SUMMARY
DNA polymerase θ (Polθ) confers resistance to chemotherapy agents that cause DNA-protein crosslinks (DPCs) at double-strand breaks (DSBs), such as topoisomerase inhibitors. This suggests Polθ might facilitate DPC repair by microhomology-mediated end-joining (MMEJ). Here, we investigate Polθ repair of DSBs carrying DPCs by monitoring MMEJ in Xenopus egg extracts. MMEJ in extracts is dependent on Polθ, exhibits the MMEJ repair signature, and efficiently repairs 5′ terminal DPCs independently of non-homologous end-joining and the replisome. We demonstrate that Polθ promotes the repair of 5′ terminal DPCs in mammalian cells by using an MMEJ reporter and find that Polθ confers resistance to formaldehyde in addition to topoisomerase inhibitors. Dual deficiency in Polθ and tyrosyl-DNA phosphodiesterase 2 (TDP2) causes severe cellular sensitivity to etoposide, which demonstrates MMEJ as an independent DPC repair pathway. These studies recapitulate MMEJ in vitro and elucidate how Polθ confers resistance to etoposide.
Graphical Abstract
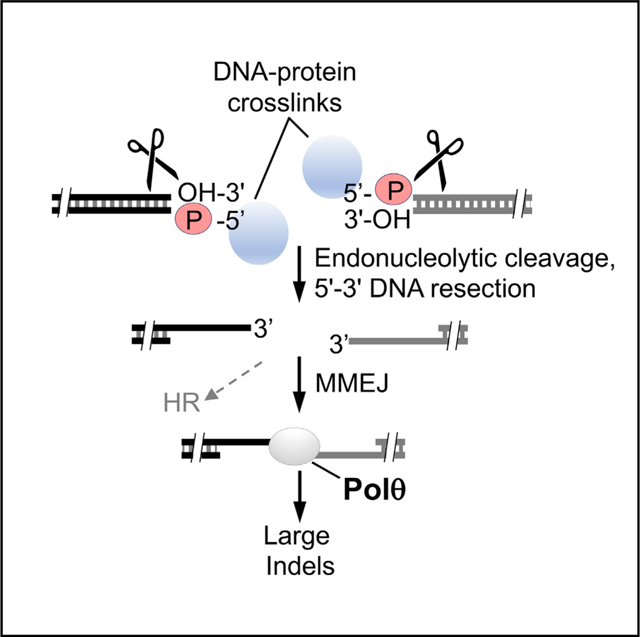
In brief
Chandramouly et al. find that Polθ protects cells from DNA-protein crosslink (DPC) agents and promotes microhomology-mediated end-joining (MMEJ) repair of DPCs occurring at double-strand breaks (DSBs) in Xenopus egg extracts as well as mammalian cells. Polθ-mediated repair of DPCs occurring at DSBs is independent of non-homologous end-joining (NHEJ) and homologous recombination (HR).
INTRODUCTION
Elucidating the mechanisms of double-strand break (DSB) repair can inform strategies for reducing cellular resistance to geno-toxic chemotherapeutics, such as those that promote DNA-protein crosslinks (DPCs). A relatively newly discovered DSB repair pathway referred to as alternative end-joining or microhomology-mediated end-joining (MMEJ) acts independently of nonhomologous end-joining (NHEJ) and homologous recombination (HR) (Kent et al., 2015; Mateos-Gomez et al., 2015; Yousefzadeh et al., 2014). The early phase of MMEJ likely requires poly-ADP ribose polymerase 1 (PARP1) that facilitates recruitment of the essential MMEJ factor DNA polymerase θ (Polθ) to sites of DNA damage (Ceccaldi et al., 2015; Mateos-Gomez et al., 2015). MMEJ functions during S/G2 cell cycle phases and acts on 3′ single-stranded DNA (ssDNA) overhangs like HR (Truong et al., 2013). Mre11 and CtIP are essential for MMEJ and initiate the resection process, resulting in 3′ ssDNA overhangs (Black et al., 2016; Sfeir and Symington, 2015). Polθ facilitates synapsis of the 3′ ssDNA termini by using minimal base pairing (≥2 bp) within microhomologous sequence tracts and then extends each overhang, resulting in stabilization of the DNA synapse (Black et al., 2016, 2019; Kent et al., 2015). Following additional end processing, DNA ligase 3 (Lig3) or ligase 1 seals the DNA (Lu et al., 2016; Sfeir and Symington, 2015).
Polθ is important for DSB repair in HR-deficient cancer cells and is therefore synthetic lethal with HR factors such as BRCA1/2 (Ceccaldi et al., 2015; Mateos-Gomez et al., 2015). Polθ also confers resistance to multiple chemotherapy agents (Higgins et al., 2010; Wang et al., 2019; Yousefzadeh et al., 2014). For example, Polθ promotes resistance to chemotherapy agents that cause DPCs at DSBs, such as toposisomerase inhibitors etoposide and camptothecin and ionizing radiation (IR) (Wang et al., 2019; Yousefzadeh et al., 2014). This suggests that Polθ plays a role in repairing toxic DPCs by the MMEJ pathway.
Previously characterized DPC repair processes include tyrosyl-DNA phsophodiesterase 1 and 2 (TDP1/2) that cleave 3′ and 5′ tyrosyl-DNA crosslinks, respectively; protease-mediated (i.e., SPRTN) pathways that act at stalled replication forks; and HR that uses MRN-CtIP to initiate endonucleolytic cleavage of DPCs at DSBs (Deshpande et al., 2016; Juarez et al., 2018; Nakano et al., 2009; Stingele et al., 2017). NHEJ also promotes DPC repair downstream of tyrosyl-DNA phosphodiesterase 2 (TDP2) (Gómez-Herreros et al., 2013). Whether Polθ contributes to these DPC repair mechanisms or acts by MMEJ to confer resistance to DNA-protein crosslinking agents has remained unknown. Here, we demonstrate that Polθ acts independently of TDP2 to confer resistance to etoposide and show that Polθ promotes MMEJ repair of DSBs harboring 5′-terminal DPCs using Xenopus egg extracts and a cellular MMEJ reporter assay. Together, these data explain how Polθ confers resistance to DPC agents such as etoposide.
RESULTS
Polθ promotes resistance to DNA-protein crosslinking agents
One of the mechanisms by which DPCs are repaired is through the MRN-CtIP complex (Stingele et al., 2017). MRN acts with CtIP to initiate 5′–3′ DNA resection during S phase and G2 that is required for HR (Syed and Tainer, 2018). The nuclease activities of Mre11 and CtIP confer resistance to etoposide and camptothecin that covalently link Top2 and Top1, respectively, onto 5′ and 3′ DNA ends (Hoa et al., 2016; Makharashvili et al., 2014; Stingele et al., 2017). MRN-CtIP and its yeast homolog complex (Mre11-Rad50-Xrs2 along with Sae2) perform endonucleolytic cleavage of DNA just upstream of DSB ends that is stimulated by 5′ biotin-streptavidin conjugation that models DPCs (Aparicio et al., 2016; Cannavo and Cejka, 2014; Deshpande et al., 2016). Additional studies have shown that BRCA1 collaborates with MRN-CtIP to facilitate DPC repair, suggesting that the HR pathway acts on DSBs containing protein adducts (Aparicio et al., 2016; Nakamura et al., 2010). Indeed, multiple studies demonstrate that HR-deficient cells are sensitive to topoisomerase inhibitors (Al Abo et al., 2014; Gómez-Herreros et al., 2013; Nakamura et al., 2010; Treszezamsky et al., 2007). Intriguingly, HR and MMEJ are thought to share the same DNA resection mechanism involving MRN and CtIP (Truong et al., 2013), which suggests that HR and MMEJ process identical DSB ends. Based on this idea, we hypothesized that Polθ-dependent MMEJ facilitates the repair of DSBs containing DPCs.
Polθ promotes cellular resistance to topoisomerase crosslinking agents etoposide and camptothecin (Wang et al., 2019; Yousefzadeh et al., 2014). To confirm this and begin to explore Polθ involvement in DPC repair, we first tested the sensitivity of isogenic Polq+/+ and Polq−/− mouse induced pluripotent cells (iPSCs) to etoposide and camptothecin. Consistent with prior reports, Polθ null cells were significantly more sensitive to these topoisomerase-DNA crosslinking agents, as revealed by clonogenic survival (Figure 1A). Polq−/− Lin−Sca1+c-Kit+ (LSK) murine stem/early progenitor bone marrow cells were also highly sensitive to etoposide, as expected (Figure 1B). To explore a more general role for Polθ in DPC repair we tested the susceptibility of Polq−/− iPSCs and Polq−/− LSK murine bone marrow cells to formaldehyde that acts as a non-specific DNA-protein crosslinking agent. Consistent with a general role for Polθ in DPC repair, we observed a significant reduction in the survival of Polq−/− versus Polq+/+ cells following formaldehyde treatment (Figure 1C). These data show that Polθ confers resistance to formaldehyde.
