Abstract
Mutual crosstalk among poly(ADP-ribose) (PAR), activated PAR polymerase 1 (PARP1) metabolites, and DNA repair machinery has emerged as a key regulatory mechanism of the DNA damage response (DDR). However, there is no conclusive evidence of how PAR precisely controls DDR. Herein, six deubiquitinating enzymes (DUBs) associated with PAR-coupled DDR were identified, and the role of USP39, an inactive DUB involved in spliceosome assembly, was characterized. USP39 rapidly localizes to DNA lesions in a PAR-dependent manner, where it regulates non-homologous end-joining (NHEJ) via a tripartite RG motif located in the N-terminus comprising 46 amino acids (N46). Furthermore, USP39 acts as a molecular trigger for liquid demixing in a PAR-coupled N46-dependent manner, thereby directly interacting with the XRCC4/LIG4 complex during NHEJ. In parallel, the USP39-associated spliceosome complex controls homologous recombination repair in a PAR-independent manner. These findings provide mechanistic insights into how PAR chains precisely control DNA repair processes in the DDR.
INTRODUCTION
The cellular response to DNA damage is a primary anti-cancer barrier. This process maintains genomic integrity by recruiting and activating assorted proteins including chromatin remodelers, DNA repair enzymes, and writer enzymes involved in post-translational modification (PTM) of diverse DNA damage-linked proteins (1–4). Upon DNA damage, histone family proteins and chromatin modifiers are the major targets of PTM writers. Indeed, PTMs contribute to the tight regulation of the DNA damage response (DDR) (5–8). A major PTM in DDR is poly(ADP-ribosyl)ation (PARylation), which is mediated by poly(ADP-ribose) polymerases (PARPs). At DNA lesion sites, PARylation is mainly initiated by PARP1 activation, resulting in the accumulation of PARylated proteins and poly(ADP-ribose) (PAR) chains. These molecules are essential factors for recruiting DNA repair-associated proteins to modulate chromatin dynamics during DNA damage.
PAR chains can be broadly divided into two types. The first type is covalently conjugated to PARylated acceptors, and the other non-covalently interacts with substrates (9–11). To date, 10 PAR-chain binding motifs have been identified: the classical PAR-binding motif ([HKR]1-X2-X3-[AIQVY]4-[KR]5-[KR]6-[AILV]7-[FILPV]8) (12), macrodomain (13–15), WWE domain (16,17), PAR-binding zinc finger (PBZ) molecules ([K/R]xxCx[F/Y]GxxCxbbxxxxHxxx[F/Y]xH) (18,19), FHA/BRCT domain (20), RNA recognition (RRM) motif (21), SR repeats and KR-rich motifs (22,23), oligonucleotide/oligosaccharide-binding (OB)-fold domain (24), PIN domain (25) and RG/RGG motif (26–28). Recent studied have shown that PAR binding activities of the FUS or EWS proteins that contain RG/RGG motifs are crucial for PAR-dependent DNA repair. Defects in this response may contribute to the progression of diseases such as amyotrophic lateral sclerosis (ALS) (29–31). In addition, crosstalk between PAR and RGG motifs of intrinsically disordered proteins (IDPs), including FUS, initiates liquid demixing and induces phase separation (32,33). These changes subsequently alter the soluble intracellular space by generating membrane-less compartments for dynamic protein assembly in the DDR (32–36).
PAR chains are directly linked to the ubiquitin-coupled DDR pathway. RNF146 (also known as Iduna) is recruited to DNA lesions upon the interaction of its WWE domain with PAR chains that are generated by hyper-activated PARP1, followed by PARP1 degradation by RNF146 activity (16). CHFR contains an N-terminal PBZ motif and localizes to DNA double-strand breaks (DSBs) in a PAR-dependent manner (37). Additionally, PAR-dependent DTX3L, also known as BAL1 macrodomain-interacting partner BBAP, selectively induces the mono-ubiquitination of histone H4K91, leading to the retention of 53BP1 and BRCA1 at DSBs (38,39). Most studies have suggested that PAR chains are required for the recruitment of PARP1-linked ubiquitin E3 ligases, which contain PAR-binding motifs or domains, and facilitate DNA repair processes during DDR (16,37–39).
In parallel, deubiquitinating enzymes (DUBs) have also been intensively studied in the chromatin context and have recently received increasing attention for understanding the basis of genomic stability and cancer development (40,41). Ubiquitin-specific peptidase 1 (USP1) was among the first ubiquitin hydrolases identified as key players that promote homologous recombination (HR) repair by deubiquitination of FANCD2 and PCNA (42,43). USP1 knockout mice display increased DDR dysfunction, which in turn elevates perinatal lethality, hypersensitivity against DNA-damaging agents, and male infertility (44–46). USP3 is a chromatin-coupled DUB that regulates histone H2A/H2B deubiquitination, and its ablation leads to DNA break accumulation, resulting in replication stress (47). Furthermore, USP4, USP5 and USP7 are potential oncogenes that regulate p53 stability in the DDR context (48–51). Intriguingly, recent studies have shown that some ubiquitin E3 ligases, including RNF169, TRIP12 and UBR5, antagonize ubiquitin signaling during DNA repair, similar to the inhibitory effect of ubiquitin signaling by DUBs on DDR. These findings indicate that the ubiquitin-mediated DNA repair process could be controlled by different methods (52–54). Although several ubiquitin E3 ligases and DUBs are linked to DDR, the crosstalk between PAR and DUBs in response to DNA damage remains unclear. To elucidate this relationship, a laser micro-irradiation (mIR) system was used to screen for novel PAR-coupled DUBs that translocate to DNA lesions. Phylogenetic analysis revealed functional DUB clustering into subgroups including the USP, JAMM, OTU and zinc-finger ubiquitin-specific protease (ZF-UBP) families. Among these, we focused on USP39, an inactive DUB, to assess its function in the DDR. We observed that USP39 strongly interacts with PAR-chains via an N-terminal 46 amino acid (N46) tripartite RG motif. This interaction initiates liquid-demixing-mediated non-homologous end-joining (NHEJ) in the DDR by recruiting the XRCC4/LIG4 complex. Moreover, we found that a USP39-coupled spliceosome complex simultaneously controls HR in a PAR-independent manner. These findings provide mechanistic insights into the functional link between PAR, liquid demixing, and DUB-regulated NHEJ repair.
MATERIALS AND METHODS
Cell lines and culture
Human osteosarcoma (U2OS) and normal MRC-5 cell lines were purchased from ATCC. And human embryonic kidney (HEK293FT) cell line was purchased from Thermo Scientific. U2OS and HEK 293FT cells were maintained in Dulbecco's modified Eagle's medium (DMEM, GIBCO) supplemented with 10% (v/v) fetal bovine serum (FBS, GIBCO). MRC-5 cells were maintained in Eagle's Minimum Essential Medium (EMEM, ATCC) supplemented with 10% (v/v) FBS. U2OS based HR and NHEJ reporter cells (U2OS-DR-GFP, U2OS-EJ5-GFP) were kindly provided from Dr. Jeremy Stark and were maintained in DMEM supplemented with 10% (v/v) FBS, 1 mg/ml puromycin (Sigma). WT MEF, PARP1 knockout MEF, and ATM knockout MEF cells were cultured with DMEM supplemented with 10% (v/v) FBS. U2OS-2-6-3 cells were kindly provided from Dr Roger A. Greenberg (University of Pennsylvania).
DUBs library cloning and plasmids
To generate the pENTRY Donor 221 vectors of human deubiquitinating enzymes (DUBs), DUBs were PCR amplified with individual primers from human liver cDNA libraries. pENTRY-DUBs were then transferred into a pDEST53 (GFP N-terminal) vector using a Gateway LR cloning system (Invitrogen). Some of GFP-DUBs (EGFP N-terminal) were kindly provided by Dr Stephen P. Jackson. Detailed information of plasmids is given in Supplementary Tables S1 and S2. To generate USP39 mutant plasmids, site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene). Each deletion mutant of USP39 was constructed using a classical PCR method. All mutation sites and deletion regions were validated by DNA sequence analysis, and each primer set used in this study is described in Supplementary Table S3.
siRNA sequences, antibodies and chemicals
siRNA sequences, antibodies and chemicals are described in Supplementary Tables S4–S6.
Live cell imaging with laser micro-irradiation
To analyze laser-induced DDR, cells were plated onto confocal glass bottom dishes (SPL) and transfected with the appropriate plasmids. After 1 day, cells were incubated with 10 μM 5-bromo-2′-deoxyuridine (BrdU, Sigma) for 30 h. Cells were treated for 1 h with the PARP inhibitor (PJ34, 5 μM; Santa Cruz Biotechnology and Olaparib, 10 μM; Avention) or the ATM inhibitor (KU55933, 10 μM; Sigma) before laser-induced DSBs were created. DSBs were induced for 3 s (32 lines/s) using a wavelength ultraviolet A laser (405 nm) in a temperature-controlled chamber (37°C, 5% CO2). The laser output was set to 100%, and 5–10 iterations were used to generate DSBs with a 60× oil objective. For live cell imaging, images were taken every second up to 300 s after laser-induced DNA damage and kinetic analysis was conducted using NIS-Elements AR software (Nikon). Each data series were normalized with respect to baseline values. For high-dose damage, DSBs were induced for 3 s (128 lines/s) using a wavelength ultraviolet A laser (405 nm) in a temperature-controlled chamber (37°C, 5% CO2).
Immunofluorescence
After laser-inducing DSBs, cells were incubated at 37°C for the indicated times. Cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 10 min at room temperature and washed three times with PBS. Subsequently, cells were permeabilized with 0.25% Triton X-100 in PBS for 15 min at room temperature and blocked with 1% BSA in PBS for 30 min. After blocking, cells were incubated with the required primary antibody for 18 h at 4°C. Cells were then washed and stained with appropriated secondary antibody in blocking solution for 1 h at room temperature. After washing, nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma) solution for 10 min, and each well was mounted with 12 mm glass slides using Vectashield mounting medium (Vector Labs).
Analysis of ionizing radiation-induced foci formation
Cells were transfected with each siRNA alone or in combination with siRNA-resistant plasmids using Lipofectamine 2000 (Invitrogen). Forty-eight hours later, cells were irradiated with a gamma cell irradiator with the appropriate IR dose. After 1 or 6 h, cells were fixed with 4% PFA in PBS and stained with the appropriate antibody. Foci were detected by confocal microscope (Nikon A1R), and representative immunofluorescence images were used for manual foci counting. The percentage of cells having >10 foci per cell was calculated.
Micronucleus assay
Cells were transfected with each siRNA using Lipofectamine RNAiMax (Invitrogen). After 48 h, cells were treated with 100 μg/ml Zeocin for 12 h and then washed with PBS. Cells were then fixed with 4% PFA in PBS and permeabilized with 0.25% triton X-100, followed by staining with anti-F-actin and anti-γH2AX antibodies. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma) solution and micronuclei were detected using a confocal microscope (Nikon A1R) with a 60× oil objective.
Purification of recombinant protein from Sf9 cells or E. coli
pENTRY vectors containing DUBs were transferred into the pDEST20 vector using a Gateway LR cloning system. To generate recombinant baculovirus, DH10Bac Escherichia coli, which contained a baculovirus shuttle vector (bacmid), was infected with pDEST20 plasmids. Purified bacmid DNA was transfected into Sf9 insect cells using Cellfectin® II Reagent (Invitrogen). After 3 days, Sf9 cells were collected and lysed with NETN buffer (25 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1% NP-40, 0.1% Triton X-100, protease inhibitor cocktail and 1 mM PMSF). Soluble protein extracts were obtained by centrifugation at 14 000 rpm and 4°C and incubated with Glutathione-Sepharose4B resin (GE Healthcare Life Science) according to the manufacturer's manual. Resin was washed with NETN buffer, and then bound proteins were eluted with elution buffer (50 mM HEPES, pH 7.5, 40 mM reduced glutathione, 100 mM NaCl, 30% glycerol and 0.03% Triton X-100). Protein concentrations were determined using the Bradford method (Bio-Rad). To purify, GST-protein, pDEST15 vector was transformed to BL21pLys and purified as we described previously (55). To obtain GST-free proteins, GST-fusion proteins derived from pDEST15-3C were incubated with HRV 3C protease (GE Healthcare) in cleavage buffer (50 mM Tris–HCl pH 7.4; 150 mM NaCl; 1 mM EDTA; 1 mM DTT) for 24 h at 4°C. The cleaved protein was purified by passing through a GST affinity column that has an affinity for the HRV 3C protease and the GST tag. The purity of the final product was examined by Coomassie brilliant blue staining following electrophoresis on an 8−16% gradient SDS-PAGE. All other proteins including DUBs were also purified in the same experimental condition. Detailed information of all proteins used in this study is described in Supplementary Table S7.
