Abstract
In plants and some animal lineages, RNA silencing is an efficient and adaptable defense mechanism against viruses. To counter it, viruses encode suppressor proteins that interfere with RNA silencing. Phloem-restricted viruses are spreading at an alarming rate and cause substantial reduction of crop yield, but how they interact with their hosts at the molecular level is still insufficiently understood. Here, we investigate the antiviral response against phloem-restricted turnip yellows virus (TuYV) in the model plant Arabidopsis thaliana. Using a combination of genetics, deep sequencing, and mechanical vasculature enrichment, we show that the main axis of silencing active against TuYV involves 22-nt vsiRNA production by DCL2, and their preferential loading into AGO1. Moreover, we identify vascular secondary siRNA produced from plant transcripts and initiated by DCL2-processed AGO1-loaded vsiRNA. Unexpectedly, and despite the viral encoded VSR P0 previously shown to mediate degradation of AGO proteins, vascular AGO1 undergoes specific post-translational stabilization during TuYV infection. Collectively, our work uncovers the complexity of antiviral RNA silencing against phloem-restricted TuYV and prompts a re-assessment of the role of its suppressor of silencing P0 during genuine infection.
INTRODUCTION
To defend themselves against pathogens, plants have developed a molecular arsenal allowing them to detect and resist the incoming threat. In turn, pathogens have adopted numerous evasion strategies and can exploit plant defenses with the resulting arms race leading to complex and ever-changing host–microbe interactions. One such focal point of plant defense and viral counter-defense is RNA silencing, with traces of such interactions evident across both plant and virus diversity (1,2). All RNA silencing pathways rest on the action of small RNA (sRNA) whose production depends on the enzymatic activity of RNAse III proteins called Dicer-like (DCLs). In the case of RNA viruses, production of viral small interfering (vsi)RNA is triggered by double-stranded (ds)RNA replication intermediates or intramolecular foldback structures in the viral genome mainly by the action of DCL4 and DCL2, generating 21- and 22-nt vsiRNA duplexes respectively (3–5). This first layer of detection and degradation is reinforced by specialized effector proteins called ARGONAUTE (AGO) that associate with the vsiRNA to form the antiviral RNA-inducted silencing complex (RISC) (6). The RISC can target RNA in a sequence-specific manner, leading to endonucleolytic cleavage (slicing) catalyzed by the AGO and/or via translational repression coupled with mRNA decay (7). The silencing signal can further be amplified through the conversion of single stranded (ss)RNA targets into dsRNA by host-encoded RNA-dependent RNA polymerase (RDR) proteins, providing new template for secondary vsiRNA production by DCLs, that are important to achieve optimal silencing for some viruses (8–11). Its layered, self-reinforcing and sequence specific mechanism makes RNA silencing a particularly potent immune system.
In order to foil this mechanism most, if not all, plant viruses deploy specialized proteins that are known as viral suppressor of RNA silencing, or VSRs. VSRs across different virus families that are highly diverse have adopted various strategies to impair different steps of RNA silencing (1,12). Their deployment usually results in the abrogation of the cell-to-cell and eventual long distance movement of vsiRNA and therefore of plant immunization (13–15), resulting in the accumulation of high amounts of the viral genome often accompanied by strong symptoms. Consequently, viruses for which VSR activity has been abrogated are impaired in their ability to move long distances and achieve systemic infection (5,11,15–19). This observation has been instrumental in deciphering the silencing components involved in plant defense, since their inactivation leads to a rescue of viral movement. However, a causal link between VSR and virus movement has only been established for comparatively few viruses, while many more known VSRs await in vivo characterization in the context of infection.
The P0 VSR of phloem-restricted poleroviruses presents an interesting case: while the intricacies of its mode of action are understood, they do not necessarily reconcile with observations made in the context of infection nor with the behaviour of natural variants of the protein. By hijacking the S phase kinase associated protein 1 (SKP1)–Cullin 1–F-box (SCF) E3 ubiquitin ligase complex and enforcing degradation of most AGO proteins, P0 impedes the formation of vsiRNA-RISC (20–22). In the case of the AGO1 protein, it has been shown that its interaction with P0 leads to its ubiquitination and vacuolar degradation (23,24). This strategy seems to be particularly effective, as in heterologous patch assays in N. benthamiana, P0 of turnip yellows virus (TuYV) is able to suppress the potent RNA silencing reaction to transgenes, enabling strong and persistent expression of GFP. Accordingly, strong dosage of P0 leads to developmental phenotypes reminiscent of AGO1 knockout plants (25,26) because P0 also disables miRNA-RISC assembly (22,27).
On the opposite spectrum, studies employing a TuYV that is unable to produce P0 show that it is dispensable for systemic infection (28) while the resulting systemic infection by the wild type (WT) virus is asymptomatic in Arabidopsis (29). Furthermore, P0 proteins from polerovirus isolates collected throughout the world display varying degrees of silencing efficiency in patch assay, ranging from strong (27,30–33) and moderate (34–36) to non-existent (31,37). Intriguingly, the start codon of ORF0 encoding P0 of TuYV has a poor initiation context (38) which results in leaky scanning by the ribosomes and therefore reduced initiation. Optimization of the 5′ context decreases viral RNA accumulation and leads to second-site mutations of the start codon that restore low translation initiation in the systemic progeny (39). Similarly, potato leafroll virus (PLRV) genomic leader sequence exerts an inhibitory effect on translation of the downstream ORF0 and ORF1 (40). These observations suggest that neither high accumulation nor strong suppression activity are traits that have been selected for during plant-polerovirus coevolution. These contrasting observations raise the question of the role of P0 during infection of the plant host, and of its interaction with the RNA silencing machinery.
We recently uncovered a suppressor of the P0-dependent developmental phenotype typically associated with AGO1 degradation, that we called ago1-57 (41). This single Gly to Asp substitution in the DUF1785 of AGO1 inhibits the SCFP0–AGO1 interaction and therefore renders the mutant AGO1 protein non-degradable by P0. We also showed that this mutation hinders sRNA duplex ‘unwinding’ by AGO1 itself, particularly for perfect duplexes, a hallmark of siRNA rather than miRNA. Accordingly, miRNA-programmed RISC activity is mostly unperturbed by the mutation, leading to a mild developmental phenotype, while endogenous siRNA-programmed RISC is strongly affected, which results in the near-complete loss of secondary siRNA. Thus, this unique allele of AGO1 allows for unprecedented decoupling of miRNA-guided pathways from siRNA guided ones, which are relied upon to perform antiviral RNA silencing.
Here, we take advantage of this unique allele to investigate the silencing components required for efficient antiviral immunity against phloem-restricted TuYV. We show that TuYV-derived vsiRNAs are largely channelled towards AGO1, and that its antiviral importance is only evident in some missense mutants. We further show that TuYV RNA is mainly cleaved into 22-nt vsiRNA by vascular DCL2 in a mechanism that is distinct from that described for turnip crinckle virus (TCV) infection. Our study also provides evidence for the production of secondary siRNA from host transcripts, a feature that is dependent on both DCL2 and AGO1. Finally, while transgenic vascular P0 can recapitulate AGO1 degradation in the phloem, AGO1 undergoes vascular post-translational stabilization during TuYV infection in a P0-independent manner.
MATERIALS AND METHODS
Plant lines
AGO1 point mutants have been described previously: ago1-57 (41), ago1-38 (42), ago1-27 (43). ago2-1 (SALK_003380), ago5-1 (SALK_063806), ago7-1 (SALK_037458), ago10-3 (SALK_019738) and the resulting combination mutants have been described previously (18). dcl2-1 (SALK_064627), dcl4-2 (GABI_160G05), dcl2-1/dcl4-2 (5), dcl2-5/dcl3-1 (3) dicer-like single and combination mutants have been described previously. rdr6-12 and sgs3-14 (SALK_001394) (44), SUC-SUL (45), SUC-SUL/ pSUC:P15-FHA (15) have been described previously. ago1-57/ago2-1, ago1-57/dcl2-1 and ago1-57/dcl4-2 plants were obtained by crossing their respective single mutants and selected by genotyping of the F2 population. SUC-SUL/ago1-57 were obtained by crossing and selected by genotyping and for Basta resistance. The triples dcl2-1/dcl4-2/ago1-57 was obtained by crossing the double dcl2-1/dcl4-2 with the double dcl2-1/ago1-57 and selected by genotyping.
Constructs
35S:CFP-AGO1 construct and its mutant derivative were previously described (41).
For overexpression constructs of P0: TuYV (21), CABYV (20), BMYV (46) and PLRV (39), all ORF0s were cloned into a pBIN vector, under a 35S promoter as described previously (39).
35S:tagRFP and 35S:tagRFP-AGO1 are in the pEAQΔP19-GG vector and were cloned as described (47). AGO1 cDNA was amplified with primers containing SapI restriction sites and combined with N-terminal tagRFP fragment in a Golden Gate reaction.
For 35S:P0-tagRFP constructs, the P0 coding sequences with T156A/T159C mutations (to avoid translation of a truncated P1 protein from the P0 vector) were amplified from the XVE:P0-myc constructs (22,41) with oligo primers containing the AttB1 and AttB2 sites and the PCR product was cloned in a pDONR221 by BP recombination. The entry clones were then recombined with the pGWB660 vector by LR gateway reaction producing 35S promoter driven P0 construct fused to tagRFP at the C terminus.
For 35S:DCL2-tagRFP and 35S:DCL4-tagRFP constructs the genomic sequence from start codon to the last codon before the stop codon was amplified with oligo primers containing the attB1 and AttB2 sites and the PCR product was subsequently cloned in a pDONR221 by BP recombination. The entry clones were then recombined with the pGWB660 vector by LR gateway reaction producing 35S promoter driven genomic constructs fused to tagRFP at their C termini.
To obtain the pCoYMV:P0-HA lines, the promoter sequence of CoYMV (1039bp) was amplified from the vector pGEM 3zf + CoYMV-Suc51 (48) with oligo primers containing AttB4 and AttB1R recombination sites and the sequences was mobilized into the pDONR P4P1r vector by BP Gateway recombination. The P0-3xHA fusions (WT or LP1) were obtained by seamless Gibson assembly (NEB) of the P0 coding sequences with T156A/T159C mutations and a 3xHA-stop fragment with overlapping primers into the pFK202 plasmid. The resulting fusion was then amplified with AttB1 and AttB2 containing primers and mobilized into the pDONR221 plasmid by gateway BP reaction. Pieces were assembled into the pK7m24GW plasmid (http://www.psb.ugent.be/gateway/) by double recombination LR reaction to obtain the final binary plasmid. The plasmid was then introduced into Agrobacterium strain GV3101 pMP90 and used to transform SUC-SUL plants (49).
To obtain the pSUC:Flag-AGO1 lines, the SUC2 promoter (943 bp) was amplified from the pEP1 plasmid (50) with oligo primers containing AttB4 and AttB1R recombination sites and the sequences was mobilized into the pDONR P4P1r vector by BP Gateway recombination. The 3xFlag sequences was amplified from a pENTRY-3xFlag vector with primers containing attB1 and attB2 sites and a 5′ Kozak consensus sequence and the PCR product was subsequently cloned in a pDONR Zeo by BP recombination. AGO1 CDS sequence was amplified with primers containing attB2R and attB3 sites and mobilized into the p2R-P3 plasmid by gateway BP reaction. All fragments were assembled by three-way LR gateway reaction into the pB7m34GW vector. The plasmid was then introduced into Agrobacterium strain GV3101 pMP90 and used to transform Col-0 plants.
For cloning PCR products, Phusion high fidelity DNA polymerase 2X master mix (Thermo Scientific) was used, except for the genomic DCL2 and DCL4 for which Platinum SuperFi DNA Polymerase (Invitrogen) was used, following the manufacturer's protocol. All clones were verified by in-house Sanger sequencing before proceeding to the next step. All primers used for cloning are available in Supplemental Table S2.
Plant growth and infection conditions
For in vitro culture, seeds were surface sterilized using ethanol, plated on growth medium (MS salts [Duchefa], 1% sucrose, and 0.8% agar, pH 5.7), stored 2 days at 4°C in the dark, and then transferred to a plant growth chamber under a 16-h-light/8-h-dark photoperiod (22°C/20°C). For standard plant growth, seeds were directly sown on soil (Hawita Fruhstorfer) in trays and kept under a 12-h-light/12-h-dark regime for 14 days, then transferred in 16-h-light/8-h-dark growth chambers, under fluorescent light (Osram Biolux 49W/965).
For infection conditions, seeds were directly sown on soil (Hawita Fruhstorfer) in trays and kept under a 12-h-light/12-h-dark regime for about 4 weeks. Agrobacterium tumefaciens GV3101 pMP90 containing binary plasmids of TuMV-GFP, TuMV-AS9-GFP, TRV-PDS (RNA1 + RNA2-PDS) recombinant viruses were grown overnight at 28°C in antibiotic-supplemented LB media, pelleted 15 min at 4000g and resuspended in infiltration medium (10 mM MgCl2/10 mM MES/200 μM Acetosyringone). The cells were then adjusted to an OD600 nm of 0.5 and agroinfiltrated into three leaves per plants with a needleless syringe. Plants post-inoculation were then grown in either 12-h-light/12-h-dark period with fluorescent light or under long day condition in greenhouse. For TuYVs81, TuYVs81P0− or the empty vector (mock), the procedure was the same with the following modifications: after pelleting of the initial cultures, cells were resuspended in 10.5 g/l K2HPO4/4.5 g/l KH2PO4/1 g/l (NH4)2SO4/0.5 g/l sodium citrate/0.1 g/l MgSO4/0.4% (v/v) glycerol/0.1 g/l MES/200 μM Acetosyringone and incubated 5–6 h in the dark. Cells were pelleted again and resuspended in infiltration medium and handled as described above. Infections were left to progress until the time indicated in the figure legends, and systemic leaves were collected from the indicated number of individuals. Plants were photographed at the indicated time using a DSLR camera mounted on a stand at a fixed distance, or for systemic TuMV-AS9-GFP propagation, using a Zeiss axio zoom equipped with GFP filter.
