ABSTRACT
Aims/Introduction
It was reported previously that N4bp2l1 expression increases in 3T3‐L1 cells in a differentiation‐dependent manner and N4bp2l1 knockdown suppresses adipocyte differentiation. However, the physiological function of N4BP2L1 in adipocytes remains unknown. This study aimed to elucidate the physiological mechanism of N4bp2l1 expression and the role of N4BP2L1 in the physiological function of adipocytes.
Materials and Methods
Analysis of gene expression levels of N4bp2l1 in adipose tissue during feeding in mice was conducted. Identification of transcription factors that regulate N4bp2l1 expression was conducted using a reporter assay. Investigation of N4BP2L1‐interacting proteins was carried out using immunoprecipitation. A GLUT4 translocation assay and a glucose uptake assay in 3T3‐L1 adipocytes were performed using N4bp2l1 overexpression and knockdown adenovirus.
Results
The results indicated that N4bp2l1 is a novel FoxO1 target gene and its expression is controlled by the insulin‐mediated regulation of FoxO1. N4BP2L1 interacts with dynactin, which binds to the microtubule motor dynein, indicating that N4BP2L1 is involved in GLUT4 trafficking and glucose uptake in 3T3‐L1 adipocytes.
Conclusions
Our results suggest that N4BP2L1 is involved in adipocyte homeostasis by interacting with dynein–dynactin and affecting GLUT4‐mediated glucose uptake and the insulin signaling pathway.
Keywords: Adipocyte, GLUT4, N4BP2L1
N4bp2l1 is a novel FoxO1 target gene that is controlled by insulin‐mediated regulation of FoxO1. N4BP2L1 interacts with dynactin, which binds to the microtubule motor dynein, suggesting that N4BP2L1 is involved in GLUT4 trafficking and glucose uptake.
INTRODUCTION
The NEDD4‐binding protein 2‐like 1 gene (N4BP2L1) is located on human chromosome 13q13.1. It encodes a 243‐amino‐acid protein that possesses several alternatively spliced isoforms. N4BP2L1 is highly expressed in nasopharyngeal carcinoma and oral squamous cell carcinoma, and its overexpression is significantly associated with nodal metastasis and poor outcomes in patients with oral squamous cell carcinoma 1 , 2 .
In our previous study 3 , N4bp2l1 was highly expressed in lipid‐rich brain and adipose mouse tissues. A significant association between the mutant allele of N4BP2L1 rs497255 and body mass index (P = 1.6 × 10−15) was observed in the Genetic Investigation of Anthropometric Traits (GIANT) UK Biobank genome‐wide association studies for height and body mass index 4 . We demonstrated that N4bp2l1 mRNA expression increased in 3T3‐L1 cells in a differentiation‐dependent manner and that N4bp2l1 knockdown suppressed adipocyte differentiation and decreased adipocyte differentiation marker genes (C/ebpα, C/ebpβ, and Pparγ) 3 . Furthermore, we found that N4bp2l1 is a novel target gene of upstream transcription factor 1 (USF1), which is involved in lipid metabolism. N4bp2l1 is transcriptionally regulated by USF1 and plays a significant role in adipocyte differentiation. This suggests that N4bp2l1 expression is associated with lipid accumulation and body mass index. However, the physiological function of N4BP2L1 in adipocytes remains unknown.
In this study, we demonstrated that N4bp2l1 is a novel forkhead box protein O1 (FoxO1) target gene controlled by the insulin‐mediated regulation of FoxO1. Furthermore, N4BP2L1 interacts with dynactin, which binds to the microtubule motor dynein, indicating that N4BP2L1 is involved in glucose transporter type 4 (GLUT4) trafficking and glucose uptake. These results show that N4BP2L1 plays an important role in GLUT4‐mediated glucose uptake and thus in the insulin signaling pathway.
MATERIALS AND METHODS
Animal experiments
Eight‐week‐old male C57BL/6 mice were obtained from CLEA Japan. The mice were maintained on a normal chow diet. Normal chow diet (CE‐2) and high‐fat diet (HFD32) were purchased from CLEA Japan. Animal experimental protocols were approved by the Jichi Medical University Animal Ethics Committee (permit number 17081‐01, March 29, 2018).
Cells and culture conditions
The 3T3‐L1 and human embryonic kidney 293T (HEK293T) cells were maintained in low‐glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum and 100 units of penicillin/streptomycin at 37°C under 5% CO2. The 3T3‐L1 cells (preadipocytes) were cultured and differentiated into 3T3‐L1 adipocytes (mature adipocytes) as described previously 5 . Differentiated 3T3‐L1 adipocytes were infected with the adenovirus at a multiplicity of infection of 30 plaque‐forming units per cell 6 . Differentiated 3T3‐L1 adipocytes were infected with adenovirus 2 days before use in the experiment.
