Abstract
microRNAs are frequently modified by addition of untemplated nucleotides to the 3′ end, but the role of this tailing is often unclear. Here we characterize the prevalence and functional consequences of microRNA tailing in vivo, using Caenorhabditis elegans. MicroRNA tailing in C. elegans consists mostly of mono-uridylation of mature microRNA species, with rarer mono-adenylation which is likely added to microRNA precursors. Through a targeted RNAi screen, we discover that the TUT4/TUT7 gene family member CID-1/CDE-1/PUP-1 is required for uridylation, whereas the GLD2 gene family member F31C3.2—here named GLD-2-related 2 (GLDR-2)—is required for adenylation. Thus, the TUT4/TUT7 and GLD2 gene families have broadly conserved roles in miRNA modification. We specifically examine the role of tailing in microRNA turnover. We determine half-lives of microRNAs after acute inactivation of microRNA biogenesis, revealing that half-lives are generally long (median = 20.7 h), as observed in other systems. Although we observe that the proportion of tailed species increases over time after biogenesis, disrupting tailing does not alter microRNA decay. Thus, tailing is not a global regulator of decay in C. elegans. Nonetheless, by identifying the responsible enzymes, this study lays the groundwork to explore whether tailing plays more specialized context- or miRNA-specific regulatory roles.
INTRODUCTION
microRNAs (miRNAs) are small non-coding RNAs that bind Argonaute (Ago) proteins to form the miRNA-induced silencing complex (miRISC), which represses complementary target mRNAs (1). miRNA-target interactions are important in sculpting gene regulation for normal physiology (2).
Canonical miRNAs are derived from longer primary transcripts (pri-miRNAs) from which a hairpin-like secondary structure is recognized and excised by the Microprocessor complex, made up of the ribonuclease Drosha and its protein cofactor DGCR8/Pasha (3). This excised hairpin, known as the miRNA precursor (or pre-miRNA), is exported to the cytoplasm where its loop is cleaved by Dicer (3). One strand of the resulting miRNA duplex (the ‘guide’ strand) is then preferentially loaded into Ago, and the passenger strand (‘star’ strand) is degraded (3).
In contrast to biogenesis, the process of miRNA turnover is poorly understood, despite turnover's equivalent importance in determining miRNA abundance. In general, miRNA half-lives are long, on the order of ∼20 h, with some miRNAs exhibiting characteristically shorter half-lives (4–8). Certain physiological signals and developmental contexts can accelerate miRNA turnover by poorly understood mechanisms, such as neuronal activity which globally destabilizes miRNAs in the retina (9).
Untemplated nucleotide additions to the 3′ end (‘tailing’) can regulate both biogenesis and decay of miRNAs through multiple mechanisms. These modifications are carried out by a class of enzymes known as terminal nucleotidyl transferases (TENTs), which also have other roles in regulating mRNAs and viral genomes (10,11).
The most well-understood examples of tailing regulating miRNAs involve uridylation of miRNA precursors. This type of tailing generally impacts atypical precursors that lack the canonical 2-nt single-stranded 3′ overhang which is generated by Microprocessor cleavage and recognized by Dicer (12). The type of uridylation and its consequence depends on the length of the 3′ overhang, where 1-nt overhangs are mono-uridylated to favor biogenesis and further recessed ends are oligo-uridylated resulting in decay of the precursor (12). Precursor uridylation can also regulate biogenesis by shifting the site of Dicer cleavage, which can result in alternative miRNA guide strand selection (arm-switching) (13). Oligo-uridylation and decay of the let-7 precursor are induced even in the context of a canonical 2-nt 3′ overhang by binding of the RNA-binding protein LIN-28 (14,15). Finally, non-canonical Microprocessor-independent miRNA precursors derived from short introns (mirtron precursors) are also highly uridylated, inhibiting their biogenesis in Drosophila (16,17).
Tailing can also occur on mature miRNAs, again with a wide variety of outcomes. During embryonic development of Drosophila, fish, and mice, terminal adenylation of maternally-contributed miRNAs induces sequence-non-specific miRNA decay at the maternal to zygotic transition (18). In contrast to the clearance of adenylated maternally-contributed miRNAs, terminal adenylation of mature miRNAs is stabilizing and activating for miR-122 and a subset of other miRNAs (19–21). Through a process known as target-directed miRNA degradation (TDMD), target RNAs destabilize miRNAs with extensive complementarity, but high rates of tailing observed in TDMD are apparently dispensable to this destabilization (22–35).
Beyond effects on stability, 3′ miRNA tailing can even alter target repertoire (36). Overall, tailing of miRNA precursors and mature miRNAs has a wide range of regulatory consequences. How tailing is coupled to these various outcomes in a miRNA- and context-dependent manner is not understood.
In this work, we set out to examine the role of tailing in miRNA turnover in a model organism in vivo. We first use deep sequencing and meta-analysis of published data to characterize miRNA tailing in C. elegans. We perform a candidate RNAi screen by deep sequencing to discover the enzymes responsible for tailing, demonstrating that terminal uridylyltransferase 4 (TUT4)/TUT7 ortholog caffeine induced death homolog 1 (CID-1) is required for U-tailing, and defective in germ line development 2 (GLD2) ortholog F31C3.2 – here named GLD-2-related 2 (GLDR-2) – is responsible for A-tailing. Finally, we use acute inactivation of miRNA biogenesis to measure miRNA turnover and show that, while U-tailing increases as miRNAs age, this is correlative not causative of miRNA turnover. Overall, this work importantly defines ancestral functions of TENT families that modify miRNAs, and—by defining the required enzymes—lays the groundwork for future studies to determine whether miRNA tailing in Caenorhabditis elegans may have context-dependent or miRNA-specific functions.
MATERIALS AND METHODS
C. elegans maintenance and RNAi
N2 (wild type) and SX1137 (pash-1(mj100) I) worms were maintained at 15°C. For RNA samples, L1s were synchronized by alkaline hypochlorite lysis of gravid adults followed by hatching in M9 with 1 mM cholesterol at 15°C for 48–72 h. L1s were plated on NGM seeded with either OP50 or RNAi bacteria and maintained at 15°C until early day one of gravid adulthood (until just a few embryos were laid on the plate, ∼76 h after plating). (This time point was chosen because it achieved the best synchrony of SX1137, which tends to develop slightly asynchronously, even at permissive temperature). This time point was harvested for N2 samples, SX1137 samples in the RNAi screen, and served as 0h in time courses involving SX1137. Additional SX1137 plates were shifted to 25°C, and samples were collected at the indicated time points after upshift for time courses on OP50 or RNAi. pup-1(tm1021), pup-2(tm4344) and pup-1/-2(om129) mutations were maintained over a balancer chromosome at 15–20°C; RNA was isolated from homozygous F2 (M-Z-) 1-day adult hermaphrodites raised at 22°C.
RNAi plates were supplemented with 1μg/ml IPTG and 1μg/ml Carbenicillin (and poured no more than two weeks prior to use and stored at 4°C). The RNAi bacteria (HT115 expressing an RNAi vector) were prepared from single clones derived from the Ahringer library (Source Bioscience), the Vidal library (Source Bioscience), or custom RNAi vectors (Supplementary Table S1). All previously-reported RNAi phenotypes were observed. Furthermore, a robust RNAi response in each sample was confirmed by the presence of abundant siRNAs mapping to the targeted locus.
Small RNA sequencing
Total RNA was isolated by Trizol extraction. Spike-ins were added at a final concentration of 0.1 pg/μl (OP50 samples) or 1 pg/μl (RNAi samples) in 100 ng/μl total RNA. See Supplementary Table S2 for spike-in information. Libraries were prepared using the NEBNext Small RNA Library Prep set for Illumina with the following modifications. The first size selection was performed after the RT reaction by excising 65–75nt cDNA from an 8% denaturing acrylamide gel. PCR products were again size selected using a Pippin prep or 6% native acrylamide gel. Input RNA and final library sample quality were assessed using a Bioanalyzer. Equimolar amounts of each library were pooled; up to 24 indexed samples were pooled together for sequencing. For pup-1(tm1021), pup-2(tm4344) and pup-1/-2(om129) libraries, ∼18–35 bp inserts were size-selected. Sequencing was performed on a NextSeq500 with a minimum of 10M single-end reads of minimum length 50 nt.
Data analysis
Deep sequencing data were analyzed using the miTRATA web interface (37) after preprocessing the data using miTRATA’s accompanying Python3-based preprocess.seq pipeline, installed on the NIH High Performance Computing Cluster in a dedicated miniconda environment. The mature C. elegans miRNAs from miRBase Release 22.1 (38) were used for miTRATA mapping. A custom C++ script reformatted the miTRATA output by consolidating read numbers for all reads meeting the following criteria into a tabular format: a tail:head ratio >0.12 (16) and a tail length ≤3nt. These reformatted tailing data were further analyzed in RStudio. (One non-unique miRNA pair was of sufficient abundance to be included in tailing analysis—mir-44/45-3p; mir-45-3p was therefore culled from tabular data to avoid plotting this data twice). To calculate reads per million, reads were normalized by the number of genome-mapping reads in the library (Supplementary Table S3). The number of genome-mapping reads was determined using Bowtie (39) to align to the C. elegans genome (WS215), with the arguments -v 3 -f -B 1 -a –best –strata. Alignments were then filtered based on the length of the reads and the number of mismatches as follows: for sequence lengths 16–17, 18–19, 20–24 or >24: zero, one, two or three mismatches were allowed, respectively. Reads passing this threshold were considered ‘genome-mapping reads’. A custom bash/R pipeline was used to calculate trimming relative to canonical miRNA length from the miTRATA output files.
