Abstract
Irradiation of cells with short-wavelength ultraviolet light (UVC) changes the program of gene expression, in part within less than 15 min. As one of the immediate-early genes in response to UV, expression of the oncogene c-fos is upregulated. This immediate induction is regulated at the transcriptional level and is transient in character, due to the autocatalyzed shutoff of transcription and the rapid turnover of c-fos mRNA. In an experiment analyzing the kinetics of c-fos mRNA expression in murine fibroblasts irradiated with UVC, we found that, in addition to the initial transient induction, c-fos mRNA accumulated in a second wave starting at 4 to 5 h after irradiation, reaching a maximum at 8 h, and persisting for several more hours. It was accompanied by an increase in Fos protein synthesis. The second peak of c-fos RNA was caused by an UV dose-dependent increase in mRNA half-life from about 10 to 60 min. With similar kinetics, the mRNAs of other UV target genes (i.e., the Kin17 gene, c-jun, IκB, and c-myc) were stabilized (e.g., Kin17 RNA from 80 min to more than 8 h). The delayed response was not due to autocrine cytokine secretion with subsequent autostimulation of the secreting cells or to UV-induced growth factor receptor activation. Cells unable to repair UVC-induced DNA damage responded to lower doses of UVC with an even greater accumulation of c-fos and Kin17 mRNAs than repair-proficient wild-type cells, suggesting that a process in which a repair protein is involved regulates mRNA stability. Although resembling the induction of p53, a DNA damage-dependent increase in p53 was not a necessary intermediate in the stabilization reaction, since cells derived from p53 knockout mice showed the same pattern of c-fos and Kin17 mRNA accumulation as wild-type cells. The data indicate that the signal flow induced by UV radiation addresses not only protein stability (p53) and transcription but also RNA stability, a hitherto-unrecognized level of UV-induced regulation.
Men and mice and derived cells in culture react to irradiation with ultraviolet light A, B, or C (UVA, -B, or -C) by initiating a new program of gene expression that has been named the UV response (reviewed in references 24, 30–32, and 66). Most of the response known so far is transcriptional, as shown by run-on analyses or promoter studies. Within 10 min, the immediate-early genes c-fos, c-jun, and junB are transcriptionally activated, preceded by activation of transcription factors such as SRF-TCF, ATF-2, and CREB (15, 33, 55). NF-κB target genes are activated with a delay of about 2 h (6, 44). Since several of the response genes carry out regulatory functions themselves (e.g., c-fos), secondary response genes are transcribed with considerable delay (6, 9, 58). Transcriptional activation of the immediate response genes is triggered through signal transduction which originates from one of several primary UV target molecules (for a review, see reference 31). UV can induce signal transduction cascades through DNA lesions introduced through irradiation (9, 46, 47, 58), through damaged ribosomal RNA (34), and through the inactivation of one of many oxidation-sensitive protein tyrosine phosphatases (29, 41).
Other levels of gene regulation have, as yet, received less attention. Stabilization of protein is responsible for the UV-DNA lesion-induced increases in p53 and E2F-1 levels (10, 11, 45, 71). Nothing is known about UV influences on pre-RNA splicing and almost nothing about the regulation of mRNA turnover. The turnover of UV-induced c-fos transcripts at its peak level is rapid, and several structural features of c-fos RNA have been made responsible for this rapid degradation (12, 36, 37, 50, 56, 63). One of the features common to rapid-turnover transcripts, sequences in the 3′ untranslated region (UTR) (reviewed in reference 16), are eliminated in Fos-expressing retroviruses (19, 20, 51). These viral fos transcripts are stable, enhancing oncogenicity.
The mammalian UV response resembles in part the bacterial SOS response (reviewed in references 31 and 69). One of the key proteins in the SOS response is the bacterial RecA protein. Eukaryotic proteins with similarities to the bacterial RecA protein have been identified. Most of them are encoded by rad genes which are induced in response to irradiation and involved in the repair of irradiation-induced DNA lesions (7, 62). By screening a murine expression library with anti-RecA antibodies, the nuclear protein Kin17 was identified (2, 3). Kin17 and RecA appear to share an epitope located in the RecA carboxy-terminal region that is involved in the regulation of DNA binding and in the SOS response in Escherichia coli, while differing in most other respects (38, 39, 43, 68). Overexpression of Kin17 seems to be toxic for mammalian cells (40). Bacterially produced Kin17 binds to DNA (64, 65). The physiologic function of Kin17 is unknown as yet.
We report here that UVC treatment of several mammalian cell lines produces two peaks of c-fos RNA abundance, at 30 to 60 min and at about 8 h. While the first peak represents the known transcriptional response, this newly discovered second peak of accumulation is, as we report here, due to UV-induced stabilization of c-fos RNA. Several other short-lived RNAs are also stabilized. The UV-induced accumulation of Kin17 RNA is exclusively caused by RNA stabilization and follows the kinetics and characteristics of the second c-fos peak. Other inducers of c-fos, such as phorbol ester or growth factors, cannot trigger the biphasic c-fos response and cannot reduce RNA turnover. Furthermore, RNA stabilization is induced in cells defective in the Xeroderma pigmentosum group A (XPA) DNA repair gene xpa at a lower UV dose than in wild-type cells. Although this is reminiscent of the induction of p53, UV-induced elevation of p53 protein levels is not an intermediate in the stabilization of c-fos and Kin17 RNA.
MATERIALS AND METHODS
Cell lines and their treatment.
The following cell lines were used: NIH 3T3 cells were obtained from Peter Gruss, Göttingen, Germany; p53−/− mouse embryo fibroblasts, p53+/+ mouse embryo fibroblasts (p53−/− and p53+/+ [22]), and CB17 cells were obtained from Michael Fritsche, Freiburg, Germany; p53−/−mdm2−/− double knockout cells were obtained from David Lane, Dundee, Scotland; primary ΔXPA NIH 3T3 (ΔXPA) and XPA+ NIH 3T3 (wild-type) cells were obtained from Harry van Steeg, Bilthoven, The Netherlands; and c-fos−/− and c-fos+/+ murine embryonic fibroblasts were obtained from Peter Angel, Heidelberg, Germany.
All cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 8% fetal calf serum (FCS), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in humidified 6% CO2.
For UVC irradiation, the culture medium was removed and the culture dishes were washed once with warm phosphate-buffered saline (PBS). The cells were irradiated (without PBS) with a germicidal lamp (254 nm and 30 J/m2, unless otherwise indicated), and the original culture medium was returned to the cells. Growth factors (epidermal growth factor [EGF], 2 ng/ml; basic fibroblast growth factor [bFGF], 10 ng/ml; and interleukin-1α [IL-1α], 2 ng/ml), and 4 μM methyl methansulfonate (MMS), or phorbol ester (12-O-tetradecanoyl-phorbol-13-acetate [TPA], 60 ng/ml) were directly added to the culture medium. Treatment with gamma irradiation was as follows: cells in culture medium were exposed to 5 Gy of radiation in a cobalt-60 gamma radiation source at a dose rate of 5.75 Gy/min. A stock solution of suramin (final concentration, 0.3 mM) was freshly prepared in H2O and diluted into the culture medium 30 min prior to treatment of cells. For the RNA stability assay, cells were UVC irradiated with a dose of 30 J/m2. After a 45-min or 4.5-h incubation period, actinomycin D-mannitol (Sigma) solubilized in water was added to the cultures to a final concentration of 5 μg/ml and incubated further, as indicated.
RNA preparation, Northern blots, and cDNA probes.
