Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the COVID-19 pandemic. Angiotensin-converting enzyme 2 (ACE2) has been identified as the host cell receptor that binds to the receptor-binding domain (RBD) of the SARS-COV-2 spike protein and mediates cell entry. Because the ACE2 proteins are widely available in mammals, it is important to investigate the interactions between the RBD and the ACE2 of other mammals. Here we analyzed the sequences of ACE2 proteins from 16 mammals, predicted the structures of ACE2-RBD complexes by homology modeling, and refined the complexes using molecular dynamics simulation. Analyses on sequence, structure, and dynamics synergistically provide valuable insights into the interactions between ACE2 and RBD. The analysis outcomes suggest that the ACE2 of bovine, cat, and panda form strong binding interactions with RBD, while in the cases of rat, least horseshoe bat, horse, pig, mouse, and civet, the ACE2 proteins interact weakly with RBD.
Keywords: ACE2, Homology modeling, Molecular dynamics, RBD, SARS-CoV-2
Graphical abstract
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel coronavirus, is responsible for the new type of severe pneumonia COVID-19 [1]. Hundreds of millions of people have tested positive for the SARS-CoV-2, and the number of infections still rapidly increases with mutant variants of the virus also noted [2]. SARS-CoV-2 utilizes the human angiotensin-converting enzyme 2 protein (ACE2) to initiate the spike protein binding and to facilitate the viral attachment to host cells [[3], [4], [5], [6], [7], [8]]. Recently, reports of other animals testing positive for SARS-CoV-2 are emerging. Studies on viral replication and susceptibility to SARS-CoV-2 suggested that the virus replicates efficiently in cats or ferrets [9]. There are reports of dog, cat and tiger testing positive for SARS-CoV-2 [[10], [11], [12]]. Therefore, it is highly desirable to study the susceptibility of those mammalian animals, which are in close contact with humans. Because ACE2 proteins exist in many mammalian animals, potentially making them susceptible to SARS-CoV-2, we gathered ACE2 sequences of 16 animals for detailed analysis (Table 1 ). By studying the interactions between the receptor binding domain (RBD) of virus spike protein and ACE2 receptors, we hope to provide information on animal susceptibility to the SARS-CoV-2. It has been established that the RBD of the SARS-CoV-2 (denoted as RBD hereafter) and the human ACE2 (hACE2) form stable complexes, as shown in recently determined crystal structures [13,14] and computer simulations [15]. This provides an opportunity to investigate the interactions between RBD and ACE2 of other mammalian animals. Although such knowledge alone may not be sufficient to accurately predict the susceptibility of animals to SARS-CoV-2, the information is valuable in understanding the interactions between RBD and ACE2.
Table 1.
ACE2 proteins selected in this study.
| Source of ACE2 | Scientific name of animals | Reason for selectiona | |
|---|---|---|---|
| human | Homo sapiens | n/a |  |
| Bovine/Cow | Bos taurus | 1 |  |
| Cat | Felis catus | 1 |  |
| Chinese Horseshoe bat | Rhinolophus sinicus | 2 |  |
| Dog | Canis lupus familiaris | 1 |  |
| Giant panda | Ailuropoda melanoleuca | 1, 4 |  |
| Horse | Equus caballus | 1 |  |
| Least Horseshoes bat | Rhinolophus pusillus | 2 |  |
| Malayan pangolin | Manis javanica | 2,4 |  |
| Mouse | Mus musculus | 1 |  |
| Palm civet | Paguma larvata | 2 |  |
| Pig | Sus scrofa | 1 |  |
| Rabbit | Oryctolagus cuniculus | 1 |  |
| Rat | Rattus norvegicus | 1 |  |
| Sheep | Ovis aries | 1 |  |
| Siberian tiger | Panthera tigris altaica | 1,3 |  |
Reasons for selection: 1 = in close contact with humans, 2 = known hosts of related coronaviruses, 3 = news reports on positive SARS-COV-2 test, 4 = endangered animal.
The conservation of ACE2 residues and structures of ACE2-RBD complexes are reported in a few studies [[16], [17], [18]], dynamics simulations were also applied to investigate the dynamical features of the ACE2-RBD interactions [19,20]. In this report, we combined sequence analysis, structure prediction, molecular dynamics to investigate the interactions between ACE2 and RBD. Using the crystal structure of SARS-CoV-2 RBD and human ACE2 complex (hACE2-RBD) as the template [21], ACE2-RBD complex structures were constructed for previously mentioned ACE2 proteins, and the dynamics of these complexes were investigated using simulations. Based on conservation in ACE2 residues, similarity in electrostatic potentials, and dynamical interactions revealed from simulations, we classified these ACE2-RBD interactions into weak, medium, and strong categories.
2. Materials and methods
2.1. Homology modeling of the ACE2-RBD complex structures
The ACE2 sequences were obtained from the NCBI and uniport databases [22,23]. Using the SWISS-MODEL interactive server [24], we modeled structures for 15 mammalian ACE2 proteins, based on the hACE2 structure (PDB ID: 6LZG [21]). Model validation and assessment were carried out using the assessment tools within the SWISS-MODEL server. Ramachandran plots were used to check the stereochemical quality of the structures by analyzing both per residue and overall geometry. Molprobity was applied to perform all-atom contact analysis and compute scores based on contacts, percentage of Ramachandran outliers and percentage of bad-side chain contact [25]. After ACE2 structure prediction, the SARS-CoV-2 RBD and ACE2 complexes were assembled by superposing the predicted homology structure of ACE2 proteins to the hACE2-RBD complex structure.
