Abstract
The inositol phosphatase SHIP binds to the FcγRIIB1 receptor and plays a critical role in FcγRIIB1-mediated inhibition of B-cell proliferation and immunoglobulin synthesis. The molecular details of SHIP function are not fully understood. While point mutations of the signature motifs in the inositol phosphatase domain abolish SHIP's ability to inhibit calcium flux in B cells, little is known about the function of the evolutionarily conserved, putative noncatalytic regions of SHIP in vivo. In this study, through a systematic mutagenesis approach, we identified the inositol phosphatase domain of SHIP between amino acids 400 and 866. Through reconstitution of a SHIP-deficient B-cell line with wild-type and mutant forms of SHIP, we demonstrate that the catalytic domain alone is not sufficient to mediate FcγRIIB1/SHIP-dependent inhibition of B-cell receptor signaling. Expression of a truncation mutant of SHIP that has intact phosphatase activity but lacks the last 190 amino acids showed that the noncatalytic region in the C terminus is essential for inhibitory signaling. Mutation of two tyrosines within this C-terminal region, previously identified as important in binding to Shc, showed a reduced inhibition of calcium flux. However, studies with an Shc-deficient B-cell line indicated that Shc-SHIP complex formation is not required and that other proteins that bind these tyrosines may be important in FcγRIIB1/SHIP-mediated calcium inhibition. Interestingly, membrane targeting of SHIP lacking the C terminus is able to restore this inhibition, suggesting a role for the C terminus in localization or stabilization of SHIP interaction at the membrane. Taken together, these data suggest that the noncatalytic carboxyl-terminal 190 amino acids of SHIP play a critical role in SHIP function in B cells and may play a similar role in several other receptor systems where SHIP functions as a negative regulator.
B-cell immune response to antigens is terminated or attenuated by surface receptors such as FcγRIIB1 and CD22 on B cells (5, 11, 34, 48). These inhibitory receptors recruit specific intracellular signaling proteins, which play a key role in attenuating the early activation events initiated by cross-linking of the B-cell receptor (BCR). FcγRIIB1 is an important mediator of the attenuation of B-cell activation by antibody-antigen immune complexes in the later phases of the immune response (49). Coengagement of FcγRIIB1 with BCR results in a potent inhibitory signal that depends on the recruitment of Src homology 2-containing inositol phosphatase (SHIP). SHIP binds to the phosphorylated immunotyrosine-based motif (ITIM) in the cytoplasmic region of FcγRIIB1 (43, 44), and SHIP-mediated dephosphorylation of specific phosphoinositide products has been implicated in terminating the BCR-induced activation events (4, 14, 53).
SHIP was initially characterized in hematopoietic cells as a 145-kDa phosphoprotein that coprecipitated with the adapter protein Shc upon stimulation of specific receptors (6–8, 37, 50, 52, 54). Molecular cloning of SHIP identified it as a 5′-inositolphosphatase (5′-IPase), based on homology with other 5′-IPases (9, 13, 29, 36, 45, 57). SHIP specifically dephosphorylates phosphatidylinositol-3,4,5-trisphosphate (PIP3), a major product of phosphoinositide-3-kinase (PI3K) enzymatic action, as well as inositoltetrakisphosphate (IP4), both in vitro (28, 36) and in vivo (53). The requirement for SHIP in FcγRIIB1-mediated inhibition of BCR signaling has been well established (4, 5, 14, 20, 32, 44, 48, 53). Recruitment of enzymatically active SHIP to the receptor complex results in potent inhibition of intracellular calcium flux (12, 30, 44), diminished activation of the serine-threonine kinase Akt (1, 3, 17, 27), inhibition of the Ras/mitogen-activated protein kinase pathway (56), and the regulation of apoptosis (2, 38, 47). Further evidence for a crucial role for SHIP in negative regulation of BCR signaling comes from studies with SHIP knockout mice as well as SHIP−/− Rag−/− chimeric mice, in which BCR-mediated responses are heightened and the FcγRIIB1-dependent inhibition of BCR responses is abolished (23, 39).
It is noteworthy that SHIP also negatively regulates histamine release in response to engagement of the immunoglobulin E (IgE) receptor and Steel factor (25, 26, 43), as well as the proliferative response to interleukin-3 and the macrophage colony-stimulating factor (36). Ex vivo studies with cells from SHIP-deficient mice have suggested that in the absence of SHIP, the myeloid progenitor cells hyperproliferate in response to cytokines and hematopoietic growth factors, with the dose-response curve being left-shifted (23). Taken together, these studies have clearly established a functional role for SHIP as a negative regulator of cytokine and antigen receptor signaling.
The 145-kDa isoform of SHIP, the predominant form expressed in hematopoietic cells, is composed of an N-terminal Src homology 2 (SH2) domain, a central, loosely defined IPase domain, and a C-terminal region which contains multiple motifs involved in protein-protein interactions (9, 13, 29, 36, 45, 57). While mutation of residues within the signature motifs of the IPase abolishes SHIP's ability to inhibit calcium flux (44), little is known about the function of noncatalytic regions of SHIP in vivo. Interestingly, multiple isoforms and cleavage products of SHIP have been detected in hematopoietic cells and have been proposed to perform specific functions (10, 15, 41). Since the putative catalytic domain of SHIP is left intact in the various isoforms, the regions outside of the catalytic domain, through interaction with other proteins, are likely to play a key role in determining SHIP function under different conditions. Thus, defining the precise boundaries of the phosphatase domain of SHIP, identifying the proteins that bind to the noncatalytic regions, and determining how they regulate SHIP function have become important issues to be resolved.
In this study, through a systematic mutagenesis approach, we determined that the boundaries of the IPase domain of SHIP exist between amino acids 400 and 866. Through reconstitution of a SHIP-deficient B-cell line with wild-type (wt) and mutant forms of SHIP, we demonstrate that the catalytic domain alone is not sufficient and that the C-terminal noncatalytic region is essential for FcγRIIB1/SHIP-mediated inhibitory signaling. Our data also suggest that the C-terminal region of SHIP may play a key role in the localization of SHIP to the membrane during inhibitory signaling.
MATERIALS AND METHODS
Plasmids.
The original murine SHIP cDNA was kindly provided by Gerald Krystal (Terry Fox Labs, Vancouver, Canada). Plasmids encoding glutathione-S-transferase (GST)-tagged SHIP were generated by cloning the wt and mutant constructs in-frame into the pEBG vector as described previously (33). SHIP versions tagged with three copies of hemagglutinin (HA) were generated in the pEBB vector as described before (33) and subcloned into the pApuro vector (55). Both the pEBG and pEBB vectors regulate protein expression under the elongation factor-1 (EF-1) promoter, while the pApuro vector expresses proteins under the chicken actin promoter. The constructs encoding partial regions of SHIP were generated by PCR and/or subcloning of the appropriate regions into the pEBG or pEBB vector. All PCR-generated products were sequenced to ensure fidelity. The plasmids encoding SHIP-SH2 fused to the linker were generated in the pApuro vector. The 18-amino-acid linker was designed based on the description of Pantoliano et al. (46). An enzymatically inactive form of SHIP was generated by incorporating three point mutations, P671A, D675A, and R676G, into the IPase signature motif as described previously (44). The plasmid encoding FcγRIIB1 in pApuro was kindly provided by Tomo Kurosaki (Osaka, Japan). FcγRIIB1 was also subcloned into the pEFneo vector, which drives expression under the EF-1 promoter. For generation of Fc receptor (FcR)-SHIP chimeric proteins, the insert encoding SHIPwt or SHIP1–900 was fused in-frame to a truncated FcγRIIB1 (Fc; truncated at residue 300 in its cytoplasmic tail and thus lacking the ITIM motif that normally binds SHIP) in the pApuro vector.
Cell lines, antibodies, and reagents.
The SHIP-deficient and wt DT40 cell lines were obtained from Tomo Kurosaki. These cells were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum, 1% chicken serum (Sigma Biochemicals, St. Louis, Mo.), penicillin, streptomycin, 2 mM l-glutamine, and 50 μM 2-mercaptoethanol. Murine A20 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, 2 mM l-glutamine, and 20 μM 2-mercaptoethanol. 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, and antibiotics. Rabbit anti-mouse IgG [F(ab′)2 fragment and intact antibody] was purchased from Jackson ImmunoResearch Laboratories (West Grove, Pa.). Murine monoclonal anti-chicken IgM (M4) antibody was obtained from Southern Biotechnology Associates (Birmingham, Ala.). Polyclonal and monoclonal anti-Shc antibodies and the horseradish peroxidase-conjugated antiphosphotyrosine RC20 antibody were purchased from Transduction Laboratories. Rabbit anti-SHIP antiserum was a generous gift from G. Krystal. Purified, constitutively active PI3K (P110*) was kindly provided by A. Klippel (Chiron Corporation, Emeryville, Calif.).
Transfections.
293T cells were transfected by the calcium phosphate precipitation method with reagents from 5′ → 3′ Inc. according to manufacturer's instructions. DT40 cells were transfected with 20 μg of linearized plasmid DNA (pApuro for SHIP constructs or pEFneo for FcγRIIB1) by electroporation at 250 V and 960 μF. Cells were cultured for 24 h before selection in medium containing puromycin (0.5 μg/ml) or G418 (2.0 mg/ml) in 96-well plates. DT40 lines expressing FcγRIIB1 were screened with the 2.4G2 antibody and by flow cytometry (see below). A representative FcγRIIB1-positive clone was used for transfection of plasmids encoding the various SHIP constructs. The clones expressing HA-SHIP were identified by Western blot with anti-HA antibody.
Stimulation, immunoprecipitations, and immunoblotting.
