Abstract
Long noncoding RNAs (lncRNAs) have emerged as prominent regulators of gene expression in eukaryotes. The identification of lncRNA orthologs is essential in efforts to decipher their roles across model organisms, as homologous genes tend to have similar molecular and biological functions. The relatively high sequence plasticity of lncRNA genes compared with protein-coding genes, makes the identification of their orthologs a challenging task. This is why comparative genomics of lncRNAs requires the development of specific and, sometimes, complex approaches. Here, we briefly review current advancements and challenges associated with four levels of lncRNA conservation: genomic sequences, splicing signals, secondary structures and syntenic transcription.
Keywords: comparative genomics, long noncoding RNAs, orthologs, sequence analysis
Introduction
Long noncoding RNAs (lncRNAs) represent an abundant class of transcripts longer than 200 bases that do not encode proteins. In ENSEMBL 102 [1], there are 82 760 human lncRNAs originating from 28 480 genes; thus, they outnumber protein-coding genes (n=22 803). Despite indisputable progress in accumulating lncRNA-related data, relatively little is known about their functions, with no more than a few thousands of them subjected to detailed functional studies. To date, lncRNAs have been linked to virtually all cellular processes, including transcription, splicing, translation, and the cell cycle [2–6], and have also been implicated in human diseases, such as malignant transformation, while a number of them represent diagnostic and prognostic biomarkers for cancer [7,8]. lncRNAs participate in different steps of gene expression. For instance, many modulate the act of transcription, e.g. through promoter modifications, creating a permissive chromatin environment or binding transport factors to inhibit the nuclear localization of specific transcription factors [9,10].
The heterogeneity in the modes of lncRNA action poses a huge challenge to functional studies and for distinguishing which are bona fide functional. Here, comparative genomics, conservation studies in particular, may be helpful. First, evolutionary conservation is often associated with functionality, while the presence of orthologs with established roles makes the predictions stronger. Second, the level and type of lncRNA conservation may be used to assign lncRNAs to a hypothetical functional domain, thus facilitating subsequent functional studies. For instance, conservation of exon sequences points to functions exerted through mature lncRNA molecules, such as lncRNA:RNA base pairings that are associated with RNA processing and stability [11,12]. On the other hand, the evidence accumulated thus far strongly suggests that for most lncRNAs it is not their sequences but the act of transcription that is associated with their biologically relevant functions. This is especially true for so-called antisense lncRNAs [13,14], whose function is to modulate the expression of sense partners. Because of the nature of the underlying lncRNA:protein interactions with low sequence specificity, there are virtually no constraints upon the conservation of sequences of these lncRNAs. They do, however, often exhibit positional conservation; i.e., they occupy the same or neighboring genomic loci in closely related species.
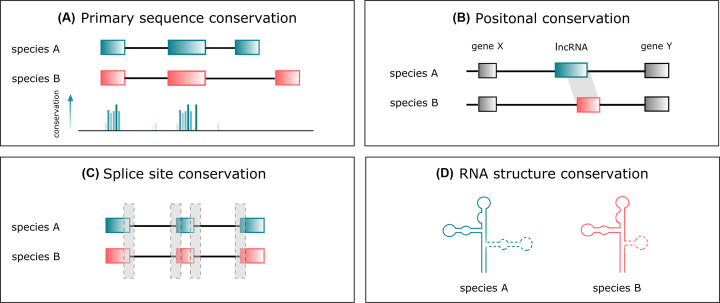
In this review, we consider four dimensions of lncRNA conservation from the perspective of the genome. The abovementioned conservation of primary sequences (Figure 1A) and positional conservation of lncRNAs (Figure 1B) are complemented by insight into their splice sites and other splicing signals (Figure 1C), and their secondary structures (Figure 1D): we consider the pros and cons of existing methodologies and their algorithmic challenges, discuss relevant studies and pinpoint some novel or emerging approaches, such as the application of alignment-free metrics and the conservation of promoter sequences.
Figure 1. Different levels of lncRNAs conservation.
Homologous lncRNAs may exhibit primary sequence similarity (A), but their positional conservation is more common (B). The primary sequence may possess, however, short, functional sequences conserved between species, in particular splicing signals (C). Finally, studies on secondary structures, though of limited usability in studying lncRNAs, are occasionally used in the search for conserved lncRNAs or their functional units (D).
We are not covering the issue of conservation of lncRNA expression, processing, and tissue-specificity but one should keep in mind this represents another layer of complexity in conservation and functional studies. For example, it has been demonstrated that positionally conserved lncRNAs in human embryonic stem cells are in most cases spliced and then exported to the cytoplasm, while their mouse counterparts are predominantly unspliced and retained in the nucleus, leading to species-specific functions of lncRNAs in pluripotency maintenance [15]. Generally, the most conserved lncRNAs tend to be broadly expressed and they display significantly higher expression levels than those with high evolutionary rates [16,17].
