Abstract
The formation of microbial biofilms enables single planktonic cells to assume a multicellular mode of growth. During dispersion, the final step of the biofilm lifecycle, single cells egress from the biofilm to resume a planktonic lifestyle. As the planktonic state is considered to be more vulnerable to antimicrobial agents and immune responses, dispersion is being considered a promising avenue for biofilm control. In this Review, we discuss conditions that lead to dispersion and the mechanisms by which native and environmental cues contribute to dispersion. We also explore recent findings on the role of matrix degradation in the dispersion process, and the distinct phenotype of dispersed cells. Last, the translational and therapeutic potential of dispersing bacteria during infection will be discussed.
Graphical Abstract
In this Review, Rumbaugh and Sauer discuss the environmental cues and microorganism-derived signals that lead to the biofilm dispersal response, recent findings of matrix-degrading enzymes required for cells to liberate themselves from the biofilm matrix, novel insight into the mechanisms and regulation of dispersal, as well as the implications of these insights for biofilm control effort.
INTRODUCTION
Bacteria exhibit two modes of growth: the free-living planktonic mode or the sessile, surface-attached mode within biofilms, which are structured communities encased in a self-produced polymeric matrix1–3. The ability to form a biofilm is not only a common trait of various microorganisms, including lower-order eukaryotes, but has also been recognized as the dominant mode of bacterial growth in nature1,3,4. The formation of biofilms is a developmental process that is initiated by planktonic (free-living) organisms aggregating and/or transitioning to a surface-associated lifestyle, and is completed when cells escape from the biofilm structure in a process referred to as dispersion to return to the single cell, planktonic mode of growth (see below). Although it is now widely accepted that most if not all bacterial species form biofilms in a cyclic process, the first and most detailed information of the biofilm developmental life cycle stems from research with Pseudomonas aeruginosa. This developmental process has been described as a sequential, highly regulated process, involving at least five phenotypically distinct stages (Fig. 1a), with each biofilm developmental stage corresponding to unique patterns of protein production and gene expression5–9. Key features relating to the sessile mode of growth include loss of flagella gene expression, production of biofilm matrix components, induction of antibiotic resistance mechanisms (including efflux-pumps even when biofilms were grown in the absence of antibiotics) and increased levels of virulence determinants10–12. As a consequence, biofilm cells display characteristics and behavior that are distinct from their planktonic counterparts, with the hallmark characteristics including innate resistance to host immune defenses and their increased tolerance to stresses, including starvation, dehydration and antimicrobials. Notably, biofilms have been reported to be 10–1000-times more tolerant to various antibiotics compared to their planktonic counterparts13.
Figure 1: Biofilm formation and dispersion.
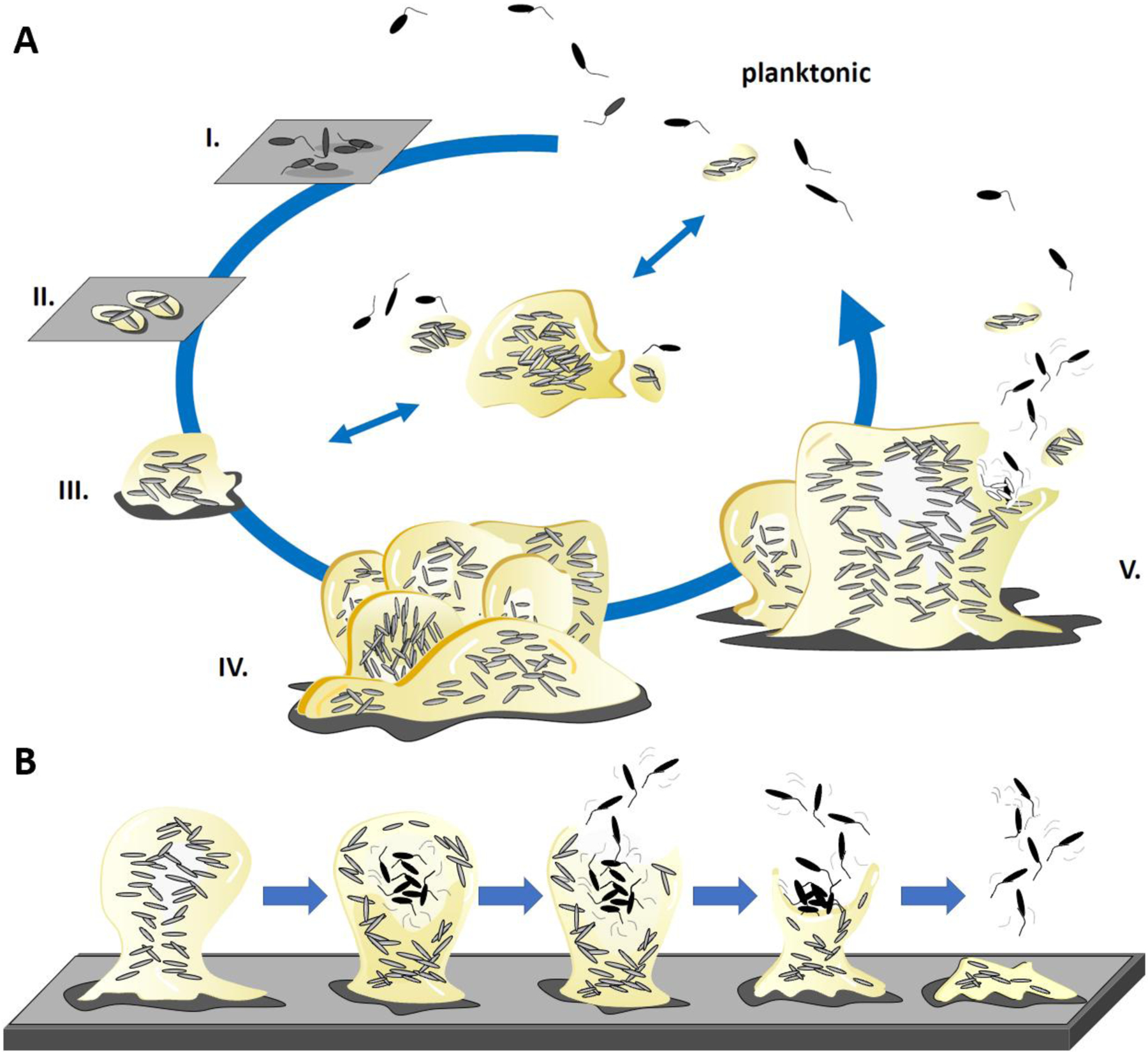
a) The schematic is based on the in vitro analysis of single species Pseudomonas aeruginosa biofilms. The formation of biofilms is a cyclic process that occurs in a stage-specific and progressive manner. The process is initiated following surface contact by single planktonic cells. Several developmental steps are discernable as reversible attachment (step I), irreversible attachment (step II) and biofilm maturation (steps III-IV)6,26. During the initial event in biofilm development, bacteria attach to substratum at the cell pole or via the flagellum (step I). Cells are cemented to the substratum and form nascent cell clusters with all cells in contact with the substratum. Transition to this stage coincides with loss of flagella gene expression and the production of biofilm matrix components (step II). Cell clusters mature and are several cells thick, embedded in the extracellular polymeric substances matrix (step III). Biofilms fully mature, which is apparent by clusters and microcolonies having reached maximum thickness (step IV). The biofilm life cycle comes full circle when biofilms disperse (step V). This stage is characterized by cells evacuating the interior portions of cell clusters, forming void spaces. Recent reports describe the formation of aggregates that exhibit biofilm-like features in the absence of surfaces (middle panel)174. Based on in vivo observations, these aggregates can attach to surfaces or disperse174 (indicated by the arrows). However, it is unclear if aggregate formation requires the same biofilm formation pathways and dispersion events. b) The schematic shows the discernable events that lead to biofilm dispersion. During dispersion, the inside of a biofilm becomes fluid, and cells within this zone begin to show signs of agitation and movement. Subsequently, cells escape the biofilm via a disruption in the microcolony wall through which cells evacuate, entering the bulk liquid as single bacteria. This leaves behind biofilms with central voids. If the dispersion response is extensive, the biofilm structure may further erode.
These features pose severe consequences in medical and industrial settings. For example, biofilms are frequently found on engineered surfaces where they cause biological fouling (biofouling). Biofouling is associated with materials deterioration such as corrosion, loss in heat-transfer efficiency and mechanical clogging in fluid transport systems (reviewed in Refs.14,15). Marine biofouling is associated with more than a 30% increase in fuel consumption by large sea-going vessels to overcome the viscous drag imposed by fouling organisms on ship hulls14. Estimates suggest that governments and industry spend more than $6 billion annually for repairs and preventive maintenance activities that result from biofouling, highlighting the economical challenge of biofilms14. However, biofilms pose the most daunting challenge to 21st century medical care1,16–22. According to the National Institutes of Health, biofilms are responsible for >80% of microbial infections and >60% of all nosocomial infections, with the Center for Disease Control estimating biofilms to be the etiologic agent in 60% of all chronic infections (program announcements PA-03–047, PA-06–537). Individuals at risk for developing biofilm-related infections include those with medical implants and medical devices, and immunocompromised patients, such as those suffering from cystic fibrosis and diabetic neuropathy1,16–22. In the Unites States, the annual incidence of biofilm-related infections is 1.96 million cases, causing an estimated 268,000 deaths, with over $18 billion in direct costs spent on the treatment of these infections23.
Detachment is believed to be the main mechanism that limits the overall biofilm biomass accumulation and refers to the passive release of biofilm particles due to mechanical or shear stress24. Distinct detachment mechanisms have been recognized based on the shear stress and/or the extent by which biofilm particles are removed by grazing, abrasion, erosion and sloughing25. Grazing refers to loss of biofilm cells by the feeding activity of eukaryotic organisms, whereas abrasion is caused by collisions of biofilm cells with particles from the environment. Erosion and sloughing refer to loss of biofilm cells by fluid frictional forces, with erosion removing small portions of the biofilm and sloughing removing intact pieces of biofilm or the entire biofilm. In contrast to detachment, dispersion is an active event in which sessile, matrix-encased biofilm cells actively escape from the biofilm, leaving behind eroded biofilms and biofilms having central voids (Fig. 1b)5,26–28. Dispersion is generally characterized as the terminal stage of biofilm development (Fig. 1), often referred to as seeding dispersal29, as dispersion is assumed to lead to the translocation of bacteria to new sites for colonization.
Relative to the initiation of biofilm formation, the mechanism or mechanisms by which bacteria disperse from the biofilm have received little attention until recently. However, given that dispersion coincides with biofilm bacteria converting to a planktonic mode of growth that is more vulnerable to antimicrobial agents and immune responses, dispersion is now being considered a promising avenue for biofilm control. Dispersion is a rapidly emerging field of biofilm research, as evidenced by the growing numbers of publications and the growing interest by pharmaceutical and start-up companies pursuing anti-biofilm products.
In this Review, we summarize our current knowledge of biofilm dispersion, including the environmental cues and microorganism-derived signals that lead to the dispersal response, recent findings of matrix-degrading enzymes required for cells to liberate themselves from the biofilm matrix, and novel insight into the mechanisms and regulation of dispersal. In addition, we discuss consequences of escape to dispersed cells and the host as well as implications of these insights for biofilm control efforts. Although we primarily focus on the dispersion response of the Gram-negative pathogen P. aeruginosa as the best-characterized dispersion system to date, examples from other microorganisms are included and parallels are drawn when possible. It is important to note that much of what will be discussed in this Review originated from in vitro studies that were conducted under a limited number of environmental conditions (for example, experiments carried out under flowing or semi-batch conditions) with a much smaller subset of biofilm experiments having been conducted using animal models. Thus, caution should be used when considering the in vivo or clinical implications of biofilm mechanisms that have only been studied in vitro.
The biofilm lifestyle
Biofilm formation is generally considered a cyclic process comprising phenotypically distinct stages. Despite the diversity of biofilm-forming species and biofilm architectures, numerous studies using single-species have revealed biofilm formation to coincide with distinct developmental stages and general features regardless of species5,30. For one, biofilm formation is initiated with surface attachment by a few planktonic cells, which occurs in two stages: reversible and irreversible attachment. Whereas reversible attachment enables first surface contact via electrostatic and hydrophobic interactions, the interaction is unstable, with cells frequently returning to the liquid phase (Fig. 1a). During this stage, first surface contact by rod-shaped flagellated cells is primarily mediated via the flagellum31. In addition, pili (such as type 1 pili and type IV pili), curli fibers and antigen 43 have been shown to mediate attachment31. Non-motile Gram-positive bacteria make surface contact via pili and adhesins31. Examples of adhesins include SagA andAcm of Enterococcus faecium, and Ace, Esp and Enterococcal biofilm pili (Ebp) of E. faecalis31. Microscopic observations suggest that once cells are irreversibly attached they cease to move and initiate matrix production, probably to ‘cement’ themselves to the surface or to each other. This is followed by clonal growth into more complex multicellular structures during the maturation stages of biofilm development31,32. Various mature biofilm structures have been reported, including unstructured and overall flat biofilms, and mushroom-like or pillar-like structures referred to as microcolonies or macrocolonies that are interspersed with fluid-filled channels (Fig. 1a). However, most of these structures are only seen in environmental biofilms or biofilms grown in vitro. By contrast, available evidence from in vivo-grown biofilms and imaging of clinical biopsies indicate much smaller and less differentiated structures or aggregates33.