Stimulation of DT40 cells was performed as previously described (44). Briefly, cells were preincubated at 37°C with either 3 μg of F(ab′)2 fragment of rabbit anti-mouse IgM per ml for BCR stimulation or 6 μg of an intact form of the same antibody per ml for BCR plus FcR cross-linking. Stimulation was initiated by adding 1 μg of mouse anti-chicken IgM (M4) per ml. For Western blotting and immunoprecipitation, cells were lysed in lysis buffer containing 50 mM Tris (pH 7.6); 150 mM NaCl; 1% NP-40; 10 mM sodium pyrophosphate; 10 μg/ml each of aprotinin, leupeptin, and pepstatin; 10mM NaF; 1 mM NaVO4; and 2 mM phenylmethylsulfonyl fluoride. Cellular debris was cleared by centrifugation, and proteins were precipitated from lysates with either glutathione-Sepharose beads or the relevant antibody plus protein A-conjugated beads. Beads were washed four times with lysis buffer, and bound proteins were analyzed by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis and developed by enhanced chemiluminescence.
IPase assays.
32P-labeled PIP3 was generated by using a constitutively active form of PI3K (p110*) (31). A mixture of 12.5 nmol of phosphatidylinositol-(4,5)-bisphosphate (PIP2) and 185 nmol of phosphatidylserine were sonicated in 10 mM HEPES (pH 7)–1 mM EGTA to generate micelles and incubated in 500 μl of a kinase buffer consisting of 0.2 mM ATP, 5 mM MgCl2, 30 mM HEPES (pH 7), 5 μM EGTA, 150 μCi of [γ32P]ATP, and 5 μg of p110* for 30 min at room temperature. The reaction was terminated by addition of EDTA, and lipids were extracted in 800 μl of chloroform-methanol (1:1) and 125 μl of 5 M HCl. After drying the organic phase, the lipids were resuspended in 50 mM Tris (pH 7.5)–0.125% NP-40 by sonication and used as the substrate for the phosphatase assays. Phosphatase assays were performed for 30 min to 1 h at 37°C after adding 100 μl of the substrate solution and 50 μl of 50 mM Tris (pH 7.5)–30 mM MgCl2 to immunoprecipitated enzyme equilibrated in Tris-MgCl2 buffer. The lipids were extracted as above and analyzed on thin-layer chromatography plates activated in potassium oxalate-methanol as described before (36). The phosphatase activity of SHIP toward IP4 was assayed with 3H-labeled IP4 (NEN Dupont). Immunoprecipitated SHIP and SHIP mutants were washed in 50 mM MES (2-N-morpholinoethanesulfonic acid)–3 mM MgCl2 and incubated for 30 min at 37°C in the same buffer supplemented with 10 μM unlabeled IP4 (a generous gift from G Prestwich, University of Utah) and 25 nM (0.05 μCi/100 μl) [3H]IP4. The reaction was stopped by adding 1 ml of cold water and analyzed by fast protein liquid chromatography (FPLC) (Pharmacia). Samples were loaded on a strong cationic Mono Q (Pharmacia,) column and eluted on a linear gradient of 0.3 to 0.7 M monobasic ammonium phosphate at 0.5 ml/min. Fractions of 0.5 ml were collected, and radioactivity was counted in a scintillation counter.
Intracellular calcium measurements.
Cells (5 × 106) were incubated in 1 μg of Indo 1 (Molecular Probes, Eugene, Oreg.) per ml in complete medium for 20 min in a humidified incubator at 39°C. Cells were washed in HEPES buffer (150 mM NaCl, 5 mM KCl, 1 mM each CaCl2 and MgCl2, 10 mM HEPES [pH 7.4], 0.1% glucose, and 1% fetal calf serum), resuspended in the same buffer, and transferred to a cuvette. Secondary antibodies, 1.5 μg of F(ab′)2 fragment of rabbit anti-mouse IgM per ml (for BCR cross-linking alone) or 3 μg of intact rabbit anti-mouse IgM per ml (for BCR plus FcR cross-linking), were added prior to measurement. Using an SLM 8100-C spectrofluorimeter (22), calcium flux was recorded upon excitation at 340 nm as the ratio of fluorescence emissions at 398 and 480 nm. The background was recorded for 20 s, followed by addition of mouse anti-chicken IgM (M4) antibody at 1 μg/ml.
FACS analysis.
For fluorescence-activated cell sorting (FACS), cells were incubated at 5 × 106 cells/ml in phosphate-buffered saline (PBS) with 1 μg of 2.4G2 (anti-FcγRIIB1) antibody per ml for 20 min at 4°C. Cells were washed twice in PBS and stained with fluorescein isothiocyanate-conjugated anti-rat IgG at 4 μg/ml for 20 min at 4°C. The cells were then washed and analyzed on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, Calif.). As a negative control, cells were stained with fluorescein isothiocyanate-conjugated anti-rat IgG without primary antibody.
RESULTS
Defining the minimal IPase domain of SHIP.
To gain a better molecular understanding of SHIP function, we undertook systematic mutagenesis to delineate the catalytic and noncatalytic regions of SHIP. To define the boundaries of the IPase domain of SHIP, we generated GST-tagged versions of wt SHIP and various truncation mutants that have deletions at the amino or carboxyl terminus (schematically shown in Fig. 1A). To test the catalytic activity of these SHIP versions, these constructs were transiently expressed in 293T cells, the proteins were precipitated by glutathione beads, and the IPase activity toward radiolabeled PIP3 was measured. To ensure that the IPase activity measured in these assays is in fact due to SHIP and not to a coprecipitating phosphatase activity from 293T cells, we expressed a catalytically inactive SHIP protein in which three residues in the signature motifs highly conserved among 5′-IPases have been mutated (see Materials and Methods) (44). No detectable phosphatase activity was associated with this mutant (data not shown). Varying the amount of SHIP protein or the time of incubation with substrates revealed that in our routine 30-min assay, most, if not all, of the substrate was dephosphorylated (data not shown). Identical experiments with HA-tagged versions of SHIP showed that the HA and GST tags have no influence on the IPase activity determined in these assays (data not shown).
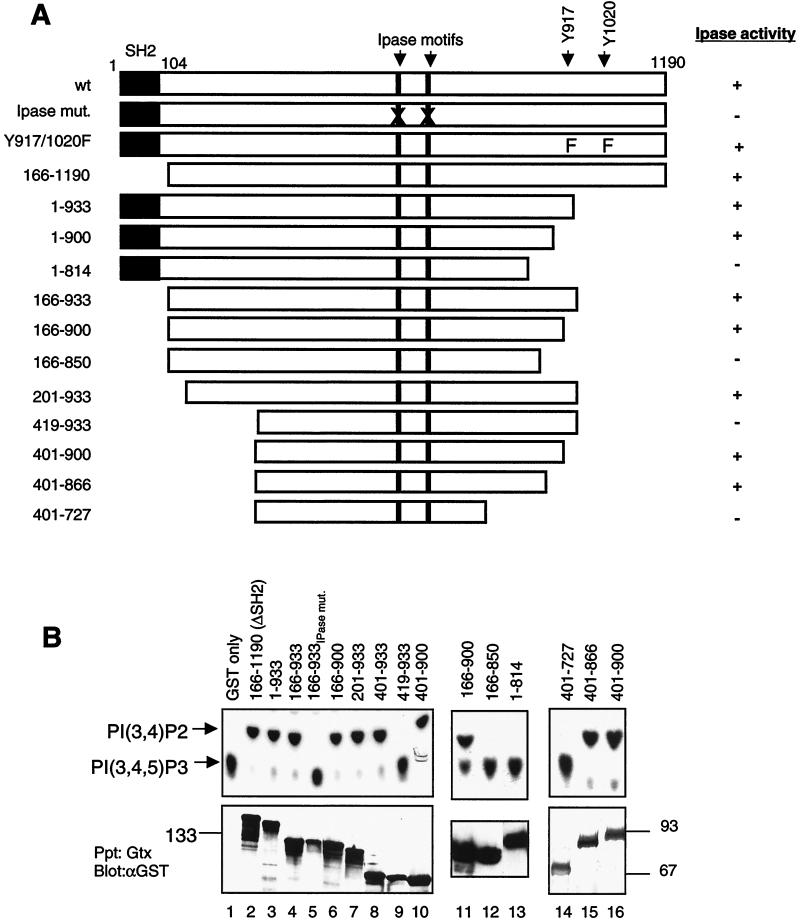
FIG. 1.
Minimal phosphatase region of SHIP. (A) Schematic diagram of the SHIP deletion and truncation mutants. All constructs were GST tagged at the N terminus. The positions of the SH2 domain, IPase signature motifs, and the two Shc-phosphotyrosine binding sites are indicated. The IPase activity of each construct, based on the data shown in panel B, is shown. (B) 293T cells were transfected with the indicated constructs, and the expressed proteins were precipitated with glutathione-Sepharose beads. One half of the beads were assayed for phosphatase activity toward 32P-labeled PIP3 and analyzed by thin-layer chromatography (upper panel). The other half was analyzed for protein expression by immunoblotting with an anti-GST antibody (lower panel). Sizes are shown in kilodaltons. (C) Phosphatase activity of GST-tagged tyrosine mutants of SHIP expressed in 293T cells and precipitated with glutathione-Sepharose beads. The lower panel shows the immunoblot of precipitated proteins with anti-GST antibody. (D) Phosphatase activity of wt and mutant SHIP proteins toward soluble IP4. Proteins precipitated, after transient expression in 293T cells, were incubated with 3H-labeled IP4, and the reaction products were analyzed by FPLC as described in the text. Arrows indicate the peaks for substrate (IP4) and the product inositol-1,3,4-trisphosphate.
We then tested deletion mutants of SHIP in this assay for their activity toward PIP3. Deletion of the SH2 domain of SHIP (construct 166-1190) had no detectable effect on the IPase activity (Fig. 1B, lane 2). Similarly, deleting much of the C terminus of SHIP through truncation after amino acid 933 (1-933) also did not affect its catalytic activity (Fig. 1B, lane 3). As would be predicted from the above results, a construct that expressed amino acids 166 to 933 of SHIP was enzymatically active (Fig. 1B, lane 4). The triple mutation within the IPase signature motifs again completely abolished the activity of the 166-933 protein (Fig. 1B, lane 5).
