Abstract
Host cell factor 1 (HCF-1; also called C1) is a 230-kDa protein which is cleaved posttranslationally into separate but associated N- and C-terminal polypeptides. These polypeptides are components of the C1 complex, along with Oct-1 and the viral protein VP16. The C1 complex is formed when herpes simplex virus (HSV) infects a cell and is responsible for transcription of the HSV immediate-early genes. A temperature-sensitive mutation in the N-terminal kelch domain of HCF-1 reversibly arrests cells in a G0-like state when grown at the nonpermissive temperature, and the same domain interacts with VP16 in the formation of the C1 complex. The form of HCF-1 in primary G0 cells was investigated by using peripheral blood mononucleocytes and serum-arrested human primary fibroblasts. A novel 50-kDa N-terminal fragment of HCF-1 encompassing the kelch domain was identified in the cytoplasm of these cells. This fragment arises by proteolysis of the full-length HCF-1 protein and is able to associate with VP16.
Upon herpes simplex virus (HSV) infection, the viral protein VP16 (also called αTIF and vmw65) is released from the virion particle and forms a complex with two cellular proteins, Oct-1 and host cell factor 1 (HCF-1; also called C1 and VCAF) (2, 7, 16, 19). This complex, termed the C1 complex, directs specific transcription from the alpha/immediate-early (α/IE) promoter element of the viral genome (8). Oct-1 is a member of the POU domain family of proteins and normally regulates transcription from octamer and related elements (5′-ATGCAAAT-3′) found in the promoters of a diverse array of cellular genes (5, 21). In contrast, the complex of Oct-1 and VP16 has specificity for the viral α gene promoters, with Oct-1 contacting the 5′ half of the α/IE promoter element (5′-ATGCTAAT-3′), and VP16 making contacts in the 3′ half (8). HCF-1 stabilizes the interaction between VP16 and Oct-1 and may also contact the DNA (8, 23).
HCF-1 consists of an array of polypeptides, varying in length from 100 to 230 kDa (6, 9, 23). All of the observed peptides are derived from the proteolysis of a single 2,035-amino-acid, 230-kDa protein (6, 23). After translation, HCF-1 is imported into the nucleus, where it is cleaved at one or more of the six near-perfect 26-amino-acid repeats found near the center of the protein. The cleavage is specific and occurs between a glutamic acid and a threonine (6, 25). Neither this 26-amino-acid sequence nor a related motif has been identified as a cleavage site in other proteins, but this repeat can direct the cleavage of a heterologous protein when inserted into its coding sequence (25). After cleavage, the N and C termini of HCF-1 remain strongly associated (6, 25). The N terminus of HCF-1 contains a series of six kelch repeats (named after the Drosophila egg chamber protein kelch, in which they were originally recognized). These repeats are found in other unrelated proteins and are predicted to fold into a barrel-like β-propeller structure (22). The first 360 amino acids of HCF-1 (HCF1–360), encompassing the kelch repeat region, are sufficient for binding to VP16 and for formation of a fast-mobility C1-like complex in vitro (12, 22). The cellular protein luman, or LZIP, a member of the basic leucine zipper family of DNA-binding proteins, has also been shown to bind HCF-1 in this region (1, 13). VP16 and luman share a short region of amino acid homology that, when mutated, greatly reduces both proteins' ability to bind HCF-1 (1, 14).
A proline-to-serine mutation in the third kelch repeat (P134S) confers a temperature-sensitive phenotype on HCF-1. BHK cells carrying this mutation in their only copy of HCF-1 arrest in a G0-like state. These cells divide for 36 h after a shift to the nonpermissive temperature, then arrest and remain highly viable for several days. If they are shifted back to the permissive temperature during this time, they will reenter the G1 phase of the cell (3). This suggests that a function of HCF-1 is required for cells to cycle. Consistent with this hypothesis, HCF-1 is most abundant in cycling cells, including fetal tissue and immortalized cell lines (6, 24). It has therefore been proposed that VP16 interacts with HCF-1 to allow the infecting virus to sense the proliferative state of the host cell (3).
At the nonpermissive temperature, the expression levels and proteolytic profiles of the P134S mutant HCF-1 are identical to those of the wild-type protein, and the mutant N terminus is still able to associate with the cleaved C terminus. However, mutant HCF-1 is unable to support VP16-mediated transcription from an α/IE reporter element in vivo and does not support C1 complex formation in vitro (3, 22). Nearly the entire N terminus of HCF-1 (HCF1–902) is required for complementation of the temperature-sensitive phenotype; deletion of as few as 66 additional amino acids from the C terminus of this fragment renders it unable to prevent G0 arrest at the nonpermissive temperature (22).
The role of HCF-1 in controlling HSV entry into either a replicative or latent state is not known. The amount of VP16 that enters the nucleus may influence the balance toward one type of infection or the other (20). La Boissiere et al. recently reported that VP16 entry into the nucleus depends on the HCF-1 nuclear localization signal (NLS), which is found within the C terminus of HCF-1. When the NLS was deleted, transfected HCF-1 remained in the cytoplasm, along with a large portion of the cotransfected VP16 (11). In extracts from dorsal root ganglia, a cell type in which HSV is able to establish latency, a faster-mobility form of the C1 complex has been observed, suggesting that an altered form of HCF-1 may be present (4). Interestingly, immunofluorescent staining of HCF-1 in trigeminal ganglia shows the protein to be sequestered in cytoplasmic granules, and conditions which activate HSV from latency rapidly induce the appearance of HCF-1 in the nucleus of these cells (10). We have investigated the structure of HCF-1 in primary cells and describe a truncated form of HCF-1 that is present in resting, or G0, cells. This N-terminal fragment is able to bind VP16 and is present in cytoplasmic extracts from these cells.
