Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has stimulated the search for effective drugs for its prevention and treatment. Natural products are an important source for new drug discovery. Here, we report that, NK007(S,R), a tylophorine malate, displays high antiviral activity against SARS-CoV-2 with an EC50 0.03 μM in vitro, which is substantially lower than that of remdesivir (EC50: 0.8 μM in vitro), the only authorized drug to date. The histopathological research revealed that NK007(S,R) (5 mg/kg/dose) displayed a protection effect in lung injury induced by SARS-CoV-2, which is better than remdesivir (25 mg/kg/dose). We also prepared two nanosized preparations of NK007(S,R), which also showed good efficacy (EC50: NP-NK007, 0.007 μM in vitro; LP-NK007, 0.014 μM in vitro). Our findings suggest that tylophora alkaloids, isolated from the traditional Chinese medicine Cynanchum komarovii AL, offer a new skeleton for the development of anticoronavirus drug candidate.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Traditional Chinese medicine, Tylophorine, Nanosized preparation
As of September 1, 2021, there have been 217 558 771 reported cases of coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the disease has led to 4 517 240 deaths in more than 200 countries.1,2 Kissler and co-workers reported that one-off blocking measures cannot prevent the spread of SARS-CoV-2, and the United States may need to continue social isolation measures until 2022.3 So far, there are currently no specific drugs or vaccines available for the treatment of SARS-CoV-2 infected patients, the only authorized drug by the United States Food and Drug Administration is the nucleotide analogue remdesivir.4,5 Related studies show that remdesivir has a strong resistance to SARS-CoV-2 in vitro.6 The COVID-19 pandemic has stimulated the search for effective drugs for prevention and treatment of the disease.
The response to a viral pandemic requires the simultaneous implementation of short-term measures and long-term planning. The use of existing drugs is the focus of short-term measures, and indeed a series of first-line drugs that have been found to have potential utility for treatment of COVID-19 are currently being explored, including ritonavir and lopinavir,7,8 ribavirin,9 chloroquine and hydroxychloroquine,10 and favipiravir and remdesivir.11 Long-term planning focuses on the identification of druggable targets and the discovery and development of new drug molecules. Progress toward target-based design of new drugs against COVID-19 will undoubtedly be facilitated by the recent determination of the structures of the main protease (Mpro)12 and the RNA-dependent RNA polymerase13 from SARS-CoV-2. The advantages of target-based drug design are the clear target and mechanism and high selectivity, but this approach is subject to the development of drug resistance.
An alternative approach is to design new drugs based on natural products, which are excellent sources of molecules with novel chemical structures and unique mechanisms of action. Natural products often act on multiple targets, which tends to hinder the development of drug resistance. However, this approach is relatively labor-intensive, and the determining the mechanism of action of natural products can be difficult and time-consuming. Nevertheless, they have been an important source of new drugs14−16 and may be useful for the identification of molecules with activity against SARS-CoV-2.