Being in a biofilm enables microorganisms to colonize competitive niches and survive stressful environments34. For example, compared to planktonic cells, being in a community improves the chances of survival when challenged by nutrient deprivation, dehydration, pH changes, bacteriophages or predators35. This protective effect is mostly due to the polymeric matrix that encases the community, which not only provides structural support but traps nutrients from the environment for metabolic utilization by the resident bacteria, efficiently retains water through hydrogen-bond interactions, and provides a protective, bunker-like shield for the biofilm residents2. During infection the biofilm provides protection from the immune system and from antimicrobial agents including biocides, oxidizing agents and antibiotics35. Current thinking is that resilience against antimicrobial agents observed in biofilm communities is the combined result of different mechanisms, including starvation-induced slow growth and reduced metabolic rates36–40, the presence of persister cells41–45, and the sequestration and restricted penetration of antimicrobials through the matrix46–56. Recent evidence further suggests that microbial biofilm communities resist the action of antimicrobial agents by expressing genes that encode various xenobiotic efflux and export systems12,57,58, activating the stringent response (signaled by the alarmones guanosine tetraphosphate and guanosine pentaphosphate (collectively referred to as (p)ppGpp) that modulate transcription of a third of all genes in the cell)40, or expressing ribosome hibernation factors11. Dispersion is an active event in which sessile, matrix-encased biofilm cells convert en-masse to the planktonic mode of growth, apparent by single cells actively escaping the biofilm. This process can leave behind eroded biofilms with central voids5,26–28, or cavities within the biofilm structure formed by the evacuation of bacteria located in the center of mature biofilm (Fig. 1b)26,29,59. Dispersion is often referred to as the final stage of biofilm development, as it marks the departure from the biofilm and the transition of formerly sessile cells to the planktonic mode of growth, which facilitates a new cycle of biofilm development at new sites of colonization29(Fig. 1).
Cues and signals of biofilm dispersal
Although dispersion enables dissemination, it also leaves the former biofilm residents vulnerable. Dispersed cells face the loss of multiple fitness advantages, including drug tolerance5,60. So why leave? Simply put, being in a biofilm is not always advantageous. Bacteria residing at different locations within the biofilm structure experience concentration gradients of nutrient resources, oxygen and waste products (such as acids produced by fermentation in oxygen-depleted zones) as well as extracellular signaling molecules11,34,61. Transcriptome analyses of drip reactor-grown P. aeruginosa biofilms and Escherichia coli colony biofilms confirmed that biofilm cells respond to these gradients by inducing various stress responses (Fig. 2). This was apparent by the presence of high mRNA levels of genes linked to hypoxia (or long-term anoxia), which is indicative of oxygen deprivation, increased expression of RpoS -regulated genes, which is indicative of general stress and stationary phase conditions, and increased expression levels of genes that are linked to nutrient stress and slow growth10,11,34 (Fig. 2a). As the biofilm grows in size, these chemical gradients grow steeper, which results in increased non-uniform access to resources by the resident community and leads to the formation of subpopulations11,34. In addition, continued biofilm growth coincides with increased cellular crowding that further exacerbates chemical gradients and leads to nutritional competition34,61. Thus, it is not surprising that the formation of chemical gradients within biofilms have been proposed as the driving force of dispersion10,34,61,62.
Figure 2: Environmental conditions initiating dispersion.
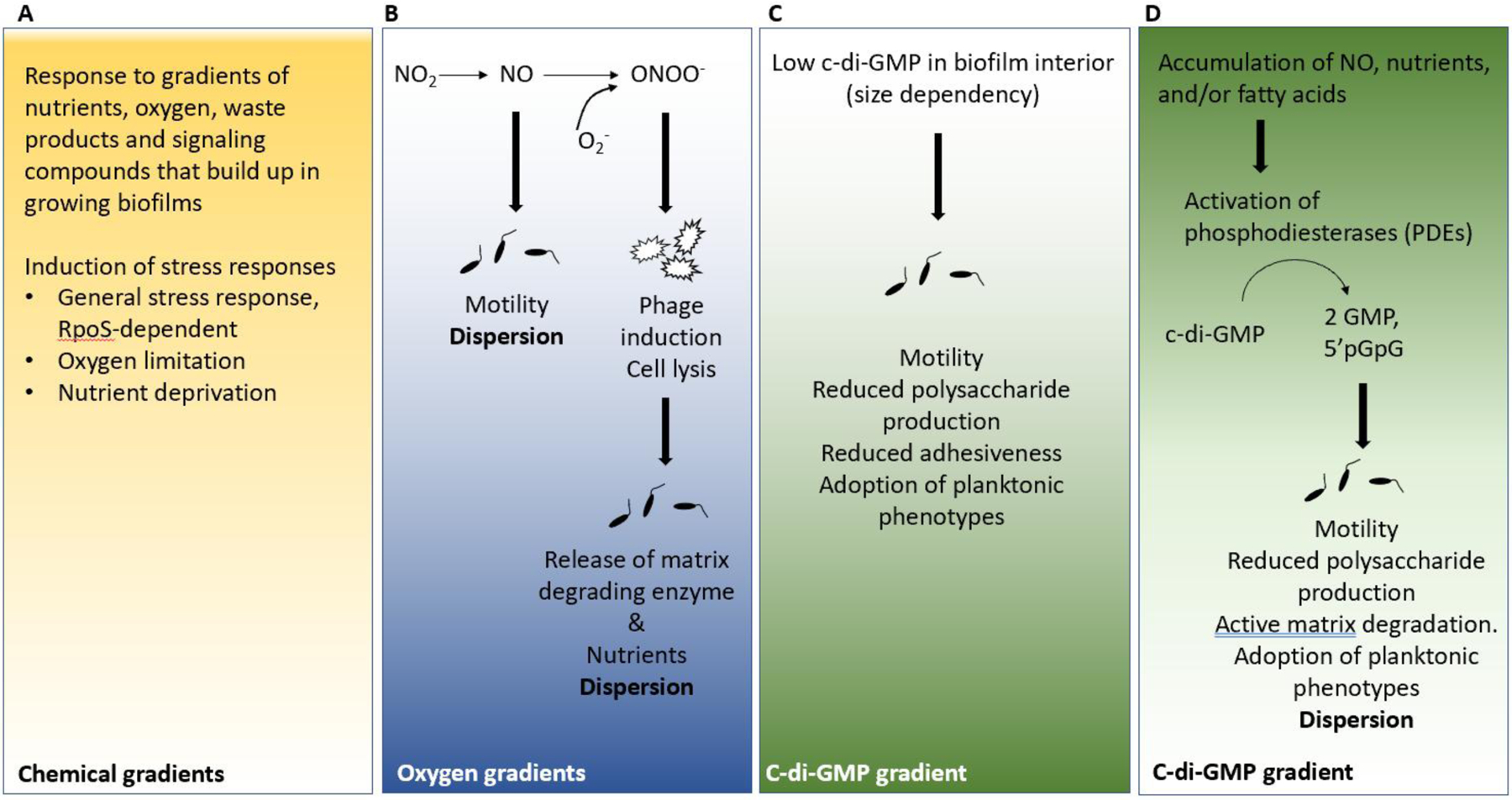
Each panel indicates various chemical gradients that are likely to be present in biofilms; low concentrations of those chemicals have been linked to dispersion. Moreover, phenotypic changes such as motility or induction of events that are triggered in response to the limited availability of compounds are shown. a) Bacteria residing at different locations within the biofilm structure experience concentration gradients of nutrient resources, oxygen, waste products and extracellular signaling molecules. Biofilm cells respond to these gradients by inducing various stress responses, such as the RpoS -dependent general stress response, as well as increased expression of genes involved in the response to oxygen limitation and nutrient deprivation. b) Under limiting oxygen conditions, such as found in the interior of the biofilm, anaerobic denitrification leads to the generation of nitric oxide (NO). Exposure to NO is linked to the reduction in cellular bis-(3’-5’)-cyclic dimeric guanosine monophosphate (c-di-GMP) levels and thus increased motility and dispersion. Furthermore, NO can react with superoxide to generate the cell-toxic radical ONOO–. ONOO– causes cellular damage, bacteriophage induction and cell lysis. Cell lysis results in the release of degradative enzymes that contribute to the enzyme-mediated breakdown of the biofilm matrix and/or loosening of the biofilm matrix. In addition, cell lysis generates nutrients for growth. c)-d) Levels of c-di-GMP vary throughout the biofilm, with the lowest levels being detected in the biofilm interior. Low c-di-GMP levels contribute to phenotypes generally associated with the planktonic mode of growth, including increased motility, increased drug susceptibility, but reduced adhesiveness and matrix production. Bacteria residing at different locations within the biofilm structure experience the accumulation of native dispersion cues including NO, cis-DA, and nutrients. Sensing of dispersion cues activates phosphodiesterases (PDEs) capable of degrading c-di-GMP, ultimately resulting in an overall reduction in the levels of c-di-GMP. Phenotypes associated with low c-di-GMP levels include increased motility, reduced adhesiveness, reduced matrix production and dispersion.
But what exactly is it about these steep chemical gradients that lead residents of the sessile community to leave their protective environment? Is there a tipping point at which resuming a solitary lifestyle outweighs the benefits of being in a biofilm and if so, what tips the scale? Two types of dispersal triggers that induce active dispersion have been elucidated: native and environmentally induced. Although native dispersion occurs in response to self-synthesized signaling molecules or cues that are likely to be the result of steep gradients within the biofilm, environmentally induced dispersion occurs in response to sensing factors that are present in the surrounding environment or the changing conditions of the surrounding environment (Table 1).
Table 1.
Cues, signals and environmental factors linked to dispersion.
| Cues, signals and factors | Species | Effector regulatory system | Source |
|---|---|---|---|
| Fatty acid signaling (DSF and BDSF) | Xanthomonas campestris | DSF and Rpf genes positively control the synthesis of manA-encoded endo-β-1,4-mannanase, which degrades the matrix; DSF negatively affects the expression of xagABC encoding a glycosyl transferase system Induction of low c-di-GMP levels | |
| Fatty acid signaling (cis-DA) | Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, Bacillus subtilis, Proteus mirabilis, Staphylococcus aureus, Streptococcus pyogenes, Candida albicans | Similar to DSF and BDSF, cis-DA may induce low c-di-GMP levels, [Au: is this entry specific for Pseudomonas aeruginosa etc? yes, specific for P. aeruginosa but could also be for other strains but has not been explored] | Correct reference is Davies and Marques, 2009, Ref #5964,109,164 |
| Oxygen depletion | P. aeruginosa | Dispersion response is dependent on PDE RbdA, probably to induce low c-di-GMP levels | 165 |
| Starvation due to cessation of flowing conditions (oxygen or nutrients) | Shewanella oneidensis | Dispersion-inducing conditions are linked to mxdB encoding a putative membrane-associated glycosyl transferase and the probable induction of a PDE to reduce c-di-GMP levels | 48 |
| Pseudomonas putida, P. aeruginosa, P. fluorescens | Dispersion-inducing conditions have been linked to low c-di-GMP levels and the release of the adhesin LapA in biofilms by P. putida and P. fluorescens. Additionally, reports suggest lack of nutrients and cell lysis but not the the accumulation of a metabolic product to be contribute to the dispersion response | 66–69 | |
| S. aureus | Quorum sensing, Agr-dependent | 134 | |
| Nutrient availability (glucose, glutamate, succinate and citrate) | P. aeruginosa | Dispersion in response to nutrients is dependent on phosphorylation events, and coincides with a decrease in c-di-GMP and increased PDE activity Factors identified to have a role include BdlA, PDEs DipA and RbdA, and DGCs NicD and GcbA Induction of matrix-degrading enzymes include endonucleases EndA and EddA, and glycosyl hydrolases PelA and PslG | 87,93,95,107,115,116,166,167 |
| Acinetobacter sp, S. pneumoniae | Dispersion response is linked to quorum sensing/Interkingdom signaling | 168 | |
| Serratia marcescens | dispersion is linked to quorum sensing | 169 | |
| Nitric oxide | P. aeruginosa | Dispersion in response to nitric oxide coincides with a decrease in c-di-GMP and increased PDE activity Factors identified to have a role include BdlA, PDEs NbdA, MucR, DipA and RbdA Induction of matrix-degrading enzymes include endonucleases EndA and EddB, and glycosyl hydrolases PelA and PslG | 97,98,115,116,170 |
| Nitrosomonas europaea Ammonia oxidizer | no effector proteins have been identified | 171 | |
| Iron | P. aeruginosa | Exposure to elevated iron concentrations repress the expression of certain genes essential for scavenging iron, with scavenging and acquisition of iron being essential for biofilm formation by P. aeruginosa. | 79,172 |
| Reduction of c-di-GMP | P. aeruginosa | Dispersion induced by overexpression of the response regulator RcsB which leads to induction of the PDE PvrR | 173,68 |
| P. putida | Dispersion induced by overexpression of E. coli-derived PDE YhjH, which might lead to LapG-dependent release of LapA | 94 |
PDE, phosphodiesterase; DGC, diguanylate cyclase, cis-DA, cis-2 decenoic acid; DSF, cis-11-methyl-2-dodecenoic acid; BDSF, cis-2-dodecenoic acid; c-di-GMP, bis-(3’-5’)-cyclic dimeric guanosine monophosphate; DGCs, diguanylate cyclases.
Native dispersion inducers and inducing conditions.
Native dispersion of biofilms grown in flow cells has been reported to be initiated by bacteria that are located in the center of mature biofilms. Cells coordinately evacuate the biofilm structure, probably through small openings in the biofilm matrix, and enter the surrounding environment, leaving behind large transparent cavities, or hollow structures that consist of non-motile cells26,29,59. These observations highlight an important characteristic of the dispersion response: dispersion rarely involves the entire biofilm (Fig. 1). Instead, only cells in select areas within a biofilm will disperse or show signs of dispersion events29. The specific subpopulation that disperses from the center of biofilms has been linked to the diameter and thickness of the biofilm (but not to biofilm age). Microscopic observations of biofilms grown in flow cells under continuous flow conditions indicated that dispersion events in the biofilm interior were only seen within biofilms that exceeded a minimum diameter of 40 μm and an overall biofilm height of 10 μm28,29. The biofilm diameter was found to increase with increasing flow rate, which suggests a relationship between dispersion and medium flow, with the latter affecting the transport of various growth resources into the biofilm structure, including nutrients, oxygen and extracellular signaling molecules. Thus transport, which determines the steepness of the chemical gradient, rather than size of the biofilm, is the determining factor of whether biofilms initiate a dispersion response28. This also suggests that an inducer is responsible for initiating biofilm dispersion, and such an inducer would be removed by diffusive and advective transport when the biofilm is small but accumulate as the biofilm grows in size.
