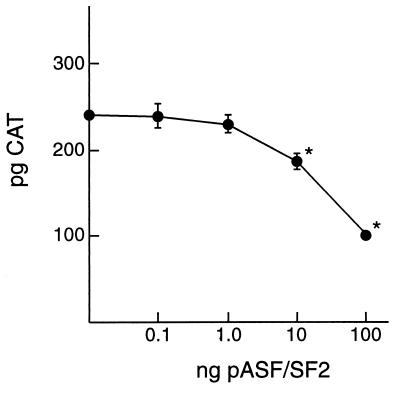
The finding that similar cis-acting sequences mediate nuclear export, RNA binding, and exon inclusion suggested that EIAV Rev directly competes with SF2/ASF for binding at a similar site on the viral pre-mRNA. If so, increasing concentrations of SF2/ASF would inhibit Rev-dependent nuclear export activity. To test this, initial studies were done to determine the linear range of Rev-dependent nuclear export activity. Based on these results, Cf2th cells were transfected with 0.5 ng of pRevWT and 10-fold-increasing concentrations of pSF2/ASF. Results indicated a significant, dose-dependent decrease in CAT levels in the presence of pSF2/ASF (Fig. 7). This suggests that SF2/ASF and Rev are mutually competitive and is consistent with the conclusion that both proteins bind to nearly identical sequences on the viral pre-mRNA. Western blot analyses confirmed that SF2/ASF protein levels increased concomitantly with increased amounts of transfected plasmid DNA (data not shown). The level of Rev produced by a transfected Rev cDNA was below the limit of detection by Western blot. Thus, we could not eliminate the possibility that the decrease in Rev-mediated nuclear export activity resulted from quantitative changes in Rev levels rather than changes in Rev binding.
FIG. 7.
SF2/ASF inhibits Rev-dependent nuclear export. pERRE-1 reporter plasmid was cotransfected with 0.5 ng of pRevWT and increasing amounts of pSF2/ASF. CAT levels were quantified by ELISA and are reported as picograms of CAT per normalized lysate. Results represent the mean of six independent transfections, and the error bars denote the standard error of the mean. Asterisks indicate values significantly different (P < 0.05) from control transfections which contained no pSF2/ASF.
DISCUSSION
In addition to its role in nuclear export of incompletely spliced viral mRNAs, EIAV Rev mediates alternative splicing of the four-exon multiply spliced EIAV mRNA (22). Gontarek and Derse (17) demonstrated that both GST-Rev and the SR protein SF2/ASF cross-link in vitro with exon 3 and proposed a model in which Rev disrupts SR protein interactions required for exon inclusion. Consistent with these previous data, we show that the purine-rich sequence is required for GST-Rev binding and for exon 3 recognition in splicing reactions in vitro. In addition, a 57-nt sequence containing the ESE was shown to act as a functional RRE in a heterologous nuclear export assay system. Mutation of GAA nucleotide repeats in the ESE reduced GST-Rev binding, exon 3 splicing in vitro, and nuclear export of ESE-containing pre-mRNA. trans-Complementation assays demonstrated that SF2/ASF inhibited Rev-dependent nuclear export in a dose-dependent manner. Therefore, both SF2/ASF-mediated exon 3 splicing and Rev-mediated RNA export have similar cis-acting RNA requirements, and EIAV Rev and SF2/ASF appear to be mutually competitive. From these data, we conclude that the purine-rich sequence within exon 3 of EIAV is both an ESE and a functional RRE. Extending the earlier model (17), we propose that Rev-mediated nuclear export requires binding at or near the ESE and that this results in skipping of exon 3 through direct inhibition of SF2/ASF-ESE interactions required for recognition of exon 3 by the host cell splicing machinery. The use of an ESE as an RRE is unprecedented among complex retroviruses.
It is interesting that mutation of the 5′ purine stretch (mutA) decreased nuclear export but appeared to have little effect on GST-Rev binding in vitro. Studies with HIV-1 Rev also indicate that sequences in the HIV-1 RRE are required for nuclear export but not for RNA binding (25, 26). While these observations have not been fully explained, it is likely that RNA secondary structure may play a role. Secondary structure is a key determinant for HIV-1 binding, multimerization, and function (7, 8, 10, 11, 24, 25, 27). No biochemical data are available to date to confirm the proposed structure (17) of the EIAV ESE/RRE, and it is not possible to predict the structural effects of the mutations used in our study. Our data would suggest that mutation of the 5′ purine stretch does not affect the primary binding site of Rev but may alter distant structures required for Rev-mediated nuclear export. It has been demonstrated that HIV-1 Rev multimerization occurs only after binding to a primary site on the RNA, and furthermore, other regions of the RNA are important for secondary binding (8, 10, 32). However, we were unable to observe Rev multimerization in our RNA binding assays, including those assays containing the ESE. Therefore, it remains unclear why mutA exhibited reduced activity with no apparent defect in RNA binding. It is also possible that binding of host cell proteins to the RRE may be required to facilitate Rev export activity. Further studies will be necessary to delineate the role of this purine region in Rev-mediated nuclear export.