Statistical analysis
Fisher's exact test for enrichment of tailing on 3p- or 5p-derived miRNAs was performed by counting miRNAs as above or below 1% tailed, and pooling these counts from three biological replicates to generate the contingency table, which was analyzed in GraphPad QuickCalcs. For the RNAi screen, GraphPad Prism v8.4.3 was used to perform one-way repeated measures ANOVA followed by Dunnett's multiple comparison test. All RNAi conditions were compared to an empty vector sample, and this comparison was repeated for each empty vector sample. RNAi conditions that were significant in comparison to each empty vector sample are highlighted, with the least significant P-value reported. All miRNAs >50 RPM in all empty vector replicates were analyzed. To determine miRNA half-lives, for each miRNA with >50 RPM at 0h in both replicates, fold change at each time point relative to 0h was calculated using spike-in-normalized reads. GraphPad Prism v8.4.3 was used to fit a one phase decay non-linear regression model with the following constraints: Y0 = 1, Plateau = 0, K > 0.
Phylogenetic analysis
MEGA-X (40) was used to build phylogenetic trees of orthologous genes (to CID-1 and PUP-2, or to GLD-2 and F31C3.2) identified in WormBase ParaSite version WBPS15 (41). Orthologs from select non-nematode species were included as shown (Supplementary Figures S7 and S9). The classification of nematode species into clades was downloaded from the same WormBase Parasite version WBPS15. Additional trees containing only C. elegans genes and common model organisms are shown in Figures 2 and 3. All alignments were generated using Clustal W (Multiple Alignment Gap Opening Penalty = 10, Gap Extension Penalty = 0.2, Negative Matrix Off, Delay Divergent Cutoff 30%). Maximum Likelihood method was used in MEGA-X, with no test of phylogeny, JTT model of substitution, uniform rates among sites, no subsetting (‘use all sites’), NNI, automatic initial tree, and no branch swap filter.
Figure 2.
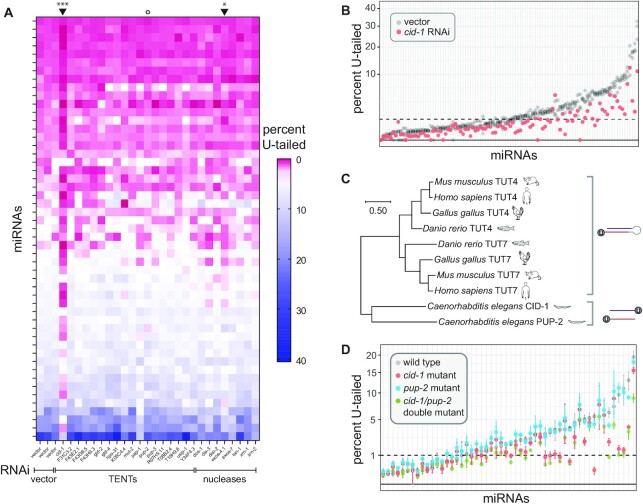
miRNA uridylation requires CID-1. (A) Heatmap summarizing percent of mono-uridylation across each miRNA (rows) in each RNAi condition (columns). All miRNAs with >50 RPM in all empty vector replicates and an average of > 1% mono-uridylation across the three empty vector replicates are shown. Arrowheads above heatmap indicate columns significantly different than vector (cid-1 and exos-4 RNAi). ***P-value = 0.0003, *P-value = 0.016. Open circle indicates pup-2 RNAi, which does not have an effect. (B) Percent mono-uridylation in vector or cid-1 RNAi. Each column is an individual miRNA. All miRNAs with > 50 RPM in all empty vector replicates are shown. Three biological replicates of empty vector are shown in gray. (C) Phylogenetic relationship of CID-1, PUP-2, TUT4 and TUT7. (Note the following alternative names for these genes CID-1/CDE-1/PUP-1, PUP-2, TUT4/TENT3A/ZCCHC11/PAPD3 and TUT7/TENT3B/ZCCHC6/PAPD6). (D) Percent mono-uridylation in indicated genotype. All miRNAs with >50 RPM in all wild type replicates are shown. Mean and standard error are shown for three biological replicates per genotype.
Figure 3.
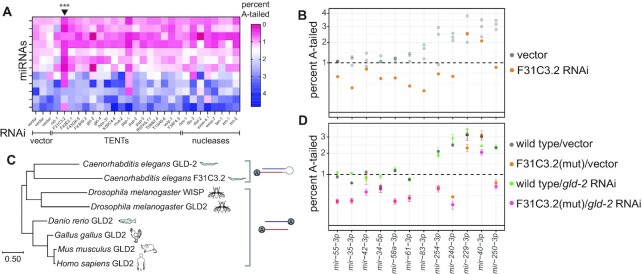
miRNA adenylation requires F31C3.2/GLDR-2. (A) Heatmap summarizing percent of mono-adenylation across each miRNA (rows) in each RNAi condition (columns). Arrowhead above heatmap indicates column significantly different from vector, corresponding to F31C3.2 RNAi, which reduces adenylation. ***P-value = 0.0003 (B) Percent mono-adenylation in vector or F31C3.2/GLDR-2 RNAi. Three biological replicates of empty vector are shown in gray. (A, B) All miRNAs with >50 RPM in all empty vector replicates and an average of >1% mono-adenylation across the three empty vector replicates are shown. (C) Phylogenetic tree of F31C3.2/GLDR-2, GLD-2 and WISP. (Note that GLD2 in humans is also known as TENT2/TUT2/PAPD4/APD4). (D) Percent mono-adenylation in indicated genotypes/RNAi. Mean and standard error for four biological replicates are shown. MiRNAs shown are the same as those in (A, B).
Genome editing
To generate the F31C3.2/gldr-2(cdb187) mutant, adult N2 hermaphrodites were injected with Cas9/gRNA RNPs to perform CRISPR. The gRNAs (gKFV05 and gKFV06) were designed to cleave within 300nt upstream and downstream of the F31C3.2/gldr-2 ORF and ordered from IDT as Alt-R crRNAs. Both guides were used at 2 μM in the injection mix. 500 ng/μl of oKFV101 (Supplementary Table S2) was used as a ssDNA oligo repair donor to produce a null deletion mutant. The final injection mix also included 2 μM IDT Cas9 as well as 2 μM of a dpy-10 gRNA and 820 nM of a dpy-10 ssDNA oligo repair donor to generate a Dpy and Rol selection marker (42). See Supplemental Supplementary Table S2 for gRNA, donor, and genotyping primer sequence details.
Transgenesis and TDMD sample preparation
Constructs for TDMD triggers were generated using pCFJ420 (myo-3p::gfp::h2b::tbb-2 terminator) as a backbone (43). For each trigger, sequences with high complementarity to the miRNAs of interest were ordered as complementary oligonucleotides, annealed at 12.5 μM each in IDT duplex buffer, digested with BsmI, and cloned into the tbb-2 3′-UTR region of pCFJ420 (also digested with BsmI). See Supplemental Supplementary Table S2 and Supplementary Figure S15 for sequence details. The resulting plasmids (pKFV18, pKFV19 and pKFV20) were then injected into adult N2 hermaphrodite gonads to generate extra-chromosomal arrays. Injection mixes included pKFV18, pKFV19, pKFV20 or pCFJ420 (no-binding site negative control) along with the following selection markers: pDD282 (for Hygromycin resistance), pRF4 (for Rol phenotype) and mCherry expression plasmids (pGH8, pCFJC90 and pCFJ104) (44,45). See Supplementary Table S2 for injection mix details. Two to three stably transmitting arrays were isolated for each injected construct. To generate samples for deep sequencing, mixed populations were bleached in sodium hypochlorite and synchronized overnight in M9 supplemented with 0.3 mg/ml Hygromycin to select for worms carrying the extra-chromosomal array, then grown at 20°C on standard NGM seeded with OP50 until early day one of gravid adulthood. miRNA-Taqman assays (ThermoFisher Scientific) were performed according to manufacturer's instructions, using synthetic miRNAs to generate a standard curve for absolute quantification. Digital droplet PCR (ddPCR) was performed as a two-step RT-qPCR by first synthesizing oligo(dT) primed cDNA using the iScript™ Select cDNA Synthesis Kit (Biorad), then running ddPCR via the standard EvaGreen program on a Biorad QX200 ddPCR system and analyzing in Quantasoft software (see Supplementary Table S2 for primer sequences).