One such inducer was identified in P. aeruginosa as the fatty acid signaling molecule cis-2-decenoic acid (cis-DA) (Table 1). Fatty acid signals are involved in intra-species, inter-species and cross-kingdom communication, and they are capable of regulating bacterial growth, virulence, motility, polymer production, biofilm development, and biofilm dispersion and persistence63. Cis-DA is no exception, as this fatty acid signaling molecule induces biofilm dispersion by a range of Gram-negative and Gram-positive bacteria and yeast59. Additional fatty acid signaling molecules include cis-11-methyl-2-dodecenoic acid (DSF), capable of disaggregating small aggregates such as flocs by Xanthomonas campesitris64, and the Burkholderia cenocepatia cis-2-dodecenoic acid (BDSF) that has been shown to induce dispersion of Francisella novicida biofilms65.
Moreover, oxygen and pyruvate have also been identified as inducers of native dispersion. There are several supportive observations of oxygen depletion as a possible trigger of dispersion. For example, dispersion is initiated in cells grown in biofilms upon cessation of flow48,66–69, dispersion originates deep within biofilm structures in areas that are subject to steep oxygen gradients (Fig. 2b). In addition, cells experiencing oxygen-limiting and electron-rich conditions70–72, referred to as ‘reductive stress’ (that is, too many NADH/electrons and not enough O2) undergo dispersal73.
Some microorganisms can cope with oxygen depletion and reductive stress by using an inorganic molecule other than oxygen such as sulfate or nitrate as a final electron acceptor for anaerobic cellular respiration. However, in the absence of alternative anaerobic respiratory pathways or alternative terminal electron acceptors, species like P. aeruginosa initiate fermentative processes, including pyruvate fermentation74,75. The pyruvate in P. aeruginosa PA14 biofilms that enable pyruvate fermentation was shown to be self-produced and released by metabolically active cells exposed to oxygen, using a pyocyanin-dependent mechanism76. Thus, pyruvate is likely to be produced mostly in the periphery of biofilms. The role of pyruvate in biofilm formation was supported by the addition of 10 mM pyruvate to the growth medium of biofilms grown in semi-batch conditions in microtiter plates, which resulted in a substantial increase in biofilm biomass accumulation by P. aeruginosa, whereas continuous addition of pyruvate dehydrogenase to deplete pyruvate prevented the formation of three-dimensional biofilm structures72. By contrast, enzymatic depletion of pyruvate from mature P. aeruginosa biofilms induced dispersion73, which suggests pyruvate availability, and thus the ability to sustain anaerobic survival, is a switch that inversely regulates biofilm formation and biofilm dispersion.
Pyruvate availability also affects biofilm formation and dispersion by Staphylococcus aureus, but the mechanism responsible has not yet been elucidated73. Pyruvate formate lyase has been reported to have a role in S. aureus biofilm formation, by functioning as a formate supplier for metabolic processes during anaerobiosis, with formate oxidation correlating with the generation of NADH/H77. These findings not only point at redox stress being a driving force of dispersion, but also at dispersion being a reversal of biofilm formation. Dispersion being a reversion of biofilm formation is further supported by the role of iron. One study78 demonstrated the formation of structured P. aeruginosa biofilms to be favored in media with low iron availability, as increasing iron concentrations resulted in increasingly unstructured biofilms. By contrast, P. aeruginosa biofilms have been reported to undergo a dispersion response when challenged with free iron in vitro79.
Dispersion is also associated with increased cell death32,80. Cell lysis has been discussed as a result of localized anaerobic regions within the biofilm, apparent by biofilms predominantly exhibiting gene expression profiles consistent with anaerobic growth11,81, and coinciding with fermentative82 or anaerobic respiratory pathways, including denitrification. Anaerobic denitrification in the interior of biofilm microcolonies generates nitric oxide (NO) that, if not reduced by NO reductase to nitrous oxide, can react with superoxide to generate the cell-toxic radical ONOO–32 (Fig. 2b). ONOO– causes cellular damage, bacteriophage induction and cell lysis32,83, with the latter resulting in the release of degradative enzymes (not thought to be transported across the cell membrane), that contribute to the enzyme-mediated breakdown of the biofilm matrix and/or loosening of the biofilm matrix32,80. In addition, cell lysis generates nutrients for growth32,80 (Fig. 2b). There is also evidence that prophages influence the pattern of cell death and lysis seen in biofilms. For P. aeruginosa, prophage-mediated cell lysis was first described for strain PAO1, which displayed characteristic voids in the center of mature microcolonies of flow cell-grown biofilms83. Cell death in the center of these microcolonies was attributed to lysis by prophages that are closely related to the filamentous phage Pf1 and reside within the genome of PAO1. Deletion of this prophage from PAO1 resulted in microcolonies devoid of hollow centers containing lysed cells. It is proposed that prophage-mediated cell death is an important mechanism of differentiation inside microcolonies that facilitates dispersal of a subpopulation of surviving cells. Prophages have since been described to contribute to microcolony maturation and stability, dispersal, virulence and the propagation of morphotypic variants84–86.
Although the mechanism or mechanisms by which (many of) these cues or factors induce dispersion is not fully understood, many of these either acerbate or eliminate chemical gradients present in biofilms depending on their concentration in the biofilm interior (Fig. 2). For instance, cell lysis may contribute to the nutrient support of surviving cells and the release of enzymes capable of degrading matrix components. Moreover, the sudden availability of nutrients may contribute to the modulation of bis-(3’-5’)-cyclic dimeric guanosine monophosphate (c-di-GMP) levels (. Likewise, NO has been linked to the reduction of c-di-GMP levels (see below, Fig. 2b). Given that these chemical gradients are the driving forces of biofilm formation by affecting gene expression and protein production, including the modulation of various stress responses10,34,61, it is apparent that some dispersion inducers function by reversing the biofilm developmental process. Moreover, dispersion is a localized event. As such, dispersion is a response by a subpopulation or subpopulations within the biofilm. For instance, cells in the interior of a P. aeruginosa biofilm convert into a motile phenotype and disperse, while non-dispersing biofilm cells are non-motile, primarily found in the biofilm periphery, and create a ‘wall phenotype’29. Although it is unclear whether biofilms differentiate into subpopulations prior to (or during) the dispersion response, it is seems that the development of dispersing subpopulations is likely to coincide with biofilm cells experiencing a multitude (or a specific combination) of chemical gradients (Fig. 2). Taken together, biofilm subpopulations capable of dispersing are likely to locate within larger, more mature biofilms, and they are characterized by hypoxia, experience cell death, free radicals and other cues that predispose the cells for dispersion.
Environmentally induced dispersion and inducers.
Dispersion in response to exposure to factors present in the surrounding environment or the changing conditions of the surrounding environment is referred to as environmentally induced dispersion. This is in contrast to native dispersion that occurs in response to signaling molecules or cues that are synthesized by the resident biofilm cells. Similar to native dispersion which is initiated in the biofilm interior, dispersion events induced by environmental factors also result in void formation. However, voids induced by environmental factors look more like eroded biofilms composed of a non-motile layer of cells that make up the base of the biofilm prior to dispersion (Fig. 1b). Similar to native dispersion, environmentally induced dispersion rarely involves the entire biofilm (Fig. 1), with no more than 80% of the biomass being removed upon induction of dispersion87. Environmentally induced dispersion cues and factors include NO, cis-DA, iron, induction of starvation inducing conditions (oxygen and/or nutrients), and a step-increase of various nutrients including sugars and amino acids (Table 1). The similarity between conditions and cues initiating environmentally induced and native dispersion is striking, which suggests similarities in dispersion cue sensing and dispersal mechanism or mechanisms between the two active dispersion responses. Moreover, environmentally induced dispersion inducers are not limited to a single species. For example, NO has been demonstrated to disperse single-species and mixed-species biofilms of clinically and industrially relevant microorganisms88. Similarly, cis-DA was found to disperse a range of Gram-negative and Gram-positive bacteria and yeast59, which suggests that cis-DA may not only be capable of dispersing single species but also mixed species biofilms.
Dispersal mechanisms
Although little is known about native dispersion, the mechanism of dispersion in response to various environmental stimuli including nutrient cues, NO and others (Table 1) has been relatively well characterized in P. aeruginosa. Sensing of dispersion cues is accomplished by a membrane-associated protein complex. Cue sensing and relay of signal is accomplished via a series of post-transcriptional modifications that ultimately result in an overall reduction in the levels of the intracellular signaling molecule c-di-GMP. Low c-di-GMP levels in turn, are likely to contribute to the phenotypes of dispersed cells associated with the planktonic mode of growth, including motility, susceptibility and enhanced phagocytosis. Moreover, it is now apparent that dispersed cells increase expression of genes that assist in the degradation of the polymeric matrix that shields and stabilizes the biofilm structure.
Modulation of c-di-GMP levels.
Although most research investigating the role of c-di-GMP has focused on Gram-negative bacteria, recent findings suggest that c-di-GMP has similar roles in Gram-positive bacteria; however, targets and regulatory mechanisms may differ89. Specifically, the intracellular signaling molecule c-di-GMP is now recognized as a near-ubiquitous second messenger that coordinates diverse aspects of bacterial growth and behavior, including motility, virulence, cell cycle progression and biofilm formation (reviewed in Ref.90). High levels of c-di-GMP foster the sessile lifestyle, whereas low levels enhance the planktonic mode of growth. Levels of c-di-GMP are modulated by the opposing activities of two sets of enzymes, diguanylate cyclases (DGCs), which generate c-di-GMP from two molecules of GTP, and phosphodiesterases (PDEs), which degrade c-di-GMP90.
Although high c-di-GMP levels are generally associated with the biofilm mode of growth, recent findings suggest levels of c-di-GMP are not uniform throughout biofilms (Fig. 2c). Using a GFP reporter for which the fluorescence intensity is directly proportional to the concentration of intracellular c-di-GMP91, a striking difference in c-di-GMP concentrations was seen across the biofilm structure. c-di-GMP was localized in relatively high amounts at the outer boundary of large, mature biofilms, whereas smaller, less developed biofilms showed a more uniform distribution of c-di-GMP92. Whether the c-di-GMP gradients noted in biofilms predispose subpopulations of biofilm cells to disperse is not known. However, several reports indicate that dispersed Gram-negative cells that have escaped the biofilm structure are characterized by reduced c-di-GMP levels compared with biofilm cells93–96. Moreover, dispersed cells display increased expression of PDEs and increased PDE activity48,93,97–99.
In turn, low c-di-GMP levels in dispersed cells enhance phenotypes associated with the planktonic mode of growth, including motility. For example, in P. aeruginosa, motility is regulated by the major flagellar gene regulator FleQ, a c-di-GMP-responsive transcriptional regulator that enhances motility but represses Pel polysaccharide production at low c-di-GMP levels100. Moreover, low c-di-GMP levels affect susceptibility to antibiotics, apparent by clinical P. aeruginosa isolates from the cystic fibrosis lung displaying aggregative phenotypes in liquid (that is, a phenotype indicative of high c-di-GMP levels) and displaying 2–8-fold higher minimal inhibitory concentration (MIC) values to several antibiotics than revertants of this aggregative phenotype that displayed low c-di-GMP levels101–105. Moreover, dispersed cells exhibit reduced production of proteins involved in resistance mediated by cationic antimicrobial peptides including colistin106 and tobramycin58.
Dispersion cue sensing.
Based on our current understanding, low c-di-GMP levels are a result of dispersion cue sensing (Fig. 3, see above). In P. aeruginosa, dispersion cue sensing involves the chemotaxis-like MCP homolog, BdlA, the c-di-GMP PDEs DipA, RbdA, MucR and NbdA, and the DGCs GcbA and NicD. These proteins form a membrane-associated complex that perceives and relays dispersion cues to promote the modulation of c-di-GMP levels107, apparent by an initial increase in c-di-GMP levels followed by a sharp decrease107. The DGC NicD and the PDE NbdA contribute to dispersion in a cue-specific manner, with NbdA sensing NO97 and NicD sensing nutrient cues107 (Fig. 3). By contrast, BdlA and the PDEs DipA and RbdA are central to the dispersion response regardless of the cue that is being sensed (Fig. 3). Sensing of dispersion cues results in the activation of BdlA by non-processive cleavage, with active BdlA recruiting and activating two PDEs, RbdA and DipA, ultimately resulting in the reduction of c-di-GMP levels97,107,108. In addition, fatty acid signaling contributing to native dispersion has been linked to the decrease of intracellular c-di-GMP levels109.
Figure 3: Sensing of dispersion cues.
