Abstract
Background
Eggshell is a bio-ceramic material comprising columnar calcite (CaCO3) crystals and organic proteinaceous matrix. The size, shape and orientation of the CaCO3 crystals influence the microstructural properties of chicken eggshells. However, the genetic architecture underlying eggshell crystal polymorphism remains to be elucidated.
Results
The integral intensity of the nine major diffraction peaks, total integral intensity and degree of orientation of the crystals were measured followed by a genome-wide association study in 839 F2 hens. The results showed that the total integral intensity was positively correlated with the eggshell strength, eggshell thickness, eggshell weight, mammillary layer thickness and effective layer thickness. The SNP-based heritabilities of total integral intensity and degree of orientation were 0.23 and 0.06, respectively. The 621 SNPs located in the range from 55.6 to 69.1 Mb in GGA1 were significantly associated with TA. PLCZ1, ABCC9, ITPR2, KCNJ8, CACNA1C and IAPP, which are involved in the biological process of regulating cytosolic calcium ion concentration, can be suggested as key genes regulating the total integral intensity.
Conclusions
The findings greatly advance the understanding of the genetic basis underlying the crystal ultrastructure of eggshell quality and thus will have practical significance in breeding programs for improving eggshell quality.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-08103-1.
Keywords: Chicken, Eggshell, Crystal structure, X-ray diffraction, GWAS
Background
In the global egg industry, the integrity of the eggshell is critical for the economic viability of poultry production. The shells must be strong enough to prevent failure during packing and transportation [1]. The eggshell also forms an embryonic chamber for the developing chick by providing mechanical protection, allowing gas exchange and preventing microbial contamination [2, 3]. The process of eggshell formation is also a classic model for studying biomineralization [4]. The avian eggshell is composed of calcium carbonate, organic matrix and other minerals [5–7]. Several factors have been shown to affect eggshell structure [5, 8, 9] with most studies still based on optical observation [10]. Because of the difficulty of measurement, few studies have focused on the crystal ultrastructure of eggshells. X-ray crystallography has shown that calcium carbonate in avian eggshell is in the form of calcite crystals [11, 12]. The microstructural characteristics influenced by the intricate interlacing of the crystal units have been reported to vary significantly from one eggshell to another, even within the same species [13–15]. The eggshell consists of ceramic material whose mechanical properties are determined by its microstructure [16]. Chicken eggshells consisting of highly-oriented crystals of abnormal sizes have been reported to be generally weaker than those consisting of smaller and less well-oriented crystals [14, 17]. A comparison of eggshells at different ages showed that aged hens produced a greater variation in grain morphology and crystallographic texture than young hens [17]. The crystallographic texture of eggshells is also correlated with the thickness and strength of the shell. The crystal size and orientation has been found to be strongly positively correlated (r = 0.65) with the thickness of the eggshell [18]. The degree of orientation of crystals has been found to explain approximately 40 % of the variance in eggshell strength [17]. All these studies have indicated that the ultrastructure of eggshell was the key factor affecting eggshell quality.
Eggshell texture and ultrastructure are regulated by organic matrix proteins, which control the mineralization process and influence the biomechanical properties of the eggshell [19]. Association studies between polymorphisms of genes encoding shell proteins and shell characteristics have revealed that certain alleles were correlated with the mechanical properties of the eggshell [20, 21]. Ovalbumin and ovo-transferrin (LTF) markers were found to be associated with crystal size, while ovocleidin-116 and ovocalyxin-32 (RARRES1) markers were associated with crystal orientation [18]. Because the crystal is the basic component of construction of the eggshell, the alleles of matrix proteins and genes known to be involved in eggshell formation have also been found to be associated with crystal size and orientation [7, 18, 21]. Therefore, genetic selection of hens with suitable eggshell ultrastructure properties is important for improving eggshell quality. However, the genetic mechanism of the eggshell crystal ultrastructure is still unclear.
In the present study, the crystal structure of chicken eggshells was determined using X-ray crystallography. To study the genetic basis of biomineralization, a genome-wide association study (GWAS) was performed in the designated F2 population by using a high-density 600k SNP chip for chickens. We aimed to explore the genetic architecture of eggshell crystal structure to identify candidate genes, which may be valuable for the genetic improvement of eggshell quality traits.
Results
Phenotypic description and genetic parameters
The descriptive statistics for nine reflection peaks (A1-A9), total integral intensity (TA) and degree of orientation (OD) were shown in Table 1. The estimates of SNP-based heritability, the genetic and phenotypic correlations between eggshell crystal structure traits were shown in Table 2. The individual integral intensity exhibited higher phenotypic variation (11.71–21.23 %) than that of TA (9.40 %). Thus, the TA could provide a good estimate of the size of the eggshell crystals. The SNP-based heritability (h2snp) of TA was 0.23. The phenotypic correlations between every individual integral intensity (A1-A9) were low (r=-0.15–0.35). The absolute values of the genetic correlations were all higher than those of the phenotypic correlations. The variation in OD was higher (23.72 %) than that of the other traits and the value of h2snp was low (0.06).
Table 1.
Descriptive statistics for individual integral intensity, total integral intensity and degree of orientation
| Traits | N | Mean | SD | CV (%) | Min | Max |
|---|---|---|---|---|---|---|
| A1 | 839 | 25335.41 | 4813.91 | 19.00 | 12953.58 | 38929.12 |
| A2 | 839 | 239384.50 | 49882.98 | 20.84 | 104335.20 | 372372.10 |
| A3 | 839 | 50264.81 | 10673.14 | 21.23 | 24546.91 | 80485.42 |
| A4 | 839 | 71896.02 | 10669.69 | 14.84 | 44550.43 | 100413.00 |
| A5 | 839 | 70718.49 | 13935.86 | 19.71 | 36597.64 | 108482.90 |
| A6 | 839 | 131559.70 | 17180.31 | 13.06 | 83325.08 | 180757.50 |
| A7 | 839 | 57379.84 | 7497.32 | 13.07 | 37028.29 | 78801.67 |
| A8 | 839 | 38877.79 | 4554.47 | 11.71 | 26827.77 | 51180.12 |
| A9 | 839 | 40565.02 | 5966.54 | 14.71 | 24762.44 | 56568.73 |
| TA | 837 | 726470.70 | 68300.65 | 9.40 | 549996.40 | 906125.60 |
| OD | 822 | 1.71 | 0.41 | 23.72 | 1.00 | 2.86 |
Abbreviations: N = number of samples; Mean = arithmetic mean; SD = standard deviation; CV = coefficient of variance; Min = minimum; Max = maximum; A1-A9 = integrate intensity of peaks of No. 1–9; TA = total intensity of all 9 peaks; OD = degree of orientation
Table 2.
