SUMMARY
Single-cell gene expression technologies are powerful tools to study cell types in the human brain, but efforts have largely focused on cortical brain regions. We therefore created a single-nucleus RNA-sequencing resource of 70,615 high-quality nuclei to generate a molecular taxonomy of cell types across five human brain regions that serve as key nodes of the human brain reward circuitry: nucleus accumbens, amygdala, subgenual anterior cingulate cortex, hippocampus, and dorsolateral prefrontal cortex. We first identified novel subpopulations of interneurons and medium spiny neurons (MSNs) in the nucleus accumbens and further characterized robust GABAergic inhibitory cell populations in the amygdala. Joint analyses across the 107 reported cell classes revealed cell-type substructure and unique patterns of transcriptomic dynamics. We identified discrete subpopulations of D1- and D2-expressing MSNs in the nucleus accumbens to which we mapped cell-type-specific enrichment for genetic risk associated with both psychiatric disease and addiction.
In brief
For this NeuroResource, >100 cell classes from five brain regions in the human reward circuitry are characterized by single-nucleus RNA sequencing, with interactive apps made available. The molecular relationships across this circuitry are described, and genetic risk for various psychiatric and substance use phenotypes is quantified across all cell classes.
Graphical Abstract
INTRODUCTION
Recent advances in single-cell and single-nucleus RNA-sequencing (scRNA-seq/snRNA-seq) technologies have facilitated the molecular characterization of diverse cell types in the postmortem human brain during development (Darmanis et al., 2015; Li et al., 2018a; Zhong et al., 2018, 2020) and have been used to assess cell-type-specific gene expression differences in the context of several brain disorders, including Alzheimer’s disease (AD), autism spectrum disorder (ASD), multiple sclerosis, and major depressive disorder (MDD) (Mathys et al., 2019; Nagy et al., 2020; Schirmer et al., 2019; Velmeshev et al., 2019). Identification of cell-type-specific gene expression signatures has contributed to understanding the relationship between molecular identity and cell function as it relates to brain health, neurological disease, and genetic risk for neuropsychiatric disorders, such as schizophrenia (SCZ; Skene et al., 2018). While substantial advancements have been made in understanding cell-type heterogeneity both within and across individual regions of the human brain, the majority of snRNA-seq reports are limited to a small number of brain areas. These primarily include the hippocampus (HPC) (Franjic et al., 2020; Habib et al., 2017) and several heavily studied subregions of the cortex (Lake et al., 2016), including the dorsolateral prefrontal cortex (DLPFC) (Li et al., 2018a; Nagy et al., 2020), medial temporal cortex (Darmanis et al., 2015; Hodge et al., 2019), entorhinal cortex (Grubman et al., 2019), and anterior cingulate cortex (Velmeshev et al., 2019). Molecular profiling of less studied cortical subregions, including the subgenual anterior cingulate cortex (sACC) and striatal and limbic brain regions (including the nucleus accumbens [NAc] and the amygdala [AMY]), is lacking in the human brain. The sACC, NAc, and AMY are interconnected within well-established circuit loops that mediate important behavioral and neurobiological functions, including signaling for reward and motivation, as well as processing emotional valence, particularly for fearful and threatening stimuli (Haber and Knutson, 2010; Janak and Tye, 2015; Russo and Nestler, 2013). Importantly, the cellular composition of individual neuronal subtypes in these regions substantially differs from previously well-profiled cortical and hippocampal regions (Saunders et al., 2018; Zeisel et al., 2018).
For example, the NAc contains dopaminoceptive populations of GABAergic medium spiny neurons (MSNs)—the principal projecting cell type that makes up to 95% of neurons in rodents—that harbor unique physiological and cellular properties (Gerfen et al., 1990; Kawaguchi, 1997; Kronman et al., 2019; Russo and Nestler, 2013). Early functional characterization of MSNs revealed two distinct classes of MSNs based on the expression of D1 versus the D2 dopamine receptors (D1-MSNs and D2-MSNs, respectively) (Lobo, 2009; Lobo et al., 2006). However, recent sc-/snRNA-seq studies in the rodent striatum, and in the NAc specifically, revealed more complex transcriptional diversity within broader D1- and D2-MSN subclasses than was previously appreciated (Gokce et al., 2016; Saunders et al., 2018; Stanley et al., 2020; Zeisel et al., 2018). Moreover, subpopulations of MSNs are differentially recruited in response to cocaine exposure and mediate divergent functional effects on behavioral responses to drugs of abuse (Savell et al., 2020). Similarly, single-cell profiling studies in the rodent AMY identified specialized populations of Cck-expressing neurons that are preferentially activated by behavioral experience, including exposure to acute stress (Wu et al., 2017). However, whether and to what extent this transcriptional diversity is conserved in these areas of the human NAc and AMY has not yet been fully explored. Given the evidence for the functional importance of specific cell types in these areas of the rodent brain, single-cell profiling of these regions in human may identify analogous cell populations, which can then be analyzed in the context of neurobiological dysfunction in human brain disorders.
Here, we used snRNA-seq to define the molecular taxonomy of distinct cell types in subcortical regions (NAc and AMY) that act as key nodes within circuits that mediate critical brain and behavioral functions, including reward signaling and emotional processing. For a subset of samples, this included NeuN enrichment to better profile neuronal diversity. We also validated molecular profiles for previously identified cell types in the HPC and DLPFC, and identified similar cell types in the sACC, an additional cortical region central to limbic system function that has been implicated in affective disorders. Furthermore, we evaluate cross-species conservation of NAc and AMY cell types between humans and rodents, specifically focusing on comparisons of MSN subpopulations identified as playing key roles in reward processing and addiction. We survey the transcriptomic architecture across 107 robust cell classes, identifying molecular relationships between cell populations and patterns of divergence within specialized MSNs. Finally, by integrating genetic studies for substance use and neuropsychiatric disorders, we show differential cell-type association, or differential expression of risk loci-associated genes, with a number of neuropsychiatric or substance use phenotypes, highlighting the clinical relevance of understanding cell-type- and region-specific expression in the human brain.
RESULTS
Identification of refined MSN subpopulations in human NAc
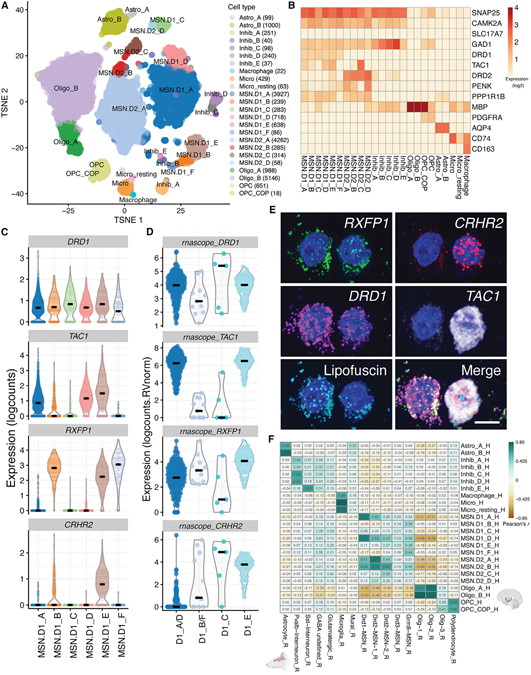
To evaluate the transcriptional landscape of MSNs and other cell populations in the human NAc, we analyzed 19,892 total nuclei from 8 donors. We performed data-driven clustering to generate 24 cell clusters across six broad cell types, including GABAergic inhibitory neurons, MSNs, oligodendrocytes, oligodendrocyte precursor cells, microglia, and astrocytes (Figure 1A). Of the 10 distinct neuronal clusters expressing established D1- and D2-MSNs markers (Figure 1B), including PPP1R1B (encoding DARPP-32), six of these MSN subclusters were enriched for DRD1 (D1_A–D1_F) and four were enriched for DRD2 (D2_A–D2_D). These MSN subclusters collectively made up between 85% and 95% of neuronal nuclei from the neuron-enriched samples (Table S3), lending human evidence that, similar to the rodent, the vast majority of nuclei in this region of the striatum are composed of MSNs (Kawaguchi, 1997). Clusters D1_A and D2_A represented the largest D1-MSN (67%) and D2-MSN (87%) subclasses, respectively. As expected, MSN subclusters showed differential enrichment of several neuropeptides, including proenkephalin (PENK), tachykinin 1 (TAC1), and prodynorphin (PDYN) (Figure S1) (Lobo, 2009; Lobo et al., 2006; Savell et al., 2020). Surprisingly, the classical D1-MSN marker, TAC1, was enriched in D2_C MSNs, while it was largely absent in the smaller D1_B, D1_C, and D1_F MSNs (Figure 1B). Similarly, the classical D2-MSN marker PENK was enriched in the large population of D2_A MSNs, in addition to D2_B and D2_D, but depleted in the smaller population of D2_C MSNs (Figure S1). Differential expression of these neuropeptides in D1- and D2-MSN subclasses was confirmed using single-molecule fluorescence in situ hybridization (smFISH) with 4-plex RNAscope technology (Maynard et al., 2020); Figure S1).
Figure 1. Distinct classes of D1- and D2-expressing MSNs in human NAc.
(A) t-distributed stochastic neighbor embedding (tSNE) plot of 19,789 nuclei (n = 8 donors) across 21 clusters, including 6 clusters of D1 MSNs and 4 clusters of D2 MSNs.
(B) Heatmap depicting log2 expression of canonical marker genes used to annotate each cluster.
(C) Violin plots for 4 genes differentially expressed (log2-normalized counts) in specific D1 classes (or class groups: CRHR2, DRD1, RXFP1, and TAC1) that were selected for validation using single-molecule fluorescence in situ hybridization (smFISH).
(D) Log2 expression of respective transcript counts per smFISH region of interest (ROI) (ROI size normalized) post lipofuscin masking (autofluorescence). Each DRD1+ ROI was classified into a Euclidean distance-predicted MSN class (or group of classes) and its/their respective expression.
(E) Multiplex smFISH in human NAc depicting a D1_C (left) and D1_E (right) MSN, side by side. Maximum intensity confocal projections showing the expression of DAPI (nuclei), CRHR2, DRD1, TAC1, and lipofuscin autofluorescence. Merged image without lipofuscin autofluorescence. Scale bar, 10 μm.
(F) Heatmap of Pearson correlation values evaluating the relationship between our human-derived NAc cell classes (rows) and reported rat NAc populations from Savell et al. (2020). Correlation was performed on the combined top 100 markers/cell populations in which annotated homology exists (here, 582 genes; see STAR Methods).
Using differential expression analyses, we identified the most preferentially expressed genes in each MSN class and found tens to hundreds of unique markers for D1- and D2-MSN classes (at false discovery rate [FDR] <1e6; Table S5). Among D1-MSNs, three classes (D1_B, D1_E, and D1_F) were enriched for the relaxin family peptide receptor 1, RXFP1, but only the TAC1− D1_F MSNs were enriched for the GABAA receptor subunit, GABRQ, and the relaxin family peptide receptor 1, RXFP1 (Figures 1C and S2). Similarly, only D1_E MSNs expressed substantial levels of CRHR2, encoding corticotropin-releasing hormone receptor 2, a protein implicated in mediating the response to stress in the brain (Figure 1D). The TAC1− D1_C MSNs could be distinguished from all other MSN classes by the elevated expression of relaxin family peptide receptor 2, RXFP2, and the depletion of both RXFP1 and GABRQ (Figure S2), although this small class of D1 MSNs was difficult to identify via smFISH. Consistent with the identification of a discrete D2-MSN class expressing Htr7 in the mouse striatum (Gokce et al., 2016; Stanley et al., 2020), we identified the enrichment of HTR7 in D2_C (TAC1+; PENK−) MSNs, but not other classes of D2 MSNs (Figure S3). Similar to D1_E MSNs, the HTR7+ D2_C cluster was the only D2-MSN class expressing CRHR2, although to a lesser degree. The existence of these novel D1- and D2-MSN classes was validated by smFISH on NAc brain sections derived from independent postmortem human brain donors (Figures 1D, 1E and S1–S3). Several other genes, including CASZ1, GPR6, and EBF1, were differentially expressed in unique D1- and/or D2-MSN classes (Figure S4). CASZ1 was highly enriched in the D1_B, D1_E, and D2_C classes, GPR6 in all D2 classes, and EBF1 in the D1_C class.