We then tested progressive truncations from the 166-933 region to narrow down the catalytic domain of SHIP. A mutant SHIP truncated at amino acid 900 still retained enzymatic activity (Fig. 1B, lane 6). With respect to the N terminus, sequences up to amino acid 400 were dispensable for in vitro activity of SHIP, whereas loss of a short 19-amino-acid stretch between residues 401 and 419 resulted in a completely inactive enzyme (Fig. 1B, lanes 7 to 10). This suggested that the N terminus of the catalytic domain of SHIP is located between amino acids 401 and 419.
We also tested larger deletions at the C terminus to further specify the carboxyl-terminal end of the catalytic domain. The mutant containing amino acids 166-850 and truncation mutant 1-814 were both inactive toward PIP3, suggesting that the C-terminal end of the IPase domain lies between residues 850 and 900 (Fig. 1B, lanes 12 and 13). It should be noted that in some experiments, we have detected residual activity with the 166-850 protein (data not shown). Interestingly, sequence homology comparisons between SHIP and nine other IPases showed that significant homology starts around amino acid 400 and ends around amino acid 727 of SHIP. However, as shown above, truncation at residue 814 results in complete loss of SHIP's enzymatic activity, while truncation at 850 severely reduced this activity. To rule out the possibility that the IPase domain may in fact end at residue 727 and that the region between residues 727 and 850 negatively influences SHIP enzymatic activity in an as yet undefined manner, we generated a construct spanning residues 401 to 727. This mutant had no catalytic activity toward PIP3 (Fig. 1B, lane 14), suggesting that the IPase domain of SHIP is longer than what the sequence homology with other 5′-IPases would predict. The only other protein in the database that had a region similar to residues 727 to 900 was the recently identified SHIP homolog SHIP-2. The homology between SHIP (also called SHIP-1) and SHIP-2 is mainly concentrated in the residues up to 870. In addition, we observed that among the non-SH2-containing 5′-phosphatases, synaptojanin shows some extended homology to SHIP, which stops at amino acid 866 of SHIP. We therefore generated a construct that covered the region between 401 and 866 and found this to be enzymatically active, similar to the 401-900 protein (Fig. 1B, lanes 15 and 16). A bacterially expressed 401-866 protein also had full catalytic activity (data not shown). Kinetic studies suggested that the 401-900 protein was comparable to the wt SHIP protein in dephosphorylating PIP3 (data not shown). Taken together, these observations narrowed the C-terminal end of the IPase domain to amino acid 866.
SHIP has two tyrosines in the C-terminal region that we have previously shown to serve as Shc-phosphotyrosine binding sites (33). As shown in Fig. 1C, SHIP proteins with a mutation of either or both sites retained enzymatic activity, suggesting that the mutation of these two sites per se does not affect the phosphatase activity. However, it must be noted that the wt SHIP expressed in 293T cells is not phosphorylated and hence is not associated with Shc or other phosphotyrosine-binding domain- or SH2-containing proteins (data not shown). Therefore, these data do not rule out a possible modulation of SHIP's enzymatic activity by molecules that may bind to these sites.
SHIP has also been demonstrated to have in vitro 5′-IPase activity toward a soluble inositol substrate, IP4 (9, 13, 29). We determined whether the minimal IPase domain defined above would also dephosphorylate IP4. The IPase activity of the above mutants toward 3H-labeled IP4 was tested, and the reaction products were analyzed by FPLC with a Mono-Q column. As shown in Fig. 1D, the 401-900 protein was capable of dephosphorylating IP4 as efficiently as the wt SHIP, whereas the 419-933 protein was completely inactive. This suggested that the regions of SHIP required for dephosphorylation of both PIP3 and IP4 are the same or essentially overlap.
Catalytic region of SHIP alone is not sufficient for FcγRIIB1-mediated inhibitory signaling in B cells.
It has been demonstrated that the recruitment of SHIP (via its SH2 domain) to the tyrosine-phosphorylated FcγRIIB1 is required for inhibition of BCR-mediated elevation of intracellular calcium (44). Since the BCR-induced calcium flux occurs downstream of PIP3 generation and activation of the tyrosine kinase Btk, SHIP-mediated dephosphorylation of PIP3 and, in turn, the termination of Btk activation leading to decreased activation of phospholipase C γ2 has been recognized as the mechanism for this inhibition (4, 14, 53). The requirement for SHIP in FcγRIIB1-mediated inhibition has been best demonstrated in the DT40 chicken B-cell line which has been made SHIP deficient through targeted gene disruption (20, 42, 44). To address the requirement for the different regions of SHIP in vivo, we chose to reconstitute this SHIP-deficient DT40 line with wt SHIP (SHIPwt) and mutant versions of SHIP and determine the FcγRIIB1-mediated inhibition of calcium flux as a readout. We generated stable cell lines expressing murine FcγRIIB1 and either wt or mutant SHIP proteins (see below). Multiple clones expressing each of the transfected proteins (at least three to five clones) were routinely analyzed. The data presented are representative of multiple experiments performed with each of these lines.
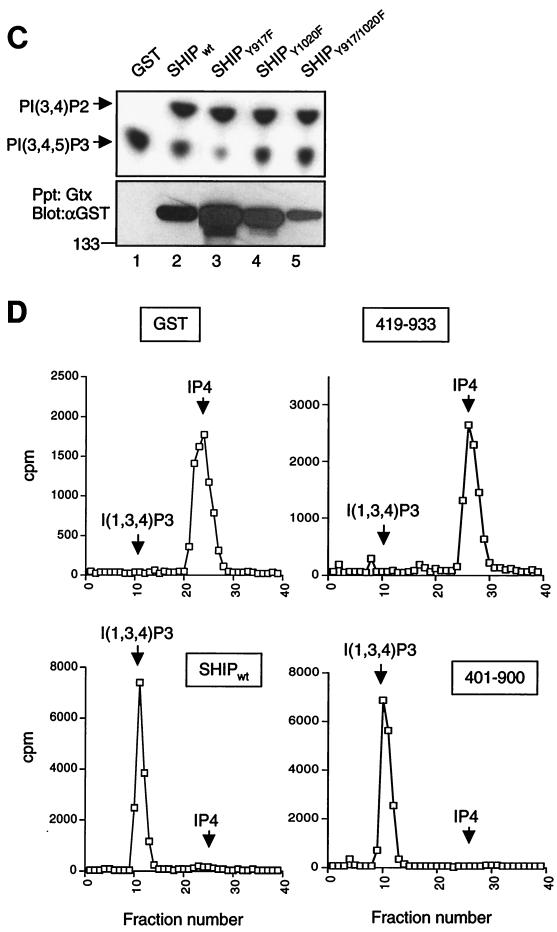
We first tested whether the minimal phosphatase region of SHIP alone would be sufficient for inhibition of calcium by FcR. Since the SHIP SH2 is required for binding to FcγRIIB1, to examine the effect of the IPase domain alone in cells, we needed to provide the SH2 domain to this protein for targeting to the FcγRIIB1. To achieve this, we generated a fusion construct in which the SHIP SH2 domain followed by an 18-amino-acid flexible linker (46) was fused to amino acids 401 to 900 of SHIP (SH2-18aa-401-900). As a control, the SHIP SH2 domain was fused to the rest of SHIP through the same linker (i.e., full-length SHIP interrupted by the linker; SH2-18aa-96-1190) (Fig. 2A). SHIP−/− DT40 cells were transfected with these constructs, and multiple clones were selected. The expression of these mutant constructs in two independent clones is shown in Fig. 2B. When analyzed for FcγRIIB1/SHIP-dependent inhibition of calcium flux, the minimal region that carries the IPase activity was unable to mediate inhibition of BCR-induced calcium flux. In contrast, clones expressing SH2-18aa-96-1190 showed the expected inhibition of BCR-mediated calcium flux upon BCR plus FcR coligation, demonstrating no apparent defect in the design of the fusion construct (Fig. 2C, right panels). Consistent with other reports, we also observed a higher level of BCR-induced calcium flux in SHIP-deficient DT40 cells and those reconstituted with nonfunctional SHIP proteins than in DT40 cells reconstituted with functional SHIP proteins (Fig. 2) (see below). The above data strongly suggested that regions other than the catalytic domain of SHIP are required for full inhibition of calcium flux through FcγRIIB1.
FIG. 2.
Minimal phosphatase region of SHIP alone is not sufficient for inhibition of calcium flux by FcγRIIB1. (A) Schematic diagram of the constructs designed for targeting the 401 to 900 and 96 to 1190 regions of SHIP to the FcR through fusion with the SHIP-SH2 domain via a flexible 18-amino-acid linker. (B) Expression of the constructs depicted in panel A in SHIP−/− DT40 stable clones. Equal numbers of cells were lysed, and total lysates were analyzed for expression of HA-tagged proteins by immunoblotting. Molecular size markers are indicated on the left (in kilodaltons). (C) Independent clones of SHIP−/− DT40 cells stably transfected with SH2-18aa-401-900 (left panels) or SH2-18aa-96-1190 (right panels) were loaded with indo-1 and analyzed for calcium flux as described in Materials and Methods. Recording of the fluorescence ratio was initiated prior to stimulation of cells with BCR cross-linking or BCR plus FcR co-cross-linking. The arrow indicates the time of addition of the stimulating antibody.
Sequences C-terminal to residue 900 are essential for SHIP function.