Sequence-level conservation
In recent years, several software programs based on sequence homology have been proposed for use in lncRNA conservation analysis [18–20]. In particular, alignment-based algorithms (e.g., BLASTN or BLAT) are widely used to search for similarities in primary sequences between lncRNAs. Such comparisons are performed utilizing solely lncRNA sequences or, additionally, genomes of interest. For example, Marques et al. compared the substitutions in mouse and rat transcriptional initiation regions and demonstrated that enhancer-associated lncRNAs generally display lower conservation than other classes of lncRNAs [21]. In another study, Hezroni et al. applied BLASTN to create a network of sequence similarities, comparing long intergenic RNAs (lincRNAs; autonomously transcribed lncRNAs that do not overlap annotated coding genes) from sea urchins and 16 vertebrates and demonstrated that most lincRNAs are lineage-specific [18]. A number of studies have revealed that only a small fraction of lncRNAs exhibit primary sequence conservation similar to that of protein-coding genes [22], with some well-studied examples among them. For example, phylogenetic analysis comparing 20 mammals showed high evolutionary conservation of MALAT1 lncRNA [23], known to be a modulator of gene expression and involved in tumor growth and metastasis. Rapid evolution of lncRNAs has resulted in weak sequence conservation not only within animals but also in plants. It has been shown that fewer than 2% of the lncRNAs in A. thaliana are conserved across plants [24]. Although the general conservation level of lncRNAs is very low, there is a special group of lncRNAs dubbed transcribed-ultraconserved regions (T-UCRs) that are encoded by a subset of ultraconserved genomic regions and are highly conserved in humans, rats, and mice [25]. Typically, they are detected as transcribed by microarray experiments and not present in most RNA-seq based reconstructions of polyadenylated transcripts [25]. Recent studies highlight the importance of T-UCRs in the regulation of genes expression. For example, uc.372 promotes the expression of genes related to lipid synthesis and uptake by binding pri-miR-195/pri-miR-4668 and by suppressing maturation of mir-195/mir-4668 [26].
Because the full sequences of many lncRNAs are poorly preserved, analyses of shorter regions have been proposed, leading to the discovery of short-sequence homologies (microhomologies) [18,27]. It has been shown that these microhomologous regions are repeated within an lncRNA and can act as protein binding sites [28,29]. However, another solution has been proposed by Noviello et al., who successfully applied alignment-free string similarity metrics for the identification of lncRNA homologs based on their promoter sequences [19]. Kirk et al. have also developed an aligment-free approach called SEEKR. They found that lncRNAs with related functions have similar k-mer profiles despite lacking linear homology [30]. Recently, Ross et al. have introduced a novel technique that complements the SEEKR algorithm, LncLOOM. This method efficiently detects both known and novel functional elements in lncRNAs [31]. Interestingly, a growing body of evidence shows that promoter regions in lncRNAs are more conserved than the remainder of their sequences [29,32]. Moreover, Derrien et al. compared the conservation level of different regions of lncRNA genes and observed that promoters have been conserved to almost the same extent as protein-coding genes [33]. In addition, a number of studies have shown that exons of lncRNAs are better conserved than introns [34,35]. On the other hand, Pegueroles and Gabaldón analyzed sets of human lncRNAs, demonstrating that although lncRNA exons are enriched in conserved elements, they are not conserved to a greater extent than introns [36].
Conservation of splicing signals
Because of the overall weak primary sequence conservation, studies of other lncRNA features, such as their gene structures, may shed light on the evolution and functional potential of these molecules. In general, the conservation of splicing patterns has been widely studied, as it refers directly to the preservation of the exon–intron structure, considered a hallmark of functional transcripts. Despite the fact that lncRNAs seem to be less efficiently spliced than pre-mRNAs [37–40], noncoding exons are reported to universally originate from alternative splicing [41], and distinct lncRNA isoforms have the potential to act in a divergent manner [42–44].
The most common approach to splicing conservation studies requires a set of high-quality splice sites in species of interest, multiple genome alignments and, finally, statistical testing to detect and measure conservation. For example, Ponjavic et al. [45] evaluated splice site conservation with a chi-square test to compare signals from bona fide splice sites against a set of background dinucleotides, not associated with splicing. They indicated that intronic terminal dinucleotides in mouse lncRNAs are significantly more conserved in humans and rats than dinucleotides that are not associated with splice-site signals. A similar strategy was applied by Chernikova et al. [46], who analyzed the evolutionary conservation of dinucleotides at 5′ and 3′ ends of introns using pairwise comparison between human and mouse lincRNAs and multiple sequence alignments for 15 animal species. Moreover, the researchers analyzed the conservation of the exon–intron structures using a parsimony approach. The analysis was performed using the DNAPARS program with positions of mouse or human introns as a reference for gene structures. Interestingly, some of them have been conserved for over 100 million years. However, a substantial fraction of lincRNAs introns are not conserved – a high turnover rate of lincRNAs introns in the Glires lineage was reported [46].