For analysis of infiltrated leaves, the same protocol was used, but five leaves per plant were infiltrated with a needleless syringe. Only the inoculated leaves were sampled at the indicated time points. To account for biological variation, several inoculated leaves were harvested per time point. For one given experiment either all the inoculated leaves were taken off a set number of plants, or one inoculated leaf from each individual was harvested per time point, with identical result.
For aphid-mediated inoculation, seeds were directly sown on soil (Hawita Fruhstorfer) in trays and kept under a 12-h-light/12-h-dark regime for 18 days. Each plant was then challenged with two Myzus persicae fed on either 20% sucrose solution, 20% sucrose solution containing 67 mg/ml TuYV virions or alternatively were left untreated. After 4 days, insecticide was applied to kill the aphids and plants were kept under a 12-h-light/12-h-dark period with fluorescent light for 25 days before sampling. Each plant was individually weighed (after removal of root system and cotyledons) and ground with liquid N2 with a mortar and pestle. For each, 100 mg (±5 mg) were kept at –80°C in Eppendorf tubes for DAS-ELISA.
Transient expression assays and local leaf infection in N. benthamiana
Binary constructs were transformed in Agrobacterium GV3101 pMP90 and infiltrated in N. benthamiana for transient expression assays. Agrobacterium cells were grown overnight at 28°C in 5 to 10 ml LB medium supplemented with antibiotics, resuspended in infiltration medium at an OD600nm of 0.1–0.3 per construct, and incubated for 2–4 h at room temperature before being infiltrated into leaves of 4-week-old plants with a needleless syringe. Plants were maintained in growth chambers under a 16-h-light/8-h-dark photoperiod with a constant temperature of 22°C. For VRC colocalization assays, 35S:B2-GFP N. benthamiana plants (51) were first screened under UV light in order to select plants with visible GFP. These plants were infiltrated with a mixture of the TuYVs81 infectious clone or the empty vector and with the construct of interest. Plants were kept 3 days in the growth chamber before imaging.
DAS-ELISA
TuYV was detected in non-inoculated leaves of A. thaliana by double-antibody sandwich enzyme-linked immunosorbent assay (52). Flat-bottomed NUNC 96-wells plates were coated with 1:400 BWYV IgG (Loewe) (53) in 1.59 g/l Na2CO3/2.93 g/l NaHCO3 pH 9.6, 100 μl per well and incubated at 37°C in the dark for 4 h. N2 chilled samples were ground in safe lock Eppendorf tubes using a Silamat S7 (Ivoclar vivadent) and extracted in 400 μl 1× PBS/0.05% (v/v) Tween 20/2% (w/v) soluble PVP 360, resuspended by shaking for 15 min and centrifuged for 5 minutes at 3000 rpm at 4°C. Coated plates were washed three times with 1× PBS/0.05% (v/v) Tween 20 and 100 μl of supernatant was loaded into each well for an overnight binding at 4°C. The plates were then washed three times in 1× PBS/0.05% (v/v) Tween 20, and 100 μl per well of 1:400 BWYV AP conjugate (Loewe) in 1× PBS/0.05% (v/v) Tween 20/2% (w/v) PVP 360 was added and plates were incubated at 37°C in the dark for 3 h. Plates were washed three time as described above and 100 μl per well of freshly prepared substate buffer was added (1 M diethanolamine/5 mM MgCl2/1 mg/ml substrate tablet [4-nitrophenyl phosphate Na2-salt]). O.D405 nm was determined after 90 min of incubation. Technical triplicates were obtained for one given sample, and each plate contained a positive control (TuYVs81 with yellow veins) and a negative control (Mock-infected individual). Blank was obtained by performing the reaction on buffer only.
Infection kinetic scoring by dot blot
A leaf disk (5 mm Ø) from young systemic leaves of each individual plant inoculated with TuYVs81 was removed at the indicated time after inoculation. Protein extracts were obtained by adding 50 μl of 2× Laemmli buffer (200 mM Tris–HCl pH 6.8/ 8% [w/v] SDS/ 40% [v/v] glycerol/ 0.05% [w/v] bromophenol blue/3% [v/v] B-mercaptoethanol) to each leaf disk in 96-well plates and grinding at maximum speed for 3 min in a Retsch mixer mill at room temperature. The plates were then centrifuged for 5 min at 4000g and the supernatant was transferred to a fresh 96-well plate to denature proteins for 5 min at 95°C. 5 μl of the denatured protein sample was manually spotted onto an activated PVDF Immobilon-P membrane (Millipore) mounted on top of a paper stack (bottom up: dry stack of paper towel, one dry whatman filter paper cut to membrane dimension, one wet whatman filter paper cut to membrane dimension in 25 mM Tris base/192 mM glycine/20% (v/v) ethanol), for a maximum of 96 samples per membrane, mirroring the plate. The membrane was left to dry at 60°C for about 10 min, reactivated in absolute ethanol, washed in deionized water for 2 min, incubated in TBST + 5% milk (20 mM Tris base/150 mM NaCl/0.1% Tween 20, pH 7.4) for about 20 min, washed in TBST without milk and hybridized overnight in 1:15000 (v/v) of anti-readthrough (RT) (54) rabbit antibody in TBST + 2.5% (w/v) BSA. Membranes were then processed as indicated for western blots, and signal was acquired on ECL films. Membranes were then colored in Coomassie blue to mark the position of the samples, dried, and film and membrane were overlayed to pinpoint infected individuals. This process was repeated several times for each inoculated plant, and individual plants were counted as positive only if at least two independent time points returned positive signals.
Meselect for vascular bundle enrichment
To obtain one sample of vascular tissues for protein or RNA analysis, the indicated amount of leaves (in the figure legends) from a given genotype or treatment were harvested fresh from different plants. Leaves were carefully laid on their abaxial side to the sticky side of a labelling tape. Another tape was added on the adaxial side of the leaf, and this tape was slowly removed, taking away the adaxial epidermal cells. During preparation of the remaining leaves, the processed leaves on tape containing the abaxial epidermis were left face down in ice cold 0.4 M mannitol/10 mM CaCl2/20 mM KCl/0.1% (w/v) BSA/20 mM MES pH 5.7. Once all were processed, they were incubated face down in the same buffer with the addition of 1% (w/v) cellulase Onozuka (Yakult) and 0.25% (w/v) macerozyme Onozuka R10 (Yakult) for 15 minutes under gentle rotation at room temperature. Mesophyll cells that were released in the protoplast buffer were collected by pipetting gently with a cut tip and centrifuged at 200g at 4°C with brakes on. The pelleted cells were extracted either in Laemmli 2× buffer for protein extraction, or in 1 ml Tri-Reagent for RNA extraction. The tapes were left face down in ice-cold wash buffer (154 mM NaCl/125 mM CaCl2/5 mM KCl/5 mM glucose/2 mM MES pH 5.7), were gently shaken, and the buffer was refreshed once for further washing. The vascular tissue network was then lifted off the leaves with tweezers, starting from the central vein, and collected into a tube with ice cold wash buffer until all were processed. All collected vascular bundles were quickly drained on paper towel, and flash frozen in a safelock Eppendorf tube containing 2mm Ø glass beads. Proteins were extracted in Laemmli 2× as described in the immunoblotting section, and RNA was extracted in tri-reagent after grinding of the frozen vasculatures in the silamat S7.
Protein analysis and immunoblotting
N2 chilled samples were ground in safe lock Eppendorf tubes containing 2 mm Ø glass beads, using a Silamat S7 (Ivoclar vivadent) and total proteins were extracted in 2X Laemmli buffer by mixing again in the Silamat S7 for 20 s. Samples were denatured for 5 min at 95°C and quantified using the amido black method. 10 μl of supernatant mixed with 190 μl of deionized water and added to 1 ml of normalized 10% (v/v) acetic acid/90% (v/v) methanol/0.05% (w/v) Amido Black (Naphtol Blue Black, Sigma N3393) buffer, mixed and centrifuged for 10 min at maximum speed. Pellets are then washed in 1 ml of 10% (v/v) acetic acid/90% (v/v) ethanol, centrifuged 5 min at maximum speed and resuspended in 0.2N NaOH. OD630 nm was determined, with NaOH solution as blank, and protein concentration is calculated using the OD = a[C] + b determined curve. 2.5–40 μg of total protein extracts were separated on SDS-PAGE gels and blotted onto PVDF Immobilon-P membrane (Millipore). AGO1 protein was detected using the anti-AGO1 antibody (rabbit polyclonal, AS09 527; Agrisera) diluted 1:10 000 (v/v). AGO2 protein was detected using the anti-AGO2 antibody (rabbit polyclonal, AS13 2682; Agrisera) diluted 1:5000 (v/v). AGO4 protein was detected using the anti-AGO4 antibody (rabbit polyclonal, AS09 617; Agrisera) diluted 1:5000 (v/v). Myc-tagged proteins were detected using anti-myc antibody (mouse monoclonal; Roche) diluted 1:5000 (v/v), or anti-myc antibody HRP-coupled (mouse monoclonal, Miltenyi Biotec) diluted 1:2000 (v/v). GFP-tagged proteins were detected using the anti-GFP antibody (JL-8, Clontech Takara) diluted 1:2000 (v/v) or anti-GFP HRP-coupled (mouse monoclonal, Miltenyi Biotec) diluted 1:5000 (v/v). Flag-tagged proteins were detected using the anti-Flag HRP-coupled antibody (mouse monoclonal, A8592; Sigma-Aldrich) diluted 1:5000 (v/v). HA-tagged proteins were detected using the anti-HA HRP-coupled antibody (mouse monoclonal, Miltenyi Biotec) diluted 1:5000 (v/v). TuYV Readthrough protein was detected as described in the dot blot section. DCL1 protein was detected using anti-DCL1 antibody (rabbit polyclonal, AS19 4307; Agrisera) diluted 1:2000 (v/v). Mouse monoclonal antibodies were detected with a goat anti-mouse IgG HRP-linked antibody (62-6520; Invitrogen) diluted 1:10 000 (v/v). Rabbit polyclonal antibody were detected with a goat anti-rabbit IgG HRP-linked antibody (65-6120; Invitrogen) diluted 1:10 000 (v/v). Hybridized membranes were reacted with Clarity or Clarity Max ECL (Biorad) and imaged using a Fusion FX (Vilbert). For signal quantification, the plot lane function of ImageJ was used to obtain the raw intensity for a signal of interest as well as from the whole Coomassie stained lane. Each signal was subsequently normalized to the total protein signal, and the values obtained from all the replicates were plotted using with(data,boxplot(value∼treat)) command in R were data contains the imputed values, separated according to treatment (∼treat).
sRNA library preparation
Total RNA samples and AGO1 IP samples were both obtained from Col-0 and ago1-57 rosette leaves from TuYVs81 infected of mock -inoculated plants 16 days after inoculation as described in RNA related methods and immunoprecipitation of AGO1 and AGO2 sections. For both genotypes, only plants with vein yellowing were included in the analysis. Plants were separated into pools of 7–8 plants per biological replicate, and the resulting mixed and ground tissue was used as the input material for the AGO1 IP and the total RNA. About 100 mg of material was used to extract total RNA using tri-reagent as described, and IPs were performed from 1 g starting material. AGO1-loaded sRNAs were then extracted by adding Tri-Reagent directly on the magnetic beads. Library preparation and sRNA sequencing (single-read, 50 bp, V4 chemistry) on Illumina HiSeq were performed by Fasteris (http://www.fasteris.com), with each sample split and sequenced in two independent lanes. FASTQ file generation, demultiplexing, and adapter removal were done by Fasteris.
Mapping, quantification and differential analysis of sRNA
Reads (18–26 nucleotides long) were aligned and quantified using Shortstack v3.8.5 with the ‘fractional’ option (55), allowing for no mismatches, with the Arabidopsis genome (TAIR10) and TuYVs81 genome as reference. Mapping statistics are provided in Supplementary Figure S2A. Total 20–24 nucleotide reads mapped per library were normalized in read per million mapped (RPM) and the obtained values per category were plotted as stacked histograms using ggplot2 in R. Alternatively, RPM values for the 21, 22 and 24-nt subsets were used for filled histograms per category. To obtain the vsiRNA distribution graphs over TuYVs81 genome, bam files were used as input in MISIS (Mapped short interfering RNA Spots Identification Software (56)) and plotted with the TuYVs81 genome on the x-axis. For coverage plots of target genes, 18–26 nucleotide reads were mapped to TAIR10 genome only, no mismatches allowed, using bowtie1.2.3 (57) and normalized in Count per million mapped reads (CPM) with a bin size = 1 using deepTools version 3.3.0 (58). Reads from minus strand were converted into negative values, and the resulting bigWig files were visualized using JBrowse (59). Raw counts for TAIR10 loci (>10 counts across conditions) were used for differential analysis between mock and TuYVs81 libraries, Col-0 mock and TuYVs81 libraries, ago1-57 mock and TuYVs81 libraries, ago1-57 and Col-0 libraries, either from total RNA of AGO1-purified sRNA using DEseq2 v1.12.4 (60). Heatmaps were generated from the top variable loci across either total RNA or AGO1 IP datasets, using the pheatmap R package. See Supplemental Table S1 for all DA loci. Deep sequencing files have been deposited to the NCBI Gene expression Omnibus (GSE176378).