Adenoviral expression vectors
Full‐length N4bp2l1‐cDNA and Glut4‐cDNA were subcloned into the pENTR/D‐TOPO vector (Thermo Fisher Scientific, Waltham, MA, USA). The HiBiT DNA sequence was fused to the first extracellular loop region of Glut4 cloned into pENTR 7 . The pENTR‐N4bp2l1 (C‐terminal FLAG‐tagged) vector and pENTR‐Glut4‐HiBiT were transferred into the pAd/CMV/V5‐DEST vector using the Gateway System (Thermo Fisher Scientific). The shRNAs of N4bp2l and LacZ were cloned into BLOCK‐iT U6 entry vector (Thermo Fisher Scientific). The shRNA sequence for N4bp2l1 shRNA#1 was 5′‐cacc GAATAACTATGAAGTTATA ttcaagaga TATAACTTCATAGTTATTC‐3′ and for N4bp2l1 shRNA#2 was cacc GAAAGAATTGGATTGAAAT ttcaagaga ATTTCAATCCAATTCTTTC. The pENTR vector inserts were transferred into the adenovirus vector pAd/PL‐DEST using the Gateway System (Thermo Fisher Scientific). Adenovirus amplification and purification were performed as described previously 8 .
Real‐time PCR (qPCR)
The extraction of total RNA, cDNA synthesis, and qPCR assays were performed as described previously 9 , 10 . The relative gene expression levels were quantified by qPCR, followed by normalization to the internal control gene ribosomal protein lateral stalk subunit P0 (Rplp0). The primers used for this analysis are listed in Table S1.
Western blot
Whole‐cell lysates were separated by SDS‐polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Proteins were detected with N4BP2L1 (HPA038971; Merck, Branchburg, NJ, USA), Akt (58295; Cell Signaling Technology, Danvers, MA, USA), phospho‐Akt (4060; Cell Signaling Technology), FLAG (F1804, Merck), V5 (46‐0705; Thermo Fisher Scientific), and β‐actin antibody (G043; abm, Richmond, Canada).
Luciferase assay
The luciferase assay was performed using the dual‐luciferase reporter assay system (Promega, Madison, WI, USA). Mouse N4bp2l1 were generated using PCR with mouse genomic DNA as a template. The 3T3‐L1 cells were cotransfected with each expression vector, mouse N4bp2l1 promoter region including exon1 and intron1 that drive firefly luciferase expression (pGL4.10 mN4bp2l1−375/+1284‐Luc) and Renilla reniformis luciferase vector (pGL4.74) for use as an internal control reporter. The cells were incubated for 24 h post‐transfection at 37°C in 5% CO2 and lysed in 100 μL 1X passive lysis buffer (Promega). Lysate was used for the luciferase assay, and luminescence was detected using a luminometer (Thermo Fisher Scientific).
Electrophoretic mobility shift assay (EMSA)
The probe (CTTGGCTCCTGTTTACATCTCTGTG) for EMSA was labeled using the biotin 3′ end DNA labeling kit (Thermo Scientific). The HEK293T cells were transfected with a FoxO1 expression vector driven by a CMV promoter. Nuclear protein was extracted 48 h post‐transfection. The biotin‐labeled probe was mixed with the FoxO1‐overexpressing nuclear protein extract and allowed to incubate at room temperature for 20 min. For the detection of the DNA‐protein bands, we used the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using the SimpleChIP enzymatic chromatin IP kit (Cell Signaling Technology). Immunoprecipitation was performed using a FoxO1 antibody (2880; Cell Signaling Technology) with mouse IgG as the negative control. After immunoprecipitation, the associated DNA was amplified with a primer pair: N4bp2l1 promoter+1067 Fwd 5′‐GTTCTATTACCTCCCCTCGCAGCTG‐3′ and N4bp2l1 promoter+1259 Rv 5′‐CGACTAGTGTGTGCCCGATAACGTT‐3′.
GLUT4‐HiBiT translocation assay
GLUT4‐HiBiT translocation was evaluated in 3T3‐L1 adipocytes cultured in 96‐well plates using the Nano Glo HiBiT extracellular detection system (Promega). The assay was performed according to the manufacturer’s protocol. Briefly, 3T3‐L1 adipocytes were pretreated with or without 50 µM LY294002 (129‐04861; Fujifilm Wako Pure Chemical Corporation,Tokyo, Japan) for 30 min, then stimulated with 1 µM insulin in serum‐free DMEM, and incubated for 1 h. The HiBiT detection reagents were then added and the cells were incubated at room temperature for 5 min prior to the detection of luminescence. Luminescence was detected using a luminometer (Thermo Fisher Scientific).
Glucose uptake assay
Glucose uptake was evaluated in 3T3‐L1 adipocytes cultured in 96‐well plates using the glucose Uptake‐Glo assay (Promega). The assay was performed according to the manufacturer’s protocol. Briefly, after an overnight incubation in serum‐free DMEM, the cells were washed with glucose‐free DMEM (042‐32255, FUJIFILM Wako Pure Chemical Corporation), and the cells were pretreated with or without 50 µM LY294002 for 30 min, and then stimulated with 1 µM insulin for 1 h. Insulin stimulation was followed by the addition of 1 mM 2‐deoxyglucose (2DG) in glucose‐free DMEM, and incubated for 10 min. The 2DG6P detection reagents were then added and the lysates were incubated at room temperature for 1 h prior to the detection of luminescence. Luminescence was detected using a luminometer (Thermo Fisher Scientific).
