Abstract
The budding yeast PLC1 gene encodes a homolog of the δ isoform of mammalian phosphoinositide-specific phospholipase C. Here, we present evidence that Plc1p associates with the kinetochore complex CBF3. This association is mediated through interactions with two established kinetochore proteins, Ndc10p and Cep3p. We show by chromatin immunoprecipitation experiments that Plc1p resides at centromeric loci in vivo. Deletion of PLC1, as well as plc1 mutations which abrogate the interaction of Plc1p with the CBF3 complex, results in a higher frequency of minichromosome loss, nocodazole sensitivity, and mitotic delay. Overexpression of Ndc10p suppresses the nocodazole sensitivity of plc1 mutants, implying that the association of Plc1p with CBF3 is important for optimal kinetochore function. Chromatin extracts from plc1Δ cells exhibit reduced microtubule binding to minichromosomes. These results suggest that Plc1p associates with kinetochores and regulates some aspect of kinetochore function and demonstrate an intranuclear function of phospholipase C in eukaryotic cells.
The hydrolysis of phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] by phospholipase C (PLC) is a key early event in the regulation of diverse cell functions by many extracellular molecules. The reaction yields two prominent eukaryotic second messengers: 1,2-diacylglycerol (DG) and inositol 1,4,5-triphosphate [Ins(1,4,5)P3] (15, 41, 44). The hydrophilic Ins(1,4,5)P3 triggers the release of calcium from internal stores and thus modulates Ca2+- and calmodulin-regulated pathways (3), while the hydrophobic DG activates the phospholipid- and Ca2+-dependent protein kinase C (50). As a result, cytoplasmic PLC plays vital roles in the signal transduction cascades which ultimately regulate nuclear events.
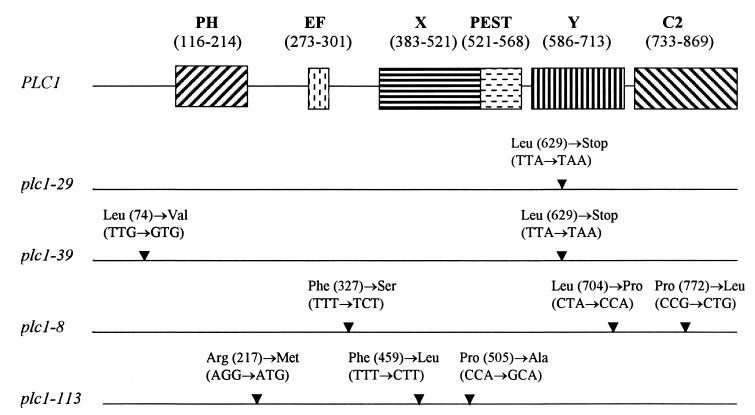
Three classes of mammalian phosphatidylinositol (PtdIns)-specific PLC isoenzymes, designated β, γ, and δ, were identified via biochemical, molecular, and immunological approaches (41). The Saccharomyces cerevisiae gene coding for PtdIns-specific PLC (PLC1) has also been cloned, and the protein product (Plc1p) is most closely related to mammalian δ isoforms, both in terms of sequence identity and in arrangements of conserved domains (23, 54, 68). All isoenzymes of the three main families of PLC, as well as the yeast Plc1p, contain a series of common modules which facilitate the enzymatic reaction. The modular structure contains an N-terminal pleckstrin homology (PH) domain, an EF-hand calcium binding domain, an active site containing two conserved regions termed X and Y boxes, and a C-terminal C2 lipid binding domain (32, 35) (schematic representation of the modular structure of Plc1p is shown in Fig. 4).
FIG. 4.
Structural features of PLC1 and mutations in plc1 alleles. The positions of the amino acid residues of the PH domain, EF-hand calcium binding domain (EF), catalytic X and Y domains, PEST motif, and C2 domain in Plc1p are shown in parentheses. The positions of mutations in individual plc1 alleles are indicated (▾), along with corresponding amino acid and nucleotide changes.
While PLC1 is not an essential gene, its deletion results in a number of phenotypes, including slow growth (progressively exacerbated at higher temperatures), osmosensitivity, defective utilization of carbon sources other than glucose, altered cell morphology, and inability to complete cytokinesis (23). Flick and Thorner (24) identified two genes which, when present in high copy number, suppress the temperature sensitivity of cells in which the PLC1 gene has been deleted (plc1Δ). One of these genes, PHO81, codes for an inhibitor of a cyclin-dependent protein kinase comprised of PHO80 and PHO85 gene products. The second suppressor is a novel gene, SPL2. In addition, Plc1p was previously shown to interact with the PtdIns kinase homolog Tor2p (43). Recent data implicate Plc1p in the regulation of several processes. Plc1p is required for synthesis of inositol hexakisphosphate, which supports efficient export of mRNA from the nucleus (70), and for oxidative-stress-induced destruction of cyclin Ume3p (12). In addition, Plc1p appears to be involved in signal transduction pathways responsible for the sensing of nutrients. Plc1p is required for glucose-induced activation of plasma membrane H+-ATPase (9) and is also a component of a nitrogen signaling pathway controlling the developmental switch from yeast-like to pseudohyphal growth (1). However, Plc1p's relationship to defined signal transduction pathways as well as the subcellular distribution of Plc1p in yeast remains unknown.
PLC1 was also identified in a genetic screen for mutants with increased frequencies of chromosome missegregation (54). The plc1-1 mutant, a temperature-sensitive mutant isolated in this screen, displayed a 32-fold increase in chromosome missegregation events. However, since plc1-1 cells appeared to have normal nuclei and spindle morphologies and were not supersensitive to the microtubule (MT)-destabilizing drug benomyl, Payne and Fitzgerald-Hayes reasoned that it was unlikely that the chromosome missegregation phenotype results from a defect in the components of the mitotic segregation apparatus (54). Rather, they concluded that plc1-1 affects chromosome segregation indirectly, possibly by interfering with proper timing of the cell cycle or by altering intracellular concentrations of calcium.
The kinetochore is a specialized organelle that mediates chromosome attachment to spindle MTs. Kinetochores are composed of centromeric DNA and a set of associated proteins. Whereas in animal cells the centromeres span several megabases, have complex organizations, and bind several MTs, in S. cerevisiae the centromere (CEN) is 125 bp long and engages only a single MT (67). CEN of S. cerevisiae includes three conserved elements, termed CDEI, CDEII, and CDEIII (22, 30). CBF3, a four-protein complex that binds the CDEIII sequence, is absolutely essential for kinetochore assembly and function in vivo (28). The four protein subunits of CBF3 have molecular masses of 110, 64, 58, and 23 kDa (11, 16, 40, 60). The genes borne by p110 (NDC10, CBF2, and CTF14) (16, 26, 33), p64 (CEP3 and CBF3b) (39, 61), p58 (CTF13) (16), and p23 (SKP1) (11, 60) have been cloned and characterized.
Traditionally, the PtdIns cycle and PLC activity have been thought to be associated with the plasma membrane. However, several reports suggest that an independently regulated PtdIns cycle operates also within the nucleus (4, 10, 13, 14, 45, 55). The regulation and downstream effectors of this nuclear PtdIns cycle are unknown. In this paper, we demonstrate that in S. cerevisiae Plc1p associates with the kinetochore and appears to affect the binding interaction between MTs and the kinetochore, which is essential for proper chromosome segregation and cell cycle progression. These results indicate a specific nuclear function for PLC in eukaryotic cells.
MATERIALS AND METHODS
Strains and media.
Yeast strains used in this study (listed in Table 1) were grown in rich medium (YPD; 1% yeast extract, 2% Bacto Peptone, 2% glucose) or under selection in synthetic minimal medium containing glucose (SD) and supplemented with appropriate nutrients. Yeasts were manipulated as described previously (8).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source and/or reference |
|---|---|---|
| W303-1a | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 ssd1-d2 can1-100 | R. Rothstein |
| YPH499 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | P. Hieter |
| PM1201 | MATα his3-Δ200 leu2-3,112 trp1 ura3-52 CSE4-HA | M. M. Smith; 46 |
| HL1-1 | W303-1a plc1::URA3 | 41 |
| PN101 | YPH499 plc1::URA3 | This study |
| CG1945 | MATa ura3-52 his3-200 lys2-801 trp1-901 ade2-101 Leu2-3,112 gal4-542 gal80-538 LYS::GAL1-HIS3 Cyhr2 URA3::GAL417-mers (×3)-CYC1-lacZ | Clontech |
| YM4271 | MATa ura3-52 his3-200 lys2-801 ade2-101 ade5 trp1-901 leu2-3,112 tyr1-501 gal4Δ gal80Δ ade5::hisG | Clontech |
| JC107 | YM4271[pHISi-1-CEN] | This study |
| JC108 | YM4271[pHISi-1-CEN M53] | This study |
Plasmids.
Plasmids pAS2-1-PLC1 and pYX232-PLC1-3HA have already been described (43). Plasmids pGBT9-PLC1 and pACT2-PLC1 were constructed by ligating a PLC1-containing BamHI fragment from pAS2-1-PLC1 into pGBT9 and pACT2, respectively. To construct plasmid pRS413-PLCP, which contains the promoter region of PLC1, a 1,100-bp fragment of the PLC1 promoter region was amplified by PCR using the oligonucleotides PLCP5 (5′-GCGCGAATTCGGGCCCACTTTTGGACGCTGGCGTCGC-3′), which hybridizes 1,107 bp upstream of the ATG start codon and introduces an ApaI site, and PLCP3 (5′-GCGCGAATTCACACTGCGTGAATGACCTTACCG-3′), which hybridizes 6 bp upstream of the ATG start codon and introduces an EcoRI site. The amplified 1.1-kb fragment was digested with EcoRI and ApaI and ligated into EcoRI- and ApaI-digested pRS413. Subsequently, an EcoRI-SacI fragment from pYX232-PLC1-3HA was ligated into EcoRI- and SacI-digested pRS413-PLCP to construct pRS413-PLC1.
