Abstract
KRAS-activating mutations are oncogenic drivers and are correlated with radioresistance of multiple cancers, including colorectal cancer, but the underlying precise molecular mechanisms remain elusive. Herein we model the radiosensitivity of isogenic HCT116 and SW48 colorectal cancer cell lines bearing wild-type or various mutant KRAS isoforms. We demonstrate that KRAS mutations indeed lead to radioresistance accompanied by reduced radiotherapy-induced mitotic catastrophe and an accelerated release from G2/M arrest. Moreover, KRAS mutations result in increased DNA damage response and upregulation of 53BP1 with associated increased non-homologous end-joining (NHEJ) repair. Remarkably, KRAS mutations lead to activation of NRF2 antioxidant signaling to increase 53BP1 gene transcription. Furthermore, genetic silencing or pharmacological inhibition of KRAS, NRF2 or 53BP1 attenuates KRAS mutation-induced radioresistance, especially in G1 phase cells. These findings reveal an important role for a KRAS-induced NRF2-53BP1 axis in the DNA repair and survival of KRAS-mutant tumor cells after radiotherapy, and indicate that targeting NRF2, 53BP1 or NHEJ may represent novel strategies to selectively abrogate KRAS mutation-mediated radioresistance.
INTRODUCTION
RAS genes encode a family of membrane-bound GTP-binding proteins whose members include HRAS, NRAS and KRAS (1). Deep sequencing studies have demonstrated that KRAS mutations are the single most common mutation in many human cancers, and these mutations result in constitutively active KRAS. KRAS mutations occur with extraordinarily high frequency in pancreatic (>90–95%), colorectal (40%) and lung cancers (20%). Under physiological conditions, KRAS functions as a binary molecular switch, cycling between an inactive GDP-bound and active GTP-bound state. In the GTP-bound state, KRAS binds and activates various downstream effectors to activate signaling pathways important for cell proliferation and survival, including mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K) pathways. Upon point mutations, most commonly at codons 12 and 13, KRAS protein becomes constitutively active and acquires oncogenic properties. Across tumor types, the presence of KRAS mutations often indicate poor prognosis and predict lack of response to targeted inhibitors of receptor tyrosine kinases, such as small molecular inhibitor of epidermal growth factor receptor (EGFR) or anti-EGFR antibody therapy (2). Therefore, therapies successfully directed against mutant KRAS activity would have broad implications in the field of oncology. However, developing clinically useful inhibitors of mutant KRAS has proven challenging (3–6). Thus, focus has been largely on downstream KRAS targets to develop a treatment that abolishes mutant KRAS activity (7,8). Nevertheless, recent preclinical and clinical data have shown that the responses of cancer cells to RAF or MEK inhibitors that act downstream of KRAS are transient, either alone or in combination, probably due to the paradoxical activation of RAS-extracellular signal-regulated kinase (ERK) and PI3K-AKT signaling (9,10). Therefore, there remains an urgent need to elucidate the mechanisms of the resistance of KRAS mutant cancer cells to current therapeutics and find novel targets to ultimately eradicate mutant KRAS activity clinically.
DNA damage response (DDR) and subsequent activation of repair pathways serve to protect cells from developing harmful mutations as a result of replicative stress and genomic instability (11–13). DDR is also responsible for protecting cancer cells from DNA damages induced by radiotherapy and genotoxic chemotherapies. DNA double strand breaks (DSBs) and single strand breaks (SSBs) are common after these therapies, with DSBs being the most lethal events to the tumor cells (14). DSB repair pathways include homologous recombination (HR) and non-homologous end-joining (NHEJ). NHEJ is especially critical for repairing DSBs after radiotherapy and operates throughout the cell cycle, compared to HR which operates chiefly in late S-G2 phases (15). Key components initiating the DDR after DSB and SSBs include ATM and ATR, as well as their downstream effectors CHK1, CHK2 and p53 (16). Radiotherapy is an important component of cancer therapy by improving local control through inducing tumor cell death and is used to treat patients prior to surgery, after surgery, or for patients who cannot have surgery due to advanced disease (17). However, radioresistance often leads to local failure and metastatic dissemination of cancer cells. Therefore, it is critical to elucidate mechanisms of radioresistance in order to engender development of novel radiosensitizers, thereby improving local control and the outcomes of cancer patients.
It has been well documented that hyperactivation of KRAS can lead to development of intrinsic radioresistance in tumor cells. A seminal preclinical study published in 1988 established that over-expression of KRAS, NRAS or HRAS induces radioresistance (18). Subsequent studies demonstrated that KRAS downregulation by siRNA or chemical inhibitors of farnesylation radiosensitizes tumor cells (19,20). Because patients are increasingly being tested for tumor mutations, there is now emerging clinical evidence that KRAS mutations are associated with resistance to radiation in colorectal and lung cancer (21–24). Most recently, a phase II clinical study of high-dose radiation for colorectal cancer metastases showed that KRAS mutations were associated with increased risk of failure after radiation (25). In addition, it has been shown that radiotherapy itself activates RAS-MAPK signaling in KRAS-mutant cells and that inhibition of MAPK signaling can attenuate cell survival after radiotherapy (26). Importantly, RAS mutations are found to be associated with increased survival of cancer cells from oxidative and other genotoxic stress possibly by promoting direct removal of oxidized nucleotides, DNA base excision repair (BER) and alternative non-homologous end-joining (alt-NHEJ) pathway (27–30). In addition, Ras-MEK signaling engages ATR-Chk1 activation in various cancers to enable their survival upon chemotherapy-induced DNA damage (31). Expression of wild-type H- and N-RAS alters mutant RAS induced oncogenic signaling and DNA damage response, further affecting tumor progression and chemosensitivity (32,33). Despite these studies, there has been a lack of mechanistic understanding of how cytosolic, mutated KRAS promotes radioresistance, and whether this occurs through increased nuclear DNA repair activity or other mechanisms. We hypothesized that KRAS mutations contribute to a poor response to radiation and certain genotoxic chemotherapies by imparting heightened DNA DSB repair capabilities to tumor cells, and that these KRAS mutant-dependent DNA repair pathways can be selectively targeted in KRAS mutant tumor cells to enhance therapeutic efficacy after radiation.
In this study, we utilized isogenic colorectal cancer HCT116 and SW48 cell lines bearing wild-type or mutant KRAS (G12C, G12D, G12V, G13D) to explore the role and underlying mechanisms of KRAS mutation in the repair of ionizing radiation-induced DSB. We found that KRAS mutation enhances DNA damage repair by promoting NHEJ. Mechanistic studies identified that 53BP1, a critical mediator of NHEJ, is upregulated in KRAS mutant tumor cells, and that 53BP1 nuclear foci are rapidly induced in KRAS-mutant cells after radiation and resolve earlier than in KRAS wild-type cells. Moreover, 53BP1 depletion preferentially radiosensitizes KRAS mutant cells in vitro, and stable depletion effectively radiosensitizes tumors in vivo. Importantly, we demonstrate that KRAS mutation upregulates NRF2 and NRF2-related transcriptional targets, and that KRAS genetic silencing or chemical inhibition reduces both 53BP1 and NRF2 expression in KRAS mutant cells. Furthermore, we show that NRF2 binds to 53BP1 promoter elements to a greater extent in KRAS mutant cells than wild-type cells. Finally, genetic or chemical suppression of NRF2 reduces 53BP1 expression and effectively radiosensitizes KRAS mutant but not KRAS wild-type tumor cells. Thus, we have identified a KRAS-NRF2-53BP1 oncogenic transcriptional regulatory pathway that imparts accelerated DNA repair and survival to KRAS mutant tumor cells after radiation. These findings suggest that targeting NRF2 and 53BP1 may represent novel strategies to selectively abrogate KRAS mutant-driven radioresistance.
MATERIALS AND METHODS
Cell culture, chemicals and antibodies
HEK293T cells were obtained from American Type Culture Collection and maintained in DMEM (Invitrogen, Carlsbad, CA) with 10% FBS. The paired human colorectal cancer cell lines HCT116 KRAS wild-type and G13D mutant cells (KRASWT/−; KRAS−/MUT) were provided by Bert Vogelstein through the Johns Hopkins Genetic Resources Core Facility (GRCF) and cultured in McCoy's 5A (Invitrogen) with 10% FBS (GE Healthcare, Chicago, IL) and 1% penicillin/streptomycin (Life Technologies, Carlsbad, CA). SW48 KRAS G13D cells (SW48WT/WT; SW48WT/MUT) were generously provided by Dr. Ching-Shih Chen and were grown in RPMI1640 (Invitrogen) with 10% FBS and 1% penicillin/streptomycin. Additional SW48 KRAS isogenic cells (G12D, G12V, G12C) were obtained from Horizon Discovery (Cambridge, UK). All cells were cultured in 37°C with 5% CO2 incubator. Typically, cells were kept in culture minimum two passages prior and maximum 20 passages when the experiments were performed. The identity of all cell lines was confirmed by STR genotyping (Identifier Kit, Applied Biosystems, Carlsbad, CA). For the detection of mycoplasma in cell culture, the Universal Mycoplasma Detection Kit (ATCC) was used. MEK inhibitor GSK1120212/trametinib (GSK) (ChemieTek, Indianapolis, IN) and NRF2 inhibitor Brusatol (Selleckchem, Houston, TX) were dissolved in DMSO (Sigma, St.Louis, MO). Anti-RAS, KRAS antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX); anti-total ERK1/2, phospho-ERK1/2 (Thr202/Tyr204), total ATM, phospho-ATM (Ser1981), total DNA-PKcs, phospho-DNA-PKcs (Ser2056), total BRCA1, NRF2, 53BP1, phospho-H2AX (Ser139), Keap1, Lamin A and GAPDH antibodies were purchased from Cell signaling Technology (Danvers, MA). Anti-β-tubulin was from Sigma (St. Louis, MO).
DNA extraction and KRAS mutation analysis
Exon 2 of KRAS gene was amplified by polymerase chain reaction (PCR) using the following primers: Forward 5′-TGA CAT GTT CTA ATA TAG TCA G-3′ and reverse 5′-ACA AGA TTT ACC TCT ATT GTT G-3′. PCR was performed as previously described (34). PCR products were purified using a DNA purification kit (Zymo Research, Irvine, CA) and direct Sanger sequencing was performed on capillary electrophoresis using Applied Biosystems 3730 DNA Analyzer (ThermoFisher, Waltham, MA).