2.2. Comparison of the electrostatic potential of ACE2 on RBD binding interface
PyMOL (https://pymol.org/2/) was utilized to compute and visualize the electrostatic potential maps at the ACE2-RBD complex interfaces. These maps depict the electrostatic potential surface rendered from the numerical solutions of the Poisson-Boltzmann equation [26]. The electrostatic potential surfaces were simplified into 2D projection images for pairwise comparison and clustering analysis. The hierarchical clustering algorithm was applied to group these 2D projection images.
2.3. Molecular dynamics simulations of ACE2-RBD complexes
GROMACS-5.1.2 [27] was used for MD simulations of ACE2-RBD complexes. All complexes were parametrized with CHARMM27 force fields [28]. Disulfide bonds were maintained as in the crystal structure of the hACE2-RBD complex. Complexes were solvated in the triclinic box with a minimum distance of 10 Å between the complex and the box boundaries. Solvated systems were neutralized by adding ions (Na+ and Cl−) to 0.15 mM. Then, these systems were subjected to steepest descent energy minimization, followed by constant volume (NVT) and constant pressure of 1 bar (NPT) equilibrations, for 1 ns and 3 ns respectively. During system equilibration, positional restraints were applied on non-hydrogen atoms of ACE2-RBD complexes. Temperature and pressure were controlled by the V-rescale method [29] and Parrinello-Rahman method [30], respectively. Finally, 50 ns production simulations were carried out at NPT conditions. VMD [31] and UCSF Chimera [32] were used to visualize and analyze simulation trajectories. The physical binding interactions comprising van der Waals and electrostatic components were calculated between each ACE2 and RBD for the structures sampled in MD simulations.
3. Results and discussions
3.1. Sequence analysis and the conservation at the RBD binding interface
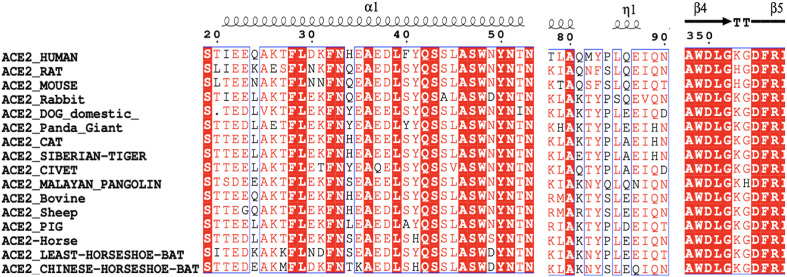
Multiple sequence alignment was carried out using CLUSTALW program [33,34], and the aligned sequences were redrawn with the human ACE2 crystal structure as the reference using the ESPript webserver [35]. All ACE2 proteins comprise amino acids from position 19 to 614, except for dog ACE2, which has a gap at position 20 (Fig. 1 and Fig. S1). In the human ACE2-RBD complex, the amino acids of ACE2 at the N-terminal helix-1 (residues 19–42), near the η1 (residues 82–83), helix-13 (residue 330) and β-hairpin-4,5 (residues 352–357), have been identified as the key residues (Fig. 1) that bind to the RBD [13,15,21].
Fig. 1.
The comparison for the key residues at the binding interfaces after multiple sequence alignment analysis.
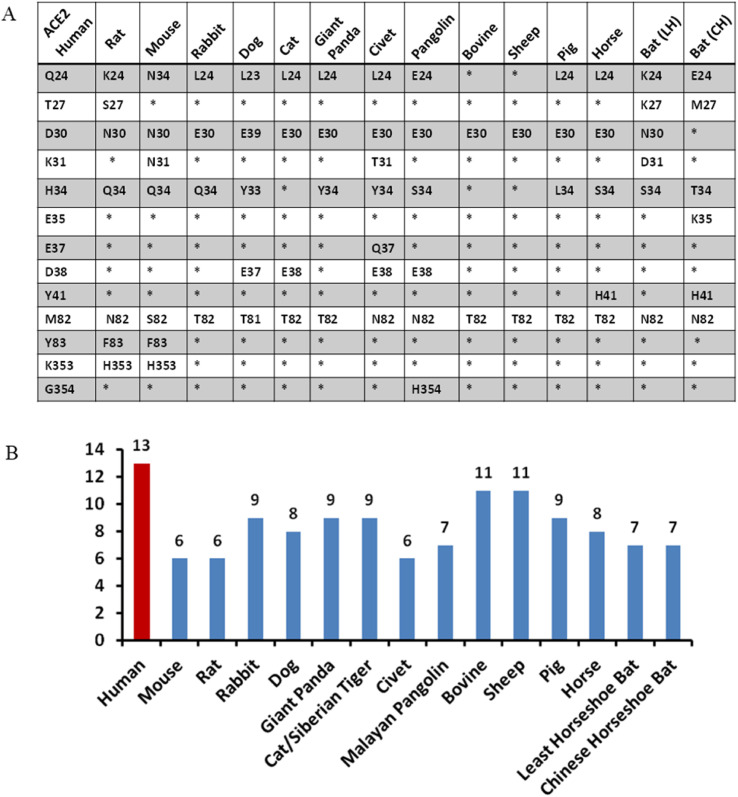
Based on the hACE2-RBD structure, we further identified 13 key residues on the RBD-interacting interface and analyzed their sequence conservations compared to hACE2 (Fig. 2 ). Siberian tiger and cat share the same ACE2 protein residues among these 13 residues, so the analysis on tiger ACE2 is inferred from cat ACE2. Taking the hACE2 sequence as the reference, substituted residues of these 13 positions are summarized in Fig. 2A and Table S1. We found that residues at positions 24, 27, 31, 34 and 82 are highly variable among these ACE2 proteins. The H34 of hACE2 has the largest variation, which is substituted by Q (Rat, Mouse and Rabbit), Y (Dog, Giant Panda and Civet), S (Pangolin, Horse and Least Horseshoe Bat), L (Pig), and T (Chinese Horseshoe Bat). The Q24 has four variations: K (Rat and Least Horseshoe Bat), N (Mouse), L (Rabbit, Dog, Cat, Giant Panda, Civet, Pangolin, Pig and Horse), and E (Chinese Horseshoe Bat). T27, K31, and M82 all have three different substitutions, while D30 and M82 have two different possible substitutions. Fig. 2B shows the number of identical residues of each ACE2 to hACE2 at these 13 positions. Bovine and sheep ACE2 proteins differ from hACE2 at only two positions, while the ACE2 proteins of mouse, rat and civet are different from hACE2 at 7 out of 13 positions.
Fig. 2.
Residue conservation analysis. (A) Comparison of 13 critical residues in binding to SARS-CoV-2 RBD. Bat (LH) stands for Least Horseshoe and Bat (CH) stands for Chinese Horseshoe Bat. Cat ACE2 is used to represent both cat and Siberian tiger ACE2 proteins, their sequences are identical at these 13 positions. (B) The number of identical residues compared to hACE2 at the 13 positions marked in (A).
3.2. Model assessment and validation
Ramachandran analysis showed that 96–99% of amino acid residues are within the energetically favored region for all the predicted ACE2 structures (Fig. S2). The Molprobity evaluation also showed that all the predicted structures are of good quality. A structure with a lower MolProbity score is considered to be better among structures at the specific resolution [25]. For reference, the structure of the hACE2-RBD complex was resolved at 2.50 Å, and the Molprobity score is 1.10 for the structure that was used as the template for modeling other ACE2 proteins. In comparison, the predicted ACE2 structures of cat, bovine, giant panda, rat, civet and least horseshoe bat have Molprobility scores of 0.91, 1.03, 1.00, 0.77, 0.93 and 0.96, respectively.
Additionally, the modeled structures were submitted to the Structural Analysis and Verification Server (SAVES) (https://saves.mbi.ucla.edu/), and the programs ERRAT and Verify3D were selected for validation. ERRAT validates the modeled structures by analyzing the statistics of non-bonded interactions between different atom types and plots the value of the error function versus position of a 9-residue sliding window, calculated by comparison with statistics from highly refined structures while the Verify3D program analyzed the compatibility of an atomic model (3D) with its amino acid sequence (1D) to assess the 3D protein structure. The results are summarized in Table S2. Both programs reiterate that all the predicted structures were of good quality and suitable for use in further studies.
3.3. Electrostatic potential surface at the binding interface
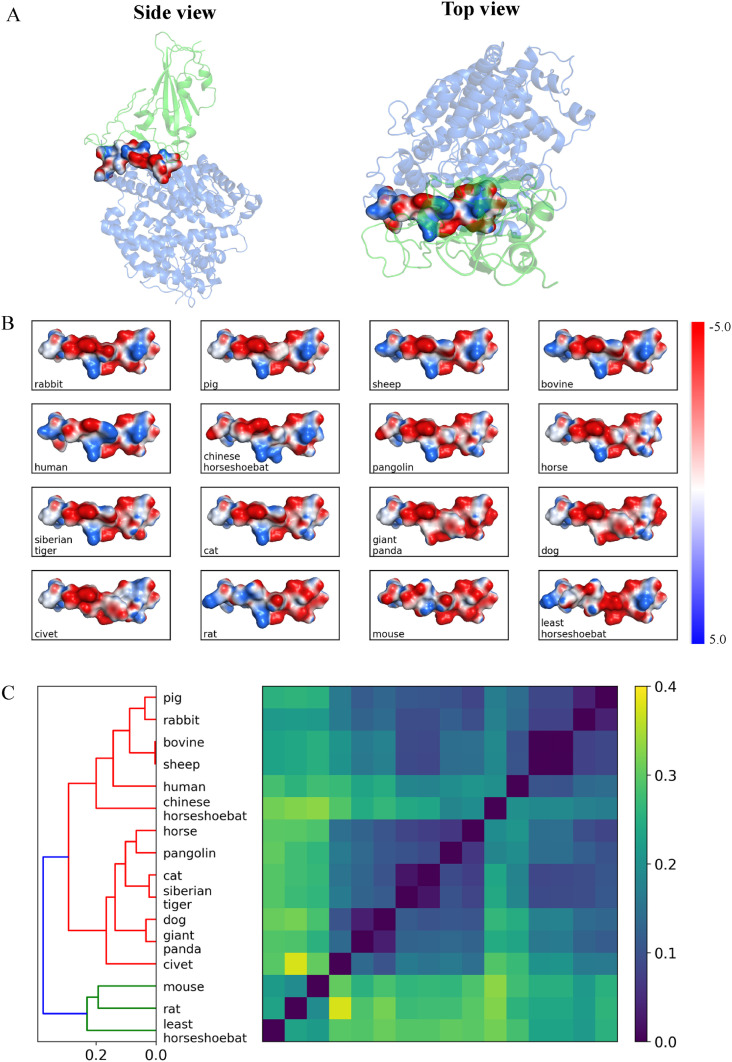
The electrostatic potential surfaces for the central region of ACE2 helix-1 (residues 30–37) are shown in Fig. 3 for all ACE2-RBD complexes. According to electrostatic potential maps, this region features a charge distribution composed of both positively and negatively charged sites in human, bovine and cat ACE2, while the electrostatic potentials are mostly negative for dog and pig ACE2 proteins. Clustering analysis on electrostatic potential surfaces showed that the bovine/sheep/pig/rabbit ACE2 proteins have similar features as hACE2 in this region (Fig. 3C). The mouse/rate/least-horseshoe-bat ACE2 show the least similarity in the electrostatic potential features in this region compared to other ACE2 proteins.