For construction of pYX242-HT-T7-NDC10 and pACT2-NDC10, the NDC10 coding sequence was generated by PCR with the oligonucleotides NDC10-5 (5′-GCGCGGATCCTGAGATCATCGATTTTGTTTCTACTAAAATTG-3′) and NDC10-3 (5′-GCGCGAGCTCTTATTGGGCCCAGTTAGATAGATATACTAACAGACCATCAAATG-3′). Primer NDC10-5 introduces a BamHI site, and NDC10-3 introduces SacI and ApaI sites. The PCR-derived NDC10 fragment was digested with BamHI and SacI and ligated into BamHI and SacI sites of the pACT2 (Clontech) or pYX242-HT-T7 vector (8) to yield pACT2-NDC10 or pYX242-HT-T7-NDC10, respectively. Plasmid pYX232-NDC10-3HA was constructed by ligating a BamHI-ApaI fragment containing NDC10 from pACT2-NDC10 into BamHI- and ApaI-digested pYX232-PLC1-3HA. Plasmids pYX242-NDC10-3HA and pAS2-1-NDC10 were constructed by ligating a BamHI-XhoI fragment from pYX232-NDC10-3HA into BamHI- and XhoI-digested pYX242 or BamHI- and SalI-digested pAS2-1.
For construction of the pACT2-NDC10(194–446) and pGEX-5X-1-NDC10(194–446) plasmids, the NDC10 sequence coding for amino acids 194 to 667 was generated by PCR with the primers NDC5 (5′-GCGCGGATCCCAGAAACTAACCATCTCTCTG-3′) and NDC3 (5′-TCCGCGAGCTCGAATTCCCGCCTTGATGTGTTAGGCCC-3′). Sequence coding for amino acids 194 to 446 was released from this fragment by digestion with BamHI (introduced by the NDC5 primer) and EcoRI (internal site within the NDC10 sequence) and ligated into similarly digested pACT2 and pGEX-5X-1 plasmids.
Plasmids pAS2-1-CTF13 and pACT2-CTF13 were built by first adding a BamHI site to the 5′ end of the coding region of CTF13 and an ApaI site to the 3′ end of the coding region by PCR with the oligonucleotides CTF5 (5′-GCTAGGATCCCCAGAAGTCCATGTCGC-3′) and CTF3 (5′-CGCGCTGCAGGGCCCTTTCACATCGATAAATCTCTACTCG-3′). The fragment was digested with BamHI and ApaI and ligated into BamHI- and ApaI-digested pAS2-1-NDC10 and pACT2-NDC10. Plasmids pAS2-1-CEP3, pACT2-CEP3, pAS2-1-SKP1, and pACT2-SKP1 were constructed in a similar way by adding a BamHI site to the 5′ end of the corresponding coding region and an ApaI site to the 3′ end of the coding region by PCR. Oligonucleotides CEP5 (5′-GCGCGGATCCGTACCACTCAACTGAAATCC-3′) and CEP3 (5′-CGCGCTGCAGGGCCCAGGAAAGTATGTCAGAAATGTTATATTCG-3′) were used to build pAS2-1-CEP3 and pACT2-CEP3 plasmids. Oligonucleotides SKP5 (5′-GCGCGGATCCTGACTTCTAATGTTGTCCTAGTGAGTGG-3′) and SKP3 (5′-CGCGCTGCAGGGCCCTACGGTCTTCAGCCCATTCATTTTC-3′) were used to construct the pAS2-1-SKP1 and pACT2-SKP1 plasmids.
One-hybrid assay.
The integrating reporter plasmid pHISi-1-CEN was constructed by annealing two 5′-end-phosphorylated oligonucleotides, CEN-5 (5′- [P]AATTCGCGCAGCTGTATTAGTGTATTTGATTTCCGAAAGTTAAAAAAGAAATAGTAAGAAATATATATTTCCC-3′) and CEN-3 (5′-[P]GGGAAATATATATTTCTTACTATTTCTTTTTTAACTTTCGGAAAT CAAATACACTAATACAGCTGCGCG-3′), and by subsequent cloning into EcoRI- and SmaI-digested pHISi-1 (Clontech, Palo Alto, Calif.). Another reporter plasmid, pHISi-1-CEN-M53, was constructed in the same way as described above except that the CDEIII sequence with single base substitutions (underlined; C→A and G→T) was used. Both plasmids were linearized with XhoI and separately transformed into the YM4271 strain to construct two reporter strains, YM4271[pHISi-1-CEN] and YM4271[pHISi-1-CEN-M53]. Both reporter strains were individually transformed with the pACT2-NDC10, pACT2-CEP3, pACT2-CTF13, pACT2-SKP1, and pACT2-PLC1 plasmids, selected on plates coated with SD–Leu-His, and subsequently streaked on plates containing SD–Leu-His plus 50 mM 3-amino-1,2,4-triazole (3-AT).
Chromatin immunoprecipitation.
In vivo chromatin cross-linking and immunoprecipitation were performed essentially as described previously (5, 47) with several minor modifications. Briefly, yeast cells were grown in 50 ml of YPD to an A600 of 1.2 to 1.5, at which point they were fixed for 2 h by the addition of formaldehyde to a concentration of 1%. Subsequently, the cells were converted to spheroplasts with Zymolyase (8). Spheroplasts were washed sequentially in 2.5 ml of ice-cold phosphate-buffered saline (10 mM NaH2PO4, 150 mM NaCl at pH 7.4), 2.5 ml of buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES at pH 6.5), and 2.5 ml of buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES at pH 6.5). Finally, the spheroplasts were resuspended in 300 μl of lysis buffer (1% sodium dodecyl sulfate, 10 mM EDTA, 50 mM Tris-HCl at pH 8.0) supplemented with protease inhibitors at the following concentrations: 10 μg/ml each for leupeptin, aprotinin, pepstatin, and antipain; 1 mM for phenylmethylsulfonyl fluoride; 2 mM for benzamidine; 0.5 mM for TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone); and 100 μg/ml for TPCK (N-tosyl-l-phenylalanine chloromethyl ketone). The suspension was then sonicated 10 times for 10 s each to fragment chromosomal DNAs to an average size of ∼500 bp. The suspension was centrifuged for 10 min at 10,000 × g, and the supernatant was diluted with 2.7 ml of dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl at pH 8.0) supplemented with protease inhibitors at the above-named concentrations. The resultant solubilized chromatin solution was divided in three aliquots (1 ml each), and each aliquot was precleared by adding 50 μl of 50% protein A-Sepharose 4B slurry and incubating for 10 min at 4°C with gentle rocking. Beads were then harvested by centrifugation, and the supernatant was incubated with 4 μg of 12CA5 antibody overnight on ice. Sonicated phage λ DNA (2 μg) and protein A-Sepharose 4B beads (50 μl of 50% slurry) were added, and the incubation continued for 1 h. Beads were then harvested and washed, and the DNA was released and extracted as described previously (5).
Total input DNA and coimmunoprecipitated DNA were analyzed by PCR (25-μl reaction mixture) for the presence of various test loci. PCRs used 1/50 of the immunoprecipitates and 1/1,200 of the total input DNA. PCR products (7 μl) were separated on 2% agarose gels and visualized with ethidium bromide. The 243-bp region encompassing CEN3 (bp 113925 to 114168 of chromosome III) was amplified with the CEN3-5 (5′-GATCAGCGCCAAACAATATGG-3′) and CEN3-3 (5′-AACTTCCACCAGTAAACGTTTC-3′) primers as described previously (47, 51). The 278-bp region 177 kb to the right of CEN3 was amplified with NCEN-R5 (5′-ATCGCATTTCTTTTCGTCCACA-3′) and NCEN-R3 (5′-TTGGACGGGGATCGTTCGTA-3′), and the 249-bp fragment of the LEU2 coding region 23 kb to the left of CEN3 was amplified with NCEN-L5 (5′-TTAAAGCTATTTCTGATGTTCGTTCC-3′) and NCEN-L3 (5′-TACATGGTCTTAAGTTGGCGTAC-3′).
Mutagenesis of PLC1 and gap repair.
