Abstract
KEOPS (Kinase, Endopeptidase and Other Proteins of Small size) is a five-subunit protein complex that is highly conserved in eukaryotes and archaea and is essential for the fitness of cells and for animal development. In humans, mutations in KEOPS genes underlie Galloway–Mowat syndrome, which manifests in severe microcephaly and renal dysfunction that lead to childhood death. The Kae1 subunit of KEOPS catalyzes the universal and essential tRNA modification N6-threonylcarbamoyl adenosine (t6A), while the auxiliary subunits Cgi121, the kinase/ATPase Bud32, Pcc1 and Gon7 play a supporting role. Kae1 orthologs are also present in bacteria and mitochondria but function in distinct complexes with proteins that are not related in structure or function to the auxiliary subunits of KEOPS. Over the past 15 years since its discovery, extensive study in the KEOPS field has provided many answers towards understanding the roles that KEOPS plays in cells and in human disease and how KEOPS carries out these functions. In this review, we provide an overview into recent advances in the study of KEOPS and illuminate exciting future directions.
INTRODUCTION
In 2006, a previously unknown five-subunit protein complex was discovered in two independent studies. This complex contained the putative protein kinase Bud32 and the putative endopeptidase Kae1 as well as the three uncharacterized proteins Gon7, Pcc1 and Cgi121. The presence of Kae1 and Bud32 led to the naming of this complex as KEOPS (Kinase, Endopeptidase and Other Proteins of Small size) (1) or EKC (Endopeptidase‐like and Kinase Chromatin‐associated) (2). For simplicity, we will use KEOPS naming throughout this review. Three key findings illuminated by these landmark studies include:
All KEOPS subunits are conserved across archaeal and eukaryotic organisms apart from Gon7 which appeared specific to eukaryotes.
The Kae1 subunit is even more well conserved representing one of only ∼60 genes that are universally found in the genomes of all living cells (3).
The presence of all subunits in KEOPS are required for its ability to support the well-being of cells as measured by growth.
Over the past 15 years, a flurry of studies resolved many mysteries of how KEOPS functions, including its core biochemical role, together with the Sua5 protein, in catalyzing the universal N6-threonylcarbamoyl adenosine (t6A) modification of transfer RNAs (tRNAs), its role in human disease and its structure. However, many critical questions remain unanswered. In this review, we will provide a comprehensive overview into how the KEOPS complex was discovered, the role of KEOPS in cells and in human disease, the atomic structure of KEOPS and how this informs on function and lastly, the pressing frontier questions in the KEOPS field that remain to be addressed.
HOW KEOPS WAS DISCOVERED
The pre-KEOPS period
Bud32 was first identified in Saccharomyces cerevisiae as a gene required for normal cell growth and polarized cell divisions, commonly known as ‘budding’ (hence its name) (4). Sequence analysis predicted that Bud32 is a member of the eukaryotic protein kinase superfamily (5). Protein kinases adopt a bi-lobal architecture with a smaller N-terminal lobe (denoted N-lobe) and a larger C-terminal lobe (denoted C-lobe). An inter-lobe cleft harbors elements projecting from the two lobes that are required for ATP binding and for the catalytic transfer of the gamma phosphate of ATP to a hydroxyl bearing sidechain in proteinaceous substrates (for a review on the structure of kinases, see (6)). Although the N-lobe of Bud32 harbors all the canonical elements characteristic of a functional protein kinase enzyme, the C-lobe appears abnormally minimalistic (see below). Despite the unusually abbreviated C-lobe, Bud32 demonstrated autophosphorylation activity in vitro (5,7–9), a property of most protein kinases (10). As expression of catalytically dead Bud32 mutants did not fully rescue the slow growth phenotype of the bud32Δ yeast strain (11), Bud32’s phospho-transfer function was inferred to be critical for its cellular function. Within the human kinome, Bud32 bears closest resemblance to the Rio kinases (12). Since the Rio kinases and Bud32 are the only two protein kinases conserved across eukaryotes and archaea (12,13), the two may represent primordial members of the eukaryotic protein kinase superfamily from which the numerous other family members in eukaryotes have evolved.
Subsequent studies identified a Bud32 ortholog in humans named Tp53 reactive kinase or p53-related protein kinase (TPRK or PRPK respectively) based on a report assigning p53 as a PRPK substrate (14), which was most likely an in vitro artifact. A yeast two-hybrid analysis using PRPK as bait identified TPRK-binding protein (TPRKB), the human ortholog of yeast Cgi121, as a binding partner (15). Cgi121 was a protein of unknown function described only to be conserved between Caenorhabditis elegans and humans in a Comparative Gene Identification study (hence its name) (16).
A separate yeast two-hybrid study with S. cerevisiae Bud32 as bait identified OSGEP, a poorly characterized protein that was erroneously reported to have O-sialo-glycoprotease activity, as a physical interactor (17). This led to the renaming of OSGEP to kinase-associated endopeptidase 1 (Kae1) (11). In some archaeal species such as Methanocaldococcus jannaschii, Kae1 and Bud32 are fused into a single polypeptide, hinting that their functions are intertwined.
As noted above, Kae1-family enzymes are part of a small group of ∼60 genes present in the genomes of all autonomous cells (3). Family members in archaea and eukaryotes are called Kae1 while the bacterial orthologs are called YgjD/TsaD (18). Similar to Kae1, TsaD is essential for bacterial growth (19). Eukaryote genomes also encode the Kae1 paralogue Qri7 (18). The open reading frame (ORF) of Qri7 contains an N-terminal mitochondrial targeting signal peptide. Accordingly, although Qri7 is encoded in the nucleus, it resides exclusively in the mitochondria (20–22). Analogous to the essential role of Kae1 and TsaD in cellular fitness, Qri7 is essential for the function of mitochondria (20,21).
Very little was known about Pcc1 and Gon7. Deletion of the GON7 gene (as well as BUD32) perturbed the transfer of mannosyl-phosphate groups to cell surface oligosaccharides in S. cerevisiae (23). Pcc1 was completely uncharacterized, in part because of a lack of annotation resulting from its coding by an ORF containing an intron with a non-canonical splice site in S. cerevisiae (24).
Converging roads to the discovery of the KEOPS complex
Two unrelated genetic screens conducted in S. cerevisiae using different strategies led to the discovery of the KEOPS complex. The first study conducted a genome-wide screen for suppressors of the growth defect of cells lacking a functional form of Cdc13, an essential protein that protects telomere ends from erosion (1). The analysis revealed that deletion of the CGI121 gene enabled growth in the absence of Cdc13. Tandem affinity purification experiments of Cgi121 from yeast cells coupled to mass spectrometry revealed that Cgi121 resides in a protein complex with Bud32, Kae1 and Gon7. The physical interaction between KEOPS proteins was further supported by a large-scale analysis of the yeast interactome (25). Supporting the notion that the newly identified subunits of KEOPS form a functional complex, knockout of any of these genes led to highly similar slow growth and shortened telomeres phenotypes (1), which are most likely an indirect result of the role KEOPS plays in translation fidelity (elaborated on later in this review).
The second study (2) utilized a different starting point, searching for suppressors of splicing defects caused by a U to A mutation at the fifth position of U1snRNP, which yields a cold-sensitive phenotype (26). The analysis revealed that the cold-sensitive phenotype could be overcome by expression of an intron-less version product of the S. cerevisiae YKR095‐A gene. This gene encoded a small uncharacterized protein that was required for the transcriptional activation of pheromone and galactose responsive genes and for polarized cell divisions. Chromatin-immunoprecipitation experiments showed the YKR095‐A protein associated with chromatin, leading to its naming as polarized growth chromatin-associated controller 1 (Pcc1) (2). Tandem affinity purification coupled to mass spectrometry revealed that Pcc1 resides within a complex with Gon7, Kae1, Bud32 and Cgi121.
THE ROLE OF KEOPS IN CELLS AND IN HUMAN DISEASE
KEOPS catalyzes the t6A modification of tRNA
The requirement for KEOPS for cell growth suggested that it fulfills an essential biochemical function. However, the precise role of KEOPS remained elusive until 2011. In this section, we provide an overview on how KEOPS activity is essential for the fidelity of translation through the role KEOPS plays in generating the universal tRNA modification N6-threonylcarbamoyl adenosine (t6A).
tRNAs undergo a complex and multi-staged biogenesis processes that includes the instalment of multiple posttranscriptional modifications, catalyzed by tRNA-modifying enzymes (27). To date, over 100 tRNA posttranscriptional modifications have been identified (28). Each tRNA is estimated to carry 5 to 15 modifications distributed throughout its structure. These modifications regulate a range of tRNA functions from degradation to roles in translation and modification dysregulation is implicated in human disease (29–32). In particular, the decoding activities of tRNAs are dependent on base modifications at positions 34 and 37 within the tRNA anticodon loop (29), which critically influence the rate and fidelity of translation and the integrity of the proteome (33). Of all tRNA modifications characterized, approximately 17% are conserved in all domains of life (34). Not unexpectedly, conserved modifications are generated by similarly conserved enzymes.
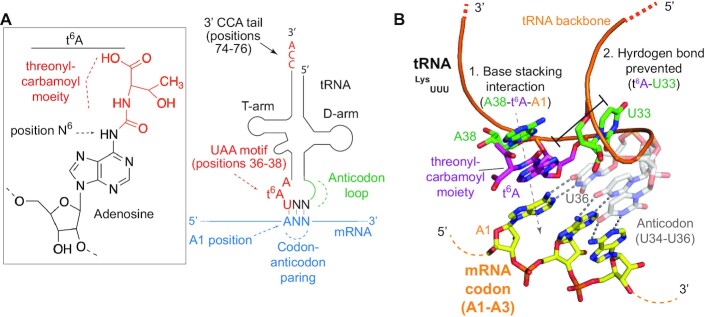
t6A is a universal tRNA modification discovered in 1969 (35) that is specifically found in tRNAs that decode ANN codons, where N denotes any nucleotide (Figure 1A) (28). t6A occurs exclusively on A37, one position 3′ to the anticodon triplet (encompassing positions 34 to 36) and remodels and stabilizes the structure of the tRNA anticodon loop by preventing an inter-loop hydrogen bond between A37 and U33 (Figure 1B) (36–40). This remodeling facilitates enhanced binding to the ribosome (41). In the ribosome, t6A reinforces A1:U36 codon-anticodon binding by inducing base stacking interactions between A37 and A38 of the tRNA and the A at the first position of the codon triplet in the mRNA (Figure 1B) (40,42,43). As a result, t6A is essential for the fidelity of translation and for the functionality of ANN-decoding tRNAs (44–47). t6A can be further modified to five different derivative forms (28). In Escherichia coli and possibly in other species, almost all t6A is cyclized (denoted ct6A) by the CsdL/TsdA enzyme (48). Crystal structures show that ct6A adopts a hydantoin structural form. Like t6A, ct6A stabilizes the structure of the anticodon loop. However, it is not clear if ct6A helps codon–anticodon binding in the ribosome (49). t6A can also be N6-methylated by TRMO (tRNA methytransferase O) to form m6t6A (50) or N2-methyl-thiolated by MtaB/Cdkla1l to form ms2t6A (51). These two specific t6A derivative modifications provide further tuning to the decoding activities of t6A modified tRNAs.
