Abstract
DNA is an ultra-high-density storage medium that could meet exponentially growing worldwide demand for archival data storage if DNA synthesis costs declined sufficiently and random access of files within exabyte-to-yottabyte-scale DNA data pools were feasible. Here we demonstrate a path to overcome the second barrier by encapsulating data-encoding DNA file sequences within impervious silica capsules that are surface-labeled with single-stranded DNA barcodes. Barcodes are chosen to represent file metadata, enabling selection of sets of files with Boolean logic directly, without use of amplification. We demonstrate random access of image files from a prototypical 2 KB image database using fluorescence sorting with selection sensitivity of 1 in 106 files, which thereby enables 1 in 106N selection capability using N optical channels. Our strategy thereby offers a scalable concept for random-access of archival files in large-scale molecular datasets.
INTRODUCTION
While DNA is the polymer selected by evolution for the storage and transmission of genetic information in biology, it can also be used for the storage of arbitrary digital information at densities far exceeding conventional data storage technologies such as flash and tape memory, at scales well beyond the capacity of the largest existing data centers1,2. Recent progress in nucleic acid synthesis and sequencing technologies continue to reduce the cost of writing and reading DNA, foreshadowing future commercially competitive DNA-based information storage1,3–5. Demonstrations of its viability as a general information storage medium include the storage and retrieval of books, images, computer programs, audio clips, works of art, and Shakespeare’s sonnets using a variety of encoding schemes6–12, with data size limited primarily by the cost of DNA synthesis. In each case, digital information was converted to DNA sequences composed of ~100–200 nucleotide (nt) data blocks for ease of chemical synthesis and sequencing. Sequence fragments were then assembled to reconstruct the original, encoded information.
While significant effort in DNA data storage has focused on increasing the scale of DNA synthesis, as well as improving encoding schemes, an additional crucial aspect of data storage systems is the ability to efficiently retrieve specific files, or arbitrary subsets of files. To date, molecular random access has largely relied on conventional polymerase chain reaction (PCR)8,10,12,13, which uses up to ~20–30 heating and cooling cycles with DNA polymerase to selectively amplify specific DNA sequences from a DNA data pool using primers. Nested addressing barcodes14–16 have also been used to uniquely identify a greater number of files, as well as biochemical affinity tags to selectively pull down oligos for targeted amplification17.
While powerful demonstrations of PCR have shown successful file retrieval from a 150 GB file system18, significant limitations include, first, the length of DNA needed to uniquely label DNA data strands for file indexing, which reduces the DNA available for data storage. For example, for an exabyte-scale data pool, each file requires at least three barcodes17, or up to sixty nucleotides in total barcode sequence length, thereby reducing the number of nucleotides that can be used for data encoding. Second, PCR-based retrieval requires an aliquot of the entire data pool to be irreversibly consumed for random access, and therefore additional PCR amplification of the entire data pool may periodically be needed to restore this loss of data. In this case, each PCR amplification may introduce stochastic variation in copy number of the file sequences, leading to up to 2% data loss per amplification19 if using ten-fold physical redundancy, as recently suggested18. Finally, avoiding spurious amplification of off-target files due to crosstalk of PCR primers with incorrect barcodes or main file sequences requires careful primer design20. While strategies exist to circumvent these preceding challenges, they generally reduce data density and might not be easily scalable to exabyte and larger file-systems. For example, data loss due to periodic PCR amplification of the entire data pool19 may be reduced by increasing the physical redundancy of the files in the main data pool, and PCR crosstalk can be mitigated by spatial segregation of data into distinct pools21 or extraction of selected DNA using biochemical affinity17,22.
As an alternative to PCR-based approaches, here we introduce a direct random access memory approach that retrieves specific files, or arbitrary subsets of files, directly using physical sorting, without a need for amplification, and without any potential for barcode-memory crosstalk, while also preserving non-selected files intact by recycling them into the original memory pool. To realize this file system, we first encapsulate DNA-based files physically within discrete, impervious silica capsules9,23,24, which we subsequently surface-label with unique single-stranded DNA barcodes that offer Boolean logic based selection on the entire data pool via simple hybridization. Downstream file selection may then be optical, physical, or biochemical, with sequencing-based read-out following de-encapsulation of the memory DNA from the silica capsule. Each “unit of information” encoded in DNA we term a file, which includes both the DNA encoding the main data as well as any additional components used for addressing, storage, and retrieval. Each file contains a file sequence, consisting of the DNA encoding the main data, and addressing barcodes, or simply barcodes, which are additional short DNA sequences used to identify the file in solution using hybridization. We refer to a collection of files as a data pool or database, and the set of procedures for storing, retrieving, and reading out files is termed a file system (see Supplementary Section S0 for a full list of terms).
As a proof-of-principle of our archival DNA file system, we encapsulated 20 image files, each composed of a ~0.1 kilobyte image file encoded in a 3,000-base-pair plasmid, within monodisperse, 6-µm silica particles that were chemically surface-labeled using up to three 25-mer single-stranded DNA (ssDNA) oligonucleotide barcodes chosen from a library of 240,000 orthogonal primers, which allows for individual selection of up to ~1015 possible distinct files using only three unique barcodes per file20 (Fig. 1). While we chose plasmids to encode DNA data in order to produce microgram quantities of DNA memory at low cost and to facilitate a renewable, closed-cycle write-store-access-read system using bacterial DNA data encoding and expression25–28, our file system is equally applicable to single-stranded DNA oligos produced using solid-phase chemical synthesis2,6,7,9–12,17 or gene-length oligos produced enzymatically29–32. Fluorescence-activated sorting (FAS) was then used to select target subsets of the complete data pool by first annealing fluorescent oligonucleotide probes that are complementary to the barcodes used to address the database33, enabling direct physical retrieval of specific, individual files from a pool of (106)N total files, where N is the number of fluorescence channels employed, without enzymatic amplification or associated loss of nucleotides available for data encoding. We also demonstrate Boolean AND, OR, NOT logic to select arbitrary subsets of files with combinations of distinct barcodes to query the data pool, similar to conventional Boolean logic applied in text and file searches on solid-state silicon devices.
Figure 1 |. Write-access-read cycle for a content-addressable molecular file system.
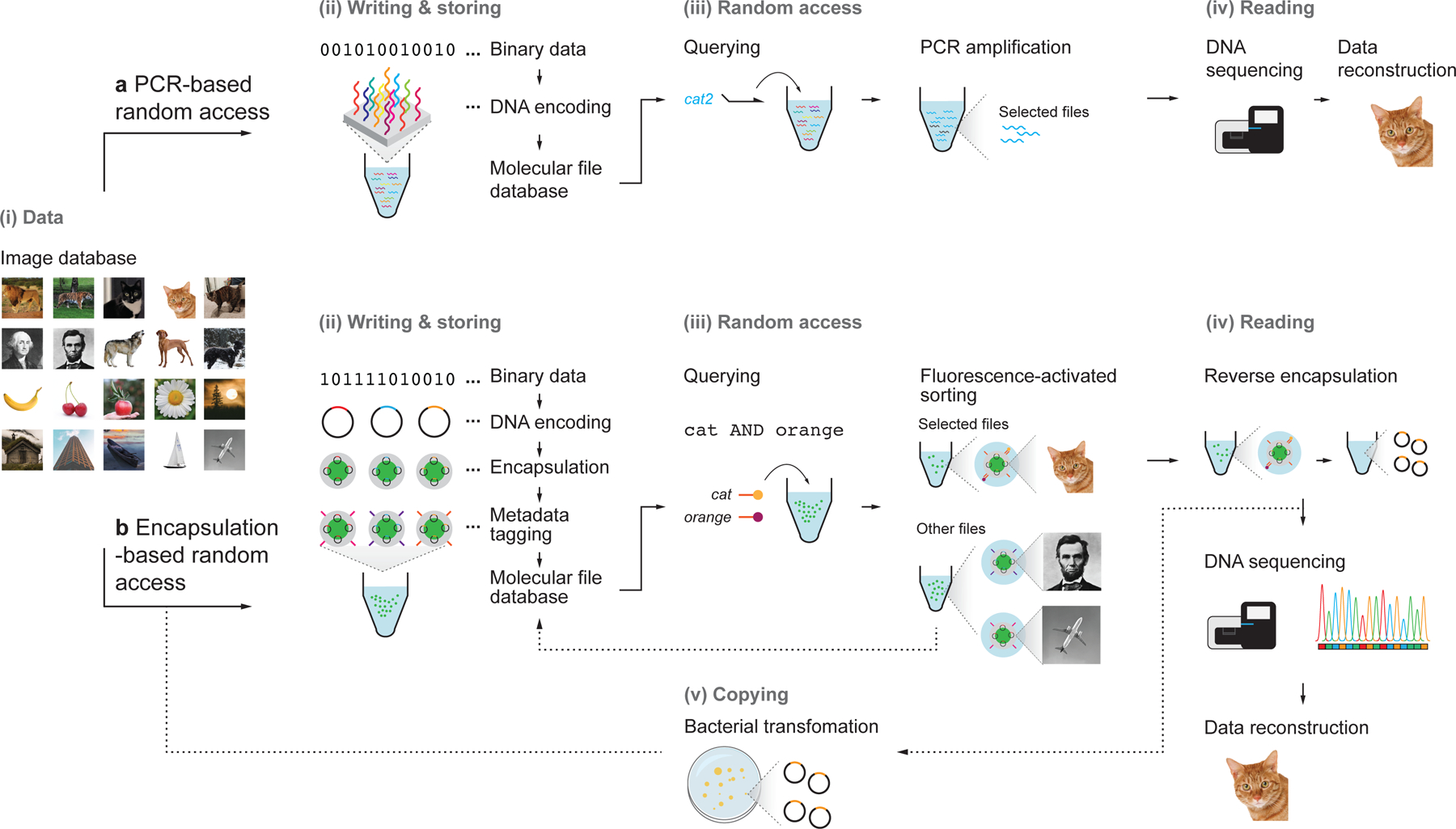
a, A general framework for DNA data storage that uses PCR-based random access and its associated challenges. b, We demonstrate here an alternative file encapsulation-based file system that allows for scalable indexing and Boolean logic selection and retrieval. Colored images were converted into 26 × 26-pixel, black-and-white icon bitmaps. The black-and-white images were then converted into DNA sequences using a custom encoding scheme (see Methods). The DNA sequences that encoded the images (file sequences) were inserted into a pUC19 plasmid vector and encapsulated into silica particles using sol-gel chemistry. Silica capsules were then addressed with content barcodes using orthogonal 25-mer single-stranded DNA strands, which were the final forms of the files. Files were pooled to form the molecular file database. To query a file or several files, fluorescently-labelled 15-mer ssDNA probes that are complementary to file barcodes were added to the data pool. Particles were then sorted with fluorescence-activated sorting (FAS) using two to four fluorescence channels simultaneously. Files that are not selected are returned to the molecular database. Addition of a chemical etching reagent into the target populations released the encapsulated DNA plasmid. Sequences for the encoded images were validated using Sanger sequencing or Illumina MiniSeq. Because plasmids were used to encode information, re-transformation of the released plasmids into bacteria to replenish the molecular file database thereby closed the write-access-read cycle.