Poly(A)+ RNA was isolated according to the method of Rahmsdorf et al. (50), resolved on a 1.4% agarose-formaldehyde gel, and transferred onto a Hybond N+ nylon membrane. Hybridization was performed as previously described (50) using radiolabelled cDNA probes comprising either an ∼1,000-bp PstI fragment of v-fos (19), an ∼1,000-bp fragment of mouse Kin17 obtained by PCR with oligonucleotides 5′-GAGCCCCAAGGCCATCGCCAAT-3′ and 5′-TTCCTGCGTCTCAACTTCCATA-3′ (39), cDNAs of c-jun, IκB, c-myc, urokinase-type plasminogen activator (u-PA), and EF-1, or an ∼1,200-bp PstI fragment of GAPDH (25).
Metabolic labelling and immunoprecipitation.
Cells were grown in DMEM without methionine and cysteine, supplemented with 8% dialyzed FCS and pulse-labelled with 150 μCi of Pro-mix (Amersham) per ml, for 2 h. Cells were washed twice in ice-cold PBS and lysed on ice in ice-cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 8], 125 mM NaCl, 0.5% IGEPAL- CA630, 0.5% sodium desoxycholate, 0.1% sodium lauryl sulfate, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride). The lysate was centrifuged at 150,000 × g and 4°C for 20 min. The lysate was precleared by incubation with 2 μl of a preimmune serum and protein A-agarose (Calbiochem). The resulting supernatant was added to 2 μl of a rabbit polyclonal anti-Fos antiserum precoupled to protein A-agarose. Immunoprecipitation reactions were incubated on a rocking platform at 4°C for 1.5 h. The agarose-antibody-Fos complexes were collected by centrifugation, washed four times in RIPA buffer, resuspended in sample buffer, and resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (8). The gel was fixed in 30% methanol–10% acetic acid for 20 min, washed in water for 20 min, and enhanced in 1 M sodium salicylate–30% methanol for 20 min. The gel was dried and put on X-ray film for exposure.
Run-on analysis.
Isolation of nuclei and in vitro transcription was performed according to the procedure outlined in reference 28. After the transcription reaction was performed, the nuclei were lysed in solution D (4 M guanidinium-isothiocyanate, 0.5% N-laurylsarcosine, 25 mM Na citrate, 143 mM β-mercapthoethanol). A 1/10 volume of 2 M Na acetate, pH 4, was added, and the reaction was vortexed. An equal volume of phenol was added, vortexed, followed by a 1/10 volume of chloroform, agitated, and incubated on ice for 15 min. Phases were separated by centrifugation at 15,000 × g for 20 min, and the upper phase was mixed with an equal volume of isopropanol and centrifuged at 15,000 × g for 15 min. The resulting pellet was solubilized in 300 μl of solution D, and the radioactive RNA was precipitated with an equal volume of isopropanol and washed with 70% ethanol.
Binding of DNA to nitrocellulose was performed as described in reference 28 using a dot blot apparatus. Filters were prehybridized at 65°C for 2 h in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M Na citrate; 450 mM NaCl and 45 mM Na citrate) supplemented with 3× Denhardt solution (0.06% Ficoll 400, 0.06% polyvinylpyrrolidone, 0.06% bovine serum albumin), 6 mM NaH2PO4, 9 mM Na2HPO4, 0.045% Na4P2O7, 1% SDS, 10 mM EDTA, and 50 μg of salmon sperm DNA/ml.
RNA was denatured at 85°C for 5 min, and hybridization was carried out with 8 × 105 cpm of each sample in 1.5 ml of hybridization solution (450 mM NaCl, 45 mM Na citrate, 0.04% Ficoll 400, 0.04% polyvinylpyrrolidone, 0.06% bovine serum albumin, 6 mM NaH2PO4, 9 mM Na2HPO4, 0.045% Na4P2O7, 0.1% SDS, 10 mM EDTA, 10 μg of salmon sperm DNA per ml) at 65°C for 36 h. The filters were washed as described for Northern blots in reference 50 and placed on X-ray films for exposure.
MTT assay.
MTT (dimethylthiazolyldiphenyltetrazolium bromide) is reduced to formazan by mitochondrial respiratory enzymes. This reaction correlates with cell viability and can be quantified spectrophotometrically.
Cells were irradiated with UVC and counted, and 103 cells/well were plated in 96-well plates in 50 μl of DMEM. After 36 h or 5 days, respectively, 50 μl of MTT freshly solubilized in DMEM at a concentration of 2 mg/ml was added, and the cells were returned to the incubator for an additional 4 h. The reaction was stopped by removing the medium and adding 50 μl of 100% isopropanol to the cells. The degree of MTT conversion was measured immediately at 590 nm.
RESULTS
Two waves of UVC-induced c-fos mRNA accumulation in murine fibroblasts.
Several UV-responsive genes, e.g., those coding for metalloproteases or plasminogen activator, are transcriptionally activated with considerable delay (31, 47). Their transcription depends on the presence of AP-1 (Fos-Jun) binding sites, and UV-induced transcription can be prevented by antisense depletion of Fos or Jun (57). Murine c-fos−/− cells do not respond to UVC with transcriptional activation of the genes encoding collagenase 1, stromelysin 1, or stromelysin 3 (59). The late requirement for AP-1 in the UVC-induced transcriptional activation of AP-1-dependent genes contrasts with the rapid and transient kinetics of c-Fos and c-Jun synthesis. Apparently, a second condition must be met for metalloprotease transcription which is not established at the time of the early Fos and Jun peak of synthesis, that is, at times when Fos and Jun of the initial expression period should be depleted by turnover. This second condition must occur later. We therefore wondered which AP-1 molecules would then act on these promoters at late time points.
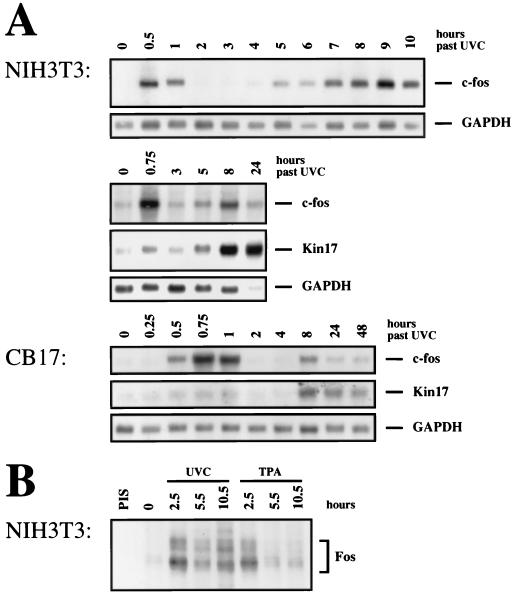
A possible solution was found by measuring the abundance of c-fos RNA over many hours after UV irradiation of cells. In NIH 3T3 cells irradiated with UVC (30 J/m2), we detected two waves of c-fos mRNA accumulation, at 30 to 60 min and at 8 to 10 h (Fig. 1A). Between these times, at 2 and 3 h, c-fos mRNA was at a nondetectable level or at a level comparable to that of noninduced cells. The second peak was less sharp, with accumulation starting at 4 to 5 h after irradiation and persisting for several hours. The synthesis of Fos protein, as shown by pulse-labelling and immunoprecipitation, corresponded to the RNA levels: a first peak at about 2.5 h was followed by a second increase at 10.5 h postirradiation (Fig. 1B). Since Fos protein is more stable than c-fos RNA, Fos abundance did not completely return to control levels between the peaks.
FIG. 1.
