Abstract
Control of the translational repressor, PHAS-I, was investigated by expressing proteins with Ser/Thr → Ala mutations in the five (S/T)P phosphorylation sites. Results of experiments with HEK293 cells reveal at least three levels of control. At one extreme is nonregulated phosphorylation, exemplified by constitutive phosphorylation of Ser82. At an intermediate level, amino acids and insulin stimulate the phosphorylation of Thr36, Thr45, and Thr69 via mTOR-dependent processes that function independently of other sites in PHAS-I. At the third level, the extent of phosphorylation of one site modulates the phosphorylation of another. This control is represented by Ser64 phosphorylation, which depends on the phosphorylation of all three TP sites. The five sites have different influences on the electrophoretic properties of PHAS-I and on the affinity of PHAS-I for eukaryotic initiation factor 4E (eIF4E). Phosphorylation of Thr45 or Ser64 results in the most dramatic decreases in eIF4E binding in vitro. However, each of the sites influences mRNA translation, either directly by modulating the binding affinity of PHAS-I and eIF4E or indirectly by affecting the phosphorylation of other sites.
PHAS-I, also known as 4E-BP1, is a 117-amino-acid (Mr ≈ 12,400) member of a family of eukaryotic initiation factor 4E (eIF4E)-binding proteins that are involved in the control of mRNA translation (19, 29, 36). In eukaryotic cells, the 5′ end of almost all mRNA is capped with m7GpppN (where N is any nucleotide) (35). The availability of eIF4E, the mRNA cap-binding protein, limits the rate of translation initiation (31, 36). To facilitate translation, eIF4E must bind eIF4G, a scaffolding protein that also binds eIF4A, an ATP-dependent helicase that allows more efficient translation of mRNAs with structured 5′ untranslated regions. The complex of eIF4E, eIF4G, and eIF4A increases translation by increasing the efficiency of binding and/or scanning by the 40S ribosomal subunit, which is linked to eIF4G by eIF3. Nonphosphorylated PHAS-I binds eIF4E and prevents eIF4E from binding to eIF4G (13). Thus, when overexpressed in cells, PHAS-I inhibits the translation of capped mRNA but does not inhibit cap-independent translation initiated from a viral internal ribosomal entry site (IRES) (27). When PHAS-I is phosphorylated in the appropriate site(s), the PHAS-I–eIF4E complex dissociates (20, 27), freeing eIF4E to bind eIF4G and thereby increasing translation initiation.
Five phosphorylation sites conforming to a (S/T)P motif have been identified in PHAS-I (8). The sites surround the residues (Arg50 to Met59) that have been implicated in eIF4E binding (23) and are conserved in the two other PHAS family members (28). Another site, Ser111, found only in PHAS-I, is phosphorylated in vitro by casein kinase II (9, 15). Phosphorylation decreases the electrophoretic mobility of PHAS-I, and three electrophoretic forms, designated α, β, and γ in order of decreasing mobility, are typically resolved by polyacrylamide gel electrophoresis (PAGE) in the presence of sodium dodecyl sulfate (SDS) (20). The sites that are most important in controlling electrophoretic mobility, eIF4E binding, and mRNA translation in cells have not been determined.
PHAS-I is controlled by the mammalian target of rapamycin (mTOR) signaling pathway. The effects of amino acids (14, 17, 39) and/or insulin (3, 21) on increasing the phosphorylation of PHAS-I are attenuated by rapamycin, which inhibits the function of TOR proteins (1). Expressing mTOR in cells increases the phosphorylation of PHAS-I, and mTOR is able to phosphorylate PHAS-I in vitro (4–6, 10, 14). It was recently proposed that the phosphorylation of PHAS-I in cells occurs by an ordered mechanism in which the phosphorylation of Thr36 and Thr45 by mTOR must occur before the other sites in PHAS-I can be phosphorylated (10).
In the present study, PHAS-I proteins having mutations in the five (S/T)P sites were overexpressed in HEK293 cells to investigate the role of the different phosphorylation sites. The results allow the identification of sites that are responsible for the gel shift and that are important in regulating eIF4E binding and mRNA translation. In addition, we demonstrate definitively that Thr36 and Thr45 do not need to be phosphorylated in cells for phosphorylation of Thr69 and Ser82 to occur.
MATERIALS AND METHODS
PHAS-I expression vectors.
Ser64 was mutated to Cys, Asp, Glu, Asn, and Thr in wild-type (WT) PHAS-I cDNA, and Ala64 in 5A PHAS-I cDNA was mutated to Glu and Asp, by oligonucleotide-directed mutagenesis as described previously for the preparation of cDNA encoding PHAS-I proteins with Ser/Thr to Ala mutations in the five (S/T)P sites (40). To disrupt eIF4E binding, Ala mutations of Leu and Met at positions 58 and 59 were created (23). The coding region of each plasmid was sequenced and found to be free of undesired mutations. For expression in cells, PHAS-I cDNA was excised from pBluescript SK(−) with HindIII and XbaI and inserted between the HindIII and XbaI sites of pCMV4 (2).
Cell culture and transfections.
Human embryonic kidney 293 (HEK293) cells (ATTC CRL 1573) were seeded at 2 × 104 cells/cm2 and cultured for 24 h in growth medium composed of 10% (vol/vol) horse serum in Dulbecco's modified Eagle's medium (DMEM). Transfections were performed using a method involving calcium phosphate precipitation (7). To obtain equal levels of expression, it was necessary to use different amounts of the respective pCMV4 constructs (0.3 μg/cm2 for WT and A64 PHAS-I, and 0.1 μg/cm2 for all others). The precipitated DNA was removed after 16 h, and the cells were incubated in growth medium for 5 h and then used in experiments as described below.
32P-labeling experiments.
Two protocols were used to label cells with 32P. Protocol 1 was used to assess the acute regulation of phosphorylation. To induce a quiescent state, transfected cells (in 35- or 60-mm-diameter dishes) were incubated without serum in DMEM supplemented with 0.2% bovine serum albumin (BSA). After 15 h, the cells were rinsed once and incubated in low-phosphate (Pi) buffer (0.2 ml/cm2) supplemented with 0.5 mCi of 32Pi (ICN Pharmaceuticals) per ml. Low-Pi buffer contained 145 mM NaCl, 5.4 mM KCl, 1.4 mM CaCl2, 1.4 mM MgSO4, 0.1 mM sodium phosphate, 10 mM sodium HEPES, 25 mM NaHCO3, 5 mM glucose, and 0.5% BSA (pH 7.4). The incubations were terminated after 3 h. Where indicated, rapamycin (final concentration, 50 nM) (Calbiochem) was added after 90 min and recombinant human insulin (final concentration, 700 nM) and/or amino acids (25-fold dilution of 50× minimal essential medium amino acids plus l-glutamine) (GIBCO BRL) were added after 150 min. To terminate the incubation, cells were rinsed with buffer A (150 mM NaCl, 10 mM EDTA, 10 mM sodium pyrophosphate, 2 mM sodium orthovanadate, 100 mM NaF, 50 mM sodium HEPES [pH 7.5]) and then lysed at 0°C in buffer A (30 μl/cm2) supplemented with 1% NP-40, 200 nM microcystin-LR, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml, and 1 mM phenylmethylsulfonyl fluoride.
Protocol 2 was used to assess the phosphorylation of PHAS-I proteins under the conditions used to investigate mRNA translation. The transfected cells were incubated for 15 h in DMEM supplemented with 0.2% BSA and 700 nM insulin. This relatively high concentration of insulin, at which the receptors for both insulin and insulinlike growth factor type 1 are activated, was used to attempt to maximally stimulate the phosphorylation of PHAS-I. The medium was then replaced with phosphate-free DMEM that was supplemented with BSA, insulin, and 0.1 mM Na32Pi. After 3 h, the incubations were terminated as described for protocol 1.
Immunoprecipitation of PHAS-I and affinity purification of PHAS-I–eIF4E complexes.