PAR overlay assay
Each of recombinant protein was subjected to SDS-PAGE and transferred onto a nitrocellulose membrane. In the case of dot blots, recombinant protein was dotted onto a nitrocellulose membrane. And the membranes were blocked with 5% Difco skimmed milk powder (BD Bioscience) in PBST (PBS plus 0.05% Tween 20) and then incubated for 1 h at room temperature with PAR polymer. After washing in PBST, PAR-binding proteins were detected with PAR antibody. Recombinant Histone H3, Iduna (positive control) and GST (negative control) were used as positive and negative controls for PAR overlay assay, respectively.
HR and NHEJ repair analysis
HR or NHEJ efficiency was measured in U2OS-based reporter cell lines (U2OS-DR-GFP, U2OS-EJ5-GFP). These cells were transfected with the indicated siRNA alone or in combination with an V5-siRNA-resistant plasmid. On the following day, I-SceI was transfected into each reporter cell. After 72 h, HR and NHEJ repair efficiency were analyzed by quantification of GFP-positive reporter cells using flow cytometry.
Neutral comet assay
To monitor the extent of DNA repair, U2OS cells were transfected with siRNA alone or in combination with an GFP-siRNA-resistant plasmid. After 48 h, each cell was treated with 40 μg/ml Zeocin for 2 h and then washed with PBS. Next, cells were further incubated for 2 h in a temperature-controlled chamber (37°C, 5% CO2). The neutral comet assay was performed according to the manufacturer's manual (Trevigen), and tail moments were measured using OpenComet V1.3 software.
Clonogenic survival assay
Clonogenic viability was examined using a colony forming assay. Cells were transfected with the appropriate siRNA. And 24 h later, cells were harvested and seeded using the appropriate number on a 6 cm dish. The following day, cells were treated with ionizing radiated with a gamma cell irradiator with the indicated IR dose (0–4 Gy). Cells were then incubated in a temperature-controlled chamber (37°C, 5% CO2) for 14 days. Resulting colonies were fixed with methanol and stained with 0.5% Crystal violet (Sigma). Colonies were counted and normalized to plating efficiencies.
Immunoprecipitation
Cells were harvested and lysed in IP lysis buffer containing 0.5% Triton X-100, 0.5% NP-40, protease inhibitor cocktail and 1 mM PMSF in PBS. The lysates were centrifuged at 12 000 g for 10 min at 4°C, and the supernatants were incubated with the appropriate antibody for 18 h at 4°C. Next, the supernatants were incubated with protein G beads for 12 h at 4°C and washed with IP lysis buffer. Bead-bound proteins were eluted with SDS sample buffer and boiled supernatants were separated by 8–16% SDS-PAGE. Proteins were detected by immunoblot with the appropriate antibodies. For IP of FLAG-tagged proteins, anti-FLAG M2 affinity gel was used.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using EZ-ChIP Kits (Millipore) according to the protocol described by Shanbhag et al. (2). Briefly, DSBs were induced in U2OS 2-6-3 cells using an mCherry-FokI-endonuclease. After 48 h, cells were crosslinked for 10 min using 1% PFA and were then quenched with glycine. Cells were then lysed in SDS buffer and sonicated. After centrifugation, the supernatants were incubated overnight with 5 μg aliquots of primary antibodies (anti-USP39, control rabbit IgG, or anti-γH2AX). Antibody bound protein/DNA complexes were then pulled down using protein G beads and were eluted with elution buffer. Eluted protein/DNA complexes were finally digested with protease K and purified DNA samples were analyzed by qRT-PCR for DSBs or normal sites; (Proximal chromosome 1 or distal chromosome 7) with the primer sets listed in Supplementary Table S3.
Cell synchronization
U2OS cells were synchronized at the G1/S boundary by a double thymidine block. Cells were treated with 2 mM thymidine (Sigma) for first thymidine block, and then cells were immediately transfected with siCtrl or siUSP39. After 16 h, cells were washed two times with PBS and incubated in normal fresh media for 8 h. Then, thymidine was added into cells for a second thymidine block and incubated for 16 h. The cells were released from the double thymidine block by washing twice with PBS and cultured in normal fresh media. Synchronized cells were collected at each indicated time following release. Asynchronized cells were harvested at 48 h after knockdown as a control. Cell cycle profile was determined using propidium iodide (PI; Sigma) staining as described below.
Cell cycle analysis
To examine cell cycle analysis in USP39 knockdown cells, cells were trypsinized following synchronization and centrifuged at 850 g for 5 min. The cells were washed once with PBS, and then fixed in ice-cold 70% ethanol in PBS for 30 min at 4°C. Fixed cells were collected by centrifugation at 850 g for 5 min at 4°C and then washed two times with PBS. For cell cycle analysis, cells were resuspended in PBS containing 50 μg/ml RNase A and incubated for 10 min at 25°C. After incubation, DNA contents were stained with PI staining solution (100 μg/ml of PI in PBS) at 37°C for 10 min. Cell cycle progression was analyzed by FACS Canto II flow cytometer (BD Bioscience). At least 10 000 cells were captured for each sample to determine the percentage of G1, S and G2/M.
In vitro aggregation assay and Transmission electron microscopy (TEM) analysis
Experiment was performed according to the protocol described by Altmeyer et al. (32). For in vitro aggregation assay, GST-free USP39 WT or RG/AA was diluted in 40 mM HEPES–KOH, 150 mM KCl, pH 7.4 to a final concentration of 0.04 mg/ml (0.615 μM), and then PAR chains (Trevigen) were added to a final concentration of 14 nM (molar ratio USP39:PAR = 44:1). For preparation of PARG-treated PAR, PAR chians (Trevigen) were pre-incubated with recombinant PARG (0.013 μg/ml; Trevigen) for 6 h at 37°C. All samples were incubated for 24 h at room temperature using a Intelli-mixer (ELMI). The samples were plated onto a carbon-coated grids and incubated for 1 min, followed by staining with UranylLess (EMS) for 3 min. TEM images were obtained using a Field Emission Scanning Electron Microscope (ZEISS, Sigma 500) operated at an acceleration voltage of 25.00 kV. At least 200 aggregate sizes per sample were analyzed and quantified using MetaMorph software.
Image quantification
Images were obtained using a Nikon A1R confocal microscope (Nikon). All images from each experiment were taken at the same exposure time. Image quantification was performed using either NIS-Elements AR software (Nikon) or ImageJ (National Institutes of Health). For laser micro-irradiation analysis, the GFP intensity of damaged areas was measured and normalized to the GFP intensity of each nucleus.
Statistical analysis
Data are presented as mean ± standard error of mean (s.e.m.). Comparisons between two groups were made with the Student's t-test. For parametric multiple comparison, one-way analysis of variance followed by the Tukey–Kramer test was employed by GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA). Significance was defined as P < 0.05. Each experiment was performed independently three times.
RESULTS
Screening for DUBs localized to DNA lesion(s) in a PAR-dependent manner
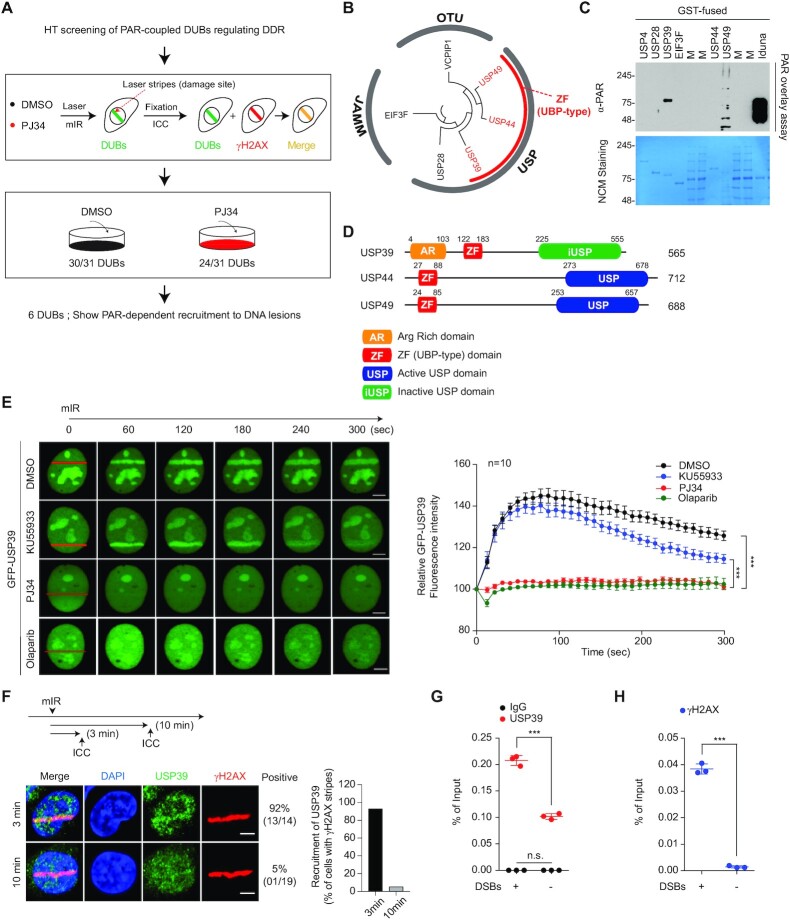
PARP1 is a master regulator of the DDR, along with ataxia telangiectasia mutated (ATM), and is linked to the ubiquitin-coupled DDR pathway. Although several ubiquitin E3 ligases are associated with PAR signaling during biological events, it remains unclear how DUBs interact with PAR chains in the DDR. To address this point, we collected 31 DUBs, which are translocated to damaged chromatin (56,57), and screened them using an mIR system in the absence or presence of PJ34, which interferes with PARP activity but not with sensing nicked DNA (Figure 1A). Among these 31 DUBs, 19% (six DUBs: USP28, USP39, USP44, USP49, VCPIP1 and EIF3F) required PARP activity for recruitment to DNA lesions (Supplementary Figure S1A and B). Phylogenic analysis revealed that these six DUBs can be functionally classified into USP, JAMM and OTU DUBs. We also observed that some of the USPs (USP39, USP44 and USP49) contain a ZF-UBP domain and can bind the free C-terminal of ubiquitin (58) (Figure 1B). These data suggest that the identified DUBs could associate with PAR chains for recruitment to DSBs, as well as PAR-coupled ubiquitin E3 ligases. To verify this hypothesis, these six recombinant purified DUBs were examined with PAR chain overlay assays. USP39 strongly bound to linear and/or branched PAR-chains under denaturing conditions, whereas USP49 exhibited mild interactions with PAR chains (Figure 1C). Using multiple sequence alignment, we observed that USP44 and USP49 contain conserved ZF-UBP and active USP domains, while USP39 possesses an inactive USP domain (iUSP) and an additional arginine-rich motif (AR) at its N-terminus (Figure 1D and Supplementary Figure S2A).
Figure 1.
Systematic screening of PARP1-dependent DUBs associated with the DDR. (A) Schematic representation of the high-throughput screening (HTS) strategy to identify PARP1-dependent DUBs. (B) Phylogenetic analysis of the six identified DUBs. The sequence-based phylogenetic tree was analyzed by Molecular Evolutionary Genetics Analysis software (MEGA6). (C) PAR overlay assay. Recombinant DUBs were separated by SDS-PAGE and then used in the PAR overlay assay. Iduna was used as a positive control. (D) Schematic illustration of functional domains of ZF-UBP DUBs. (E) GFP-USP39 was transfected into U2OS cells and then treated with either an ATM inhibitor (KU55933) or a PARP inhibitors (PJ34 and Olaparib). Stripe formation by USP39 was analyzed in living cells. Data represent the mean ± standard error of the mean (s.e.m.) from ten cells or more. For parametric multiple comparison, one-way analysis of variance followed by the Tukey-Kramer test was used. (***P ≤ 0.001). Each experiment was performed independently three times. (F–H) Endogenous USP39 is accumulated at the DNA lesions. DNA lesions were induced by mIR in U2OS cells and the translocation of endogenous USP39 to the lesions was analyzed using immunostaining as indicated (F). Each experiment was performed independently three times. ChIP-qPCR analysis for the distribution of endogenous USP39 (G) and γH2AX (H) in FokI-induced DSBs (G and H). Data represent mean ± s.e.m. from three independent ChIP-qPCR analyses. Each dot represents the mean of one experiment. Statistical significance was determined using the Student's t-test (***P ≤ 0.01). IgG was used as negative control. Scale bars, 5 μm.
