Abstract
The stable insertion of the retroviral genome into the host chromosomes requires the association between integration complexes and cellular chromatin via the interaction between retroviral integrase and the nucleosomal target DNA. This final association may involve the chromatin-binding properties of both the retroviral integrase and its cellular cofactor LEDGF/p75. To investigate this and better understand the LEDGF/p75-mediated chromatin tethering of HIV-1 integrase, we used a combination of biochemical and chromosome-binding assays. Our study revealed that retroviral integrase has an intrinsic ability to bind and recognize specific chromatin regions in metaphase even in the absence of its cofactor. Furthermore, this integrase chromatin-binding property was modulated by the interaction with its cofactor LEDGF/p75, which redirected the enzyme to alternative chromosome regions. We also better determined the chromatin features recognized by each partner alone or within the functional intasome, as well as the chronology of efficient LEDGF/p75-mediated targeting of HIV-1 integrase to chromatin. Our data support a new chromatin-binding function of integrase acting in concert with LEDGF/p75 for the optimal association with the nucleosomal substrate. This work also provides additional information about the behavior of retroviral integration complexes in metaphase chromatin and the mechanism of action of LEDGF/p75 in this specific context.
INTRODUCTION
Retroviral infection requires the stable insertion of the viral DNA genome into the chromosomes of the host (for a review on integration, see (1)). This interaction between the incoming integration complex and the host chromosome thus constitutes the first contact between the viral and cell genomes that will modulate all the further steps of the infection. This multistep process is governed by the targeting of viral DNA/integrase complexes, called intasomes, toward suitable chromatin regions and their subsequent functional association with nucleosomal DNA. Several cellular factors have been shown to play a role in this targeting mechanism, such as LEDGF/p75 and CPSF6 for lentiviruses (2,3) and BET proteins for gammaretroviruses (4,5). Nevertheless, the integrase protein (IN) by itself has been shown to be a key determinant of integration site selection and to interact directly with chromatin components such as remodeling factors and histones (6–9). Interestingly, the carboxy-terminal domain (CTD) of HIV-1 IN (residues 220–270) displays a strong structural similarity to Tudor domains (10) which are known to bind histone tails (11,12). These data make histone tails good candidates for potential receptors of incoming intasomes. This idea is further supported by findings showing that retroviral IN can bind, via its CTD, directly to histone tails (8,9,13). HIV-1 IN mutagenesis studies have shown that amino acid substitutions that affect IN binding to nucleosomes and histone tails also affect viral infectivity (8). Furthermore, mutations in the CTD were reported to affect insertion site selection based on chromatin density (8,14). Indeed, CTD mutations such as R231A/H, which were shown to decrease IN affinity for histone tails, also decrease viral infectivity and retarget integration sites in regions with lower nucleosome density. In contrast, mutations which increased the affinity for the nucleosome, e.g. D253H, were shown to improve viral infectivity (8). Similar results were obtained in the case of the prototype foamy virus (PFV) intasome, for which mutations impairing the interactions of the intasome with the nucleosome also impaired integration, as observed for HIV-1 (9). Taken together, these data suggest that retroviral INs may have intrinsic chromatin-binding properties that enable their participation, in concert with tethering cofactors, in binding to the final chromatin insertion site.
Structure of the chromatin varies during the cell cycle. Lentiviruses can infect non-dividing cells since they do not need nuclear membrane breakdown to reach the chromatin, in contrast to other retroviruses requiring mitosis for efficient infection. Furthermore, dividing cells, as activated T-cells, have been also shown to be more susceptible to infection (15). This suggests that the HIV-1 integration complexes can meet various types of chromatin structures including, in some situations, metaphases where chromatin condensation has been reported to delay HIV-1 integration (15). This raises the question whether incoming intasomes reaching this specific chromatin structure may bind but not integrate. Noticeably, it has been reported that the chromatin structure surrounding the insertion site modulates retroviral integration into nucleosomes (16). In particular, chromatin compaction was reported to prevent HIV-1 integration, while chromatin remodeling promoted viral DNA insertion, suggesting that additional contacts between incoming intasomes and nucleosomal substrates must be established for optimal integration (6,7,16). Interestingly, the lentiviral cofactor LEDGF/p75 has been reported to strongly bind metaphase chromosomes (17,18) and its role in targeting the integration complex toward chromatin has been clearly demonstrated extensively in the literature. LEDGF/p75 has been shown to stimulate in vitro HIV-1 integration onto nucleosomes (19), and the binding of the LEDGF/p75 PWWP domain to mononucleosomes and its preference for H3K36me3 have been reported in vitro (19,20). Despite these data, how integration complexes behave depending on the chromatin structures met in the context of chromosomes, remains poorly understood and documented. In particular, the molecular basis for how LEDGF/p75 engages chromatin in the context of its complex with IN and how IN chromatin-binding properties may influence this targeting remain mainly unclear, especially in the context of metaphase chromosomes.
The lack of experimental systems allowing monitoring of the direct association between proteins and chromatin in the absence of other cofactors prevented further characterization of these molecular processes. Using a combination of biochemical approaches and metaphase chromosome-spreads imaging we investigated the LEDGF/p75-mediated chromatin tethering of the HIV-1 IN in this chromosome context. Our study revealed that this retroviral IN has an unedited intrinsic property enabling it to bind and recognize specific chromatin regions even in the absence of its cofactor. We also found that this function is carried in the IN CTD. Furthermore, we show here that this IN chromatin-binding property is modulated by the IN interaction with LEDGF/p75, which modifies the efficiency of IN binding to chromosomes and redirects the enzyme to alternative/different chromatin regions, highlighting its tethering function. The chronology of the LEDGF/p75-mediated targeting of HIV-1 IN to chromatin has been further investigated, shedding light on the mechanism of chromatin tethering function of LEDGF/p75. Finally, we used this metaphase model to better determine the functional anchoring of integration complexes on this specific structure and better depict the chromatin tethering process in different retroviral intasome models.
MATERIALS AND METHODS
Proteins purification and antibodies
The HIV-1 IN was expressed in E.coli (Rosetta) and the cells were lysed in buffer containing 50 mM HEPES pH 7.5, 5 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF). The lysate was centrifuged and IN extracted from the pellet in buffer containing 1 M NaCl, 50 mM HEPES pH 7.5, 1 mM EDTA, 1 mM DTT, 7 mM CHAPS. The protein was then purified on butyl column equilibrated with 50 mM HEPES pH 7.5, 200 mM NaCl, 1 M ammonium sulfate, 100 mM EDTA, 1 mM DTT, 7 mM CHAPS, 10% glycerol. The protein was further purified on heparin column equilibrated with 50 mM HEPES pH 7.5, 200 mM NaCl, 100 mM EDTA, 1 mM DTT, 7 mM CHAPS, 10% glycerol. LEDGF/p75 was expressed in PC2 bacteria and the cells were lysed in lysis buffer containing 20 mM Tris pH 7.5, 1 M NaCl, 1mM PMSF added lysozyme and protease inhibitors. The protein was purified by nickel-affinity chromatography and the His-tag was removed with 3C protease, 4°C over night. After dilution down to 150 mM NaCl, the protein was further purified on SP column equilibrated with 25 mM Tris pH 7.5, 150 mM NaCl (gradient from 150 mM to 1M NaCl), then DTT was added to 2 mM final and the protein was concentrated for size exclusion chromatography. Gel filtration was performed on a superdex 200 column (GE Healthcare) equilibrated with 25 mM Tris pH 7.5, 500 mM NaCl. Two mM DTT were added to eluted protein that was then concentrated to about 10 mg/ml. The PFV IN was expressed in E.coli in the presence of 50 μM ZnCl2 and the cells were lysed in buffer containing 25 mM Tris pH 7.4, 500 mM NaCl, 1 mM PMSF. The lysate was centrifuged and the protein was purified by nickel-affinity chromatography. The His-tag was removed with 3C protease, 4°C over night. After dilution down to 100mM NaCl, the protein was further purified on SP column equilibrated with 25 mM Tris pH 7.4, 100 mM NaCl (gradient from 100 mM to 1 M NaCl), then DTT 2 mM final was added to the protein that was concentrated for size exclusion chromatography. Gel filtration was performed on a superdex 200 column (GE Healthcare) equilibrated with 25 mM Tris pH 7.4, 500 mM NaCl. Five mM of DTT were added to the eluted protein that was concentrated to about 15 mg/ml.
Polyclonal anti-HIV-1 IN antibody was purchased from Bioproducts MD (Middletown, MD, USA). Monoclonal anti-HIS was purchased from Abcam (ref ab18184). HRP-conjugated secondary anti-rabbit antibody was purchased from Sigma (ref A8275) and anti-mouse from Jackson ImmunoResearch (ref 715-035-151). Monoclonal Anti H4 was purchased from active motif (ref 61521, 1/400), polyclonal anti H3 from abcam (ref ab1791, 1/50), polyclonal anti H3K27ac from abcam (ref ab4729, 1/400), monoclonal anti H3K27me3 from abcam (ref ab6002, 1/400, polyclonal anti H3K36me3 from cell signaling (ref 9762, 1/50), monoclonal Anti LEDGF/p75 from BDbiosciences (ref 61175, 1/600). Polyclonal Anti-SSRP1 was purchased from Abcam (ref 21584 (1/140)) and monoclonal Anti-SSRP1 was purchased from Abcam (ref 26212 (1/400)). Polyclonal anti-Rabbit IgG Secondary Antibody, Alexa Fluor 488 was purchased from FISHER SCIENTIFIC (1/400). Polyclonal anti-Rabbit IgG Secondary Antibody, Alexa Fluor 594 was purchased from Fisher Scientific (1/400). Polyclonal anti-Mouse IgG Secondary Antibody, Alexa Fluor 546 was purchased from Fisher Scientific (1/400).