PHAS-I–eIF4E complexes were partially purified by using m7GTP-Sepharose beads, and PHAS-I was immunoprecipitated as described previously (20). Samples were subjected to SDS-PAGE (18). PHAS-I and eIF4E were identified by immunoblotting (22). Binding of PHAS-I proteins to eIF4E in vitro was assessed by far Western analyses using a 32P-labeled FLAG-eIF4E fusion protein (27). 32P-labeled proteins were detected by autoradiography or phosphorimaging.
Phosphopeptide mapping of PHAS-I.
PHAS-I immune complexes were suspended in 50 μl of 20 mM EDTA–0.1% 2-mercaptoethanol–50 mM Tris-HCl (pH 6.8) and incubated at 100°C for 15 min. PHAS-I proteins, which are relatively heat stable, were recovered in the supernatants after centrifuging the samples at 16,000 × g for 10 min. Samples were incubated at 37°C with lysyl endopeptidase (20 μg/ml) for 15 h, and the resulting phosphopeptides were resolved by high-pressure liquid chromatography (HPLC) as described previously (8).
Preparation and phosphorylation of recombinant PHAS-I proteins.
PHAS-I proteins were expressed in bacteria and purified (40). The PHAS-I proteins (20 μg/ml) were incubated with recombinant mitogen-activated protein (MAP) kinase (7 μg/ml), activated as described previously (8), in 1 mM dithiothreitol–7.5 mM MgCl2–40 mM sodium HEPES (pH 7.4) with either 0.5 mM ATP (for FLAG-eIF4E-binding experiments) or 0.5 mM [γ-32P]ATP (to measure phosphorylation).
Measurements of cap-dependent and -independent mRNA translation.
A bicistronic reporter, designated pRLIRESFL, encoding Renilla luciferase and firefly luciferase was used to investigate cap-dependent mRNA translation. To make pRLIRESFL, a cDNA fragment including the Renilla luciferase coding region and stop codon was generated using PCR with pRL-CMV (Promega) as template and primers that introduced EcoRI sites in both ends. After digestion with EcoRI, the fragment was inserted into the EcoRI cloning site of p2332 (provided by John Majors, Washington University). The p2332 plasmid contains the promoter from cytomegalovirus upstream of the EcoRI site and contains sequences encoding the IRES from encephalomyocarditis virus followed by firefly luciferase and the simian virus 40 t-intron and polyadenylation signal downstream of the EcoRI site. pRLIRESFL directs the synthesis of an mRNA from which Renilla luciferase is translated in a manner dependent on the 5′ cap and firefly luciferase is translated in a cap-independent manner through the IRES.
For expression studies, HEK293 cells in 22-mm-diameter dishes were cotransfected with pRLIRESFL (0.05 μg/dish) and the pCMV4–PHAS-I constructs. DNA precipitates were removed after 16 h, and the cells were incubated in growth medium for 5 h. The medium was then replaced with DMEM supplemented with 0.2% BSA, and the cells were incubated for 15 h with 700 nM insulin. To terminate the incubation, the cells were rinsed with phosphate-buffered saline and incubated at 0°C for 15 min in 250 μl of passive lysis buffer (Promega) supplemented with 10 μg of leupeptin per ml, 10 μg of aprotinin per ml, 1 mM PMSF, and 200 nM microcystin-LR. Lysates were centrifuged at 16,000 × g for 15 min at 4°C. The activities of the two luciferases were measured in triplicate samples of the supernatants by using the dual-luciferase reporter assay system (Promega).
RESULTS
Influence of phosphorylation site mutations on electrophoretic mobility and phosphorylation.
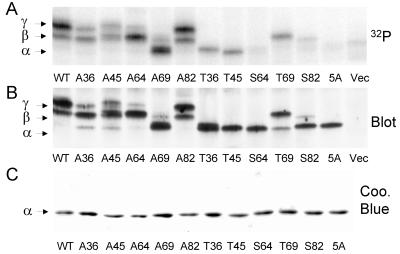
To investigate the roles of different phosphorylation sites in controlling PHAS-I, Ser/Thr→Ala mutations were introduced into the five (S/T)P sites (Fig. 1). HEK293 cells expressing PHAS-I proteins were incubated with 32Pi and a maximally effective concentration of insulin. The proteins were immunoprecipitated and subjected to SDS-PAGE. After transfer to a membrane, an autoradiogram (Fig. 2A) and PHAS-I immunoblot (Fig. 2B) were prepared. No PHAS-I was detected in extracts from cells that had been transfected with the pCMV4 vector alone (Fig. 2B). Therefore, we were able to use PHAS-I antibody to identify the overexpressed proteins, obviating the need for an epitope tag, which could affect the properties of the relatively small PHAS-I protein. In both the autoradiogram and immunoblot, WT PHAS-I appeared as two bands, corresponding to the β and γ forms. No 32P was incorporated into 5A PHAS-I (Fig. 2A), and 5A PHAS-I migrated as a single band corresponding to the α form (Fig. 2B), as did all of the nonphosphorylated mutant proteins purified from bacteria (Fig. 2C).
FIG. 1.
Mutant PHAS-I proteins. Ser/Thr → Ala or other mutations in the five (S/T)P phosphorylation sites in PHAS-I are indicated by bold type.
FIG. 2.
Effect of mutations on phosphorylation and electrophoretic mobility. PHAS-I proteins were expressed in HEK293 cells, which were incubated with 32Pi and insulin (see protocol 2 in Materials and Methods). The proteins were immunoprecipitated, subjected to SDS-PAGE, and transferred to an Immobilon membrane. (A) Phosphorimage showing 32P-labeled PHAS-I proteins. (B) PHAS-I immunoblot of the membrane. (C) Coomassie blue stain of recombinant PHAS-I proteins purified from bacteria.
The mobilities of the mutant proteins from HEK293 cells differed markedly, due to differences in the sites of phosphorylation. The 32P content in A36 PHAS-I (Fig. 2A) and the relative amount of the protein found in the γ form (Fig. 2B) were lower than in WT PHAS-I. The mobility of A36 PHAS-Iβ was slightly higher than the mobility of WT PHAS-Iβ (Fig. 2A). As might be expected, phosphorylation of T36 PHAS-I slightly decreased its mobility (Fig. 2A). However, phosphorylation of T36 did not result in the appearance of β or γ forms, indicating that phosphorylation of sites other than Thr36 are needed to generate these forms. The effects of mutating Thr45 were similar to those of mutating Thr36. However, the mobility of A45 PHAS-Iβ was indistinguishable from that of WT PHAS-Iβ. T45 PHAS-I was phosphorylated in cells, but the protein remained in the α form. Thus, phosphorylation of Thr45 appears insufficient to affect the mobility of PHAS-I. A64 PHAS-I was phosphorylated when expressed in HEK293 cells. Interestingly, almost all of the protein was found in the β form, although a small amount of γ form was detected. These findings suggest that Ser64 phosphorylation is needed for the optimum generation of the fully retarded electrophoretic form of PHAS-I. Mutating Thr69 had the most pronounced influence on the electrophoretic mobility of PHAS-I. A69 PHAS-I was phosphorylated in HEK293 cells, but almost all of it remained in the α form. T69 PHAS-I not only was phosphorylated but also accumulated in the β form, indicating that the α-to-β shift is due to phosphorylation of T69. The fact that the mobility of nearly all of T69 PHAS-I was retarded indicates that Thr69 was almost completely phosphorylated. Thus, it is clear that phosphorylation of Thr69 is not dependent on the phosphorylation of Thr36 and/or Thr45. The 32P content and electrophoretic pattern of A82 PHAS-I were very similar to those of WT PHAS-I. Thus, Ser82 does not appear to be phosphorylated as highly as the other sites. As might be expected, the phosphorylation of S82 PHAS-I was also relatively low in HEK293 cells. The phosphorylated form of S82 PHAS-I exhibited reduced electrophoretic mobility resembling that of recombinant S82 PHAS-I phosphorylated by MAP kinase in vitro (40). However, the influence of Ser82 phosphorylation on mobility was less than that of Thr69 phosphorylation.