Summary of genetic analysis between individual integral intensity, total integral intensity and degree of orientation
| Traits | A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | TA | OD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 |
0.05 (0.05) |
0.22 (0.52) |
0.94 (0.5) |
0.63 (0.48) |
NA |
0.45 (0.48) |
0.87 (0.42) |
1.00 (0.66) |
NA |
0.87 (0.44) |
0.41 (0.65) |
| A2 |
0.02 (0.03) |
0.10 (0.05) |
-0.50 (0.47) |
0.46 (0.27) |
0.17 (0.53) |
1.00 (0.18) |
0.59 (0.3) |
0.73 (0.23) |
0.36 (0.44) |
0.90 (0.07) |
NA |
| A3 |
0.09** (0.03) |
-0.05 (0.03) |
0.06 (0.05) |
0.68 (0.25) |
0.83 (0.39) |
-0.43 (0.38) |
0.84 (0.22) |
0.51 (0.36) |
0.27 (0.53) |
0.17 (0.33) |
1.00 (0.47) |
| A4 |
0.07 (0.03) |
0.07** (0.03) |
0.26** (0.03) |
0.21 (0.06) |
0.84 (0.32) |
0.07 (0.24) |
0.80 (0.15) |
0.74 (0.18) |
0.45 (0.31) |
0.71 (0.14) |
0.56 (0.24) |
| A5 |
0.02 (0.03) |
0 (0.03) |
0.27** (0.03) |
0.17** (0.03) |
0.05 (0.05) |
0.64 (0.53) |
0.63 (0.35) |
0.83 (0.34) |
1.00 (0.84) |
0.85 (0.38) |
0.64 (0.5) |
| A6 |
0.07* (0.03) |
0.41** (0.03) |
-0.15** (0.03) |
0 (0.03) |
-0.06 (0.03) |
0.15 (0.05) |
0.38 (0.25) |
0.74 (0.21) |
0.46 (0.37) |
0.88 (0.11) |
-0.88 (0.29) |
| A7 |
0.16** (0.03) |
-0.04 (0.03) |
0.35** (0.03) |
0.32** (0.03) |
0.32** (0.03) |
-0.11** (0.03) |
0.24 (0.06) |
0.75 (0.18) |
0.61 (0.31) |
0.79 (0.15) |
0.44 (0.3) |
| A8 |
0.11* (0.03) |
0.29** (0.03) |
0.20** (0.03) |
0.29** (0.03) |
0.20** (0.03) |
0.23** (0.03) |
0.28** (0.03) |
0.15 (0.05) |
0.94 (0.34) |
0.94 (0.10) |
0.06 (0.38) |
| A9 |
0.08* (0.03) |
0.10* (0.03) |
0.14** (0.03) |
0.20** (0.03) |
0.07* (0.03) |
0.13* (0.03) |
0.26** (0.03) |
0.31** (0.03) |
0.07 (0.05) |
0.61 (0.31) |
0.37 (0.51) |
| TA |
0.15** (0.03) |
0.84** (0.01) |
0.25** (0.03) |
0.34** (0.03) |
0.31** (0.03) |
0.52** (0.02) |
0.26** (0.03) |
0.50** (0.02) |
0.31** (0.03) |
0.23 (0.06) |
-0.12 (0.30) |
| OD |
0.04 (0.03) |
-0.78** (0.02) |
0.18** (0.03) |
0.52** (0.02) |
0.10** (0.03) |
-0.34** (0.03) |
0.23** (0.03) |
-0.04 (0.03) |
0.03 (0.03) |
-0.48** (0.02) |
0.06 (0.05) |
Lower triangle: phenotypic correlations; Diagonal: heritability estimates based on genome; Upper triangle: genetic correlations; Standard errors of the estimates are in parentheses; A1-A9 = integrate intensity of peaks of No. 1–9; TA = total intensity of all 9 peaks; OD = degree of orientation; *Significant linkage at P < 0.05. **Significant linkage at P < 0.01. NA: the genetic correlation value did not converge when we calculated the genetic correlation by the GCTA
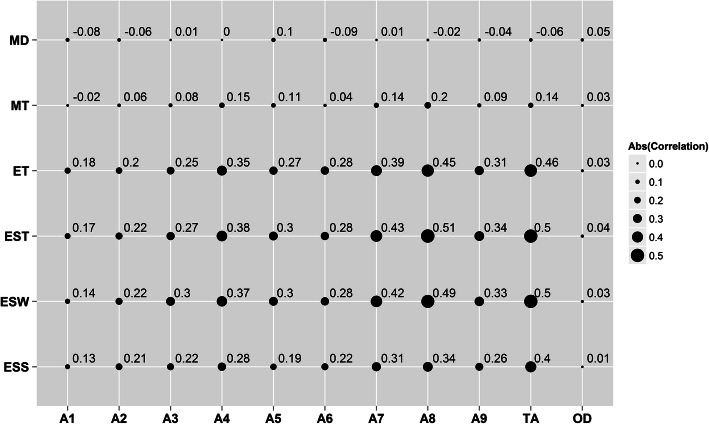
In a previous study, we also measured the ultrastructure of these eggshells [22]. The phenotypic correlations between the crystal structure and eggshell ultrastructure are shown in Fig. 1. TA was positively correlated with eggshell strength (ESS), eggshell thickness (EST), eggshell weight (ESW), mammillary layer thickness (MT) and effective layer thickness (ET) (r = 0.14–0.50), but OD was not significantly correlated with these eggshell quality traits (p < 0.05). There was a high genetic correlation between the total peak area intensity and eggshell ultrastructure (r = 0.52), and a medium genetic correlation between the OD and the eggshell ultrastructure (r = 0.31) (Table 3). However, the genetic correlation of crystal structure with the eggshell ultrastructure all exhibited a large standard error (Table 3).
Fig. 1.
The phenotypic correlations between the crystal structure and eggshell quality. Abbreviations: A1-A9 = integrate intensity of peaks of No. 1–9; TA = total intensity of all 9 peaks; OD = degree of orientation; ESS = eggshell strength; ESW = eggshell weight; EST = eggshell thickness; ET = effective layer thickness; MT = mammillary layer thickness; MD = mammillary density. Abs (Correlation) = Absolute Correlation
Table 3.
The genetic correlations between the crystal structure and eggshell quality traits
| Traits | MD | MT | ET | EST | ESW | ESS |
|---|---|---|---|---|---|---|
| A1 | -0.38(0.39) | 0.01(0.39) | 0.66(0.36) | 0.53(0.32) | 0.77(0.32) | 0.95(0.55) |
| A2 | -0.82(0.19) | 0.62(0.23) | 0.45(0.22) | 0.62(0.19) | 0.46(0.19) | 0.13(0.25) |
| A3 | 0.22(0.35) | -0.08(0.33) | 0.79(0.20) | 0.69(0.23) | 0.63(0.20) | 0.48(0.28) |
| A4 | 0.01(0.21) | 0.14(0.20) | 0.82(0.11) | 0.73(0.11) | 0.63(0.11) | 0.55(0.14) |
| A5 | -0.33(0.33) | 0.57(0.37) | 0.82(0.28) | 0.92(0.30) | 0.80(0.20) | 0.27(0.29) |
| A6 | -0.30(0.24) | 0.30(0.23) | 0.64(0.15) | 0.67(0.14) | 0.45(0.16) | 0.44(0.18) |
| A7 | -0.21(0.20) | 0.16(0.21) | 0.97(0.10) | 0.97(0.07) | 0.90(0.08) | 0.80(0.12) |
| A8 | -0.24(0.22) | 0.46(0.20) | 0.99(0.09) | 0.99(0.08) | 0.76(0.11) | 0.45(0.18) |
| A9 | -0.56(0.28) | 0.21(0.29) | 0.68(0.24) | 0.60(0.24) | 0.49(0.23) | 0.50(0.25) |
| TA | -0.61(0.17) | 0.53(0.17) | 0.83(0.09) | 0.88(0.07) | 0.75(0.09) | 0.52(0.14) |
| OD | 0.87(0.31) | -0.42(0.30) | 0.48(0.29) | 0.39(0.28) | 0.31(0.26) | 0.65(0.28) |
Standard errors of the estimates are in parentheses; A1-A9 = integrate intensity of peaks of No. 1–9; TA = total intensity of all 9 peaks; OD = degree of orientation; ESS = eggshell strength; ESW = eggshell weight; EST = eggshell thickness; ET = effective layer thickness; MD = mammillary density; MT = mammillary layer thickness
Genome-wide association study (GWAS)
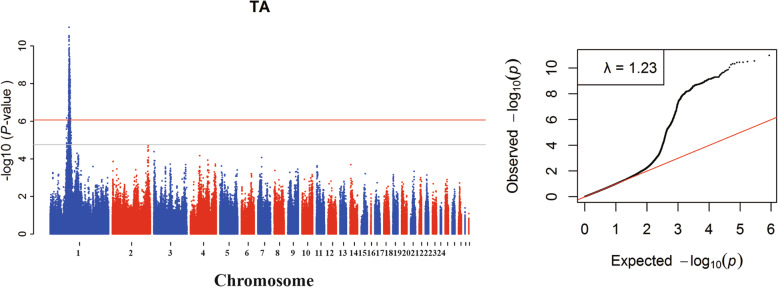
The association analyses revealed a total of 621 genome-wide significant SNPs associated with TA (Additional file 1: Table S2). No genome-wide significant locus was associated with OD. In addition, 1096 and 1 SNPs exhibited associations with TA and OD under suggestive levels of significance, respectively. The Manhattan and quantile-quantile (QQ) plots in Fig. 2 showed a global view of the P-values for all SNPs affecting the ultrastructure of the eggshell crystals. All significant SNPs associated with TA were in a 13.5-Mb region between 55.6 and 69.1 Mb on chromosome 1 (GGA1).