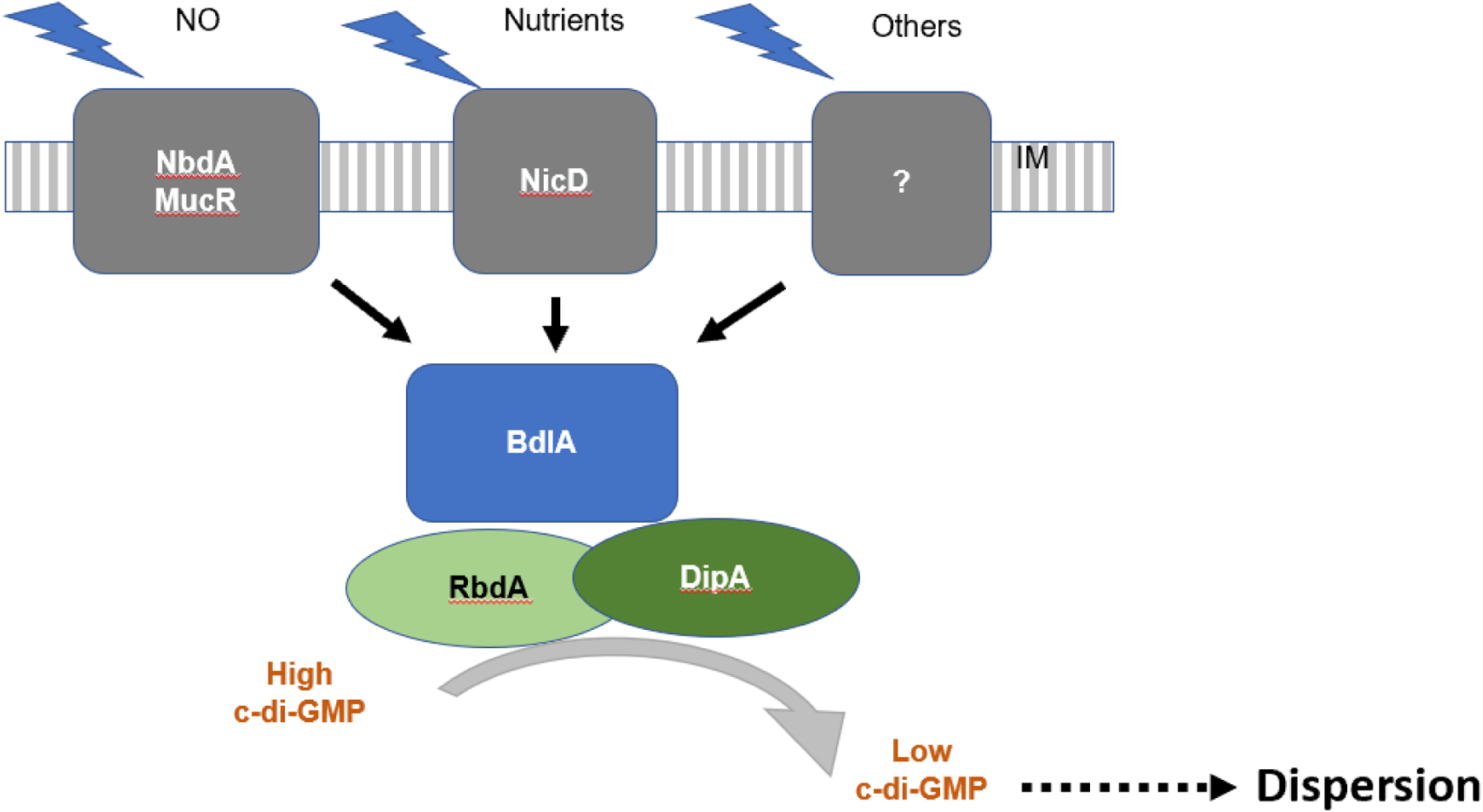
Model of dispersion cue sensing and modulation of bis-(3’-5’)-cyclic dimeric guanosine monophosphate (c-di-GMP) levels to induce dispersion. In Pseudomonas aeruginosa, dispersion cue sensing involves the chemotaxis-like MCP homolog, BdlA, the c-di-GMP phosphodiesterases (PDEs) DipA, RbdA, MucR and NbdA, and the diguanylate cyclases (DGCs) GcbA and NicD. The membrane-associated PDEs MucR and NbdA and the DGC NicD are involved in perceiving and relaying dispersion cues to promote the modulation of c-di-GMP levels. The DGC NicD and the PDE NbdA contribute to dispersion in a cue-specific manner, with NbdA sensing NO and NicD sensing nutrient cues. Receptors for other dispersion cues such as fatty acids have not yet been elucidated (indicated by the question mark). BdlA and the PDEs DipA and RbdA are central to the dispersion response regardless of the cue that is being sensed and contribute to the overall reduction of c-di-GMP levels post dispersion cue sensing.
Dispersion induced by fatty acids such as cis-DA has also been linked to the modulation of c-di-GMP64,110. Previous work on X. campestris demonstrated that fatty acid sensing requires a two-component system, RpfC–RpfG, whereas in B. cenocepacia fatty acids directly bind to the receptor protein RpfR, with binding stimulating the PDE activity of RpfR111. Although detailed investigations of the dispersion response by Gram-positive bacteria are limited112, it is likely that c-di-GMP has a similar role in the dispersion response by Gram-positive bacteria, albeit signaling may involve a new conformation of c-di-GMP and if so, likely novel receptors and novel binding motifs for c-di-GMP relative to Gram-negative bacteria113.
Disintegration of the biofilm matrix.
Events subsequent to cue sensing and reduction of c-di-GMP levels are less defined but support the notion that dispersion coincides with the disintegration of the biofilm matrix. Although the matrix composition is highly diverse amongst bacterial species, it is generally composed of extracellular polysaccharides, proteinaceous factors such as carbohydrate-binding proteins, adhesins, or amyloid fibers and extracellular DNA (eDNA) (Box 1; reviewed in Ref.114). The matrix, which accounts for ~90% of the biofilm biomass, is a viscoelastic structure that not only tethers cells within the biofilm and functions as a protective shield, but also functions as a scaffold for the three-dimensional biofilm structure114.
Textbox 1. Effectors leading to biofilm disassembly rather than dispersion.
Biofilms disperse via several mechanisms, and this commonly involves extensive regulatory and metabolic changes that a biofilm undergoes to enable the evacuation of dispersed cells. Moreover, dispersion coincides with extensive structural changes, apparent by the substantial loss of biofilm biomass, indicating that a key aspect of the dispersion response is the partial breakdown of the biofilm matrix composed of extracellular DNA (eDNA), polysaccharides and proteins.
In recent years, several effectors have been described that target the biofilm matrix. The effectors that are capable of matrix dissolution often have enzymatic activity and include polysaccharide-degrading enzymes, chitinases, proteases and nucleases (reviewed in Ref.127). These enzymes are similar to enzymes that are produced by the dispersing biofilm subpopulation. However, although the production of dispersal-induced matrix-degrading enzymes are the result of regulatory and metabolic changes that are induced during the dispersion response, these enzymes can be exogenously added to biofilms to induce a passive biofilm disassembly without inducing regulatory and metabolic changes by the resident biofilm population. Thus, induced biofilm disassembly may result in an egressing cell population that retains the biofilm phenotype rather than adopts the dispersal phenotype. Notable phenotypes associated with dispersion but likely not during biofilm disassembly include increased susceptibility to antimicrobials, shift towards an acute virulence phenotype, and susceptibility to immune cells. Moreover, biofilm disassembly may coincide with the release of cell aggregates rather than single planktonic cells.
One such effector is dispersin B, a poly-N-acetylglucosaminidase capable of degrading matrix polysaccharides131–133. Originally identified in a transposon screen of Aggregatibacter actinomycetemcomitans for genes that are essential for dispersal, dispersin B was later found to induce biofilm disassembly in a range of bacterial species, including Staphylococcus epidermidis, when exogenously added to biofilms131–133. Additional glycoside hydrolases, enzymes that hydrolyze the glycosidic linkages between two or more carbohydrates175, have been demonstrated to disassemble biofilms. Exposure of purified P. aeruginosa glycoside hydrolases PelA and PslG that are not only required for the synthesis their respective exopolysaccharides Pel and Psl but also contribute to their degradation129, has been shown to disassemble established P. aeruginosa biofilms129. Likewise, a cocktail of glycoside hydrolases (cellulase, α-amylase) that target glycosidic linkages commonly seen within the exopolysaccharides secreted by a wide range of pathogens including β-1,4 bond present in cellulose, and the α-1,4 bond in amylose, has been shown to lead to the disassembly of S. aureus and P. aeruginosa monoculture and co-culture biofilms176. In addition to eDNA being a source of nutrients, phosphorus and nitrogen177, eDNA was described in 2002 to have a role in biofilms123. Since then, eDNA has been shown to be deposited on the stalk of biofilms enabling the formation of mushroom-shaped microcolonies, to enhance biofilm formation probably by enabling direct or indirect interactions with the bacterial cell surface, and to cross-link matrix components178. Given eDNA is a structural element of the biofilm matrix, eDNA degradation by DNase I has likewise been shown to result in the release of large amounts of biomass in various biofilm-forming species, including P. putida, S. aureus, S. oneidensis and Bacillus licheniformis127. However, DNase treatment leading to biofilm disassembly seems to be limited to young, less structured biofilms123, probably due to mature biofilms harboring increasing amounts of matrix material other than eDNA, and eDNA being more centrally located within the biofilm structure.
In addition to matrix-degrading enzymes, amphipathic molecules that reduce surface tension have been shown to induce biofilm disassembly. For instance, exogenous addition of rhamnolipids triggers detachment by P. aeruginosa biofilms, whereas the biosurfactant putisolvins induces detachment by P. putida biofilms detachment127. It is of interest to note that rhamnolipids failed to disassemble filamentous biofilms by S. marcescens str. MG1169.
Matrix composition has been extensively investigated in P. aeruginosa and found to be composed of the Psl, Pel and alginate exopolysaccharides, eDNA and matrix stabilizing proteins such as the adhesin CdrA114 (Fig. 4a). Alginate is a linear unbranched polysaccharide that contains different amounts of (1→4′)-linked β-d-mannuronic acid and α-l-guluronic acid residues, Psl is a neutral pentasaccharide subunit that contains mannose, rhamnose and glucose, and Pel is a glucose-rich cellulose-like polysaccharide that can cross-link to eDNA. The level of these polysaccharides within the matrix varies across P. aeruginosa strains. Given that dispersion coincides with cells evacuating the protective biofilm environment, it is not surprising that dispersion has been linked to the increased expression and production of matrix-degrading enzymes. Specifically, NO- and nutrient-induced dispersion have been linked to increased expression of genes encoding endonucleases such as endA115 (EndA degrades DNA present in the matrix) and glycoside hydrolases such as pslG and pelA116 (PslG and PelA degrade Psl and Pel matrix polysaccharides116), as well as the increased production of matrix degrading enzymes, compared to planktonic and biofilm cells96. Below we summarize the events that lead to the enzymatic disintegration of the biofilm matrix in response to dispersion cue sensing.
Figure 4. Mechanisms resulting in biofilm dispersal.
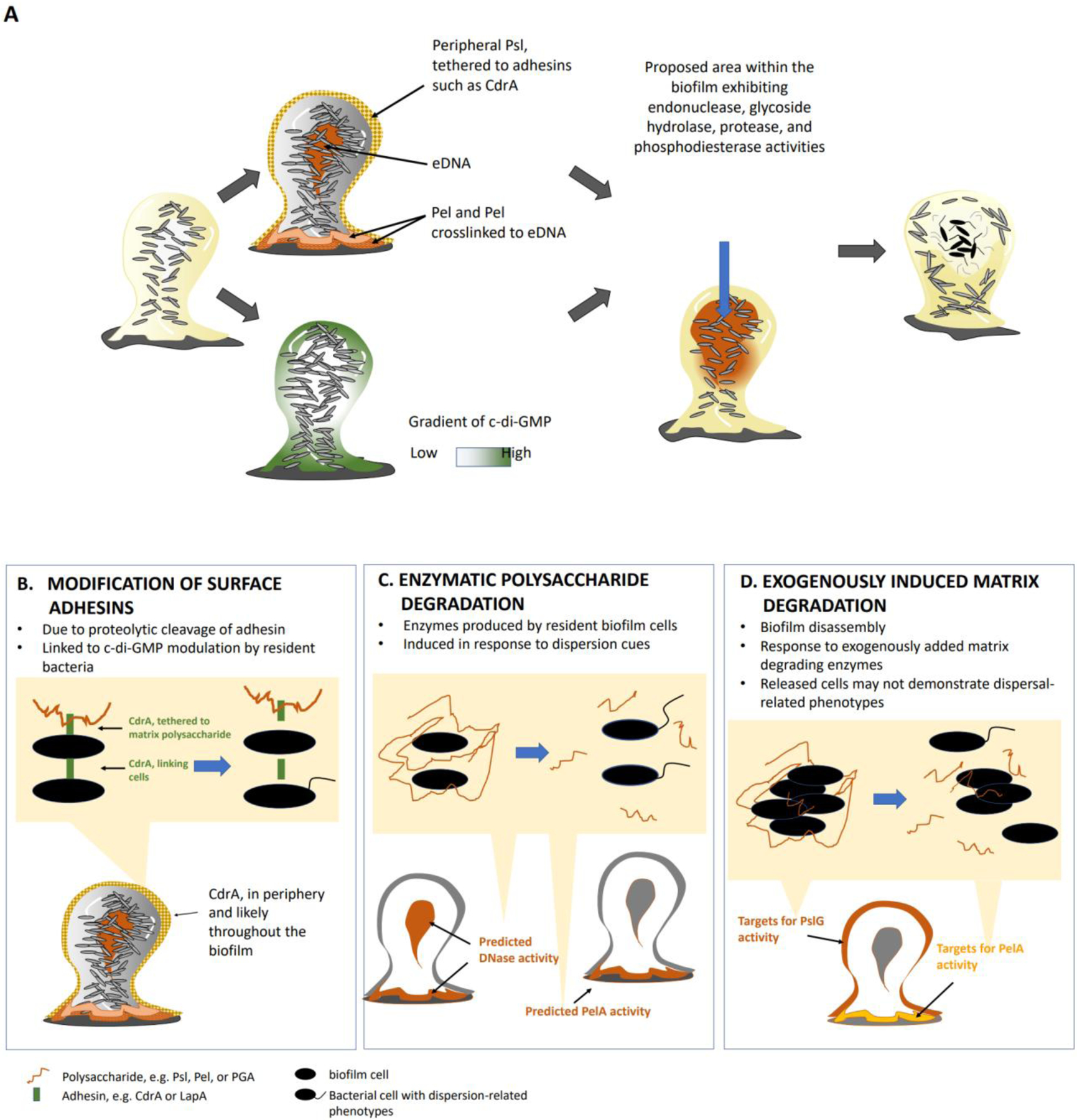
(a) Model of spatial localization of biofilm matrix components and bis-(3’-5’)-cyclic dimeric guanosine monophosphate (c-di-GMP), and proposed biofilm areas with active matrix- and c-di-GMP-degrading activities, that lead to biofilm dispersion. In Pseudomonas aeruginosa PAO1, the matrix polysaccharide Psl is localized at the periphery and the base of biofilms. Psl interacts with the adhesin protein CdrA and forms a protective but non-rigid structure in between cells and around the biofilm structure. The Pel polysaccharide is primarily located at the base of the biofilm and crosslinked to extracellular DNA (eDNA). However, eDNA is not limited to the biofilm base but has also been detected in the biofilm interior. Mature biofilms are characterized by low c-di-GMP levels in the interior of the biofilm whereas c-di-GMP levels in immature or less structured biofilms is more uniform. Considering the link between dispersion, low c-di-GMP levels, and matrix degradation, the biofilm periphery and biofilm interior are the most likely to be the locations within the biofilm that experiecne increased matrix- and c-di-GMP-degrading activities. Moreover, these locations coincide with observed dispersion events such as void formation and erosion of the biofilm structure. b) Surface adhesins such as CdrA or LapA are cleaved to untether the polysaccharide matrix and break cell-cell interactions. (c) Matrix components are enzymatically degraded (by intrinsic DNases or the hydrolase PelA) in response to native or environmental dispersion cues (nitric oxide, nutrients or cis-2-decenoic acid). d) Biofilm disassembly can be induced by exogenously added matrix-degrading enzymes. As enzymes are exogenously added, matrix degradation is expected to primarily target the periphery of the biofilm structure (see Box 1).