Effect of mutations on the phosphorylation of Ser64 and Thr69.
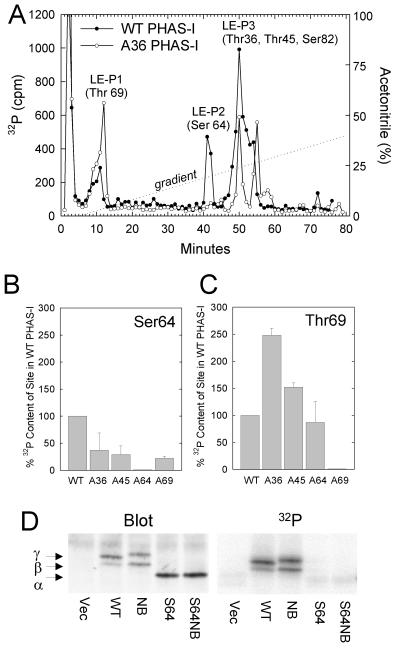
S64 PHAS-I was not appreciably phosphorylated (Fig. 2A). To determine whether HEK293 cells lacked the kinase to phosphorylate Ser64 or whether the phosphorylation of Ser64 was dependent on the phosphorylation of other sites, WT PHAS-I was immunoprecipitated from 32P-labeled cells and digested with lysyl endopeptidase. Three major peaks, labeled LE-P1, LE-P2, and LE-P3, were observed after fractionating the digest by reverse-phase HPLC (Fig. 3A). The phosphopeptides found in these peaks were previously identified by amino acid sequencing (8). LE-P1 contains the Thr69 peptide, LE-P2 contains the Ser64 peptide, and LE-P3 contains a mixture of peptides including one with Ser82 and another with both Thr36 and Thr45. The presence of 32P in LE-P2 indicated that Ser64 was phosphorylated in wild-type PHAS-I.
FIG. 3.
Evidence of interactions among phosphorylation sites. HEK293 cells expressing WT PHAS-I, A36 PHAS-I, A45 PHAS-I, or A69 PHAS-I were incubated with 32Pi and insulin (protocol 2). PHAS-I proteins were immunoprecipitated and digested with lysyl endopeptidase. The digests were fractionated by HPLC. (A) Elution profile of 32P-labeled peptides derived from WT PHAS-I and A36 PHAS-I. The three major peaks of 32P are designated LE-P1, LE-P2, and LE-P3. The phosphorylation sites previously identified (8) in these peaks are shown in parentheses. (B) Decreased phosphorylation of Ser64 in mutants lacking any one of the three TP sites. The amounts of 32P in the Ser64 peaks from the mutant proteins are expressed as percentages of the 32P content of the Ser64 peak from WT PHAS-I. The results are mean values plus half the range from two experiments. (C) Enhanced phosphorylation of Thr69 in mutants lacking Thr36 and Thr45. The 32P contents of the Thr69 peaks from the mutant proteins were determined and are expressed relative to that from WT PHAS-I. (D) Effect of disrupting the eIF4E-binding domain on PHAS-I phosphorylation. Cells were transfected with pCMV4 alone (Vec) or with constructs encoding WT PHAS-I, NBPHAS-I, S64 PHAS-I, or S64 NBPHAS-I and incubated with 32P and insulin, and the PHAS-I proteins were immunoprecipitated. A PHAS-I blot (Blot) and phosphorimage (32P) are presented.
Experiments with single Ala mutants of PHAS-I were performed to determine which of the other sites contribute to the control of Ser64 phosphorylation. The initial fractions in LE-P3 contain the Ser82 peptide and the doubly phosphorylated Thr36 and Thr45 peptide. Mutation of either Thr36 (Fig. 3A) or Thr45 (results not shown) allows the resolution of two peaks in LE-P3: the phosphorylated Ser82 peptide, which elutes at 50 min, and the singly phosphorylated Thr36 or Thr45 peptide, which elutes 5 min later. The phosphorylation of Ser64 in A36 PHAS-I was much lower than in WT PHAS-I (Fig. 3A). However, mutation of either Thr45 or Thr69 also decreased Ser64 phosphorylation (Fig. 3B), indicating that phosphorylation of all three TP sites is needed for optimal phosphorylation of Ser64. Interestingly, Thr69 in A36 PHAS-I contained over twice as much 32P as the site in WT PHAS-I does (Fig. 3A and C). Thr69 phosphorylation was also increased after Thr45 was mutated, although the effect was smaller than that of mutating Thr36 (Fig. 3C). An implication is that phosphorylation of Thr36 and/or Thr45 enhances the phosphorylation of Thr69.
In vitro MAP kinase rapidly phosphorylates Ser64 in free PHAS-I (8); however, binding to eIF4E dramatically inhibits phosphorylation by MAP kinase (21). Thus, when PHAS-I is complexed with eIF4E, the Ser64 site appears to be less accessible, at least to certain kinases. To investigate the possibility that phosphorylation of Ser64 in cells occurred secondarily to dissociation of the PHAS-I–eIF4E complex, we performed experiments with a S64 NBPHAS-I. This protein has Leu58 → Ala and Met59 → Ala mutations, which have been shown to abolish high-affinity binding of WT PHAS-I to eIF4E (24). Control experiments confirmed that the mutations inhibited the binding of S64 PHAS-I to eIF4E (I. Mothe-Satney and J. C. Lawrence, Jr., unpublished data). Disrupting eIF4E binding per se did not prevent phosphorylation of PHAS-I, since the nonbinding form of the protein having the five (S/T)P sites was phosphorylated as well as WT PHAS-I (Fig. 3D). The nonbinding form of S64 PHAS-I was not phosphorylated, and its electrophoretic mobility was identical to that of S64 PHAS-I (Fig. 3D).
Control of PHAS-I phosphorylation by amino acids, insulin, and rapamycin.
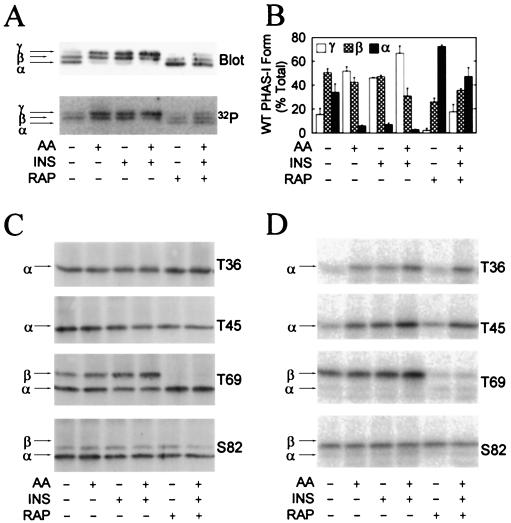
To investigate hormonal control of PHAS-I, HEK293 cells were incubated without amino acids or serum to induce quiescence. Under these conditions, most of the WT PHAS-I was found in the α and β forms (Fig. 4A). Supplying amino acids or insulin decreased the amount of the α form and increased that of the γ form. The combination of amino acids plus insulin almost abolished the α form, and more than 60% of PHAS-I was present in the γ form (Fig. 4B). Rapamycin decreased the levels of the β and γ forms and markedly increased that of the α form. The mobility changes are consistent with the established effects of insulin and amino acids on increasing the phosphorylation of PHAS-I and of rapamycin on decreasing the phosphorylation of the protein (19). The results of experiments in which the 32P content of PHAS-I was determined were also consistent (Fig. 4A and 5A). However, the effects of insulin and rapamycin, as well as those of amino acids, on 32P labeling of PHAS-I (Fig. 5A) were smaller than suggested by the dramatic changes in the electrophoretic mobility of the protein (Fig. 4A). These findings indicated that the sites having the most pronounced influence on electrophoretic mobility were affected to a greater extent than the sites that do not affect mobility. To investigate this hypothesis, 32P-labeling experiments were conducted with cells expressing T36 PHAS-I, T45 PHAS-I, T69 PHAS-I, or S82 PHAS-I. The phosphorylation of T36 PHAS-I was increased approximately 2.5-fold by incubating cells with either insulin or amino acids (Fig. 4D). The effect of the combination was approximately equal to the additive effects of the two treatments (Fig. 5B). Rapamycin attenuated the effects of insulin plus amino acids on increasing the phosphorylation of T36 PHAS-I but had no effect on the basal level of phosphorylation. The effects of insulin, amino acids, and rapamycin on the phosphorylation of T45 PHAS-I closely resembled the effects of these agents on T36 PHAS-I (Fig. 5B and C).