SARS-CoV-2 is an enveloped single-stranded RNA virus. The spike protein on the viral envelope binds to the ACE2 receptors of host cells, and a viral protease can assist virus invasion. After the virus enters the cell, its genes are released, the viral replicase and transcriptase are synthesized, and RNA replication and transcription are accomplished by an RNA-dependent RNA polymerase. Then structural proteins are synthesized, and finally new virus particles are assembled and released. All these steps of the virus life cycle are potential targets for drug therapy,6,17 and a number of small-molecule drugs have been shown to inhibit various viral processes at micromolar concentrations.12,18
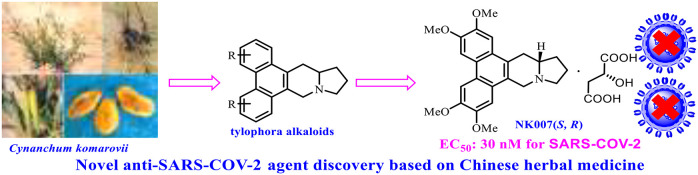
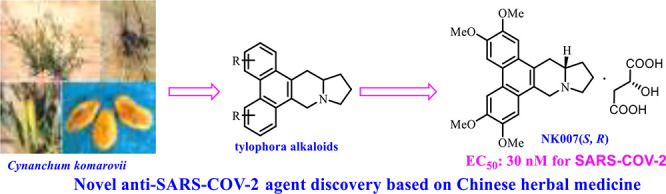
Traditional Chinese medicines are a potential source of therapeutic small molecules, and the use of such medicines for the treatment of COVID-19 patients has recently been reviewed.19 One important traditional Chinese medicine is Cynanchum komarovii AL, which has been used to treat fever, diarrhea, and cholecystitis.20 This herbaceous plant, which is widely distributed in arid sandy areas of northwest China, belongs to the genus of Cynanchum Linn. in the family Asclepiadaceae.21
In work aimed at generating active lead compounds based on traditional Chinese medicines, we discovered that Cynanchum komarovii AL has good antiviral activity against tobacco mosaic virus (TMV) and that the active components were tylophora alkaloids, such as tylophorine.22 However, the tylophorine content of the plants is very low, and the molecule is light-sensitive and poorly soluble. In efforts to overcome these disadvantages, we synthesized various tylophora alkaloids (Figure 1),23 and we designed and synthesized a series of derivatives and determined their anti-TMV activities.24−27 In addition, our evaluation of the bioactivity spectrum of these compounds showed that they have good anticancer,28 anti-HIV,29 and immunomodulatory activities.30,31 As an optimized compound, a racemate of tylophorine malate (designated NK007) is being industrialized as an anti-TMV candidate. Herein, we report that we have prepared optically pure NK007(S,R) (Figure 1) and evaluated its ability to inhibit SARS-CoV-2.
Figure 1.
Discovery of NK007(S,R).
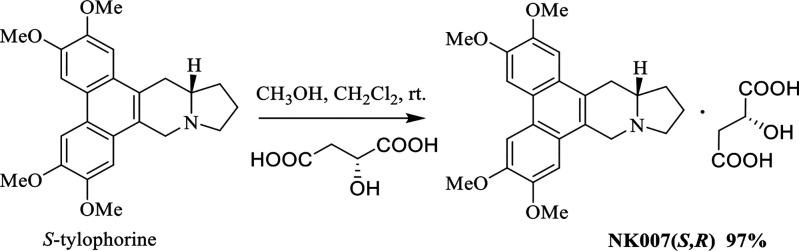
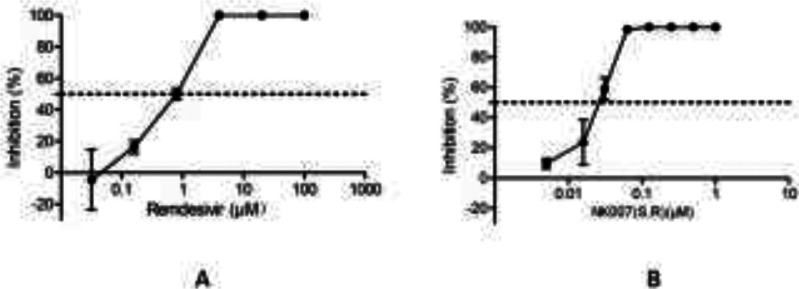
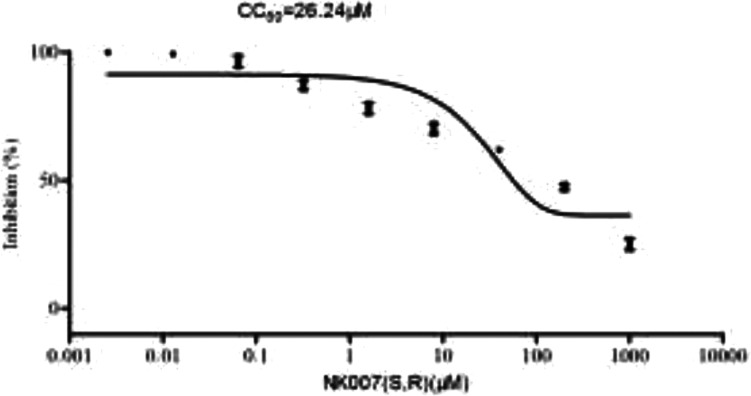
NK007(S,R) was synthesized from S-tylophorine24 in 97% yield (Scheme 1), and its in vitro activity against SARS-CoV-2 was tested by means of a previously reported method10 with remdesivir as a control. Under our test conditions, the EC50 of remdesivir is 0.8 μM (Figure 2A), whereas that of NK007(S,R) is substantially lower at 0.03 μM (Figure 2B). Furthermore, safety tests showed that NK007(S,R) is safe in the test state (CC50: 26.24 μM); the safety index is 868 (Figure 3).