In addition to describing transcriptional diversity in D1- and D2-MSNs, we also identified 5 clusters of GABAergic inhibitory neurons expressing the marker genes GAD1 and GAD2, but depleted for MSN marker genes (Figures 1B and S5). These clusters contained different transcriptionally defined classes, including GABAergic neurons expressing somatostatin (SST; Inhib_E), neuropeptide Y (NPY; Inhib_E), prepronociceptin (PNOC; Inhib_E), vasoactive intestinal peptide (VIP; Inhib_B), and tachykinin 3 (TAC3; Inhib_A; Figure S5; Table S5). While we did not observe robust expression of parvalbumin (PVALB) in any cluster, classes Inhib_C and Inhib_D showed high expression of KIT, encoding the protein c-Kit, which is frequently co-expressed in mouse Pvalb/PV+ GABAergic neurons (Enterría-Morales et al., 2020). smFISH for PVALB and other top marker genes for Inhib_C/_D (PTHLH, KIT, GAD1) confirmed that these GABAergic clusters likely represent unique PV-expressing interneuron classes (Figure S5).
We next evaluated the conservation of NAc cell types across species by comparing our cluster-level transcriptional profiles with those generated in a previous snRNA-seq study of the NAc following cocaine experience in a rat model system, which analyzed a total of 16 subclusters across 15,631 rat NAc nuclei (Savell et al., 2020). Correlation analyses between our NAc cell classes with those derived from rat NAc revealed that glial populations, including astrocytes, microglia, oligodendrocytes, and oligodendrocyte progenitor cells, were highly conserved (Figure 1F). GABAergic inhibitory neuron populations were also well correlated across species as rat Sst-expressing and likely Pvalb-expressing clusters overlapped with human Inhib_E and Inhib_C/_D classes, respectively (Pearson’s r = 0.63, 0.63, and 0.62, respectively). We also observed substantial correlation between rat and human D1- and D2-MSNs, especially between rat Drd1-expressing MSNs and human D1_A/_D1_D MSNs (r = 0.74, 0.74, respectively). Beyond the overlap of rat Drd2-expressing MSNs in the human D2_A and D2_B MSN classes (r = 0.77, 0.70, respectively), we additionally saw positive correlations across D1- and D2-MSN subtypes, such that rat Drd2-expressing MSNs also showed enrichment in our human D1_A/_D MSNs. This result is not likely fully explained by the co-expression of DRD1 and DRD2 in the same nucleus because, while we we did find that 11.2% of all MSNs expressed both DRD1 and DRD2 to some degree, these dual-expressing nuclei were by far the most enriched in the D1_E class (43.4% of D1_E nuclei expressing both DRD1/DRD2) (Figures 1B and S1). In addition, many of the top markers for either the D1_A or D2_A classes were highly expressed in both MSN clusters (Figure S4), suggesting that the majority of canonically dichotomous D1- or D2-MSNs may be more molecularly similar than previously appreciated. This is supported by the observation (data not shown) that D2_A exhibited the least number of differentially expressed genes compared to D1_A (776 genes at FDR < 0.05; see STAR Methods) than with any other D1 (range: 791–2,296 genes) or D2 class (range: 1,146–2,310 genes). We did not observe strong enrichment for rat Drd3- and Grm8-expressing MSNs in any human MSN subclusters. Likewise, a few human D1-(D1_B, _C, _E_ and _F) and D2- (D2_C and _D) MSN classes did not appear to be convincingly represented in rat MSN subtypes (Figure 1F; see discussion). While these data suggest overall conservation between rat and human NAc cell types, there appear to be transcriptional features that are unique among specialized subpopulations of rodent and human MSNs.
Atlas of molecularly defined cell types in AMY
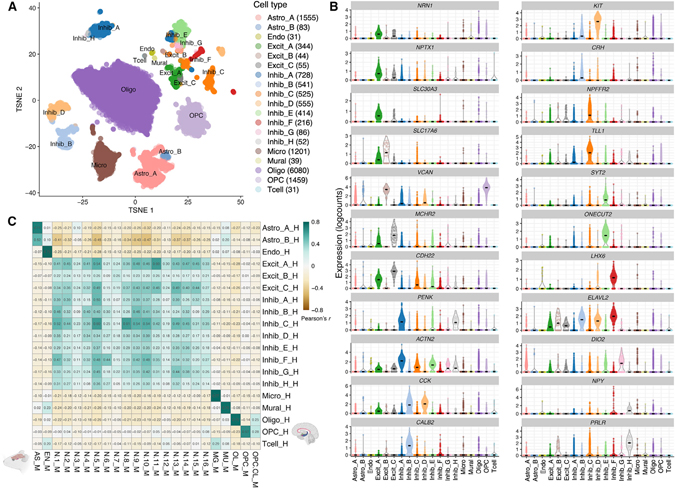
The AMY, a medial structure of the temporal lobe, is noted for its role in processing emotional valence, particularly for both fear and reward (Janak and Tye, 2015; Wassum and Izquierdo, 2015). Dysfunction in amygdalar signaling is implicated in MDD, bipolar disorder (BIP), and posttraumatic stress disorder (PTSD) (Fenster et al., 2018; Garrett and Chang, 2008; Murray et al., 2011). The human AMY can be subdivided into a number of distinct regions based on histology, immunohistochemical classifications, connectivity, and neural activation patterns, as revealed by functional magnetic resonance imaging (fMRI) of the brain (Barger et al., 2012; Schumann and Amaral, 2005; Sorvari et al., 1995; Tyszka and Pauli, 2016; Zhang et al., 2018). Studies in the rodent and non-human primate AMY have identified different cell compositions across the AMY, which likely correspond to differential patterns of synaptic connections between cell types across amygdalar subregions, and with extraamygdalar brain regions (Chareyron et al., 2011). Hence, it is likely that various cell types with unique molecular signatures also exist within the human AMY, which can be surveyed by snRNA-seq. We analyzed 14,039 nuclei from the AMY of 5 adult neurotypical donors to create a molecular taxonomy of cell types in this brain region. We identified 19 clusters that corresponded to 4 broad glial cell types (Oligo, Astro, Micro, and oligodendrocyte progenitor cell [OPC]), stromal (endothelial [Endo]; and mural cells) or immune cell populations (macrophages and T cells), and 11 neuronal classes (Figures 2A and S6). Glial cell populations were present at similar proportions between the non-NeuN-enriched donors (mean 54.4% Oligo; 12.3% Astro; 10.7% Micro; 11.5% OPC), but we observed a varied distribution of neuronal classes between donors, and the stromal/immune cell classes, as these were rarer (see discussion; Table S3). Despite this, after correcting for donor batch effects, we identified hundreds of genes enriched in each broad glial, stromal, and neuronal classes at FDR < 1–6 (top markers shown in Table S5).
Figure 2. Atlas of molecularly defined cell types in human AMY.
(A) tSNE plot of 14,039 nuclei across 19 clusters, including 3 clusters of excitatory neurons and 8 clusters of GABAergic inhibitory neurons.
(B) Expression violin plots for the top genes for each of the neuronal classes (log2-normalized counts).
(C) Heatmap of Pearson correlation values evaluating the relationship between our human-derived AMY cell classes (rows) versus the cell populations reported in Chen et al. (2019), derived from mouse medial amygdala (MeA). Correlation was performed on the combined top 100 markers/cell populations in which annotated homology exists (here, 480 genes; see STAR Methods).
Within the 11 neuronal classes expressing the pan neuronal marker gene SNAP25, 3 clusters were enriched for excitatory neuronal markers (SLC17A7, SLC17A6) and 8 clusters were enriched for inhibitory GABAergic markers (GAD1, GAD2; Figure S6). The three excitatory classes comprised different functional classes of neurons (referred to as “Excit_A” to “_C”), with top markers including NRN1, NPTX1, and SLC30A3 (encoding neuritin, neuronal pentraxin 1, and zinc transporter 3, respectively) for Excit_A, and SLC17A6 and VCAN (Versican, typically associated with OPCs) for Excit_B (Figure 2B). NRN1, NPTX1, SLC30A3, and VCAN have been implicated in modulation of synaptic plasticity and memory (Figueiro-Silva et al., 2015; Horii-Hayashi et al., 2008; Sindreu and Storm, 2011; Yao et al., 2018). Top markers for class Excit_C included MCHR2 (melanin-concentrating hormone receptor 2) and CDH22 (pituitary/brain cadherin). In addition, Excit_A is a large class made up of four subpopulations (see discussion).
Compared to the excitatory neuron classes, we identified a greater diversity of inhibitory GABAergic classes and subclasses (see STAR Methods). Those classes of note include cholecystokinin (CCK)-containing regular-spiking interneurons (Inhib_B, Inhib_D) evident by high expression of CCK (Figure 2B). Of these CCK-expressing GABAergic classes, Inhib_B was also enriched in VIP and CALB2 (calretinin), whereas Inhib_D showed enrichment for KIT. NPY was specific for the smaller of two PENK+ classes, Inhib_A and Inhib_H (Figure 2B), whereas SST and TAC3 were enriched in specific subclusters of some of these interneuron classes (data not shown). More functional characterization includes Inhib_B enrichment for CRH (corticotropin-release hormone/factor)-expressing subpopulations. CRH is a key regulator of the hypothalamic-pituitary-adrenal (HPA) axis, which is critical for both the acute stress response and adaptation to chronic stress. Finally, NPFFR2 and TLL1, additional genes associated with HPA axis regulation, were selectively expressed in Inhib_C (Lin et al., 2016; Tamura et al., 2005). These classes of neurons correspond to neuronal subclasses previously linked to anxiety in the AMY, as reviewed in Babaev et al. (2018), and also better clarify some expected patterns of molecular identity, such as that of SST and PRKCD (which characterize striatal-like GABAergic neurons of the centrolateral AMY) are not necessarily mutually exclusive (data not shown).
We then compared our cluster-level transcriptomic profiles to those of a previously published single-cell dataset derived from the mouse medial amygdala (MeA) (Chen et al., 2019) to evaluate the conservation of amygdalar cell types between humans and rodents (Figure 2C). Across the top shared homologous genes (see STAR Methods), we observed a substantial correlation between several mouse and human AMY cell types. For example, our human glutamatergic class Excit_A (SLC17A6+, SLC17A7+) most closely correlated with the mouse MeA glutamatergic subcluster “N.11” (Pearson correlation: r = 0.60). The marker genes that were most highly conserved between these subclusters included SLC30A3, NPTX1, and NRN1. Another notable pair of cell classes conserved between species was human inhibitory neuronal class, Inhib_C, and mouse inhibitory subcluster MeA “N.8” (r = 0.61). The top shared genes between these clusters included NPFFR2, GRM8, and FOXP2. Although we observed selective co-expression of NPFFR2 and TLL1 in human Inhib_C, we note the absence of orthologous Tll1 expression in all mouse MeA neuronal subclusters (Figure S6), including the corresponding cluster “N.8,” suggesting species differences in the molecular characteristics of neuronal subpopulations. Evidence supporting this is that MCHR2 (a top marker for Excit_C) is restricted to higher-order mammals, whereas rodent genomes only encode the related Mchr1 (Tan et al., 2002). In assessing the extent to which marker genes (per human AMY cell class) overlapped with this shared homology gene space, an average of 10.4% cell-type markers appeared to be human specific (data not shown). Importantly, several neuronal subpopulations in the mouse and human datasets lacked strong correlation across species (e.g., human Excit_B, mouse “N.3,” “N.7,” and “N.12”, with all r ≤ 0.30), either suggesting possible molecular divergence between species, or unique differences between the cell-type makeup of amygdalar subregions, such that all subpopulations may not be fully represented in our human AMY samples compared to mouse MeA samples. Our cross-species analysis demonstrates the conservation of neuronal subtypes between human AMY and mouse MeA, but highlights potential differences in the cellular distribution and transcriptomic profiles across neuronal subtypes.