Figure 1. Polθ confers resistance to DNA-protein crosslinking agents.
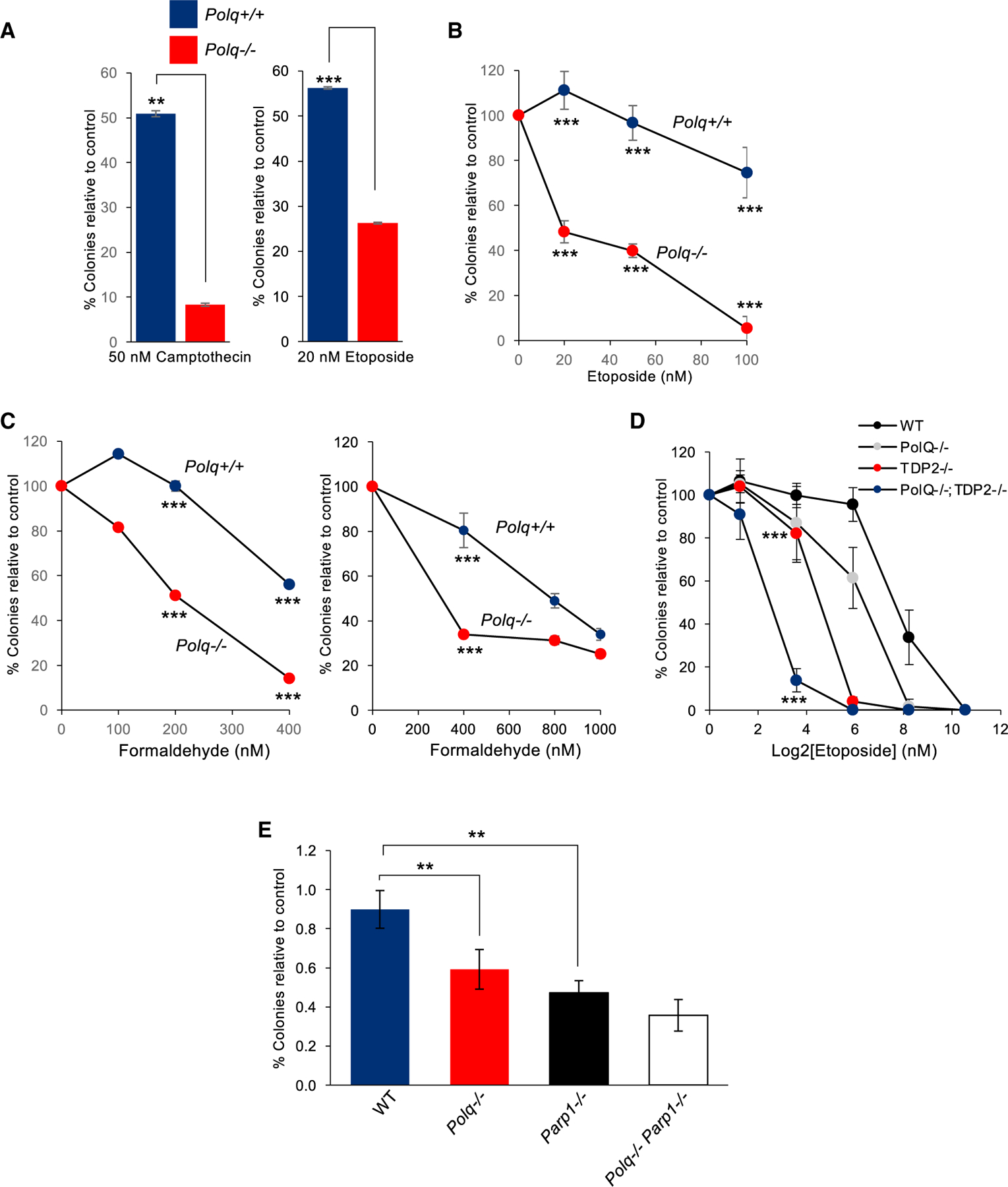
(A) Bar plots showing percentage of colonies relative to control after treatment with the indicated topoisomerase inhibitors in Polq+/+ and Polq−/− mESCs. Data represent mean. n = 3 ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance from two-sample t test: p = 0.004 for camptothecin and p = 0.001 for etoposide.
(B) Plot showing percentage of colonies relative to control after treatment with the indicated concentrations of etoposide in Polq+/+ and Polq−/− mouse bone marrow cells. Data represent mean. n = 3 ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance from two sample t test: p = 0.00017 for 20 nM, p = 0.0001 for 50 nM, p = 0.0003 for 100 nM.
(C) Plot showing percentage of colonies relative to control after treatment with indicated concentrations of formaldehyde in Polq+/+ and Polq−/− mESCs (left) and bone marrow cells (right). Data represent mean (n = 3) ± SEM (left), SD. (right). *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance from two-sample t test: p = 0.001 for 200 nM formaldehyde and p = 0.00001 for 400nM formaldehyde (left). Statistical significance from two-sample t test: p = 0.0002 for 400 nM (right).
(D) Plot showing percentage of colonies relative to control after treatment with indicated concentrations of etoposide in HCT116 cells with indicated gene knockouts. *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance from two-sample t test: p < 0.00001 for 3.8 nm.
(E) Plot showing colonies normalized to control after treatment with indicated concentrations of etoposide in Polq+/+, Polq−/−, Parp1 −/−, and Polq−/− Parp1−/− mouse bone marrow cells. *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance from two-sample t test for 50 nm: p = 0.009 for wild-type versus Polq−/−, p = 0.0013 for wild-type versus Parp1−/−, p = not significant for Parp1 −/− versus double null.
Importantly, Polθ-dependent MMEJ occurs independently of NHEJ. This infers that Polθ promotes resistance to DNA-protein crosslinking agents independently of NHEJ. TDP2 acts upstream of NHEJ to cleave 5′ DPCs, such as 5′-phosphotyrosine adducts, by its 5′-tyrosyl DNA phosphodiesterase activity (Pommier et al., 2014; Stingele et al., 2017). Thus, we envisaged that Polθ acts independently of TDP2 to confer resistance to etoposide. As a control, we found that TDP2−/− human HCT116 cells are highly sensitive to etoposide as expected (Figure 1D). POLQ−/− HCT116 cells were also significantly more sensitive to etoposide than WT HCT116 cells (Figure 1D). Dual deficiency in Polθ and TDP2, however, further increased cellular sensitivity to etoposide (Figure 1D). Hence, these genetic data demonstrate that Polθ plays a major role in 5′ DPC repair that is independent of TDP2-mediated NHEJ. To gain insight into whether other MMEJ factors contribute to etoposide resistance, we compared the effects of etoposide in mouse bone marrow cells null for Polq versus those null for Parp1, which also promotes MMEJ and facilitates Polθ recruitment to DNA breaks (Ceccaldi et al., 2015; Mateos-Gomez et al., 2015). The results demonstrate a similar sensitivity to etoposide in Polq−/− and Parp1−/− cells, and no significant increase in sensitivity is observed in Polq−/− Parp1−/− double-knockout cells (Figure 1E). These data indicate that Polθ and PARP1 act within the same pathway (e.g., MMEJ) to confer etoposide resistance.