RNA related methods
Total RNA extraction was performed in Tri-Reagent (MRC) and RNA blots were performed as described previously (41) with starting material between 10 and 20 μg total RNA. DNA oligo sequences used for [γ-32P]ATP labeling and DNA oligos used to generate the PCR products for [α-32P]CTP-labeled Klenow products are available in Supplemental Table S2. For quantitative RT-PCR, 1–2 μg of total RNA treated with DNase I (Thermo Fisher Scientific) was reverse transcribed using either the High-capacity cDNA reverse transcription kit (Applied Biosystems) or the SuperScript IV Reverse Transcriptase (Invitrogen). All RT reactions performed from TuYVs81-infected samples and their control counterparts contained 2.5 μM random hexamers and 0.05 μM Tu-4942-rev primer. Quantitative PCR reactions were performed in a total volume of 10 μl of SYBR Green master mix I (Roche) in 384-wells plates on a Lightcycler LC480 apparatus (Roche) with the following settings: initial denaturation: 95°C 5 min. Cycling: 95°C 10 s, 60°C 15 s, 72°C 15 s single acquisition mode. Each cycle was repeated 45 times. Melting curve: 95°C 5 s, 55°C 1 min, 95°C in continuous acquisition mode with a ramp rate of 0.11°C/s, 5 acquisition per °C. All primers were designed using either Quant prime qPCR primer design tool (https://quantprime.mpimp-golm.mpg.de/) or Light cycler design software 2.0 with the following settings: min amplicon size 60, primer Tm 64, reaction conditions LC DNA master SYBR. For each cDNA, technical triplicates were obtained from the same plate, and expression data was normalized using the SAND (AT2G28390) and EF1a (AT5G60390) genes as internal controls for adult leaves and the TIP41 and EXP (AT4G26410) for seedlings. Relative quantification was done using the ΔΔCt method using a in house excel macro.
Immunoprecipitation of AGO1 and AGO2
Immunoprecipitation of endogenous AGO1 were performed as described previously (41), using the anti-AGO1 and anti-AGO2 antibodies, described in the immunoblotting section. In both cases, 5 μg of water-resuspended antibody was used to bind to 30 μl of PureProteome Protein A magnetic beads (Millipore).
Confocal microscopy
All confocal imaging was done using a Leica SP8 CLSM. For agro-infiltrated tobacco leaves, abaxial epidermal cells were imaged from leaf-disks (at least two disks per leaf, with at least three different leaves per combination, on separate plants). Leaf disks were mounted between a microscope slide and a coverslip, with a tape in between to account for the thickness and the coverslip was further taped to the slide before adding deionized water. Leaf disks were vacuum infiltrated before microscopy. Usual excitation/detection range parameters for GFP and tagRFP were 488 nm/505–520 nm and 561 nm/570–630 nm. Emissions were collected using the system's hybrid (Hyd) detectors and sequential scanning was employed for all acquisitions. Images were processed using FIJI (61).
RESULTS
siRNA-RISC-deficient point mutation in AGO1 highlights its crucial role in antiviral defense against TuYV
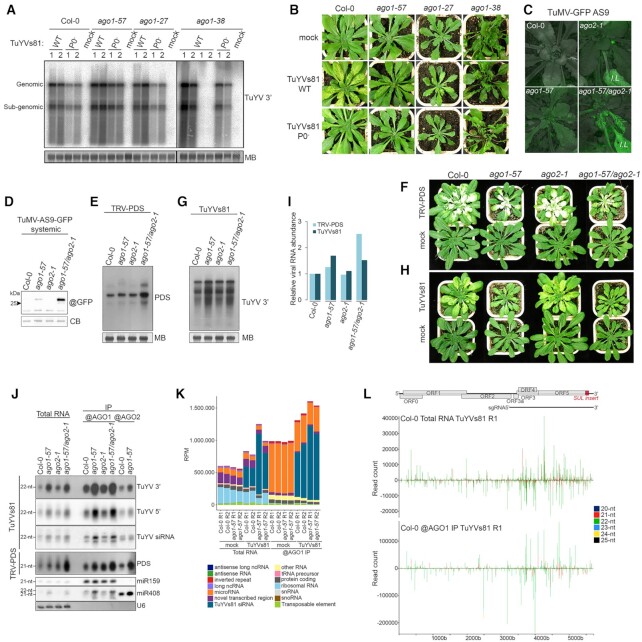
We have previously demonstrated that the G371D mutation, carried by the ago1-57 allele of AGO1, allows evasion from the P0 VSR while simultaneously causing retention of perfect siRNA duplexes, leading to the inhibition of secondary siRNA production (41). We therefore addressed the implications of this dual phenotype during TuYV infection in the model plant Arabidopsis thaliana (hereafter Arabidopsis), using an infectious clone of TuYV containing an 81 bp insertion of the AtCHLI1 gene in the 3′ non-coding region of the genome (29), referred to as TuYVs81. Since the systemic viral propagation is limited to the phloem and can act as a trigger to produce vsiRNA that move cell-to-cell, silencing of the CHLI1 gene in the neighbouring cells through virus-induced gene silencing (VIGS) typically leads to yellowing around the veins. WT (Col-0) and different ago1 mutant alleles were inoculated with either a WT TuYVs81 or the P0-deficient clone TuYVs81P0– and viral RNA accumulation was assessed in systemic tissues. Intriguingly, while absence of P0 consistently resulted in less viral RNA accumulation relative to the WT TuYVs81, none of the tested ago1 alleles exhibited the same phenotype. ago1-27 plants (that contain a point mutation in the PIWI domain) accumulated a similar amount of TuYVs81 RNA to the WT plants. The ago1-57 and ago1-38 plants (the latter containing a point mutation in the N domain) accumulated moderately more viral RNA (Figure 1A). While Col-0 and ago1-27 plants display clear vein yellowing, ago1-57 plants presented reduced yellowing despite elevated viral RNA levels, indicating that this mutant allele of AGO1 is impaired in its ability to establish VIGS (Figure 1B). None of the ago1-38 plants displayed any vein yellowing, despite the presence of the viral RNA in the systemic leaves. Any VIGS defect observed in presence of the WT virus was exacerbated when infected with TuYVs81P0–, resulting in even less vein yellowing in ago1-57. We conclude that TuYVs81-based VIGS is sensitive to the amount of trigger RNA, relies on AGO1 for its completion, and that different AGO1 point mutations result in varying VIGS as well as antiviral RNA silencing efficacy.
Figure 1.
AGO1 is the main Argonaute protein involved in defense against turnip yellows virus. (A) Accumulation of TuYVs81 RNA in systemic leaves of Col-0, ago1-57, ago1-27 and ago1-38 plants at 19 days post-infiltration (dpi) with either WT or P0-less (P0–) virus. Mock stands for mock-inoculated plants. Each lane represents a pool of four to seven individuals from which RNA was extracted from either the youngest rosette leaves (1) or older rosette leaves with yellow veins (2). For ago1-38 that did not display any vein yellowing, leaves were sampled in a similar fashion from plants displaying reddened leaves (left) or not (right, no virus accumulation). Viral RNA abundance was measured by RNA gel blot; loading control is obtained by staining the membrane with methylene blue (MB). ‘@’ indicates hybridization with DNA probe against the 3′ part of the TuYV genome. Samples were run on the same gel on two levels. Samples that ran on the bottom row are thus separated by a line. (B) Representative image of the infected genotypes analyzed in (A). (C) Representative image of the TuMV-AS9-GFP infected plants at 9 dpi. Successful systemic movement is achieved only in ago1-57 and the double mutant while expression of viral-derived GFP in the inoculated leaves (IL) is clearly visible. (D) Rescue of the TuMV-AS9-GFP systemic movement in the single ago1-57 and the double ago1-57/ago2-1 genetic backgrounds. Systemic leaves of TuMV-AS9-GFP inoculated plants were harvested at 13 dpi (n = 5 plants), and GFP protein content was measured by immunoblot. ‘@’ indicates hybridization with GFP antibody, loading control is obtained by post-staining the membrane with coomassie blue (CB). (E) Accumulation of TRV-PDS RNA in systemic leaves in the indicated genotypes at 20 dpi (n = 5 plants) measured by RNA gel blot. ‘@’ indicates hybridization with DNA probe against the PDS insert and loading control is obtained by staining the membrane with methylene blue (MB). (F) Representative individuals infected with TRV-PDS displaying systemic leaf whitening due to the silencing of the PDS gene. (G) Accumulation of TuYVs81 RNA in systemic leaves in the indicated genotypes at 20 dpi (n = 5 plants) measured by RNA gel blot. ‘@’ indicates hybridization with DNA probe and loading control is obtained by staining the membrane with methylene blue (MB). (H) Representative individuals infected with TuYVs81 displaying systemic vein yellowing. (I) Quantification of viral RNA signal in (E) and (G) relative to Col-0 and normalized to MB signal. (J) vsiRNA abundance in total RNA, AGO1 and AGO2 immunoprecipitates in TuYVs81 and TRV-PDS infected leaves. ‘@’ indicates hybridization with DNA probe or use of a specific antibody for immunoprecipitation. (K) Global quantification of 21-nt to 24-nt small RNA reads aligned to the reference Arabidopsis genome and TuYVs81 genome per functional categories (araport11), expressed as (RPM) reads per million ([category count × 1 000 000]/total mapped reads). Libraries were obtained from total RNA and AGO1 IP in mock-inoculated (mock) or infected (TuYVs81) systemic leaves at 16 dpi (n = 7 or 8 individual plants per replicate). R1 = replicate 1, R2 = replicate 2. (L) Distribution of TuYVs81-derived sRNA reads (20-nt to 25-nt) along the TuYVs81 genome in Col-0 total RNA and AGO1 IP replicate 1 (R1), with MISIS. Bars indicate the position of the 5′ (+strand) and 3′ (–strand) extremity of each mapped sRNA. Y-axis represents read counts, and each size category is represented in the indicated color.
Other AGO proteins have been shown to be involved in the silencing of several RNA viruses, sometimes in combinations (6). We therefore tested the contribution of different AGOs to TuYV silencing by infecting single or multiple knock out mutants for AGO5, AGO10, AGO2 and AGO7, in addition to ago1-27, which is a missense allele. Quantification of the viral RNA in systemic leaves revealed that only the ago1-57 mutation caused enhanced accumulation of TuYV (Supplementary Figure S1A), further demonstrating the importance of AGO1 in TuYV antiviral defense. Here too, the ago1-27 mutant did not exhibit any difference, neither did the combinations that contained that allele.
Because AGO2 has been described in several studies as the main antiviral AGO (62–64) or to function in tandem with AGO1 (18,65), we introgressed the ago2-1 mutation into the ago1-57 background and tested for antiviral performance against several well-studied RNA viruses. Plants were inoculated with an infectious clone of turnip mosaic virus (TuMV) containing a GFP reporter and a point mutation inactivating the VSR activity of HC-Pro, referred to as TuMV-AS9-GFP (11). While GFP was clearly visible in inoculated leaves of ago2-1 plants, systemic spread of the virus did not occur (Figure 1C and D). By contrast, ago1-57 containing plants allowed spread of TuMV-AS9-GFP into the systemic leaves. This reveals a previously unreported role for AGO1 in defense against TuMV and shows that both fully functional AGO1 and AGO2 are needed to mount efficient defense against TuMV-AS9-GFP. We then infected the same genotypes with tobacco rattle virus (TRV) carrying a PHYTOENE DESATURASE (PDS) fragment, referred to as TRV-PDS (66), which triggers potent VIGS against this gene resulting in a bleached leaf phenotype. Both AGO1 and AGO2 have been reported as necessary for efficient antiviral silencing during TRV infection (65). Accordingly, we observed increased viral RNA and impaired PDS silencing in the double ago1-57/ago2-1 mutant, while single mutants behaved like Col-0 (Figure 1E and F). In contrast, ago2-1 single mutation did not affect TuYVs81 RNA accumulation, while the single ago1-57 and the double mutant showed enhanced accumulation and an identical impairment in VIGS (Figure 1G–I). This suggests that, as opposed to TuMV and TRV, efficient antiviral silencing of TuYV by the plant does not rely on the coordinated action of AGO1 and AGO2, but rather that AGO1 alone is necessary to mount an efficient response.
Since AGO proteins can bind overlapping cohorts of sRNA and act redundantly on an RNA target, impairment of a particular AGO can result in compensatory loading by another AGO. We reasoned that if AGO2 can act as a surrogate to AGO1, then compromising AGO1 function should lead to an increase in AGO2 loading with vsiRNA. We therefore performed immunoprecipitations of both AGO1 and AGO2 (Supplementary Figure S1B) and analyzed the associated sRNA in plants systemically infected by TRV-PDS and TuYVs81. TRV-PDS-derived vsiRNA were loaded into AGO1 and AGO2, and in the ago1-57 background we observed increased loading into AGO2 (Figure 1J). TuYVs81 vsiRNA were found to be predominantly associated with AGO1, with increased production of vsiRNA in ago1-57 plants likely leading to increased loading of AGO1-57. Little or no compensatory loading of AGO2 was observed, further establishing the pivotal role of AGO1 in TuYVs81 antiviral defense. We also compared loading abilities of AGO1-57 and AGO1-27 (Supplementary Figure S1C) and found increased AGO2 loading with vsiRNA only in ago1-27, concomitant with impaired loading of the AGO1-27 protein of both vsiRNA and miRNA. This can be explained by the increased AGO2 protein level observed in ago1-27 (Supplementary Figure S1D) caused by lessened miR403-AGO1 complex assembly (Supplementary Figure S1C), a feature not observed in the ago1-57 mutant. Like for TuYVs81, we also found that loading of AGO1 and AGO2 with SUL siRNA was identical in WT and ago1-57 plants (Supplementary Figure S1E and F), indicating that the mutant protein is not affected in siRNA loading, as we have previously shown (41). Taken together, these results suggest that ago1-57 presents a unique opportunity for deciphering the antiviral role of AGO1 in planta.