Time course of c-fos and Kin17 mRNA induction by UV. (A) NIH 3T3 and CB17 cells were irradiated with UVC (30 J/m2) and harvested at the indicated time points. Poly(A)+ RNA was prepared, and 7.5 μg of each sample was resolved on a 1.4% agarose-formaldehyde gel. The RNA was transferred to a Hybond N+ nylon membrane and sequentially probed with 32P-labelled cDNAs coding for v-fos (hybridizing to c-fos mRNA), Kin17, and GAPDH and placed on X-ray films. (B) NIH 3T3 cells were either irradiated with UVC (30 J/m2) or treated with 60 ng of TPA per ml and harvested 2.5 to 10.5 h after irradiation as indicated. Before harvest, cells were pulse-labelled with 150 μCi of Pro-mix (Amersham) per ml for 2 h and lysed in RIPA buffer. The lysates were precleared by incubation with preimmune serum (PIS) and protein A-agarose. Fos was immunoprecipitated with a polyclonal anti-Fos antibody and protein A-agarose and loaded onto a SDS–10% polyacrylamide gel. The gel was fixed in acetic acid-methanol, enhanced with sodium salicylate, and dried, and X-ray film was exposed for 2 weeks. The bands enclosed within the bracket represent differentially phosphorylated forms of Fos protein.
Several cell lines responded to UV irradiation with two waves of c-fos RNA accumulation, although the height of the second peak was variable: the height of the peak was most prominent in BALB/c 3T3 cells, NIH 3T3 fibroblasts, and JB6 epidermal cells and less pronounced in murine NIH 3T3-like fibroblasts derived from CB17 or SCID mice and in several human cell lines (Fig. 1A and data not shown). A possible explanation for this variability between cell lines will become clear below.
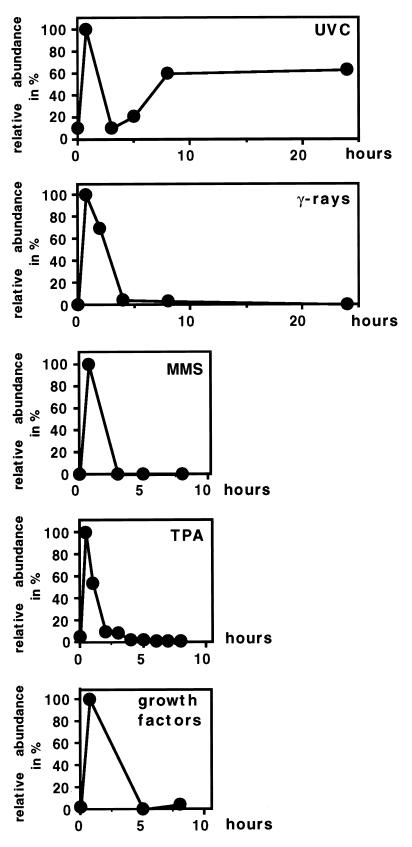
Of all c-fos transcription-inducing agents examined, only UVC irradiation caused two waves of c-fos mRNA accumulation, while all agents triggered the immediate-early transcription in NIH 3T3 cells. Agents examined included a combination of growth factors (EGF, bFGF, and IL-1α), TPA, gamma irradiation, and MMS. Representative Northern blots were scanned with a laser densitometer and plotted (Fig. 2).
FIG. 2.
The biphasic induction of c-fos mRNA is specific for UVC irradiation. NIH 3T3 cells were irradiated with UVC (30 J/m2) or gamma rays (50 Gy) or treated with 4 μM MMS, TPA, (60 ng/ml), or a mixture of growth factors (EGF, [2 ng/ml], bFGF [10 ng/ml], and IL-1α [2 ng/ml]). At the indicated time points, poly(A)+ RNA was prepared, and 5 μg of each sample was resolved on a 1.4% agarose-formaldehyde gel. Hybridizations were carried out as described in the legend to Fig. 1. Relative levels normalized for GAPDH were determined by phosphorimager and densitometry.
Kin17 RNA accumulation was induced with kinetics resembling those during the late phase of c-fos RNA (Fig. 1A). It could only be induced by UVC, not by TPA or growth factors (results not shown), and the relative induction in different cell lines corresponded to the heights of the second c-fos peak, suggesting that the late accumulation of c-fos RNA and that of Kin17 followed the same mechanism.
The second wave of c-fos mRNA abundance and the accumulation of Kin17 mRNA are due to UV-induced transcript stabilization.
The immediate-early induction of c-fos is transcriptional. To investigate whether the 8-h peak of c-fos mRNA accumulation also depended on an elevated transcriptional rate, we performed nuclear run-on analyses at early and late time points after UVC irradiation. Only an early increase in rate was detected in these analyses (Fig. 3). Obviously, transcriptional rate is not significantly increased late after UV irradiation. Kin17-promoter-luciferase constructs failed to show UV-induced transcription above basal levels (data not shown), also suggesting that UV did not increase the transcriptional rate. The observed increases in RNA abundance (Fig. 1A) must therefore be regulated on a posttranscriptional level.
FIG. 3.
Run-on analysis of c-fos transcription after UV irradiation. NIH 3T3 cells were irradiated with UVC (30 J/m2). At the indicated time points, nuclei were isolated and elongation of transcripts was performed in vitro in the presence of 32P-labelled nucleotide triphosphates. A total of 8 × 105 cpm at each time point were hybridized to plasmids encoding c-fos, c-jun, or GAPDH.
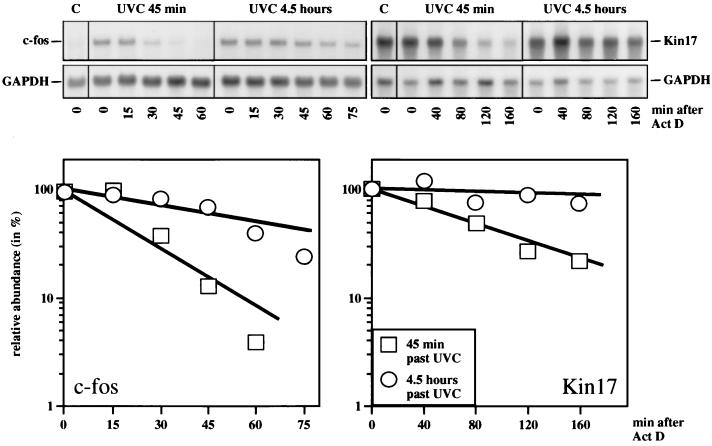
The slow increase in rate and lasting abundance (Fig. 1A) are compatible with a change in the turnover of RNA. The magnitude of this response would thus depend on the basal transcriptional rate, which may indeed differ between cell types. To measure RNA stability, we harvested cells at various times after the addition of the transcriptional inhibitor actinomycin D (Act D). Act D was added at 45 min and at 4.5 h after irradiation with UVC, and RNA was isolated and probed for c-fos and Kin17 RNA by Northern blot analysis. At 45 min after UVC irradiation, c-fos mRNA decayed with a half-life of 15 to 20 min. In contrast, the half-life of c-fos mRNA at 4.5 h was 60 to 70 min (Fig. 4). Since Kin17 RNA abundance was low 45 min after UVC irradiation, long exposure times were required to measure turnover. The half-life was determined to be about 80 min (Fig. 4), similar to that in mock-treated cells (data not shown). Within 4.5 h, UVC irradiation prolonged the half-life of Kin17 mRNA to more than 8 h (extrapolated from Fig. 4). Thus, the relatively stable Kin17 RNA was nevertheless further stabilized by a factor of more than 5.
FIG. 4.
UV induces c-fos and Kin17 mRNA stabilization. NIH 3T3 cells were irradiated with UVC (30 J/m2). At 45 min or 4.5 h after irradiation, Act D (5 μg/ml) was added, and the cells were harvested immediately (0) and every 15 min (for determining the decay of c-fos mRNA) or every 40 min (for determining the decay of Kin17 mRNA), respectively (top). Poly(A)+ RNA was prepared, and 15 μg of each sample was resolved and hybridized as described in the legends to Fig. 1 and 2. The specific hybridization signals for c-fos and Kin17 mRNA were corrected for GAPDH mRNA, which was used as a loading control, and plotted in the graphs (bottom). The relative abundance of c-fos and Kin17 mRNA, respectively, at the time of Act D addition was set at 100%.