Statistical analysis
Experimental studies were performed in triplicates. Statistical significance was tested using the unpaired two‐tailed Student’s t‐test or one‐way ANOVA with Bonferroni test for multiple comparisons. Data were expressed as mean ± SEM. Statistical significance was set at P < 0.05.
RESULTS
Expression levels of N4bp2l1 mRNA in adipocytes
Our previous study demonstrated that N4bp2l1 expression increases in 3T3‐L1 cells during adipocyte differentiation and that N4bp2l1 knockdown suppresses adipocyte differentiation, suggesting that the regulation of N4bp2l1 expression is important for the development of adipocyte and physiological function 3 . However, the molecular mechanisms by which N4bp2l1 expression is regulated in adipose tissue and by which N4BP2L1 functions intracellularly and is involved in adipocyte differentiation are largely unknown. Therefore, to first examine how N4bp2l1 expression is regulated in adipose tissue, we determined whether it is influenced by feeding. After 24 h of fasting, we examined N4bp2l1 expression in epididymal adipose tissue (eWAT) of mice fed either a normal or a high‐fat (HF) diet and found that it was significantly reduced after feeding with both normal and HF diets (Figure 1a). Then, we examined how the reduction of N4bp2l1 mRNA in eWAT is related to feeding. To determine whether feeding‐induced insulin influences N4bp2l1 downregulation, we investigated the N4bp2l1 expression levels in insulin‐treated 3T3‐L1 adipocytes. The results revealed that 2 h after insulin treatment, the N4bp2l1 expression levels were significantly decreased and remained so for 8 h (Figure 1b). After 4 h of insulin treatment, the N4BP2L1 protein levels decreased (Figure 1c). To confirm whether insulin signaling is responsible for the decrease in N4BP2L1 protein expression, we blocked insulin signaling using a PI3 kinase inhibitor, namely, LY294002. Insulin reduced the N4BP2L1 protein levels, which is in agreement with Figure 1c. However, interestingly, LY294002 further reduced the N4BP2L1 protein levels and did not counteract the insulin‐induced reduction in N4BP2L1 protein (Figure 1d).
Figure 1.
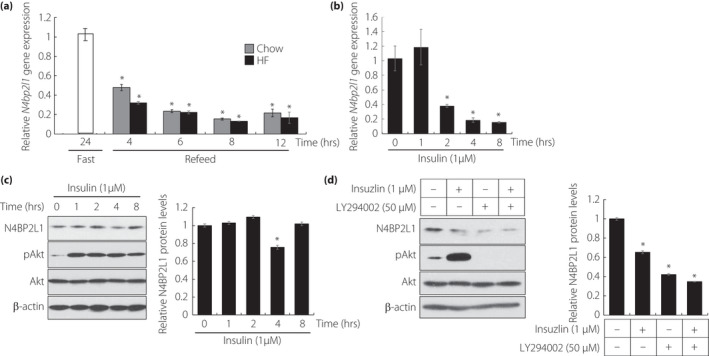
Insulin decreases N4bp2l1 mRNA expression levels. (a) The expression levels of N4bp2l1 under various feeding states in epididymal adipose tissue. Mice fasted for 24 h or fasted (white bar) for 24 h and were refed for 4, 6, 8, and 12 h. Wild‐type mice were fed normal chow diet (gray bar) or HF diet (black bar) during the feeding period; n = 3 per group, *P < 0.01 vs 24 h fasting. (b) The effect of N4bp2l1 expression on insulin stimulation in 3T3‐L1 adipocytes; n = 3 per group, *P < 0.01 vs 0 h. (c) The effect of N4BP2L1 protein expression on insulin stimulation in 3T3‐L1 adipocytes. Whole‐cell lysates were extracted from 3T3‐L1 adipocytes at several time points following insulin treatment (left panel). Quantification of relative N4BP2L1 protein levels via western blot. The data were normalized to β‐actin levels (right panel); n = 3 per group, *P < 0.01 vs 0 h. (d) Whole‐cell lysates from 3T3‐L1 adipocytes with or without pretreatment with LY294002 (50 μM) for 30 min followed by exposure to insulin (1 μM) for a further 4 h were subjected to western blot analysis using anti‐N4BP2L1, anti‐phosphorylated Akt, anti‐Akt, and anti‐β‐actin (left panel). Quantification of relative N4BP2L1 protein levels via western blot. The data were normalized to β‐actin levels (right panel); n = 3 per group, *P < 0.01 vs. 0 h.