MATERIALS AND METHODS
Preparation of whole-cell lysates.
Peripheral blood mononucleocytes (PBMCs) from 10 ml of whole blood were isolated immediately after donation as described below, then boiled in 200 μl of 2× sodium dodecyl sulfate (SDS) loading buffer, and sonicated for 1 min. For 293 and HeLa cells, the cells from one confluent 10-cm plate were boiled in 300 μl of 2× SDS buffer and sonicated for 1 min. A 20-μl amount of each preparation was separated by SDS–4 to 20% polyacrylamide gel electrophoresis (PAGE) Ready Gel (Bio-Rad) and transferred to nitrocellulose, and Western blots were done as described below.
Preparation of PBMC whole-cell extracts.
PBMC whole-cell extracts were prepared as described previously (15). Leukopacks were obtained from the Dana Farber Cancer Institute blood bank. Cell suspension (30 ml) was placed in each of four sterile 50-ml tubes, underlaid with 15 ml of Ficoll-Paque Plus, and centrifuged at 1,500 rpm (400 × g) at room temperature for 30 min. The interface, which contains the PBMCs, was recovered with a sterile pipette. Isolated PBMCs were washed five times with phosphate-buffered saline (PBS) and resuspended at 4 × 106 cells/ml in RPMI 1640 containing 10% heat-inactivated calf serum. A total of 3 × 107 cells were split into six 125-ml cultures. The T0 sample was harvested immediately, phytohemagglutinin (PHA) was added to each of the remaining cultures to a concentration of 1 mg/ml, and the cultures were placed in a 37°C incubator. At 0, 24, 48, 72, 96, and 120 h, one culture was harvested as follows: 35 ml of cell suspension was washed twice in PBS, resuspended in 350 μl of WCE buffer (100 mM HEPES [pH 7.9], 35% glycerol, 500 mM KCl, 5 mM MgCl2, 5 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], 5 mg leupeptin of leupeptin per ml, 5 mg of aprotinin per ml), then flash frozen in liquid N2, and immediately thawed at 30°C. The resulting extract was cleared by ultracentrifugation for 45 min at 45,000 rpm. The protein concentration of each extract was measured and diluted to 3.2 mg/ml with WCE buffer. Whole-cell extracts were stored at −80°C. A 20-μl amount of each extract was run on duplicate SDS–4 to 20% PAGE Ready Gels (Bio-Rad). Duplicate gels were transferred to nitrocellulose, and Western blots were performed with either antiserum 2159, raised against the N terminus of HCF-1 (C1) (T. Kristie), or αrHCF antiserum, raised against a recombinant C-terminal polypeptide of HCF-1 (W. Herr).
Mixing experiments.
Ten microliters of T0 or T120 or a mixture of 10 μl each of T0 and T120 PBMC whole-cell extracts was brought to 50 μl with buffer D and then dialyzed against buffer D (20 mM HEPES [pH 7.9], 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT) for 3 h at 4°C. The dialyzed samples were then incubated for 1 h at 37°C, and an equal volume of 2× SDS loading buffer was added. Samples were analyzed by Western blotting with antisera 2159 and αrHCF. Alternatively, 30 μl of 35S-labeled HA-HCF1–1019 (prepared by in vitro translation) was incubated with 10 μl of either T0 or T96 PBMC whole-cell extract and incubated at 37°C for 1 h. The proteins were immunoprecipitated with polyclonal antihemagglutinin (αHA) antiserum (Y-11; Santa Cruz), separated on an SDS–4 to 20% PAGE Ready Gel, and detected by autoradiography.
GST pull-down.
A total of 50 ng of a glutathione-S-transferase (GST)-VP16 protein or an equivalent volume of IP buffer (100 mM NaCl, 50 mM Tris [pH 8], 2 mM MgCl2, 0.5 mM PMSF, 0.5 mM DTT, 0.05% NP-40) was added to 12 μl of 35S-labeled HCF1–1019 or HCF1–460 (prepared by in vitro translation), and the total volume was brought to 200 μl with IP buffer. After incubation at 4°C for 40 min, 20 μl of glutathione-Sepharose was added, and binding was allowed to proceed at 4°C for 40 min. The reaction mixtures were then washed twice with cold IP buffer, separated on an SDS–4 to 20% PAGE Ready Gel, and detected by autoradiography.
EMSA with PBMC whole-cell extracts.
For the electrophoretic mobility shift assay (EMSA), protein A-VP16 (PA-VP16) fusion protein was prepared as described previously (17). pRB608 probe was digested with EcoRI and HindIII and then labeled with Klenow fragment in the presence of 100 μCi of [α-32P]dTTP, 100 μCi of [α-32P]dATP, 1.4 mM dGTP, and 1.4 mM dCTP. Binding reaction mixtures contained 20 mM HEPES (pH 7.9), 60 mM KCl, 4% glycerol, 0.5 mM EDTA, 0.75 mM DTT, 50,000 cpm of probe, 1 mg of poly(dIdC), 1 mg of sonicated salmon sperm DNA, 15 ng of PA-VP16, and 2 μl of whole-cell extract. Reaction mixtures were incubated at room temperature for 5 min and then separated on a 4% native polyacrylamide gel, and the complexes were detected by autoradiography.
Serum starvation of WI38 cells.