The increase in mRNA stability may well account for the second wave of c-fos mRNA and for the induction by UV of Kin17 mRNA accumulation, although a minor contribution by transcriptional activation cannot be completely ruled out because of the low basal transcriptional rate of both genes.
Several mRNAs with relatively short half-lives are stabilized by UV.
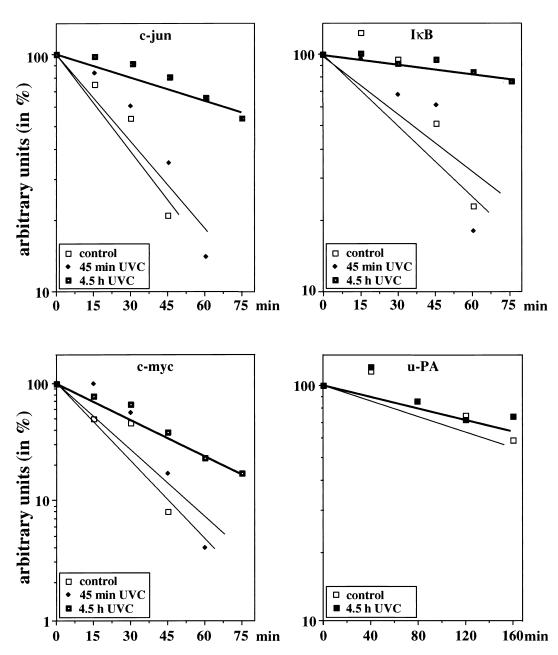
UV-induced stabilization addresses two species of RNA with rather different half-lives. An obvious question is how specific is this stabilization process? We examined a series of other UV-responsive genes and found that they fall into two classes: c-jun, c-myc, and IκB RNAs were also stabilized (Fig. 5). These RNAs have in common a relatively short spontaneous half-life of 15 to 30 min (Fig. 5). u-PA RNA (Fig. 5) and the RNA for the translation elongation factor 1 (EF-1) (data not shown) were not stabilized. We conclude that UV causes an increase in lifetime for several, but not all, mRNAs.
FIG. 5.
Stabilization of several short-lived mRNAs after UV irradiation. NIH 3T3 cells were irradiated with UVC (30 J/m2) or left unirradiated for control and treated as described in the legend to Fig. 4. Hybond N+ nylon membrane was sequentially probed with 32P-labelled cDNAs encoding c-jun, IκB, c-myc, u-PA, and GAPDH, respectively. Specific hybridization signals were quantified by evaluating scanned X-ray film using NIH Image software, corrected for GAPDH, and plotted. The relative abundance of the particular mRNAs at the time of Act D addition was set at 100%.
UVC-induced RNA stabilization is independent of secreted growth factors or of growth factor receptors.
The immediate-early transcriptional activation of c-fos by UVC has been shown to depend, at least in part, on the ligand-independent activation of receptor tyrosine kinases and other receptors (4, 18, 33, 52, 55, 61). Experimental evidence included (i) UV-induced tyrosine autophosphorylation of several receptor tyrosine kinases (41, 55) and receptor clustering (4, 52), (ii) inhibition of the UV response by the growth factor receptor poison suramin, (iii) growth factor-induced downregulation of growth factor receptors and subsequent transient refractoriness either to the same growth factor or to UV (33, 55), and (iv) the inhibition of the UV response upon introduction of dominant negative versions of certain receptor tyrosine kinases (55). We aimed at obtaining similar types of evidence for the UV-induced stabilization.
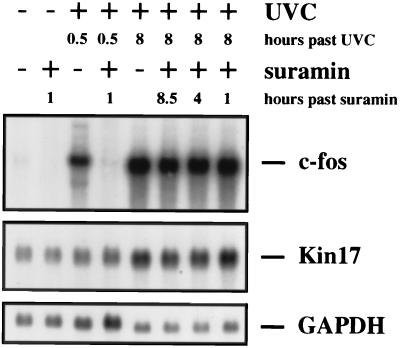
As expected, pretreatment of cells with suramin for 30 min completely inhibited early c-fos mRNA accumulation (Fig. 6). This is in agreement with the previous finding that growth factor receptors are activated by UVC. Suramin could not, however, prevent UV-induced stabilization of c-fos mRNA (the second peak of RNA abundance) or the basal synthesis or the induction of Kin17 RNA at 8 h after UVC irradiation (Fig. 6). Thus, while clearly the early transcriptional induction of c-fos was suramin sensitive, RNA stabilization did not depend on activation of a suramin-sensitive receptor tyrosine kinase. At the same time, the suramin experiment rules out the possibility that the late stabilization of RNA was caused by the induced secretion of a growth factor, since its action would also be poisoned by suramin; this would have been a realistic possibility, since growth factor secretion can indeed be triggered by irradiation of cells with UVC (6, 42, 54). Participation of a secreted growth factor was also made unlikely by an experiment with serum-starved cells and hourly medium changes (see Fig. 8, lower panel). Late c-fos RNA accumulation was barely affected by medium exchange.
FIG. 6.
Pretreatment of cells with suramin does not interfere with c-fos or Kin17 mRNA stabilization. NIH 3T3 cells were grown in DMEM supplemented with 0.5% FCS for 24 h prior to UVC irradiation. The cells were irradiated with UVC (30 J/m2) where indicated or left unirradiated for control. Where marked in the figure, suramin was added to a final concentration of 0.3 mM 30 min prior to UVC irradiation or 4 h (4 h after suramin) or 7 h (1 h past suramin) after irradiation. The cells were harvested at 0.5 h or 8 h after UVC irradiation as shown, poly(A)+ RNA was prepared, and 5 μg of each sample was resolved and hybridized as described in the legends to Fig. 1 and 2.
FIG. 8.
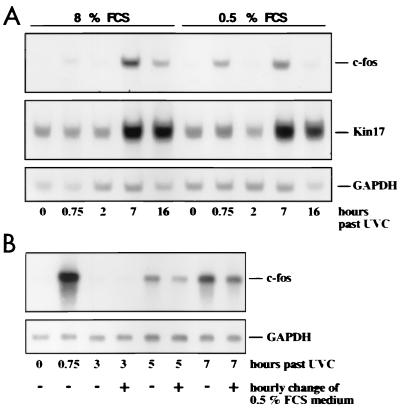
Influence of serum on RNA stabilization by UV. (A) NIH 3T3 cells were either grown in DMEM supplemented with 8% FCS or were serum starved in DMEM plus 0.5% FCS prior to UVC irradiation for 24 h. Poly(A)+ RNA was analyzed as described in the legends to Fig. 1 and 2. (B) NIH 3T3 cells were serum starved in DMEM supplemented with 0.5% FCS for 24 h prior to UVC irradiation. Cells were irradiated with 30 J/m2 UVC and, where marked in the figure key (+), the culture medium was replaced every hour by fresh DMEM plus 0.5% FCS. Poly(A)+ RNA was prepared and analyzed as described in the legends to Fig. 1 and 2.
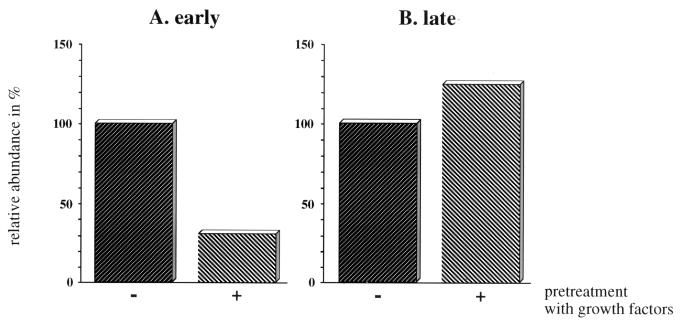
Cells whose relevant receptor tyrosine kinases had been made refractory to their ligands by pretreatment with these ligands no longer show the early c-fos response to UV (Fig. 7A) (33, 55), as UV addresses these same receptors in a ligand-independent fashion. The receptors for EGF, IL-1α, and bFGF have previously been shown to be relevant for the UV response in cultured HeLa cells (55). This is also the case for the early response in NIH 3T3 cells (Fig. 7A). The late response was, however, not altered in cells pretreated with EGF, IL-1α, and bFGF (Fig. 7B), suggesting either that another secreted (and suramin-resistant) cytokine was responsible or that receptor-dependent pathways were not involved in UV-induced RNA stabilization. Only the latter possibility is compatible with the suramin experiment.