Figure 1.
t6A is a posttranscriptional modification of A37 in the anticodon loop of ANN-decoding tRNAs. (A) (left) Chemical structure of N6-threonycarbamoyladenosine (t6A). The threonylcarbamoyl moiety is shown in red and the adenosine is shown in black. (right) A schematic structure of an ANN-decoding tRNA engaged with an ANN codon within mRNA. Codon–anticodon pairing is shown in blue. The 34-UAA-36 motif and the position of t6A as well as the CCA tail are shown in red. (B) Structure of the anticodon loop of t6A modified Thermus thermophilus tRNALysUUU (PDB 1XM). The UUU anticodon triplet is shown in grey engaged with an AAA codon in yellow. For simplicity, the ribosome is not shown. A38 (green) and t6A (purple) of the tRNA form base stacking interactions with A1 of the mRNA (yellow). This stabilizes the binding of U36 (grey) with A1. The presence of t6A prevents base pairing with U33 (green). This stabilizes anticodon loop structure.
In 2009, 40 years after the discovery of t6A, the first enzyme in the t6A biosynthesis pathway was discovered through a comparative genomics approach that surveyed universally conserved proteins of unknown function (47) (for a detailed review of the conservation and diversity of t6A biosynthesis enzymes, see (18)). This analysis revealed that Sua5 (Suppressor of upstream AUG 5) (52) is directly involved in t6A biosynthesis. However, since Sua5 in isolation was not sufficient to t6A modify tRNA in vitro (47), the t6A biosynthesis pathway was predicted to involve additional gene products. Interestingly, some bacterial and archaeal genomes encode proteins that contain a Sua5-like domain fused to a Kae1-like domain (53) hinting at a linked functional role between these distinct classes of enzymes. Rhizobium fredii NolO, a Kae1-Sua5 like fusion protein, functions as a carbamoyl-transferase (54), which raised the possibility that Sua5 and Kae1 could participate in catalyzing reactions with similar chemistry (53), which ultimately proved to be the case.
In 2011, three breakthrough studies identified the Kae1/TsaD/Qri7 family of enzymes as the missing component in t6A biosynthesis (46,55,56). All three studies noted the similarity between Kae1 to the Kae1-Sua5 like fusion proteins as an indication that both enzymes might co-operate to generate t6A. The first study (56) noted that the extraordinary conservation of the Kae1 protein indicated that it would be involved in a universal cellular process. This led to a search for a potential role Kae1 might play in the synthesis of the major macromolecules in cells. While global DNA and RNA levels were only mildly affected in S. cerevisiae cells in the absence of Kae1 activity, protein synthesis was dramatically reduced, implicating Kae1 in translation. This finding led to the hypothesis that Kae1 works with Sua5 to t6A modify tRNAs. The second study (46) used a comparative genomics approach, reasoning that since t6A is universally conserved the candidate enzymes involved in its biosynthesis would have a universal phylogenetic distribution and an unknown biochemical function. Remarkably, one of the few enzymes that met both criteria were the Kae1/TsaD/Qri7 family of enzymes (57) hinting that Kae1 would collaborate with Sua5 in the t6A biosynthesis pathway. Both the first and second studies proved their working hypotheses by showing that mutations in Kae1 in S. cerevisiae resulted in a loss of t6A. The third study (55) revealed that the knockout of KEOPS genes leads to transcriptional activation of target genes of the transcription factor Gcn4 in a Gcn2-independent manner. Gcn4 protein expression is normally suppressed by upstream ORFs that contain AUG start codons. The Gcn4 protein is synthesized when the upstream kinase Gcn2 phosphorylates eIF2α, suppressing the inhibitory effect of the upstream ORFs (for an extensive review of Gcn4 translational regulation see (58)). Therefore, increased Gcn4 expression in the absence of KEOPS activity hinted that KEOPS would influence the selection of AUG start sites. Because Sua5 (45) and its downstream product t6A (47) are important for translation initiation, this led the authors to hypothesize that KEOPS is involved in generating the t6A modification.
Together, these breakthrough studies unequivocally proved that Sua5 and Kae1/TsaD/Qri7 are needed for the t6A modification of tRNA in vivo (46,47,55,56). Subsequent studies with purified proteins (together with their respective binding partners) demonstrated the sufficiency of these enzymes to reconstitute the t6A modification of tRNA in vitro (59–61). The specific role each enzyme plays in this process and how these roles were assigned is described later in this review.
KEOPS activity is essential for diverse aspects of cell biology and mutations in KEOPS underlie Galloway–Mowat syndrome
Genetic studies conducted in the eukaryotic species yeast, zebra fish, fruit flies and mice as well as the archaeon Haloferax volcanaii have established an essential role for KEOPS in normal cell growth and in animal development.
KEOPS function has been most extensively characterized in vivo in budding yeast. Knockout of any individual KEOPS gene leads to pleiotropic phenotypes, ranging from slow growth to shortened telomeres and to defects in transcription (1,2,44,55,62,63). These phenotypes are most likely an indirect result of the role KEOPS plays in tRNA modification and translation and of the far-reaching downstream effects that the perturbation of translation has on virtually all cellular processes. The impact of KEOPS activity on translation has been studied using reporter based assays, which showed that the absence of the t6A modification leads to an increase in translation mis-initiation and in ± 1 frame-shift events (46,47,55) as well as defects in stop codon recognition (45,64). A higher resolution study using ribosome profiling revealed that the role t6A has in translation varies considerably from one ANN codon to another. The translation rates of ANN codons that are decoded by rare tRNAs or tRNAs with weak G34:U3 pairing at the wobble position decreased in the absence of t6A. In contrast, the translation rates of ANN codons that are dependent on high abundance tRNAs or tRNA with strong I34:C3 pairing at the wobble position were in fact increased by the absence of t6A (44). Both types of changes in translation rate likely have a major role in the perturbed translation observed in the absence of t6A.
The importance of KEOPS activity for the well-being of cells has been confirmed in a variety of other model organisms. Briefly, in the archaeon H. volcanaii, the cgi121 and kae1-bud32 fusion genes are essential for cell growth, while knockout of pcc1 yields a less severe growth defect (65). In Drosophila melanogaster, only Pcc1, Kae1 and Bud32 (termed Prpk) orthologs have been identified thus far (18,66). Knockout or knockdown of Kae1, Bud32, Pcc1 or Sua5 in Drosophila leads to reduced larval size and transformation to pupa (66–69). The sensitivity to Kae1 knockdown varies between different tissues, with mitotic tissues especially sensitive while non-proliferating tissues much less sensitive (68). Interestingly, the rare larvae that manage to transform to pupa despite knockdown of Prpk develop into flies with reduced cell and organ size but without extreme defects in body patterning (70). This result is reminiscent of mutants in the TOR pathway (71) and hints at a potential role for KEOPS in regulating TOR signaling (67,72). In agreement with this possibility, the growth defects induced by Prpk knockdown are reversed by expression of an activated mutant of the S6 kinase (70), a downstream effector of the TOR pathway (73). In Danio rerio, osgep and tprkb knockout (Kae1 and Cgi121 orthologs respectively) leads to microcephaly and to a shortened life span (74,75). Similarly, knockout of either of the osgep, lage3, prpk or tprkb (Kae1, Pcc1, Bud32 and Cgi121 orthologs respectively) genes in Mus musculus leads to severe microcephaly and early lethality (74). Why the developmental defects resulting from KEOPS dysregulation localize with great frequency to the brain remains an open question.
In line with studies in model organisms, genetic analysis in humans have identified pathogenic mutations that underlie Galloway–Mowat syndrome (GAMOS) in each of the five KEOPS genes and in Sua5 (74,76,77). GAMOS is a recessive genetic disease that manifests in renal and neurological dysfunctions with a considerable variety of severity (74,76–78). As a result, GAMOS patients die at very early stages of life. Since GAMOS inducing mutations are common to KEOPS and Sua5 encoding genes, it is highly likely that reduced t6A levels underpins the GAMOS disease phenotype. Consistent with this notion, introduction of GAMOS disease mutations into yeast Kae1 inhibits the modification of tRNA in vivo (74,76). KEOPS pathogenic mutations elicit complex cellular phenotypes that likely stem from perturbed translation including activation of the DNA-damage and unfolded protein response, ER stress and defects in actin regulation (74,76). Some pathogenic mutations in KEOPS exert their effect by disrupting the formation of KEOPS, while others are pathogenic for unknown reasons (74,77). Further studies are required for a complete understanding of the molecular mechanisms connecting KEOPS mutations to GAMOS.
t6A derivative modifications are also important for cellular processes and their dysregulation is implicated in human disease. Mammalian Cdkal1 for example catalyzes the methythiolation of t6A to ms2t6A (51). In the absence of Cdkal1 activity, insulin synthesis is inhibited (51) due to misreading of lysine codons in pro-insulin (79) leading to development of type 2 diabetes.
HOW KEOPS FUNCTIONS TO MODIFY tRNA
Sua5 makes the t6A modification reaction intermediate threonlycarbamoyl adenylate, which Kae1 uses to modify tRNA
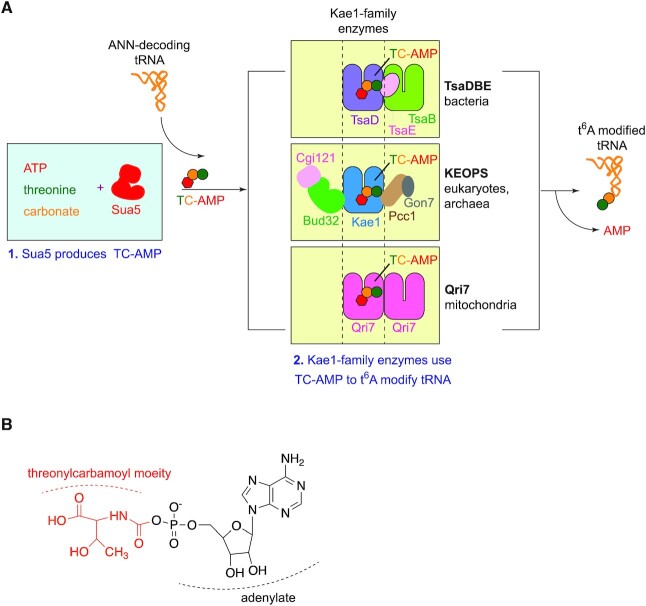
The mechanism by which Sua5 and Kae1 collaborate to modify tRNA with t6A was delineated through a series of elegant biochemical and structural studies described below. Briefly, the t6A modification of tRNA occurs in a reaction with two major steps. In the first step, Sua5 utilizes threonine, CO2/HCO3− and ATP precursors to generate the diffusible intermediate molecule threonylcarbamoyl adenylate (TC-AMP) while releasing PPi from ATP. In the second step, TC-AMP and tRNA are bound by Kae1/TsaD/Qri7, which then transfer the threonylcarbamoyl moiety from TC-AMP to the N6 acceptor site of A37 in tRNA in a reaction that generates AMP (Figure 2A).
Figure 2.
t6A is universally biosynthesized in a two-step reaction by Sua5 and Kae1/TsaD/Qri7 enzymes. (A) Schematic of the universal t6A biosynthesis pathway. Step 1- Sua5 utilizes ATP, threonine and HCO3−/CO2 to catalyze the formation of threonylcarbamoyl adenylate (TC-AMP). Step 2- TC-AMP is used by Kae1/TsaD/Qri7 enzymes to t6A modify ANN-decoding tRNA substrates. (top) TsaD functions with the TsaB and TsaE subunits in bacteria. (middle) Kae1 functions with Cgi121, Bud32, Pcc1 and Gon7 subunits in the cytoplasm in eukaryotes and in archaea. (Bottom) Qri7 functions either as a homo-dimer as shown in figure or as a monomer. (B) Chemical structure of threonylcarbamoyl adenylate (TC-AMP). The threonylcarbamoyl moiety is shown in red and the adenylate is shown in black.