We employed the PCR procedure for localized mutagenesis and gap repair as described previously (49). PLC1 was first amplified by PCR with a slightly error-prone Taq polymerase, using the primers PLC-M5 (5′-CCTTGACATGATTTTGAAAATGG-3′) and PLC-M3 (5′-TAGAGGTGTGGTCAATAAGAGCG-3′) and with pGBT9-PLC1 as a template. The PCR product (100 ng) was cotransformed with 100 ng of EcoRI- and BamHI-digested pGBT9 into the CG1945 strain harboring the pACT2-NDC10 plasmid. About 105 transformants were selected on SD–Leu-Trp plates and subsequently replica plated onto plates containing SD–Leu-Trp-His plus 2.5 mM 3-AT. The transformants that were unable to grow on these plates were identified, and pGBT9-PLC1 mutant plasmids were isolated and transformed back into the CG1945 strain containing pACT2-NDC10 to verify the inability of the transformants to grow on plates containing SD–Leu-Trp-His plus 2.5 mM 3-AT. Twenty pGBT9-PLC1 plasmids were isolated in this way and transformed in plc1Δ cells. We took advantage of the fact that the pGBT9-PLC1 plasmid can suppress the temperature sensitivity, osmosensitivity, and nocodazole sensitivity of plc1Δ cells. We eliminated those pGBT9-PLC1 mutant plasmids, which suppressed none of these phenotypes and were not phenotypically distinguishable from plc1Δ cells. These plasmids probably encode mutated Plc1p with grossly reduced ability to function in vivo. All other transformants (plc1Δ cells harboring the mutated pGBT9-PLC1 plasmid) displayed temperature sensitivity and nocodazole sensitivity but osmoresistance (ability to grow in YPD medium containing 0.7 M NaCl). We selected three mutant plasmids with the tightest phenotypes and subcloned the coding regions of PLC1 as EcoRI-SalI fragments in the pRS413-PLCP plasmid (see above) to yield pRS413-PLC1-8, pRS413-PLC1-29, and pRS413-PLC1-39 (these plasmids express PLC1 from its natural promoter). The resultant plasmids were transformed in plc1Δ cells, and the temperature sensitivity, nocodazole sensitivity, and osmoresistance of the transformants were confirmed. Hereafter, these mutants are referred to as the plc1-8, plc1-29, and plc1-39 mutants.
The temperature-sensitive plc1 mutants were prepared in a similar way by PCR mutagenesis and gap repair (49). PLC1 was amplified with Taq polymerase by using the primers PLCP-M5 (5′-GAAGGGCGTTTAGATAAAGCCCC-3′) and PLCP-M3 (5′-CGAGCGCAGCGAGTCAGTGAG-3′). The PCR product (100 ng) was cotransformed with 100 ng of EcoRI- and XhoI-digested pRS413-PLC1 (the PLC1 insert is released by the digest) into the HL1-1 strain (plc1Δ). About 5 × 104 transformants were selected on plates containing synthetic complete medium (SC) lacking His at 28°C and subsequently replica plated onto YPD plates at 37°C and YPD plates containing 0.7 M NaCl at 28°C. The transformants that were unable to grow at 37°C but grew well at 28°C on YPD plates with 0.7 M NaCl were identified. Mutagenized pRS413-PLC1 plasmids were isolated and retested in the HL1-1 strain. Four plasmids were temperature sensitive but osmoresistant on both minimal and rich medium upon retesting.
Minichromosome stability assay.
Mitotic minichromosome stability was measured as a fraction of cells that retained the plasmid after growth in nonselective medium (38, 46). Briefly, wild-type or plc1Δ cells containing the empty pRS413 plasmid, or plc1Δ cells containing PLC1, plc1-8, plc1-29, or plc1-39 in pRS413 (CEN HIS3) were transformed with the pRS414 plasmid (CEN TRP1). For each transformant, five single colonies were inoculated separately into medium nonselective for the pRS414 plasmid and grown for about 24 h at 28°C. At the end of the growth, the cultures were still in the exponential phase, as was determined by counting the cells with a hemacytometer and by measuring A600. For each culture, the frequency of Trp+ cells was determined at the time of inoculation (F0) and at the end of nonselective growth (Fend) by plating appropriately diluted cultures on YPD plates and subsequently replica plating them onto SC lacking tryptophan. The rate of plasmid loss per generation was determined as described previously (38, 46), according to the equation Fend = F0(FD)G, where FD is the fraction of cells in each generation that retain the minichromosome and G is the number of generations. The fraction of cells that lose the minichromosome per generation is (1 − FD). The number of cell doublings was calculated by counting the total cell numbers at the beginning and at the end of nonselective growth.
In addition, the minichromosome stability was also determined by the color colony assay (29). Wild-type yeast cells are white. Mutation in the ADE2 gene, which is required for purine biosynthesis, causes an accumulation of a red intermediate in the purine biosynthetic pathway, and therefore the ade2 colonies are red. The red phenotype of cells containing an ochre mutation in the ADE2 gene (ade2-101) can be suppressed by an ochre-suppressing tRNA, SUP11. We determined the stability of the SUP11-marked minichromosome pUN20 (17) in the haploid plc1Δ strain. Deletion of PLC1 (plc1::URA3) was introduced into the YPH499 strain (ade2-101) as described previously (54), and the resultant strain, PN101, was transformed with the pUN20 and pRS413 plasmids containing a PLC1, plc1-8, plc1-29, or plc1-39 allele. For each transformant, five single white colonies (haploid ade2-101 cells with a single copy of SUP11 are white) from color plates were inoculated separately into medium nonselective for the pUN20 plasmid and grown for three generations to eliminate the cells containing more than one copy of the pUN20 plasmid. Subsequently, the cells were spread onto color medium plates to yield a density of about 100 colonies per plate and incubated for 4 to 5 days at 28°C and then overnight at 4°C. The number of half-sectored red-white colonies (which represent missegregation events that occurred at the first division after plating) divided by the total number of mostly white colonies plated represents the missegregation frequency of the pUN20 minichromosome.
Minichromosome-MT binding assay.
Preparation of yeast lysates containing minichromosomes and the minichromosome-MT binding assay were done as described previously (36, 37). Briefly, yeast cultures were grown to an A600 of ∼0.6 after the cells were maintained in exponential growth for several generations. In experiments with nocodazole-arrested cells, nocodazole was added to the cultures to 15 μg/ml for 6 h and nocodazole was also present during the preparation of spheroplasts (36). Cells were spheroplasted by glusulase and osmotically lysed in EBB buffer (10 mM Tris-Cl [pH 7.4], 10 mM MgCl2, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.1 mM dithiothreitol). Minichromosomes were eluted from nuclei by adding 0.3 M NaCl. After 5 min of incubation, the extracts were diluted threefold by adding EBB buffer and subjected to two subsequent centrifugations, each at 15,000 × g for 20 min. The clear supernatant was removed, supplemented with 10 μM taxol, and used for the MT binding assay. Purified tubulin was polymerized in vitro into MTs with an average length of 2 μm as directed by the manufacturer (ICN Biochemicals, Inc.) and stabilized by addition of the MT-stabilizing drug taxol (10 μM final concentration). Different amounts of stabilized MTs were added to 500-μl aliquots of the cleared extracts to initiate the MT binding assay. After 15 min of incubation at room temperature, the reaction mixtures were centrifuged at 15,000 × g for 8 min. The supernatant and pellet fractions were separated, and the amount of minichromosomes in each fraction was determined by Southern blot using a TRP1 probe. The calibration of the band intensities was performed by loading different amounts of the TRP1 DNA fragment on the gels, and the band intensities of the scanned images were quantified using UN-SCAN-IT software (Silk Scientific).
Other methods.
Two-hybrid screening, expression and purification of the glutathione S-transferase (GST)–Ndc10(194–446) fusion protein, Western blotting, and coprecipitation assays were carried out as described previously (8, 43). The DNA contents of cells were determined by flow cytometry of propidium iodide-stained cells (64).
RESULTS
Plc1p interacts with two components of the kinetochore complex.
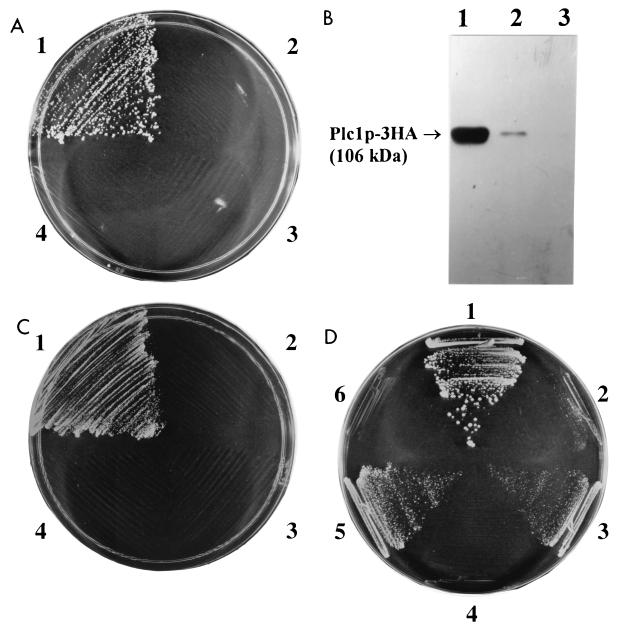
Because the gene encoding PtdIns-specific PLC, PLC1, was isolated in a genetic screen of mutants that exhibit defects in chromosome segregation (54), it seemed to us that Plc1p might be involved in kinetochore function. To test this hypothesis, we used the two-hybrid assay (21) to determine whether Plc1p interacts with Ndc10p, a component of the CBF3 complex of the kinetochore. We found that Plc1p interacts with full-length Ndc10p (Fig. 1A). We also constructed several truncation alleles of Ndc10p and identified amino acids 194 to 446 as the essential domain required for interaction with Plc1p (data not shown). To confirm the interaction between Plc1p and Ndc10p by an independent biochemical approach, we expressed the domain of Ndc10p encompassing amino acids 194 to 446 in Escherichia coli as a fusion with GST, purified the fusion protein on glutathione (GSH)-Sepharose 4B, and tested its ability to interact with Plc1p. Because Plc1p is expressed at a very low level in S. cerevisiae (23), we overexpressed Plc1p from a high-copy-number vector with a strong, constitutive triose phosphate isomerase promoter as a C-terminal fusion with three copies of the hemagglutinin (HA) epitope (plasmid pYX232-PLC-3HA) in a protease-deficient strain, BJ5465 (43). Cell lysate prepared from this strain was incubated with GST-Ndc10p(194–446) fusion protein or with GST protein as the control. The resulting protein complexes were isolated using GSH-Sepharose 4B beads. GST-Ndc10p(194–446) fusion protein (but not GST protein) was able to precipitate Plc1p-3HA from the yeast cell lysate (Fig. 1B), confirming that Plc1p can interact with Ndc10p.