FIG. 7.
Differential effect of growth factor pretreatment on early and late c-fos induction. NIH 3T3 cells were grown in DMEM supplemented with 0.5% FCS for 24 h prior to UVC irradiation and irradiated with 30 J/m2 UVC. Where marked in the figure, cells were treated with a cocktail of growth factors (EGF, 2 ng/ml; bFGF, 10 ng/ml; IL-1α, 2 ng/ml) at 30 min prior to irradiation. At 45 min (early) (A) and 8 h (late) (B), irradiated cells were harvested. Poly(A)+ RNA was prepared, and 5 μg of each sample was analyzed as described in the legend to Fig. 1. The relative abundance of c-fos mRNA was determined densitometrically and plotted after correction for GAPDH mRNA. The relative abundance of c-fos mRNA in cells that had been irradiated but not treated with growth factor was set at 100%.
Interestingly, a constant supply of 8% FCS enhanced the late c-fos RNA peak (compared to cells starved at 0.5% FCS [Fig. 8]), perhaps reflecting a higher basal rate of c-fos transcription under serum-enhanced conditions. With Kin17, this effect was marginal (Fig. 8).
Different UVC dose requirements for c-fos and Kin17 mRNA stabilization in cells deficient or proficient in the xpa gene.
UVC affects several primary target molecules relevant for gene regulation. We ruled out receptor tyrosine kinases by the experiments described in the previous paragraph. Several late cellular reactions to UVC irradiation observed earlier appear to depend on UVC-induced DNA damage: e.g., the stabilization of p53 protein (45, 71), the release of cytokines (6), and the late transcriptional activation of several genes encoding proteases and of the metallothionein 2A gene (9, 58). That these reactions require prior UVC-induced DNA damage has been deduced from the finding that human cells deficient in the repair of UVC-induced DNA damage, such as cells derived from patients with XPA or Cockayne's syndrome, activated these functions at much lower UVC doses than did normal wild-type cells. After UV irradiation, cells from normal human individuals or mice remove by nucleotide excision repair UV-induced 6-4 photoproducts and cyclobutane pyrimidine dimers from DNA with half-lives of 90 min and 4 h, respectively (26). In particular, cells from patients with a deficiency in the xpa gene or from mice with a disruption of this gene do not repair at any measurable rate. After the initially identical deposit of UV-induced DNA damage, the density of lesions will progressively divert between normal and repair-deficient cells. To reach the same density of lesions at a given time after irradiation, a higher dose must be applied to normal cells than to repair-deficient cells. Thus, photoproduct-dependent processes will occur at a lower dose in repair-deficient cells than in wild-type cells (9).
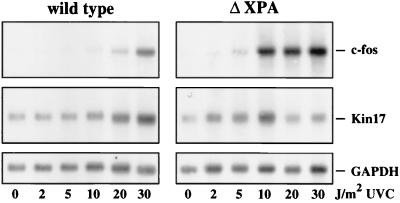
To investigate whether the late wave of c-fos mRNA accumulation and the induction of Kin17 mRNA, like the stabilization of p53 protein (45), depended on the capacity of cells to repair UVC-induced DNA damage, we made use of 3T3 cells (designated ΔXPA) derived from mice in which the murine homologue of the human xpa gene had been knocked out by recombinational inactivation (21), and 3T3 cells from their XPA+/+ siblings. We first confirmed that UVC induced the second wave of c-fos mRNA induction and Kin17 mRNA induction in ΔXPA cells and then tested the dose dependence for RNA stabilization (Fig. 9). Doses above 20 to 30 J/m2 were needed for stabilization of both c-fos and Kin17 RNA in repair-proficient wild-type cells. Only 5 to 10 J/m2 were required for half-maximal induction in the repair-deficient ΔXPA cells (Fig. 9). At UVC doses above 10 J/m2, Kin17 mRNA disappeared in preference over c-fos RNA. We have no definitive explanation for this difference. Possibly the primary transcript of Kin17 is much larger than that of c-fos, and thus higher UV doses may preferentially affect basal Kin17 transcription by a DNA lesion-dependent block of elongation. From the dose-response curves shown in Fig. 9, we can conclude either that UVC-induced DNA lesions are intermediates in the induced stabilization of c-fos and Kin17 RNAs or that components of the XPA complex affect RNA stability.
FIG. 9.
UV dose dependence of RNA stabilization in cells from XPA knockout mice and from wild-type cells. Fibroblast cell lines from XPA knockout mice (ΔXPA) and from the respective parental mice (wild type) were irradiated with 2, 5, 10, 20, or 30 J/m2 UVC or left unirradiated for control. At 16 h postirradiation, poly(A)+ RNA was prepared and processed as described in the legends to Fig. 1 through 3.
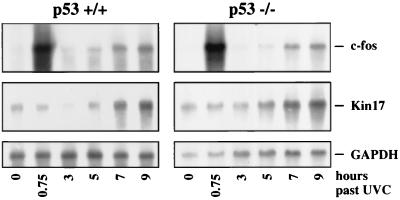
As the UV dose dependence of RNA stabilization resembled that of p53 protein stabilization, we considered whether p53 could mediate stabilization of RNA (see Discussion). However, as shown in Fig. 10, the stabilization of both c-fos and Kin17 RNA was identical in isogenic embryonic mouse fibroblasts differing only in p53 (p53+/+ versus p53−/− [22]) (Fig. 10), excluding a role for p53 in UVC-induced c-fos and Kin17 RNA stabilization. Also, cells doubly negative for p53 and Mdm2 responded like wild-type cells, with two peaks of c-fos RNA (results not shown), indicating that Mdm2 also has no role in the stabilization process.
FIG. 10.
Equal induction of RNA stability by UV both in p53+/+ and p53−/− cells. Fibroblast cell lines from p53 knockout mice (p53−/−) and from the wild-type mice (p53+/+) were irradiated with UVC (30 J/m2). The cells were harvested at the indicated time points, and poly(A)+ RNA was prepared and analyzed as described in the legends to Fig. 1 through 3.
Delayed death of c-fos−/− cells.
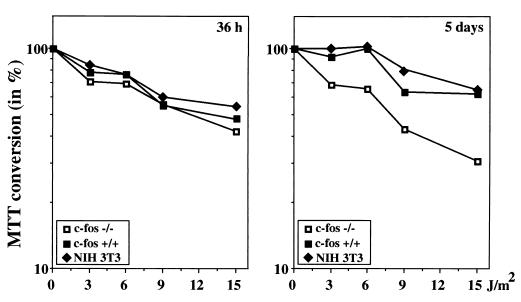
Fos is the subunit of a transcription factor, AP-1, which is decisive for a large number of genes. In its absence, the UV response of several genes is hampered (59). The induction of c-fos transcription and of RNA stabilization could establish a survival advantage. Along these lines of thought, c-fos−/− cells have been examined for survival after UV irradiation. Indeed, c-fos−/− cells were found to be less resistant to UV (measured by lactic dehydrogenase release), an effect not accounted for by a difference of DNA repair (59). We have confirmed this result using a different survival assay, the MTT conversion assay, which measures viability instantaneously (Fig. 11). Interestingly, viability of c-fos−/− cells was decreased with an enormous delay: at 36 h, MTT counts were still normal, while decreasing progressively thereafter.