Scheme 1. Synthesis of NK007(S,R).
Figure 2.
Determination of the EC50 values of remdesivir and NK007(S,R).
Figure 3.
Determination of the CC50 value of NK007(S,R).
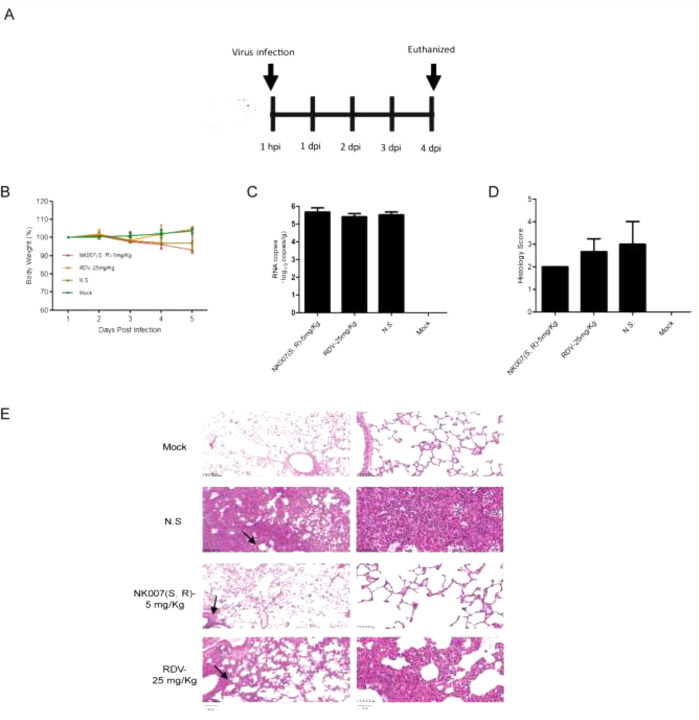
NK007(S,R) was further used to study the therapy outcome in the COVID-19 rat model. A COVID-19 golden hamster rat model was generated according to a recently published paper.32 Briefly, golden hamster rats were randomly divided into four groups. Rats were then infected intranasally with SARS-CoV-2 (1 × 105 PFU). One hour after SARS-CoV-2 infection, rats in the NS (normal saline, 0.9% NaCl) group, the remdesivir group, and the NK007(S,R) group received normal saline, remdesivir, or NK007(S,R) intranasally. The Mock group is mock-infected rats. After virus infection at day 0, rats received NK007(S,R) or remdesivir or NS treatment 3 times as shown in Figure 4A. All rats were euthanized at day 4, and several parameters were measured, including body weight loss, SARS-CoV-2 RNA copies in the lungs, and pathological change in lung tissues.
Figure 4.
NK007(S,R) and remdesivir (RDV) inhibit virus replication in lungs of COVID-19 hamster rats. (A) Schematic diagram of NK007(S,R) or remdesivir treated COVID-19 hamster rats. (B) Weight loss of infected rats treated with NK007 or remdesivir. (C) Viral RNA copies in rat lungs at day 4 (unpaired t test). (D) Histological scores of lung inflammation of NK007(S,R) or remdesivir treated SARS-CoV-2 infected rats (unpaired t test). (E) Representative hematoxylin-eosin (HE) staining of lungs from rats harvested at day 4. Bar: 100 μm.