Fig. 3.
Electrostatic potential surface analysis. (A) ACE2 binding interface to RBD at two orientations. (B) The top view of the electrostatic potential surfaces for the central binding region of between ACE2 and SARS-CoV-2-RBD. In humans, cat and bovine ACE2, positions 30–37 comprise both positive and negatively charged residues. The residue substitutions in ACE2 of dog and civet at the same region lead to negatively charged patches. (C) Hierarchical clustering results are based on the similarity between electrostatic potential surfaces.
3.4. Binding interactions assessed from MD refinement data
Computational predictions of 3D structure models of protein are often complemented with crucial refinement to improve the structures for accurate models [36]. In this study, MD simulation was used to refine the predicted ACE2-RBD complexes assembled by superposing the predicted ACE2 structures to the hACE2 of the hACE2-RBD complex crystal structure. Based on the template, we assumed that besides diversity in RBD interacting residues of ACE2 among 15 mammals, the ACE2-RBD interface would be similar to the crystal structure of the hACE2-RBD complex. Moreover, attempts to build the ACE2-RBD complexes by molecular docking using ZDOCK (https://zdock.umassmed.edu) led to inconsistent complex structures (Fig. S3).
The structural integrity and stability during the refinement process for all complexes were quantified using the root mean square deviation (RMSD) of the Cα with respect to the initial structure and their fluctuations during the refinement trajectories (Fig. S4). For all ACE2-RBD complexes, the RMSD of the spike RBD remained stable during the refinement process with an average value of 1.7 Å. For hACE2 and all modeled ACE2 of the mammals, small fluctuations in the RMSD were observed at the beginning of the refinement simulation. The most significant conformational changes are the movement of the β1-β2 loop (residues 130–143) of the ACE2 as evidenced by the RMSF of their Cα (Fig. S4). The loop is located away from the binding interface which remained stable during the refinement process.
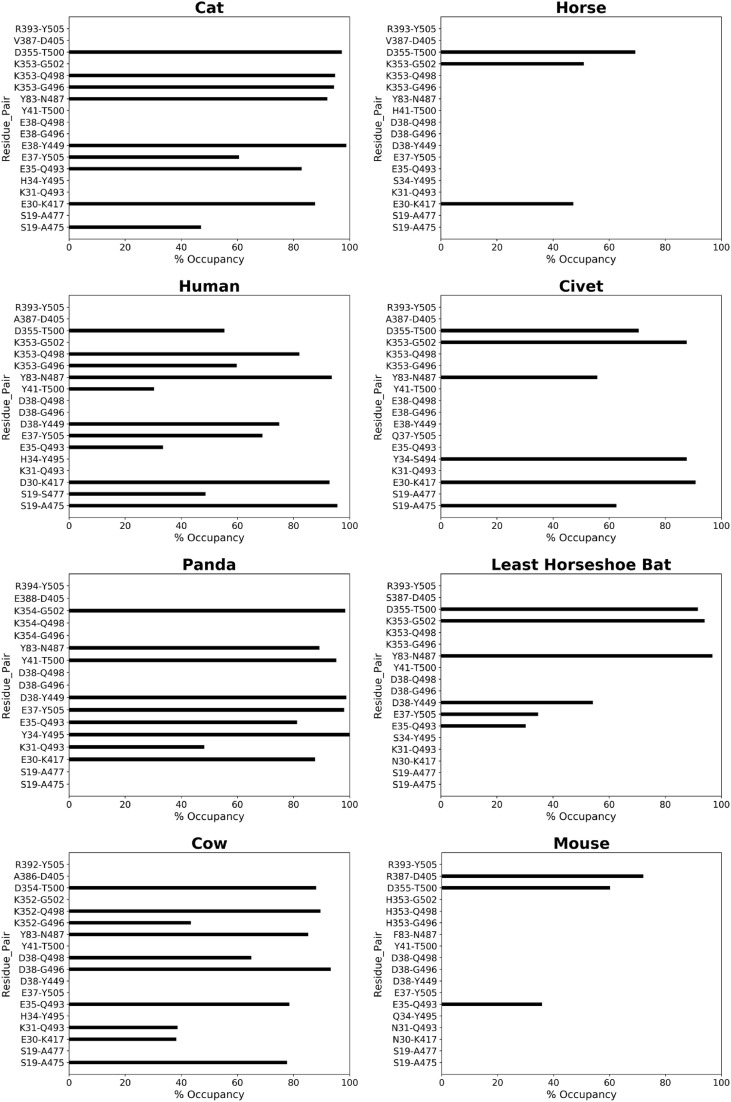
Refinement MD trajectories of 16 ACE2-RBD complexes were analyzed with a focus on the ACE2 residues at the binding interface (Figs. S5–S7). We focused on the analysis of interfacing hydrogen bonds and contacts between ACE2 and RBD, which are directly involved in the binding interactions. The occupancies of hydrogen bonds and contacts were calculated from MD refinement trajectories and only the residues with occupancy greater than 30% are were considered. For these ACE2-RBD complexes, five frequently observed hydrogen bonds are D30:K417, E35:Q493, Y83:N487, K353:G502, and D355:T500 (ACE2 residues are placed on the left of the colon, and RBD residues on the right). In the following, detailed discussions are grouped based on the number of substitutions among the 13 key residues.