While only twenty icon-resolution images were chosen as our image database, representing diverse subject matter including animals, plants, transportation, and buildings (Supplementary Fig. 1), our file system may in principle be scaled to considerably larger sets of images, limited primarily by the cost of DNA synthesis and the need to develop strategies for high-throughput silica encapsulation of distinct file sequences and surface-based DNA labeling for barcoding (Supplementary Fig. 1). Because physical encapsulation separates file sequences from external barcodes that are used to describe the encapsulated information, our file system offers long-term environmental protection of encoded file sequences via silica encapsulation for permanent archival storage9,23,24, where external barcodes may be renewed periodically, further protected with secondary encapsulation, or data pools may simply be stored using methods implemented in PCR-based random access, such as dehydrating the data pool and immersing the dried molecular database in oil21.
File Synthesis
Digital information in the form of 20 icon-resolution images was stored in a data pool, with each image encoded into DNA and synthesized on a plasmid. We selected images of broad diversity, representative of distinct and shared subject categories, which included several domestic and wild cats and dogs, US presidents, and several human-made objects such as an airplane, boats, and buildings (Fig. 1 and Supplementary Fig. 1). To implement this image database, the images were substituted with black-and-white, 26 × 26-pixel images to minimize synthesis costs, compressed using run-length encoding, and converted to DNA (Supplementary Fig. 1, 2). Following synthesis, bacterial amplification, and sequencing validation (Supplementary Fig. 3), each plasmid DNA was separately encapsulated into silica particles containing a fluorescein dye core and a positively charged surface23,24. Because the negatively charged phosphate groups of the DNA interact with positively charged silica particles, plasmid DNA condensed on the silica surface, after which N-[3-(trimethoxysilyl)propyl]-N,N,N-trimethylammonium chloride (TMAPS) was co-condensed with tetraethoxysilane to form an encapsulation shell after four days of incubation at room-temperature9,23 (Fig. 2a) to form discrete silica capsules containing the file sequence that encodes for the image file. Quantitative PCR (qPCR) of the reaction supernatant after encapsulation (Supplementary Fig. 4) showed full encapsulation of plasmids without residual DNA in solution. To investigate the fraction of capsules that contained plasmid DNA, we compared the fluorescence intensity of the intercalating dye TO-PRO when added pre- versus post-encapsulation (Fig. 2). All capsules synthesized in the presence of both DNA and TO-PRO showed a distinct fluorescence signal, consistent with the presence of plasmid DNA in the majority of capsules, compared with a silica particle negative control that contained no DNA. In order to test whether plasmid DNA was fully encapsulated versus partially exposed at the surface of capsules, capsules were also stained separately with TO-PRO post-encapsulation (Fig. 2b). Using qPCR to measure released DNA after de-encapsulation, we estimated 106 plasmids per capsule (Supplementary Fig. 5 and Supplementary Table 3). Because encapsulation of the DNA file sequence relies only on electrostatic interactions between positively-charged silica and the phosphate backbone of DNA, our approach can equally encapsulate any molecular weight of DNA molecule applicable to MB and larger file sizes, as demonstrated previously23, and is compatible with alternative DNA file compositions such as 100–200-mer oligonucleotides that are commonly used2,6,7,11,12,17 or multiple layers of encapsulated DNA to increase the data density per capsule34.
Figure 2 |. Encapsulation of DNA plasmids into silica and surface barcoding.
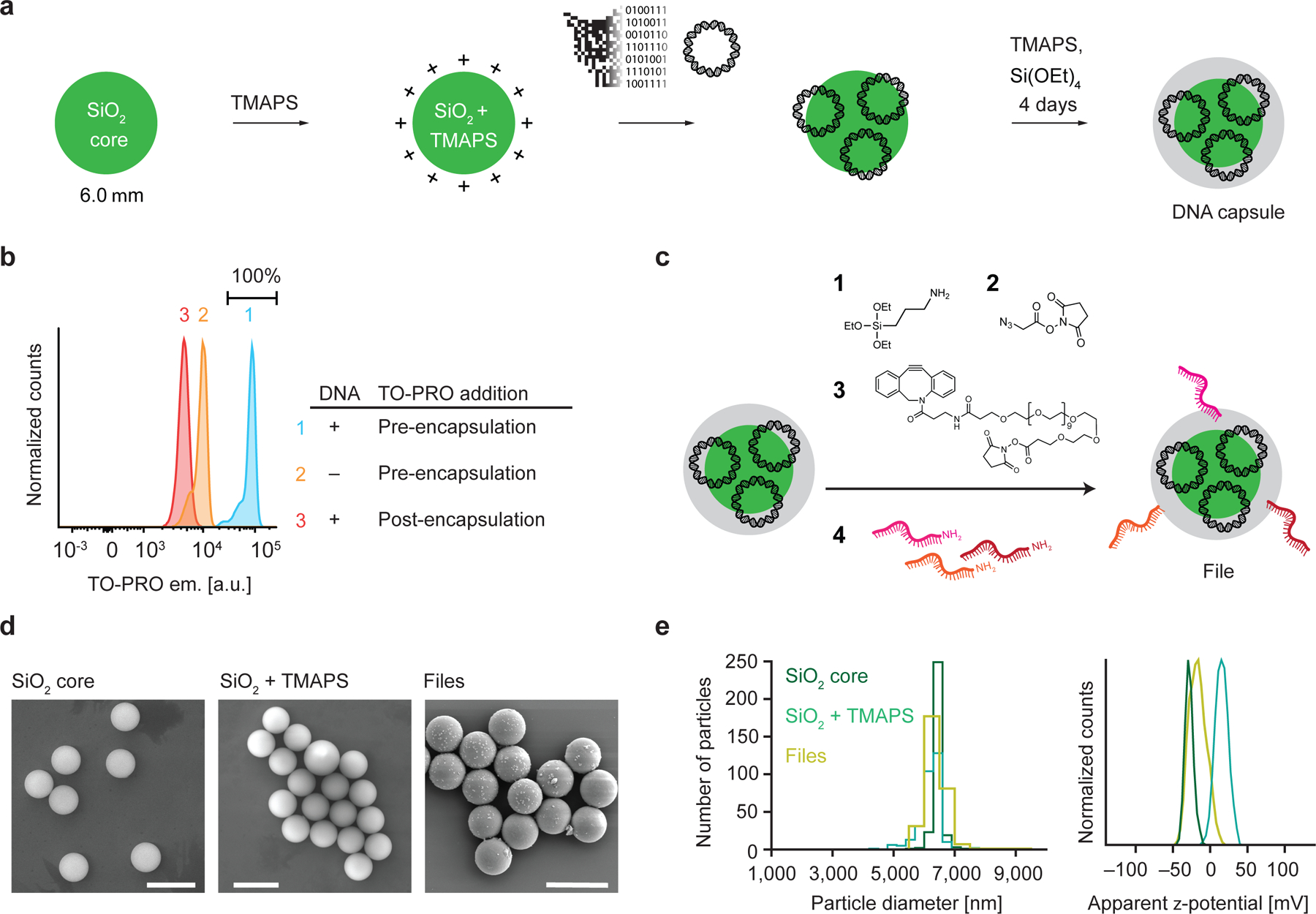
a, Workflow of silica encapsulation 23. b, Raw fluorescence data from FAS experiments to detect DNA staining of TO-PRO during or after encapsulation. c, Functionalization of encapsulated DNA particles. d, Representative scanning electron microscopy images of bare silica particles, silica particles functionalized with TMAPS, and the file. Scale bar: 10 µm. e, Distribution of particle sizes determined from microscopy data determined from particle size measurements taken from ten different fields of view for each sample (left) and zeta potential analyses of silica particles and files.