FIG. 11.
c-fos is required for cell survival. c-fos+/+, c-fos−/−, and NIH 3T3 cells were irradiated with the indicated UVC dose, counted, and plated in quadruplicate at a density of 103 cells/well in 96-well plates. After 36 h or 5 days, respectively, MTT was added at a final concentration of 1 mg/ml, and the cells were returned to the incubator for additional 4 h. The medium was replaced by isopropanol, and MTT conversion was immediately measured at 590 nm. The mean values of three independent experiments were calculated and plotted as percentages of nonirradiated cells.
DISCUSSION
This paper describes a novel feature of c-fos regulation, UV-induced stabilization of its RNA—surprising in view of the fact that many laboratories (including one of ours) have studied c-fos for years. UV-induced stabilization of RNA appears to be selective: the RNAs for c-fos, c-jun, c-myc, IκBα, and Kin17 (this paper) are stabilized, whereas actin, GAPDH (see Fig. 4), u-PA, and EF-1 RNA turnover was not changed (data not shown and this paper). Stabilization of RNA can, of course, only occur with RNA that is intrinsically unstable. Nevertheless, both Kin17 and u-PA show moderately long half-lives but are discriminated by the stabilization process. The magnitude and time course of stabilization depend on the rate of transcription and therefore vary with different genes. The spontaneous transcription of c-fos and c-jun is a function of growth conditions and likely differs between cell types. A low transcriptional rate in several human cell lines may be the reason that UVC-induced c-fos RNA stabilization in these cells was barely detectable.
UVC-induced RNA stabilization varied with the capacity of cells to repair photoproducts in nuclear DNA. In repair (XPA)-deficient fibroblasts, a two- to three-times-lower UV dose was required for RNA stabilization than in repair-proficient cells. This observation supports our hypothesis that the density of DNA lesions (and of protein complexes assembled at these lesions) is an intermediate in induced RNA stabilization, as lesion density remains constant in XPA cells, while wild-type cells remove lesions with a half-life of 90 min to 4 h (26). To retain the same lesion density in repair-proficient cells, the initial dose applied must be higher in repair-proficient cells. Alternative explanations for the different dose responses in XPA and wild-type cells need to be considered (see below).
Following the DNA damage hypothesis, it is obvious that additional steps are required between the introduction of DNA lesions and the activation or inactivation of critical proteins affecting RNA stability. A hypothetical scheme could involve a lesion-dependent arrest of transcription which is converted in an unknown fashion into a signaling pathway to the RNA-degrading exosome (67). Indeed, transcriptional induction of genes and stabilization of p53 correlate best with DNA lesions in transcribed regions of the genome (9, 71). To identify components of a signal transduction chain involved in the UV-dependent stabilization of RNA, we tested numerous inhibitors. None, however, prevented the stabilization: vanadate, SB203580 (Jun N-terminal protein kinases [JNKs] and p38), PD98059 and U0126 (MEKs), Ly294002 (phosphatidylinositol 3-kinases, ATM, and DNA-dependent protein kinase), wortmannin, rapamycin (S6 kinase), roscovitin (CDKs), okadaic acid (protein phosphatase 2A), H7 (protein kinase C), trichostatin (histone deacetylase), and several inhibitors of protein synthesis. Tentatively, this also excludes the involvement of known protein kinases recognizing DNA strand breaks.
RNAs regulated by changes in turnover carry specific cis-acting elements. One of these elements, consisting of several repeats of the sequence AUUUA, is found in the 3′ UTR (16, 36, 53). Indeed, c-fos, c-jun, c-myc, and Kin17 RNAs carry AUUUA repeats in their 3′ UTRs. These AU-rich sequences bind several specific proteins, one of which clearly increases RNA stability (49). Also, cis-acting elements in other sites of the mature RNA likely bind specific proteins, affecting RNA stability. We hypothesize that a signal transduction cascade originating from a DNA photoproduct leads to activation of RNA binding proteins involved in RNA stability. Interestingly, JNK can prolong the lifetime of IL-2 RNA (17). If JNK were activated by DNA damage (which is still debated [1]), UV-induced JNK activity would be a good candidate as a member of the signal transduction cascade. Inhibitor studies, however, speak against a role by JNK.
Could cells defective in XPA lack repair of some other cellular component damaged by UV? We are not aware of another XPA-dependent repair process but cannot exclude its existence. For instance, double-stranded RNA, e.g., in ribosomes, could be subjected to XPA-dependent lesion repair. Damage to ribosomal RNA has been reported to cause signal transduction to JNK (34). One could also argue that the increased response in XPA cells reflects release from an RNA turnover process, suggesting that XPA and partner proteins were actively involved in mediating high rates of RNA turnover. This involvement would need to occur only after UV irradiation. UV irradiation could then be absorbed by a relevant non-DNA and/or non-RNA target. But we find difficult to explain, in this hypothetical scheme, the dose difference of UV-induced RNA stabilization between XPA and wild-type cells.
Could a transcriptional factor be involved in RNA stabilization? It is puzzling that the activation of several transcription factors not only affects transcription but also the turnover of their target transcripts: e.g., the estrogen receptor (13), glucocorticoid receptor (48), and p53 (27). To our knowledge, the mechanisms of how transcription factors could achieve RNA stabilization are not clear. One could argue that, at the same time, they transcribe genes whose products participate in RNA metabolism.
p53 is an interesting candidate for the stabilization of c-fos and Kin17 RNA as well as other stress-induced RNAs (35), since p53 is rescued from rapid proteasome-dependent degradation by a DNA damage-dependent mechanism (5, 14). Furthermore, p53 has been reported to induce RNA stability of its transcriptional target gene, p21WAF-1 (27). Moreover, c-fos (but not Kin17) is also a target gene of the transcription factor p53 (23 and our unpublished data). However, several arguments speak against a participation of p53 in UV-dependent c-fos and Kin17 RNA stabilization: (i) p53 levels in cells are increased by both UV and X-ray treatment, whereas c-fos and Kin17 RNA stability is induced only by UV. (ii) p21 RNA turnover regulation by p53 is under tyrosine kinase-phosphatase control in the absence of p53, as shown by the effect of vanadate (27). Vanadate had no effect on UVC-induced c-fos and Kin17 RNA stabilization (as discussed above). (iii) UV-induced RNA stabilization occurs in both p53+/+ and p53−/− cells.
Although stabilization of c-fos RNA does not involve p53, p53 does interact in an interesting way with the transcription factor AP-1, which contains Fos and Jun as subunits. As shown by the comparison of Jun+/+ and Jun−/− cells, p53 synthesis is repressed by a Jun-containing member of the AP-1 family (60). p53, in turn, induces immediate-early c-fos transcription by binding to an intronic p53 binding site (23), detected by activating a temperature-sensitive p53 mutant protein.
A simple interpretation of the UV effect on RNA stability could be that the overall inhibition of transcription and translation by UVC preferentially affected the availability of an RNA-degrading enzyme. In fact, the inhibition of translation by cycloheximide indeed stabilizes c-fos RNA (50), and UVC treatment of cells leads to the disappearance of specific proteins, e.g., the oncoprotein Mdm2 (10, 70). The stabilization of c-fos and Kin17 RNA occurs under irradiation conditions which reduce overall transcription, due to the arrest of elongation at sites of DNA lesions. One could speculate that the physiological significance of RNA stabilization represents a rescue mechanism for important survival functions which are (for example) AP-1 and NF-κB (6) dependent. This is compatible with the observed survival-promoting action of Fos (59 and this paper).
ACKNOWLEDGMENTS
We are grateful to Helmut Ponta and Martin Göttlicher for text and figure criticism.
This work was supported by the European Community (grant F14P-CT96-0052) and by the Association pour la Recherche sur le Cancer (contract no. 6060) and Electricité de France (contract no. 8702). P. Kannouche received a fellowship from the Institut National de la Science et la Technologie Nucléaire (INSTN) du CEA and from EDF.