N4bp2l1 is a novel target gene of FoxO1
Insulin decreased N4bp2l1 expression in 3T3‐L1 adipocytes (Figure 1b), suggesting that insulin signaling‐mediated transcription factors are involved in the regulation of N4bp2l1 expression. We reported previously that N4bp2l1 is a novel target gene of USF1 by N4bp2l1 promoter analysis 3 ; however, we did not find any transcription factors that were associated with insulin signaling. Therefore, we examined the presence of insulin signaling‐related transcription factors in the N4bp2l1 promoter region using LASAGNA‐Search 2.0. We searched the N4bp2l1 promoter region, including intron 1, and identified the elements that may bind C/EBP, FoxO1, and FoxA2 (Figure 2a), in addition to the previously identified USF1 3 .
Figure 2.
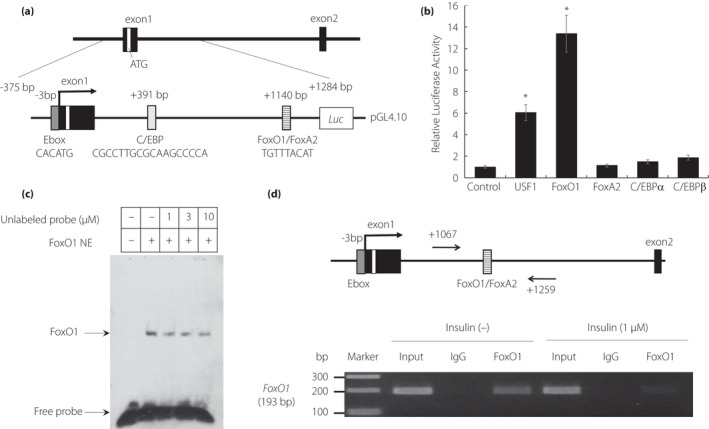
The activation of the N4bp2l1 promoter by FoxO1. (a) A schematic diagram of the mouse N4bp2l1 promoter with the potential binding sites of lipid metabolism‐related transcription factors. (b) The mouse N4bp2l1 promoter region including exon 1 and intron 1 was fused to a luciferase reporter gene (pGL4.10 mN4bp2l1−375/+1284‐Luc). The 3T3‐L1 cells were cotransfected with pGL4.10 mN4bp2l1−375/+1284‐Luc as a reporter gene. Renilla reniformis luciferase vector (pGL4.74) served as an internal control reporter, and the control vector was pcDNA3.1. n = 3 per group, *P < 0.01 vs. control. (c) Electrophoretic mobility shift assay (EMSA) for FoxO1 binding to the +1140 bp region of the N4bp2l1 gene. Nuclear extracts of FoxO1 protein were incubated with the biotin‐labeled +1140 bp region of the N4bp2l1 gene in the presence or absence of unlabeled probes. The competition was performed using unlabeled probes as competitors from 1‐ to 10‐fold molar excesses. (d) The ChIP assay for FoxO1 binding to the +1140 bp region of the N4bp2l1 gene using 3T3‐L1 adipocytes with/without insulin (1 μM). The cells were harvested 4 h following insulin treatment. The extracted genomic DNA was subjected to immunoprecipitation, which was performed using an anti‐FoxO1 antibody, with IgG as a negative control. For comparison, amplification derived from unprecipitated chromatin is also demonstrated (input).
Subsequently, we performed a reporter assay to determine whether these transcription factors bind to the N4bp2l1 promoter region, including intron 1, and increase the transcriptional activity of N4bp2l1. Consistent with our previous study 3 , USF1 significantly increased the transcriptional activity of N4bp2l1. We identified a new transcription factor, FoxO1, which increased the transcriptional activity of N4bp2l1 by 13‐fold compared with the control (Figure 2b). To examine whether FoxO1 directly binds to intron 1 of the N4bp2l1 gene, EMSA was performed. FoxO1 directly bound to intron 1 (the TGTTTACAT sequence) of the N4bp2l1 gene, and the protein binding specificity of the TGTTTACAT sequence probe was suppressed by competition with an excess of the unlabeled probe (Figure 2c). Furthermore, to elucidate whether FoxO1 physically binds to the endogenous N4bp2l1 promoter, we conducted a ChIP assay of genomic DNA using 3T3‐L1 adipocytes. The results confirmed that FoxO1 directly binds to the FoxO1 binding site in 3T3‐L1 adipocytes without insulin, whereas FoxO1 does not bind to the FoxO1 binding site of N4bp2l1 in insulin‐treated 3T3‐L1 adipocytes (Figure 2d).
N4BP2L1 forms a complex with dynactin
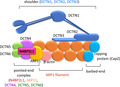
To investigate the functions of N4BP2L1, we examined the proteins that interact with N4BP2L1 using the BioGRID interaction database. We found that dynactin subunit proteins (ARP11 and DCTN3–6) interact with N4BP2L1 (https://thebiogrid.org/, last accessed on January 25, 2021). As ARP11 and DCTN4–6 constitute the pointed‐end complex domain of dynactin 11 , N4BP2L1 is predicted to represent a novel member of the pointed‐end complex domain. Thus, we subsequently conducted immunoprecipitation studies to confirm whether N4BP2L1 binds to dynactin subunits in the cells. The co‐immunoprecipitation assay with N4BP2L1 was performed using Dctn4, Dctn5, and Dctn6 vectors, which are the point‐end complexes of dynactin. We demonstrated that N4BP2L1 binds to DCTN3–6 (Figure 3a), indicating that N4BP2L1 represents a novel member of the dynactin complex (Figure 3b).