Intasomes assembly
HIV intasomes were purified essentially using the method described in (21) with some modifications. Briefly recombinant HIV IN (7 μM) was incubated with LEDGF/p75 (8 μM) and it's cognate pre-processed vDNA (4 μM) coupled to FITC at 37°C for 13 min in a buffer containing 25 mM HEPES pH 7.0, 40 mM KAc, 3 mM CaCl2, 10 μM ZnCl2, 2 mM DTT and 80 mM NaCl in a final volume of 200 μl. After the incubation, the mixture was adjusted at 310 mM NaCl and placed on ice for 5 min. Intasomes were purified with a Superose 6 10/300 24 ml equilibrated with 310 mM NaCl, 25 mM HEPES pH 7.0 and 3 mM CaCl2. Intasomes fractions (eluted around 10.5 ml) were pooled and concentrated with a 30 kDa cut-off vivaspin column. The sequence of the ODN used for the intasome assembly are reported in S1. PFV intasomes were purified essentially using the method described in (22). Briefly, recombinant PFV IN (135 μM) and it's cognate pre-processed vDNA (80 μM) labeled with FITC were dialyzed over night at 18°C against a buffer containing 20 mM BTP pH 7.5, 200 mM NaCl, 25 μM ZnCl2, 2 mM DTT using a 3.5 kDa cut-off dialysis cassette. The mixture was then loaded on a SuperDex200 10/300 24 ml equilibrated with 320 mM NaCl and 20 mM BTP pH 7.5. Intasomes fractions (eluted around 10.5 ml) were pooled and concentrated with a 30 kDa cut-off vivaspin column. The sequence of the ODN used for the intasome assembly are reported in S1.
Chromosomes spreads
Chromosome spreads were obtained after culture of peripheral blood T-lymphocytes, in accordance with standard procedures used for conventional karyotyping on peripheral blood cells. Briefly, 0.5 ml of peripheral blood was added to 5 ml of specific culture medium (ChromoSynchroPTM kit, EuroClone, Pero, Italy), T-lymphocytes were thereby cultured for 72 h. After 48 h of culture, reagents for lymphocytes culture synchronization (ChromoSynchroPTM kit, EuroClone, Pero, Italy) were added, according to the manufacturer's recommendations. Mitotic cells were arrested at the prometaphase stage by adding to the culture 50 μl of Colcemid at 10 μg/ml (KaryoMAX ColcemidTM, Thermo Fisher Scientific, Waltham, MA, USA) for 30 min, then cells undergo hypotonic shock for 15 mn before fixation with a solution of methanol and acetic acid (3/1, v/v). Finally, chromosome spreading was performed on Superfrost Plus™ slides (Thermo Fisher Scientific, Waltham, MA, USA).
Chromosomes interaction assays
Chromosomes spreads slides were permeabilized with Triton X‐100 1%, saponin 0.4% for 15 min and blocked with 2% bovine serum albumin (BSA), saponin 0.1%, SVF 2% for 1 h. All incubations were carried out at room temperature and were followed by three PBS-saponin 0.1% washes. Chromosomes spreads were incubated with IN or LEDGF/p75 diluted in blocking buffer for 1 h. Then, chromosomes spreads were incubated with primary antibodies for 1 h and secondary antibodies for 45 min. Antibodies were diluted in blocking buffer too. DNA were stained with Dapi (100 ng/ml). PBS washes then water wash are performed. Finally, chromosomes spreads were mounted onto glass slides with Prolong Diamond (Life Technologies). Epifluorescence microscopy was carried out on a Zeiss AxioImager Z1 using a 63× objective. Images were acquired in FIJI.
Analysis of chromatin binding
The interaction with chromosomes was monitored by immunofluorescence using antibodies directed against the studied factor and chromatin was monitored by the DAPI staining. The fluorescence signal among selected chromosomes was quantified with ImageJ software after subtracting the background computed as average pixel value in 50 pixels rolling ball radius. The different channels were merged and a segmented line of 4 units width along chromosomes was drawn from both sides of the analyzed chromosomes previously identified by inverted DAPI staining and oriented by centromere positioning (see S2). The DAPI staining was used to precisely determine the exact shape and size of the chromosome and the distribution profile of the protein fluorescence signal along each chromosome. The intensity of binding, as well as the distribution of the protein, were then compiled from at least 10 independent spreads and up to 10 chromosomes which could be unambiguously identified. Both registration of intensity profiles using DAPI stained-chromatin as reference (see S2), and correlation between the DAPI stained-chromatin and the protein distribution was performed using MATLAB. Profiles presented in the figures of this manuscripts are obtained after computing means and standard deviation along available profiles. Pearson correlation for each pair of curves was computed and the proportion of significant correlation (P value) was also provided for each analysis. In that analysis the Pearson correlation will be >0 if correlation, =0 if no correlation, <0 if anti-correlation. The P-value indicates if the correlation (or anti-correlation) is significant. The mean correlation ± standard deviation is provided among all the comparison and a positive mean correlation indicates that most of the time, the curve correlates together, a close to 0 mean can indicate that curves do not correlate together. Another comparison based on P-value (ranksum test) computed at each position along the chromosome was also performed in order to identify regions where there is a significant difference while taking into account the distribution profile variability among all the measurements. In this analysis a P-value <0.01 in one region indicates there is a significant difference and a P-value >0.05 indicates no significant difference meaning either that the compared profiles are highly different in the two groups or the compared profiles are highly the same in the two groups.
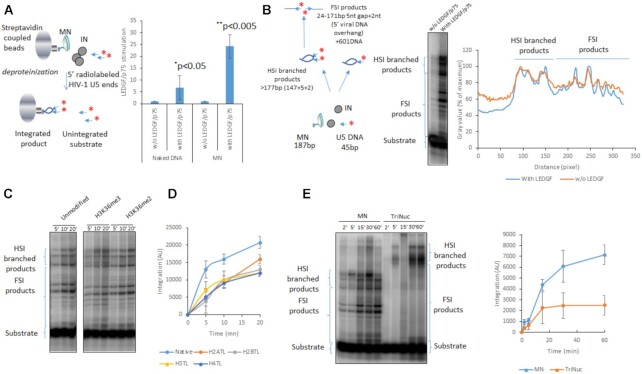
Integration assays
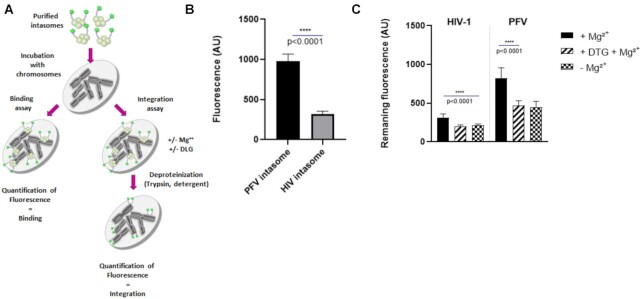
In vitro HIV-1 concerted integration assays were performed as previously reported (8) using recombinant purified IN or IN•LEDGF/p75 complex (200 nM in IN monomers) and biotinylated native or modified 601 mono-nucleosome or tri-nucleosomes assembled as previously reported (8). Under typical conditions IN/viral DNA complexes were pre-assembled 20 min at 4°C in the presence of the 17/19 bp U5 viral ends DNA hybrid radiolabeled in 5’ (10 nM) and LEDGF/p75. The pre-assembled complex was then incubated with MN (50 ng DNA) for 5 min in 20 mM HEPES pH 7, 10 mM MgCl2, 20 μM ZnCl2, 100 mM NaCl, 10 mM DTT final concentrations. After reaction products are treated with proteinase K and phenol/chloroform/isoamyl alcohol (25/24/1 v/v/v) before salt precipitation. Reaction samples are then loaded on 6–15% native polyacrylamide gel and run for 5 h at 200 V. The gel was then dried and submitted to autoradiography before analysis using ImageJ software. For quantitative assay the acceptor MN substrates were immobilized on streptavidin-coupled beads before reaction and the reaction products were deproteinized as described above and the integration was quantified by counting the remaining radioactivity bound to magnetized beads. Integration assays using purified assembled intasomes were performed as followed: 200 ng of mononucleosome were incubated with 30nm final concentration purified intasome in 100 mM NaCl, 20 mM BTP pH 7, 12.5 mM MgSO4 in a final volume of 40 μl for PFV intasome, and for HIV intasome in 20 mM HEPES pH 7, 7.5% DMSO, 8% PEG, 10 mM MgCl2, 20 μM ZnCl2, 100 mM NaCl, 5 mM DTT final concentration. The mix was then incubated at 37°C for 15 min and 1 h, respectively. Then reaction was stopped by the addition of 5.5 μl of a mix containing 5% SDS and 0.25 M EDTA and deproteinized with proteinase K (Promega) for 1 h at 37°C. Nucleic acids were then precipitated with 150 μl of ethanol overnight at −20°C. Samples were then spun at top speed for 1 h at 4°C, and the pellets were dried and then re-suspended with DNA loading buffer. Integration products were separated on an 8% native polyacrylamide gel. For integration assays onto chromosomes, each intasome was incubated 1 h in specific optimized reaction conditions (see above) at 37°C. Chromosomes were then treated 10 min with 5 μg/ml of trypsin at RT and washed extensively with Triton X‐100 1%, saponin 0.4%. Integration efficiency was evaluated by measuring the intensity of fluorescence remaining on chromosomes compared to conditions without Mg2+ or with the IN active site inhibitor dolutegravir 1 μM final concentration.
Cellular Integration site selectivity and chromatin features analyzes
K562 cell line (human immortalized myelogenous leukemia cell line) purchased from ATCC company were transduced as previously described by lentiviral vector (23). An optimized multiplicity of infection (MOI) of 1 was used, which resulted in 25–35% of the cells containing one copy of proviral DNA as determined before. Removing of non-integrated viral DNA and library generation were performed as previously reported (23). The 200–500 bp size range of the indexed libraries were agarose gel-isolated and mixed equimolarly for 100 base, single-end Illumina sequencing on a HiSeq 2000 instrument using 40% PhiX DNA spike-in at Genewiz, USA. The raw reads starting with condition-specific indexes were grouped and filtered for the presence of the virus-specific nested primer followed by LTR sequences at the tip of the LTR. The rest of the reads were quality-trimmed as soon as 2 out of 5 bases had quality scores less than a Phred score of 20. We used bowtie (24) with the TAPDANCE tool (25) to map the reads to the hg19 human genome assembly in cycles with decreasing read length of 60, 55, 50, 45, 40, 35 allowing 3, 3, 3, 2, 1, 0 mismatches, respectively, with the following bowtie parameters in the mapping cycles: [–quiet -a -v < nu. mismatches allowed> -m 1 –suppress 5,6,7 –f]. Any insertion site was considered valid if there were at least five independent reads supporting it. All read pre-processing and follow-up analyses were done in R (R Development Core Team (2008). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3–900051-07-0, URL http://www.R-project.org.). BEDTools (26) and genomation were used to analyze the representation of ISs in chromatin state segment datasets making use of a consensus merge of the segmentations produced by the ChromHMM and Segway software (27).