NS-treated COVID-19 hamsters had a significant body weight loss compared with Mock hamsters on day 4 post infection (p value = 0.0093), while NK007(S,R) or remdesivir treated hamsters were with no significant body weight loss when compared to the Mock hamsters (Figure 4B). We found that the average number of viral RNA copies in the lungs of NK007(S,R) (∼105.7) or remdesivir (∼105.4) treated rats have no significant difference with NS-treated infected rats (∼105.5) (Figure 4C). And the histopathological changes in rat lung tissues are a key index to assess the therapy effects of NK007(S,R) or remdesivir in COVID-19 golden hamster rats. The lungs of rats were assessed by grading the injuries in accordance with the International Harmonization of Nomenclature and Diagnostic Criteria (INHAND) scoring standard. As shown in Figure 4D,E, the average histopathological score of virus-infected rats in NS group was approximately 3, along with an infiltration of lymphocytes and neutrophils, and the alveolar septum, bronchus, and perivascular interstitium were significantly widened. For rats infected by SARS-CoV-2, treatment of remdesivir at a dose of 25 mg/kg got histopathological scores, approximately 2.7 where the alveolar septum, bronchus, and perivascular interstitium were obviously widened, along with an infiltration of some of lymphocytes and neutrophils. Treatment of NK007(S,R) at a dose of 5 mg/kg got pathological scores, approximately 2.0. NK007(S,R) treatment significantly decreased lung inflammation in SARS-CoV-2 infected rats. The local alveolar septum, bronchi, and perivascular interstitial widening were significantly decreased in the remdesivir treatment group compared with NS-treated COVID-19 hamsters. And the NK007(S,R) treated group showed the lowest lung inflammation. According to histopathological results, NK007(S,R) shows protection from lung injury induced by SARS-CoV-2 inflammation, and the therapy outcome of NK007(S,R) at a dose of 5 mg/kg is better than that of remdesivir at a dose of 25 mg/kg.
The good pharmacological activity of NK007(S,R) lays a foundation for its application in the treatment of COVID-19. We further developed two nanosized preparations of NK007(S,R) and evaluated their anti-SARS-CoV-2 activities in vitro.
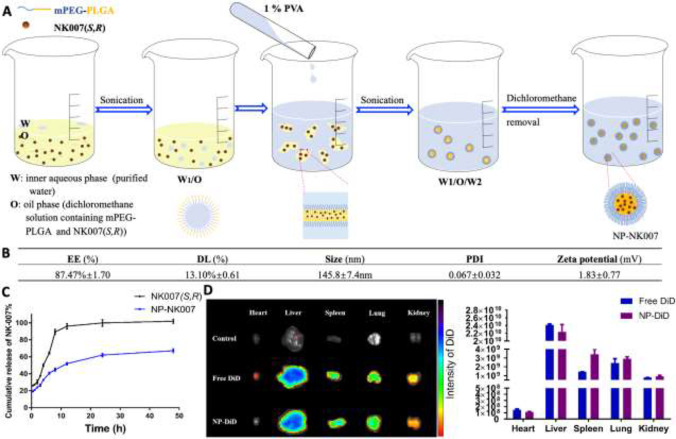
The self-assembled PEG–PLGA nanoparticle loaded with NK007(S,R) (NP-NK007) was prepared using a double-emulsion (W1/O/W2) solvent evaporation method (Figure 5A).33 The mean particle size of NP-NK007 was 145.8 ± 7.4 nm with a PDI of 0.067 ± 0.032 (Figure 5B) and a slightly positive zeta potential of 1.83 ± 0.77 mV (Figure S4). The encapsulation efficiency and drug loading of nanoparticles was 87.47% ± 1.70 and 13.10% ± 0.61 (n = 3), respectively. The release profile of NK007(S,R) out of NP-NK007 was performed (Figure 5C). NP-NK007 released NK-007(S,R) gradually. Within a 48 h period, only 66.51% ± 2.00% of NK-007(S,R) was released from NP-NK007. All the results illustrated that NK007(S,R) was efficiently entrapped in the nanoparticles and might achieve a sustained release in vivo. The biodistribution of nanoparticles was studied in healthy C57BL/6 mice. As shown in Figure 5D, NP-DiD, the self-assembled PEG–PLGA nanoparticle loaded with a lipophilic fluorescent dye DiD prepared in the same method of NP-NK007, displayed enhanced distribution in the lung compared to the free DiD. It indicated that NP-NK007 might have the potential to achieve higher antivirus activity in vivo than NK-007(S,R) because of the improved accumulation of NK-007(S,R) delivered by the nanoparticles in the lung.
Figure 5.
Preparation and characterization of NP-NK007. (A) The schematic diagram of NP-NK007 preparation processes. (B) Physical-chemical properties of NP-NK007. (C) The release profile of NK007(S,R) out of NP-NK007 in 0.5% Tween 80 PBS phosphate buffer (pH7.4). (D) The representative fluorescent image and quantitative analysis of different vital organs at 2 h after intravenous injection of NP-DiD. *p < 0.05, compared to the free DiD group.