REFERENCES
- 1.Adler V, Fuchs S Y, Kim J, Kraft A, King M P, Pelling J, Ronai Z. Jun-NH2-terminal kinase activation mediated by UV-induced DNA lesions in melanoma and fibroblast cells. Cell Growth Differ. 1995;6:1437–1446. [PubMed] [Google Scholar]
- 2.Angulo J F, Moreau P L, Maunoury R, Laporte J, Hill A M, Bertolotti R, Devoret R. KIN, a mammalian nuclear protein immunologically related to E. coli RecA protein. Mutat Res. 1989;217:123–134. doi: 10.1016/0921-8777(89)90064-5. [DOI] [PubMed] [Google Scholar]
- 3.Angulo J F, Rouer E, Mazin A, Mattei M G, Tissier A, Horellou P, Benarous R, Devoret R. Identification and expression of the cDNA of KIN17, a zinc-finger gene located on mouse chromosome 2, encoding a new DNA-binding protein. Nucleic Acids Res. 1991;19:5117–5123. doi: 10.1093/nar/19.19.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aragane Y, Kulms D, Metze D, Wilkes G, Pöppelmann B, Luger T A, Schwarz T. Ultraviolet light induces apoptosis via direct activation of CD95 (Fas/Apo-1) independently of its ligand CD95L. J Cell Biol. 1998;140:171–182. doi: 10.1083/jcb.140.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates S, Vousden K H. Mechanisms of p53-mediated apoptosis. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender K, Göttlicher M, Whiteside S, Rahmsdorf H-J, Herrlich P. Sequential DNA damage-independent and -dependent activation of NF-kappaB by UV. EMBO J. 1998;17:5170–5181. doi: 10.1093/emboj/17.17.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson F E, Stasiak A, West S C. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner C, Knebel A, Radler-Pohl A, Sachsenmaier C, Herrlich P, Rahmsdorf H-J. DNA damaging agents and growth factors induce changes in the program of expressed gene products through common routes. Environ Mol Mutagen. 1994;24:3–10. doi: 10.1002/em.2850240103. [DOI] [PubMed] [Google Scholar]
- 9.Blattner C, Bender K, Herrlich P, Rahmsdorf H-J. Photoproducts in transcriptionally active DNA induce signal transduction to the delayed U.V.-responsive genes for collagenase and metallothionein. Oncogene. 1998;16:2827–2834. doi: 10.1038/sj.onc.1201827. [DOI] [PubMed] [Google Scholar]
- 10.Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol. 1999;19:3704–3713. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blattner C, Tobiasch E, Litfin M, Rahmsdorf H-J, Herrlich P. DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene. 1999;18:1723–1732. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- 12.Brawerman G. mRNA degradation in eukaryotic cells: an overview. In: Belasco J, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press, Inc.; 1993. pp. 150–159. [Google Scholar]
- 13.Brock M L, Shapiro D J. Estrogen stabilizes vitellogenin mRNA against cytoplasmic degradation. Cell. 1983;34:207–214. doi: 10.1016/0092-8674(83)90151-4. [DOI] [PubMed] [Google Scholar]
- 14.Brown J P, Pagano M. Mechanism of p53 degradation. Biochim Biophys Acta. 1997;1332:O1–O6. doi: 10.1016/s0304-419x(96)00048-0. [DOI] [PubMed] [Google Scholar]
- 15.Büscher M, Rahmsdorf H-J, Litfin M, Karin M, Herrlich P. Activation of the c-fos gene by UV and phorbol ester: different signal transduction pathways converge to the same enhancer element. Oncogene. 1988;3:301–311. [PubMed] [Google Scholar]
- 16.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 17.Chen C Y, Del Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 18.Coffer P J, Burgering B M, Peppelenbosch M P, Bos J L, Kruijer W. UV activation of RTK activity. Oncogene. 1995;11:561–569. [PubMed] [Google Scholar]
- 19.Curran T, Peters G, Van Beveren C, Teich N M, Verma I M. FBJ murine osteosarcoma virus: identification and molecular cloning of biologically active proviral DNA. J Virol. 1982;44:674–682. doi: 10.1128/jvi.44.2.674-682.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curran T, MacConnell W P, van Straaten F, Verma I M. Structure of the FBJ murine osteosarcoma virus genome: molecular cloning of its associated helper virus and the cellular homolog of the v-fos gene from mouse and human cells. Mol Cell Biol. 1983;3:914–921. doi: 10.1128/mcb.3.5.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Vries A, van Oostrom C T, Hofhuis F M, Dortant P M, Berg R J, de Gruijl F R, Wester P W, van Kreijl C F, Capel P J, van Steeg H, Verbeek S J. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature. 1995;377:169–173. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- 22.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 23.Elkeles A, Juven-Gershon T, Israeli D, Wilder S, Zalcenstein A, Oren M. The c-fos proto-oncogene is a target for transactivation by the p53 tumor suppressor. Mol Cell Biol. 1999;19:2594–2600. doi: 10.1128/mcb.19.4.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fornace A J., Jr DNA-damage inducible genes in mammalian cells. Annu Rev Genet. 1992;26:505–524. [Google Scholar]
- 25.Fort P, Marty L, Piecharzyk M, el Sabrouty S, Dani C, Jeanteur P, Blanchard J M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 27.Gorospe M, Wang X, Holbrook H J. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol Cell Biol. 1998;18:1400–1407. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenberg M E, Bender T P. Preparation and analysis of RNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley; 1987. [Google Scholar]
- 29.Herrlich P, Böhmer F-D. Redox regulation of signal transduction in mammalian cells. Biochem Pharmacol. 2000;59:35–41. doi: 10.1016/s0006-2952(99)00298-1. [DOI] [PubMed] [Google Scholar]
- 30.Herrlich P, Rahmsdorf H-J. Transcriptional and post-transcriptional responses to DNA-damaging agents. Curr Opin Cell Biol. 1994;6:425–431. doi: 10.1016/0955-0674(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 31.Herrlich P, Blattner C, Knebel A, Bender K, Rahmsdorf H-J. Nuclear and non-nuclear targets of genotoxic agents in the induction of gene expression. Shared principles in yeast, rodents, man and plants. Biol Chem. 1997;378:1217–1229. doi: 10.1515/bchm.1997.378.11.1217. [DOI] [PubMed] [Google Scholar]
- 32.Herrlich P, Rahmsdorf H-J, Bender K, Blattner C, Knebel A. Signal transduction induced by adverse agents: activation by inhibition. The UV response 1997. In: Puga A, Wallace K B, editors. Molecular biology of the toxic response. Philadelphia, Pa: Taylor & Francis; 1998. pp. 479–492. [Google Scholar]
- 33.Iordanov M, Bender K, Ade T, Schmid W, Sachsenmaier C, Engel K, Gaestel M, Rahmsdorf H-J, Herrlich P. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 1997;16:1009–1022. doi: 10.1093/emboj/16.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iordanov M S, Pribnow D, Magun J L, Dinh T H, Pearson J A, Magun B E. Ultraviolet radiation triggers the ribotoxic stress response in mammalian cells. J Biol Chem. 1998;273:15794–15803. doi: 10.1074/jbc.273.25.15794. [DOI] [PubMed] [Google Scholar]
- 35.Jackman J, Alamo I, Jr, Fornace A J., Jr Genotoxic stress confers preferential and coordinate messenger RNA stability on the five gadd genes. Cancer Res. 1994;54:5656–5662. [PubMed] [Google Scholar]