Polθ promotes DPC repair in Xenopus egg extracts
To directly examine Polθ DPC repair, we recapitulated MMEJ by using membrane-free extracts from unfertilized interphase Xenopus eggs that are incapable of initiating replication (Liao et al., 2016). Because MRN-CtIP cleaves 5′ DNA-streptavidin linkages by Mre11 endonuclease activity (Deshpande et al., 2016) and MRN-CtIP is essential for the resection initiation step of MMEJ (Truong et al., 2013; Zhang and Jasin, 2011), we used a similar model system that uses 5′ avidin-biotin linkages (Figure 2A, top right). Mre11 and CtIP are essential for the 5′–3′ resection of DNA containing 5′ biotin-avidin adducts in Xenopus egg extracts (Liao et al., 2016). Mre11 is also essential for resecting DNA possessing 5′-phosphotyrosine adducts that model 5′ DNA-Top2 covalent lesions following etoposide treatment (Liao et al., 2016). To assess whether MMEJ is active in Xenopus egg extracts and capable of repairing DSBs with 5′ DPCs, uniformly 32P-labeled 5′ avidin-conjugated DNA 5.7 kb in length was incubated with extracts during a time course in HEPES buffer containing ATP and a ATP regenerating system as in previous studies (Liao et al., 2016). Reactions were terminated by the addition of EDTA and proteinase K that degrades protein but leaves DNA intact. Radiolabeled DNA was then resolved in native agarose gels and visualized by a phosphorimager. Consistent with previous studies (Liao et al., 2016), we observed the formation of high-molecular-weight products that were due to intermolecular DSB repair and thus concatemer formation (Figure 2A, right). The repair junction sequencing analysis below confirmed intermolecular DSB repair (Figure 3). Dimers were also formed that were subsequently converted to concatemers during the time course (Figure 2A, right). Minor degradation of the DNA was also detected during the earlier time points, which has been shown to be due to 5′–3′ resection in prior studies (Liao et al., 2016). In the absence of 5′ DNA-protein adducts (clean ends; 5′ phosphate), the majority of products resembled circular supercoiled DNA monomers that have previously been shown to be due to NHEJ and occur independently of Mre11 (Figure 2A, left; Di Virgilio and Gautier, 2005; Labhart, 1999). Concatemer products were still formed in the absence of 5′-avidin, albeit to a lesser extent (Figure 2A, left).
Figure 2. Polq promotes the repair of 5′ DPCs in Xenopus extracts.

(A) Non-denaturing gels showing a time course of DSB repair of the indicated DNA substrates in Xenopus egg extracts. % concatemer products indicated.
(B) Western blots showing the presence and absence of Polθ in mock-depleted (control) and Polθ-depleted Xenopus egg extracts. % extracts loaded indicated at right.
(C) SDS gel of purified recombinant full-length human Polθ.
(D) Non-denaturing gels showing a time course of DSB repair of the indicated DNA substrates in Xenopus egg extracts either mock depleted (control, lanes 2–5, left), Polθ depleted (lanes 6–9, left), or Polθ depleted with recombinant Polθ added (right). % concatemer products indicated.
(E) Western blots showing the presence and absence of Ku70 in mock-depleted (control) and Ku70-depleted Xenopus egg extracts. % extracts loaded indicated at right.
(F) Non-denaturing gel showing a time course of DSB repair of the indicated DNA substrate in Xenopus egg extracts either mock depleted (control, left) or Ku70 depleted (right). % concatemer products indicated.
Figure 3. Sequence analysis of 5′ DPC repair junctions.
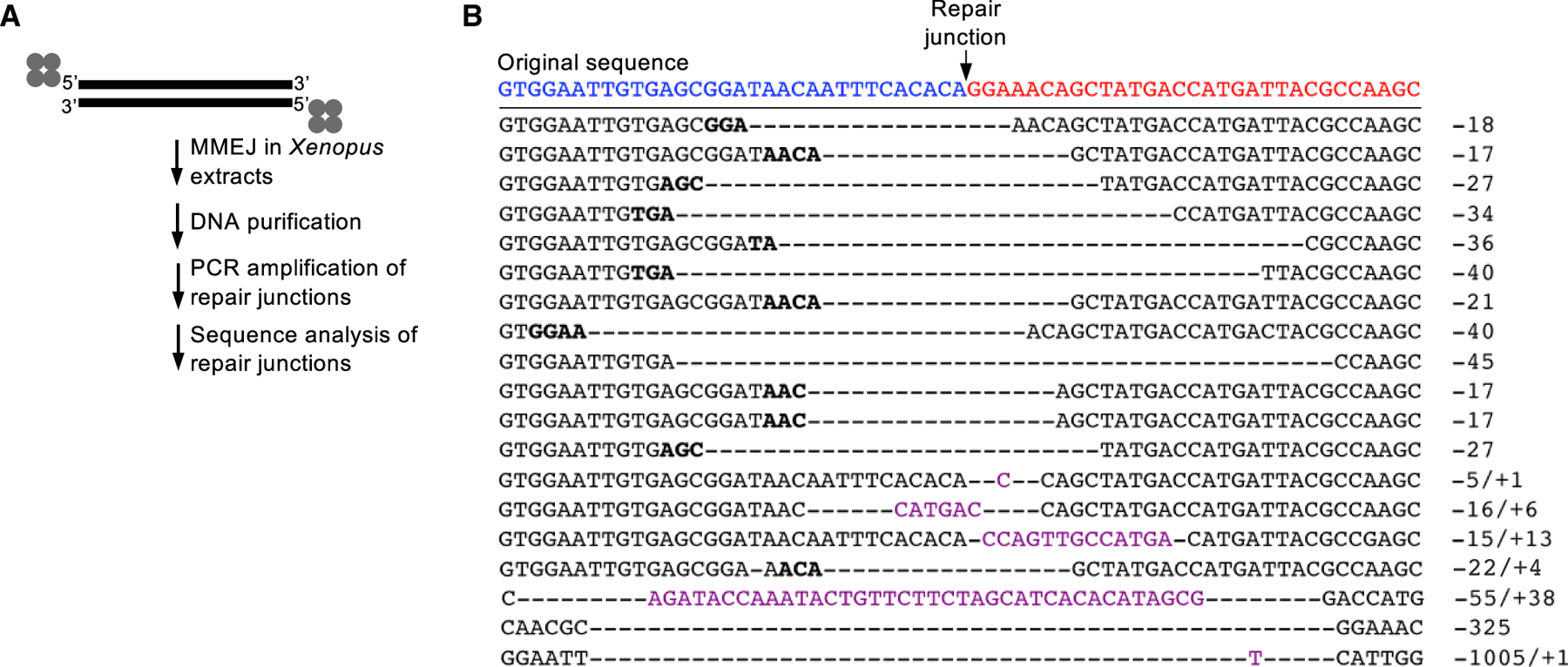
(A) Schematic of methods for sequencing DPC repair junctions.
(B) Sequences of DPC repair junctions. Sequence representative of accurate end-joining of original sequences (red and blue) indicated at top. Bold type represents microhomology. Hyphens represent deleted nucleotides. Magenta type represents insertions. Deletions and insertion sizes are indicated by negative and positive numbers, respectively, at right.
To determine whether Polθ promotes repair of the DSBs containing 5′ avidin-biotin linkages, the polymerase was depleted from extracts by using a polyclonal antibody generated against the polymerase domain of Xenopus Polθ (495 C-terminal residues). Our data demonstrate efficient (>90%) depletion of Polθ from extracts (Figure 2B). The DSB repair time course assay was repeated using Polθ-depleted versus mock-depleted extracts. Polθ-depleted extracts caused a dramatic reduction in the formation of high-molecular-weight concatemer DSB repair products versus mock-depleted extracts even after 3 h, indicating that Polθ promotes 5′ DPC repair during DNA end-joining (Figure 2D, left). The majority of 5′-avidin conjugated DNA in Polθ-depleted extracts were resected, and a smaller fraction was converted into dimers, likely due to residual Polθ (Figure 2D, left, lanes 6–9). To validate Polθ involvement in this process, we purified full-length human Polθ by using previously described methods (Figure 2C; Black et al., 2019) and then tested whether adding back recombinant Polθ to the depleted extracts rescues concatemer formation. Our recent biochemical studies demonstrate that full-length Polθ is fully active in MMEJ even at low 1- to 3-nM concentrations (Black et al., 2019). Consistent with this result, adding even small amounts of recombinant Polθ to the Polθ-depleted extracts resulted in partial rescue of concatemer DSB repair products (Figure 2D, right). We note that we were unable to further concentrate full-length recombinant Polθ due to precipitation issues. Therefore, only a modest rescue of concatemer formation was observed due to a limited concentration of the large 290-kDa enzyme. Nevertheless, our data clearly demonstrate that depletion and replenishment of Polθ abolishes and rescues, respectively, DSB repair of DNA containing 5′ DPCs.