Fig. 2.
Manhattan plots (left) and quantile–quantile plots (right) of the observed P-values for TA. The Manhattan plots indicate -log10 (observed P-values) for genome-wide SNPs (y-axis) plotted against their respective positions on each chromosome (x-axis), and the horizontal gray and red lines depict the genome-wide suggestive (1.73 × 10− 5) and significant (8.64 × 10− 7) thresholds, respectively. For quantile-quantile plots, the x-axis shows the expected -log10-transformed P-values, and the y-axis represents the observed -log10-transformed P-values. The genomic inflation factors (λ) are shown on the top left in the QQ plot. TA represents the total integral intensity of all peaks
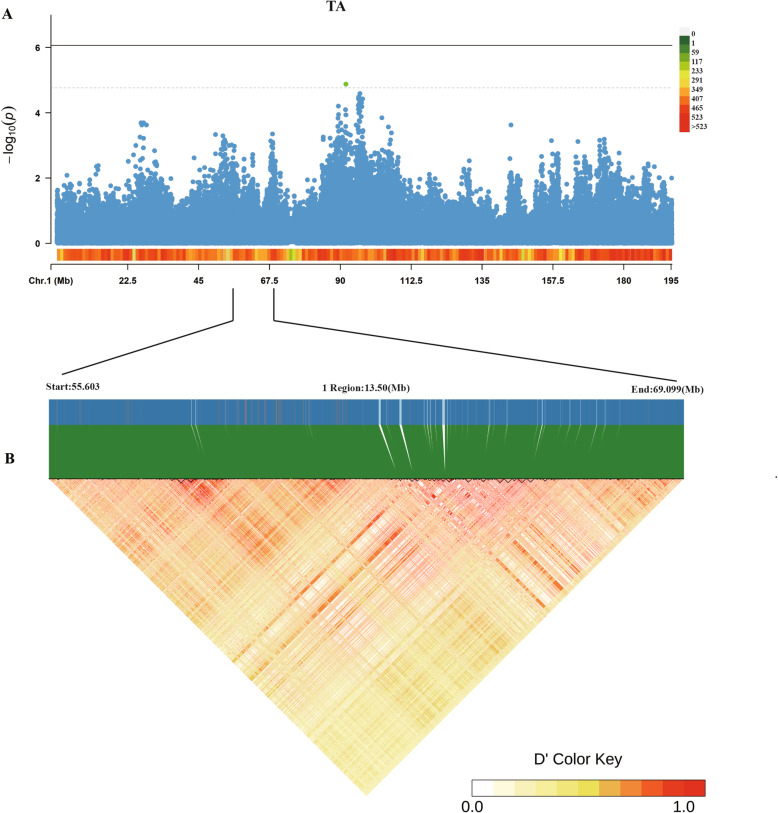
To identify independent SNPs, a further series stepwise conditional analyses were performed and the locus rs315989578, that was found to be significantly associated with TA was fitted into the model to examine these associations. The levels of significance for all loci around the most significant SNP decreased substantially (Fig. 3A) during the stepwise conditional Genome-wide Association (GWA) analysis, with no other independent loci being detected. The Fig. 3B illustrated the linkage disequilibrium between the regions associated with TA traits. A region showing a relatively strong linkage disequilibrium was in chr1: 55.6 Mb and 69.1 Mb. We also conducted a GWAS analysis on A1-A9 (Additional file 2: Table S1). The result showed that no significant SNPs were associated with the A1, A2, A3, A4, A5, A7 and A8. 163 and 330 genome-wide significant SNPs were associated with the A6 and A9, respectively. These significant SNPs almost coincided with the significant SNPs for TA.
Fig. 3.
Conditional association analyses (A) and linkage disequilibrium (LD) analysis (B) for TA. The genotype of rs315989578 was added to the multivariate model as a covariate for the conditional analysis. The LD plot of significant SNPs with the total intensity of all peaks in the region ranged from 55.6 to 69.1 Mb. Haplotypes are indicated by sample symbols connected by a solid line. The color represents Hedrick’s multiallelic D’, which indicates the degree of LD between two blocks. TA represents the total integral intensity of all peaks
SNP annotation and promising genes associated with eggshell ultrastructure
The annotation of significant SNPs using the Variant Effect Predictor (VEP) tool supplied by Ensembl could help identify promising genes associated with eggshell crystal structure. Detailed information about the genes is summarized in Additional file 1: Table S2. In total, 74 potential candidate genes all with significant quantitative trait loci (QTLs) were identified. The results of enrichment analysis were listed in Additional file 1: Table S3. We found that six genes were significantly enriched in the blood circulation (GO:0008015): CACNA1C (voltage-dependent L-type calcium channel subunit alpha-1 C), IAPP (islet amyloid polypeptide), ITPR2 (inositol 1,4,5-trisphosphate receptor type 2), PLCZ1 (1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1), ABCC9 (ATP-binding cassette sub-family C member 9) and KCNJ8 (ATP-sensitive inward rectifier potassium channel 8). Among them, the CACNA1C, IAPP, ITPR2 and PLCZ1 also played considerable roles in the biological process of regulating cytosolic calcium ion concentration. We also found 26 genes that were regulated by the octamer transcription factor 1 (Oct-1). However, no information on the interaction network is available for these candidate genes.