CdrA is a c-di-GMP regulated adhesin that reinforces the biofilm matrix, probably by tethering Psl117,118. High c-di-GMP levels have been linked to a cell surface localization of CdrA, whereas low c-di-GMP levels coincide with the release of CdrA from the outer membrane via cleavage by the periplasmic cysteine protease LapG. The first evidence of CdrA contributing to dispersion was inferred by the finding that lapG mutants displayed increased attachment to plastic and glass surfaces117, whereas a cdrA deletion mutant displayed decreased attachment relative to the parental strain upon extended incubation. The role of CdrA in dispersion has recently been confirmed by the observation that a cdrA deletion mutantwas ‘hyper’-dispersive relative to P. aeruginosa strains with an inactivated lapG, which exhibited reduced dispersion115. These findings suggest that the presence of the surface-associated matrix protein CdrA impedes the dispersion response and that CdrA release, and thus untethering of the matrix, is likely to be a first step that enables matrix degradation. (Fig. 4b)
A similar mechanism was noted to contribute to dispersion by P. putida and P. fluorescens. In these two species, the adhesin LapA, similar to CdrA, is localized at the outer membrane at high c-di-GMP levels, but the adhesin is cleaved from the cell surface by the protease LapG in the presence of low levels of c-di-GMP119. Starvation‐induced dispersal of P. putida biofilms was dependent on LapG-dependent proteolytic cleavage of LapA66,94. Similarly, LapG-dependent proteolytic cleavage of LapA at low c-di-GMP levels was found to contribute to phosphate limitation‐induced dispersal of P. fluorescens biofilms119. However, it is likely that other proteases in addition to LapG have a role. This is supported by CdrA release being sensitive to self-produced proteases other than LapG120, proteinase K being capable of disassembling P. putida biofilms94, and the finding that lowering the c-di-GMP level in P. aeruginosa by overexpressing the phosphodiesterase PA2133 was not sufficient to induce dispersion116.
It is now widely acknowledged that eDNA is a ubiquitous and pivotal structural component of biofilms by diverse group of microorganisms, including P. aeruginosa, uropathogenic. coli, nontypeable Haemophilus influenzae and Staphylococcus epidermidis121,122. As a structural component, eDNA may also have a role in dispersion (Fig. 4c). Although exogenously added DNAses were shown to be effective in disassembling young P. aeruginosa biofilms123 and biofilms produced by P. putida, S. aureus, Shewanella oneidensis and Bacillus licheniformis124–127, DNAses were mostly ineffective in disassembling mature biofilms. The difference in the efficacy of DNAse to disassemble biofilm structures is likely to be due to the fact that eDNA being predominately found in the iinterior of mature biofilms (Figs. 4a, 4c). However, dispersed P. aeruginosa cells exhibit increased expression of genes encoding the secreted nucleases EndA, EddA and EddB, which are not only capable of degrading eDNA present in the biofilm matrix but are essential for the dispersion response115. Dispersed cells also have increased expression of pelA but not pslG116, which encode glycoside hydrolases, with purified PelA and PslG having been shown to degrade Pel and Psl, respectively, and contribute to biofilm disassembly128–130. Interestingly, overexpression of pelA but not pslG expression resulted in dispersion of P. aeruginosa biofilms116, and pslG-induced dispersion was only detectable in the absence of CdrA116.
Polysaccharide degradation has been linked to dispersion of biofilms of various Gram-positive and Gram-negative bacteria, including Aggregatibacter actinomycetemcomitans, Aggregatibacter pleuropneumoniae, E. coli and S. epidermidis, harbor linear polymers of N-acetyl-D-glucosamines referred to as polysaccharide intercellular adhesin (PIA); poly-N-acetyl glucosamine (PNAG, β-(1–6)-linked N-acetylglucosamine), and polysaccharide adhesin (PGA, β-1,6-N-acetyl-D-glucosamine)(Fig. 4d). The enzyme responsible for the degradation has been identified as dispersin B, a secreted glycoside hydrolase131–133 (Box 1). Moreover, the E. coli CsrA protein has been shown to repress the synthesis of PGA, , thus aiding in dispersion31. Likewise, the agr system of S. aureus that contributes to the expression of matrix polysaccharides and an extracellular protease that degrades proteins such as adhesins, has been shown to contribute to dispersal134.
These findings indicate that dispersion coincides with the expression of genes encoding matrix-degrading enzymes, and requires several matrix-degradative enzymes that work together to degrade multiple matrix components, including eDNA, polysaccharides and adhesins (Fig. 4). It is of interest to note that the regulatory mechanisms driving matrix degradation are not well understood. More specifically, it is unclear whether matrix degradation is the direct result of c-di-GMP modulation or if additional factors or regulatory proteins are required to induce the expression of genes encoding matrix-degrading enzymes. Although it has been suggested that dispersed cells can be generated by simply reducing the intracellular c-di-GMP content through modulation of PDEs135, recent evidence suggests that molecular mechanisms regulating the dispersion response are likely to be more complex, requiring both c-di-GMP-dependent and -independent processes58,116. This is supported by recent findings of dispersion being induced upon overproduction of the endonuclease 1 EndA 115, and the glycosyl hydrolase PelA 116, whereas the adhesin CdrA was found to impede the dispersion response 116. endA, pelA, and cdrA are directly regulated by AmrZ, the central regulator of biofilm formation by P. aeruginosa136–138. Given the contribution of CdrA and matrix-degrading proteins in the P. aeruginosa dispersion response, it is likely that the central regulator of biofilm formation AmrZ has a role in the regulation of the dispersion response.
Escape and consequences
Although it is widely assumed that dispersed cells return to their planktonic state, studies with many different microbial species have shown that dispersed cells display distinct phenotypes. A common hallmark of dispersed cells is their high level of phenotypic heterogeneity139. This is not surprising considering that cells within the biofilm itself have high mutation frequency, presumably due to the close association of a high concentration of cells, as well as the adaptive pressure of different microenvironments present within the biofilm140. Dispersing cells with diverse phenotypes into the environment are likely to represent an important ecological adaptation mechanism that increases the chance some cells will survive and create new biofilm communities.
Many reports characterize dispersed cells as having phenotypes somewhere between that of biofilm cells and planktonic cells, but distinct from both57,87,139,141–143, and the heterogeneity within the dispersed subpopulation can be vast. For example, Klebsiella pneumoniae cells that spontaneously detached due to shear stress in a flow-cell biofilm displayed transcriptomes that were distinct from planktonic cells and sessile biofilm cells144. Cells representing five different physiological states were documented (that is, exponential planktonic growth, stationary planktonic growth, a 7 hr-old biofilm, a 13 hour-old biofilm and a dispersed biofilm), and 40 distinct transcriptional signatures were reported. Although the transcriptomes of dispersed cells resembled those of cells in a 7 hr-old biofilm more than the other cell subpopulations, they still displayed distinct transcriptional profiles. The most representative clusters of orthologous group (COG) affiliations of dispersed cells were translation, ribosomal structure and biogenesis and amino acid transport and metabolism, which suggests that these cells possess distinct metabolic capabilities compared to both biofilm and planktonic cells.
Dispersed cells are typically more motile, virulent and adherent, display different metabolic signatures and altered antimicrobial susceptibilities compared to their biofilm and planktonic counterparts (see below)57,87,141–143,145. Although these phenotypes can be short-lived as the dispersed cells transition to a more planktonic phenotype over time, this suggest that detached or dispersed cells are primed for recolonization at new sites57,142. For example, RNA sequencing analysis of Candida albicans grown in flow-cells demonstrated that the dispersed yeast cells exhibited enhanced adhesion, invasion and biofilm formation in comparison to their age-matched planktonic counterparts142. The dispersed cells also seemed to be transcriptionally reprogrammed to adopt different metabolic capabilities, which enables them to acquire specific nutrients and amino acids, and to metabolize alternative carbon sources in comparison to their biofilm-associated counterparts, despite being grown in the same nutritional environment. This reprogramming was manifested in the significant up-regulation of select high-affinity transporters and increased expression of genes involved in gluconeogenesis in dispersed versus biofilm-associated cells142. In another study, dispersal of P. aeruginosa was induced by the reduction of intracellular c-di-GMP, and transcriptomic analysis was performed on the resulting cell subpopulations57. Planktonic P. aeruginosa cells exhibited higher expression levels of genes involved in quorum sensing compared to biofilm and dispersed cells. Not surprisingly, biofilm cells exhibited higher expression of the genes involved in extracellular matrix synthesis, and lower expression of the genes involved in motility and chemotaxis compared with planktonic and dispersed cells. By contrast, dispersed cells exhibited higher expression of virulence genes and lower expression of the genes involved in iron uptake in comparison to planktonic cells and biofilm cells.
It is intuitive that dispersed cells express high levels of genes encoding virulence factors and genes important for motility and adhesion as they leave the protective environment of the biofilm and enter a much harsher environment, and the consequence to the host could mean a subsequently worse infection. In a C. albicans jugular vein catheter mouse model, genetic induction of dispersal from biofilms growing in the lumen of catheters resulted in enhanced disseminated infection, whereas repressing dispersal resulted in a 15-fold decrease in C. albicans infection of distal organs142. Likewise, dispersed P. aeruginosa cells have also displayed enhanced virulence in vivo. It was shown that P. aeruginosa cells that have been dispersed from in vitro grown biofilms by reduction of intracellular c-di-GMP killed significantly more macrophages and C. elegans than their planktonic counterparts57. Moreover, dispersing P. aeruginosa cells from a chronic wound infection with glycoside hydrolases caused systemic spread of the bacteria and death by sepsis in a mouse model146 (Fig. 5). Notably, these three examples include two different kingdoms of microorganisms, three different dispersal methods and completely different in vivo models, yet all the dispersed cells displayed enhanced virulence. Although clearly more studies are needed, existing data suggest that some dispersal phenotypes, such as heightened virulence, are conserved regardless of dispersal method, and the consequences of dispersal on the host or environment must be thoroughly considered.
Figure 5. Bacterial dissemination triggered by glycoside hydrolase treatment of Pseudomonas aeruginosa biofilms.
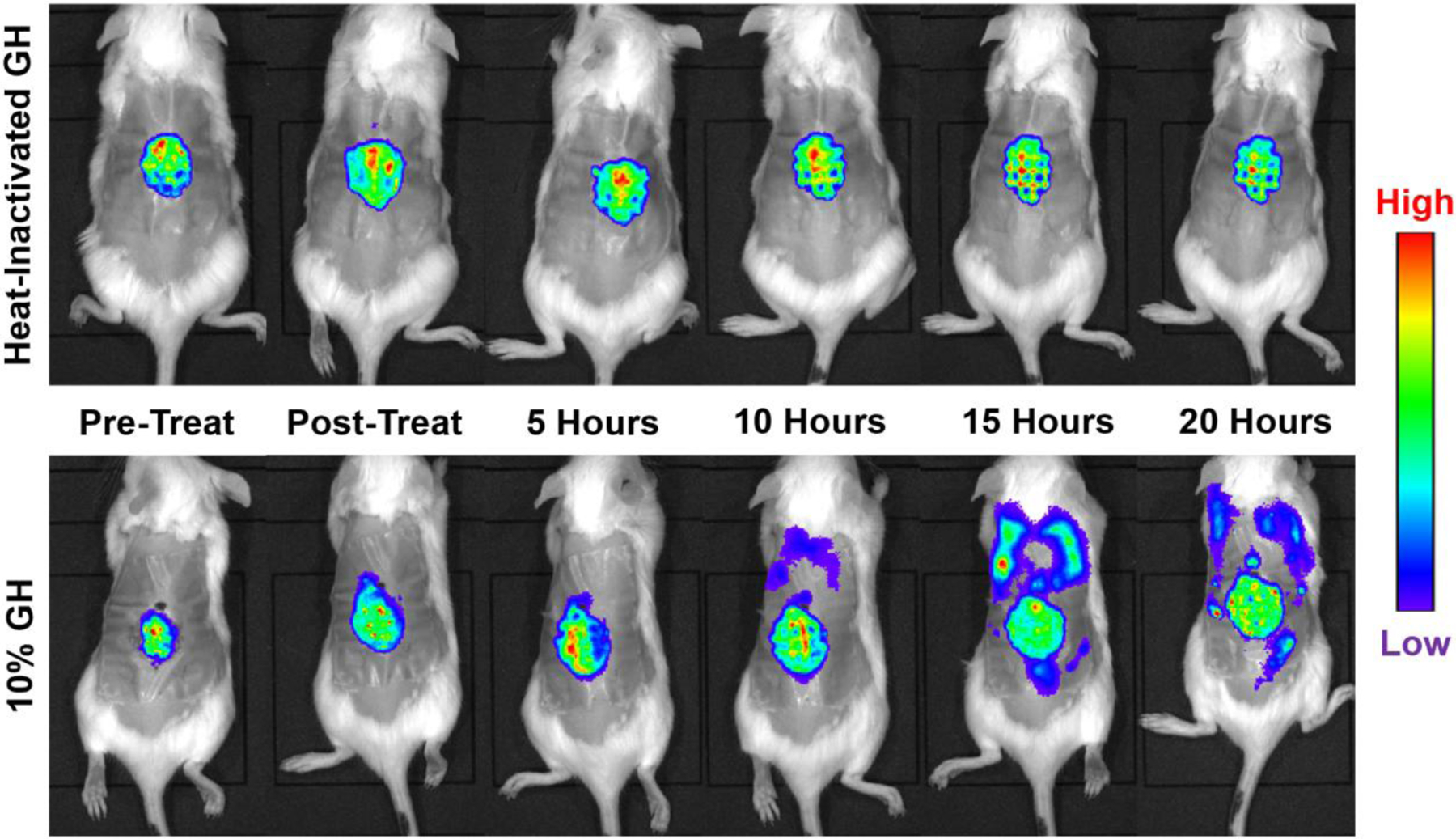
In vitro image system (IVIS) imaging shows that treatment of 48-hour-old mouse chronic wounds, infected with bioluminescent P. aeruginosa, with α-amylase and cellulase (glycoside hydrolase treatment), resulted in systemic spread of the infection (bottom panles). By contrast, systemic spread was not obeserved after treatment with heat-inactivated enzymes (top panels). Modified from Ref.146.