RESULTS
Patterns of miRNA tailing in C. elegans suggest different substrates than in flies and vertebrates
To determine the nature and prevalence of miRNA tailing in C. elegans, we first cloned and deep sequenced small RNAs from wild type animals. To ensure reliable measurements of tailing, only miRNAs with >50 reads per million were included in tailing analysis. To determine the level of background in our sequencing and computational pipeline, ten synthetic miRNAs (whose sequences are endogenous to plants and not present in C. elegans—Supplementary Table S2) were spiked in to purified total RNA before preparation for deep sequencing. Although these RNA species were never present in the context of cellular lysate, up to 1% tailing was detected; therefore, tailing levels measured <1% (delineated by dashed line on all tailing plots) are low confidence and could arise due to PCR or sequencing errors.
Most tailing consisted of a single nucleotide addition (Figure 1A). Tails of two or more nucleotides were much rarer, and so we focused our analysis on further characterization of the single-nucleotide tails. Three replicates of adult animals showed strong correlation in proportions of tailing (Supplementary Figure S1). Modest levels of miRNA tailing were observed, and tailing in C. elegans consisted primarily of uridylation with a much lower frequency of adenylation (Figure 1B, Supplementary Table S4). This is in contrast to humans and flies, where both adenylation and uridylation are prevalent (46–49). In addition, a single miRNA, mir-83-3p, was highly cytidylated in all replicates (Figure 1B, Supplementary Figure S1), a modification that is rarely observed in animals, but was recently reported as a frequent addition to plant miRNA precursors (50).
Figure 1.
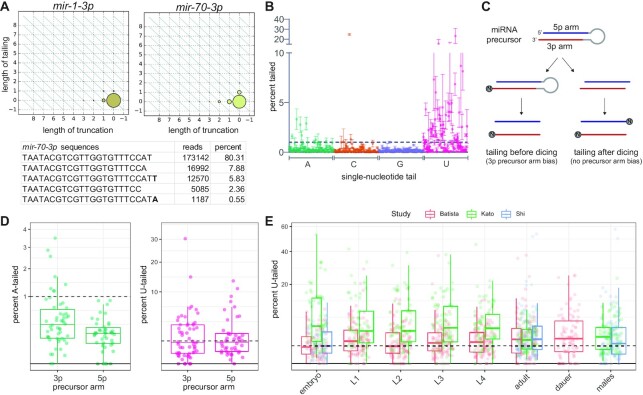
Characterization of miRNA tailing in C. elegans. (A) 2D matrix representing the truncation and tailing status of the indicated miRNAs. The sizes of the dots represent the proportion of reads, with the canonical sequence at zero on both axes, and increasingly trimmed or tailed species plotted to the left on the x-axis or upward on the y-axis, respectively. Bottom: Number of reads of most abundant mir-70-3p species in one wild type adult library, with the canonical sequence being the most abundant. (B) Mean and standard deviation of prevalence of single nucleotide 3′ terminal additions of each indicated nucleotide from three biological replicates. (C) Schematic of miRNA tailing preceding or following Dicer-mediated cleavage of the miRNA precursor. (D) Comparison of prevalence of mono-adenylation and mono-uridylation on 3p- versus 5p-derived miRNAs. One biological replicate is shown. (E) Meta-analysis of three published datasets shows tailing across C. elegans developmental stages. Prevalence of mono-uridylation of miRNAs across development is shown. Only miRNAs with >50 RPM in the indicated library were analyzed. (B, D) Only miRNAs with >50 RPM in all biological replicates were analyzed. (B, D, E) Each dot represents an individual miRNA.
Either the mature miRNA species or the miRNA precursor may be the substrate of the observed tailing. If the vast majority of observed tailing events occur on miRNA precursors, then miRNAs derived from the 3′ arm of the precursor hairpin (3p) would be expected to bear more terminal additions than those derived from the 5′ arm of the hairpin (5p) since the 3′ end of the 5p arm is not accessible for tailing prior to Dicer-mediated cleavage (Figure 1C). In vertebrates and Drosophila, miRNA precursors are the primary substrate of uridylation, though mature miRNAs can also be uridylated (25,46–48,51). In contrast, 3p and 5p miRNAs displayed similar levels of uridylation in C. elegans (Figure 1D), indicating that U-tailing likely predominantly occurs on mature miRNA species. Untemplated adenylation generally occurs on mature miRNAs in vertebrates, and a modest 3p bias suggests that both precursor and mature miRNAs are adenylation substrates in Drosophila (46–48). In contrast, A-tailing in C. elegans shows a strong bias, occurring primarily on 3p-derived miRNAs (Figure 1D); miRNAs displaying above 1% A-tailing were significantly enriched in the 3p-derived class of miRNAs (Fisher's exact test P-value < 0.0001). This suggests that A-tailing in C. elegans occurs predominantly on miRNA precursors.
Because global developmentally-regulated tailing has been demonstrated in other models (18), we examined published datasets from developmental time courses to determine whether tailing of C. elegans miRNAs is correlated with any particular developmental stage (52–54). No consistent global differences in rates of uridylation or other types of tailing were observed (Figure 1E and Supplementary Figure S2A, Supplementary Table S5). We were especially interested in oocyte and early embryo samples since miRNA adenylation is highly prevalent in these time points and leads to turnover of maternally-contributed miRNAs in other species (18), but neither adenylation nor uridylation was globally dynamic during the course of early embryonic development in C. elegans (Supplementary Figure S2B, Supplementary Table S5) (55). When examining the most highly tailed miRNAs from our data (Figure 1B), these miRNAs were largely expressed fairly evenly across development, with the exception of the embryo-enriched mir-35-42 family of miRNAs (of which one member was among the top 12 U-tailed miRNAs and three were among the top 12 A-tailed miRNAs) (Supplementary Figure S3). When examining the tailing of the highly tailed miRNAs across development, tailing was approximately equally prevalent across development (Supplementary Figure S4).
miRNA uridylation requires CID-1
To determine what role tailing may play in regulating miRNAs in C. elegans, we next set out to identify the enzymes responsible for these modifications. To this end, we performed RNAi against a list of candidate terminal nucleotidyl transferases (TENTs). For each knockdown, we performed deep sequencing of small RNAs, and compared the proportion of tailed isoforms to those in three biological replicates of empty vector. Of RNAi against 18 candidate tailing enzymes, only the RNAi targeting cid-1 significantly abrogated miRNA U-tailing (P-value = 0.0003, Figure 2A-B, Supplementary Figure S5, Supplementary Table S6), and this was reproduced in independent experiments (Supplementary Figure S6A). Knockdown of cid-1 decreased U-tailing of both 3p- and 5p-derived miRNAs equally, suggesting that CID-1 acts on mature miRNAs (Supplementary Figure S6B). Though cid-1(lf) was previously reported to reduce miRNA U-tailing (56), our study allows for the comparison of the relative contributions of all putative TENTs, demonstrating that other TENTs are not required. In particular, knockdown of pup-2, a paralog of cid-1, does not affect miRNA terminal uridylation (Supplementary Figure S5).
Both CID-1 (also known as PUP-1 or CDE-1) and PUP-2 are orthologs of TUT4 and TUT7, which act redundantly, along with TENT2/TUT2/GLD2, to uridylate miRNA precursors in mammals (12,15,57). The common ancestor of nematodes and vertebrates likely contains only one TUT4/7 family gene, and a recent duplication occurred in each lineage to give rise to two paralogs (Figure 2C) (10). The duplication in nematodes likely occurred in the common ancestor of Rhabditina or earlier (Supplementary Figure S7). In contrast, the Drosophila lineage has lost this gene family (10). Overall, the role of CID-1 demonstrates that the function of the TUT4/7 gene family in uridylation of miRNA species is deeply conserved, though the substrate specificity shows a different bias (preferentially targeting mature miRNAs rather than precursors in C. elegans) (Figure 2C).
To evaluate any redundancy between CID-1 and PUP-2, we assessed the U-tailing of miRNAs in individual knock-out mutants of cid-1 and pup-2 and a double cid-1/pup-2 mutant. Three biological replicates of wild type and the single and double mutants were profiled by deep sequencing. The data from single mutant animals recapitulated the results from our RNAi screen. U-tailing was strongly disrupted in the cid-1(tm1021) mutant but not in the pup-2(tm4344) mutant (Figure 2D, Supplementary Table S7). In the cid-1/pup-2(om129) double mutant, we observe a decrease in U-tailing that in most cases is comparable to that of the single cid-1(tm1021) mutant (Figure 2D, Supplementary Table S7). This reaffirms the role of cid-1 in U-tailing and suggests very little redundancy in its function with pup-2. Although CID-1 and PUP-2 are redundant in their biological function of protecting germline fate (58), they do not appear to be redundant for this molecular function. CID-1 and PUP-2 are much less similar (17.4%) than human TUT4 and TUT7 (44.5%), consistent with the lack of redundancy of CID-1 and PUP-2 for miRNA uridylation in C. elegans.
miRNA adenylation requires F31C3.2/GLDR-2
We also analyzed the RNAi screen of putative TENTs to identify enzymes required for adenylated miRNA species, with the goal of assessing whether adenylation affects processing or stability of miRNAs. Because of the generally low level of A tailing and the noise inherent in tailing levels <1%, we highlight those miRNAs that were >1% adenylated in empty vector samples in Figure 3. Knockdown of F31C3.2 reduced A-tailing of these miRNAs (P-value = 0.0003, Figure 3A-B, Supplementary Figure S8) in multiple replicates (Supplementary Figure S6C).