Sua5 generates TC-AMP
As early as 1974, t6A biosynthesis was known to require the precursors threonine, ATP and HCO3− (80). Knowing that Sua5 was involved in the t6A modification process and that Sua5 has ATP binding properties (81,82), Deutsch et al. and Perrochia et al. examined the effect of threonine and tRNA on its ATP hydrolysis activity. They found that ATP consumption by Escherichia coli, Pyrococcus abyysi and S. cerevisiae Sua5 proteins was dependent on threonine (but not tRNA). This hinted that Sua5 produces an adenylated threonine containing molecule for use in the t6A modification reaction (59,60). Subsequent studies using Bacillus subtilis and S. cerevisiae Sua5 proteins showed this molecule to be TC-AMP (Figure 2B) (83). TC-AMP is a relatively short-lived molecule that breaks down to threonylcarbamoyl and AMP moieties. Its stability is strongly affected by pH and temperature, with a half-life of ∼20 seconds at pH 7.5 and at 37°C (83). TC-AMP can be stabilized by its binding to Sua5, but not by its binding to Kae1/TsaD/Qri7 enzymes (84). The structural basis by which Sua5 generates TC-AMP was delineated by X-ray co-crystal and NMR structures of Sua5 with threonine, ATP and HCO3− precursors (81,85–87). After the discovery that Sua5 produces TC-AMP, re-analysis of a previously determined crystal structure of Sulfolobus tokodaii Sua5 bound to ATP, revealed serendipitously that the ATP moiety bound in the Sua5 active site was actually TC-AMP (81,88).
Kae1/TsaD/Qri7 uses TC-AMP to make t6A
The discovery that Sua5 generates the precursor TC-AMP suggested that the Kae1/TsaD/Qri7 enzymes would employ TC-AMP and tRNA to catalyze the remainder of the t6A modification reaction. In support, P. abyssi KEOPS binds tRNA in vitro and co-purifies with tRNA when expressed in E. coli cells, suggesting that this complex directly acts on tRNA (60). Evidence that the Kae1/TsaD/Qri7 enzymes carry out the final step of the t6A modification reaction came from the analysis of Qri7, which together with Sua5 are sufficient to carry out the t6A modification of tRNA in vitro (61). tRNA modification could be achieved with Sua5 and Qri7 in chambers separated by a 2 kDa size cut off membrane but only when tRNA was in the same chamber as Qri7 (61). In vivo, it is currently unclear how the unstable TC-AMP is efficiently shuttled to the active site of the Kae1/TsaD/Qri7 enzymes before its breakdown.
Remarkably, although all three Kae1 family orthologs perform the same biochemical reaction, each ortholog functions in a completely different protein complex (Figure 2A). The mitochondrial Qri7 is the most simplified member of the family and catalyzes tRNA modification without any auxiliary proteins (61,89). The bacterial TsaD protein depends on its binding to YaeZ/TsaB, which is a structural mimic of Kae1 enzymes (90–92). TsaD bound to TsaB can bind tRNA but has limited catalytic capacity and likely catalyzes only a single round of tRNA modification (93). For multiple rounds of tRNA modification, TsaD-TsaB depend on the activity of the ATPase subunit YjeE/TsaE (59,92–95) for reasons that are not entirely understood. Notably, TsaB and TsaE are not similar in structure and apparent function to any KEOPS subunits. The significance of the different complex compositions is one of the most exciting mysteries in the field that remain unresolved (see ‘CONCLUDING REMARKS’ section).
The tRNA modification activity of Sua5 and Kae1/TsaD/Qri7 enzymes extends to tRNA modifications other than t6A. Sua5 can generate adenylated variants of TC-AMP using serine or hydroxy norvaline (hn) in place of threonine. In turn, the bacterial TsaD protein can utilize hn-carbamoyl adenylate to generate an hn6A modification on substrate tRNAs (96). Since the hn6A modification is also observed in archaeal species (97,98), this would suggest that KEOPS in both archaea and eukaryotes may similarly utilize hn-carbamoyl adenylate to modify tRNA. However, the functional relevance of this modification remains in question.
Structure of the individual subunits
Bud32 is an atypical protein kinase that lacks the infrastructure for canonical protein substrate recognition
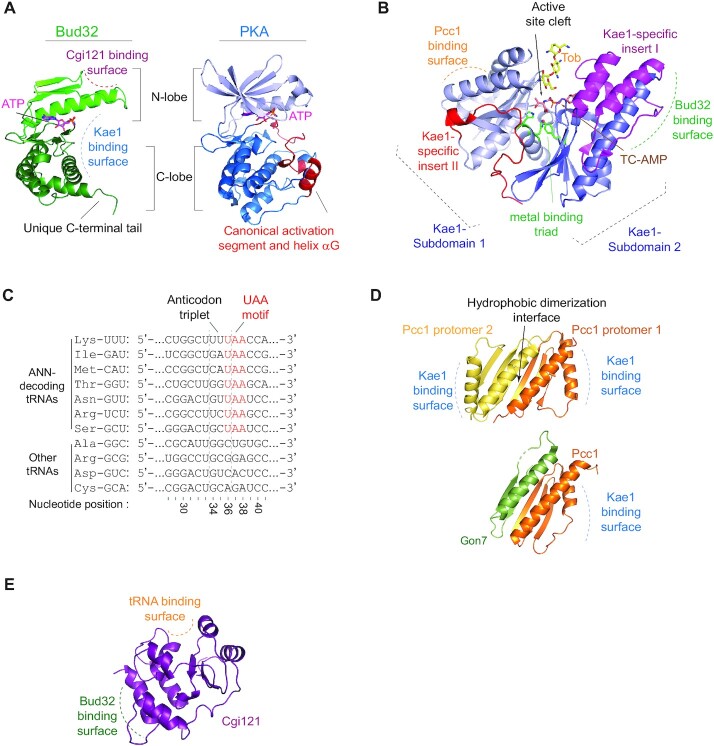
Eight crystal structures of complexes containing Bud32 have been deposited to the PDB (for a list of the structures of proteins that take part in t6A modifying complexes, see Table 1). These include structures of Bud32 orthologs from archaea, yeast and human in complex with either Kae1, Cgi121 or both. These structures reveal a bi-lobal domain organization characteristic of the eukaryotic protein kinase family as exemplified by PKA, a well-studied prototypical kinase (Figure 3A) (for review on the composition of the kinase domain, see (6)). The kinase N-lobe of Bud32 possesses the canonical antiparallel β−strands, buttressed against a helix αC (Figure 3A) (8,9,99). The C-lobe, which is connected to the N-lobe through a flexible hinge region, is predominantly α-helical and greatly abbreviated (i.e. of smaller size) relative to the C-lobes of other eukaryotic protein kinases. One peculiar feature of the abbreviated kinase C-lobe is the absence of all infrastructure typically required for protein substrate recognition, including helix-αG and the activation segment (100) (Figure 3A). A second unique feature of the kinase C-lobe is the presence of a basic C-terminal tail with no apparent homology in other members of the kinome (5).
Table 1.
Structures of KEOPS proteins and sub-complexes. All structures reported are crystal structures unless stated otherwise. f = fusion protein. car-AMP = carboxy-adenylate. BK951 = TC-AMP mimetic.
| t6A modifying complex | Proteins | Species | Ligand | PDB code |
|---|---|---|---|---|
| Eukaryotes (cytoplasm) and archaea | Bud32-Cgi121 | S. cerevisiae | - | 4WWA |
| Bud32-Cgi121 | S. cerevisiae | AMP | 4WW7 | |
| Bud32-Cgi121 | S. cerevisiae | ADP | 4WW9 | |
| Bud32-Cgi121 | S. cerevisiae | AMP-PnP | 4WW9 | |
| Bud32-Cgi121 | H. sapiens | AMP-PnP | 6WQX | |
| Kae1-Bud32(f) | M. jannaschii | AMP-PnP | 2VWB | |
| Kae1-Bud32(f) | M. jannaschii | - | 3EN9 | |
| Kae1 | P. abyssi | - | 2IVO | |
| Kae1 | P. abyssi | ATP | 2IVP | |
| Kae1 | P. abyssi | AMP-PnP | 2IVN | |
| Kae1-Pcc1 | T. acidophilum, P. furiosus | - | 3ENO | |
| Kae1-Pcc1 | M. jannaschii, P. furiosus | AMP | 5JMV | |
| Kae1-Bud32(f)-Cgi121 | M. jannaschii | - | 3ENH | |
| Pcc1-Pcc1 dimer | P. furiosus | - | 3ENC | |
| Pcc1-Gon7 | S. cerevisiae | - | 4WXA | |
| Pcc1-Gon7 | S. cerevisiae | - | 4WX8 | |
| Gon7-Pcc1-Kae1 | H. sapiens | - | 6GWJ | |
| Cgi121 | S. cerevisiae | - | 4XAH | |
| Cgi121 | H. sapiens | - | 3ENP | |
| Cgi121 (NMR) | M. jannaschii | - | 2K8Y | |
| Cgi121 | M. jannaschii | tRNA | 7KJT | |
| Mitochondria | Qri7-Qri7 dimer | S. cerevisiae | 4K25 | |
| Qri7 | S. cerevisiae | AMP | 4WUH | |
| Bacteria | TsaB dimer | E. coli | - | 1OKJ |
| TsaB dimer | S. enterica | - | 2GEM, 2GEL | |
| TsaB dimer | V. parahaemolyticus | - | 3R6M | |
| TsaB dimer | P. aeruginosa | - | 4Y0W, 5BR9 | |
| TsaB dimer | T. maritima | - | 2A6A | |
| TsaE | H. influenzae | - | 1FL9 | |
| TsaE | H. influenzae | ADP | 1HTW | |
| TsaE | B. subtilus | ADP | 5MVR, 5NP9 | |
| TsaD-TsaB | E. coli | BK951, AMP | 6Z81 | |
| TsaD-TsaB | E. coli | ADP | 4YDU | |
| TsaD(E12A mutant)-TsaB | E. coli | ATP | 4WQ4 | |
| TsaD(V85E mutant)-TsaB | E. coli | ATP | 4WQ5 | |
| TsaD-TsaB | S. typhimurium | AMP | 3ZET | |
| TsaD-TsaB | S. enterica | ATP-γ-S | 3ZEU | |
| TsaD-TsaB-TsaE | T. maritima | ATP, car-AMP | 6N9A | |
| TsaD-TsaB-TsaE | T. maritima | AMPcPP | 6S84 |
Figure 3.
Structural characteristics of the individual KEOPS subunits. (A) Bud32 is an atypical protein kinase. (left) The structure of M. jannaschii Bud32 (PDB 2VWB) is shown with AMP-PnP in the active site (depicted as ATP for simplicity). Highlighted is the unique C-terminal tail and the location of the binding surfaces for Cgi121 and Kae1. (Right) Structure of the kinase domain of M. musculus PKA (PDB 1ATP) is shown with ATP in the active site. Highlighted are the canonical activation segment and helix-αG (red) that are distinctively absent from the structure of Bud32. (B) Kae1 is an ASKHA fold enzyme with two unique insertions. The structure of M. Jannaschii Kae1 (PDB 2VWB) is shown with the threonylcarbamoyl adenylate (TC-AMP) mimetic BK951 (PDB 6Z81) and substrate analogue tobramycin (tob, PDB 3VET) superimposed on its active site. Highlighted are the Kae1-specific inserts I and II (purple and red, respectively) and the location of the binding surfaces for Bud32 and Pcc1. (C) Kae1/TsaD/Qri7 enzymes substrate tRNAs harbor a universal UAA motif in the anticodon loop. A sequence alignment of representative ANN-decoding and non-ANN-decoding tRNAs from M. jannaschii is shown highlighting that ANN-decoding tRNAs harbor a 36-UAA-38 motif (red) within the anticodon loop that partially overlaps with the anticodon triplet (positions 34–36). Note that none of the non-ANN-decoding tRNAs have this motif. (D) Pcc1 can homo-dimerize or hetero-dimerize with Gon7. (top) The structure of P. furiosus Pcc1 homo-dimer is shown (PDB 3ENC). The dimerization interface and the location of the Kae1 binding surface are highlighted. (bottom) The structure of human Pcc1-Gon7 hetero-dimer is shown (PDB 6GWJ). Note the similarity between the Pcc1 homo-dimer and Pcc1-Gon7 hetero-dimer configuration. (E) Cgi121 is a single domain protein. The structure of M. jannaschii Cgi121 (PDB 3ENH) is shown. The location of the binding surfaces for Bud32 and tRNA are highlighted.
