Abstract
Large indels greatly impact the observable phenotypes in different organisms including plants and human. Hence, extracting large indels with high precision and sensitivity is important. Here, we developed IndelEnsembler to detect large indels in 1047 Arabidopsis whole-genome sequencing data. IndelEnsembler identified 34 093 deletions, 12 913 tandem duplications and 9773 insertions. Our large indel dataset was more comprehensive and accurate compared with the previous dataset of AthCNV (1). We captured nearly twice of the ground truth deletions and on average 27% more ground truth duplications compared with AthCNV, though our dataset has less number of large indels compared with AthCNV. Our large indels were positively correlated with transposon elements across the Arabidopsis genome. The non-homologous recombination events were the major formation mechanism of deletions in Arabidopsis genome. The Neighbor joining (NJ) tree constructed based on IndelEnsembler's deletions clearly divided the geographic subgroups of 1047 Arabidopsis. More importantly, our large indels represent a previously unassessed source of genetic variation. Approximately 49% of the deletions have low linkage disequilibrium (LD) with surrounding single nucleotide polymorphisms. Some of them could affect trait performance. For instance, using deletion-based genome-wide association study (DEL-GWAS), the accessions containing a 182-bp deletion in AT1G11520 had delayed flowering time and all accessions in north Sweden had the 182-bp deletion. We also found the accessions with 65-bp deletion in the first exon of AT4G00650 (FRI) flowered earlier than those without it. These two deletions cannot be detected in AthCNV and, interestingly, they do not co-occur in any Arabidopsis thaliana accession. By SNP-GWAS, surrounding SNPs of these two deletions do not correlate with flowering time. This example demonstrated that existing large indel datasets miss phenotypic variations and our large indel dataset filled in the gap.
INTRODUCTION
Genomic variations include single-nucleotide polymorphisms (SNPs), small indels, and structural variations (SVs). In plant, one important challenge is to identify genomic variations that affect observable phenotypes. Previous studies mostly focused on SNPs and small indels. Some interesting discoveries have been made. Fang et al. (2) reported that some loci associated with lint yield and fiber quality in cotton. SNPs in the promoter regions of FLOWERING LOCUS T and FLOWERING LOCUS C orthologs correspond to the different rapeseed ecotype groups (3).
However, recent studies revealed that single-nucleotide polymorphisms (SNPs) and small indels cannot completely explain the phenotypic differences (4,5). Abundant evidence from genetics and molecular biology has clearly demonstrated large indels (insertions or deletions of size > 50 bp), in particular, play a major role to explain the phenotypic variances that affecting a series of important agronomic and quality traits in crops. For example, Wang et al. (6) reported that an insertion in the promoter region of the ZmVPP1 gene, encoding a vacuolar H+ pyrophosphatase, in maize associated with enhancement of photosynthetic efficiency and root development. Wang et al. (7) reported that a tandem duplication at GL7 (Grain Length on Chromosome 7) locus in rice, which increased grain length and improved grain appearance. Elevated copy numbers of Ppd-B1 (Photoperiod-B1) gene in wheat were found to be related to early flowering and day neutral (8). A 30.2-kb duplicated segment at the Female locus led to the development of gynoecy in cucumber (9).
Previous studies also demonstrated that large indels associate with ecogeographical adaptations. For example, Maron et al. (10) reported that the tandem triplication of MATE1 (malate transporter) gene in maize associated with superior aluminum tolerance and these maize lines originate from regions of highly acidic soils. A copy number variation (CNV) in ZmWAK (wall-associated kinase) gene is conferred resistance to head smut in maize. The ZmWAK gene was absent in many modern maize lines but present in wild relatives (11). The insertion of a 1-kb sequence upstream of HvAACT1 (Al-activated citrate transporter) gene enables barley to adapt to acidic soils (12).
Although large indels are important, the number of discovered phenotypic large indels are far smaller than that of phenotypic SNPs. The main hurdle is that existing methods cannot accurately identify large indels from whole genome sequencing data. No single method can call large indels with high accuracy. Different methods have different strengths and weakness, depending on the applied strategies. To improve sensitivity and specificity, one approach is to combine the results from multiple methods (13). A number of previous studies used this approach (14–16). Through ensembling different methods, like DELLY (17), BreakDancer (18), Pindel (19), Manta (20) and Lumpy (21), these studies predicted variations with relatively high accuracy (22). Fuentes et al. (23) combined multiple approaches and developed a SV prediction pipeline in 3000 rice genomes. The sensitivity of their pipeline is 60% for deletions and is very low for tandem duplications. The specificity of their pipeline for deletions, tandem duplications were 86% and 60%, respectively. Brandler et al. (24) utilized three complementary algorithms to detect SVs in 235 autism spectrum disorder (ASD) genomes. They assessed the sensitivity of deletion by applying their methods to 27 individuals in the 1000G Project. The sensitivity for detecting deletion was 75%, 61% and 25% for lengths >1000 bp, 100–1000 bp and <100 bp, respectively.
Here, we present a sensitive and accurate large indel caller IndelEnsembler that enables us to call large indels appearing in >1% of Arabidopsis samples. We applied IndelEnsembler on 1047 Arabidopsis genomes and called large indels against the TAIR10 (Arabidopsis Col-0) reference genome. We showed that IndelEnsembler is highly accurate and it can discover novel phenotypic indels of size > 50 bp that cannot be found in previous studies.
MATERIALS AND METHODS
Overview of the discovery pipeline IndelEnsembler
We independently ran Lumpy (21), Manta (20), SurVIndel (25) and TranSurVeyor (26) on the short paired-end reads of each of the 1047 samples. Lumpy is a general probabilistic framework that calls variants by integrating multiple sequence alignments signals, including read-pair and split-read. Manta is a fast assembly-based caller that can accurately discover genomic variants. It calls a large fraction of variants to base-pair resolution. SurVIndel is a statistical method that focuses on detecting deletions and tandem duplications using discordant paired reads, clipped reads as well as insert size distribution. It is effective in calling deletions and tandem duplications that are in repetitive regions. TranSurVeyor proposed a realignment strategy to resolve the problem of transposition calling from high-throughput next-generation sequencing. TranSurVeyor shows at least 3.1-fold improvement of F1-score compared with existing database-free methods. We ran the above four tools using default parameters (see supplementary method for detail).
In this study, we focused to call three types of SVs: deletions (DELs), tandem duplications (DUPs) and insertions (INSs). Lumpy provided DELs, Manta provided all three types, SurVIndel provided DELs and DUPs while TranSurVeyor provided INSs (due to transposition). We ignored DUPs called by Lumpy because, when evaluated on our 7 benchmark datasets (as elaborated in Evaluation of large indel callsets), they had unusually low precision (average precision = 0.23, compared to 0.49 for Manta and 0.52 for SurVIndel). For each sample, we merged the large indels provided by the four callers, in order to generate a final single-sample large indel callset. The merging procedure was as follows. Initially, the final set is made of all the calls from SurVIndel. Next, we use our large indel comparison routine to identify large indels in Manta not in our current final set, and we add them. Finally, we repeat for Lumpy and TranSurVeyor. Given the final callset for each sample, we cluster large indels across samples as described in ‘Large indel clustering’. The output is a set of unique large indels across the 1047 samples, where each large indel is associated with a list of samples that display it.
Large indel clustering
We developed a flexible and efficient algorithm for clustering of large indels (i.e. DELs/DUPs/INSs) based on graph theory. The algorithm requires a compatibility criterion that defines whether any two large indels are compatible, i.e. they can be assigned to the same cluster. Our goal is to divide the indels into clusters so that every indel is compatible with every other indel in the same cluster, while at the same time minimizing the number of clusters we generated.
First, we define a simple compatibility criterion that determines whether two large indels can be clustered together. In order for two indels to be compatible, they must be of the same type (i.e. both are DELs, both are DUPs or both are INSs), they must alter the same chromosome and the sum of the distance between their left breakpoints and the distance between their right breakpoints must be at most 200 bp.
Next, we define the graph used by our clustering algorithm. We represent each indel as a vertex in a graph. Two vertices are connected by an edge if the corresponding indels are compatible. Our clustering algorithm wants to partition the vertices into a set of clusters such that every cluster is a clique (i.e. a set of pairwise connected vertices), and we want the number of clusters to be as small as possible. This problem is known in graph theory as minimum clique cover problem. Finding an optimal minimum clique cover is computationally unfeasible for large graphs, so we use a simple heuristic. The detailed algorithm is shown as a pseudocode in supplementary method. Let the neighborhood of a vertex v be the set of vertices connected to v. We sort the vertices of the graph by neighborhood size, in descending order. For each vertex v, we check its neighbors. For each neighbor that already belongs to a clique, we test whether v can be added to that clique. Finally, among all the cliques that v can join, we choose the largest. If v cannot join any clique, we create a new clique with v as its sole element.
Finally, for each clique, we output a consensus indel by choosing the median start coordinate and the median end coordinate among the indel belonging to the clique. We also output the list of the samples that display that indel.
DEL/DUP/INS comparison routine
In order to compare DELs/DUPs/INSs, we used a simple repeat-aware routine introduced in Rajaby and Sung (25). Given two DELs/DUPs/INSs, the routine reports whether they are equivalent. First of all, the two DELs/DUPs/INSs must alter the same chromosome and be of the same type, otherwise they are considered as different large indels.