Figure 3.
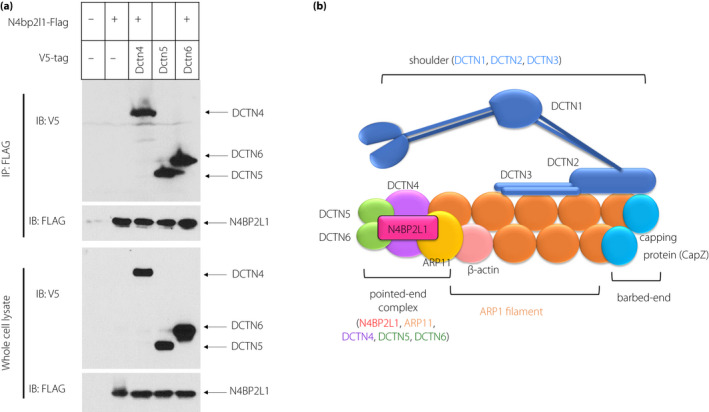
N4BP2L1 interacts with dynactin. (a) FLAG‐tagged N4bp2l1 and V5‐tagged dynactin subunit (Dctn4–6) vectors were cotransfected into HEK293T cells, and immunoprecipitation of the FLAG‐tagged proteins was performed using anti‐FLAG antibodies. Immunoblot analysis was conducted using anti‐V5 or anti‐FLAG antibodies. (b) Schematic models of the dynactin‐N4BP2L1 complex. N4BP2L1 binds to DCTN4–6 and possibly forms a complex with the pointed‐end complex domain.
N4BP2L1 affects GLUT4 translocation and is associated with glucose uptake in 3T3‐L1 adipocytes
The microtubule motor protein dynein interacts with dynactin and is involved in GLUT4 trafficking 12 . To determine whether N4BP2L1, which binds to dynactin, is involved in GLUT4 trafficking via dynein–dynactin interactions, we conducted GLUT4‐HiBiT translocation assays using N4bp2l1 overexpression and knockdown 3T3‐L1 adipocytes (Figure 4a). In N4bp2l1‐overexpressing 3T3‐L1 adipocytes, insulin‐induced GLUT4 translocation to the membrane was significantly increased compared with the GFP controls (Figure 4b). In N4bp2l1‐knockdown 3T3‐L1 adipocytes, GLUT4 translocation to the membrane was significantly decreased after 1 h of insulin treatment compared with shLacZ controls, whereas GLUT4 translocation to the membrane was increased, except after 1 h of insulin treatment (Figure 4c). Next, we examined glucose uptake in N4bp2l1‐overexpressing and knockdown adipocytes. In N4bp2l1‐overexpressing 3T3‐L1 adipocytes, glucose uptake was increased, which is consistent with insulin‐induced GLUT4 translocation to the plasma membrane. Blocking insulin signaling using a PI3 kinase inhibitor, namely, LY294002, also increased glucose uptake in N4bp2l1‐overexpressing 3T3‐L1 adipocytes compared with GFP controls (Figure 4d). This result is consistent with the results of GLUT4 translocation to the membrane (Figure 4b). Conversely, in N4bp2l1‐knockdown 3T3‐L1 adipocytes, GLUT4 translocation to the membrane was increased (Figure 4c), whereas glucose uptake was decreased (Figure 4e).
Figure 4.
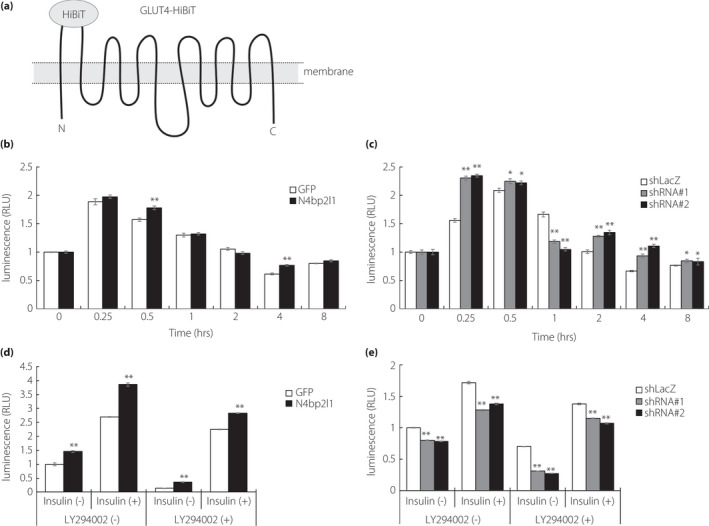
N4BP2L1 affects GLUT4 translocation and is involved in glucose uptake. (a) A schematic diagram of GLUT4‐HiBiT. The N‐ and C‐termini of GLUT4 are in the cytoplasm, and HiBiT is fused to the first extracellular loop of GLUT4. In the presence of insulin, GLUT4‐HiBiT translocates to the cell membrane to promote glucose uptake. GLUT4‐HiBiT can regulate the translocation of the GLUT4 in live cells in real time. (b and c) N4BP2L1 influences GLUT4 translocation in 3T3‐L1 adipocytes. 3T3‐L1 adipocytes were co‐infected with GLUT4‐HiBiT and either GFP or N4bp2l1 adenovirus or either shLacZ or N4bp2l1 shRNA adenovirus. After 48 h, the cells were incubated with or without insulin (1 μM) for each time point. GLUT4‐HiBiT translocation to the cell membrane was measured using a detection reagent containing LgBiT and NanoLuc substrate. n = 3 per group, *P < 0.05, **P < 0.01 vs GFP or shLacZ. (d and e) N4BP2L1 influences glucose uptake in 3T3‐L1 adipocytes. 3T3‐L1 adipocytes were infected with either GFP or N4bp2l1 adenovirus or either shLacZ or N4bp2l1 shRNA adenovirus. After 48 h, the cells were preincubated with or without LY294002 (50 μM) for 30 min followed by exposure to insulin (1 μM) for a further 1 h. Afterward, glucose uptake assay was performed. n = 3 per group, *P < 0.05, **P < 0.01 vs GFP or shLacZ.