KRAS activity assay
Ras GTP was detected using a Ras Activation ELISA assay kit (Millipore) according to manufacturer's instruction. GST-Raf-RBD was used to pull-down RAS-GTP from 50 μg of cell lysate prepared the same way as for immunoblotting, and then a primary antibody for RAS (detecting pan-RAS) or KRAS was added, followed by incubation with an HRP-conjugated secondary antibody. After addition of developing reagent, chemiluminescent reaction was determined with a Fluoroskan Ascent FL luminometer.
Neutral comet assay
KRAS isogenic cells were treated with 10 Gy ionizing radiation to induce DNA double strand breaks. At different time points, cells were trypsinized and subjected to neutral comet assay using the Trevigen Comet assay kit (Trevigen, USA) following the manufacturer’s protocol. Cells were imaged using a Zeiss Axios Observer Z1 fluorescence microscope (Carl Zeiss, White Plains, NY). Comet tail moment were measured and quantified using CometScore software (TriTek Corp, Sumerduck, VA). At least 50 cells were counted and imaged per time point.
Immunofluorescence for nuclear foci and mitotic catastrophe
Nuclear foci and mitotic catastrophe were analyzed by immunofluorescence staining as previously described (35). Briefly, cells were cultured and treated on glass coverslips, washed with PBS twice, and fixed with 2% paraformaldehyde, permeabilized with 1% Triton X-100 and blocked with 3% bovine serum albumin (BSA) in phosphate buffer saline (PBS). For nuclear foci analysis, cells were incubated with anti-γH2AX, anti-53BP1, anti-RAD51, anti-DNA-PKcs, or anti-BRCA1 antibody for 2 h in a humidified chamber at room temperature. For mitotic catastrophe assay, the cells were stained with anti-tubulin antibody. After rinsed with PBS, the cells were stained with an Alexa Fluor 488-conjugated secondary antibody (Biotium, Hayward, CA), along with DAPI for 1 h. Cells were then rinsed, mounted with coverslips, and sealed until visualization with Zeiss Axio Observer Z1 fluorescence microscope (Carl Zeiss). For each experiment, the number of foci per cell and mitotic catastrophe (>2 nuclear lobes, or several micronuclei) was determined in at least 100 cells.
Bromodeoxyuridine (BrdU) pulse-chase assay
Cells were pulsed with 10 μM BrdU for 30 min, washed with medium containing 10 μM thymidine, irradiated, and collected at different time points. The cells were processed and analyzed as previously described (36) using anti-BrdU (Pharmingen) and FITC-conjugated anti-mouse (Biotium) antibodies in conjunction with propidium iodide staining. Samples were analyzed on a LSRII flow cytometer (Becton Dickinson) with FlowJo software (Tree Star).
In vivo tumor growth studies
All animal studies were conducted according to our IACUC approved protocol. Six- to eight-week-old nude mice (Taconic Farms Inc., NY) were housed in a pathogen-free facility. Five million tumor cells were suspended in a 1:1 mixture of PBS/Matrigel (BD Biosciences) and injected subcutaneously into the left flank of a mouse. When the average tumor volume reached 100–200 mm3, ionizing radiation was initiated, with 2 or 4 Gy per day, for 5 continuous days with a RS2000 radiator. Radiation was targeted to the tumor mass in the left flank with custom lead shielding to block the rest of the mouse. To obtain a tumor growth curve, perpendicular diameter measurements of each tumor were measured 2–3 times/week with digital calipers, and volumes were calculated using the formula (L × W × W)/2. At the end of experiments, the mice were sacrificed, and tumors were removed.
HR and NHEJ repair analysis
HR and NHEJ were measured in cells as previously described (37,38). Briefly, cells were transiently transfected with NHEJ-GFP-PEM1, or HR-GFP-PEM1 plasmids, together with siRNA, and mCherry plasmids as a transfection control for GFP-positive cells. Twenty-four hours later, cells were infected with adenovirus expressing I-SceI restriction enzyme that induces DSBs in the recognition sequence within the reporter construct. Eighteen hours later, virus-containing medium was replaced by normal cell culture medium. After 24 h, GFP-positive and mCherry-positive cells were measured on LSRII flow cytometer. HR and NHEJ activity were calculated as the ratio of GFP/mCherry.
Real-time quantitative PCR analysis
Total cellular RNA was isolated using Trizol (Invitrogen), and one microgram of total RNA was reverse-transcribed using Supercript reverse transcriptase (Bio-Rad). PCR was performed on iCYCLER real-time PCR machine (Bio-Rad) using SYBR-Green chemistry (Bio-Rad). The gene expression levels were normalized to housekeeping gene GAPDH. The sequences of the used primers are shown in Supplementary Table S1.
Chromatin immunoprecipitation (ChIP) assay
Cells were fixed with 1% formaldehyde in PBS, sonicated to shear chromatin and subjected to ChIP as previously described (39). The cell lysate was incubated with rabbit anti-NRF2 antibody or normal rabbit IgG (Cell Signaling Technology) in combination with ChIP-Grade Protein G Magnetic Beads (Cell Signaling Technology). The beads-bound protein/DNA complexes were recovered by incubating in elution buffer (1% SDS and 0.1 M NaHCO3) at room temperature followed by incubating in 0.2 M NaCl for 5 h at 65°C to reverse formaldehyde crosslinking. After proteinase K treatment, the immunoprecipitated and input DNA were purified with phenol/chloroform precipitation and analyzed in triplicate by real-time qPCR. Primers for the AREs of 53BP1 promoter region are described in Supplementary Table S2. ChIP-qPCR was repeated twice for confirmation.
Statistical analysis
The statistical significance of difference was determined by Student’s t test. All values were expressed as mean ± standard deviation (SD). All analyses were two-sided, and the difference with a P-value of <0.05 were considered statistically significant in all analyses. Statistical correlations between expression, protein abundance, or KRAS mutation status tested with a linear model. Survival analysis performed with cox proportional hazard test and visualized with Kaplan–Meier plots.
RESULTS
KRAS oncogenic mutations increase KRAS activation and induce radioresistance
We assessed the effects of various KRAS mutations on radiation sensitivity, using homologous-recombined, isogenic HCT116 or SW48 cell lines bearing wild-type (WT) or G13D mutant (MUT) KRAS. To confirm KRAS mutation in these cell lines, Sanger sequencing of KRAS exon 2 was performed in genomic DNA isolated from HCT116 KRAS WT/- (‘HCT116 WT/-’), HCT116 KRAS -/MUT (‘HCT116-/MUT’), SW48 KRAS WT/WT (‘SW48 WT/WT’) and SW48 KRAS WT/MUT (‘SW48 WT/MUT’) cells. In HCT116 isogenic cells, a single G to A transition at codon 13 was detected in the HCT116 -/MUT cells. In SW48 isogenic cells, a G and A were both detected at codon 13 in SW48 WT/MUT cells suggesting heterozygosity of the mutant allele (Supplementary Figure S1A). To biologically characterize the resulting KRAS G13D mutation in these isogenic pairs, we assessed RAS activity in HCT116 WT/- and HCT116 -/MUT cells. We confirmed that HCT116 -/MUT cells with G13D mutation demonstrated both increased Pan-RAS and KRAS-specific activity comparing with HCT116 WT/- cells (Supplementary Figure S1B). Immunoblotting confirmed there was increased phosphorylation of ERK1/2 at Thr202 and Tyr204 (pERK) in HCT116 -/MUT and SW48 WT/MUT cells in comparison to the corresponding isogenic KRAS wild-type cells. In addition, transient exogenous expression of KRAS G13D in HEK293T cells resulted in increased pERK levels confirming RAS-RAF-MEK-ERK activation (Supplementary Figure S1C).
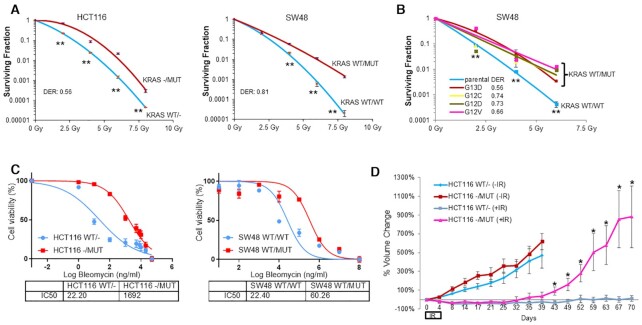
To determine whether KRAS mutation contributes to radioresistance, KRAS wild-type (HCT116 WT/- and SW48 WT/WT) and KRAS G13D mutant (HCT116 -/MUT and SW48 WT/MUT cells) isogenic cells were treated with increasing doses of ionizing radiation (IR) followed by standard radiation clonogenic assay. Compared to KRAS WT cell lines, there was a higher surviving fraction of cells with KRAS G13D in response to IR (Figure 1A). To determine if this effect is specific for the G13D mutation, we also performed radiation clonogenic assays using a set of isogenic SW48 cell lines bearing G12C, G12D or G12V allelic mutations of KRAS. In comparison to KRAS WT cells, all KRAS MUT cell lines showed significant radioresistance (Figure 1B). We confirmed that KRAS mutation also induced profound resistance to bleomycin, a radiomimetic chemotherapy that induces DSBs similar to radiation (40) (Figure 1C). Additionally, we examined radiation sensitivity data from a collection of over 500 Cancer Cell Line Encyclopedia (CCLE) cell lines from Yard et al. (41), and pooled radiation sensitivity data for each of the top two tumor types with KRAS mutations (colorectal cancer and lung adenocarcinoma). Compared to KRAS WT cell lines, although KRAS MUT cells showed slight increases in radioresistance, the differences were not statistically significant (Supplementary Figure S2). Next, we tested whether KRAS mutation alters radiosensitivity in vivo using mouse xenografts. When tumors of HCT116 -/MUT and HCT116 WT/- cell lines reached ∼100–150 mm3, the tumors were treated with or without 4 Gy radiation every day for five consecutive days (total 20 Gy). As shown in Figure 1D, radiation treatment led to durable control of HCT116 WT/- tumors, but HCT116 -/MUT tumors demonstrated rapid and aggressive re-growth indicating radioresistance in vivo, which corroborated our in vitro findings. Taken together, these results support that KRAS mutation contributes to radioresistance, in accordance with clinical observations.
Figure 1.