SNP contribution to phenotypic variation
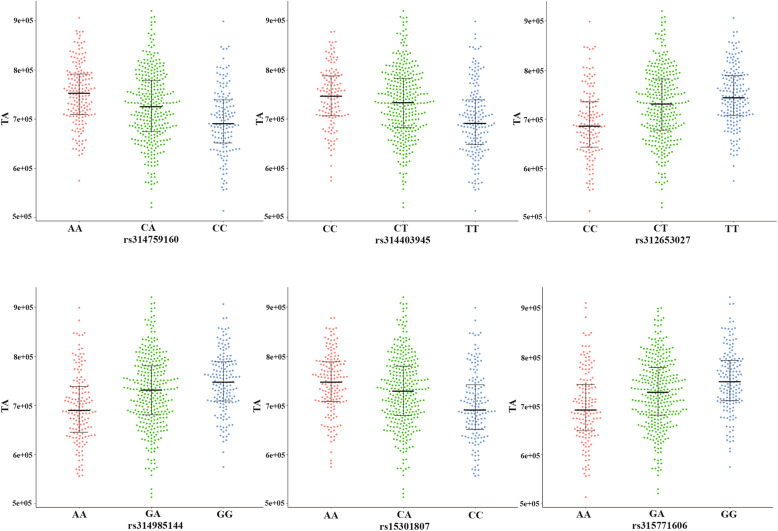
Table 4 showed six meaningful loci extracted which reached genome-wide significance in the univariate GWAS for analysis: rs314759160, rs314985144, rs314403945, rs15301807, rs315771606 and rs312653027 in PLCZ1, ABCC9, ITPR2, KCNJ8, CACNA1C and IAPP, respectively. These six SNPs exerted contributions of between 3.93 % and 5.90 % to the phenotypic variation in TA. Notably, rs314759160 exhibited the largest allelic substitution effect on TA with the substitution of one copy of the effect allele at the rs314759160 site causing a 0.345 SD/allele decrease in TA. The phenotypic differences between the genotypes of those SNPs were shown in boxplots (Fig. 4), revealing that homozygotes of the effect allele and of the alternative alleles possessed the lowest and highest phenotypic values, respectively, whereas the values for the heterozygotes were intermediate. Data also showed that the most significant QTL associated with TA was the rs315989578 at the position 63.8 Mb on chromosome 1 (p-value 1.01e-11). However, this SNP located in the intergenic region, explained 6.45 % of the phenotypic variation (Additional file 1: Table S2).
Table 4.
Contributions of six SNPs in genes potentially related to the total integral intensity
| SNP | Gene | Consequence | Chr. | Position (bp) | EA/AA | P | MAF | Beta (SE) | CPV (%) |
|---|---|---|---|---|---|---|---|---|---|
| rs314759160 | PLCZ1 | intron_variant | 1 | 64,223,356 | A/C | 4.96E-10 | 0.454 | -0.345(-0.055) | 5.90 |
| rs314403945 | ITPR2 | intron_variant | 1 | 67,660,408 | T/C | 9.18E-09 | 0.475 | 0.320(-0.055) | 5.11 |
| rs312653027 | IAPP | downstream_gene_variant | 1 | 65,271,009 | C/T | 5.42E-09 | 0.443 | -0.391(-0.054) | 5.02 |
| rs314985144 | ABCC9 | intron_variant | 1 | 66,908,211 | G/A | 2.75E-08 | 0.468 | -0.305(-0.054) | 4.63 |
| rs15301807 | KCNJ8 | intron_variant | 1 | 66,959,507 | A/C | 3.52E-07 | 0.470 | -0.285(-0.056) | 4.05 |
| rs315771606 | CACNA1C | intron_variant | 1 | 61,199,051 | G/A | 5.13E-07 | 0.468 | -0.281(-0.056) | 3.93 |
Abbreviations: EA/AA: effect allele (minor allele)/alternative allele (major allele); EA = effect allele (minor allele); AA = alternative allele (major allele); MAF = minor allele frequency; CPV = contribution to phenotypic variance. Beta is the estimated allelic substitution effect per copy of the effect allele (EA) based on an inverse-normal transformed scale under an additive model, expressed in SD unit/allele; SE = standard error of the beta
Fig. 4.
Boxplots of the SNP effects on the TA. The figure shows the genotypes of six representative SNPs (x-axis) versus the phenotypic values of the corresponding traits (y-axis). TA represents the total integral intensity of all peaks
Discussion
The eggshell of chicken is a bio-ceramic material comprising columnar calcite (CaCO3) crystals and organic proteinaceous matrix [7]. Previous studies have found that the size and orientation of calcium carbonate crystals influenced the eggshell quality [17, 18]. In the present study, we used X-ray diffraction to measure the traits of calcium carbonate crystals in the eggshell. This method was based on the analysis of X-ray diffraction patterns formed by intact eggshells, with the intensity of the spots displayed in these patterns being directly related to the size of crystals in the eggshells [12]. Our results showed that the ESS, ESW, EST, ET and MT increased as the size of the crystals increased (Fig. 1), which was consistent with previous studies [18, 21]. We speculated that in eggshells with larger crystals a faster crystal growth rate had occurred, causing the deposition of more calcites in the eggshell and thus increasing its strength, weight and thickness [23]. Moreover, the palisade layer was mainly composed of packed calcite crystals, being governed by the geometry of the nucleation sites, crystal growth rate and interactions between the growing crystals [24]. The thickness and strength of the eggshell would increase as the size of the calcite crystal increases[18]. Interestingly, our results have shown that the TA was highly negatively correlated with the OD (r = − 0.48, Table 2). This indicated that the crystal structure of the larger crystals was more randomly oriented, which was consistent with a previous study [18]. Table 3 showed that TA was highly genetically correlated with the microstructure traits ET, MT, ESS and EST, results consistent with those of Dunn’s study [18]. We also noted that the genetic correlations were high between the A7, A8, ET and EST. However, this high correlation has not been reported in other studies. X-ray diffraction studies have showed that the ET was oriented to a few crystal planes [5]. So, we hypothesized that the arrangement of some specific crystal planes (such as A7 and A8) may have an important effect on the eggshell structure. However, further study is needed for the crystallographic structure mechanism. In the present study, OD was not correlated with the quality of the chicken eggshells. The estimated heritability of OD (h2 = 0.06) was also low with a large estimation error, but was smaller than previous estimates [18], partly because of the higher variation coefficient for OD (Table 2). The genetic correlations between OD and ET in the present study were high (r = 0.48) (Table 3), and Dunn et al. also found a similar genetic correlation (r = 0.32) between OD and ET[18], but the estimation errors were large in both studies. To determine whether this estimate is reliable, our experiments will need to be repeated with a larger sample. Therefore, further studies are needed to explore the mechanical aspects of crystallographic texture.
The eggshell is composed of four layers, with ET accounting for more than 70 % of the total thickness [25]. Therefore, changes in the effective layer of eggshells can significantly affect their properties [26]. In a previous study, the phenotypic correlation between the EST and ET of eggshells was 0.92, while the genetic correlation was 0.96 [22]. The association analysis of QTLs in the region between 57.3 and 71.4 Mb of GGA1 was significantly associated with ESS, ESW and EST [27]. We have also identified a 9-Mb QTL (GGA1: 59.4–68.5 Mb) in this region which was associated with the ET [22]. These results have indicated that this region of chromosome 1 had a great influence on the microstructure of eggshells, but the details are unclear. Interestingly, the present study has found that this region was also closely related to the TA. One reason may be that there was a strong correlation (r = 0.5) between the ET and TA (Fig. 1). However, no QTLs were identified as being associated with crystal growth orientation. We speculated that the size of the crystal was the dominant influence on the structure of the eggshell, which was independent of the orientation of crystal growth. Our results have also shown that GGA1 was closely related to the ultrastructure of the chicken eggshell.