FIG. 4.
Stimulation of PHAS-I phosphorylation by amino acids and insulin. Cells expressing WT PHAS-I, T36 PHAS-I, T45 PHAS-I, T69 PHAS-I, or S82 PHAS-I were incubated with 32Pi (see protocol 1 in Materials and Methods) for 3 h before extracts were prepared. Prior to terminating the incubations, the cells were treated as follows: no additions, amino acids [AA] and/or insulin [INS] for 30 min, rapamycin [RAP] for 90 min, and rapamycin for 60 min followed by rapamycin plus insulin and amino acids for 30 min. PHAS-I proteins were immunoprecipitated from extracts, subjected to SDS-PAGE, and transferred to Immobilon membranes. (A) Immunoblot depicting the α, β, and γ forms of WT PHAS-I (Blot) and a phosphorimage showing the 32P-labeled protein (32P). (B) Relative proportions of α, β, and γ of WT PHAS-I. The optical density of each of the forms was determined by scanning laser densitometry and is expressed as a percentage of the total. Means and standard errors from three experiments are presented. (C) Immunoblots showing T36 PHAS-I, T45 PHAS-I, T69 PHAS-I, and S82 PHAS-I. (D) Phosphorimages showing the 32P-labeled mutant proteins.
FIG. 5.
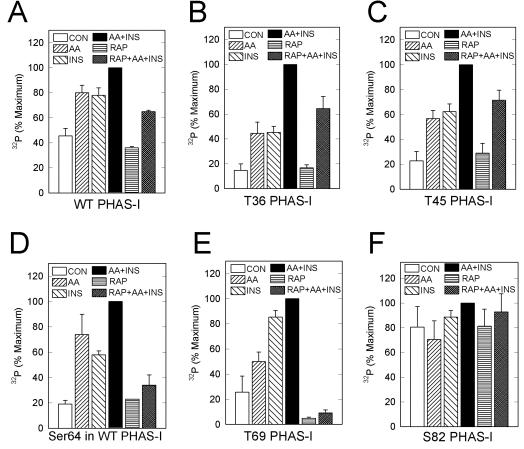
Regulated phosphorylation of Thr36, Thr45, and Thr69 does not require phosphorylation of other sites. Experiments were performed as described in the legend to Fig. 4. In addition, WT PHAS-I was immunoprecipitated and incubated with lysyl endopeptidase. The digests were fractionated by reverse-phase HPLC as described in the legend to Fig. 3, and the amounts of 32P in the peak fractions containing the Ser64 phosphopeptide were determined. The relative amounts of 32P associated with WT PHAS-I (A), T36 PHAS-I (B), T45 PHAS-I (C), Ser64 in WT PHAS-I (D), T69 PHAS-I (E), and S82 PHAS-I (F) were determined after correcting for the amounts of the proteins estimated by immunoblotting. The results are expressed as a percentages of the respective maximum values and are mean values and standard errors from three experiments.
Both insulin and amino acids increased the phosphorylation of T69 PHAS-I, although the effect of insulin was about twice that of amino acids (Fig. 4D and 5E). As expected, the increases in T69 phosphorylation were associated with increases in the amount of the protein found in the β form (Fig. 4C). Rapamycin markedly decreased the basal phosphorylation of T69 PHAS-I and essentially abolished the effect of insulin plus amino acids on increasing the phosphorylation of PHAS-I (Fig. 4D and 5E). These effects of rapamycin were associated with a total loss of the β form and a corresponding increase in the level of the α form (Fig. 4C). Thus, the phosphorylation of T69 was particularly sensitive to rapamycin.
The phosphorylation of Ser82 was distinctly different from that of the other sites (Fig. 4C). Neither insulin nor amino acids changed the 32P content of Ser82 PHAS-I or the electrophoretic mobility of the protein (Fig. 4D). The phosphorylation of S82 was also insensitive to rapamycin (Fig. 5F).
Because the phosphorylation of Ser64 was markedly reduced by mutation of the three TP sites, S64 PHAS-I could not be used to investigate the control of Ser64 phosphorylation. The effects of amino acids and insulin on this site were investigated by peptide mapping of WT PHAS-I (Fig. 5D). Lysyl endopeptidase C digests of protein that had been immunoprecipitated from 32P-labeled cells were subjected to HPLC, and the amounts of 32P in the peak fractions containing the Ser64 peptide were determined. Insulin and amino acids increased the 32P content of Ser64 3- and 3.5-fold, respectively, and the combination increased phosphorylation of the site 5-fold (Fig. 5D). Rapamycin alone had little, if any, effect on the 32P content of Ser64. However, rapamycin markedly inhibited the phosphorylation of this site in response to insulin and amino acids.
Effect of phosphorylation site mutations on eIF4E binding.
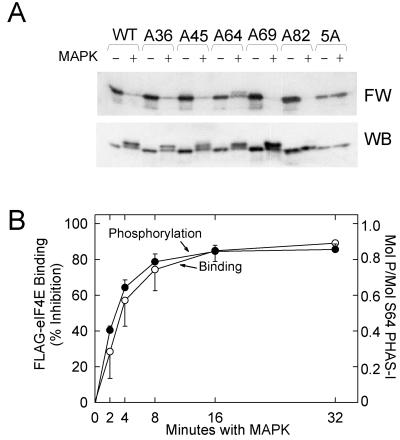
To investigate the role of the five (S/T)P sites in controlling eIF4E binding, mutant PHAS-I proteins were phosphorylated in vitro with purified MAP kinase. Samples of the phosphorylated proteins were subjected to SDS-PAGE, transferred to Immobilon membranes, and probed with a 32P-labeled FLAG-eIF4E fusion protein. Phosphorylation of WT PHAS-I almost abolished binding to FLAG-eIF4E (Fig. 6A). None of the individual site mutations prevented the loss of FLAG-eIF4E binding, indicating that phosphorylation of no single site is essential for the loss of eIF4E binding. The effects of phosphorylating T36 PHAS-I, T45 PHAS-I, and S82 PHAS-I on FLAG-eIF4E binding were recently described (40). To investigate the effect of Ser64 phosphorylation, S64 PHAS-I was incubated for increasing times with MAP kinase. FLAG-eIF4E binding decreased with a time course essentially identical to that of phosphate incorporation (Fig. 6B).
FIG. 6.
Effect of phosphorylating mutant PHAS-I proteins in vitro on eIF4E binding. (A) Proteins were incubated with ATP in the absence (−) or presence (+) of MAP kinase (MAPK) for 5 h. Samples were subjected to SDS-PAGE before the proteins were transferred to an Immobilon membrane. A far Western blot (FW) prepared using 32P-labeled FLAG-eIF4E and a PHAS-I immunoblot are shown. (B) S64 PHAS-I was incubated for increasing times with recombinant MAP kinase and either [γ-32P]ATP or unlabeled ATP before samples were subjected to SDS-PAGE. Gels containing 32P-labeled samples were stained with Coomassie blue. The S64 PHAS-I bands were excised, and the 32P contents were determined by scintillation counting. The results are expressed as moles of phosphate incorporated per mole of S64 PHAS-I. S64 PHAS-I that had been phosphorylated with unlabeled ATP was transferred to an Immobilon membrane, which was probed with 32P-labeled FLAG-eIF4E. The results represent the inhibition of binding due to MAP kinase. The results for phosphorylation (●) and FLAG-eIF4E binding (○) are mean values and standard errors from three experiments.