KRAS mutation leads to radiation resistance in vitro and in vivo. (A) HCT116 and SW48 isogenic cells bearing either WT or MUT KRAS (G13D) allele were subjected to radiation clonogenic assays with the indicated radiation doses (2, 4, 6 and 8 Gy). (B) Radiation clonogenic assays showed that SW48 isogenic lines bearing G12C, G12D, G12V or G13D mutation all demonstrated radioresistance compared to the KRAS WT parental cells. Data shown as mean ± SD. Experiments were performed three times with triplicates; DER, dose enhancement ratio. (C) HCT116 and SW48 KRAS G13D isogenic cells were treated with increasing doses of bleomycin (0–50 000 ng/ml) for 72 h and cytotoxicity was measured with alamarBlue assay. IC50 was much higher in KRAS mutant cells than in KRAS wild-type cells, indicating KRAS mutation induces resistance to bleomycin. Data shown as mean ± SD. Experiments were performed three times with four replicates. (D) HCT116 WT and G13D MUT isogenic cells were implanted s.c. into the left flanks of athymic nude mice and treated with sham radiation or radiation (IR, 4 Gy daily × 5 days during Days 0–4); *P < 0.05, **P < 0.001.
KRAS-mutated cells demonstrate enhanced DNA damage response and repair ability in response to radiation
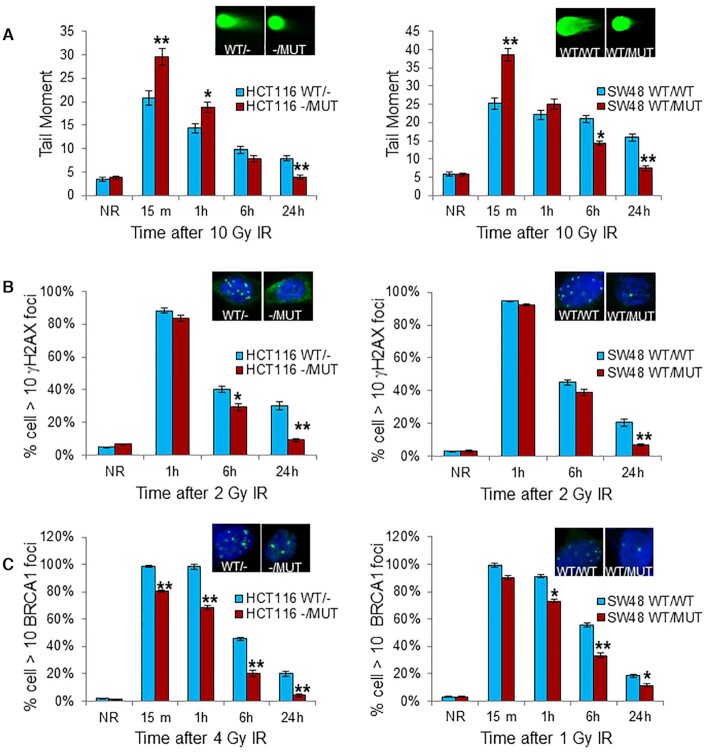
A major mechanism of cell death induced by radiation is through induction of DSBs and subsequent post-mitotic death. Efficient and faithful DNA damage repair is critical to normal cell survival but is also a mechanism contributing to tumor cell therapeutic resistance. To test if KRAS mutation alters the global DNA repair process in irradiated cells, we performed neutral comet assay, a sensitive technique for detecting DNA damage, particularly DSBs, at the level of individual cells (42). At 15 min and 1 h after 10 Gy, there was significantly higher tail moment in HCT116 -/MUT and SW48 WT/MUT cells compared to HCT116 WT/- and SW48 WT/WT cells, suggesting altered DNA damage response in KRAS MUT cancer cells (Figure 2A). However, after 6 and 24 h post-IR, there was reduced tail moment in HCT116 -/MUT and SW48 WT/MUT compared to HCT116 WT/- and SW48 WT/WT cells, suggesting accelerated DNA damage repair in cancer cells with KRAS mutation. Phosphorylation of histone H2AX (γH2AX) status also reflects the kinetics of DNA damage production and DSB repair. We performed immunofluorescence analysis of γH2AX nuclear foci of these cells after IR. As expected, IR induced dramatic γH2AX foci in the nuclei of all cells at 1 h after IR regardless of KRAS mutation status (Figure 2B). However, at 6 and 24 h post-IR there were significantly fewer γH2AX foci in KRAS MUT cells compared to KRAS WT cells. Interestingly, there were significantly fewer BRCA1 nuclear foci in KRAS MUT cells compared to KRAS WT cells both early during the radiation response (15–60 min) and during recovery from IR (6 and 24 h) (Figure 2C), suggesting that double-strand breaks may be repaired more quickly, or at least that a KRAS mutation is altering BRCA1’s participation (or need for involvement) in the DNA repair process.
Figure 2.
KRAS mutation accelerates DNA damage repair after radiation. (A) KRAS G13D isogenic cells received 10 Gy, followed by neutral comet assay at the indicated time points after radiation. The graph showed the mean tail moment of minimum of 50 cells. (B) Immunofluorescence of γH2AX nuclear foci at the indicated time points following IR (2 Gy). Mean percentage of cells with >10 γH2AX foci at each time point after IR from two experiments of no <100 cells per condition are shown. (C) Immunofluorescence of BRCA1 nuclear foci of the cells treated as in (B), but with 4 Gy (HCT116) or 1 Gy (SW48) radiation, followed by indicated repair time. The graph showed the quantitation of γH2AX and BRCA1 foci. Left: HCT116 isogenic cells, Right: SW48 isogenic cells. Data shown as mean ± SD. Experiments were performed three times. Inset: representative pictures from comet assay (A), and foci staining (B and C) at 24 h after IR; *P < 0.05, **P < 0.001.
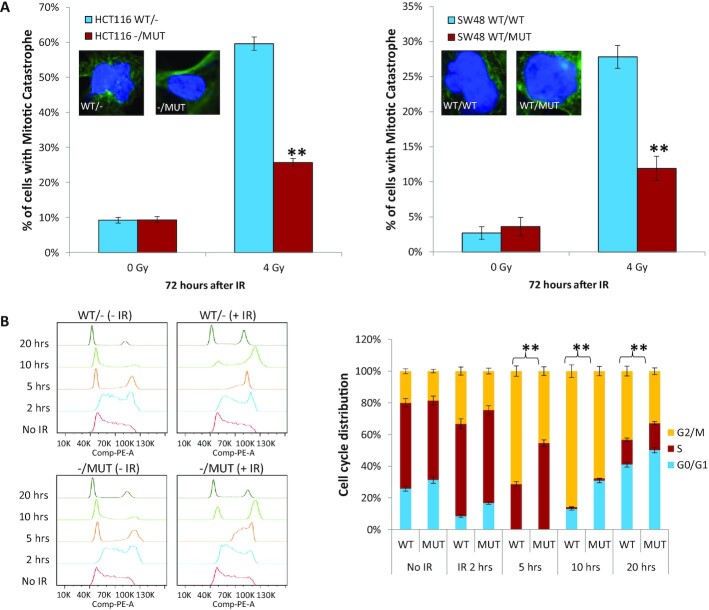
Mitotic catastrophe occurs if a cell cannot sufficiently repair DNA damage, therefore resulting in aberrant chromosome segregation in mitosis, and ultimately post-mitotic cell death. Mitotic catastrophe is the predominant form of cell death induced by IR, and these events can be scored by the presence of cells with > 2 nuclear lobes. To further assess the consequences of DNA damage to cells with or without KRAS mutation, we measured the incidence of mitotic catastrophe at 72 h after IR. We found the presence of a KRAS mutation was associated with lower levels of mitotic catastrophe in both HCT116 and SW48 cells (Figure 3A). Furthermore, cell cycle regulation is an important determinant of IR sensitivity, with cells being most radiosensitive in the G2/M phase, but less sensitive in the G1 phase (43,44). We first compared cell cycle distribution of two pairs of isogenic asynchronously growing cells. We found that KRAS mutant cells had slightly higher proportions of cells in G1 phase, and significantly less G2/M cells compared to wild-type KRAS cells (Supplementary Figure S3), supporting that KRAS mutant cells may also be more radioresistant by virtue of different inherent cell cycle distribution. A common cellular response to IR is the activation of G2/M arrest within 24 h, providing time for tumor cells to repair the DNA damage after IR. Once repaired, the tumor cells can re-enter the cell cycle. Using BrdU-labeling to track cell cycle progression of S phase labeled HCT116 tumor cells after IR, we also found that a KRAS mutation resulted in earlier recovery from IR-induced G2/M arrest at 10 and 20 h after IR, resulting in accelerated G2/M progression and re-entry of tumor cells back into the G0/G1 phase of the cell cycle (Figure 3B).
Figure 3.
KRAS mutation attenuates mitotic catastrophe and relieves cell cycle arrest after radiation. (A) KRAS G13D isogenic cells were irradiated with 4 Gy. Seventy-two hours later, cells were co-stained with tubulin and DAPI to quantitate mitotic catastrophe events (multinucleated cells as shown in inserts). The graph showed percentage of mitotic catastrophe cells in minimum of 100 cells. Experiments were performed three independent times. (B) Cell cycle distribution was analyzed by BrdU incorporation assay. Left, BrdU pulsed HCT116 isogenic cells were treated with 4 Gy and collected for flow cytometry at the indicated time points. Right, proportion of cells in the G0/G1, S or G2/M phases of the cell cycle were displayed by flow cytometry histograms. Note higher proportion of cells re-entering G0/G1 at 10 h after radiation in KRAS MUT cells. Data shown as mean ± SD. Experiments were performed three times; **P < 0.001.