Convergent and divergent cell classes with unique molecular signatures across brain regions
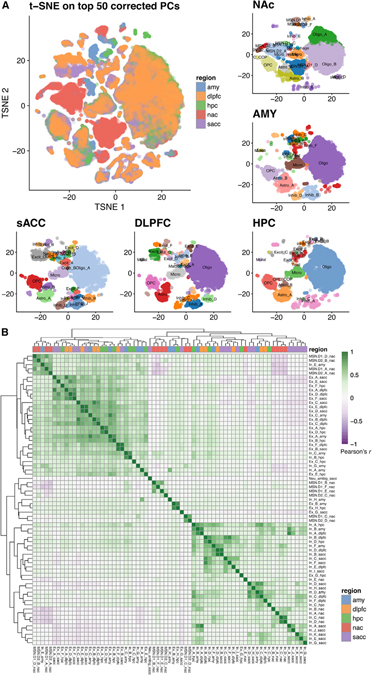
To complement the cell class populations described in the previous sections for the NAc and AMY, we additionally defined the catalog of cell-type clusters and cluster-specific genes within the other brain regions in our dataset (sACC, DLPFC, and HPC), separately (Figures 3A and S7). We further benchmarked our transcriptomic profiles against other published datasets that profiled similar regions in the postmortem human brain. Overall, our HPC subpopulations correlated well with the broad cell classes as reported in Habib et al. (2017 (Figure S8). We additionally observed strong overlap between our DLPFC to the reported PFC profiles from Velmeshev et al. (2019) (Figure S9), or similarly, sACC to the ACC set (Figure S10). Interestingly, our sACC subpopulations did not correlate more strongly with the ACC subpopulation profiles than their co-reported PFC profiles from Velmeshev et al. (2019), whereas our DLPFC subclusters generally correlated only slightly more strongly to the reported PFC than ACC subpopulations. This suggests that these cortical regions share a high degree of overlap in their nuclear transcriptomic profiles. The strength of correlation to these benchmark datasets demonstrates the robustness and utility of our pipeline, and the presented data significantly expand the existing repository of postmortem human brain snRNA-seq datasets.
Figure 3. Across-regions analyses reveal whole-brain transcriptomic architecture and neuronal subtype similarities across regions.
(A) tSNE array of a total of 70,615 nuclei, paneled by each brain region and their regionally defined cell classes (a total of 107 cell classes).
(B) Pairwise correlation of t-statistics, comparing the top cell class marker genes of the 107 classes (total of 3,715 genes). Here, only the 69 neuronal classes are shown. Regions are colored and labeled in lowercase as the suffix (e.g., as “_hpc” for HPC); “Excit_” is abbreviated as “Ex_” and “Inhib_” as “In_.” Scale values are of Pearson correlation coefficient (r), which are printed in each cell in the version shown in Figure S11.
To generate a global view of the transcriptomic architecture across the five brain regions, we compared gene expression patterns across all 107 regionally defined cell classes. Overall, each glial or stromal cell class (Oligos, Astros, OPCs, Micros, Endothelial cells, Mural cells, Macrophages, and T cells) showed broadly consistent gene expression patterns across all five brain regions (Figure S12), in line with previous analyses of broad non-neuronal cell populations using DNA methylation data (Rizzardi et al., 2019). However, in cases that yielded multiple classes of glial cells, such as astrocytes, there were unique blocks of shared transcriptional programs between subclasses. For example, the sACC “Astro_B,” HPC “Astro_B,” and NAc “Astro_A” are tightly correlated and more distinct from the other astrocyte subclasses, whereas the “Astro_B” from the AMY seem to be most distinct, in that it does not cluster with any of the rest. Among the annotated “Astro” classes, these are characterized by the highest expression of DST, COL19A1, and MACF1 (Figure S13). Although most of the other astrocyte classes (aside from the sACC “Astro_B”) themselves are characterized by specific or higher expression of unique markers, the AMY “Astro_B” showed a distinctly lower transcriptional activity state (Figure S13), likely related to this somewhat divergent astrocyte class from the amygdala, as seen with “Micro_resting,” or putative resting or dormant microglia, found in the NAc (Figures 1A and S13). Finally, we highlight that most of these reported classes of glial cell populations comprise a greater diversity of subclusters (see discussion; Table S4), the characterization of which is beyond the scope of this article.
Within the neuronal set of region-specific annotations, totaling 69 neuronal classes across 28,150 nuclei, most of the inhibitory or excitatory populations showed transcriptional patterns that clustered these broad classes together across brain regions, as expected (Figures 3B; and S11), although this was not exclusively the case in all instances. For example, the AMY Inhib_A, _C, _E, and _G, in addition to HPC Inhib_B, clustered with the excitatory branch, although they distinctly express more canonically inhibitory-classifying markers (Figure S7). We also observed strong similarities between unique pairs of neuronal classes across regions, such as between AMY and DLPFC (“In_B_amy” and “In_A_dlpfc”; r = 0.86). This DLPFC inhibitory subpopulation shares many top markers with its AMY counterpart (Table S5), including CALB2 and VIP, in addition to selective expression of CRH. In addition, we saw a variety of strongly correlating DLPFC-sACC neuronal class pairs, such as “Ex_F_dlpfc” and “Ex_B_sacc” (r = 0.94), and “Ex_E_dlpfc” and “Ex_D_sacc” (r = 0.94). This suggests potentially overlapping layer-specific identities in these regions, as evident, for example, by “Ex_F_dlpfc” and “Ex_B_sacc” most highly correlating with the corresponding and reported “L5/6” cluster from (Velmeshev et al., 2019) (r = 0.84, 0.83, respectively; Figures S9 and S10).
Integrating these neuronal classes across regions also suggested an excitatory transcriptomic signature in the NAc-specific MSN.D1_A/_D classes, in addition to D2_A/_B MSNs, as their profiles clustered with the broad excitatory branch of neuronal classes (Figures 3B and S11), whereas the remaining, less abundant MSN classes (MSN.D1_B/_C/_E/_F, and D2_C/_D) clustered in the general inhibitory branch. Strikingly, the former MSN classes negatively correlate with most other MSN and GABAergic inhibitory populations within the NAc, suggesting potentially divergent transcriptional programs across NAc neuronal classes. This observation was supported by hierarchical NAc cluster relationships, where the four D1/D2 MSN classes carrying more excitatory signature are seen to be more related than the remaining six (Figure S14), and further by separation between these groupings by the top principal components (PCs) describing variance across all NAc nuclei (Figure S14). Further investigation will be needed to assess this divergent feature of MSN classes and identify what unique roles these dopaminoceptive neurons, with respect to their general D1/D2 classification, play in reward and emotional valence processing. These analyses illustrate the utility of integrating cell-type profiles from across datasets or related tissue types/regions, to better understand cell-type identity at a full transcriptome level.
Enrichment of region-specific cell subtypes in psychiatric disease and substance use
Genome-wide association studies (GWAS) have identified a plethora of genetic risk variants or loci (segregating variants in linkage disequilibrium [LD]) for common psychiatric disorders, including SCZ (Pardiñas et al., 2018; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), ASD (Grove et al., 2019), BIP (Stahl et al., 2019), MDD (Howard et al., 2019; Wray et al., 2018), PTSD (Nievergelt et al., 2019), AD (Jansen et al., 2019), and attention-deficit/hyperactivity disorder (ADHD; Demontis et al., 2019). In addition, a large GWAS was recently performed with 1.2 million individuals to identify the genetic risk and correlations for alcohol and tobacco use (Liu et al., 2019). Approaches have been developed to identify the biological context or relevance of the hundreds of risk loci that are often identified for a given disorder or phenotype, such as LD score regression (Finucane et al., 2015), which assesses the heritability of complex phenotypes across input categories/genomic regions and their measured LD with single-nucleotide polymorphism (SNP)-level variants. Multi-marker analysis of genomic annotation (MAGMA) (de Leeuw et al., 2015) is an alternative approach that defines gene-level localization of GWAS risk, then integrates this with gene set observations, affording flexibility to assess a variety of marker lists, such as for brain region-specific snRNA-seq subcluster profiles, in two separable analyses.
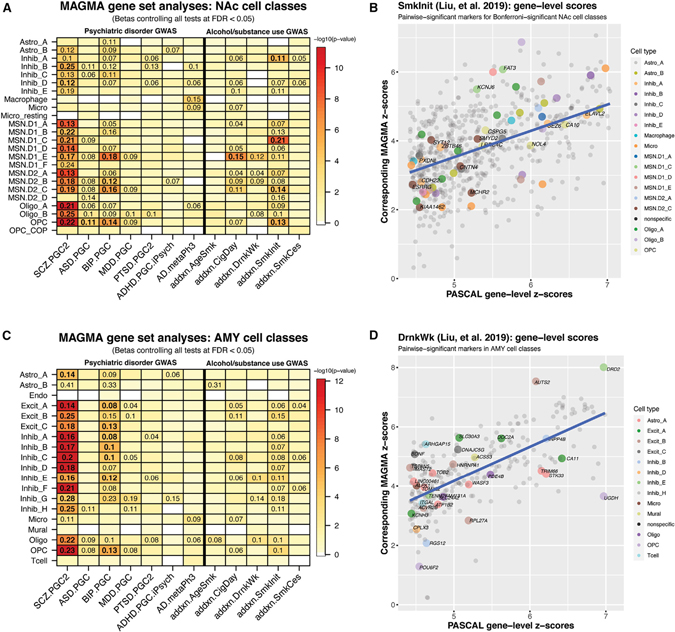
We used MAGMA to identify which cell subtypes in this study harbored aggregated genetic risk for psychiatric disorders and found robust signals across many nuclear transcriptomic profiles in each of the five profiled brain regions. As expected, many DLPFC and HPC neuronal subtypes exhibited significant effect sizes for both SCZ and BIP GWAS, spanning both excitatory and inhibitory classes (Figure S15), extending and strengthening previous findings in Bryois et al. (2020) and Skene et al. (2018). After controlling with the strict Bonferroni multiple test correction across all MAGMA gene set tests (threshold p < 3.89e–5), 9 of 10 DLPFC neuronal cell classes, which were FDR significant (controlling for FDR < 0.05), retained significant association for both SCZ and BIP. This near-uniform pattern of SCZ and BIP risk association to neuronal DLPFC cell classes was similar in the sACC (Figure S15), although only one inhibitory cell class in the sACC retained Bonferroni significance. This suggests potential regional differences in inhibitory cell classes between the two cortical brain regions, in their manifestation of genetic risk for bipolar disorder. Contrary to these patterns of risk association in the cortex, the HPC showed disorder-specific patterns of Bonferroni-significant risk (Figure S15). For example, HPC Excit_D significantly associated with only BIP, at this threshold (p = 2.96e–5, β = 0.17), whereas Excit_H associated with only SCZ (p = 4.48e–13). This rare excitatory class (Table S3) additionally showed the strongest effect size (β = 0.34) for SCZ among all GWAS tested for the cell classes of these 3 regions. Interestingly, this small, hippocampal population was most enriched for VCAN, SLC17A6 (VGLUT2), and both SoxC transcription factors, SOX4 and SOX11. None of the cell class profiles in these cortical or hippocampal regions retained significant signal for aggregated GWAS risk in MDD, PTSD, ADHD, AD, or ASD (other than in DLPFC OPCs), after Bonferroni correction, although there were some differential FDR-significant signals across certain cell classes for these disorders (Table S6).
It has been shown that broad mouse striatal neuronal populations (GABAergic inhibitory neurons, Drd1, and Drd2-expressing MSNs) additionally associated with SCZ (Skene et al., 2018) and BIP (Bryois et al., 2020) genetic risk. We demonstrate that most of our refined MSN classes in the human NAc exhibited strong associations with SCZ risk with variable effect sizes, even at the Bonferroni correction threshold (Figure 4A), aside from D1_F and D2_D. Classes MSN.D1_E, MSN.D2_B, and MSN.D2_C additionally associated with BIP at this threshold, and interestingly, the smallest D2 class, MSN.D2_D, which showed associations with neither SCZ nor BIP, was FDR significant for association with ASD (p = 0.0076, β = 0.14). NAc GABAergic inhibitory neuron classes Inhib_B and Inhib_D showed strong SCZ associations, but none to BIP, at the Bonferroni threshold. Within AMY, we observed significant associations in most of our neuronal classes to SCZ and BIP (Figure 4C). As with all of the other regions (other than DLPFC and NAc OPCs to ASD), our AMY cell classes exhibited differential, albeit weaker, FDR-significant associations to the other psychiatric disorders assessed. In summary, the NAc analyses showed complementary findings of Drd1- and Drd2-expressing striatal MSN associations with SCZ and BIP. We also dissected these mouse association signals with more relevant human GABAergic inhibitory neuron and MSN subpopulations in the NAc, and further extended this analysis to human AMY snRNA-seq-defined cell classes.
Figure 4. Genetic associations of NAc and AMY cell populations with psychiatric disease and addiction phenotypes.
(A) MAGMA associations of 12 GWAS for each of 24 cell classes profiled in human NAc.