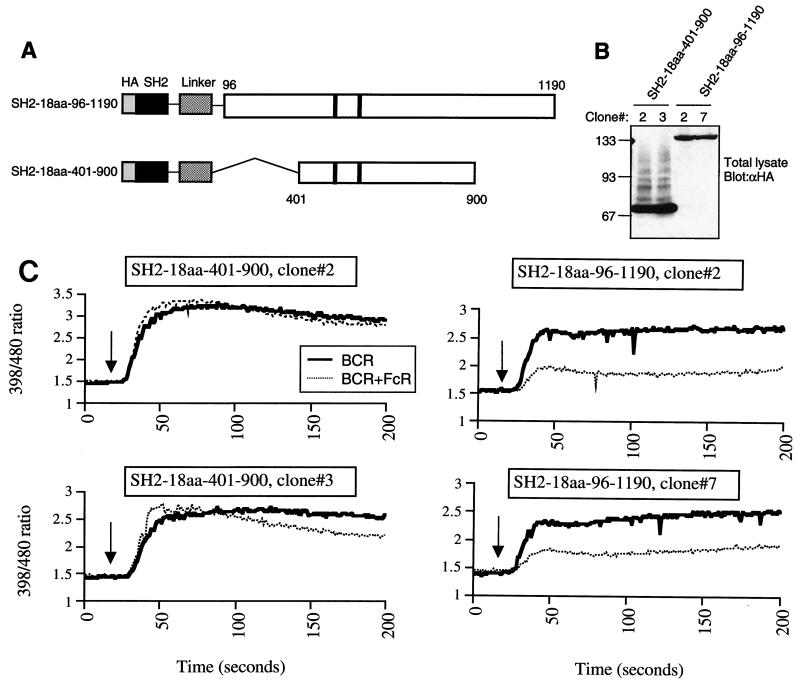
Two noncatalytic regions, amino acids 100 to 400 between the SH2 domain and the beginning of the IPase domain and the C-terminal region beyond residue 900, are missing from the SH2-18aa-401-900 protein. To test the role of the C-terminal region, we generated DT40 clones stably expressing a SHIP protein with the truncation at residue 900 (SHIP1–900) in SHIP−/− DT40 cells. Cross-linking of the BCR with the FcγRIIB1 showed much reduced inhibition of calcium flux in SHIP1–900-expressing cells compared with that in cells transfected with SHIPwt (Fig. 3A). The expression levels of both the SHIPwt and SHIP1–900 proteins were comparable in these clones (Fig. 3B). Surface expression of FcγRIIB1 on cells transfected with SHIP1–900 was equal to or slightly higher than that on cells transfected with SHIPwt, ruling out differences in FcR expression levels as a reason for diminished inhibition by SHIP1–900 (Fig. 3C). As shown in Fig. 3D, SHIP1–900 protein precipitated from these DT40 cells had readily detectable in vitro enzymatic activity toward PIP3. We also considered the possibility that SHIP1–900 may not bind FcR as efficiently as SHIPwt, resulting in this phenotype. However, as shown in Fig. 3E, both the SHIP1–900 and SHIPwt proteins could coprecipitate equivalent amount of phosphorylated FcγRIIB1. These data clearly demonstrated that the C-terminal region beyond amino acid 900 is required for the efficient function of SHIP in FcγRIIB1-mediated inhibitory signaling.
FIG. 3.
C terminus of SHIP required for inhibition of calcium flux by FcγRIIB1. (A) SHIP−/− DT40 cells and transfectants expressing HA-tagged wt SHIP or SHIP truncated at amino acid 900 (SHIP1–900) were analyzed for calcium flux upon BCR cross-linking (heavy solid line) or BCR plus FcR co-cross-linking (dotted line) as described in the legend to Fig. 2. (B) Expression of SHIPwt and SHIP1–900 in the DT40 clones used for calcium analysis in panel A was analyzed by immunoblotting. (C) Expression of FcγRIIB1 on DT40 transfectants used in the above experiments was analyzed by flow cytometry. FcR-negative parental DT40 cells were used as a negative control (dashed line). The SHIPwt transfectant (left panel) and the two SHIP1–900 clones (right panel) are shown as solid lines. (D) Phosphatase activity of SHIPwt and SHIP1–900 proteins immunoprecipitated (IP) from DT40 clones with anti-HA antibody analyzed with PIP3 as the substrate. The lower panel shows the immunoblot of precipitated proteins with anti-HA antibody. Sizes are shown in kilodaltons. (E) Coprecipitation of phosphorylated FcγRIIB1 with SHIPwt and SHIP1–900 proteins in DT40 clones. Cells were either left unstimulated or activated by coligation with BCR plus FcR for 5 min and lysed, and SHIP proteins were immunoprecipitated with anti-HA antibody and analyzed by immunoblotting with antiphosphotyrosine (γpTyr) antibody (upper panel) or anti-HA antibody (lower panel).
Tyrosines 917 and 1020 of SHIP play a role in FcγRIIB1-dependent inhibition of calcium flux.
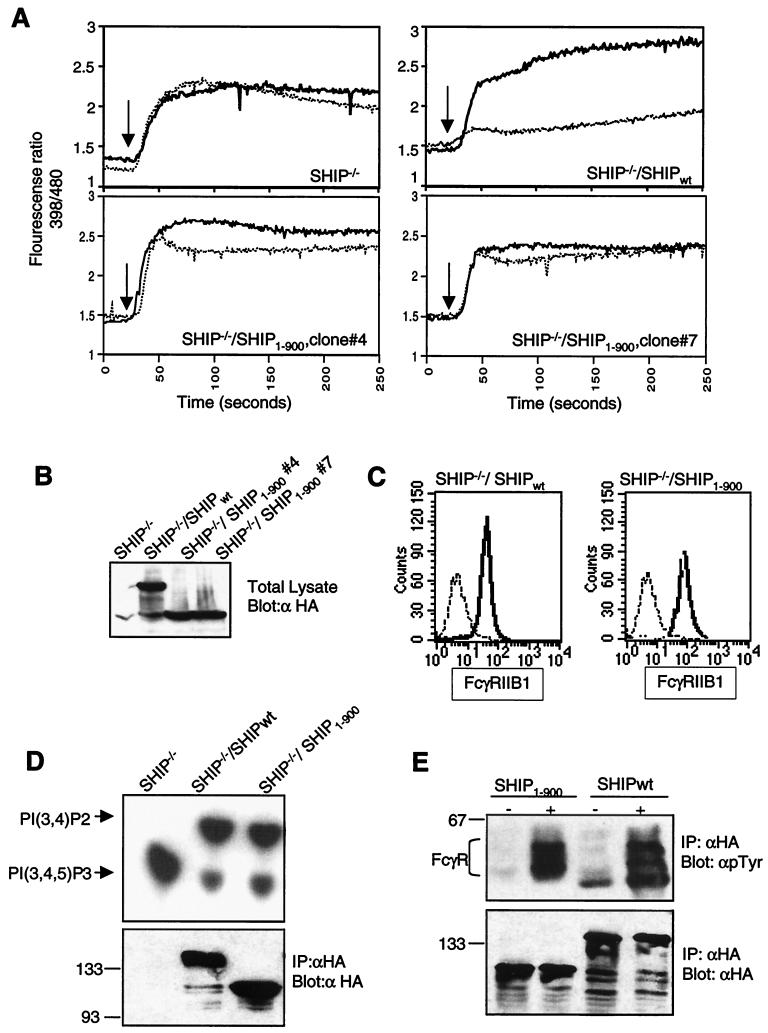
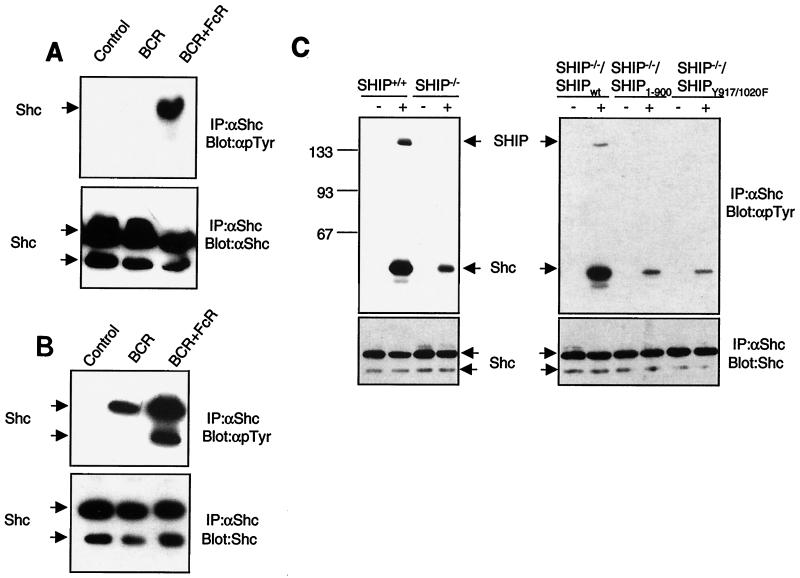
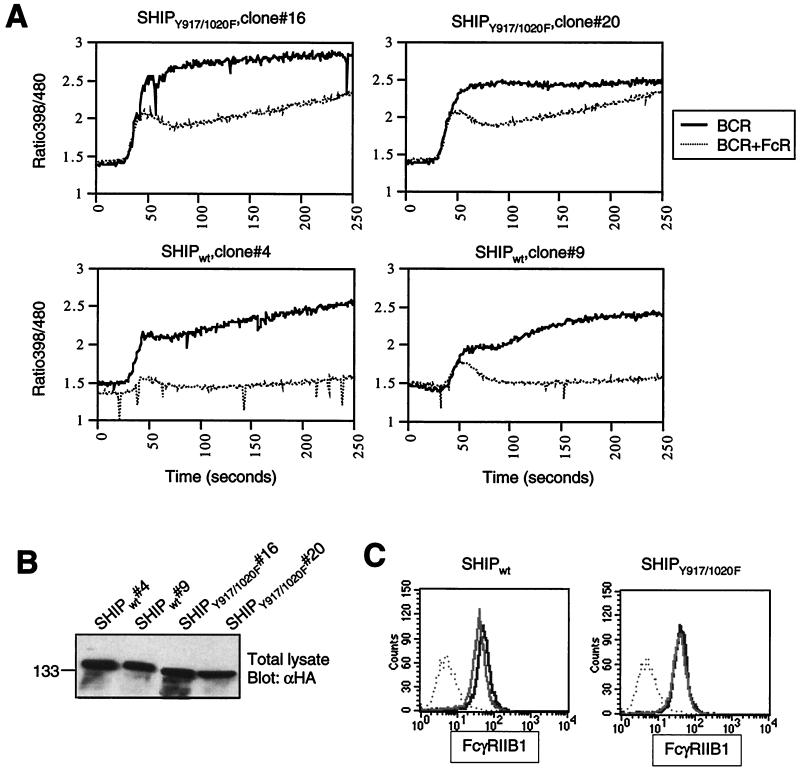
The C-terminal region of SHIP contains two tyrosines (Y917 and Y1020) that we have previously identified as binding sites for the Shc-phosphotyrosine binding domain (33). To test whether the absence of Shc binding sites may have contributed to the diminished function of the SHIP1–900 mutant, we transfected the SHIP-deficient DT40 cell line with a plasmid encoding full-length SHIP in which both Y917 and Y1020 have been mutated to phenylalanine (SHIPY917F/Y1020F). We have previously demonstrated that SHIPY917F/Y1020F fails to interact with Shc (33). As expected, SHIPY917F/Y1020F expressed in these DT40 clones also failed to associate with Shc upon BCR plus FcR co-cross-linking (data not shown and Fig. 5C). We generated multiple clones expressing different levels of SHIPY917F/Y1020F protein and analyzed those that closely matched the expression level of SHIPwt protein. The calcium flux profile of two of the clones after BCR alone and BCR plus FcR co-cross-linking is shown in Fig. 4A. We found that FcγRIIB1-dependent inhibition of the BCR-mediated calcium flux was much less efficient in SHIPY917F/Y1020F-expressing cells than in SHIPwt-reconstituted cells. This difference was most pronounced in the inhibition of sustained calcium levels (Fig. 4A). While the sustained rise in calcium levels was essentially abrogated upon BCR plus FcR co-cross-linking in SHIPwt-expressing cells, this was not the case in SHIPY917F/Y1020F-expressing cells. As shown in Fig. 4B and C, the cell lines used in these experiments express comparable levels of both SHIP proteins as well as FcγRIIB1. It is noteworthy that during the analysis of multiple clones expressing SHIPY917F/Y1020F and repeated analyses of the same clone, we observed a greater variability in the extent of inhibition compared with SHIP1–900. The reason for this variability is unclear. Nevertheless, we consistently observed that the profile of the calcium inhibition in SHIPY917F/Y1020F-expressing cells was clearly different from that with the SHIPwt clones. These data suggested a role for the two tyrosines and potentially for Shc binding to these two sites as being important in SHIP-mediated regulation of the calcium flux.