F31C3.2 is in the TENT2/GLD2 gene family of non-canonical cytoplasmic poly(A) polymerases, and we therefore name it GLD-2-related 2 (GLDR-2) (Figure 3C). The common ancestor of Bilateria likely had a single GLD2 gene, and duplications have occurred in the Nematoda lineage (GLD-2 and F31C3.2/ GLDR-2) and the Drosophila lineage (GLD2 and WISPY) (10). The nematode duplication likely occurred in the common ancestor of clades III-V or earlier (Supplementary Figure S9). The single GLD2 gene in mammals and the paralog WISPY in Drosophila both catalyze adenylation of mature miRNAs (18,19,21,47).
To determine whether F31C3.2/GLDR-2 has any functional redundancy with its paralog GLD-2, we assessed A-tailing in conditions where both genes are disrupted. We generated a F31C3.2/gldr-2 knockout mutant, F31C3.2/gldr-2(cdb187), using CRISPR-Cas9, deleting the entire ORF of the gene. The mutant progeny were viable and superficially wild type. Because disruption of gld-2 disrupts germline development and generates infertile animals (59), we used RNAi to knock down gld-2 in a wild type or F31C3.2/gldr-2(cdb187) mutant background. Four biological replicates of each genotype/RNAi condition were deep sequenced. Results of single gene disruptions recapitulated those of the RNAi screen. A-tailing was significantly disrupted in the F31C3.2/gldr-2(cdb187) mutant on empty RNAi vector, but not in gld-2 RNAi of wild type worms (Figure 3D, Supplementary Figure S6D, Supplementary Table S8). When gld-2 RNAi was performed in the F31C3.2/gldr-2(cdb187) background, the disruption of A-tailing was comparable to that in the F31C3.2/gldr-2(cdb187) mutant on empty RNAi vector (Figure 3D, Supplementary Figure S6D, Supplementary Table S8). (The efficiency of gld-2 knockdown was evidenced by the induction of the germline defective phenotype and strong induction of siRNAs at the gld-2 locus in all gld-2 RNAi samples). This suggests that there is little to no redundancy between F31C3.2/GLDR-2 and GLD-2 in miRNA tailing in C. elegans.
Here we show that the paralog F31C3.2/GLDR-2—rather than GLD-2 itself—adenylates miRNAs in C. elegans, though the enrichment of this modification on 3p-derived miRNAs (Figure 1D) suggests that precursors rather than mature miRNAs are the substrate of F31C3.2/GLDR-2. Although F31C3.2/GLDR-2 previously had no known function in vivo, a high throughput assay in yeast showed that the enzyme is indiscriminate with respect to substrate nucleotide (60). However, knockdown of F31C3.2/GLDR-2 only affects the prevalence of A tailing (and not C, G or U additions) in our in vivo experiments. Thus, additional factors in the C. elegans cellular context may confer nucleotide substrate specificity to F31C3.2/GLDR-2.
RNAi screen does not identify nucleases targeting tailed miRNAs
In many previous studies, tailed miRNA precursors or tailed mature miRNAs are targeted to specific nucleases (61–64). To determine whether tailed miRNAs are the substrate of a specific nuclease, we also performed RNAi against a panel of nucleases previously implicated in miRNA turnover (4,61,62,65–68). We observed only subtle effects on tailing (Figure 2A, 3A, Supplementary Figures S5 and S8). RNAi against exosome component exos-4.1 showed statistically significant differences in U-tailing compared to the vector control (P-value = 0.016). However, the effect was a slight decrease in tailed species, suggesting an indirect effect of the RNAi. (The direct effect of knocking down an exonuclease that targets tailed miRNAs would be an increase in the prevalence of tailing). Overall, these data suggest that tailed miRNAs may not be specifically targeted to any of the examined nucleases. (Note that PARN-1 was not included since previous studies showed no effect of parn-1(lf) on miRNA tailing or stability in C. elegans (69)). The knockdowns of nucleases displayed both a robust small RNA response mapping to each targeted locus and the expected deleterious phenotypes based on previous studies (70–73). Together this suggests that knockdown was sufficiently potent to impair activity of the enzymes. Since the strong pleiotropic phenotypes induced by these knockdowns introduce concerns of sample synchrony and staging, we did not analyze the impacts of these knockdowns on miRNA abundance. Overall, we have not observed evidence for the role of any of these nucleases in the trimming of tailed miRNAs.
miRNAs are relatively long-lived
Having identified the enzymes required for miRNA tailing in C. elegans, we sought to examine the potential involvement of tailing in regulating miRNA turnover. To this end, we first characterized miRNA turnover in C. elegans by measuring miRNA half-lives using a system in which miRNA biogenesis can be acutely inactivated: a temperature-sensitive allele of the DGCR8 homolog pash-1 (74). Upon upshift to restrictive temperature, pash-1(ts) no longer processes new miRNAs, and the decay of existing miRNAs can be observed over time (Figure 4A) (74,75). To measure miRNA decay, we collected time course samples 0, 6, 24 and 48 h after upshift to restrictive temperature. Because miRNAs comprise a large portion of the small RNA complement and global miRNA levels will change over the time course after pash-1 inactivation, synthetic miRNAs not present in the C. elegans genome were spiked in to the samples at a constant concentration relative to total RNA to allow for robust normalization. Adult animals shifted to restrictive temperature produced a few non-viable progeny and did not proliferate further, so dilution of miRNAs by increasing amounts of total RNA is minimal in this system. After normalization to spike-ins, global decay of miRNAs was observed over the 48 h time course in two biological replicates (Figure 4B, Supplementary Table S9).
Figure 4.
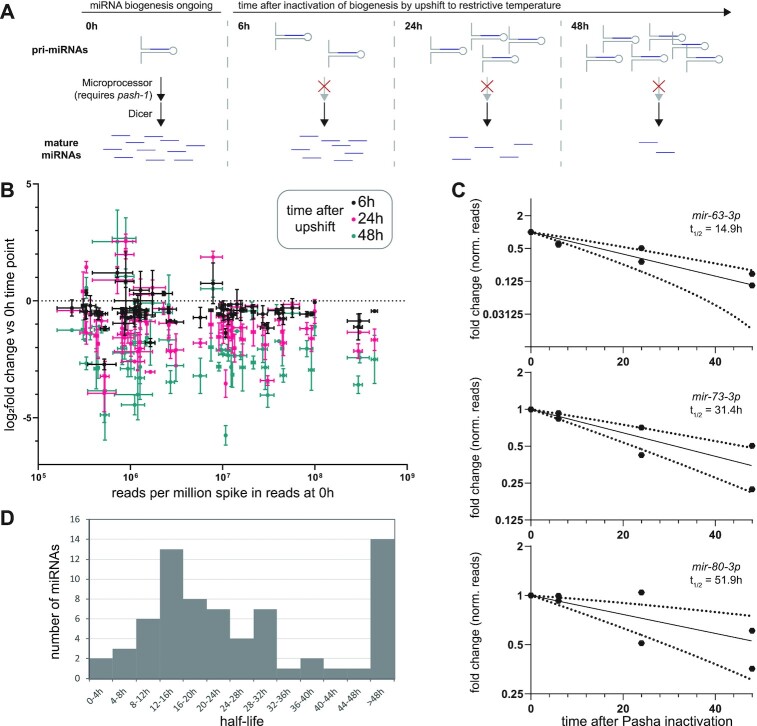
Characterization of miRNA half-lives in C. elegans. (A) Schematic of time courses after pash-1 inactivation used for measuring miRNA decay. (B) Log2 fold change of miRNA reads compared to 0 h after PASH-1 inactivation (y-axis) versus abundance at 0 h (x-axis). Reads were normalized to spike-ins. Mean and standard error of two biological replicates are shown. (C) Representative time course data plotted as fold change, with exponential decay model and 95% prediction bands (dashed lines). Data for two biological replicates is shown. (D) Distribution of miRNA half-lives modeled from pash-1(ts) time course data. (B–D) Only miRNAs with >50 RPM in both replicates at the 0 h timepoint were included in the analysis.