(B) MAGMA-computed, gene-level Z scores, compared to their reported significant PASCAL scores, for “SmkInit” from Liu et al. (2019). Genes are colored if they were statistically significant for pairwise marker tests, for the corresponding NAc cell class, and additionally labeled if that cell class was Bonferroni significant in MAGMA association with the phenotype.
(C) MAGMA associations for each of 16 cell classes profiled in human AMY.
(D) Same as (B) but for “DrnkWk” and colored/labeled by AMY pairwise cell class markers (no MAGMA-gene set analysis result restriction).
For the MAGMA heatmaps: displayed numbers are the effect size (b) for significant associations (controlled for false discovery rate [FDR] < 0.05), on a Z (standard normal) distribution. Bolded numbers are those that additionally satisfy a strict Bonferroni correction threshold of p < 3.89e–5. Heatmap is colored by empirical log10(p value) for each association test.
AD, Alzheimer’s disease; ADHD, attention-deficit/hyperactivity disorder; ASD, autism spectrum disorder; BIP, bipolar disorder; MDD, major depressive disorder; PTSD, posttraumatic stress disorder; SCZ, schizophrenia. The suffix for these (e.g., “.PGC2”) reference the specific study (see STAR Methods). For the Liu et al. (2019) phenotypes: “addxn.,” addiction; “AgeSmk,” age of initiation of regular smoking; “CigDay,” number of cigarettes per day; “DrnkWk,” number of drinks per week; “SmkInit,” whether regular smoking was ever reported (binary variable); “SmkCes,” if so, had an individual stopped smoking (binary variable).
We further tested for alcohol and tobacco use GWAS (Liu et al., 2019) genetic risk associations across subcluster profiles from each brain region, focusing on the subcortical regions centered in reward circuitry, the NAc and AMY, and their subcluster profiles described above. This highlighted various MSN and inhibitory classes in the NAc differentially associated with genetic risk for regular smoking behavior (“SmkInit”) and heaviness of smoking “CigDay” at the Bonferroni correction threshold (Figure 4A). In addition to their gene set markers collectively contributing to risk associations to these substance use phenotypes, we saw that many genes with strong gene-level risk scores for a given phenotype were themselves also markers for those and additional cell classes (e.g., “SmkInit” in Figure 4B; see STAR Methods). Although no AMY cell classes associated with any of the five substance use phenotypes at the strict Bonferroni correction threshold, they were still differentially associated with the non-”SmkInit” phenotypes. One of these classes was DRD2-expressing Inhib_E (Figure 4C), which was the strongest AMY cell class associated with the heaviness of drinking (“DrnkWk”, p = 0.00031, β = 0.10). As with the NAc, many other cell classes were characterized by selectively enriched genes harboring local, gene-level risk (e.g., “DrnkWk” in Figure 4D), even though the gene set analyses did not assign strict Bonferroni-significant cell class association. Collectively, these results provide complementary human findings for genetic risk associations to those previously described for psychiatric disease, further identifying subpopulations in the NAc and AMY harboring aggregated risk for substance use behaviors.
DISCUSSION
In this study, we used snRNA-seq to profile five human brain regions within the ventral striatum (NAc), limbic system (AMY and HPC), and two cortical subregions (sACC and DLPFC) that are interconnected within the larger reward circuitry. While efforts to generate a diverse human cell type atlas at the single-cell level are under way (Han et al., 2020), the landscape of specialized molecular cell types across the complex human brain remains largely unexplored at this level of number of samples and regional diversity. This study is the first, to our knowledge, to systematically profile and compare across multiple interconnected cortical and subcortical human brain areas, selected for their function and association with risk for neuropsychiatric disorders and addiction. We placed special emphasis on analyses in the NAc and the AMY, given their roles in emotional processing and reward signaling and the lack of any current human snRNA-seq reference data in these regions. While this study was performed in neurotypical donors, the strong cell-type-specific associations to genetic risk for these disorders provide important information about disease etiology. This link to genetic risk is important, given that differential gene expression identified in case-control studies of postmortem tissue is difficult to interpret as signals may more likely represent consequences, rather than causes, of these disorders (Collado-Torres et al., 2019; Jaffe et al., 2020). More generally, understanding the transcriptomic architecture and cell-type composition across the normal human brain is crucial to understanding the etiology of disease and the molecular pathology observed in postmortem tissues to identify and prioritize potential novel disease targets. Our study is a significant contribution as it demonstrates differential enrichment of disease risk in snRNA-seq-defined cell populations across multiple brain regions, including the NAc and AMY, which have not yet been profiled at the single-cell/single-nucleus level in the human brain.
The NAc is a central hub for reward signaling, and altered function in circuits encompassing the NAc is implicated in a number of psychiatric disorders as well as drug addiction. Consistent with studies that used single-cell sequencing to profile the mouse striatum, including ~1,000 striatal cells in each study (Gokce et al., 2016; Stanley et al., 2020), we identified several discrete classes of D1- and D2-expressing MSNs in human NAc. We also observed co-expression of DRD1 and DRD2 in a small subset of MSNs. While these dual-expressing neurons did not emerge as their own cluster, they were largely found in the D1_E class (Figures 1B and S1). Interestingly, this cluster showed the strongest enrichment of genes associated with psychiatric and substance use phenotypes, indicating that this particular subpopulation may be especially vulnerable to dysfunction in these disorders. Among D1 subtypes, D1_E MSNs (along with SST+, NPY+ interneuron class Inhib_E, and to some degree, MSN.D2_D) show selective expression of CRHR2, a gene encoding corticotropin-releasing hormone receptor 2, suggesting that they may be particularly susceptible to the effects of CRH, which is released and mediates the physiological and behavioral response to stress, modulating several neurotransmitter systems, including dopamine release (Bonfiglio et al., 2011; Payer et al., 2017). Given that dysfunction of the CRH system has been associated with many psychiatric disorders, including depression, anxiety, and PTSD (Claes, 2004), understanding which cell types express CRH receptors may aid in more specific targeting of the stress axis for therapeutic developments.
Similar to Gokce et al. (2016), we also observed promiscuous expression of “typical” D1 and D2 neuropeptide marker genes (TAC1 and PENK, respectively) in both D1- and D2-MSN classes, providing further evidence that these classic markers may not be as selectively expressed as previously understood. Future studies using spatial transcriptomic approaches will be important to clarify whether TAC1-expressing D1- and D2-MSN classes show topographical organization in the NAc core versus shell. Anatomical location may explain differences in TAC1 and PENK expression in specific MSN classes, as it is well established that specific neuropeptides are expressed in a spatial gradient across the core and shell (Prensa et al., 2003; Salgado and Kaplitt, 2015; Stanley et al., 2020; Voorn et al., 1989). To better interpret clinical implications of studies that focus on circuitry encompassing the NAc in animal models, further understanding of similarities and differences across species for cell types that contribute to NAc function are important. While many cell populations were conserved between rat and human NAc (Savell et al., 2020), we did observe differences in specific MSN classes, which may indicate unique molecular features between analogous MSN classes and/or the existence of divergent MSN subclasses, as exemplified by the lack of a specific human MSN classes with strong correlation to the rat “Grm8-MSN” subpopulation (Figure 1F). However, given the small positive correlations measured with human D2_B (r = 0.40) and various D1 classes, it is possible that this Grm8-expressing population encompasses a broader species equivalent of these less abundant D1/D2 classes, which express variable GRM8 (Figure S4). We also were unable to identify a population of cholinergic interneurons. While these are thought to be more abundant in the human neostriatum compared to the rodent, where they account for only ~0.3% of neurons (Graveland et al., 1985; Rymar et al., 2004; Tepper and Bolam, 2004), it is likely that the relatively low rate of sampling and the relative rarity of this population’s accounts for this lack of observation.
In addition to profiling NAc cell types, we generated a molecular taxonomy of human amygdala cell types. Among the more diverse set of inhibitory (8 of 11, total) neuronal classes in our AMY profiles, the stress modulator CRH was specifically enriched in Inhib_B. Top markers in the AMY Inhib_C subcluster included NPFFR2 (Neuropeptide FF Receptor 2) and TLL1 (Tolloid-Like 1), which are both associated with glucocorticoid signaling and the response to stress (Lin et al., 2016, 2017; Tamura et al., 2005). Comparing human AMY subcluster profiles to data from the mouse MeA (Chen et al., 2019), we found that Inhib_C and its corresponding population in mouse (MeA “N.8” subcluster; Figure 2C) were of the most strongly correlated neuronal classes (r = 0.61). While Tll1 expression was notably absent in mouse MeA, Npffr2 and other top MeA “N.8” marker genes were shared with Inhib_C (Figure S6). We emphasize the comparisons to single-nucleus/single-cell datasets from animal models in the present study as a quick method for comparing annotated profiles, whereas other more computationally heavy algorithms exist to more definitively answer questions about conservation and species specificity (Butler et al., 2018; Haghverdi et al., 2018). Nevertheless, these insights highlight the importance of deriving reference snRNA-seq datasets across the human brain, as molecular gene markers may not be shared across species between analogous neuronal subpopulations.
Integrating the transcriptomic profiles across our total of 107 reported cell classes in the NAc, AMY, sACC, DLPFC, and HPC showed patterns of expected similarity, particularly among glial cell classes. However, this analysis also revealed some within-cell-type substructure that highlight unique relationships between these regionally defined classes. As an example, we noted a cluster of strongly correlated astrocyte classes from each of the five brain regions (Figure S12), and a small cluster made up of the sACC “Astro_B,” HPC “Astro_B,” and NAc “Astro_A.” The most unique astrocyte population, “Astro_B” from AMY (83 nuclei), appeared to be a metabolically low class of astrocytes, suggested by its low distribution of total unique molecular identifier (UMI) capture (Figure S13) and specific depletion of central transcription factors (TCF4, SOX6, NFIA, and NFIB) and various signaling/growth regulators (AKT3, APC, and PREX2; data not shown). These observations complement recent work focused on surveying astrocyte diversity in the mammalian brain (Batiuk et al., 2020) or across other glial cell types and their developmental origin (Chamling et al., 2021; Masuda et al., 2020). We additionally observed a variety of shared neuronal cell classes across regions. Most strikingly, this revealed that despite their broad D1/D2 classification, MSNs separate into divergent groups that exhibit a more excitatory versus a more inhibitory signature, and these respective groups of MSN classes are generally negatively correlated (Figures S11 and S14). A comprehensive characterization of the molecular pathways driving this divergence at the transcriptome level, in addition to how this influences their potential roles within the existing micro- and macrocircuitry, is beyond the scope of this work. However, this full integration of single-nucleus profiles across regions demonstrates a practical method of assessing cell-type relationships and elucidating patterns across the cell-type manifold, while maintaining the molecular resolution of transcriptomic signatures related to their tissues of origin.
We finally used the snRNA-seq data from the five profiled regions to ask whether specific cell classes harbored aggregate genetic risk for various neuropsychiatric disorders and/or features of substance use. We confirmed previous findings by identifying strong associations for neuronal classes in the DLPFC and HPC with both SCZ and BIP (Bryois et al., 2020; Skene et al., 2018) and significantly extended these findings by providing associations with specific sACC excitatory and inhibitory populations (Figure S15). We not only confirmed previously observed associations to broad striatal populations defined in the mouse but also showed, for the first time, that individual populations of dopaminoceptive (DRD1/2) neurons in the human NAc may be differentially associated with SCZ and BIP (Figure 4). We also found that specific classes of GABAergic inhibitory neurons in the human AMY were preferentially associated with SCZ that were not significantly associated with BIP. These observations highlight a potential role for these subcortical brain regions in mediating genetic risk for SCZ and BIP.
As both the NAc and AMY play critical roles in reward signaling, we also evaluated the enrichment of genetic risk for addiction or substance use behaviors (Liu et al., 2019). Intriguingly, the genetic risk for adopting regular smoking associated more broadly across most neuronal populations, whereas other phenotypes assessed in this addiction GWAS showed more preferential associations to certain classes. This suggests that the risk for adopting addictive-like behaviors may affect these brain regions more broadly than specific features of addiction (Figure 4A and 4C). With regard to the other features, the MSN.D1_E class significantly associated with genetic risk for heaviness of smoking (“CigDay”) and drinking (“DrnkWk”). As a top marker for this class was CRHR2, this may be a key population in understanding these features of addiction. Many rodent studies have implicated CRH receptors in alcohol consumption and alcohol dependence (Heilig and Koob, 2007; Yong et al., 2014). In the AMY, two neuronal classes drew our attention, due to their association with multiple phenotypes, including “Inhib_C” and “Inhib_E.” Due to their marker expression of NPFFR2/TLL1 and serotonin receptors HTR4/HTR2C, respectively (data not shown; Table S5), these GABAergic classes may be of interest in understanding amygdalar circuits underlying genetic risk for substance use. From these analyses, we surveyed our diversity of neuronal classes profiled in the NAc and AMY for their clinical relevance in psychiatric disease and addiction behaviors. In addition, we have extended such analyses for these regions, which have formerly only been performed on cell-type profiles defined in murine models (Bryois et al., 2020; Skene et al., 2018) to their relevant human context, and with increased resolution of molecularly defined classes. Finally, we narrowed down those classes manifesting the greatest genetic risk, potentially highlighting some neuronal subclasses mediating certain substance use behaviors.