Confluent WI38 cells were split 1:8 into Dulbecco's modified Eagle's medium containing either 10 or 0.1% heat-inactivated calf serum. The cells in 10% heat-inactivated calf-serum were split 1:8 after 8 days. On day 14, two plates of each serum condition were trypsinized, fixed, and stained with propidium iodide, and their DNA content was analyzed by fluorescence-activated cell sorting (FACS) to confirm that the cells grown under serum starvation were arrested. Twelve plates from each serum condition were washed twice with cold PBS and harvested by scraping. One-third of the cells were resuspended in 1 ml of RIPA lysis buffer (50 mM Tris [pH 8], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 1 mM EDTA), and 2/3 were resuspended in 1 ml of NP-40 lysis buffer (10 mM HEPES [pH 7.9], 250 mM KCl, 5% glycerol, 0.25% NP-40, 0.2 mM EDTA, 5 mg of leupeptin per ml, 5 mg of aprotinin per ml, 1 mM PMSF), and the nuclei and cytoplasms were fractionated. Fractions were analyzed by Western blotting with antiserum 2159.
Immunofluorescence of HA-HCF-1, HA-HCF1–460, and VP16.
COS-1 cells were grown on coverslips in six-well culture dishes and transfected with LipofectAMINE (Gibco-BRL) with either 1 μg of HA-HCF-1, 1 μg of HA-HCF1–460, 1 μg of HA-HCF-1 plus 50 ng of VP16, or 1 μg of HA-HCF1–460 plus 50 ng of VP16. Cells were washed with PBS at 48 h posttransfection and fixed with 4% paraformaldehyde for 15 min. Cells were washed three times in wash buffer (PBS containing 0.2% Tween 20) and permeabilized for 7 min in 0.2% SDS in PBS. Cells were washed three times in wash buffer and incubated at 37°C in blocking buffer (1 × PBS, 5% goat serum, 0.2% Tween 20, 0.2% fish skin gelatin) for 30 min. Cells were immunostained with monoclonal antibody 12CA5 and polyclonal αVP16 (Clontech) in blocking buffer for 1 h at 37°C. Cells were washed three times for 5 min each with wash buffer, blocked for 5 min, and then incubated at 37°C with fluorescein isothiocyanate-labeled anti-rabbit immunoglobulin (Ig) antibody and Texas red-labeled anti-mouse Ig antibody for 30 min in blocking buffer. Cells were then washed three times for 5 min each, with DAPI (4′,6′-diamidino-2-phenylindole) present in the final wash, and mounted on slides.
RESULTS
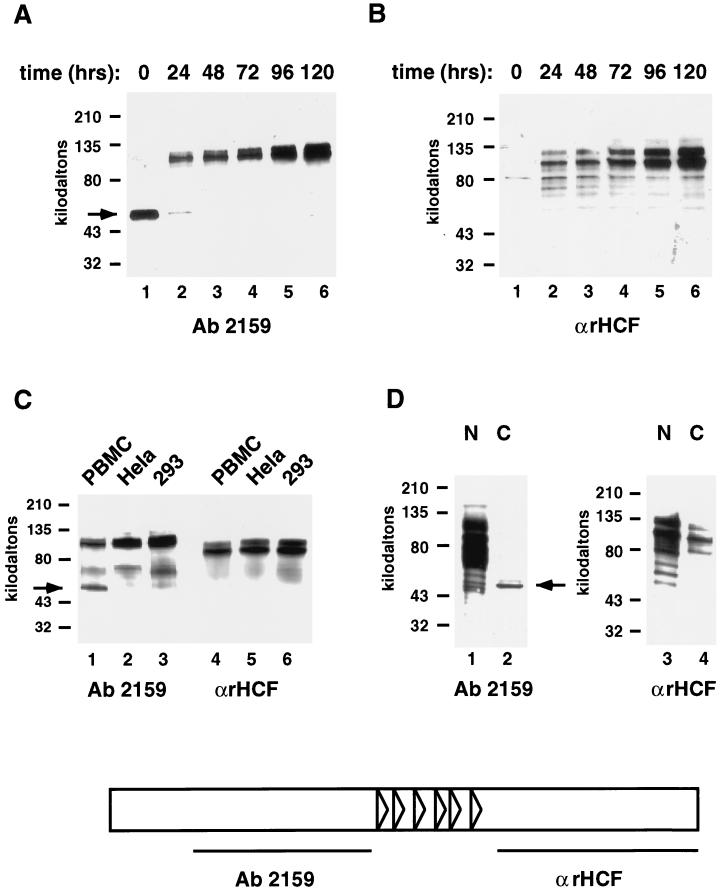
HCF-1 is abundant in rapidly cycling cell types, such as fetal tissue and immortalized cell lines, and a single mutation in the only copy of HCF-1 in one of these lines, BHK, causes the cells to arrest in a G0-like state. To investigate the state of HCF-1 in wild-type G0 cells, PBMCs from whole blood were cultured in the presence and absence of the mitogen PHA. Whole-cell extracts were made from the cultures at a series of time points following mitogenic stimulation and probed by Western blot for the presence of HCF-1 (Fig. 1A and B). Extracts from unstimulated PBMCs contained only a 50-kDa protein that cross-reacted with HCF-1 antiserum 2159 (HCFp50) (Fig. 1A, lane 1). Upon mitogenic stimulation, HCFp50 disappeared from extracts within 24 h, and the characteristic higher-molecular-weight forms of HCF-1 appeared (Fig. 1A, lanes 2 to 6). HCFp50 is derived from the N terminus of HCF-1, as it is detected by antiserum 2159, which was raised against amino acids 384 to 1015 of the N terminus, and not by αrHCF, antiserum prepared against the C terminus of HCF-1 (Fig. 1B, lane 1). The typical set of HCF-1 C-terminal polypeptides were present in extracts by 24 h after mitogenic stimulation of PBMCs, along with the higher-molecular-weight N-terminal polypeptides (Fig. 1A and B, lanes 2 to 6). HCFp50 apparently constitutes a unique form of HCF-1 that is only present in whole-cell extracts from unstimulated PBMCs which are in G0.