Because MMEJ is known to act independently of NHEJ, the observed 5′ DPC repair process is expected to occur independently of essential NHEJ factors such as Ku70/80. Indeed, we found that depletion of Ku70 had no significant effect on 5′ DPC repair, as indicated by concatemer formation (Figure 2F). Controls showed efficient depletion of Ku70 from Xenopus extracts (Figure 2E) and that Ku70 depletion has no effect on concatemer formation in the absence of 5′ DPCs (Figure S1A). Taken together, the data in Figure 2 demonstrate that Polθ promotes the repair of DSBs carrying 5′ DPCs independently of NHEJ. Furthermore, because these extracts are not competent in replication initiation, the observed Polθ-mediated DPC repair also occurs independently of the replisome that can activate proteolysis-dependent DPC repair (Larsen et al., 2019).
Xenopus DPC repair reveals the MMEJ signature
To confirm that the 5′ avidin-biotin linkages are removed during Polθ-dependent MMEJ, we sequenced the repair junctions. The presence of deletions at repair junctions would confirm endonucleolytic cleavage of the DSB ends carrying avidin-biotin linkages. On the other hand, the lack of deletions would suggest the unlikely possibility that the DPCs remain intact following MMEJ. The repair signature of MMEJ is generally described as large (>5–10 bp) deletions and/or large (>2 nucleotides [nt]) insertions often flanked by microhomology tracts ≥2–4 bp in length (Chan et al., 2010; Mateos-Gomez et al., 2015; Schimmel et al., 2017; van Schendel et al., 2015; Yousefzadeh et al., 2014). This repair signature is used to designate MMEJ activity and is observed in HR-deficient cancer cells that rely on Polθ for their survival (Ahrabi et al., 2016; Ceccaldi et al., 2015; Kamp et al., 2020; Mateos-Gomez et al., 2015).
We evaluated the sequences of junctions formed by the repair of DSBs harboring 5′ avidin-biotin conjugates by purifying DNA from the Xenopus extract DSB repair reactions and then amplifying the repair junctions by PCR, followed by sequencing individual PCR products after subcloning into plasmid vectors (Figure 3A). Consistent with Polθ-dependent MMEJ activity, 100% of junctions contained large (≥5 bp) deletions and 21% of junctions exhibited large (4–38 bp) insertions (Figure 3B). Furthermore, microhomology 2–4 bp in length flanked 63% of the repair junctions (Figure 3B). Importantly, 17 out of 19 repair junctions showed that deletions occurred at both DSB ends, which unequivocally demonstrates removal of DNA termini carrying 5′ avidin-biotin linkages (Figure 3B). The overwhelming presence of the MMEJ signature in conjunction with the requirement for Polθ in efficient DSB repair of 5′ avidin-conjugated DNA shown in Figure 2 strongly supports MMEJ as the major repair mechanism at work. For example, in contrast to NHEJ, which typically results in no insertions or in very few cases a 1-bp insertion, a relatively high proportion of MMEJ junctions result in large (≥2 bp) insertions, which is due to the ability of Polθ to efficiently add nucleotides to the 3′-terminal ends of ssDNA overhangs (Kent et al., 2016). Aside from Polθ, the only other DNA polymerase capable of promoting relatively large insertions by efficient terminal transferase activity is terminal deoxynucleotidyl transferase (TdT) that exclusively functions during antibody maturation in pre-B and pre-T lymphoid cells (Loc’h and Delarue, 2018). Taken together, the observed MMEJ signature strongly supports Polθ-dependent MMEJ repair of DSBs carrying 5′ DPCs.
Polθ promotes DPC repair in mammalian cells
To confirm the ability of Polθ to promote DPC repair in mammalian cells, we developed a reporter assay that detects MMEJ of a split green fluorescent protein (GFP) expression construct conjugated with 5′-terminal DPCs (Figures 4A and 4B). Here, the upstream and downstream portions of a GFP expression vector were synthesized by PCR with a non-specific AT-rich sequence adjacent to a 6-bp overlapping sequence tract as microhomology. The DNA constructs were prepared with either 5′-biotin-streptavidin linkages on both ends (Figure 4A) or with 5′-phosphotyrosine adducts on the ends proximal to the microhomology tracts along with 5′-biotin-streptavidin linkages on the opposite ends (Figure 4B). The 5′-phosphotyrosine adducts are formed following partial proteolytic degradation of Top2 after etoposide induces covalent crosslinking of a Top2 active site tyrosine residue to the 5′-phosphate at DNA ends (Gao et al., 2012; Shi et al., 2012). Controls showed highly efficient conjugation of the 5′-biotinylated DNA constructs with streptavidin prior to transfection (Figure S1B). MMEJ of the left and right 5′-adducted DNA constructs in cells following co-transfection is expected to use the 6-bp microhomology tract and thus activate GFP expression (Figures 4A and 4B). This is based on the fact that MMEJ typically results in relatively large deletions flanked by microhomology. For example, recent studies demonstrate that microhomology tracts (≥3 bp) are efficiently used to repair CRISPR-Cas9-induced DSBs in human cells by MMEJ, resulting in predictable microhomology-mediated deletions (Grajcarek et al., 2019).
Figure 4. Polq promotes the repair 5′ of DPCs in cells.
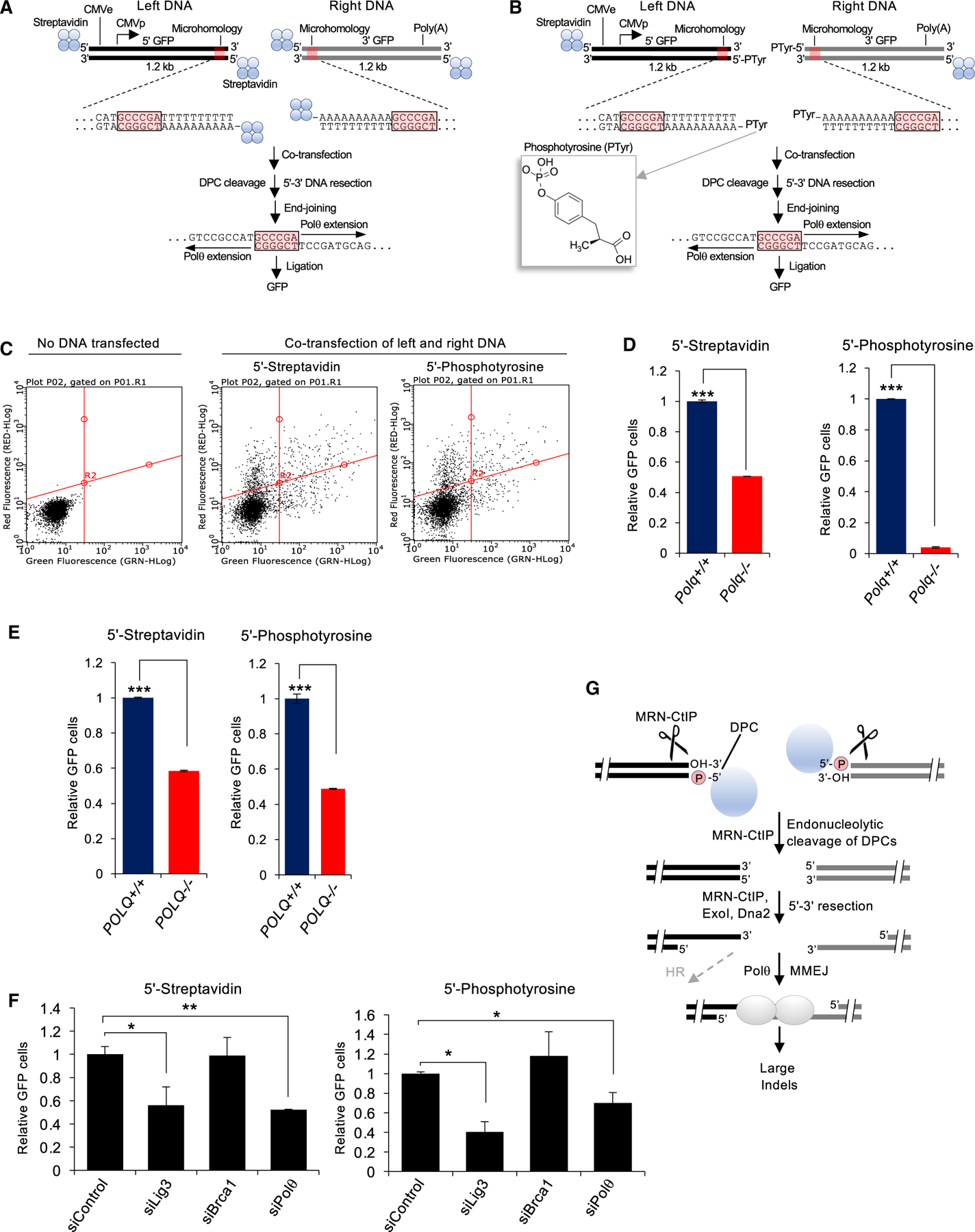
(A) Schematic of MMEJ reporter containing 5′-streptavidin-biotin linkages. Middle inset highlights internal termini of left and right MMEJ reporter DNA constructs. Schematic of MMEJ reporter assay (bottom).