While we identified and characterized a diversity of robust neuronal classes with our analytical pipelines for this study, we recognize that our sample sizes may not fully capture all cell types or subpopulations, such as the striatal cholinergic interneurons mentioned above. The most direct evidence for this is that there remains some bias in the donor makeup of certain classes (Figure S16; Table S3), keeping NeuN enrichment for a subset of samples in mind. However, despite steps to mitigate the impact of the small input for our sample processing protocol (see STAR Methods), we expect some degree of sampling bias since cell-type makeup is not expected to be homogeneous within a single region. For example, the NAc core or shell has different functional properties and differs in regard to their afferent and efferent connections, and, thus, differences in cell composition across these two subregions is expected (Heimer et al., 1991; Li et al., 2018b; Zahm and Heimer, 1993). Integration of spatial transcriptomic technologies with snRNA-seq data in these regions (Maynard et al., 2021) will help resolve expected heterogeneity across these adjacent subregions. Furthermore, while many groups have recently begun to identify sex-specific differences in specific roles or hormonal responses of neuronal subpopulations (Cao et al., 2018; Chen et al., 2019), we believe our study remains underpowered to potentially recapitulate these observations. It is noted that the capture of certain non-neuronal cell classes was observed in only select brain regions, namely endothelial cells, as these were only identified in the AMY. However, we identified mural cells (comprising pericytes and vascular smooth muscle cells) in the AMY, DLPFC, and HPC. With these sample sizes still limited and an emphasis on NeuN enrichment in a subset of our data, we believe that these smaller, stromal populations were inherently not captured by our protocols in some regions. Nevertheless, we highlight that most of the cell classes we report are made up of their own set of subclusters (Table S4; see method details and data and code availability).
Another caveat to these snRNA-seq data is the lack of gene expression information from the cytosolic compartment, such as the neuropil. This is an important caveat, given that synaptic signaling is implicated in neuropsychiatric disorders, and gene products localized to the synapse are enriched for SCZ genetic risk (Skene et al., 2018). In addition, mRNA from some expected marker genes (e.g., PVALB) may be preferentially localized to the cytosol, as demonstrated with smFISH for the GAD1+ interneuron “Inhib_C/_D” classes in the NAc (Figure S5). However, this seems to be cell population and/or area specific, in regard to the transcriptional and nuclear-export dynamics of the respective cell population, as PVALB was highly expressed in some DLPFC subpopulations (data not shown; see data and code availability). These observations and those by others thus emphasize that snRNA-seq will not capture the full transcriptomic profile of cell populations, including activation-induced or disease-associated molecular changes restricted to the cytosol (Thrupp et al., 2020). However, as we previously demonstrated (Maynard et al., 2021), snRNA-seq-defined cell populations can be registered to spatial transcriptomic data, which does retain such information, for further characterization of transcriptomic profiles.
In summary, we used snRNA-seq to profile five human brain regions with roles in the reward circuitry. We defined transcriptomic profiles for 107 regionally defined cell-type classes and characterized the architecture of molecular relationships across these brain regions. We finally identified associations with genetic risk for neuropsychiatric disorders and substance use phenotypes in unique neuronal subpopulations in the NAc and AMY. This study provides a significant step toward constructing a single-nucleus transcriptomic atlas of the human brain and illustrates the utility of this type of data in understanding the diversity of cell populations, as well as their roles in biology and disease.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources not provided below (see data and code availability) should be directed to the lead contact, Keri Martinowich (keri.martinowich@libd.org).
Materials availability
This study did not generate new reagents or materials.
Data and code availability
Raw single-nucleus RNA-seq read data is publicly available from the Globus endpoint ‘jhpce#tran2021’, linked from https://research.libd.org/globus. Data files containing de-identified and processed SingleCellExperiment objects are hosted on Amazon S3, and the links are available on the README.md of the GitHub repository for this project (https://github.com/LieberInstitute/10xPilot_snRNAseq-human). RNAscope data generated in this study will be shared by the lead contact upon request.
All code for processing and analyzing the data has been archived at the time of submission at Zenodo: https://doi.org/10.5281/zenodo.5149046.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Post-mortem human tissue
Post-mortem human brain tissue from eight neurotypical donors of European ancestry from age 40 to 69 (Table S1) was obtained by autopsy from the Office of the Chief Medical Examiner for the State of Maryland under State of Maryland Department of Health and Mental Hygiene Protocol 12–24. Clinical characterization, diagnoses, and macro- and micro-scopic neuropathological examinations were performed on all samples using a standardized paradigm, and subjects with evidence of macro- or micro-scopic neuropathology were excluded. Details of tissue acquisition, handling, processing, dissection, clinical characterization, diagnoses, neuropathological examinations, RNA extraction and quality control measures have been described previously (Lipska et al., 2006). Dorsolateral prefrontal cortex (DLPFC, n = 3) and hippocampus (HPC, n = 3) tissue was microdissected using a hand-held dental drill as previously described (Collado-Torres et al., 2019). The subgenual anterior cingulate cortex (sACC, n = 5) was dissected under visual guidance from the medial aspect of the forebrain at the level of the rostrum of the corpus callosum. Dissections were performed ventral to the corpus callosum, and dorsal to the orbital frontal cortex (BA11). Medially it was bounded by the interhemispheric fissure, while laterally it was bounded by the corona radiata/centrum semiovale. For the amygdala (AMY, n = 5), a block containing the structure was dissected under visual guidance at the level of its maximal size, taken from a 1 cm thick slab of one hemisphere, and sectioned in the coronal plane. The amygdala block was chosen by visual inspection at a level that contained the maximal number of subnuclei. Landmarks for selection of the amygdala block included presence of the internal and external segments of the globus pallidus, the anterior commissure, and optic tract. The block containing the nucleus accumbens was taken from a 1 cm thick slab of one hemisphere, and sectioned in the coronal plane. The nucleus accumbens (NAc, n = 8) block was chosen at a level where the putamen and caudate are joined by the accumbens at the ventral aspect of the striatum, with clear striations separating the putamen from the caudate. Additional landmarks include the presence of the anterior aspect of the temporal lobe and the claustrum.
Given the low number of female donors (n = 2) and inclusion in only three (NAc, AMY, and sACC) of the regions in the present study, in addition to the variable enrichment (DAPI/PI, alone, or with NeuN enrichment) scheme across samples (see Table S2), our study is underpowered to assess effects or influences due to sex, such as cell class proportion or regulation/effect on gene expression.
METHOD DETAILS
snRNaseq data generation
We performed single-nucleus RNA-seq (snRNA-seq) on 24 samples from 3–8 individual donors, per region (n = 3 DLPFC, n = 3 HPC, n = 5 AMY, n = 5 sACC, n = 8 NAc), using 10x Genomics Chromium Single Cell Gene Expression V3 technology (Zheng et al., 2017). Nuclei were isolated using a “Frankenstein” nuclei isolation protocol developed by Martelotto et al. for frozen tissues (Habib et al., 2016, 2017; Hu et al., 2017; Lacar et al., 2016; Lake et al., 2016). Briefly, ~40mg of frozen, ground tissue was homogenized in chilled Nuclei EZ Lysis Buffer (MilliporeSigma #NUC101) using a glass dounce with ~15 strokes per pestle. Homogenate was filtered using a 70 μm-strainer mesh and centrifuged at 500 × g for 5 minutes at 4°C in a benchtop centrifuge. Nuclei were resuspended in the EZ lysis buffer, centrifuged again, and equilibrated to nuclei wash/resuspension buffer (1x PBS, 1% BSA, 0.2U/μL RNase Inhibitor). Nuclei were washed and centrifuged in this nuclei wash/resuspension buffer three times, before labeling with DAPI (10 μg/mL) or propidium iodide (PI) (depending on processing batch). For 3 NAc, 2 sACC, and 2 AMY samples from individual donors, nuclei were additionally labeled with Alexa Fluor 488-conjugated anti-NeuN (MilliporeSigma cat. #MAB377X), at 1:1000 in the same wash/resuspension buffer, for 30 minutes on ice, to facilitate enrichment of neurons during fluorescent activated cell sorting (FACS). Samples were then filtered through a 35 μm-cell strainer and sorted on a BD FACS Aria II Flow Cytometer (Becton Dickinson) at the Johns Hopkins University Sidney Kimmel Comprehensive Cancer Center (SKCCC) Flow Cytometry Core, or Bio-Rad S3e Cell Sorter (depending on processing batch) into 10X Genomics reverse transcription reagents. Gating criteria hierarchically selected for whole, singlet nuclei (by forward/side scatter), G0/G1 nuclei (by DAPI or PI fluorescence), and NeuN-positive cells for the respective NeuN-enriched samples. A “null” sort of nuclei into the wash buffer was additionally performed from the same preparation, for quantification of nuclei concentration and to ensure that sorted nuclei were intact and free of debris. For each sample, approximately 8,500 single nuclei were sorted directly into 25.1 μL of reverse transcription reagents from the 10x Genomics Single Cell 3′ Reagents kit (without enzyme). The 10x Chromium process was performed and libraries prepared, according to manufacturer’s instructions (10x Genomics), and finally sequenced on the Next-seq (Illumina) at the Johns Hopkins University Transcriptomics and Deep Sequencing Core.
RNAscope single molecule fluorescent in situ hybridization (smFISH)
Fresh frozen NAc from two independent donors was sectioned at 10 μm and stored at −80°C. In situ hybridization assays were performed with RNAscope technology utilizing the RNAscope Fluorescent Multiplex Kit V2 and 4-plex Ancillary Kit (Cat # 323100, 323120 ACD, Hayward, California) according to the manufacturer’s instructions. Briefly, tissue sections were fixed with a 10% neutral buffered formalin solution (Cat # HT501128 Sigma-Aldrich, St. Louis, Missouri) for 30 minutes at room temperature (RT), series dehydrated in ethanol, pretreated with hydrogen peroxide for 10 minutes at RT, and treated with protease IV for 30 minutes. Sections were incubated with 5 different probe combinations to assess MSN and inhibitory neuron subtypes: 1) “Square”: DRD1, TAC1, RXFP2, GABRQ (Cat 524991-C4, 310711-C3, 452201, 483171-C2, ACD, Hayward, California); 2) “Circle”: DRD1, TAC1, CRHR2, RXFP1 (Cat 524991-C4, 310711-C3, 469621, 422821-C2); 3) “Triangle”: DRD1, DRD2, TAC1, PENK (Cat 524991-C4, 553991, 310711-C2, 548301-C3); 4) “Star”: DRD1, DRD2, CRHR2, HTR7 (Cat 524991-C4, 553991-C3, 469621, 413041-C2). 5) “Swirl”: PVALB, GAD1, PTHLH, KIT (Cat 422181-C4, 404031-C3, PTHLH, 606401-C2). Following probe labeling, sections were stored over-night in 4x SSC (saline-sodium citrate) buffer. After amplification steps (AMP1–3), probes were fluorescently labeled with Opal Dyes (Perkin Elmer, Waltham, MA; 1:500) and stained with DAPI (4′,6-diamidino-2-phenylindole) to label the nucleus. Lambda stacks were acquired in z-series using a Zeiss LSM780 confocal microscope equipped with a 63x × 1.4NA objective, a GaAsP spectral detector, and 405, 488, 555, and 647 lasers as previously described (Maynard et al., 2020). All lambda stacks were acquired with the same imaging settings and laser power intensities. For each subject, high magnification 63x images were randomly acquired in the NAc (n = 2 subjects, n = 2 sections per subject, n = 12 images per section).