FIG. 1.
Plc1p associates with the kinetochore complex CBF3. (A) Two-hybrid assay of the Plc1p and Ndc10p interaction. Cells of strain CG1945 harboring the following plasmids were streaked on plates containing SD–Trp-Leu-His plus 2.5 mM 3-AT and incubated for 4 days at 30°C: pAS2-1-PLC and pACT2-NDC10 (sector 1), pAS2-1-PLC and pACT2 (sector 2), pAS2-1 and pACT2-NDC10 (sector 3), and pAS2-1 and pACT2 (sector 4). The above-named strains grew equally well on SC–Leu-Trp plates. (B) GST-Ndc10p specifically coprecipitates Plc1p. Plc1p was expressed in yeast as a C-terminal fusion with three copies of the HA epitope (Plc1p-3HA), and the total cell extract (lane 1) was incubated with GST-Ndc10p(194–446) fusion protein (lane 2) or with GST protein (lane 3). The protein complexes were isolated on GSH-Sepharose 4B, and the samples were analyzed by Western blotting with 12CA5 antibody recognizing the HA epitope. The amounts of samples loaded onto all three lanes correspond to the amount of the total cell extract (1%). Thus, the difference in the intensities of the Plc1p-3HA bands in lanes 1 and 2 reflects recovery of Plc1p-3HA. The amounts of Plc1p-3HA in the cell lysate and in the GST-Ndc10p(194–446)-precipitated sample were compared quantitatively by Western blot analysis using several different amounts of both samples (data not shown). From the band intensities of scanned images, we estimate that under the conditions of the experiment, GST-Ndc10p(194–446) precipitates approximately 10% of Plc1p-3HA in the cell lysate. (C) Two-hybrid assay of the Plc1p and Cep3p interaction. Cells of strain CG1945 harboring the following plasmids were streaked on plates containing SD–Trp-Leu-His plus 10 mM 3-AT and incubated for 4 days at 30°C: pAS2-1-PLC and pACT2-CEP3 (sector 1), pAS2-1-PLC and pACT2 (sector 2), pAS2-1 and pACT2-CEP3 (sector 3), and pAS2-1 and pACT2 (sector 4). The above-named strains grew equally well on SC–Leu-Trp plates. (D) Plc1p associates with the assembled CBF3 complex. Strain YM4271 was transformed with either pHISi-1-CEN or pHISi-1-CENM53 integrative plasmids to create two reporter strains, JC107 and JC108, respectively. The JC107 strain (sectors 1, 3, and 5) and the JC108 strain (sectors 2, 4, and 6) harboring the following plasmids were streaked on plates containing SD–Leu-His plus 50 mM 3-AT and incubated for 4 days at 30°C: pACT2-NDC10 (sectors 1 and 2), pACT2-PLC1 (sectors 3 and 4), and pACT2-CEP3 (sectors 5 and 6). The strains grew equally well on SC-Leu plates.
These results prompted us to test whether Plc1p also interacts with additional protein components of the kinetochore. We fused coding regions of CEP3, CTF13, and SKP1 to a GAL4 activation domain (AD) and tested the interaction of the corresponding fusion proteins with Plc1p fused to a Gal4 DNA binding domain (BD). Coexpression of the Cep3p-Gal4 AD fusion with the Plc1p-Gal4 BD results in the expression of the HIS3 marker in the CG1945 reporter strain (Fig. 1C). Cep3p, therefore, is an additional kinetochore component that interacts with Plc1p. No interaction between Plc1p and Ctf13p or Skp1p was detectable by the two-hybrid assay.
Plc1p associates with the assembled CBF3 complex of the kinetochore.
The interaction of Plc1p with Ndc10p and Cep3p can be interpreted in two ways: (i) Plc1p interacts with Ndc10p and Cep3p prior to assembly into the kinetochore or (ii) Plc1p contacts the assembled kinetochore via Ndc10p and Cep3p. Additionally, it is also conceivable that the domains of Ndc10p and Cep3p responsible for the interaction with Plc1p are hidden within the CBF3 complex after assembly of the kinetochore and are not available for interaction with Plc1p. To resolve these issues, we determined whether the assembled CBF3 complex interacts with Plc1p. To this end, we used a modified one-hybrid assay. The one-hybrid system is an in vivo assay used to test binding of a protein to short target DNA sequences. When a DNA binding protein is fused to the Gal4 AD and expressed in yeast, it can bind to its specific recognition sequence inserted in the promoter region of a reporter gene (such as HIS3) and activate its transcription. We used a one-hybrid assay to test the binding of CBF3 proteins to the CDEIII sequence. The rationale was that reporter gene transcription would be activated only if the CBF3 complex was assembled on the CDEIII sequence. Conversely, cis-acting mutations in CDEIII would prevent proper assembly of the CBF3 complex (56), resulting in a failure to activate the transcription of the reporter gene. A similar one-hybrid assay was used previously by Ortiz et al. (51) in a screen which resulted in the identification of Ctf19p, Mcm21p, and Okp1p as components of the kinetochore.
We constructed two new derivatives of the YM4271 reporter strain by inserting either the wild-type CDEIII sequence (strain JC107) or the CDEIII sequence with a point mutation in the conserved CCG motif (strain JC108) upstream in the promoter of the HIS3 reporter gene. JC107 and JC108 cells were then transformed with plasmids for expression of Gal4 AD fusions with Ndc10p, Cep3p, Ctf13p, and Skp1p, and the transformants were streaked on plates containing 50 mM 3-AT. JC107 cells expressing Gal4 AD fusions of Ndc10p and Cep3p were able to grow on plates containing SD–Leu-His plus 50 mM 3-AT. JC108 cells expressing the same fusions, but with the point mutation in the CDEIII sequence, were not able to grow on the same medium (Fig. 1D). JC107 cells expressing the Gal4 AD fusion with Ctf13p grew very weakly, and cells expressing the Skp1p fusion were unable to grow on plates containing SD–Leu-His plus 50 mM 3-AT (data not shown). Interestingly, the JC107 cells expressing the Gal4 AD-Ndc10p fusion were able to grow even at 90 mM 3-AT. Thus, in the JC107 strain, the CBF3 complex can assemble on the CDEIII sequence, and this results in the activation of transcription of the reporter HIS3 gene. A point mutation in the conserved CCG motif of the CDEIII sequence in strain JC108 disrupts assembly of the CBF3 complex and prevents the transcriptional activation of HIS3.
When strains JC107 and JC108 were transformed with pACT2-PLC1, Gal4 AD-Plc1p fusion protein was able to activate transcription of HIS3 in the JC107 strain but not in the JC108 strain (Fig. 1D). All four subunits of CBF3 are needed for assembly of CBF3 on CDEIII, and mutations in any one of the four CBF3 subunits abolish the CDEIII binding activity of CBF3 (19, 34, 58). The above results indicate an interaction between Plc1p and the assembled CBF3 complex. However, we cannot exclude the possibility that Plc1p also interacts with the individual Cep3p and Ndc10p components prior to their assembly in CBF3.
Plc1p localizes to CEN DNA in vivo.
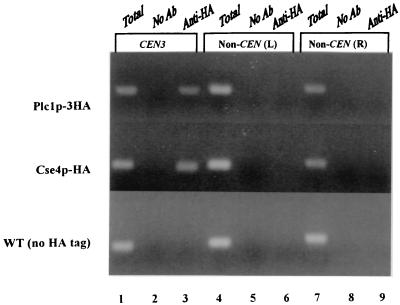
To test whether the interaction between Plc1p and the CBF3 complex occurs in vivo at the CEN locus, we used in vivo cross-linking followed by chromatin immunoprecipitation. Cells expressing Plc1p-3HA were first treated with formaldehyde to cross-link the cellular structures and then lysed and sonicated to shear the chromatin. Subsequently, Plc1p-3HA was immunoprecipitated using anti-HA (12CA5) antibody and protein A-Sepharose. As a positive control, we also used a strain expressing an epitope-tagged allele of CSE4, CSE4-HA. Cse4p, a histone H3 variant, is a structural component of the yeast kinetochore (48). To exclude the possibility that CEN chromatin is preferentially precipitated with anti-HA antibody or protein A-Sepharose, we also performed the chromatin immunoprecipitation with extracts that contained no HA-tagged proteins. The immunoprecipitated DNA was analyzed by PCR for the presence of the CEN3 fragment and two noncentromeric control fragments. The CEN3 DNA fragment specifically coimmunoprecipitated with Plc1p-3HA and Cse4-HAp (Fig. 2). Immunoprecipitates from mock-treated extracts and from extracts of an untagged strain did not contain detectable amounts of the CEN3 fragment. In addition, noncentromeric loci were not also detectable in any immunoprecipitate. Thus, Plc1p localizes to the centromere in vivo, presumably by interactions with the CBF3 components Cep3p and Ndc10p.
