Abstract
The tumor suppressor function of Rb is intimately related to its ability to interact with E2F and repress the transcription of E2F target genes. Here we describe a novel E2F product that specifically interacts with Rb in quiescent cells. This novel E2F, which we term E2F3b, is encoded by a unique mRNA transcribed from an intronic promoter within the E2F3 locus. The E2F3b RNA differs from the previously characterized E2F3 RNA, which we now term E2F3a, by the utilization of a unique coding exon. In contrast to the E2F3a product that is tightly regulated by cell growth, the E2F3b product is expressed equivalently in quiescent and proliferating cells. But, unlike the E2F4 and E2F5 proteins, which are also expressed in quiescent cells and form complexes with the p130 protein, the E2F3b protein associates with Rb and represents the predominant E2F-Rb complex in quiescent cells. Thus, the previously described specificity of Rb function as a transcriptional repressor in quiescent cells coincides with the association of Rb with this novel E2F product.
Pathways controlling the progression of cells out of quiescence, through G1, and into S phase have now been elucidated in considerable detail. A variety of studies have demonstrated the role of G1 cyclin-dependent kinases (cdk) that phosphorylate and control the activity of Rb and related proteins, leading to the accumulation of E2F transcription factor activity (for reviews, see references 6, 9, 17, 21, 26, 27, and 30). It is also clear that the disruption of this pathway, either through the activation of positive-acting components such as the G1 cyclins or the inactivation of negative components such as p53, Rb, and the cyclin kinase inhibitors, can lead to a loss of cell growth control that underlies the development of various forms of human cancer (10, 30).
E2F activity is comprised of a complex array of DNA binding activities involving various members of a family of related proteins that include six distinct E2F members and at least two heterodimer partners, DP1 and DP2 (4, 22). The complexity of the E2F activity suggests a complexity of function whereby the individual family members might play distinct roles in cellular growth control. Indeed, emerging evidence indicates that distinct roles can be ascribed to the individual E2F activities. For instance, it has been suggested that the E2F4 and E2F5 proteins, which specifically associate with the Rb-related p130 protein in quiescent cells (28), function to repress transcription of various genes encoding proteins important for cell growth. In contrast, E2F3 has been suggested to play a positive role in transcription control during the cell cycle and appears to be important for efficient induction of S phase in cycling cells (18). E2F1, on the other hand, appears to function in the induction of apoptosis (2, 14, 23, 25, 31), accumulating at the initial G1/S transition as cells re-enter the cell cycle (18).
Additional insight into the role of E2F and Rb family proteins in the control of transcription has come from the analysis of expression of various E2F target genes in cells derived from mouse embryos deficient in Rb family members (7, 11). These studies have provided evidence for deregulation of various E2F target genes in cells lacking Rb or Rb family members. Particularly striking is the specificity in the control of E2F-regulated genes exhibited by individual Rb family members (11). The absence of Rb function coincided with partial derepression of cyclin E and p107 expression but normal control of various other E2F targets. In contrast, the combined absence of the p130 and p107 activities led to the deregulation of a number of E2F targets but no change in cyclin E nor p107. These results thus suggest that the individual Rb family members play distinct roles in the control of transcription in quiescent cells.
We have now further explored the role of E2F activities in the control of gene expression as a function of cell proliferation. Because of recent work that highlights an apparent important role of E2F3 activity in regulating transitions during the cell cycle (18), we have further analyzed the control of E2F3 accumulation in quiescent and growing cells. In so doing, we have revealed a complexity in the organization of the E2F3 locus whereby two independent but overlapping transcription units direct the synthesis of two distinct mRNAs that differ with respect to the initial exon utilized. These two RNAs encode related but distinct proteins that differ with respect to expression. In particular, we find that an E2F3 product that is expressed in quiescent cells specifically associates with Rb and thus may be critical for the specificity of Rb transcription control in quiescent cells.
MATERIALS AND METHODS
Cell culture.
REF52 cells were grown in Dulbecco modified Eagle medium (DMEM) containing 10% serum (5% fetal bovine serum and 5% calf serum). To bring cells to quiescence, cells were plated at approximately 3,500 cells/cm2 and incubated overnight. The next day, the cells were washed once with DMEM, and the culture medium was replaced with DMEM containing 0.25% serum; cells were then further incubated for 48 h prior to replacement with culture medium containing 10% serum.
Isolation of E2F3 genomic DNA.
A mouse BAC library (Genome Systems) was screened with a 1.5-kb human E2F3 cDNA probe isolated by HindIII/BamHI digestion of pcDNA3-E2F3. Two positive clones, 305 G72 and 146 J18, were identified and isolated. EcoRI, HindIII, and KpnI digestion of the two clones demonstrated similar restriction patterns.