(B) Schematic of MMEJ reporter containing 5′-phosphotyrosine adducts on the ends proximal to the microhomology tracts along with 5′-biotin-streptavidin linkages on the opposite ends. Middle inset highlights internal termini of left and right MMEJ reporter DNA constructs. Structure of phosphotyrosine is shown on the left. Schematic of MMEJ reporter assay (bottom).
(C) FACS plots showing green fluorescent protein (GFP) and red fluorescent protein (RFP) in Polq+/+ mESCs following no transfection (left), co-transfection of the left and right streptavidin-biotin GFP reporter DNA constructs along with mCherry (middle), and co-transfection of the left and right phosphotyrosine GFP reporter DNA constructs along with mCherry (right).
(D) Bar plot showing relative GFP following co-transfection of left and right MMEJ reporter DNA constructs conjugated with streptavidin (left) and following co-transfection of left and right MMEJ reporter DNA constructs conjugated with phosphotyrosine (right) in Polq+/+ and Polq−/− mESCs. GFP+ frequencies are normalized to transfection efficiency and shown relative to Polq+/+ cells (Polq+/+ = 1). Data were pooled from two separate experiments and performed in triplicate for each condition. ±SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance from two-sample t test between Polq+/+ versus Polq−/−. p = 0.0003 (left). p = 0.001 (right).
(E) Same as in (D) in POLQ+/+ and POLQ−/− HEK293T cells. GFP+ frequencies are normalized to transfection efficiency and shown relative to Polq+/+ cells (Polq+/+ = 1). Data are pooled from two separate experiments and performed in triplicate for each condition. ±SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance from two-sample t test between POLQ+/+ versus POLQ−/−. p = 0.00006 (left). p = 0.0001 (right)
(F) Bar plot showing relative GFP following co-transfection of left and right MMEJ reporter DNA constructs conjugated with streptavidin (left), following co-transfection of left and right MMEJ reporter DNA constructs conjugated with phosphotyrosine (right) in Polq+/+ mESCs and following co-transfection of siRNA control, siRNA against LIG3, siRNA against BRCA1, and siRNA against Polθ in Polq+/+ mESCs. GFP+ frequencies are normalized to transfection efficiency and shown relative to non-targeting siRNA (siControl = 1). Data were pooled from two separate experiments and performed in triplicate for each condition. ±SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance from two-sample t test between siControl versus siLig3, p = 0.048; siControl versus siPolθ, p = 0.009 (left); between siControl versus siLig3, p = 0.01; siControl versus siPolθ, p = 0.02 (right).
(G) Model of Polθ MMEJ repair of 5′ DPCs.
We co-transfected the left and right DNA constructs containing either 5′-streptavidin or 5′-phosphotyrosine adducts proximal to the microhomology tracts, along with a mCherry expression vector as an internal transfection control, into Polq+/+ and Polq−/− mouse embryonic stem cells (mESCs) (Mateos-Gomez et al., 2017) and POLQ+/+ and POLQ−/− HEK293T cells generated by CRISPR-Cas9 engineering (Figure S1C). At 72-h post-transfection, %GFP and %mCherry were measured by fluorescence activated cell sorting. Transfection efficiency was measured using mCherry. GFP+ frequencies were normalized to both transfection efficiency and parallel control samples. Representative fluorescence-activated cell sorting (FACS) plots demonstrate activation of GFP following co-transfection of the left and right GFP DNA reporter constructs with 5′-streptavidin and 5′-phosphotyrosine adducts, indicating MMEJ (Figure 4C). We note that the left and right PCR DNA constructs were treated with DpnI prior to their purification in order to degrade the CMV-GFP plasmid PCR template and thus prevent any possible background GFP expression. The absence of Polθ in both mESCs and 293T cells resulted in significantly lower GFP following transfection of DNA constructs with 5′-streptavidin and 5′-phosphotyrosine (Figures 4D and 4E; Figures S2A and S2B). Hence, Polθ promotes the repair of DSBs carrying 5′-streptavidin and 5′-phosphotyrosine adducts, which requires deletion of the DNA termini and use of the 6-bp microhomology. As a control, we found that a similar reduction in GFP was observed following siRNA suppression of Polθ (Figure 4F; Figures S2C and S2F). siRNA knockdown of the MMEJ factor Lig3 also significantly reduced MMEJ of both 5′-adducted DNA constructs, whereas knockdown of BRCA1 had no effect, as expected (Figure 4F; Figures S2C and S2F). Taken together, these data confirm that both 5′-adducted GFP reporters are repaired by MMEJ. Additionally, we found that expression of Polθ WT, but not a polymerase mutant, rescues MMEJ of the 5′-adducted DNA substrates (Figures S2D and S2E). The results of these MMEJ reporter assays are consistent with those obtained from Xenopus egg extracts in which similar DNA substrates with 5′ protein (avidin) adducts are repaired by Polθ-dependent MMEJ. Furthermore, because replication initiation requires specific replication origin protein loading and strict cell-cycle control, and exogenous DNA is not replicated in mammalian cells in the absence of large T-antigen and its replication origin, our GFP assay reports on MMEJ events that occur independently of the replication fork, similar to the Xenopus MMEJ system. Hence, our data indicate that Polθ-dependent MMEJ is capable of promoting 5′ DPC repair independently of the replisome.
DISCUSSION
Here, we discover the ability of Polθ-dependent MMEJ to repair DSBs possessing 5′ DPCs. Consistent with these findings, we discover that Polθ confers cellular resistance to formaldehyde and confirm Polθ’s ability to promote cellular tolerance to etoposide and camptothecin. We also find that Polθ acts independently of TDP2 to promote resistance to etoposide, which indicates MMEJ as an independent mechanism of 5′ DPC repair.
Separate studies have shown that MRN along with CtIP promotes endonucleolytic cleavage of DSBs carrying 5′ DPCs (Deshpande et al., 2016; Liao et al., 2016; Figure 4G). In fact, 5′-streptavidin stimulates endonucleolytic activity of MRN-CtIP and the yeast ortholog MRX-Sae2 complex (Cannavo and Cejka, 2014; Deshpande et al., 2016). Because MRN-CtIP promotes MMEJ, and MMEJ and HR share the same resection initiation process (Truong et al., 2013), we propose a model whereby MRN-CtIP endonucleolytic activity cleaves DPCs at DSB ends (Figure 4G). This model is consistent with the requirement for Mre11 and CtIP in resection of DSBs containing 5′ DPCs in Xenopus egg extracts (Liao et al., 2016) and biochemical studies of MRN-CtIP and MRX-Sae2 (Cannavo and Cejka, 2014; Deshpande et al., 2016). Once the DNA-protein adduct is cleaved, the resection machinery can complete its process along with additional nucleases and helicases (i.e., Dna2, BLM, and ExoI) that ultimately leads to MMEJ or HR (Figure 4G).
Previous studies demonstrated that the BRCA1-CtIP interaction is needed for MRN-CtIP repair of 5′-Top2 DNA adducts in replication-competent Xenopus egg extracts (Aparicio and Gautier, 2016). MMEJ repair of 5′ DPCs in the replication-incompetent extracts used here, however, shows no dependency on BRCA1 (Figure S3). We reconcile the difference in the requirement for BRCA1 in 5′ DPC repair in the prior report due to replication fork coupling. For example, replication fork collision with Top2-DNA crosslinks was implicated in triggering BRCA1-dependent MRN-CtIP 5′ DPC repair in replication-competent Xenopus extracts (Aparicio and Gautier, 2016). Thus, in the context of the replication fork, BRCA1 likely plays a primary role in facilitating DPC repair by HR, whereas Polθ MMEJ probably acts as a backup mechanism. This is consistent with the synthetic lethal relationship between BRCA1 and Polθ. Despite the ability of Polθ to promote DPC repair independently from the replication fork in our assays, MMEJ is likely to be activated following replication-DPC collisions because Polθ confers resistance to topoisomerase inhibitors. Future studies will be needed to elucidate the upstream signaling mechanisms responsible for coordinating Polθ MMEJ repair of DPCs.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact: Richard T. Pomerantz (Richard.Pomerantz@jefferson.edu).