Far Western blotting was also used to investigate the role of phosphorylation sites in controlling eIF4E binding of PHAS-I derived from cells. WT PHAS-I and mutant proteins were expressed in HEK293 cells, which were incubated with amino acids and a maximally effective concentration of insulin before 32P-labeled FLAG-eIF4E binding was measured. Little, if any, of the probe bound to WT PHAS-I (Fig. 7A), although a small amount of binding to WT PHAS-I was detected in some experiments. These differences among experiments are presumably related to differences in the extent of insulin-stimulated phosphorylation of PHAS-I. An immunoblot of the same membrane used for the far Western blot revealed that essentially all of the WT PHAS-I was present in the phosphorylated β and γ forms (Fig. 7B). The immunoblot also confirmed that the levels of expression of WT PHAS-I and the mutant proteins were similar.
FIG. 7.
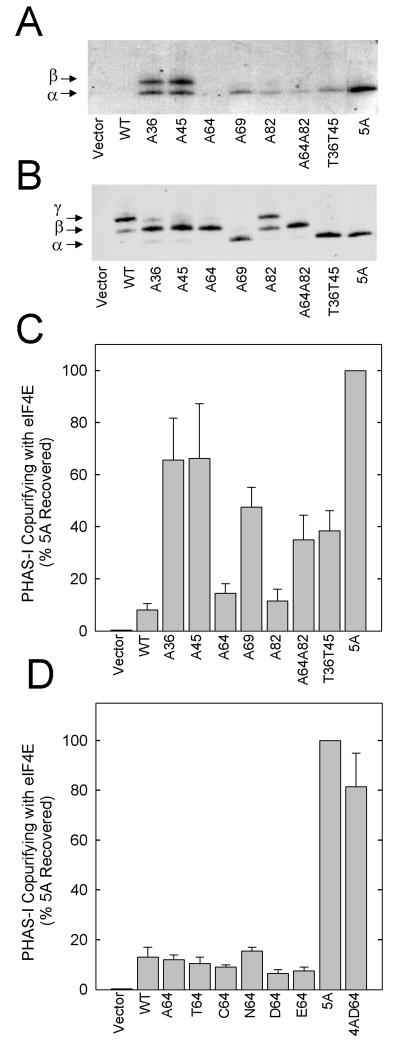
Effect of mutations on eIF-4E binding. HEK293 cells were transfected and incubated as described in Materials and Methods (protocol 2). PHAS-I proteins were immunoprecipitated from extracts and subjected to SDS-PAGE. (A) Far Western blot showing binding to 32P-labeled FLAG-eIF4E. (B) PHAS-I immunoblot of the membrane used for the far Western blot. (C) Copurification of mutant proteins with endogenous eIF-4E. PHAS-I–eIF4E complexes were isolated from extracts by using m7GTP-Sepharose, and the relative levels of PHAS-I and eIF4E were determined by immunoblotting. The results represent the amounts of the different PHAS-I proteins that were isolated with the cap affinity resin expressed as a percentage of the 5A PHAS-I that was recovered and are mean values and standard errors from three experiments. (D) Copurification with eIF4E of PHAS-I proteins with different substitutions at position 64. Mean values plus half the range from two experiments are presented.
Mutating either Thr36 or Thr45 to Ala markedly increased 32P-labeled FLAG-eIF4E binding, suggesting that these sites contribute to the control of the association state of PHAS-I and eIF4E (Fig. 7A). Mutating Thr69 also increased binding but to a lesser extent than that caused by mutations in the other two TP sites. In contrast, mutating Ser64 to Ala had little, if any, effect on increasing FLAG-eIF4E binding. Mutating Ser82 only slightly increased binding. FLAG-eIF4E binding to the mutant lacking both SP sites was similar to that observed with A82 PHAS-I. FLAG-eIF4E binding was markedly increased by mutating all five S/TP sites. This finding was not surprising, since 5A PHAS-I is not phosphorylated in cells. None of the proteins having mutations in four of the five available sites exhibited lower binding to eIF4E than did 5A PHAS-I, except for Thr69 PHAS-I, whose binding was approximately equal to that of A36 PHAS-I and A45 PHAS-I (Mothe-Satney and Lawrence, unpublished).
The conditions used to assess binding by far Western analysis differ from those used for intact cells. Moreover, it is possible that phosphorylation of certain sites might be sufficient to inhibit the association of PHAS-I and eIF4E, but insufficient to promote dissociation of eIF4E once the PHAS-I–eIF4E complex has formed. To address these issues, the amounts of the mutant proteins bound to eIF4E in cells were determined by isolating PHAS-I–eIF4E complexes with m7GTP-Sepharose. The results are expressed relative to the amount of 5A PHAS-I that copurified with eIF4E (Fig. 7C). Binding of WT PHAS-I to endogenous eIF4E was only 8% of that of 5A PHAS-I, consistent with the relatively high level of phosphorylation of WT PHAS-I. The Thr36 and Thr45 mutations had the largest impact of any of the single-site mutations, increasing binding by approximately eightfold. Mutating Thr69 increased binding by sixfold. The recovery of A64 PHAS-I and A82 PHAS-I with eIF4E was approximately twice that of WT PHAS-I, although the differences were within the range of experimental variability. Thus, the effects of mutating individual SP sites on binding were smaller than the effects of mutating the TP sites. The binding of the double mutant lacking both Ser64 and Ser82 was approximately four times that of the wild-type protein.
The lack of effect of mutating Ser64 on eIF4E binding was surprising, in view of the striking effect of phosphorylating Ser64 on eIF4E binding (Fig. 6). Therefore, we investigated the effect of expressing proteins having other substitutions in position 64 (Fig. 7D). The binding of C64 PHAS-I and N64 PHAS-I (Fig. 1), which cannot be phosphorylated at this position, to endogenous eIF4E was not significantly different from that of A64 PHAS-I (Fig. 7D). T64 PHAS-I also exhibited very low binding to eIF4E. To investigate the effect of introducing a negative charge at position 64, we expressed mutant proteins having Glu or Asp substitutions. The effects of D64 PHAS-I and E64 PHAS-I on binding were no different from those of A64 PHAS-I. However, because Ser64 was highly phosphorylated under these conditions, it would have been difficult to detect an effect of the acidic substitutions. For this reason, we also introduced Asp into position 64 in the 5A mutant. 5A PHAS-I and 4AD64 exhibited almost the same binding to endogenous eIF4E.
Effect of phosphorylation site mutations on cap-dependent mRNA translation.