Next, we performed sRNA deep sequencing of total RNA and AGO1 IPs from mock-inoculated and TuYVs81 infected Col-0 and ago1-57 plants (Supplementary Figure S2A). After verifying successful immunoprecipitation of AGO1 in all replicates and samples (Supplementary Figure S2B), sRNA reads of 18–26nt were mapped to the reference Arabidopsis genome (67) and the TuYVs81 genome and normalized 21–24nt reads mapping to the different categories are shown in Figure 1K. As expected, AGO1 IPs of mock samples contained abundant reads mapping to miRNA, that were mostly of 21-nt (Supplementary Figure S2C). In infected samples, a large amount of vsiRNA were recovered from both the total RNA and the AGO1 IP, indicating that a large amount of the produced vsiRNA is channelled towards AGO1, as observed by northern blot. These vsiRNA were found to originate from both strands and to completely cover the TuYVs81 genome, with some hotspots found across the replicates and samples (Figure 1L and Supplementary Figure S2D). TuYVs81-mappers were enriched in AGO1 IP with some reaching a read count of over 400 000 in AGO1-57 IPs. Intriguingly, most reads mapping to the TuYVs81 genome were of 22-nt (Figure 1L and Supplementary Figure S2C) suggesting that DCL2, among the four Arabidopsis DCL proteins, is predominantly responsible for TuYVs81 processing. We also observed an impaired 5′U bias in AGO1 IPs from the ago1-57 plants (Supplementary Figure S2E). This is explained by the molecular phenotype of the allele, which retains both the guide strand and the passenger strand (41), the latter having a 5′ extremity that can be any nucleotide, artificially raising the amount of 5′ C, G and A reads. Altogether our results show that AGO1 plays a key role in orchestrating antiviral defense against TuYV, by loading a vast population of vsiRNA which are mostly 22nt-long.
Non-degradable AGO1-57 impairs establishment of systemic TuYVs81 infection
After numerous TuYVs81 infections using agro-inoculation we noticed that about only half of the initially inoculated ago1-57 plants developed VIGS, in contrast to Col-0 inoculated plants that showed up to 100% systemic infection rate (Supplementary Figure S3A). Since ago1-57 is impaired in VIGS, we chose to monitor the presence of the viral readthrough protein (RT) in young leaves of the inoculated plant population during the progression of infection. This showed a similar kinetic to that of VIGS, with only 43.3% of the ago1-57 plants exhibiting infection, while infection of ago1-57 with TuMV-GFP and TRV-PDS reached 100% (Supplementary Figure S3A). We hypothesized that this observation could be explained by the evasion of AGO1-57 from P0-mediated degradation or alternatively, that the ago1-57 mutation leads to constitutive activation of defense-related genes leading to enhanced resistance, as observed for TRV-infected ago1-27 (65), and regulated by miRNA and phasiRNA in multiple plant species (68–70). To discriminate between these two hypotheses, we performed similar infection kinetics in the three ago1 hypomorphic mutants used previously. We found that only the ago1-57 plant population exhibited a 50% loss of systemic infection by TuYVs81, while ago1-27 and ago1-38 both reached 100% infection, albeit with very dissimilar kinetics (Figure 2A and B). Importantly, this loss was not observed during establishment of TuMV-GFP infection (Figure 2C and D), indicating that it is unique to the ago1-57-TuYVs81 interaction, and likely results from the undegradable nature of the mutant protein.
Figure 2.
ago1-57 uniquely affects systemic movement of TuYVs81 due to delayed viral RNA accumulation after agrobacterium-mediated inoculation but fails to provide any protective effect in vector-mediated infection. (A) Kinetic of systemic TuYVs81 infection in Col-0, ago1-57, ago1-27 and ago1-38 represented as the cumulated percentage of infected plants in the inoculated population (n = 14 individuals per genotype). To avoid any confounding effect introduced by VIGS deficiency in the mutants, infected individuals were scored by the detection of the TuYVs81 readthrough protein (RT, ORF5) in leaf patch from young systemic leaves at 6, 7, 9, 12, 15 and 21 dpi using either western or dot blot. Plants that exhibit systemic VIGS in between these sampling times are also counted as infected at the time point at which the VIGS was first observed. (B) TuYVs81 viral RNA abundance in systemic leaves of the indicated mutants at 15 dpi measured by RNA blot. Each sample represents a mix of leaf patches for all infected individuals at that time point in the kinetic. ‘@’ indicates hybridization with DNA probe and loading control is obtained by staining the membrane with methylene blue (MB). (C) Kinetic of systemic TuMV-GFP infection in Col-0, ago1-57, ago1-27 and ago1-38 represented as the cumulated percentage of infected plants in the inoculated population (n = 6 individuals per genotype). Infected individuals were scored by the detection of GFP in systemic leaves at the indicated day. (D) Detection of TuMV-GFP in systemic leaves of the indicated mutants at 9 dpi measured by immunoblot. ‘@’ indicates hybridization with GFP antibody, loading control is obtained by post-staining the membrane with Coomassie blue (CB). (E) ago1-57 displays delayed viral RNA accumulation after inoculation. Measurement of TuYVs81 RNA in inoculated leaves at 0, 49, 95 and 120 h post infiltration (hpi) measured by RT-qPCR in the indicated genetic backgrounds represented as bar graph relative to Col-0 49hpi. Represented values are means of technical triplicates, error bars represent the SEM. For each time point, five infiltrated leaves from four individuals were harvested for each genotype. *P < 0.05, **P < 0.01, ***P < 0.001 with Student's t-test, one-tailed, paired. (F) Accumulation of TuYVs81 readthrough protein (RT) at 0, 49, 95 and 120 h post infiltration (hpi) measured by immunoblot. Samples are from the same tissues as in (E). ‘@’ indicates hybridization with RT antibody, loading control is obtained by post-staining the membrane with Coomassie blue (CB). (G) G371D mutation does not confer undegradability to AGO1 in presence of P0 from diverse polerovirus species. Left panel: Schematic representation of the P0 protein, with the region containing the F-box motif highlighted in red and the corresponding alignment of P0 from turnip yellows virus (TuYV), cucurbit aphid-borne yellows virus (CABYV), beet mild yellowing virus (BMYV) and potato leafroll virus (PLRV) shown below. The minimal F-box motif is boxed in red, and the sequence logo for the alignment is shown below. Right panel: AGO1 degradation test in N. benthamiana. Both versions of CFP-AGO1 (WT or G371D) were expressed either without (ø) or with the indicated P0 proteins in two separate leaves and an equivalent amount of leaf patches were collected at 4 dpi. All infiltrated patches contain P19. Fusion protein levels were assessed by immunoblot on two different membranes (GFP corresponds to Coomassie stain CB1 and cMyc to CB2) and ‘@’ indicates hybridization with the corresponding specific antibody. Expression of the untagged P0 constructs was verified by RNA blot using DNA probes specific for each sequence (@ P0) and equal loading was assessed by staining the corresponding membrane with methylene blue (MB1 and MB2). (H) Detection and quantification of aphid-transmitted WT TuYV virions in systemic leaves of Col-0 or ago1-57 individual plants by DAS-ELISA. Eighteen-day-old plants were individually challenged with two Myzus persicae fed on either 20% sucrose solution (+Suc) or 20% sucrose solution containing 67mg/ml TuYV virions (+TuYV) or alternatively were left untreated (N.I). Each bar represents the mean OD at 405 nm (technical triplicate measurements) for a single individual within the considered category, and error bars represent SD. Individuals for which the OD was ≤ to those of the NI and +Suc control plants are considered as non-infected and are colored in black. Difference between the Col-0 and ago1-57 populations is statistically different (Kruskal and Wallis test). (I) Fresh weight measurement of all the analyzed plants in (H) after removal of the non-infected plants, expressed in mg. ***P < 0.001 (two-way ANOVA followed by Tukey honest significant differences test to compare both genotypes and treatments).
P0-less mutants of TuYV show lower viral replication in protoplasts (28), and PLRV P0 is essential for viral multiplication in inoculated tissues (71), suggesting that suppression of RNA silencing is important during early steps of infections. Since AGO1-57 is resistant to P0-mediated degradation, it should exert a similar effect to the loss of viral P0 during the local establishment of infection. We therefore assayed viral RNA abundance in Arabidopsis leaves agro-inoculated with TuYVs81 in ago1-57 compared to Col-0 and ago1-27. In all experiments, ago1-57 leaves contained significantly less viral RNA than Col-0 (Figure 2E and Supplementary Figure S3B) and less viral RT protein (Figure 2F). This slower build-up of infection in ago1-57 was not shared by the ago1-27 mutant, that is sensitive to P0-mediated degradation (41), and is thus the consequence of resistance to P0.
Because P0 proteins from different poleroviruses contain the minimal F-box consensus motif (LPxxL/I), we tested if the G371D mutation enables AGO1 evasion from a range of P0 proteins. WT and CFP-AGO1G371D were co-infiltrated with P0 proteins from TuYV, PLRV, cucurbit aphid-borne yellows virus (CABYV) and beet mild yellowing virus (BMYV), and CFP-AGO1 degradation was assayed as previously described (21). Surprisingly, while the G371D mutation protected AGO1 from P0Tu-mediated degradation, the mutation failed to prevent degradation by the three other P0 proteins tested (Figure 2G). This highlights the existence of different modes of AGO1 recognition employed by viral P0s and indicates that the strategy employed by TuYV to target AGO1 is not a conserved feature of poleroviruses.
Finally, we assayed the performance of ago1-57 when TuYV is delivered by its natural aphid vector, that directly injects virions into phloem cells. When infection was monitored 25 days after transmission we found that the number of plants that scored positive for the presence of the virus was comparable between Col-0 (89.5%) and ago1-57 (87.5%) (Figure 2H). Not only did the ago1-57 plants not display the reduced systemic infection as observed for agro-inoculation, but all the plants contained significantly more virion than their WT counterparts. Intriguingly, while TuYV infection normally leads to a symptomless infection in Arabidopsis, we observed a statistically significant reduction in fresh weight for the TuYV-infected ago1-57 plants (Figure 2I), suggesting that impairment of AGO1 function leads to symptomatic infection. Taken together, our analysis shows that evasion of degradation by the mutant AGO1 causes delay in the establishment of local infection, leading to a decrease in systemic infection. However, this apparent resistance phenotype is overruled by direct inoculation into phloem cells.
TuYV RNA is mostly processed by DCL2, but both DCL2 and DCL4 are necessary to mount an effective antiviral defense
To test the contribution of DCLs to vsiRNA production, we performed northern blots from tissues systemically infected by TuYVs81, in single and multiple mutants containing the dcl2-1 or dcl2-5, dcl4-2, dcl3-1 and ago1-57 alleles. In dcl2-1 infected plants, the main 22-nt TuYV vsiRNA band present in wild-type was replaced by a 21-nt band, indicating that the bulk of vsiRNA is indeed the product of DCL2, while loss of DCL4 did not affect the main vsiRNA population (Figure 3A). Loss of both DCL2 and DCL4 resulted in the sole accumulation of a 24-nt signal that is otherwise minimal in WT plants, while the dcl2-5/dcl3-1 combination led to a single 21-nt signal. Loss of 22-nt vsiRNA in dcl2-1 was consistently accompanied by reduced silencing signal spread, a feature that was not observed in the dcl4-2 plant (Figure 3B and Supplementary Figure S4A), mirroring their relative contribution to vsiRNA production in the phloem cells. VIGS signal was abolished in dcl2-1/dcl4-2 plants, while dcl2-5/dcl3-1 were identical to single dcl2-1, indicating that the residual 24-nt do not constitute a mobile silencing signal sufficient to initiate VIGS. Identical vsiRNA patterns were observed when the ago1-57 mutation was added to each combination (Figure 3A). Combining dcl2-1 and ago1-57, that both exhibit weak VIGS, led to a complete loss of cell-to-cell silencing spread, which was not the case in dcl4-2/ago1-57, that produces abundant vsiRNA. Like ago1-57, single dcl2-1 and dcl4-2 mutations lead to a two-fold increase of TuYV RNA in systemic tissues, that was not exacerbated when introduced in the ago1-57 background (Figure 3C). Only the double dcl2-1/dcl4-2 lead to a dramatic increase in the amount of TuYV RNA accompanied by severe symptoms (Figure 3B). Intriguingly, the triple mutant combination led to an even greater viral RNA accumulation, supporting that the 24-nt vsiRNA are able to exert antiviral activity through AGO1.
Figure 3.