Materials availability
Cell lines generated in this study are available upon request from the lead contact.
Data and code availability
This study did not generate any unique datasets or code. The datasets supporting this study have not been deposited in a public repository.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Lin-cKIT+ primary cells, Polq+/+, Polq−/− mESCs and POLQ−/− HEK293T cells were cultured in appropriate media (more information are provided in Method details) and incubated at 37C with 5% CO2.
METHOD DETAILS
Cell lines
The following oligonucleotides (IDT, IA) were used for constructing plasmids that express PolQ or TDP2 gRNAs (PAM sequences are in parenthesis and not part of oligos): TTCATATAGGAGTTCATCA(TGG)/TGATGAACTCCTATATGAA (PolQ); ATATAACTGTAGC TGACTC(TGG)/GAGTCAGCTACAGTTATAT (TDP2). Mutant cells were isolated by the HPRT co-targeting method as previously described (Liao et al., 2015). PolQ status was determined by amplifying the target region from wild-type and mutant cells and then Sanger sequencing (Genewiz, NJ). TPD2 status was determined by western blot with an anti-TDP2 antibody (Bethyl Laboratories, TX) against samples prepared from wild-type and mutant cells. Primary murine bone marrow cells were isolated from mice. Polq+/+ and Polq−/− mice were obtained by breeding Polq−/− mice (JAX #006194). Parp1−/− mice were provided by Roberto Caricchio (Temple University). They were crossed with Polq−/− mice to make the Polq−/−Parp1−/− mice. Lin-cKIT+ primary cells were isolated by magnetic sorting using the EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit (StemCell) followed by EasySep Mouse CD117 (cKIT) Positive Selection Kit (StemCell), and were subsequently cultured in IMDM + 10% FBS supplemented with a cocktail of growth factors (3 ng/mL IL3, 3 ng/mL IL6, 5 ng/mL SCF). Polq+/+, Polq−/− mESCs and iPSCs were generated in prior studies as described (Mateos-Gomez et al., 2015, 2017). POLQ−/− HEK293T cells were generated by CRISPR-Cas9 engineering and were purchased from Genscript, Piscataway, NJ. HEK293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum, 2mM L-glutamine, non-essential amino acids, and penicillin/streptomycin. mES and iPS cells were cultured in DMEM supplemented with 15% fetal bovine serum (GIBCO), 2 mM L-glutamine (Sigma), 100 U/ml penicillin (Sigma), 0.1 μg/ml streptomycin (Sigma), 0.1 mM non-essential amino acids (Invitrogen), leukemia inhibitory factor (LIF) and 2-Betamercaptoethanol (GIBCO 21985). All cell lines were cultured at 37°C in a humidified incubator containing 5% CO2.
Clonogenic survival assays
Polq+/+ and Polq−/− iPSCs from a stock culture were plated on six well plates at 500 cells/well. Cells were treated with the indicated concentrations of camptothecin, etoposide or formaldehyde 24 h after plating. Medium was replaced every three days with the chemicals until the colonies were ready for staining in 8–10 days. Medium was removed from plates, cells were rinsed with PBS. Fixation was carried out with Acetic acid/methanol (1:7) for 30 minutes followed by staining of colonies with 0.5% crystal violet for 2 hours at room temp. Dishes were rinsed with water and left for drying overnight at room temp. Counting of colonies was performed with ImageJ software. Polq+/+, Polq−/−, Parp1−/− and Polq−/− Parp1−/− Lin−Sca1+c-Kit+ (LSK) murine stem/early progenitor bone marrow cells were plated at 104 cells per well in a 96 well plate and treated with etoposide or formaldehyde at the indicated concentrations. After 48 h, cells were counted via trypan blue exclusion and immediately plated in MethoCult (StemCell) containing threshold level (1/10X) of growth factors. Colonies were counted after approximately 7 days. HCT116 WT and mutant cells were grown in DMEM supplemented with 10% fetal bovine serum, 2mM L-glutamine, non-essential amino acids, and penicillin/streptomycin at 37°C in a humidified incubator containing 5% CO2. Wild-type HCT116 cells and the indicated mutant HCT116 cells were seeded in 6 well plates at 500 cells/well. After two days, etoposide was added to each well at the indicated concentrations. Plates were incubated for 9 more days and then stained with crystal violet to visualize colonies. Colonies were counted and the averages and standard deviations of surviving colonies as percentages of no drug controls were calculated and plotted. For comparisons of averages, a one-tailed t test was conducted at 95% confidence interval (C.I.).
Preparation of Xenopus egg extracts and immunodepletion
Membrane-free extracts derived from unfertilized interphase Xenopus eggs were prepared as described (Liao et al., 2016). For immunodepletions, extracts (40μl + 20μl ELB (10mM HEPES (pH7.5), 250mM sucrose, 2.5mM MgCl2, 50mM KCl, 1mM DTT)) were incubated with 20 μl Protein A Sepharose beads pre-coated with 80 μl of the rabbit serum or no serum at 4°C for 1.5 h. After two rounds of depletion, extracts were saved as 5 μl aliquots at −80°C. Rabbit antibodies against Xenopus Ku and Polθ were raised against the Ku70 subunit and the C-terminal 495 amino acids of Polθ. Recombinant Xenopus Ku and Polθ (C-terminal 495 residues) were over-expressed as GST-fusion proteins in E. coli, purified as inclusion bodies, and injected into rabbits following standard procedures.
DSB repair of 5′ avidin conjugated DNA in Xenopus egg extracts
The DNA substrates were prepared by amplifying a 5.7 kb plasmid using Pfu DNA polymerase (Promega, WI) and oligonucleotides carrying 5′-biotin (Midland, TX) in the presence of 32P-α-dATP (Perkin Elmer). PCR products were purified by gel-filtration with Sepharose CL-2B beads (Sigma-Aldrich, MO). Peak fractions were pooled and concentrated to 37.5 ng/μl. 5′ avidin DNA was prepared by pre-incubating 5′ biotin DNA (20 ng/μl) with Neutravidin (4 μg/μl) (Pierce/ThermoScientific, IL) for 10 minutes. A typical repair assay contained 5 μl non-depleted extracts, mock depleted extracts, or extracts depleted of Polθ, Ku70 or BRCA1. 0.5 μl 10x ATP mix (20 mM ATP/200 mM phosphocreatine/0.5 mg/ml creatine kinase/50 mM DTT), 1.5 ng/μl DNA, and ELB buffer (total volume = 7.5 μl). Reactions were incubated at room temp and samples taken at the indicated times were mixed with an equal volume of 2% SDS/25 mM EDTA. At the end, samples were brought up to 10 μl with H2O and treated with 1 μl proteinase K (10 mg/ml) at room temp for 2 hours. Products were separated on 1% TAE/agarose gels by electrophoresis and gels were dried and exposed to X-ray films. For analysis of repair junctions, the 2 kb fragment bordering the junction was amplified by PCR, subcloned into a pUC vector, and sequenced by the Sanger method (Genewiz, NJ).
Construction of mutant cells
The following oligonucleotides (IDT, IA) were used for constructing plasmids that express PolQ or TDP2 gRNAs (PAM sequences are in parenthesis and not part of oligos): TTCATATAGGAGTTCATCA(TGG)/TGATGAACTCCTATATGAA(PolQ); ATATAACTGTAGCT GACTC(TGG)/GAGTCAGCTACAGTTATAT (TDP2).
Mutant cells were isolated by the HPRT co-targeting method as previously described (Liao et al., 2015). PolQ status was determined by amplifying the target region from wild-type and mutant cells and then Sanger sequencing (Genewiz, NJ). TPD2 status was determined by western blot with an anti-TDP2 antibody (Bethyl Laboratories, TX) against samples prepared from wild-type and mutant cells. Primary murine bone marrow cells were isolated from Polq+/+ and Polq−/− mice, which were obtained by breeding Polq+/− mice (JAX #006194). Lin-cKIT+ primary cells were isolated by magnetic sorting using the EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit (StemCell) followed by EasySep Mouse CD117 (cKIT) Positive Selection Kit (StemCell), and were subsequently cultured in IMDM + 10% FBS supplemented with a cocktail of growth factors (3 ng/mL IL3, 3 ng/mL IL6, 5 ng/mL SCF). Polq+/+ and Polq−/− mESCs and iPSCs were generated in prior studies as described (Mateos-Gomez et al., 2015, 2017).