For comparing DELs and DUPs, we use two different criteria: a strict criterion, and a relaxed criterion. Two DELs/DUPs are the same according to the strict criterion if (a) the sum of the distances between their left breakpoints and the distance between their right breakpoints is at most 200 bp and (b) the smallest of the two is covered for at least 90% by the largest one. If all conditions are satisfied, the two DELs/DUPs are deemed to be the equivalent. When the strict criterion fails and both DELs/DUPs are located in the same repetitive region (according to TRF (27) annotations of the reference genome), we apply the relaxed criterion. Two DELs/DUPs are the same according to the relaxed criterion if (a) the difference in length is at most 200 bp and (b) the two deleted or duplicated sequences have high similarity score, i.e. when aligned with an affine gap penalty scoring scheme (+2 match, −−2 mismatch, −4 gap open, −1 gap extend), the score is greater than or equal to the length of the smaller event. When comparing two INSs, we require that the distance between the two insertion sites is at most 200 bp. If both INSs report an inserted sequence, we also require that they have high similarity score.
Evaluation of large indel callsets
We evaluated the performance of IndelEnsembler using 7 publicly available Arabidopsis thaliana genomes (https://1001genomes.org/data/MPIPZ/MPIPZJiao2020/releases/current/strains/). For each sample, short reads and an assembled genome are provided. We produced a catalogue of indels using IndelEnsembler on short reads, and we used Assemblytics (28) to generate a ground truth catalogue to assess recall (Supplementary Table S1). Given the ground truth G and the set of called large indels C for the same sample, we compared each large indel in G with every large indel in C using our large indel comparison routine. If at least one match was found, we considered the large indel in G to be correctly called. Recall was the number of correctly called large indels divided by the size of G.
By aligning the assembled genomes to the reference genome using Minimap2 (29), a state-of-the-art tool for alignment of long sequences, we noticed that some of the calls were not reported by Assemblytics but were clearly supported by the aligned contigs. Therefore, we employed a different strategy for assessing precision. For a given sample, we evaluated the precision of the called DELs/DUPs/INSs as follows. We aligned its assembled genome to the reference using Minimap2. Calls that fall in regions of the reference not covered by any assembled contigs could not be validated and were discarded from the comparison. For each remaining called DEL, we extracted the portion of assembled contig aligned to putative deleted region: if the alignment displayed a deletion of size at least 50 bp and within 200 bp of the size of the called DEL, we considered the called DEL to be a true positive. Called INSs and DUPs were validated similarly: a DUP/INS must be displayed at most 200 bp away from the called DUP/INS site, respectively, and the size of the DUP/INS must be at least 50 bp and within 200 bp of the size of the called DUP/INS. For both DELs and INSs/DUPs, we also employed a repeat-aware relaxed criterion, identical to what we introduced in ‘large indel comparison routine’. Precision was computed as the number of true positives divided by the size of the called catalogue, after excluding DELs/DUPs/INSs that could not be validated. F1-score of precision and recall provides a weighted averaging of both precision and recall. It is defined as the harmonic mean between precision and recall, i.e. F1-score = 2 × (recall × precision)/(recall + precision).
Finally, we estimated the recall of the full catalogue of large indels called on the 1047 Arabidopsis samples, both for our method and for AthCNV. We used the same Assemblytics ground truth sets for the seven samples that we used to assess single-sample recall, and we followed the same procedure. Interestingly, AthCNV does not report whether a CNV deletes or inserts copies, therefore we had to consider each CNV as both deletion and tandem duplication, potentially overestimating its recall.
SNPs/large indels annotation
SNP data (1001 genomes_snp-short-indel_only_ACGTN.vcf.gz) were downloaded from the 1001 Genomes Project server. From 1047 accessions for which we used to call large indels, we obtained ∼12.9 million SNPs. By VCFtools (0.1.17) (30), we filtered SNPs with parameters ‘—maf 0.05 –max-missing 0.9’. Then, an in-house Python script further filtered the SNPs based on hybrid rate lower than 0.05. As a result, 845,188 SNPs were kept. The annotations and effects of SNPs and large indels on gene function were predicted using SnpEff software (31). The centromere positions were defined as described previously (32).
LD analysis of DEL
For each common DEL (MAF > 5%) in the population, the nearest flanking 150 SNPs upstream and 150 SNPs downstream were selected for LD (r2 values) calculated. For every SNP-SNP pair, r2 values were computed. Then, the pairs are ranked by decreasing r2 values and a median SNP-SNP rank was calculated. We also calculated the r2 values for the 300 SNP–DEL pairs. The relative LD metric of DEL to SNP is denoted as the number of times the r2 values of the SNP-DEL pairs was greater than the SNP-SNP median rank. DEL variants with the relative LD metric less than 100 were classified as low-LD with flanking SNPs. DEL variants with the relative LD metric between 100 and 200 were classified as mid-LD with flanking SNPs, while DEL variants with the relative LD metric greater than 200 were classified as high-LD with flanking SNPs.
Mechanism of DEL formation and Deleted genes enrichment analysis
This study used BreakSeq deletion formation mechanism analysis pipeline (33) to infer the deletion formation mechanisms. Note that IndelEnsembler called the nucleotide resolution breakpoints of all deletions. DEL-genes that were covered by DELs for ≥90% of their length were used to perform functional enrichment analysis. Enrichment analysis was performed with Panther Tools (Panther database v.16.0; Mi et al. (34)). The classification of the gene duplication types (tandem versus segmental) were conducted based on information retrieved from the Plaza v.4.0 database (35).
GWAS using the SNP and DEL datasets
SNP/DEL-GWAS was performed for flowering time under 10°C and 16°C using a mixed linear model (MLM) in EMMAX (36), using a kinship matrix. The kinship matrix was computed by the EMMAX ‘-kin’ command with default parameters. We plotted Manhattan and QQ plots using the R package ‘CMplot’. Circos v0.69 (37) was used to plotted large indels and the distribution of deletion formation mechanisms in the genome.
Neighbor-joining cluster analysis
TagSNPs were selected using PLINK v.1.90 (38) with parameter ‘–blocks’ to construct Neighbor joining (NJ) tree of SNPs. We take individual DEL calls as alleles of genetic markers to construct NJ tree of DELs. The NJ tree was constructed using TreeBeST v1.9.2 (39) software with 1000 replicates of bootstrap. An online tool Interactive tree of life (iTOL) v3 (40) was used to display the NJ tree. Principal component analysis of all tagSNPs was performed using Genome-wide Complex Trait Analysis (GCTA) v1.91.7 (41) software with default parameters.
RESULTS
Discovery pipeline IndelEnsembler
IndelEnsembler is an ensemble method for identifying deletions (DELs), tandem duplications (DUPs) and insertions (INSs) (either novel or due to transposition) from next generation sequencing data. It merges calls from different callers: Lumpy (21), Manta (20), SurVIndel (25) and TranSurVeyor (26). These callers were chosen because they use different approaches that complement each other: Lumpy relies on discordant pairs and split reads, Manta uses an assembly-based approach, SurVIndel offers better recall in repetitive regions by using statistical methods to detect the large indels, and TranSurVeyor specialises in insertions due to transposition. More details are reported in Materials and Methods.
To illustrate the goodness of IndelEnsembler, we applied IndelEnsembler to call large indels from the genome resequencing dataset of 1047 Arabidopsis thaliana accessions (NCBI SRA with id SRP056687), which is used in the study of (42). The paired-end reads in this dataset were mapped to the reference genome Tair10 (Arabidopsis Col-0) using BWA-MEM 0.7.10. Indels were called independently on each sample using IndelEnsembler. Similar or identical calls were clustered across samples using a novel in-house algorithm (see supplementary methods), retaining calls detected in at least 1% samples. Finally, we remove calls longer than 500 kb as well.
The final callset consists of 34 093 DELs, 12 913 DUPs and 9773 INSs, with size ranges of 50–494 176 bp (median 309 bp) for DELs, 50–494 500 bp (median 182 bp) for DUPs and 50–1762 bp (median 311 bp) for INSs (Figure 1A and Supplementary Table S5-7). We observe that duplication and insertion events are rarer than deletion events. There are several reasons for this imbalance. First, insertion and deletion are a relativity concept. When we compare one thousand Arabidopsis genomes to the reference genome Tair10 (which is Col-0), deletions (insertions, resp.) in Col-0 are insertions (deletions, resp.) in other Arabidopsis genomes. Because the Arabidopsis genomes are affected by the expansion of transposable elements (TEs), all TEs insert in other Arabidopsis thaliana genomes will be predicted as deletions in Col-0 (56). Second, insertion and duplication in Arabidopsis genomes are much harder to detect using short next generation sequencing (NGS) reads compared to the deletion events (23). Therefore, a large proportion of nondeletion events may go undetected.