Using spike-in-normalized reads, time course data were fit to exponential decay curves to calculate miRNA half-lives. Half-lives determined using data from only one replicate were well correlated between replicates (Supplementary Figure S10A). Therefore, we used data from both replicates together to fit half-life values with greater confidence. The fit of the decay curves was best when deep sequencing data were normalized to spike-ins, as expected, significantly outperforming normalization to total mapped reads (Supplementary Figure S10B). Data for miRNAs with half-lives longer than the 48 h course of the experiment (14 out of 68 analyzed miRNAs) generally did not fit the exponential decay curve well (Supplementary Figure S10C, Supplementary Table S10). In contrast, shorter-lived miRNAs fit the curve well, with 40 out of 54 miRNAs having a fit with R2 > 0.8 (Figure 4C, Supplementary Figure S10C, Supplementary Table S10). Overall, miRNAs were fairly stable, with a median half-life of 20.7 h (Figure 4D, Supplementary Table S10), similar to most observations in mammalian and Drosophila cells (4,6–8,76). These results using spike-in normalization and deep sequencing yield longer half-lives than a previous study using pash-1(ts) (Supplementary Figure S10D) (74). The previous study employed microarray miRNA quantification and normalization to non-miRNA small RNAs (siRNA and mirtron); when our data were normalized to mirtron reads, we observed a similar range of half-lives to their study (Supplementary Figure S10E). This suggests that mirtron or siRNA normalization does not effectively control for library-wide changes. Nonetheless, both studies highlighted similar sets of fast-decaying miRNAs, with mir-61, mir-71, mir-253 and mir-250 among the six lowest half-lives in both datasets (Supplementary Table S10) (74).
The proportion of tailed and trimmed miRNAs increases over time after biogenesis
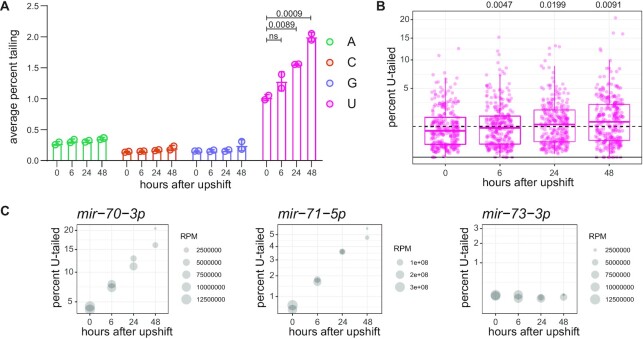
Previous studies have demonstrated that terminal modifications correlate with miRNA turnover (22,23,30,34,35). These studies demonstrated that miRNAs that are targeted for decay display high levels of 3′ tailing. In our datasets, as miRNAs turn over during the time course after PASH-1 inactivation, the ensemble of miRNAs becomes increasingly skewed towards older miRNAs (approaching turnover). To determine whether miRNA terminal modifications are associated (correlatively or causatively) with miRNA turnover in our dataset, we quantified tailing across the time course. We observed that indeed later time points displayed higher levels of uridylation (Figure 5A, B, Supplementary Table S9). This global trend included a large increase in tailing for mir-70–3p and many other miRNAs, although not all miRNAs displayed increased tailing over the time course (Figure 5C, Supplementary Table S9). The increase of U-tailing on aging miRNAs is consistent with the mature miRNA being the substrate of U-tailing as suggested above (Figure 1C-D). No change was observed in adenylation of miRNAs over the time course (Figure 5A), consistent with its substrate being miRNA precursors.
Figure 5.
The proportion of tailed and trimmed miRNAs increases over time after biogenesis. (A) Average percent of reads bearing each indicated single-nucleotide tail. (B) Prevalence of mono-uridylation of miRNAs across time course after PASH-1 inactivation in one biological replicate. Each dot represents an individual miRNA. (A, B) One-way ANOVA was used to compare all time points to 0 h; when ANOVA was significant, Dunnett's multiple comparison test was performed, and P-values are shown above graph. (C) Representative plots for indicated miRNAs, where y-axis indicates percent mono-uridylation, and size of dot represents read number. Note that reads here are reads per million spike-in reads, an arbitrary unit.
Like tailing, 3′ trimming is also often observed concomitant with tailing (22,30). Therefore, we also analyzed trimming over the time course after PASH-1 inactivation. Like tailing, global 3′ trimming increased in later time points, as indicated by a lower proportion of canonical-length reads and increased proportion of shorter genome-matching reads (Supplementary Figure S11). Like tailing, this subtle global trend was more apparent when examining individual miRNAs (Supplementary Figure S11). Thus, both tailing and trimming increase in older miRNAs that are approaching turnover.
If miRNA tailing regulates turnover, two models may explain the increase in U-tailing observed over the time course after PASH-1 inactivation. On one hand, if tailing triggers turnover, then older miRNAs would be tailed as they approach decay, increasing the relative abundance of tailed species over time. On the other hand, if tailing protects miRNAs from decay, this could also result in the relative accumulation of tailed miRNAs at later time points, as non-tailed miRNAs are decayed more rapidly.
We therefore asked whether tailing is correlated (or anti-correlated) with the rate of miRNA turnover. We did not observe an overall correlation between initial prevalence of the U-tailed isoform and half-life for a given miRNA (Supplementary Figure S12A), nor between rate of increase in tailing and half-life (Supplementary Figure S12B). While this indicates that terminal uridylation is unlikely to be the primary determinant of turnover, uridylation might play a modulatory role that is not apparent in bulk analysis due to larger effects of other unknown parameters.
Abrogation of miRNA tailing does not affect miRNA turnover
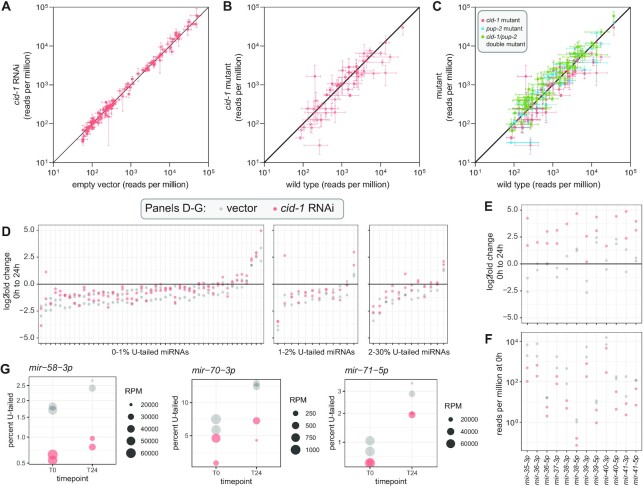
To determine whether uridylation may play a modulatory role - either stimulating or impairing turnover - we sought to disrupt miRNA tailing and then measure rates of miRNA turnover in its absence. In wild type animals, disrupting U-tailing in the context of cid-1 RNAi, cid-1 knockout or cid-1/pup-2 double knockout did not significantly change abundance of miRNAs, suggesting that turnover is unchanged (Figure 6A–C). To further examine the effect of tailing on miRNA decay, we profiled in cid-1 RNAi after PASH-1 inactivation in the pash-1(ts) background (0 and 24 h after upshift). RNAi against cid-1 reduced U-tailing at both time points (Supplementary Figure S13A, Supplementary Table S11). However, when we examined the change in miRNA abundance from zero to 24 h after PASH-1 inactivation, cid-1 RNAi did not have a global effect. We divided the miRNAs into subsets based on the percent of tailed species and saw no effect on turnover in miRNAs that were tailed 0–1%, 1–2% or 2–30% (Figure 6D). One exception to this was miRNAs of the embryo-enriched mir-35–41 cluster, which showed a higher log2fold change in cid-1 RNAi than vector control (Figure 6E). However, this is likely due to a lower starting amount of these miRNAs in T0 cid-1 RNAi samples (Figure 6F); indeed, these miRNAs are the only ones with less than half the abundance in cid-1 RNAi T0 samples as compared to vector T0 samples. Furthermore, both strands of the miRNA duplex derived from these precursors behaved similarly, supporting an effect of cid-1 RNAi on transcription or biogenesis rather than on decay rate of the mature miRNA species (Figure 6E, F). Because these miRNAs are largely germline and embryo-specific (53,77), the difference in their abundance most likely corresponds to slight differences in staging of cid-1 RNAi T0 samples due to pleiotropic effects of cid-1(lf), which negatively impacts germline function and embryo production (58). Finally, we also specifically examined miRNAs that show high increases in uridylation after pash-1 inactivation to determine whether these cases might be particularly sensitive to disrupting uridylation by cid-1 RNAi. For these miRNAs, tailing was disrupted, but turnover was not reproducibly altered (Figure 6G). Taken together, these data show that miRNA uridylation in C. elegans is not a global regulator of miRNA decay.
Figure 6.
Uridylation does not globally regulate miRNA abundance or decay. (A) Abundance of miRNAs in cid-1 RNAi versus empty vector. Mean and standard error are plotted. (B, C) Abundance of miRNAs in indicated mutant versus wild type. (A–C) Except for cid-1 RNAi (two biological replicates), mean and standard error of three biological replicates are plotted. All miRNAs > 50 RPM in all empty vector or wild type replicates, respectively, are shown. (D) Log2fold change in reads per million from zero to 24 h after PASH-1 inactivation. miRNAs are divided into three plots by the average percent mono-uridylation in empty vector at 0 h. All miRNAs with >50 RPM at 0 h in both empty vector replicates are shown, except for mir-35–41. (E) Log2fold change from zero to 24 h after PASH-1 inactivation for mir-35–41 guide (3p) strands and star (5p) strands. All miRNAs from this group are shown, regardless of abundance, except for mir-35-5p and mir-37-5p which had ∼0 reads in all samples. (F) Abundance of mir-35–41 guide and star strands at 0 h (prior to PASH-1 inactivation). (G) Plots showing percent mono-uridylation and miRNA abundance at zero and 24 h after PASH-1 inactivation. Size of dot represents abundance, and y-axis is percent mono-uridylation.