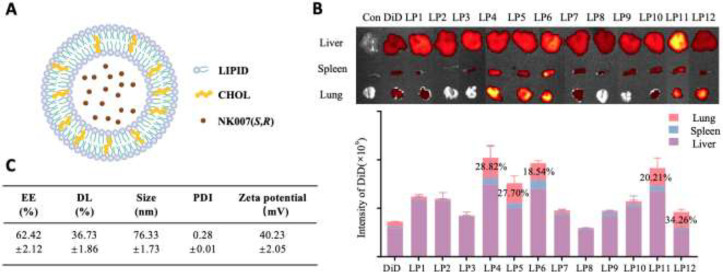
The liposome loaded with NK007(S,R) (LP-NK007) was prepared by reverse-phase evaporation method. As presented in Figure 6A, we designed another platform for lung targeted delivery of NK007(S,R) encapsulating into a lipid bilayer envelope. First, the in vivo distribution test was used to screen the lipid with the best lung targeting ability. Similarly, DiD was employed as the fluorescence probe to characterize the in vivo distribution profiles of different liposomes. As seen in Figure 6B, the relative fluorescence intensity of LP12 (DOTAP-based LP-DiD) was the strongest in the lung and was similar to free DiD in the spleen and liver. This implied that NK007(S,R) could be enriched in the lung at most when delivered by LP12 and had the least impact on other organs. Therefore, LP-NK007 containing DOTAP was selected for the subsequent experiments. The characteristics of the optimal LP-NK007 were shown in Figure 6C including EE%, DL%, particle size, and PDI. LP-NK007 had average diameters around 75 nm with narrow PDI. The encapsulation efficiency and drug loading of LP-NK007 was 62.4% and 36.7%, respectively.
Figure 6.
Structure and characterization of LP-NK007. (A) The schematic view of the LP-NK007. NK007(S,R) is encapsulated into liposomes. (B) Accumulation of LP-DiD in lungs. Representative ex vivo fluorescent images and quantitative analysis of organs at 2 h after intravenous injection of LP-DiD. (C) Characteristics of LP-NK007.
Under our test conditions, the in vitro EC50 values of NP-NK007 and LP-NK007 were 0.007 and 0.014 μM respectively, and the CC50 values of NP-NK007 and LP-NK007 were 20 and 1 μM respectively, which indicated that these two nanosized preparations could be used as drug sustained-release agents.
Starting from a traditional Chinese medicine, we obtained an anticoronavirus drug candidate with a novel skeleton. The compound, a tylophorine malate designated NK007(S,R), showed substantially higher activity (EC50: 0.03 μM) against SARS-CoV-2 than remdesivir (EC50: 0.8 μM). NK007(S,R) showed protection in lung injury induced by SARS-CoV-2 inflammation, and the therapy outcome of NK007(S,R) at a dose of 5 mg/kg was better than that of remdesivir at a dose of 25 mg/kg. We further developed two nanosized preparations of NK007(S,R) and evaluated their anti-SARS-CoV-2 activities, which indicated that these two nanosized preparations could be used as drug sustained-release agents. The findings reported herein are encouraging, and we hope they will contribute to the ongoing fight against the COVID-19 pandemic.
Glossary
ABBREVIATIONS
- COVID-19
the coronavirus disease 2019
- SARS-CoV-2
severe acute respiratory syndrome coronavirus
- ACE2
angiotensin-converting enzyme 2
- INHAND
International Harmonization of Nomenclature and Diagnostic Criteria
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.1c00481.
Experimental and synthetic methods, spectral characterization of compounds, tables and figures of EC50 and CC50 values, size distribution and ζ potential graphs (PDF)
Author Contributions
# Ziwen Wang and Fei Ye contributed equally to this work.
Author Contributions
W., W.T., and X.S. conceived the project. Z.W., H.S., Y. Liu, Y. Li, and Y.Z. are responsible for proposal, compound design, synthesis and structure determination. F.Y., L.Z., R.L., B.H., and W.W. are responsible for the test of activity. Y.F. and W.X. are responsible for the preparation of two nanosized preparations of NK007(S,R). Y.D. is responsible for providing sample.
This study was supported by Natural Science Fund of China (21732002, 21772145) and Frontiers Science Center for New Organic Matter, Nankai University (63181206). Dedicated to the 100th anniversary of Chemistry at Nankai University.
The authors declare no competing financial interest.
This article is made available via the ACS COVID-19 subset for unrestricted RESEARCH re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic.