Next, we chemically attached unique content addresses on the surfaces of silica capsules using orthogonal 25-mer ssDNA barcodes (Supplementary Fig. 6) describing selected features of the underlying image for file selection. For example, the image of an orange tabby house cat (Supplementary Fig. 1) was described with cat, orange, and domestic, whereas the image of a tiger was described with cat, orange, and wild (Supplementary Fig. 1 and Supplementary Table 2). To attach the barcodes, we activated the surface of the silica capsules through a series of chemical steps. Condensation of 3-aminopropyltriethoxysilane with the hydroxy-terminated surface of the encapsulated plasmid DNA provided a primary amine chemical handle that supported further conjugation reactions (Fig. 2c).
We modified the amino-modified surface of the silica capsules with 2-azidoacetic acid N-hydroxysuccinimide (NHS) ester followed by an oligo(ethylene glycol) that contained two chemically orthogonal functional groups: the dibenzocyclooctyne functional group reacted with the surface-attached azide through strain-promoted azide-alkyne cycloaddition while the NHS ester functional group was available for subsequent conjugation with a primary amine. Each of the associated barcodes contained a 5’-amino modification that could react with the NHS-ester groups on the surface of the silica capsules, thereby producing the complete form of our file. Notably, the sizes of bare, hydroxy-terminated silica particles representing capsules without barcodes were comparable with complete files consisting of capsules with barcodes attached, confirmed using scanning electron microscopy (Fig. 2d and 2e, left). These results were anticipated given that the encapsulation thickness was only on the order of 10 nm23 and that additional steps to attach functional groups minimally increases the capsule diameter. We also observed systematic shifts in the surface charge of the silica particles as different functional groups were introduced onto their surfaces (Fig. 2e). Using hybridization assays with fluorescently-labelled probes35–37, we estimated the number of barcodes available for hybridization on each file to be on the order of 108 (Supplementary Fig. 7). Following synthesis, files were pooled and stored together for subsequent retrieval. The data pool contains at least ~106 copies of each file based on the mass of silica used during encapsulation (Supplementary Section 5). Short-read sequencing was used to read each file sequence and reconstruct the encoded image following selection and de-encapsulation, in order to validate the complete process of image file encoding, encapsulation, barcoding, selection, de-encapsulation, sequencing, and image file reconstruction (Supplementary Figs. 9 and 10).
File Selection
Following file synthesis and pooling, we used FAS to select specific targeted files from the complete data pool through the reversible binding of fluorescent probe molecules to the file barcodes (Supplementary Figs. 11 and 12). In FAS, files in solution are hydrodynamically focused into a single stream of droplets. At sufficiently low concentrations of files, each droplet contains a single file and passes through a laser beam. All files contained a fluorescent dye, fluorescein, in their core as a marker to distinguish files from other particulates such as spurious silica particles that nucleated in the absence of a core or insoluble salts that may have formed during the sorting process. Each detected fluorescein event was therefore interpreted to indicate the presence of a single file during FAS (Supplementary Fig. 11). To apply a query such as flying to the image database, the corresponding fluorescently labeled ssDNA probe was added, which hybridized to the complementary barcode displayed externally on the surface of a silica capsule for FAS selection (Fig. 3a). Fluorescence originating from fluorescein and/or fluorescently-labelled ssDNA are used to separate fractions using sorting gates, which are drawn based on specific ranges of fluorescence intensities.
Figure 3 |. Single-barcode sorting.
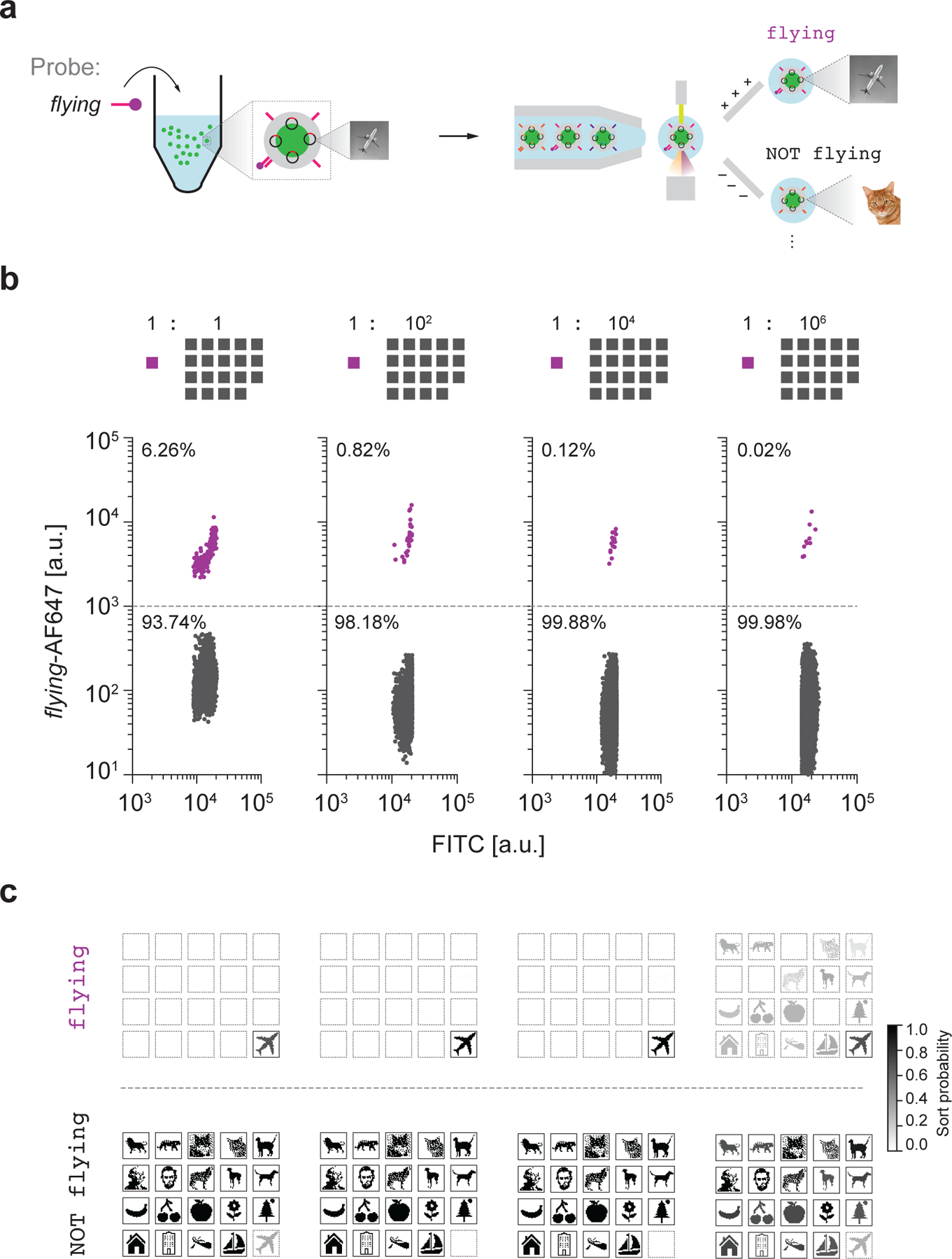
a, Schematic diagram of file sorting using FAS. b, Sorting of Airplane from varying relative abundance of the other nineteen files as background. Percentages represent the numbers of particles that were sorted in the gate. Colored traces in each of the sorting plots indicate the target population. c, Sequencing validation using Illumina MiniSeq. Sort probability is the probability that a file is sorted into one gated population over the other gated populations. We note that the sort probability is influenced by errors in FAS sorting and sequencing. Boxes with solid outlines indicate files that should be sorted into the specified gate. Other files have dashed outlines.
We subjected the entire data pool to a series of experiments to test selection sensitivity of target subsets using distinct queries. First, we evaluated single-barcode selection of an individual file, specifically Airplane, out of a pool of varying concentrations of the nineteen other files as background (Fig. 3b, Supplementary Fig. 13). To select the Airplane file, we hybridized an AFDye 647-labelled ssDNA probe that is complementary to the barcode flying, which is unique to Airplane. We were able to detect and select the desired Airplane file through FAS even at a relative abundance of 10−6 compared with each other file (Fig. 3c). While comparable in sensitivity to a nested PCR barcoding data indexing approach17, unlike PCR that requires 20–30 of rounds of heating and cooling to selectively amplify the selected sequence, our approach selects files directly without need for thermal cycling and amplification. This strategy also applies to gating of N barcodes simultaneously in parallel optical channels, which offers file selection sensitivity of 1 in 106N total files, where common commercial FAS systems offer up to N = 17 channels38,39. For example, comparison of the retrieved sequences between the flying gate and the NOT flying gate after chemical release of the file sequences from silica encapsulation revealed that 60–95% of the Airplane files were sorted into the flying gate (Supplementary Figs. 18–21), where we note that any sort probability above 50% indicates enrichment of Airplane within the correct population subset (flying) relative to the incorrect subset (NOT flying). In general, a file with sort probability p has sort error rate 1–p if it is a target file and error rate p if it is an off-target file. In this case, for example, a sort probability of 100% for Airplane into the flying subset would indicate ideal performance. Besides single file selection, our approach allows for repeated rounds of FAS selection, as well as Boolean logic, described below.