The formation of eggshells is mainly achieved by biomineralization in the chicken uterus [16, 28, 29]. Two systems have been proposed as playing an important role in the biomineralization of eggshells: the ion transport system and the matrix protein system [30]. The ion transport system provides sufficient ions for eggshell formation and matrix proteins play a regulatory role in the process of eggshell mineralization [31, 32]. In a previous study, ABCC9, ITPR2, KCNJ8 and WNK1 have been mapped as being associated with ET and EST [22], and all these genes are also involved in ion transport with their expression detected in the chicken uterus. Many studies have also reported that the eggshell matrix proteins were associated with EST [7, 21]. Genetic associations for crystal measurements showed that OVAL and ovo-transferrin were associated with TA, while OC116 and OCX-32 were associated with OD [18]. In the present study, we have identified 76 candidate genes including ABCC9, ITPR2, KCNJ8 as being associated with crystal size and six genes with the process of blood circulation (Additional file 1: Table S3). ABCC9 and KCNJ8 encoded membrane-associated receptors that combined with the potassium channel have been reported to become smooth muscle ATP-sensitive potassium channels in the heart [33]. Inositol 1,4,5 trisphosphate receptors (ITPRs), a family of endoplasmic reticulum Ca2+ channels, are essential for controlling intracellular Ca2+ levels in virtually every type of mammalian cell [34]. ITPR2 has been found to be highly expressed in cardiomyocytes and to be associated with human heart failure [35, 36]. As important ion transmembrane channels, these genes are reported to play a vital role in cardiac contraction [37]. It has been reported that the calcium required for eggshell calcification mainly depends on blood circulation, and more efficient calcium transport could promote the calcification of eggshells [38]. Our results thus suggested that the strength of cardiac contractility may affect the mineralization of eggshells.
Several genes related to intracellular calcium concentration were also identified. ITPR2 was also directly involved in the process of Ca2+ signal transduction and released during bone growth [39]. Ca2+ oscillation is mediated by the inositol 1,4,5-trisphosphate receptors 2 and 3 (ITPR2 and ITPR3) in the osteoclast differentiation [40]. Except for previously reported major candidate genes, we have also identified several new genes associated with crystal size, CACNA1C, IAPP and PLCZ1, which influence the function of regulating cytosolic calcium ion concentration. CACNA1C encoded the protein, voltage-dependent L-type calcium channel subunit alpha-1 C, which is essential for normal blood pressure regulation through the contraction of arterial smooth muscle cells [41]. CACNA1C is highly expressed in the brain, heart, jejunum, ovary, pancreatic beta-cells and vascular smooth muscle [41–43]. Ca2+ influx through CACNA1C has also been reported to activate osteogenic transcriptional programs and promote mineralization [44]. We therefore propose that CACNA1C might play an important role in regulating the concentration of calcium in uterine fluid. Amylin is encoded by IAPP and is a member of the calcitonin family of hormones that is co-secreted with insulin by the pancreatic beta cells [45]. Amylin has been reported to use the calcitonin receptor to inhibit osteoclastogenesis in vivo [45]. There was no direct evidence that PLCZ1 participated in the release of calcium but it has been reported that it indirectly affected the transmembrane transport of calcium [46, 47]. Interestingly, we also found that 22 candidate genes, including ITPR2, PLCZ1 and CACNA1C, were regulated by the transcription factor, Oct-1, which acted as downstream of notch signaling during radial glia formation. Oct-1 has been reported as forming a novel complex with the transcription factor (Runx2) to regulate the expression of the mammary gland-specific gene, beta-casein. Runx2 is essential for expressing several bone-specific genes and is primarily considered as a master regulator of bone development [48]. This suggested that Oct-1 might also be related to the development of bone. The present study has shown that several genes related to bone growth were significantly associated with the strength of eggshell crystals. Except for eggshell formation, the biomineralization process in birds occurs in the bone calcification [49]. Some eggshell matrix proteins such as OC-116 have been found to be expressed in bone [50], which indicated that there may be similar biological pathways in the two mineralization processes. Our results further support this hypothesis. It is also worth noting that no known eggshell matrix protein was found in these candidate genes, because some matrix proteins such as ovocleidin-17 have not yet been annotated in the reference genome. It will thus be necessary to obtain more information on the crystallization system for eggshells.
Conclusions
The integral intensity and degree of orientation of eggshell crystal were determined, then used for a GWA analysis with a 600 K high-density SNP array. The result showed that 621 genome-wide significant SNPs ranging from 55.6 to 69.1 Mb in GGA1 were associated with TA. Ultimately, six genes, PLCZ1, ABCC9, ITPR2, KCNJ8, CACNA1C and IAPP, were considered as promising genes that were implicated in the integral intensity of eggshell crystals and could improve eggshell quality. The present study has revealed the genetic basis of the crystallographic ultrastructure of eggshells and improved our understanding of its mechanical properties. These new findings will also provide a useful theoretical basis on biomineralization in chicken eggshells.
Methods and materials
Experimental population
White Leghorn (WL) and Dongxiang (DX) chickens, representing a standard breed and a Chinese indigenous strain, respectively, were used to construct an F2 resource population for the present study. Six WL and six DX males were mated with 133 DX and 80 WL females, respectively. Then, 25 cocks and 407 hens from the WL/DX cross and 24 cocks and 235 hens from DX/WL cross in the F1 generation were selected to produce the F2 generation. A total of 3749 chickens from the F2 population were reared in individual cages with ad libitum access to feed. Finally, 839 hens from 49 half-sib families and 365 full-sib families with sufficient phenotypic and pedigree information were chosen for SNP genotyping. All the experimental animals were hens and came from the same batch.
Phenotypic measurements
Eggs were collected from hens at 66 weeks of age on three successive days to ensure at least one egg per hen. The egg contents were removed then the eggshells were cleaned with tap water and dried naturally at room temperature. Pieces (approximately 3 × 5 mm) of eggshell were taken at the equator. The shells were measured using X-ray diffraction (R-Axis Spider XRD, Rigaku Corp., Tokyo, Japan). The measurement conditions were as follows: Mo Kα (λ = 0.71075 Å), 50 KV, 80 mA, a pin-hole collimator (0.8 mm in diameter) and an exposure time of 30 s per frame. The samples were mounted with the inner surface of the eggshell facing the incident X-ray beam. The beam passed through the sample and the transmission diffraction pattern was recorded on the detector. The software THCLXPD attached to the XRD was used to analyze the diffraction patterns by measuring the intensity of the reflection spots.
The integrated intensities were determined from each eggshell sample using nine different rings. These rings were oriented with a set of (hkl) crystallographic planes: 113, 104, 012, 110, 202, 018, 122, 208 and 300. The data from the nine reflection peaks (A1-A9) were added to give the TA to lower the data variability. The crystal size was estimated from the intensity of the reflection spots [51]. To determine the OD, a calcite powder standard (Joint Committee on Powder Diffraction File, PDF 05-0586) having a completely random orientation was used as a reference pattern for the crystals. The ratio between the integrated intensities of the calcite reflections from the eggshell and those of the standard sample (PDF 05-0586) provided a measure of the relative orientation of the eggshell crystals [9]. In the present study, the strongest integrated intensities of the standard sample (PDF 05-0586) were 113 and 104 (Additional file 1: Table S1), indicating that the crystals were preferentially oriented to these planes. The ratio (Rstd) of the calcite powder standard sample (A113/A104) was 0.18. Thus, the OD of eggshell crystals was calculated using Eq. (1):
| 1 |
The value of OD represented the degree of the preferential orientation of the crystals. For a value of one, the sample consists of randomly oriented crystals and for a value > 1, the sample consists of highly-oriented crystals [17].