Human USP39 encodes a well-conserved protein that is homologous to Sad1 in Saccharomyces cerevisiae, except for an additional 67 amino acids that are only conserved in mammals (Supplementary Figure S2B). Both yeast Sad1 and human USP39 are spliceosome components and are classified as DUBs, but lack DUB activity because their core protease active sites are mutated (59,60). However, the functional role of USP39 in DDR remains unclear. Therefore, we focused on investigating the functional effect of USP39 in the DNA-damaged state. To determine the role of USP39 in PARP-coupled DDR, we monitored whether USP39 localizes to damaged chromatin in the presence of an ATM or a PARP inhibitor. Consistent with the primary DUB screening, USP39 was not enriched in laser-induced DSB-containing stripes in the presence of PARP inhibitors including PJ34 or Olaparib (Figure 1E and Supplementary Figure S3A). The ATM inhibitor, KU55933, failed to prevent USP39 enrichment in laser-induced DSB-containing stripes (Figure 1E and Supplementary Figure S3A and B). In addition, we detected endogenous USP39 accumulation in mIR- and FokI-induced DNA lesions (Figure 1F–H).
USP39 recruitment to laser-induced DSB-containing stripes was also examined in ATM or PARP1 knockout mouse embryonic fibroblast (MEF) cells. We found that PARP1 is a key factor for USP39 recruitment to DSB-containing DNA lesions, consistent with the inhibitor experiments (Supplementary Figure S3C). Next, the relative retention kinetics between USP39 and PAR were assessed at laser-induced DSB-containing stripes. We observed that both proteins behaved similarly in DNA lesions, suggesting that PARP1 activity is directly linked to USP39 recruitment to DNA lesions (Supplementary Figure S3D). As expected, RNF168, an ATM pathway-coupled ubiquitin E3 ligase, was recruited to DNA lesions in an RNF8-dependent and RNF20-independent manner. In contrast, the ATM-dependent pathway was unnecessary for USP39 recruitment (Supplementary Figure S3E–G). These data suggest that USP39 is linked to the DDR, and that USP39 requires PARP1 activity. Finally, these results suggest that USP39 in the DDR relies on the hierarchy of PARP1 activation.
USP39 is an essential DUB for the regulation of DSB repair
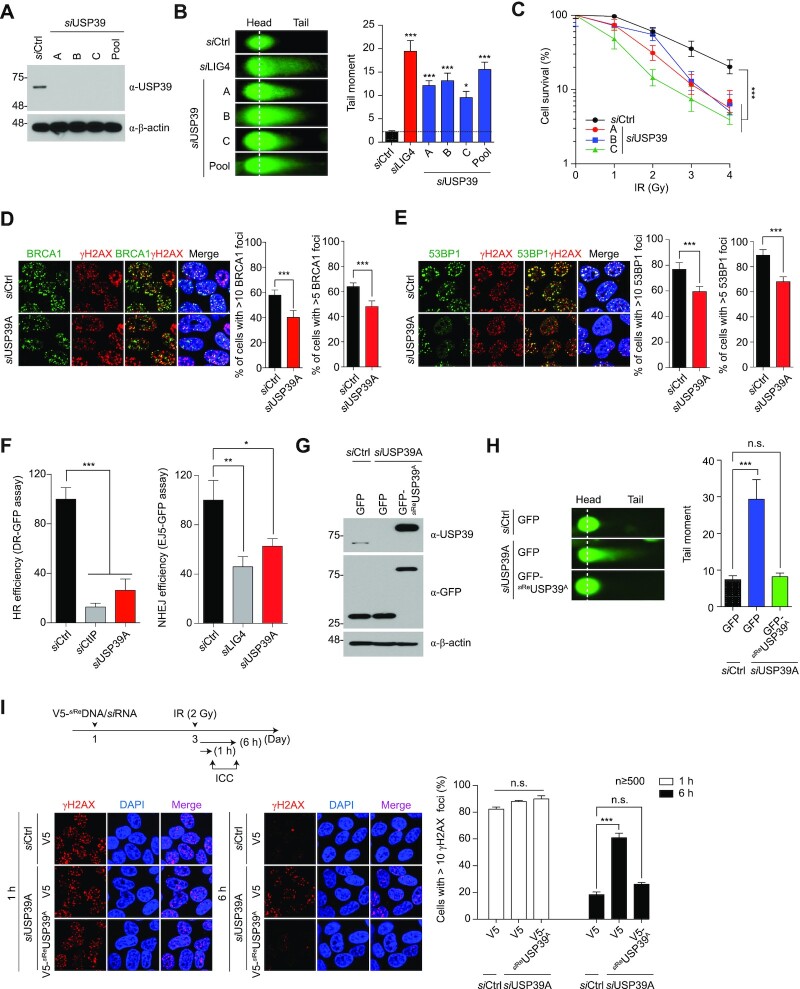
Next, we investigated the physiological roles of USP39 in DNA repair processes. To ascertain whether USP39 modulates DSB repair, we knocked down USP39 with individual or pooled siRNAs targeting three different USP39 mRNA regions. Then, we performed a neutral comet assay after treatment with zeocin, a radiomimetic chemical drug (Figure 2A, B and Supplementary Figure S4A). USP39 ablation induced severe genomic instability similar to LIG4 knockdown, which is a key regulator of NHEJ (Figure 2B and Supplementary Figure S4A). We also found that micronuclei were significantly increased by USP39 knockdown, indicating that the USP39-coupled DSB repair process is directly linked to the maintenance of chromosomal stability (Supplementary Figure S4B). A clonogenic survival assay revealed that cell viability was significantly decreased by USP39 knockdown when compared to that in the siRNA control, and this decrease was dependent on the radiation dose (Figure 2C).
Figure 2.
USP39 is a key factor for maintenance of genomic integrity and cell survival. (A) The knockdown efficacy of each USP39 siRNA or its pool (siUSP39-A, -B and -C mixture) was tested in U2OS cells. (B) Ablation of endogenous USP39 leads to genomic instability. USP39-depletion induced genomic instability, which was monitored by a neutral comet assay (left panel), and the level of genomic fragmentation was quantified as indicated (right panel). siRNA-targeting LIG4 was used as a positive control for the comet assay. Statistical significance was determined using one-way ANOVA followed by the Tukey Kramer test (C) Clonogenic cell survival assay. Statistical significance was determined using one-way ANOVA followed by the Tukey Kramer test. (D and E) Analysis of BRCA1 and 53BP1 IRIF formation. U2OS cells were transfected with control or USP39 siRNAs. Cells were fixed and stained with BRCA1 (D) or 53BP1 (E) antibodies, and nuclei were counterstained with DAPI as indicated by experimental conditions (left panel). Foci formation per cell ≥ 100 was calculated as indicated (right panel). Statistical significance was determined by the Student's t-test (F) Comparative analysis of HR and NHEJ repair activities in USP39 knockdown cells. siCtIP and siLIG4 were used as a positive control for HR and NHEJ repair assay, respectively. Statistical significance was determined using one-way ANOVA followed by the Tukey Kramer test (G) GFP-siReUSP39 was generated, and its resistance against USP39 siRNA was tested. (H) GFP-siReUSP39 was reintroduced into cells with ablated endogenous USP39 and USP39-dependent regulation of genomic stability was monitored and quantified as described above. Statistical significance was determined using one-way ANOVA followed by the Tukey Kramer test (I) USP39 is involved in the early response to DNA damage. USP39 siRNA and V5-siReUSP39 were transfected into U2OS cells and 48 h later, cells were treated with γ-irradiation (2 Gy). The number of γH2AX foci at DSB sites was monitored (left panel) and quantified (right panel). Statistical significance was determined using one-way ANOVA followed by the Tukey Kramer test. Data represent the mean ± s.e.m. from three hundred cells or more. All results represent at least three independent experiments. ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05. n.s., not significant.
DSBs are mainly repaired by HR and NHEJ, and BRCA1 and 53BP1 play decisive roles in which repair process is used. In addition, BRCA1 and 53BP1 antagonistically control the temporal choice between NHEJ and HR (61,62). To determine whether USP39 is a crucial factor in HR or NHEJ repair processes, we observed BRCA1- and 53BP1-irradiation-induced foci (IRIF) formation, their respective IRIF formations representing each repair pathway. Remarkably, we found that both BRCA1 and 53BP1 IRIF require USP39 for efficient repair, suggesting that USP39 may simultaneously control HR and NHEJ (Figure 2D and E). To further confirm the function of USP39 in the DNA repair pathway, we performed DNA repair analysis using a GFP reporter system to measure the efficiency of HR or NHEJ during USP39 knockdown. Consistent with previous results, we observed that USP39 depletion reduced both HR and NHEJ repair (Figure 2F). To obtain corroborating evidence, the same experiment was conducted using alternative USP39 siRNAs (siUSP39-B and -C), and similar results were obtained (Supplementary Figure S5A–C).
Next, to verify that genomic instability is directly caused by USP39 knockdown, we generated an siRNA-resistant USP39 construct (siReUSP39) and introduced this construct into cells with ablated endogenous USP39. As expected, the increased DNA fragmentation caused by USP39 depletion was dramatically reversed by siReUSP39 overexpression, indicating that endogenous USP39 is directly involved in the maintenance of genomic integrity (Figure 2G and H). ATM and MDC1 recruitment-mediated Ser139 phosphorylation in H2AX (γH2AX) is required for ionizing IRIF formation at DNA break sites within chromatin. Thus, the formation and resolution of γH2AX IRIFs comprise the DSB repair process. To examine whether USP39 is a key factor in this DNA repair process, cells with ablated USP39 or siReUSP39 overexpression were γ-irradiated (2 Gy), and γH2AX IRIFs were counted at different time points. One hour after irradiation, the number of γH2AX IRIFs in the USP39 knockdown cells was comparable to the foci numbers in cells rescued by siReUSP39. However, 6 h after irradiation, the siReUSP39-rescued cells displayed a significantly lower number of γH2AX IRIFs than that of USP39 knockdown cells (Figure 2I and Supplementary Figure S5D). Interestingly, we observed that the number of 53BP1 IRIFs in the USP39 knockdown cells was significantly reduced 6 h post irradiation, suggesting that USP39 is involved in 53BP1 recruitment to DNA lesions (Supplementary Figure S5E). In parallel, as shown Figure 2D, we also observed that USP39 depletion leads to defective BRCA1 foci formation. Increasing evidence states that BRCA1 is part of several complexes with distinct roles in DSB signaling and repair (63). To ensure the role of USP39 in BRCA1-mediated HR repair, we further monitored the formation of RAD51 foci since RAD51 is known to be a downstream marker for DNA end section and HR repair. As expected, we found that RAD51 foci were significantly reduced by USP39 knockdown, suggesting that USP39 could act as an upstream modulator for BRCA1-mediated HR repair (Supplementary Figure S5F).
In part, the choice of DNA repair pathway is determined by the stage of the cell cycle. Recent studies have shown that silencing of USP39 could lead to cell cycle arrest at G0/G1 (64), G1 (65) or G2/M phases (66) in different cell types. To clarify whether USP39 depletion alters the cell cycle distribution in U2OS cells, we analyzed the cell cycle of USP39-depleted cells under double thymidine blocking using flow cytometry, which showed a significant increase in the population of cells in the G1 phase, and a simultaneous decrease in the G2/M phase of the cell cycle, compared with those in the control. Consistently, USP39 knockdown led to G1 arrest in asynchronized U2OS cells, suggesting that USP39 may be required for the G1/S transition (Supplementary Figure S6). The preference for NHEJ in G1 phase and HR in S and G2 phases implies that the modes of DSB repair are regulated during the cell cycle. At the same time, cell cycle checkpoints play a critical role in delaying the onset of mitosis until DSB repair is completed. Although, it is unclear how the regulation of cell cycle progression by USP39 affects DSB repair, our results indicate that USP39 is an indispensable factor in the regulation of the cell cycle-coupled DNA repair pathway (Supplementary Figure S6). Taken together, these data show that USP39 plays a key role in NHEJ and HR repair processes, thereby regulating cell viability after IR-induced DNA damage in human cells.