Biofilm control
It has been proposed that removing cells from the protective shield of a biofilm will render them more susceptible to antimicrobials and the host immune response. Given our current understanding about the mechanisms of dispersal and the broad phenotypes dispersed cells can adopt, one would not necessarily predict that dispersion would be an effective method for biofilm control. It is possible that dispersed cells would be more tolerant to antimicrobials than MIC testing on planktonic cells would predict, be highly virulent and adept at forming new biofilms at other locations. Thus, dispersing cells, without the capability of efficiently killing them, could result in a substantially larger ecological problem or more deadly infection. However, despite these perils, dispersal agents have gained traction over the past decade as a viable therapeutic option, and many published studies have demonstrated proof-of-principal for this strategy.
With a fundamental understanding of what constitutes the extracellular polymeric substance (EPS) of biofilms, enzymes that degrade these components gained enthusiasm early on as potential dispersal agents. Dozens of enzymes, including many nucleases, glycoside hydrolases and proteases, have been tested for their ability to degrade biofilms and disperse cells (reviewed in Ref.147). As microorganisms themselves produce and use these enzymes for biofilm degradation and dispersal, their exploitation is an intuitive strategy. Most dispersal attempts have been early-stage, pre-clinical studies, which applied purified enzymes to in vitro-grown biofilms and determined the reduction in biomass or percentage of cells dispersed. Biofilms formed by various bacteria and fungi were shown to be susceptible to degradation and some enzymes have also demonstrated efficacy on polymicrobial biofilms (reviewed in Ref.147).
Although previous studies have widely affirmed the potential of EPS-degrading enzymes to disrupt biofilms in vitro, far fewer have examined the efficacy of dispersal agents in vivo. Glycoside hydrolases represent one class of EPS-degrading enzymes that have moved into animal studies and the commercialization pipeline. The most well studied of these enzymes is dispersin B from A. actinomycetemcomitans. Dispersin B degrades PNAG by hydrolyzing β(1,6) glycosidic linkages. This enzyme is effective at disrupting the biofilms of multiple Gram-negative and Gram-positive bacterial species, including many pathogens132,148–151. Dispersin B has also shown efficacy in some in vivo models152–154 and commercialization of the agent is being pursued, although currently only for the treatment of animals. Effective treatment with other glycoside hydrolases, including amylase and cellulase, has also been demonstrated in vivo in recent reports128,146, which supports the rationale for their use in treating of biofilm-associated infections.
As investigators have learned more about what chemical cues lead to dispersal they have exploited these processes for therapeutic purposes. The standouts in this category are cis-DA acid and NO63,155. As indicated above, cis-DA acid is a fatty acid signal that prevents biofilm development and induces biofilm dispersion for a number of microorganisms in vitro59,156–158. Moreover, cis-DA has also shown some promise in a mouse model of periprosthetic joint infection159. NO is a ubiquitously produced biological signaling molecule that induces biofilm dispersal for a number of different microorganisms in vitro (reviewed in Ref.155); however, it has also shown efficacy in a number of animal models. For example, applying NO-releasing chitosan dressings to methicillin-resistant Staphylococcus aureus -infected wounds in mice resulted in enhanced biofilm dispersal, wound size reduction, epithelialization rates and collagen deposition in comparison to control groups160. A liposomal formulation of an NO donor, applied as a topical sinus wash, effectively reduced S. aureus biofilms in a sheep model of rhinosinusitis161, and NO-releasing catheters have shown effective at reducing biofilm burden in vivo162,163. Excitingly, NO-based therapeutic approaches have entered human clinical trials. Interventional studies testing the efficacy of NO to treat recalcitrant chronic rhinosinusitis, chronic non-healing wounds, prevent urinary tract infections, and reduce antibiotic tolerance in patients with cystic fibrosis have all entered clinical trials.
In many ways a biofilm is analogous to a cancerous tumor. Biofilms are tolerant and difficult to eradicate and have the potential to metastasize to other locations. As with cancer, the goal is to kill the cells in the biofilm with minimal collateral damage to the host. There are many potential options to chemically induce, enzymatically induce or mechanically break up biofilm and cause dispersion, but the successful agents will cause low levels of collateral damage, lack toxicity and demonstrate successful clinical trials. Importantly, biofilm dispersal agents have the capacity to potentiate the efficacy of antimicrobials. Ideally, they would be used as adjunctive agents with broad application for many biofilm-related infections, and because most are not bactericidal they are unlikely to cause resistance.
Conclusions
Dispersion is an active event in which sessile, matrix-encased biofilm cells convert en-masse to the planktonic mode of growth, apparent by single cells that are actively escaping the biofilm. The active escape from the protective biofilm environment seems to be driven by steepening chemical concentration gradients of nutrient resources, oxygen and waste products over the course of biofilm development that lead to the accumulation of molecules that are not only capable of inducing dispersion (that is, NO) but furthermore contribute to conditions that foster the formation of subpopulations in the interior of biofilms. These subpopulations are either more susceptible to dispersion cues or are being exposed to dispersion conditions to egress from the biofilm. The review of native and environmental dispersion cues suggests not only a striking similarity between cues initiating environmentally induced and native dispersion, but also similarities in cue sensing and the pathways between the two active dispersion responses. Based on current knowledge, the dispersion response involves dispersion cue sensing which subsequently results in an overall reduction of the c-di-GMP pool. Although subsequent events are less defined, dispersion coincides with the enzymatic disintegration of the biofilm matrix and subsequent egress from the biofilm. The dispersed subpopulation is typically more motile, virulent and adherent, displays different metabolic signatures and altered antimicrobial susceptibilities, which has resulted in dispersed cells being considered phenotypically between that of biofilm cells and planktonic cells, but distinct from both.
Based on a number of recent studies summarized here, it is apparent that dispersion has the potential to be exploited for therapeutic purposes to treat biofilm-related infections. However, although we have gained a deeper knowledge of the dispersion response in recent years, there remain numerous gaps in our knowledge. For instance, how is the sensing of dispersion cues linked to matrix degradation and subsequent dispersion? Is the relay simply based on the modulation of c-di-GMP or are additional non-ci-di-GMP regulated mechanisms involved? Given that dispersion in P. aeruginosa is linked to motility and matrix degradation, it is likely that FleQ is involved. Likewise, AmrZ, the central regulator of biofilm formation by P. aeruginosa, may have a role as the genes encoding endonucleases such as endA and glycoside hydrolases such pelA and cdrA, are directly controlled by AmrZ. Additionally, we know little about how the biofilm environment influences subpopulations to disperse. Is it possible that these subpopulations are already primed to disperse and if so, what pushes them over the edge to leave the biofilm? And what regulatory mechanisms contribute to the distinct phenotypes displayed by dispersed cells? When considering dispersion as a treatment strategy, we also know little about how efficient the dispersion response has to be for it to be an effective therapeutic agent, or whether enzymatic disassembly the biofilm structure is as effective as dispersion. But more importantly, our current understanding of dispersion is based on experimental evidence obtained under limited in vitro conditions with few microorganisms grown as single species biofilms. To close these gaps in our knowledge, it will not only be necessary to explore dispersion by a larger number of mono-species and mixed-species biofilm pathogens, but also to expand dispersion research from basic to translational studies, by exploring dispersion and dispersion inducers in pre-clinical and clinical studies.
Acknowledgements
This work was supported by grants from the National Institutes of Health (R21 AI137462-01A1 to KPR, 2R01 AI080710 to KS) and the Ted Nash Long Life Foundation to KPR.
GLOSSARY
- Abrasion
Loss of biofilm cells by collisions with particles from the environment; form of detachment
- Dispersion
Generally characterized as the terminal stage of biofilm development, dispersion is an active regulated event during which cells actively escape from the biofilm, leaving behind eroded biofilms and biofilms having central voids. Often referred to as seeding dispersal or dispersal29, as dispersion is assumed to lead to the translocation of bacteria to new sites for colonization.
- Disassembly
Describes the egress from biofilms and/or the disintegration of the biofilm structure in in response to exogenously added matrix-degrading enzymes. The term is frequently used when it is unclear whether the released cell subpopulation retains the biofilm phenotype or adopts the dispersal phenotype.
- Detachment
Characterized as the passive release or loss of biofilm cells or biofilm particles due to mechanical, physical or frictional forces. Another term used to refer to detachment is dissolution.
- Dissemination
The translocation of dispersed biofilm cells to new sites.
- Diffusive and advective transport
Processes that move nutrients, waste, gases or other compounds through the biofilm and the surrounding environment. Advection refers to transport of compounds with fluid flow whereas diffusion eliminates sharp discontinuities of compounds through the action of random motions.
- Native dispersion (also referred to as native dispersion)
Refers to dispersion in response to self-synthesized signaling molecules or cues that are likely to be the result of steep gradients within the biofilm.
- Quorum sensing
also referred to as cell-to-cell signaling, refers to the regulation of gene expression in response to the production and release of chemical signal molecules called autoinducers that increase in concentration as a function of cell density.
- RpoS
also referred to as sigma-38 (σ38), primary regulator of stationary phase genes and central regulator of the general stress response
- Shear
strain in the structure of a substance produced by pressure
Footnotes
Competing interests
The authors declare no competing interests.
Further links
CDC program announcement PA-03–047: https://grants.nih.gov/grants/guide/pa-files/pa-03-047.html
CDC programme announcement: PA-06–537: https://grants.nih.gov/grants/guide/pa-files/PA-06-537.html
REFERENCES
- 1.Costerton JW, Stewart PS & Greenberg EP Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Flemming H-C et al. Biofilms: an emergent form of bacterial life. Nature Reviews Microbiology 14, 563, doi: 10.1038/nrmicro.2016.94 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Geesey GG, Richardson WT, Yeomans HG, Irvin RT & Costerton JW Microscopic examination of natural sessile bacterial populations from an alpine stream. Can. J. Microbiol 23, 1733–1736 (1977). [DOI] [PubMed] [Google Scholar]
- 4.Costerton JW et al. Bacterial biofilms in nature and disease. Annual Reviews in Microbiology 41, 435–464 (1987). [DOI] [PubMed] [Google Scholar]
- 5.Stoodley P, Sauer K, Davies DG & Costerton JW Biofilms as complex differentiated communities. Annual Reviews in Microbiology 56, 187–209, doi:doi: 10.1146/annurev.micro.56.012302.160705 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Petrova OE & Sauer K A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathogens 5, e1000668 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrova OE, Gupta K, Liao J, Goodwine JS & Sauer K Divide and conquer: the Pseudomonas aeruginosa two-component hybrid SagS enables biofilm formation and recalcitrance of biofilm cells to antimicrobial agents via distinct regulatory circuits. Environmental Microbiology 19, 2005–2024, doi: 10.1111/1462-2920.13719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Toole GA & Kolter R Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Molecular Microbiology 28, 449–461, doi:doi: 10.1046/j.1365-2958.1998.00797.x (1998). [DOI] [PubMed] [Google Scholar]
- 9.Davey ME & O’Toole GA Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev 64, 847–867 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heacock-Kang Y et al. Spatial transcriptomes within the Pseudomonas aeruginosa biofilm architecture. Molecular Microbiology 106, 976–985, doi: 10.1111/mmi.13863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williamson KS et al. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. Journal of Bacteriology 194, 2062–2073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao J, Schurr MJ & Sauer K The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug-efflux pumps in Pseudomonas aeruginosa biofilms. Journal of Bacteriology 195, 3352–3363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies D Understanding biofilm resistance to antibacterial agents. Nature Reviews Drug Discovery 2, 114, doi: 10.1038/nrd1008 (2003). [DOI] [PubMed] [Google Scholar]
- 14.de Carvalho CCCR Marine Biofilms: a successful microbial strategy with economic implications. Frontiers in Marine Science 5, doi: 10.3389/fmars.2018.00126 (2018). [DOI] [Google Scholar]
- 15.Fitridge I, Dempster T, Guenther J & de Nys R The impact and control of biofouling in marine aquaculture: a review. Biofouling 28, 649–669, doi: 10.1080/08927014.2012.700478 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Hauser AR, Jain M, Bar-Meir M & McColley SA Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin. Microbiol. Rev 24, 29–70, doi: 10.1128/CMR.00036-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerr KG & Snelling AM Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect 73, 338–344, doi: 10.1016/j.jhin.2009.04.020 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Lyczak JB, Cannon CL & Pier GB Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev 15, 194–222 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pirnay JP et al. Pseudomonas aeruginosa population structure revisited. PLoS One 4, e7740, doi: 10.1371/journal.pone.0007740 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberg AL et al. The importance of bacterial sepsis in intensive care unit patients with acquired immunodeficiency syndrome: implications for future care in the age of increasing antiretroviral resistance. Crit Care Med 29, 548–556 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Rabello LS et al. Clinical outcomes and microbiological characteristics of severe pneumonia in cancer patients: a prospective cohort study. PLoS One 10, e0120544, doi: 10.1371/journal.pone.0120544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassett DJ, Borchers MT & Panos RJ Chronic obstructive pulmonary disease (COPD): evaluation from clinical, immunological and bacterial pathogenesis perspectives. J Microbiol 52, 211–226, doi: 10.1007/s12275-014-4068-2 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Omar A, Wright JB, Schultz G, Burrell R & Nadworny P Microbial biofilms and chronic wounds. Microorganisms 5, 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Loosdrecht MCM, Picioreanu C & Heijnen JJ A more unifying hypothesis for the structure of microbial biofilms. FEMS Microbial Ecology 24, 181–183 (1997). [Google Scholar]