FIG. 1.
Presence of a unique 50-kDa fragment of HCF-1, HCFp50, in unstimulated PBMC extracts. (A and B) PBMCs were mitogen stimulated for the times shown, and whole-cell extracts were prepared, separated by SDS-PAGE, and blotted with HCF-1 antiserum 2159 (A) or αrHCF (B). (C) PBMC, HeLa, and 293 cells were boiled immediately after harvest in 2× SDS loading buffer, separated by SDS-PAGE, and blotted with HCF-1 antisera 2159 and αrHCF. (D) PBMCs were separated into nuclear and cytoplasmic fractions, separated by SDS-PAGE, and blotted with HCF-1 antisera 2159 and αrHCF. Arrows in all panels show the position of HCFp50. Schematic at bottom indicates the regions of HCF-1 against which antisera 2159 and αrHCF were raised.
Interestingly, a Western blot of unstimulated PBMC cells which were isolated and boiled immediately after donation in SDS loading buffer revealed that full-length N-terminal and C-terminal fragments of HCF-1 were present in the cells, along with HCFp50 (Fig. 1C, lanes 1 and 4). In the previous experiment, apparently either these high-molecular-weight proteins were not extracted from the cells, or the polypeptides were extracted and then converted to HCFp50 in the extract. When cycling HeLa and 293 cells were boiled directly in SDS loading buffer, only high-molecular-weight HCF-1 N- and C-terminal polypeptides were detected (Fig. 1C, lanes 2 and 3 and lanes 5 and 6, respectively). When unstimulated PBMCs were separated into nuclear and cytoplasmic fractions, HCFp50 was detected primarily in the cytoplasmic fraction (Fig. 1D, lane 2), while the larger N-terminal polypeptides were present exclusively in the nuclear fraction (lane 1). The majority of C-terminal polypeptides were retained in the nucleus, although a small portion were extracted into the cytoplasmic fraction (Fig. 1D, lanes 3 and 4). The detection of some C-terminal polypeptides in the cytoplasm (which almost certainly results from limitations in the fractionation protocol), but none of the large N-terminal polypeptides, most likely reflects a difference in the sensitivities of the two antibodies used for the Western blots. The important conclusion arises from the finding that a large majority of the HCFp50 fractionates in the cytoplasm, suggesting that HCFp50 is in a different subcellular location or state than the full-length HCF-1 polypeptides.
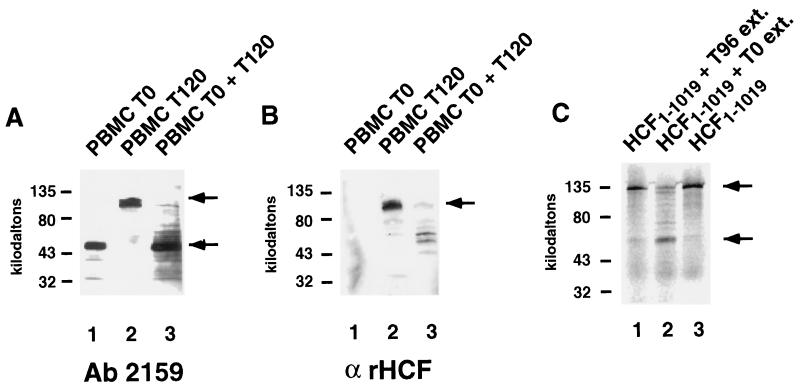
We were interested in determining whether the 50-kDa fragment of HCF-1 observed in unstimulated PBMCs could have arisen via proteolysis of the larger HCF-1 polypeptides. A mixing experiment was performed in which PBMC whole-cell extract that had been mitogen stimulated for 120 h (T120), and thus contained the larger HCF-1 polypeptides, was mixed with an equal volume of unstimulated PBMC extract (T0), which contained only HCFp50. After incubation for 1 h at 37°C, the larger HCF-1 N-terminal polypeptides from the T120 extract had been degraded, as they were undetectable by Western blot with antiserum 2159 (Fig. 2A, lanes 1 to 3). Similarly, the HCF-1 C-terminal polypeptides, which were present in the T120 extract but not in the T0 extract, were also degraded upon incubation with the T0 extract (Fig. 2B, lanes 1 to 3). Whether the 50-kDa fragment detected by antiserum 2159 was produced during degradation of the larger HCF-1 N-terminal polypeptides could not be determined due to the presence of HCFp50 in the T0 PBMC extract. To address this question, an HA-tagged 110-kDa N-terminal fragment of HCF-1 (HA-HCF1–1019)was synthesized in the presence of 35S-labeled methionine, using in vitro translation. A small amount of the lysate containing HA-HCF1–1019 was incubated with either the PBMC G0 extract or the PBMC mitogen-stimulated cycling extract (Fig. 2C). Each mixture was immunoprecipitated with an anti-HA antibody and resolved by SDS-PAGE. A 50-kDa band was immunoprecipitated following incubation with the G0 PBMC extract (Fig. 2C, lane 2). The full-length radiolabeled N terminus was immunoprecipitated following incubation with a PBMC mitogen-stimulated cycling extract (Fig. 2C, lane 1). Because the 50-kDa fragment retained the HA tag from the in vitro-translated HA-HCF1–1019, it was derived from the N terminus of the protein. These results indicate that G0 PBMC extracts contain a proteolytic activity that can generate HCFp50 from larger N-terminal polypeptides.