The tripartite RG motif of USP39 is crucial for its recruitment to DNA lesions during NHEJ repair
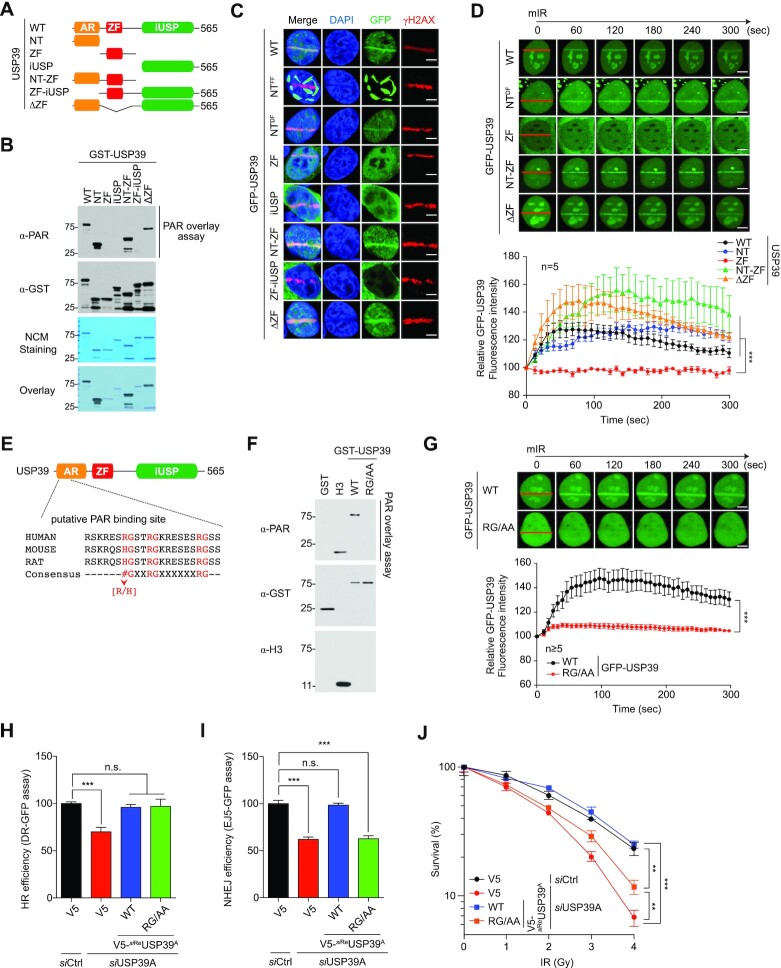
Given that USP39 strongly binds to PAR chains, and that USP39 recruitment to DNA lesions depends on PARP1 activity in the DDR, thereby regulating the DNA repair process for cell survival, various USP39 mutants were generated to identify the USP39 region involved in its recruitment to DNA lesions (Figure 3A and Supplementary Figure S2B). These mutations were: N-terminal (NT; 1–103), UBP-type zinc finger (ZF; 104–224), inactive USP domain (iUSP; 225–565), NT-zinc finger (NT-ZF; 1–224), ZF-iUSP (104–565) and ΔZF (Δ; 104–224). To identify which USP39 region is required for PAR chain binding, we performed PAR chain overlay assays using WT and USP39 mutants. Interestingly, we observed that the NT region containing an AR-rich motif is responsible for interacting with PAR chains (Figure 3B). We also observed that all mutants containing the USP39 NT region were localized to the nucleus, whereas the others were predominantly expressed in the cytosol, suggesting that amino acids 1–103 are necessary for nuclear localization. Strikingly, we found that the nuclear expression patterns of NT were clearly separated into a dispersed form (DF, hereafter named NTDF) or a filament-like form (FF, hereafter referred to as NTFF), which is presumably caused by increased USP39 aggregation under certain conditions, such as increased protein expression level. To investigate how PAR chain binding ability affects USP39 recruitment, the behavior of all mutants was examined with the mIR system. Remarkably, all mutants containing NT and NTDF localized to DSBs, whereas NTFF failed to translocate to damaged chromatin, suggesting that the NT region is indispensable for sensing DSBs and recruiting USP39 to DNA lesions (Figure 3C). To further validate the role of NT in USP39 recruitment, the translocation rate of WT and mutant USP39 was monitored in living cells using the mIR system. As expected, all mutants containing NT moved rapidly to laser-induced stripes, whereas the other mutants did not, indicating that NT is a bona fide region facilitating USP39 recruitment to DNA lesions (Figure 3D). To narrow down the PAR chain target sequences, we performed domain mapping by generating more specific NT deletion mutants: NT1-98, NT1-46 and NT47-98. These mutants were assessed using the PAR chain overlay assay and the mIR system. NT1-46 (hereafter referred to as N46), the first half of NT, contains critical amino acids for binding to PAR chains and for translocation to DNA lesions (Supplementary Figure S7A–C). These data suggest that N46 of USP39 is required for PAR chain binding and DSB sensing. Intriguingly, N46 contains a tripartite RG motif, a conserved PAR chain binding sequence composed of an RG-X2-RG-X6-RG layer, suggesting that this motif might be important for connecting PAR chains to USP39 (Figure 3E). To further test whether the RG repeats in N46 are critical for regulating USP39-mediated DNA repair and cell survival, we generated RG repeat point mutants with mutations of all RG residues to alanine (hereafter referred to as RG/AA). As expected, mutating RG motifs in N46 decreased USP39 PAR chain binding (Figure 3F and Supplementary Figure S7D). We also found that RG/AA mutants do not translocate to DNA lesions, suggesting that PAR chain binding is crucial for USP39 localization to DSBs (Figure 3F and G). Remarkably, we observed that the frequency of NTFF generation was significantly reduced by NT46 RG/AA overexpression, indicating that USP39-mediated generation of NTFF is directly linked to either PARP1 activity or PAR (Supplementary Figure S7E). In addition, we also found that reintroducing either WT USP39 or the RG/AA mutant rescued USP39 depletion-mediated HR failure, but the RG/AA mutant failed to rescue the NHEJ defect caused by USP39 ablation. These results indicate that USP39 regulates NHEJ but not HR in a PAR-binding dependent manner (Figure 3H and I). Using clonogenic analysis, we also confirmed that the RG/AA mutant failed to rescue USP39 knockdown-mediated cell death (Figure 3J). Taken together, these data suggest that the tripartite RG motif of USP39 is crucial for USP39-coupled NHEJ repair in a PAR-dependent manner.
Figure 3.
The RG motif of USP39 is not only involved in interaction with PAR-chains but also essential for its recruitment to DNA lesions. (A) Schematic illustration of USP39 mutants. (B) Analysis of PAR-binding activity of USP39 WT or each deletion mutant. Indicated proteins were purified from insect cells and then used in PAR overlay assays. PAR-binding activity was monitored by immunoblot with the indicated antibody. (C and D) The N-terminal region of USP39 is critical for its recruitment to mIR-induced DSBs. Indicated mutants of GFP-tagged USP39 were transfected to U2OS cells, and stripe formation by USP39 WT or deletion mutants was analyzed from fixed (C) or living cells (D) as indicated. Data represent the mean ± s.e.m. from five cells. Statistical significance was determined using one-way ANOVA followed by the Tukey Kramer test (E) Identification of tripartite RG motifs as the putative PAR-binding sites in USP39. (F) Recombinant USP39 WT and RG/AA mutant were subjected to the PAR overlay assay as indicated. GST or H3 was used as negative or positive controls, respectively. (G) Tripartite RG motifs are critical for translocation of USP39 to mIR-induced DSBs. GFP-USP39 WT or RG/AA mutant were transfected into U2OS cells and mIR-induced stripe formation was monitored in living cells (upper panel). The efficacy of translocation was quantified as indicated (lower panel). Data represent the mean ± s.e.m. from five cells or more. Statistical significance was determined by the Student's t-test. (H and I) Comparative analysis of HR (H) and NHEJ (I) repair activities in USP39 knockdowns or cells rescued by reintroduction of siReUSP39 or RG/AA as indicated. Statistical significance was determined using one-way ANOVA followed by the Tukey–Kramer test (J) Clonogenic cell survival assay, which was performed using the indicated experimental conditions. Statistical significance was determined using one-way ANOVA followed by the Tukey Kramer test. Scale bars, 5 μm. Data represent the mean ± s.e.m. of three independent experiments. ***P ≤ 0.001, **P ≤ 0.01. n.s., not significant.
The tripartite RG motif of USP39 is crucial for PAR chain-mediated liquid demixing at DNA lesions
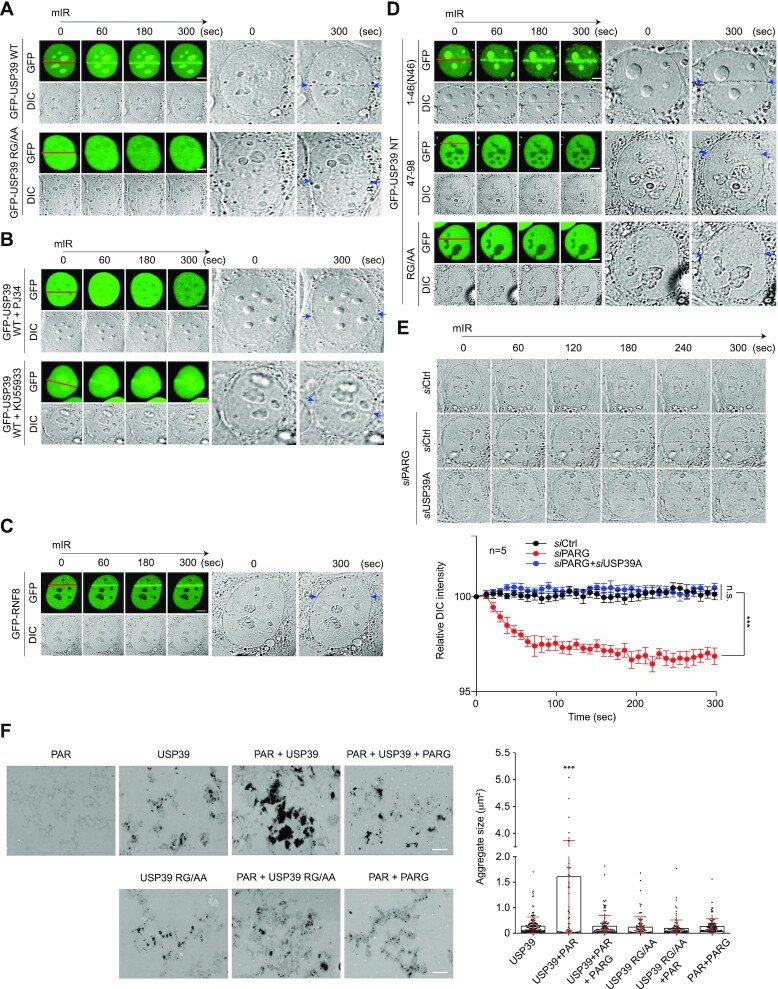
Recently, it was proposed that intrinsically disordered proteins containing RG/RGG motifs strongly drive the generation of membrane-less compartments at DNA lesions by a phase transition called liquid demixing, which in turn dynamically orchestrates the DDR (32). To determine whether these RG repeats are involved in PAR chain binding and liquid demixing, we performed mIR assay with WT USP39 and RG/AA mutant. Surprisingly, we observed that WT USP39 forms transient, distinct light-diffracting dark stripes exactly at USP39 accumulation regions, whereas RG/AA mutants failed to induce liquid demixing-mediated dark stripe formation (Figure 4A). Consistent with these results, PJ34, a PARP inhibitor, completely eliminated USP39-mediated light-diffracting dark stripe formation (Figure 4B). Additionally, USP39-mediated liquid demixing during the DDR was found to only depend on PARP1 activity, since the ATM inhibitor KU55993 failed to prevent USP39-mediated dark stripe formation (Figure 4B). Moreover, ATM-dependent GFP-RNF8 moved to mIR-induced DNA lesions but was not accompanied by liquid demixing, suggesting that the ATM pathway is not directly linked to liquid demixing (Figure 4C). To verify whether USP39 N46 is an essential region for facilitating PAR-mediated liquid demixing, we monitored liquid-demixing-mediated dark stripe formation in cells expressing the N46 RG/AA mutant. As expected, N46 itself induced dark stripe formation (Figure 4D). To further confirm whether endogenous USP39 is physiologically linked to PAR-seeded liquid demixing, we monitored mIR-induced dark stripe formation in cells depleted of endogenous PAR glycohydrolase (PARG) and/or USP39. Strikingly, we observed that PARG knockdown-mediated dark stripe formation was completely abolished by USP39 knockdown, indicating that USP39 is a major factor in the regulation of PAR-mediated liquid demixing (Figure 4E).
Figure 4.
USP39 drives PAR-seeded liquid demixing via its RG repeats in the N46 region. (A–C) USP39 WT leads to transient formation of distinct light-diffracting dark stripes via its RG motif. (A) USP39 WT and RG/AA mutant were transfected into U2OS cells and then monitored for translocation to DSBs using a mIR system. (B) The formation of USP39-mediated dark stripes was monitored in presence of a PARP inhibitor. (C) ATM-dependent RNF8 was used as negative control. mIR-induced dark stripe formation was monitored in bright-field. Each right panel show a 12× magnification of the last DIC images. Red lines indicate a micro-irradiated area and blue arrows point to light-diffracting dark stripes. Scale bars, 5 μm. (D) The N46 of USP39 is sufficient for phase separation. The N-terminal regions of GFP-USP39 (1–46, 47–98, RG/AA) were transfected into U2OS cells that were then monitored for the formation of dark stripes after mIR-induced DNA damage. Scale bars, 5 μm. Each right panel shows a 12× magnification of the last DIC image. Red lines indicate a micro-irradiated area and blue arrows point to light-diffracting dark stripes. (E) Endogenous USP39 regulates PAR-seeded phase separation. siUSP39 or siPARG were transfected into U2OS cells and mIR-induced dark stripe formation was then monitored using bright-field microscopy. Representative images are shown in the upper panel and quantification results are shown in the lower panel. Scale bars, 5 μm. Statistical significance was determined by the Student's t-test. (F) USP39 WT and RG/AA mutant were incubated at RT for 24 h with or without PAR or PARG-treated PAR. Aggregate sizes were analyzed by TEM (left panel) and quantified (right panel). Statistical significance was determined using one-way ANOVA followed by the Tukey Kramer test. Scale bars, 2 μm. Data represent the mean ± s.e.m. of three independent experiments and quantification results represent the mean ± s.e.m. from five cells. ***P ≤ 0.001, n.s., not significant.