RESULTS
Analysis of intrinsic HIV-1 IN and LEDGF/p75 chromatin-binding properties
To monitor the direct binding of proteins onto chromatin, we used chromosomes isolated from blood cells and spread on glass (see S2 and a full description in the methods section). Chromosome spreads provide important sources of biological material in which individual chromosomes can be identified. In metaphase chromatin structures, little to no transcription occurs (28), demonstrating that the cellular machinery has been mainly eliminated and not available to interfere with the direct binding of our proteins of interest to complicate the study of the intrinsic chromatin-binding properties of proteins. This choice also removes a major source of variability between non-metaphasic chromosomes, which restrains the analysis to the intrinsic chromatin-binding properties of our candidates. Furthermore, most of the chromatin features were found to be conserved, as DNA content genes and epigenetic marks positions (29), when compared to interphase chromosomes, allowing the specific study of the influence of these parameters on proteins binding. We thus first adapted the (pro)metaphase chromosome spreading procedure described previously (29) to maintain chromatin and nucleosome integrity and deplete the material associated with chromatin-binding proteins. For this purpose, we used synchronized peripheral T lymphocytes containing HIV-1-susceptible cells and a fixation buffer adjusted to 3/1 (v/v) methanol/acetic acid to maintain chromosome integrity for further binding assays. Then, the chromosomes were treated with a detergent-containing permeation buffer allowing the smooth release of the proteins associated with the chromatin except for the nucleosome components. The integrity of the chromosome was then checked by DAPI staining and immunostaining using antibodies directed against histones as previously shown (29) (S3). We also ensured the absence of contamination with chromatin-associated proteins, which would have biased our analysis. In particular, we controlled for the lack of cellular factors previously reported to modulate HIV-1 integration, including FACT and LEDGF/p75. As reported in S3B, immunostaining, as well as western blots, did not detect the presence of these factors, indicating that they were not present, at least not in significant amounts, in the chromosome preparations.
The methodology for analyzing the chromatin-binding property of a protein candidate was established as described in S2. Briefly, the protein of interest was incubated with the chromosome spreads under conditions described in the materials and methods section. The interaction with chromosomes was then monitored by immunofluorescence using antibodies directed against the studied factor, and the chromatin was visualized by DAPI staining. The protein interaction was evaluated after quantification of the fluorescence signal among selected chromosomes with ImageJ software. DAPI staining was used to determine the exact shape and size of the chromosome (identified by inverted DAPI staining) and the distribution profile of the protein fluorescence signal along each selected chromosome. The correlation between the DAPI stained chromatin and the protein distribution was determined using a MATLAB pipeline developed in-house for this work (see materials and methods section). The intensity of binding, as well as the distribution of the protein, were then compiled from at least ten independent spreads and up to ten chromosomes that could be unambiguously identified, oriented by inverted DAPI staining. The means and standard deviations of the data were calculated. Using this method, the data on both the efficiency of the interaction and the preference for chromosomal regions were assessed by measuring the fluorescence signal along the different chromosomes. Analyses were performed mostly on chromosomes 1, 2 and 3 since (i) they could be identified without additional staining (i.a. Giemsa banding) which would not be compatible with our assays and (ii) they sustain HIV-1 integration both in vivo and in cellulo in various studies as confirmed in our analyses of integration sites distribution among chromosomes in K562 lymphoid cells transduced with VSV-G pseudotyped lentiviral vector (S4 and insertion sites positions in the Supplementary File S1).
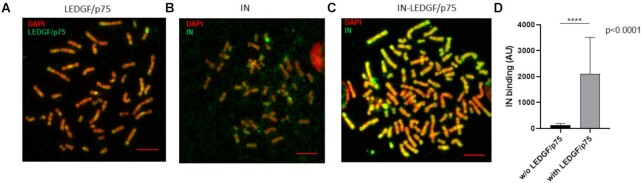
When incubated separately with metaphase chromosomes, both LEDGF/p75 and IN showed chromatin binding. Indeed, as reported in Figure 1A, LEDGF/p75 showed strong binding to chromatin which appears consistent with the previously observed capability of this protein to bind metaphases in cells (18). HIV-1 IN also bound chromosomes with distinct preferences for chromatin regions (Figure 1B). In HIV-1-infected cells, IN and LEDGF/p75 are assumed to form a complex that allows efficient targeting of the intasome toward targeted chromatin sites. Thus, to investigate the impact of LEDGF/p75 on the intrinsic chromatin-binding property of HIV-1 IN, we compared the binding efficiency of IN alone or in the presence of LEDGF/p75. As reported in Figure 1C, IN was found to strongly bind the chromosome in the presence of its cofactor LEDGF/p75. Quantification of the intensity of binding of IN to chromatin in each condition (total IN fluorescence signal) along the chromosomes analyzed from the 7 to 11 spreads showed that the presence of LEDGF/p75 strongly and significantly improved the amount of IN bound to chromatin (see quantification data in Figure 1D).
Figure 1.
Binding of human LEDGF/p75 and HIV-1 IN to chromosomes spreads. Either recombinant purified LEDGF/p75 or HIV-1 IN (4nM) were incubated alone (respectively A and B) or together (C) with chromosomes spreads under the conditions described in materials and methods section. Monitoring of their interaction with the chromatin was performed by immunofluorescence with the corresponding antibody coupled to ALEXA 594 (green signal) and DAPI staining (red signal). The intensity of total IF signal has been quantified in each conditions using ImageJ software and the IN binding without or with LEDGF/p75 were reported in (D) as mean from the quantification of 7–11 chromosomes in each condition ± standard deviation (SD), P-value was calculated by student test. Scale bar = 10 μM.
LEDGF/p75 modulates both the efficiency and specificity of IN binding to chromatin
In order to get information about the binding specificity of HIV-1 IN and LEDGF/p75 we next analyzed the distribution of each protein alone or together. As reported in Figure 2, while both proteins bound chromosomes, significant differences were detected in their preference for chromosomal regions.
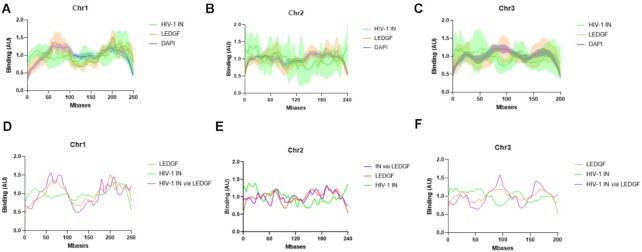
Figure 2.
Comparison between the distribution of HIV-1 IN alone or in the presence of LEDGF/p75. Either recombinant purified LEDGF/p75 or HIV-1 IN (4 nM) was incubated with chromosomes spreads under the conditions described in materials and methods section alone or together. The protein binding to chromatin was monitored by immunofluorescence as performed in Figure 1. The distribution profile of IN and LEDGF/p75 along chromosome 1, 2 and 3 was analyzed and reported respectively in (A), (B) and (C). The distribution profile of IN incubated with chromosome spreads in the presence of LEDGF/p75 has been monitored and quantified and the distribution curve has been reported in (D) for the chromosome 1, (E) for chromosome 2 and (F) for the chromosome 3. The data are reported as means from 7 to 11 serials of quantifications ± SD. In (D)–(F), the SD were omitted to improve readability.
Indeed, the analysis of the LEDGF/p75 distribution performed on chromosomes which were unambiguously identified and oriented on the basis of DAPI staining showed that similar regions of each chromosome were apparently enriched with the factor (Figure 2A–C). LEDGF/p75 localization and DAPI staining were then compared, and statistical analysis, including Pearson correlation and P-value (percentage of comparisons with high correlations) calculation, was applied to determine the similarities and the differences between the distribution curves. The mean and standard deviation of the Pearson correlation calculation (>0 for a correlation, =0 for no correlation and <0 for a negative correlation) performed between all paired LEDGF/p75 and DAPI curves unambiguously showed a high correlation (Pearson correlation for chromosome 1 = 0.53 ± 0.21 and P < 0.01 for 84% of pairs, for chromosome 2 = 0.51 ± 0.18 and P < 0.01 for 90% of pairs, for chromosome 3 = 0.39 ± 0.22 and P < 0.01 for 92% of pairs).
Analysis of HIV-1 IN chromatin-binding properties also revealed a reproducible interaction of the retroviral enzyme with specific regions of the chromosomes. Regions bound by IN in the context of a specific chromosome were reproducibly enriched with the retroviral enzyme (Figure 2A–C). Strikingly, in contrast to the observation for LEDGF/p75, the comparison between IN localization and DAPI staining showed only low, or no, correlation (Pearson correlation for chromosome 1 = 0.14 ± 0.21, P < 0.01 for 29% of pairs, for chromosome 2 = 0.16 ± 0.26, P < 0.01 for 50% of pairs, and for chromosome 3 = 0.083 ± 0.22, P < 0.01 for 15% of pairs). Merging of the LEDGF/p75 and IN chromatin interaction signal confirmed that the distribution profile of each protein showed only partial overlap (see the merged curved in Figure 2A–C). Direct statistical comparison of the distribution of HIV-1 IN and LEDGF/p75 showed no or low correlation between these proteins on chromosome 1 (Pearson correlation = 0.19 ± 0.3, P < 0.01 for 57% of pairs), chromosome 2 (Pearson correlation = 0.09 ± 0.24, P < 0.01 for 40%) and chromosome 3 (Pearson correlation = 0.04 ± 0.31, P < 0.01 for 32% of pairs). These data confirmed that both proteins bind chromatin but showed different affinities for chromosome regions.