KRAS mutation upregulates 53BP1 to promote NHEJ repair
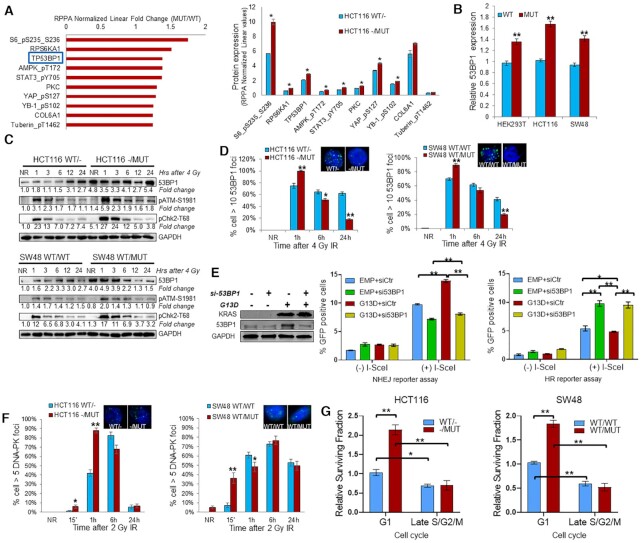
In order to assess how a KRAS mutation promotes heightened DNA repair in the nuclei of cells, we performed reverse phase protein array (RPPA) on lysates from untreated HCT116 KRAS isogenic cells. Of the proteins significantly upregulated in KRAS MUT cells, the most upregulated DNA repair protein was 53BP1 (Figure 4A). Since 53BP1 is a nuclear protein important for the response to DSB damage, and important for mediating genomic integrity, we focused our efforts on this protein (45,46). Analyses of 53BP1 in publicly available RNA expression databases also demonstrated 53BP1 is upregulated in colorectal cancer tissues compared with normal tissues and has a trend for association with KRAS mutation (Supplementary Figure S4). Furthermore, analysis of TCGA data shows that higher 53BP1 expression is associated with significantly worse disease-free survival (DFS, log-rank P < 0.05) and trend toward worse overall survival (log-rank P = 0.068) (Supplementary Figure S5A). We further investigated whether 53BP1 expression was associated with outcomes based on KRAS mutation status and found that higher 53BP1 expression is associated with worse DFS in KRAS MUT tumors but not KRAS WT tumors (Supplementary Figure S5B). These results suggest that 53BP1 might be an overall biomarker of poor prognosis, particularly in KRAS MUT tumors. We also assessed 53BP1 protein expression in a tissue microarray (TMA) of patients with rectal cancer treated preoperatively with neoadjuvant chemoradiation (CRT) prior to surgical resection. The TMA consisted of pre-therapy tumor biopsies and tumor tissue obtained post-CRT (at the time of surgery) along with adjacent normal rectal tissues that also received CRT. Interestingly, 53BP1 expression was significantly upregulated in tumors after CRT compared to (i) adjacent normal rectal epithelial cells (that received CRT) and (ii) tumor tissue pre-CRT (Supplementary Figure S6). In addition, higher tumor 53BP1 expression in post-CRT was correlated with adverse pathologic features including lymphovascular invasion (P = 0.003) and perineural invasion (P = 0.10).
Figure 4.
KRAS mutation upregulates 53BP1 to influence DNA double strand break repair pathway choice. (A) TP53BP1 is one of the top 10 upregulated proteins in HCT116–/MUT relative to HCT116WT/- by RPPA analysis. Left panel: RPPA analysis displaying the fold change of top 10 upregulated protein expression or phosphorylation in KRAS mutant HCT116 cells relative to KRAS wild-type HCT116 cells. Right panel: RPPA analysis displaying the protein expression or phosphorylation levels in HCT116 isogenic cells. (B) Exponentially growing HCT116 and SW48 KRAS G13D isogenic cells, and HEK293T cells transfected with a plasmid expressing KRAS G13D or empty vector were subjected to qRT-PCR analysis of TP53BP1 mRNA. (C) Western blot and densitometric quantification of proteins at the indicated hours post 4 Gy IR (NR = no radiation, time zero hours). Fold change of protein are compared to KRAS wild-type cells without IR. (D) At the indicated hours post 4 Gy, isogenic HCT116 and SW48 cells were prepared for immunofluorescence of 53BP1 nuclear foci. Inset: representative pictures of foci at 24 h after IR. (E) HEK293T cells were transfected with KRAS G13D plasmid (G13D), empty vector, 53BP1 siRNA (si53BP1) or scrambled control siRNA (siCtr) for 48 h and subjected to immunoblotting. DNA damage reporter assays were performed by transfecting HEK293T cells with NHEJ-GFP or HR-GFP reporter plasmid, together with KRAS G13D, empty vector (EMP), si53BP1 or siCtr plasmid for 24 h, followed by addition of adenovirus containing I-SceI to induce DNA DSB in the reporter plasmid consensus sequence. After 24 h, the cells were analyzed for GFP-positive cells by flow cytometry to measure NHEJ or HR repair proficiency. The values represent the mean ± SD from triplicate dishes, and at least three independent experiments were performed. mCherry plasmid was used to normalize transfection efficiency. (F) At the indicated time post 4 Gy, isogenic HCT116 and SW48 cells were prepared for immunofluorescence of DNA-PKcs nuclear foci. Data shown as mean ± SD. Experiments were performed three times. (G) Clonogenic assay using cells enriched in G1 phase and late S/G2/M phase. Surviving fraction at 2 Gy was normalized to KRAS wild-type G1 cells; *P < 0.05, **P < 0.001
In our isogenic cell lines, qRT-PCR confirmed increased mRNA levels of 53BP1 in HCT116 -/MUT and SW48 WT/MUT cells in comparison to the KRAS wild-type cells (Figure 4B). In addition, transient exogenous expression of KRAS G13D in HEK293T cells resulted in upregulation of 53BP1 mRNA level. At the protein level, 53BP1 was markedly upregulated in KRAS MUT isogenic tumor cells at baseline (no radiation), confirming RPPA findings (Figure 4C). 53BP1 was also noted to be induced by radiation, similar to our above findings in the TMA. 53BP1 has been shown to be required for optimal activation of ATM-Chk2 checkpoint, suggesting that over-expression of 53BP1 may heighten and prolong an ATM-Chk2 activation response (47). Indeed, we noted increased activation of ATM and Chk2 early after IR in KRAS mutant cells, but at subsequent time points, we did not see prolongation of ATM or Chk2 activation (Figure 4C). To further evaluate 53BP1 recruitment to DSBs, we irradiated cells and performed immunofluorescence for 53BP1 nuclear foci. We found accelerated formation and resolution of 53BP1 nuclear foci after IR in KRAS-mutant cells (Figure 4D), suggesting that KRAS-mutant cells are ‘primed’ for a 53BP1-mediated response following IR.
53BP1 is recruited to sites of DSBs and promotes NHEJ-mediated repair (46). To more directly determine whether KRAS G13D mutation maintains cell survival by increasing 53BP1 expression to alter HR and/or NHEJ repair, we applied fluorescent reporter constructs for sensitive and quantitative measurement of HR (DR-GFP) or NHEJ repair (Pem-1-NHEJ) in HEK293T cells transfected with a plasmid-expressing KRAS G13D in the presence or absence of co-transfection of 53BP1 siRNA. NHEJ and HR reporter constructs are based on an engineered GFP gene containing recognition sites for I-SceI endonuclease for induction of DSBs. The starting constructs are GFP negative as the GFP gene is inactivated by additional exon, or by mutations. Successful repair of I-SceI induced DSBs by NHEJ or HR restores the functional GFP gene. The percentage of GFP positive cells counted by flow cytometry provides a quantitative measure of NHEJ or HR efficiency (38). We found that expression of KRAS MUT significantly enhanced NHEJ but had minimal effect on HR (Figure 4E). Knockdown of 53BP1 by siRNA significantly reduced the upregulation of NHEJ activity by KRAS G13D, whereas knockdown of 53BP1 significantly increased HR. Furthermore, assessment of nuclear foci kinetics of DNA-PKcs (critical effector enzyme mediating NHEJ repair) after 2 Gy of IR showed that KRAS MUT cells have increased DNA-PK foci formation within the first 15 min after radiation (Figure 4F), indicating a rapid NHEJ-mediated response to DSB formation. Conversely, no consistent differences in the kinetics of RAD51 foci were noted in the KRAS WT vs MUT isogenic cell lines for both HCT116 and SW48 paired cells (Supplementary Figure S7A).
Since NHEJ repair can operate throughout cell cycle phases, while HR repair occurs predominantly in S and G2 phases, we also explored the relative radiosensitivity to 2 Gy of cells at different cell cycle phases using an ES-Fucci reporter plasmid to sort cells into G1 or late S/G2/M phases. As Figure 4G shows, G1 phase sorted cells were more radioresistant compared to late S/G2/M phase cells. With regard to KRAS mutation, in G1 enriched cells, KRAS mutation caused significant radiation resistance, resulting in 2.1- and 1.8-fold increased survival in HCT116 and SW48 KRAS MUT isogenic cells respectively (Figure 4G). However, in late S/G2/M phase cells there was no difference between KRAS WT and MUT cells. Taken together, these results suggest that KRAS G13D mutation enhances NHEJ repair activity at least in part via upregulating 53BP1, which ‘primes’ the cells for an NHEJ repair response.
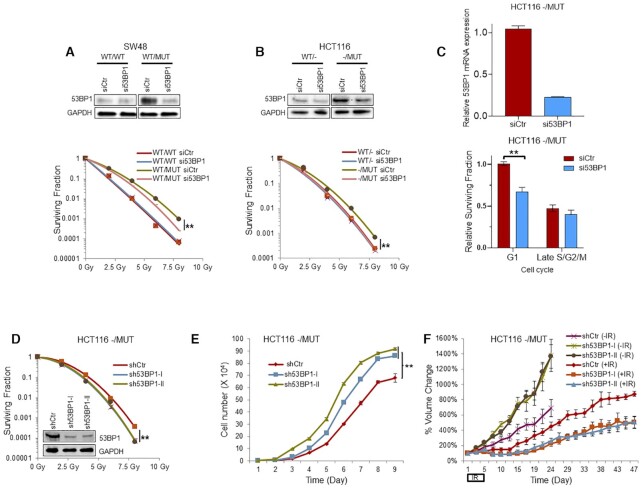
We next determined whether upregulation of 53BP1 contributes to the radioresistance of tumor cells with a KRAS G13D mutation. We performed radiation clonogenic assay in our KRAS isogenic cells following genetic depletion of 53BP1 by siRNA. We found that 53BP1 knockdown did not affect the radiosensitivity of SW48 WT/WT cells, whereas significantly radiosensitized SW48 WT/MUT cells (Figure 5A). Similar results were obtained in HCT116 cells (Figure 5B). Furthermore, cell cycle enriched cell radiation clonogenic assay revealed that 53BP1 genetic silencing significantly sensitized HCT116 KRAS mutant G1 phase cells to radiation treatment (Figure 5C). To test these results in vivo, we established two stable HCT116 -/MUT cell lines, each expressing an independent 53BP1 shRNA, as well as a scrambled shRNA control stable cell line. Partial silencing was confirmed by immunoblotting (Figure 5D). Similar to the transient knockdown of 53BP1, stable 53BP1 knockdown sensitized HCT116 -/MUT cells to radiation (Figure 5D). Interestingly, stable 53BP1 knockdown cells demonstrated a proliferative advantage over the control cells under normal growth conditions in vitro (Figure 5E). In vivo, we tested the ability of 53BP1 depletion to radiosensitize radioresistant HCT116 -/MUT cells compared to shCtr HCT116 -/MUT tumors using xenograft modeling. Interestingly, unirradiated stable 53BP1 knockdown tumors grew faster than the unirradiated shCtr tumors, corroborating the in vitro cell data (Figure 5F). However, stable 53BP1 knockdown tumors demonstrated enhanced radiosensitivity compared with the HCT116 -/MUT shCtr tumors. Immunoblotting of tumor lysates confirmed the stable knockdown of 53BP1 in the HCT116/MUT tumors (Supplementary Figure S7B).