FIG. 5.
SHIP required for maximal phosphorylation of Shc in response to BCR plus FcR co-cross-linking. (A and B) DT40 cells (A) or A20 cells (B) were left unstimulated or stimulated by BCR cross-linking alone or BCR plus FcR coligation. Cells were lysed, and Shc was immunoprecipitated (IP) with polyclonal anti-Shc antibody. Immunoprecipitates were analyzed by immunoblotting with antiphosphotyrosine antibody (dpTyr) (upper panels) or anti-Shc antibody (lower panels). The p52 and p46 isoforms of Shc are indicated by arrows. (C) Parental DT40 cells, SHIP−/− DT40 cells, or SHIP−/− DT40 cells reconstituted with SHIPwt, SHIP1–900, or SHIPY917/1020F were left unstimulated (lanes —) or stimulated by BCR plus FcR co-cross-linking (lanes +), and Shc phosphorylation was analyzed as described above. Sizes are shown in kilodaltons.
FIG. 4.
Two tyrosines in the C terminus of SHIP are required for inhibition of calcium flux by FcγRIIB1. (A) SHIP−/− DT40 cells and transfectants expressing HA-tagged SHIPwt or SHIPY917/1020F were analyzed for calcium flux upon BCR cross-linking alone (heavy solid line) or BCR plus FcR co-cross-linking (dotted line) as described in the legend to Fig. 2. (B) Expression of SHIPwt and SHIPY917/1020F in the stable clones used in panel A was determined by anti-HA immunoblotting. (C) Expression of FcγRIIB1 on DT40 transfectants used in the above experiments was analyzed by flow cytometry with the 2.4G2 antibody. FcR-negative parental DT40 cells were used as a negative control (dotted line). The SHIPwt transfectants (left panel) and SHIPY917/1020F clones (right panel) are shown as solid lines.
The best known readout for Shc involvement in signaling has been its tyrosine phosphorylation. We therefore determined whether Shc phosphorylation might be altered in cells expressing wt and mutant forms of SHIP. We consistently observed that the tyrosine phosphorylation of Shc in response to BCR plus FcR coligation is much higher than that after BCR cross-linking alone in chicken DT40 cells (Fig. 5A) and the murine A20 B-cell line (Fig. 5B), consistent with a previous report (30). In the DT40 cells, BCR-induced Shc phosphorylation is very weak (detectable after long exposure of the film) but is significantly enhanced by BCR plus FcR co-cross-linking (Fig. 5A). Interestingly, when we compared Shc phosphorylation after BCR plus FcR coligation in parental DT40 cells and SHIP-deficient DT40 cells, we detected a significantly diminished level of Shc phosphorylation in the SHIP-deficient cells (Fig. 5C, left panel). When the SHIP-deficient cells were reconstituted with SHIPwt, we could again detect a higher level of Shc phosphorylation as well as its association with SHIP in response to BCR plus FcR coligation (Fig. 5C, right panel). In contrast, Shc was poorly phosphorylated in cells reconstituted with SHIP mutants that cannot interact with Shc, i.e., SHIP1–900 and SHIPY917F/Y1020F (Fig. 5C, right panel). These data suggested that interaction of Shc with SHIP is critical for efficient tyrosine phosphorylation of Shc.
Shc binding to SHIP is not required for FcγRIIB1-mediated calcium inhibition.
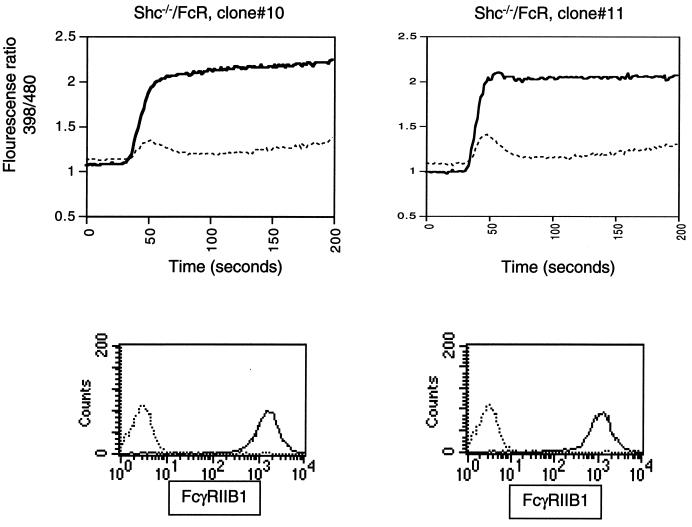
Although there is no direct evidence to date that tyrosine phosphorylation of Shc influences calcium levels in B cells, the above data suggested that the failure of SHIP to complex with Shc and the subsequent failure of efficient Shc tyrosine phosphorylation may have contributed to the diminished function of the SHIPY917F/Y1020F and SHIP1–900 proteins. To directly test the requirement for Shc in FcγRIIB1/SHIP-dependent inhibition of BCR-induced calcium flux, we made use of a DT40 cell line in which Shc expression has been abolished by targeted gene disruption (21). The Shc-deficient DT40 cell line was stably transfected with a plasmid encoding murine FcγRIIB1, and multiple clones expressing FcγRIIB1 on their surface were established. Analysis of calcium flux in these cells showed that coligation of FcR with BCR efficiently inhibited BCR-induced calcium flux in the absence of Shc expression. Data for two independent clones are shown in Fig. 6. In addition, we could not detect a significant difference in the enzymatic activity of Shc-bound and Shc-free SHIP in our in vitro IPase assays with PIP3 as a substrate (data not shown). These data suggested that Shc-SHIP complex formation is not required for FcγRIIB1-mediated inhibition of calcium flux and that the diminished function of SHIPY917F/Y1020F may be due to the failure of the tyrosine mutants to interact with another molecule(s) besides Shc.
FIG. 6.
Shc not required for SHIP-mediated inhibition of calcium flux by FcγRIIB1. Shc−/− DT40 cells stably transfected with FcγRIIB1 were analyzed for calcium flux upon BCR cross-linking alone (heavy solid line) or BCR plus FcR co-cross-linking (dotted line). Lower panels show FcγRIIB1 expression in these clones analyzed by flow cytometry.
Membrane targeting of SHIP1–900 restores inhibition of calcium flux.
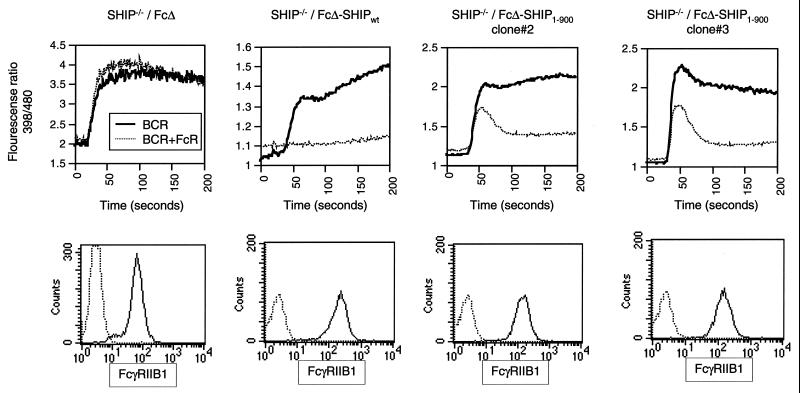
We then tested the possibility that the C terminus of SHIP may play a role in stabilizing the membrane localization of SHIP during FcγRIIB1-mediated inhibitory signaling. We fused either SHIPwt or SHIP1–900 directly to the cytoplasmic domain of a truncated FcγRIIB1 (that lacks the ITIM motif that would normally recruit SHIP). SHIP−/− DT40 cells were stably transfected with plasmids encoding a truncated FcγRIIB1 (FcΔ), Fc fused to wt SHIP (FcΔ-SHIPwt), or Fc fused to SHIP1–900 (FcΔ-SHIP1–900). The calcium fluxes in these cells were analyzed after BCR cross-linking alone or BCR plus FcR cross-linking. As shown in Fig. 7, FcΔ-SHIP1–900 was very efficient in inhibiting BCR-mediated calcium flux. It is noteworthy that FcΔ-SHIPwt was more efficient than Fc-SHIP1–900 in inhibiting the calcium flux. Despite this, FcΔ-SHIP1–900 showed far greater inhibition than the cytoplasmic version of SHIP1–900 (Fig. 3). This was seen with multiple FcΔ-SHIP1–900-expressing clones. As expected, the control Fc clones did not show an inhibition of calcium flux after BCR plus FcR co-cross-linking. As shown in the bottom panel of Fig. 7, FcΔ, FcΔ-SHIPwt, and FcΔ-SHIP1–900 were expressed comparably on the cell surface. These data suggested that membrane targeting significantly restores SHIP1–900 function and that the C-terminal noncatalytic region of SHIP may play a key role in stabilizing SHIP interaction at the membrane.