The data on eggshell quality traits (ESS, EST, ESW, MT and ET) from our previous publications was used in the correlation analysis[22, 52]. The descriptive statistics of all the phenotypic data were calculated using R (version 3.3.1) (https://www.rproject.org/).
Genotyping and quality control (QC)
All blood samples were collected from brachial veins of chickens by standard venipuncture. Genomic DNA was extracted from whole blood samples using standard phenol-chloroform method and the 839 chickens were genotyped using the Affymetrix 600 K chicken SNP chip (Affymetrix Inc. Santa Clara, CA, USA). Array Power Tools (APT) (version 1.16.0; Affymetrix Inc) software was used for quality control and genotype calling. SNPs with a minor allele frequency of < 5 % and a Hardy-Weinberg equilibrium test with P < 1 × 10− 6 were removed using the PLINK version 1.90 package. The BEAGLE version 4.0 procedure [53] was used to impute sporadic missing genotypes, and SNPs with an imputation quality score of R2 > 0.5 were retained for the next analysis step. A total of 839 samples and 435,867 SNPs finally remained for the subsequent GWAS.
GWA analysis
The loci-based analysis was performed using the generalized linear mixed model implemented in GEMMA [54], where the kinship matrix is calculated using the standardization method. The mixed model was based mainly on the additive effect of sites:
| 2 |
Where: Y is the vector phenotypes of eggshell; µ the overall mean; X the covariance matrix (containing the first ten PCA principal components obtained from analysis of the population substructure); b the estimator vector of fixed effects; Z is an incidence matrix associating u with y. u the additive polygenic effect (0, GVg), with G the genomic kinship matrix, and the additive effect variance Vg; S the design matrix containing the corresponding SNP sites; α the substitution effect size corresponding to each site; and e the vector of random residual effects (N (0, IVe)), with I the identity matrix and the residual variance (Ve).
The genomic inflation factor (λ) was calculated using the R package, qqman (Version 0.1.2) [55]. Multiple test thresholds were calculated using the simpleM method [56]. Stepwise conditional analyses were conducted using the generalized linear mixed model of the R package with the strongest SNP identified being added at each step as a covariate. A total of 58,358 valid inspections was obtained. The genome-wide significance level was 8.57 × 10− 7 (0.05/58,358), with a suggestive significance level of 1.71 × 10− 5 (1.00/58,358). Principal component analysis was performed using the EIGENSOFT software (v.2.04) with the results showing that there was stratification in the experimental population. In order to correct the population stratification, we selected the top ten PCA [57] components as covariates in the GWAS analysis.
The SNP-based heritability of eggshell crystal traits, calculated using the residual maximum likelihood (REML) method in GCTA (version 1.24) [58], was estimated from the same mixed model as used in the GWAS analysis. The genetic correlations were estimated from a standard bivariate linear mixed model [59] using the Bivariate GREML analysis of GCTA [60]. We used Visscher’s method [61] to calculate the power of genetic correlation. We calculated the phenotypic variance contribution (CPV) of those genome-wide significant SNPs based on the equation , with allele frequencies p and q, β was corresponding effect size of SNP identified in association study, was the phenotypic variance [62].
Linkage disequilibrium analysis and gene identification
We performed a series of linkage disequilibrium (LD) analyses to characterize the causative SNPs within strong LD regions by applying the solid spine algorithm in Haploview software version 4.2 [63]. The significant SNPs were functionally annotated, then the candidate genes related to the significant SNPs or genomic regions were identified using the Variant Effect Predictor [64]. The Biomart tools were supported by Ensembl based on the Galgal6 assembly.
Functional annotation
Gene Ontology annotation and KEGG pathway were completed using the Metascape website to assign genes with corresponding terms [65, 66]. The identities of all the candidate genes were converted into those of the homologous genes in humans.
Supplementary information
Acknowledgements
We gratefully acknowledge the Jiangsu Institute of Poultry Science for their assistance with sample collection.
Abbreviations
- TA
total integral intensity
- OD
degree of orientation
- ESS
eggshell strength
- EST
eggshell thickness
- ESW
eggshell weight
- MT
mammillary layer thickness
- ET
effective layer thickness
- GWAS
Genome-wide association study
- GWA
Genome-wide association
- SNP
Single nucleotide polymorphism
- QTL
Quantitative trait loci
- LD
Linkage disequilibrium
- REML
Restricted maximum likelihood
- MAF
Minor allele frequency
- HWE
Hardy-Weinberg equilibrium
- PCA
Principal component analysis
- VEP
Variant effect predictor
Authors’ contributions
N.Y. conceived the study and designed the project. Q.L., and Z.D. performed genetic and bioinformatics analyses. Z.D., C.S., J.Z., and G.X. contributed to collecting samples and measuring phenotypic data. Q.L., and Z.D. wrote the manuscript. C.S., and N.Y. revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the China Agriculture Research Systems (CARS-40) and by the Programs for Changjiang Scholars and Innovative Research in University (IRT15R62).
Availability of data and materials
The datasets generated for this study can be found in figshare repository, 10.6084/m9.figshare.16627099.v1 .
Declarations
Ethics approval and consent to participate
We confirm that standard guidelines were followed for all the experimental protocols related to animal experimentation in this study were reviewed and approved by the Animal Welfare Committee of China Agricultural University. The study was carried out in compliance with the ARRIVE guidelines.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Quanlin Li and Zhongyi Duan contributed equally to this work.
References
- 1.Bain MM, Nys Y, Dunn IC. Increasing persistency in lay and stabilising egg quality in longer laying cycles. What are the challenges? Br. Poult. Sci. 2016;57(3):330–338. doi: 10.1080/00071668.2016.1161727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onagbesan O, Bruggeman V, Smit LD, Debonne M, Witters A, Tona K, Everaert N, Decuypere E. Gas exchange during storage and incubation of Avian eggs: effects on embryogenesis, hatchability, chick quality and post-hatch growth. World’s Poultry Science Journal. 2007;63(4):553–573. doi: 10.1017/S0043933907001614. [DOI] [Google Scholar]
- 3.Chien YC, Hincke MT. McKee. MD. Ultrastructure of avian eggshell during resorption following egg fertilization. J Journal of Structural Biology. 2009;168(3):527–538. doi: 10.1016/j.jsb.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Leach RM. Biochemistry of the Organic Matrix of the Eggshell. J Poultry Ence. 1982;61(10):2040–2047. doi: 10.3382/ps.0612040. [DOI] [Google Scholar]
- 5.Cain CJ, Heyn AN. X-Ray Diffraction Studies of the Crystalline Structure of the Avian Egg Shell. Biophys J. 1964;4(1):23–39. doi: 10.1016/S0006-3495(64)86767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien YC, Hincke MT, Vali H, McKee MD. Ultrastructural matrix-mineral relationships in avian eggshell, and effects of osteopontin on calcite growth in vitro. J Struct Biol. 2008;163(1):84–99. doi: 10.1016/j.jsb.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Nys Y, Gautron J, Garcia-Ruiz JM, Hincke MT. Avian eggshell mineralization: biochemical and functional characterization of matrix proteins. Comptes rendus - Palevol. 2004;3(6):549–562. doi: 10.1016/j.crpv.2004.08.002. [DOI] [Google Scholar]
- 8.Hamilton RMG. Methods and Factors That Affect the Measurement of Egg Shell Quality. J Poult. Sci. 1982;61(10):2022–2039. doi: 10.3382/ps.0612022. [DOI] [Google Scholar]
- 9.Sharp RM, Silyn-Roberts H. Development of preferred orientation in the eggshell of the domestic fowl. J Biophysical Journal. 1984;46(2):175–179. doi: 10.1016/S0006-3495(84)84010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunton P. Research on eggshell structure and quality: an historical overview. Rev. Bras. Ciênc. Avícola. 2005;7(2):67–71. doi: 10.1590/S1516-635X2005000200001. [DOI] [Google Scholar]
- 11.Foulkes. RH, Parsons. J, Schuknecht. HF. X-Ray Diffraction of Otoliths and Egg Shells of Bird and Reptile. J American Naturalist. 1958; 92(866):319.