Boolean Search
Beyond direct selection of 1 in 106N individual random files, without thermal cycling or loss of fidelity due to primer crosstalk, our system offers the ability to apply Boolean logic to select random file subsets from the data pool. AND, OR, and NOT logical operations were applied by first adding to the data pool fluorescently labeled ssDNA probes that were complementary to the barcodes (Fig. 4, left). This hybridization reaction was used to distinguish one or several files in the data pool, which were then sorted using FAS. We used two to four fluorescence channels simultaneously to create the FAS gates that executed the target Boolean logic queries (Fig. 4, middle). To demonstrate a NOT query, we added to the data pool an AFDye 647-labelled ssDNA probe that hybridized to files that contained the cat barcode. Files that did not show AFDye 647 signal were sorted into the NOT cat subset (Fig. 4a, Supplementary Fig. 14a). An example of an OR gate was applied to the data pool by simultaneously adding dog and building probes that both had the TAMRA label (Fig. 4b, Supplementary Fig. 14b). All files that showed TAMRA signal were sorted into the dog OR building subset by the FAS. Finally, an example of an AND gate was achieved by adding fruit and yellow probes that were labelled with AFDye 647 and TAMRA, respectively. Files showing signal for both AFDye 647 and TAMRA were sorted into the fruit AND yellow subset in the FAS (Fig. 4c, Supplementary Fig. 14c). For each example query, we validated our sorting experiments by releasing the file sequence from silica encapsulation and sequencing the released DNA with Illumina MiniSeq (Fig. 4, right). Sort probabilities of each file for each search query are shown in Supplementary Figs. 22–24.
Figure 4 |. Fundamental Boolean logic gates.
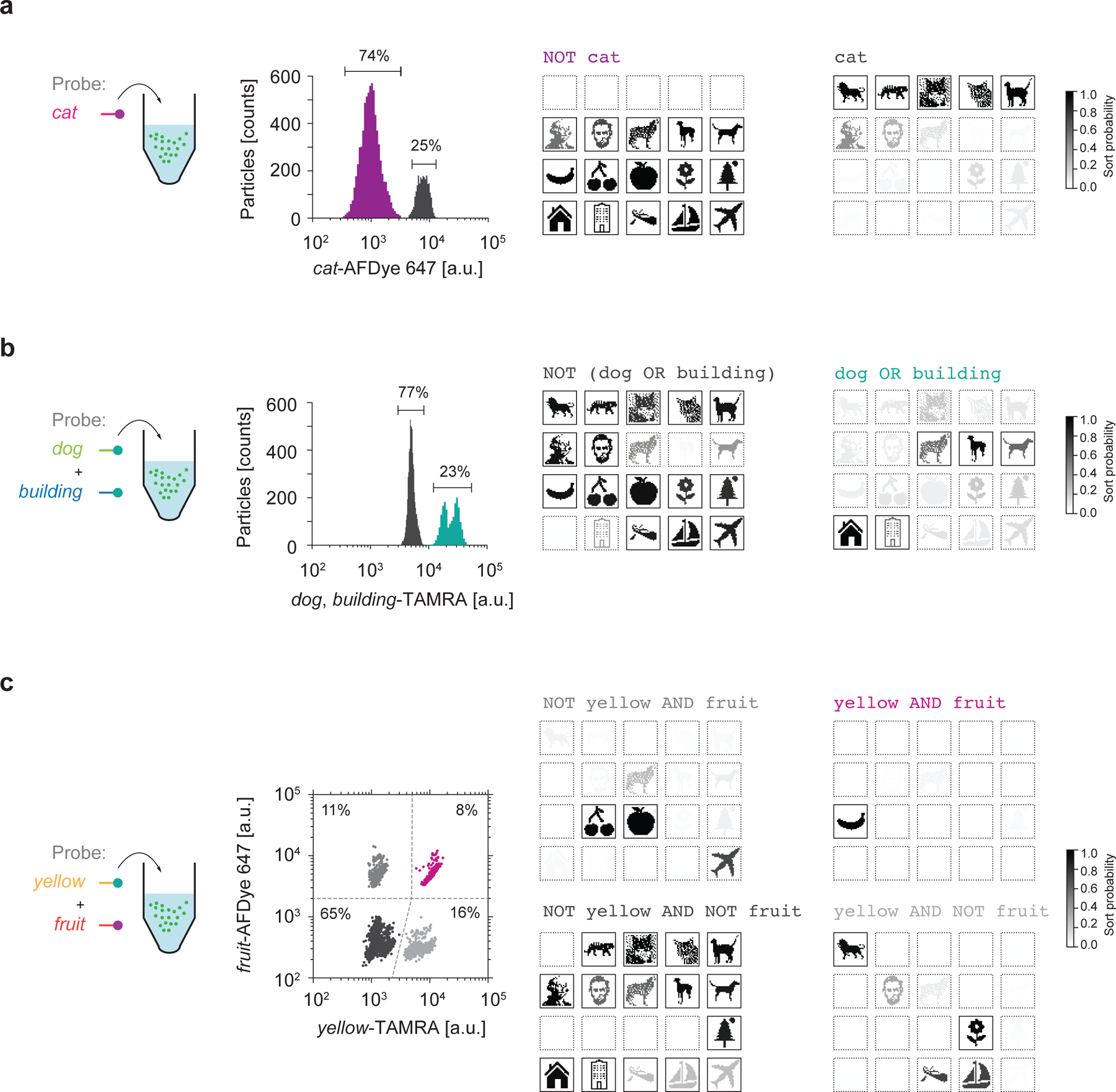
a, NOT cat selection. Raw fluorescence trace from the FAS system (left) plotted on a 1D sorting plot showing the percent of particles that were sorted in each gate. Sequencing using Illumina MiniSeq tested selection specificity (right). b, dog OR building selection. Raw fluorescence trace from the FAS system (left) plotted on a 1D sorting plot showing the percent of particles that were sorted in each gate. Sequencing using Illumina MiniSeq evaluated sorting using the OR gate (right). c, A 2D sorting plot to perform a yellow AND fruit gate. Percentages in each quadrant show the percentages of particles that were sorted in each gate. Colored traces in all of the sorting plots indicate the target populations. Sort probability is the probability that a file is sorted into one gated population versus the other gated populations. We note that the sort probability is influenced by errors in FAS sorting and sequencing. Boxes with solid outlines indicate files that were intended to sort into the specified gate. Other files have dashed outlines.
The preceding demonstrations of Boolean logic gates enable file sorting with varying specificity of selection criteria for the retrieval of different subsets of the data pool. FAS can also be used to create multiple gating conditions simultaneously, thereby increasing the complexity of target file selection operations, as noted above. To demonstrate increasingly complex Boolean search queries, we selected the file containing the image of Abraham Lincoln from the data pool, which included images of two presidents, George Washington and Abraham Lincoln. The president ssDNA probe, fluorescently labeled with TAMRA, selected both Lincoln and Washington files from the data pool. The simultaneous addition of the 18th century ssDNA probe, fluorescently labeled with AFDye 647 (Fig. 5a, left), discriminated Washington, which contained the 18th century barcode, from the Lincoln file (Fig. 5a, middle, Supplementary Fig. 15a). The combination of these two ssDNA probes permitted the complex search query president AND (NOT 18th century). Sequencing analysis of the gated populations after reverse encapsulation validated that the sorted populations matched search queries for president AND (NOT 18th century), president AND 18th century, and NOT president (Fig. 5a, right; Supplementary Fig. 25).
Figure 5 |. Arbitrary logic searching.