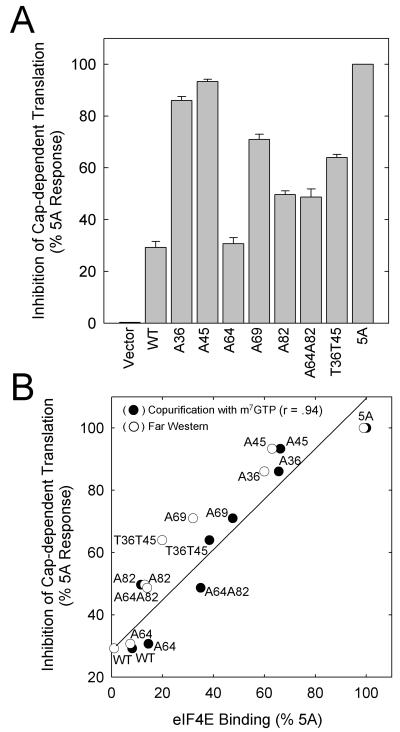
mRNA translation was investigated using a dual luciferase reporter, which allowed assessment of the effects of the PHAS-I proteins on cap-dependent and cap-independent translation. Measuring cap-independent translation driven by the IRES element should allow the detection of nonspecific inhibitory effects of the PHAS-I proteins on the translation machinery (27). None of the constructs used inhibited cap-independent translation (results not presented). Thus, the inhibitory effects of expressing the PHAS-I proteins on cap-dependent translation were presumably related to inhibition of the function of eIF4E. In these experiments, 5A PHAS-I inhibited translation of the reporter by approximately 60% and WT PHAS-I inhibited translation by only 20%. Nevertheless, we were concerned that the effects of some of the mutations might have been limited by the availability of eIF4E. If this were the case, it would not have been possible to detect differences in translation among the mutants that bound more tightly to eIF4E. This did not appear to be a problem, since the same relationships among the mutants with respect to translation inhibition were observed when cells were transfected with 25-fold less cDNA (Mothe-Satney and Lawrence, unpublished). A64 PHAS-I did not inhibit cap-dependent translation (Fig. 8A); however, all of the other single mutations generated proteins that had greater inhibitory effects on translation than that of WT PHAS-I. Mutating Thr36 or Thr45 had the most dramatic effects, approaching the inhibition produced by 5A PHAS-I. The Thr69 → Ala mutation had the next largest effect, followed by the Ser82 → Ala mutation.
FIG. 8.
Effect of mutations on cap-dependent mRNA translation. HEK293 cells were cotransfected with pRLIRESFL and the PHAS-I expression vectors. The cells were incubated as described in the legend to Fig. 7 before the activities of the two luciferases were measured. None of the PHAS-I proteins decreased firefly luciferase activity, which was used to correct for small differences in the transfection efficiency of the reporter construct among samples. Approximately equal amounts of the different PHAS-I proteins were expressed (see Fig. 7B, for example). (A) Inhibition of cap-dependent translation resulting from PHAS-I proteins, expressed relative to that produced by 5A PHAS-I (mean values and standard errors from three experiments). (B) Relationship between eIF4E binding and inhibition of cap-dependent mRNA translation. The relative amounts of PHAS-I proteins copurifying with eIF-4E (●) or the relative amounts of FLAG–eIF4E binding (○) from Fig. 7 are plotted against the inhibition of cap-dependent mRNA translation. The line and correlation coefficient (r) were generated by linear regression analysis of translation inhibition versus PHAS-I protein recovered with m7GTP-Sepharose.
As shown in Fig. 8B, there was a direct correlation between translation inhibition and eIF4E binding, assessed by either far Western analysis or copurification of PHAS-I proteins. This relationship provides additional evidence that the inhibition of translation by the mutant proteins is due to specific inhibition of the function of eIF4E. As indicated by the y intercept on the plot in Fig. 8B, significant inhibition of translation can occur in the absence of detectable binding. Presumably, this inhibition is due to lower-affinity interactions that are not detected by the available methods to assess the binding of PHAS-I to eIF4E in cells.
DISCUSSION
The role of the five (S/T)P sites in controlling PHAS-I was investigated by expressing mutant PHAS-I proteins in HEK293 cells. As with all overexpression studies, there is the issue of whether the effects observed are representative of events that occur with the endogenous proteins. The present finding that phosphorylation was confined to the same five (S/T)P sites previously identified in endogenous PHAS-I in rat adipocytes indicates that ectopic phosphorylation of the proteins expressed in HEK293 cells did not occur (8). Furthermore, insulin stimulated the phosphorylation of PHAS-I expressed in HEK293 cells in the same four sites that were phosphorylated in response to the hormone in adipocytes (8), and phosphorylation occurred via a rapamycin-sensitive pathway, as with the endogenous PHAS-I proteins in other cell types (3, 12). These findings argue that regulation of the phosphorylation of the expressed PHAS-I proteins occurs by normal signaling mechanisms. The results obtained with these proteins have important implications with respect to the mechanisms involved in the regulation of phosphorylation and the roles of the different sites in controlling the function of the PHAS-I protein.
It has become common practice to use a gel shift assay to evaluate the phosphorylation of PHAS-I. A caveat in using this assay to evaluate functional control of PHAS-I is that sites having the largest influence on electrophoretic mobility do not necessarily have the largest effects on eIF4E binding. Phosphorylation of either Thr45 (40) or Ser64 alone did not affect mobility, although phosphorylation of either of these two sites abolished eIF4E binding in vitro. PHAS-1β is generated by phosphorylation of Thr69 or Ser82. PHAS-Iγ results from the phosphorylation of Thr69 and either Ser64 or Ser82. Phosphorylated Thr36 and Thr45 may appear in the α, β, and γ forms, since these two sites do not significantly influence electrophoretic mobility.
With multisite phosphorylation, the potential exists for interactions among phosphorylation sites. It was recently proposed that PHAS-I phosphorylation involves a two-step mechanism in which phosphorylation of Thr36 and Thr45, occurring when PHAS-I is bound to eIF4E, is required for the phosphorylation of the other sites (10). We have confirmed that mutating Thr36 or Thr45 decreases the phosphorylation of Ser64 in HEK293 cells. However, other findings are clearly incompatible with certain aspects of the two-step model (10). For example, mutating Thr69 also markedly decreased the phosphorylation of Ser64, indicating that Ser64 phosphorylation depends on the phosphorylation of all three TP sites. Also, PHAS-I proteins lacking Thr36 and Thr45 could be phosphorylated in both Thr69 and Ser82. Indeed, the absence of Thr36 or Thr45 actually enhanced the phosphorylation of Thr69, and the phosphorylation of Thr69 was regulated in a rapamycin-sensitive manner by amino acids and insulin in the mutant lacking the four other sites. The finding that the three PHAS-I proteins having an individual TP site with mutations in the four remaining sites were phosphorylated implies that there is not an obligate order in the phosphorylation of Thr36, Thr45, and Thr69. S64 NBPHAS-I was not phosphorylated, indicating that dissociation of the PHAS-I–eIF4E complex, which occurs in response to phosphorylation of the TP sites, is not sufficient for Ser64 phosphorylation. Moreover, since phosphorylation of Ser64 still required the TP sites when PHAS-I was rendered incapable of high-affinity binding to eIF4E, the ordered phosphorylation of Ser64 does not have to occur when PHAS-I is bound to eIF4E as proposed in the two-step model (10).
Hierarchal phosphorylation, a mechanism in which phosphorylation of one site creates a consensus site for phosphorylation of a second site (32), might explain the dependence of Ser64 on prior phosphorylation of the TP sites. Phosphorylated Ser or Thr residues ([S/T]*) are found in the consensus motifs, [S/T]XXX[S/T]* and [S/T]*XX[S/T], for phosphorylation by glycogen synthase kinase 3 and casein kinase I, respectively (32). If such a mechanism is involved in the control of Ser64 phosphorylation, the consensus motif is likely to be complex, since three sites were required for the phosphorylation of Ser64. As suggested previously (10), phosphorylation might create a recognition motif that recruits the Ser64 kinase and/or other regulatory factors to PHAS-I. However, the ordered-phosphorylation model is based on studies in intact cells, and there are other potential explanations of the data. For example, phosphorylation could appear to be ordered if phosphorylation of the TP sites protected the Ser64 site from dephosphorylation. Interestingly, evidence for such a mechanism was recently obtained for the control of nPKCδ, another downstream protein in the mTOR signaling pathway (26). In this case, phosphorylation of Thr505 in the activation loop of the kinase appears to markedly decrease the rate of Ser662 dephosphorylation.