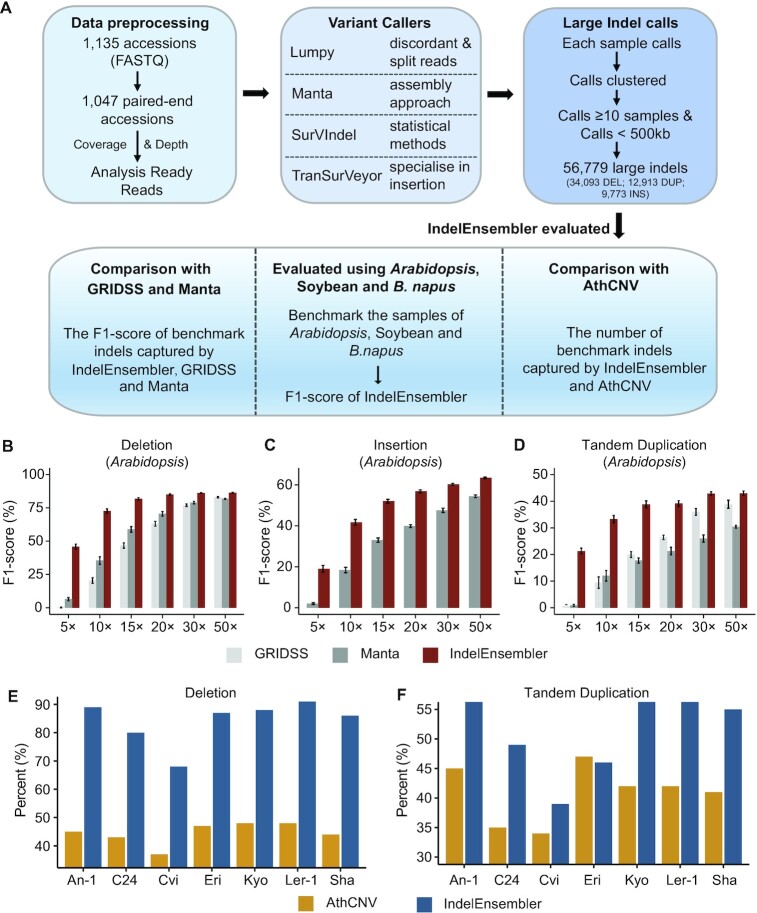
Figure 1.
Summary and Evaluation the performance of IndelEnsembler. (A) Large indel discovery pipeline IndelEnsembler in 1047 Arabidopsis. The software comparison of GRIDSS, Manta and IndelEnsembler in detecting deletions (B), insertions (C) and duplications (D) in Arabidopsis. (E) The percent of ground truth deletions captured by AthCNV and IndelEnsembler. (F) The percent of ground truth duplications captured by AthCNV and IndelEnsembler.
Evaluation of IndelEnsembler
We evaluated the performance of IndelEnsembler using seven Arabidopsis thaliana samples, for which short paired-end reads and assembled genomes are provided in (43). IndelEnsembler was run using the short reads of each sample independently. For assessing recall, a benchmark catalogue of large indels (Supplementary Table S1) is created by aligning Tair10 reference with the assembled genome of each sample using Assemblytics (28); then, the detected indels are compared with the ground truth to compute the recall. For precision, since the assemblies are incomplete, precision was only assessed using the detected indels that fall within an assembled contig. More details are given in Methods.
Supplementary Figure S1A–C shows the recall and precision of IndelEnsembler for DEL, INS and DUP, respectively, for the seven samples. As expected, DELs are the easiest class of variations to detect, and the mean recall of IndelEnsembler across the seven samples is 0.93, while the mean precision 0.8. INSs were more challenging to predict, with the mean recall 0.55 and the mean precision 0.79. DUPs are by far the most difficult class of indels to detect (26), and the mean recall is 0.4 while the mean precision 0.47. It must be noted that tandem repeats, where DUPs primarily occur, are notoriously difficult to capture in assembles and therefore the statistics for DUPs may be underestimated, especially precision. This is also supported by the fact that Assemblytics reports an unusually low number of DUPs (Supplementary Table S2). We examined how the sequencing depth affects the performance of IndelEnsembler in terms of F1-score. We generated datasets with different sequencing depths (5×, 10×, 15×, 20×, 30× and 50×) in Arabidopsis, Soybean and B. napus (see Supplementary Methods) for performance evaluation. For the performance comparison, we also evaluated the performance of GRIDSS (44) and Manta since GRIDSS and Manta are the current best methods for calling indels (45). Figure 1B–D shows the F1-score of GRIDSS, Manta and IndelEnsembler in detecting deletions, insertions and tandem duplications in Arabidopsis. As expected, the F1-score of IndelEnsembler increases as the sequencing depth increases. In particular, when the sequencing depth is low, IndelEnsembler performs much better than GRIDSS and Manta. We also observed that the improvement of F1-score of IndelEnsembler reach the plateau when the sequencing depth is above 15×. Same tendency was observed in Soybean and B. napus (Supplementary Figure S1D–I). These results suggested that IndelEnsembler performs better than or on par with GRIDSS and Manta.
AthCNV (1) is the previously published catalogue of large indels in the 1047 A. thaliana. It was also compiled using an ensemble of SV callers, although the specific callers used were different from the ones used in IndelEnsembler. The complete catalogue of accepted large indels in AthCNV consists of 89 140 entries. However, AthCNV does not report the list of large indels for the seven benchmark samples, so a direct comparison is not possible. We compare AthCNV and IndelEnsembler indirectly. We expect that most of the ground truth large indels appearing in the seven benchmark samples will also appear in the 1047 Arabidopsis thaliana. Therefore, we estimated the completeness of IndelEnsembler and AthCNV by checking the number of ground truth large indels that are captured by each method in the 1047 Arabidopsis thaliana. Note that AthCNV does not distinguish whether the events are DELs or DUPs, therefore we matched each AthCNV event with both benchmark DELs and DUPs, which could lead to an overestimation of AthCNV completeness. Furthermore, AthCNV does not report clear INSs, so we could not compare them. Despite IndelEnsembler reporting less large indels (56 779), we consistently capture nearly twice of the ground truth DELs (on average 84% for IndelEnsembler and 44% for AthCNV). We also capture on average 27% more DUPs (Figure 1E, F). This suggests that the callset of IndelEnsembler is considerably more complete.
Zmienko et al. (1) partitioned the 89 140 DELs/DUPs in AthCNV into 19 003 events of length >500 bp (called LargeCNVAthCNV) and 70 137 events of length between 50 and 500 bp (called SmallCNVAthCNV). Zmienko et al. (1) mentioned that LargeCNVAthCNV was more accurate and they focused their analysis to LargeCNVAthCNV only. To compare AthCNV and IndelEnsembler, we also partitioned the 56,779 large indels in IndelEnsembler into 19 296 events of length >500 bp (called LargeCNVIndelEnsembler) and 37 483 events of length between 50 and 500 bp (called SmallCNVIndelEnsembler). We observed that LargeCNVIndelEnsembler and LargeCNVAthCNV have good overlap (Supplementary Figure S2A and Supplementary Table S3). 17 134 events in LargeCNVAthCNV overlapped by at least 1 bp with that in LargeCNVIndelEnsembler. Therefore, IndelEnsembler and AthCNV have similar performance for calling large indels of size >500 bp.
Next, we compare SmallCNVIndelEnsembler and SmallCNVAthCNV. They do not have a good overlap (Supplementary Figure S2B). Only 34 538 events in SmallCNVAthCNV have at least 1 bp overlap with that in SmallCNVIndelEnsembler. 35 599 events appeared in SmallCNVAthCNV–only and 20 383 events appeared in SmallCNVIndelEnsembler–only. Although SmallCNVIndelEnsembler–only contained less events, it robustly captured nearly thrice of the ground truth DELs compared with SmallCNVAthCNV–only (Supplementary Table S4). SmallCNVIndelEnsembler–only also captured more DUPs compared with SmallCNVAthCNV–only (Supplementary Table S4). This indicates that SmallCNVAthCNV has a lot of false positives.
Statistics of large indels for 1047 Arabidopsis genomes
As the indels of size 50–500 bp predicted by AthCNV have high false positive rate, AthCNV (1) focused the subsequent analysis on large indels of size >500 bp only. However, large indels of length between 50 and 500 bp also greatly impact on the observable phenotypes (6,46–48). Since our large indel dataset is more complete compared with AthCNVs, this paper provides a comprehensive analyze for all large indels of size >50 bp. First, Figure 2A shows the frequency distributions for DELs, DUPs and INSs. Their frequency distributions follow the power law, consistent with the expectation from the neutral theory of evolution (49).
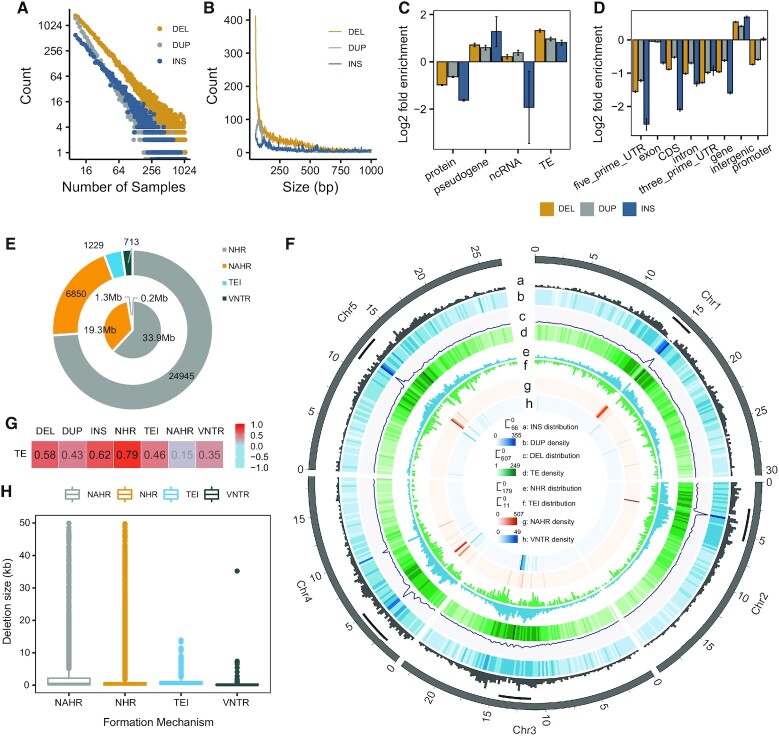
Figure 2.