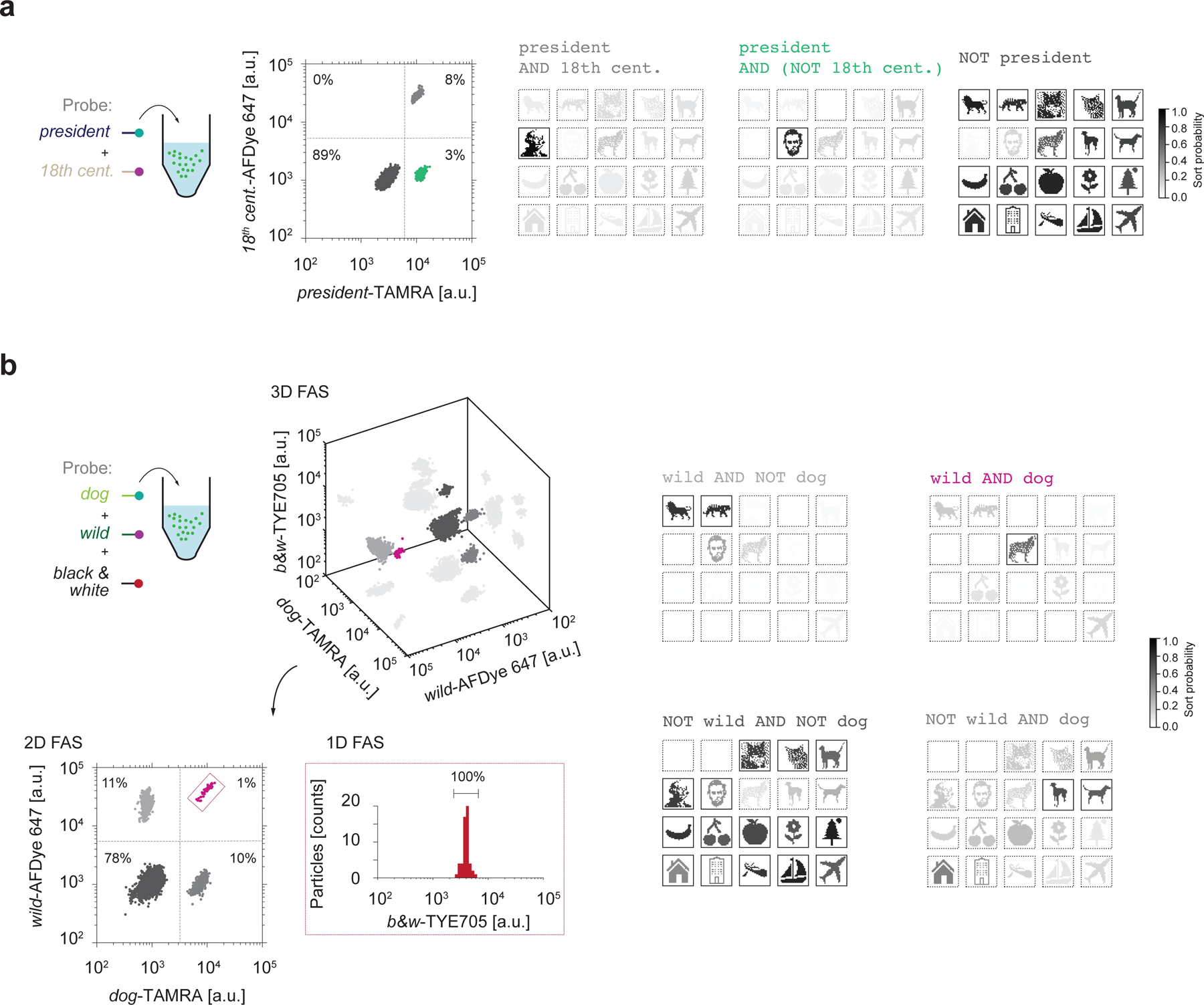
a, president AND (NOT 18th century) sorting. A 2D sorting plot (middle) was used to sort Lincoln by selecting a population that has high TAMRA fluorescence but low AFDye 647 fluorescence. Sequencing using MiniSeq offered quantitative evaluation of the sorted populations. b, Multiple fluorescence channels were projected into a 3D FAS plot (left and top). There are three possible 2D plots that can be used for sorting. To select the Wolf image using the query wild AND dog, a 2D plot of wild versus dog was first selected and then populations selected using quadrant gates (left and bottom). One of the quadrants were then selected where the Wolf image should belong based on the wild AND dog query in order to test whether only a single population was present in the TYE705 fluorescence channel. Sequencing quantified the sorted populations (right) using Illumina MiniSeq. Sort probability is the probability that a file was sorted into one gated population over the other gated populations. We note that the sort probability is influenced by errors in FAS sorting and sequencing. Boxes with solid outlines indicate files that would ideally be sorted into the specified gate. Other files have dashed outlines.
To demonstrate the feasibility of performing Boolean search using more than three fluorescence channels for sorting, we also selected the Wolf file from the data pool using the query dog AND wild, and used the black & white probe to validate the selected file (Fig. 5b, left). Because conventional FAS software is only capable of sorting using 1D and 2D gates, we first selected one out of the three possible 2D plots (Fig. 5b, left and bottom, Supplementary Figure 15b): dog-TAMRA against wild-AFDye 647. We examined the black & white-TYE705 channel on members of the dog AND wild subset (Fig. 5b, left and bottom, Supplementary Figure 15b). Release of the encapsulated file sequence and subsequent sequencing of each gated population from the dog versus wild 2D plot validated sorting (Fig. 5b, right; Supplementary Fig. 26).
Our use of plasmids as a substrate for encoding information offered the ability to restore selected files back into the data pool after retrieval. In cases where single images were selected (Figs. 4c, 5a), we were able to transform competent bacteria from each search query that resulted in a single file (Supplementary Fig. 27). Amplified material was pure and ready for re-encapsulation into silica particles, which could be re-introduced directly back into the data pool and thereby represents a complete write-store-access-read cycle that in principle may be applied larger-scale datasets, with periodic renewal of single-stranded DNA barcodes and bacterial replication of DNA data following reading25–27. Notably, in a practical implementation of our file system, only the selected files must be restored via amplification, encapsulation, and barcoding, because files that are not selected can be returned to the data pool immediately following FAS. While sort probabilities were typically below the optimal 100% targeted for a specific file or file subset query, future work may characterize sources of error that could be due to sample contamination or random FAS errors. The latter type of error may be mitigated through repeated cycles of file selection in series.
Discussion & Outlook
We introduced a direct random access molecular file system for the retrieval of arbitrary files and file-subsets from an archival DNA data store. Our file system overcomes several challenges associated with pre-existing PCR-based file systems, including obviating the need for numerous heating and cooling cycles and enzymatic synthesis, and eliminating non-specific crosstalk between file sequences and barcodes17,19,21, while enabling arbitrary Boolean logical search queries. Notwithstanding, several technical limitations also exist for our system.
First, while our file system supports theoretical storage densities that are orders of magnitude higher than archival file storage systems based on hard disk, magnetic tape, or similar archival media1,40 commonly used today, silica encapsulation of DNA file sequences lowers overall data storage density compared with bare DNA memory (Supplementary Section 6). Second, latency associated with DNA-based barcoding of silica capsules may limit its application to archival data storage and retrieval applications for the foreseeable future. Specifically, advanced liquid handling or microfluidics will be needed to manipulate large numbers of distinct files during the encapsulation and barcoding steps, and latency associated with the encapsulation process itself might need to be reduced from days, as currently implemented, to several hours or less, for example, using encapsulant alternatives such as alginates41 or synthetic polymers42, in order to realize a practical DNA data storage device (Supplementary Section 13). This is in contrast to existing file systems, such as PCR and nested file addressing8,11,12,17,22, wherein barcodes are instead written directly with the DNA file sequences using solid-phase phosphoramidite synthesis, g., using DNA microarrays. Third, retrieval time is currently limiting for our system in an exabyte-scale data pool because FAS scales linearly with the size of the pool. For example, in our proof-of-concept system, data reconstruction was successful with 100 copies of a file (Figure 3 and Supplementary Figure 13), so that our file size of 0.1 kB and search rate of 1,000 files per second yield a final sorting rate of only ~1 kB per second. However, because our proof-of-concept system did not make use of the vast majority of DNA stored in each capsule, the data stored in each capsule can in theory be significantly increased to yield a search rate of ~1 GB per second, or ~15 days to search through 1 PB of data (Supplementary Section 10). Additional technology development such as custom flow nozzles to accommodate high flow rates close to the limit of a typical FAS instrument (~104 particles per second or ~10 GB per second), parallel flow cytometry to achieve search rates of ~106 particles per second per device43 (~1 TB per second), or physical sorting strategies such as direct biochemical pulldown using magnetic extraction17,44,45 may therefore be required to implement a practical exabyte- or larger-scale memory storage and retrieval device with our system.
Finally, while DNA file protection by silica encapsulation offers millennium-scale protection and storage9 of the encapsulated file, unprotected DNA barcodes and their covalent linkers are susceptible to hydrolysis and will therefore require either periodic renewal or additional protection, as noted at the outset of this article. As also noted there, similar to PCR-based approaches, barcodes can be protected by dehydrating the data pool or immersing files in hydrophobic liquids21. Alternatively, secondary encapsulation with chemistries orthogonal to silica de-encapsulation, such as cleavable synthetic polymers46 or salts47, may be explored.
The latency times associated with file selection and recovery of DNA from files renders our file system ideally suited to archival data storage in which data are written once, stored for decades or longer, and accessed rarely, file protection by silica encapsulation offers millennium-scale storage9 of immutable data, such as astronomical image databases48, high-energy physics datasets49, or high-resolution deep ocean floor mapping50. Finally, because our system is not limited to synthetic DNA, it also applies to compact and energy-efficient long-term archival storage of bacterial, human, and other genomes for archival sample preservation and retrieval.
Methods
Generating file sequences.
Twenty 26 × 26-pixel, black-and-white icon bitmaps were generated as representations of 20 high-resolution color images (Supplementary Fig. 1) encompassing a broad range of subject matters. Each black-and-white icon was converted to a length-676 bitstring in a column-first order, with each black or white pixel encoded as a 0 or 1, respectively. This bitstring was compressed via run length encoding, replacing each stretch of consecutive 0s or 1s with a 2-tuple (value, length) to generate a list of 2-tuples describing the entire bitstring. The maximum length is 15; runs longer than 15 bits are encoded as multiple consecutive runs of the same value. This run length encoding was converted to a sequence of base-4 digits as follows:
Begin with an empty string, and set the current run to the first run of the list.
Append the value of the current run (0 or 1).
Using 2 base-4 digits, append the length of the current run.
If the length of this run was 15, encode the next run starting with Step (2). Otherwise, encode starting at Step (3). If no runs remain, the process is complete.