Machine learning or statistical models are implemented in conservation studies as well. For instance, Rose et al. trained a support vector machine model to detect and rate donor and acceptor splice site candidates in multiple sequence alignments [47]. They also used MaxEntScan [48], a tool that scores splice site sequences in terms of their similarity to canonical splice sites. MaxEntScan is based on maximum entropy distribution and is considered to offer the most unbiased approximation for modeling short sequence motifs [49]. Remarkably, it considers dependencies between both nonadjacent and adjacent positions and is used extensively in analyses of lncRNA splice-site conservation [50–52]. One such study was performed on vertebrate lncRNAs [50], in which multiple sequence alignment together with transcriptomics data were utilized to determine the homologous positions in splice sites, leading to the construction of a comparative map of splice sites. The authors showed that in conserved human transcripts, 87% of splice sites are present in other species, including mice, rats, cows, and dogs, yet most of the novel splice sites originated during primate evolution. MaxEntScan has also been used in recent studies on lncRNA splicing conservation in 16 Brassica species [52]. These studies revealed that nearly 18% of lincRNAs display splicing conservation in at least one exon in Brassicaceae plant family members. In comparison with that of vertebrates, the level of lincRNA conservation measured by gene structure is significantly lower in plants.
The splicing conservation may also involve short sequence motifs recognized by proteins associated with splicing regulation. Examples of this motif are purine-rich hexamers that are recognized by serine arginine-rich proteins (SR proteins). These so-called exonic splice enhancers (ESEs) were explored, inter alia, by Schüler et al. [53]; these authors showed that experimentally confirmed human hexamers (ESEs) evolve considerably slower than nonenhancer sequences in lncRNAs. Another group implemented the RESCUE-ESE algorithm [54] to identify hexamers that are significantly enriched or depleted within exonic sequences relative to their flanking intronic sequences [55]. The authors reported that evolutionary constraints are more concentrated near intron–exon boundaries. Moreover, these regions in lncRNAs contain a high density of ESEs, which are conserved across mammals [55]. Despite the fact that both of the aforementioned studies indicate the action of purifying selection to preserve exonic splicing enhancers within lncRNAs, the biological interpretation is difficult, particularly because splicing of lncRNAs is generally inefficient [37,39,40]. More in-depth insight is provided by experimental evidence showing that lncRNAs are unable to bind SR proteins securely [56] and by studies showing the diverse dynamics of intron excision across lncRNAs [38].
Secondary structure conservation
Diversified functions of lncRNAs, including participation in post-transcriptional regulation, are often associated with their ability to recognize and bind molecular targets, where the secondary and tertiary structures of related RNA molecules may play pivotal roles. As a result, studying the structural architecture of lncRNAs may help to determine their exact modes of action. The possibility of these molecules to maintain certain structures was reported, for example by Smith et al. in a genome-wide study of mammalian evolutionarily conserved structures. Smith et al. identified millions of putative functional RNAs, some of which overlap with annotated ncRNAs [57]. Another report pointed to a set of conserved brain lncRNAs being significantly enriched with predicted RNA secondary structures [58]. Furthermore, Pegueroles and Gabaldón [36] observed that positions in paired lncRNA regions that participate in the formation of folds show lower probabilities of having SNPs. Thus secondary structures impose limits on changes to lncRNA sequences (which may lead to degeneration of the structural motifs), suggesting their functional relevance. Yang and Zhang showed, using data from a parallel analysis of RNA structure (PARS), that lncRNAs are substantially less folded than mRNAs [59]. These results were surprising as it is assumed that lncRNAs often require a secondary structure to perform their functions, while the mRNAs do not. However, the authors pointed out that it is difficult to clearly explain the observations since analyses may be biased by insufficient representation of functional lncRNAs, for example. On the other hand, strong secondary structures may not be preferred as they could hinder base-pairings that are at the core of many RNAs functions.
Although the secondary structure conservation of lncRNAs is lower than that of messenger RNAs or ribosomal RNAs [20,59], some functional lncRNAs with experimentally confirmed structural motifs, including Cyrano [60], Xist [61,62], MALAT1 [63,64], SRA [65] and GAS5 [66], have been found. For instance, recently published studies on MALAT1 in vertebrates revealed an evolutionarily conserved core consisting of numerous helices [64]. At the center of the functional core of Cyrano is a cloverleaf structure maintained for more than 400 million years, enriched in several protein binding sites and masking a target site for miR-7 [60]. The aforementioned studies were based on a similar approach: secondary structures are experimentally determined by chemical and/or enzymatic probing and then used in comparative sequence analyses. Interestingly, resolved secondary structures may be used in the identification of lncRNAs in other species, as shown in studies on COOLAIR [67]; its secondary structure was successfully used to predict the corresponding exons in evolutionarily divergent Brassicaceae species.
Computational methods for the detection of conserved secondary structures are quite well established, such as those based on covariance models (CMs), which are able to automatically learn statistical models derived from multiple RNA alignments. CMs are implemented with popular homology search tools, e.g., Infernal [68] or CMfinder [69]. CMfinder was applied in research on vertebrate genomes, which were screened for conserved RNA structures (CRSs) [70]. The authors pinpointed some structurally conserved lncRNAs and noted that CRS density decreases from the 5′ to the 3′ end of lncRNAs. Another tool based on covariation models is R-scape, which initially showed limited utility on eukaryotic lncRNAs, as it found no statistically significant support for the proposed secondary structures in some of the lncRNAs with experimentally verified structures [71]. However, recent studies have shown that R-scape efficiently detects conserved secondary structures when appropriately parameterized [72]. Notably, since CMs need to be trained on sets of homologous sequences that are not always available, attempts have been made to utilize algorithms comparing structures without sequence alignments, e.g., BEAGLE [73], which was used to successfully study the secondary structure similarities in four different Caenorhabditis worm species [20].