FIG. 7.
Membrane-targeted SHIP1–900 can mediate FcγRIIB1-mediated calcium inhibition. SHIP−/− DT40 cells were stably transfected with plasmids encoding FcγRIIB1 truncated at its cytoplasmic tail (FcΔ), FcΔ fused to wt SHIP (FcΔ-SHIPwt), or FcΔ fused to SHIP1–900) The cells were analyzed for calcium flux after cross-linking with BCR alone or BCR plus FcR (top panels). The increase in BCR-induced calcium flux seen at later time points is unique to the FcΔ-SHIP clones, and the reason for this is currently unclear. The surface expression of the FcΔ and FcΔ-SHIP fusion proteins was determined by flow cytometry with the anti-FcR antibody 2.4G2 (bottom panel). The data are representative of at least two independent experiments. Multiple clones that were analyzed showed similar results.
DISCUSSION
In the past few years since its original discovery as an IPase, SHIP has been demonstrated to play a key role as a negative regulator of multiple receptor systems. While SHIP's role as the mediator of the potent negative signal delivered through FcγRIIB1 is established, the molecular mechanism(s) involved is less well defined. In this report, by defining the boundaries of the catalytic and noncatalytic regions of SHIP, we demonstrate that the recruitment of an enzymatically active SHIP to the FcγRIIB1 alone is not sufficient for FcγRIIB1/SHIP-mediated inhibition of the BCR signals. We show that the noncatalytic region in the C terminus of SHIP plays an essential role in SHIP function in vivo. While the two tyrosines Y917 and Y1020 appear to be important in SHIP function, our studies with Shc-deficient cells suggest that the binding of Shc to these sites is not required for SHIP-mediated inhibition of calcium flux. Studies with membrane-targeted SHIP proteins indicate that the C terminus of SHIP, at least in part, functions through stabilizing SHIP localization at the membrane. Taken together, these data suggest that the C-terminal 190 amino acids of SHIP play a critical role in SHIP function in B cells and may also be required in various other systems where SHIP functions as a negative regulator of signaling.
Interestingly, the noncatalytic regions of the 145-kDa form of human and murine SHIP are highly conserved (greater than 90% amino acid identity), suggesting an evolutionarily conserved role for these regions. Different isoforms of SHIP and cleavage products have been detected in hematopoietic cells and have been implicated to perform specific functions (10, 15, 41). Lucas and Rohrschneider have reported the regulated expression of a 135-kDa form of SHIP that appears during myeloid lineage development, carrying an internal deletion between residues 920 and 980 (41). Since our data indicate that the IPase domain of SHIP exists between residues 400 and 866, essentially all of the isoforms detected thus far must carry an intact IPase domain but differ in the noncatalytic regions. Given our finding that the noncatalytic regions of SHIP play a critical role in FcR signaling, the differences in the regions outside the enzymatic domain could determine the specific function for the different isoforms.
It is also noteworthy that the noncatalytic regions of SHIP are not conserved in other non-SH2-containing 5′-IPases. The phenotype of SHIP-deficient mice, in which other 5′-IPases are apparently expressed in hematopoietic cells but are unable to substitute for SHIP function, suggests an essential role for these noncatalytic regions. In this regard, we found that expression of other 5′-IPases is unable to reconstitute a SHIP-deficient B-cell line even when targeted to the FcR (M. J. Aman et al., unpublished data). As mentioned earlier, the IPase domain of SHIP is larger than what is predicted for other 5′-IPases. Whether the failure of these other 5′-IPases to substitute for SHIP is due to the unique role played by SHIP's noncatalytic regions or the unique IPase domain of SHIP or both remains to be determined. We also observed that the murine SHIP, while quite capable of reconstituting the SHIP deficiency, may be less efficient than the endogenous chicken SHIP in the DT40 cells. This stems from the diminished phosphorylation of HA-tagged murine SHIP that we observed compared with endogenous SHIP, despite higher expression of the tagged protein. The cause for this difference is unclear and awaits the cloning of chicken SHIP cDNA.
While our data demonstrate a requirement for the C terminus of SHIP in FcγRIIB1/SHIP-mediated inhibition of calcium signaling, precisely how this region contributes to SHIP function is not clear. Several, not mutually exclusive, possibilities exist. For example, the C terminus could help stabilize localization of SHIP at the membrane. Although the SH2 domain of SHIP has been shown to bind to the ITIM motif in the cytoplasmic tail of the FcγRIIB1 receptor, the kinetics or stability of this interaction at the membrane has not yet been determined. The C terminus of SHIP may stabilize the FcR-SHIP complex either directly or indirectly through interacting with other proteins. Alternatively, C-terminal interactions may be required for efficient localization to specific membrane compartments within cells, where the calcium signals may be initiated. While a role for the C terminus in stabilizing SHIP interaction at the membrane is suggested by the FcΔ-SHIP1–900 construct, it is noteworthy that the constitutive presence of this protein on the membrane or the topology or turnover rate of the Fc-SHIP protein may have also contributed to this effect.
Protein secondary-structure predictions of the region beyond residue 866 suggest a predominantly unstructured region. This is not unexpected, given the numerous prolines throughout this region. This implies that the regulation of SHIP function by the C terminus is most likely mediated through proteins that interact with this region. In this regard, we looked at the significance of the best-characterized protein known to bind to SHIP, Shc. While the mutations of the tyrosines previously identified as Shc binding sites appeared to play a role in SHIP function, Shc-deficient cells showed efficient FcγRIIB1-mediated inhibition of calcium flux. This suggested that Shc-SHIP complex formation is not required for this inhibition. However, it is noteworthy that we observed an unexpected role for Shc-SHIP complex formation in efficient Shc phosphorylation. The precise role of Shc-SHIP complex formation in regulating FcγRIIB1-mediated signaling is unclear. Although a SHIP-mediated sequestration of Shc away from Grb2, and subsequent inhibition of the Ras/mitogen-activated protein kinase pathway was proposed initially (56), this has not been confirmed by others (19; our unpublished observations). Nevertheless, it appears that Shc phosphorylation is regulated through its interaction with SHIP and may have functional consequences that remain to be elucidated.
A recent report by Gupta et al. has suggested that the p85 subunit of PI3K (via its SH2 domain) can interact with the Y917 site on SHIP (18). The failure to interact with p85 or other SH2- or phosphotyrosine binding domain-containing proteins might also have contributed to the deficiency in function of the SHIPY917F/1020F mutant. In addition, p62 Dok, originally identified as a Ras-GAP binding protein, has also been shown to interact via its phosphotyrosine binding domain with the two tyrosines in the C terminus of SHIP (J. C. Cambier, personal communication). The significance of p85 and p62 Dok interactions with SHIP in regulating SHIP function remains to be established. Besides the two tyrosines Y917 and Y1020, there are also four proline-rich motifs in the C terminus of SHIP which can bind Grb2 (9, 29) or other SH3 domain-containing proteins. Grb2 has been shown to interact via its SH3 domain with proline-rich regions of SHIP and concurrently via its SH2 domain with phosphorylated Shc (19). Although we have failed to see a role for Shc-SHIP complex formation in inhibition of calcium flux, the direct binding of Grb2 and its regulation of SHIP function are still possible. The precise role of the proline-rich regions and the involvement of Grb2 are currently under investigation.
Another potential role for the C terminus could be in facilitating substrate accessibility for SHIP. The calcium inhibition profile of the membrane-targeted FcΔ-SHIP1–900 protein is not the same as that of Fc-SHIPwt. We consistently observed that the membrane-targeted Fc-SHIP is more potent in inhibiting BCR-mediated calcium flux than wt SHIP, possibly due to a greater fraction of SHIP molecules being present on the membrane (data not shown). This may also be due to the topology of the Fc-SHIP proteins on the membrane or to lower turnover of Fc-SHIP from the membrane. If the sole function of the C terminus were to stabilize SHIP interaction at the membrane, we would have seen comparable inhibition with FcΔ-SHIP1–900 and FcΔ-SHIPwt. Perhaps another role for the C terminus of SHIP may be to provide better accessibility to the substrate PIP3. A study of the phospholipid contents of the NHK96 and LS5 cell lines showed that PIP3 makes up less than 0.01% of the total cellular phospholipids (59). Gold and Aebersold observed only a modest increase in the level of PIP3 over the background upon BCR stimulation, showing that even after activation this phospholipid is present at very low concentrations (16). While better accessibility to PIP3 through the C terminus is an attractive possibility, we have not seen a difference in in vitro enzymatic activity between SHIPwt and SHIP1–900. However, the phosphatidylserine/PIP3 micelles used in these experiments very likely do not adequately represent the complex composition of plasma membrane. It is conceivable that in the context of the natural plasma membrane, the C terminus may facilitate SHIP's access to substrate by interaction with certain membrane components or mediate SHIP's localization to specialized membrane microdomains where PIP3 is present or calcium signaling is initiated. Further detailed studies are required to address this issue.