QUANTIFICATION AND STATISTICAL ANALYSIS
snRNaseq raw data processing
We processed the sequencing data with the 10x Genomics’ Cell Ranger v3.0 pipeline, aligning to the human reference genome GRCh38, with a reconfigured GTF such that intronic alignments were additionally counted given the nuclear context, to generate UMI/feature-barcode matrices (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/advanced/references#premrna). Per the output metrics of Cell Ranger, each sample was sequenced to a median depth of 284.3M reads (IQR: 253.7M-419.0M). We started with raw feature-barcode matrices from this output for analysis with the Bioconductor suite of R packages for single-cell RNA-seq analysis (Amezquita et al., 2020) using Bioconductor (Huber et al., 2015) version 3.12. For quality control (QC) and nuclei calling, we first used a Monte Carlo simulation-based approach to assess and exclude empty droplets or those with random ambient transcriptional noise, such as from debris (Griffiths et al., 2018; Lun et al., 2019). This was then followed by mitochondrial rate adaptive thresholding, which, though expected to be near-zero in this nuclear context, we applied a 3x median absolute deviation (MAD) threshold, to allow for flexibility in output/purity of nuclear enrichment by FACS using scater’s isOutlier (Lun et al., 2016). Finally, within each sample, we computed doublet scores implemented with R package scDblFinder’s computeDoubletDensity function (Dahlin et al., 2018), to assess putative doublet-driven clustering (see below). This QC pipeline yielded 11,202 high-quality nuclei from the DLPFC, 10,268 nuclei from HPC, 15,669 nuclei from AMY, 15,669 nuclei from sACC, and 20,571 nuclei from NAc. Collectively, these exhibited a median unique molecular identifier (UMI) count of 9,450(interquartile range, IQR: 5,513–23,078 UMIs) per nucleus, and a median detected gene count of 3,225 (IQR: 2,292–5,739) genes captured per nucleus. These feature-barcode gene counts were then rescaled across all nuclear libraries, using batchelor’s multiBatchNorm (Haghverdi et al., 2018). Finally, these rescaled counts were log2-transformed for identification of highly-variable genes (HVGs) with scran’s modelGeneVar (Lun et al., 2016), taking all genes with a greater variance than the fitted trend.
Dimensionality reduction and clustering
Principal components analysis (PCA) was then performed on the HVGs to reduce the high dimensionality of nuclear transcriptomic data for each region, implementing batchelor’s fastMNN PC coordinate correction to remove batch effects at the donor (highest variance-contributing)-level (Amezquita et al., 2020; Haghverdi et al., 2018). The optimal principal component (PC) space was defined with iterative graph-based clustering to determine the d PCs where resulting n clusters stabilize, with the constraint that n clusters < / = (d + 1) PCs (Lun et al., 2016), resulting in a chosen d between 59–99 PCs. In this PC-reduced space, graph-based clustering was performed to identify what we classified as preliminary clusters; specifically, k-nearest neighbors with k = 20 neighbors and the Walk-trap method from R package igraph (Csardi and Nepusz, 2006) for community detection. We then took all feature counts for these assignments and pseudo-bulked counts (Crowell et al., 2019; Kang et al., 2018; Lun and Marioni, 2017) across these preliminary nuclear clusters, rescaling for combined library size and log-transformed normalized counts, using scater’s librarySizeFactors (Lun et al., 2016). With the pseudo-bulked count profiles, we then performed hierarchical clustering to identify preliminary cluster relationships, and finally merged with the cutreeDynamic function of R package dynamicTreeCut (Langfelder et al., 2016), or keeping split clusters at the preliminary resolution, if generally well-represented across donors, as this suggested biologically valid subpopulations (for example, neuronal subtypes) as opposed to more likely batch-driven preliminary clusters. However, in some cases, cluster marker identification (see below) suggested sample bias in true, biological subpopulations (see discussion). The final clusters merged at the appropriate tree height were then annotated for broad cell type identity with well-established cell type markers (Mathys et al., 2019), and with a letter suffix where multiple broad cell class populations were defined (‘Excit_A’, ‘Excit_B’, etc.). We also used Bioconductor package scater’s (McCarthy et al., 2017) implementation of non-linear dimensionality reduction techniques, t-SNE (van der Maaten and Hinton, 2008) and UMAP (McInnes et al., 2018), with default parameters and within the aforementioned optimal PC space, simply for visualization of the high-dimensional structure in the data, which generally complemented the clustering results. Additionally, in the HPC, AMY, sACC, and NAc analyses, we flagged clusters that were driven by low transcript capture or doublets (suggested by ‘dual’ cell-type marker expression, but confirmed with high doublet scores), and these were removed prior to down-stream analyses and from the t-SNE display, resulting in a final n nuclei analyzed per region of: 11,202 from the DLPFC, 10,139 nuclei from HPC, 14,039 nuclei from AMY, 15,343 nuclei from sACC, and 19,892 nuclei from NAc (an average of 96.8% nuclei kept post-QC, above). These final numbers of nuclei analyzed per regionally-defined cell class (and subcluster) and by donor can be found in Tables S3 and S4.
Cluster marker identification
For marker identification with our final clusters defined in each brain region, we utilized scran’s findMarkers (Lun et al., 2016) function for two sets of statistics:
Pairwise t tests, to identify differences between each cluster, or
Implementing the function findMarkers to perform a cluster-versus-all-other-nuclei t test iteration
In both cases, we re-computed non-scale-matched log2-transformed counts (from logNormCounts), including a donor covariate to properly model linearly (in the design = parameter) on this unwanted batch effect (as multiBatchNorm, which is preferred as the input to fastMNN, above, removes much of the sequencing depth differences being modeled). The latter approach, 2), we consider a lessstringent marker test for enriched genes in a given cluster, but which would not necessarily differentiate between said cluster and all others. In addition to these statistics, for each cell class, we computed a Boolean parameter for non-0 median expression of each gene, to differentiate between noise-driven statistics. We used the results from both tests to interpret cell type identity beyond the broad classes (excitatory versus inhibitory neuron), and to identify markers to probe via smFISH (below). The top 40 markers from each test result (including the respective non-0-median expression filter applied) are provided for each regionally-defined cell class in Table S5 (regions separated by worksheet), where the ‘_pw’ suffix corresponds to the pairwise tests (set 1), and ‘_1vAll’ to the enriched expression test (set 2).
Importantly, 2) can be used to return a statistic, Cohen’s D, or the standardized log-fold change, which we used to back-compute a single t-statistic for each cluster per gene, using:
t = std.logFC * sqrt(n), where n = the total n nuclei (per region/dataset)
* Back-computing a single t-statistic cannot be generated with the result of 1) due to pairwise testing.
Comparing cell class conservation between datasets or across species
The t-statistics, described above, can then be used to compare such ‘transcriptomic profiles’ to those we computed for publicly-available postmortem human datasets, using the provided cell type annotations (or across our 5 regions), and compute the Pearson correlation coefficient (r), as was done in the spatial registration approaches in spatialLIBD (Maynard et al., 2021), across all shared expressed genes.
To perform cross-species conservation analyses, we generated these t-statistics (from marker test 2., above) per gene, per reported cell annotation, subsetting on shared homologous genes between our human data and rat or mouse, using the ‘DB Class Key’ identifier provided by http://www.informatics.jax.org/downloads/reports/HOM_AllOrganism.rpt, before computing the pairwise correlations. In the case of “many-to-many” orthalog scenarios, we took the highest-expressing paralog as the surrogate for each homologous pair, though these were small sets of genes in both rat and mouse cases. Correlation Pearson’s r for both the human-versus-rat NAc and human AMY versus mouse MeA sections were performed in the gene space defined by the combined top-100 markers per cell class/subcluster (whether markers for the human cell classes or the reported subpopulations in each respective rodent model), where species homology information was known. In the NAc comparison, this was across 582 homologous marker genes, and for the AMY, 480 homologous genes.
GWAS association analyses with MAGMA
The latest version (v1.08) of Multi-marker Analysis of GenoMic Annotation (MAGMA; de Leeuw et al., 2015) was used to test for genetic risk association of our 107 regionally-defined cell classes with schizophrenia (SCZ: Pardiñas et al., 2018; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), autism spectrum disorder (ASD: Grove et al., 2019), bipolar disorder (BIP: Stahl et al., 2019), major depressive disorder (MDD: Wray et al., 2018), posttraumatic stress disorder (PTSD: Nievergelt et al., 2019); Alzheimer’s disease (AD: Jansen et al., 2019); attention deficit/hyperactivity disorder (ADHD: Demontis et al., 2019); and for alcohol and tobacco use phenotypes (Liu et al., 2019). For the marker gene sets, we used any genes defined as enriched per subpopulation (using marker test set 2, from above), at the Benjamini & Hochberg false discovery rate (FDR) < 1e-6 (Benjamini and Hochberg, 1995) and a restriction that the median expression of putative marker gene per cell class be > 0. SNPs were first annotated to genes, using window sizes from −10kb to +35kb of each gene, with the 1000 Genomes EUR reference panel, and gene-level analyses were performed, using provided summary statistics from each of the above listed GWAS (via https://www.med.unc.edu/pgc/download-results/ or https://genome.psych.umn.edu/index.php/GSCAN for results from Liu et al. (2019) and the snp-wise = mean model, to test whether there was enrichment of genetic risk for disease/phenotype in each gene. Following this, we performed the default competitive gene set analysis with the 107 regionally-defined marker sets, testing for association of gene-level risk and whether genes were enriched/specific to each subpopulation. From the empirical p-value of the gene set analysis, we performed multiple test correction with both false-discovery rate (FDR) and the stricter Bonferroni procedure (threshold p < 3.89e-5) across all 1284 (107 regionally-defined subpopulations and 12 GWAS phenotypes tested) tests. All genetic association test results are provided in Table S6.
RNAscope data analysis
Following image acquisition, lambda stacks in z-series were linearly unmixed in Zen software (weighted; no autoscale) using reference emission spectral profiles previously created in Zen (Maynard et al., 2020) and saved as Carl Zeiss Image “.czi” files. Images were segmented and quantitatively analyzed in MATLAB using dotdotdot software (Maynard et al., 2020) and statistical analyses were performed in R v4.0.4: For each of the five experiments (see below for elaboration), we combined DAPI-defined region of interest (ROI)-level data from all respective images, and used data-driven cutoffs based on distributional overlap to determine binary expression levels (i.e., expressed or unexpressed) for each gene/channel, for cell class [group] prediction. In each experiment, each GAD1+ (or DRD1+ or DRD1+/DRD2+, pending on the experiment) ROI was classified into a Euclidean distance-predicted neuronal cell class (or group of classes, as accordingly, due to 4-plex limitations), based on the lowest distance. Probe counts were then quantified as the number of dots per 10,000 ROI pixels, post-lipofuscin masking, then log2-transformed (annotated as ‘rnascope_[GENE]’ in Figures 1D, S1, S2, S3, and S5).
Experiment-specific information
- Circle
1033 ROIs were quantified across 48 images taken from 4 tissue sections across from 2 donors (two sections/donor). 251 ROIs were classified as DRD1+ with > 3 dots post lipofuscin masking, and among these ROIs, RXFP1 and CRHR2 binarized expression, for prediction only, was classified as > 3 dots and TAC1 expression was classified as > 6 dots. Corresponds to experiment shown in Figure 1D.
- Square
1126 ROIs were quantified across 48 images taken from 4 tissue sections across from 2 donors (two sections/donor). 341 ROIs were classified as DRD1+ with > 3 dots post lipofuscin masking, and among these ROIs, RXFP2, GABRQ, and TAC1 binarized expression, for prediction 2nly, were each classified as > 6 dots. Corresponds to experiment shown in Figure S2.
- Triangle
1039 ROIs were quantified across 47 images taken from 4 tissue sections across from 2 donors (two sections/donor). 271 ROIs were classified as either DRD1+ or DRD2+ with > 3 dots post lipofuscin masking in either gene, and among these ROIs, TAC1 and PENK binarized expression, for prediction only, were classified as > 6 dots. Corresponds to experiment shown in Figure S1.
- Star
1003 ROIs were quantified across 44 images taken from 4 tissue sections across from 2 donors (two sections/donor). 482 ROIs were classified as either DRD1+ or DRD2+ with > 3 dots (post lipofuscin masking) in either gene, and among these ROIs, HTR7 and CRHR21 binarized expression, for prediction only, were classified as > 6 dots. Corresponds to experiment shown in Figure S3.
- Swirl
989 ROIs were quantified across 44 images taken from 4 tissue sections across from 2 donors (two sections/donor). 212 ROIs were classified as GAD1+ inhibitory neurons with > 6 dots post lipofuscin masking, and among these ROIs, PVALB, KIT and PTHLH binarized expression, for prediction only, were classified as > 6 dots. Corresponds to experiment shown in Figure S5.