- 36.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 37.Kabnick K S, Housman D E. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988;8:3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kannouche P, Pinon-Lataillade G, Mauffrey P, Faucher C, Biard D S, Angulo J F. Overexpression of Kin17 protein forms intranuclear foci in mammalian cells. Biochimie. 1997;79:599–606. doi: 10.1016/s0300-9084(97)82009-3. [DOI] [PubMed] [Google Scholar]
- 39.Kannouche P, Pinon-Lataillade G, Tissier A, Chevalier-Lagente O, Sarasin A, Mezzina M, Angulo J F. The nuclear concentration of Kin17, a mouse protein that binds to curved DNA, increases during cell proliferation and after UV irradiation. Carcinogenesis. 1998;19:781–789. doi: 10.1093/carcin/19.5.781. [DOI] [PubMed] [Google Scholar]
- 40.Kannouche P, Angulo J F. Overexpression of Kin17 protein disrupts nuclear morphology and inhibits the growth of mammalian cells. J Cell Sci. 1999;112:3215–3224. doi: 10.1242/jcs.112.19.3215. [DOI] [PubMed] [Google Scholar]
- 41.Knebel A, Rahmsdorf H-J, Ullrich A, Herrlich P. Dephosphorylation of receptor tyrosine kinases as target of regulation by radiation, oxidants or alkylating agents. EMBO J. 1996;15:5314–5325. [PMC free article] [PubMed] [Google Scholar]
- 42.Krämer M, Sachsenmaier C, Herrlich P, Rahmsdorf H-J. UV irradiation-induced interleukin-1 and basic fibroblast growth factor synthesis and release mediate part of the UV response. J Biol Chem. 1993;268:6734–6741. [PubMed] [Google Scholar]
- 43.Kurumizaka H, Aihara H, Ikawa S, Kashima T, Bazemore L R, Kawasaki K, Sarai A, Radding C M, Shibata T. A possible role of the C-terminal domain of the RecA protein. A gateway model for double-stranded DNA binding. J Biol Chem. 1996;271:33515–33524. doi: 10.1074/jbc.271.52.33515. [DOI] [PubMed] [Google Scholar]
- 44.Li N, Karin M. Ionizing radiation and short wavelength UV activate NF-kappaB through two distinct mechanisms. Proc Natl Acad Sci USA. 1998;95:13012–13017. doi: 10.1073/pnas.95.22.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maltzman W, Czyzyk L. UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miskin R, Reich E. Plasminogen activator: induction of synthesis by DNA damage. Cell. 1980;19:217–224. doi: 10.1016/0092-8674(80)90403-1. [DOI] [PubMed] [Google Scholar]
- 47.Miskin R, Benishai R. Induction of plasminogen activator by UV light in normal and xeroderma pigmentosum fibroblasts. Proc Natl Acad Sci USA. 1981;78:6236–6240. doi: 10.1073/pnas.78.10.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paek I, Axel R. Glucocorticoids enhance stability of human growth hormone mRNA. Mol Cell Biol. 1987;7:1496–1507. doi: 10.1128/mcb.7.4.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng S, Chen C, Xu N H, Shyu A B. RNA stabilization by the AU-rich element binding protein HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahmsdorf H-J, Schönthal A, Angel P, Litfin M, Rüther U, Herrlich P. Posttranscriptional regulation of c-fos mRNA expression. Nucleic Acids Res. 1987;15:1643–1659. doi: 10.1093/nar/15.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raymond V, Atwater J A, Verma I M. Removal of a messenger-RNA destabilizing element correlates with the increased oncogenicity of proto-oncogene fos. Oncogene Res. 1989;5:1–12. [PubMed] [Google Scholar]
- 52.Rosette C, Karin M. Ultraviolet light and osmotic stress: activation of the JNK cascade through multiple growth factor and cytokine receptors. Science. 1996;274:1194–1197. doi: 10.1126/science.274.5290.1194. [DOI] [PubMed] [Google Scholar]
- 53.Ross J. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 1996;12:171–175. doi: 10.1016/0168-9525(96)10016-0. [DOI] [PubMed] [Google Scholar]
- 54.Rotem N, Axelrod J H, Miskin R. Induction of urokinase-type plasminogen activator by UV light in human fetal fibroblasts is mediated through a UV-induced secreted protein. Mol Cell Biol. 1987;7:622–631. doi: 10.1128/mcb.7.2.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sachsenmaier C, Radler-Pohl A, Zinck R, Nordheim A, Herrlich P, Rahmsdorf H-J. Involvement of growth factor receptors in the mammalian UVC response. Cell. 1994;78:963–972. doi: 10.1016/0092-8674(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 56.Schiavi S C, Wellington C L, Shyu A B, Chen C, Greenberg M E, Belasco J B. Multiple elements in the c-fos protein-coding region facilitate messenger-RNA deadenylation and decay by a mechanism coupled to translation. J Biol Chem. 1994;269:3441–3448. [PubMed] [Google Scholar]
- 57.Schönthal A, Herrlich P, Rahmsdorf H-J, Ponta H. Requirement for fos gene expression in the transcriptional activation of collagenase by other oncogenes and phorbol esters. Cell. 1988;54:325–334. doi: 10.1016/0092-8674(88)90195-x. [DOI] [PubMed] [Google Scholar]
- 58.Schorpp M, Mallick U, Rahmsdorf H-J, Herrlich P. UV-induced extracellular factor from human fibroblasts communicates the UV response to non-irradiated cells. Cell. 1984;37:861–868. doi: 10.1016/0092-8674(84)90421-5. [DOI] [PubMed] [Google Scholar]
- 59.Schreiber M, Baumann B, Cotten M, Angel P, Wagner E F. Fos is an essential component of the mammalian UV response. EMBO J. 1995;14:5338–5349. doi: 10.1002/j.1460-2075.1995.tb00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner E F. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheikh M S, Antinore M J, Huang Y, Fornace A J., Jr Ultraviolet-irradiation-induced apoptosis is mediated via ligand independent activation of tumor necrosis factor receptor 1. Oncogene. 1998;17:2555–2563. doi: 10.1038/sj.onc.1202292. [DOI] [PubMed] [Google Scholar]
- 62.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 63.Shyu A B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid messenger-RNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 64.Timchenko T, Bailone A, Devoret R. Btcd, a mouse protein that binds to curved DNA, can substitute in Escherichia coli for H-NS, a bacterial nucleoid protein. EMBO J. 1996;15:3986–3992. [PMC free article] [PubMed] [Google Scholar]
- 65.Tissier A, Kannouche P, Mauffrey P, Allemand I, Frelat G, Devoret R, Angulo J F. Molecular cloning and characterization of the mouse Kin17 gene coding for a Zn-finger protein that preferentially recognizes bent DNA. Genomics. 1996;38:238–242. doi: 10.1006/geno.1996.0623. [DOI] [PubMed] [Google Scholar]
- 66.Tyrrell R M. Oxidant, antioxidant status and photocarcinogenesis: the role of gene activation. Photochem Photobiol. 1996;63:380–383. doi: 10.1111/j.1751-1097.1996.tb03049.x. [DOI] [PubMed] [Google Scholar]
- 67.Van Hoof A, Parker R. The exosome: a proteasome for RNA. Cell. 1999;99:347–350. doi: 10.1016/s0092-8674(00)81520-2. [DOI] [PubMed] [Google Scholar]
- 68.Voloshin O N, Wang L J, Camerini-Otero R D. Homologous DNA pairing promoted by a 20-amino-acid peptide derived from RecA. Science. 1996;272:868–872. doi: 10.1126/science.272.5263.868. [DOI] [PubMed] [Google Scholar]
- 69.Walker G C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- 70.Wu L, Levine A J. Differential regulation of the p21/WAF-1 and mdm2 genes after high-dose UV irradiation: p53-dependent and p53-independent regulation of the mdm2 gene. Mol Med. 1997;3:441–451. [PMC free article] [PubMed] [Google Scholar]
- 71.Yamaizumi M, Sugano T. UV-induced nuclear accumulation of p53 is evoked through DNA damage in actively transcribed genes independent of the cell cycle. Oncogene. 1994;9:2775–2784. [PubMed] [Google Scholar]