Interactions of Rev-like proteins with SR proteins have been demonstrated in other complex retroviruses. SR proteins have been shown to bind the HIV-1 RRE in a Rev-dependent manner (28). The same study also demonstrated that excess exogenous SF2/ASF could produce a dose-dependent inhibition of HIV-1 Rev function in vivo. We have previously reported an inhibition of EIAV replication in activated macrophages associated with a delay in the appearance of incompletely spliced RNAs (31). The data presented here suggest that this inhibition may be a result of competition of SF2/ASF with Rev for binding at the ESE. This hypothesis is supported by our data showing that excess SF2/ASF provided in trans can inhibit Rev function in transient-transfection assays. Together, these results suggest that the inhibition in activated macrophages may be due to an increase in the level of SR proteins. It is known that expression of SR proteins varies in cells at different states of activation and differentiation (16, 30, 33), including an increased expression of the SR protein SRp30c in activated T cells (30). However, little is yet known about the phenotype of SR proteins in monocyte cells. Also, our data cannot rule out the possibility that in addition to competing for binding at the RRE, Rev may also inhibit function via protein-protein interactions.
Previous reports have demonstrated that mutations in the NES do not affect the alternative splicing activity of EIAV Rev (4, 18). To date, no laboratory has identified a Rev protein which is competent in nuclear export but deficient in alternative splicing. Therefore, it is not clear whether alternative splicing of exon 3 is merely a consequence of Rev-mediated nuclear export or if it plays a separate role in virus replication. The RREs of most complex retroviruses are located near the SU-TM cleavage site or in the 3′ end of env. The location of EIAV RRE in the 5′ env is unique and may be explained by the requirement of the ESE for exon 3 inclusion. The env mRNA is spliced with the exon 2 splice acceptor (Fig. 1). A singly spliced mRNA using the exon 3 splice acceptor has not been observed in infected cells and would encode a truncated Env protein lacking the signal peptide. A singly spliced mRNA using the exon 4 splice acceptor is observed, which produces a truncated transmembrane protein from the alternate start codon present in exon 1. Therefore, the use of an ESE as an RRE may function to silence recognition of exon 3 to eliminate another singly spliced transcript. Although a number of retroviruses utilize cis-acting sequences such as ESEs to take advantage of cellular mechanisms of alternative splicing, EIAV appears to be the only retrovirus to encode a trans-acting protein that directly modulates SR-mediated alternative splicing. EIAV was the first lentivirus described and is smaller and genetically less complex than the other lentiviruses. It is possible that the EIAV Rev-ESE interaction represents a transitional step in the evolution of the Rev-Rex pathway utilized by most complex retroviruses. Interestingly, previous work in our laboratory and others suggested that EIAV may possess two separate RREs (4, 22). In the current study, reporter constructs containing the 57-nt ESE region showed significantly reduced activity compared with the ERRE-All reporter construct, containing a larger portion of the env gene (Fig. 6A). However, a second RRE has not been identified, nor is it clear that such an element can function independently of the ESE to mediate export of viral pre-mRNAs. Additional studies will help to fully understand the biological and evolutionary significance of the EIAV Rev-mediated export pathway.
ACKNOWLEDGMENTS
We thank Yvonne Wannemuehler for technical assistance; Tom Hope for plasmids pERRE-All, pERRE-1, and pDM138; and Sean Murphy and Prasith Baccam for statistical analysis.
This work was supported by funds from the Carver Grant Trust (S.C.), an Iowa State University-University of Iowa interinstitutional grant in Biomedical Sciences (S.C. and C.M.S.), USDA grant 96-358204-3847 (S.C.), PHS grant CA 28951 from the National Cancer Institute (C.M.S.), and PHS grant AI36073 from the National Institute of Allergy and Infectious Disease (C.M.S.).