However, although PAR, generated by activated PARP1, and endogenous USP39 were predicted to localize to the mIR-damaged chromatin, mIR-induced dark stripes were not observed under endogenous conditions (Figure 4C and E). This result suggests that the amount of PAR synthesized on the damaged chromatin is not sufficient to produce dark stripes caused by liquid demixing under endogenous conditions. To address this point, further experiments were performed using a high-powered laser that increased PAR production from damaged chromatin, and the formation of dark stripes was monitored under endogenous conditions (Supplementary Figure S8). Indeed, we observed that dark stripes were produced by mIR with high-powered laser under endogenous conditions. Moreover, we also observed that these dark stripes disappeared significantly with USP39 knockdown (Supplementary Figure S8). These data clearly support that endogenous USP39 is also involved in PAR-dependent liquid demixing upon laser-induced DNA damage.
Previously, Altmeyer et al. reported that PAR chains facilitate liquid demixing-mediated IDP assembly via protein RG motifs in a cell-free system (32). This leads to the hypothesis that PAR chains could induce the assembly of USP39 at the end of liquid demixing in a similar way. To address this hypothesis, we monitored USP39 assembly using transmission electron microscopy (TEM) in a cell-free system. Remarkably, we observed that co-incubation of USP39 with PAR chains at physiological pH significantly enhanced USP39 aggregation, resulting in larger or more condensed aggregates (Figure 4F). No comparable aggregate structures were found in samples incubated with PAR alone (Figure 4F). TEM-based quantification of USP39 aggregates confirmed that larger aggregates were significantly increased in samples incubated with PAR (Figure 4F). Importantly, the increased aggregate size was completely lost when PAR was degraded by recombinant PARG prior to the addition of USP39 (Figure 4F). Consistent with the role of the USP39 RG tripartite motif as a PAR sensor, the USP39 RG/AA mutant failed to create aggregates when incubated with high concentrations of PAR chains (Figure 4F). Combined with the in vivo analysis, these data provide evidence for the intrinsic ability of PAR chains to induce USP39 condensation and aggregation. Taken together, these results indicate that the USP39 tripartite RG motif is important for interactions with PAR chains and is critical for specific NHEJ regulation in the DDR by PAR-USP39-seeded liquid demixing.
USP39 directly drives XRCC4/LIG4 dependent NHEJ repair
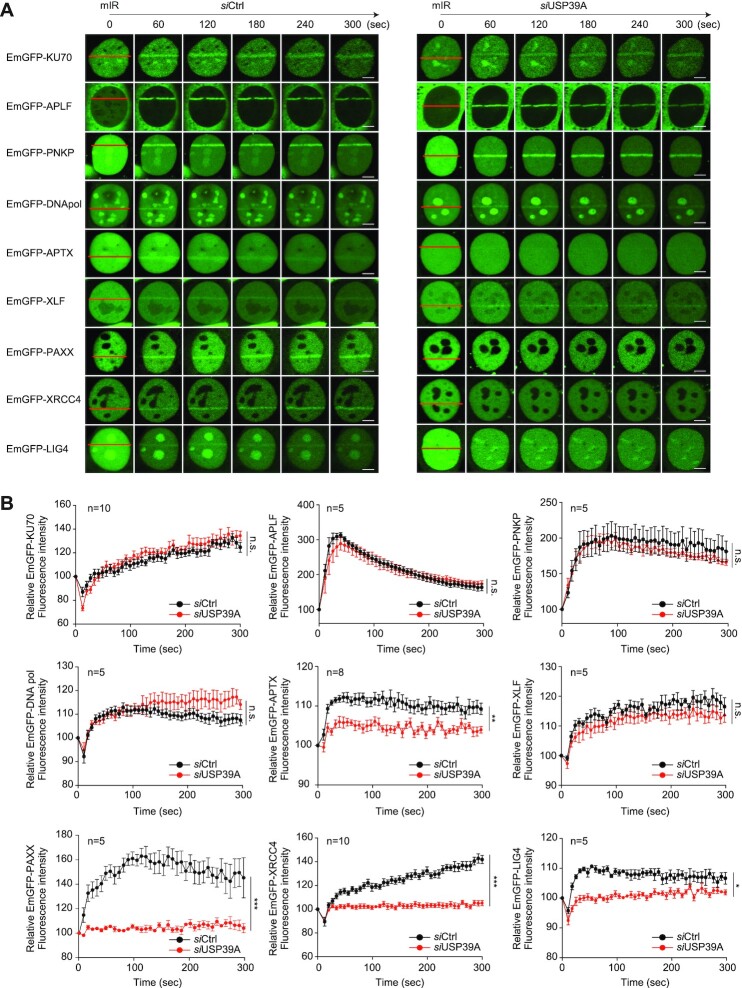
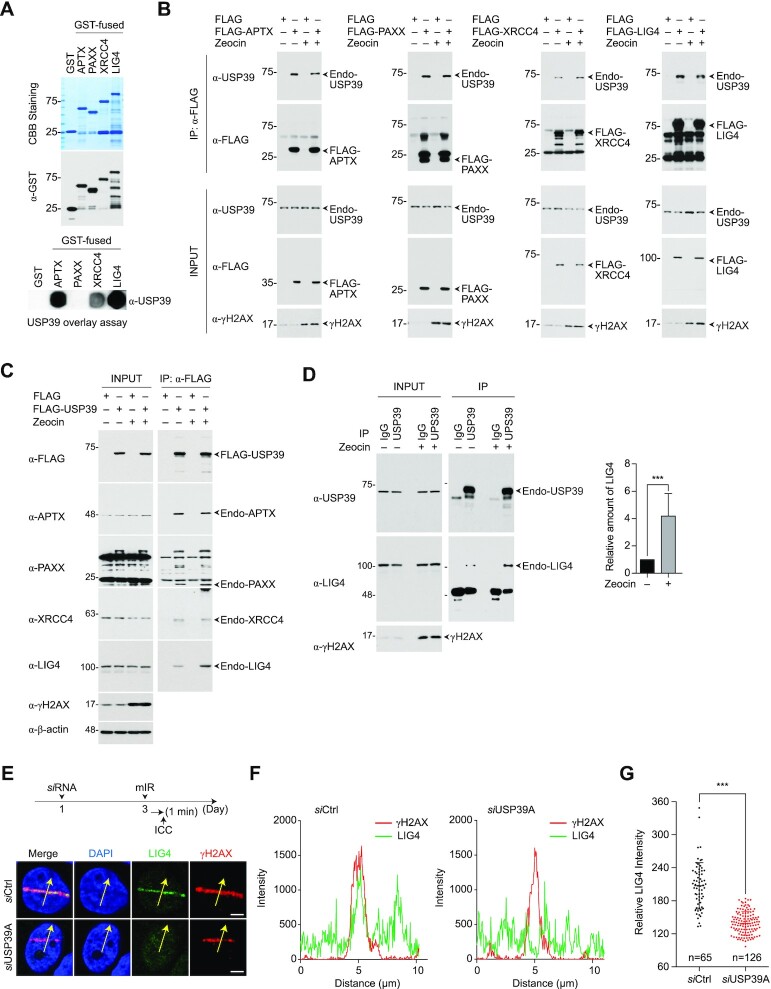
We next explored the molecular mechanism underlying NHEJ regulation by USP39. To determine where USP39 physically exerts its influence on the NHEJ cascade, we knocked down USP39 and monitored the recruitment of nine major regulatory factors involved in NHEJ using the mIR system. Among them, we found that APTX, PAXX, XRCC4 and LIG4 (hereafter collectively referred to as APXL proteins) exhibit USP39-dependent recruitment to mIR-induced DNA lesions, indicating that USP39 acts as an upstream regulator in the NHEJ cascade (Figure 5A and B). To further elucidate the functional role of USP39, we analyzed the physical interactions between USP39 and its downstream factors. Indeed, GST-free USP39 directly interacts with recombinant APTX, XRCC4 and LIG4, but not with PAXX (Figure 6A). Then, using an immunoprecipitation assay, we validated that endogenous USP39 interacts with exogenously expressed APXL proteins and vice versa in the presence of a DNA-damaging reagent, suggesting that USP39 is an indispensable factor in driving APXL protein-mediated NHEJ repair (Figure 6B and C).
Figure 5.
USP39 controls the recruitment of XRCC4/LIG4 to DSBs for NHEJ repair. (A and B) USP39 is key regulator of the NHEJ repair pathway by controlling the recruitment of APXL proteins (APTX, PAXX, XRCC4, and LIG4). The indicated NHEJ regulatory factors were transfected into U2OS cells along with controls or USP39-targeted siRNA. After 48 h, cells were analyzed in the mIR system and translocation efficacy was monitored (A) and quantified (B). Statistical significance was determined by the Student's t-test. Scale bars, 5 μm. Data represent the mean ± s.e.m. from five cells or more. ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05. n.s., not significant.
Figure 6.
USP39 is a key factor in creating NHEJ complexes by interplay with PAXX, APTX, XRCC4 and LIG4. (A) GST-free USP39 directly interacts with recombinant GST-APTX, GST-XRCC4 and GST-LIG4, but not GST-PAXX. Indicated recombinant proteins were purified from insect cells and used in a USP39 overlay assay. GST was used as negative control. (B and C) Cell extracts from HEK293FT-expressing FLAG-tagged NHEJ factors (B) or FLAG-USP39 (C) were immunoprecipitated with anti-FLAG antibody and then analyzed by immunoblotting. (D) Endogenous USP39 interacts more strongly with endogenous LIG4 in the DNA-damaged state. The cell lysates derived from normal or DNA-damaged cells were immunoprecipitated with anti-USP39 antibody, and then analyzed for interaction between endogenous USP39 and endogenous LIG4. (E–G) Depletion of USP39 leads to failure of proper accumulation of endogenous LIG4 at DSBs. siRNA-targeting USP39 was transfected into U2OS cells and then stripe formation of endogenous LIG4 was analyzed. Yellow arrows indicate the area of signal intensity measurement (E). The accumulated level of γH2AX or LIG4 at the DNA lesions was analyzed by Nikon NIS software (F). Relative intensity of endogenous LIG4 accumulated at DSBs was monitored in cells expressing either siCtrl or siUSP39. n indicates the total cell number used in quantification (G). Statistical significance was determined by the Student's t-test. Scale bars, 5 μm. Data represent the mean ± s.e.m. of three independent experiments. ***P ≤ 0.001.
Moreover, we confirmed that endogenous USP39 interacts with endogenous LIG4 and observed using IP that LIG4 is more enriched with USP39 in the DNA-damaged condition (Figure 6D). Subsequently, we assessed the recruitment of endogenous LIG4 to DNA lesions in USP39 knockdown cells. As expected, LIG4 failed to translocate into the stripes (Figure 6E-G). Using human non-cancer lung fibroblasts (MRC-5 cells), we also demonstrated that the recruitment of both GFP-XRCC4 and GFP-LIG4 to damaged chromatin was drastically inhibited by USP39 knockdown (Supplementary Figures S9A-C). These results indicate that USP39 is an essential factor for LIG4 translocation to DSBs and NHEJ in the DDR.
Recently, PAXX and XLF were shown to have overlapping functions in the KU80-dependent NHEJ pathway (67–69). To clarify their function in USP39-PAR-coupled NHEJ, we investigated PAR-dependent PAXX and XLF recruitment. Interestingly, we found that the PARP inhibitor, PJ34, completely abolishes PAXX recruitment to DSBs, but not XLF, indicating that XLF and PAXX play distinct roles in PAR-mediated DNA repair (Supplementary Figure S10A and B). Emerging evidence revealed that Ku70, Ku80, XRCC4 and LIG4 are essential components for DSB repair via NHEJ and that XRCCR and LIG4 form a tight complex for the end-joining process (70). Furthermore, XRCC4 is translocated to DSBs in a PARP1-dependent manner (71). Because USP39 directly interacts with the XRCC4/LIG4 complex and is involved in the translocation to DSBs, we examined whether PARP1 regulates XRCC4/LIG4 complex translocation to DSBs in a USP39-dependent manner. We monitored the kinetics of XRCC4/LIG4 complex accumulation in mIR-induced DSBs in PARP1 knockout MEF cells. Consistent with the results of a previous study, XRCC4 and LIG4 require PARP1 to move into DSBs (Supplementary Figure S10C and D). Taken together, these observations show that USP39 may play a role as a regulator of the APXL cascade in NHEJ repair by generating a bridge between Ku family proteins and the XRCC4/LIG4 complex.