The analysis of the distribution of IN alone or in the presence of LEDGF/p75 was then performed and reported in Figure 2D–F. The global distribution of IN along the chromosomes was found modified by the presence of its cofactor. Indeed, when comparing the correlation between IN alone and LEDGF/p75 alone to the correlation between IN in the presence of LEDGF/p75 and LEDGF/p75 alone the Pearson correlation shifted from 0.19 ± 0.3, P < 0.01 = 0.57, to 0.45 ± 0.16, P < 0.01 = 0.86, in the presence of LEDGF/p75 for chromosome 1 and from 0.04 ± 0.31, P < 0.01 = 0.32, without LEDGF/p75 to 0.23 ± 0.25, P < 0.01 = 0.37, in the presence of LEDGF/p75 for chromosome 3. A calculation of the p-value (Wilcoxon rank sum test) performed in each position of the chromosome (see S5) allowed us to more precisely define several regions of the chromosomes preferred or excluded in each condition. These statistical and correlative analyses of the IN distribution showed that in the presence of LEDGF/p75, HIV-1 IN was found to be mainly retargeted toward alternative regions of chromatin that were mainly similar to those recognized by LEDGF/p75 alone.
Taken together these data indicate that LEDGF/p75 modified IN localization along chromosomes demonstrating its IN retargeting function in our model and its capability to modulate intrinsic IN chromatin binding properties. We next further investigated the determinants of these distinct chromatin binding processes.
HIV-1 IN and LEDGF/p75 determinants of chromatin tethering and targeting processes
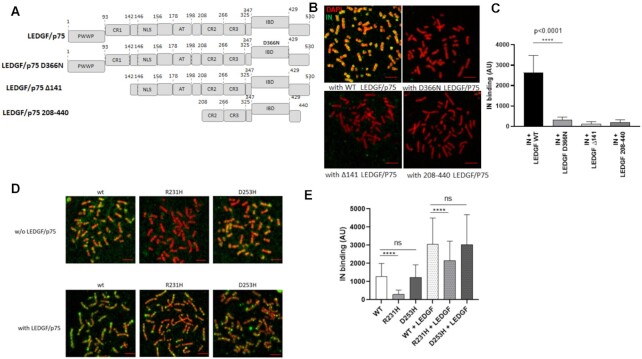
The LEDGF/p75-IN interaction sites have been precisely mapped on both partners, and the IN-binding domain (IBD) of LEDGF/p75 and the catalytic core domain of IN (CCD) have been identified as interacting regions (see Figure 3A). The IBD D366N mutation has been shown to prevent the ability of LEDGF/p75 to interact with IN (30), and extensive studies have shown that the PWWP domain and AT hook domains of LEDGF/p75 mediate its chromatin-binding properties (18,20,31). The association of LEDGF/p75 with AT-enriched regions stained with DAPI found in Figure 2 correlates finely with the presence of the AT-Hook domain in LEDGF/p75 and may account for the capability of the protein to bind high AT-contains DNAs (17). This was further supported by the data obtained with LEDGF/p75 mutants lacking the AT-Hook. Indeed, deletion of the PWWP and AT hook domains (mutant 208–440 and mutant Δ141, respectively) prevented IN chromatin targeting by LEDGF/p75 (Figure 3B-C). We also showed that the D366N mutation preventing the LEDGF/p75 binding to IN is also associated with a decrease in the capability of LEDGF/p75 to target IN toward chromatin. To note, treatments with LEDGins or ALLINIs drugs that block the IN-LEDGF/p75 association also prevented the IN-LEDGF/p75 binding to chromosomes confirming the importance of the IN interaction to LEDGF/p75 in the anchoring mechanism (data not shown).
Figure 3.
Effect of LEDGF/p75 and IN carboxyterminal domain mutants on the modulation of HIV-1 IN chromatin binding property. The different constructions used in the experiments are reported in (A). Full length, D366N LEDGF/p75 punctual mutant impaired for binding to HIV-1 IN and truncations deleted for the PWWP chromatin binding domain and AT hook of LEDGF/p75 were compared for their effect on HIV-1 IN binding to chromosomes. The purified LEDGF/p75 mutants were added to HIV-1 IN in the chromatin binding assay and their effects on the anchoring of IN were compared to the one showed by WT protein (B). The total IN binding signal was quantified along chromosomes (here chromosome 1) in the different conditions and are reported in the histogram as means of eight independent set of quantification (between 738 and 954 quantification points) ± SD. Statistics were performed by Student's t-test and the calculated P is reported on the figure (C). WT, R231H and D253D IN proteins were incubated with chromosome spreads in the absence or in the presence of LEDGF/p75 (4nM) and the IN binding to chromatin was monitored by immunofluorescence using anti-IN antibody (D). The intensity of total IF signal has been quantified in each condition using ImageJ software on several chromosomes and reported as IN binding. The data are reported as mean of the quantification of ten chromosomes 1 (1026–1122 quantification points) ± SD. Statistics were performed by Student's t-test and the calculated p is reported on the figure (E). Scale bar = 10 μM.
Since IN alone shows affinity for chromatin, we also investigated the molecular determinants of this interaction and their roles in tethering the viral enzyme in the presence of LEDGF/p75 to chromatin. Target DNA and histone-binding properties have been described for the CTD of several retroviral INs, including HIV-1 IN (8,13,22). Thus, we analyzed the importance of this domain in binding to chromosomes. Several amino acids have been identified in HIV-1 IN as important for the interaction to both the target DNA and histone tail (8,13,14,32). These positions include the R231 and D253 residues. To test the chromatin-binding properties of HIV-1 IN, we used recombinant INs mutated in these positions and showed that these mutations affected their association with nucleosomes and histone tails (see extended analysis of the CTD mutants in (8) and S6). As reported in Figure 3D and quantified in Figure 3E, the R231H mutant IN was found to poorly bind chromatin, correlating closely with its poor ability to bind nucleosomes in vitro. In contrast, D253H mutant of IN, which presented similar, or even better, affinity for nucleosomal DNA than the WT enzyme, bound chromatin with efficiency close to WT. We next investigated whether this intrinsic chromatin-binding property of the IN CTD also contributes to the interaction of the viral protein in the presence of LEDGF/p75 with the chromosome. The R231H and D253H IN proteins binding to chromosome were next analyzed in the presence of LEDGF/p75. In this context, the differences found in the association of the mutants with chromatin compared to that of the wild-type enzyme were drastically smoothed, confirming that LEDGF/p75 is a strong modulator of the chromatin-binding property of IN (Figure 3D, E). However, in the presence of LEDGF/p75, the R231H chromatin-binding efficiency was still significantly lower than that of the wild-type and D253H mutant proteins (8). Taken together, our results indicate that the CTD domain of HIV-1 IN participates in the recognition and binding of the viral protein to the chromatin regions of the chromosomes, even in the presence of its cofactor.
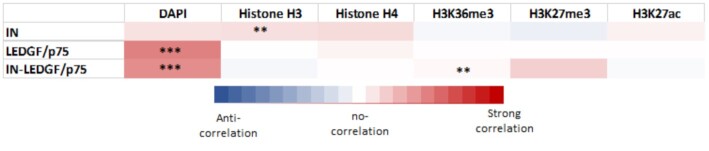
Since IN and LEDGF/p75 are expected to bind both DNA and nucleosomes, their association with chromosomes regions may also be modulated by the access to these components of the chromatin. To investigate this, we probed the DNA content by DAPI staining and nucleosome accessibility by histone distribution (see S7). We then performed a correlative analysis with the distribution of our protein candidates. As reported in the heat maps shown in Figure 4, a significant correlation was found between the HIV-1 IN and histone distributions on chromosomes (Pearson correlation = 0.135 ± 0.013 and 40–55% of pairs with P < 0.01). IN distribution correlated poorly with DAPI with pearson correlation = 0.12 ± 0.02 and 22–29% of pairs with P < 0.01. In contrast, no correlation was found between LEDGF/p75 and histones distribution (pearson correlation = 0.24 ± 0.039 and 36–40% of pairs with P < 0.01) while a strong correlation was found with DAPI staining (pearson correlation = 0.495 ± 0.049 and 84–88% of pairs with P < 0.01). These data confirmed that both proteins behaved differently on chromatin and suggested a significant link between the IN distribution and chromatin accessibility via mostly histones, while LEDGF/p75 distribution on metaphase chromatin depends mostly on DNA content. Analyses performed with the IN-LEDGF/p75 complex also showed a high correlation with DAPI staining (Pearson correlation = 0.450 ± 0.010 and 82–86% of pairs with P < 0.01) and no correlation with histone distribution (Pearson correlation = –0.019 ± 0.033 and 30–35% of pairs with P < 0.01) confirming the retargeting process and LEDGF/p75-mediated IN anchoring. Additional comparison of the protein distribution with histone marks including H3K36me3, associated with gene bodies, H3K27me7, associated with heterochromatin and H3K27ac associated with active transcription showed poor correlation with IN and LEDGF/p75 (see histone modifications distributions in S7 and heat maps in Figure 4).
Figure 4.
Correlation analysis between the protein distributions and chromatin features. Distributions of IN, LEDGF/p75 and IN-LEDGF/p75 were compared to DAPI staining and histones distribution. The mean and standard deviation of the pearson correlation calculation (>0 for a correlation, =0 for no correlation and < 0 for a negative correlation) were performed between all paired protein distribution and DAPI/histone curves. The p-value (Wilcoxon rank sum test) at each position and the percentage of pairs with P < 0.01 was also calculated and were reported as *** if > 0.80 and ** if > 0.40).
In summary, over all conditions tested, the best correlations were found for LEDGF/p75 versus DAPI and IN versus histones. These data suggest that the first major process for LEDGF/p75 and LEDGF/p75-mediated IN anchoring to chromosomes was the accessibility to chromatin, AT-rich regions and nucleosome without strong preference for specific histone modifications in the metaphase context.
Catalysis of integration requires the formation of a nucleocomplex formed between the IN protein, LEDGF/p75 and the viral DNA. To further determine the property of these functional complexes we next performed integration assays.