In view of the complexities in the control of PHAS-I, the simple hypothesis that the five (S/T)P sites in PHAS-I are regulated by a common mechanism can be eliminated. Ser82 was not phosphorylated in response to insulin or amino acids, indicating that Ser82 is not subject to the same control as the other sites whose phosphorylation was increased by these agents. Insulin and amino acids increased the phosphorylation of T36 PHAS-I, T45 PHAS-I, and T69 PHAS-I, indicating that the stimulatory effects of these agents on the phosphorylation of Thr36, Thr45, and Thr69 occurred independently of other sites. Although the accumulation of phosphate in Ser64 is complicated by the dependence on prior phosphorylation of the TP sites, the finding that amino acids and insulin increased the phosphorylation of the same four sites is consistent with the hypothesis that these agents act via a common upstream effector. The effect of insulin on increasing the phosphorylation of PHAS-I is mediated by the protein kinase B (PKB) signaling pathway (11, 33, 37); however, PKB is not activated by amino acids (14, 39). mTOR is a more likely common effector, since the effects of both insulin and amino acids are attenuated by rapamycin. The PHAS-I kinase activity of mTOR is increased in response to insulin (33, 34). Activation of mTOR occurs by a PKB-dependent pathway, which leads to an increase in phosphorylation of Ser2448 in the COOH-terminal region of mTOR (25, 33). Amino acids appear to have a permissive effect on the phosphorylation of mTOR by PKB (25).
The marked effects of rapamycin on decreasing the phosphorylation of Thr69 suggest that this site might be directly phosphorylated in cells by mTOR, which is able to phosphorylate Thr69 in vitro (4). Decreasing Thr69 phosphorylation by rapamycin would be expected to reduce the phosphorylation of Ser64. Nevertheless, it is paradoxical that phosphorylation of Thr36 and Thr45, the sites preferred by mTOR in vitro (6, 10, 40), is less sensitive to rapamycin than is the phosphorylation of Thr69 and Ser64. A kinase that associates with mTOR and that can be released upon incubation with an mTOR antiserum was recently described (16). This enzyme was reported to specifically phosphorylate Ser64 and to promote the dissociation of the PHAS-I–eIF4E complex, but the kinase phosphorylated PHAS-I only when it was bound to eIF4E, a finding that would seem to exclude a role in phosphorylating Ser64 in the free PHAS-I protein. It was recently concluded that MAP kinase participates in the control of PHAS-I (30), as was proposed several years ago (20). However, it has been argued that MAP kinase activation is neither necessary nor sufficient for the phosphorylation of PHAS-I (38). Additional work is needed to identify the kinases that phosphorylate PHAS-I in cells.
Far Western analysis was used to investigate the effect of phosphorylating PHAS-I in vitro on eIF4E binding. An advantage of this method is that it allows a direct assessment of eIF4E binding to purified PHAS-I proteins phosphorylated in defined sites. We recently demonstrated that phosphorylation of Thr45 in T45 PHAS-I markedly decreased FLAG-eIF4E binding whereas phosphorylating Thr36 in T36 PHAS-I or Ser82 in S82 PHAS-I had less pronounced inhibitory effects on binding (40). In the present study, phosphorylation of S64 PHAS-I by MAP kinase in vitro was found to abolish FLAG-eIF4E binding. Interestingly, introducing Asp at position 64 in 5A PHAS-I did not decrease eIF4E binding, indicating that acidic substitutions do not mimic the effect of phosphorylating the mutant protein in Ser64. The excellent correlation between the loss of eIF4E binding and the stoichiometry of S64 PHAS-I phosphorylation leaves little doubt that Ser64 phosphorylation inhibits binding. In an earlier study in which WT PHAS-I was phosphorylated by MAP kinase, the loss of binding did not appear to correlate with the extent of phosphorylation of Ser64, and it was concluded that phosphorylating Ser64 did not inhibit eIF4E binding (8). Imprecision in measuring the stoichiometry of phosphorylation of Ser64, which was complicated by the presence of other sites in this previous study, is the likely reason for the erroneous conclusion. We have been unable to efficiently phosphorylate Thr69 in vitro; however, Thr69 in T69 PHAS-I may be almost completely phosphorylated in cells, as evidenced by the accumulation of most of the protein in the β electrophoretic form. FLAG-eIF4E binding to this phosphorylated form was attenuated (Mothe-Satney and Lawrence, unpublished), indicating that phosphorylation of Thr69 decreases the affinity of PHAS-I for eIF4E. Taken together, the results of far Western analyses of the phosphorylated forms of T36, T45, S64, T69, and S82 PHAS-I indicate the following order for the influence of phosphorylation on eIF4E binding in vitro: Ser64 > Thr45 > Thr69 > Thr36 > Ser82. Thr45 and Ser64 flank the eIF4E-binding motif (23), which may explain their greater influence relative to the other sites.
The effects of mutating the different sites on eIF4E binding of PHAS-I proteins in cells were assessed by both far Western analyses and copurification of mutant proteins with endogenous eIF4E. Presumably, the presence of other endogenous proteins, such as eIF4G, that influence the binding of PHAS-I to eIF4E in cells could lead to differences between binding assessed by the two methods. It is reassuring that when expressed relative to the binding of 5A PHAS-I, there was reasonably good agreement between binding results obtained by far Western analysis and by copurification with eIF4E. In contrast, the relative influence of in vitro phosphorylation of the different sites on eIF4E binding did not correlate with the relative effects of mutating the sites on eIF4E binding of PHAS-I proteins expressed in cells. For example, mutating Ser64 had little, if any, effect on the amount of PHAS-I bound to eIF4E in cells. To investigate the possibility that the Ala mutation itself might have decreased binding, we expressed proteins with Asn, Cys, and Thr substitutions at position 64. Our choice of these substitutions was influenced by the recent discussion of unpublished structural studies in which Ser64 was placed in close proximity to Glu70 in eIF4E. If they are positioned appropriately, a hydrogen bond could form between these two residues. If this were the case, introducing an Ala mutation in place of Ser64 would reduce the binding affinity, since Ala cannot participate in hydrogen bonding. The three alternative substitutions have the potential to participate in hydrogen bonding but (except for Thr) cannot be phosphorylated. The result that binding of C64 PHAS-I and N64 PHAS-I to eIF4E in cells was very similar to that of A64 PHAS-I supports the conclusion that phosphorylation of Ser64 is not necessary for the dissociation of the PHAS-I–eIF4E complex. Whether Ser64 phosphorylation is sufficient to inhibit binding in cells is still not clear.
We also found that mutating Ala36 increased eIF4E binding much more than expected on the basis of the modest effect of Thr36 phosphorylation on eIF4E binding in vitro observed previously (40). The fact that the Ala36 mutation not only ablates the Thr36 site but also decreases the phosphorylation of Ser64 in cells is a potential explanation. The result that binding of eIF4E to PHAS-Iγ, which may be generated by the phosphorylation of Ser64 in combination with Thr69, is never observed supports the concept that phosphorylation of Ser64 may act in combination with the phosphorylation of other sites to modulate the affinity of PHAS-I for eIF4E. While such interactions also complicate the interpretation of the findings with A45 PHAS-I and A69 PHAS-I, they provide an elegant mechanism through which the phosphorylation of the TP sites can control eIF4E binding by facilitating the accumulation of phosphate in Ser64.
Mutating any individual site except Ser64 increased eIF4E binding, and there was a very good correlation between the amount of PHAS-I bound to eIF4E and the inhibition of cap-dependent mRNA translation. Thus, each of the TP sites appears to be able to influence mRNA translation, either directly by modulating the binding affinity of PHAS-I and eIF4E or indirectly by affecting the phosphorylation of Ser64. The equivalents of Thr36, Thr45, Ser64, and Thr69 are found in all members of the PHAS family thus far discovered in species ranging from slime mold to humans on the evolutionary scale. Presumably, the functional importance explains why the sites are so highly conserved.
ACKNOWLEDGMENTS
This research was supported in part by National Institutes of Health grants DK52753 and DK28312 (to J.C.L.), NIH grants HL19242 and DK52378 (to T.A.J.H.), and a fellowship from the Juvenile Diabetes Foundation (to I.M.-S.).
We thank John Majors for p2332, Kevin Bowman for expert technical assistance, and Gregory Brunn and Angus Scrimgeour for their critical reading of the manuscript.