ATP binds to the catalytic cleft of Bud32 situated between the N and C- lobes. The ATP binding mechanism in Bud32 is conserved with that of other protein kinases (6,99,101). Specifically, the adenine base of ATP is wedged towards the kinase hinge. The ribose and phosphate groups are coordinated by the conserved β−3 Lys, αC-helix Glu, Gly rich loop, and Asp side chain of the conserved Asp-Phe-Gly (DFG) motif through a bridging Mg/Mn ion. Phospho-transfer function of protein kinases is mediated in part by a catalytic Asp residue within a conserved His-Arg-Asp (HRD) motif in a region termed the catalytic loop (6). This Asp residue functions as a catalytic base to extract a proton from the target hydroxyl group and is conserved in Bud32, suggesting that Bud32 also shares the ability to catalyze phosphate transfer. Accordingly, Bud32 proteins manifest autophosphorylation activity (5,8,9) and its phospho-transfer function is essential for biological function (9,11). However, the absence of structural elements normally required for protein phosphorylation as well as experiments showing that the autophosphorylation acceptor-sites are not required for physiological activity (7,102) suggests an alternate role for its phospho-transfer function. In line with this prediction, the chief function of Bud32 appears directed at ATP hydrolysis (i.e. transfer of phosphate to water) (102).
Kae1 has a bi-lobal ASKHA fold
Nine crystal structures of Kae1 alone or in complex with different KEOPS proteins have been deposited to the PDB (Table 1). These include structures of Kae1 orthologs from archaea, yeast and humans either alone, in complex with Pcc1, in complex with Pcc1 and Gon7, in complex with Bud32 or in complex with Bud32 and Cgi121. Likewise, multiple structures of the Kae1 orthologs Qri7 and TsaD have been solved in their respective complexes. These structures reveal that Kae1 adopts a bi-lobal domain organization characteristic of the ASKHA (acetate and sugar kinases/Hsc70/actin) family of enzymes (Figure 3B) (103,104). The two subdomains of Kae1 bear considerable similarity to each other and contain a five stranded β-sheet core flanked by α-helices. In the cavity between the two subdomains, lies a conserved metal binding triad, which co-ordinates a Zn, Mg or Fe ion. Kae1 enzymes have two unique inserts relative to other ASKHA fold enzymes, termed Kae1 specific insert I and II, which are tightly associated with sub-domain 2 and 1 respectively (Figure 3B) (9,104). Unique inserts in ASKHA-fold enzymes commonly contribute to the unique sub-family functions (103), which also holds true for Kae1 proteins (see below).
How does the active site of Kae1 bind its substrates TC-AMP and tRNA? The unstable nature of TC-AMP hindered the ability to obtain atomic level insight into how it is bound by Kae1-family enzymes. However, recently, Kopina et al. developed a non-hydrolysable TC-AMP analogue (termed BK951) and reported its structure bound to the E. coli TsaD-TsaB heterodimer (84). The structure revealed that the adenine base portion of TC-AMP is ‘sandwiched’ between subdomain 2 and Kae1 specific insert I, similar to the nucleotide binding mechanism of Kae1 enzymes reported in previous structures (8,84,91,95,105). A cation bound by the metal binding triad serves to coordinate the carboxylate group of the threonine moiety of TC-AMP (Figure 3B). This binding mode directs the threonylcarbamoyl group towards a cavity in the active site cleft, presumably guiding it to the binding site for tRNA. How the threonylcarbamoyl group is activated by Kae1 for the transfer reaction to N6 of A37 remains unclear. To date, there is no structure of a Kae1 family enzyme with a tRNA substrate. The expected position of A37 can at best be inferred from the co-structure of TobZ, a NodU family carbamoyl transferase, with its substrate the antibiotic tobramycin (53,88). Since A37 and tobramycin should occupy a similar position in their respective enzyme's active site, superimposing the structure of the TobZ-tobramycin complex onto Kae1 places A37 in close proximity to TC-AMP and the metal binding triad (Figure 3B).
What features of substrate tRNAs dictates the substrate selectivity of Kae1 enzymes? ANN-decoding tRNAs universally harbor a conserved 36-UAA-38 motif within the anticodon loop with the A37 site at its center (Figure 3C). This motif partially overlaps with the anticodon at positions 34 to 36. The presence of the 36-UAA-38 motif can be explained in part through the roles these nucleotides play in translation (elaborated on above and shown in the structure presented in Figure 1B). However, the UAA motif also plays a role in the ability of the tRNA to be modified by Kae1. Mutation of U36 or A38 disrupts t6A modification, demonstrating the necessity of the UAA motif for tRNA recognition by Kae1. Insertion of a 36-UAA-38 motif into the anticodon loop of non-substrate tRNAs can lead to their modification (106–108) demonstrating sufficiency in some instances. For example, mutations of either U36 or A38 in tRNAIle disrupts its t6A modification in Xenopus laevis oocytes (107). In the same system, converting the 36-CAC-38 motif of tRNAVal, which is normally not t6A modified, to a 36-UAA-38 motif was sufficient for inducing its t6A modification (107). Highlighting biological significance, mutation of the UAA motif in substrate tRNAs can contribute to human disease. For example, pathogenic mutations that cause myoclonus epilepsy associated with ragged red fibers (MERRF) disease alters the 36-UAA-38 motif of mitochondrial (m) tRNAThr to UAG. This mutation inhibits its t6A modification by the human Qri7 orthologue OSGEPL1 (106,108). In contrast, mtRNAIle naturally harbors a non-canonical 36-UAG-38 motif and is therefore normally not a substrate for t6A modification. However, a mtRNAIle G38A pathogenic mutation that leads to Leigh syndrome reconstitutes a canonical 36-UAA-38 motif, inducing the unnatural t6A modification of this tRNA (106). As noted below, there are other substrate determinants that are distal to anticodon region (see below).
Pcc1 has homo-dimerization capabilities
Six structures of Pcc1 from archaeal, yeast and human species have been deposited to the PDB (Table 1). These structures include those of the Pcc1 homo-dimer and of the Pcc1-Kae1, Pcc1-Gon7 and Pcc1-Kae1-Gon7 complexes. These structures reveal that Pcc1 is a small protein (∼8 kDa in Pyrococcus furiosus for example) composed of a three-strand antiparallel β-sheet flanked on one side by two α-helices (Figure 3D). Pcc1 alone crystalizes as a homo-dimer with an antiparallel configuration of the two monomers. The dimerization of Pcc1 is mediated through a hydrophobic interface, which is generated mainly by the longer of the two helices in each monomer. Mutations in this interface in P. furiosus Pcc1 yields insoluble proteins, suggesting that archaeal Pcc1 is an obligatory dimer (9). The Pcc1 dimer generates a continuous six anti-parallel β-sheet on one face of the dimer packed against four continuous helices of the other face. Since there are no structures of a eukaryotic Pcc1 protein in its apo form, it is unclear if all Pcc1 proteins are obligatory dimers, but the ability to dimerize has been reported for the human Pcc1 ortholog Lage3 (109).
Gon7 is an intrinsically disordered protein that becomes partially structured in binding to Pcc1
Gon7 alone is an intrinsically disordered protein (109). However, a portion of Gon7 assumes a stable structure upon binding to Pcc1 (77,99). Three structures of Gon7 from yeast and human species have been deposited to the PDB (Table 1). These structures include those of Gon7-Pcc1 and Gon7-Pcc1-Kae1 complexes. The structured region of Gon7 (residues 1 to 20 and residues 25–50, human nomenclature used) form two anti-parallel β-sheets and a single α-helix (Figure 3D). The C-terminus of Gon7 (residues 51–100) is enriched with acidic residues, a feature that is conserved among Gon7 orthologs (77,99).
Cgi121 structure
Nine crystal structures and 1 solution structure solved by NMR of Cgi121 alone or in complex with other KEOPS proteins or substrate tRNA have been deposited to the PDB (Table 1). These include the structures of Cgi121 orthologs from archaea, yeast, and human either alone, in complex with Bud32, in complex with Bud32-Kae1 or in complex with tRNALysUUU. These structures revealed that Cgi121 is a single domain protein containing a core antiparallel β-sheet of 3 to 5 strands sandwiched between α-helices on either side (9) (Figure 3E).
Basis for KEOPS subunit interaction with each other and with substrate tRNA
Overview: KEOPS subunits adopt a linear binding topology
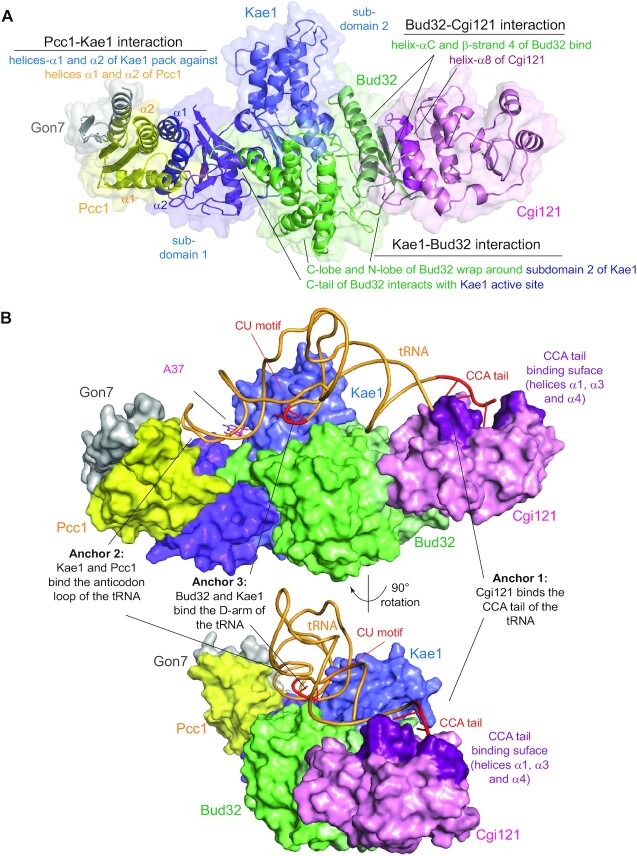
The architecture and tRNA binding mechanism of KEOPS have been elucidated by solving a series of binary and trinary structures of its subunits (Table 1). In brief, KEOPS adopts a linear binding architecture (Figure 4A). The two enzymes Kae1 and Bud32 bind directly to each other at the center of KEOPS. On opposing ends, Bud32 binds to Cgi121 and Pcc1 binds to Kae1 (Figure 4A). Bound to Kae1, Pcc1 exists as a homo-dimer or as a hetero-dimer with Gon7 (Figure 3D).
Figure 4.