FIG. 2.
Localization of Plc1p to the CEN3 DNA locus. Cross-linked chromatin was prepared from cells expressing Plc1p-3HA (HL1-1 strain harboring plasmid pRS413-PLC1) or Cse4-HAp (strain PM1201) and wild-type cells (WT) (W303-1a) expressing no tagged protein. Chromatin was immunoprecipitated with the anti-HA antibody (Ab) 12CA5 (lanes 3, 6, and 9) or mock treated (lanes 2, 5, and 8), and aliquots of total input material (lanes 1, 4, and 7; in an amount corresponding to 0.8 μl of the chromatin solution) and coimmunoprecipitated DNA (in an amount corresponding to 20 μl of the chromatin solution) were analyzed by PCR with primers specific for CEN3 (244 bp) and two noncentromeric loci on yeast chromosome III: non-CEN (L) (249 bp) and non-CEN (R) (278 bp).
plc1Δ cells are hypersensitive to nocodazole and display an increased rate of minichromosome loss.
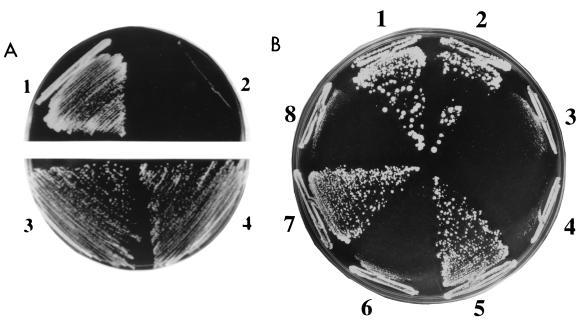
Nocodazole is an MT-depolymerizing drug that arrests cells at mitosis (52). Recent studies suggest that low concentrations of nocodazole alter MT dynamics and cause unstable attachment of the kinetochore to spindle MTs (63, 66). Since mutations in kinetochore-associated proteins can result in nocodazole hypersensitivity (25), we determined the resistance of plc1Δ cells to nocodazole. Disruption of PLC1 resulted in nocodazole hypersensitivity (Fig. 3A). When considered together with the previous report that mutation in PLC1 causes a chromosome segregation defect (54), our results suggest that Plc1p may affect the fidelity of chromosome segregation. Therefore, we tested the stability of minichromosomes in plc1Δ cells. The stability of the pRS413 minichromosome was measured as a fraction of cells that retained the minichromosome after growth in nonselective medium (38). The stability of the pUN20 minichromosome (17) was measured using the half-sectored colony color assay (29). The results of both methods were in good agreement and showed that the minichromosome loss rate in plc1Δ cells was about fivefold higher than in the wild-type cells (Table 2). Nocodazole (2 μg/ml) increased the minichromosome loss rate twofold in the wild-type strain but almost fivefold in plc1Δ cells (Table 2). This indicates that the deletion of PLC1 acts synergistically with nocodazole in destabilizing minichromosomes. It should be noted, however, that this high rate of loss (32%; Table 2) for plc1Δ cells in the presence of nocodazole is at the high limit of the plasmid loss rate measurable by this assay and corresponds to the loss rate of an acentric minichromosome or YRp plasmid (36, 38). For the same reason, we were unable to determine the plasmid loss rate of plc1Δ cells in the presence of nocodazole by the half-sectored colony assay. In this case, frequent sectoring made identification of half-sectored colonies impossible.
FIG. 3.
Nocodazole sensitivity of PLC1 mutants can be suppressed by NDC10 overexpression. (A) plc1Δ cells (HL1-1) are hypersensitive to nocodazole. Wild-type W303-1a (sectors 1 and 3) and plc1Δ (sectors 2 and 4) cells were streaked on a YPD plate containing 4 μg of nocodazole per ml (sectors 1 and 2; nocodazole was added to the medium from a stock solution of 3.3 mg/ml in dimethyl sulfoxide) or a YPD plate containing only a corresponding amount of the solvent (0.12% dimethyl sulfoxide; sectors 3 and 4). The plates were incubated for 3 days at 28°C. (B) Suppression of the nocodazole sensitivity of plc1-29 and plc1-39 mutants by NDC10 overexpression. plc1Δ cells (HL1-1) harboring the following plasmids were streaked on a YPD plate containing 4 μg of nocodazole per ml and incubated at 28°C for 3 days: pRS413-PLC1 and pYX242-HT-T7-NDC10 (sector 1), pRS413-PLC1 and pYX242-HT-T7 (sector 2), pRS413 and pYX242-HT-T7-NDC10 (sector 3), pRS413 and pYX242-HT-T7 (sector 4), pRS413-plc1-29 and pYX242-HT-T7-NDC10 (sector 5), pRS413-plc1-29 and pYX242-HT-T7 (sector 6), pRS413-plc1-39 and pYX242-HT-T7-NDC10 (sector 7), and pRS413-plc1-39 and pYX242-HT-T7 (sector 8).
TABLE 2.
Effect of PLC1 mutations on the mitotic stability of minichromosomes
| Straine | Generation timed | Avg loss per generation (%)a with:
|
||
|---|---|---|---|---|
| pRS314b plus nocodazole at:
|
pUN20c | |||
| 0 μg/ml | 2 μg/ml | |||
| PLC1 | 1.5 | 1.0 ± 0.1 | 1.9 ± 0.2 | 1.4 ± 0.1 |
| plc1Δ | 5.0 | 6.1 ± 0.8 | 32.4 ± 4.5 | 6.1 ± 0.6 |
| plc1-8 | 4.0 | 3.1 ± 0.4 | 12.7 ± 1.6 | 3.5 ± 0.4 |
| plc1-29 | 2.5 | 2.1 ± 0.3 | 4.1 ± 0.4 | 1.9 ± 0.2 |
| plc1-39 | 3.0 | 2.5 ± 0.3 | 5.9 ± 0.7 | 1.3 ± 0.1 |
The minichromosome stability of pRS414 or pUN20 was measured for at least five independent transformants and is reported as the percentage of minichromosome loss per generation at 28°C (standard deviations are also indicated).
The mitotic stability of the pRS413 minichromosome was measured as a fraction of cells that retained the minichromosome after growth in nonselective medium (35, 43) as described in Materials and Methods. All strains used for this assay were derived from W303-1a.
The mitotic stability of the pUN20 minichromosome was measured by using the half-sectored colony assay (27) as described in Materials and Methods. All strains used for this assay were derived from YPH499.
Generation time (in hours) was measured in YPD medium at 28°C for strains derived from W303-1a.
PLC1, plc1Δ, plc1-8, plc1-29, and plc1-39 denote plc1Δ strains harboring the pRS413-PLC1, pRS413, pRS413-plc1-8, pRS413-plc1-29, and pRS413-plc1-39 plasmids, respectively.
Disruption of the interaction between Plc1p and Ndc10p results in temperature sensitivity, nocodazole sensitivity, and an increased rate of minichromosome loss.
What is the biological significance of the interaction between Plc1p and the kinetochore? To answer this question, we generated three plc1 mutants (the plc1-8, plc1-29, and plc1-39 mutants) that do not interact with Ndc10p (see Materials and Methods). In addition, none of these mutants interacted in the two-hybrid assay with Cep3p or with the assembled kinetochore in the one-hybrid assay described above. All three plc1 mutants displayed slower growth at the permissive temperature (28°C), temperature sensitivity (inability to grow at temperatures higher than 36°C), nocodazole sensitivity (inability to grow in YPD medium containing more than 4 μg of nocodazole per ml), and a slightly higher rate of minichromosome loss (Table 2). All three mutants displayed the osmoresistant phenotype (ability to grow in YPD medium containing 0.7 M NaCl), indicating that a subset of Plc1p functions is not altered in these mutants. To determine if Ndc10p overexpression can suppress the temperature sensitivity and/or nocodazole sensitivity in these mutants, Ndc10p was overexpressed either as an N-terminal fusion with the His tag and T7 epitope (HT-T7) or as a C-terminal fusion with three HA tags. Overexpression of both fusion proteins suppressed the nocodazole sensitivity of the plc1-29 and plc1-39 mutants (Fig. 3B) and weakly suppressed the temperature sensitivity of the plc1-39 mutant (data not shown). These results provide genetic evidence for an interaction between Plc1p and Ndc10p.
To determine whether the enzymatic activities [the hydrolysis of PtdIns(4,5)P2] of the three mutants were likely to be affected, we sequenced the plc1-29, plc1-39, and plc1-8 alleles (Fig. 4). Since the mutations in all three plc1 alleles map to conserved regions which are presumably important for PtdIns(4,5)P2 hydrolysis (7, 18, 20), it is very likely that the enzymatic activities of the plc1 alleles are severely diminished or perhaps completely absent. Therefore, the resulting phenotypes of the three plc1 alleles (temperature sensitivity, nocodazole sensitivity, and minichromosome missegregation) are due to either the kinetochore binding defect, the diminished ability to hydrolyze PtdIns(4,5)P2, or both.