Synthesis of DNA substrates for cellular MMEJ reporter assay
PCR preparation followed recommended conditions for the Phusion® High-Fidelity DNA Polymerase (New England BioLabs®) with 10ng of the 3kb pCMV-GFP plasmid in 1x Phusion HF Buffer. PCR for the left-flank DNA with 6bp of internal microhomology (PCR1.6I.SA2.) was performed with forward primer RP500B and RP506B. PCR for the right-flank DNA with 6bp of internal microhomology (PCR2.6I.SA2.) was performed with primers RP507B and RP503B. Following PCR, left or right flank DNA constructs were pooled together and digested with DpnI (New England Biolabs®) in 1X CutSmart® buffer and then purified via QIAGEN® QIAquick PCR Purification Kit. Purified PCR was then conjugated to Strepatividin (Sigma) at 110 ng/μl of PCR and 0.8 μg/μl Streptavidin in 10 mM Tris-HCl 7.5, 100 mM NaCl at 37°C for 1 hr. Conjugation was confirmed by resolution in a 0.8% agarose gel stained with ethidium bromide. To synthesize DNA substrates with 6 bp microhomology and a 5′-phosphotyrosine, this procedure was repeated using different left-flank DNA and right-flank DNA for the PCR. PCR for the left-flank DNA was performed with forward primer RP500B and reverse primer RP506-5′-phosphotyrosine (The Midland Certified Reagent Co.). PCR for the right-flank DNA was performed with forward primer RP507–5′-phosphotyrosine (The Midland Certified Reagent Co.) and reverse primer RP503B.
Cellular MMEJ reporter assay
2 × 105 mouse embryonic stem cells were transfected with 0.25 μg each of left and the right flank of GFP in suspension along with 100 ng of pCAGGS-mcherry (https://www.addgene.org/41583/) using Lipofectamine 2000 (Invitrogen). As a negative control, buffer that was used to re-suspend DNA in experimental wells was used for transfection in control wells. For HEK293T, 1 × 105 cells were plated and 24hours later, 0.25 μg each of left and the right flank of GFP in along with 100 ng of pCAGGS-mcherry was transfected using Lipofectamine 2000. For overexpression of POLQWT (https://www.addgene.org/64875/) as well as POLQ-DY2230AA polymerase mutant (https://www.addgene.org/64878/), 1 × 105 cells HEK293T cells were plated and after 24 hours later, 200 ng of either POLQWT or POLQ-DY2230AA polymerase mutant was transfected using lipofectamine 2000. 24 hours after transfection of the POLQ plasmids, 0.25 μg each of left and the right flank of GFP along with 100 ng of pCAGGS-mcherry was transfected using Lipofectamine 2000. For siRNA experiments, cells were transfected with 20 pmol siRNA along with 0.25 ug each of left and right flank of GFP, 100 ng of mCherry. Positive GFP and RFP cell frequencies were measured 3 days post transfection by flow cytometry using GUAVA easycyte 5-HT (Luminex corp) in independent replicates. For all the MMEJ reporter assays, the frequency of repair events was normalized using percentage of red fluorescence signal generated by simultaneous transfection with mcherry expression vector (pCAGGS-mcherry). For comparisons Polq+/+ versus Polq−/−, each repair value normalized to transfection efficiency is expressed relative to Polq+/+ (Polq+/+ = 1). In the case of siRNA experiments, each repair value normalized to transfection efficiency is expressed relative to non-targeting siRNA (siControl = 1). Data is represented as the mean and standard error of the mean of two independent experiments, with triplicates per condition per experiment. Statistical analysis was by two sample t test.
RT-qPCR
A portion of mES cells from MMEJ reporter assays performed with siRNA was used for RNA extraction. RNA was extracted using High Capacity cDNA Reverse Transcription Kit (Thermofisher Scientific Catalog # 4368814). Analysis of first-strand cDNA was by Power SYBR Green PCR master mix (Applied Biosystems Catalog # 4367659). An Applied Biosystem StepOne plus PCR System was used for RT-qPCR. We used conventional SYBR green RT-qPCR assays of Gapdh and the siRNA-targeted gene. Primers used for RT–qPCR:
Mouse BRCA1—sense: CGAGGAAATGGCAACTTGCCTAG;
Mouse BRCA1—antisense: TCACTCTGCGAGCAGTCTTCAG;
Mouse POLQ—sense: GTCGAGAGGAGCTTGTTTGC;
Mouse POLQ—antisense: CGCTTGTTTGTTCCTGTCCC;
Mouse LIG3—sense: AAG GCA GAC TTT GCT GTG GT
Mouse LIG3—antisense: AAT GCT TTG GAA TCG GTT TG
Mouse GAPDH—sense: CATCACTGCCACCCAGAAGACTG;
Mouse GAPDH—antisense: ATGCCAGTGAGCTTCCCGTTCAG.
mRNA was measured in triplicates with a standard curve generated for each gene using cDNA obtained from each sample. The expression level of target genes was normalized to internal Gapdh.
Immunoblotting
A portion of 293T cells from the MMEJ reporter assays performed after overexpression of POLQWT or POLQ-DY2230AA polymerase mutant was used for western blotting analysis. Cells were resuspended in IP lysis buffer (Cat. No: 87787, Thermo scientific, USA) and laemmli buffer was used to make whole-cell protein extracts. Equal amounts (20 μg) whole-cell protein lysates were separated on 4–20% bis tris gels (GenScript) by electrophoresis then transferred onto Protran BA85 nitrocellulose membrane (Whatman, Germany) and immunoblotted with antibodies against Actin (MA-5-11869,1:20000, Invitrogen) or POLQ (PA5-69577,1:500, Invitrogen) overnight followed by secondary antibodies IRDye 800CW (926-32210, 1:10000) or IRDye 680CW (926-68073, 1:10000). Blots were scanned using ODYSSEY software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Two sample t test was used in Figures 1, 4, and S2. Mean, SEM and SD values are shown, p < 0.05 was considered statistically significant. Statistically significant p values and number of replicates are indicated in the Figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PolQ polyclonal antibody | This paper | N/A |
| PolQ polyclonal antibody | Invitrogen | PA5-69577 |
| Ku70 polyclonal antibody | Yan lab | N/A |
| Actin antibody | Invitrogen | MA-5-11869 |
| Chemicals, peptides, and recombinant proteins | ||
| Etoposide | Sigma-Aldrich | E1383 |
| Camptothecin | Sigma-Aldrich | PHL89593 |
| Formaldehyde | Sigma-Aldrich | 252549 |
| Streptavadin | Sigma-Aldrich | S3762 |
| Avidin | Sigma-Aldrich | A8706 |
| POLQ | Pometantz Lab | N/A |
| Phusion® High-Fidelity PCR Kit | New England Biolabs | E0553S |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher | 11668030 |
| Pfu DNA polymerase | Promega | M7741 |
| DpnI | New England Biolabs | R0176S |
| QIAquick PCR Purification Kit | QIAGEN | 28104 |
| Experimental models: cell lines | ||
| HCT116 | Fox Chase Cancer Center | N/A |
| HCT116 POLQ−/− | This paper | N/A |
| HCT116 TDP2−/− | This paper | N/A |
| HCT116 POLQ−/−, TDP2−/− | This paper | N/A |
| mESCs | Sfeir laboratory | N/A |
| mESCs Polq−/− | Sfeir laboratory | N/A |
| iPSCs | Sfeir laboratory | N/A |
| iPSCs Polq−/− | Sfeir laboratory | N/A |
| Polq+/+ Lin−Sca1 +c-Kit+ bone marrow cells | This paper | N/A |
| Polq−/− Lin−Sca1+c-Kit+ bone marrow cells | This paper | N/A |
| Parpl−/− Lin−Sca1+c-Kit+ bone marrow cells | This paper | N/A |
| Polq−/− Parpl−/− Lin−Sca1+c-Kit+ bone marrow cells | This paper | N/A |
| POLQ−/− HEK293T cells | This paper | N/A |
| Oligonucleotides | ||
| RP 500B | IDT | N/A |
| RP 503B | IDT | N/A |
| RP506B | IDT | N/A |
| RP507B | IDT | N/A |
| RP506PT | IDT | N/A |
| RP507PT | IDT | N/A |
| Recombinant DNA | ||
| pCMV-GFP | Addgene | 11153 |
Highlights.