- 25.Breyers JD Modeling biofilm accumulation. In: Physiology Models in Microbiology. Bazin MJ Prosser JI, (eds.) Boca Raton, FL: 2, 109–144 (1988). [Google Scholar]
- 26.Sauer K, Camper AK, Ehrlich GD, Costerton JW & Davies DG Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. Journal of Bacteriology 184, 1140–1154 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrova OE & Sauer K Escaping the biofilm in more than one way: desorption, detachment or dispersion. Current Opinion in Microbiology 30, 67–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies DG Biofilm Dispersion. In Biofilm Highlights; Springer: Berlin, 1–28 (2011). [Google Scholar]
- 29.Purevdorj-Gage B, Costerton WJ & Stoodley P Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151, 1569–1576, doi:doi: 10.1099/mic.0.27536-0 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Otto M Staphylococcal biofilms. Curr Top Microbiol Immunol 322, 207–228, doi: 10.1007/978-3-540-75418-3_10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kostakioti M, Hadjifrangiskou M & Hultgren SJ Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med 3, a010306–a010306, doi: 10.1101/cshperspect.a010306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb JS Differentiation and dispersal in biofilms. The biofilm mode of life: mechanisms and adaptations, 165–174 (2007). [Google Scholar]
- 33.Bjarnsholt T et al. The in vivo biofilm. Trends in Microbiology 21, 466–474 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Serra DO & Hengge R Stress responses go three dimensional–the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environmental Microbiology 16, 1455–1471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR & Lappin-Scott HM Microbial biofilms. Annual Reviews in Microbiology 49, 711–745 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Sternberg C et al. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol 65, 4108–4117 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anwar H, Strap JL, Chen K & Costerton JW Dynamic interactions of biofilms of mucoid Pseudomonas aeruginosa with tobramycin and piperacillin. Antimicrob. Agents Chemother 36, 1208–1214, doi: 10.1128/aac. (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderl JN, Zahller J, Roe F & Stewart PS Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother 47, 1251–1256, doi: 10.1128/aac.47.4.1251-1256.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fux CA, Wilson S & Stoodley P Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. Journal of Bacteriology 186, 4486–4491, doi: 10.1128/jb.186.14.4486-4491.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen D et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986, doi: 10.1126/science.1211037 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spoering AL & Lewis K Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. Journal of Bacteriology 183, 6746–6751, doi: 10.1128/jb.183.23.6746-6751.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keren I, Kaldalu N, Spoering A, Wang Y & Lewis K Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett . 230, 13–18 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Brooun A, Liu S & Lewis K A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother 44, 640–646, doi: 10.1128/aac.44.3.640-646.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keren I, Shah D, Spoering A, Kaldalu N & Lewis K Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. Journal of Bacteriology 186, 8172–8180, doi: 10.1128/jb.186.24.8172-8180.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah D et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6, 53 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campanac C, Pineau L, Payard A, Baziard-Mouysset G & Roques C Interactions between biocide cationic agents and bacterial biofilms. Antimicrob. Agents Chemother 46, 1469–1474, doi: 10.1128/aac.46.5.1469-1474.2002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Picioreanu C, van Loosdrecht MCM & Heijnen JJ Two-dimensional model of biofilm detachment caused by internal stress from liquid flow. Biotechnol. Bioeng 72, 205–218 (2001). [PubMed] [Google Scholar]
- 48.Thormann KM et al. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. Journal of Bacteriology 188, 2681–2691, doi: 10.1128/jb.188.7.2681-2691.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderl JN, Franklin MJ & Stewart PS Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother 44, 1818–1824, doi: 10.1128/aac.44.7.1818-1824.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis K Riddle of biofilm resistance. Antimicrob. Agents Chemother 45, 999–1007, doi: 10.1128/aac.45.4.999-1007.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart PS & Costerton JW Antibiotic resistance of bacteria in biofilms. The Lancet 358, 135–138 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Stewart PS Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother 40, 2517–2522 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drenkard E Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5, 1213–1219, doi: 10.1016/j.micinf.2003.08.009 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Reichhardt C & Parsek MR Confocal laser scanning microscopy for analysis of Pseudomonas aeruginosa biofilm architecture and matrix localization. Frontiers in Microbiology 10, doi: 10.3389/fmicb.2019.00677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doroshenko N et al. Extracellular DNA impedes the transport of vancomycin in Staphylococcus epidermidis biofilms preexposed to subinhibitory concentrations of vancomycin. Antimicrobial Agents and Chemotherapy 58, 7273–7282, doi: 10.1128/aac.03132-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tseng BS et al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environmental Microbiology 15, 2865–2878, doi: 10.1111/1462-2920.12155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chua SL et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyle. Nature Communications 5, 4462, doi: 10.1038/ncomms5462 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Chambers JR, Cherny KE & Sauer K Susceptibility of Pseudomonas aeruginosa dispersed cells to antimicrobial agents is dependent on the dispersion cue and class of the antimicrobial agent used. Antimicrobial Agents and Chemotherapy 61, e00846–00817, doi: 10.1128/aac.00846-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies DG & Marques CNH A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. Journal of Bacteriology 191, 1393–1403, doi: 10.1128/jb.01214-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costerton J Introduction to biofilm. Int J Antimicrob Agents 11, 217–221 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Stewart PS & Franklin MJ Physiological heterogeneity in biofilms. Nature Reviews Microbiology 6, 199 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Haussler S & Fuqua C Biofilms 2012: new discoveries and significant wrinkles in a dynamic field. Journal of Bacteriology 195, 2947–2958 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marques CN, Davies DG & Sauer K Control of Biofilms with the Fatty Acid Signaling Molecule cis-2-Decenoic Acid. Pharmaceuticals 8, 816–835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dow JM et al. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proceedings of the National Academy of Sciences 100, 10995–11000, doi: 10.1073/pnas.1833360100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dean SN, Chung M-C & van Hoek ML Burkholderia diffusible signal factor signals to Francisella novicida to disperse biofilm and increase siderophore production. Applied and Environmental Microbiology 81, 7057–7066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gjermansen M, Ragas P, Sternberg C, Molin S & Tolker-Nielsen T Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol 7, 894–904, doi:doi: 10.1111/j.1462-2920.2005.00775.x (2005). [DOI] [PubMed] [Google Scholar]
- 67.Delille A, Quiles F & Humbert F In situ monitoring of the nascent Pseudomonas fluorescens biofilm response to variations in the dissolved organic carbon level in low-nutrient water by attenuated total reflectance-Fourier transform infrared spectroscopy. Applied and Environmental Microbiology 73, 5782–5788 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schleheck D et al. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. (2009). [DOI] [PMC free article] [PubMed]
- 69.Hunt SM, Werner EM, Huang B, Hamilton MA & Stewart PS Hypothesis for the role of nutrient starvation in biofilm detachment. Applied and Environmental Microbiology 70, 7418–7425 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoodley P, deBeer D & Lewandowski Z Liquid flow in biofilm systems. Appl. Environ. Microbiol 60, 2711–2716 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rasmussen K & Lewandowski Z Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol. Bioeng 59, 302–309, doi: (1998). [DOI] [PubMed] [Google Scholar]
- 72.Petrova OE, Schurr JR, Schurr MJ & Sauer K Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Molecular Microbiology 86, 819–835 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goodwine J et al. Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo. Scientific Reports 9, 3763 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eschbach M et al. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. Journal of Bacteriology 186, 4596–4604, doi: 10.1128/jb.186.14.4596-4604.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schreiber K et al. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. Journal of Bacteriology 188, 659–668, doi: 10.1128/jb.188.2.659-668.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Price-Whelan A, Dietrich LEP & Newman DK Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. Journal of Bacteriology 189, 6372–6381, doi: 10.1128/jb.00505-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leibig M et al. Pyruvate formate lyase acts as a formate supplier for metabolic processes during anaerobiosis in Staphylococcus aureus. Journal of Bacteriology 193, 952–962 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang L et al. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153, 1318–1328, doi: 10.1099/mic.0.2006/004911-0 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Lanter BB, Sauer K & Davies DG Bacteria present in carotid arterial plaques are found as biofilm deposits which may contribute to enhanced risk of plaque rupture. mBio 5, 01206–01214, doi: 10.1128/mBio.01206-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas VC & Hancock LE Suicide and fratricide in bacterial biofilms. The International journal of artificial organs 32, 537–544 (2009). [DOI] [PubMed] [Google Scholar]
- 81.Hentzer M, Eberl L & Givskov M Transcriptome analysis of Pseudomonas aeruginosa biofilm development: anaerobic respiration and iron limitation Biofilms 2, 37–61 (2005). [Google Scholar]
- 82.Sriramulu DD, Lünsdorf H, Lam JS & Römling U Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol 54, 667–676, doi: 10.1099/jmm.0.45969-0 (2005). [DOI] [PubMed] [Google Scholar]
- 83.Webb JS et al. Cell death in Pseudomonas aeruginosa biofilm development. Journal of Bacteriology 185, 4585–4592, doi: 10.1128/jb.185.15.4585-4592.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rice SA et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. Isme J. 3, 271–282 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sutherland IW, Hughes KA, Skillman LC & Tait K The interaction of phage and biofilms. FEMS Microbiology Letters 232, 1–6, doi: 10.1016/s0378-1097(04)00041-2 (2004). [DOI] [PubMed] [Google Scholar]
- 86.Secor PR et al. Biofilm assembly becomes crystal clear–filamentous bacteriophage organize the Pseudomonas aeruginosa biofilm matrix into a liquid crystal. Microbial Cell 3, 49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sauer K et al. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. Journal of Bacteriology 186, 7312–7326, doi: 10.1128/jb.186.21.7312-7326.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barraud N et al. Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms. Microbial biotechnology 2, 370–378 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Purcell EB & Tamayo R Cyclic diguanylate signaling in Gram-positive bacteria. FEMS Microbiology Reviews 40, 753–773, doi: 10.1093/femsre/fuw013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hengge R Principles of c-di-GMP signalling in bacteria. Nature Reviews Microbiology 7, 263–273 (2009). [DOI] [PubMed] [Google Scholar]
- 91.Rybtke MT et al. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Applied and Environmental Microbiology 78, 5060–5069, doi: 10.1128/aem.00414-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nair HA, Periasamy S, Yang L, Kjelleberg S & Rice SA Real time, spatial, and temporal mapping of the distribution of c-di-GMP during biofilm development. Journal of Biological Chemistry 292, 477–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Basu Roy A, Petrova OE & Sauer K The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. Journal of Bacteriology 194, 2904–2915, doi: 10.1128/jb.05346-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gjermansen M, Nilsson M, Yang L & Tolker-Nielsen T Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Molecular Microbiology 75, 815–826, doi: 10.1111/j.1365-2958.2009.06793.x (2010). [DOI] [PubMed] [Google Scholar]
- 95.Morgan R, Kohn S, Hwang S-H, Hassett DJ & Sauer K BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. Journal of Bacteriology 188, 7335–7343 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Y et al. BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathogens 10, e1004168, doi: 10.1371/journal.ppat.1004168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Y, Heine S, Entian M, Sauer K & Frankenberg-Dinkel N NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by a MHYT-domain coupled phosphodiesterase. Journal of Bacteriology 195, 3531–3542, doi: 10.1128/jb.01156-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barraud N et al. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. Journal of Bacteriology 191, 7333–7342, doi: 10.1128/jb.00975-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Christensen LD et al. Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic di-GMP level in the bacteria. Infection and Immunity 81, 2705–2713, doi: 10.1128/iai.00332-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baraquet C & Harwood CS FleQ DNA Binding Consensus Sequence Revealed by Studies of FleQ-Dependent Regulation of Biofilm Gene Expression in Pseudomonas aeruginosa. Journal of Bacteriology 198, 178–186, doi: 10.1128/jb.00539-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Häußler S, Tümmler B, Weißbrodt H, Rohde M & Steinmetz I Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis 29, 621–625, doi: 10.1086/598644 (1999). [DOI] [PubMed] [Google Scholar]
- 102.Häußler S et al. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol 52, 295–301, doi: 10.1099/jmm.0.05069-0 (2003). [DOI] [PubMed] [Google Scholar]
- 103.Proctor RA et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nature Reviews Microbiology 4, 295–305 (2006). [DOI] [PubMed] [Google Scholar]
- 104.Proctor RA, van Langevelde P, Kristjansson M, Maslow JN & Arbeit RD Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis 20, 95–102, doi: 10.1093/clinids/20.1.95 (1995). [DOI] [PubMed] [Google Scholar]
- 105.Drenkard E & Ausubel FM Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416, 740–743 (2002). [DOI] [PubMed] [Google Scholar]