Bovine ACE2-RBD shows a highly similar hydrogen-bonding pattern as hACE2-RBD (Fig. 4 ). However, the refined structure shows slightly weaker binding interactions compared to hACE2. Similarly, experimental binding affinity for Bovine ACE2 to the RBD has been reported to be 3–4 fold weaker compared to hACE2 [37]. Cat ACE2 shows stronger hydrogen bonding interactions with RBD than hACE2, but the hydrogen bond (H-bond) between Y41 and T500 is absent (Fig. 4). Cat ACE2-RBD also exhibits the highest interaction energy among 16 ACE2-RBD complexes (see Table 2 ). This is consistent with recent reports on domestic cats being infected by SARS-CoV-2 [9,10]. A recently resolved complex structure of cat ACE2-RBD reveals similar binding as the hACE2-RBD complex [37]. When compared to the experimental structure, our model deviates by 0.6 Å at the ACE2-RBD interface mainly contributed by the terminal residue orientation. Furthermore, our model correctly predicts and depicts all the hydrogen bonds observed in the experimental structure. Experimental data also show that cats are efficient in replicating SARS-CoV-2 [37], suggesting that four substitutions do not inhibit RBD binding. Interestingly, terminal residues’ orientation and movements also contributed to the differences between cat-ACE2 and Siberian tiger ACE2-RBD complexes, despite the highest sequence similarity between the two animals. A difference at the terminal residue S19 in tiger lead to the loss of interaction with RBD residue A475 (Fig. S8).
Fig. 4.
Occupancies of hydrogen bonds at ACE2-RBD interface. The left panels are hydrogen bonding patterns in strong binding cases (as labeled above each plot); the right panels correspond to the weak binding cases. Each hydrogen bond comprises one residue from ACE2 and one from RBD, shown on the left and right of the hyphen respectively.
Table 2.
Molecular interaction energies between ACE2 and RBD. 250 structures from the simulations were used to compute interaction energies. Strong interactions are highlighted in bold font.
| Complex | Interaction energy of homology models (kcal/mol) | No. of H-bonds having >30% occupancy | Average interaction energy (kcal/mol) |
|---|---|---|---|
| Human | −169.56 | 11 | −170.08 ± 10.79 |
| Rat | −135.33 | 5 | −125.69 ± 15.52 |
| Mouse | −127.39 | 3 | −107.31 ± 9.16 |
| Rabbit | −165.99 | 9 | −151.48 ± 15.56 |
| Dog | −168.71 | 7 | −147.79 ± 14.23 |
| Siberian Tiger | −155.13 | 7 | −144.82 ± 13.43 |
| Cat | −170.80 | 9 | −175.77 ± 12.20 |
| Civet | −155.95 | 6 | −115.67 ± 12.16 |
| Bovine | −182.96 | 10 | −162.83 ± 14.04 |
| Least Horseshoe Bat | −157.98 | 6 | −114.12 ± 17.08 |
| Malayan Pangolin | −179.45 | 9 | −155.46 ± 10.94 |
| Pig | −172.54 | 4 | −127.67 ± 13.74 |
| Chinese Horseshoe Bat | −176.70 | 5 | −140.40 ± 13.55 |
| Horse | −148.84 | 3 | −119.62 ± 11.46 |
| Sheep | −181.49 | 9 | −146.63 ± 15.32 |
| Panda | −187.17 | 9 | −167.30 ± 9.92 |
Panda and pig ACE2 proteins both differ from hACE2 at four positions, but their interactions with RBD are quite different. Panda ACE2 forms 9 strong hydrogen bonds with RBD (Fig. 4). The pig ACE2 interacts with RBD more weakly than panda ACE2, in line with experimental studies showing that SARS-CoV-2 infection was not detectable in pigs or their cell lines [38]. Although experimental binding affinity for Panda ACE2 to RBD is not available, for pig ACE2, the binding affinity has been reported to be 2-fold weaker compared to hACE2 [37]. The difference in the interaction profiles of panda and pig may be due to H34Y and H34L substitutions respectively, increasing electrostatic interactions of the panda ACE2-RBD interface.