Modulation of the in vitro nucleosomal integration by LEDGF/p75
To further investigate the effect of LEDGF/p75 on functional integration complexes and determine whether the LEDGF/p75-mediated increase in IN chromatin-binding was related to the nucleosomal features of chromosomes, we first performed typical in vitro integration assays using biotinylated nucleosomes (MNs) immobilized on streptavidin beads as previously performed (8,13). As shown in Figure 5A, LEDGF/p75 strongly stimulated HIV-1 IN activity on MNs, while integration onto naked DNA was only slightly improved in the presence of the cofactor. These data were in agreement with data from the literature (19) and suggested that LEDGF/p75 modulated the functional interfaces between the HIV-1 intasome and the nucleosome in addition to its modulation function on IN chromatin association found above.
Figure 5.
Effect of LEDGF/P75 on in vitro integration catalyzed by HIV-1 IN onto nucleosomes. Integration was performed using radiolabelled viral DNA, HIV-1 IN with or without LEDGF/p75 using either biotinylated naked 601 DNA or MN assembled on 601 sequence. The integration was quantified by measuring the remaining radioactivity on streptavidin beads after hammering the target DNA (A) or by loading the integration products on polyacrylamid 6–12% gradient gel (B). Data are reported in (A) as stimulation effect of LEDGF/p75 according to the control experiment performed without the cellular factor on either naked of MN targets. Data were obtained from at least three independent experiments and are reported as means ± SD. Statistics were performed by student test and the calculated p is reported on the figure. Integration product structures were analyzed on native polyacrylamide 6–12% gel as reported in (B). The half site integration (HSI) and full site integration (FSI) products are schematized in the figure. The positions of the integration sites on the nucleosomal DNA in the absence or in the presence of LEDGF/p75 were compared by scanning the gel migration profile using ImageJ software and reporting the radioactivity signal intensity in function of the migration distance from the top of the gel (representative analysis reported in the figure). Integration performed in the presence of LEDGF/p75 with MNs carrying histone modifications H3K36me2/me3 (C), tailless MNs (D) and tri-nucleosome (E) were performed under similar reaction conditions and products were quantified as described previously. Data were obtained from three to four independent experiments and are reported as means ± SD.
Since we showed that LEDGF/p75 could also modify the IN preference for chromatin regions we investigated whether it could affect the integration selectivity toward the nucleosomal structure. The positions of the integration products generated on MNs were thus analyzed using native polyacrylamide gels. As shown in Figure 5B, the integration products formed in the presence of IN alone were distributed along the MNs with preferred sites. While the stimulation of the integration by LEDGF/p75 was confirmed, the distribution profiles of the integration sites did not appear drastically changed, as indicated by the insertion site position analysis shown in the left panel of Figure 5B. These results suggest that LEDGF/p75 improves nucleosomal integration catalyzed by IN without affecting the global anchoring position for the integration complex onto MN. MN assembled with histone octamers carrying histone modifications H3K36me3, known to be recognized by LEDGF/p75, showed similar integration profiles without significant difference in the insertion efficiency (Figure 5C). Similar results were obtained with H3K36me2 MNs. This suggested that these modifications were not required for optimal in vitro integration nor involved in the local insertion selectivity at the surface of the targeted nucleosome. However, deletion of the histone tails significantly decreased the integration efficiency indicating their importance for optimal viral DNA insertion (Figure 5D). Taken together, these biochemical data agree closely with the findings showing the stimulation of IN binding to chromatin by LEDGF/p75 observed on chromosomes, indicating that the cellular factor potentiates IN functional interactions with nucleosomes and modulate the chromatin binding properties of the retroviral enzyme. The similar integration profile observed in in vitro integration assays performed either in the presence or in the absence of LEDGF/p75, either using modified histone tails additionally suggests that the differences observed in IN binding to chromatin in the presence of LEDGF/p75 were not due to altered specificity for the nucleosome structure but were mainly related to distinct preference for local surrounding chromatin features, suggesting a role of chromatin neighboring the interaction site. To better investigate the influence of the neighboring chromatin, we performed integration assay using polynucleosomes. Data reported in Figure 5E indicated that, while the isolated nucleosome is a good substrate for integration, the tri-nucleosomes was poorly efficient as integration template. This result confirmed that the neighboring nucleosomes modulate the access to targeted nucleosomes for efficient integration.
Taken together, all these data indicate that the main determinant for efficient functional association between the HIV-1 intasome and the nucleosome is the accessibility of the nucleosome at the final insertion site that is determined by the chromatin structure of the surrounding chromosomal region. In concert with the binding data obtained using chromosome spreads, our results suggest that the association to chromosome may be a prerequisite step before integration that may then require additional processes such as local chromatin remodeling which could improve the nucleosome accessibility.
Optimal IN targeting toward chromatin requires preformed IN-LEDGF/p75 complex
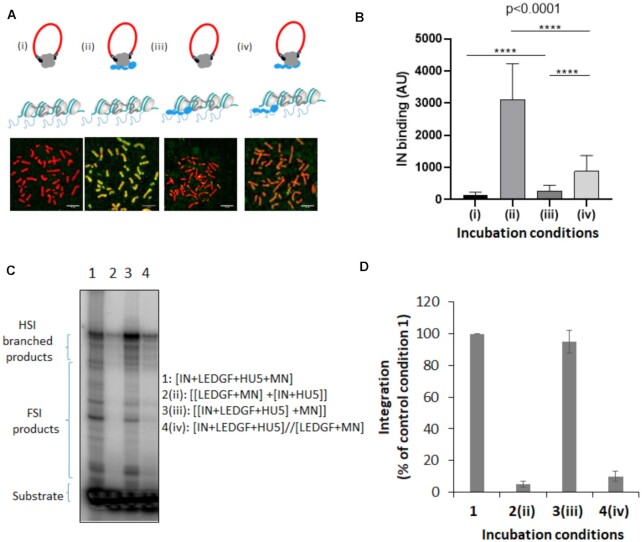
Since the chromosomes binding assay performed in this work recapitulated the LEDGF/p75 targeting of IN toward chromatin, we used this model to depict the targeting process better. The time-of-addition experimental scheme presented in Figure 6A shows that, although incubation of LEDGF/p75 with chromosomes before adding IN (iii) slightly improved IN binding to chromatin, the best IN chromatin anchoring by LEDGF/p75 was observed when both proteins were preincubated before their addition to the chromosome spreads (Figure 6B, condition (ii)).
Figure 6.
Time of addition analysis of the LEDGF/p75-mediated chromatin tethering of HIV-1 IN. HIV-1 IN was incubated with chromosome spreads in the presence or in the absence of LEDGF/p75 following different time of addition conditions reported in (A): (i) IN alone, (ii) IN and LEDGF/p75 were preincubated before being added to the chromosome spreads, (iii) LEDGF/p75 was pre-incubated with chromosome spreads before adding IN and (iv) IN and LEDGF/p75 were preincubated together and added to chromosomes spreads previously precincubated with LEDGF/p75. The total amount of LEDGF was similar in all conditions. The intensity of total IN IF signal has been quantified in each condition using ImageJ software on several chromosomes and reported as IN binding. The data are reported in (B) as mean of the quantification from 12 chromosomes (here chromosome 3, 1135–1220 quantification points per condition) ± standard deviation (SD). Statistics were performed by student test and the calculated p is reported on the figure. Integration assay onto MN was performed in difference incubation conditions using recombinant IN with or without LEDGF/p75 as done in Figure 5. A representative gel is reported in (C) and quantification data obtained from at least three independent experiments are reported IN in the graphic (D) as means ± SD. Integration product structures are reported as half site integration (HSI) or full site integration (FSI). Condition 1: IN, LEDGF/p75, viral donor U5 DNA and target nucleosomal DNA were pre-incubated altogether, condition 2: [LEDGF/p75/target nucleosomal DNA] and [IN/viral U5 donor DNA] were preincubated separately before reaction, condition 3: IN, LEDGF/p75 and the viral donor U5 DNA were preincubated together before adding the target nucleosomal DNA, condition 4: [IN, LEDGF/p75, viral donor U5 DNA] and [LEDGF/p75, target nucleosomal DNA] were incubated separately before reaction.
Interestingly, preincubation of LEDGF/p75 with chromosomes before adding the preformed IN•LEDGF/p75 complex (conditions (iii) and (iv)) decreased the IN-binding efficiency compared to conditions without LEDGF/p75 bound to chromosomes. These data suggest, first, that the efficient retargeting of the incoming IN requires the formation of the complex before interacting with chromatin, and second, the presence of LEDGF/p75 on the final anchoring sites may prevent optimal binding. We further investigated this process with a functional integration assay to determine whether the integration process followed similar chronological rules. As reported in Figure 6C and quantification in Figure 6D, preincubation of IN with LEDGF/p75 led to maximal integration efficiency in vitro, while the incubation of LEDGF/p75 with nucleosomes before adding IN or the preformed IN•LEDGF/p75 complex profoundly prevented optimal integration. These data are in agreement with the chromatin binding of IN observed in the chromosomes spreads, supporting the functional relevance of our system and strengthening our conclusions.
Altogether, these results indicate that without preliminary interaction between both factors, IN binding to chromatin was relatively inefficient, and IN alone does not appear efficient in finding LEDGF/p75-enriched sites on chromosomes. In addition to the data obtained in the integration assays, this finding further supports the hypothesis that incoming intasomes form functional complexes with LEDGF/p75 before reaching chromatin sites. We further investigated this finding by analyzing the behavior of fully functionally pre-assembled retroviral intasomes which constitute the biological relevant species.
Chromatin-binding and integration properties of functional intasomes
A native HIV-1 intasome has been assembled and purified using HIV-1 IN, LEDGF/p75 recombinant protein and short ODN containing a viral DNA end sequence tagged with FITC following previously reported conditions (21). As shown in S8, HIV-1 intasome showed expected elution profile from size exclusion chromatography confirming the correct assembly. Enough complex could be obtained to perform further functional integration assays. Typical in vitro integration assays shown in S8 confirmed that the purified complex was catalytically active allowing its characterization.