Large indels distribution and spatial distribution of deletions based on the different formation mechanisms. (A) Frequency spectrum of deletions (yellow) duplications (grey) and insertions (blue) amongst 1047 Arabidopsis thaliana accessions. (B) Size distribution of large indels in our discovery set. (C) Enrichment/depletion of large indels in various gene types. (D) Enrichment/depletion of large indels in various genomic regions. (E) Distribution of different DEL formation mechanisms. Outer circle represents number of DELs per mechanism. Inner circle represents cumulative genomic size of these events. (F) Tracks (outer to inner circles) indicate the following: a–d, insertions, duplications, deletions and transposable elements (TEs) per 200 kb (red color indicates more); e–h, deletions per 200 kb per mechanism (NHR, TEI, NAHR, VNTR), range max: 179, 11, 507, 49. (G) Pearson's correlation of deletion, duplication, insertion and four mechanism with TEs. Pearson's correlation coefficients (r) of DEL, DUP, INS, NHR, TEI, NAHR and VNTR with TE were 0.58, 0.43, 0.79, 0.46, 0.15, 0.35, respectively; P< 2.2 × 10–16, P< 2.2 × 10–16, P< 2.2 × 10–16, P< 2.2 × 10–16, P= 6.6 × 10–16, P= 1.6 × 10–5, P= 5.7 × 10–14, respectively. (H) Sizes of deletions formed by different mechanisms.
Figure 2B shows the size distribution for each type of large indels between 50 and 1000 bp. Unlike DEL and INS, DUPs show a peak around 107 bp. We observed that the DUPs in the peak are enriched with DNA/MuDR and LTR/Gypsy TEs but severely depleted in RC/Helitron TEs (Supplementary Figure S2C). Supplementary Table S8 presents statistics of DELs, DUPs and INSs on different chromosomes. The result indicated that the number of DELs (r = 0.91, P = 3.24 × 10–2, Pearson's correlation), DUPs (r = 0.89, P = 4.33 × 10–2, Pearson's correlation) and INSs (r = 0.97, P = 5.76 × 10–3, Pearson's correlation) were positively correlated with chromosome size. The TAIR10 release contains 27 416 protein coding genes, 924 pseudogenes, 3,903 transposable element genes and 1359 ncRNAs. Large indels occur more often in transposable element genes and pseudogenes (Figure 2C) and intergenic regions (Figure 2D); however, large indels depleted in protein genes (Figure 2C) and genic regions (Figure 2D).
Mechanisms of DEL formation and spatial distribution of large indels
We tried to infer the deletion formation mechanisms for the 34 093 DELs (see Methods) by the BreakSeq deletion formation mechanism analysis pipeline (33). There are four possible deletion formation mechanisms: (i) non-allelic homologous recombination (NAHR) (50); (ii) nonhomologous recombination (NHR) (51), involving nonhomologous end-joining (NHEJ), fork stalling and template switching (FoSTeS) or microhomology-mediated break-induced repair (MMBIR); (iii) mobile element insertion, involving retrotransposons or DNA transposons (TEI) and (iv) expansion or shrinkage of a variable numbers of tandem repeats (VNTR). We successfully inferred the formation mechanisms of 33 737 deletions (Figure 2E). NHR was found to be the major formation mechanism of DELs, determined either by deletion count (73.94%) or total deletion length (61.98%). In addition, we determined that NAHR accounted for 20.3% deletion events, the remaining 3.64% and 2.11% deletion events were attributed to TEI and VNTR, respectively (Figure 2E).
We next analyzed the spatial distribution of large indels and the four different classes of deletions formation mechanisms (Figure 2F). We observed a strong enrichment of these four types of deletions on centromeres. Their occurrences have high correlation with that of TEs (Pearson's correlation coefficient r = 0.58, 0.43, 0.62, 0.79, 0.46, 0.15, 0.35, respectively; P < 2.2 × 10–16, P < 2.2 × 10–16, P < 2.2 × 10–16, P < 2.2 × 10–16, P = 6.6 × 10–16, P = 1.6 × 10–5, P = 5.7 × 10–14, respectively), especially for the NHR formation mechanisms (Figure 2G). This observation suggested that TE enriched regions represent an important source of deletions. This result is also consistent with the previous study that the number of presence/absence variation (PAV) gene was positively correlated with transposon elements density in Arabidopsis thaliana (52). By relating the formation mechanisms to deletion sizes, we observed a broad size range in NAHR, NHR and TEI, whereas there was a relatively small range of deletion sizes in VNTR (Figure 2H). These results were consistent with the previous study in Oryza (53).
Genomic content in DEL/DUP/INS regions
We observed uneven genome coverage by large indels called by IndelEnsembler (Table 1). In particular, 80–96% of the chromosome centromeric regions were covered by DEL/DUP/INSs. Also, a very large number of genes (13 102) overlapped with large indels (Figure 3A and Supplementary Table S9). (As a comparison, though AthCNV called more large indels, only 7712 genes are covered by AthCNV-based large indels.) We also observed a large number of TEs overlapped with large indels. They constituted 76.95% of all TEs. We hereafter refer to genes and TEs covered by DEL/DUP by at least 1 bp as DEL/DUP-genes and DEL/DUP-TEs, respectively. INS inserted within 2kb upstream of genes and TEs as INS-genes and INS-TEs, respectively.
Table 1.
Summary of genome coverage by DEL/DUP/INSs
| Region type | No. of variants | Mean coverage (%) of the given region typea |
|---|---|---|
| Genome | 56 878 | 40.50 |
| Centromeres | 12 101 | 87.37 |
| Outside centromeres | 44 777 | 36.37 |
| Overlapping protein-coding genes | 15 240 | 26.77 |
| Overlapping pseudogenes | 2108 | 68.29 |
| Overlapping TEs | 27 155 | 78.57 |
aCalculated from the following formula: coverage in individual region of a given type = number of bases overlapped by any DEL/DUP/INS/number of all bases in this region × 100%; average value is reported in the table.
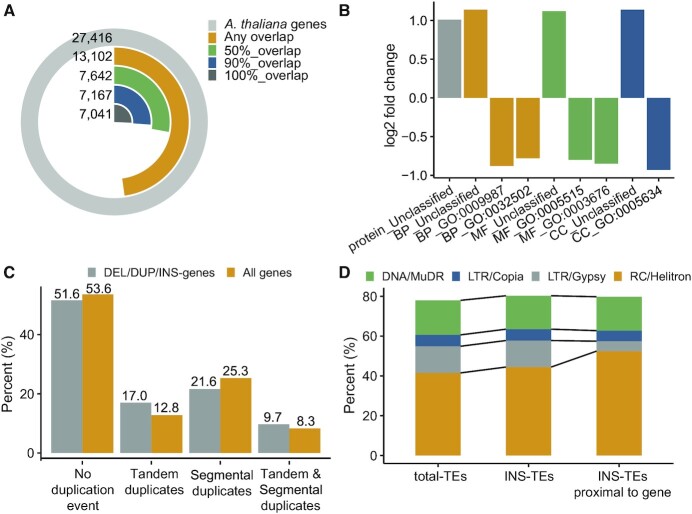
Figure 3.
Genomic Content in Regions Overlapped by DEL/DUP/INSs. (A) Fractions of annotated Arabidopsis genes with various degrees of overlap with DEL/DUP/INSs. (B) Over- and underrepresented protein types and GO terms among the DEL/DUP/INS-genes. All terms are either significantly enriched or depleted (binomial test with Bonferroni-corrected P-value < 0.05). (GO:0009987: cellular process; GO:0032502: developmental process; GO:0005515: protein binding; GO:0003676: nucleic acid binding; GO:0005634: nucleus). (C) Percentages of DEL/DUP/INS-genes and all genes that are overlapped with tandem duplicated and/or segmental duplicated regions. (D) Repeat families composition for all Arabidopsis TEs, all INS-TEs and all gene-proximal INS-TEs (located within ±2 kb distance).
We further studied 7167 DEL/DUP/INS-genes that were covered by DEL/DUP/INSs for ≥90% of their length (Figure 3A). These genes were significantly overrepresented in protein-coding genes of unclassified type (binomial test with Bonferroni-corrected P-value < 0.05; Figure 3B) and unclassified based on the Molecular Function, Biological Process, and Cellular Component Gene Ontology (GO) terms. Terms related to cellular process, developmental process and nucleus were significantly underrepresented in the DEL/DUP/INS-genes data set (Supplementary Table S10). This indicated that DEL/DUP/INS-genes are not core genes of Arabidopsis.