A model for the KEOPS-tRNA holo-enzyme-substrate complex. (A) Model of KEOPS reveals the basis for the interactions between its subunits and shows that they adopt a linear binding. A cartoon representation with semi-transparent surface of a model of the KEOPS complex. The model was generated using the structures of the Cgi121-Bud32-Kae1, Kae1-Pcc1 and Gon7-Pcc1-Kae1 sub-complexes (PDB 3ENH, 3ENO and 6GWJ respectively, see Table 1). Gon7 (gray) binds to Pcc1 (yellow), which binds to Kae1 (blue). Kae1 binds also to Bud32 (green), which binds Cgi121 (purple). Bud32 (green) binds to subdomain 2 of Kae1 (light blue) through an extensive surface that is composed of the N and C lobes. The C-tail of Bud32 interacts with the active site cleft of Kae1. Pcc1 (yellow) binds to helices α1 and α2 of subdomain 1 of Kae1 (dark blue) in vicinity of the active site cleft of Kae1. Cgi121 (purple) binds to helix-αC and β-strand 4 of the N-lobe of Bud32 (dark green) through helix α8. (B) A model of KEOPS-tRNA holoenzyme substrate complex shows the basis for substrate interaction by KEOPS. For clarity, proteins are shown in surface representation and tRNA in cartoon representation and complex is shown in two different orientations. According to the model, Cgi121 (pink) binds the tRNA CCA tail (red) via helices α1, α3 and α4 (purple). Kae1 (blue) and Pcc1 (yellow) interact with the tRNA anticodon loop and Bud32 (green) and Kae1 interact with the tRNA D-arm. Gon7 (gray) does not participate in tRNA binding. This binding mode directs A37 of tRNA (magenta) into the active site of Kae1 and positions the CU motif of tRNA (red) to the interface between Bud32 and Kae1.
The kinase domain of Bud32 ‘wraps’ around subdomain 2 of Kae1
Consistent with the initial yeast two hybrid analysis (11), Bud32 binds directly to Kae1. The Kae1 binding surface in Bud32 is composed of regions from both N and C lobes (Figure 4A). The reciprocal binding surface on Kae1 is composed of subdomain 2, including the Kae1-specific insert I. The highly conserved C-terminal tail of Bud32 interacts with the active site of Kae1 (Figure 4A) (8,9). While not essential for binding to Kae1, the C-terminal tail of Bud32 is required the ability of KEOPS to modify tRNA in vitro and for KEOPS functionality in vivo as assessed in yeast (9,102).
Pcc1 binds to subdomain 1 of Kae1 and can control KEOPS dimerization
The Pcc1 binding surface on Kae1 is composed primarily by helices α1 and α2, which pack against helices α1 and α2 of Pcc1 (Figure 4A). The Kae1-Pcc1 binding mechanism is similar to that of TsaD-TsaB and to the dimerization mechanism of Qri7. The significance of this convergent binding mode is currently unclear (see CONCLUDING REMARKS section). The Pcc1 homo-dimer presents two non-overlapping Kae1-binding surfaces, raising the enticing possibility that KEOPS might dimerize through Pcc1. In support, archaeal KEOPS with wild-type Pcc1 forms dimers in solution (60,110). In contrast, KEOPS reconstituted with a synthetic asymmetric Pcc1-Pcc1 fusion protein with disruptive mutations on one of two Kae1 binding surfaces is monomeric (110). Since monomeric and dimeric KEOPS complexes have comparable tRNA modification activity in vitro, the role of this dimerization is unclear (110) (for a model of a KEOPS dimer see (9)). Gon7 binds to Pcc1 through the surface that mediates the dimerization of archaeal Pcc1 (Figure 3D), Gon7 was therefore proposed to regulate the dimerization function of Pcc1 and thus KEOPS dimerization (99). In support, yeast KEOPS that includes Gon7 is monomeric in solution with a 1:1 binding stoichiometry between its all five subunits (99). Furthermore, 4-subunit human KEOPS (without hGon7) forms dimers in solution while 5-subunit human KEOPS is monomeric (109,111). Lastly hGon7 appears to enhance or stabilize the expression levels of KEOPS in vivo since in the absence of hGon7 overall expression levels of KEOPS subunits are repressed (77). These findings might hint indirectly that KEOPS dimerization could regulate its degradation in eukaryotes, however this possibility requires further exploration.
Cgi121 binds and regulates Bud32 and recruits tRNA to KEOPS
Consistent with the initial yeast two-hybrid analysis (15), Cgi121 binds directly to Bud32. In archaeal protein structures, the Cgi121 binding surface on Bud32 is composed primarily of N-lobe helix-αC and β-strand 4 (Figure 4A). The reciprocal binding surface on archaeal Cgi121 is composed primarily of helix α8. The Cgi121 binding mode in the eukaryotic proteins is essentially the same with slight elaborations. The N-lobe of eukaryotic Bud32 is slightly larger than its archaeal ortholog with an extra helix (termed αA-helix) that participates in binding eukaryotic Cgi121. The binding mode between Bud32 and Cgi121 is reminiscent of cyclin-dependent kinases binding to cyclins (112). Thus, not unexpectedly, Cgi121 potentiates both the ATPase and low level autophosphorylation activity of Bud32 (note that the autophosphorylation activity is unlikely to be physiologically relevant) (9,101,102). Structures of Bud32 in complex with Cgi121 reveal an active-like conformation with productively positioned catalytic elements (99,101). To date, no structures of Bud32 alone have been reported for comparison.
Apart from of its ability to bind Bud32, Cgi121 has intrinsic ability to bind tRNA. This ability is essential for the efficient recruitment of tRNA to KEOPS. A co-crystal structure of a Cgi121-tRNA complex revealed that tRNA binding by Cgi121 is directed almost exclusively towards the 3' CCA tail (positions 74–76, 5'-C74-C75-A76-3') (Figure 4B) (102). The binding surface on Cgi121 for tRNA is composed primarily of α−helices 1, 3 and 4. The CCA tail is a universal element present in all mature tRNAs that plays multiple essential roles in the functionality and regulation of tRNAs. Perhaps most importantly, the terminal nucleotide of the CCA tail serves for the attachment of amino acids (aminoacylation) by aminoacyl tRNA synthetases (113). The CCA tail also plays an important role in quality control of tRNAs and translation. For example, the CCA tail is excised by ANKZF1 to help the recycling of stalled ribosomes (114) and a second CCA tail is added by the CCA-adding enzyme to structurally unstable tRNAs as a signal for their destruction by the tRNA degradation machinery (115). Cgi121 tRNA binding activity is dependent on the presence of an intact and non-aminoacylated CCA tail. Specifically, deletion of the CCA tail, addition of a second CCA or attachment of bulky groups to the 3′ of A76 inhibits tRNA binding by Cgi121 (102). This dependency renders KEOPS activity highly sensitive to the status of the CCA tail of tRNA substrates. The biological role of this dependency is still not understood (see CONCLUDING REMARKS section).
A model for the KEOPS-tRNA holo-enzyme-substrate complex
A model for the KEOPS-tRNA holo-enzyme substrate complex was generated using the Cgi121-tRNA structure and structures of KEOPS sub-complexes (Figure 4B) (102). According to this model, all four core KEOPS subunits (i.e. Cgi121, Bud32, Kae1 and Pcc1) form an extended tRNA binding surface that is highly complementary to the structure of the tRNA. The tRNA is anchored on both ends to the KEOPS complex with the CCA end bound by Cgi121 and the anticodon loop interacting with Kae1 and Pcc1. At its center, the tRNA D-arm is predicted to contact Bud32 and Kae1 (Figure 4B). Therefore, the tRNA binding mode of KEOPS seems to interrogate the entire ‘inner side’ of the L shape of tRNA. Although all subunits are predicted to take part in contacting tRNA, only mutations on Bud32 and Cgi121 adversely affect tRNA binding affinity. Bud32 does not bind tRNA independently but serves to potentiate the tRNA binding activity of Cgi121. Bud32 achieves this likely by forming secondary contacts with tRNA and by allosterically inducing a high affinity state in the CCA tail binding surface of Cgi121 (102). Despite not contributing to binding affinity, the predicted tRNA binding surface in Kae1 and Pcc1 are none-the-less vital for tRNA modification activity (102).
The ATPase activity of Bud32 is regulated by tRNA binding to KEOPS
The ATPase activity of Bud32 is essential for the ability of KEOPS to modify tRNA substrates in vitro and is required for Bud32’s essential function in vivo (9,102). Interestingly, activation of Bud32 ATPase activity is strongly potentiated by tRNA, but only in the context of the Cgi121-Bud32-Kae1 sub-complex or the full KEOPS complex (102). Mutations in any of the KEOPS subunits on the predicted tRNA binding surfaces inhibit the ability of tRNA to activate Bud32 ATPase. How tRNA substrates regulate Bud32 ATPase activity remains an unresolved question. However, in archaea this ability is not correlated with the anticodon loop UAA motif but rather the presence of a di-nucleotide CU motif located at positions 10–11 within the tRNA D-arm (102). This motif is almost completely conserved in ANN-decoding tRNAs and absent from almost all non-ANN-decoding tRNAs with the sole exception of tRNAAla. In the KEOPS-tRNA complex, the CU motif is predicted to localize at the interface between Bud32 and Kae1 in proximity to the active site of Bud32 (Figure 4B) (102). The role this element has in the activation of Bud32 and in the ability of KEOPS to modify tRNA awaits further investigation.
CONCLUDING REMARKS AND QUESTIONS REMAINING TO BE ANSWERED
Although many questions about how KEOPS functions have been answered over the past 15 years, many more remain to be addressed, as outlined below.
Why has KEOPS evolved added complexity?
Fifteen years of studying the structure and function of KEOPS have revealed an intricate mechanism of action. Unlike Qri7, which functions without binding partners, the activity of Kae1 is dependent on at least three binding partners. This raises the question of what function does this added complexity serve when in principle Kae1 could function like Qri7?
We consider two tantalizing possibilities. First, the machinery built around Kae1 could enable the regulation of its tRNA modifying activity. What benefit could there be from KEOPS downregulation? Although the total absence of t6A is harmful for cells, temporal control of t6A levels could be beneficial for cells as a mean to control translation under specific cellular conditions. For example, Ser and Arg can be coded by t6A-dependent (AGT/C and AGG/A respectively) and t6A-independent codons (TCT/C/A/G and CG/T/C/A/G respectively). As such, KEOPS downregulation would favor the translation of mRNA transcripts enriched with Ser and Arg codons that are not dependent on t6A modification. Thaiville et al. reported that in yeast mRNAs encoding ∼300 proteins exclusively contain Arg codons that are t6A-dependent (44). These would be prime candidates for downregulation in response to KEOPS inhibition.
Translation is regulated by modulation of other types of tRNA modifications at positions 34 and 37 (for examples, see references (116–119)) and mRNAs that are differentially translated due to the presence or absence of tRNA modifications have been termed modification tunable transcripts (MoTTs) (120). If the temporal regulation of t6A is exploited for regulatory means, the added complexity of KEOPS provides multiple opportunities to achieve this end. For example, the tRNA binding mechanism of KEOPS renders its function sensitive to Cgi121 expression levels. The short half-life of Cgi121 in yeast (∼30 min) (9) and its sub-stoichiometric level relative to the other subunits (121), make it an excellent restriction point for regulation.