REFERENCES
- 1.Ali S A, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. Biotechniques. 1995;18:746–750. [PubMed] [Google Scholar]
- 2.Amendt B A, Hesslein D, Chang L-J, Stoltzfus C M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beisel C E, Edwards J F, Dunn L L, Rice N R. Analysis of multiple mRNAs from pathogenic equine infectious anemia virus (EIAV) in an acutely infected horse reveals a novel protein, ttm, derived from the carboxy terminus of the EIAV transmembrane protein. J Virol. 1993;67:832–842. doi: 10.1128/jvi.67.2.832-842.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belshan M, Harris M E, Shoemaker A E, Hope T J, Carpenter S. Biological characterization of Rev variation in equine infectious anemia virus. J Virol. 1998;72:4421–4426. doi: 10.1128/jvi.72.5.4421-4426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter S, Alexandersen S, Long M J, Perryman S, Chesebro B. Identification of a hypervariable region in the long terminal repeat of equine infectious anemia virus. J Virol. 1991;65:1605–1610. doi: 10.1128/jvi.65.3.1605-1610.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll R, Derse D. Translation of equine infectious anemia virus bicistronic tat-rev mRNA requires leaky ribosome scanning of the tat CTG initiation codon. J Virol. 1993;67:1433–1440. doi: 10.1128/jvi.67.3.1433-1440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochrane A W, Chen C-H, Rosen C A. Specific interaction of the human immunodeficiency virus rev protein with a structured region in the env mRNA. Proc Natl Acad Sci USA. 1990;87:1198–1202. doi: 10.1073/pnas.87.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook K S, Fisk G J, Hauber J, Usman N, Daly T J, Rusche J R. Characterization of human immunodeficiency virus type 1 protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 1991;19:1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen B R. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly T J, Doten R C, Rennert P, Auer M, Jaksche H, Donner A, Fisk G, Rusche J R. Biochemical characterization of binding of multiple HIV-1 Rev monomeric proteins to the Rev responsive element. Biochemistry. 1993;32:10497–10505. doi: 10.1021/bi00090a028. [DOI] [PubMed] [Google Scholar]
- 11.Dayton E T, Konings D A M, Powell D M, Shapiro B A, Butini L, Maizel J V, Dayton A I. Extensive sequence-specific information throughout the CAR/RRE, the target sequence of the human immunodeficiency virus type 1 Rev protein. J Virol. 1992;66:1139–1151. doi: 10.1128/jvi.66.2.1139-1151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer U, Huber J, Boelens W C, Mattal I W, Luhrmann R. The HIV-1 rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 14.Fridell R A, Bogerd H P, Cullen B R. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fridell R A, Partin K M, Carpenter S, Cullen B R. Identification of the activation domain of equine infectious anemia virus Rev. J Virol. 1993;67:7317–7323. doi: 10.1128/jvi.67.12.7317-7323.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 17.Gontarek R R, Derse D. Interactions among SR proteins, an exonic splicing enhancer, and a lentivirus Rev protein regulate alternative splicing. Mol Cell Biol. 1996;16:2325–2331. doi: 10.1128/mcb.16.5.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris M E, Gontarek R R, Derse D, Hope T J. Differential requirements for alternative splicing and nuclear export functions of equine infectious anemia virus Rev protein. Mol Cell Biol. 1998;18:3889–3899. doi: 10.1128/mcb.18.7.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacks T, Madhani H D, Masiarz F R, Varmus H E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakami T, Sherman L, Dahlbert J, Gazit A, Yaniv A, Tronick S R, Aaronson S A. Nucleotide sequence analysis of equine infectious anemia proviral DNA. Virology. 1987;158:300–312. doi: 10.1016/0042-6822(87)90202-9. [DOI] [PubMed] [Google Scholar]
- 21.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 22.Martarano L, Stephens R, Rice N, Derse D. Equine infectious anemia virus trans-regulatory protein Rev controls viral mRNA stability, accumulation, and alternative splicing. J Virol. 1994;68:3102–3111. doi: 10.1128/jvi.68.5.3102-3111.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayeda A, Screaton G R, Chandler S D, Fu X-D, Krainer A R. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen H, Cochrane A, Dillon P, Nalin C, Rosen C. Interaction of the human immunodeficiency virus type 1 rev protein with a structured region in env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 1990;4:1357–1364. doi: 10.1101/gad.4.8.1357. [DOI] [PubMed] [Google Scholar]
- 25.Olsen H S, Nelbock P, Cochrane A W, Rosen C A. Secondary structure is the major determinant for interaction of HIV rev protein with RNA. Science. 1990;247:845–848. doi: 10.1126/science.2406903. [DOI] [PubMed] [Google Scholar]
- 26.Olsen H S, Beidas S, Dillon P, Rosen C A, Cochrane A W. Mutational analysis of the HIV-1 Rev protein and its target sequence, the Rev responsive element. J Acquir Immune Defic Syndr. 1991;4:558–567. [PubMed] [Google Scholar]
- 27.Powell D M, Zhang M J, Konings D A M, Wingfield P T, Stahl S J, Dayton E T, Dayton A I. Sequence specificity in the higher-order interaction of the rev protein of HIV-1 with its target sequence, the RRE. J Acquir Immune Defic Syndr. 1995;10:317–323. [PubMed] [Google Scholar]
- 28.Powell D M, Amaral M C, Wu J Y, Maniatis T, Greene W C. HIV Rev-dependent binding of SF2/ASF to the Rev response element: possible role in Rev-mediated inhibition of HIV RNA splicing. Proc Natl Acad Sci USA. 1997;94:973–978. doi: 10.1073/pnas.94.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz S, Felber B K, Benko D M, Fenyo E M, Pavlakis G N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Screaton G R, Caceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith T A, Davis E, Carpenter S. Endotoxin treatment of EIAV-infected horse macrophage cultures decreases production of infectious virus. J Gen Virol. 1998;79:747–755. doi: 10.1099/0022-1317-79-4-747. [DOI] [PubMed] [Google Scholar]
- 32.Van Rck D I, Venkatesan S. Real-time kinetics of HIV-1 Rev-Rev response element interactions. J Biol Chem. 1999;274:17452–17463. doi: 10.1074/jbc.274.25.17452. [DOI] [PubMed] [Google Scholar]
- 33.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 34.Zapp M, Hope T, Parslow T, Green M. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus rev protein: a dual function for an arginine-rich motif. Proc Natl Acad Sci USA. 1988;88:7734–7738. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zapp M L, Green M R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]