Dog and horse ACE2 have five substitutions (four are at positions 24,30,34,82, and one occurs at position 38 for dog and position 41 for horse). A dog was the first domestic animal reported testing positive with a low level of SARS-CoV-2 infection [39,40]. The dog ACE2 (dACE2) contains a notable variation at N-terminal-helix 1 which results in gapping (deletion) at position 20, revealed in the sequence alignment. While this deletion does not appear to affect the complex structure revealed in the homology model, it slightly differs from the crystal structure of dACE2-RBD that has been solved [41]. In the case of our homology model of dACE2-RBD (Fig. S6), the occupancy of hydrogen bonds (Fig. S8) between RBD and E37, Y40, Q41 of dACE2 is lower than 30%. When superposed together focusing on the residues at the ACE2-RBD interface, our model and the crystal structure differ by 0.93 Å. This can be explained by the missing N-terminal residues (M, Q, S, T which are solved in the crystal structure) in the predicted model, the position and orientation of E22 and D23 (as the terminal residues in the predicted model) were affected during the MD refinement. However, these weakened interactions are also in line with the reported experimental binding affinities of dACE2 (K D = 123 nM) to RBD as compared to the hACE2 (K D = 18.5 nM) [41]. Horse ACE2 forms only 3 hydrogen bonds with RBD (Fig. 4), this alludes to weaker interactions which are in agreement with the reported 6–7 folds weaker interactions between horse ACE2 and RBD [37].
With respect to hACE2, pangolin, CH-bat, and LH-bat ACE2 differ at 6 positions. Due to the co-evolution with other coronaviruses, pangolin and bats were speculated to be intermediate hosts of SARS-CoV-2. Despite having 6 substitutions, pangolin ACE2 forms strong interactions with RBD in the homology model (Table 2). However, upon refinement by MD, the complex showed diminished binding interactions. Published experimental cells assays reported a 3–4 fold weaker binding of pangolin ACE2 to RBD [37]. Previously study on viral determinants of adaptation to hACE2 has shown that the Q24K mutant of ACE2 revealed a slight inhibition effect on the binding to the RBD of SARS-CoV spike protein, and the binding is abolished for K31D mutant [42]. The mutant may exert a similar effect to the SARS-CoV-2 infection. LH-bat ACE2 has both substitutions, leading to weak interactions with only 6 hydrogen bonds (Fig. 4).
Compared to hACE2, civet, mouse, and rat have 7 substitutions in their ACE2 at the interface. Studies have shown civets can be infected by coronaviruses in natural environments [43]. Our models show that in civet ACE2, the important hydrogen bond D30:K417 in hACE2-RBD is not formed between E30 of civet ACE2 and K417 of RBD (Fig. 4). Mouse ACE2 shows the weakest interaction with RBD, with only 3 hydrogen bonds. The mutation Y83F in both mice/rats results in the loss of the hydroxyl moiety of tyrosine (in hACE2), losing a hydrogen bond with the N487 of the RBD. This Y83F mutation has been reported to inhibit interaction with SARS-CoV spike RBD [42]. Another noticeable substitution occurs at the highly conserved K353, which is replaced by histidine in both mouse and rat ACE2 (Fig. 2). The K353H substitution eliminated hydrogen bonds with N501 of RBD, exerting a significant impact on RBD binding. These structural observations from the models are in line with both reported computational studies and cells assays which reported a more than 10-fold increase in K D for the bats' species to RBD [37]. The hydrogen bonds occupancy and contacts for all the animals studied are summarized in Fig. 4, Fig. S8, Table 4, and Table S3.
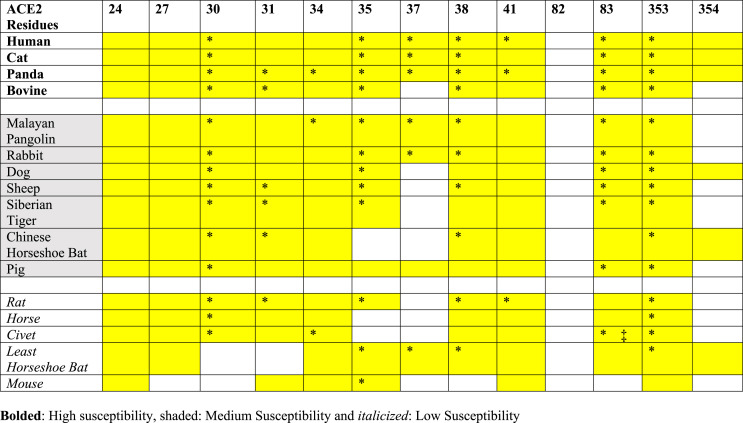
Table 4.