SHIP has also been shown to function as a negative regulator in many other systems, such as stimulation via colony-stimulating factor-1, granulocyte-macrophage colony-stimulating factor, and several other cytokines, growth hormone, and thrombin (24, 35, 51, 58). While there is involvement of calcium in some cases, there is no calcium flux in others. Interestingly, SHIP-deficient mice exhibit defects in a number of different hematopoietic lineages, and studies with these mice clearly suggest a role for SHIP in setting thresholds during cytokine stimulation and proliferation in the myeloid lineage (23, 39, 40). Whether the C terminus is also required for SHIP-mediated regulation of these events remains to be determined. Given the crucial role played by SHIP in multiple receptor systems, the data presented in this report provide the initial steps toward a better molecular understanding of negative regulation through SHIP.
ACKNOWLEDGMENTS
We thank Tomo Kurosaki for providing us with the DT40 cell lines and the FcγRIIB1 cDNA and Gerald Krystal for the original SHIP cDNA. We thank Anke Klippel for purified p110* protein and Glen Prestwich and Andrew Morris for IP4 and PIP2, respectively. We thank Ulrike Lorenz for critical reading of the manuscript.
This work was supported by an RO1 grant from the National Institutes of Health and by a grant from the Jeffress Gwathmy Memorial Trust (to K.S.R.). M.J.A. was supported by an NIH Immunology Training Grant.
REFERENCES
- 1.Aman M J, Lamkin T D, Okada H, Kurosaki T, Ravichandran K S. The inositol phosphatase SHIP inhibits Akt/PKB activation in B cells. J Biol Chem. 1998;273:33922–33928. doi: 10.1074/jbc.273.51.33922. [DOI] [PubMed] [Google Scholar]
- 2.Ashman R F, Peckham D, Stunz L L. Fc receptor off-signal in the B cell involves apoptosis. J Immunol. 1996;157:5–11. [PubMed] [Google Scholar]
- 3.Astoul E, Watton S, Cantrell D. The dynamics of protein kinase B regulation during B cell antigen receptor engagement. J Cell Biol. 1999;145:1511–1520. doi: 10.1083/jcb.145.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolland S, Pearse R N, Kurosaki T, Ravetch J V. SHIP modulates immune receptor responses by regulating membrane association of Btk. Immunity. 1998;8:509–516. doi: 10.1016/s1074-7613(00)80555-5. [DOI] [PubMed] [Google Scholar]
- 5.Bolland S, Ravetch J V. Inhibitory pathways triggered by ITIM-containing receptors. Adv Immunol. 1999;72:149–177. doi: 10.1016/s0065-2776(08)60019-x. [DOI] [PubMed] [Google Scholar]
- 6.Chacko G W, Tridandapani S, Damen J E, Liu L, Krystal G, Coggeshall K M. Negative signaling in B lymphocytes induces tyrosine phosphorylation of the 145-kDa inositol polyphosphate 5-phosphatase, SHIP. J Immunol. 1996;157:2234–2238. [PubMed] [Google Scholar]
- 7.Crowley M T, Harmer S L, DeFranco A L. Activation-induced association of a 145-kDa tyrosine-phosphorylated protein with Shc and Syk in B lymphocytes and macrophages. J Biol Chem. 1996;271:1145–1152. doi: 10.1074/jbc.271.2.1145. [DOI] [PubMed] [Google Scholar]
- 8.Damen J E, Liu L, Cutler R L, Krystal G. Erythropoietin stimulates the tyrosine phosphorylation of Shc and its association with Grb2 and a 145-Kd tyrosine phosphorylated protein. Blood. 1993;82:2296–2303. [PubMed] [Google Scholar]
- 9.Damen J E, Liu L, Rosten P, Humphries R K, Jefferson A B, Majerus P W, Krystal G. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5-triphosphate 5-phosphatase. Proc Natl Acad Sci USA. 1996;93:1689–1693. doi: 10.1073/pnas.93.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damen J E, Liu L, Ware M D, Ermolaeva M, Majerus P W, Krystal G. Multiple forms of the SH2-containing inositol phosphatase, SHIP, are generated by C-terminal truncation. Blood. 1998;92:1199–1205. [PubMed] [Google Scholar]
- 11.DeFranco A L, Chan V W, Lowell C A. Positive and negative roles of the tyrosine kinase Lyn in B cell function. Semin Immunol. 1998;10:299–307. doi: 10.1006/smim.1998.0122. [DOI] [PubMed] [Google Scholar]
- 12.Diegel M L, Rankin B M, Bolen J B, Dubois P M, Kiener P A. Cross-linking of Fc gamma receptor to surface immunoglobulin on B cells provides an inhibitory signal that closes the plasma membrane calcium channel. J Biol Chem. 1994;269:11409–11416. [PubMed] [Google Scholar]
- 13.Drayer A L, Pesesse X, De Smedt F, Woscholski R, Parker P, Erneux C. Cloning and expression of a human placenta inositol 1,3,4,5-tetrakisphosphate and phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase. Biochem Biophys Res Commun. 1996;225:243–249. doi: 10.1006/bbrc.1996.1161. [DOI] [PubMed] [Google Scholar]
- 14.Fluckiger A C, Li Z, Kato R M, Wahl M I, Ochs H D, Longnecker R, Kinet J P, Witte O N, Scharenberg A M, Rawlings D J. Btk/Tec kinases regulate sustained increases in intracellular Ca2+ following B-cell receptor activation. EMBO J. 1998;17:1973–1985. doi: 10.1093/emboj/17.7.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geier S J, Algate P A, Carlberg K, Flowers D, Friedman C, Trask B, Rohrschneider L R. The human SHIP gene is differentially expressed in cell lineages of the bone marrow and blood. Blood. 1997;89:1876–1885. [PubMed] [Google Scholar]
- 16.Gold M R, Aebersold R. Both phosphatidylinositol 3-kinase and phosphatidylinositol 4-kinase products are increased by antigen receptor signaling in B cells. J Immunol. 1994;152:42–50. [PubMed] [Google Scholar]
- 17.Gold M R, Scheid M P, Santos L, Dang-Lawson M, Roth R A, Matsuuchi L, Duronio V, Krebs D L. The B cell antigen receptor activates the Akt (protein kinase B)/glycogen synthase kinase-3 signaling pathway via phosphatidylinositol 3-kinase. J Immunol. 1999;163:1894–1905. [PubMed] [Google Scholar]
- 18.Gupta N, Scharenberg A M, Fruman D A, Cantley L C, Kinet J P, Long E O. The SH2 domain-containing inositol 5′-phosphatase (SHIP) recruits the p85 subunit of phosphoinositide 3-kinase during FcgammaRIIb1-mediated inhibition of B cell receptor signaling. J Biol Chem. 1999;274:7489–7494. doi: 10.1074/jbc.274.11.7489. [DOI] [PubMed] [Google Scholar]
- 19.Harmer S L, DeFranco A L. The src homology domain 2-containing inositol phosphatase SHIP forms a ternary complex with Shc and Grb2 in antigen receptor-stimulated B lymphocytes. J Biol Chem. 1999;274:12183–12191. doi: 10.1074/jbc.274.17.12183. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto A, Hirose K, Okada H, Kurosaki T, Iino M. Inhibitory modulation of B cell receptor-mediated Ca2+ mobilization by Src homology 2 domain-containing inositol 5′-phosphatase (SHIP) J Biol Chem. 1999;274:11203–11208. doi: 10.1074/jbc.274.16.11203. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto A, Okada H, Jiang A, Kurosaki M, Greenberg S, Clark E A, Kurosaki T. Involvement of guanosine triphosphatases and phospholipase C-gamma2 in extracellular signal-regulated kinase, c-Jun NH2-terminal kinase, and p38 mitogen-activated protein kinase activation by the B cell antigen receptor. J Exp Med. 1998;188:1287–1295. doi: 10.1084/jem.188.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haverstick D M, Gray L S. Increased intracellular Ca2+ induces Ca2+ influx in human T lymphocytes. Mol Biol Cell. 1993;4:173–184. doi: 10.1091/mbc.4.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helgason C D, Damen J E, Rosten P, Grewal R, Sorensen P, Chappel S M, Borowski A, Jirik F, Krystal G, Humphries R K. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hibi M, Hirano T. Signal transduction through cytokine receptors. Int Rev Immunol. 1998;17:75–102. doi: 10.3109/08830189809084488. [DOI] [PubMed] [Google Scholar]
- 25.Huber M, Helgason C D, Damen J E, Liu L, Humphries R K, Krystal G. The src homology 2-containing inositol phosphatase (SHIP) is the gatekeeper of mast cell degranulation. Proc Natl Acad Sci USA. 1998;95:11330–11335. doi: 10.1073/pnas.95.19.11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber M, Helgason C D, Scheid M P, Duronio V, Humphries R K, Krystal G. Targeted disruption of SHIP leads to steel factor-induced degranulation of mast cells. EMBO J. 1998;17:7311–7319. doi: 10.1093/emboj/17.24.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacob A, Cooney D, Tridandapani S, Kelley T, Coggeshall K M. FcgammaRIIb modulation of surface immunoglobulin-induced Akt activation in murine B cells. J Biol Chem. 1999;274:13704–13710. doi: 10.1074/jbc.274.19.13704. [DOI] [PubMed] [Google Scholar]