A second possibility is that the complexity of KEOPS addresses a specific challenge or need of modifying cytoplasmic tRNAs that is not required for mitochondrial tRNAs. The cytoplasm contains a much larger number of tRNAs, both ANN and non-ANN-decoding, which synthesize a far larger number of proteins relative to mitochondria. The human mitochondrial genome transcribes only 22 different tRNAs (122), of which 5 are substrates for t6A modification (28,123). These tRNAs are needed for the synthesis of only 13 mitochondrial proteins (122). The human nuclear genome in contrast contains approximately 500 tRNA genes, including hundreds that are not KEOPS substrates, and these participate in the synthesis of 20 000–30 000 different proteins. Perhaps Kae1 requires its auxiliary subunits to deal with the greater diversity of tRNAs and the greater demand for modified tRNAs in the cytoplasm relative to those observed in mitochondria. In support of this possibility, while Qri7 and KEOPS display comparable catalytic rates in purified systems, massive overexpression of cytoplasmic Qri7 is required for its ability to rescue the growth defects resulting from the knockout of KEOPS (61). This in turn raises the question of what specific advantage the added complexity of KEOPS actually provides relative to Qri7? Possible advantages include the ability to readout tRNA features beyond the anticodon loop (102). This enhancement of substrate binding mechanism could serve as a substrate quality control mechanism that would help discriminate between functional and non-functional tRNA substrates, which might perturb the translation machinery. A second advantage may be the ability to coordinate t6A modification with other tRNA modification and processing events. tRNAs undergo many processing steps from their initial transcription to their degradation, and many of these steps occur in a hierarchic manner (124,125). In support of this possibility, since a subset of tRNAs contain an intron that is located between U36 and A37, the t6A modification by KEOPS can only occur after splicing. Likewise, the tRNA binding mechanism entails that t6A modification will occur after CCA tailing and before aminoacylation (102). How the activity of KEOPS is affected by other tRNA modifications and processing events and what role this might play awaits further study.
How does the ATPase activity of Bud32 promote the t6A modification of tRNA by Kae1?
The ATPase activity of Bud32 is required for KEOPS activity in vitro (102) and in vivo (9). However, it is still unclear how this activity exerts a specific effect on the tRNA t6A modification reaction. Since the ATPase activity of Bud32 precedes the tRNA modification activity of Kae1 (102) it is reasonable to infer that it serves to activate Kae1. A candidate region that could mediate an activating effect on Kae1 is the conserved C-terminal tail of Bud32. This region is dispensable for the activation of ATPase activity by tRNA (and thus exerts its essential effect downstream of the ATPase activating event) but is required for the tRNA modification activity of Kae1. How could this region of Bud32 mediate the activation of Kae1? Structures of Kae1-Bud32 complexes reveal that the C-terminus of Bud32 resides near the Kae1 active site and the Pcc1 binding surface on Kae1 (Figure 4A). From this perch, the C-terminal tail of Bud32 is well placed to help remodel the active site of Kae1 into a productive conformation. Alternatively, its position could serve in some capacity to collaborate with Pcc1. Either way, since ATP binding and hydrolysis by protein kinases is known to influence inter-lobe closure (126), Bud32’s catalytic function could generate movements of the C-tail of Bud32 to enable its supporting role in Kae1 catalytic function.
What are the precise roles of Pcc1 and Gon7 in the activity of KEOPS?
One of the main unanswered issues in the KEOPS field is the precise role of Pcc1. Pcc1 binds close to the active site of Kae1. Interestingly, the absence of Pcc1 has the same effect on KEOPS as the deletion of the C-tail of Bud32, namely the absence of both has minimal effect on the tRNA binding activity of KEOPS or the activation of Bud32 ATPase activity in response to binding tRNA (102). However, both are necessary for the tRNA modification activity of KEOPS in vitro and in vivo (9,102). The proximity of Pcc1 to the active site of Kae1 indicates that Pcc1 might function to activate Kae1, similar to the C-tail of Bud32. Providing an initial clue of Pcc1’s involvement in the regulation of the t6A catalytic function of KEOPS, tRNA binding to KEOPS induces perturbations to the binding interface between the Kae1 and Pcc1 subunits as measured by hydrogen deuterium exchange mass-spectrometry (102).
Another possible role for Pcc1 in t6A modification could be to guide the anticodon loop of the tRNA into the active site of Kae1. Pcc1 is predicted to directly contact tRNA in close vicinity of the anticodon loop harbouring the A37 modification site (Figure 4B). Whatever the exact role of Pcc1, mutation of an invariant Arg residue on its inferred tRNA binding surface (Arg 63 in P. furiosus Pcc1) disrupts t6A modification function (102).
The role of the Gon7 subunit in KEOPS also remains elusive. Since Gon7 proteins have not been identified in archaeal species (18) they appear to be an adaptation specific to eukaryotic KEOPS. Thus, Gon7 does not likely fulfill an essential role in the biochemical activity of KEOPS. In support, human KEOPS is active in vitro in the absence of Gon7 (102,109). In vivo however, the absence of Gon7 yields a strong phenotype that is similar to the absence of other KEOPS subunits (1) and in humans mutations in Gon7 can cause GAMOS (77). Interestingly, Gon7 possesses a highly conserved acidic C-terminus (residues 51–100 in human Gon7) that does not participate in binding to Pcc1 and remains unstructured in the Pcc1-Gon7 complex. Uncovering what this region does could provide insight into the essential role of Gon7 in cells.
Conclusion and relation to other t6A modifying complexes
A great amount has been learned over the past 15 years of intensive KEOPS study. While not covered here in detail, parallel research efforts on the structure (Table 1) and function of the orthologous tRNA modification machinery in bacteria (for examples, see references (59,93–95) by the Van Tilbeurgh, Crecy-Lagard, Iwata-Reuyl and Swairjo labs) have been similarly informative. These efforts have shed light into functions shared across the Kae1/TsaD/Qri7 family, and functions specific to the divergent protein complexes each enzyme operates in (see Figure 2A for schematic representations of the complexes). Comparing KEOPS with these orthologous systems raises two interesting points for consideration.
First, as noted above, a striking common feature of the three different t6A modifying enzymes is the conserved binding mode between Kae1/TsaD/Qri7 and their respective binding partners Pcc1/TsaB/Qri7. Each of these interactions (i.e. Kae1-Pcc1, TsaD-TsaB and Qri7-Qri7) is essential for tRNA modification. The precise function of this conserved binding mode remains elusive, but once assigned for Kae1, TsaD or Qri7, it will likely be informative for all three systems. A second striking feature is the dependency of Kae1 and TsaD on the ATPase subunits Bud32 and TsaE respectively for efficient tRNA modification. Despite this apparent similarity, Bud32 and TsaE play distinct roles in their respective complexes. In KEOPS, Bud32 promotes tRNA binding and its ATPase activity is required for tRNA modification (102) while in the TsaD-TsaB-TsaE complex, TsaE competes with tRNA binding (93) but its ATPase activity is required for maximal tRNA modification (92,93,95). How precisely the ATPase activities of Bud32 and TsaE exert their effects on Kae1 and TsaD respectively and if there are similarities in these effects remains to be determined.
No doubt, future studies will illuminate answers to these remaining questions and others that unify and differentiate the mitochondrial, bacterial and eukaryotic/archaeal t6A modification machineries and their important biological relevance.
DATA AVAILABILITY
The structures used for generating Figures 1B, 3A, 3B, 3D, 3E and 4A can be found on the PDB website. The model presented in Figure 4B can be provided upon request.
ACKNOWLEDGEMENTS
We wish to thank Ms. Susan Kelso, Dr Salima Daou, professor Ute Kothe, professor Mark Bayfield, Ms. Jennifer Porat and professor Daniel Durocher for their critical reading of this manuscript and their useful comments. We wish to thank Ms. Samara Ona for her help in generating the KEOPS-tRNA model presented in Figure 4B.
Contributor Information
Jonah Beenstock, The Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Ontario, M5G 1X5, Canada.
Frank Sicheri, The Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Ontario, M5G 1X5, Canada; Department of Molecular Genetics, University of Toronto, Ontario, M5S 1A8, Canada; Department of Biochemistry, University of Toronto, Ontario, M5S 1A8, Canada.
FUNDING
CIHR [FDN 143277 to F.S.]. Funding for open access charge: CIHR [FDN 143277].
Conflict of interest statement. None declared.
REFERENCES
- 1. Downey M., Houlsworth R., Maringele L., Rollie A., Brehme M., Galicia S., Guillard S., Partington M., Zubko M.K., Krogan N.J.et al.. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006; 124:1155–1168. [DOI] [PubMed] [Google Scholar]
- 2. Kisseleva-Romanova E., Lopreiato R., Baudin-Baillieu A., Rousselle J.C., Ilan L., Hofmann K., Namane A., Mann C., Libri D.. Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J. 2006; 25:3576–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koonin E.V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003; 1:127–136. [DOI] [PubMed] [Google Scholar]
- 4. Ni L., Snyder M.. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001; 12:2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stocchetto S., Marin O., Carignani G., Pinna L.A.. Biochemical evidence that Saccharomyces cerevisiae YGR262c gene, required for normal growth, encodes a novel Ser/Thr-specific protein kinase. FEBS Lett. 1997; 414:171–175. [DOI] [PubMed] [Google Scholar]
- 6. Kornev A.P., Taylor S.S.. Dynamics-driven allostery in protein kinases. Trends Biochem. Sci. 2015; 40:628–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Facchin S., Lopreiato R., Stocchetto S., Arrigoni G., Cesaro L., Marin O., Carignani G., Pinna L.A.. Structure-function analysis of yeast piD261/Bud32, an atypical protein kinase essential for normal cell life. Biochem. J. 2002; 364:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hecker A., Lopreiato R., Graille M., Collinet B., Forterre P., Libri D., van Tilbeurgh H.. Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: a model for the eukaryotic EKC/KEOPS subcomplex. EMBO J. 2008; 27:2340–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mao D.Y., Neculai D., Downey M., Orlicky S., Haffani Y.Z., Ceccarelli D.F., Ho J.S., Szilard R.K., Zhang W., Ho C.S.et al.. Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol. Cell. 2008; 32:259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beenstock J., Mooshayef N., Engelberg D.. How do protein kinases take a selfie (autophosphorylate). Trends Biochem. Sci. 2016; 41:938–953. [DOI] [PubMed] [Google Scholar]
- 11. Lopreiato R., Facchin S., Sartori G., Arrigoni G., Casonato S., Ruzzene M., Pinna L.A., Carignani G.. Analysis of the interaction between piD261/Bud32, an evolutionarily conserved protein kinase of Saccharomyces cerevisiae, and the Grx4 glutaredoxin. Biochem. J. 2004; 377:395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kannan N., Taylor S.S., Zhai Y., Venter J.C., Manning G.. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007; 5:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kennelly P.J. Protein Ser/Thr/Tyr phosphorylation in the Archaea. J. Biol. Chem. 2014; 289:9480–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abe Y., Matsumoto S., Wei S., Nezu K., Miyoshi A., Kito K., Ueda N., Shigemoto K., Hitsumoto Y., Nikawa J.et al.. Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. J. Biol. Chem. 2001; 276:44003–44011. [DOI] [PubMed] [Google Scholar]
- 15. Miyoshi A., Kito K., Aramoto T., Abe Y., Kobayashi N., Ueda N.. Identification of CGI-121, a novel PRPK (p53-related protein kinase)-binding protein. Biochem. Biophys. Res. Commun. 2003; 303:399–405. [DOI] [PubMed] [Google Scholar]