TuYvs81 RNA is mainly processed by DCL2 into 22-nt vsiRNA, yet both DCL2 and DCL4 are necessary to efficiently silence the virus. (A) Analysis of vsiRNA size in infected TuYVs81 infected plants at 17 dpi using RNA gel blot. Most of the vsiRNA populations is of 22-nt and is lost in a dcl2-1 single or combination mutant. ‘@’ indicates hybridization with the indicated DNA probe. Endogenous siRNA are used to control for the proper identity of the dcl mutants and U6 signal is the loading control. (B) Representative image of the infected genotypes analyzed in (A). Left panel: whole plant view. Right panel: inset of young systemic leaves. White arrow indicate very feint vein yellowing. (C) TuYVs81 viral RNA abundance in systemic leaves of the indicated mutants at 17 dpi measured by RT-qPCR. Levels are displayed relative to infected Col-0. Each sample represents a pool of several individuals from the indicated genotype. Only individuals that scored positive for the presence of systemic TuYVs81 (via detection of the RT protein in leaf patches) were harvested. P value above each sample is for pairwise comparison of the sample to Col-0. *P < 0.05, **P < 0.01, ***P < 0.001 with Student's t-test, two-tailed, unequal variance. (D) Both DCL2 and DCL4 are redirected to cytosolic viral replication complexes (VRC) during TuYVs81 infection and colocalize with viral double stranded RNA. Representative single plane confocal images of transiently expressed 35S:tRFP, 35S:DCL2genomic-tRFP and 35S:DCL4genomic-tRFP with (TuYVs81) or without (mock) the virus in leaves of transgenic N. benthamiana stably expressing the double-stranded RNA-binding B2-GFP protein. Observations are from leaf discs of 3–5 days post-infiltration. Inset scale bar is 10 μm. See also Supplementary Figure S5 for additional images.
In order to test the contribution of secondary siRNA to antiviral silencing of TuYVs81, we infected loss of function mutants for RDR6 and SGS3, that are both required for production of dsRNA and silencing signal amplification from AGO1 cleavage products, a mechanism that participates in antiviral silencing for some plant RNA viruses (8,10,11,72). This revealed that neither VIGS (Supplementary Figure S4A), vsiRNA profiles (Supplementary Figure S4B) nor TuYV RNA accumulation (Supplementary Figure S4C) were affected in those mutants. The same mutants presented systemic infection kinetics that were comparable to those of WT and dcl mutant plants (Supplementary Figure S4D). This shows that silencing signal amplification via RDR6/SGS3 is dispensable for antiviral silencing during infection by TuYVs81.
Since both DCL2 and DCL4 have antiviral activity against TuYVs81, but DCL2 rather than DCL4 is the major contributor of vsiRNA, we sought to establish their localization relative to viral replication complexes (VRCs). We took advantage of the recently established stable N. benthamiana line expressing the eGFP-tagged dsRNA binding domain of flock house virus (FHV) B2 protein (B2-GFP) that allows in vivo visualization of dsRNA, the minimal component of the VRCs (51). Consistent with their ability to produce vsiRNA from double stranded TuYV RNA, both AtDCL2-tRFP and AtDCL4-tRFP were found to equally re-localize from the nucleus to the B2-GFP labeled VRCs (Figure 3D and Supplementary Figure S5), which was not the case for the nucleo-cytoplasmic tRFP control. In summary, our results show that both DCL2 and DCL4 are directed to VRCs upon infection to initiate antiviral defense against TuYVs81, but DCL2 is the major source of vsiRNA in systemically infected tissues.
DCL4 and DCL2 operate normally in TuYV-infected vasculature
To characterize more specifically the relative roles of DCL2 and DCL4 in infected vasculature, we employed MeSelect (mechanical separation of leaf compound tissues), a technique which enables isolation of vascular bundle cells (73). As expected, TuYVs81 RNA was enriched when compared to whole leaf extracts (Figure 4A and Supplementary Figure S6A), and the loss of DCL2 lead to an increased accumulation of TuYVs81 RNA, as observed in whole leaves (Figure 3C). Vascular vsiRNA were 22-nt in size (Figure 4B and Supplementary Figure S6B), confirming that their presence in whole leaf arises from the processing of TuYVs81 RNA by DCL2 in the vasculature. We also observed a smearing pattern above the 22-nt signal only in the enriched vasculature (Supplementary Figure S6B) suggesting persistence of precursor RNA species/replication intermediates.
Figure 4.
DCL2 processing is not a consequence of detectable viral manipulation in the infected vasculature. (A) Measurement of TuYVs81 RNA in systemic whole leaves of Col-0 and dcl2-1 plants (n = 8–10 individuals) or enriched vascular bundles of the equivalent plants (n = 20 leaves) at 17 dpi by RT-qPCR represented as bar graph relative to infected Col-0 leaves. Represented values are means of technical triplicates, error bars represent the SEM. **P < 0.01, with Student's t-test, one-tailed, paired. (B) Analysis of sRNA abundance in total RNA extracted from the whole leaf or the vasculature of mock (−) or TuYVs81 infected (+) Col-0 and ago1-57 plants by RNA gel blot. For leaf tissue, a mixture of infected leaves from several individual was used (n = 8) and enriched vascular bundles (n = 24 leaves) were obtained from the equivalent plants at 16 dpi. ‘@’ indicates hybridization with DNA probe and U6 signal is the loading control. (C) Detection of the DCL1 and viral RT proteins in total protein extracted from the whole leaf or the vasculature of mock (−) or TuYVs81 infected (+) Col-0 and ago1-57 plants by immunoblot. Samples were prepared from the same material as in (B). ‘@’ indicates hybridization antibodies, loading control is obtained by post-staining the membrane with Coomassie blue (DCL1 corresponds to Coomassie stain CB1 and RT to CB2). (D) Measurement of DCL2 and DCL4 mRNA abundance in the same samples as (A) by RT-qPCR, represented as bar graph relative to - Col-0 vasculatures. Note that loss-of-function allele dcl2-1 contains WT level of DCL2 messenger RNA. Represented values are means of technical triplicates, error bars represent the SEM. **P < 0.01, ns P > 0.05 with Student's t-test, two-tailed, paired. See Supplementary Figure S6C for an additional experiment. (E) Analysis of TRV-PDS (+) infected whole leaves and vasculature of Col-0 plants (n = 9 individuals, 8 leaves for vasculature) at 16 dpi by RNA gel blot. Top panel: sRNA blot shows equivalent profiles of vsiRNA made against the PDS insert that are mostly processed by DCL4 into 21-nt. An oligoprobe recognizing the conserved 3′ of RNA1 and 2 of TRV shows that this portion of the viral RNA is mostly processed by DCL2 into 22-nt, irrespective of the tissue. Bottom panel: TRV RNA is present in both tissue types and is not enriched in the vasculature. ‘@’ indicates hybridization with DNA probe, U6 signal and methylene blue staining are the loading control. Note that chloroplastic 16S and 23S rRNA are only visible in whole leaf. (F) Measurement of TuYVs81 RNA in systemic Col-0 whole leaves (n = 8–10 individuals) or enriched vascular bundles of the equivalent plants (n = 20 leaves) at 17 dpi by RT-qPCR represented as bar graph relative to infected leaves. Plants were inoculated either with the empty vector (mock), with the WT TuYVs81 or with a P0-less mutant. Represented values are means of technical triplicates, error bars represent the SEM. **P < 0.01, ***P < 0.001 with Student's t-test, one-tailed, paired. (G) Analysis of vsiRNA profile from whole leaf of Col-0 plants infected with either WT TuYVs81 or the P0-less TuYVs81 by RNA gel blot. Samples are the same as in (F), infected dcl2-1 is used a control for the absence of 22-nt. ‘@’ indicates hybridization with DNA probe and U6 signal is the loading control.
Similarly to TuYV, turnip crinkle virus (TCV) is also processed mainly into 22-nt-long vsiRNA in Arabidopsis, its VSR P38 inhibiting the AGO1-miR162 regulation of DCL1 (74) which itself represses DCL4 and DCL3 via an unknown mechanism (8). We observed a mild decrease in the miR162 signal in both Col-0 and ago1-57 infected vasculature (Figure 4B), but no appreciable difference in the abundance of the DCL1 protein in the same tissue (Figure 4C), ruling out a mechanism similar to TCV regulation of DCL1 in the case of TuYV. Measurement of DCL2 and DCL4 mRNA abundance showed that neither are affected by the presence of the virus in the vasculature (Figure 4D and Supplementary Figure S6C). To test if the activity of DCL4 is impaired in the presence of TuYVs81, we monitored the accumulation of DCL4-dependent endogenous sRNA. While miR822 is barely produced in the vasculature, siR255 (TAS1) abundance was unperturbed by the presence of the virus in the vasculature (Figure 4B), suggesting that DCL4 functionality is intact. Similarly, IR71 22-nt siRNA are unaffected by the virus, demonstrating that DCL2 is functioning normally. Because DCL4 and DCL2 usually exhibit hierarchical activity towards transgenes and RNA viruses, with DCL4 being the prime source of siRNA over DCL2, we wondered whether in the vasculature this hierarchy could be inverted. We thus compared sRNA profiles in whole leaf to those in vasculature recovered from TRV-PDS infected plants, an RNA virus that is primarily processed into 21-nt by DCL4 in WT plants (5) and found that the vsiRNA profiles were identical both for the PDS reporter and the 3′ of RNA1 and RNA2 (Figure 4E), demonstrating that the vascular DCL hierarchy is identical to that of the leaves.
Collectively, these results show that processing of TuYVs81 by DCL2 is not a consequence of attenuated DCL4 expression and/or activity, nor of increased expression or overall activity of DCL2 in infected vasculature. Neither DCL2 nor DCL4 show vasculature-specific antiviral activity when challenged with an unrelated RNA virus, suggesting in turn manipulation of the DCL balance by the TuYV. To test whether P0 could affect the balance between DCL2 and DCL4 in TuYV processing, we compared the vsiRNA profiles in systemic tissues infected with either WT TuYVs81 or TuYVs81P0–. As observed previously in whole leaves (Figure 1A), TuYVs81P0– was less abundant than its WT counterpart in the isolated vasculatures (Figure 4F) but was nonetheless enriched comparatively to the leaf. Although the signal was weaker, TuYVs81P0– infection still resulted in a major 22-nt signal (Figure 4G), indicating that P0 is not responsible for preponderant processing of TuYV by DCL2.
Vascular AGO1 is sensitive to P0 but is unexpectedly stabilized in presence of TuYV
Next, we took advantage of the MeSelect method to observe viral P0-driven degradation of AGO1, a feature that is obscured in whole leaf tissues, and habitually observed with over-expression of P0 in Arabidopsis (22,24,41) or in transient co-expression assays (Figure 2G). We therefore separated vasculature from Col-0 and ago1-57 infected plants, with both genotypes presenting a significant enrichment of TuYVs81 RNA in the vasculature (Figure 5A and Supplementary Figure S7A). Strikingly, rather than a depletion, we observed an increased accumulation of vascular AGO1 protein in the presence of the virus, irrespective of the background used (Figure 5B and Supplementary Figure S7B). This enrichment relative to the mock inoculated vasculature was very robust and observed in vasculatures collected at different days post inoculation as quantified in Figure 5C (n = 5 separate infection experiments), and was not observed for either AGO2 and AGO4 (Figure 5B and Supplementary Figure S7B).
Figure 5.