This process produces a quaternary string describing the entire run length encoding of the image. To avoid homopolymer runs and repeated subsequences in the final nucleotide sequence, each digit is offset by a random number generated from a linear congruential random number generator (LCG) beginning with a random seed (i.e. the ith value generated by the LCG is added, modulo 4, to the ith base-4 digit of the quaternary string). We used an LCG of multiplier 5, modulus 231-1, and increment 0, although any LCG parameters with period longer than the length of the sequence would be suitable.
The final “randomized” quaternary string is converted to nucleotides by a direct mapping (0 = G, 1 = A, 2 = T, 3 = C). The number used to seed the LCG is prepended to this sequence by converting it into a ternary string of length 20, whose digits are encoded in nucleotides via a base transition table, as done previously by Goldman et al.7: (0 = GA, AT, TC, CG; 1 = GT, AC, TG, CA; 2 = GC, AG, TA, CT). The first digit is encoded directly (0 = A, 1 = T, 2 = C).
This sequence was modified with additional flanking sequences added to the beginning and end. A 64-bit wavelet hash of each bitmap was calculated using the whash function provided by the ImageHash Python package, available through the Python Package Index (https://pypi.org/project/ImageHash/). The 64-bit hash was split into two 32-bit halves, each of which was represented in a length-24 ternary string that was subsequently converted to nucleotides through the same process as applied to the seed. The two 24-nt regions were appended to the beginning and end of the sequence (Supplementary Table 1). The sequence containing image hash, seed, and image encoding, was additionally flanked on the 5’ and 3’ ends by sequences (5′-CGTCGTCGTCCCCTCAAACT-3′ and 5′-GCTGAAAAGGTGGCATCAAT-3′, respectively) that allow amplification from a “master primer” pair that would amplify every sequence in the molecular plasmid database (Supplementary Table 1 and Supplementary Fig. 2).
The final sequence was checked for problematic subsequences, specifically, GGGG, CCCC, AAAAA, TTTTT, and the restriction enzyme recognition sites GAATTC and CTGCAG. If any of these subsequences were found outside of expected occurrences in the constant flanking regions, the entire sequence was regenerated with a new random seed until no such subsequences appeared.
The generated sequences were cloned on a pUC19-based vector. The software for sequence encoding and decoding is publicly available on GitHub at https://github.com/lcbb/DNA-Memory-Blocks/51 and the plasmids are publicly available from AddGene (https://www.addgene.org/browse/article/28206796/). Each master primer and hash barcode pairs were verified by PCR and agarose gel analysis and PCR bias was checked by qPCR (Supplementary Fig. 3).
Generated DNA sequences were ordered as custom genes synthesized, cloned, sequenced, and validated by Integrated DNA Technologies into pUC19-based vector. Each clone was designed with flanking single EcoRI and PstI cut sites, sequences which had been designed against in the inserts, enabling future sub-cloning to alternative vectors using standard digestion and ligation. All plasmids were amplified in bacterial cultures and purified using Qiagen (Venlo, Netherlands) HiSpeed Midi or Maxi kits following the protocol provided by the manufacturer. Briefly, DH5α cells were made chemically competent by growing 100 mL of cells to 0.5 and 0.6 at OD600. The cells were gently pelleted and incubated on ice in 100 mM CaCl2 for 30 minutes. The pelleted cells were then brought up in 100 mM CaCl2 with 10% glycerol. Transformations from each plasmid were accomplished by incubation of 20 µL chemically competent cells with 1 ng of plasmid DNA for 30 minutes on ice, followed by heating to 42 °C for 45 seconds, placing on ice for 2 minutes, and then addition of 1 mL of pre-warmed SOC media (Thermo Fisher; catalog number: 15544034). The mixture was shaken for 1 hour at 250 rpm at 37 °C. A volume of 20 µL of transformed cells were streaked to single colonies on agar plates that were supplemented with 100 µg mL−1 ampicillin. Single colonies were chosen for growth in 100 mL LB with 100 µg mL−1 ampicillin (Millipore Sigma; catalog number: A5354) overnight in disposable baffled flasks and then followed by pelleting cells from the culture. Pelleted cells were lysed and treated with Qiagen purification buffers. Plasmid DNA was then purified on the provided silica support matrix and eluted in 1 mL of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Plasmid preparations were verified by PCR, Sanger sequencing, and short-read sequencing using Illumina MiniSeq for quality control (Supplementary Fig. 3).
DNA encapsulation.
A volume of 1.0 mL of 50 mg mL−1 fluorescein-core 5 µm silica particles (Creative Diagnostics; catalog number: DNG-L034) was added into a 2.0 mL DNA/RNA LoBind Eppendorf tube. The particles were centrifuged at 1,000 rpm for 10 seconds using a benchtop centrifuge. The particles were redispersed in 1.0 mL anhydrous ethanol with vigorous vortexing. The particles were centrifuged and redispersed in ethanol five times. We then added 50 µL of 50% TMAPS in methanol (Alfa Aesar; catalog number: H66414) to the dispersed 5 µm silica particles (50 mg mL−1 in ethanol). The mixture was stirred overnight at room temperature using a thermal mixer (Thermo Fisher; catalog number: 13687711) at 1,200 rpm. The mixture was centrifuged at 1,000 rpm and washed with ethanol five times to remove any unreacted TMAPS. The functionalized particles were finally redispersed in 1.0 mL DNAse/RNAse-free water (Thermo Fisher; catalog number: 10977015). The particles were stored at room-temperature until further use.
For each data encoding plasmid, a mass of 1.0 mg of TMAPS-functionalized, fluorescent 5 µm particle was added into a 2 mL LoBind Eppendorf tube containing 15 µg of plasmid DNA dissolved in 1 mL of water. The mixture was mixed gently using a tube revolver (Thermo Fisher; catalog number: 88881001) at 30 rpm and at room-temperature for 5 minutes. A volume of 10 µL of 50% TMAPS in methanol was then added to mixture and stirred for 10 minutes, at 1,000-rpm, and at 25 °C using a thermal mixer. After 10 minutes, a volume of 2 µL of tetraethoxysilane (TEOS; Millpore Sigma; catalog number: 333859) was added and the mixture is stirred for 24 hours, at 1000 rpm, and at 25 °C using a thermal mixer (Thermo Fisher). An additional 5 µL of TEOS was then added and the mixture is stirred for 4 days, at 1,000 rpm, and at 25 °C which forms our encapsulated DNA particles or DNA capsules. The mixture is centrifuged at 2,000 × g for 3 minutes to sediment the DNA capsules then the supernatant was removed without disturbing the particle sediment pellet. The particles were washed repeatedly for 5 times by re-dispersing the particles with 1 mL of water, sedimenting the particles with a centrifuge at 2,000 × g for 3 minutes, and removing the supernatant. After the final wash, the DNA capsules were re-dispersed in 1 mL of ethanol with 30 seconds of vortex mixing. A volume of 20 µL of 2-aminopropyltriethoxysilane (Millipore Sigma; catalog number: 440140), was then added and the mixture was stirred for 18 hours, at 1,000 rpm, and at 25 °C using a thermal mixer. The mixture is centrifuged at 2,000 × g for 3 minutes to sediment the amino-modified DNA capsules then the supernatant was removed without disturbing the particle sediment pellet. The particles were washed repeatedly for 5 times by re-dispersing the particles with 1 mL of N-methyl-2-pyrrolidone, sedimenting the particles with a centrifuge at 2,000 × g for 3 minutes, and removing the supernatant. After the final wash, the DNA capsules were re-dispersed in 1 mL of N-methyl-2-pyrrolidone with 30 seconds of vortex mixing and the resulting colloidal suspension was then transferred into a clean 2-mL Eppendorf LoBind tube.
Barcoding DNA capsules.
Each encoded image was annotated with three semantic metadata descriptors (Supplementary Table 2) associated with the original image (Supplementary Fig. 1). A table was then generated to associate each descriptor with a unique sequence chosen from a list of 240,000 orthogonal barcode sequences20. Hexylamine-modified single-stranded DNA oligonucleotides that represent each descriptor were purchased from IDT and dissolved in nuclease-free water with a final concentration of 500 µM upon receipt.