- 16. Lai C.H., Chou C.Y., Ch’ang L.Y., Liu C.S., Lin W.. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 2000; 10:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abdullah K.M., Lo R.Y., Mellors A.. Cloning, nucleotide sequence, and expression of the Pasteurella haemolytica A1 glycoprotease gene. J. Bacteriol. 1991; 173:5597–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thiaville P.C., Iwata-Reuyl D., de Crecy-Lagard V.. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t(6)A), a universal modification of tRNA. RNA Biol. 2014; 11:1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arigoni F., Talabot F., Peitsch M., Edgerton M.D., Meldrum E., Allet E., Fish R., Jamotte T., Curchod M.L., Loferer H.. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 1998; 16:851–856. [DOI] [PubMed] [Google Scholar]
- 20. Oberto J., Breuil N., Hecker A., Farina F., Brochier-Armanet C., Culetto E., Forterre P.. Qri7/OSGEPL, the mitochondrial version of the universal Kae1/YgjD protein, is essential for mitochondrial genome maintenance. Nucleic Acids Res. 2009; 37:5343–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haussuehl K., Huesgen P.F., Meier M., Dessi P., Glaser E., Adamski J., Adamska I.. Eukaryotic GCP1 is a conserved mitochondrial protein required for progression of embryo development beyond the globular stage in Arabidopsis thaliana. Biochem. J. 2009; 423:333–341. [DOI] [PubMed] [Google Scholar]
- 22. Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K.. Global analysis of protein localization in budding yeast. Nature. 2003; 425:686–691. [DOI] [PubMed] [Google Scholar]
- 23. Corbacho I., Olivero I., Hernandez L.M.. Identification of low-dye-binding (ldb) mutants of Saccharomyces cerevisiae. FEMS Yeast Res. 2004; 4:437–444. [DOI] [PubMed] [Google Scholar]
- 24. Brachat S., Dietrich F.S., Voegeli S., Zhang Z., Stuart L., Lerch A., Gates K., Gaffney T., Philippsen P.. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 2003; 4:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P.et al.. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006; 440:637–643. [DOI] [PubMed] [Google Scholar]
- 26. Seraphin B., Kretzner L., Rosbash M.. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988; 7:2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El Yacoubi B., Bailly M., de Crecy-Lagard V.. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012; 46:69–95. [DOI] [PubMed] [Google Scholar]
- 28. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A.et al.. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gustilo E.M., Vendeix F.A., Agris P.F.. tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol. 2008; 11:134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lyons S.M., Fay M.M., Ivanov P.. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett. 2018; 592:2828–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021; 22:375–392. [DOI] [PubMed] [Google Scholar]
- 32. Vare V.Y., Eruysal E.R., Narendran A., Sarachan K.L., Agris P.F.. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules. 2017; 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nedialkova D.D., Leidel S.A.. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell. 2015; 161:1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lorenz C., Lunse C.E., Morl M.. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules. 2017; 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schweizer M.P., Chheda G.B., Baczynskyj L., Hall R.H.. Aminoacyl nucleosides. VII. N-(Purin-6-ylcarbamoyl)threonine. A new component of transfer ribonucleic acid. Biochemistry. 1969; 8:3283–3289. [DOI] [PubMed] [Google Scholar]
- 36. Durant P.C., Bajji A.C., Sundaram M., Kumar R.K., Davis D.R.. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry. 2005; 44:8078–8089. [DOI] [PubMed] [Google Scholar]
- 37. Lescrinier E., Nauwelaerts K., Zanier K., Poesen K., Sattler M., Herdewijn P.. The naturally occurring N6-threonyl adenine in anticodon loop of Schizosaccharomyces pombe tRNAi causes formation of a unique U-turn motif. Nucleic Acids Res. 2006; 34:2878–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stuart J.W., Gdaniec Z., Guenther R., Marszalek M., Sochacka E., Malkiewicz A., Agris P.F.. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry. 2000; 39:13396–13404. [DOI] [PubMed] [Google Scholar]
- 39. Seelam Prabhakar P., Takyi N.A., Wetmore S.D.. Posttranscriptional modifications at the 37th position in the anticodon stem-loop of tRNA: structural insights from MD simulations. RNA. 2021; 27:202–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murphy F.V.t., Ramakrishnan V., Malkiewicz A., Agris P.F.. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004; 11:1186–1191. [DOI] [PubMed] [Google Scholar]
- 41. Yarian C., Marszalek M., Sochacka E., Malkiewicz A., Guenther R., Miskiewicz A., Agris P.F.. Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry. 2000; 39:13390–13395. [DOI] [PubMed] [Google Scholar]
- 42. Weissenbach J., Grosjean H.. Effect of threonylcarbamoyl modification (t6A) in yeast tRNA Arg III on codon-anticodon and anticodon-anticodon interactions. A thermodynamic and kinetic evaluation. Eur. J. Biochem. 1981; 116:207–213. [DOI] [PubMed] [Google Scholar]
- 43. Grosjean H., Westhof E.. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016; 44:8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thiaville P.C., Legendre R., Rojas-Benitez D., Baudin-Baillieu A., Hatin I., Chalancon G., Glavic A., Namy O., de Crecy-Lagard V.. Global translational impacts of the loss of the tRNA modification t(6)A in yeast. Microb Cell. 2016; 3:29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin C.A., Ellis S.R., True H.L.. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell. Biol. 2010; 30:354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. El Yacoubi B., Hatin I., Deutsch C., Kahveci T., Rousset J.P., Iwata-Reuyl D., Murzin A.G., de Crecy-Lagard V.. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011; 30:882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. El Yacoubi B., Lyons B., Cruz Y., Reddy R., Nordin B., Agnelli F., Williamson J.R., Schimmel P., Swairjo M.A., de Crecy-Lagard V.. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009; 37:2894–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miyauchi K., Kimura S., Suzuki T.. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013; 9:105–111. [DOI] [PubMed] [Google Scholar]
- 49. Matuszewski M., Wojciechowski J., Miyauchi K., Gdaniec Z., Wolf W.M., Suzuki T., Sochacka E.. A hydantoin isoform of cyclic N6-threonylcarbamoyladenosine (ct6A) is present in tRNAs. Nucleic Acids Res. 2017; 45:2137–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kimura S., Miyauchi K., Ikeuchi Y., Thiaville P.C., Crecy-Lagard V., Suzuki T.. Discovery of the beta-barrel-type RNA methyltransferase responsible for N6-methylation of N6-threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res. 2014; 42:9350–9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wei F.Y., Suzuki T., Watanabe S., Kimura S., Kaitsuka T., Fujimura A., Matsui H., Atta M., Michiue H., Fontecave M.et al.. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 2011; 121:3598–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Na J.G., Pinto I., Hampsey M.. Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics. 1992; 131:791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aravind L., Koonin E.V.. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 1999; 287:1023–1040. [DOI] [PubMed] [Google Scholar]
- 54. Jabbouri S., Relic B., Hanin M., Kamalaprija P., Burger U., Prome D., Prome J.C., Broughton W.J.. nolO and noeI (HsnIII) of Rhizobium sp. NGR234 are involved in 3-O-carbamoylation and 2-O-methylation of Nod factors. J. Biol. Chem. 1998; 273:12047–12055. [DOI] [PubMed] [Google Scholar]
- 55. Daugeron M.C., Lenstra T.L., Frizzarin M., El Yacoubi B., Liu X., Baudin-Baillieu A., Lijnzaad P., Decourty L., Saveanu C., Jacquier A.et al.. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res. 2011; 39:6148–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Srinivasan M., Mehta P., Yu Y., Prugar E., Koonin E.V., Karzai A.W., Sternglanz R.. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 2011; 30:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Galperin M.Y., Koonin E.V.. Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004; 32:5452–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005; 59:407–450. [DOI] [PubMed] [Google Scholar]
- 59. Deutsch C., El Yacoubi B., de Crecy-Lagard V., Iwata-Reuyl D.. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J. Biol. Chem. 2012; 287:13666–13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Perrochia L., Crozat E., Hecker A., Zhang W., Bareille J., Collinet B., van Tilbeurgh H., Forterre P., Basta T.. In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya. Nucleic Acids Res. 2013; 41:1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wan L.C., Mao D.Y., Neculai D., Strecker J., Chiovitti D., Kurinov I., Poda G., Thevakumaran N., Yuan F., Szilard R.K.et al.. Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system. Nucleic Acids Res. 2013; 41:6332–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pollo-Oliveira L., Klassen R., Davis N., Ciftci A., Bacusmo J.M., Martinelli M., DeMott M.S., Begley T.J., Dedon P.C., Schaffrath R.et al.. Loss of Elongator- and KEOPS-Dependent tRNA Modifications Leads to Severe Growth Phenotypes and Protein Aggregation in Yeast. Biomolecules. 2020; 10:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Peng J., He M.H., Duan Y.M., Liu Y.T., Zhou J.Q.. Inhibition of telomere recombination by inactivation of KEOPS subunit Cgi121 promotes cell longevity. PLos Genet. 2015; 11:e1005071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Joshi K., Bhatt M.J., Farabaugh P.J.. Codon-specific effects of tRNA anticodon loop modifications on translational misreading errors in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2018; 46:10331–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Naor A., Thiaville P.C., Altman-Price N., Cohen-Or I., Allers T., de Crecy-Lagard V., Gophna U.. A genetic investigation of the KEOPS complex in halophilic Archaea. PLoS One. 2012; 7:e43013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rojas-Benitez D., Eggers C., Glavic A.. Modulation of the proteostasis machinery to overcome stress caused by diminished levels of t6A-modified tRNAs in Drosophila. Biomolecules. 2017; 7:25. [Google Scholar]
- 67. Rojas-Benitez D., Ibar C., Glavic A.. The Drosophila EKC/KEOPS complex: roles in protein synthesis homeostasis and animal growth. Fly (Austin). 2013; 7:168–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin C.J., Smibert P., Zhao X., Hu J.F., Ramroop J., Kellner S.M., Benton M.A., Govind S., Dedon P.C., Sternglanz R.et al.. An extensive allelic series of Drosophila kae1 mutants reveals diverse and tissue-specific requirements for t6A biogenesis. RNA. 2015; 21:2103–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rojas-Benitez D., Thiaville P.C., de Crecy-Lagard V., Glavic A.. The Levels of a Universally Conserved tRNA Modification Regulate Cell Growth. J. Biol. Chem. 2015; 290:18699–18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ibar C., Cataldo V.F., Vasquez-Doorman C., Olguin P., Glavic A.. Drosophila p53-related protein kinase is required for PI3K/TOR pathway-dependent growth. Development. 2013; 140:1282–1291. [DOI] [PubMed] [Google Scholar]
- 71. Zhang H., Stallock J.P., Ng J.C., Reinhard C., Neufeld T.P.. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000; 14:2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Thiaville P.C., de Crecy-Lagard V.. The emerging role of complex modifications of tRNA(Lys)UUU in signaling pathways. Microb Cell. 2015; 2:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Magnuson B., Ekim B., Fingar D.C.. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 2012; 441:1–21. [DOI] [PubMed] [Google Scholar]