We also examined if these 7167 DEL/DUP/INS-genes are enriched in tandem duplication regions. We found that 17.04% of DEL/DUP/INS-genes are in tandem duplication regions (additionally, 9.74% underwent both segmental and tandem duplications; Figure 3C). On the other hand, only 12.87% of all Arabidopsis genes are in tandem duplicated regions. Consistent with the results in Zmienko et al. (1), these observations indicate that the regions of tandem duplications are sites that accumulate rearrangements and consequently, show high structural diversity.
Next, we studied the DEL/DUP/INS-TEs. We have 19 984 DEL-TEs, 13 459 DUP-TEs and 7595 INS-TEs. Supplementary Table S11 classified these DEL/DUP/INS-TEs by the TE superfamilies of Arabidopsis. ∼70% of DEL/DUP/INS-TEs belong to the four main TE superfamilies: DNA/MuDR, LTR/Copia, LTR/Gypsy and RC/Helitron TEs. Supplementary Tables S12–S14 summarized the proportions of TE superfamilies that are covered by DEL-TEs, DUP-TEs and INS-TEs. We observed that DEL/DUP-TEs were slightly depleted in RC/Helitron TEs and enriched in LTR/Gypsy TEs (Supplementary Figure S3A, B). However, for DEL/DUP-TEs that were proximal to genes, they were slightly enriched in RC/Helitron TEs but severely depleted in LTR/Gypsy TEs. They were also moderately enriched in DNA/MuDR elements. These results were consistent with that in Zmienko et al. (1). INS-TEs showed a different distribution. INS-TEs were slightly enriched in RC/Helitron TEs. For INS-TEs that are proximal to genes, they are even more significantly enriched in RC/Helitron TEs while severely depleted in LTR/Gypsy TEs (Figure 3D and Supplementary Table S13). Previous studies showed that Helitron TEs tends to be inserted into AT-rich regions while the promoter regions were usually enriched with ATs. Therefore, Helitron may affect gene expression through insertion into the promoter region (54–57). INS-TEs were slightly enriched in RC/Helitron TEs, probably because 64.45% INS-TEs were proximal to genes, which was higher than 55.49% for DEL-TEs and 52.52% for DUP-TEs.
Interplay between the DEL/DUP /INS-related genes and TEs
To investigate the relationship between genes and TEs, we compared the genomic distributions of DEL-genes, DEL-TEs, NONDEL-genes and NONDEL-TEs (NONDEL-genes and NONDEL-TEs are genes and TEs, respectively, that do not overlap with any DEL). Both DEL-genes and DEL-TEs were located closer to the chromosome centromeres than their NONDEL-counterparts and this tendency was much stronger for TEs than for genes (Supplementary Figure S4A). We also repeated the same analysis for DUP and INS. Same tendency was observed in DUPs (Supplementary Figure S4B) and INSs (Supplementary Figure S4C). This result is consistent with the finding in Zmienko et al. (1).
We analyzed the relative positions of DEL/DUP/INS-TEs to their nearby genes. In Fig. 5E of Zmienko et al. (1), it is observed that DEL/DUP/INS-TEs are mostly overlap with the genes or on the upstream of their nearby genes. Unlike Zmienko et al. (1), we showed that significantly more TEs called by IndelEnsembler are inserted in the upstream flanking regions of their nearby genes, but not significantly overlap with genes (Figure 4A–C). This result tells the importance of predicting the base-level breakpoints of the indels, which improves the resolution of the figures. Figure 4A–C also indicated that the gene–TE pairs with the same variation statuses were located closer to each other than pairs with the opposite statuses.
Figure 4.
Links between genes and TEs variation and localization. (A) Distances of proximal TEs around DEL-genes. (B) Distances of proximal TEs around DUP-genes. (C) Distances of proximal TEs around INS-genes. For each gene, a proximal TE was defined as each TE that overlaps with this gene (distance = 0) or locate within 2 kb upstream from the gene's 5' untranslated region (distance < 0) or locate within 2 kb downstream from 3' untranslated region (distance > 0). (D) Distance between gene and centromere for every gene-TE pair classified by variation status (Wilcoxon rank sum test with continuity correction for the difference between DEL-DEL and DEL-NONDEL groups, P-value < 2.2e–16). (E) Distance between gene and centromere for every gene-TE pairs classified by variation status (Wilcoxon rank sum test with continuity correction for the difference between INS-INS and INS-NONINS groups, P-value < 2.2e-16). (F) Distance between gene and centromere for every gene-TE pairs by variation status (Wilcoxon rank sum test with continuity correction for the difference between DUP-DUP and DUP-NONDUP groups, P-value > 0.05). Boxplots in (D), (E) and (F) show median (inner line) and inner quartiles (box). Whiskers extend to the highest and lowest values no greater than 1.5 times the inner quartile range.
We also analyzed the locations of gene-TE pairs. Unlike Zmienko et al. (1), the localization of DEL gene-DEL TE pairs and DEL gene-NONDEL TE pairs were biased to centromeres (Figure 4D), while the localization of other statuses of gene-TE pairs were biased toward non-centromeres (Wilcoxon rank sum test with continuity correction for the difference between DEL gene-DEL TE pairs and NONDEL gene-DEL/NONDEL TE pairs, P-value < 2.2e–16; Wilcoxon rank sum test with continuity correction for the difference between DEL gene-NONDEL TE pairs and NONDEL gene-DEL/NONDEL TE pairs, P-value < 2.2e–16). Same tendency was observed in DUPs (Figure 4E) and INSs (Figure 4F). Our observations confirmed the presence of selective constraints reciprocally imposed on genes and TEs, which is an important factor contributing to their present variation and genomic distribution patterns.
A genome-wide association study (GWAS) of flowering time
Although large indels have been recognized as the causative mutations for many traits (23), previous studies mainly link phenotypic diversity to SNPs only. If large indels have a strong linkage disequilibrium (LD) with the adjacent SNPs, the effect of large indels have been assessed by those studies; otherwise, the effect of large indels is unassessed. To determine whether large indels represent a previously unassessed source of genetic variation, we checked how frequently the common deletions (minor allele frequency (MAF) > 5%) were linked to adjacent SNPs (58,59). Surprisingly, 48.91% of the common deletions had low LD with nearby SNPs in our deletion sets, suggesting that they represent a source of genetic diversity not assessed by SNPs (see methods and Figure 5A, B and Supplementary Figure S5 and Supplementary Table S15). We found a positive correlation between MAF and LD states of deletions. Deletions with high MAF are more often classified as high linkage disequilibrium (Figure 5B), indicating that some deletions were under adaptive selection.
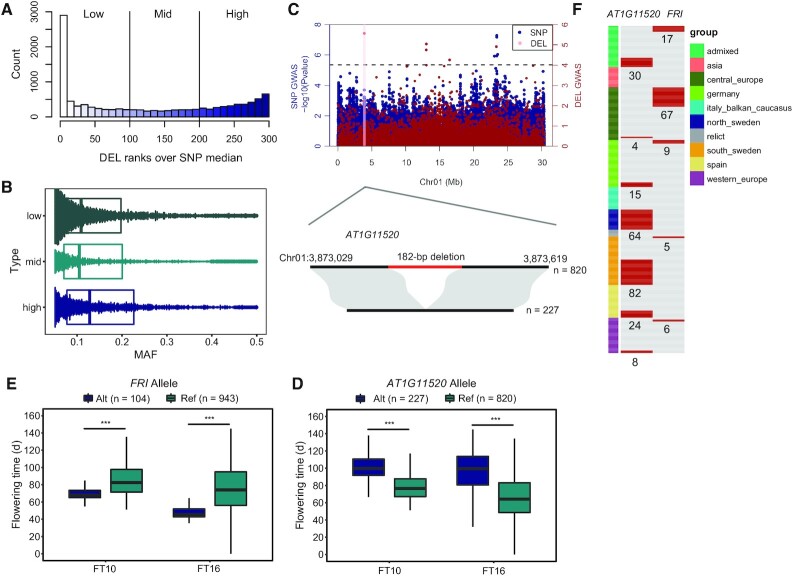
Figure 5.
An overview of significant deletions. (A) Histogram of the relative LD metrices for common DELs. (B) Boxplots showing distribution of minor allele frequencies for each LD category. (C) Top, Manhattan plot of SNPs (with ∼12.9 million SNPs obtained from 1001 Genomes) and DELs genome-wide association studies for flowering time under 16°C. The red line represents the candidate gene AT1G11520 on Chromosome 1. Bottom, A 182-bp deletion that present in 227 Arabidopsis thaliana accessions and not present in 820 Arabidopsis thaliana accessions. (D) The boxplots that show the flowering time of accessions with different AT1G11520 alleles. (E) The boxplots that show the flowering time of accessions with different AT4G00650 (FRI) alleles (*** P< 0.001, P values were determined using two-tailed Student's t-tests). (F) The distribution of individuals with deletions in AT1G11520 and AT4G00650 (FRI). Different geographic groups are represented by different color. The numbers below the red bars indicate the number of accessions with the corresponding deletions.