Figure 5.
Depletion of 53BP1 sensitizes KRAS mutant cells to radiotherapy. (A and B) SW48 and HCT116 KRAS G13D isogenic cells were transfected with si53BP1 or control scrambled siRNA (siCtr) and subjected to immunoblotting to confirm 53BP1 knockdown (top panel), and clonogenic assay to evaluate effects of 53BP1 depletion on radiation sensitivity (bottom panel). (C) Cell cycle enriched clonogenic assay with 2 Gy (bottom panel) in HCT116 KRAS mutant cells after 53BP1 genetic silencing confirmed by qPCR (top panel). (D) Clonogenic assays shows 53BP1 stable knockdown radiosensitizes tumor cells in both two 53BP1 stable knockdown cell lines. Inset: Immunoblotting confirmed 53BP1 stable knockdown in HCT116 -/MUT cells. (E) Cell proliferation assay shows 53BP1 depletion increases cell proliferation rates. (F) 53BP1 stable knockdown cells were subcutaneously implanted into left flanks of athymic mice for in vivo assessment of radiosensitivity. Once tumors reached 100–150 mm3, mice were randomized to no radiation or radiation (2 Gy per day, days 1–5); **P < 0.001.
KRAS mutation increases NRF2 expression that subsequently enhances 53BP1 gene transcription
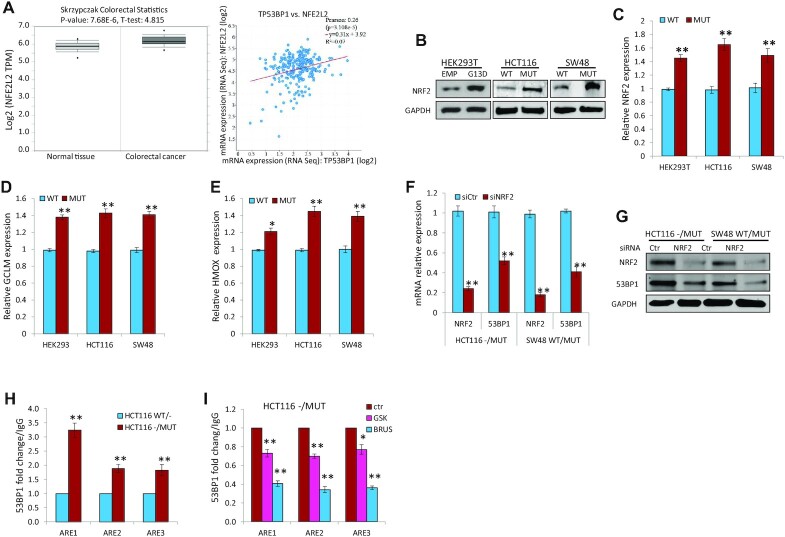
NRF2 is an anti-oxidant response gene encoding for the NRF2 transcription factor that has been implicated in promoting radiation resistance. The 53BP1 gene promoter contains three antioxidant responsive elements (ARE1, 2, 3) to which NRF2 directly binds, and it has been reported that NRF2 protects cancer cells from IR by inducing 53BP1-mediated DSB repair (48). Our analyses of DNA damage and repair gene expression in the publicly available databases revealed that NRF2 is significantly upregulated in colorectal cancer tissues compared with that in normal tissues, and that NRF2 expression directly correlates with 53BP1 expression (Figure 6A). We hypothesized that KRAS mutation might increase 53BP1 expression via NRF2. To test this hypothesis, we first assessed NRF2 protein levels in isogenic HCT116 and SW48 cells, and HEK293T cells transiently expressing exogenous KRAS G13D by immunoblotting and found there was dramatic increase of NRF2 expression in the presence of KRAS mutation (Figure 6B). qRT-PCR confirmed that there are increased mRNA levels of NRF2 (Figure 6C) and its well-established transcriptional targets, GCLM (Figure 6D) and HMOX (Figure 6E), in HCT116-/MUT, SW48 WT/MUT and 293T cells with ectopically expressed KRAS G13D. In addition, silencing NRF2 by siRNA markedly decreased both the mRNA and protein levels of 53BP1 in HCT116-/MUT and SW48 WT/MUT cells (Figure 6F and G). Moreover, ChIP assay identified that NRF2 binds to ARE1, ARE2 and ARE3 within the 53BP1 promoter to a greater extent in KRAS MUT HCT116 cells versus HCT116 WT/- cells (Figure 6H). To confirm that this observation is dependent on KRAS activation and NRF2, we treated HCT116 -/MUT cells with either MEK1/2 inhibitor trametinib (downstream of KRAS activation) or NRF2 inhibitor brusatol. We found that pharmacologic inhibition of either MEK1/2 or NRF2 significantly reduced the NRF2 binding to ARE1, ARE2 and ARE3 within the 53BP1 promoter (Figure 6I).
Figure 6.
KRAS mutation increases NRF2 expression and subsequent TP53BP1 gene transcription. (A) NRF2 is upregulated in colorectal cancer and correlates with 53BP1 expression. Left: Analysis of RNA expression datasets reveals upregulation of NRF2 in Oncomine datasets (Skrzypczak colorectal) using www.oncomine.org; Right: NRF2 (NFE2L2) and 53BP1 RNA expression are directly correlated (TCGA COAD). (B) Immunoblotting demonstrates upregulation of NRF2 in exogenously expressed KRAS G13D (HEK293T cells), or endogenous KRAS G13D (HCT116 and SW48 isogenic cells). (C–E) Exponentially growing HCT116 and SW48 isogenic cells, and HEK293T cells transfected with a plasmid expressing KRAS G13D or empty vector were subjected to qRT-PCR analysis of NRF2 mRNA (C), and its downstream transcriptional targets GCLM (D) and HMOX (E). (F and G) HCT116 and SW48 isogenic cells were transfected with NRF2 siRNA (siNRF2) or control siRNA (siCtr). After 48 h, RNA (F) and protein (G) expression levels of NRF2 and 53BP1 were assessed by qRT-PCR and immunoblotting, respectively. (H and I) ChIP assays demonstrate that NRF2 binds to 53BP1 promoter in a greater extent in KRAS mutant cells (H), which was prevented by pharmacological inhibition of either MEK1/2 with trametinib (GSK, 100 nM) or Nrf2 with brusatol (BRUS, 100 nM) (I); *P < 0.05, **P < 0.001.
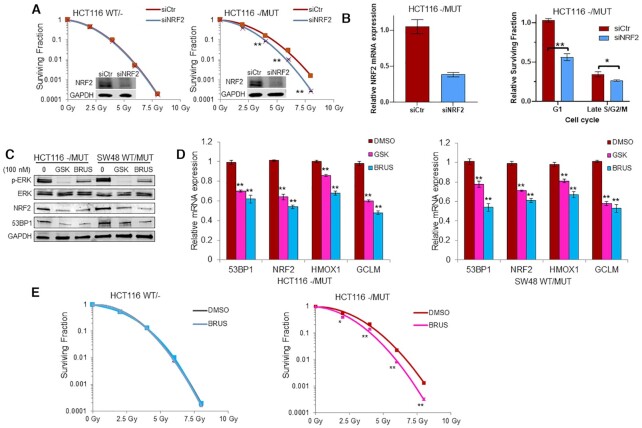
In order to assess whether NRF2 could promote KRAS-associated radioresistance, radiation clonogenic assays were performed in isogenic HCT116 and SW48 cells following knockdown of NRF2 by siRNA (Figure 7A and Supplementary Figure S7C). We found that NRF2 knockdown did not affect the radiosensitivity of HCT116 WT/- cells but significantly radiosensitized HCT116 -/MUT cells. Furthermore, cell cycle enriched clonogenic assay indicated NRF2 gene silencing sensitized KRAS mutant cells to IR predominantly in G1 and a small amount in late S/G2/M phases (Figure 7B). To extend our observations, we treated both HCT116 and SW48 KRAS mutant cells with 100 nM trametinib or brusatol. Immunoblotting showed that MEK1/2 or NRF2 inhibition suppressed the phosphorylation of ERK1/2 (pERK) as well as NRF2 and 53BP1 levels (Figure 7C). qRT-PCR demonstrated that inhibition of either MEK1/2 or NRF2 also suppressed the mRNA levels of 53BP1, NRF2 and NRF2 targets GCLM and HMOX (Figure 7D). Moreover, radiation clonogenic survival revealed that pharmacologic NRF2 inhibition with 10 nM brusatol preferentially sensitized HCT116 -/MUT but not HCT116 WT/- cells to radiation (Figure 7E).
Figure 7.
Inhibition of NRF2 sensitizes KRAS mutant tumor cells to radiotherapy. (A) HCT116 G13D isogenic cells were transfected with NRF2 specific siRNA (siNRF2) or control siRNA (siCtr) for 48 h before IR and subjected to radiation clonogenic assay. Inset: Immunoblotting confirmed NRF2 knockdown in HCT116 isogenic cells. (B) Cell cycle enriched clonogenic assay with 2 Gy (right panel) in HCT116 KRAS mutant cells after NRF2 genetic silencing (left panel). (C and D) HCT116 and SW48 isogenic cells were treated with 100 nM MEK1/2 inhibitor trametinib (GSK) or 100 nM NRF2 inhibitor brusatol (BRUS) for 24 h. Total proteins were extracted for immunoblotting of the indicated proteins (C). mRNA expression levels of 53BP1, NRF2, HMOX1 and GCLM were determined by qRT-PCR (D). (E) Isogenic HCT116 cells were cultured in media containing 10 nM brusatol 3 h prior IR and subjected to radiation clonogenic survival assays; *P < 0.05, **P < 0.001.