Next, we examined whether A-tailing altered miRNA abundance or turnover. In wild type, although F31C3.2/gldr-2 RNAi reduces A-tailing, RNAi did not alter miRNA abundance (Supplementary Figure S14A-B). F31C3.2/gldr-2 knockout and F31C3.2/gldr-2 knockout with gld-2 RNAi also showed minimal effects on miRNA abundance (Supplementary Figure S14C–F). Small changes in the abundance of three mir-35–42 family members appeared to be due to changes in synthesis, rather than changes in turnover of the miRNAs, as indicated by similar changes in star strand abundance (Supplementary Figure S14G); as noted above, this is likely due to slight staging differences due to pleiotropic effects. In the pash-1(ts) setting, fewer miRNAs could be examined with high confidence due to lower abundance and/or lower A-tailing (lower miRNA abundance may be the result of reduction of pash-1 function even at permissive temperature, and likewise, a reduced lifetime of miRNA precursors in this setting could lead to reduced A-tailing). Nonetheless, we still observed that F31C3.2/gldr-2 RNAi abrogated A-tailing in the 24 h time point (Supplementary Figure S14H, Supplementary Table S11). When examining the change in miRNA abundance from zero to 24 h after temperature shift, F31C3.2/gldr-2 RNAi did not affect the rate of miRNA decay (Supplementary Figure S14I). Therefore, the adenylation mediated by F31C3.2/GLDR-2 does not regulate miRNA stability.
High levels of mirtron uridylation do not affect abundance
Mirtrons are non-canonical miRNAs that derive from small introns; debranching of these intron lariats results in a miRNA precursor-like hairpin structure, bypassing the requirement for Microprocessor-mediated cleavage (78,79). Previous work showed that mirtron precursors are highly uridylated and that, in Drosophila, this uridylation is carried out by the Drosophilidae-specific TUTase6-related enzyme Tailor and negatively regulates mirtron abundance (10,16,17,80). In wild type sequencing libraries, only one mirtron, mir-62, was abundant enough for reliable tailing analysis. However, in pash-1(ts) libraries at restrictive temperature, mirtrons are relatively elevated in sequencing libraries since they do not depend on Microprocessor activity, while canonical miRNAs are globally depleted. This allowed for the analysis of tailing of multiple mirtrons. We observed that (as previously reported from aggregate sequencing data), 3p-derived mirtrons are highly uridylated to a much greater degree than canonical miRNAs, with the exception of mir-62 (Supplementary Figure S13B) (80). Not only is mir-62 atypical in its proportion of tailing, but mir-62 is also much more abundant than the other mirtrons (Supplementary Figure S13B). Like uridylation of canonical miRNAs, uridylation of mirtrons also required CID-1 (Supplementary Figure S13B). Surprisingly, although knockdown of cid-1 strongly abrogated mirtron tailing, the abundance of the mirtrons was not elevated as was observed in Drosophila (upon knockdown of Tailor) (Supplementary Figure S13B). Therefore, although the high rate of U-tailing is conserved for most C. elegans mirtrons, this modification does not regulate mirtron abundance.
Standard over-expression transgenes are too lowly expressed to induce detectable TDMD in C. elegans
Recent work provided evidence that some C. elegans miRNAs may be targeted by TDMD, whereas another previous study described target-mediated miRNA protection (TMMP) of miRNAs in C. elegans (27,81). To further interrogate the relationship between target binding and miRNA stability in C. elegans, we attempted to induce TDMD of specific miRNAs using exogenous transgenic triggers. We modified a ubiquitously expressed GFP construct (Peft-3::GFP::tbb-2 3′UTR) to contain two highly complementary binding sites for a miRNA of interest in the transgene's 3′UTR (Supplementary Figure S15A). Because TDMD triggers are generally very highly expressed, we chose a promoter and 3′UTR from very highly expressed genes (eft-3 encodes a translation elongation factor, and tbb-2 encodes β-tubulin). These elements are commonly used in the C. elegans field when the highest levels of over-expression are desired (43,82–84). We also used extra-chromosomal arrays to express these constructs; extra-chromosomal arrays are used for high amplitude over-expression because they encode hundreds of copies of the transgene (85,86). We chose to target lin-4, mir-237 and mir-85 because of their moderate expression level, moderate to slow decay rate, and low proportion of tailing. Also, the biology of lin-4 is well studied, and it shares a seed sequence with mir-237 (87). Extra-chromosomal arrays expressing these transgenes did not induce any phenotype (such as a lin-4 loss-of-function phenotype). Deep sequencing showed that the miRNAs were not altered in abundance in the presence of the transgene expressing their cognate putative TDMD trigger (Supplementary Figure S15B). The TDMD triggers also did not alter tailing of the targeted miRNAs (Supplementary Figure S15C). Most previously described triggers of TDMD are expressed at the same or higher levels than the targeted miRNA (23,28,33); the most efficient known TDMD trigger, the lncRNA Cyrano, is an extreme case which effects multiple turnover TDMD when expressed at a 17-fold lower level than its target miRNA (35). To determine if our transgenes are expressed at a similar stoichiometry with respect to the targeted miRNAs, we performed absolute quantification of the transgene mRNA and the targeted miRNAs using digital droplet PCR (ddPCR) and miRNA Taqman assays, respectively. Despite being expressed from a high-copy transgene with an efficient promoter, each transgene mRNA was >300-fold lower in abundance than the targeted miRNA (Supplementary Figure S15D). Thus, the stoichiometry renders these transgenes unlikely to induce TDMD. Because we have already applied all possible strategies to highly overexpress the transgene, the only option to examine a more equimolar transgene:miRNA pair would be to target a very lowly expressed miRNA. We estimated that, because the average ratio of the TDMD triggers described in Supplementary Figure S15 to their respective miRNA is 1116-fold higher, a miRNA would need to be 1116-fold lower expressed than lin-4, mir-237 and mir-85 to be equimolar. These three miRNAs have an average of 1726 reads per million in our sequencing libraries, so to be equimolar with the average TDMD trigger, a miRNA would need to be expressed on the order of 1.5 reads per million. At this level of expression, measuring tailing or decreases in abundance by deep sequencing would not be possible. Therefore, using traditional protein-coding transgenes as synthetic triggers of TDMD for moderately-expressed miRNAs is not feasible. In the long term, discovering endogenous triggers of TDMD will be more likely to serve as an effective platform for dissecting the features of TDMD in C. elegans.
DISCUSSION
In this work, we characterize the nature of miRNA tailing in C. elegans, with the broad view of determining ancestral functions of this class of post-transcriptional modification. We show that uridylation is far more prevalent than adenylation in C. elegans, unlike mammals and Drosophila in which both modifications are common (46–49).
We show that the substrate of these additions is shifted in different lineages. Based on its even distribution between 3p- and 5p-derived miRNAs, we infer that uridylation targets mature miRNAs in C. elegans, whereas precursors are the predominant substrate in mammals and Drosophila (25,46–48,51). In contrast, adenylation is skewed towards 3p-derived miRNAs in C. elegans, indicating that miRNA precursors are its likely substrate, unlike in vertebrates where mature miRNAs are more often adenylated and Drosophila where both precursors and mature miRNAs are likely substrates (46–48).
Despite the shifts in the apparent substrate of the modification, the enzymes required for these modifications are largely conserved. MiRNA uridylation requires CID-1, which is in the same TUT-4/7 gene family that carries out this modification (along with TUT2/GLD2) in mammals (12,15,57). MiRNA adenylation requires F31C3.2/GLDR-2. F31C3.2/GLDR-2 is in the TUT2/GLD2/PAPD-4 gene family which carries out miRNA adenylation in mammals and Drosophila (18,19,21,47). Importantly, the enzymatic activities of these enzymes in uridylation/adenylation has been previously demonstrated in vitro or in a heterologous tethering assay (56,60).
We characterize rates of miRNA decay in C. elegans, which are largely similar to those in other organisms (4,6–8,76). The median half-life – 20.7h – is relatively long. Half-lives in this study may even be slightly underestimated due to slight expansion of the culture after upshift to restrictive temperature. Nonetheless, certain miRNAs (especially mir-61, mir-71, mir-253 and mir-250) are reproducibly short-lived across replicates in our study and previous studies (74). How these fast decay rates are specified will be an important area of ongoing study. Moreover, we have only analyzed a single condition; decay may be differentially regulated for subsets of miRNAs in distinct developmental stages or under stress conditions. Notably, a study conducted in L1 stage larvae highlighted different fast-decaying miRNAs than our study in adults (88), suggesting that miRNA turnover may be developmentally regulated.