To check if some of these deletions are phenotypic, we used our deletions (and SNPs obtained from The 1001 Genomes Consortium (42)) to perform a genome-wide association study for flowering time under 10°C and 16°C, respectively. We indeed found two significant loci for flowering time under 16°C on chromosome 1 and chromosome 4 that could not be represented by local SNPs (Figure 5C and Supplementary Figure S6 and Supplementary Figure S7). The locus on Chr1 is a 182 bp deletion located in the exon of AT1G11520, which encodes spliceosome associated protein-like protein. Meaningfully, the 227 Arabidopsis thaliana individuals containing the 182 bp deletion had delayed flowering time than those not containing the deletion (Figure 5D). The locus on Chr4 was a 377 bp deletion and resulted in a 65 bp deletion in the first exon of AT4G00650 (FRI), which encodes FRIGIDA-like protein that was a major determinant of natural variation in Arabidopsis flowering time and activates expression of the floral repressor FLOWERING LOCUS C (FLC, AT5G10140). Many early flowering accessions carry this loss-of-function FRI alleles (60). The 104 accessions with this deletion flowered earlier than those without it, consistent with the observation above (Figure 5E and Supplementary Figure S7). The deletions on AT1G11520 and AT4G00650 had mid and low LD with nearby SNPs, respectively. This indicated that these two deletions were unassessed genetic diversity, which are important genetic variants for studying Arabidopsis thaliana flowering time. In addition, those two deletions do not appear in the previous results of AthCNV dataset (1). This suggested that there are still some genomic variations that were undetected and our large indel dataset can complement the existing catalog of know phenotype-related genomic variations present in Arabidopsis genome.
Interestingly, the two deletions on AT1G11520 and AT4G00650 cannot co-occur in the same Arabidopsis thaliana individual (Figure 5F). The distribution of accessions with deletion on AT1G11520 are mainly in north Sweden (64 accessions) and south Sweden (82 accessions) (Supplementary Table S16). Significantly, all accessions in north Sweden have the 182 bp deletion on AT1G11520. This may be due to the accessions in north Sweden located in the area of low temperature, and the delay of flowering time of accessions with deletion on AT1G11520 can improve survival. The distribution of accessions with deletion on AT4G00650 are mainly in central Europe (68 accessions) (Supplementary Table S16).
We also detected three deletions that significant associated with flowering time under 10°C or 16°C (Supplementary Table S17). A 321 bp deletion on Chr1 which resulted in a 135 bp delete on AT1G35360 (Supplementary Figure S8A), a transposable element gene, belong to copia-like retrotransposon family, the 33 individuals with this deletion flowered later than those without it (Supplementary Figure S8B). Another locus occurring on Chr1 was a 3,572 bp deletion that resulted in a 1,086 bp delete on AT1G63070 (Supplementary Figure S8C). AT1G63070 is a pentatricopeptide (PPR) repeat-containing protein. The 26 accessions with this deletion flowered later than those without it (Supplementary Figure S8D). We also found a 3105 bp deletion that resulted in the deletion of three genes (Supplementary Figure S8E). AT3G06990 and AT3G07000 are Cysteine/Histidine-rich C1 domain family protein. The 21 accessions with this deletion flowered later than those without it (Supplementary Figure S8F).
We also used our insertions to perform a genome-wide association study for flowering time under 10°C and 16°C, respectively (Supplementary Figure S9). We detected some insertions that significant associate with flowering time under 10°C or 16°C (Supplementary Table S18). There are 12 accessions with a 433 bp insertion located on the second exon of AT1G26570 (Supplementary Figure S10A). These 12 accessions flowered later than the other accessions (Supplementary Figure S10B). We also found a 1030 bp insertion located on the intron of AT4G37235 (Supplementary Figure S10C). The 22 accessions with this insertion flowered later than those without it (Supplementary Figure S10D). Lastly, we found a 302 bp insertion located on the seventh exon of AT5G40230 (Supplementary Figure S10E). The 18 accessions with this insertion flowered later than those without it (Supplementary Figure S10F). The above examples showed that our large indel callset is useful. They provided us an opportunity to investigate the causative agent of the observed phenotypic variation in Arabidopsis thaliana.
Neighbor-joining cluster analysis of 1047 Arabidopsis thaliana
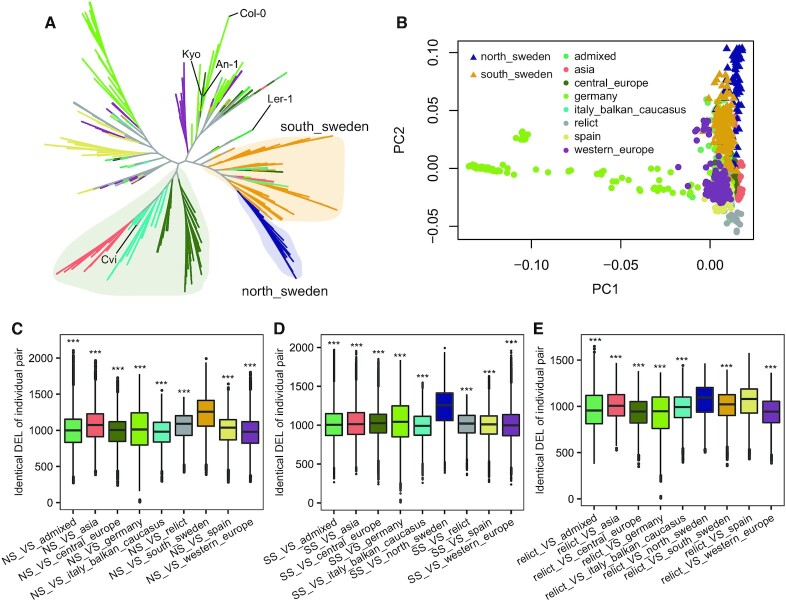
Although ongoing work demonstrates the importance of large indels as a source of genetic variations during evolution (61), little evolution works have been performed using large indels. To explore the evolution of 1047 A. thaliana, we took common individual DEL calls (MAF ≥ 0.05) as genetic markers to perform neighbor-joining cluster analysis based on Nei's genetic distances (Figure 6A) (62). Similarly, we also took genome-wide SNP calls to infer a NJ tree for the same set of A. thaliana (Supplementary Figure S11A). The NJ tree inferred from both DELs and SNPs are consistent. The 1047 A. thaliana accessions were classified into different subclades that corresponded to their geographic groups (42). Interestingly, the common ancestor of the accessions of north Sweden was a descendant of the common ancestor of south Sweden in both of DEL-based and SNP-based NJ trees, suggesting that the north Sweden might derive from south Sweden. We also check the average number of shared DELs in accession pairs between south Sweden and north Sweden. This number is significantly larger than the average number of shared DELs in both (i) accession pairs between north Sweden and other groups (Figure 6C) and (ii) accession pairs between south Sweden and other groups (Figure 6D). This is another evident that accessions of north Sweden are derived from south Sweden.
Figure 6.
Neighbor-joining cluster analysis of deletions in 1047 Arabidopsis thaliana. (A) The NJ tree is constructed based on deletions of 1047 Arabidopsis thaliana accessions. Different colors on the NJ tree correspond different groups. The group of north Sweden, south Sweden, Asia, Italy-Balkan-Caucasus and central Europe were shaded. Reference genome Col-0 and 4 accessions used for evaluating the performance of IndelEnsembler were marked. (B) The PCA plot of deletions of 1047 Arabidopsis thaliana. (C) Boxplots that show the number of shared DELs between accessions of north Sweden and accessions of all other groups (differences between north Sweden versus south Sweden relative to north Sweden with other groups were statistically analyzed based on two-tailed Student's t-tests, *** P-value < 0.001). (D) Boxplots that show the number of shared DELs between accessions of south Sweden and accessions of all other groups. (Differences between south Sweden versus north Sweden relative to south Sweden with other groups were statistically analyzed based on two-tailed Student's t-tests, *** P-value < 0.001). (E) Boxplots that show the number of shared DELs between accessions of relict and accessions of all other groups. (Differences between relict versus north Sweden relative to relict with other groups were statistically analyzed based on two-tailed Student's t-tests; Differences between relict versus spain relative to relict with other groups were statistically analyzed based on two-tailed Student's t-tests, *** P-value < 0.001).
Similarly, the DEL-based principal component analysis (PCA) of 1047 Arabidopsis thaliana accessions also showed that the accessions of north Sweden were evolutionary closer to that of south Sweden, supporting the results of NJ tree (Figure 6B). Our DEL-based PCA were clear than CNV-based PCA in Zmienko et al. (1) to reflected the global distribution of the accessions from east (Asia) to west (Germany and western Europe) for PC1 and north (north Sweden) to south (Spain and relict) for PC2.
Significantly, our results were in agreement with the previous proposed two-wave expansion model of Arabidopsis thaliana across Eurasia, through the analysis of the extent of relict introgression in the non-relict genomes (63). The first wave started from the populations of different glacial refugia and expanded northwards at the end of last ice age. Subsequently, a second wave started from a population of central Europe, which expands to Italy, Balkan and Caucasus, and finally to Asia along the east-west axis. The second wave is supported by our DEL-based NJ tree (Figure 6A), where the accessions of Asia, Italy-Balkan-Caucasus and central Europe are mostly in the same branch of the tree. The three groups are separated into three parts and we can clearly see that central Europe derives Italy-Balkan-Caucasus, which derives Asia. Note that the SNP-based NJ tree (Supplementary Figure S11A) also show similar observation, but it is less clear. The footprint of the first wave after the two-wave expansions is that the relict accessions are mainly found in the south and north of species range (63). This footprint is supported by the fact that the average number of shared DELs in both (i) accession pairs between relict and north Sweden and (ii) accession pairs between relict and Spain are significantly higher than that between relict and other groups (Figure 6E). The above results indicated that our DELs not only have a significant influence on traits (58,61,64) but also have important roles in evolutionary analysis.