Vascular AGO1 is post-translationally stabilized in presence of TuYVs81 and loaded viral 22-nt siRNA promotes production of secondary siRNA. (A) Measurement of TuYVs81 RNA in systemic whole leaves of Col-0 and ago1-57 plants (n = 8) or enriched vascular bundles of the equivalent plants (n = 24 leaves) at 16 dpi by RT-qPCR represented as bar graph relative to infected Col-0 leaves. Represented values are means of technical triplicates, error bars represent the SEM. *P < 0.05, ***P < 0.001 with Student's t-test, one-tailed, paired between tissues, unequal variance between genotypes. (B) Representative immunoblot of AGO1 and AGO2 accumulation in systemic whole leaves (n = 12 individuals) and in enriched vascular bundles of the equivalent plants (n = 18 leaves) at 21 dpi in Col-0 and ago1-57, in the absence (−) or presence (+) of TuYVs81. ‘@’ indicates hybridization with the indicated antibodies, and loading control is obtained by post-staining the membrane with Coomassie blue (AGO1 on CB1 and AGO2 on CB2). (C) Quantification of the AGO1 signal normalized to total protein signal (Coomassie blue stain, whole lane) in Col-0 vasculatures without (mock) or in the presence of TuYVs81. Collected values represent 5 biological replicates in which vasculatures were enriched at either 15, 16, 17 or 21 dpi from different infection experiments. P-value = 0.001196 with Student's t-test, one-tailed, paired. (D) Representative single plane confocal images of transiently expressed 35S:tRFP-AGO1 with (TuYVs81) or without (mock) the virus in leaves of transgenic N. benthamiana stably expressing the double-stranded RNA-binding B2-GFP protein. Observations are from leaf discs at 3 dpi. Inset scale bar is 10 μm. See also Supplementary Figure S8A for additional images. (E) P0 colocalizes with viral double stranded RNA. Representative confocal images of transiently expressed 35S:P0-tRFPwith (TuYVs81) or without (mock) the virus in leaves of transgenic N. benthamiana stably expressing the double-stranded RNA-binding B2-GFP protein. Observations are from leaf discs at 3 dpi. Inset scale bar is 10 μm. See also Supplementary Figure S8B for additional images. (F) Levels of AGO1 and AGO2 mRNA are not significantly affected by the presence of TuYVs81 in the plant vasculature. mRNA abundance in the same samples as (A) by RT-qPCR, represented as bar graph relative to Col-0 vasculatures. Represented values are means of technical triplicates, error bars represent the SEM. *P < 0.05, ns P > 0.05 with Student's t-test, two-tailed, paired. See Supplementary Figure S7E for an additional experiment. (G) Analysis of AGO1 and AGO2 protein level in enriched vascular bundles of Col-0 or dcl2-1 plants (n = 20 leaves from 8 to 10 individuals per genotype and treatment) at 17 dpi after inoculation with E.V (−), TuYVs81 WT or TuYVs81 P0–. ‘@’ indicates hybridization with the indicated antibodies, and loading control is obtained by post-staining the membrane with Coomassie blue (AGO1 on CB1 and AGO2 on CB2). (H) Persistent companion cell expression of P0 enhances spreading of SUL siRNA. Image of representative individual adult plants of WT, pCoYMV:P0-HA WT and LP1 in the SS background. (I) Vascular AGO1 is degraded in presence of WT P0-HA. Immunoblot of AGO1 and P0-HA accumulation in whole leaves (n = 4 individuals) and in enriched vascular bundles of the equivalent plants (n = 10–16 leaves) in the indicated genotypes. Plants are the same as in (H). ‘@’ indicates hybridization with the indicated antibodies, and loading control is obtained by post-staining the membrane with Coomassie blue (CB). (J) Analysis of sRNA abundance in total RNA extracted from the whole leaf or vasculature by RNA gel blot. RNA were obtained from the same samples as in (I). ‘@’ indicates hybridization with DNA probe and U6 signal is the loading control. (K) Degradation of AGO1 by vascular P0 leads to increased abundance of the AGO1 mRNA in the vasculature and decrease of the total amount of CHLI1 and CHLI2 mRNA. mRNA abundance in the same samples as (J) by RT-qPCR, represented as bar graph relative to SS leaf. Represented values are means of technical triplicates, error bars represent the SEM. ***P < 0.001, **P < 0.01, ns P > 0.05 with Student's t-test, two-tailed, equal variance. (L) Heatmap of annotation units showing the most variation in small RNA abundance across the eight AGO1 IP libraries. (M) Top panel: Schematic representation of the TuYVs81 genomeSUL insert is represented as a red square in 3′ of the viral sequence. Bottom panel: Browser view of normalized sRNA reads (CPM count per million of mapped reads) mapping on both strands of CHLI1 and CHLI2 genes (0mm). Red squares represent the regions in the transcript that are identical to the SUL insert of TuYVs81. sRNA reads present within the pink highlighted area are directly produced from the viral RNA and trigger the production of the secondary siRNA population found in 3′ of both transcripts. Production of secondary siRNA is impaired in ago1-57 plants.
Because this observation was unexpected, given that SCFP0 enables vacuolar degradation of ARGONAUTE proteins (23,24), we explored their localization relative to TuYVs81 VRCs using the B2-GFP assay in N. benthamiana. In contrast to what was observed for DCL proteins, tRFP-AGO1 did not fully colocalize with the dsRNA but rather re-localized to the immediate vicinity of TuYVs81 VRCs (Figure 5D and Supplementary Figure S8A). This localization was reminiscent of that of the ssRNA and CP of potato virus X (PVX) relative to that of B2-GFP labelled dsRNA, that are partitioned within larger ‘viral factories’ (51) also referred to as X-bodies (75). This suggests that AGO1 does not fully access TuYV VRCs, which contain dsRNA, but rather peripheral viral structures that could be rich in viral ssRNA. On the contrary P0-tRFP, which presented bright punctate cytosolic structures in non-inoculated leaves, colocalized with the VRCs in infected leaves (Figure 5E and Supplementary Figure S8B). Thus, AGO1 and P0 localizations only partially overlap during TuYVs81 infection, suggesting that only a fraction of AGO1 would be available for ubiquitination by viral P0 at any given time.
Since AGO1 mRNA level is under tight control by the AGO1-miR168 feedback loop (76), we monitored miR168 abundance in TuYVs81-infected Arabidopsis vasculature (Figure 4B and Supplementary Figure S7C) and found it to be unchanged by TuYV. Accordingly, AGO1 and AGO2 mRNA abundance was identical in mock and TuYVs81-infected vasculature (Figure 5F, Supplementary Figure S7D and S7E), ruling out a transcriptional or post-transcriptional regulation of AGO1 caused by the virus. We also observed stabilization of a vascular-restricted Flag-AGO1 protein (pSUC:Flag-AGO1, two independent lines) in TuYVs81 infected whole leaves (Supplementary Figure S7F) that itself did not result from transcriptional activation of the transgene (Supplementary Figure S7G). We also tested the possible involvement of the viral P0 in AGO1 stabilization, or a correlation between this phenomenon and TuYVs81 processing by DCL2 by analyzing AGO1 abundance in vasculature isolated from TuYVs81P0– infected plants, as well as in dcl2-1 plants. In both cases, AGO1 overaccumulation was still evident (Figure 5G).
Finally, we tested the ability of P0 to degrade AGO1 and its consequences on silencing in vascular cells by engineering a P0-HA stable line under the control of the commelina yellow mottle virus promoter (pCoYMV) that drives specific expression in companion cells (77). This transgene was introduced into the SUC-SUL background (45) to monitor silencing. Lines expressing the transgene were confirmed by RT-qPCR in in vitro grown seedlings and exhibited vein yellowing (Supplementary Figure S9), in contrast to a pSUC:P15-FHA lines that prevented movement of SUL siRNA to the neighbouring cells (15). Strikingly, older plants containing the WT P0-HA exhibited a strong enhancement of leaf yellowing that was absent from plants of the same age containing the LP1 version of P0-HA (Figure 5H). Isolation of vasculature revealed the enrichment of the P0-HA protein compared to whole leaves and the degradation of vascular AGO1 by P0-HA, that was dependent on an intact F-box motif (Figure 5I), demonstrating that vascular cells are a suitable environment for P0-mediated degradation. Degradation of AGO1 was accompanied by stabilization of miR160c*, a hallmark of miR160 duplex accumulation, as well as destabilization of guide miRNA (Figure 5J). SUL siRNA quantity was not changed by P0-HA presence in the vasculature, indicating that movement rather than biogenesis was impaired. Overall CHLI1 and CHLI2 mRNA were decreased in the pCoYMV:P0-HA-WT plants, due to SUL siRNA accessing transcripts far removed from the vascular initiation site (Figure 5K). Vascular AGO1 homeostasis was perturbed (Figure 5K), most likely as a consequence of disturbance in the miR168-AGO1 feedback loop. All in all, we demonstrate that vascular P0, when produced by a transgene, is competent in degrading AGO1, and the resulting depletion of AGO1 potentiates the cell-to-cell movement of siRNA to the surrounding tissues. Nevertheless, during genuine TuYVs81 infection, vascular AGO1 protein is stabilized via a mechanism that is not caused by a post-transcriptional regulation and depends neither on P0 nor DCL2.
AGO1-loaded 22-nt vsiRNA promote production of secondary siRNA from Arabidopsis transcripts
Given that 22-nt sRNA can initiate secondary siRNA cascade from AGO-targeted RNA (78,79), we wondered if the abundant TuYV-derived 22-nt vsiRNA could lead to production of secondary siRNA from Arabidopsis transcripts. We therefore performed pairwise differential enrichment analysis of sRNA reads mapping to the Arabidopsis genome between infected and mock samples and found very little change apart from a few loci. Figure 5L shows the 20 most deregulated loci across all IP datasets, with the two bottom-most clusters corresponding to MIR genes and phasiRNA producing loci that we have previously shown to be affected by the ago1-57 mutation (41). The top-most cluster contains nine loci that accumulate sRNA reads only in the presence of TuYVs81. Analysis of the total RNA datasets showed a similar behaviour for five out of nine loci and uncovered an additional gene following this pattern (Supplementary Figure S10A). Additional quantitative analysis of the AGO1 IPs revealed very few loci presenting sRNA enrichment with an adjusted P-value < 0.05 that did not overlap with pre-existing sRNA, namely AT1G18480, AT5G45930, AT5G11000 and AT4G31540 (Supplementary Figure S10B). The fact that these sRNA cohorts are bound to AGO1 and less abundant in AGO1-57 IPs suggests that they are part of a functional RISC, and that their biogenesis is partly affected by the ago1-57 passenger strand retention phenotype.
The two top variable loci in all cases were CHLI1 (AT1G18480) and CHLI2 (AT5G45930). Since TuYVs81 contains the 81 bp reporter fragment of CHLI1 and that 62 bp of that insert present an almost 100% sequence identity with CHLI2, we checked the siRNA distribution along these two loci. As expected, the regions overlapping with the s81 insert contained abundant siRNAs, but an additional population of siRNA was found corresponding to the 3′ half of the two transcripts, that cannot be directly attributed to TuYVs81 processing, and was mostly lost in AGO1-57 IP (Figure 5M). We interpret this observation as a primary source of abundant 22-nt vsiRNAs originating from the viral genome loaded into AGO1 and initiating production of secondary siRNA downstream of the targeting site. Since these primary vsiRNA are made from a largely double stranded RNA precursor, they form perfectly matched duplexes that are retained into AGO1-57 (41), thus disabling production of transitive siRNA in the mutant. Next, we analyzed CHLI1-derived siRNA (SUL 3′) by northern blot (Supplementary Figure S10C). This revealed that TuYVs81-triggered secondary siRNA are 21-nt in length, are greatly diminished in both ago1-57 and ago1-27, are entirely lost in dcl2-1, rdr6-12 and sgs3-14 mutants, and that DCL2 can replace DCL4 for their biogenesis. Their production is specific to TuYV containing the s81 insert, as neither WT TuYV nor the unrelated TuMV-GFP elicited their production. Regarding the cause of siRNA production from the other unrelated loci, we hypothesize the existence of discrete TuYV regions that present micro-homologies with the candidate genes that support production of 22-nt vsiRNA sufficient to guide de novo transitivity. Whether vsiRNA targeting of host transcripts is merely accidental or contributes to the infection and/or the host response will need further investigations.
DISCUSSION
Antiviral RNA silencing mechanism against phloem-restricted TuYV
In this study, we dissected the molecular machinery mediating antiviral silencing of the phloem-restricted polerovirus, TuYV, which revealed several unusual characteristics. First, we find that DCL2, rather than DCL4, is the major supplier of antiviral sRNA, which results in accumulation of large quantities of 22-nt vsiRNA produced in the vasculature, in accordance with the tropism of poleroviruses. This is in stark contrast with most viruses that have been described and form compatible interactions with Arabidopsis, which usually relies on DCL4 to produce the bulk of antiviral siRNA (4,5,11,18,80,81), or for which processing by DCL2 is a consequence of virus manipulation, like in the case of TCV (8,74). For TuYV, we do not observe any perturbation of the activity of DCL2 and DCL4 in infected vasculature, nor does loss of viral P0 affect the pattern of vsiRNA production. Nevertheless, both DCL2 and DCL4 are required to mount a full antiviral response, as loss of both results in a dramatic accumulation of TuYVs81 RNA. Accordingly, we show that both DCL2 and DCL4 are able to re-localize to TuYV VRCs in N. benthamiana infected leaves, showing that both retain their ability to access viral dsRNA, likely via their dsRNA-binding motifs and/or their cofactors. Although the reason for which DCL2 over DCL4 cleaves TuYV dsRNA remains to be investigated, a recent work suggests that in vivo DCL4 enzymatic activity is sensitive to the intrinsic properties of target RNA, and that its processivity might be more tunable than that of other DCL proteins (82). Thus, although both can access the dsRNA, perhaps TuYV dsRNA structural properties are less compatible with efficient processing by DCL4, leaving DCL2 free to cleave it into 22-nt. One might also ponder if dependence on DCL2 might be common in nature and in diverse pathosystems, in other words, if Arabidopsis Col-0 apparent low reliance on DCL2 represents the exception rather than the norm. Interestingly, abundant 22-nt vsiRNA were detected in cotton plants infected with cotton leafroll dwarf virus (CLRDV) (83), in maize leaves containing a novel polerovirus (33) and in wheat infected with barley yellow dwarf virus (BYDV) (84), suggesting that DCL2 is active against Luteoviridae in general.
Another unexpected feature observed in our study is the simultaneous reliance on AGO1 for silencing, with its specific stabilization in infected vasculature. Effective antiviral silencing requires the function of AGO–vsiRNA effector complexes and for most well-described RNA viruses, AGO2 seems to play a major antiviral role, alongside AGO1 (18,62–65,85). The situation is quite different in the case of TuYVs81, for which the ago1-57 mutation was sufficient to induce a two-fold increase in the amount of viral RNA and the addition of the ago2-1 mutation did not lead to further increase. We also found that the vast majority of vsiRNA were loaded into AGO1, further underscoring its importance. Remarkably, none of the defects observed in ago1-57 were observed in ago1-27, which is the most commonly used hypomorphic ago1 allele. It is therefore possible that the contribution of AGO1 to antiviral silencing in many plant-virus interactions has been underestimated. Use of the ago1-57 allele, that uncouples AGO1 activity in siRNA pathways from miRNA pathways and shows little developmental phenotype will no doubt prove valuable to future investigations.
Finally, we observed that some of the TuYVs81-derived vsiRNA can trigger the production of siRNA from host encoded transcripts. The production of these vsiRNA-primed secondary siRNA is made possible by the fact that TuYVs81 RNA is cleaved by DCL2 over DCL4 to generate a cohort of 22-nt vsiRNA, which can initiate secondary siRNA cascades while 21-nt isoforms cannot (78,79). Moreover, TuYVs81 siRNA are preferentially loaded into AGO1 rather than AGO2. This fact also converges towards the production of secondary siRNA, as AGO1 initiates tasiRNA biogenesis (86,87). By contrast, AGO2 loaded with a 22-nt siRNA can direct cleavage of the matching RNA but cannot stimulate production of secondary siRNA (88). To our knowledge, this is the first report of vsiRNA-directed silencing amplification.