The USP39 ZF domain is important for XRCC4/LIG4 complex recruitment
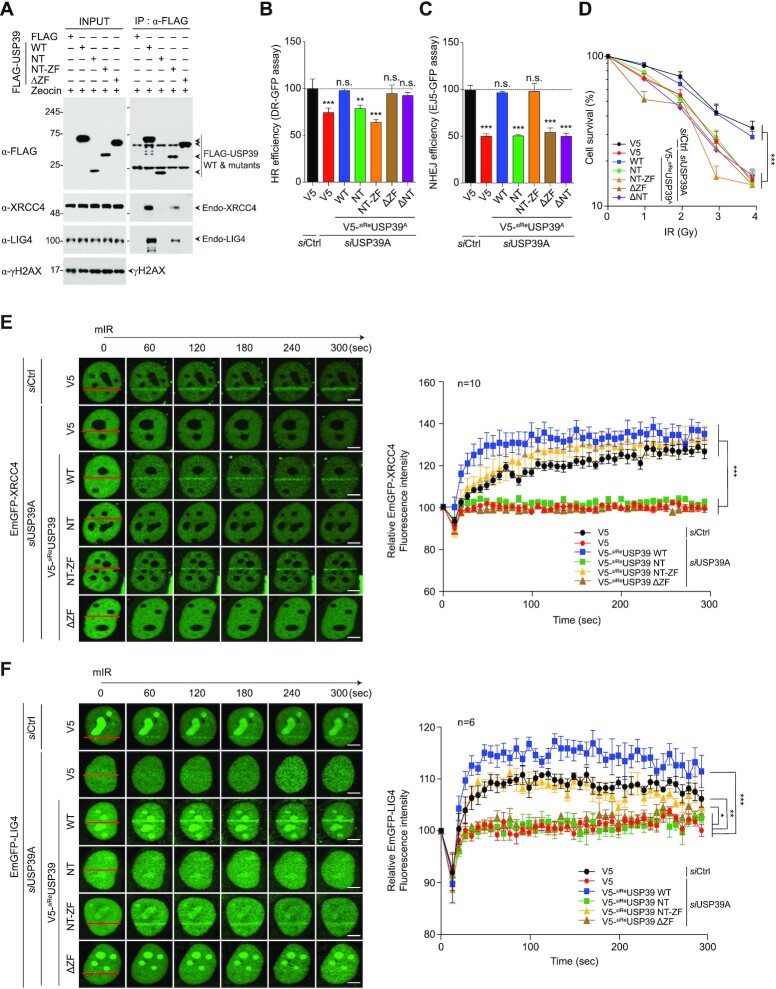
To obtain a more detailed view of the interaction between USP39 and the XRCC4/LIG4 complex, we investigated which USP39 domain is responsible for interaction with both XRCC4 and LIG4. We used immunoprecipitation analysis with cells expressing USP39 deletion mutants (WT, NT, NT-ZF and ΔZF). We observed that both USP39 WT and NT-ZF strongly interacted with endogenous LIG4, whereas NT and ΔZF mutants failed to bind, indicating that the USP39 ZF domain is critical for its interaction with the XRCC4/LIG4 complex (Figure 7A). Next, we monitored the effect of the ZF domain in USP39-mediated HR or NHEJ repair. Surprisingly, we found that USP39-linked HR repair does not require the NT or ZF domains. However, both domains are necessary for NHEJ repair. Thus, USP39 specifically regulates NHEJ via its ZF domain, which itself regulates the interaction with XRCC4/LIG4 complex (Figure 7B and C). In parallel, we examined whether the ZF domain is important for USP39-mediated NHEJ repair or cell survival. Consistently, we observed that the ZF domain is essential for USP39-mediated cell survival, together with the N-terminal RG motifs (Figure 7D). Subsequently, we evaluated the recruitment of the XRCC4/LIG4 complex to DSBs in cells expressing siReUSP39 or USP39 deletion mutants (siReNT, siReNT-ZF, siReZF and siReΔZF), which are resistant to USP39 siRNA. As expected, both XRCC4 and LIG4 translocated to DNA lesions in a ZF domain-dependent manner (Figure 7E, F and Supplementary Figure S11). These data raise the question as to whether the USP39 ZF domain is involved in liquid demixing, along with the tripartite RG motif in N46. To clarify this question, we performed an mIR experiment with WT USP39 and its ΔZF mutant and monitored liquid-demixing-mediated dark stripe formation at DNA lesions. Intriguingly, we found that the USP39 ZF domain was not required for the transient formation of the distinct light-diffracting dark stripes (Supplementary Figure S12). These data indicate that the USP39 ZF domain is a major determinant of its interaction with the XRCC4/LIG4 complex at DNA lesions but not for USP39-mediated phase separation by liquid demixing.
Figure 7.
The ZF domain of USP39 is an essential region for interaction with XRCC4 and LIG4. (A) The ZF domain of USP39 serves as a docking site for the interaction with XRCC4/LIG4 complex. Cells were transfected with FLAG-tagged WT or mutant of USP39 and were subjected to IP with FLAG antibody and then analyzed by immunoblotting. (B and C) Comparative analysis of HR (B) and NHEJ (C) repair activities in USP39 knockdown cells or cells rescued by reintroducing siReUSP39 or deletion mutants. Statistical significance was determined using one-way ANOVA followed by the Tukey Kramer test. (D) A clonogenic cell survival assay was performed using the indicated experimental conditions. Statistical significance was determined using one-way ANOVA followed by the Tukey Kramer test. (E and F) The ZF domain of USP39 is important for recruitment of XRCC4 and LIG4. GFP-tagged XRCC4 or LIG4 were transfected into USP39 knockdown cells along with the indicated siRNA-resistant V5-fused WT or USP39 mutants. Stripe formation by each deletion mutant was monitored (left panels in E and F) and the efficacy of translocation was quantified by Nikon NIS software as indicated (right panels in E and F). Statistical significance was determined using one-way ANOVA followed by the Tukey–Kramer test. Scale bars, 5 μm. Data represent the mean ± s.e.m. of three independent experiments and quantification results represent the mean ± s.e.m. from five cells. ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05. n.s., not significant.
USP39 regulates HR repair in a spliceosome complex-dependent manner
USP39 was first identified as a component of a spliceosome complex that modulates tri-snRNP assembly (72). More recently, this spliceosome was shown to promote HR repair during DDR (73). To determine whether USP39-mediated DNA repair processes are linked to spliceosome activity, we conducted a DNA repair assay using cells depleted of SART1, SART3 or snRNP27, which are critical factors for tri-snRNP complex assembly. Surprisingly, HR repair was significantly diminished by SART1, SART3 and snRNP27 knockdown (73). However, NHEJ was only affected by USP39 knockdown, indicating its spliceosome-independent regulation (Supplementary Figure S13A and B). Notably, USP39 is involved in the initial stages of spliceosome assembly, which regulates RNA splicing and controls the HR repair process. To confirm that USP39 regulates the NHEJ pathway in a spliceosome-independent manner, we monitored XRCC4 recruitment to DSBs in SART1, SART3, and snRNP27 knockdown cells. We observed that XRCC4 accumulation in DNA lesions was significantly reduced only in USP39-depleted cells but not in SART1, SART3, or snRNP27 knockdown cells, indicating that USP39 has a spliceosome-independent function in the regulation of XRCC4/LIG4-linked NHEJ (Supplementary Figure S13C). The function of USP39 in the spliceosome is essential for efficient splicing of a large number of transcripts. To rule out that a decrease in transcript levels of USP39-linked factors causes HR or NHEJ defects in USP39 depleted cells, we performed qPCR for USP39-linked HR or NHEJ factors and found that none of the NHEJ factors identified in this study were downregulated by USP39 depletion (Supplementary Figure S13D). However, we found that BRCA1 mRNA was significantly reduced by USP39 depletion. Recently, Wang et al. reported that USP39 is an oncogenic splicing factor in ovarian cancer cells, and its depletion led to global impairment of splicing (74). Interestingly, similar to our results, they also found that the level of BRCA1 mRNA is significantly decreased by USP39 depletion (74). These results strongly support that downregulation of BRCA1 expression may be the result of impaired splicing caused by USP39 depletion. In parallel, we also monitored the protein levels of 53BP1 and BRCA1 in USP39-depleted cells. Intriguingly, no quantitative change of 53BP1 protein was observed, but we found that the amount of BRCA1 protein was significantly reduced, as shown in the Supplementary Figure S13E. Although it is still unclear how the USP39-coupled spliceosome specifically regulates the expression of BRCA1 mRNA, our data strongly support that USP39 could control BRCA1-linked HR process in a PARP1-independent and spliceosome-dependent manner.
DISCUSSION
Protein function depends on the associated amino acid sequence, including its domains and motifs. Comparative proteome analysis of five complete eukaryotic genomes revealed differences in individual and multiple amino acid sequences that seem evolutionarily associated with species-specific cellular responses (75). Herein, we determined that human USP39 is an indispensable factor in mediating PARP1-coupled NHEJ. Further, we demonstrated that the USP39 tripartite RG motif located in the N-terminal region, which is only conserved in mammals, is an essential region for binding PAR chains and facilitating liquid demixing. In contrast, the USP39 ZF domain mediates recruitment of NHEJ factors, including APTX, PAXX, XRCC4 and LIG4, to DNA lesions. In parallel, we showed that USP39 regulates HR via its iUSP domain, which is conserved from yeast to humans, indicating that this domain facilitates HR but not the NHEJ pathway. Intriguingly, emerging evidence shows that the USP39 iUSP domain is indispensable for spliceosome assembly, initiation, and maturation in yeast and human cells (60). Herein, we showed that the iUSP domain is also critical for USP39-mediated HR repair. Remarkably, the iUSP domain and many other residues within USP39 are conserved from yeast to humans, whereas the NT 1–46 region is only conserved in mammals. Indeed, we showed that NT 1–46 is important for interaction with PAR chains, and that this domain is crucial for USP39-mediated regulation of the NHEJ process in mammals.
As previously noted, yeast Sad1, unlike USP39, lacks the N46 region, suggesting that it cannot bind PAR chains or participate in NHEJ in yeast. Notably, the yeast genome does not encode any PARP1 homologs, suggesting that PARP1-coupled NHEJ repair is compromised in yeast. Early studies demonstrated that DNA damage induced by radiation or radiomimetic chemicals is predominantly repaired by HR in yeast because of a limited ability to perform NHEJ.
In contrast, higher eukaryotes efficiently utilize imprecise NHEJ. It remains unclear why compared to mammals, yeast exhibits a limited capacity for imprecise NHEJ. Multiple explanations have been proposed from different perspectives. First, NHEJ in mammals may be related to the difficulty of searching for or locating a homologous template for HR repair in larger genomes, especially because both yeast and humans have defined chromosome territories in the nucleus. Second, mammals possess a more highly programmed DNA-breaking process. For example, PARP1 is involved in B cell processes, including V(D)J recombination that generates diverse antibodies in the immune system (76–78). In higher eukaryotes, imprecise NHEJ appears to be essential for preserving genomic integrity and promoting survival. Lastly, mammals have at least three major factors, DNA-PKs, BRCA1, and Artemis that regulate NHEJ, but these proteins are absent in yeast (79–81). A recent study showed that BRCA1 controls HR and NHEJ in a cell cycle-dependent manner (82). Remarkably, they found that BRCA1 facilitates the fidelity of NHEJ repair during the G1 phase by interacting with KU80. Furthermore, DNA-PKs and BRCA1 are spatially or temporally connected with PARP1 or PAR chains to facilitate DNA repair processes in mammals but not yeast (79,83). Thus, the N46 region of USP39 may have been added in mammals during evolution for its adaptor function to connect PAR signaling and specificity for DDR-linked USP39 function. Although Sad1 does not contain a PAR chain binding motif, it shares 31.86% sequence similarity with human USP39, including two conserved ZF (UBP-type) and iUSP domains, suggesting that these domains may be responsible for the shared function between yeast and humans.