In a complementary approach, several temperature-sensitive plc1 mutants were generated (see Materials and Methods). We selected mutants which were unable to grow at a restrictive temperature (37°C) but that were osmoresistant and grew at nearly wild-type rates at the permissive temperature (28°C). All of these mutants were nocodazole sensitive (5 μg/ml) and failed to interact with Ndc10p and Cep3p in the two-hybrid assay and with the CBF3 complex in the one-hybrid assay even at the permissive temperature. Thus, the spectrum of phenotypes of these mutants is identical to that of the plc1-29, plc1-39, and plc1-8 mutants, which were isolated on the basis of their inability to interact with Ndc10p. The sequencing of plc1-113, the allele with the tightest temperature-sensitive phenotype, revealed three point mutations (Fig. 4). Two of these mutations change the conserved amino acid residues within the catalytic X domain and are therefore likely to affect the enzymatic activity of Plc1p (7, 18, 20). Thus, none of the isolated plc1 alleles can bind kinetochores, and all of the alleles acquired mutations in conserved amino acid residues within the catalytic region (7, 18, 20).
We attempted to separate the two activities of Plc1p [i.e., to bind kinetochores and to hydrolyze PtdIns(4,5)P2] by mapping the Plc1p domain required for interaction with kinetochores. Neither the C-terminal domain (amino acids 334 to 869), which includes the catalytic X and Y domains, nor the N-terminal domain (amino acids 1 to 360), which includes the PH domain and EF-hand calcium binding domain, interacted with Ndc10p or Cep3p in a two-hybrid assay or with the CBF3 complex in a one-hybrid assay (data not shown).
Collectively, the above data suggest that both the catalytic activity and the binding to kinetochores may require correct folding of Plc1p and that the interaction between Plc1p and the CBF3 complex occurs over a relatively large surface area of the Plc1p molecule and requires the presence of at least part of the catalytic domain. It is possible that the same mutation of Plc1p may affect both enzymatic activity and the interaction with kinetochores. Perhaps the surface area of the Plc1p molecule involved in the interaction with kinetochores also includes the catalytic region.
plc1Δ cells exhibit mitotic delay.
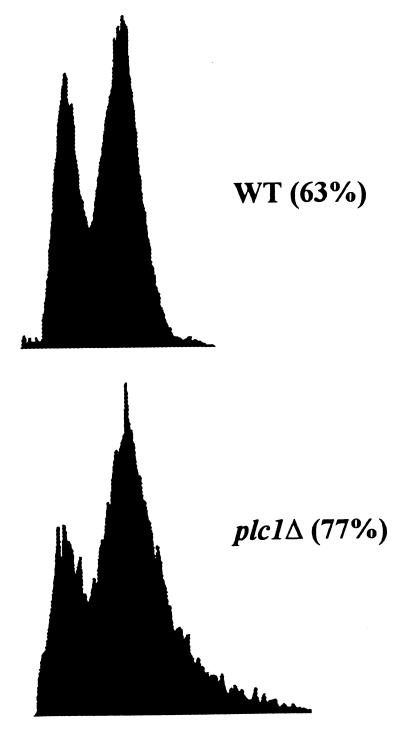
In addition to the kinetochore-MT interactions, the fidelity of chromosome transmission is also controlled by the action of at least one of the kinetochore-based mitotic checkpoint control systems (27, 31, 42, 56). Mutations that weaken a single centromere or mutations in genes that encode centromere DNA binding proteins can delay the cell cycle in the G2/M stage, suggesting a role for the kinetochore in checkpoint control systems in yeast cells (16, 53, 59, 64, 65). Despite the good potential for being the signal-transducing molecule of the kinetochore, Plc1p did not appear to be involved in mitotic checkpoint control. After treatment with nocodazole (15 μg/ml), the plc1Δ cells properly arrested as large-budded cells and maintained viability for at least 6 h (data not shown). However, if Plc1p is involved in attachment of an MT to the kinetochore, then plc1Δ cells would be expected to linger in the G2/M stage of the cell cycle, as was observed for ctf13, cep3, and skp1 mutants (2, 11, 16, 61). We examined the cell and nuclear morphologies of wild-type and plc1Δ cells during exponential growth at 28°C (Table 3). The frequencies of large-budded cells (with the diameter of the bud being at least 75% of the diameter of the mother cell) with a single nucleus positioned within 50% of the mother cell proximal to the neck (53, 59) were 12% for the wild-type strain and 26% for the plc1Δ strain. Exponentially growing plc1Δ cells also displayed a larger amount of cells with a 2N DNA content than the wild-type strain (Fig. 5). Taken together, these results demonstrate that plc1Δ cells exhibit a relatively mild delay at the G2/M stage of the cell cycle. About 8% of plc1Δ cells also displayed a two-budded cell phenotype (Table 3). Depending on the size of the second bud, it is either with or without a nucleus. The appearance of cells with two buds in the plc1Δ population suggests an additional partial defect or delay in cytokinesis. This observation is supported by the previous work of Flick and Thorner (23), who described the appearance of multibudded plc1Δ cells after 10 h of incubation at 37°C (restrictive temperature for the plc1Δ mutant). Perhaps in addition to its role in G2/M transition, Plc1p has an additional role in cytokinesis, which is more pronounced during suboptimal growth conditions, such as high temperature.
TABLE 3.
plc1Δ cells display mitotic-delay morphology
| Strain | % of cells with cell morphologya:
|
||||
|---|---|---|---|---|---|
| UB | SB | LB
|
DB | ||
| Single nucleus | Two separate nuclei | ||||
| Wild type | 37 ± 4.2 | 32 ± 4.0 | 12 ± 1.9 | 19 ± 2.5 | 0 |
| plc1Δ | 25 ± 3.5 | 19 ± 2.1 | 26 ± 3.1 | 22 ± 2.7 | 8 ± 1.5 |
Logarithmic asynchronous cultures of wild-type (W303-1a) and plc1Δ (Hl1-1) cells growing in YPD at 28°C were stained with 4′,6-diamidino-2-phenylindole, and nuclear and bud morphologies were scored. At least 300 separated cells were scored for each sample in each experiment. The experiment was performed three times, and standard deviations are indicated. UB, unbudded cells; SB, small-budded cells (with the diameter of the bud being less than 75% of the diameter of the mother cell); LB, large-budded cells with single nuclei (cells with single nuclei positioned within 50% of the mother cell proximal to the neck and with the diameter of the bud being at least 75% of the diameter of the mother cell); LB, large-budded cells with two separate nuclei (one nucleus in the mother cell, one nucleus in the bud); DB, cells with two buds (occasionally cells with more than two buds were observed and were also scored as DB).
FIG. 5.
Flow cytometry of wild-type W303-1a (WT) and plc1Δ (HL1-1) cells exponentially growing in YPD at 28°C. Cells were fixed with ethanol and stained with propidium iodide. The fractions of cells in G2/M are indicated in parentheses.
Minichromosomes in plc1Δ extract exhibit reduced MT binding.
Previous work suggested that kinetochore function can be perturbed by a failure either to form the core CEN DNA-protein complex or to assemble components that interact with the MTs to ensure faithful chromosome segregation (36). We used an assay developed by Kingsbury and Koshland (36, 37) to measure the ability of minichromosomes formed in vivo to bind MTs in vitro. The wild-type and plc1Δ mutant strains were transformed with the centromeric plasmid (minichromosome) pRS414, which was then assayed in clarified extracts of these cultures for binding to taxol-stabilized MTs. In the control lysate without MTs, about 2 to 3% of the plasmid pelleted when it was subjected to centrifugation. However, in the presence of a saturating concentration of MTs, about 24% of the plasmid pelleted with MTs in the wild-type extract and about 13% pelleted in the plc1Δ extract (Table 4). When the same experiment was performed with the pRS424 plasmid, which does not contain the CEN sequence and which is segregated by a kinetochore-independent mechanism, the amounts of plasmid precipitated from the wild-type and plc1Δ lysates were nearly identical and increased from only 2% (control experiment without MTs) to about 4% (saturating concentration of MTs). To improve the sensitivity of the assay and to exclude the possibility that the observed difference in MT binding activities between the two strains was merely the result of different cell cycle profiles, the assay was also performed with lysates prepared from nocodazole-arrested cells (Table 4). The G2/M transition is the phase when the kinetochore is required for progression of the cell cycle through mitosis. It is also when the kinetochore is in its most active state. Kinetochores of cells arrested with nocodazole at the G2/M transition exhibit the highest MT binding activity (36). Our data for the wild-type strain correspond to these previous results. The saturating concentration of MTs pelleted about 48% of the plasmid in the wild-type strain but only 28% in the plc1Δ strain (Table 4). The amount of 2μm plasmid (pRS424) precipitated from nocodazole-arrested wild-type and plc1Δ cells did not significantly differ from the amounts precipitated from asynchronous cultures and ranged from 2% in experiments without MTs to about 4% in experiments with a saturating concentration of MTs (data not shown). Although the difference in MT binding between the wild-type and plc1Δ strains is relatively small, it is reproducible in both asynchronous and mitotic extracts and suggests that in plc1Δ cells the kinetochore function is indeed partially impaired.