FIG. 2.
HCFp50 can be formed by proteolysis. (A and B) Western blots with antisera 2159 (A) and αrHCF (B) of PBMC whole-cell extracts which were mitogen stimulated for 0 h (lane 1) or 120 h (lane 2) or with a mixture of the two (lane 3). (C) In vitro-translated 35S-labeled HCF p120 was mixed with whole-cell extract from PBMCs that had been mitogen stimulated for 96 h (lane 1) or mixed with unstimulated PBMC whole-cell extract (lane 2). Lane 3, HCF-1 N terminus translation product alone. The upper arrows show the position of the larger N-terminal polypeptides of HCF-1 (A), the C-terminal polypeptides of HCF-1 (B), and the HCF1–1019 in vitro translation product (C). The lower arrows show the position of HCFp50.
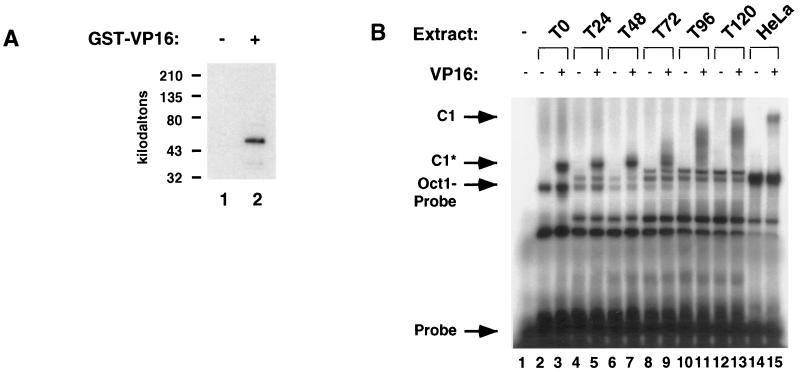
The high-affinity site of interaction between HCF-1 and VP16 has been mapped to the N-terminal kelch domain, which HCFp50 should encompass. To confirm that HCFp50 is capable of binding VP16, a GST pull-down experiment was performed (Fig. 3A). PBMC G0 extract was incubated in the presence or absence of GST-VP16, glutathione-Sepharose was added to the reaction mixture, and the fraction bound to the beads was separated by SDS-PAGE. A Western blot performed with antiserum 2159 detected a single 50-kDa species (Fig. 3A, lane 2). When GST-VP16 was left out of the mixture, no proteins were detected with the 2159 antiserum (Fig. 3A, lane 1). This confirms the anticipated interaction between HCFp50 and VP16.
FIG. 3.
Association of HCFp50 with VP16, and formation of C1 complex with VP16, Oct-1, and DNA. (A) Pull-down by glutathione-Sepharose of HCFp50 from unstimulated PBMC extracts in the absence (lane 1) and presence (lane 2) of GST-VP16. (B) PBMC extracts that were mitogen stimulated for the times indicated were incubated with radiolabeled probe in the presence and absence of VP16. C1, C1 complex formed by full-length HCF-1, Oct-1, VP16, and probe. C1*, faster-mobility form of the complex formed in unstimulated and early stimulated PBMC extracts. Lanes 14 and 15 show C1 complex formation from HeLa cell extract.
Because HCFp50 contains the kelch repeats of HCF-1 and is able to bind VP16, it should also be active for C1 complex formation. To test if this was the case, extracts of unstimulated PBMCs from each time point following mitogen stimulation were assayed by EMSA in the presence of VP16, and a radiolabeled oligonucleotide containing the α/IE promoter element (Fig. 3B). A faster-migrating form of the C1 complex (designated C1*) was observed when the binding reaction mixture contained extract from unstimulated PBMCs (Fig. 3B, lane 3). The C1 complexes formed with extracts from each successive time point were of a slower mobility than the one immediately preceding it (Fig. 3B, lanes 5, 7, 9, 11, and 13), approaching the mobility of the C1 complex formed from a HeLa extract (lane 15). Both the C1* complex and the other C1 complexes were VP16 dependent (even-numbered lanes) and could be supershifted with 2159 antibody (data not shown), indicating the presence of HCF-1 N-terminal polypeptides, as would be expected in the VP16-dependent complexes. Interestingly, the difference in mobility of the C1* complexes formed from extracts of mitogen-stimulated PBMCs at different time points did not correlate simply with the size of the HCF-1 detected in these extracts (see Fig. 1A and B). For instance, the T24 extract contained N- and C-terminal fragments of HCF-1 that appeared to be full-length, but the C1 complex formed from this extract was of a faster mobility than the C1 complex from HeLa cells. The mobilities of the C1 complexes formed from mitogen-stimulated PBMC extracts decreased at successive time points, as the level of full-length HCF-1 increased. HCF-1 purifies from HeLa cells as a large complex, probably consisting of oligomers of the protein. In addition, reconstitution of the C1 complex with material prepared from insect cells suggests that other proteins in addition to HCF-1, Oct-1, and VP16 may associate with the core C1 complex (8).