Recently, the Lehner group demonstrated that dosage-sensitive proteins in yeast share physiological properties with proteins known to be involved in liquid demixing (84). In fact, PARPs and their metabolites, PAR chains, are not found in prokaryotes or yeast, indicating that PAR-mediated liquid demixing does not occur in yeast. Intriguingly, Sad1, the yeast homolog of USP39, lacks the human USP39 N46 region, which contains the tripartite RG motif, suggesting that this motif may have evolved in mammals to modulate very complex intracellular signaling pathways, including DNA repair processes.
The PAR sensors FUS and EWS are well characterized by Mastrocola et al. and Wang et al. (29,30). These groups were the first to show RGG-dependent PAR binding and FUS recruitment to DNA lesions. Further, these groups characterized PAR-dependent functions in NHEJ and HR repair in cancer and neuronal cells. Rulten et al. suggested that defects in the PAR binding activity of FUS may contribute to ALS (31). These studies strongly indicate that RBP PAR binding activity occurs via RGG motifs and are required for physiological DNA repair. In addition, RGG motifs in FUS or EWS are essential for inducing liquid demixing to generate membrane-less compartments, which act as filtrate barriers to control the transit of DNA repair-associated factors to DNA lesions (32,33). Although these studies showed that FUS and EWS proteins regulate PAR-dependent liquid demixing, they did not explain how PAR-associated FUS or EWS control the recruitment of other factors involved in DNA repair. In the present study, we show that USP39 has a tripartite RG motif and a ZF domain, which are required for USP39-mediated, PAR-dependent liquid demixing and XRCC4 and LIG4 recruitment, respectively. These findings suggest that FUS and USP39 simultaneously induce PAR-dependent liquid demixing in DNA lesions but interact with different repair factors via their specific domains, such as the USP39 ZF domain. Taken together, multiple repair factors may be recruited to DNA lesions in a PAR-dependent manner. However, selective recruitment of repair factors to damaged chromatin may be spatiotemporally controlled by PAR binding proteins, including FUS and USP39, depending on the type of DNA repair and its complexity. It was previously speculated that the PAR-induced phase separation could provide a flexible tether required to keep the DNA ends in DSBs together, which in turn could facilitate NHEJ (85). Recently, Singatulina et al. reported that interaction between PAR and FUS strongly enhanced assembly of the damaged DNA-rich compartment (86). Although, these data were derived from an in vitro reconstitution experiment, it seems to be reasonable to expect that PAR-induced liquid demixing may help keep the DSB DNA ends together to facilitate NHEJ. In a similar manner, USP39 may simultaneously participate in this process with FUS or other NHEJ factors to help in the assembly of a damaged DNA-rich compartment in vivo. In fact, mIR-induced dark stripe formation was dramatically diminished in U2OS cells by endogenous USP39 depletion even when the endogenous FUS was active. These data strongly support that USP39 is also an indispensable factor for PAR-mediated liquid demixing to facilitate NHEJ. Nonetheless, it remains unclear how liquid demixing is induced by PAR-coupled USP39 and how this phenomenon is linked to DNA repair. Future structural analyses of PAR-binding USP39 mutants will help to validate this model.
Recently, it was shown that USP4, a spliceosomal deubiquitinase, regulates and promotes end resection and HR repair via interplay with the CtIP/MRN complex in a spliceosome-independent manner. Remarkably, our study implies that USP39 also promotes HR via mechanisms shared with its splicing function. It is noteworthy that both USP4 and USP39 have similar functions with respect to HR, while the DUB activity of USP4, but not of USP39, plays a role in HR repair. To date, 93 DUBs have been identified and classified into five different groups: USP, UCH, Josephine, OTU and JAMM/MPN. Among these, 13% of DUBs were predicted to be inactive based on their USP-domain activity, indicating the importance of ubiquitin-independent functions of DUBs in regulating numerous fundamental biological processes.
In the present study, we demonstrated that USP39 has dual functions in controlling NHEJ and HR in a PAR-dependent and spliceosome-dependent manner, respectively. In terms of HR, however, it remains largely unclear how USP39 selectively and/or coordinately regulates its function. Recently, it was shown that USP39 controls the stability of CHK2 protein via its DUB activity in DDR (87). They also found that depletion of USP39 leads to deregulation of CHK2 stability, which results in compromising the DDR-induced G2/M check point, decreasing apoptosis, and conferring cancer cell resistance to radiation treatment (87). In addition, it is also well documented that BRCA1 is phosphorylated by CHK2 kinase in DDR, and abrogation of CHK2 phosphorylation on BRCA1 delays end resection and the dispersion of BRCA1 from DSBs (88–90). Although the aforementioned study showing the DUB activity of USP39 is controversial compared to that in previous studies (59,60), it is possible that USP39 modulates BRCA1-coupled HR in a PAR-independent fashion in various ways.
Lastly, a recent study showed that the recruitment of 53BP1 to DNA lesions was not impaired after PARP inhibition (91). However, we observed that depletion of USP39 regulated by PARP1 activity causes a defect in 53BP1 foci formation. Taken together, this suggests that USP39 regulates 53BP1 foci formation in a PARP1-independent manner and that it is not mechanistically related to PAR-dependent NHEJ, which is regulated by USP39/XRCC4/LIG4 axis-mediated liquid demixing. Similarly, it is increasingly clear that 53BP1 and its binding partners block excessive DNA-end resection, thereby directing DNA repair through the NHEJ pathway (92,93). Therefore, it can be speculated that USP39 may have a PARP1-independent function in regulating the 53BP1-mediated blocking of DNA-end resection in DDR. In parallel, we have also shown that USP39 depletion leads to decreased mRNA expression of BRCA1, which might affect the BRCA1 foci formation. Although, how USP39 modulates BRCA1 mRNA expression as a spliceosome component remains unclear, it is clear that it is not linked to the PAR-dependent USP39 function in DDR. Therefore, future studies will be required to understand in detail how USP39 controls the NHEJ and HR repair processes at multiple levels.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Ho Chul Kang, Ph.D; Email: hckang@ajou.ac.kr
Supplementary Material
ACKNOWLEDGEMENTS
We thank SP. Jackson for providing GFP-DUBs clones, J. Stark for providing GFP-reporter cell lines for repair assays.
Author contributions: J.J.K., S.Y.L., H.K., V.L.D. and H.C.K. conceived and designed the experiments. J.J.K. and S.Y.L. performed the majority of the experiments. J.M.C. and S.K. contributed to PAR binding experiments. Y.H. performed the TEM experiment. J.Y. and H.Y. performed the FACS analysis. S.P., D.P., J.H.J. and S.C. contributed to additional analysis of immunofluorescence data. H.C., C.G.K. and T.M.D. contributed to deep discussion and comments. H.K., V.L.D. and H.C.K. interpreted the data and wrote the paper.
Contributor Information
Jae Jin Kim, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Physiology, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Biomedical Sciences, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Life Science, Hallym University, Chuncheon 24252, Republic of Korea.
Seo Yun Lee, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Physiology, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Biomedical Sciences, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea.
Yiseul Hwang, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Physiology, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Biomedical Sciences, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea.
Soyeon Kim, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Physiology, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Biomedical Sciences, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea.
Jee Min Chung, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Physiology, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Biomedical Sciences, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea.
Sangwook Park, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Physiology, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Biomedical Sciences, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea.
Junghyun Yoon, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Physiology, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Biomedical Sciences, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea.
Hansol Yun, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Physiology, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Biomedical Sciences, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea.
Jae-Hoon Ji, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Biochemistry & Structural Biology, University of Texas Health Science Center, San Antonio, TX, USA.
Sunyoung Chae, Institute of Medical Science, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea.
Hyeseong Cho, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Biochemistry and Molecular Biology, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Biomedical Sciences, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea.
Chan Gil Kim, Department of Biotechnology, Konkuk University, Chungju 380-701, Republic of Korea.
Ted M Dawson, Neuroregeneration and Stem Cell Programs, Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Solomon H. Snyder Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Hongtae Kim, Center for Genomic Integrity Institute for Basic Science (IBS), Ulsan National Institute of Science and Technology, Ulsan, Republic of Korea; School of Life Sciences, Ulsan National Institute of Science and Technology, Ulsan, Republic of Korea.
Valina L Dawson, Neuroregeneration and Stem Cell Programs, Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Solomon H. Snyder Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Physiology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Ho Chul Kang, Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Gyeonggi, 16499, Republic of Korea; Department of Physiology, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea; Department of Biomedical Sciences, Ajou University School of Medicine, Suwon, Gyeonggi 16499, Republic of Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea grants funded by the Korean government (MSIP) [NRF-2017R1A2B4011925, 2020R1A2C2004988]; Brain Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning [NRF-2015M3C7A1029038, in part]; This research was also supported by a grant for the Korea Health Technology R&D project of the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea [HI16C0992]; T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases. Funding for open access charge: National Research Foundation of Korea [NRF2020R1A2C2004988].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shanbhag N.M., Rafalska-Metcalf I.U., Balane-Bolivar C., Janicki S.M., Greenberg R.A.. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010; 141:970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lord C.J., Ashworth A.. The DNA damage response and cancer therapy. Nature. 2012; 481:287–294. [DOI] [PubMed] [Google Scholar]
- 4. Jackson S.P., Durocher D.. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 2013; 49:795–807. [DOI] [PubMed] [Google Scholar]
- 5. van Attikum H., Gasser S.M.. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009; 19:207–217. [DOI] [PubMed] [Google Scholar]
- 6. Sulli G., Di Micco R., d’Adda di Fagagna F.. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat. Rev. Cancer. 2012; 12:709–720. [DOI] [PubMed] [Google Scholar]
- 7. Panier S., Boulton S.J.. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014; 15:7–18. [DOI] [PubMed] [Google Scholar]
- 8. Dantuma N.P., van Attikum H.. Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J. 2016; 35:6–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo X., Kraus W.L.. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012; 26:417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tallis M., Morra R., Barkauskaite E., Ahel I.. Poly(ADP-ribosyl)ation in regulation of chromatin structure and the DNA damage response. Chromosoma. 2014; 123:79–90. [DOI] [PubMed] [Google Scholar]
- 11. Li M., Yu X.. The role of poly(ADP-ribosyl)ation in DNA damage response and cancer chemotherapy. Oncogene. 2015; 34:3349–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pleschke J.M., Kleczkowska H.E., Strohm M., Althaus F.R.. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000; 275:40974–40980. [DOI] [PubMed] [Google Scholar]
- 13. Ahel D., Horejsi Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S.C., Jackson S.P.et al.. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009; 325:1240–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gottschalk A.J., Timinszky G., Kong S.E., Jin J., Cai Y., Swanson S.K., Washburn M.P., Florens L., Ladurner A.G., Conaway J.W.et al.. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:13770–13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feijs K.L., Forst A.H., Verheugd P., Luscher B.. Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat. Rev. Mol. Cell Biol. 2013; 14:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang H.C., Lee Y.I., Shin J.H., Andrabi S.A., Chi Z., Gagne J.P., Lee Y., Ko H.S., Lee B.D., Poirier G.G.et al.. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14103–14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z., Michaud G.A., Cheng Z., Zhang Y., Hinds T.R., Fan E., Cong F., Xu W.. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012; 26:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C.. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008; 451:81–85. [DOI] [PubMed] [Google Scholar]
- 19. Li G.Y., McCulloch R.D., Fenton A.L., Cheung M., Meng L., Ikura M., Koch C.A.. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:9129–9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li M., Lu L.Y., Yang C.Y., Wang S., Yu X.. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013; 27:1752–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krietsch J., Caron M.C., Gagne J.P., Ethier C., Vignard J., Vincent M., Rouleau M., Hendzel M.J., Poirier G.G., Masson J.Y.. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012; 40:10287–10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malanga M., Czubaty A., Girstun A., Staron K., Althaus F.R.. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J. Biol. Chem. 2008; 283:19991–19998. [DOI] [PubMed] [Google Scholar]