Polθ confers resistance to DNA-protein crosslink (DPC) agents
Polθ promotes MMEJ repair of 5′-DPCs in Xenopus egg extracts
Polθ promotes MMEJ repair of 5′-DPCs in mammalian cells
Polθ acts independently of NHEJ and HR to repair 5′-DPCs
ACKNOWLEDGMENTS
The research was funded by the following National Institutes of Health grants 1R01GM115472-01 and 1R01GM130889-01 to R.T.P., R01GM57962 to H.Y., R01CA186238 and R01CA244179 to T.S., and 5 T32 CA009009035-43 to K.S.-R. We are grateful to Dr. Sfeir for providing Polq+/+ and Polq−/− iPSCs and mESCs.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108820.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ahrabi S, Sarkar S, Pfister SX, Pirovano G, Higgins GS, Porter AC, and Humphrey TC (2016). A role for human homologous recombination factors in suppressing microhomology-mediated end joining. Nucleic Acids Res. 44, 5743–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Abo M, Dejsuphong D, Hirota K, Yonetani Y, Yamazoe M, Kurumizaka H, and Takeda S (2014). Compensatory functions and interdependency of the DNA-binding domain of BRCA2 with the BRCA1-PALB2-BRCA2 complex. Cancer Res. 74, 797–807. [DOI] [PubMed] [Google Scholar]
- Aparicio T, and Gautier J (2016). BRCA1-CtIP interaction in the repair of DNA double-strand breaks. Mol. Cell. Oncol 3, e1169343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio T, Baer R, Gottesman M, and Gautier J (2016). MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J. Cell Biol 212, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black SJ, Kashkina E, Kent T, and Pomerantz RT (2016). DNA Polymerase θ: A Unique Multifunctional End-Joining Machine. Genes (Basel) 7, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black SJ, Ozdemir AY, Kashkina E, Kent T, Rusanov T, Ristic D, Shin Y, Suma A, Hoang T, Chandramouly G, et al. (2019). Molecular basis of microhomology-mediated end-joining by purified full-length Polθ. Nat. Commun 10, 4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, and Cejka P (2014). Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514, 122–125. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, O’Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, et al. (2015). Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SH, Yu AM, and McVey M (2010). Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6, e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Lee JH, Arora S, and Paull TT (2016). Nbs1 Converts the Human Mre11/Rad50 Nuclease Complex into an Endo/Exonuclease Machine Specific for Protein-DNA Adducts. Mol. Cell 64, 593–606. [DOI] [PubMed] [Google Scholar]
- Di Virgilio M, and Gautier J (2005). Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J. Cell Biol 171, 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Huang SY, Marchand C, and Pommier Y (2012). Biochemical characterization of human tyrosyl-DNA phosphodiesterase 2 (TDP2/TTRAP): a Mg(2+)/Mn(2+)-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes. J. Biol. Chem 287, 30842–30852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Herreros F, Romero-Granados R, Zeng Z, Alvarez-Quilón A, Quintero C, Ju L, Umans L, Vermeire L, Huylebroeck D, Caldecott KW, and Cortés-Ledesma F (2013). TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 9, e1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajcarek J, Monlong J, Nishinaka-Arai Y, Nakamura M, Nagai M, Matsuo S, Lougheed D, Sakurai H, Saito MK, Bourque G, and Woltjen K (2019). Genome-wide microhomologies enable precise template-free editing of biologically relevant deletion mutations. Nat. Commun 10, 4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins GS, Prevo R, Lee YF, Helleday T, Muschel RJ, Taylor S, Yoshimura M, Hickson ID, Bernhard EJ, and McKenna WG (2010). A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 70, 2984–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa NN, Shimizu T, Zhou ZW, Wang ZQ, Deshpande RA, Paull TT, Akter S, Tsuda M, Furuta R, Tsutsui K, et al. (2016). Mre11 Is Essential for the Removal of Lethal Topoisomerase 2 Covalent Cleavage Complexes. Mol. Cell 64, 580–592. [DOI] [PubMed] [Google Scholar]
- Juarez E, Chambwe N, Tang W, Mitchell AD, Owen N, Kumari A, Monnat RJ Jr., and McCullough AK (2018). An RNAi screen in human cell lines reveals conserved DNA damage repair pathways that mitigate formaldehyde sensitivity. DNA Repair (Amst.) 72, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp JA, van Schendel R, Dilweg IW, and Tijsterman M (2020). BRCA1-associated structural variations are a consequence of polymerase theta-mediated end-joining. Nat. Commun 11, 3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, and Pomerantz RT (2015). Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol 22, 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent T, Mateos-Gomez PA, Sfeir A, and Pomerantz RT (2016). Polymerase θ is a robust terminal transferase that oscillates between three different mechanisms during end-joining. eLife 5, e13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P (1999). Ku-dependent nonhomologous DNA end joining in Xenopus egg extracts. Mol. Cell. Biol 19, 2585–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NB, Gao AO, Sparks JL, Gallina I, Wu RA, Mann M, Räschle M, Walter JC, and Duxin JP (2019). Replication-Coupled DNA-Protein Crosslink Repair by SPRTN and the Proteasome in Xenopus Egg Extracts. Mol. Cell 73, 574–588.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Tammaro M, and Yan H (2015). Enriching CRISPR-Cas9 targeted cells by co-targeting the HPRT gene. Nucleic Acids Res. 43, e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Tammaro M, and Yan H (2016). The structure of ends determines the pathway choice and Mre11 nuclease dependency of DNA double-strand break repair. Nucleic Acids Res. 44, 5689–5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loc’h J, and Delarue M (2018). Terminal deoxynucleotidyltransferase: the story of an untemplated DNA polymerase capable of DNA bridging and templated synthesis across strands. Curr. Opin. Struct. Biol 53, 22–31. [DOI] [PubMed] [Google Scholar]
- Lu G, Duan J, Shu S, Wang X, Gao L, Guo J, and Zhang Y (2016). Ligase I and ligase III mediate the DNA double-strand break ligation in alternative end-joining. Proc. Natl. Acad. Sci. USA 113, 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, et al. (2014). Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol. Cell 54, 1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, and Sfeir A (2015). Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez PA, Kent T, Deng SK, McDevitt S, Kashkina E, Hoang TM, Pomerantz RT, and Sfeir A (2017). The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol 24, 1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui KM, Tsutsui K, Hartsuiker E, et al. (2010). Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 6, e1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, et al. (2009). Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J. Biol. Chem 284, 27065–27076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Huang SY, Gao R, Das BB, Murai J, and Marchand C (2014). Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst.) 19, 114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J, Kool H, van Schendel R, and Tijsterman M (2017). Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 36, 3634–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, and Symington LS (2015). Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem. Sci 40, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K, Kurahashi K, Gao R, Tsutakawa SE, Tainer JA, Pommier Y, and Aihara H (2012). Structural basis for recognition of 5′-phosphotyrosine adducts by Tdp2. Nat. Struct. Mol. Biol 19, 1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J, Bellelli R, and Boulton SJ (2017). Mechanisms of DNA-protein crosslink repair. Nat. Rev. Mol. Cell Biol 18, 563–573. [DOI] [PubMed] [Google Scholar]
- Syed A, and Tainer JA (2018). The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu. Rev. Biochem 87, 263–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treszezamsky AD, Kachnic LA, Feng Z, Zhang J, Tokadjian C, and Powell SN (2007). BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res. 67, 7078–7081. [DOI] [PubMed] [Google Scholar]
- Truong LN, Li Y, Shi LZ, Hwang PY, He J, Wang H, Razavian N, Berns MW, and Wu X (2013). Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA 110, 7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schendel R, Roerink SF, Portegijs V, van den Heuvel S, and Tijsterman M (2015). Polymerase Θ is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat. Commun 6, 7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song Y, Li S, Kurian S, Xiang R, Chiba T, and Wu X (2019). DNA polymerase θ (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J. Biol. Chem 294, 3909–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefzadeh MJ, Wyatt DW, Takata K, Mu Y, Hensley SC, Tomida J, Bylund GO, Doublié S, Johansson E, Ramsden DA, et al. (2014). Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 10, e1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, and Jasin M (2011). An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol 18, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code. The datasets supporting this study have not been deposited in a public repository.