Residues of ACE2 which form H-bonds (* = >30% Occupancy) and vdw contacts (yellow color = > 30% Occupancy) with SARS-CoV-2 RBD.
We have quantified the interaction between ACE2 and RBD by molecular mechanics energy comprising van der Waals and electrostatic interaction terms. As summarized in Table 2, the ACE2 of cat, panda, bovine, and human form strong interactions with the RBD, while the ACE2-RBD interactions are much weaker in the cases of mouse, least horseshoe bat, civet, horse, rat, and pig. The sequence conservation, molecular interactions are correlated to experimental results (Table 3 ), providing insights on the interactions at molecular levels.
Table 3.
Binding interactions are classified based on sequence identity and interactions.
| Mammals | Experimental results | Sequence identity | Interaction energy |
|---|---|---|---|
| Human | High [1,21,46,47] | High | High |
| Bovine | Medium [37] | High (11/13) | High |
| Sheep | Medium [37] | High (11/13) | Medium |
| Cat | High [9,10,37,48] | Medium (9/13) | High |
| Tiger | Medium [12] | Medium (9/13) | Medium |
| Panda | Not available | Medium (9/13) | High |
| Pig | Medium [37,38] | Medium (9/13) | Low |
| Rabbit | Low [48] | Medium (9/13) | Medium |
| Dog | Medium [11,41] | Medium (8/13) | Medium |
| Horse | Medium [37] | Medium (8/13) | Low |
| Pangolin | Medium [48] | Medium (7/13) | Medium |
| Bat (LH) | Not susceptible [37] | Medium (7/13) | Low |
| Bat (CH) | Not susceptible [37] | Medium (7/13) | Medium |
| Civet | Not susceptible [48] | Low (6/13) | Low |
| Mouse | Not susceptible [8,44,48] | Low (6/13) | Low |
| Rat | Not susceptible [48] | Low (6/13) | Low |
Classification criteria: Sequence identity of 13 residues compared to hACE2 (Low, 0–50%; Medium, 50–75%; High, 75%–100%); Interaction energy (Low, −100 ∼ −130 kcal/mol; Medium, −130 ∼ −160 kcal/mol; and High, −160 ∼ −190 kcal/mol).
To identify key amino acid residues for the viral-host infusion, we considered both the hydrogen bonds (Table 4, Fig. 4 and Fig. S8) and contacts between ACE2 and RBD (Table 4 and Table S3) with occupancy of >30% during the refinement trajectories. From our analysis, we identified residues at positions 30 and 83 (D30 and Y83 in hACE2) do not form H-bonds or contacts in ACE2-RBD complexes of mouse and least horseshoe bat (Table 4 ). Salt-bridge between residues D/E30 of ACE2 and K417 of the SARS-CoV-2 RBD has been reported to be very crucial for ACE2- SARS-CoV-2 RBD interaction [13,44,45]. Multiple studies have shown that K353 is crucial for host viral interactions. A single humanizing mutation H353K in mouse ACE2 has been reported to enhance viral entry, infection efficiency, thusly rendering mice susceptible to SARS-CoV-2 [44]. Moreover, our results show that residue at position 353 interacts with RBD through H-bond formation in all mammals except mice. In addition to residues at position 353, residue in positions 34 and 41 also form contacts with SARS-CoV-2 RBD in all mammals studied.
4. Conclusions
Sequences, homology models, and refined conformations are analyzed for 16 ACE2 proteins in complex with the RBD of SARS-CoV-2 spike protein. The ACE2 of bovine and sheep exhibit high sequence identities to human ACE2. MD refinement simulation reveals that bovine, cat, and panda ACE2 proteins show strong binding interactions with the RBD. ACE2 of dog, Siberian tiger, Malayan pangolin, rabbit, sheep, and rabbit show relatively weaker interactions. This study provides a molecular basis for differential interactions between ACE2 and RBD in 16 mammals and will be useful in predicting the host range of the SARS-CoV-2.
Declaration of competing interest
None Declared.
Acknowledgment
The work is supported by Beijing Computational Science Research Center (CSRC) via a director discretionary grant. The research is supported by the National natural science foundation (NSFC Grant number: U1930402) to Haiguang Liu.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.105017.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. January 2020 doi: 10.1056/NEJMoa2001017. Published online. NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease (COVID-19). Accessed February 20, 2021. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 3.Li W., Moore M.J., Vasllieva N., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (80-) 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 6.Kuba K., Imai Y., Ohto-Nakanishi T., Penninger J.M. Trilogy of ACE2: a peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol. Ther. 2010;128(1):119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao L., Deng W., Huang B., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. Published online. [DOI] [PubMed] [Google Scholar]
- 9.Shi J., Wen Z., Zhong G., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science (80-) 2020 doi: 10.1126/science.abb7015. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coronavirus: Belgian cat infected by owner. Accessed April 7, 2020. https://www.brusselstimes.com/all-news/belgium-all-news/103003/coronavirus-belgian-woman-infected-her-cat/.