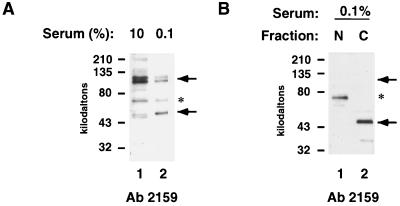
Finally, we investigated whether the generation of HCFp50 is unique to PBMCs. The human secondary cell line WI38 was cultured for 14 days in either high serum, in which they will continue to cycle, or low serum, which causes them to arrest in G0. FACS analysis showed that the WI38 cells grown in high serum contained 23% S-phase cells, while those grown in low serum contained only 2% S-phase cells, indicating that the WI38 cells grown in low serum were in fact arrested (data not shown). Extracts from these cells were analyzed for HCF-1 by SDS-PAGE. Probing of a Western blot with antiserum 2159 revealed that the serum-starved WI38 cells also contain a 50-kDa species, similar to the fragment found in unstimulated PBMCs (Fig. 4A, lane 2). The serum-starved WI38 extracts also contain very low levels of C-terminal polypeptides from HCF-1, also similar to unstimulated PBMCs (data not shown). When nuclear and cytoplasmic extracts from the serum-starved WI38 cells were analyzed, the 50-kDa species was found in the cytoplasm (Fig. 4B, lane 2), while the larger HCF-1 N-terminal polypeptides were predominantly nuclear (data not shown). These data suggest that G0 cells contain a unique 50-kDa form of HCF-1, HCFp50, which is derived from the N terminus of the protein, and that this protein appears to be present in the cytoplasm of these cells, in contrast to the nuclear localization of the larger HCF-1 N-terminal polypeptides.
FIG. 4.
Appearance of HCFp50 in serum-starved WI38 cells. (A) Western blot of WI38 cells grown in 10% (lane 1) or 0.1% (lane 2) serum with antiserum 2159. (B) Western blot of nuclear (lane 1) and cytoplasmic (lane 2) fractions of WI38 cells grown in 0.1% serum with antiserum 2159. The upper arrows show the position of the larger N-terminal polypeptides of HCF-1; the lower arrows show the position of HCFp50. The asterisk indicates a nonspecific band that cross-reacts with the 2159 antibody.
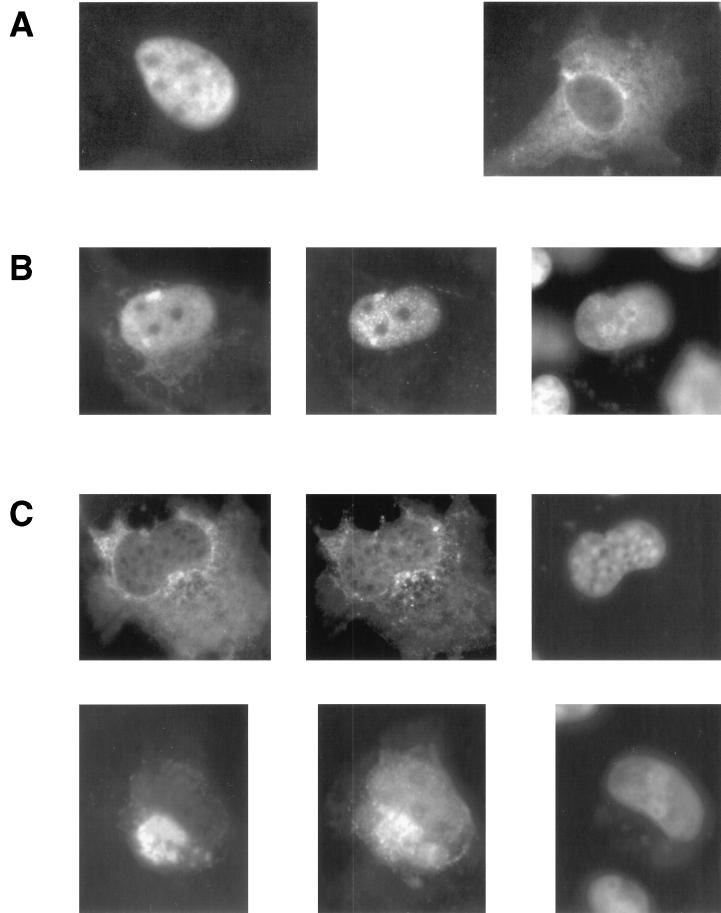
It has previously been reported that deletion of the nuclear localization signal of HCF-1 causes the protein to localize to the cytoplasm. Expression of this form of HCF-1 also sequestered coexpressed VP16 in the cytoplasm (11). To test whether HCFp50 would have a similar activity, COS-1 cells were transfected with HA-tagged HCF-1 or with HA-tagged HCF1–460 and stained with monoclonal antibody 12CA5 (Fig. 5A). This N-terminal fragment of HCF-1 was chosen because of its close approximation to the size of HCFp50 (as determined by SDS-PAGE) and corresponding location within HCF-1. Cells transfected with a plasmid encoding HA-HCF-1 and immunostained with monoclonal antibody 12CA5, which recognizes the HA tag, showed an almost exclusively nuclear localization, with occasional staining of both the nucleus and cytoplasm (Fig. 5A, left panel). In contrast, cells transfected with a plasmid encoding HA-HCF1–460 showed staining in the cytoplasm, with little or no staining in the nucleus (Fig. 5A, right panel). When a plasmid encoding VP16 was cotransfected with full-length HA-HCF-1 (Fig. 5B), VP16 protein accumulated in the nucleus (middle panel) along with HA-HCF-1 (left panel; DAPI staining shown in the right panel). In contrast, when a plasmid encoding VP16 was cotransfected with a plasmid encoding HA-HCF1–460 (Fig. 5C), VP16 was distributed between the cytoplasm and nucleus (middle panels, top and bottom). The cytoplasmic VP16 was accumulated in a pocket of very bright staining which was coincident with a very strong HA-HCF1–460 signal (left panels, top and bottom; DAPI staining shown in the right panels), suggesting that the two proteins interact in the cytoplasm. The VP16 that is visible in the nucleus is likely to have been imported by the endogenous HCF-1 present in these cells.