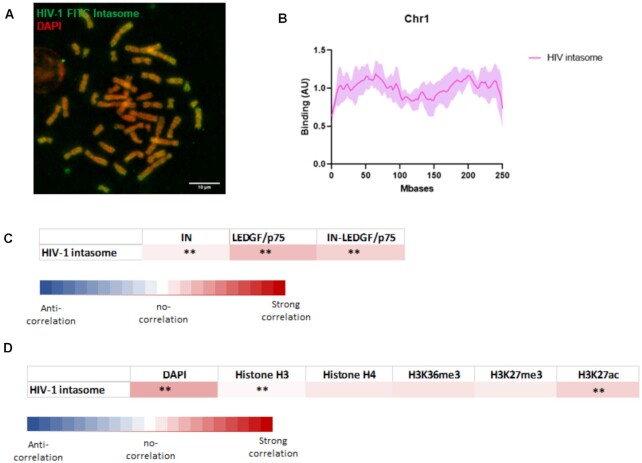
The analysis of the HIV-1 intasome chromatin binding properties showed an efficient association with the chromosomes (Figure 7A). The distribution analysis also revealed preferences for some chromatin regions as reported for the chromosome 1 in Figure 7B and chromosome 2/3 in S9. Comparison with the binding property of IN, LEDGF/p75 or IN-LEDGF/p75 showed that the HIV-1 intasome behaved mostly as LEDGF/p75 and IN-LEDGF/p75 (see the heat map in Figure 7C). Especially, pearson correlation between intasome and IN-LEDGF/p75 distributions = 0.25 ± 0.21 with P < 0.01 for 51% of pairs) suggests that the structuration with the viral DNA did not change the overall chromatin association property of the complex and indicating that LEDGF/p75 remained the major chromatin targeting motor in the functional complex. However, the differences between IN and IN-LEDGF/p75 distribution were lowered when compared to the differences between IN and the intasome. This suggests that the IN chromatin binding determinants are more prone to be expressed within the functional complex that in the IN-LEDGF/p75 complex leading, in these conditions, to a more intermediate distribution profile of the intasome between IN, LEDGF/p75 and IN-LEDGF/p75. More detailed analysis of the correlation between intasome distribution and chromatin features confirmed that the intasome recognized metaphase chromatin regions mostly similar to the IN-LEDGF/p75 complex with a strong correlation found with DAPI staining (see Figure 7D).
Figure 7.
Analysis of the chromatin binding property of HIV-1 intasome. HIV-1 intasome was assembled using IN, LEDGF/p75 and the viral short DNA sequence corresponding to the viral end tagged with FITC. The intasome was purified by size exclusion chromatohraphy (see S8). After verification of their functionality in in vitro concerted integration assays the intasome (4nM) were incubated with chromosomes spreads. Interaction profile was determine by FITC-epifluorescence acquisitions (A). The distribution profile onto chromosome 1 was compared determined as reported in materials and methods section (an example of chromosome 1 is reported in (B), see the profile of chromosome 2 and 3 in S9). The data are reported as means from the quantification of eight to nine chromosomes ± standard deviation (SD). Scale bar = 10 μM. Correlation between the distributions of the intasome and the distribution of IN alone, LEDGF/p75 alone or IN-LEDGF/p75 complex (C) as well as DAPI staining, H3/H4, H3K36me3, K3K27me3 and H3K27ac (D) was compared. The mean and standard deviation of the Pearson correlation calculation (>0 for a positive correlation, =0 for no correlation and <0 for negative correlation) were performed between all paired protein distribution and DAPI/histone curves. The P-value (Wilcoxon rank sum test) at each position and the percentage of pairs with P < 0.01 was also calculated and were reported as *** if > 0.80 and ** if > 0.40).
To determine whether this profile was specific for HIV-1 intasome, we analyzed the chromatin-binding properties of an additional functional intasome: the prototype foamy virus (PFV). Following indications from the literature (22), we assembled a PFV intasome exhibiting the proper integration activity (S8). Analysis of the chromatin-binding properties of the purified intasome as determined with the chromosomes spreads binding assay (Figure 8A) also indicated that this complex binds chromatin more efficiently than HIV-1 intasome (see the chromosome binding imaging in S10 and quantification of binding in Figure 8B). Distribution analysis on the different chromosomes also showed a non-uniform binding of PFV intasome (see S10). Direct comparison of HIV-1 and PFV intasome distribution showed only a partial correlation (Pearson correlation = 0.28 ± 0.20 with P < 0.01 for 0.55% of pairs). Noticeably, the PFV intasome chromatin binding signal was more homogeneous along chromosomes than for HIV-1 intasome or HIV-1 IN•LEDGF/p75 complex. A control experiment performed with PFV in the presence of LEDGF/p75 showed no effect of the cellular factor, as expected from the specificity of the cellular protein for lentiviral INs (S10). This result confirmed that the differences found between the two intasomes were not due to LEDGF/p75 but to intrinsic chromatin-binding properties specific for each complex, further validating the specificity of our assay.
Figure 8.
Analysis of the chromatin binding and integration efficiency of HIV-1 and PFV intasomes. Chromosomes spreads were used either to quantify the direct intasome binding or the integration efficiency (A). After incubation with intasome (5 nM) under binding conditions the fluorescence was quantified on nine chromosomes and reported as means ± SD (B). Exposition was adjusted to 2000 ms for HIV-1 and 1500 ms for PFV. For quantification of the integration efficiency, after incubation with intasomes under conditions allowing integration, samples were then treated with trypsin and washed extensively with Triton X-100. The remaining fluorescence was then quantified by epifluorescence in each conditions ± Mg2+ or dolutegravir 1 μM (DTG). Data are reported in (C) as mean of fluorescence from eight chromosomes ± standard deviation (SD).
To determine whether the HIV-1 intasome could integrate into the metaphase chromosomes we set up an integration assay as described in materials and methods section and Figure 8A). Briefly, the intasome was incubated with the chromosomes spreads with, or without Mg2+ or integrase inhibitor dolutegravir for 1 h at 37°C. Samples were then treated with trypsin and washed extensively with detergent to eliminate proteins, the remaining fluorescence was then quantified by epifluorescence. The integration efficiency was evaluated as the difference between the fluorescence signal quantified in Mg2+ conditions and conditions without Mg2+ or with dolutegravir. In these conditions the HIV-1 intasome was found to be poorly efficient in catalyzing integration into chromosomes (Figure 8C). Experiments performed with the PFV intasome showed that this complex was 3–4 times more efficient in catalyzing integration compared to the HIV-1 orthologue, confirming its different behavior onto metaphase chromatin and its higher capability to accommodate the metaphase chromatin.
Taken together all these data indicate that chromosome binding and integration properties may differ from intasome to intasome. These data, also confirm that while HIV-1 intasome can bind metaphase chromatin, efficient integration may require additional cell activities in contrast to PFV intasome that accommodates more easily with this chromatin structure as suggested by previous biochemical data (16).
DISCUSSION
During the retroviral integration process, the association between the incoming intasome and host chromatin constitutes a point of no return and a platform for molecular exchanges between the virus and the cell. While the early targeting processes of the intasome toward chromatin have been investigated for a long time, its final association with the nucleosome is not well characterized especially with regard to chromatin structure.
Lentiviruses can infect dividing and non-dividing cells suggesting that the incoming integration complexes can meet different types of chromatin structures including metaphases. However, whether incoming intasomes can bind and integrate in these different structures remained obscure.
Recent advances have shown that after nuclear entry and targeting toward a suitable region of the genome by cellular IN or CA cofactors, such as CPSF6 and LEDGF/p75 for lentiviruses, the intasome and the histone component of the nucleosome directly interact (8,13,22). Furthermore, IN domains, such as the CTD, have been shown to directly engage the target DNA and participate in insertion site selection in chromatin (14,32). These data indicate that the CTD of INs is the central domain for its association with the target substrate via its direct DNA- and histone-binding properties. In the specific case of HIV-1, previous data showed that HIV-1 IN association with histone tails, especially the H4 tail, modulated its functional binding to the nucleosome, leading to structural changes and the stabilization of the intasome/nucleosome complex (8,13). Mutations within the CTD that affect retroviral IN-histone binding were also shown to affect both the functional association with the nucleosome integration site selectivity in vitro and in vivo (8,22). Taken together, these data support the presence of a chromatin-binding function in the CTD of IN that may participate in the functional association with chromatin and insertion site selection. However, to date, no model for validating this function or investigating its molecular determinants is available. Here, using metaphase chromosomes spreads as a tool, in addition to biochemical approaches, we provided new data about the chromatin-binding properties of functional retroviral intasomes and their cellular and viral components.
Indeed, the analysis of the chromatin-binding property of HIV-1 IN alone showed that the protein was able to selectively bind regions of chromatin with a certain specificity. Since no endogenous FACT or LEDGF/p75 was detected (cf. S3), IN binding to chromatin was more likely an intrinsic property of the retroviral enzyme and not due to contamination by IN cofactors, which have the potential to interact both with chromatin and IN. Both the specificity and efficiency of IN binding to chromosomes were affected by the presence of the IN cofactor LEDGF/p75. A similar integration profile observed in integration assays performed on mononucleosomes in vitro, additionally suggested that the differences observed in IN binding to chromatin in the presence of LEDGF/p75 were not due to altered nucleosome specificity but were mainly related to distinct preferences of the IN•LEDGF/p75 complex for local chromatin features. This outcome suggests a role for nucleosomes neighboring the final interaction site selected within chromatin. In vitro integration assays performed with polynucleosomes confirmed that the neighboring nucleosomes modulate the access to the insertion site supporting the hypothesis that the local chromatin structure affects the functional binding of the incoming intasome. The analysis of the LEDGF/p75 distribution along the chromosomes showed that it appeared to correlate closely with chromatin DAPI staining. In contrast, the poor correlation found between IN distribution and DAPI staining suggested that the chromatin features recognized by IN were mainly independent of DNA structure and composition. The partial correlation found between IN distribution and nucleosome and histones accessibility confirmed that the access to nucleosome constitutes an important factor for IN interaction with the chromatin. Strikingly, no significant correlation could be found between protein distribution and histone marks. Especially, the H3K36me3 assumed to be recognized by LEDGF/p75, appeared not recognized in the metaphase chromosome context of our assay. This may be due to the low amount of this modification in the metaphase state or the requirement of additional cellular activities, or specific chromatin structures, for the LEDGF/p75 recognition. These new questions, provided by this work, about the still poorly documented process of LEDGF/p75 binding to histone marks in a chromosomal context remain to be investigated further.