TABLE 4.
plc1Δ extracts exhibit reduced MT binding to minichromosomes
| MT concn (MTs/ml) | Avg % of minichromosomes in the MT pelleta
|
|||||
|---|---|---|---|---|---|---|
| Asynchronous cells
|
Nocodazole-arrested cells
|
|||||
| Wild type
|
plc1Δ
|
Wild-type CEN | plc1Δ CEN | |||
| CEN | 2μm | CEN | 2μm | |||
| 0 | 2.1 ± 0.3 | 2.0 ± 0.4 | 2.5 ± 0.6 | 2.6 ± 0.6 | 2.0 ± 0.3 | 1.9 ± 0.3 |
| 4 × 107 | 8.7 ± 1.5 | 2.5 ± 0.5 | 5.1 ± 1.0 | 2.3 ± 0.4 | 19.9 ± 2.7 | 12.6 ± 2.1 |
| 4 × 108 | 21.3 ± 2.9 | 3.1 ± 0.5 | 10.9 ± 2.0 | 2.9 ± 0.5 | 43.5 ± 6.9 | 25.2 ± 4.6 |
| 4 × 109 | 24.2 ± 3.8 | 3.8 ± 0.6 | 13.4 ± 2.1 | 3.7 ± 0.6 | 47.7 ± 7.8 | 28.1 ± 4.9 |
Cleared lysates were prepared from W303-1a and plc1Δ cells (HL1-1) transformed with the centromeric plasmid pRS414 or the 2μm plasmid pRS424. Cells were arrested at the G2/M stage of the cell cycle by incubating them for 6 h in the presence of 15 μg of nocodazole per ml. Under these conditions, 90% of both wild-type and plc1Δ cells arrested as large-budded cells. Different amounts of bovine MTs were added to the lysates and incubated for 15 min at 20°C. MTs were pelleted, and the percentage of minichromosomes that cosedimented with the MTs was determined. The experiment was performed five times, and the standard deviations are indicated. The amount of the 2μm plasmid cosedimenting from lysates of nocodazole-arrested cells did not significantly differ from the amount cosedimenting from lysates of asynchronous cells (data not shown).
DISCUSSION
It is generally accepted that PtdIns signaling pathways are present in nuclei (4, 14), and enzymes responsible for PtdIns turnover and signaling, such as PLC and protein kinase C, have been identified in the nuclear interior, associated with nonmembrane nuclear structures (10, 13, 45, 55). The nuclear PtdIns cycle is regulated independently of the PtdIns cycle at the plasma membrane (14). Nuclear PtdIns(4,5)P2, for instance, decreases as cells progress through the S phase of the cell cycle (69), and nuclear PLC activity and DG levels increase at the G2/M transition (62). However, the processes regulated by the nuclear PtdIns cycle have not been identified and the biological function of the nuclear PtdIns cycle remains unknown. Our data demonstrate that in S. cerevisiae Plc1p associates with kinetochores and appears to affect the binding of kinetochores to MTs.
Plc1p exhibits all three characteristics expected of a kinetochore protein (61). First, Plc1p associates with centromeric DNA through interaction with the CBF3 complex. Second, plc1Δ cells delay in the G2/M stage of the cell cycle, are hypersensitive to nocodazole, and display a greater instability of minichromosomes. In addition, minichromosomes isolated from plc1Δ cells have a reduced ability to bind MTs. Third, in comparison with the wild-type cells, plc1-1 mutant cells display a 32-fold-higher frequency of missegregation of a chromosome fragment (cen3X69) carrying a destabilizing mutation in the CDEIII region of the centromere (54).
The CBF3 complex is essential for binding of kinetochores to MTs in vitro. However, CBF3 does not mediate efficient MT attachment on its own (57). Attachment requires the presence of additional cellular factors. It was proposed that CBF3 forms a DNA-binding scaffold onto which additional kinetochore components assemble, including proteins that directly bind to MTs (57). However, the identities of these proteins remain unknown. Our data suggest that Plc1p may be one of these accessory proteins of the kinetochore involved in binding MTs.
Since Plc1p is not essential for cell viability and does not contain a recognizable MT binding motif, it is more likely that Plc1p has a regulatory rather than a structural role in the MT-kinetochore interaction. An alternative explanation may be that CBF3 subunits bind MTs directly but that Plc1p is required to achieve an active MT binding conformation of the CBF3 subunits. Using in vivo cross-linking and immunoprecipitation, Meluh and Koshland (47) demonstrated the existence of centromeric subcomplexes which probably correspond to kinetochore assembly intermediates. In their model, kinetochore assembly initiates with recruitment of the CBF3 complex, and possibly Mif2p, to CDEIII. The resultant subcomplex then serves as the nucleation site around which a fully functional kinetochore is assembled if CDEI and CDEII are adjacent. Conceivably, Plc1p may function in this folding process, which may affect the ability of kinetochores to bind MTs.
The MT binding activity of kinetochore complexes is high in mitotic extracts and low in interphase extracts. However, the DNA binding activity of CBF3 is high in both stages (57). These results suggest that CBF3 is assembled on the centromeric DNA throughout the cell cycle but that the MT attachment is controlled by a cell cycle-dependent interaction of additional factors with a constitutively bound CBF3 scaffold. At present we do not know whether Plc1p remains stably localized at the CEN locus throughout the cell cycle or whether it localizes to the CEN locus only during a particular stage(s) of the cell cycle.
Currently, it is difficult to determine whether only the ability of Plc1p to bind kinetochores or this binding ability and Plc1p's enzymatic activity are required for the apparent mitotic functions of Plc1p. The sequencing of the plc1-29, plc1-39, plc1-8, and plc1-113 alleles revealed that they all acquired mutations which likely affect the enzymatic activity. Since all of these mutants failed to interact with Ndc10p and Cep3p in a two-hybrid assay and with the CBF3 complex in a one-hybrid assay, we believe that they do not interact with kinetochores. Therefore, the corresponding phenotypes can be attributed to either a loss of binding to kinetochores, a decrease (or loss) of enzymatic activity, or both. Since plc1Δ cells display only a mild mitotic delay and only a sixfold-increased rate of minichromosome loss, it seems likely that Plc1p is not a direct structural component of the kinetochore but rather a regulatory factor affecting some aspects of the kinetochore activity.
Since kinetochores are unlikely to contain any PtdIns(4,5)P2 component which could be used as a substrate for Plc1p, it seems unlikely that Plc1p would hydrolyze PtdIns(4,5)P2 at the kinetochore. It is possible that the interaction of Plc1p with the kinetochore is spatially and perhaps also temporally separated from its enzymatic activity. It remains to be determined which of these two events [binding to kinetochore and hydrolysis of PtdIns(4,5)P2] is relevant for the observed mitotic functions of Plc1p. However, since nuclei of mammalian cells contain a pool of PtdIns(4,5)P2 which is not associated with nuclear envelope (4, 6, 71), the possibility that the kinetochore-bound Plc1p hydrolyzes PtdIns(4,5)P2 cannot be completely discarded. It will be important to determine whether Plc1p remains localized to the CEN locus throughout the cell cycle or whether it localizes to the CEN locus only during a particular stage(s) of the cycle and whether this localization and interaction with the CBF3 complex regulates the Plc1p-dependent hydrolysis of PtdIns(4,5)P2. The possibility that PLC is regulated in a cell cycle-specific manner is supported by the observation that in mammalian cells nuclear PLC activity and DG levels are increased at the G2/M transition (62). In this context, it will be important to explore further the role of PLC during mitosis in both yeast and mammalian cells.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (1R15 GM/OD55937-01) and the American Cancer Society (BE-239) to A.V.
We are grateful to D. Burke, M. Fitzgerald-Hayes, M. Hall, P. Hieter, D. Koshland, M. Smith, F. Spencer, and J. Thorner for providing plasmids and strains and to P. Paine and I. Vancurova for critical readings of the manuscript and helpful comments. We thank the staff of the Flow Cytometry Laboratory of the State University of New York at Stony Brook for performing the FACS analysis and the DNA sequencing facility of the Michigan State University for sequencing the plc1 mutants.