REFERENCES
- 1.Abraham R T. Mammalian target of rapamycin: immunosuppressive drugs uncover a novel pathway of cytokine receptor signaling. Curr Opin Immunol. 1998;10:330–336. doi: 10.1016/s0952-7915(98)80172-6. [DOI] [PubMed] [Google Scholar]
- 2.Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 3.Beretta L, Gingras A-C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 4.Brunn G J, Fadden P, Haystead T A J, Lawrence J C., Jr The mammalian target of rapamycin phosphorylates sites having a (Ser/Thr)-Pro motif and is activated by antibodies to a region near its COOH-terminus. J Biol Chem. 1997;272:32547–32550. doi: 10.1074/jbc.272.51.32547. [DOI] [PubMed] [Google Scholar]
- 5.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H, Houghton P J, Lawrence J C, Jr, Abraham R T. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 6.Burnett P E, Barrow R K, Cohen N A, Snyder S H, Sabatini D M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fadden P, Haystead T A J, Lawrence J C., Jr Identification of phosphorylation sites in the translational regulator, PHAS-I, that are controlled by insulin and rapamycin in rat adipocytes. J Biol Chem. 1997;272:10240–10247. doi: 10.1074/jbc.272.15.10240. [DOI] [PubMed] [Google Scholar]
- 9.Fadden P, Haystead T A J, Lawrence J C., Jr Phosphorylation of the translational regulator, PHAS-I, by protein kinase CK2. FEBS Lett. 1998;435:105–109. doi: 10.1016/s0014-5793(98)01047-3. [DOI] [PubMed] [Google Scholar]
- 10.Gingras A-C, Gygi S P, Raught B, Polakiewicz R D, Abraham R T, Hoekstra M F, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gingras A-C, Kennedy S G, O'Leary M A, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves L M, Bornfeldt K E, Argast G M, Krebs E G, Kong X, Lin T-A, Lawrence J C., Jr cAMP- and rapamycin-sensitive regulation of the association of eukaryotic initiation factor 4E and the translational regulator PHAS-I in aortic smooth muscle cells. Proc Natl Acad Sci USA. 1995;92:7222–7226. doi: 10.1073/pnas.92.16.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara K, Yonezawa K, Weng Q-P, Kozlowski M T, Belham C M, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 15.Heesom K J, Avison M B, Diggle T A, Denton R M. Insulin-stimulated kinase from rat fat cells that phosphorylates initiation factor 4E-binding protein 1 on the rapamycin-insensitive site (serine-111) Biochem J. 1998;336:39–48. doi: 10.1042/bj3360039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heesom K J, Denton R M. Dissociation of the eukaryotic initiation factor-4E/4E-BP1 complex involves phosphorylation of 4E-BP1 by an mTOR-associated kinase. FEBS Lett. 1999;457:489–493. doi: 10.1016/s0014-5793(99)01094-7. [DOI] [PubMed] [Google Scholar]
- 17.Kimball S R, Shantz L M, Horetsky R L, Jefferson L. Leucine regulates translation of specific mRNAs in L6 myoblasts through mTOR-mediated changes in availability of eIF4E and phosphorylation of ribosomal protein S6. J Biol Chem. 1999;274:11647–11652. doi: 10.1074/jbc.274.17.11647. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence J C, Jr, Abraham R T. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem Sci. 1997;22:345–349. doi: 10.1016/s0968-0004(97)01101-8. [DOI] [PubMed] [Google Scholar]
- 20.Lin T-A, Kong X, Haystead T A J, Pause A, Belsham G, Sonenberg N, Lawrence J C., Jr PHAS-I as a link between mitogen activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 21.Lin T-A, Kong X, Saltiel A R, Blackshear P J, Lawrence J C., Jr Control of PHAS-I by insulin in 3T3-L1 adipocytes: synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated protein kinase-independent pathway. J Biol Chem. 1995;270:18531–18538. doi: 10.1074/jbc.270.31.18531. [DOI] [PubMed] [Google Scholar]
- 22.Lin T-A, Lawrence J C., Jr Control of the translational regulators, PHAS-I and PHAS-II, by insulin and cAMP in 3T3-L1 adipocytes. J Biol Chem. 1996;271:30199–30204. doi: 10.1074/jbc.271.47.30199. [DOI] [PubMed] [Google Scholar]
- 23.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4G and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mader S, Sonenberg N. Cap binding complexes and cellular growth control. Biochimie. 1995;77:40–44. doi: 10.1016/0300-9084(96)88102-8. [DOI] [PubMed] [Google Scholar]
- 25.Nave B T, Ouwens D M, Withers D J, Alessi D R, Shepherd P R. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino acid deficiency on protein translation. Biochem J. 1999;344:427–431. [PMC free article] [PubMed] [Google Scholar]
- 26.Parekh D, Ziegler W, Yonezawa K, Hara K, Parker P J. Mammalian TOR controls one of two kinase pathways acting upon nPKCδ and nPKCɛ. J Biol Chem. 1999;274:34758–34764. doi: 10.1074/jbc.274.49.34758. [DOI] [PubMed] [Google Scholar]
- 27.Pause A, Belsham G J, Gingras A-C, Donze O, Lin T-A, Lawrence J C, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 28.Poulin F, Gingras A-C, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem. 1998;273:14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- 29.Proud C G, Denton R M. Molecular mechanisms for the control of translation by insulin. Biochem J. 1997;328:329–341. doi: 10.1042/bj3280329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao G N, Madamanchi N R, Lele M, Gadiparthi L, Gingras A-C, Eling T E, Sonenberg N. A potential role for extracellular signal-regulated kinases in prostaglandin F2α-induced protein synthesis in smooth muscle cells. J Biol Chem. 1999;274:12925–12932. doi: 10.1074/jbc.274.18.12925. [DOI] [PubMed] [Google Scholar]
- 31.Rhoads R E. Signal transduction pathways that regulate eukaryotic protein synthesis. J Biol Chem. 1999;274:30337–30340. doi: 10.1074/jbc.274.43.30337. [DOI] [PubMed] [Google Scholar]
- 32.Roach P J. Control of glycogen synthase by hierarchal protein phosphorylation. FASEB J. 1990;4:2961–2968. [PubMed] [Google Scholar]
- 33.Scott P H, Brunn G J, Kohn A D, Roth R A, Lawrence J C., Jr Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proc Natl Acad Sci USA. 1998;95:7772–7777. doi: 10.1073/pnas.95.13.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott P H, Lawrence J C., Jr Attenuation of mammalian target of rapamycin activity by increased cAMP in 3T3-L1 adipocytes. J Biol Chem. 1998;273:34496–34501. doi: 10.1074/jbc.273.51.34496. [DOI] [PubMed] [Google Scholar]
- 35.Shatkin A J. mRNA caps—old and newer hats. Bioessays. 1987;7:275–277. doi: 10.1002/bies.950070611. [DOI] [PubMed] [Google Scholar]
- 36.Sonenberg N, Gingras A-C. The mRNA 5′-cap binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 37.Ueki K, Yamamoto-Honda R, Kaburagi Y, Yamauchi T, Tobe K, Burgering B M T, Coffer P J, Komuro I, Akanuma Y, Yazaki Y, Kadowaki T. Potential role of protein kinase B in insulin-induced glucose transport, glycogen synthesis, and protein synthesis. J Biol Chem. 1998;273:5315–5322. doi: 10.1074/jbc.273.9.5315. [DOI] [PubMed] [Google Scholar]
- 38.von Manteuffel S R, Gingras A-C, Ming X-F, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70S6K pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Campbell L E, Miller C M, Proud C G. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem J. 1998;334:261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang D, Brunn G J, Lawrence J C., Jr Mutational analysis of sites in the translational regulator, PHAS-I, that are selectively phosphorylated by mTOR. FEBS Lett. 1999;453:387–390. doi: 10.1016/s0014-5793(99)00762-0. [DOI] [PubMed] [Google Scholar]