DISCUSSION
This study integrated different methods into one pipeline IndelEnsembler to call large indels. We generated a database of large indels from the sequencing data of 1047 Arabidopsis obtained from the 1001 Genomes. We detected ∼34 000 nucleotide resolution deletions, ∼13 000 tandem duplications and ∼10 000 insertions longer than 50 bp that are distributed across the Tair10 reference genome. Our large indel dataset enabled us to investigate their functional impact. Consistent with previous studies (23,65), we observed that large indels are not enriched in coding sequences. In total, 52.02% of the deletions and 48.23% of the tandem duplications were 1 bp overlapping with transposable elements, 56.02% of the insertions were overlapped with TEs or were located within 2-kb upstream regions of the TEs, which significantly contribute to genomic variation in plants (66). Large indels tend to enrich in centromeres and locate in regions with high density of transposable elements, consistent with the result reported in cucumber (1,9).
Our large indel datasets enabled us to examine the distribution of deletion formation mechanisms in Arabidopsis. We observed that NHR is the most frequent formation mechanism. The formation mechanisms of >3.59% deletions are related to TEI, indicated that the importance of transposable and retrotransposable elements in shaping the Arabidopsis genome. Four formation mechanisms of deletions were positively correlated with TEs, which suggested that TE enriched regions represent an important source of DELs (9).
Among the identified common deletions, 48.91% had low linkage disequilibrium with nearby SNPs, indicated that they represent genetic variants currently overlooked. We further demonstrated the utility of our deletion datasets for Neighbor-joining cluster analysis. Results indicated that the DEL-based NJ tree is similar to the SNP-based tree. Moreover, DEL-based tree separates different subspecies more clearly.
More importantly, our large indel dataset enriches phenotypic variants. We demonstrated the discovery of phenotypic large indels by performing genome-wide association analysis to screen candidate genes for flowering time in Arabidopsis. This technique yielded fewer significant associations than traditional SNP-GWAS but could focus on absent loci in the reference genome and obtain accurate positions. We directly detected two deletions that have significant association with flowering time. First, the deletion of the gene AT1G11520 increases the flowering time, and all north Sweden accessions contain this deletion. On the contrary, another deleted gene FRI decreases the flowering time. These two deletions cannot present in the same accession. We also identified a few other deletions and insertions that are associated with flowering time. These phenotypic loci were misses in AthCNV (1). Further experiments will be needed to establish a causal link between these indels and the associated phenotype. We foresee that GWAS analyses based on structural variations should become routinely performed on the population level projects, with the goal to identify much more gene-trait associations. Our large indel resource will be of important value for the Arabidopsis research and enables the discovery of phenotypic diversity related genes. Although short-reads originating from SVs are often aligned poorly (especially for the complex genome), population scale SV callings are mostly based on short-read sequencing (9,23,58,67,68) since short read sequencing is cheaper than third-generation sequencing (69). On the other hand, long read sequencing technologies (PacBio single molecule, real-time (SMRT) sequencing and Oxford Nanopore Technology (ONT) sequencing) enable the detection of SVs at single base pair resolution even in repetitive regions. As the cost of long read sequencing is decreasing, the development of long read analysis methods provides opportunities to improve SV calling in population level (70).
DATA AVAILABILITY
The information of the accessions used for large indels discovery are provided in Supplementary Table S19. The genomic coordinates of large indels identified are listed in Supplementary Table S5-7. We also provided a web interface at http://yanglab.hzau.edu.cn/IndelEnsembler to accessed the large indels. Users could fetch the information of large indels by entering the gene ID or gene region, the results included basic description of the large indels and their overlapped genes in Arabidopsis. IndelEnsembler has been released as free and open source software under a GNU General Public License (GPL version 3). The latest source code are available at https://github.com/kensung-lab/IndelEnsembler.
Supplementary Material
ACKNOWLEDGEMENTS
The computations in this paper were run on the bioinformatics computing platform of the National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University.
Author contributions: W.-K.S. and Q.-Y.Y. conceived and designed the study. D.-X.L., R.R., L.Z. and Z.-Q.Y. performed data analyses. R.R. and D.-X.L. contributed IndelEnsembler for large indels detection. L.-L.W. prepared the web interface for data visualization. D.-X.L. and R.R. wrote the manuscript. W.-K.S. and Q.-Y.Y. revised the manuscript. All the authors read and approved the manuscript.
Contributor Information
Dong-Xu Liu, National Key Laboratory of Crop Genetic Improvement, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Ramesh Rajaby, School of Computing, National University of Singapore, 117417 Singapore; NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, 117456, Singapore.
Lu-Lu Wei, National Key Laboratory of Crop Genetic Improvement, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Lei Zhang, Precision Medical Laboratory, Wuhan Children's Hospital (Wuhan Maternal and Child Healthcare Hospital), Tongji Medical College, Huazhong University of Science & Technology, Wuhan 430016, China.
Zhi-Quan Yang, National Key Laboratory of Crop Genetic Improvement, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Qing-Yong Yang, National Key Laboratory of Crop Genetic Improvement, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China; School of Computing, National University of Singapore, 117417 Singapore.
Wing-Kin Sung, National Key Laboratory of Crop Genetic Improvement, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China; School of Computing, National University of Singapore, 117417 Singapore; Genome Institute of Singapore, Genome, 138672 Singapore.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Plan of China [2017YFE0104800]; National Natural Science Foundation of China [32070559]; Fundamental Research Funds for the Central University HZAU [2662018PY068]. Funding for open access charge: Fundamental Research Funds for the Central University HZAU.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zmienko A., Marszalek-Zenczak M., Wojciechowski P., Samelak-Czajka A., Luczak M., Kozlowski P., Karlowski W.M., Figlerowicz M.. AthCNV: a map of DNA copy number variations in the Arabidopsis genome. Plant Cell. 2020; 32:1797–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fang L., Wang Q., Hu Y., Jia Y., Chen J., Liu B., Zhang Z., Guan X., Chen S., Zhou B.et al.. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 2017; 49:1089–1098. [DOI] [PubMed] [Google Scholar]
- 3. Wu D., Liang Z., Yan T., Xu Y., Xuan L., Tang J., Zhou G., Lohwasser U., Hua S., Wang H.et al.. Whole-genome resequencing of a worldwide collection of rapeseed accessions reveals the genetic basis of ecotype divergence. Molecular plant. 2019; 12:30–43. [DOI] [PubMed] [Google Scholar]
- 4. Springer N.M., Ying K., Fu Y., Ji T., Yeh C.T., Jia Y., Wu W., Richmond T., Kitzman J., Rosenbaum H.et al.. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 2009; 5:e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saxena R.K., Edwards D., Varshney R.K.. Structural variations in plant genomes. Brief. Funct. Genomics. 2014; 13:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang X., Wang H., Liu S., Ferjani A., Li J., Yan J., Yang X., Qin F.. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016; 48:1233–1241. [DOI] [PubMed] [Google Scholar]
- 7. Wang Y., Xiong G., Hu J., Jiang L., Yu H., Xu J., Fang Y., Zeng L., Xu E., Xu J.et al.. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015; 47:944–948. [DOI] [PubMed] [Google Scholar]
- 8. Díaz A., Zikhali M., Turner A., Isaac P., Laurie D. Copy Number Variation Affecting the Photoperiod-B1 and Vernalization-A1 Genes Is Associated with Altered Flowering Time in Wheat (Triticum aestivum). PLoS One. 2012; 7:e33234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Z., Mao L., Chen H., Bu F., Li G., Sun J., Li S., Sun H., Jiao C., Blakely R.et al.. Genome-wide mapping of structural variations reveals a copy number variant that determines reproductive morphology in cucumber. Plant Cell. 2015; 27:1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maron L.G., Guimaraes C.T., Kirst M., Albert P.S., Birchler J.A., Bradbury P.J., Buckler E.S., Coluccio A.E., Danilova T.V., Kudrna D.et al.. Aluminum tolerance in maize is associated with higher MATE1 gene copy number. PNAS. 2013; 110:5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zuo W., Chao Q., Zhang N., Ye J., Tan G., Li B., Xing Y., Zhang B., Liu H., Fengler K.A.et al.. A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 2015; 47:151–157. [DOI] [PubMed] [Google Scholar]
- 12. Fujii M., Yokosho K., Yamaji N., Saisho D., Yamane M., Takahashi H., Sato K., Nakazono M., Ma J.F.. Acquisition of aluminium tolerance by modification of a single gene in barley. Nat. Commun. 2012; 3:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alkan C., Coe B.P., Eichler E.E.. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011; 12:363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Genome of the Netherlands C. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014; 46:818–825. [DOI] [PubMed] [Google Scholar]