REFERENCES
- 1.Ansari K, Martin S, Farkasovsky M, Ehbrecht I M, Kuntzel H. Phospholipase C binds to the receptor-like GPR1 protein and controls pseudohyphal differentiation in Saccharomyces cerevisiae. J Biol Chem. 1999;274:30052–30058. doi: 10.1074/jbc.274.42.30052. [DOI] [PubMed] [Google Scholar]
- 2.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 3.Berridge M J. Inositol triphosphate and calcium signaling. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 4.Boronenkov I V, Loijens J C, Umeda M, Anderson R A. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol Biol Cell. 1998;9:3547–3560. doi: 10.1091/mbc.9.12.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 6.Caramelli E, Matteucci A, Zini N, Carini C, Guidotti L, Ricci D, Maraldi N M, Capitani S. Nuclear phosphoinositide-specific phospholipase C, phosphatidylinositol 4,5-bisphosphate and protein kinase C during rat spermatogenesis. Eur J Cell Biol. 1996;71:154–164. [PubMed] [Google Scholar]
- 7.Cheng H-F, Jiang M-J, Chen C-L, Liu S-M, Wong L-P, Lomasney J W, King K. Cloning and identification of amino acid residues of human phospholipase Cδ1 essential for catalysis. J Biol Chem. 1995;270:5495–5505. doi: 10.1074/jbc.270.10.5495. [DOI] [PubMed] [Google Scholar]
- 8.Choi J H, Lou W, Vancura A. A novel membrane-bound glutathione S-transferase functions in the stationary phase of the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:29915–29922. doi: 10.1074/jbc.273.45.29915. [DOI] [PubMed] [Google Scholar]
- 9.Coccetti P, Tisi R, Martegani E, Teixeira L S, Brandao R L, Castro I M, Thevelein J M. The PLC1 encoded phospholipase C in the yeast Saccharomyces cerevisiae is essential for glucose-induced phosphatidylinositol turnover and activation of plasma membrane H+-ATPase. Biochim Biophys Acta. 1998;1405:147–154. doi: 10.1016/s0167-4889(98)00099-8. [DOI] [PubMed] [Google Scholar]
- 10.Cocco L, Gilmour R S, Ognibene A, Letcher A J, Manzoli F A, Irvine R F. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem J. 1987;248:765–770. doi: 10.1042/bj2480765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper K F, Mallory M J, Strich R. Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol Cell Biol. 1999;19:3338–3348. doi: 10.1128/mcb.19.5.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Divecha N, Banfic H, Irvine R F. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. EMBO J. 1991;10:3207–3214. doi: 10.1002/j.1460-2075.1991.tb04883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Divecha N, Banfic H, Irvine R F. Inositides and the nucleus and inositides in the nucleus. Cell. 1993;74:405–407. doi: 10.1016/0092-8674(93)80041-c. [DOI] [PubMed] [Google Scholar]
- 15.Divecha N, Irvine R F. Phospholipid signaling. Cell. 1995;80:269–278. doi: 10.1016/0092-8674(95)90409-3. [DOI] [PubMed] [Google Scholar]
- 16.Doheny K F, Sorger P K, Hyman A A, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the Saccharomyces cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elledge S J, Davis R W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988;70:303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- 18.Ellis M V, James S R, Perisic O, Downes C P, Williams R L, Katan M. Catalytic domain of phosphoinositide-specific phospholipase C (PLC). Mutational analysis of residues within the active site and hydrophobic ridge of PLCδ1. J Biol Chem. 1998;273:11650–11659. doi: 10.1074/jbc.273.19.11650. [DOI] [PubMed] [Google Scholar]
- 19.Espelin C W, Kaplan K B, Sorger P K. Probing the architecture of a simple kinetochore using DNA-protein crossing. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Essen L-O, Perisic O, Katan M, Wu Y, Roberts M F, Williams R L. Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C. Biochemistry. 1997;36:1704–1718. doi: 10.1021/bi962512p. [DOI] [PubMed] [Google Scholar]
- 21.Fields S, Song O K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 23.Flick J, Thorner J. Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5861–5876. doi: 10.1128/mcb.13.9.5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flick J, Thorner J. An essential function of a phosphoinositide-specific phospholipase C is relieved by inhibition of a cyclin-dependent protein kinase in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:33–47. doi: 10.1093/genetics/148.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foreman P K, Davis R W. CDP1, a novel Saccharomyces cerevisiae gene required for proper nuclear division and chromosome segregation. Genetics. 1996;144:1387–1397. doi: 10.1093/genetics/144.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goh P Y, Kilmartin J V. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorbsky G J. Cell cycle checkpoints: arresting progress in mitosis. Bioessays. 1997;19:193–197. doi: 10.1002/bies.950190303. [DOI] [PubMed] [Google Scholar]
- 28.Hegemann J H, Shero J H, Cottarel G, Philippsen P, Hieter P. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2523–2535. doi: 10.1128/mcb.8.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hieter P, Mann C, Snyder M, Davis R W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 30.Hieter P, Pridmore D, Hegemann J H, Thomas M, Davis R W, Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985;42:913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- 31.Hoyt M A, Totis L, Roberts B T S. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 32.James S. The structure of phospholipase C isoforms and the regulation of phosphoinositide hydrolysis. Biochem Soc Trans. 1998;26:354–359. doi: 10.1042/bst0260354. [DOI] [PubMed] [Google Scholar]
- 33.Jiang W, Lechner J, Carbon J. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J Cell Biol. 1993;121:513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan K B, Hyman A A, Sorger P K. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 35.Katan M. Families of phosphoinositide-specific phospholipase C: structure and function. Biochim Biophys Acta. 1998;1436:5–17. doi: 10.1016/s0005-2760(98)00125-8. [DOI] [PubMed] [Google Scholar]
- 36.Kingsbury J, Koshland D. Centromere-dependent binding of yeast minichromosomes to microtubules in vitro. Cell. 1991;66:483–495. doi: 10.1016/0092-8674(81)90012-x. [DOI] [PubMed] [Google Scholar]
- 37.Kingsbury J, Koshland D. Centromere function on minichromosomes isolated from budding yeast. Mol Biol Cell. 1993;4:859–870. doi: 10.1091/mbc.4.8.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koshland D, Rutledge L, Fitzgerald-Hayes M, Hartwell L H. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 1987;48:801–812. doi: 10.1016/0092-8674(87)90077-8. [DOI] [PubMed] [Google Scholar]
- 39.Lechner J. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, Cbf3. EMBO J. 1994;13:5203–5211. doi: 10.1002/j.1460-2075.1994.tb06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 41.Lee S B, Rhee S G. Significance of PIP2 hydrolysis and regulation of phospholipase C isozymes. Curr Opin Cell Biol. 1995;7:183–189. doi: 10.1016/0955-0674(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 42.Li R, Murray A W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 43.Lin H, Choi J H, Vancura A. Phosphoinositide-specific phospholipase C interacts with phosphatidylinositol kinase homolog TOR2. Biochem Biophys Res Commun. 1998;252:285–289. doi: 10.1006/bbrc.1998.9639. [DOI] [PubMed] [Google Scholar]
- 44.Majerus P W. Inositol phosphate biochemistry. Annu Rev Biochem. 1992;61:225–250. doi: 10.1146/annurev.bi.61.070192.001301. [DOI] [PubMed] [Google Scholar]
- 45.Martelli A M, Gilmour R S, Bertagnolo V, Neri L M, Manzoli L, Cocco L. Nuclear localization and signalling activity of phosphoinositidase C beta in Swiss 3T3 cells. Nature. 1992;358:242–245. doi: 10.1038/358242a0. [DOI] [PubMed] [Google Scholar]
- 46.Meluh P B, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meluh P B, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meluh P B, Yang P, Glowczewski L, Koshland D, Smith M M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 49.Muhlrad D, Hunter R, Parker R A. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 50.Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 51.Ortiz J, Stemman O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Owellen R J, Hartke C A, Dickerson R M, Hains F O. Inhibition of tubulin-microtubule polymerization by drugs of the vinca alkaloid class. Cancer Res. 1976;36:1499–1502. [PubMed] [Google Scholar]
- 53.Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Biol Cell. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Payne W E, Fitzgerald-Hayes M. A mutation in PLC1, a candidate phosphainositide-specific phospholipase C gene from Saccharomyces cerevisiae, causes aberrant mitotic chromosome segregation. Mol Cell Biol. 1993;13:4351–4364. doi: 10.1128/mcb.13.7.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Payrastre B, Nievers M, Boonstra J, Breton M, Verkleij A J, Van Bergen en Henegouwen P M. A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. J Biol Chem. 1992;267:5078–5084. [PubMed] [Google Scholar]
- 56.Rudner A D, Murray A W. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- 57.Sorger P K, Severin F F, Hyman A A. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorger P K, Doheny K F, Hieter P, Kopski K M, Huffaker T C, Hyman A A. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc Natl Acad Sci USA. 1995;92:12026–12030. doi: 10.1073/pnas.92.26.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spencer F, Hieter P. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:8908–8912. doi: 10.1073/pnas.89.19.8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stemmann O, Lechner J. The Saccharomyces cerevisiae kinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO J. 1996;15:3611–3620. [PMC free article] [PubMed] [Google Scholar]
- 61.Strunnikov A V, Kingsbury J, Koshland D. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J Cell Biol. 1995;128:749–760. doi: 10.1083/jcb.128.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun B, Murray N R, Fields A P. A role for nuclear phosphatidylinositol-specific phospholipase C in the G2/M phase transition. J Biol Chem. 1997;272:26313–26317. doi: 10.1074/jbc.272.42.26313. [DOI] [PubMed] [Google Scholar]
- 63.Vasquez R J, Howell B, Yvon A M C, Wadsworth P, Cassimeris L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol Biol Cell. 1997;8:973–985. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Burke D J. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wells W A E, Murray A W. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wendell K L, Wilson L, Jordan M A. Mitotic block in HeLa cells by vinblastine: ultrastructural changes in kinetochore-microtubule attachment and centrosomes. J Cell Sci. 1993;104:261–274. doi: 10.1242/jcs.104.2.261. [DOI] [PubMed] [Google Scholar]
- 67.Winey M, Mamay C L, O'Toole E T, Mastronarde D N, Giddings T H, Jr, McDonald K L, McIntosh J R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoko-o T, Matsui Y, Yagisawa H, Nojima H, Uno I, Toh-e A. The putative phosphoinositide-specific phospholipase C gene, PLC1, of the yeast Saccharomyces cerevisiae is important for cell growth. Proc Natl Acad Sci USA. 1993;90:1804–1808. doi: 10.1073/pnas.90.5.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.York J D, Majerus P W. Nuclear phosphatidylinositols decrease during S-phase of the cell cycle in HeLa cells. J Biol Chem. 1994;269:7847–7850. [PubMed] [Google Scholar]
- 70.York J D, Odom A R, Murphy R, Ives E B, Wente S R. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 71.Zini N, Neri L M, Ognibene A, Scotlandi K, Baldini N, Maraldi N M. Increase of nuclear phosphatidylinositol 4,5-bisphosphate and phospholipase C β1 is not associated to variations of protein kinase C in multidrug-resistant saos-2 cells. Microsc Res Tech. 1997;36:172–178. doi: 10.1002/(SICI)1097-0029(19970201)36:3<172::AID-JEMT5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]