Using all the DNA capsules from the previous step, a mass of 5 mg of 2-azido acetic acid N-hydroxysuccinimide ester (Click Chemistry Tools; catalog number: 1070) and 5 µL N,N-diisopropylethylamine (Millipore Sigma; catalog number: D125806) as catalyst were added and the mixture was stirred for 2 hours, at 1,000 rpm, and at 25 °C using a thermal mixer. The azide-modified DNA capsules were washed repeatedly for 5 times by re-dispersing the particles with 1 mL of N-methyl-2-pyrrolidone (Millipore Sigma; catalog number: 270458), sedimenting the azide-modified DNA capsules with a centrifuge at 2,000 × g for 3 minutes, and removing the supernatant. After the final wash, the azide-modified DNA capsules were re-dispersed in 1 mL of N-methyl-2-pyrrolidone. A mass of 2 mg of DBCO-PEG13-NHS ester (Click Chemistry Tool; catalog number: 1015) was added and the mixture was stirred for 30 minutes, at 1,000 rpm, and at °C using a thermal mixer. The particles were washed repeatedly for 5 times by re-dispersing the PEG-modified DNA capsules with 1 mL of N-methyl-2-pyrrolidone, sedimenting the PEG-modified DNA capsules with a centrifuge at 2,000 × g for 3 minutes, and removing the supernatant. After the final wash, the PEG-modified DNA capsules were re-dispersed in 200 µL of N-methyl-2-pyrrolidone with 30-seconds of vortex mixing and 1 minute sonication. A volume 10 µL of each ssDNA barcode (500 µM in nuclease-free water from IDT) and 200 µL of PEG-modified DNA capsules were added to 770 µL of 0.1 M bicarbonate buffer (pH 9.2; Alfa Aesar; catalog number: AAJ67384AE) in a 1.5 mL Eppendorf LoBind tube. The mixture was stirred for 2 hours, at 1,000 rpm, and at 25 °C using a thermal mixer to produce the final form of our data blocks or “files”. The files were washed repeatedly for 5 times by re-dispersing the particles with 1 mL of saline TAE buffer (40 mM Tris, 20 mM acetate, 2 mM EDTA, 1.0% Tween-20, 1.0% sodium dodecyl sulfate, and 500 mM NaCl), sedimenting the particles with a centrifuge at 2,000 × g for 3 minutes, and removing the supernatant. After the final wash, the particles were re-dispersed in 500 µL of saline TAE buffer with 30 seconds of vortex mixing and 1 minute sonication. All the files were then pooled together which forms the file pool or molecular file database with an estimated final concentration of 2.0 mg mL−1 in 10.0 mL of saline TAE buffer.
Silica particle characterization.
Surface zeta-potentials were measured using a Malvern Zetasizer Nano ZSP. All samples for surface zeta-potentials were prepared and measured in a standard fluorescence quartz cuvette (catalog number: 3-Q-10) from Starna Cells, Inc. at a concentration of 0.1 mg mL−1 with a volume of 700 µL. A universal ‘dip’ probe (Malvern Panalytical; catalog number: ZEN1002) was used to measure zeta potential of particles. Scanning electron microscopy of the particles were performed using a Zeiss Gemini 2 Field Emission Scanning Electron Microscope. Samples were mounted on silicon substrates for non-DNA modified particles and glass for DNA-modified particles.
Querying molecular file database using fluorescently-labelled probes.
The molecular file database was vortexed for 10 seconds, sonicated for two minutes, and re-vortexed for another 10 seconds to re-disperse the settled particles. For all sorting experiments except Figure 3, a volume of 100-µL of the molecular database (2 mg mL−1) is added into a 1.5 mL Eppendorf LoBind tube. Dye-labelled probes (Supplementary Section 7) for querying the molecular file database were added such that the final concentration of the DNA-dye single-stranded DNA in solution is 5 µM. The resulting mixtures were mixed at 70 °C at 1,200-rpm using a thermal mixer for 5 minutes. The mixtures were then cooled to 20 °C at 1,200 rpm using a thermal mixer over 20 minutes and then centrifuged at 10,000 × g for 1 minute. The supernatant was discarded, and the pelleted particles were washed with 500-µL of saline TAE buffer. The sedimentation and washing process was repeated for five additional times to remove non-specifically bound dye-DNA. The particles are finally re-suspended in 500 µL of saline TAE buffer.
For sorting experiments performed in Figure 3 and Supplementary Fig. 13, the workflow for querying the molecular file database remains the same except the composition of the molecular file database:
| Ratio of Airplane:19 other files |
Volume of Airplane stock solution [µL] |
Volume of stock solution of 19 other files [µL] [a] |
|---|---|---|
| 1:1 | 500 [a] | 0 |
| 1:102 | 5 [a] | 495 |
| 1:104 | 5 [b] | 495 |
| 1:106 | 5 [c] | 495 |
Concentration of stock solution is 2 mg mL−1
Concentration of stock solution is 2 × 10−2 mg mL−1 obtained from serial dilution
Concentration of stock solution is 2 × 10−4 mg mL−1 obtained from serial dilution
Fluorescence-activated sorting.
All fluorescence-activated sorting (FAS) experiments were performed on a BD FACSAria III flow cytometer. Samples were filtered through a Corning® 70-µm cell strainer (Fisher Scientific; catalog number: 07–201-431) prior to particle sorts. Samples are flowed into the instrument with 1× PBS as sheath fluid at a flow rate that maintains an events detection rate of 1,200 events per second and below. We found that performing sorting at a flow rate that exceeds this events rate clogged the FAS instrument intermittently. All sorts were accomplished with a standard 70 µm nozzle. The sample was held at room-temperature and agitated periodically every 5 minutes by pausing the sort and vortexing the sample vigorously with a vortex mixer. We note that the internal agitator in the flow cytometer with a 300 rpm agitation speed was not sufficient to prevent the silica particles from sedimenting over time and we found that periodically agitating the sample tube every 5 minutes with a vortex mixer was more effective. Since all files must contain a fluorescein core, all particles were gated by default using the ‘FITC’ laser and detector settings, which is defined by gating the majority population in the ‘FITC-A’ channel histogram, in addition to standard FSC and SSC gates to minimize sorting of doublets (Supplementary Fig. 11). All FAS experiments were performed at room-temperature and at a 500 µL constant volume given that sorting rates, detection rates, and flow rates can vary significantly across different flow cytometers.
Release of DNA from sorted files.
Sorted populations were centrifuged at 10,000 × g for 1 minute. The supernatant was carefully removed with a pipette to avoid disturbing the silica pellets. A volume of 45 µL of CMOS-grade 5:1 buffered oxide etch (VWR; catalog number: JT5192–3) was then added. The mixture was vortexed for 5 seconds to re-suspend the pellet and the mixture was statically incubated at room temperature for 5 minutes. A volume of 5 µL of 1 M phosphate buffer (0.75 M Na2HPO4; 0.25 M NaH2PO4; pH 7.5 at 0.1 M) was then added, vortexed for 1 second, and desalted twice through an Illustra MicroSpin S-200 HR column (Millipore Sigma; GE27–5120-01).
Sequencing of retrieved files.
For Illumina MiniSeq and MiSeq sequencing, the master primer pair with 5′ extensions matching Illumina Nextera sequencing adapters were used to amplify all plasmids simultaneously (Supplementary Fig. 9). Template amounts were adjusted based on concentrations determined with Qubit fluorescence assay (Thermo Fisher) or qPCR. If required, the amplification was simultaneously followed by qPCR and enough cycles were used to rise above the cycle threshold, or alternatively obtain a final concentration of 2 ng µL−1 . Dual sequence indices were then added to the adaptor-modified inserts at the 5′ and 3′ ends, associating the sequencing lane with a particular selection experiment, which was followed by SPRI bead cleanup (Beckman Coulter). A 25 µL PCR reaction amplified the material over 8–10 cycles using KAPA HiFi polymerase (Roche) with 1 ng of template and 1 µM forward and reverse primers. After amplification, the PCR mixture was combined with 20 µL of SPRIselect beads (Beckman Coulter), mixed, and let stand for 5 min. The mix was then separated by magnetic plates, and washed twice with 150 µL 80% ethanol, dried for 2 min, and eluted in 20 µL Qiagen TE buffer. Samples were quantified using the Qubit fluorescence assay with the provided high-sensitivity buffer and standards. A sequencing pool was generated to approximately equimolar amount per index pair. Illumina MiniSeq with 150 × 150 read lengths was used to read out the start and end of each sequence. Sequences were demultiplexed, and sequence clustering was used to count the number of occurrences of each image (Supplementary Section 9). The sort probability for a file into a particular fraction was calculated as the count associated with that file divided by the sum of the counts for that file over all fractions generated from an initial sample. This metric is also referred to as enrichment throughout the text. In Figs. 3–5 in the main text, the enrichment of each file is indicated by the percent opacity of the images displayed on the grid.
Supplementary Material
Acknowledgments.
We gratefully acknowledge fruitful discussions with Charles Leiserson and Tao B. Schardl on the scalability and generalizability of our barcoding approach. We thank Glenn Paradis, Michael Jennings, and Michele Griffin of the Flow Cytometry Core at the Koch Institute in MIT and Patricia Rogers of the Flow Cytometry Facility at the Broad Institute of Harvard and MIT for assistance and fruitful discussions in developing the flow cytometry workflow. We also thank David Mankus of the Nanotechnology Materials Core Facility at the Koch Institute in MIT for assistance in the imaging of the particles using the scanning electron microscope and Alla Leshinsky of the Biopolymer and Proteomics Core at the Koch Institute at MIT for assistance in mass spectrometry characterization.
Funding. M.B., J.L.B., T.R.S., and J.B. gratefully acknowledge funding from the Office of Naval Research N00014–17-1–2609, N00014–16-1–2506, N00014–12-1–0621, and N00014–18-1–2290 and the National Science Foundation CCF-1564025, CCF-1956054, HDR OAC-1940231, and CBET-1729397. Research was sponsored by the U.S. Army Research Office and accomplished under cooperative agreement W911NF-19–2-0026 for the Institute for Collaborative Biotechnologies. Additional funding to J.B. was provided through an NSF Graduate Research Fellowship (Grant # 1122374). P.C.B. was supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. C.M.A. was supported by NIH grant F32CA236425.