FIG. 5.
(A) Immunofluorescence of transfected HA-HCF-1 (left panel) and HA-HCF1–460 (right panel) in COS-1 cells with monoclonal antibody 12CA5. (B) Immunofluorescence of HA-HCF-1 cotransfected with VP16 in COS-1 cells; HA-HCF-1 was detected with antibody 12CA5 (left panel), VP16 was detected with polyclonal αVP16 (middle panel), and the nucleus was stained with DAPI (right panel). (C) Immunofluorescence of HA-HCF1–460 and VP16 cotransfected in COS-1 cells; HA-HCF1–460 was detected with antibody 12CA5 (left panels, top and bottom), VP16 was detected with polyclonal αVP16 (middle panels, top and bottom), and the nucleus was stained with DAPI (right panels, top and bottom).
DISCUSSION
A novel 50-kDa fragment of HCF-1 is found in resting PBMCs and serum-starved WI38 cells. HCFp50 appears to be generated by proteolysis of the larger N-terminal polypeptides of HCF-1 in resting cells. A proteolytic activity was present in extracts from resting PBMCs that produced HCFp50 from in vitro-translated HA-HCF1–1019. This activity was not detectable in extracts from mitogen-stimulated PBMCs, indicating that it is G0 specific.
Resting cells accumulate an N-terminal fragment of HCF-1 (p50) that is not present at similar levels in cycling cells. This p50 fragment contains the kelch domain of HCF-1, appears to be localized to the cytoplasm, and is readily extractable from G0 PBMCs. A proteolytic activity present in resting PBMC extract is sufficient to process higher-molecular-weight HCF-1 polypeptides to HCFp50. Whether these proteolytic activities are involved in the generation of HCFp50 in vivo is not clear. It is unlikely that HCFp50 is only generated during extraction, since immediate boiling of cells in SDS buffer reveals the presence of HCFp50. Although only a 50-kDa N-terminal fragment of HCF-1 was present in whole-cell extracts of resting PBMCs, larger polypeptides from both the N and C termini were clearly present in the nuclei of these cells. There are several possible explanations for the absence of high-molecular-weight HCF-1 polypeptides in extracts from resting PBMCs. It is possible that the higher-molecular-weight fragments of HCF-1 in these cells are not extractable by the conditions used for making whole-cell extracts. This explanation would imply that upon mitogenic stimulation, the state of HCF-1 changes, allowing it to become more extractable. Alternatively, the proteolytic activity present in resting cells could degrade the higher-molecular-weight forms of HCF-1 in the whole-cell extract, precluding their detection.
Previous studies have shown that the kelch repeats of HCF-1 are necessary and sufficient for association with VP16 and formation of a complex with VP16, Oct-1, and DNA (12, 22). As expected, HCFp50 is able to associate with VP16 and mediate a fast-mobility form of the C1 complex, termed C1*. Following mitogen stimulation of PBMCs, the mobility of this complex slowed and approached the mobility of a C1 complex formed from cycling HeLa cell extracts. The migration rate of the C1-like complex probably reflects the size of the complex of HCF-1 proteins that bind to VP16 and Oct-1. The mobilities of the C1-type complex generated from extracts of resting cells and the early time points following mitogen stimulation did not change dramatically even though the molecular weight of the extracted HCF-1 protein doubled, as determined by SDS-PAGE. The most likely explanation for this apparent inconsistency is that the mobility of the C1-type complex is primarily determined by the oligomeric state of HCF-1 and not solely by the molecular weight of its fragments. The form of HCF-1 extracted from HeLa cells has an apparent molecular size in excess of 106 Da when assayed by chromatographic exclusion (9). Furthermore, N-terminal and C-terminal fragments of HCF-1 are bound in these complexes (23). Thus, the mobility of the C1-type complex prepared from extracts of stimulated cells probably reflects the formation of multimers of HCF-1. This is consistent with the observation that following mitogenic stimulation of PBMCs, the level of larger HCF-1 polypeptides continues to increase for at least 120 h. In addition, other factors may accumulate in stimulated cells which associate with the C1 complex.
By cell fractionation and immunofluorescence, HCFp50 appears to be localized in the cytoplasm. Previous data also support a cytoplasmic localization for a protein similar to HCFp50. First, a consensus NLS has been identified in the C terminus of HCF-1, a region not contained within HCFp50 (11). When a mutant form of HCF-1 lacking this consensus NLS was transfected into COS-1 cells, it accumulated in the cytoplasm, whereas the full-length protein accumulated in the nucleus. Transfection of a plasmid encoding the kelch repeats alone also produced a cytoplasmic localization (11). Coexpression of full-length HCF-1 with VP16 causes both proteins to be localized to the nucleus of COS-1 cells (11; this report). Expression of HA-HCF1–460 with VP16 results in a significant cytoplasmic accumulation of VP16.
HCF-1 is clearly required for progression of the cell cycle, as a single-amino-acid change results in the temperature-sensitive arrest of a BHK cell line (3). It has therefore been suggested that HSV uses HCF-1 to sense the cell cycle state of the cell (3). La Boissiere et al. (11) suggest a model to explain a possible mechanism for this action of HCF-1 in HSV infection. Because it appears that HCF-1 is required for nuclear import of the transactivating factor VP16, the association of VP16 with HCFp50 in G0 cells could result in VP16's being sequestered in the cytoplasm of an infected cell (11). It has previously been postulated that the decision by the virus to enter either a replicative or latent life cycle may be influenced in part by the amount of VP16 that enters the nucleus (20). Thus, sequestering VP16 in the cytoplasm of a cell through association with HCFp50 would suppress the activation of the IE genes and promote a latent infection.