Syntenic lncRNAs
Since it is difficult to identify orthologous lncRNAs by sequence similarity [74,75] or the conservation of secondary structures [71], the evolutionarily conserved position in the genome (synteny) may be particularly useful. Syntenic lncRNAs are found either in the same genomic region across compared species, as determined by whole genome alignment (WGA), or are found between syntenic protein-coding genes. Syntenic lncRNAs, also referred to as syntologs, represent loci with conserved transcription that typically exhibit little or no sequence similarity. In other words, syntenic transcription may generate RNAs whose expression itself, rather than the sequence, is required for lncRNA function. Indeed, a growing body of evidence shows that for most human lncRNAs, their transcription alone, rather than the production of the mature RNA molecules, is of biological importance. Good examples are natural antisense transcripts (NATs), which originate from the opposite genomic strand compared with their sense partners. It is estimated that as many as 70% of human genes display antisense transcription [13,14], with NATs modulating their expression and processing in a number of ways [76,77]. The prevailing mechanism is the recruitment of complex epigenetic machinery that results in histone modifications and subsequent transcriptional deregulation of target genes [78]. Antisense lncRNAs may also affect expression of their sense counterparts through transcriptional intereference (TI) mechanisms. One such example is yeast SER3 protein coding gene being overlapped by the SRG1 lncRNA, whose transcription increases nucleosome density at the SER3 promoter [79]. Antisense lncRNAs may also trigger methylation at GC-rich genomic regions that are often associated with vertebrate genomes. An example is α-thalassemia, where antisense RNA LUC7L represses expression of HBA2 (α-Globin) by triggering methylation of its CpG islands [80]. More functions played by NATs are reviewed elsewhere [76,77]. Examples of syntologs with relatively degenerate sequences are those referred to as topological anchor point RNAs (tapRNAs), whose roles are linked to chromatin looping and topology, especially in the neighborhood of developmental genes [81]. They act by binding the DNA in cis; hence, they maintain complementarity irrespective of how much the counterpart DNA sequence is changed, indicating little or no evolutionary constraints upon their sequences. It should be noted, however, that particular fragments of these transcripts might be under stricter evolutionary constraints. tapRNAs contain conserved sequence domains recognized by transcription factors and RNA-binding proteins with zinc finger domains, leading to their elevated overall sequence conservation compared with most lncRNAs [81].
Although the search for syntenic lncRNAs proved to be an effective way of obtaining orthologous lncRNAs [82,83], there are substantial issues when calling syntologs. In general, whether two lncRNAs found within syntenic genomic blocks are orthologous remains in question, as they tend to occupy nonoverlapping conserved fragments of genomic DNA or are even found in a relatively large, nonconserved genomic region between two syntenic blocks (or two orthologous protein-coding genes). The syntenic lncRNAs might also show conservation limited to short parts of their sequences (e.g. within one exon out of many), which further sows the seeds of doubt whether they should be called orthologs. Inference pipelines, such as slncky [82], also fail to unanimously call lncRNA homologs in cases where more than one lncRNA can be found at neighboring genomic loci or when lncRNA and its genomic neighborhood are enriched with repetitive elements. In these cases, a single orthologous pair is selected based on statistical analysis but with no guarantee that the best solution is provided. For example, to reduce reporting alignments that may be driven by repetitive elements, slncky aligns each lncRNA to the shuffled intergenic regions and seeks to establish a null distribution and to determine the empirical 5% threshold for significant alignment scores. Second, the search for syntologs relies heavily on the quality of WGAs, which project expressed lncRNA loci that correspond to loci in other species. As a result, although the approach is less dependent on evolutionary distance than other sequence-based analyses, it works best with closely related species. Nevertheless, Herrera-Ubeda et al. managed to find syntenic lncRNAs between humans and lancelets using LincOFinder, a new pipeline that identifies conserved lincRNAs between evolutionarily distant species by means of microsynteny analyses [83]. The pipeline considers only the clusters formed by one upstream gene, the lincRNA and one downstream gene and led to the discovery of 32 clusters, with only 16 found to have bona fide orthologous lincRNA after manual inspection.
Notably, an efficient search for syntologous lncRNAs requires high-quality annotations for both species of interest, which are not always available but are essential in different algorithm steps. In particular, protein-coding gene annotations are used to resolve spurious conservation relationships, but the ability to detect lncRNA orthologs relies mostly on extensive annotations of lncRNAs in a given species. As mentioned earlier, available lncRNA datasets are scarce for most species, while model organisms possess multiple and rather poorly overlapping catalogs of lncRNAs, as they are populated with fundamentally different experimental and in silico methods. Keeping this in mind, in our recent research on conserved lncRNAs across primates, we used our custom pipeline built on the basis of slncky, and we assembled custom lncRNA annotations based on data from publicly available RNA-Seq repositories [17]. Our analysis yielded over 78 000 expressed human lncRNAs, and from 2054 to 18 226 were conserved across individual primate species. Interestingly, lncRNAs showing only positional conservation represent as many as 66.78% of the conserved human lncRNAs in the great apes, while syntologs make up only 42.37% of the ultraconserved lncRNAs, defined as those found in all eleven primate species considered in the study.