Footnotes
Competing Interests Statement. J.L.B., T.R.S., J.B. & M.B. have filed provisional patents (17/029,948 and 16/012,583) related to this work. M.B. is the founder of Cache DNA LLC and is a member of its Scientific Advisory Board. P.C.B. is a member of the Scientific Advisory Board of Cache DNA LLC. H.H., M.R. & C.M.A. declare that they have no competing interests.
Data Availability. Gene sequences and plasmid maps are available from AddGene (https://www.addgene.org/browse/article/28206796/). Insert sequences and barcoding sequences are given in Supplementary Tables 1 and 2. All the data files used to generate the plots in this manuscript are available from M.B. upon request.
Code Availability. Software for sequence encoding and decoding is publicly available on GitHub (https://github.com/lcbb/DNA-Memory-Blocks/).
Online Content. Any methods, additional references, and supplementary information are available at https://doi.org/10.1038/s41563-021-01021-3.
References
- 1.Zhirnov V, Zadegan RM, Sandhu GS, Church GM & Hughes WL Nucleic acid memory. Nature Materials 15, 366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ceze L, Nivala J & Strauss K Molecular digital data storage using DNA. Nature Reviews Genetics 20, 456–466, (2019). [DOI] [PubMed] [Google Scholar]
- 3.Kosuri S & Church GM Large-scale de novo DNA synthesis: technologies and applications. Nature Methods 11, 499–507, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palluk S et al. De novo DNA synthesis using polymerase-nucleotide conjugates. Nature Biotechnology 36, 645–650, (2018). [DOI] [PubMed] [Google Scholar]
- 5.Lee HH, Kalhor R, Goela N, Bolot J & Church GM Terminator-free template-independent enzymatic DNA synthesis for digital information storage. Nature Communications 10, 2383, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church GM, Gao Y & Kosuri S Next-generation digital information storage in DNA. Science 337, 1628–1628 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Goldman N et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yazdi SMHT, Yuan Y, Ma J, Zhao H & Milenkovic O A rewritable, random-access DNA-based storage system. Scientific Reports 5, 14138, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grass RN, Heckel R, Puddu M, Paunescu D & Stark WJ Robust chemical preservation of digital information on DNA in silica with error‐correcting codes. Angewandte Chemie International Edition 54, 2552–2555 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Yazdi SMHT, Gabrys R & Milenkovic O Portable and error-free DNA-based data storage. Scientific Reports 7, 5011, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erlich Y & Zielinski D DNA Fountain enables a robust and efficient storage architecture. Science 355, 950–954 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Organick L et al. Random access in large-scale DNA data storage. Nature Biotechnology 36, 242 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Ranu N, Villani A-C, Hacohen N & Blainey PC Targeting individual cells by barcode in pooled sequence libraries. Nucleic Acids Research 47, e4–e4, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashiwamura S, Yamamoto M, Kameda A, Shiba T & Ohuchi A in 8th International Workshop on DNA-Based Computers (DNA8). 112–123 (Springer). [Google Scholar]
- 15.Yamamoto M, Kashiwamura S, Ohuchi A & Furukawa M Large-scale DNA memory based on the nested PCR. Natural Computing 7, 335–346 (2008). [Google Scholar]
- 16.Yamamoto M, Kashiwamura S & Ohuchi A in 13th International Meeting on DNA Computing (DNA13). 99–108 (Springer). [Google Scholar]
- 17.Tomek KJ et al. Driving the scalability of DNA-based information storage systems. ACS Synthetic Biology 8, 1241–1248, (2019). [DOI] [PubMed] [Google Scholar]
- 18.Organick L et al. Probing the physical limits of reliable DNA data retrieval. Nature Communications 11, 616, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y-J et al. Quantifying molecular bias in DNA data storage. Nature Communications 11, 3264, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Q, Schlabach MR, Hannon GJ & Elledge SJ Design of 240,000 orthogonal 25mer DNA barcode probes. Proceedings of the National Academy of Sciences 106, 2289–2294, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman S et al. High density DNA data storage library via dehydration with digital microfluidic retrieval. Nature Communications 10, 1706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin KN, Volkel K, Tuck JM & Keung AJ Dynamic and scalable DNA-based information storage. Nature Communications 11, 2981, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paunescu D, Puddu M, Soellner JOB, Stoessel PR & Grass RN Reversible DNA encapsulation in silica to produce ROS-resistant and heat-resistant synthetic DNA “fossils”. Nature Protocols 8, 2440, (2013). [DOI] [PubMed] [Google Scholar]
- 24.Paunescu D, Fuhrer R & Grass RN Protection and deprotection of DNA--high-temperature stability of nucleic acid barcodes for polymer labeling. Angewandte Chemie International Edition 52, 4269–4272, (2013). [DOI] [PubMed] [Google Scholar]
- 25.Farzadfard F et al. Single-nucleotide-resolution computing and memory in living cells. Molecular Cell 75, 769–780. e764 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farzadfard F & Lu TK Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farzadfard F & Lu TK Emerging applications for DNA writers and molecular recorders. Science 361, 870–875, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen HH et al. Long-term stability and integrity of plasmid-based DNA data storage. Polymers 10, 28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plesa C, Sidore AM, Lubock NB, Zhang D & Kosuri S Multiplexed gene synthesis in emulsions for exploring protein functional landscapes. Science 359, 343–347, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shepherd TR, Du RR, Huang H, Wamhoff E-C & Bathe M Bioproduction of pure, kilobase-scale single-stranded DNA. Scientific Reports 9, 6121, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veneziano R et al. In vitro synthesis of gene-length single-stranded DNA. Scientific Reports 8, 1–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minev D et al. Rapid in vitro production of single-stranded DNA. Nucleic Acids Research 47, 11956–11962, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reif JH et al. in 7th International Workshop on DNA-Based Computers (DNA7). 231–247 (Springer Berlin Heidelberg). [Google Scholar]
- 34.Chen WD et al. Combining data longevity with high storage capacity—layer-by-layer DNA encapsulated in magnetic nanoparticles. Advanced Functional Materials 29, 1901672, (2019). [Google Scholar]
- 35.Pillai PP, Reisewitz S, Schroeder H & Niemeyer CM Quantum-dot-encoded silica nanospheres for nucleic acid hybridization. Small 6, 2130–2134, (2010). [DOI] [PubMed] [Google Scholar]
- 36.Leidner A et al. Biopebbles: DNA-functionalized core–shell silica nanospheres for cellular uptake and cell guidance studies. Advanced Functional Materials 28, 1707572, (2018). [Google Scholar]
- 37.Sun P et al. Biopebble containers: DNA-directed surface assembly of mesoporous silica nanoparticles for cell studies. Small 15, 1900083, (2019). [DOI] [PubMed] [Google Scholar]
- 38.Perfetto SP, Chattopadhyay PK & Roederer M Seventeen-colour flow cytometry: unravelling the immune system. Nature Reviews Immunology 4, 648–655, (2004). [DOI] [PubMed] [Google Scholar]
- 39.Chattopadhyay PK et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nature Medicine 12, 972–977, (2006). [DOI] [PubMed] [Google Scholar]
- 40.Fontana RE Jr. & Decad GM Moore’s law realities for recording systems and memory storage components: HDD, tape, NAND, and optical. AIP Advances 8, 056506, (2018). [Google Scholar]
- 41.Machado AHE et al. Encapsulation of DNA in macroscopic and nanosized calcium alginate gel particles. Langmuir 29, 15926–15935, (2013). [DOI] [PubMed] [Google Scholar]
- 42.Zelikin AN et al. A general approach for DNA encapsulation in degradable polymer microcapsules. ACS Nano 1, 63–69, (2007). [DOI] [PubMed] [Google Scholar]
- 43.Hur SC, Tse HTK & Di Carlo D Sheathless inertial cell ordering for extreme throughput flow cytometry. Lab on a Chip 10, 274–280, (2010). [DOI] [PubMed] [Google Scholar]
- 44.Lee H, Kim J, Kim H, Kim J & Kwon S Colour-barcoded magnetic microparticles for multiplexed bioassays. Nature Materials 9, 745–749, (2010). [DOI] [PubMed] [Google Scholar]
- 45.Stewart K et al. in 24th International Conference on DNA Computing and Molecular Programming (DNA 24). 55–70 (Springer). [Google Scholar]
- 46.Shieh P et al. Cleavable comonomers enable degradable, recyclable thermoset plastics. Nature 583, 542–547, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohll AX et al. Stabilizing synthetic DNA for long-term data storage with earth alkaline salts. Chemical Communications 56, 3613–3616, (2020). [DOI] [PubMed] [Google Scholar]
- 48.Broekema PC, Nieuwpoort R. V. v. & Bal HE in Proceedings of the 2012 workshop on High-Performance Computing for Astronomy Date 9–16 (Association for Computing Machinery, Delft, The Netherlands, 2012). [Google Scholar]
- 49.Gaillard M & Pandolfi S CERN Data Centre passes the 200-petabyte milestone, <https://cds.cern.ch/record/2276551> (2017).
- 50.Mayer L et al. The Nippon Foundation—GEBCO seabed 2030 project: The quest to see the world’s oceans completely mapped by 2030. Geosciences 8, 63 (2018). [Google Scholar]
- 51.Banal JL et al. , lcbb/DNA-Memory-Blocks: Publication release, Zenodo (DNA-Memory-Blocks-v2.0), (2021) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