- 15. Nagasaki M., Yasuda J., Katsuoka F., Nariai N., Kojima K., Kawai Y., Yamaguchi-Kabata Y., Yokozawa J., Danjoh I., Saito S.et al.. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat. Commun. 2015; 6:8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H.et al.. An integrated map of structural variation in 2,504 human genomes. Nature. 2015; 526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rausch T., Zichner T., Schlattl A., Stutz A.M., Benes V., Korbel J.O.. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012; 28:i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan X., Abbott T.E., Larson D., Chen K.. BreakDancer: identification of genomic structural variation from paired-end read mapping. Curr. Protoc. Bioinformatics. 2014; 45:15.6.1–15.6.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z.. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009; 25:2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Kallberg M., Cox A.J., Kruglyak S., Saunders C.T.. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016; 32:1220–1222. [DOI] [PubMed] [Google Scholar]
- 21. Layer R.M., Chiang C., Quinlan A.R., Hall I.M.. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014; 15:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kosugi S., Momozawa Y., Liu X., Terao C., Kubo M., Kamatani Y.. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019; 20:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fuentes R.R., Chebotarov D., Duitama J., Smith S., De la Hoz J.F., Mohiyuddin M., Wing R.A., McNally K.L., Tatarinova T., Grigoriev A.et al.. Structural variants in 3000 rice genomes. Genome Res. 2019; 29:870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brandler W.M., Antaki D., Gujral M., Noor A., Rosanio G., Chapman T.R., Barrera D.J., Lin G.N., Malhotra D., Watts A.C.et al.. Frequency and complexity of de novo structural mutation in autism. Am. J. Hum. Genet. 2016; 98:667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rajaby R., Sung W.K.. SurVIndel: improving CNV calling from high-throughput sequencing data through statistical testing. Bioinformatics. 2019; 37:1497–1505. [DOI] [PubMed] [Google Scholar]
- 26. Rajaby R., Sung W.K.. TranSurVeyor: an improved database-free algorithm for finding non-reference transpositions in high-throughput sequencing data. Nucleic Acids Res. 2018; 46:e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999; 27 2:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nattestad M., Schatz M.C.. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics. 2016; 32:3021–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T.et al.. The variant call format and VCFtools. Bioinformatics. 2011; 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Underwood C.J., Choi K., Lambing C., Zhao X., Serra H., Borges F., Simorowski J., Ernst E., Jacob Y., Henderson I.R.et al.. Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 2018; 28:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lam H.Y., Mu X.J., Stutz A.M., Tanzer A., Cayting P.D., Snyder M., Kim P.M., Korbel J.O., Gerstein M.B.. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat. Biotechnol. 2010; 28:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mi H., Muruganujan A., Casagrande J.T., Thomas P.D.. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013; 8:1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Bel M., Diels T., Vancaester E., Kreft L., Botzki A., Van de Peer Y., Coppens F., Vandepoele K.. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018; 46:D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.Y., Freimer N.B., Sabatti C., Eskin E.. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010; 42:348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.. Circos: an information aesthetic for comparative genomics. Genome Res. 2009; 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J.et al.. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nandi T., Ong C., Singh A.P., Boddey J., Atkins T., Sarkar-Tyson M., Essex-Lopresti A.E., Chua H.H., Pearson T., Kreisberg J.F.et al.. A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence. PLoS Pathog. 2010; 6:e1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Letunic I., Bork P.. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016; 44:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang J., Lee S.H., Goddard M.E., Visscher P.M.. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011; 88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. The 1001 Genomes Consortium 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016; 166:481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiao W.B., Schneeberger K.. Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat. Commun. 2020; 11:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cameron D.L., Schroder J., Penington J.S., Do H., Molania R., Dobrovic A., Speed T.P., Papenfuss A.T.. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 2017; 27:2050–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cameron D.L., Di Stefano L., Papenfuss A.T.. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat. Commun. 2019; 10:3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pearce S., Saville R., Vaughan S.P., Chandler P.M., Wilhelm E.P., Sparks C.A., Al-Kaff N., Korolev A., Boulton M.I., Phillips A.L.et al.. Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiol. 2011; 157:1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uga Y., Sugimoto K., Ogawa S., Rane J., Ishitani M., Hara N., Kitomi Y., Inukai Y., Ono K., Kanno N.et al.. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013; 45:1097–1102. [DOI] [PubMed] [Google Scholar]
- 48. Guo J., Cao K., Deng C., Li Y., Zhu G., Fang W., Chen C., Wang X., Wu J., Guan L.et al.. An integrated peach genome structural variation map uncovers genes associated with fruit traits. Genome Biol. 2020; 21:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fu Y. Statistical properties of segregating sites. Theor. Popul. Biol. 1995; 48:172–197. [DOI] [PubMed] [Google Scholar]
- 50. Gu W., Zhang F., Lupski J.R.. Mechanisms for human genomic rearrangements. PathoGenetics. 2008; 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weckselblatt B., Rudd M.K.. Human structural variation: mechanisms of chromosome rearrangements. Trends Genet.: TIG. 2015; 31:587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bush S.J., Castillo-Morales A., Tovar-Corona J.M., Chen L., Kover P.X., Urrutia A.O.. Presence-absence variation in A. thaliana is primarily associated with genomic signatures consistent with relaxed selective constraints. Mol. Biol. Evol. 2014; 31:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bai Z., Chen J., Liao Y., Wang M., Liu R., Ge S., Wing R.A., Chen M.. The impact and origin of copy number variations in the Oryza species. BMC Genomics. 2016; 17:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gupta S., Gallavotti A., Stryker G.A., Schmidt R.J., Lal S.K.. A novel class of Helitron-related transposable elements in maize contain portions of multiple pseudogenes. Plant Mol. Biol. 2005; 57:115–127. [DOI] [PubMed] [Google Scholar]
- 55. Brunner S., Pea G., Rafalski A.. Origins, genetic organization and transcription of a family of non-autonomous helitron elements in maize. Plant J. 2005; 43:799–810. [DOI] [PubMed] [Google Scholar]
- 56. Cultrone A., Dominguez Y.R., Drevet C., Scazzocchio C., Fernandez-Martin R.. The tightly regulated promoter of the xanA gene of Aspergillus nidulans is included in a helitron. Mol. Microbiol. 2007; 63:1577–1587. [DOI] [PubMed] [Google Scholar]
- 57. Lei M., Zhang H., Julian R., Tang K., Xie S., Zhu J.K.. Regulatory link between DNA methylation and active demethylation in Arabidopsis. PNAS. 2015; 112:3553–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang N., Liu J., Gao Q., Gui S., Chen L., Yang L., Huang J., Deng T., Luo J., He L.et al.. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat. Genet. 2019; 51:1052–1059. [DOI] [PubMed] [Google Scholar]
- 59. Stuart T., Eichten S.R., Cahn J., Karpievitch Y.V., Borevitz J., Lister R.. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. eLife. 2016; 5:e20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schmalenbach I., Zhang L., Ryngajllo M., Jimenez-Gomez J.M.. Functional analysis of the Landsberg erecta allele of FRIGIDA. BMC Plant Biol. 2014; 14:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lye Z.N., Purugganan M.D.. Copy number variation in domestication. Trends Plant Sci. 2019; 24:352–365. [DOI] [PubMed] [Google Scholar]
- 62. Saitou N. The neighbor-joining methods: a new method for reconstructing phylogenetic trees. Mol.Biol.Evol. 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
- 63. Lee C.R., Svardal H., Farlow A., Exposito-Alonso M., Ding W., Novikova P., Alonso-Blanco C., Weigel D., Nordborg M.. On the post-glacial spread of human commensal Arabidopsis thaliana. Nat. Commun. 2017; 8:14458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gaut B.S., Seymour D.K., Liu Q., Zhou Y.. Demography and its effects on genomic variation in crop domestication. Nature plants. 2018; 4:512–520. [DOI] [PubMed] [Google Scholar]
- 65. Zichner T., Garfield D.A., Rausch T., Stutz A.M., Cannavo E., Braun M., Furlong E.E., Korbel J.O.. Impact of genomic structural variation in Drosophila melanogaster based on population-scale sequencing. Genome Res. 2013; 23:568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wendel J.F., Jackson S.A., Meyers B.C., Wing R.A.. Evolution of plant genome architecture. Genome Biol. 2016; 17:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K.et al.. Mapping copy number variation by population-scale genome sequencing. Nature. 2011; 470:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhou Y., Minio A., Massonnet M., Solares E., Lv Y., Beridze T., Cantu D., Gaut B.S.. The population genetics of structural variants in grapevine domestication. Nature plants. 2019; 5:965–979. [DOI] [PubMed] [Google Scholar]
- 69. Goodwin S., McPherson J.D., McCombie W.R.. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016; 17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M.C.. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018; 15:461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The information of the accessions used for large indels discovery are provided in Supplementary Table S19. The genomic coordinates of large indels identified are listed in Supplementary Table S5-7. We also provided a web interface at http://yanglab.hzau.edu.cn/IndelEnsembler to accessed the large indels. Users could fetch the information of large indels by entering the gene ID or gene region, the results included basic description of the large indels and their overlapped genes in Arabidopsis. IndelEnsembler has been released as free and open source software under a GNU General Public License (GPL version 3). The latest source code are available at https://github.com/kensung-lab/IndelEnsembler.