Construction of plasmids.
Mouse BAC clone 305 G72 was digested with EcoRI and HindIII. A 532-bp N-terminal HindIII/DraI human cDNA probe isolated a 6.5-kb EcoRI fragment and a 3-kb HindIII fragment. A 943-bp N-terminal HindIII/HindII human cDNA probe isolated both a 6.5- and a 15-kb EcoRI fragment. The 3-kb HindIII fragment and the 6.5- and 15-kb EcoRI fragments were subcloned into pBluescript SK(+/−) creating the constructs 3kbH3pBS, 6.5kbpBS, and 15kbpBS. SacI/HaeII digestion of 3kbH3pBS excised the 2-kb promoter fragment of E2F3a, which was directionally subcloned into SacI/HindIII-digested pGL2Basic (Promega). The primers GAGAGAGATCTTCCGAAAGCAGCCTGG and GAGAGAGATCTAATACCCTCCTCAGCG containing BglII sites were used to generate a 1.1-kb E2F3b promoter fragment via PCR. The PCR product was digested with BglII, subcloned into BglII-digested pGL2Basic, and sequenced.
RACE (rapid amplification of cDNA ends) analysis.
A cDNA library was generated from quiescent mouse embryo fibroblasts (MEF) using the protocol described in the Marathon cDNA Amplification Kit (Clontech). In accordance with the kit, the first gene specific primer (GSP1), which anneals in exon 5, used to amplify 5′E2F3b sequence was ACTTCAAGTCTCGTTTCTGGAAGGGCTTTCACAAC. The product of the first round of amplification was used as the template for a second round of amplification. A more upstream primer (GSP2) that anneals in exon 4 and contains a nested NotI site was used in the second amplification: GAGAGAGCGGCCGCAATCCTCGGTTAACAGTTTGAGGTCCAG. The amplified product was digested with NotI, ligated into NotI-digested pBS, and sequenced.
Primer extension analysis.
The transcription start sites for the murine E2F3a and E2F3b transcripts were determined by primer extension analysis as described previously (5). Poly(A) containing RNA was prepared from MEF that were synchronized at G1/S by hydroxyurea arrest, and 5 μg was used for primer extension. The primer for E2F3a derived from 5′ untranslated region (UTR) sequence within exon 1a (5′-CTCGTCCTCGCTCTCTACTCTTTCCCC-3′); the primer for E2F3b derived from exon 1b sequence (5′-CGGTAGTCATGGAGAGTCTACC-3′). The products of primer extension were analyzed in an 8% polyacrylamide gel containing 7 M urea along with products of DNA sequencing reactions generated with an exon 1a primer (5′-CCTCCGCTCTTCCTCTCTGAACC-3′).
Viruses.
The construction of Ad-Con and Ad-Myc have been previously described (2).
Northern analysis.
Poly(A)+ RNA was isolated from an equal amount of total RNA and processed for Northern analysis as described previously (1).
E2F DNA binding assays.
E2F assays were performed as previously described (12). Supershift analysis was carried out as previously described (12) using antibodies specific for E2F1 (SC-251x), E2F3 C terminus (SC-878), or E2F3 N terminus (SC-879). The E2F3 C-terminal antibody was generated against an 18-amino-acid peptide from the C terminus of E2F3 (DLFDAYDLEKLPLVEDFMCS), which is common to both the E2F3a and the E2F3b proteins. The E2F3 N-terminal antibody was raised against a 20-amino-acid peptide from the N terminus of the E2F3a sequence (MRKGIQPALEQYLVTAGGGE). Immunoglobulin G (IgG) was used as a control (Santa Cruz). Immunoprecipitation followed by release with deoxycholate (DOC) were performed as described previously (12).
Promoter transfection assays.
REF52 cells were transfected with either E2F3a-Luc or E2F3b-Luc reporter vectors and cytomegalovirus (CMV)–β-galactosidase as an internal control as described previously (24). After transfection, cells were brought to quiescence by serum starvation and then stimulated to grow by the addition of fresh medium with serum. Cells were harvested at various times after stimulation and luciferase, and β-galactosidase assays were performed as described earlier (24).
RESULTS
Expression of the E2F3 locus following stimulation of cell growth.