- 12.Rodriguez-Navarro AB, Yebra A, Nys Y, Jimenez-Lopez C, Garcia-Ruiz JM. Analysis of avian eggshell microstructure using X-ray area detectors. European Journal of Mineralogy. 2007;19(3):391–398. doi: 10.1127/0935-1221/2007/0019-1724. [DOI] [Google Scholar]
- 13.Lammie D, Bain MM, Solomon SE. Scanning Microfocus Small Angle X-ray Scattering Study of the Avian Eggshell. Journal of Bionic Engineering. 2006;3(1):11–18. doi: 10.1016/S1672-6529(06)60002-4. [DOI] [Google Scholar]
- 14.Ahmed AM, Rodriguez-Navarro AB, Vidal ML, Gautron J, Garcia-Ruiz JM, Nys Y. Changes in eggshell mechanical properties, crystallographic texture and in matrix proteins induced by moult in hens. Br Poult Sci. 2005;46(3):268–279. doi: 10.1080/00071660500065425. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez-Hernandez A, Gomez-Morales J, Rodrı´guez-Navarro A, Gautron J, Nys Y, Garcia-Ruiz J. Identification of some active proteins in the process of hen eggshell formation. Crystal Growth & Design. 2008;8(12):4330–4339. doi: 10.1021/cg800786s. [DOI] [Google Scholar]
- 16.Nys YM, Hincke MT, Arias JL, Garcia-Ruiz JM, Solomon SE. Avian eggshell mineralization. J Avian Poultry Biology Reviews. 1999;10(3):143–166. [Google Scholar]
- 17.Rodriguez-Navarro A, Kalin O, Nys Y, Garcia-Ruiz JM. Influence of the microstructure on the shell strength of eggs laid by hens of different ages. Br Poult Sci. 2002;43(3):395–403. doi: 10.1080/00071660120103675. [DOI] [PubMed] [Google Scholar]
- 18.Dunn IC, Rodríguez-Navarro AB, Mcdade K, Schmutz M, Preisinger R, Waddington D, Wilson PW, Bain MM. Genetic variation in eggshell crystal size and orientation is large and these traits are correlated with shell thickness and are associated with eggshell matrix protein markers. Animal Genetics. 2012;43(4):410–418. doi: 10.1111/j.1365-2052.2011.02280.x. [DOI] [PubMed] [Google Scholar]
- 19.Brionne A, Nys Y, Hennequet-Antier C, Gautron J. Hen uterine gene expression profiling during eggshell formation reveals putative proteins involved in the supply of minerals or in the shell mineralization process. BMC Genomics. 2014;15:220. doi: 10.1186/1471-2164-15-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belcher AM, Christensen RJ, Hansma PK. Control of crystal phase switching and orientation by soluble mollusc-shell proteins. [J]. Nature. 1996;381(6577):56–58. doi: 10.1038/381056a0. [DOI] [Google Scholar]
- 21.Dunn IC, Joseph NT, Bain M, Edmond A, Wilson PW, Milona P, Nys Y, Gautron J, Schmutz M, Preisinger R, et al. Polymorphisms in eggshell organic matrix genes are associated with eggshell quality measurements in pedigree Rhode Island Red hens. Anim Genet. 2009;40(1):110–114. doi: 10.1111/j.1365-2052.2008.01794.x. [DOI] [PubMed] [Google Scholar]
- 22.Duan Z, Sun C, Shen M, Wang K, Yang N, Zheng J, Xu G. Genetic architecture dissection by genome-wide association analysis reveals avian eggshell ultrastructure traits. Sci Rep. 2016;6:28836. doi: 10.1038/srep28836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Navarro A, Garcia-Ruiz JM. Model of textural development of layered crystal aggregates. European Journal of Mineralogy. 2000;12(3):609–614. doi: 10.1127/ejm/12/3/0609. [DOI] [Google Scholar]
- 24.Hahn EN, Sherman VR, Pissarenko A, Rohrbach SD, Fernandes DJ, Meyers MA. Nature’s technical ceramic: the avian eggshell. Journal of the Royal Society Interface. 2017;14(126):20160804. doi: 10.1098/rsif.2016.0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maxwell TH. The eggshell: structure, composition and mineralization. J Frontiers in Bioence. 2012;17(1):1266–1280. doi: 10.2741/3985. [DOI] [PubMed] [Google Scholar]
- 26.Sun C, Duan Z, Qu L, Yang N, Xu G. Expression analysis for candidate genes associated with eggshell mechanical property. J. Integr. Agric. 2016;15:397–402. doi: 10.1016/S2095-3119(14)60969-2. [DOI] [Google Scholar]
- 27.Sun C, Lu J, Yi G, Yuan J, Duan Z, Qu L, Xu G, Wang K, Yang N. Promising Loci and Genes for Yolk and Ovary Weight in Chickens Revealed by a Genome-Wide Association Study. Plos One. 2015;10(9):e0137145. doi: 10.1371/journal.pone.0137145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwasawa A, Rahman MA, Roy TK, Moriyama A, Yoshizaki N. Morphological and histochemical changes in the uterus epithelium during eggshell formation in quail. Journal of Poultry Ence. 2010;47(2):183–189. doi: 10.2141/jpsa.009112. [DOI] [Google Scholar]
- 29.Jonchere V, Brionne A, Gautron J, Nys Y. Identification of uterine ion transporters for mineralisation precursors of the avian eggshell. BMC. Physiol. 2012;12:10. doi: 10.1186/1472-6793-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonchere V, Rehault-Godbert S, Hennequet-Antier C, Cabau C, Sibut V, Cogburn LA, Nys Y, Gautron J. Gene expression profiling to identify eggshell proteins involved in physical defense of the chicken egg. BMC Genomics. 2010;11:57. doi: 10.1186/1471-2164-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson TW, Goldner AM. Calcium transport across avian uterus. I. Effects of electrolyte substitution. J American Journal of Physiology. 1973;225(6):1508. doi: 10.1152/ajplegacy.1973.225.6.1508. [DOI] [PubMed] [Google Scholar]
- 32.Lavelin I, Meiri N, Genina O, Alexiev R, Pines M. Na + – K + –ATPase gene expression in the avian eggshell gland: distinct regulation in different cell types. J Physiol Regul Integr Comp Physiol. 2001;281(4):R1169–R1176. doi: 10.1152/ajpregu.2001.281.4.R1169. [DOI] [PubMed] [Google Scholar]
- 33.Ellis JA, Lamantia A, Chavez R, Scurrah KJ, Nichols CG, Harrap SB. Genes controlling postural changes in blood pressure: comprehensive association analysis of ATP-sensitive potassium channel genes KCNJ8 and ABCC9. Physiol Genomics. 2010;40(3):184–188. doi: 10.1152/physiolgenomics.00173.2009. [DOI] [PubMed] [Google Scholar]
- 34.Mangla A, Guerra MT, Nathanson MH. Type 3 inositol 1,4,5-trisphosphate receptor: A calcium channel for all seasons. Cell Calcium. 2019; 85:10.1016/j.ceca.2019.102132. [DOI] [PMC free article] [PubMed]
- 35.Futatsugi A, Kuwajima G, Mikoshiba K. Muscle-specific mRNA isoform encodes a protein composed mainly of the N-terminal 175 residues of type 2 Ins(1,4,5)P3 receptor. J Biochem. 1998;334:559–563. doi: 10.1042/bj3340559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutstein DE, Marks AR. Role of inositol 1,4,5-trisphosphate receptors in regulating apoptotic signaling and heart failure. Heart Vessels. 1997;12:53–57. doi: 10.1007/BF02820867. [DOI] [PubMed] [Google Scholar]