N4BP2L1 affects the lipid metabolism‐related genes in 3T3‐L1 adipocytes
To investigate the expression levels of Glut4, lipid metabolism‐related genes, and several genes of dynactin and dynein in 3T3‐L1 adipocytes by N4bp2l1 knockdown and overexpression, we performed qPCR. In N4bp2l1‐overexpressing 3T3‐L1 adipocytes, Glut4 expression was significantly increased. The genes involved in adipocyte differentiation (Pparγ and aP2) and triglyceride synthesis (Srebp1c and Fas) were upregulated in N4bp2l1 overexpression. The components of dynactin (Dctn1–6) and dynein (Dync1h1, Dync1i1, Dync1li1, Dync1li2, and Dynll1) were increased in N4bp2l1‐overexpressing 3T3‐L1 adipocytes. Contrarily, the expression of these genes was decreased in N4bp2l1‐knockdown 3T3‐L1 adipocytes (Figure 5a,b).
Figure 5.
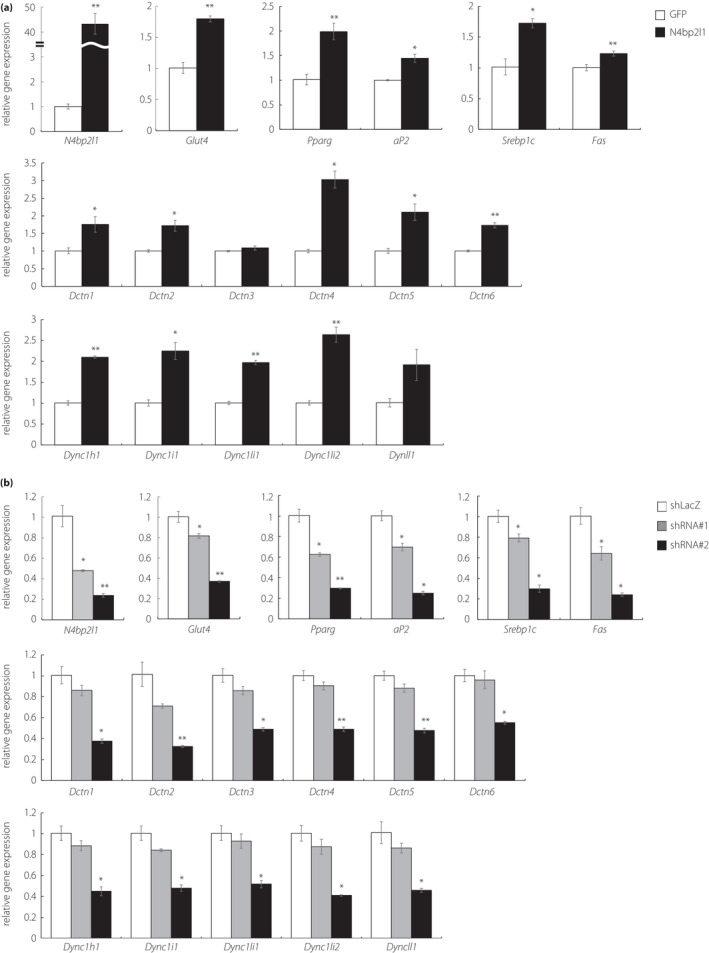
The effect of N4BP2L1 on gene expression levels in 3T3‐L1 adipocytes. (a and b) 3T3‐L1 adipocytes were infected with either GFP or N4bp2l1 adenovirus or either shLacZ or N4bp2l1 shRNA adenovirus. After 48 h, total RNA was extracted and subjected to qPCR. n = 3 per group, *P < 0.05, **P < 0.01 vs GFP or shLacZ.