Current challenges in comparative genomics of lncRNAs
When deciphering the evolution of lncRNAs, the use of comparative genomics is hindered by the poor conservation of sequences and secondary structures. Therefore, specialized computational approaches tailored to the specific characteristics of lncRNA evolution are needed. Some of the most frequently used approaches are briefly described above. The genomic context of lncRNAs represents another layer of complexity. As many of lncRNAs are expressed in antisense to protein-coding loci, are located in introns or represent alternative isoforms of protein-coding genes, it is virtually impossible to investigate their evolution independent of their associated protein-coding genes. This is why most studies are focusing on lincRNAs, but their evolution does not necessarily reflect that of all lncRNAs. Another major obstacle is extensive bias in the quality of annotations between compared species. First, in addition to model organisms, available lncRNA catalogs are quite scarce (compare 82 760 lncRNAs for humans and 1778 for chimps in ENSEMBL 102), which is often coupled with the lack of fully assembled genomes and diversified RNA-Seq data, both of which are typically required to build comprehensive sets of expressed lncRNAs. On the other hand, model organisms, such as humans, possess highly divergent datasets of lncRNAs across public resources, which largely stems from the various computational criteria being used, in some situations leading to barely overlapping sets of lncRNAs based on the same input data [84]. Additionally, since lncRNAs are most often identified from RNA-Seq data, some lncRNAs with highly specific spatiotemporal expression patterns might remain undiscovered; similarly, nonpolyadenylated lncRNAs that prevailingly remain elusive when oligo(dT) protocols for reverse transcription are applied. Reliable analysis of lncRNA conservation, orthologs across species and evolution in general depend on the establishment of standard criteria for their search and annotation, along with the growth of genomic and transcriptomic data for nonmodel organisms.
Summary
Comparative genomics of lncRNAs typically involves the formulation of dedicated algorithms and the use of specialized software, in addition to those used in previous studies on protein-coding genes.
From the perspective of genomics, there are four different levels of lncRNA conservation, and in general, lncRNA orthologs are not required to show sequence similarities.
Comparative genomics of lncRNAs is severely hampered by limited availability of quality annotations, and further progress in the field is dependent on expansion of lncRNA catalogs across species.
Abbreviations
- CM
covariance model
- CRS
conserved RNA structure
- ESE
exonic splice enhancer
- lincRNA
long intergenic RNA
- lncRNA
long noncoding RNA
- NAT
natural antisense transcript
- T-UCR
transcribed-ultraconserved region
- WGA
whole genome alignment
Contributor Information
Michał Wojciech Szcześniak, Email: miszcz@amu.edu.pl.
Izabela Makałowska, Email: izabel@amu.edu.pl.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Science Centre [grant number 2017/27/N/NZ1/00417 (to E.W.)].
Author Contribution
All authors wrote and edited the manuscript. E.W. and M.R.K. prepared the figure. All authors read and approved the final manuscript.
References
- 1.Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R.et al. (2020) Ensembl 2021. Nucleic Acids Res. 49, D884–D891 10.1093/nar/gkaa942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flintoft L. (2013) Structure and function for lncRNAs. Nat. Rev. Genet. 14, 598–598 10.1038/nrg3561 [DOI] [PubMed] [Google Scholar]
- 3.Perry R.B.-T. and Ulitsky I. (2016) The functions of long noncoding RNAs in development and stem cells. Development 143, 3882–3894 10.1242/dev.140962 [DOI] [PubMed] [Google Scholar]
- 4.Marchese F.P., Raimondi I. and Huarte M. (2017) The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 18, 206 10.1186/s13059-017-1348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., Wang W., Zhu W., Dong J., Cheng Y., Yin Z.et al. (2019) Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 20, 5573 10.3390/ijms20225573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J., Wang Y., Wang C., Hu J.-F. and Li W. (2020) LncRNA functions as a new emerging epigenetic factor in determining the fate of stem cells. Front. Genet. 11, 10.3389/fgene.2020.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolha L., Ravnik-Glavač M. and Glavač D. (2017) Long noncoding RNAs as biomarkers in cancer. Dis. Markers 2017, 10.1155/2017/7243968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarfi M., Abbastabar M. and Khalili E. (2019) Long noncoding RNAs biomarker-based cancer assessment. J. Cell. Physiol. 234, 16971–16986 10.1002/jcp.28417 [DOI] [PubMed] [Google Scholar]
- 9.Kugel J.F. and Goodrich J.A. (2012) Non-coding RNAs: key regulators of mammalian transcription. Trends Biochem. Sci. 37, 144–151 10.1016/j.tibs.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakagawa S. and Kageyama Y. (2014) Nuclear lncRNAs as epigenetic regulators—beyond skepticism. Biochim. Biophys. Acta Gene Regul. Mech. 1839, 215–222 10.1016/j.bbagrm.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 11.Hu S., Wang X. and Shan G. (2016) Insertion of an Alu element in a lncRNA leads to primate-specific modulation of alternative splicing. Nat. Struct. Mol. Biol. 23, 1011–1019 10.1038/nsmb.3302 [DOI] [PubMed] [Google Scholar]