- 23. Murawska M., Hassler M., Renkawitz-Pohl R., Ladurner A., Brehm A.. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLos Genet. 2011; 7:e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang F., Chen Y., Li M., Yu X.. The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:7278–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang F., Shi J., Chen S.H., Bian C., Yu X.. The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res. 2015; 43:10782–10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krietsch J., Rouleau M., Pic E., Ethier C., Dawson T.M., Dawson V.L., Masson J.Y., Poirier G.G., Gagne J.P.. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med. 2013; 34:1066–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thandapani P., O’Connor T.R., Bailey T.L., Richard S.. Defining the RGG/RG motif. Mol. Cell. 2013; 50:613–623. [DOI] [PubMed] [Google Scholar]
- 28. Teloni F., Altmeyer M.. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016; 44:993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mastrocola A.S., Kim S.H., Trinh A.T., Rodenkirch L.A., Tibbetts R.S.. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem. 2013; 288:24731–24741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang W.Y., Pan L., Su S.C., Quinn E.J., Sasaki M., Jimenez J.C., Mackenzie I.R., Huang E.J., Tsai L.H.. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 2013; 16:1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rulten S.L., Rotheray A., Green R.L., Grundy G.J., Moore D.A., Gomez-Herreros F., Hafezparast M., Caldecott K.W.. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014; 42:307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altmeyer M., Neelsen K.J., Teloni F., Pozdnyakova I., Pellegrino S., Grofte M., Rask M.B., Streicher W., Jungmichel S., Nielsen M.L.et al.. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 2015; 6:8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M.et al.. A liquid-to-solid phase transition of the ALS Protein FUS accelerated by disease mutation. Cell. 2015; 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- 34. Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G.et al.. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012; 149:768–779. [DOI] [PubMed] [Google Scholar]
- 35. Hyman A.A., Simons K.. Cell biology. Beyond oil and water–phase transitions in cells. Science. 2012; 337:1047–1049. [DOI] [PubMed] [Google Scholar]
- 36. Hyman A.A., Weber C.A., Julicher F.. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014; 30:39–58. [DOI] [PubMed] [Google Scholar]
- 37. Liu C., Wu J., Paudyal S.C., You Z., Yu X.. CHFR is important for the first wave of ubiquitination at DNA damage sites. Nucleic Acids Res. 2013; 41:1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan Q., Dutt S., Xu R., Graves K., Juszczynski P., Manis J.P., Shipp M.A.. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol. Cell. 2009; 36:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yan Q., Xu R., Zhu L., Cheng X., Wang Z., Manis J., Shipp M.A.. BAL1 and its partner E3 ligase, BBAP, link Poly(ADP-ribose) activation, ubiquitylation, and double-strand DNA repair independent of ATM, MDC1, and RNF8. Mol. Cell. Biol. 2013; 33:845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jacq X., Kemp M., Martin N.M., Jackson S.P.. Deubiquitylating enzymes and DNA damage response pathways. Cell Biochem. Biophys. 2013; 67:25–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McClurg U.L., Robson C.N.. Deubiquitinating enzymes as oncotargets. Oncotarget. 2015; 6:9657–9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nijman S.M., Huang T.T., Dirac A.M., Brummelkamp T.R., Kerkhoven R.M., D’Andrea A.D., Bernards R.. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005; 17:331–339. [DOI] [PubMed] [Google Scholar]
- 43. Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., D’Andrea A.D.. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006; 8:339–347. [DOI] [PubMed] [Google Scholar]
- 44. Oestergaard V.H., Langevin F., Kuiken H.J., Pace P., Niedzwiedz W., Simpson L.J., Ohzeki M., Takata M., Sale J.E., Patel K.J.. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol. Cell. 2007; 28:798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim J.M., Parmar K., Huang M., Weinstock D.M., Ruit C.A., Kutok J.L., D’Andrea A.D.. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev. Cell. 2009; 16:314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murai J., Yang K., Dejsuphong D., Hirota K., Takeda S., D’Andrea A.D.. The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol. Cell. Biol. 2011; 31:2462–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nicassio F., Corrado N., Vissers J.H., Areces L.B., Bergink S., Marteijn J.A., Geverts B., Houtsmuller A.B., Vermeulen W., Di Fiore P.P.et al.. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 2007; 17:1972–1977. [DOI] [PubMed] [Google Scholar]
- 48. Li M., Chen D., Shiloh A., Luo J., Nikolaev A.Y., Qin J., Gu W.. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002; 416:648–653. [DOI] [PubMed] [Google Scholar]
- 49. Dayal S., Sparks A., Jacob J., Allende-Vega N., Lane D.P., Saville M.K.. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J. Biol. Chem. 2009; 284:5030–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang X., Berger F.G., Yang J., Lu X.. USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J. 2011; 30:2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khoronenkova S.V., Dianova I.I., Ternette N., Kessler B.M., Parsons J.L., Dianov G.L.. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol. Cell. 2012; 45:801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen J., Feng W., Jiang J., Deng Y., Huen M.S.. Ring finger protein RNF169 antagonizes the ubiquitin-dependent signaling cascade at sites of DNA damage. J. Biol. Chem. 2012; 287:27715–27722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grofte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S.et al.. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012; 150:697–709. [DOI] [PubMed] [Google Scholar]
- 54. Poulsen M., Lukas C., Lukas J., Bekker-Jensen S., Mailand N.. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J. Cell Biol. 2012; 197:189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim J.J., Lee S.Y., Kim S., Chung J.M., Kwon M., Yoon J.H., Park S., Hwang Y., Park D., Lee J.S.et al.. A novel reciprocal crosstalk between RNF168 and PARP1 to regulate DNA repair processes. Mol. Cells. 2018; 41:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nishi R., Wijnhoven P., le Sage C., Tjeertes J., Galanty Y., Forment J.V., Clague M.J., Urbe S., Jackson S.P.. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat. Cell Biol. 2014; 16:1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Izhar L., Adamson B., Ciccia A., Lewis J., Pontano-Vaites L., Leng Y., Liang A.C., Westbrook T.F., Harper J.W., Elledge S.J.. A systematic analysis of factors localized to damaged chromatin reveals PARP-Dependent recruitment of transcription factors. Cell Rep. 2015; 11:1486–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Reyes-Turcu F.E., Horton J.R., Mullally J.E., Heroux A., Cheng X., Wilkinson K.D.. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006; 124:1197–1208. [DOI] [PubMed] [Google Scholar]
- 59. van Leuken R.J., Luna-Vargas M.P., Sixma T.K., Wolthuis R.M., Medema R.H.. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell Cycle. 2008; 7:2710–2719. [DOI] [PubMed] [Google Scholar]
- 60. Hadjivassiliou H., Rosenberg O.S., Guthrie C.. The crystal structure of S. cerevisiae Sad1, a catalytically inactive deubiquitinase that is broadly required for pre-mRNA splicing. RNA. 2014; 20:656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q.et al.. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010; 17:688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bunting S.F., Callen E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L.et al.. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010; 141:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Roy R., Chun J., Powell S.N.. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2011; 12:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao Y., Zhang B., Lei Y., Sun J., Zhang Y., Yang S., Zhang X.. Knockdown of USP39 induces cell cycle arrest and apoptosis in melanoma. Tumour Biol. 2016; 37:13167–13176. [DOI] [PubMed] [Google Scholar]
- 65. Li K.Y., Zhang J., Jiang L.C., Zhang B., Xia C.P., Xu K., Chen H.Y., Yang Q.Z., Liu S.W., Zhu H.. Knockdown of USP39 by lentivirus-mediated RNA interference suppresses the growth of oral squamous cell carcinoma. Cancer Biomark. 2016; 16:137–144. [DOI] [PubMed] [Google Scholar]
- 66. Gan Z., Han K., Lin S., Hu H., Shen Z., Min D.. Knockdown of ubiquitin-specific peptidase 39 inhibited the growth of osteosarcoma cells and induced apoptosis in vitro. Biol. Res. 2017; 50:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yano K., Morotomi-Yano K., Wang S.Y., Uematsu N., Lee K.J., Asaithamby A., Weterings E., Chen D.J.. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008; 9:91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ochi T., Blackford A.N., Coates J., Jhujh S., Mehmood S., Tamura N., Travers J., Wu Q., Draviam V.M., Robinson C.V.et al.. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015; 347:185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tadi S.K., Tellier-Lebegue C., Nemoz C., Drevet P., Audebert S., Roy S., Meek K., Charbonnier J.B., Modesti M.. PAXX is an accessory c-NHEJ factor that associates with Ku70 and has overlapping functions with XLF. Cell Rep. 2016; 17:541–555. [DOI] [PubMed] [Google Scholar]
- 70. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Luijsterburg M.S., de Krijger I., Wiegant W.W., Shah R.G., Smeenk G., de Groot A.J., Pines A., Vertegaal A.C., Jacobs J.J., Shah G.M.et al.. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell. 2016; 61:547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Makarova O.V., Makarov E.M., Luhrmann R.. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J. 2001; 20:2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Savage K.I., Gorski J.J., Barros E.M., Irwin G.W., Manti L., Powell A.J., Pellagatti A., Lukashchuk N., McCance D.J., McCluggage W.G.et al.. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol. Cell. 2014; 54:445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang S., Wang Z., Li J., Qin J., Song J., Li Y., Zhao L., Zhang X., Guo H., Shao C.et al.. Splicing factor USP39 promotes ovarian cancer malignancy through maintaining efficient splicing of oncogenic HMGA2. Cell Death. Dis. 2021; 12:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Karlin S., Brocchieri L., Bergman A., Mrazek J., Gentles A.J.. Amino acid runs in eukaryotic proteomes and disease associations. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Morrison C., Smith G.C., Stingl L., Jackson S.P., Wagner E.F., Wang Z.Q.. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat. Genet. 1997; 17:479–482. [DOI] [PubMed] [Google Scholar]
- 77. Brown M.L., Franco D., Burkle A., Chang Y.. Role of poly(ADP-ribosyl)ation in DNA-PKcs- independent V(D)J recombination. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:4532–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Galindo-Campos M.A., Bedora-Faure M., Farres J., Lescale C., Moreno-Lama L., Martinez C., Martin-Caballero J., Ampurdanes C., Aparicio P., Dantzer F.et al.. Coordinated signals from the DNA repair enzymes PARP-1 and PARP-2 promotes B-cell development and function. Cell Death Differ. 2019; 26:2667–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hu Y., Petit S.A., Ficarro S.B., Toomire K.J., Xie A., Lim E., Cao S.A., Park E., Eck M.J., Scully R.et al.. PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov. 2014; 4:1430–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jiang W., Crowe J.L., Liu X., Nakajima S., Wang Y., Li C., Lee B.J., Dubois R.L., Liu C., Yu X.et al.. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol. Cell. 2015; 58:172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Saha J., Davis A.J.. Unsolved mystery: the role of BRCA1 in DNA end-joining. J. Radiat. Res. 2016; 57:i18–i24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jiang G., Plo I., Wang T., Rahman M., Cho J.H., Yang E., Lopez B.S., Xia F.. BRCA1-Ku80 protein interaction enhances end-joining fidelity of chromosomal double-strand breaks in the G1 phase of the cell cycle. J. Biol. Chem. 2013; 288:8966–8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wei H., Yu X.. Functions of PARylation in DNA damage repair pathways. Genomics Proteomics Bioinformatics. 2016; 14:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bolognesi B., Lorenzo Gotor N., Dhar R., Cirillo D., Baldrighi M., Tartaglia G.G., Lehner B.. A Concentration-Dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 2016; 16:222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Aguzzi A., Altmeyer M.. Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 2016; 26:547–558. [DOI] [PubMed] [Google Scholar]
- 86. Singatulina A.S., Hamon L., Sukhanova M.V., Desforges B., Joshi V., Bouhss A., Lavrik O.I., Pastre D.. PARP-1 activation directs FUS to DNA damage sites to form PARG-Reversible compartments enriched in damaged DNA. Cell Rep. 2019; 27:1809–1821. [DOI] [PubMed] [Google Scholar]
- 87. Wu J., Chen Y., Geng G., Li L., Yin P., Nowsheen S., Li Y., Wu C., Liu J., Zhao F.et al.. USP39 regulates DNA damage response and chemo-radiation resistance by deubiquitinating and stabilizing CHK2. Cancer Lett. 2019; 449:114–124. [DOI] [PubMed] [Google Scholar]
- 88. Lee J.S., Collins K.M., Brown A.L., Lee C.H., Chung J.H.. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000; 404:201–204. [DOI] [PubMed] [Google Scholar]
- 89. Zhang J., Willers H., Feng Z., Ghosh J.C., Kim S., Weaver D.T., Chung J.H., Powell S.N., Xia F.. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol. Cell. Biol. 2004; 24:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Parameswaran B., Chiang H.C., Lu Y., Coates J., Deng C.X., Baer R., Lin H.K., Li R., Paull T.T., Hu Y.. Damage-induced BRCA1 phosphorylation by Chk2 contributes to the timing of end resection. Cell Cycle. 2015; 14:437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Aleksandrov R., Dotchev A., Poser I., Krastev D., Georgiev G., Panova G., Babukov Y., Danovski G., Dyankova T., Hubatsch L.et al.. Protein dynamics in complex DNA lesions. Mol. Cell. 2018; 69:1046–1061. [DOI] [PubMed] [Google Scholar]
- 92. Ochs F., Somyajit K., Altmeyer M., Rask M.B., Lukas J., Lukas C.. 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol. 2016; 23:714–721. [DOI] [PubMed] [Google Scholar]
- 93. Rother M.B., Pellegrino S., Smith R., Gatti M., Meisenberg C., Wiegant W.W., Luijsterburg M.S., Imhof R., Downs J.A., Vertegaal A.C.O.et al.. CHD7 and 53BP1 regulate distinct pathways for the re-ligation of DNA double-strand breaks. Nat. Commun. 2020; 11:5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.