KRAS mutation promotes NRF2 nuclear expression and KRAS depletion inhibits NRF2-53BP1 expression leading to radiosensitization
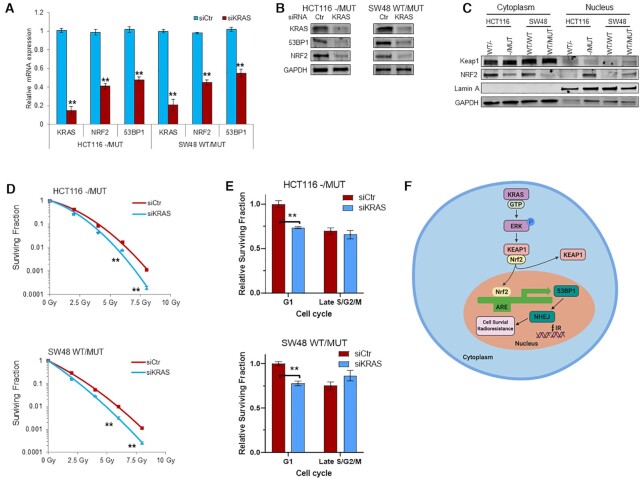
Previous studies have shown that RAS-RAF signaling controls NRF2 gene transcription via Jun and Myc transcription factors (49,50). To assess the importance of KRAS in the regulation of NRF2 gene transcription and 53BP1 expression, we knocked down KRAS by siRNA in HCT116-/MUT and SW48 WT/MUT cells. Silencing KRAS was accompanied by decrease of NRF2 and 53BP1 at both RNA and protein levels (Figure 8A and B). Under normal growth conditions, NRF2 is sequestrated by Keap1 in the cytoplasm. In response to stressful conditions, such as oxidative stress, NRF2 dissociates from Keap1 via multiple mechanisms and translocates to nucleus to stimulate transcription of genes important for the anti-oxidant response (49). RAS-MAPK signaling also regulates NRF2 intracellular localization via phosphorylating NRF2 and/or Keap1 (49). Therefore, we assessed the intracellular distribution of NRF2 by sub-cellular fractionation and found there was increased NRF2 in the nuclear fraction of KRAS mutant HCT116 and SW48 cells, consistent with activated NRF2 (Figure 8C). Finally, consistent with the critical role of NRF2 antioxidant signaling in the balance of oncogene-induced reactive oxygen species (ROS) and subsequent survival of cancer cells (51), we found that silencing of KRAS sensitized KRAS mutant HCT116 and SW48 cells to radiation (Figure 8D). Finally, cell cycle enriched clonogenic assay indicated KRAS gene silencing sensitized KRAS mutant cells to IR in the G1 phase, with minimal effect in late S/G2/M phases (Figure 8E), similar to previous findings with 53BP1 silencing.
Figure 8.
KRAS mutation promotes NRF2 nuclear expression and KRAS depletion inhibits NRF2-53BP1 expression leading to radiosensitization. (A and B) HCT116 and SW48 G13D isogenic cells were transfected with KRAS siRNA (siKRAS) or control scrambled siRNA (siCtr) for 48 h. The mRNA (A) and protein (B) expression levels of 53BP1, NRF2 and KRAS were determined by qRT-PCR and immunoblotting, respectively. (C) Subcellular fractionation of HCT116 and SW48 isogenic cells reveal higher levels of NRF2 in the nucleus in KRAS MUT cells. (D) Effects of KRAS knockdown by siRNA on radiation sensitivity of HCT116 and SW48 KRAS G13D mutant cells was evaluated by radiation clonogenic assay. (E) Cell cycle enriched clonogenic assay with 2 Gy in HCT116 KRAS mutant (top) and SW48 KRAS mutant (bottom) cells after KRAS genetic silencing. (F) Schematic model of the regulation of DSB repair via KRAS-NRF2-53BP1 signaling axis. Activated (oncogenic) KRAS increases NRF2 translocation into the nucleus, leading to increased binding and transcription of anti-oxidant response element genes, including 53BP1. Increased 53BP1 expression promotes NHEJ repair after ionizing radiation, promoting cell survival and radioresistance; **P < 0.001.
DISCUSSION
In this study, we demonstrated an oncogenic KRAS mutation induces accelerated DNA damage response and repair after radiation. Our results indicate that an activating KRAS mutation promotes gene transcription and nuclear translocation of NRF2, which in turn stimulates 53BP1 transcription to prime the cell for NHEJ-mediated repair activity (model shown in Figure 8F). Additionally, our findings that targeting KRAS, NRF2, or 53BP1 preferentially sensitizes tumor cells in the G1 phase of the cell cycle further implicates an NHEJ-mediated repair mechanism over HR repair in contributing to KRAS-mediated radioresistance. Through this study, we have identified molecular evidence of how an oncogenic KRAS mutation induces tumor cell radioresistance by detailing a KRAS-NRF2-53BP1 signaling axis which promotes NHEJ-mediated repair of radiation-induced DNA damage. Taken together, our study suggests that targeting 53BP1, NRF2 or NHEJ could represent independent novel therapeutic strategies to improve radiation therapy efficacy in KRAS-mutated cancers.
The frequency of KRAS, NRAS and HRAS mutations are ∼40–45%, 5–10% and 2–4% respectively in colorectal cancer (52,53). Detection of KRAS mutation status is routine clinical practice in the diagnosis and management of colorectal cancer, since KRAS mutations are correlated to the resistance to anti-EGFR therapy consisting of cetuximab or panitumumab (54,55). Moreover, accumulating data have shown that RAS mutations contribute to radiotherapy resistance of various cancer types (1,18–23). Most recently, a seminal study of patients with colorectal cancer metastases treated with high-dose radiation demonstrated that the presence of a KRAS mutation was associated with increased risk of tumor recurrence after radiation (25). Available data indicate that the role of KRAS mutation in tumor response to cytotoxic chemotherapies seems cancer-type dependent (3,56–58). Our phase I rectal cancer trial recently found that higher pERK levels in the tumor pre-therapy were correlated with resistance to chemoradiation (59). Preclinical data also show that elevated signaling of components upstream of ERK, such as receptor tyrosine kinases (EGFR, HER2), Raf and Mek are correlated with radioresistance (41). It has been reported that KRAS mutant cancer cells have enhanced DNA base excision repair (BER) (27), which is a principal DNA repair system to maintain genome stability and cancer cell survival due to the increased intracellular oxidative stress experienced by proliferating cancer cells. Ramdzan et al. (2014) reported that KRAS G12V mutation is synthetic lethal with CUX1, an important accessory factor of OGG1 in BER (28). In addition, MTH1, an enzyme critical for suppressing oxidative damage of nucleotides, was found to be upregulated by KRAS and proposed to contribute to the evasion of oxidative DNA damage in lung cancer cells (29). It was also reported that KRAS mutation in human lung cancer caused radiation resistance by osteopontin-EGFR pathway-enhanced cancer stem cell properties (60,61). Interestingly, it has been shown that KRAS-mutated, but not KRAS wild-type cells, rely on the alternative non-homologous end-joining (alt-NHEJ) pathway on genotoxic stress by increasing the expression of DNA ligase 3α, poly(ADP-ribose) polymerase 1 (PARP1), and X-ray repair cross-complementing protein 1 (XRCC1), all essential components of the error-prone alt-NHEJ pathway (30).
In the present study, we found various KRAS mutations led to radioresistance in isogenic colorectal cancer cells both in vitro and in preclinical animal models. Further functional investigation revealed that KRAS G13D enhanced classic NHEJ, a major DNA double-strand break repair mechanism operating throughout the cell cycle. Therefore, upregulation of DNA damage repair pathways appears to play an important role in the radioresistance of KRAS mutation-driving cancers as a KRAS mutation may harness distinct DNA repair machinery to maintain cancer cell survival after radiation treatment. The heightened dependency of RAS-transformed cells on various DNA repair systems may provide a therapeutic window that could be exploited with drugs that specifically target these pathways.
53BP1 plays an important role in the maintenance of genome stability (45,46). In agreement with a previous study showing that 53BP1 over-expression enhanced ATM-Chk2 checkpoint activation (47), we observed increased ATM and Chk2 phosphorylation after IR in KRAS mutant cells restricted to early timepoints. In addition, we found upregulation of 53BP1 in KRAS mutant cells and accelerated formation and resolution of 53BP1 nuclear foci after IR in KRAS mutant cells. Our findings suggest that KRAS mutant cells may be ‘primed’ for a 53BP1-mediated DDR and DNA repair response after radiation or genotoxins. Furthermore, we revealed that 53BP1 knockdown by RNA interference preferentially radiosensitized KRAS mutant cells both in vitro and in mouse xenograft models. Since 53BP1 translocates to sites of DSBs and promotes NHEJ, the principal repair mechanism for DSBs (46), our results suggest that 53BP1 may be a promising target for the development of radiosensitizers for colorectal cancer cells.
Ionizing radiation induces various kinds of DNA lesions, as well as reactive oxygen species (ROS), to overwhelm the repair capacity of cancer cells (62). There is growing evidence that the NRF2 antioxidant pathway, which protects cells from ROS, also plays an important role in cancer cell radioresistance (63–65). In support of this, it has been shown that RAS mutation results in increased production of ROS and that KRAS-mutant cancers cells depend on NRF2 antioxidant response and subsequent ROS detoxification for survival (49,50). In the present study, we revealed that KRAS mutant cells displayed increased NRF2 expression and elevated binding of NRF2 to the 53BP1 promoter. More importantly, we showed that silencing by siRNA or chemical suppression of NRF2 by brusatol reduced 53BP1 expression and effectively radiosensitized KRAS-mutant (but not KRAS wild-type) tumor cells. Furthermore, radiotherapy activates RAS-MAPK signaling in KRAS-mutant cells (26). Interestingly, we revealed that brusatol also inhibited the activation of RAS-ERK signaling, indicating the potential existence of a feedback signaling loop between RAS-ERK and NRF2 antioxidant response, which warrants further investigation. Therefore, targeting NRF2 may also be a novel strategy for radiosensitization of KRAS mutant cancer cells, in addition to 53BP1. Additional strategies to increase radiation response in KRAS-mutant cells are currently being tested, including targeting MEK-1/2 downstream of KRAS, based on preclinical and more recent clinical data (37,59,66,67).