ADDITIONAL RESOURCES
For each of the five brain regions in this study, we created an interactive website with the data using iSEE (Rue-Albrecht et al., 2018) and deployed at the LIBD shinyapps.io account at URLs such as https://libd.shinyapps.io/tran2021_NAc/ (and accordingly, /tran2021_sACC, /tran2021_DLPFC, /tran2021_AMY, and /tran2021_HPC).
Supplementary Material
KEY RESOURCES TABLE.
(Tran, Maynard, et al. 2021 submission)
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-NeuN (clone A60), Alexa Fluor 488-conjugated | MilliporeSigma | #MAB377X |
| Biological samples | ||
| Human postmortem brain tissue (NAc, AMY, sACC, DLPFC, HPC) | Lieber Institute for Brain Development; see Tables S1 and S2 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant RNasin ribonuclease inhibitor | Promega | N2515 |
| 4’,6-diamidino-2-phenylindole, dihydrochloride (DAPI) | Invitrogen | D1306 |
| Propidium iodide solution (PI), 1mg/mL | Invitrogen | P3566 |
| Bovine serum albumin (BSA) | VWR | 102643-516 |
| 10% neutral buffered formalin | Sigma-Aldrich | HT501128 |
| Critical commercial assays | ||
| Nuclei EZ Prep (lysis buffer) | Sigma-Aldrich | NUC101-1KT |
| Chromium Single Cell 3’ GEM, Library & Gel Bead Kit, v3 | 10x Genomics | PN-1000075 |
| Chromium Single Cell B Chip Kit | 10x Genomics | PN-1000073 |
| RNAscope Multiplex Fluorescent Kit, v2 | ACD | 323100 |
| RNAscope 4-Plex Ancillary Kit | ACD | 323120 |
| Deposited data | ||
| Raw sequencing read data (FASTQ) for all n=24 samples reported in this paper | This paper | https://research.libd.org/globus/jhpce_tran2021/index.html |
| Processed data as ‘SingleCellExperiment’ R objects, for each region (NAc, AMY, sACC, DLPFC, HPC) | This paper | Linked from: https://github.com/LieberInstitute/10xPilot_snRNAseq-human#readme |
| Summary statistics from various Psychiatric Genomics Consortium GWAS | (Various author lists) | https://www.med.unc.edu/pgc/download-results/ |
| Summary statistics for substance use phenotypes GWAS | Liu, et al. 2019 | https://genome.psych.umn.edu/index.php/GSCAN |
| Software and algorithms | ||
| Cell Ranger, v3.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/3.0/installation |
| R, v4.0.4 | R Core Team | https://www.r-project.org/ |
| Orchestrating single-cell analysis with Bioconductor (v3.12 methods/R packages) | Amezquita, et al. 2019 | https://doi.org/10.1038/s41592-019-0654-x |
| MAGMA, v1.08 | De Leeuw, et al. 2015 | https://ctg.cncr.nl/software/magma |
| Other | ||
| Code for analyses in the present study | This paper | https://doi.org/10.5281/zenodo.5149046 |
Highlights.
snRNA sequencing (70,615 nuclei) of 5 human brain regions with roles in reward
Characterization of the transcriptomic architecture across 107 cell classes
Genetic risk association for substance use phenotypes in specific cell populations
Interactive app for each brain region to explore genes of interest in cell classes
ACKNOWLEDGMENTS
The authors would like to express their gratitude to our colleagues, whose efforts have supported the collection of postmortem brain tissue to advance these studies, including at the Office of the Chief Medical Examiner of the State of Maryland, Baltimore. We also would like to acknowledge the contributions of Llewellyn B. Bigelow, MD, and Amy Deep-Soboslay for their diagnostic expertise and Daniel R. Weinberger for providing constructive commentary and editing of the manuscript. Finally, we are indebted to the generosity of the families of the decedents, who donated the brain tissue used in these studies. We thank the Johns Hopkins University Sidney Kimmel Comprehensive Cancer Center (SKCCC) Flow Cytometry Core, especially Jessica Gucwa, and the Johns Hopkins University Transcriptomics and Deep Sequencing Core, especially Linda Orzolek and Haiping Hao, for supporting snRNA-seq data generation. We also thank members of the laboratories of Dr. Weizhe Hong (University of California, Los Angeles) and Dr. Jeremy Day (University of Alabama) for facilitating data sharing and helpful conversations about cross-species comparisons. We also thank Nick Clifton for assisting our group with the initial implementation of MAGMA analyses. Funding for this project was provided by the Lieber Institute for Brain Development. B.K.B., D.B.H., K.M., and A.E.J. were partially supported by R01DA042090; K.R.M., A.S., L.C.-T., K.M., and A.E.J. were partially supported by 1R01MH123183; and S.C.H. was partially supported by R00HG009007.
Footnotes
DECLARATION OF INTERESTS
A.E.J. is employed by a for-profit biotechnology startup company (company name pending), which is unrelated to the content of this manuscript. The remaining authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.neuron.2021.09.001.
REFERENCES
- Amezquita RA, Lun ATL, Becht E, Carey VJ, Carpp LN, Geistlinger L, Marini F, Rue-Albrecht K, Risso D, Soneson C, et al. (2020). Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaev O, Piletti Chatain C, and Krueger-Burg D (2018). Inhibition in the amygdala anxiety circuitry. Exp. Mol. Med. 50, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger N, Stefanacci L, Schumann CM, Sherwood CC, Annese J, Allman JM, Buckwalter JA, Hof PR, and Semendeferi K (2012). Neuronal populations in the basolateral nuclei of the amygdala are differentially increased in humans compared with apes: a stereological study. J. Comp. Neurol. 520, 3035–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiuk MY, Martirosyan A, Wahis J, de Vin F, Marneffe C, Kusserow C, Koeppen J, Viana JF, Oliveira JF, Voet T, et al. (2020). Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 11, 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- Bonfiglio JJ, Inda C, Refojo D, Holsboer F, Arzt E, and Silberstein S (2011). The corticotropin-releasing hormone network and the hypothalamic-pituitary-adrenal axis: molecular and cellular mechanisms involved. Neuroendocrinology 94, 12–20. [DOI] [PubMed] [Google Scholar]
- Bryois J, Skene NG, Hansen TF, Kogelman LJA, Watson HJ, Liu Z, Brueggeman L, Breen G, Bulik CM, Arenas E, et al. ; Eating Disorders Working Group of the Psychiatric Genomics Consortium; International Headache Genetics Consortium; 23andMe Research Team (2020). Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat. Genet. 52, 482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Willett JA, Dorris DM, and Meitzen J (2018). Sex Differences in Medium Spiny Neuron Excitability and Glutamatergic Synaptic Input: Heterogeneity Across Striatal Regions and Evidence for Estradiol-Dependent Sexual Differentiation. Front. Endocrinol. (Lausanne) 9, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamling X, Kallman A, Fang W, Berlinicke CA, Mertz JL, Devkota P, Pantoja IEM, Smith MD, Ji Z, Chang C, et al. (2021). Single-cell transcriptomic reveals molecular diversity and developmental heterogeneity of human stem cell-derived oligodendrocyte lineage cells. Nat. Commun. 12, 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chareyron LJ, Banta Lavenex P, Amaral DG, and Lavenex P (2011). Stereological analysis of the rat and monkey amygdala. J. Comp. Neurol. 519, 3218–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PB, Hu RK, Wu YE, Pan L, Huang S, Micevych PE, and Hong W (2019). Sexually dimorphic control of parenting behavior by the medial amygdala. Cell 176, 1206–1221.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes SJ (2004). Corticotropin-releasing hormone (CRH) in psychiatry: from stress to psychopathology. Ann. Med. 36, 50–61. [DOI] [PubMed] [Google Scholar]
- Collado-Torres L, Burke EE, Peterson A, Shin J, Straub RE, Rajpurohit A, Semick SA, Ulrich WS, Price AJ, Valencia C, et al. ; BrainSeq Consortium (2019). Regional Heterogeneity in Gene Expression, Regulation, and Coherence in the Frontal Cortex and Hippocampus across Development and Schizophrenia. Neuron 103, 203–216.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell HL, Soneson C, Germain P-L, Calini D, Collin L, Raposo C, Malhotra D, and Robinson MD (2019). On the discovery of population-specific state transitions from multi-sample multi-condition single-cell RNA sequencing data. bioRxiv. 10.1101/713412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, and Nepusz T (2006). The igraph software package for complex network research. http://www.interjournal.org/manuscript_abstract.php?361100992.
- Dahlin JS, Hamey FK, Pijuan-Sala B, Shepherd M, Lau WWY, Nestorowa S, Weinreb C, Wolock S, Hannah R, Diamanti E, et al. (2018). A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood 131, e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, and Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, and Posthuma D (2015). MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, Baldursson G, Belliveau R, Bybjerg-Grauholm J, Bækvad-Hansen M, et al. ; ADHD Working Group of the Psychiatric Genomics Consortium (PGC); Early Lifecourse & Genetic Epidemiology (EAGLE) Consortium; 23andMe Research Team (2019). Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enterría-Morales D, Del Rey NL-G, Blesa J, López-López I, Gallet S, Prévot V, López-Barneo J, and d’Anglemont de Tassigny X (2020). Molecular targets for endogenous glial cell line-derived neurotrophic factor modulation in striatal parvalbumin interneurons. Brain Commun. 2, fcaa105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster RJ, Lebois LAM, Ressler KJ, and Suh J (2018). Brain circuit dysfunction in post-traumatic stress disorder: from mouse to man. Nat. Rev. Neurosci. 19, 535–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiro-Silva J, Gruart A, Clayton KB, Podlesniy P, Abad MA, Gasull X, Delgado-García JM, and Trullas R (2015). Neuronal pentraxin 1 negatively regulates excitatory synapse density and synaptic plasticity. J. Neurosci. 35, 5504–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K, et al. ; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium (2015). Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franjic D, Choi J, Skarica M, Xu C, Li Q, Ma S, Tebbenkamp ATN, Santpere G, Arellano JI, Gudelj I, et al. (2020). Molecular diversity among adult hippocampal and entorhinal cells. bioRxiv. 10.1101/2019.12.31.889139. [DOI] [Google Scholar]
- Garrett A, and Chang K (2008). The role of the amygdala in bipolar disorder development. Dev. Psychopathol. 20, 1285–1296. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ Jr., and Sibley DR (1990). D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science 250, 1429–1432. [DOI] [PubMed] [Google Scholar]
- Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Südhof TC, and Quake SR (2016). Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep. 16, 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveland GA, Williams RS, and DiFiglia M (1985). A Golgi study of the human neostriatum: neurons and afferent fibers. J. Comp. Neurol. 234, 317–333. [DOI] [PubMed] [Google Scholar]
- Griffiths JA, Richard AC, Bach K, Lun ATL, and Marioni JC (2018). Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun. 9, 2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, Pallesen J, Agerbo E, Andreassen OA, Anney R, et al. ; Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium; BUPGEN; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; 23andMe Research Team (2019). Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubman A, Chew G, Ouyang JF, Sun G, Choo XY, McLean C, Simmons RK, Buckberry S, Vargas-Landin DB, Poppe D, et al. (2019). A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22, 2087–2097. [DOI] [PubMed] [Google Scholar]
- Haber SN, and Knutson B (2010). The reward circuit: linking primate anatomy and human imaging. Neuropsychopharmacology 35, 4–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, and Regev A (2016). Div-Seq: single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. (2017). Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Zhou Z, Fei L, Sun H, Wang R, Chen Y, Chen H, Wang J, Tang H, Ge W, et al. (2020). Construction of a human cell landscape at single-cell level. Nature 581, 303–309. [DOI] [PubMed] [Google Scholar]
- Heilig M, and Koob GF (2007). A key role for corticotropin-releasing factor in alcohol dependence. Trends Neurosci. 30, 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimer L, Zahm DS, Churchill L, Kalivas PW, and Wohltmann C (1991). Specificity in the projection patterns of accumbal core and shell in the rat. Neuroscience 41, 89–125. [DOI] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii-Hayashi N, Okuda H, Tatsumi K, Ishizaka S, Yoshikawa M, and Wanaka A (2008). Localization of chondroitin sulfate proteoglycan versican in adult brain with special reference to large projection neurons. Cell Tissue Res. 334, 163–177. [DOI] [PubMed] [Google Scholar]
- Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, Coleman JRI, Hagenaars SP, Ward J, Wigmore EM, et al. ; 23andMe Research Team; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (2019). Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Fabyanic E, Kwon DY, Tang S, Zhou Z, and Wu H (2017). Dissecting Cell-Type Composition and Activity-Dependent Transcriptional State in Mammalian Brains by Massively Parallel Single-Nucleus RNA-Seq. Mol. Cell 68, 1006–1015.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Hoeppner DJ, Saito T, Blanpain L, Ukaigwe J, Burke EE, Collado-Torres L, Tao R, Tajinda K, Maynard KR, et al. (2020). Profiling gene expression in the human dentate gyrus granule cell layer reveals insights into schizophrenia and its genetic risk. Nat. Neurosci. 23, 510–519. [DOI] [PubMed] [Google Scholar]
- Janak PH, and Tye KM (2015). From circuits to behaviour in the amygdala. Nature 517, 284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, Sealock J, Karlsson IK, Hägg S, Athanasiu L, et al. (2019). Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, et al. (2018). Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36, 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y (1997). Neostriatal cell subtypes and their functional roles. Neurosci. Res. 27, 1–8. [DOI] [PubMed] [Google Scholar]
- Kronman H, Richter F, Labonté B, Chandra R, Zhao S, Hoffman G, Lobo MK, Schadt EE, and Nestler EJ (2019). Biology and Bias in Cell Type-Specific RNAseq of Nucleus Accumbens Medium Spiny Neurons. Sci. Rep. 9, 8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B, Linker SB, Jaeger BN, Krishnaswami SR, Barron JJ, Kelder MJE, Parylak SL, Paquola ACM, Venepally P, Novotny M, et al. (2016). Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 7, 11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung H-L, Chen S, et al. (2016). Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, and Horvath S (2016). dynamicTreeCut: Methods for Detection of Clusters in Hierarchical Clustering Dendrograms. https://rdrr.io/cran/dynamicTreeCut/. [Google Scholar]
- Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y, et al. ; BrainSpan Consortium; PsychENCODE Consortium; PsychENCODE Developmental Subgroup (2018a). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, eaat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen Z, Fan G, Li A, Yuan J, and Xu T (2018b). Cell-Type-Specific Afferent Innervation of the Nucleus Accumbens Core and Shell. Front. Neuroanat. 12, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-T, Liu T-Y, Yang C-Y, Yu Y-L, Chen T-C, Day Y-J, Chang C-C, Huang G-J, and Chen J-C (2016). Chronic activation of NPFFR2 stimulates the stress-related depressive behaviors through HPA axis modulation. Psychoneuroendocrinology 71, 73–85. [DOI] [PubMed] [Google Scholar]
- Lin Y-T, Yu Y-L, Hong W-C, Yeh T-S, Chen T-C, and Chen J-C (2017). NPFFR2 activates the HPA axis and induces anxiogenic effects in rodents. Int. J. Mol. Sci. 18, 1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, and Kleinman JE (2006). Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol. Psychiatry 60, 650–658. [DOI] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, et al. ; 23andMe Research Team; HUNT All-In Psychiatry (2019). Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK (2009). Molecular profiling of striatonigral and striatopallidal medium spiny neurons past, present, and future. Int. Rev. Neurobiol. 89, 1–35. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, and Yang XW (2006). FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat. Neurosci. 9, 443–452. [DOI] [PubMed] [Google Scholar]
- Lun ATL, and Marioni JC (2017). Overcoming confounding plate effects in differential expression analyses of single-cell RNA-seq data. Biostatistics 18, 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, McCarthy DJ, and Marioni JC (2016). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 5, 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, and Marioni JC; Participants in the 1st Human Cell Atlas Jamboree (2019). EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Sankowski R, Staszewski O, and Prinz M (2020). Microglia Heterogeneity in the Single-Cell Era. Cell Rep. 30, 1271–1281. [DOI] [PubMed] [Google Scholar]
- Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X, et al. (2019). Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard KR, Tippani M, Takahashi Y, Phan BN, Hyde TM, Jaffe AE, and Martinowich K (2020). dotdotdot: an automated approach to quantify multiplex single molecule fluorescent in situ hybridization (smFISH) images in complex tissues. Nucleic Acids Res. 48, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard KR, Collado-Torres L, Weber LM, Uytingco C, Barry BK, Williams SR, Catallini JL 2nd, Tran MN, Besich Z, Tippani M, et al. (2021). Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun ATL, and Wills QF (2017). Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, and Melville J (2018). UMAP: Uniform Manifold Approximation and Projection. arXiv, 1802.03426v3 http://arxiv.org/abs/1802.03426v3. [Google Scholar]
- Murray EA, Wise SP, and Drevets WC (2011). Localization of dysfunction in major depressive disorder: prefrontal cortex and amygdala. Biol. Psychiatry 69, e43–e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy C, Maitra M, Tanti A, Suderman M, Théroux J-F, Davoli MA, Perlman K, Yerko V, Wang YC, Tripathy SJ, et al. (2020). Single-nucleus transcriptomics of the prefrontal cortex in major depressive disorder implicates oligodendrocyte precursor cells and excitatory neurons. Nat. Neurosci. 23, 771–781. [DOI] [PubMed] [Google Scholar]
- Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen C-Y, Choi KW, Coleman JRI, Dalvie S, Duncan LE, Gelernter J, et al. (2019). International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat. Commun. 10, 4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, et al. ; GERAD1 Consortium; CRESTAR Consortium (2018). Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer D, Williams B, Mansouri E, Stevanovski S, Nakajima S, Le Foll B, Kish S, Houle S, Mizrahi R, George SR, et al. (2017). Corticotropin-releasing hormone and dopamine release in healthy individuals. Psychoneuroendocrinology 76, 192–196. [DOI] [PubMed] [Google Scholar]
- Prensa L, Richard S, and Parent A (2003). Chemical anatomy of the human ventral striatum and adjacent basal forebrain structures. J. Comp. Neurol. 460, 345–367. [DOI] [PubMed] [Google Scholar]
- Rizzardi LF, Hickey PF, Rodriguez DiBlasi V, Tryggvadóttir R, Callahan CM, Idrizi A, Hansen KD, and Feinberg AP (2019). Neuronal brain-region-specific DNA methylation and chromatin accessibility are associated with neuropsychiatric trait heritability. Nat. Neurosci. 22, 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue-Albrecht K, Marini F, Soneson C, and Lun ATL (2018). iSEE: Interactive SummarizedExperiment Explorer. F1000Res. 7, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, and Nestler EJ (2013). The brain reward circuitry in mood disorders. Nat. Rev. Neurosci. 14, 609–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymar VV, Sasseville R, Luk KC, and Sadikot AF (2004). Neurogenesis and stereological morphometry of calretinin-immunoreactive GABAergic interneurons of the neostriatum. J. Comp. Neurol. 469, 325–339. [DOI] [PubMed] [Google Scholar]
- Salgado S, and Kaplitt MG (2015). The nucleus accumbens: a comprehensive review. Stereotact. Funct. Neurosurg. 93, 75–93. [DOI] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savell KE, Tuscher JJ, Zipperly ME, Duke CG, Phillips RA 3rd, Bauman AJ, Thukral S, Sultan FA, Goska NA, Ianov L, and Day JJ (2020). A dopamine-induced gene expression signature regulates neuronal function and cocaine response. Sci. Adv. 6, eaba4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer L, Velmeshev D, Holmqvist S, Kaufmann M, Werneburg S, Jung D, Vistnes S, Stockley JH, Young A, Steindel M, et al. (2019). Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann CM, and Amaral DG (2005). Stereological estimation of the number of neurons in the human amygdaloid complex. J. Comp. Neurol. 491, 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindreu C, and Storm DR (2011). Modulation of neuronal signal transduction and memory formation by synaptic zinc. Front. Behav. Neurosci. 5, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, Giusti-Rodriguez P, Hodge RD, Miller JA, Muñoz-Manchado AB, et al. ; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (2018). Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 50, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorvari H, Soininen H, Paljärvi L, Karkola K, and Pitkänen A (1995). Distribution of parvalbumin-immunoreactive cells and fibers in the human amygdaloid complex. J. Comp. Neurol. 360, 185–212. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, Mattheisen M, Wang Y, Coleman JRI, Gaspar HA, et al. ; eQTLGen Consortium; BIOS Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium (2019). Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley G, Gokce O, Malenka RC, Südhof TC, and Quake SR (2020). Continuous and discrete neuron types of the adult murine striatum. Neuron 105, 688–699.e8. [DOI] [PubMed] [Google Scholar]
- Tamura G, Olson D, Miron J, and Clark TG (2005). Tolloid-like 1 is negatively regulated by stress and glucocorticoids. Brain Res. Mol. Brain Res. 142, 81–90. [DOI] [PubMed] [Google Scholar]
- Tan CP, Sano H, Iwaasa H, Pan J, Sailer AW, Hreniuk DL, Feighner SD, Palyha OC, Pong S-S, Figueroa DJ, et al. (2002). Melanin-concentrating hormone receptor subtypes 1 and 2: species-specific gene expression. Genomics 79, 785–792. [DOI] [PubMed] [Google Scholar]
- Tepper JM, and Bolam JP (2004). Functional diversity and specificity of neostriatal interneurons. Curr. Opin. Neurobiol. 14, 685–692. [DOI] [PubMed] [Google Scholar]
- Thrupp N, Sala Frigerio C, Wolfs L, Skene NG, Fattorelli N, Poovathingal S, Fourne Y, Matthews PM, Theys T, Mancuso R, et al. (2020). Single-Nucleus RNA-Seq Is Not Suitable for Detection of Microglial Activation Genes in Humans. Cell Rep. 32, 108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyszka JM, and Pauli WM (2016). In vivo delineation of subdivisions of the human amygdaloid complex in a high-resolution group template. Hum. Brain Mapp. 37, 3979–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton G (2008). Visualizing Data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605. [Google Scholar]
- Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, Bhaduri A, Goyal N, Rowitch DH, and Kriegstein AR (2019). Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorn P, Gerfen CR, and Groenewegen HJ (1989). Compartmental organization of the ventral striatum of the rat: immunohistochemical distribution of enkephalin, substance P, dopamine, and calcium-binding protein. J. Comp. Neurol. 289, 189–201. [DOI] [PubMed] [Google Scholar]
- Wassum KM, and Izquierdo A (2015). The basolateral amygdala in reward learning and addiction. Neurosci. Biobehav. Rev. 57, 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, et al. ; eQTLGen; 23andMe; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YE, Pan L, Zuo Y, Li X, and Hong W (2017). Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron 96, 313–329.e6. [DOI] [PubMed] [Google Scholar]
- Yao J-J, Zhao Q-R, Lu J-M, and Mei Y-A (2018). Functions and the related signaling pathways of the neurotrophic factor neuritin. Acta Pharmacol. Sin. 39, 1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong W, Spence JP, Eskay R, Fitz SD, Damadzic R, Lai D, Foroud T, Carr LG, Shekhar A, Chester JA, et al. (2014). Alcohol-preferring rats show decreased corticotropin-releasing hormone-2 receptor expression and differences in HPA activation compared to alcohol-nonpreferring rats. Alcohol. Clin. Exp. Res. 38, 1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahm DS, and Heimer L (1993). Specificity in the efferent projections of the nucleus accumbens in the rat: comparison of the rostral pole projection patterns with those of the core and shell. J. Comp. Neurol. 327, 220–232. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cheng H, Zuo Z, Zhou K, Cong F, Wang B, Zhuo Y, Chen L, Xue R, and Fan Y (2018). Individualized Functional Parcellation of the Human Amygdala Using a Semi-supervised Clustering Method: A 7T Resting State fMRI Study. Front. Neurosci. 12, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
- Zhong S, Ding W, Sun L, Lu Y, Dong H, Fan X, Liu Z, Chen R, Zhang S, Ma Q, et al. (2020). Decoding the development of the human hippocampus. Nature 577, 531–536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw single-nucleus RNA-seq read data is publicly available from the Globus endpoint ‘jhpce#tran2021’, linked from https://research.libd.org/globus. Data files containing de-identified and processed SingleCellExperiment objects are hosted on Amazon S3, and the links are available on the README.md of the GitHub repository for this project (https://github.com/LieberInstitute/10xPilot_snRNAseq-human). RNAscope data generated in this study will be shared by the lead contact upon request.
All code for processing and analyzing the data has been archived at the time of submission at Zenodo: https://doi.org/10.5281/zenodo.5149046.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.