DISCUSSION
We reported previously that N4bp2l1 is upregulated during adipocyte differentiation and N4bp2l1 knockdown suppresses adipocyte differentiation 3 . In the present study, we demonstrated that N4bp2l1 is a novel target gene of FoxO1 (Figure 2), indicating that insulin‐induced translocation of FoxO1 from the nucleus to the cytoplasm results in a decrease in the N4bp2l1 mRNA levels and N4BP2L1 protein levels. These data are consistent with the feeding‐induced N4bp2l1 downregulation in mouse eWAT and reduced N4BP2L1 protein in insulin‐treated 3T3‐L1 adipocytes (Figure 1). Interestingly, despite the presence of FoxO1 in the nuclei of LY294002‐treated 3T3‐L1 adipocytes (Figure S1), N4BP2L1 protein level was reduced compared with untreated 3T3‐L1 adipocytes (Figure 1d). These data suggest that LY294002‐induced inhibition of the insulin signaling pathway is involved in something other than FoxO1‐mediated N4bp2l1 transcriptional regulation, such as the inhibition of N4BP2L1 synthesis and degradation of N4BP2L1 protein. To further explore the functions of N4BP2L1, we examined the interacting proteins and found that N4BP2L1 binds to dynactin.
Dynactin is a ˜ 1 MDa complex composed of 23 subunits of 11 distinct proteins, with three structural domains, namely, actin‐related protein‐1 (ARP1) filament subunit, pointed‐end complex, and sidearm/shoulder 13 . The ARP1 filament is the core of the dynactin structure and is composed of eight ARP1 units, one β‐actin unit, and one actin‐capping protein (CapZ) that forms the barbed‐end. The sidearm/shoulder domain, which binds to CapZ, consists of DCTN1–3. The pointed‐end complex domain forms a complex with the four proteins DCTN4–6 and ARP11 and binds to the ARP1 filament 11 , 14 , 15 . N4BP2L1 was found to bind to DCTN4–6 (Figure 3a) and may have formed a complex with the pointed‐end complex domain, thus playing a role in the function of dynactin (Figure 3b). Dynactin binds to the microtubule motor dynein, which is important for most cellular functions, including vesicular trafficking, organelle transport, and mitotic spindle assembly 12 , 16 . It has been suggested previously that the microtubule motor dynein, which transports cargo toward the minus end of microtubules, plays a role in GLUT4 vesicle localization 12 . In addition, dynactin, which was first identified as an activator of dynein, acts as an adaptor between dynein and the cargo and has been shown to enhance the organelle transport capacity of dynein 15 , 17 . However, the mechanisms involved in dynein activation remain unknown.
As N4BP2L1 binds to dynactin (Figure 3), we hypothesized that the N4BP2L1–dynactin interaction is involved in the activation of dynein and determined whether it influences the localization of GLUT4, one of the functions of the dynein–dynactin complex 12 . The GLUT4 translocation assay revealed that N4bp2l1 overexpression increased GLUT4 translocation (Figure 4b) and consequent glucose uptake (Figure 4d) as well as the expressions of Glut4 and lipid metabolism‐related genes in adipocytes (Figure 5a). Conversely, we expected N4bp2l1 knockdown to decrease GLUT4 translocation. However, contrary to our expectation, GLUT4 translocation was significantly decreased only 1 h following insulin treatment but was significantly increased at other times (Figure 4c). Although insulin treatment increased the amount of GLUT4 localized at the cell membrane in N4bp2l1‐knockdown adipocytes, glucose uptake (Figure 4e) and the expressions of Glut4 and lipid metabolism‐related genes were significantly reduced (Figure 5b). As the motor protein dynein regulates the internalization of GLUT4 from the cell membrane 11 , N4BP2L1 may be involved in not only GLUT4 translocation to the membrane but also the internalization of GLUT4 (i.e., GLUT4 trafficking). This suggests that N4BP2L1 affects glucose uptake by influencing the localization of GLUT4 in the cells, as the decrease in N4BP2L1 did not unidirectionally affect GLUT4 translocation to the membrane but affected its fluctuation. In addition, the dynactin and dynein genes were altered when N4bp2l1 was overexpressed and knocked down (Figure 5), which may have also affected GLUT4 trafficking between the cytoplasm and the membrane. N4BP2L1 is likely important for adipocyte homeostasis and physiological roles because changes in N4BP2L1 expression not only alter glucose uptake and the expressions of Glut4 and lipid metabolism‐related genes, but also the expression of dynactin and dynein genes in adipocytes. However, the physiological role of adipocytes in N4BP2L1 reduction and mechanism by which decreased N4BP2L1 reduces glucose uptake remain unclear. Although not yet clarified, it is possible that insulin‐mediated reduction of N4BP2L1 suppresses excessive glucose uptake.
In summary, we demonstrated that N4bp2l1 is a novel target gene of FoxO1 and that N4BP2L1 binds to dynactin. This suggests that N4BP2L1 is involved in adipocyte homeostasis by interacting with dynein–dynactin and affecting GLUT4 trafficking and glucose uptake. How insulin‐induced decreases in N4BP2L1 levels involve GLUT4 trafficking and glucose uptake remains unknown; however, we shall investigate these issues in the future using N4BP2L1‐knockout mice.