Supplementary Material
References
- World Health Organization . WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/, 2021.
- Burki T. Coronavirus in China. Lancet Respir. Med. 2020, 8, 238. 10.1016/S2213-2600(20)30056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissler S.; Tedijanto C.; Goldstein E.; Grad Y.; Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science 2020, 368, 860–868. 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indari O.; Jakhmola S.; Manivannan E.; Jha H. An update on antiviral therapy against SARS-CoV-2: How far have we come?. Front. Pharmacol. 2021, 12, 632–677. 10.3389/fphar.2021.632677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus (COVID-19) Update: FDA issues emergency use authorization for potential covid-19 treatment: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment.
- Sanders J.; Monogue M.; Jodlowski T.; Cutrell J. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. JAMA 2020, 323, 1824–1836. 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Chen N.; Zhou M.; Dong X.; Qu J.; Gong F.; Han Y.; Qiu Y.; Wang J.; Liu Y.; Wei Y.; Xia J.; Yu T.; Zhang X.; Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020, 395, 507–513. 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibzadeh P.; Stoneman E. The novel coronavirus: A bird’s eye view. Int. J. Occup. Environ. Med. 2020, 11, 65–71. 10.15171/ijoem.2020.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.; Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discovery 2020, 19, 149–150. 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Yao X.; Ye F.; Zhang M.; Cui C.; Huang B.; Niu P.; Liu X.; Zhao L.; Dong E.; Song C.; Zhan S.; Lu R.; Li H.; Tan W.; Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020, 71, 732–739. 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Cao R.; Zhang L.; Yang X.; Liu J.; Xu M.; Shi Z.; Hu Z.; Zhong W.; Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z.; Du X.; Xu Y.; Deng Y.; Liu M.; Zhao Y.; Zhang B.; Li X.; Zhang L.; Peng C.; Duan Y.; Yu J.; Wang L.; Yang K.; Liu F.; Jiang R.; Yang X.; You T.; Liu X.; Yang X.; Bai F.; Liu H.; Liu X.; Guddat L.; Xu W.; Xiao G.; Qin C.; Shi Z.; Jiang H.; Rao Z.; Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature 2020, 582, 289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Yan L.; Huang Y.; Liu F.; Zhao Y.; Cao L.; Wang T.; Sun Q.; Ming Z.; Zhang L.; Ge J.; Zheng L.; Zhang Y.; Wang H.; Zhu Y.; Zhu C.; Hu T.; Hua T.; Zhang B.; Yang X.; Li J.; Yang H.; Liu Z.; Xu W.; Guddat L.; Wang Q.; Lou Z.; Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues T.; Reker D.; Schneider P.; Schneider G. Counting on natural products for drug design. Nat. Chem. 2016, 8, 531–541. 10.1038/nchem.2479. [DOI] [PubMed] [Google Scholar]
- Atanasov A.; Waltenberger B.; Pferschy-Wenzig E.; Linder T.; Wawrosch C.; Uhrin P.; Temml V.; Wang L.; Schwaiger S.; Heiss E.; Rollinger J.; Schuster D.; Breuss J.; Bochkov V.; Mihovilovic M.; Kopp B.; Bauer R.; Dirsch V.; Stuppner H. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Dong G.; Sheng C. Structural simplification of natural products. Chem. Rev. 2019, 119, 4180–4220. 10.1021/acs.chemrev.8b00504. [DOI] [PubMed] [Google Scholar]
- Boopathi S.; Poma A.; Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2021, 39, 3409–3418. 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W.; Zhang B.; Jiang X.; Su H.; Li J.; Zhao Y.; Xie X.; Jin Z.; Peng J.; Liu F.; Li C.; Li Y.; Bai F.; Wang H.; Cheng X.; Cen X.; Hu S.; Yang X.; Wang J.; Liu X.; Xiao G.; Jiang H.; Rao Z.; Zhang L.; Xu Y.; Yang H.; Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335. 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Islam M.; Wang J.; Li Y.; Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): A review and perspective. Int. J. Biol. Sci. 2020, 16, 1708–1717. 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiangsu New Medical College. Dictionary of Chinese Crude Drugs; Shanghai Scientific Technologic Publisher: Shanghai, 1977; p 789. [Google Scholar]