- 28.Jefferson A B, Auethavekiat V, Pot D A, Williams L T, Majerus P W. Signaling inositol polyphosphate-5-phosphatase. Characterization of activity and effect of GRB2 association. J Biol Chem. 1997;272:5983–5988. doi: 10.1074/jbc.272.9.5983. [DOI] [PubMed] [Google Scholar]
- 29.Kavanaugh W M, Pot D A, Chin S M, Deuter-Reinhard M, Jefferson A B, Norris F A, Masiarz F R, Cousens L S, Majerus P W, Williams L T. Multiple forms of an inositol polyphosphate 5-phosphatase form signaling complexes with Shc and Grb2. Curr Biol. 1996;6:438–445. doi: 10.1016/s0960-9822(02)00511-0. [DOI] [PubMed] [Google Scholar]
- 30.Kiener P A, Lioubin M N, Rohrschneider L R, Ledbetter J A, Nadler S G, Diegel M L. Co-ligation of the antigen and Fc receptors gives rise to the selective modulation of intracellular signaling in B cells. Regulation of the association of phosphatidylinositol 3-kinase and inositol 5′-phosphatase with the antigen receptor complex. J Biol Chem. 1997;272:3838–3844. doi: 10.1074/jbc.272.6.3838. [DOI] [PubMed] [Google Scholar]
- 31.Klippel A, Kavanaugh W M, Pot D, Williams L T. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurosaki T. Genetic analysis of B cell antigen receptor signaling. Annu Rev Immunol. 1999;17:555–592. doi: 10.1146/annurev.immunol.17.1.555. [DOI] [PubMed] [Google Scholar]
- 33.Lamkin T D, Walk S F, Liu L, Damen J E, Krystal G, Ravichandran K S. Shc interaction with Src homology 2 domain containing inositol phosphatase (SHIP) in vivo requires the Shc-phosphotyrosine binding domain and two specific phosphotyrosines on SHIP. J Biol Chem. 1997;272:10396–10401. doi: 10.1074/jbc.272.16.10396. [DOI] [PubMed] [Google Scholar]
- 34.Law C L, Sidorenko S P, Clark E A. Regulation of lymphocyte activation by the cell-surface molecule CD22. Immunol Today. 1994;15:442–449. doi: 10.1016/0167-5699(94)90275-5. [DOI] [PubMed] [Google Scholar]
- 35.Lin J X, Leonard W J. Signaling from the IL-2 receptor to the nucleus. Cytokine Growth Factor Rev. 1997;8:313–332. doi: 10.1016/s1359-6101(97)00021-x. [DOI] [PubMed] [Google Scholar]
- 36.Lioubin M N, Algate P A, Tsai S, Carlberg K, Aebersold A, Rohrschneider L R. p150Ship, a signal transduction molecule with inositol polyphosphate-5-phosphatase activity. Genes Dev. 1996;10:1084–1095. doi: 10.1101/gad.10.9.1084. [DOI] [PubMed] [Google Scholar]
- 37.Liu L, Damen J E, Cutler R L, Krystal G. Multiple cytokines stimulate the binding of a common 145-kilodalton protein to Shc at the Grb2 recognition site of Shc. Mol Cell Biol. 1994;14:6926–6935. doi: 10.1128/mcb.14.10.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L, Damen J E, Hughes M R, Babic I, Jirik F R, Krystal G. The Src homology 2 (SH2) domain of SH2-containing inositol phosphatase (SHIP) is essential for tyrosine phosphorylation of SHIP, its association with Shc, and its induction of apoptosis. J Biol Chem. 1997;272:8983–8988. doi: 10.1074/jbc.272.14.8983. [DOI] [PubMed] [Google Scholar]
- 39.Liu Q, Oliveira-Dos-Santos A J, Mariathasan S, Bouchard D, Jones J, Sarao R, Kozieradzki I, Ohashi P S, Penninger J M, Dumont D J. The inositol polyphosphate 5-phosphatase ship is a crucial negative regulator of B cell antigen receptor signaling. J Exp Med. 1998;188:1333–1342. doi: 10.1084/jem.188.7.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q, Sasaki T, Kozieradzki I, Wakeham A, Itie A, Dumont D J, Penninger J M. SHIP is a negative regulator of growth factor receptor-mediated PKB/Akt activation and myeloid cell survival. Genes Dev. 1999;13:786–791. doi: 10.1101/gad.13.7.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas D M, Rohrschneider L R. A novel spliced form of SH2-containing inositol phosphatase is expressed during myeloid development. Blood. 1999;93:1922–1933. [PubMed] [Google Scholar]
- 42.Okada H, Bolland S, Hashimoto A, Kurosaki M, Kabuyama Y, Iino M, Ravetch J V, Kurosaki T. Role of the inositol phosphatase SHIP in B cell receptor-induced Ca2+ oscillatory response. J Immunol. 1998;161:5129–5132. [PubMed] [Google Scholar]
- 43.Ono M, Bolland S, Tempst P, Ravetch J V. Role of the inositol phosphatase SHIP in negative regulation of the immune system by the receptor Fc(gamma)RIIB. Nature. 1996;383:263–266. doi: 10.1038/383263a0. [DOI] [PubMed] [Google Scholar]
- 44.Ono M, Okada H, Bolland S, Yanagi S, Kurosaki T, Ravetch J V. Deletion of SHIP or SHP-1 reveals two distinct pathways for inhibitory signaling. Cell. 1997;90:293–301. doi: 10.1016/s0092-8674(00)80337-2. [DOI] [PubMed] [Google Scholar]
- 45.Osborne M A, Zenner G, Lubinus M, Zhang X, Songyang Z, Cantley L C, Majerus P, Burn P, Kochan J P. The inositol 5′-phosphatase SHIP binds to immunoreceptor signaling motifs and responds to high affinity IgE receptor aggregation. J Biol Chem. 1996;271:29271–29278. doi: 10.1074/jbc.271.46.29271. [DOI] [PubMed] [Google Scholar]
- 46.Pantoliano M W, Bird R E, Johnson S, Asel E D, Dodd S W, Wood J F, Hardman K D. Conformational stability, folding, and ligand-binding affinity of single-chain Fv immunoglobulin fragments expressed in Escherichia coli. Biochemistry. 1991;30:10117–10125. doi: 10.1021/bi00106a007. [DOI] [PubMed] [Google Scholar]
- 47.Pearse R N, Kawabe T, Bolland S, Guinamard R, Kurosaki T, Ravetch J V. SHIP recruitment attenuates Fc gamma RIIB-induced B cell apoptosis. Immunity. 1999;10:753–60. doi: 10.1016/s1074-7613(00)80074-6. [DOI] [PubMed] [Google Scholar]
- 48.Ravetch J V. Fc receptors. Curr Opin Immunol. 1997;9:121–125. doi: 10.1016/s0952-7915(97)80168-9. [DOI] [PubMed] [Google Scholar]
- 49.Ravetch J V, Clynes R A. Divergent roles for Fc receptors and complement in vivo. Annu Rev Immunol. 1998;16:421–432. doi: 10.1146/annurev.immunol.16.1.421. [DOI] [PubMed] [Google Scholar]
- 50.Ravichandran K S, Lee K K, Songyang Z, Cantley L C, Burn P, Burakoff S J. Interaction of Shc with the zeta chain of the T cell receptor upon T cell activation. Science. 1993;262:902–905. doi: 10.1126/science.8235613. [DOI] [PubMed] [Google Scholar]
- 51.Rohrschneider L R, Bourette R P, Lioubin M N, Algate P A, Myles G M, Carlberg K. Growth and differentiation signals regulated by the M-CSF receptor. Mol Reprod Dev. 1997;46:96–103. doi: 10.1002/(SICI)1098-2795(199701)46:1<96::AID-MRD15>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 52.Saxton T M, van Oostveen I, Bowtell D, Aebersold R, Gold M R. B cell antigen receptor cross-linking induces phosphorylation of the p21ras oncoprotein activators SHC and mSOS1 as well as assembly of complexes containing SHC, GRB-2, mSOS1, and a 145-kDa tyrosine-phosphorylated protein. J Immunol. 1994;153:623–636. [PubMed] [Google Scholar]
- 53.Scharenberg A M, El-Hillal O, Fruman D A, Beitz L O, Li Z, Lin S, Gout I, Cantley L C, Rawlings D J, Kinet J P. Phosphatidylinositol-3,4,5-trisphosphate (PtdIns-3,4,5-P3)/Tec kinase-dependent calcium signaling pathway: a target for SHIP-mediated inhibitory signals. EMBO J. 1998;17:1961–1972. doi: 10.1093/emboj/17.7.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smit L, de Vries-Smits A M, Bos J L, Borst J. B cell antigen receptor stimulation induces formation of a Shc-Grb2 complex containing multiple tyrosine-phosphorylated proteins. J Biol Chem. 1994;269:20209–20212. [PubMed] [Google Scholar]
- 55.Takata M, Sabe H, Hata A, Inazu T, Homma Y, Nukada T, Yamamura H, Kurosaki T. Tyrosine kinases Lyn and Syk regulate B cell receptor-coupled Ca2+ mobilization through distinct pathways. EMBO J. 1994;13:1341–1349. doi: 10.1002/j.1460-2075.1994.tb06387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tridandapani S, Chacko G W, Van Brocklyn J R, Coggeshall K M. Negative signaling in B cells causes reduced Ras activity by reducing Shc-Grb2 interactions. J Immunol. 1997;158:1125–1132. [PubMed] [Google Scholar]
- 57.Ware M D, Rosten P, Damen J E, Liu L, Humphries R K, Krystal G. Cloning and characterization of human SHIP, the 145-kD inositol 5-phosphatase that associates with SHC after cytokine stimulation. Blood. 1996;88:2833–2840. [PubMed] [Google Scholar]
- 58.Yamauchi T, Ueki K, Tobe K, Tamemoto H, Sekine N, Wada M, Honjo M, Takahashi M, Takahashi T, Hirai H, Tsushima T, Akanuma Y, Fujita T, Komuro I, Yazaki Y, Kadowaki T. Growth hormone-induced tyrosine phosphorylation of EGF receptor as an essential element leading to MAP kinase activation and gene expression. Endocr J. 1998;45(Suppl.):S27–S31. doi: 10.1507/endocrj.45.suppl_s27. [DOI] [PubMed] [Google Scholar]
- 59.Zhang X, Hartz P A, Philip E, Racusen L C, Majerus P W. Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1998;273:1574–1582. doi: 10.1074/jbc.273.3.1574. [DOI] [PubMed] [Google Scholar]