- 12.Szcześniak M.W. and Makałowska I. (2016) lncRNA-RNA interactions across the human transcriptome. PLoS ONE 11, e0150353 10.1371/journal.pone.0150353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehner B., Williams G., Campbell R.D. and Sanderson C.M. (2002) Antisense transcripts in the human genome. Trends Genet. 18, 63–65 10.1016/S0168-9525(02)02598-2 [DOI] [PubMed] [Google Scholar]
- 14.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M.et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566 10.1126/science.1112009 [DOI] [PubMed] [Google Scholar]
- 15.Guo C.-J., Ma X.-K., Xing Y.-H., Zheng C.-C., Xu Y.-F., Shan L.et al. (2020) Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell 181, 621.e22–636.e22 10.1016/j.cell.2020.03.006 [DOI] [PubMed] [Google Scholar]
- 16.Managadze D., Rogozin I.B., Chernikova D., Shabalina S.A. and Koonin E.V. (2011) Negative correlation between expression level and evolutionary rate of long intergenic noncoding RNAs. Genome Biol. Evol. 3, 1390–1404 10.1093/gbe/evr116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryzghalov O., Szcześniak M.W. and Makałowska I. (2020) SyntDB: defining orthologues of human long noncoding RNAs across primates. Nucleic Acids Res. 48, D238–D245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hezroni H., Koppstein D., Schwartz M.G., Avrutin A., Bartel D.P. and Ulitsky I. (2015) Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 11, 1110–1122 10.1016/j.celrep.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noviello T.M.R., Di Liddo A., Ventola G.M., Spagnuolo A., D'Aniello S., Ceccarelli M.et al. (2018) Detection of long non–coding RNA homology, a comparative study on alignment and alignment–free metrics. BMC Bioinformatics 19, 407 10.1186/s12859-018-2441-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pegueroles C., Iraola-Guzmán S., Chorostecki U., Ksiezopolska E., Saus E. and Gabaldón T. (2019) Transcriptomic analyses reveal groups of co-expressed, syntenic lncRNAs in four species of the genus Caenorhabditis. RNA Biol. 16, 320–329 10.1080/15476286.2019.1572438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marques A.C., Hughes J., Graham B., Kowalczyk M.S., Higgs D.R. and Ponting C.P. (2013) Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol. 14, R131 10.1186/gb-2013-14-11-r131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D. and Yang M.Q. (2017) Identification and characterization of conserved lncRNAs in human and rat brain. BMC Bioinformatics 18, 489 10.1186/s12859-017-1890-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma X.-Y., Wang J.-H., Wang J.-L., Ma C.X., Wang X.-C. and Liu F.-S. (2015) Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genomics 16, 676 10.1186/s12864-015-1881-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L.et al. (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24, 4333–4345 10.1105/tpc.112.102855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng J.C., Shen J. and Ran Z.H. (2013) Transcribed ultraconserved region in human cancers. RNA Biol. 10, 1771–1777 10.4161/rna.26995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo J., Fang W., Sun L., Lu Y., Dou L., Huang X.et al. (2018) Ultraconserved element uc.372 drives hepatic lipid accumulation by suppressing miR-195/miR4668 maturation. Nat. Commun. 9, 612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ang C.E., Ma Q., Wapinski O.L., Fan S., Flynn R.A., Lee Q.Y.et al. (2019) The novel lncRNA lnc-NR2F1 is pro-neurogenic and mutated in human neurodevelopmental disorders. eLife 8, e41770 10.7554/eLife.41770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinn J.J., Zhang Q.C., Georgiev P., Ilik I.A., Akhtar A. and Chang H.Y. (2016) Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes. Dev. 30, 191–207 10.1101/gad.272187.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz-Orera J. and Albà M.M. (2019) Conserved regions in long non-coding RNAs contain abundant translation and protein–RNA interaction signatures. NAR Genom. Bioinformatics 1, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirk J.M., Kim S.O., Inoue K., Smola M.J., Lee D.M., Schertzer M.D.et al. (2018) Functional classification of long non-coding RNAs by k -mer content. Nat. Genet. 50, 1474–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross C.J., Rom A., Spinrad A., Gelbard-Solodkin D., Degani N. and Ulitsky I. (2021) Uncovering deeply conserved motif combinations in rapidly evolving noncoding sequences. Genome Biol. 22, 29 10.1186/s13059-020-02247-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N.et al. (2005) The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- 33.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H.et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H., Zhang Z. and Krause H.M. (2019) Long noncoding RNAs and repetitive elements: junk or intimate evolutionary partners? Trends Genet. 35, 892–902 10.1016/j.tig.2019.09.006 [DOI] [PubMed] [Google Scholar]
- 35.Ulitsky I. (2016) Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 17, 601–614 10.1038/nrg.2016.85 [DOI] [PubMed] [Google Scholar]
- 36.Pegueroles C. and Gabaldón T. (2016) Secondary structure impacts patterns of selection in human lncRNAs. BMC Biol. 14, 10.1186/s12915-016-0283-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tilgner H., Knowles D.G., Johnson R., Davis C.A., Chakrabortty S., Djebali S.et al. (2012) Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 22, 1616–1625 10.1101/gr.134445.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee N., Calviello L., Hirsekorn A., de Pretis S., Pelizzola M. and Ohler U. (2017) Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 24, 86–96 10.1038/nsmb.3325 [DOI] [PubMed] [Google Scholar]