Disclosure
No conflicts of interest to declare.
Author contributions
K.W. designed and conceived the experiments; K.W. conducted the experiments; K.W. wrote the manuscript. A.M., T.H., and S.I. provided critical advice on various aspects of this project.
Supporting information
Figure S1 | 3T3‐L1 adipocytes were pretreated with LY294002 (50 μM) for 30 min followed by exposure to insulin (1 μM) for a further 4 h. Cell nuclear extracts were subjected to western blot analysis using anti‐FoxO1 (2880, Cell Signaling Technology) and anti‐Histone H3 (4620, Cell Signaling Technology).
Table S1 | Forward and reverse primer sequences of target genes
Acknowledgments
This study was supported by Grants‐in‐Aid for Scientific Research (15K19523, 17K16153, and 20K08914) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and Jichi Medical University young investigator award. We would also like to thank Ms Kayo Nagashima and Ms Yukiko Ohashi for their excellent technical assistance. We thank Enago (http://www.enago.jp) for the English language review.
J Diabetes Investig. 2021; 11: 1958–1966
References
- 1. Sasahira T, Kurihara M, Nishiguchi Y, et al. Nedd 4 binding protein 2‐like 1 promotes cancer cell invasion in oral squamous cell carcinoma. Virchows Arch 2016; 469: 163–172. [DOI] [PubMed] [Google Scholar]
- 2. Sasahira T, Kirita T. Hallmarks of cancer‐related newly prognostic factors of oral squamous cell carcinoma. Int J Mol Sci 2018; 19: 2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Watanabe K, Yokota K, Yoshida K, et al. A novel upstream transcription factor 1 target gene n4bp2l1 that regulates adipogenesis. Biochem Biophys Rep 2019; 20: 100676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yengo L, Sidorenko J, Kemper KE, et al. Meta‐analysis of genome‐wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet 2018; 27: 3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watanabe K, Yoshida K, Iwamoto S. Kbtbd11 gene expression in adipose tissue increases in response to feeding and affects adipocyte differentiation. J Diabetes Investig 2019; 10: 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watanabe K, Yokota K, Yoshida K, et al. Kbtbd11 contributes to adipocyte homeostasis through the activation of upstream stimulatory factor 1. Heliyon 2019; 5: e02777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kanai F, Nishioka Y, Hayashi H, et al. Direct demonstration of insulin‐induced glut4 translocation to the surface of intact cells by insertion of a c‐myc epitope into an exofacial glut4 domain. J Biol Chem 1993; 268: 14523–14526. [PubMed] [Google Scholar]
- 8. Watanabe K, Watson E, Cremona ML, et al. Ildr2: an endoplasmic reticulum resident molecule mediating hepatic lipid homeostasis. PLoS One 2013; 8: e67234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Watanabe K, Nakayama K, Ohta S, et al. Znf70, a novel ildr2‐interacting protein, contributes to the regulation of hes1 gene expression. Biochem Biophys Res Commun 2016; 477: 712–716. [DOI] [PubMed] [Google Scholar]
- 10. Watanabe K, Nakayama K, Ohta S, et al. Ildr2 stabilization is regulated by its interaction with grp78. Sci Rep 2021; 11: 8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Urnavicius L, Zhang K, Diamant AG, et al. The structure of the dynactin complex and its interaction with dynein. Science (New York, N.Y.) 2015; 347: 1441‐1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang J, Imamura T, Olefsky JM. Insulin can regulate glut4 internalization by signaling to rab5 and the motor protein dynein. Proc Natl Acad Sci USA 2001; 98: 13084–13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yeh TY, Quintyne NJ, Scipioni BR, et al. Dynactin's pointed‐end complex is a cargo‐targeting module. Mol Biol Cell 2012; 23: 3827–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eckley DM, Gill SR, Melkonian KA, et al. Analysis of dynactin subcomplexes reveals a novel actin‐related protein associated with the arp1 minifilament pointed end. J Cell Biol 1999; 147: 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hammesfahr B, Kollmar M. Evolution of the eukaryotic dynactin complex, the activator of cytoplasmic dynein. BMC Evol Biol 2012; 12: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reck‐Peterson SL, Redwine WB, Vale RD, et al. The cytoplasmic dynein transport machinery and its many cargoes. Nat Rev Mol Cell Biol 2018; 19: 382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ayloo S, Lazarus JE, Dodda A, et al. Dynactin functions as both a dynamic tether and brake during dynein‐driven motility. Nat Commun 2014; 5: 4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 | 3T3‐L1 adipocytes were pretreated with LY294002 (50 μM) for 30 min followed by exposure to insulin (1 μM) for a further 4 h. Cell nuclear extracts were subjected to western blot analysis using anti‐FoxO1 (2880, Cell Signaling Technology) and anti‐Histone H3 (4620, Cell Signaling Technology).
Table S1 | Forward and reverse primer sequences of target genes