The DDR determines both carcinogenesis (through improper surveillance and repair of genotoxic insults) and cancer control outcomes after radiotherapy and chemotherapy. Much of our knowledge of DDR machinery comes from studies of human diseases derived from hereditary defects in DNA damage signaling and repair pathways. Alterations of DSB repair pathways affect the sensitivity of cancer cells to IR and can be exploited for effective radiation therapy. For example, cancers with ATM loss are hypersensitive to IR (68,69). However, nonhereditary forms of BRCA1/2-deficient tumors develop resistance to PARP inhibitors due to genetic reversion of DDR defects and rewiring of DDR pathways (70,71). Paradoxically, early studies showed that, although ATM patients are cancer prone and hypersensitive to IR, the DNA is not hypermutated and the cells are proficient in the repair of IR induced DNA damage (72,73). In addition, the rationale for the sensitivity of DDR-mutant cancer cells to DNA-damaging agents has been poorly understood and is often unsatisfactorily attributed to the more rapid growth of tumor cells (69). Recent deep sequencing has actually shown that most DNA damage response genes are overexpressed in most cancer types (TCGA database, unpublished). Our results suggest that, in response to IR, a KRAS activating mutation may produce higher levels of initial DNA damage at early timepoints (see 15 min at Figure 2A) perhaps through higher baseline oncogene-induced replication stress and reactive oxygen species, but that through more efficient DNA repair (i.e. over-active NHEJ) the damage can be more efficiently resolved in KRAS mutant cancer cells to allow cells to survive and continue to grow (see 6–24 h, Figure 2A). It is also possible that this excessive level of initial DNA damage may lead to enhanced activation of ATM and DNA-PK in the initial period after IR in KRAS mutant cells, contributing to a rapid response from radiation damage (Figure 4C and F). Taken together, we found that KRAS mutation resulted in enhanced DNA damage response and heightening of NHEJ-mediated repair activity of DNA strand breaks, perhaps partly mediated by increased cell cycle re-assortment to G1. However, other mechanisms may also contribute to KRAS mutation mediated radioresistance, including, but not limited to, DNA replication stress, base excision repair (BER), alt-NHEJ, chromatin condensation, and the stem cell phenotype which should be further explored (30,60,61,74). Moreover, it should be noted that a significant portion of colon cancer with mutant KRAS also have concurrent TP53 or APC mutation (75), accounting for 50–60% of cases with KRAS mutation (TCGA data). Since both HCT116 and SW48 cells have wild-type TP53, further evaluation of these mechanisms in KRAS mutant cells with concurrent TP53 mutation should be performed in colorectal as well as other cancer types.
In summary, we have elucidated an important mechanistic role for a KRAS-NRF2-53BP1 pathway in facilitating DNA damage repair and survival of KRAS-mutant colorectal cancer cells following radiotherapy. Our findings suggest that targeting NRF2, 53BP1 or NHEJ are novel therapeutic strategies to preferentially radiosensitize KRAS mutant cancers.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Analytical Cytometry at OSU for assistance with flow cytometry experiments. We thank Megan Halloran with assistance in figure generation of the model.
Contributor Information
Linlin Yang, Department of Radiation Oncology, Beckman Research Institute, City of Hope National Medical Center, Duarte, CA 91010, USA.
Changxian Shen, Department of Radiation Oncology, Beckman Research Institute, City of Hope National Medical Center, Duarte, CA 91010, USA.
Adriana Estrada-Bernal, The Ohio State University Wexner Medical Center, Arthur G. James Comprehensive Cancer Center and Richard J. Solove Research Institute, Columbus, OH 43210, USA.
Ryan Robb, The Ohio State University Wexner Medical Center, Arthur G. James Comprehensive Cancer Center and Richard J. Solove Research Institute, Columbus, OH 43210, USA.
Moumita Chatterjee, The Ohio State University Wexner Medical Center, Arthur G. James Comprehensive Cancer Center and Richard J. Solove Research Institute, Columbus, OH 43210, USA.
Nikhil Sebastian, The Ohio State University Wexner Medical Center, Arthur G. James Comprehensive Cancer Center and Richard J. Solove Research Institute, Columbus, OH 43210, USA.
Amy Webb, The Ohio State University Wexner Medical Center, Arthur G. James Comprehensive Cancer Center and Richard J. Solove Research Institute, Columbus, OH 43210, USA.
Xiaokui Mo, The Ohio State University Wexner Medical Center, Arthur G. James Comprehensive Cancer Center and Richard J. Solove Research Institute, Columbus, OH 43210, USA.
Wei Chen, The Ohio State University Wexner Medical Center, Arthur G. James Comprehensive Cancer Center and Richard J. Solove Research Institute, Columbus, OH 43210, USA.
Sunil Krishnan, Mayo Clinic Cancer Center, Jacksonville, FL 32224, USA.
Terence M Williams, Department of Radiation Oncology, Beckman Research Institute, City of Hope National Medical Center, Duarte, CA 91010, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Cancer Society [RSG-17-221-01-TBG]; National Center for Advancing Translational Sciences [KL2TR001068]; NIH [R01 CA198128 to T.M.W.]; NIH [P30 CA016058]. Funding for open access charge: American Cancer Society [RSG-17-221-01-TBG].
Conflict of interest statement. None declared.
REFERENCES
- 1. Russo M., Di Nicolantonio F., Bardelli A.. Climbing RAS, the everest of oncogenes. Cancer Discov. 2014; 4:19–21. [DOI] [PubMed] [Google Scholar]
- 2. Eser S., Schnieke A., Schneider G., Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br. J. Cancer. 2014; 111:817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baines A.T., Xu D., Der C.J.. Inhibition of Ras for cancer treatment: the search continues. Future Med Chem. 2011; 3:1787–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dang C.V., Reddy E.P., Shokat K.M., Soucek L.. Drugging the ‘undruggable’ cancer targets. Nat. Rev. Cancer. 2017; 17:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ostrem J.M., Shokat K.M.. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat. Rev. Drug Discov. 2016; 15:771–785. [DOI] [PubMed] [Google Scholar]
- 6. Samatar A.A., Poulikakos P.I.. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat. Rev. Drug Discov. 2014; 13:928–942. [DOI] [PubMed] [Google Scholar]
- 7. Roberts P.J., Der C.J.. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007; 26:3291–3310. [DOI] [PubMed] [Google Scholar]
- 8. Caunt C.J., Sale M.J., Smith P.D., Cook S.J.. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat. Rev. Cancer. 2015; 15:577–592. [DOI] [PubMed] [Google Scholar]
- 9. Karoulia Z., Gavathiotis E., Poulikakos P.I.. New perspectives for targeting RAF kinase in human cancer. Nat. Rev. Cancer. 2017; 17:676–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fiskus W., Mitsiades N.. B-Raf inhibition in the clinic: present and future. Annu. Rev. Med. 2016; 67:29–43. [DOI] [PubMed] [Google Scholar]
- 11. Bartkova J., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C.et al.. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005; 434:864–870. [DOI] [PubMed] [Google Scholar]
- 12. Brown J.S., O’Carrigan B., Jackson S.P., Yap T.A.. Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov. 2017; 7:20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R.A.. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008; 8:193–204. [DOI] [PubMed] [Google Scholar]
- 14. Lord C.J., Ashworth A.. The DNA damage response and cancer therapy. Nature. 2012; 481:287–294. [DOI] [PubMed] [Google Scholar]
- 15. O’Connor M.J. Targeting the DNA Damage Response in Cancer. Mol. Cell. 2015; 60:547–560. [DOI] [PubMed] [Google Scholar]
- 16. Kastan M.B., Lim D.S.. The many substrates and functions of ATM. Nat. Rev. Mol. Cell Biol. 2000; 1:179–186. [DOI] [PubMed] [Google Scholar]
- 17. Ben-Josef E., Lawrence T.S.. Radiotherapy: the importance of local control in pancreatic cancer. Nat. Rev. Clin. Oncol. 2011; 9:9–10. [DOI] [PubMed] [Google Scholar]
- 18. Sklar M.D. The ras oncogenes increase the intrinsic resistance of NIH 3T3 cells to ionizing radiation. Science. 1988; 239:645–647. [DOI] [PubMed] [Google Scholar]
- 19. Bernhard E.J., McKenna W.G., Hamilton A.D., Sebti S.M., Qian Y., Wu J.M., Muschel R.J.. Inhibiting Ras prenylation increases the radiosensitivity of human tumor cell lines with activating mutations of ras oncogenes. Cancer Res. 1998; 58:1754–1761. [PubMed] [Google Scholar]
- 20. Brunner T.B., Cengel K.A., Hahn S.M., Wu J., Fraker D.L., McKenna W.G., Bernhard E.J.. Pancreatic cancer cell radiation survival and prenyltransferase inhibition: the role of K-Ras. Cancer Res. 2005; 65:8433–8441. [DOI] [PubMed] [Google Scholar]
- 21. Duldulao M.P., Lee W., Nelson R.A., Li W., Chen Z., Kim J., Garcia-Aguilar J.. Mutations in specific codons of the KRAS oncogene are associated with variable resistance to neoadjuvant chemoradiation therapy in patients with rectal adenocarcinoma. Ann. Surg. Oncol. 2013; 20:2166–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mak R.H., Hermann G., Lewis J.H., Aerts H.J., Baldini E.H., Chen A.B., Colson Y.L., Hacker F.H., Kozono D., Wee J.O.et al.. Outcomes by tumor histology and KRAS mutation status after lung stereotactic body radiation therapy for early-stage non-small-cell lung cancer. Clin. Lung Cancer. 2015; 16:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grimminger P.P., Danenberg P., Dellas K., Arnold D., Rodel C., Machiels J.P., Haustermans K., Debucquoy A., Velenik V., Sempoux C.et al.. Biomarkers for cetuximab-based neoadjuvant radiochemotherapy in locally advanced rectal cancer. Clin. Cancer Res. 2011; 17:3469–3477. [DOI] [PubMed] [Google Scholar]
- 24. Russo A.L., Ryan D.P., Borger D.R., Wo J.Y., Szymonifka J., Liang W.Y., Kwak E.L., Blaszkowsky L.S., Clark J.W., Allen J.N.et al.. Mutational and clinical predictors of pathologic complete response in the treatment of locally advanced rectal cancer. J. Gastrointest. Cancer. 2014; 45:34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hong T.S., Wo J.Y., Borger D.R., Yeap B.Y., McDonnell E.I., Willers H., Blaszkowsky L.S., Kwak E.L., Allen J.N., Clark J.W.et al.. Phase II Study of Proton-Based Stereotactic Body Radiation Therapy for Liver Metastases: Importance of Tumor Genotype. J. Natl. Cancer Inst. 2017; 109: 10.1093/jnci/djx031. [DOI] [PubMed] [Google Scholar]