- 39.Schlackow M., Nojima T., Gomes T., Dhir A., Carmo-Fonseca M. and Proudfoot N.J. (2017) Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol. Cell 65, 25–38 10.1016/j.molcel.2016.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melé M., Mattioli K., Mallard W., Shechner D.M., Gerhardinger C. and Rinn J.L. (2017) Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 27, 27–37 10.1101/gr.214205.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deveson I.W., Brunck M.E., Blackburn J., Tseng E., Hon T., Clark T.A.et al. (2018) Universal alternative splicing of noncoding exons. Cells 6, 245.e5–255.e5 10.1016/j.cels.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 42.Bozgeyik E., Igci Y.Z., Sami Jacksi M.F., Arman K., Gurses S.A., Bozgeyik I.et al. (2016) A novel variable exonic region and differential expression of LINC00663 non-coding RNA in various cancer cell lines and normal human tissue samples. Tumour Biol. 37, 8791–8798 10.1007/s13277-015-4782-3 [DOI] [PubMed] [Google Scholar]
- 43.Knutsen E., Lellahi S.M., Aure M.R., Nord S., Fismen S., Larsen K.B.et al. (2020) The expression of the long NEAT1_2 isoform is associated with human epidermal growth factor receptor 2-positive breast cancers. Sci. Rep. 10, 1277 10.1038/s41598-020-57759-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma X., Zhang W., Zhang R., Li J., Li S., Ma Y.et al. (2019) Overexpressed long noncoding RNA CRNDE with distinct alternatively spliced isoforms in multiple cancers. Front. Med. 13, 330–343 10.1007/s11684-017-0557-0 [DOI] [PubMed] [Google Scholar]
- 45.Ponjavic J., Ponting C.P. and Lunter G. (2007) Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs Genome Res. 17, 556–565 10.1101/gr.6036807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chernikova D., Managadze D., Glazko G.V., Makalowski W. and Rogozin I.B. (2016) Conservation of the exon-intron structure of long intergenic non-coding RNA genes in eutherian mammals. Life 6, 27 10.3390/life6030027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rose D., Hiller M., Schutt K., Hackermüller J., Backofen R. and Stadler P.F. (2011) Computational discovery of human coding and non-coding transcripts with conserved splice sites. Bioinformatics 27, 1894–1900 10.1093/bioinformatics/btr314 [DOI] [PubMed] [Google Scholar]
- 48.Yeo G. and Burge C.B. (2004) Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11, 377–394 10.1089/1066527041410418 [DOI] [PubMed] [Google Scholar]
- 49.Jian X., Boerwinkle E. and Liu X. (2014) In silico tools for splicing defect prediction - a survey from the viewpoint of end-users. Genet. Med. 16, 497 10.1038/gim.2013.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nitsche A., Rose D., Fasold M., Reiche K. and Stadler P.F. (2015) Comparison of splice sites reveals that long noncoding RNAs are evolutionarily well conserved. RNA 21, 801 10.1261/rna.046342.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alezz M.A., Celli L., Belotti G., Lisa A. and Bione S. (2020) GC-AG introns features in long non-coding and protein-coding genes suggest their role in gene expression regulation. Front. Genet. 11, 488 10.3389/fgene.2020.00488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corona-Gomez J.A., Garcia-Lopez I.J., Stadler P.F. and Fernandez-Valverde S.L. (2020) Splicing conservation signals in plant long noncoding RNAs. RNA 26, 784–793 10.1261/rna.074393.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schüler A., Ghanbarian A.T. and Hurst L.D. (2014) Purifying selection on splice-related motifs, not expression level nor RNA folding, explains nearly all constraint on human lincRNAs. Mol. Biol. Evol. 31, 3164 10.1093/molbev/msu249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fairbrother W.G., Yeo G.W., Yeh R., Goldstein P., Mawson M., Sharp P.A.et al. (2004) RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 32, W187–W190 10.1093/nar/gkh393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haerty W. and Ponting C.P. (2015) Unexpected selection to retain high GC content and splicing enhancers within exons of multiexonic lncRNA loci. RNA 21, 320–332 10.1261/rna.047324.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krchňáková Z., Thakur P.K., Krausová M., Bieberstein N., Haberman N., Müller-McNicoll M.et al. (2019) Splicing of long non-coding RNAs primarily depends on polypyrimidine tract and 5′ splice-site sequences due to weak interactions with SR proteins. Nucleic Acids Res. 47, 911–928 10.1093/nar/gky1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith M.A., Gesell T., Stadler P.F. and Mattick J.S. (2013) Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res. 41, 8220–8236 10.1093/nar/gkt596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ponjavic J., Oliver P.L., Lunter G. and Ponting C.P. (2009) Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 5, e1000617 10.1371/journal.pgen.1000617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J.-R. and Zhang J. (2015) Human long noncoding RNAs are substantially less folded than messenger RNAs. Mol. Biol. Evol. 32, 970–977 10.1093/molbev/msu402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones A.N., Pisignano G., Pavelitz T., White J., Kinisu M., Forino N.et al. (2020) An evolutionarily conserved RNA structure in the functional core of the lincRNA Cyrano. RNA 26, 1234–1246 10.1261/rna.076117.