Modulation of the activity of VP16 during infection of cells is probably not the sole determinant in establishing the viral genome in a latent state. It has been shown that HSV can enter latency when VP16 is expressed ectopically in the sensory neurons of mice. Sequestration of VP16 in the cytoplasm, however, may contribute to viral latency by decreasing the activation of HSV IE genes (18). Recent evidence suggests that HCF-1 has an unusual structure in the trigeminal ganglia cells targeted by HSV for latency. In these cells, HCF-1 cannot be detected as a nuclear antigen using antisera to several regions of the protein, but appears to be sequestered in granules in the cytoplasm (10). Trigeminal ganglia were unique in the loss of nuclear staining of HCF-1 compared with cells from the liver and kidney. Conditions that activate the virus from these cells result in the rapid appearance of nuclear HCF-1 antigens (10). It is not clear whether this phenomenon, detected with specific antisera, is related to the accumulation of HCFp50 in resting PBMCs and WI38 cells. However, extracts of dorsal root ganglia cells have been reported to generate faster-migrating C1-type complexes (4), similar to those seen here in the presence of HCFp50.
ACKNOWLEDGMENTS
We thank T. Kristie and W. Herr for generously providing antisera. We thank D. Tantin and B. Panning for critical reading of the manuscript and helpful discussions.
This work was supported by U.S. Public Health Service grants P01-CA42063 to P.A.S. and partially by Cancer Center Support (core) grant P30-CA14051 from the National Institutes of Health.
REFERENCES
- 1.Freiman R N, Herr W. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerster T, Roeder R G. A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc Natl Acad Sci USA. 1988;85:6347–6351. doi: 10.1073/pnas.85.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 4.Hagmann M, Georgiev O, Schaffner W, Douville P. Transcription factors interacting with herpes simplex virus alpha gene promoters in sensory neurons. Nucleic Acids Res. 1995;23:4978–4985. doi: 10.1093/nar/23.24.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herr W, Sturm R A, Clerc R G, Corcoran L M, Baltimore D, Sharp P A, Ingraham H A, Rosenfeld M G, Finney M, Ruvkun G, et al. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 6.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 7.Kristie T M, LeBowitz J H, Sharp P A. The octamer-binding proteins form multi-protein--DNA complexes with the HSV alpha TIF regulatory protein. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kristie T M, Sharp P A. Interactions of the Oct-1 POU subdomains with specific DNA sequences and with the HSV alpha-trans-activator protein. Genes Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- 9.Kristie T M, Sharp P A. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus alpha- trans-induction factor (VP16) J Biol Chem. 1993;268:6525–6534. [PubMed] [Google Scholar]
- 10.Kristie T M, Vogel J L, Sears A E. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc Natl Acad Sci USA. 1999;96:1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.La Boissiere S, Hughes T, O'Hare P. HCF-dependent nuclear import of VP16. EMBO J. 1999;18:480–489. doi: 10.1093/emboj/18.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.La Boissiere S, Walker S, O'Hare P. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol Cell Biol. 1997;17:7108–118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu R, Yang P, O'Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R, Yang P, Padmakumar S, Misra V. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J Virol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Hare P, Goding C R. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell. 1988;52:435–445. doi: 10.1016/s0092-8674(88)80036-9. [DOI] [PubMed] [Google Scholar]
- 17.Pomerantz J L, Pabo C O, Sharp P A. Analysis of homeodomain function by structure-based design of a transcription factor. Proc Natl Acad Sci USA. 1995;92:9752–9756. doi: 10.1073/pnas.92.21.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sears A E, Hukkanen V, Labow M A, Levine A J, Roizman B. Expression of the herpes simplex virus 1 alpha trans-inducing factor (VP16) does not induce reactivation of latent virus or prevent the establishment of latency in mice. J Virol. 1991;65:2929–2935. doi: 10.1128/jvi.65.6.2929-2935.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stern S, Tanaka M, Herr W. The Oct-1 homeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989;341:624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 20.Valyi-Nagy T, Deshmane S L, Spivack J G, Steiner I, Ace C I, Preston C M, Fraser N W. Investigation of herpes simplex virus type 1 (HSV-1) gene expression and DNA synthesis during the establishment of latent infection by an HSV-1 mutant, in 1814, that does not replicate in mouse trigeminal ganglia. J Gen Virol. 1991;72:641–649. doi: 10.1099/0022-1317-72-3-641. [DOI] [PubMed] [Google Scholar]
- 21.Verrijzer C P, Van der Vliet P C. POU domain transcription factors. Biochim Biophys Acta. 1993;1173:1–21. doi: 10.1016/0167-4781(93)90237-8. [DOI] [PubMed] [Google Scholar]
- 22.Wilson A C, Freiman R N, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson A C, LaMarco K, Peterson M G, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 24.Wilson A C, Parrish J E, Massa H F, Nelson D L, Trask B J, Herr W. The gene encoding the VP16-accessory protein HCF (HCFC1) resides in human Xq28 and is highly expressed in fetal tissues and the adult kidney. Genomics. 1995;25:462–468. doi: 10.1016/0888-7543(95)80046-o. [DOI] [PubMed] [Google Scholar]
- 25.Wilson A C, Peterson M G, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]