- 11.Coronavirus: Hong Kong confirms a second dog is infected | South China Morning Post. Accessed April 7, 2020. https://www.scmp.com/news/hong-kong/health-environment/article/3075993/coronavirus-hong-kong-confirms-second-dog.
- 12.Tiger at NYC's Bronx Zoo tests positive for coronavirus - ABC News. Accessed April 7, 2020. https://abcnews.go.com/Health/wireStory/tiger-nycs-bronx-zoo-tests-positive-coronavirus-69989185.
- 13.Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. Published online. [DOI] [PubMed] [Google Scholar]
- 14.Shang J., Ye G., Shi K., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupala C.S., Li X., Lei J., et al. Computational simulations reveal the binding dynamics between human ACE2 and the receptor binding domain of SARS-CoV-2 spike protein. Quant. Biol. 2021;9(1):61–72. doi: 10.15302/J-QB-020-0231. [DOI] [Google Scholar]
- 16.Damas J., Hughes G.M., Keough K.C., et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. Unit. States Am. 2020 doi: 10.1073/pnas.2010146117. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.03.047. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai X., Sun J., Yan Z., et al. Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020 doi: 10.1128/jvi.00831-20. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Xu X., Zhou X., et al. Molecular simulation of SARS-CoV-2 spike protein binding to pangolin ACE2 or human ACE2 natural variants reveals altered susceptibility to infection. J. Gen. Virol. 2020 doi: 10.1099/jgv.0.001452. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luan J., Jin X., Lu Y., Zhang L. SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J. Med. Virol. 2020 doi: 10.1002/jmv.25817. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., Zhang Y., Wu L., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acland A., Agarwala R., Barrett T., et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2013 doi: 10.1093/nar/gks1189. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bateman A. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019 doi: 10.1093/nar/gky1049. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterhouse A., Bertoni M., Bienert S., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.VB C., WB A., JJ H., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inc CCG . 2015. Molecular Operating Environment (MOE), 2015.01. 1010 Sherbooke StWest, Suite #910, Montr QC, Canada, H3A 2R7. Published online. [Google Scholar]
- 27.Abraham M.J., Murtola T., Schulz R., et al. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015 doi: 10.1016/j.softx.2015.06.001. Published online. [DOI] [Google Scholar]
- 28.MacKerel A.D., Jr., Brooks C.L., III, Nilsson L., Roux B., Won Y., Karplus M. The Encyclopedia of Computational Chemistry. 1998. CHARMM: the energy function and its parameterization with an overview of the program. [Google Scholar]
- 29.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007 doi: 10.1063/1.2408420. Published online. [DOI] [PubMed] [Google Scholar]
- 30.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981 doi: 10.1063/1.328693. Published online. [DOI] [Google Scholar]
- 31.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996 doi: 10.1016/0263-7855(96)00018-5. Published online. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen E.F., Goddard T.D., Huang C.C., et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004 doi: 10.1002/jcc.20084. Published online. [DOI] [PubMed] [Google Scholar]
- 33.Multiple sequence alignment - CLUSTALW. Accessed April 8, 2020. https://www.genome.jp/tools-bin/clustalw.
- 34.Thompson J.D., Higgins D.G., Gibson T.J., Clustal W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 doi: 10.1093/nar/22.22.4673. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku316. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A R., M L.J. Methods for the refinement of protein structure 3D models. Int. J. Mol. Sci. 2019;20(9) doi: 10.3390/IJMS20092301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu L., Chen Q., Liu K., et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6(1):1–12. doi: 10.1038/s41421-020-00210-9. 2020 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meekins D.A., Morozov I., Trujillo J.D., et al. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg. Microb. Infect. 2020 doi: 10.1080/22221751.2020.1831405. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Low-level of infection with COVID-19 in pet dog. Accessed April 8, 2020. https://www.info.gov.hk/gia/general/202003/04/P2020030400658.htm.
- 40.Sit T.H.C., Brackman C.J., Ip S.M., et al. Infection of dogs with SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2334-5. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z., Zhang Y., Liu K., et al. The molecular basis for SARS-CoV-2 binding to dog ACE2. Nat. Commun. 2021;12(1):1–10. doi: 10.1038/s41467-021-24326-y. 2021 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W., Zhang C., Sui J., et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005 doi: 10.1038/sj.emboj.7600640. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W., Wong S.-K., Li F., et al. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Virol. 2006;80(9):4211–4219. doi: 10.1128/jvi.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren W., Zhu Y., Wang Y., et al. Comparative analysis reveals the species-specific genetic determinants of ACE2 required for SARS-CoV-2 entry. PLoS Pathog. 2021;17(3) doi: 10.1371/JOURNAL.PPAT.1009392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexander M.R., Schoeder C.T., Brown J.A., et al. Predicting susceptibility to SARS-CoV-2 infection based on structural differences in ACE2 across species. Faseb. J. 2020;34(12):15946–15960. doi: 10.1096/FJ.202001808R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu F., Zhao S., Yu B., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X., Chen D., Szabla R., et al. Broad and differential animal ACE2 receptor usage by SARS-CoV-2. J. Virol. 2020 doi: 10.1128/JVI.00940-20. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.