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smola M.J., Christy T.W., Inoue K., Nicholson C.O., Friedersdorf M., Keene J.D.et al. (2016) SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. U.S.A. 113, 10322–10327 10.1073/pnas.1600008113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Z., Guo J.K., Wei Y., Dou D.R., Zarnegar B., Ma Q.et al. (2020) Structural modularity of the XIST ribonucleoprotein complex. Nat. Commun. 11, 6163 10.1038/s41467-020-20040-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang B., Mao Y.S., Diermeier S.D., Novikova I.V., Nawrocki E.P., Jones T.A.et al. (2017) Identification and characterization of a class of MALAT1-like genomic loci. Cell Rep. 19, 1723–1738 10.1016/j.celrep.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCown P.J., Wang M.C., Jaeger L. and Brown J.A. (2019) Secondary structural model of human MALAT1 reveals multiple structure–function relationships. Int. J. Mol. Sci. 20, 5610 10.3390/ijms20225610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novikova I.V., Hennelly S.P. and Sanbonmatsu K.Y. (2012) Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 40, 5034–5051 10.1093/nar/gks071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hudson W.H., Pickard M.R., de Vera I.M.S., Kuiper E.G., Mourtada-Maarabouni M., Conn G.L.et al. (2014) Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate. Nat. Commun. 5, 5395 10.1038/ncomms6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hawkes E.J., Hennelly S.P., Novikova I.V., Irwin J.A., Dean C. and Sanbonmatsu K.Y. (2016) COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 16, 3087–3096 10.1016/j.celrep.2016.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eddy S.R. (2002) A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics 3, 18 10.1186/1471-2105-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao Z., Weinberg Z. and Ruzzo W.L. (2006) CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics 22, 445–452 10.1093/bioinformatics/btk008 [DOI] [PubMed] [Google Scholar]
- 70.Seemann S.E., Mirza A.H., Hansen C., Bang-Berthelsen C.H., Garde C., Christensen-Dalsgaard M.et al. (2017) The identification and functional annotation of RNA structures conserved in vertebrates. Genome Res. 27, 1371–1383 10.1101/gr.208652.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rivas E., Clements J. and Eddy S.R. (2017) A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods 14, 45–48 10.1038/nmeth.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tavares R.C.A., Pyle A.M. and Somarowthu S. (2019) Phylogenetic analysis with improved parameters reveals conservation in lncRNA structures. J. Mol. Biol. 431, 1592–1603 10.1016/j.jmb.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mattei E., Ausiello G., Ferrè F. and Helmer-Citterich M. (2014) A novel approach to represent and compare RNA secondary structures. Nucleic Acids Res. 42, 6146–6157 10.1093/nar/gku283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Diederichs S. (2014) The four dimensions of noncoding RNA conservation. Trends Genet. 30, 121–123 10.1016/j.tig.2014.01.004 [DOI] [PubMed] [Google Scholar]
- 75.Jathar S., Kumar V., Srivastava J. and Tripathi V. (2017) Technological developments in lncRNA biology. Adv. Exp. Med. Biol. 1008, 283–323 10.1007/978-981-10-5203-3_10 [DOI] [PubMed] [Google Scholar]
- 76.Wanowska E., Kubiak M.R., Rosikiewicz W., Makałowska I. and Szcześniak M.W. (2018) Natural antisense transcripts in diseases: from modes of action to targeted therapies. Wiley Interdiscip Rev. RNA 9, e1461 10.1002/wrna.1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rosikiewicz W. and Makałowska I. (2016) Biological functions of natural antisense transcripts. Acta Biochim. Pol. 63, 665–673 [DOI] [PubMed] [Google Scholar]
- 78.Kaikkonen M.U., Lam M.T.Y. and Glass C.K. (2011) Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 90, 430–440 10.1093/cvr/cvr097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martens J.A., Laprade L. and Winston F. (2004) Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429, 571–574 10.1038/nature02538 [DOI] [PubMed] [Google Scholar]
- 80.Tufarelli C., Stanley J.A.S., Garrick D., Sharpe J.A., Ayyub H., Wood W.G.et al. (2003) Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 34, 157–165 10.1038/ng1157 [DOI] [PubMed] [Google Scholar]
- 81.Amaral P.P., Leonardi T., Han N., Viré E., Gascoigne D.K., Arias-Carrasco R.et al. (2018) Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol. 19, 32, 10.1186/s13059-018-1405-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen J., Shishkin A.A., Zhu X., Kadri S., Maza I., Guttman M.et al. (2016) Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol. 17, 19 10.1186/s13059-016-0880-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Herrera-Úbeda C., Marín-Barba M., Navas-Pérez E., Gravemeyer J., Albuixech-Crespo B., Wheeler G.N.et al. (2019) Microsyntenic clusters reveal conservation of lncRNAs in chordates despite absence of sequence conservation. Biology 8, 61 10.3390/biology8030061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu J., Bai J., Zhang X., Lv Y., Gong Y., Liu L.et al. (2017) A comprehensive overview of lncRNA annotation resources. Brief. Bioinform. 18, 236–249 [DOI] [PubMed] [Google Scholar]