- 37.Medeiros-Domingo A, Tan BH, Crotti L, Tester DJ, Eckhardt L, Cuoretti A, Kroboth SL, Song C, Zhou Q, Kopp D, et al. Gain of function mutation S422L in the KCNJ8-encoded cardiac K(ATP) channel Kir6.1 as a pathogenic substrate for J-wave syndromes. Heart Rhythm. 2010;7(10):1466–1471. doi: 10.1016/j.hrthm.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matos DR. Calcium metabolism in birds. Vet Clin Exot Anim. 2008;11(1):59–82. doi: 10.1016/j.cvex.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto-Hino M, Sugiyama T, Hikichi K, Mattei MG, Hasegawa K, Sekine S, Sakurada K, Miyawaki A, Furuichi T, Hasegawa M. Cloning and characterization of human type 2 and type 3 inositol 1,4,5-trisphosphate receptors. Receptors Channels. 1994;2(1):9–22. [PubMed] [Google Scholar]
- 40.Tohmonda T, Yoda M, Iwawaki T, Matsumoto M, Horiuchi K. IRE1α/XBP1-mediated branch of the unfolded protein response regulates osteoclastogenesis. Journal of Clinical Investigation. 2015;125(8):3269–3279. doi: 10.1172/JCI76765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nystoriak MA, Nieves-Cintron M, Patriarchi T, Buonarati OR, Prada MP, Morotti S, Grandi E, Fernandes JD, Forbush K, Hofmann F, et al. Ser1928 phosphorylation by PKA stimulates the L-type Ca2 + channel CaV1.2 and vasoconstriction during acute hyperglycemia and diabetes. Science Signaling. 2017;10(463):eaaf9647. doi: 10.1126/scisignal.aaf9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz D, Mikala G, Yatani A, Engle DB, Iles DE, Segers B, Sinke RJ, Weghuis DO, Klockner U, Wakamori M. Cloning, chromosomal localization, and functional expression of the alpha 1 subunit of the L-type voltage-dependent calcium channel from normal human heart. Proc Natl Acad Sci. 1993;90(13):6228–6232. doi: 10.1073/pnas.90.13.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyford GL, Strege PR, Shepard A, Ou Y, Ermilov L, Miller SM, Gibbons SJ, Rae JL, Szurszewski JH, Farrugia G. alpha(1 C) (Ca(V)1.2) L-type calcium channel mediates mechanosensitive calcium regulation. Am J Physiol Cell Physiol. 2002;283(3):C1001-1008. doi: 10.1152/ajpcell.00140.2002. [DOI] [PubMed] [Google Scholar]
- 44.Cao C, Ren YS, Mirando AJ, Barnett A, Rouse D, Mun SH, Park-Min KH, McNulty AL, Guilak F, Karner CM, et al. Increased Ca2 + signaling through altered CaV1.2 L-type Ca2 + channel activity promotes bone formation and prevents estrogen deficiency-induced bone loss. JCI Insight. 2017;2(22):e95512. doi: 10.1172/jci.insight.95512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dacquin R, Davey RA, Laplace C, Levasseur R, Morris HA, Goldring SR, Gebre-Medhin S, Galson DL, Zajac JD, Karsenty G. Amylin inhibits bone resorption while the calcitonin receptor controls bone formation in vivo. J Cell Biol. 2004;164(4):509–514. doi: 10.1083/jcb.200312135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu W, Ruangkittisakul A, MacTavish D, Shi JY, Ballanyi K, Jhamandas JH. Amyloid beta (Abeta) peptide directly activates amylin-3 receptor subtype by triggering multiple intracellular signaling pathways. J Biol Chem. 2012;287(22):18820–18830. doi: 10.1074/jbc.M111.331181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Escoffier J, Lee HC, Yassine S, Zouari R, Martinez G, Karaouzene T, Coutton C, Kherraf ZE, Halouani L, Triki C, et al. Homozygous mutation of PLCZ1 leads to defective human oocyte activation and infertility that is not rescued by the WW-binding protein PAWP. Hum Mol Genet. 2016;25(5):878–891. doi: 10.1093/hmg/ddv617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inman CK, Li N, Shore P. Oct-1 counteracts autoinhibition of Runx2 DNA binding to form a novel Runx2/Oct-1 complex on the promoter of the mammary gland-specific gene beta-casein. Mol Cell Biol. 2005;25(8):3182–3193. doi: 10.1128/MCB.25.8.3182-3193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nys Y, Le Roy N. Calcium Homeostasis and Eggshell Biomineralization in Female Chicken. In: Feldman D, editor. Vitamin D. 4. London: Academic Press; 2018. pp. 361–382. [Google Scholar]
- 50.Rowe PS. Regulation of bone-renal mineral and energy metabolism: the PHEX, FGF23, DMP1, MEPE ASARM pathway. Crit Rev Eukaryot Gene Expr. 2012;22(1):61–86. doi: 10.1615/CritRevEukarGeneExpr.v22.i1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alejandro B, Rodriguez-Navarro XRD2DScan: new software for polycrystalline materials characterization using two-dimensional X-ray diffraction. Journal of Applied Crystallography. 2006;39(6):905–909. doi: 10.1107/S0021889806042488. [DOI] [Google Scholar]
- 52.Sun C, Liang Q, Yi G, Yuan J, Ning Y. Genome-wide association study revealed a promising region and candidate genes for eggshell quality in an F2 resource population. BMC Genomics. 2015;16(1):565. doi: 10.1186/s12864-015-1795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J of Hum Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44(7):821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. Journal of Open Source Software. 2014;3(25):731. doi: 10.21105/joss.00731. [DOI] [Google Scholar]
- 56.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32(4):361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 57.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 58.Yang JA, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am J Human Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson R. The estimation of variance and covariance components withan application when records are subject to culling. Biometrics. 1973;29:527–550. doi: 10.2307/2529174. [DOI] [Google Scholar]
- 60.Lee HC, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28(19):2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Visscher PM, Hemani G, Vinkhuyzen A, Chen GB, Lee SH, Wray NR, Goddard ME, Yang J, Barsh GS. Statistical Power to Detect Genetic (Co)Variance of Complex Traits Using SNP Data in Unrelated Samples. Plos Genetics. 2014; 10:DOI: 10.1371/journal.pgen.1004269. [DOI] [PMC free article] [PubMed]
- 62.Jocelyn Plassais, Jaemin, Kim Brian, Davis, Danielle, Karyadi. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nature Communications. 2019;10:1489. doi: 10.1038/s41467-019-09373-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 64.McLaren W, Pritchard B, Rios D, Chen YA, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minoru K, Susumu G, Yoko S, Miho F, Mao T. KEGG for integration and interpretation of large-scale molecular data sets. J Nucleic Acids Research. 2012;40(D1):D109-D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in figshare repository, 10.6084/m9.figshare.16627099.v1 .