- Gellert E.Structure and synthesis of phenanthroindolizidine alkaloids and some related compounds. In The Alkaloids: Chemical and Biological Perspectives; Pelletier S., Ed.; Academic Press: New York, 1987; pp 55–132. [Google Scholar]
- An T.; Huang R.; Yang Z.; Zhang D.; Li G.; Yao Y.; Gao J. Alkaloids from Cynanchum komarovii with inhibitory activity against the tobacco mosaic virus. Phytochemistry 2001, 58, 1267–1269. 10.1016/S0031-9422(01)00382-X. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Li Z.; Wang K.; Wang Q. Efficient and chirally specific synthesis of phenanthro-indolizidine alkaloids by Parham-Type cycloacylation. Eur. J. Org. Chem. 2010, 2010, 292–299. 10.1002/ejoc.200900920. [DOI] [Google Scholar]
- Wang K.; Su B.; Wang Z.; Wu M.; Li Z.; Hu Y.; Fan Z.; Mi N.; Wang Q. Synthesis and antiviral activities of phenanthroindolizidine alkaloids and their derivatives. J. Agric. Food Chem. 2010, 58, 2703–2709. 10.1021/jf902543r. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Wang L.; Ma S.; Liu Y.; Wang L.; Wang Q. Design, synthesis, antiviral activity, and SARs of 14-aminophenanthroindolizidines. J. Agric. Food Chem. 2012, 60, 5825–5831. 10.1021/jf3013376. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Wei P.; Xu X.; Liu Y.; Wang L.; Wang Q. Design, synthesis, and antiviral activity evaluation of phenanthrene-based antofine derivatives. J. Agric. Food Chem. 2012, 60, 8544–8551. 10.1021/jf302746m. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Wei P.; Liu Y.; Wang Q. D and E rings may not be indispensable for antofine: discovery of phenanthrene and alkylamine chain containing antofine derivatives as novel antiviral agents against tobacco mosaic virus (TMV) based on interaction of antofine and TMV RNA. J. Agric. Food Chem. 2014, 62, 10393–10404. 10.1021/jf5028894. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Wu M.; Wang Y.; Li Z.; Wang L.; Han G.; Chen F.; Liu Y.; Wang K.; Zhang A.; Meng L.; Wang Q. Synthesis and SAR studies of phenanthroindolizidine and phenanthroquinolizidine alkaloids as potent antitumor agents. Eur. J. Med. Chem. 2012, 51, 250–258. 10.1016/j.ejmech.2012.02.048. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Wang M.; Wang L.; Yao X.; Li Y.; Tan J.; Qiao W.; Geng Y.; Liu Y.; Wang Q. First discovery of tylophora alkaloids as HIV inhibitors. Lett. Drug Des. Discovery 2015, 12, 277–283. 10.2174/1570180811666141009235124. [DOI] [Google Scholar]
- Wen T.; Li Y.; Wu M.; Sun X.; Bao X.; Lin Y.; Hao J.; Han L.; Cao G.; Wang Z.; Liu Y.; Wu Z.; Hong Z.; Wang P.; Zhao L.; Li Z.; Wang Q.; Yin Z. Therapeutic effects of a novel tylophorine analog, NK-007, on collagen-induced arthritis through suppressing tumor necrosis factor α production and Th17 cell differentiation. Arthritis Rheum. 2012, 64, 2896–2906. 10.1002/art.34528. [DOI] [PubMed] [Google Scholar]
- Wen T.; Li Y.; Wu M.; Chen X.; Han L.; Bao X.; Wang Z.; Wang K.; Hu Y.; Zhou X.; Wu Z.; Wang P.; Hong Z.; Zhao L.; Wang Q.; Yin Z. A novel tylophorine analog NK-007 ameliorates colitis through inhibition of innate immune response. Int. Immunopharmacol. 2012, 14, 487–494. 10.1016/j.intimp.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Sia S.; Yan L.; Chin A.; Fung K.; Choy K.; Wong A.; Kaewpreedee P.; Perera R.; Poon L.; Nicholls J.; Peiris M.; Yen H. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.; Li J.; Zhou Y.; Huang L.; Li Y.; Xu J.; Fu C.; Guo X.; Yang J. Lipid nanoparticles-encapsulated YF4: A potential therapeutic oral peptide delivery system for hypertension treatment. Front. Pharmacol. 2019, 10, 102. 10.3389/fphar.2019.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.