While the significance of our observation in natural infection conditions needs to be elucidated further, our data provide additional information about the chromatin binding property of both retroviral IN and LEDGF/p75 alone or in complex. Additionally, the observations reported here may highlight the LEDGF/p75 mediated docking of the retroviral intasome in this specific metaphase structure. Although, the exact nature of the recognized chromatin regions remains to be determined, our data provide evidence that the intrinsic HIV-1 IN protein chromatin-binding properties are modulated by its cofactor, LEDGF/p75. This intrinsic chromatin-binding function of IN appeared to be carried by its CTD since mutations in this domain affected the efficiency of its chromosome interaction. In particular, mutations that were shown to affect the interaction of the protein with nucleosomal DNA and histone tails (8,13) affected the interaction with chromosomes in a similar manner. These results correlate closely with cellular data previously reported, i.e. a decrease in the integration efficiency of the R231H mutant and a close or better integration efficiency of the D253H mutant, with a redistribution of integration sites in both cases (8). However, despite its higher capability to bind nucleosome in vitro, the D253H mutant was not found to significantly bind chromosomes better than the WT enzyme. This may be due to differences in the association with isolated MNs and chromatin and this may be correlated to the lack of difference observed in cells for the insertion sites of this mutant versus the nucleosome density. Indeed, while the D253H mutated virus showed an increased infectivity in cells, it has been showed to behave mostly as the WT in term of preference for nucleosome density but with differences regarding the preference for transcriptional regions (8). Thus, the mutation may affect the behavior of the integration complex regarding the dynamic of the chromatin, which is not recapitulated in our model system, more than the nucleosomal structure. These data point out that, while the efficiency of cellular integration is directly related to the efficient association between the incoming integration complex, as shown in the chromosomes binding assay, the selection of the final integration site may depend in part on the intrinsic capability of the enzyme to prefer chromatin structures, even in the presence of LEDGF/p75, as demonstrated here.
Furthermore, comparisons of both the IN distribution profile and chromatin-binding efficiency in the presence or absence of LEDGF/p75 clearly showed that cellular factors modulate IN binding properties. Indeed, LEDGF/p75 greatly enhanced IN binding to chromosomes and modified its localization, targeting it mostly toward LEDGF/p75-preferred sites. We showed that both the PWWP chromatin binding and IN-binding domains in LEDGF/p75 were required for IN targeting toward chromatin. Our data indicated that LEDGF/p75 potentiated the IN functional interaction with nucleosomes and, thus, modulated the retroviral enzyme chromatin-binding property by affecting its behavior on nucleosomes. Interestingly, the R231H mutation in the IN CTD was shown to affect the enzyme chromatin-binding property, even in the presence of LEDGF/p75, suggesting that the IN CTD may participate in the final association and insertion site selection. This may provide the molecular basis for previously reported cellular selectivity analyses of viruses carrying these CTD mutations (8). Interestingly, all these data appear to corroborate of previous works showing that HIV-1 IN may modulate the interaction of LEDGF/p75 with chromatin (33). Indeed, the efficiency of viral integration into cells is directly related to the efficiently established association between the incoming integration complex, as shown on chromosomes, while the selection of the final integration site may be mediated in part by the intrinsic ability of the enzyme to prefer chromatin structures, even in the presence of LEDGF/p75, as demonstrated herein. Further characterization of the IN targeting process via LEDGF/p75 showed that maximal efficiency was observed when IN was pre-assembled with its cofactor. This finding supports a model where the presence of LEDGF/p75 complexed to the incoming integration complexes allows efficient anchoring to the final insertion sites, revealing a longstanding issue in the field. This supposition was confirmed by analyzing purified functional HIV-1 lentiviral intasomes assembled with LEDGF/p75, which showed efficient binding to chromosomes. Analysis of the HIV-1 intasome distribution showed that the differences found between IN and LEDGF/p75 or IN-LEDGF/p75 were smoothed since the functional complex showed a more intermediate profile between IN, LEDGF/p75 and IN-LEDGF/p75. These results suggest that the IN chromatin binding determinants within the functional complex could be more efficient in directing the binding site that in the IN-LEDGF/p75 complex. This could be due to specific structures adopted within the intasome. The comparison with the PFV intasome profile also suggested that the distribution profiles were intasome-specific, indicating that the integration complexes carry the main determinants for local chromatin anchoring through their nucleosome binding sites.
Further functional analysis of the capability of the HIV-1 intasome to catalyze integration into metaphase chromosomes showed that, while it can bind this chromatin structure in our assay, it has a poor capability to integrate on it, in contrast to PFV. This suggests that coupling with additional cellular activities, as chromatin remodeling as previously reported (6,7,34), may be required for efficient integration. This was confirmed by in vitro integration assays using polynucleosomes showing that while the isolated nucleosome is a good substrate for integration when it is encompassed into a polynucleosome it became poorly efficient for integration. In addition to these results, our data support the modulation of nucleosomal integration by contacts with neighboring nucleosomes.
The efficient anchoring of integration complexes by LEDGF/p75 on metaphase chromosomes may be related to the previous metaphase binding property of this factor (18). This anchoring capability may constitute an evolutive advantage for lentiviral integration complexes allowing them to bind this chromatin structure and, thus, stabilize their association to chromatin through the cell cycle for waiting for suitable integration conditions. It is currently assumed that dividing T cells display increased permissiveness to HIV-1 infection, the influence of major mitotic events such as NE breakdown and chromosome condensation/decondensation on viral DNA docking and subsequent integration remained poorly characterized. Our data show that retroviral integration complexes can bind metaphase chromosomes while integration remains poorly efficient. This observation may provide a molecular explanation to earlier observation which indicates that nuclear envelop breakdown during mitosis increases the integration complexes association to metaphase chromatin while its condensation delays the integration process (15). These data also complement previous data showing that compacted chromatin is refractory to HIV-1 integration in vitro (6,16) suggesting that the integration catalysis may require a dynamic chromatin as supported by the link between HIV-1 integration and chromatin remodeling activities (7). However, these previous results did not indicate whether retroviral intasomes can reach their integration site in metaphase chromatin or whether they bind but cannot catalyze integration and, thus, wait for next cell cycle steps as previously reported for MLV and PFV viruses. Interestingly, as mentioned above, LEDGF/p75, has been shown to bind strongly metaphases, suggesting that this factor could dock the incoming integration complex on these specific chromatin structures when encountered. Thus, LEDGF/p75 could play this role in allowing the intasome to stably seat on mitotic chromosomes waiting for suitable integration conditions. Strikingly, the viral Gag protein has been proposed to carry this docking function in the foamy virus models (35). This requirement for stable chromatin tethering process prior to integration may constitute a more general mechanism in retroviral integration that need to be further studied.
Indeed, whether the binding sites identified on the chromosomes spreads may correlate with the final cellular integration sites remains difficult to determine. In infected cells, additional pathways and factors participate in the insertion site selection including CPSF6 targeting toward highly spliced region and the spatial organization of the chromosomes within the nucleus. However, a global comparison between the chromosome intasomes association reported in this work and the distribution of integration sites along some chromosomes, as chromosome 1 shown in S11, revealed striking close profiles. This may suggest that after binding to their association sites the functional intasome may diffuse locally during the next cell cycle steps to move toward regions suitable for integration as previously reported in vitro for the PFV intasome (36,37). A first binding step to chromatin followed by a diffusion of the functional complexes toward the optimal integration sites could also be expected for other integration complexes as HIV-1. In the cells, these preferred HIV-1 integration sites are actively transcribed regions enriched in H3K36me3 histone marks known to be recognized by LEDGF/p75. Our data indicate no strong correlation between this histone modification and LEDGF/p75, IN-LEDGF/p75 or intasome distribution, at least in the metaphase context. This may be due to the low amount of this modification found in metaphase chromosomes (29). This could also suggest that the correlation between cellular integration sites and H3K36me3 may be due to a process independent from the initial binding to chromatin but to a possible coupling between the integration complexes and H3K36me3 deposit during the next stages of the cell cycle. The exact role of this modification during the integration process remains to be fully established since no impact on the integration catalysis has been detected to date.
Taken together all these data indicate that the main determinant for efficient functional association between the HIV-1 intasome and chromatin is the accessibility of the nucleosome that is determined by the structure of the surrounding chromosomal region. In concert with the binding data obtained using the chromosome spreads, our results support the hypothesis that the stable anchoring to chromosome may be a prerequisite step before integration that would require additional processes leading to an improved nucleosome accessibility. In addition to providing additional mechanistic information about the chromatin targeting process by the incoming integration complexes, the chromosomes spreads binding model used here paves the way for broad and deep analyses of these chromatin-binding mechanisms. Indeed, data obtained with functional intasomes indicated that this model could recapitulate the final step of chromatin targeting by the integration complexes in a chromosome context. Consequently, the chromosomes spreads model may constitute a suitable tool for further depicting all the parameters of this mechanism, such as the roles of additional cofactors or the kinetics of the docking process. Additionally, this assay can also be used in structure/function studies to analyze the role of specific amino acids and domains of the different integration actors in the anchoring mechanism, as shown here for the analysis of IN CTD mutants. Testing small molecules that may affect the chromatin-binding function of the enzymes should also be possible with this model, which may become complementary to the current panoply of biochemical and cellular assays used for studying the regulation of retroviral integration by chromatin components as well as for studying any protein or nucleocomplex candidate that may have chromatin-binding properties.
DATA AVAILABILITY
All additional data are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank N. Landrein and M. Bonhivers (MFP laboratory) for support in microscopy imaging. English editing has been performed by Nature Rearch Editing Service.
Contributor Information
Delphine Lapaillerie, Fundamental Microbiology and Pathogenicity Lab (MFP), UMR 5234 CNRS-University of Bordeaux, SFR TransBioMed. Bordeaux, France.
Benoît Lelandais, Imaging and modeling unit, Computational Biology Department, Institut Pasteur, Paris, France.
Eric Mauro, Fundamental Microbiology and Pathogenicity Lab (MFP), UMR 5234 CNRS-University of Bordeaux, SFR TransBioMed. Bordeaux, France.
Floriane Lagadec, Fundamental Microbiology and Pathogenicity Lab (MFP), UMR 5234 CNRS-University of Bordeaux, SFR TransBioMed. Bordeaux, France.