- 26. Williams T.M., Flecha A.R., Keller P., Ram A., Karnak D., Galban S., Galban C.J., Ross B.D., Lawrence T.S., Rehemtulla A.et al.. Cotargeting MAPK and PI3K signaling with concurrent radiotherapy as a strategy for the treatment of pancreatic cancer. Mol. Cancer Ther. 2012; 11:1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Caiola E., Salles D., Frapolli R., Lupi M., Rotella G., Ronchi A., Garassino M.C., Mattschas N., Colavecchio S., Broggini M.et al.. Base excision repair-mediated resistance to cisplatin in KRAS(G12C) mutant NSCLC cells. Oncotarget. 2015; 6:30072–30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramdzan Z.M., Vadnais C., Pal R., Vandal G., Cadieux C., Leduy L., Davoudi S., Hulea L., Yao L., Karnezis A.N.et al.. RAS transformation requires CUX1-dependent repair of oxidative DNA damage. PLoS Biol. 2014; 12:e1001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel A., Burton D.G., Halvorsen K., Balkan W., Reiner T., Perez-Stable C., Cohen A., Munoz A., Giribaldi M.G., Singh S.et al.. MutT Homolog 1 (MTH1) maintains multiple KRAS-driven pro-malignant pathways. Oncogene. 2015; 34:2586–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hahnel P.S., Enders B., Sasca D., Roos W.P., Kaina B., Bullinger L., Theobald M., Kindler T.. Targeting components of the alternative NHEJ pathway sensitizes KRAS mutant leukemic cells to chemotherapy. Blood. 2014; 123:2355–2366. [DOI] [PubMed] [Google Scholar]
- 31. Lee H.J., Cao Y., Pham V., Blackwood E., Wilson C., Evangelista M., Klijn C., Stokoe D., Settleman J.. Ras-MEK signaling mediates a critical Chk1-dependent DNA damage response in cancer cells. Mol. Cancer Ther. 2017; 16:694–704. [DOI] [PubMed] [Google Scholar]
- 32. Anastassiadis T., Brown E.J.. Wild-type RAS: keeping mutant RAS in CHK. Cancer Cell. 2014; 25:137–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grabocka E., Pylayeva-Gupta Y., Jones M.J., Lubkov V., Yemanaberhan E., Taylor L., Jeng H.H., Bar-Sagi D. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell. 2014; 25:243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monzon F.A., Ogino S., Hammond M.E.H., Halling K.C., Bloom K.J., Nikiforova M.N.. The role of KRAS mutation testing in the management of patients with metastatic colorectal cancer. Arch. Pathol. Lab. Med. 2009; 133:1600–1606. [DOI] [PubMed] [Google Scholar]
- 35. Yang L., Shen C., Pettit C.J., Li T., Hu A.J., Miller E.D., Zhang J., Lin S.H., Williams T.M.. Wee1 kinase inhibitor AZD1775 effectively sensitizes esophageal cancer to radiotherapy. Clin. Cancer Res. 2020; 26:3740–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoy C.A., Seamer L.C., Schimke R.T.. Thermal denaturation of DNA for immunochemical staining of incorporated bromodeoxyuridine (BrdUrd): critical factors that affect the amount of fluorescence and the shape of BrdUrd/DNA histogram. Cytometry. 1989; 10:718–725. [DOI] [PubMed] [Google Scholar]
- 37. Estrada-Bernal A., Chatterjee M., Haque S.J., Yang L., Morgan M.A., Kotian S., Morrell D., Chakravarti A., Williams T.M.. MEK inhibitor GSK1120212-mediated radiosensitization of pancreatic cancer cells involves inhibition of DNA double-strand break repair pathways. Cell Cycle. 2015; 14:3713–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seluanov A., Mao Z., Gorbunova V.. Analysis of DNA double-strand break (DSB) repair in mammalian cells. J. Vis. Exp. 2010; 43:e2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Han C., Srivastava A.K., Cui T., Wang Q.E., Wani A.A.. Differential DNA lesion formation and repair in heterochromatin and euchromatin. Carcinogenesis. 2016; 37:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R.A.. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008; 8:193–204. [DOI] [PubMed] [Google Scholar]
- 41. Yard B.D., Adams D.J., Chie E.K., Tamayo P., Battaglia J.S., Gopal P., Rogacki K., Pearson B.E., Phillips J., Raymond D.P.et al.. A genetic basis for the variation in the vulnerability of cancer to DNA damage. Nat. Commun. 2016; 7:11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu Y., Liu Y., Yang C.. Evaluating in vitro DNA damage using comet assay. J. Vis. Exp. 2017; 128:e6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pawlik T.M., Keyomarsi K.. Role of cell cycle in mediating sensitivity to radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2004; 59:928–942. [DOI] [PubMed] [Google Scholar]
- 44. Sinclair W.K. Cyclic X-ray responses in mammalian cells in vitro. 1968. Radiat. Res. 2012; 178:AV112–AV124. [DOI] [PubMed] [Google Scholar]
- 45. Zimmermann M., de Lange T.. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014; 24:108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goodwin J.F., Knudsen K.E.. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov. 2014; 4:1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ward I.M., Minn K., van Deursen J., Chen J.. Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 2003; 23:2556–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim S.B., Pandita R.K., Eskiocak U., Ly P., Kaisani A., Kumar R., Cornelius C., Wright W.E., Pandita T.K., Shay J.W.. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2949–E2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hayes J.D., Dinkova-Kostova A.T.. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014; 39:199–218. [DOI] [PubMed] [Google Scholar]
- 50. DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S.et al.. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011; 475:106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perera R.M., Bardeesy N.. Cancer: when antioxidants are bad. Nature. 2011; 475:43–44. [DOI] [PubMed] [Google Scholar]
- 52. Chang Y.Y., Lin P.C., Lin H.H., Lin J.K., Chen W.S., Jiang J.K., Yang S.H., Liang W.Y., Chang S.C.. Mutation spectra of RAS gene family in colorectal cancer. Am. J. Surg. 2016; 212:537–544. [DOI] [PubMed] [Google Scholar]
- 53. Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. De Roock W., Claes B., Bernasconi D., De Schutter J., Biesmans B., Fountzilas G., Kalogeras K.T., Kotoula V., Papamichael D., Laurent-Puig P.et al.. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010; 11:753–762. [DOI] [PubMed] [Google Scholar]
- 55. Cunningham D., Humblet Y., Siena S., Khayat D., Bleiberg H., Santoro A., Bets D., Mueser M., Harstrick A., Verslype C.et al.. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N. Engl. J. Med. 2004; 351:337–345. [DOI] [PubMed] [Google Scholar]
- 56. Rosell R., Gonzalez-Larriba J.L., Alberola V., Molina F., Monzo M., Benito D., Perez J.M., de Anta J.M.. Single-agent paclitaxel by 3-hour infusion in the treatment of non-small cell lung cancer: links between p53 and K-ras gene status and chemosensitivity. Semin. Oncol. 1995; 22:12–18. [PubMed] [Google Scholar]
- 57. Samouelian V., Maugard C.M., Jolicoeur M., Bertrand R., Arcand S.L., Tonin P.N., Provencher D.M., Mes-Masson A.M.. Chemosensitivity and radiosensitivity profiles of four new human epithelial ovarian cancer cell lines exhibiting genetic alterations in BRCA2, TGFbeta-RII, KRAS2, TP53 and/or CDNK2A. Cancer Chemother. Pharmacol. 2004; 54:497–504. [DOI] [PubMed] [Google Scholar]
- 58. Wang M., Han J., Marcar L., Black J., Liu Q., Li X., Nagulapalli K., Sequist L.V., Mak R.H., Benes C.H.et al.. Radiation resistance in KRAS-mutated lung cancer is enabled by stem-like properties mediated by an Osteopontin-EGFR pathway. Cancer Res. 2017; 77:2018–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu C., Williams T.M., Robb R., Webb A., Wei L., Chen W., Mikhail S., Ciombor K.K., Cardin D.B., Timmers C.et al.. Phase I trial of trametinib with neoadjuvant chemoradiation in patients with locally advanced rectal cancer. Clin. Cancer Res. 2020; 26:3117–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang M., Han J., Marcar L., Black J., Liu Q., Li X.Y., Nagulapalli K., Sequist L.V., Mak R.H., Benes C.H.et al.. Radiation resistance in KRAS-mutated lung cancer is enabled by stem-like properties mediated by an Osteopontin-EGFR pathway. Cancer Res. 2017; 77:2018–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang M., Kern A.M., Hulskotter M., Greninger P., Singh A., Pan Y.F., Chowdhury D., Krause M., Baumann M., Benes C.H.et al.. EGFR-mediated chromatin condensation protects KRAS-mutant cancer cells against ionizing radiation. Cancer Res. 2014; 74:2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bala M Concerted action of Nrf2-ARE pathway, MRN complex, HMGB1 and inflammatory cytokines - implication in modification of radiation damage. Redox. Biol. 2014; 2:832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jayakumar S., Pal D., Sandur S.K.. Nrf2 facilitates repair of radiation induced DNA damage through homologous recombination repair pathway in a ROS independent manner in cancer cells. Mutat. Res. 2015; 779:33–45. [DOI] [PubMed] [Google Scholar]
- 65. Sekhar K.R., Freeman M.L.. Nrf2 promotes survival following exposure to ionizing radiation. Free Radic. Biol. Med. 2015; 88:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chung E.J., Brown A.P., Asano H., Mandler M., Burgan W.E., Carter D., Camphausen K., Citrin D. In vitro and in vivo radiosensitization with AZD6244 (ARRY-142886), an inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase 1/2 kinase. Clin. Cancer Res. 2009; 15:3050–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Urick M.E., Chung E.J., Shield W.P. 3rd, Gerber N., White A., Sowers A., Thetford A., Camphausen K., Mitchell J., Citrin D.E. Enhancement of 5-fluorouracil-induced in vitro and in vivo radiosensitization with MEK inhibition. Clin. Cancer Res. 2011; 17:5038–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Goldstein M., Kastan M.B.. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 2015; 66:129–143. [DOI] [PubMed] [Google Scholar]
- 69. Jeggo P.A., Pearl L.H., Carr A.M.. DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer. 2016; 16:35–42. [DOI] [PubMed] [Google Scholar]
- 70. Swisher E.M., Sakai W., Karlan B.Y., Wurz K., Urban N., Taniguchi T.. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008; 68:2581–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Edwards S.L., Brough R., Lord C.J., Natrajan R., Vatcheva R., Levine D.A., Boyd J., Reis-Filho J.S., Ashworth A.. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008; 451:1111–1115. [DOI] [PubMed] [Google Scholar]
- 72. Taylor A.M., Harnden D.G., Arlett C.F., Harcourt S.A., Lehmann A.R., Stevens S., Bridges B.A.. Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature. 1975; 258:427–429. [DOI] [PubMed] [Google Scholar]
- 73. Shiloh Y., Tabor E., Becker Y.. Cellular hypersensitivity to neocarzinostatin in ataxia-telangiectasia skin fibroblasts. Cancer Res. 1982; 42:2247–2249. [PubMed] [Google Scholar]
- 74. Al Zubaidi T., Gehrisch O.H.F., Genois M.M., Liu Q., Lu S., Kung J., Xie Y., Schuemann J., Lu H.M., Hata A.N.et al.. Targeting the DNA replication stress phenotype of KRAS mutant cancer cells. Sci. Rep. 2021; 11:3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nakayama M., Oshima M.. Mutant p53 in colon cancer. J. Mol. Cell Biol. 2019; 11:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.