Curtailed role of P0 in the context of TuYV infection
Along with the sole reliance on AGO1, we clearly observe increased accumulation of the protein, only in infected vasculature, a feature that did not extend to AGO2 or AGO4. This is counterintuitive, as the experimentally validated action of the VSR P0 is the degradation of unloaded AGO molecules to prevent formation of vsiRNA-RISC. We further show that this increase in vascular AGO1 protein is not a response to the viral P0, nor a consequence of the accumulation of 22-nt vsiRNA. The differences between the results obtained with P0 in heterologous systems and the results reported here obtained during genuine virus infection are stark. This contrast highlights how expression of viral proteins as overexpressed transgenes, while often necessary to study their molecular features in vivo, does not always adequately recapitulate their true activity within the viral lifecycle.
When reconstituting the P0 degradation system in the vasculature, by expressing P0 under a companion cell promoter, we show that P0 leads to AGO1 degradation in the vasculature and that this degradation depends on its F-box motif. Thus, the local environment of the vasculature does not hinder the function of P0, nor is vascular AGO1 insensitive to P0. Interestingly, we observe an increase of leaf chlorosis in SUC-SUL plants that contain WT vascular P0, a phenotype opposite to that of a phloem-restricted VSR that sequesters sRNA duplexes and leads to suppression of SUL siRNA cell-to-cell movement (15). We interpret this observation as a direct consequence of the decrease of AGO1 (and possibly additional AGOs) in the incipient cells, which allows an increased population of mobile siRNA duplexes to exit the companion cells. This leads to increased reach of the silencing signal to additional recipient cells, where these siRNAs are loaded in turn and cause downregulation of CHLI1/2. Our observation is consistent with the ‘consumption’ model proposed by Devers et al. (89) in which cell-autonomous AGO proteins consume mobile sRNAs as they travel cell-to-cell, leading to a modulation of the traveling silencing signal. Our results show that this holds true at the initiation site - provided it contains an active AGO1 population - as exemplified here in the vasculature.
As commented above, however, these results are in contrast with our data from TuYV infections. Given that AGO1 over-accumulates in TuYV-infected vasculature, it is unlikely that the production of the P0 protein from the viral genome is sufficient to cause measurable AGO1 decay. As underlined in the introduction, several observations suggest that strong suppression activity is not necessarily favoured by poleroviruses at large. The observations gathered here support the notion that the activity of TuYV P0 is mostly curtailed during infection. First: vascular AGO1 is stabilized during TuYV infection (although this does not rule out that a fraction of the AGO1 molecules can undergo P0-directed degradation). Second: non-degradable AGO1-57 offers no advantage to the host in the context of the natural mode of infection. Third: P0 and AGO1 have contrasting localizations relative to the VRCs, with AGO1 localizing in the proximity of the VRCs, while P0 signal overlaps with VRCs. The specific localization of P0 at what is most likely the replicating viral genome contrasts with its described localization when overexpressed in absence of the TuYV RNA and suggests that P0’s main activity during infection is restricted to the VRCs rather than spread throughout the cell. We therefore propose that viral P0 is mostly acting at the site of TuYV replication and causes degradation of a relatively small pool of AGO1 at contact sites between viral factories and bona fide replication complexes.
Tolerating a level of antiviral silencing and therefore minimizing perturbation to the host miRNA pathways is expected to increase host fitness during infection and could be a driving mechanism for adaptation, likely to be favoured over time. Tolerance could be achieved by temporally and/or spatially restricting silencing suppression, as we propose for TuYV P0. Furthermore, this would theoretically allow re-routing of the host RNAi pathways for the benefit of the pathogen, as discussed above. In accordance with this notion, several recent examples of viruses dampening their VSRs have emerged. The coat protein of Cucumber mosaic virus (CMV Fny strain) inhibits the translation of the 2b suppressor, which allows a degree of antiviral silencing, promoting self-attenuation and symptom recovery in the infected plants (90). Similarly, βC1 protein of Cotton leaf curl Multan Betasatellite, a virulence factor for begomoviruses that exhibits silencing suppression activity (91), seems to be both an activator and a target of autophagy, effectively driving its own downregulation, leading to milder symptoms (92,93). We expect that more examples of viruses exercising ‘self-control’ via different strategies will emerge in the coming years.
DATA AVAILABILITY
All raw and processed data are available on the zenodo platform : https://doi.org/10.5281/zenodo.5218515. Deep sequencing files have been deposited to the NCBI Gene expression Omnibus (GSE176378).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the following individuals: Herve Vaucheret for the gift of ago7-1, ago10-3 and multiple combinations containing ago2-1 and ago1-27 seeds, Todd Blevins and Christophe Himber at the IBMP for the gift of ago5-1 and dcl2-5/dcl3-1 seeds, Christophe Ritzenthaler at the IBMP for providing the Benthamiana B2-GFP plants, Brian G. Ayre at the University of north Texas for the pCoymv-containing plasmid. We are grateful to all members of the Genschik lab, Benoit Derrien, Thibaut Hacquard, Nicolas Baumberger and Todd Blevins for fruitful discussions and Jerome Mutterer for assistance with microscopy.
Author contributions: Conceptualisation and design: Marion Clavel, Pascal Genschik. Data gathering, formal analysis, validation: Marion Clavel, Esther Lechner, Marco Incarbone, Timothée Vincent, Valerie Cognat, Ekaterina Smirnova, Maxime Lecorbeiller, Véronique Brault. Writing – original draft: Marion Clavel. Writing – review and editing: Marion Clavel, Esther Lechner, Véronique Ziegler-Graff and Pascal Genschik.
Notes
Present address: Marion Clavel and Marco Incarbone, Gregor Mendel Institute (GMI), Austrian Academy of Sciences, Vienna BioCenter (VBC), Vienna, Austria.
Present address: Ekaterina Smirnova, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Integrated Structural Biology Department, Equipe labellisée Ligue Contre le Cancer, Illkirch, France.
Contributor Information
Marion Clavel, Institut de Biologie Moléculaire des Plantes, CNRS, Université de Strasbourg, Strasbourg, France.
Esther Lechner, Institut de Biologie Moléculaire des Plantes, CNRS, Université de Strasbourg, Strasbourg, France.
Marco Incarbone, Institut de Biologie Moléculaire des Plantes, CNRS, Université de Strasbourg, Strasbourg, France.
Timothée Vincent, Institut de Biologie Moléculaire des Plantes, CNRS, Université de Strasbourg, Strasbourg, France.
Valerie Cognat, Institut de Biologie Moléculaire des Plantes, CNRS, Université de Strasbourg, Strasbourg, France.
Ekaterina Smirnova, Institut de Biologie Moléculaire des Plantes, CNRS, Université de Strasbourg, Strasbourg, France.
Maxime Lecorbeiller, Institut de Biologie Moléculaire des Plantes, CNRS, Université de Strasbourg, Strasbourg, France.
Véronique Brault, SVQV, INRA UMR1131, Université de Strasbourg, Colmar, France.
Véronique Ziegler-Graff, Institut de Biologie Moléculaire des Plantes, CNRS, Université de Strasbourg, Strasbourg, France.
Pascal Genschik, Institut de Biologie Moléculaire des Plantes, CNRS, Université de Strasbourg, Strasbourg, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Pascal Genschik acknowledges support from the European Research Council under the European Union's Seventh Framework Programme [FP7/2007–2013]; ERC advanced grant [338904], IdEx Unistra [ANR-10-IDEX-0002]; SFRI-STRAT’US project [ANR 20-SFRI-0012]; EUR IMCBio [IMCBio ANR-17-EURE-0023]; Labex NetRNA [ANR-10-LABX-0036]. Funding for open access charge: ERC advanced Grant [338904].
Conflict of interest statement. None declared.
REFERENCES
- 1. Pumplin N., Voinnet O.. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013; 11:745–760. [DOI] [PubMed] [Google Scholar]
- 2. Yang Z., Li Y.. Dissection of RNAi-based antiviral immunity in plants. Curr. Opin. Virol. 2018; 32:88–99. [DOI] [PubMed] [Google Scholar]
- 3. Blevins T., Rajeswaran R., Shivaprasad P. V., Beknazariants D., Si-Ammour A., Park H.S., Vazquez F., Robertson D., Meins F., Hohn T.et al.. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006; 34:6233–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouché N., Lauressergues D., Gasciolli V., Vaucheret H.. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006; 25:3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deleris A., Gallago-Bartolome J., Bao J., Kasschau K.D., Carrington J.C., Voinnet O.. Hierarchical action and inhibition of plant dicer-like proteins in antiviral defense. Science (80-.). 2006; 313:68–71. [DOI] [PubMed] [Google Scholar]
- 6. Carbonell A., Carrington J.C.. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015; 27:111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poulsen C., Vaucheret H., Brodersen P.. Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures. Plant Cell. 2013; 25:22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qu F., Ye X., Morris T.J.. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Donaire L., Barajas D., Martinez-Garcia B., Martinez-Priego L., Pagan I., Llave C.. Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J. Virol. 2008; 82:5167–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang X.B., Wu Q., Ito T., Cillo F., Li W.X., Chen X., Yu J.L., Ding S.W.. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garcia-Ruiz H., Takeda A., Chapman E.J., Sullivan C.M., Fahlgren N., Brempelis K.J., Carrington J.C.. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell. 2010; 22:481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Incarbone M., Dunoyer P.. RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 2013; 18:382–392. [DOI] [PubMed] [Google Scholar]
- 13. Guo H.S., Ding S.W.. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 2002; 21:398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schott G., Mari-Ordonez A., Himber C., Alioua A., Voinnet O., Dunoyer P.. Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. EMBO J. 2012; 31:2553–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Incarbone M., Zimmermann A., Hammann P., Erhardt M., Michel F., Dunoyer P.. Neutralization of mobile antiviral small RNA through peroxisomal import. Nat. Plants. 2017; 3:17094. [DOI] [PubMed] [Google Scholar]
- 16. Havelda Z., Hornyik C., Crescenzi A., Burgyan J.. In situ characterization of cymbidium ringspot tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J. Virol. 2003; 77:6082–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bayne E.H., Rakitina D.V., Morozov S.Y., Baulcombe D.C.. Cell-to-cell movement of potato potexvirus X is dependent on suppression of RNA silencing. Plant J. 2005; 44:471–482. [DOI] [PubMed] [Google Scholar]
- 18. Wang X.-B., Jovel J., Udomporn P., Wang Y., Wu Q., Li W.-X., Gasciolli V., Vaucheret H., Ding S.-W.. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell. 2011; 23:1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiba S., Hleibieh K., Delbianco A., Klein E., Ratti C., Ziegler-Graff V., Bouzoubaa S., Gilmer D.. The Benyvirus RNA silencing suppressor is essential for long-distance movement, requires both zinc-finger and nols basic residues but not a nucleolar localization for its silencing-suppression activity. Mol. Plant-Microbe Interact. 2013; 26:168–181. [DOI] [PubMed] [Google Scholar]
- 20. Pazhouhandeh M., Dieterle M., Marrocco K., Lechner E., Berry B., Brault V., Hemmer O., Kretsch T., Richards K.E., Genschik P.et al.. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baumberger N., Tsai C.-H., Lie M., Havecker E., Baulcombe D.C.. The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 2007; 17:1609–1614. [DOI] [PubMed] [Google Scholar]
- 22. Bortolamiol D., Pazhouhandeh M., Marrocco K., Genschik P., Ziegler-Graff V.. The polerovirus F Box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 2007; 17:1615–1621. [DOI] [PubMed] [Google Scholar]
- 23. Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., Isono E., Schumacher K., Genschik P.. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Michaeli S., Clavel M., Lechner E., Viotti C., Wu J., Dubois M., Hacquard T., Derrien B., Izquierdo E., Lecorbeiller M.et al.. The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1. Proc. Natl. Acad. Sci. 2019; 116:22872–22883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bohmert K., Camus I., Bellini C., Bouchez D., Caboche M., Benning C., Banning C.. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998; 17:170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trolet A., Baldrich P., Criqui M.-C., Dubois M., Clavel M., Meyers B.C., Genschik P.. Cell cycle-dependent regulation and function of ARGONAUTE1 in plants. Plant Cell. 2019; 31:1734–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fusaro A.F., Correa R.L., Nakasugi K., Jackson C., Kawchuk L., Vaslin M.F.S., Waterhouse P.M.. The Enamovirus P0 protein is a silencing suppressor which inhibits local and systemic RNA silencing through AGO1 degradation. Virology. 2012; 426:178–187. [DOI] [PubMed] [Google Scholar]
- 28. Ziegler-Graff V., Brault V., Mutterer J., Simonis M.-T., Herrbach E., Guilley H., Richards K.E., Jonard G.. The coat protein of beet western yellows luteovirus is essential for systemic infection but the viral gene products P29 and P19 are dispensable for systemic infection and aphid transmission. Mol. Plant-Microbe Interact. 1996; 9:501–510. [Google Scholar]