Although we observed that the proportion of tailed species increased over time after biogenesis, disrupting tailing had no impact on turnover. This is consistent with recent kinetic analysis which demonstrated that tailing is faster than 3′ trimming, so miRNAs are more likely to carry a tail as they approach decay (8). TDMD is the most frequently-studied context in which tailing correlates with decay, yet recent work on TDMD suggests that tailing is correlative – not causative – of TDMD, and that conformational changes that promote decay of the Argonaute-miRNA complex also expose the miRNA 3′ end to TENTs (24,26,27,35). However, in specific developmental contexts, tailing does promote wholesale miRNA turnover (18). Therefore, cell-type or developmental contexts which we have not yet examined may also couple decay to tailing in C. elegans. By identifying the enzymes required for miRNA tailing, this study lays the groundwork for discovering and dissecting such regulation.
DATA AVAILABILITY
All raw sequence data has been deposited in NCBI Sequence Read Archive (BioProject Number PRJNA703306).
Supplementary Material
ACKNOWLEDGEMENTS
We thank WormBase, the NIDDK Genomics Core, the NCI CCR Genomics Core, NIH Medical Arts, and NIH High Performance Computing. Strains are regularly received from the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Thank you to Anna Zinovyeva and members of the McJunkin lab for helpful discussions.
Contributor Information
Karl-Frédéric Vieux, National Institutes of Diabetes and Digestive and Kidney Diseases Intramural Research Program, Bethesda, MD 20815, USA.
Katherine P Prothro, National Institutes of Diabetes and Digestive and Kidney Diseases Intramural Research Program, Bethesda, MD 20815, USA; Stanford University School of Medicine, Stanford, CA 94305, USA.
Leanne H Kelley, Department of Biology, Syracuse University, Syracuse, NY 13244, USA.
Cameron Palmer, National Institutes of Diabetes and Digestive and Kidney Diseases Intramural Research Program, Bethesda, MD 20815, USA.
Eleanor M Maine, Department of Biology, Syracuse University, Syracuse, NY 13244, USA.
Isana Veksler-Lublinsky, Ben Gurion University of the Negev, Beer Sheva 8410501, Israel.
Katherine McJunkin, National Institutes of Diabetes and Digestive and Kidney Diseases Intramural Research Program, Bethesda, MD 20815, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIDDK Intramural Research Program [ZIA DK075147]. Funding for open access charge: NIDDK [ZIA DK075147].
Conflict of interest statement. None declared.
REFERENCES
- 1. Dexheimer P.J., Cochella L.. MicroRNAs: from mechanism to organism. Front. Cell Dev. Biol. 2020; 8:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alberti C., Cochella L.. A framework for understanding the roles of miRNAs in animal development. Dev. 2017; 144:2548–2559. [DOI] [PubMed] [Google Scholar]
- 3. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 4. Bail S., Swerdel M., Liu H., Jiao X., Goff L.A., Hart R.P., Kiledjian M.. Differential regulation of microRNA stability. RNA. 2010; 16:1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rissland O.S., Hong S.-J., Bartel D.P.. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol. Cell. 2011; 43:993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marzi M.J., Ghini F., Cerruti B., De Pretis S., Bonetti P., Giacomelli C., Gorski M.M., Kress T., Pelizzola M., Muller H.et al.. Degradation dynamics of micrornas revealed by a novel pulse-chase approach. Genome Res. 2016; 26:554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reichholf B., Herzog V.A., Fasching N., Manzenreither R.A., Sowemimo I., Ameres S.L.. Time-resolved small RNA sequencing unravels the molecular principles of microRNA homeostasis. Mol. Cell. 2019; 75:756–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kingston E.R., Bartel D.P.. Global analyses of the dynamics of mammalian microRNA metabolism. Genome Res. 2019; 29:1777–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krol J., Busskamp V., Markiewicz I., Stadler M.B., Ribi S., Richter J., Duebel J., Bicker S., Fehling H.J., Schübeler D.et al.. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010; 141:618–631. [DOI] [PubMed] [Google Scholar]
- 10. Modepalli V., Moran Y.. Evolution of miRNA tailing by 3′ terminal uridylyl transferases in metazoa. Genome Biol. Evol. 2017; 9:1547–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Almeida C., Scheer H., Zuber H., Gagliardi D.. RNA uridylation: a key posttranscriptional modification shaping the coding and noncoding transcriptome. Wiley Interdiscip. Rev. RNA. 2018; 9:e1440. [DOI] [PubMed] [Google Scholar]
- 12. Kim B., Ha M., Loeff L., Chang H., Simanshu D.K., Li S., Fareh M., Patel D.J., Joo C., Kim V.N.. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J. 2015; 34:1801–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim H., Kim J., Yu S., Lee Y.Y., Park J., Choi R.J., Yoon S.J., Kang S.G., Kim V.N.. A mechanism for microRNA Arm switching regulated by uridylation. Mol. Cell. 2020; 78:1224–1236. [DOI] [PubMed] [Google Scholar]
- 14. Heo I., Joo C., Cho J., Ha M., Han J., Kim V.N.. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008; 32:276–284. [DOI] [PubMed] [Google Scholar]
- 15. Heo I., Joo C., Kim Y.-K.K., Ha M., Yoon M.-J.J., Cho J., Yeom K.-H.H., Han J., Kim V.N.. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009; 138:696–708. [DOI] [PubMed] [Google Scholar]
- 16. Reimão-Pinto M.M., Ignatova V., Burkard T.R., Hung J.H., Manzenreither R.A., Sowemimo I., Herzog V.A., Reichholf B., Fariña-Lopez S., Ameres S.L.. Uridylation of RNA hairpins by tailor confines the emergence of microRNAs in Drosophila. Mol. Cell. 2015; 59:203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bortolamiol-Becet D., Hu F., Jee D., Wen J., Okamura K., Lin C.J., Ameres S.L., Lai E.C.. Selective suppression of the splicing-mediated microRNA pathway by the terminal uridyltransferase tailor. Mol. Cell. 2015; 59:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee M., Choi Y., Kim K., Jin H., Lim J., Nguyen T.A.A., Yang J., Jeong M., Giraldez A.J.J., Yang H.et al.. Adenylation of maternally inherited microRNAs by Wispy. Mol. Cell. 2014; 56:696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katoh T., Sakaguchi Y., Miyauchi K., Suzuki T., Kashiwabara S.-I., Baba T., Suzuki T.. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009; 23:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burns D.M., D’Ambrogio A., Nottrott S., Richter J.D. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011; 473:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. D’Ambrogio A., Gu W., Udagawa T., Mello C.C., Richter J.D.. Specific miRNA stabilization by Gld2-catalyzed monoadenylation. Cell Rep. 2012; 2:1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ameres S.L., Horwich M.D., Hung J.-H., Xu J., Ghildiyal M., Weng Z., Zamore P.D.. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010; 328:1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baccarini A., Chauhan H., Gardner T.J., Jayaprakash A.D., Sachidanandam R., Brown B.D.. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr. Biol. 2011; 21:369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sheu-Gruttadauria J., Pawlica P., Klum S.M., Wang S., Yario T.A., Schirle Oakdale N.T., Steitz J.A., MacRae I.J.. Structural basis for target-directed microRNA degradation. Mol. Cell. 2019; 75:1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang A., Shao T.J., Bofill-De Ros X., Lian C., Villanueva P., Dai L., Gu S.. AGO-bound mature miRNAs are oligouridylated by TUTs and subsequently degraded by DIS3L2. Nat. Commun. 2020; 11:2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han J., LaVigne C.A., Jones B.T., Zhang H., Gillett F., Mendell J.T.. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science. 2020; 370:eabc9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi C.Y., Kingston E.R., Kleaveland B., Lin D.H., Stubna M.W., Bartel D.P.. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science. 2020; 370:eabc9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de la Mata M., Gaidatzis D., Vitanescu M., Stadler M.B., Wentzel C., Scheiffele P., Filipowicz W., Grosshans H.. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 2015; 16:500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cazalla D., Yario T., Steitz J.A., Steitz J.. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010; 328:1563–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marcinowski L., Tanguy M., Krmpotic A., Rädle B., Lisnić V.J., Tuddenham L., Chane-Woon-Ming B., Ruzsics Z., Erhard F., Benkartek C.et al.. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012; 8:e1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Libri V., Helwak A., Miesen P., Santhakumar D., Borger J.G., Kudla G., Grey F., Tollervey D., Buck A.H.. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Piwecka M., Glažar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P.et al.. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017; 357:eaam8526. [DOI] [PubMed] [Google Scholar]
- 33. Ghini F., Rubolino C., Climent M., Simeone I., Marzi M.J., Nicassio F.. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat. Commun. 2018; 9:3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bitetti A., Mallory A.C., Golini E., Carrieri C., Carreño Gutiérrez H., Perlas E., Pérez-Rico Y.A., Tocchini-Valentini G.P., Enright A.J., Norton W.H.J.et al.. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol. 2018; 25:244–251. [DOI] [PubMed] [Google Scholar]
- 35. Kleaveland B., Shi C.Y., Stefano J., Bartel D.P.. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018; 174:350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang A., Bofill-De Ros X., Shao T.J., Jiang M., Li K., Villanueva P., Dai L., Gu S.. 3′ Uridylation confers miRNAs with non-canonical target repertoires. Mol. Cell. 2019; 75:511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Patel P., Ramachandruni S.D., Kakrana A., Nakano M., Meyers B.C.. miTRATA: a web-based tool for microRNA truncation and tailing analysis. Bioinformatics. 2016; 32:450–452. [DOI] [PubMed] [Google Scholar]