- 106.Chua SL et al. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy 57, 2066–2075, doi: 10.1128/aac.02499-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Basu Roy A & Sauer K Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Molecular Microbiology 94, 771–793, doi: 10.1111/mmi.12802 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cutruzzolà F & Frankenberg-Dinkel N Origin and Impact of Nitric Oxide in Pseudomonas aeruginosa Biofilms. Journal of Bacteriology 198, 55–65, doi: 10.1128/jb.00371-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou L, Zhang L-H, Cámara M & He Y-W The DSF family of quorum sensing signals: diversity, biosynthesis, and turnover. Trends in Microbiology 25, 293–303 (2017). [DOI] [PubMed] [Google Scholar]
- 110.Andrade MO et al. The HD-GYP domain of RpfG mediates a direct linkage between the Rpf quorum-sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv citri. Molecular Microbiology 62, 537–551, doi:doi: 10.1111/j.1365-2958.2006.05386.x (2006). [DOI] [PubMed] [Google Scholar]
- 111.Deng Y et al. Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proceedings of the National Academy of Sciences 109, 15479–15484, doi: 10.1073/pnas.1205037109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Abee T, Kovács ÁT, Kuipers OP & Van der Veen S Biofilm formation and dispersal in Gram-positive bacteria. Current Opinion in Biotechnology 22, 172–179 (2011). [DOI] [PubMed] [Google Scholar]
- 113.Chou S-H & Galperin MY Cyclic di-GMP in Streptomycetes: a new conformation, new binding mode, new receptor, and a new mechanism to control cell development. Molecular cell 77, 443–445, doi: 10.1016/j.molcel.2020.01.018 (2020). [DOI] [PubMed] [Google Scholar]
- 114.Flemming HC & Wingender J The biofilm matrix. Nature Reviews Microbiology 8, 623–633 (2010). [DOI] [PubMed] [Google Scholar]
- 115.Cherny KE & Sauer K Pseudomonas aeruginosa requires the DNA-specific endonuclease EndA to degrade eDNA to disperse from the biofilm. Journal of Bacteriology, JB.00059–00019, doi: 10.1128/jb.00059-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cherny KE & Sauer K Untethering and degradation of the polysaccharide matrix are essential steps in the dispersion response of Pseudomonas aeruginosa biofilms. Journal of Bacteriology 202, e00575–00519, doi: 10.1128/jb.00575-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rybtke M et al. The LapG protein plays a role in Pseudomonas aeruginosa biofilm formation by controlling the presence of the CdrA adhesin on the cell surface. MicrobiologyOpen (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Borlee BR et al. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Molecular Microbiology 75, 827–842, doi: 10.1111/j.1365-2958.2009.06991.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Monds RD, Newell PD, Gross RH & O’Toole GA Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Molecular Microbiology 63, 656–679, doi:doi: 10.1111/j.1365-2958.2006.05539.x (2007). [DOI] [PubMed] [Google Scholar]
- 120.Reichhardt C, Wong C, Passos da Silva D, Wozniak DJ & Parsek MR CdrA Interactions within the Pseudomonas aeruginosa Biofilm Matrix Safeguard It from Proteolysis and Promote Cellular Packing. mBio 9, e01376–01318, doi: 10.1128/mBio.01376-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Devaraj A et al. The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday junction recombination intermediates. Proceedings of the National Academy of Sciences 116, 25068–25077, doi: 10.1073/pnas.1909017116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Steinberger RE & Holden PA Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol 71, 5404–5410, doi: 10.1128/aem.71.9.5404-5410.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Whitchurch CB, Tolker-Nielsen T, Ragas PC & Mattick JS Extracellular DNA required for bacterial biofilm formation. Science 295, 1487, doi: 10.1126/science.295.5559.1487 (2002). [DOI] [PubMed] [Google Scholar]
- 124.Nijland R, Hall MJ & Burgess JG Dispersal of biofilms by secreted, matrix degrading, bacterial DNase. PLoS ONE 5, e15668, doi: 10.1371/journal.pone.0015668 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Boles BR & Horswill AR Staphylococcal biofilm disassembly. Trends in Microbiology 19, 449–455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gödeke J, Heun M, Bubendorfer S, Paul K & Thormann KM Roles of two Shewanella oneidensis MR-1 extracellular endonucleases. Applied and Environmental Microbiology 77, 5342–5351, doi: 10.1128/aem.00643-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McDougald D, Rice SA, Barraud N, Steinberg PD & Kjelleberg S Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nature Reviews Microbiology 10, 39–50 (2012). [DOI] [PubMed] [Google Scholar]
- 128.Pestrak MJ et al. Treatment with the Pseudomonas aeruginosa glycoside hydrolase PslG combats wound infection by improving antibiotic efficacy and host innate immune activity. Antimicrobial Agents and Chemotherapy 63, e00234–00219, doi: 10.1128/aac.00234-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baker P et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Science Advances 2, e1501632, doi: 10.1126/sciadv.1501632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yu S et al. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Research 25, 1352–1367, doi: 10.1038/cr.2015.129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kaplan JB, Ragunath C, Ramasubbu N & Fine DH Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. Journal of Bacteriology 185, 4693–4698 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kaplan JB, Ragunath C, Velliyagounder K, Fine DH & Ramasubbu N Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother 48, 2633–2636, doi: 10.1128/aac.48.7.2633-2636.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chaignon P et al. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Applied Microbiology and Biotechnology 75, 125–132, doi: 10.1007/s00253-006-0790-y (2007). [DOI] [PubMed] [Google Scholar]
- 134.Boles BR & Horswill AR agr mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathogens 4, e1000052, doi: 10.1371/journal.ppat.1000052 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chua SL et al. In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation. Nature Protocols 10, 1165–1180, doi: 10.1038/nprot.2015.067 (2015). [DOI] [PubMed] [Google Scholar]
- 136.Jones CJ et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathogens 10, e1003984, doi: 10.1371/journal.ppat.1003984 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu B et al. The Pseudomonas aeruginosa AmrZ C-terminal domain mediates tetramerization and is required for its activator and repressor functions. Environmental Microbiology Reports 8, 85–90, doi:doi: 10.1111/1758-2229.12354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jones CJ AmrZ is a central regulator of biofilm formation in Pseudomonas aeruginosa. The Ohio State University, ProQuest Dissertations Publishing, Dissertation. (2013). [Google Scholar]
- 139.Woo JK, Webb JS, Kirov SM, Kjelleberg S & Rice SA Biofilm dispersal cells of a cystic fibrosis Pseudomonas aeruginosa isolate exhibit variability in functional traits likely to contribute to persistent infection. FEMS Immunol Med Microbiol 66, 251–264, doi: 10.1111/j.1574-695X.2012.01006.x (2012). [DOI] [PubMed] [Google Scholar]
- 140.Hall-Stoodley L & Stoodley P Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol 13, 7–10, doi: 10.1016/j.tim.2004.11.004 (2005). [DOI] [PubMed] [Google Scholar]
- 141.Rollet C, Gal L & Guzzo J Biofilm-detached cells, a transition from a sessile to a planktonic phenotype: a comparative study of adhesion and physiological characteristics in Pseudomonas aeruginosa. FEMS Microbiol Lett 290, 135–142, doi: 10.1111/j.1574-6968.2008.01415.x (2009). [DOI] [PubMed] [Google Scholar]
- 142.Uppuluri P et al. Candida albicans Dispersed Cells Are Developmentally Distinct from Biofilm and Planktonic Cells. MBio 9, doi: 10.1128/mBio.01338-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu J, Ling JQ, Zhang K & Wu CD Physiological properties of Streptococcus mutans UA159 biofilm-detached cells. FEMS Microbiol Lett 340, 11–18, doi: 10.1111/1574-6968.12066 (2013). [DOI] [PubMed] [Google Scholar]
- 144.Guilhen C et al. Transcriptional profiling of Klebsiella pneumoniae defines signatures for planktonic, sessile and biofilm-dispersed cells. BMC Genomics 17, 237, doi: 10.1186/s12864-016-2557-x (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vaysse PJ, Sivadon P, Goulas P & Grimaud R Cells dispersed from Marinobacter hydrocarbonoclasticus SP17 biofilm exhibit a specific protein profile associated with a higher ability to reinitiate biofilm development at the hexadecane-water interface. Environ Microbiol 13, 737–746, doi: 10.1111/j.1462-2920.2010.02377.x (2011). [DOI] [PubMed] [Google Scholar]
- 146.Fleming D & Rumbaugh K The Consequences of Biofilm Dispersal on the Host. Scientific Reports 8, 10738, doi: 10.1038/s41598-018-29121-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fleming D & Rumbaugh K Approaches to Dispersing Medical Biofilms. Microorganisms 5, 15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Waryah CB et al. In Vitro Antimicrobial Efficacy of Tobramycin Against Staphylococcus aureus Biofilms in Combination With or Without DNase I and/or Dispersin B: A Preliminary Investigation. Microb Drug Resist 23, 384–390, doi: 10.1089/mdr.2016.0100 (2017). [DOI] [PubMed] [Google Scholar]
- 149.Izano EA, Wang H, Ragunath C, Ramasubbu N & Kaplan JB Detachment and killing of Aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J Dent Res 86, 618–622, doi: 10.1177/154405910708600707 (2007). [DOI] [PubMed] [Google Scholar]
- 150.Gawande PV, Leung KP & Madhyastha S Antibiofilm and antimicrobial efficacy of DispersinB(R)-KSL-W peptide-based wound gel against chronic wound infection associated bacteria. Curr Microbiol 68, 635–641, doi: 10.1007/s00284-014-0519-6 (2014). [DOI] [PubMed] [Google Scholar]
- 151.Itoh Y, Wang X, Hinnebusch BJ, Preston JF 3rd & Romeo T Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol 187, 382–387, doi: 10.1128/JB.187.1.382-387.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gawande PV et al. Antibiofilm Efficacy of DispersinB((R)) Wound Spray Used in Combination with a Silver Wound Dressing. Microbiol Insights 7, 9–13, doi: 10.4137/MBI.S13914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kaplan JB et al. Extracellular polymeric substance (EPS)-degrading enzymes reduce staphylococcal surface attachment and biocide resistance on pig skin in vivo. PLoS One 13, e0205526, doi: 10.1371/journal.pone.0205526 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Darouiche RO, Mansouri MD, Gawande PV & Madhyastha S Antimicrobial and antibiofilm efficacy of triclosan and DispersinB combination. J Antimicrob Chemother 64, 88–93, doi: 10.1093/jac/dkp158 (2009). [DOI] [PubMed] [Google Scholar]
- 155.Barraud N, J Kelso M, A Rice S & Kjelleberg S Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Current pharmaceutical design 21, 31–42 (2015). [DOI] [PubMed] [Google Scholar]
- 156.Sepehr S, Rahmani-Badi A, Babaie-Naiej H & Soudi MR Unsaturated fatty acid, cis-2-decenoic acid, in combination with disinfectants or antibiotics removes pre-established biofilms formed by food-related bacteria. PLoS One 9, e101677, doi: 10.1371/journal.pone.0101677 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rahmani-Badi A et al. A combination of cis-2-decenoic acid and antibiotics eradicates pre-established catheter-associated biofilms. J Med Microbiol 63, 1509–1516, doi: 10.1099/jmm.0.075374-0 (2014). [DOI] [PubMed] [Google Scholar]
- 158.Rahmani-Badi A, Sepehr S & Babaie-Naiej H A combination of cis-2-decenoic acid and chlorhexidine removes dental plaque. Arch Oral Biol 60, 1655–1661, doi: 10.1016/j.archoralbio.2015.08.006 (2015). [DOI] [PubMed] [Google Scholar]
- 159.Harris MA, Beenken KE, Smeltzer MS, Haggard WO & Jennings JA Phosphatidylcholine Coatings Deliver Local Antimicrobials and Reduce Infection in a Murine Model: A Preliminary Study. Clin Orthop Relat Res 475, 1847–1853, doi: 10.1007/s11999-016-5211-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Choi M et al. Chitosan-based nitric oxide-releasing dressing for anti-biofilm and in vivo healing activities in MRSA biofilm-infected wounds. Int J Biol Macromol, doi: 10.1016/j.ijbiomac.2019.10.009 (2019). [DOI] [PubMed] [Google Scholar]
- 161.Jardeleza C et al. An in vivo safety and efficacy demonstration of a topical liposomal nitric oxide donor treatment for Staphylococcus aureus biofilm-associated rhinosinusitis. Transl Res 166, 683–692, doi: 10.1016/j.trsl.2015.06.009 (2015). [DOI] [PubMed] [Google Scholar]
- 162.Wo Y et al. Reduction of Thrombosis and Bacterial Infection via Controlled Nitric Oxide (NO) Release from S-Nitroso-N-acetylpenicillamine (SNAP) Impregnated CarboSil Intravascular Catheters. ACS Biomater Sci Eng 3, 349–359, doi: 10.1021/acsbiomaterials.6b00622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mihu MR et al. Sustained Nitric Oxide-Releasing Nanoparticles Interfere with Methicillin-Resistant Staphylococcus aureus Adhesion and Biofilm Formation in a Rat Central Venous Catheter Model. Antimicrob Agents Chemother 61, doi: 10.1128/AAC.02020-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tao F, Swarup S & Zhang LH Quorum sensing modulation of a putative glycosyltransferase gene cluster essential for Xanthomonas campestris biofilm formation. Environmental Microbiology 12, 3159–3170 (2010). [DOI] [PubMed] [Google Scholar]
- 165.An S, Wu J. e. & Zhang L-H Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl. Environ. Microbiol 76, 8160–8173, doi: 10.1128/aem.01233-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Petrova OE, Cherny KE & Sauer K The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. Journal of Bacteriology 197.1, 174–187, doi: 10.1128/jb.02244-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Petrova OE & Sauer K Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proceedings of the National Academy of Sciences 109 16690–16695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Marks LR, Davidson BA, Knight PR & Hakansson AP Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 4, e00438–00413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rice S et al. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. Journal of Bacteriology 187, 3477–3485 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Barraud N et al. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. Journal of Bacteriology 188, 7344–7353, doi: 10.1128/jb.00779-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schmidt I, Steenbakkers PJ, op den Camp HJ, Schmidt K & Jetten MS Physiologic and proteomic evidence for a role of nitric oxide in biofilm formation by Nitrosomonas europaea and other ammonia oxidizers. Journal of Bacteriology 186, 2781–2788 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Musk DJ, Banko DA & Hergenrother PJ Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chemistry & biology 12, 789–796 (2005). [DOI] [PubMed] [Google Scholar]
- 173.Mikkelsen H, Hui K, Barraud N & Filloux A The pathogenicity island encoded PvrSR/RcsCB regulatory network controls biofilm formation and dispersal in Pseudomonas aeruginosa PA14. Molecular Microbiology 89, 450–463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kragh KN et al. Role of multicellular aggregates in biofilm formation. mBio 7, e00237–00216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Naumoff D Hierarchical classification of glycoside hydrolases. Biochemistry (Moscow) 76, 622–635 (2011). [DOI] [PubMed] [Google Scholar]
- 176.Fleming D, Chahin L & Rumbaugh K Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrobial Agents and Chemotherapy 61, e01998–01916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mulcahy H, Charron-Mazenod L & Lewenza S Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environmental Microbiology 12, 1621–1629, doi:doi: 10.1111/j.1462-2920.2010.02208.x (2010). [DOI] [PubMed] [Google Scholar]
- 178.Klausen M, Aaes-Jørgensen A, Molin S & Tolker-Nielsen T Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Molecular Microbiology 50, 61–68 (2003). [DOI] [PubMed] [Google Scholar]