- 74. Braun D.A., Rao J., Mollet G., Schapiro D., Daugeron M.C., Tan W., Gribouval O., Boyer O., Revy P., Jobst-Schwan T.et al.. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet. 2017; 49:1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jobst-Schwan T., Schmidt J.M., Schneider R., Hoogstraten C.A., Ullmann J.F.P., Schapiro D., Majmundar A.J., Kolb A., Eddy K., Shril S.et al.. Acute multi-sgRNA knockdown of KEOPS complex genes reproduces the microcephaly phenotype of the stable knockout zebrafish model. PLoS One. 2018; 13:e0191503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Edvardson S., Prunetti L., Arraf A., Haas D., Bacusmo J.M., Hu J.F., Ta-Shma A., Dedon P.C., de Crecy-Lagard V., Elpeleg O.. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet. 2017; 25:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Arrondel C., Missoury S., Snoek R., Patat J., Menara G., Collinet B., Liger D., Durand D., Gribouval O., Boyer O.et al.. Defects in t(6)A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat. Commun. 2019; 10:3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Galloway W.H., Mowat A.P.. Congenital microcephaly with hiatus hernia and nephrotic syndrome in two sibs. J. Med. Genet. 1968; 5:319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Santos M., Anderson C.P., Neschen S., Zumbrennen-Bullough K.B., Romney S.J., Kahle-Stephan M., Rathkolb B., Gailus-Durner V., Fuchs H., Wolf E.et al.. Irp2 regulates insulin production through iron-mediated Cdkal1-catalyzed tRNA modification. Nat. Commun. 2020; 11:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Elkins B.N., Keller E.B.. The enzymatic synthesis of N-(purin-6-ylcarbamoyl)threonine, an anticodon-adjacent base in transfer ribonucleic acid. Biochemistry. 1974; 13:4622–4628. [DOI] [PubMed] [Google Scholar]
- 81. Agari Y., Sato S., Wakamatsu T., Bessho Y., Ebihara A., Yokoyama S., Kuramitsu S., Shinkai A.. X-ray crystal structure of a hypothetical Sua5 protein from Sulfolobus tokodaii strain 7. Proteins. 2008; 70:1108–1111. [DOI] [PubMed] [Google Scholar]
- 82. Meng F.L., Hu Y., Shen N., Tong X.J., Wang J., Ding J., Zhou J.Q.. Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication. EMBO J. 2009; 28:1466–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lauhon C.T. Mechanism of N6-threonylcarbamoyladenonsine (t(6)A) biosynthesis: isolation and characterization of the intermediate threonylcarbamoyl-AMP. Biochemistry. 2012; 51:8950–8963. [DOI] [PubMed] [Google Scholar]
- 84. Kopina B.J., Missoury S., Collinet B., Fulton M.G., Cirio C., van Tilbeurgh H., Lauhon C.T.. Structure of a reaction intermediate mimic in t6A biosynthesis bound in the active site of the TsaBD heterodimer from Escherichia coli. Nucleic Acids Res. 2021; 49:2141–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Harris K.A., Bobay B.G., Sarachan K.L., Sims A.F., Bilbille Y., Deutsch C., Iwata-Reuyl D., Agris P.F.. NMR-based Structural Analysis of Threonylcarbamoyl-AMP Synthase and Its Substrate Interactions. J. Biol. Chem. 2015; 290:20032–20043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kuratani M., Kasai T., Akasaka R., Higashijima K., Terada T., Kigawa T., Shinkai A., Bessho Y., Yokoyama S.. Crystal structure of Sulfolobus tokodaii Sua5 complexed with L-threonine and AMPPNP. Proteins. 2011; 79:2065–2075. [DOI] [PubMed] [Google Scholar]
- 87. Pichard-Kostuch A., Zhang W., Liger D., Daugeron M.C., Letoquart J., Li de la Sierra-Gallay I., Forterre P., Collinet B., van Tilbeurgh H., Basta T.. Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t(6)A tRNA modification. RNA. 2018; 24:926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Parthier C., Gorlich S., Jaenecke F., Breithaupt C., Brauer U., Fandrich U., Clausnitzer D., Wehmeier U.F., Bottcher C., Scheel D.et al.. The O-carbamoyltransferase TobZ catalyzes an ancient enzymatic reaction. Angew. Chem. Int. Ed. Engl. 2012; 51:4046–4052. [DOI] [PubMed] [Google Scholar]
- 89. Zhou J.B., Wang Y., Zeng Q.Y., Meng S.X., Wang E.D., Zhou X.L.. Molecular basis for t6A modification in human mitochondria. Nucleic Acids Res. 2020; 48:3181–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nichols C.E., Johnson C., Lockyer M., Charles I.G., Lamb H.K., Hawkins A.R., Stammers D.K.. Structural characterization of Salmonella typhimurium YeaZ, an M22 O-sialoglycoprotein endopeptidase homolog. Proteins. 2006; 64:111–123. [DOI] [PubMed] [Google Scholar]
- 91. Nichols C.E., Lamb H.K., Thompson P., El Omari K., Lockyer M., Charles I., Hawkins A.R., Stammers D.K.. Crystal structure of the dimer of two essential Salmonella typhimurium proteins, YgjD & YeaZ and calorimetric evidence for the formation of a ternary YgjD-YeaZ-YjeE complex. Protein Sci. 2013; 22:628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang W., Collinet B., Perrochia L., Durand D., van Tilbeurgh H.. The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli. Nucleic Acids Res. 2015; 43:1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Luthra A., Swinehart W., Bayooz S., Phan P., Stec B., Iwata-Reuyl D., Swairjo M.A.. Structure and mechanism of a bacterial t6A biosynthesis system. Nucleic Acids Res. 2018; 46:1395–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Missoury S., Plancqueel S., Li de la Sierra-Gallay I., Zhang W., Liger D., Durand D., Dammak R., Collinet B., van Tilbeurgh H.. The structure of the TsaB/TsaD/TsaE complex reveals an unexpected mechanism for the bacterial t6A tRNA-modification. Nucleic Acids Res. 2018; 46:5850–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Luthra A., Paranagama N., Swinehart W., Bayooz S., Phan P., Quach V., Schiffer J.M., Stec B., Iwata-Reuyl D., Swairjo M.A.. Conformational communication mediates the reset step in t6A biosynthesis. Nucleic Acids Res. 2019; 47:6551–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Swinehart W., Deutsch C., Sarachan K.L., Luthra A., Bacusmo J.M., de Crecy-Lagard V., Swairjo M.A., Agris P.F., Iwata-Reuyl D.. Specificity in the biosynthesis of the universal tRNA nucleoside N (6)-threonylcarbamoyl adenosine (t(6)A)-TsaD is the gatekeeper. RNA. 2020; 26:1094–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wolff P., Villette C., Zumsteg J., Heintz D., Antoine L., Chane-Woon-Ming B., Droogmans L., Grosjean H., Westhof E.. Comparative patterns of modified nucleotides in individual tRNA species from a mesophilic and two thermophilic archaea. RNA. 2020; 26:1957–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yu N., Jora M., Solivio B., Thakur P., Acevedo-Rocha C.G., Randau L., de Crecy-Lagard V., Addepalli B., Limbach P.A.. tRNA Modification Profiles and Codon-Decoding Strategies in Methanocaldococcus jannaschii. J. Bacteriol. 2019; 201:e00690-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang W., Collinet B., Graille M., Daugeron M.C., Lazar N., Libri D., Durand D., van Tilbeurgh H.. Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex. Nucleic Acids Res. 2015; 43:3358–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dar A.C., Dever T.E., Sicheri F.. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005; 122:887–900. [DOI] [PubMed] [Google Scholar]
- 101. Li J., Ma X., Banerjee S., Chen H., Ma W., Bode A.M., Dong Z.. Crystal structure of the human PRPK-TPRKB complex. Commun Biol. 2021; 4:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Beenstock J., Ona S.M., Porat J., Orlicky S., Wan L.C.K., Ceccarelli D.F., Maisonneuve P., Szilard R.K., Yin Z., Setiaputra D.et al.. A substrate binding model for the KEOPS tRNA modifying complex. Nat. Commun. 2020; 11:6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hurley J.H. The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu. Rev. Biophys. Biomol. Struct. 1996; 25:137–162. [DOI] [PubMed] [Google Scholar]
- 104. Hecker A., Leulliot N., Gadelle D., Graille M., Justome A., Dorlet P., Brochier C., Quevillon-Cheruel S., Le Cam E., van Tilbeurgh H.et al.. An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res. 2007; 35:6042–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tominaga T., Kobayashi K., Ishii R., Ishitani R., Nureki O.. Structure of Saccharomyces cerevisiae mitochondrial Qri7 in complex with AMP. Acta Crystallogr. F Struct. Biol. Commun. 2014; 70:1009–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lin H., Miyauchi K., Harada T., Okita R., Takeshita E., Komaki H., Fujioka K., Yagasaki H., Goto Y.I., Yanaka K.et al.. CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat. Commun. 2018; 9:1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Morin A., Auxilien S., Senger B., Tewari R., Grosjean H.. Structural requirements for enzymatic formation of threonylcarbamoyladenosine (t6A) in tRNA: an in vivo study with Xenopus laevis oocytes. RNA. 1998; 4:24–37. [PMC free article] [PubMed] [Google Scholar]
- 108. Wang Y., Zeng Q.Y., Zheng W.Q., Ji Q.Q., Zhou X.L., Wang E.D.. A natural non-Watson-Crick base pair in human mitochondrial tRNAThr causes structural and functional susceptibility to local mutations. Nucleic Acids Res. 2018; 46:4662–4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wan L.C., Maisonneuve P., Szilard R.K., Lambert J.P., Ng T.F., Manczyk N., Huang H., Laister R., Caudy A.A., Gingras A.C.et al.. Proteomic analysis of the human KEOPS complex identifies C14ORF142 as a core subunit homologous to yeast Gon7. Nucleic Acids Res. 2017; 45:805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wan L.C., Pillon M.C., Thevakumaran N., Sun Y., Chakrabartty A., Guarne A., Kurinov I., Durocher D., Sicheri F.. Structural and functional characterization of KEOPS dimerization by Pcc1 and its role in t6A biosynthesis. Nucleic Acids Res. 2016; 44:6971–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Costessi A., Mahrour N., Sharma V., Stunnenberg R., Stoel M.A., Tijchon E., Conaway J.W., Conaway R.C., Stunnenberg H.G.. The human EKC/KEOPS complex is recruited to Cullin2 ubiquitin ligases by the human tumour antigen PRAME. PLoS One. 2012; 7:e42822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jeffrey P.D., Russo A.A., Polyak K., Gibbs E., Hurwitz J., Massague J., Pavletich N.P.. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995; 376:313–320. [DOI] [PubMed] [Google Scholar]
- 113. Ibba M., Soll D.. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000; 69:617–650. [DOI] [PubMed] [Google Scholar]
- 114. Yip M.C.J., Keszei A.F.A., Feng Q., Chu V., McKenna M.J., Shao S.. Mechanism for recycling tRNAs on stalled ribosomes. Nat. Struct. Mol. Biol. 2019; 26:343–349. [DOI] [PubMed] [Google Scholar]
- 115. Wilusz J.E., Whipple J.M., Phizicky E.M., Sharp P.A.. tRNAs marked with CCACCA are targeted for degradation. Science. 2011; 334:817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Begley U., Dyavaiah M., Patil A., Rooney J.P., DiRenzo D., Young C.M., Conklin D.S., Zitomer R.S., Begley T.J.. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007; 28:860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Chan C.T., Pang Y.L., Deng W., Babu I.R., Dyavaiah M., Begley T.J., Dedon P.C.. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012; 3:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chionh Y.H., McBee M., Babu I.R., Hia F., Lin W., Zhao W., Cao J., Dziergowska A., Malkiewicz A., Begley T.J.et al.. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 2016; 7:13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gall A.R., Datsenko K.A., Figueroa-Bossi N., Bossi L., Masuda I., Hou Y.M., Csonka L.N.. Mg2+ regulates transcription of mgtA in Salmonella Typhimurium via translation of proline codons during synthesis of the MgtL peptide. Proc. Natl. Acad. Sci. USA. 2016; 113:15096–15101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Gu C., Begley T.J., Dedon P.C.. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014; 588:4287–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S.. Global analysis of protein expression in yeast. Nature. 2003; 425:737–741. [DOI] [PubMed] [Google Scholar]
- 122. D'Souza A.R., Minczuk M.. Mitochondrial transcription and translation: overview. Essays Biochem. 2018; 62:309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Suzuki T., Yashiro Y., Kikuchi I., Ishigami Y., Saito H., Matsuzawa I., Okada S., Mito M., Iwasaki S., Ma D.et al.. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020; 11:4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Han L., Phizicky E.M.. A rationale for tRNA modification circuits in the anticodon loop. RNA. 2018; 24:1277–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Barraud P., Gato A., Heiss M., Catala M., Kellner S., Tisne C.. Time-resolved NMR monitoring of tRNA maturation. Nat. Commun. 2019; 10:3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Masterson L.R., Cheng C., Yu T., Tonelli M., Kornev A., Taylor S.S., Veglia G.. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat. Chem. Biol. 2010; 6:821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The structures used for generating Figures 1B, 3A, 3B, 3D, 3E and 4A can be found on the PDB website. The model presented in Figure 4B can be provided upon request.