- 38. Kozomara A., Griffiths-Jones S.. MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kumar S., Stecher G., Li M., Knyaz C., Tamura K.. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018; 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bolt B.J., Rodgers F.H., Shafie M., Kersey P.J., Berriman M., Howe K.L.. Using WormBase ParaSite: an integrated platform for exploring helminth genomic data. Methods in Molecular Biology. 2018; 1757:Humana Press Inc; 471–491. [DOI] [PubMed] [Google Scholar]
- 42. Arribere J.A., Bell R.T., Fu B.X.H., Artiles K.L., Hartman P.S., Fire A.Z.. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014; 198:837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frøkjær-Jensen C., Davis M.W., Ailion M., Jorgensen E.M.. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods. 2012; 9:117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dickinson D.J., Pani A.M., Heppert J.K., Higgins C.D., Goldstein B.. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 2015; 200:1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.-P., Grunnet M., Jorgensen E.M.. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008; 40:1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chiang H.R., Schoenfeld L.W., Ruby J.G., Auyeung V.C., Spies N., Baek D., Johnston W.K., Russ C., Luo S., Babiarz J.E.et al.. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010; 24:992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burroughs A.M., Ando Y., de Hoon M.J., Tomaru Y., Nishibu T., Ukekawa R., Funakoshi T., Kurokawa T., Suzuki H., Hayashizaki Y.et al.. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010; 20:1398–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berezikov E., Robine N., Samsonova A., Westholm J.O., Naqvi A., Hung J.H., Okamura K., Dai Q., Bortolamiol-Becet D., Martin R.et al.. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011; 21:203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wyman S.K., Knouf E.C., Parkin R.K., Fritz B.R., Lin D.W., Dennis L.M., Krouse M.A., Webster P.J., Tewari M.. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011; 21:1450–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Song J., Wang X., Song B., Gao L., Mo X., Yue L., Yang H., Lu J., Ren G., Mo B.et al.. Prevalent cytidylation and uridylation of precursor miRNAs in Arabidopsis. Nat. Plants. 2019; 5:1260–1272. [DOI] [PubMed] [Google Scholar]
- 51. Thornton J.E., Du P., Jing L., Sjekloca L., Lin S., Grossi E., Sliz P., Zon L.I., Gregory R.I.. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res. 2014; 42:11777–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Batista P.J., Ruby J.G., Claycomb J.M., Chiang R., Fahlgren N., Kasschau K.D., Chaves D.A., Gu W., Vasale J.J., Duan S.et al.. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell. 2008; 31:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kato M., de Lencastre A., Pincus Z., Slack F.J.. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009; 10:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi Z., Montgomery T.A., Qi Y., Ruvkun G.. High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res. 2013; 23:497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stoeckius M., Maaskola J., Colombo T., Rahn H.-P., Friedländer M.R., Li N., Chen W., Piano F., Rajewsky N.. Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nat. Methods. 2009; 6:745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van Wolfswinkel J.C., Claycomb J.M., Batista P.J., Mello C.C., Berezikov E., Ketting R.F.. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009; 139:135–148. [DOI] [PubMed] [Google Scholar]
- 57. Heo I., Ha M., Lim J., Yoon M.-J., Park J.-E., Kwon S.C., Chang H., Kim V.N.. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012; 151:521–532. [DOI] [PubMed] [Google Scholar]
- 58. Li Y., Maine E.M.. The balance of poly(U) polymerase activity ensures germline identity, survival and development in Caenorhabditis elegans. Dev. 2018; 145:dev165944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kadyk L.C., Kimble J.. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998; 125:1803–1813. [DOI] [PubMed] [Google Scholar]
- 60. Preston M.A., Porter D.F., Chen F., Buter N., Lapointe C.P., Keles S., Kimble J., Wickens M.. Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods. 2019; 16:437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chang H.M., Triboulet R., Thornton J.E., Gregory R.I.. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013; 497:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ustianenko D., Hrossova D., Potesil D., Chalupnikova K., Hrazdilova K., Pachernik J., Cetkovska K., Uldrijan S., Zdrahal Z., Vanacova A.. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013; 19:1632–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y.. Poly(A)-specific ribonuclease mediates 3′-end trimming of argonaute2-cleaved precursor micrornas. Cell Rep. 2013; 5:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Katoh T., Hojo H., Suzuki T.. Destabilization of microRNAs in human cells by 3′ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res. 2015; 43:7521–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chatterjee S., Grosshans H., Großhans H.. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009; 461:546–549. [DOI] [PubMed] [Google Scholar]
- 66. Bossé G.D., Rüegger S., Ow M.C., Vasquez-Rifo A., Rondeau E.L., Ambros V.R., Simard M.J.. The decapping scavenger enzyme DCS-1 controls microRNA levels in Caenorhabditis elegans. Mol. Cell. 2013; 50:281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Haas G., Cetin S., Messmer M., Chane-Woon-Ming B., Terenzi O., Chicher J., Kuhn L., Hammann P., Pfeffer S.. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res. 2016; 44:2873–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Elbarbary R.A., Miyoshi K., Myers J.R., Du P., Ashton J.M., Tian B., Maquat L.E.. Tudor-SN–mediated endonucleolytic decay of human cell microRNAs promotes G 1 /S phase transition. Science. 2017; 356:859–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tang W., Tu S., Lee H.C., Weng Z., Mello C.C.. The RNase PARN-1 Trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell. 2016; 164:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kamath R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., Ahringer J.. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001; 2:RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Maeda I., Kohara Y., Yamamoto M., Sugimoto A.. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 2001; 11:171–176. [DOI] [PubMed] [Google Scholar]
- 72. Sönnichsen B., Koski L.B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A.M., Artelt J., Bettencourt P., Cassin E.et al.. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005; 434:462–469. [DOI] [PubMed] [Google Scholar]
- 73. Simmer F., Moorman C., Van Der Linden A.M., Kuijk E., Van Den Berghe P.V.E., Kamath R.S., Fraser A.G., Ahringer J., Plasterk R.H.A.. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003; 1:E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lehrbach N.J., Castro C., Murfitt K.J., Abreu-Goodger C., Griffin J.L., Miska E.A.. Post-developmental microRNA expression is required for normal physiology, and regulates aging in parallel to insulin/IGF-1 signaling in C. elegans. RNA. 2012; 18:2220–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dexheimer P.J., Wang J., Cochella L.. Two MicroRNAs are sufficient for embryonic patterning in C. elegans. Curr. Biol. 2020; 30:5058–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gantier M.P., McCoy C.E., Rusinova I., Saulep D., Wang D., Xu D., Irving A.T., Behlke M.A., Hertzog P.J., Mackay F.et al.. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011; 39:5692–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Alvarez-Saavedra E., Horvitz H.R.. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 2010; 20:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ruby J.G., Jan C.H., Bartel D.P.. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007; 448:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chung W.-J., Agius P., Westholm J.O., Chen M., Okamura K., Robine N., Leslie C.S., Lai E.C.. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res. 2011; 21:286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Westholm J.O., Ladewig E., Okamura K., Nicolas R., Lai E.C.. Common and distinct patterns of terminal modifications to mirtrons and canonical microRNAs. RNA. 2012; 18:177–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chatterjee S., Fasler M., Büssing I., Grosshans H.. Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell. 2011; 20:388–396. [DOI] [PubMed] [Google Scholar]
- 82. Merritt C., Rasoloson D., Ko D., Seydoux G.. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 2008; 18:1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Friedland A.E., Tzur Y.B., Esvelt K.M., Colaiácovo M.P., Church G.M., Calarco J.A.. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods. 2013; 10:741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ashley G.E., Duong T., Levenson M.T., Martinez M.A.Q., Johnson L.C., Hibshman J.D., Saeger H.N., Palmisano N.J., Doonan R., Martinez-Mendez R.et al.. An expanded auxin-inducible degron toolkit for Caenorhabditis elegans. Genetics. 2021; 217:iyab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Stinchcomb D.T., Shaw J.E., Carr S.H., Hirsh d. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 1985; 5:3484–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mello C.C., Kramer J.M., Stinchcomb D., Ambros V.. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991; 10:3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ambros V MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 2011; 21:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Miki T.S., Rüegger S., Gaidatzis D., Stadler M.B., Großhans H.. Engineering of a conditional allele reveals multiple roles of XRN2 in Caenorhabditis elegans development and substrate specificity in microRNA turnover. Nucleic Acids Res. 2014; 42:4056–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequence data has been deposited in NCBI Sequence Read Archive (BioProject Number PRJNA703306).