- 29. Bortolamiol-Bécet D., Monsion B., Chapuis S., Hleibieh K., Scheidecker D., Alioua A., Bogaert F., Revers F., Brault V., Ziegler-Graff V.. Phloem-triggered virus-induced gene silencing using a recombinant polerovirus. Front. Microbiol. 2018; 9:2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mangwende T., Wang M.L., Borth W., Hu J., Moore P.H., Mirkov T.E., Albert H.H.. The P0 gene of sugarcane yellow leaf virus encodes an RNA silencing suppressor with unique activities. Virology. 2009; 384:38–50. [DOI] [PubMed] [Google Scholar]
- 31. Han Y.H., Xiang H.Y., Wang Q., Li Y.Y., Wu W.Q., Han C.G., Li D.W., Yu J.L.. Ring structure amino acids affect the suppressor activity of melon aphid-borne yellows virus P0 protein. Virology. 2010; 406:21–27. [DOI] [PubMed] [Google Scholar]
- 32. Liu Y., Zhai H., Zhao K., Wu B., Wang X.. Two suppressors of RNA silencing encoded by cereal-infecting members of the family Luteoviridae. J. Gen. Virol. 2012; 93:1825–1830. [DOI] [PubMed] [Google Scholar]
- 33. Chen S., Jiang G., Wu J., Liu Y., Qian Y., Zhou X.. Characterization of a novel polerovirus infecting maize in China. Viruses. 2016; 8:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Delfosse V.C., Agrofoglio Y.C., Casse M.F., Kresic I.B., Hopp H.E., Ziegler-Graff V., Distéfano A.J.. The P0 protein encoded by cotton leafroll dwarf virus (CLRDV) inhibits local but not systemic RNA silencing. Virus Res. 2014; 180:70–75. [DOI] [PubMed] [Google Scholar]
- 35. Cascardo R.S., Arantes I.L.G., Silva T.F., Sachetto-Martins G., Vaslin M.F.S., Corrêa R.L.. Function and diversity of P0 proteins among cotton leafroll dwarf virus isolates. Virol. J. 2015; 12:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Almasi R., Miller W.A., Ziegler-Graff V.. Mild and severe cereal yellow dwarf viruses differ in silencing suppressor efficiency of the P0 protein. Virus Res. 2015; 208:199–206. [DOI] [PubMed] [Google Scholar]
- 37. Kozlowska-Makulska A., Guilley H., Szyndel M.S., Beuve M., Lemaire O., Herrbach E., Bouzoubaa S.. P0 proteins of European beet-infecting poleroviruses display variable RNA silencing suppression activity. J. Gen. Virol. 2010; 91:1082–1091. [DOI] [PubMed] [Google Scholar]
- 38. Mayo M.A., Ziegler-Graff V.. Molecular biology of luteoviruses. Adv. Virus Res. 1996; 46:413–460. [DOI] [PubMed] [Google Scholar]
- 39. Pfeffer S., Dunoyer P., Heim F., Richards K.E., Jonard G.. P0 of beet western yellows virus is a suppressor of posttranscriptional gene silencing. J. Virol. 2002; 76:6815–6824. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40. Juszczuk M., Paczkowska E., Sadowy E., Zagórski W., Hulanicka D.M.. Effect of genomic and subgenomic leader sequences of potato leafroll virus on gene expression. FEBS Lett. 2000; 484:33–36. [DOI] [PubMed] [Google Scholar]
- 41. Derrien B., Clavel M., Baumberger N., Iki T., Sarazin A., Hacquard T., Ponce M.R., Ziegler-Graff V., Vaucheret H., Micol J.L.et al.. A suppressor screen for AGO1 degradation by the viral F-box P0 protein uncovers a role for AGO DUF1785 in sRNA duplex unwinding. Plant Cell. 2018; 30:1353–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gregory B.D., O’Malley R.C., Lister R., Urich M.A., Tonti-Filippini J., Chen H., Millar A.H., Ecker J.R.. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008; 14:854–866. [DOI] [PubMed] [Google Scholar]
- 43. Morel J., Godon C., Mourrain P., Feuerbach F., Proux F.. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002; 14:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peragine A. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004; 18:2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Himber C., Dunoyer P., Moissiard G., Ritzenthaler C., Voinnet O.. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003; 22:4523–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klein E., Brault V., Klein D., Weyens G., Lefèbvre M., Ziegler-Graff V., Gilmer D.. Divergence of host range and biological properties between natural isolate and full-length infectious cDNA clone of the Beet mild yellowing virus 2ITB. Mol. Plant Pathol. 2014; 15:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Incarbone M., Clavel M., Monsion B., Kuhn L., Scheer H., Poignavent V., Dunoyer P., Genschik P., Ritzenthaler C.. Immunocapture of dsRNA-bound proteins provides insight into tobacco rattle virus replication complexes and reveals Arabidopsis DRB2 to be a wide-spectrum antiviral effector. Plant Cell. 2021; 10.1093/plcell/koab214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Srivastava A.C., Ganesan S., Ismail I.O., Ayre B.G.. Effective carbon partitioning driven by exotic phloem-specific regulatory elements fused to the Arabidopsis thaliana AtSUC2 sucrose-proton symporter gene. BMC Plant Biol. 2009; 9:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Clough S.J., Bent A.F.. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- 50. Imlau A., Truernit E., Sauer N.. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell. 1999; 11:309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Monsion B., Incarbone M., Hleibieh K., Poignavent V., Ghannam A., Dunoyer P., Daeffler L., Tilsner J., Ritzenthaler C.. Efficient detection of long dsRNA in vitro and in vivo using the dsRNA binding domain from FHV B2 protein. Front. Plant Sci. 2018; 9:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Clark M.F., Adams A.N.. Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 1977; 34:475–483. [DOI] [PubMed] [Google Scholar]
- 53. Herrbach E., Lemaire O., Ziegler-Graff V., Lot H., Rabenstein F., Bouchery Y.. Detection of BMYV and BWYV isolates using monoclonal antibodies and radioactive RNA probes, and relationships among luteoviruses. Ann. Appl. Biol. 1991; 118:127–138. [Google Scholar]
- 54. Reutenauer A., Ziegler-Graff V., Lot H., Scheidecker D., Guilley H., Richards K., Jonard G.. Identification of beet western yellows luteovirus genes implicated in viral replication and particle morphogenesis. Virology. 1993; 195:692–699. [DOI] [PubMed] [Google Scholar]
- 55. Johnson N.R., Yeoh J.M., Coruh C., Axtell M.J.. Improved placement of multi-mapping small RNAs. G3. 2016; 6:2103–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Seguin J., Otten P., Baerlocher L., Farinelli L., Pooggin M.M.. MISIS: a bioinformatics tool to view and analyze maps of small RNAs derived from viruses and genomic loci generating multiple small RNAs. J. Virol. Methods. 2014; 195:120–122. [DOI] [PubMed] [Google Scholar]
- 57. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L.et al.. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Harvey J.J.W., Lewsey M.G., Patel K., Westwood J., Heimstädt S., Carr J.P., Baulcombe D.C.. An antiviral defense role of AGO2 in plants. PLoS One. 2011; 6:e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jaubert M., Bhattacharjee S., Mello A.F.S., Perry K.L., Moffett P.. ARGONAUTE2 mediates RNA-silencing antiviral defenses against potato virus X in Arabidopsis. Plant Physiol. 2011; 156:1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Garcia-Ruiz H., Carbonell A., Hoyer J.S., Fahlgren N., Gilbert K.B., Takeda A., Giampetruzzi A., Garcia Ruiz M.T., McGinn M.G., Lowery N.et al.. Roles and programming of Arabidopsis ARGONAUTE proteins during turnip mosaic virus infection. PLoS Pathog. 2015; 11:e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ma X., Nicole M.C., Meteignier L.V., Hong N., Wang G., Moffett P.. Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. J. Exp. Bot. 2015; 66:919–932. [DOI] [PubMed] [Google Scholar]
- 66. Ratcliff F., Martin-Hernandez A.M., Baulcombe D.C.. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001; 25:237–245. [DOI] [PubMed] [Google Scholar]
- 67. Cheng C.-Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D.. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017; 89:789–804. [DOI] [PubMed] [Google Scholar]
- 68. Li F., Pignatta D., Bendix C., Brunkard J.O., Cohn M.M., Tung J., Sun H., Kumar P., Baker B.. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shivaprasad P. V., Chen H.-M., Patel K., Bond D.M., Santos B.A.C.M., Baulcombe D.C.. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012; 24:859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Deng Y., Wang J., Tung J., Liu D., Zhou Y., He S., Du Y., Baker B., Li F.. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018; 14:e1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sadowy E., Maasen A., Juszczuk M., David C., Zagórski-Ostoja W., Gronenborn B., Hulanicka M.D.. The ORF0 product of Potato leafroll virus is indispensable for virus accumulation. J. Gen. Virol. 2001; 82:1529–1532. [DOI] [PubMed] [Google Scholar]
- 72. Mourrain P., Bé C., Elmayan T., Feuerbach F., Godon C., Morel J.-B., Jouette D., Lacombe A.-M., Nikic S., Picault N.et al.. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance gene required for PTGS, a possible mechanistic link. Cell. 2000; 101:533–542. [DOI] [PubMed] [Google Scholar]
- 73. Svozil J., Gruissem W., Baerenfaller K.. Proteasome targeting of proteins in Arabidopsis leaf mesophyll, epidermal and vascular tissues. Front. Plant Sci. 2015; 6:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Azevedo J., Garcia D., Pontier D., Ohnesorge S., Yu A., Garcia S., Braun L., Bergdoll M., Hakimi M.A., Lagrange T.et al.. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 2010; 24:904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tilsner J., Linnik O., Wright K.M., Bell K., Roberts A.G., Lacomme C., Santa Cruz S., Oparka K.J.. The TGB1 movement protein of potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory. Plant Physiol. 2012; 158:1359–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mallory A.C., Vaucheret H.. ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 2009; 10:521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Matsuda Y., Liang G., Zhu Y., Ma F., Nelson R.S., Ding B.. The Commelina yellow mottle virus promoter drives companion-cell-specific gene expression in multiple organs of transgenic tobacco. Protoplasma. 2002; 220:51–58. [DOI] [PubMed] [Google Scholar]
- 78. Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H.. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C.. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 2010; 17:997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Diaz-Pendon J.A., Li F., Li W.-X., Ding S.-W.. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007; 19:2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Donaire L., Wang Y., Gonzalez-Ibeas D., Mayer K.F., Aranda M.A., Llave C.. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology. 2009; 392:203–214. [DOI] [PubMed] [Google Scholar]
- 82. Montavon T., Kwon Y., Zimmermann A., Hammann P., Vincent T., Cognat V., Bergdoll M., Michel F., Dunoyer P.. Characterization of DCL4 missense alleles provides insights into its ability to process distinct classes of dsRNA substrates. Plant J. 2018; 95:204–218. [DOI] [PubMed] [Google Scholar]
- 83. Silva T.F., Romanel E.A., Andrade R.R., Farinelli L., Østerås M., Deluen C., Corrêa R.L., Schrago C.E., Vaslin M.F.. Profile of small interfering RNAs from cotton plants infected with the polerovirus Cotton leafroll dwarf virus. BMC Mol. Biol. 2011; 12:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shen C., Wei C., Li J., Zhang X., Zhong Q., Li Y., Bai B., Wu Y.. Barley yellow dwarf virus-GAV-derived vsiRNAs are involved in the production of wheat leaf yellowing symptoms by targeting chlorophyll synthase. Virol. J. 2020; 17:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Brosseau C., Moffett P.. Functional and genetic analysis identify a role for Arabidopsis ARGONAUTE5 in antiviral RNA silencing. Plant Cell. 2015; 27:1742–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.-L., Bartel D.P., Crété P.. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004; 16:69–79. [DOI] [PubMed] [Google Scholar]
- 87. Arribas-Hernández L., Marchais A., Poulsen C., Haase B., Hauptmann J., Benes V., Meister G., Brodersen P.. The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell. 2016; 28:tpc.00121.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Carbonell A., Fahlgren N., Garcia-Ruiz H., Gilbert K.B., Montgomery T.A., Nguyen T., Cuperus J.T., Carrington J.C.. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell. 2012; 24:3613–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Devers E.A., Brosnan C.A., Sarazin A., Albertini D., Amsler A.C., Brioudes F., Jullien P.E., Lim P., Schott G., Voinnet O.. Movement and differential consumption of short interfering RNA duplexes underlie mobile RNA interference. Nat. Plants. 2020; 6:789–799. [DOI] [PubMed] [Google Scholar]
- 90. Zhang X.P., Liu D.S., Yan T., Fang X.D., Dong K., Xu J., Wang Y., Yu J.L., Wang X.B.. Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta. PLoS Pathog. 2017; 13:e1006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Amin I., Hussain K., Akbergenov R., Yadav J.S., Qazi J., Mansoor S., Hohn T., Fauquet C.M., Briddon R.W.. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-BetaSatellite complex. Mol. Plant-Microbe Interact. 2011; 24:973–983. [DOI] [PubMed] [Google Scholar]
- 92. Haxim Y., Ismayil A., Jia Q., Wang Y., Zheng X., Chen T., Qian L., Liu N., Wang Y., Han S.et al.. Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife. 2017; 6:e23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ismayil A., Yang M., Haxim Y., Wang Y., Li J., Han L., Wang Y., Zheng X., Wei X., Nagalakshmi U.et al.. Cotton leaf curl multan virus βC1 protein induces autophagy by disrupting the interaction of autophagy-related protein 3 with glyceraldehyde-3-phosphate dehydrogenases. Plant Cell. 2020; 32:1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed data are available on the zenodo platform : https://doi.org/10.5281/zenodo.5218515. Deep sequencing files have been deposited to the NCBI Gene expression Omnibus (GSE176378).