Camille Tumiotto, Fundamental Microbiology and Pathogenicity Lab (MFP), UMR 5234 CNRS-University of Bordeaux, SFR TransBioMed. Bordeaux, France.
Csaba Miskey, Paul-Ehrlich-Institute, division of medical biotechnology, Langen, Germany.
Guillaume Ferran, CHU de Bordeaux, Service de Génétique Médicale, Bordeaux France.
Natacha Kuschner, CHU de Bordeaux, Service de Génétique Médicale, Bordeaux France.
Christina Calmels, Fundamental Microbiology and Pathogenicity Lab (MFP), UMR 5234 CNRS-University of Bordeaux, SFR TransBioMed. Bordeaux, France.
Mathieu Métifiot, Fundamental Microbiology and Pathogenicity Lab (MFP), UMR 5234 CNRS-University of Bordeaux, SFR TransBioMed. Bordeaux, France.
Caroline Rooryck, CHU de Bordeaux, Service de Génétique Médicale, Bordeaux France.
Zoltan Ivics, Paul-Ehrlich-Institute, division of medical biotechnology, Langen, Germany.
Marc Ruff, IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Département de Biologie Structurale intégrative, UDS, U596 INSERM, UMR7104, CNRS, Strasbourg, France.
Christophe Zimmer, Imaging and modeling unit, Computational Biology Department, Institut Pasteur, Paris, France.
Paul Lesbats, Fundamental Microbiology and Pathogenicity Lab (MFP), UMR 5234 CNRS-University of Bordeaux, SFR TransBioMed. Bordeaux, France.
Jérôme Toutain, CHU de Bordeaux, Service de Génétique Médicale, Bordeaux France.
Vincent Parissi, Fundamental Microbiology and Pathogenicity Lab (MFP), UMR 5234 CNRS-University of Bordeaux, SFR TransBioMed. Bordeaux, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French ANRS research agency [ECTZ115893, SIDACTION 16-1AEQ-10465]. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lesbats P., Engelman A.N., Cherepanov P.. Retroviral DNA integration. Chem. Rev. 2016; 116:12730–12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kvaratskhelia M., Sharma A., Larue R.C., Serrao E., Engelman A.. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 2014; 42:10209–10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sowd G.A., Serrao E., Wang H., Wang W., Fadel H.J., Poeschla E.M., Engelman A.N.. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1054–E1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharma A., Larue R.C., Plumb M.R., Malani N., Male F., Slaughter A., Kessl J.J., Shkriabai N., Coward E., Aiyer S.S.et al.. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:12036–12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Rijck J., de Kogel C., Demeulemeester J., Vets S., El Ashkar S., Malani N., Bushman F.D., Landuyt B., Husson S.J., Busschots K.et al.. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep. 2013; 5:886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lesbats P., Botbol Y., Chevereau G., Vaillant C., Calmels C., Arneodo A., Andreola M.L., Lavigne M., Parissi V.. Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLoS Pathog. 2011; 7:e1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matysiak J., Lesbats P., Mauro E., Lapaillerie D., Dupuy J.-W., Lopez A.P., Benleulmi M.S., Calmels C., Andreola M.-L., Ruff M.et al.. Modulation of chromatin structure by the FACT histone chaperone complex regulates HIV-1 integration. Retrovirology. 2017; 14:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benleulmi M.S., Matysiak J., Robert X., Miskey C., Mauro E., Lapaillerie D., Lesbats P., Chaignepain S., Henriquez D.R., Calmels C.et al.. Modulation of the functional association between the HIV-1 intasome and the nucleosome by histone amino-terminal tails. Retrovirology. 2017; 14:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maskell D.P., Renault L., Serrao E., Lesbats P., Matadeen R., Hare S., Lindemann D., Engelman A.N., Costa A., Cherepanov P.. Structural basis for retroviral integration into nucleosomes. Nature. 215AD; 523:366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lodi P.J., Ernst J.A., Kuszewski J., Hickman A.B., Engelman A., Craigie R., Clore G.M., Gronenborn A.M.. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry (Mosc.). 1995; 34:9826–9833. [DOI] [PubMed] [Google Scholar]
- 11. Lu R., Wang G.G.. Tudor: a versatile family of histone methylation ‘readers’. Trends Biochem. Sci. 2013; 38:546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adams-Cioaba M.A., Guo Y., Bian C., Amaya M.F., Lam R., Wasney G.A., Vedadi M., Xu C., Min J.. Structural studies of the tandem Tudor domains of fragile X mental retardation related proteins FXR1 and FXR2. PLoS One. 2010; 5:e13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mauro E., Lesbats P., Lapaillerie D., Chaignepain S., Maillot B., Oladosu O., Robert X., Fiorini F., Kieffer B., Bouaziz S.et al.. Human H4 tail stimulates HIV-1 integration through binding to the carboxy-terminal domain of integrase. Nucleic Acids Res. 2019; 47:3607–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Demeulemeester J., Vets S., Schrijvers R., Madlala P., De Maeyer M., De Rijck J., Ndung’u T., Debyser Z., Gijsbers R.. HIV-1 integrase variants retarget viral integration and are associated with disease progression in a chronic infection cohort. Cell Host Microbe. 2014; 16:651–662. [DOI] [PubMed] [Google Scholar]
- 15. Mannioui A., Schiffer C., Felix N., Nelson E., Brussel A., Sonigo P., Gluckman J.C., Canque B.. Cell cycle regulation of human immunodeficiency virus type 1 integration in T cells: antagonistic effects of nuclear envelope breakdown and chromatin condensation. Virology. 2004; 329:77–88. [DOI] [PubMed] [Google Scholar]
- 16. Benleulmi M.S., Matysiak J., Henriquez D.R., Vaillant C., Lesbats P., Calmels C., Naughtin M., Leon O., Skalka A.M., Ruff M.et al.. Intasome architecture and chromatin density modulate retroviral integration into nucleosome. Retrovirology. 2015; 12:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meehan A.M., Poeschla E.M.. Chromatin tethering and retroviral integration: recent discoveries and parallels with DNA viruses. Biochim. Biophys. Acta. 2010; 1799:182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Llano M., Vanegas M., Hutchins N., Thompson D., Delgado S., Poeschla E.M.. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006; 360:760–773. [DOI] [PubMed] [Google Scholar]
- 19. Botbol Y., Raghavendra N.K., Rahman S., Engelman A., Lavigne M.. Chromatinized templates reveal the requirement for the LEDGF/p75 PWWP domain during HIV-1 integration in vitro. Nucleic Acids Res. 2008; 36:1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eidahl J.O., Crowe B.L., North J.A., McKee C.J., Shkriabai N., Feng L., Plumb M., Graham R.L., Gorelick R.J., Hess S.et al.. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013; 41:3924–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ballandras-Colas A., Maskell D.P., Serrao E., Locke J., Swuec P., Jónsson S.R., Kotecha A., Cook N.J., Pye V.E., Taylor I.A.et al.. A supramolecular assembly mediates lentiviral DNA integration. Science. 2017; 355:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maskell D.P., Renault L., Serrao E., Lesbats P., Matadeen R., Hare S., Lindemann D., Engelman A.N., Costa A., Cherepanov P.. Structural basis for retroviral integration into nucleosomes. Nature. 2015; 523:366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cosnefroy O., Tocco A., Lesbats P., Thierry S., Calmels C., Wiktorowicz T., Reigadas S., Kwon Y., De Cian A., Desfarges S.et al.. Stimulation of the human RAD51 nucleofilament restricts HIV-1 integration in vitro and in infected cells. J. Virol. 2012; 86:513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sarver A.L., Erdman J., Starr T., Largaespada D.A., Silverstein K.A.T.. TAPDANCE: An automated tool to identify and annotate transposon insertion CISs and associations between CISs from next generation sequence data. BMC Bioinformatics. 2012; 13:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoffman M.M., Buske O.J., Wang J., Weng Z., Bilmes J.A., Noble W.S.. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012; 9:473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. John S., Workman J.L.. Bookmarking genes for activation in condensed mitotic chromosomes. BioEssays News Rev. Mol. Cell. Dev. Biol. 1998; 20:275–279. [DOI] [PubMed] [Google Scholar]
- 29. Terrenoire E., McRonald F., Halsall J.A., Page P., Illingworth R.S., Taylor A.M.R., Davison V., O’Neill L.P., Turner B.M. Immunostaining of modified histones defines high-level features of the human metaphase epigenome. Genome Biol. 2010; 11:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007; 35:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shun M.C., Botbol Y., Li X., Di Nunzio F., Daigle J.E., Yan N., Lieberman J., Lavigne M., Engelman A.. Identification and characterization of PWWP domain residues critical for LEDGF/p75 chromatin binding and human immunodeficiency virus type 1 infectivity. J. Virol. 2008; 82:11555–11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Serrao E., Krishnan L., Shun M.C., Li X., Cherepanov P., Engelman A., Maertens G.N.. Integrase residues that determine nucleotide preferences at sites of HIV-1 integration: implications for the mechanism of target DNA binding. Nucleic Acids Res. 2014; 42:5164–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Astiazaran P., Bueno M.T., Morales E., Kugelman J.R., Garcia-Rivera J.A., Llano M.. HIV-1 integrase modulates the interaction of the HIV-1 cellular cofactor LEDGF/p75 with chromatin. Retrovirology. 2011; 8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lesbats P., Lavigne M., Parissi V.. HIV-1 integration into chromatin: new insights and future perspective. Future Virol. 2011; 6:1035–1043. [Google Scholar]
- 35. Lesbats P., Serrao E., Maskell D.P., Pye V.E., O’Reilly N., Lindemann D., Engelman A.N., Cherepanov P. Structural basis for spumavirus GAG tethering to chromatin. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:5509–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kotlar R.M., Jones N.D., Senavirathne G., Gardner A.M., Messer R.K., Tan Y.Y., Rabe A.J., Fishel R., Yoder K.E.. Retroviral prototype foamy virus intasome binding to a nucleosome target does not determine integration efficiency. J. Biol. Chem. 2021; 296:100550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones N.D., Lopez M.A. Jr, Hanne J., Peake M.B., Lee J.-B., Fishel R., Yoder K.E. Retroviral intasomes search for a target DNA by 1D diffusion which rarely results in integration. Nat. Commun. 2016; 7:11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All additional data are available upon request.