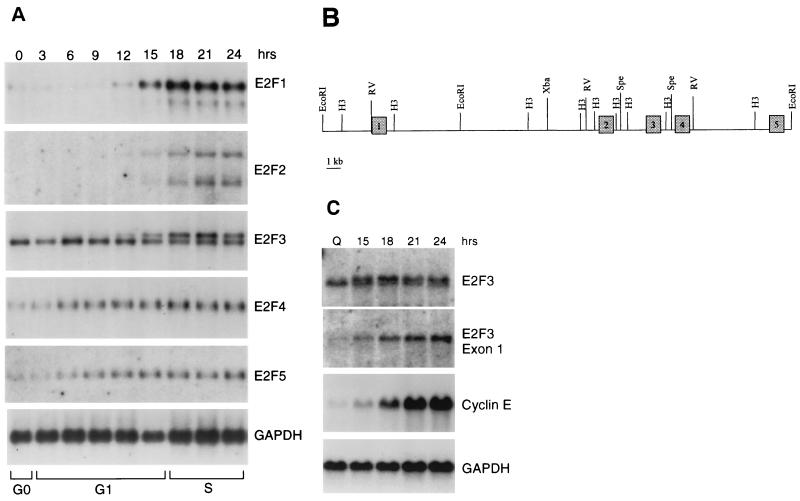
Previous studies have detailed the distinct pattern of expression of the various E2F family genes (4, 22); an example of this, as measured by Northern blot assays, is shown in Fig. 1A. Particularly striking is the distinct pattern of expression of the E2F1 and E2F2 genes compared to that of E2F4 and E2F5. Whereas the expression of the latter two genes is relatively constant as cells progress out of quiescence and into a cell cycle, the accumulation of the E2F1 and E2F2 RNAs is tightly regulated with little or no RNA evident in quiescent cells and then a large induction as cells pass through G1. Previous work has shown that this is largely a function of negative autoregulatory control of these genes in quiescent cells (8, 13, 20, 24). That is, the presence of E2F binding elements within the E2F1 and E2F2 promoters allows repression of transcription in quiescent cells, presumably mediated by an E2F-Rb family member complex.
FIG. 1.
Expression of E2F family members following stimulation of cell growth. (A) Expression of E2F family RNAs following stimulation of REF52 cell growth. Cells were brought to quiescence by serum starvation and then stimulated to grow by the addition of fresh media with serum. Samples were taken at the indicated times, and RNA was prepared and analyzed by Northern blotting as described in Materials and Methods. The transition from G1 to S phase, as indicated at the bottom of the figure, was determined by flow cytometry. (B) Genomic organization of the E2F3 locus. The exon-intron structure that includes the initial five exons of the E2F3 locus is shown in relation to restriction maps. (C) Northern analysis with E2F3 exon-specific probes. REF52 cells were brought to quiescence and then stimulated by the addition of serum. Aliquots were taken at the indicated times, and RNA was analyzed as described in Fig. 1. E2F3 RNA was detected with either a full-length cDNA probe (E2F3) or an exon 1-specific probe (E2F3 exon 1a [see Fig. 2]). For comparison, the same blots were probed for expression of the cyclin E gene as well as GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as a loading control.
The expression of E2F3 is more complex and, in fact, appears to combine the patterns for the other E2F species. In particular, two E2F3 RNAs are detected in these assays; the faster-migrating RNA remains constant through the growth stimulation, similar to the pattern of E2F4 and E2F5 expression, whereas the slower-migrating RNA accumulates with kinetics similar to those of E2F1 and E2F2. This is not a species-specific event since analysis of both MEF and human diploid fibroblasts yielded essentially the same result (data not shown).
Analysis of the E2F3 genomic locus reveals two distinct transcription units.
To explore the mechanistic basis for the generation of two E2F3 RNAs that differ in their expression, we have analyzed the genomic organization of the E2F3 locus. E2F3 mouse genomic clones were isolated by screening a mouse BAC genomic library with a radiolabeled probe derived from the full-length, 1.5-kb human E2F3 cDNA. Two positive clones were identified and isolated as described in Materials and Methods. Analysis of the clones by restriction mapping and Southern blot analysis revealed that they were overlapping clones, each containing the E2F3 gene locus. Three fragments, a 3-kb HindIII fragment, a 6.5-kb EcoRI fragment, and a 15-kb EcoRI fragment from the initial BAC clone were determined by Southern blot analysis to contain a portion of the E2F3 gene. The fragments were isolated and subcloned into pBluescript SK(+/−). Exon-intron boundaries for the first five exons were identified by sequencing using primers derived from human cDNA. Lack of homology to the human cDNA as well as the presence of consensus splice sites resolved exon-intron borders (Fig. 1B). Intron length was determined by both PCR analysis, using primers complementary to the adjacent exons, and by direct sequencing.
To further analyze the expression of the E2F3 locus, we repeated the Northern analysis using either the complete cDNA probe or a probe specific to the exon 1 sequences. As shown in Fig. 1C, the cDNA probe again detected both E2F3 transcripts. In contrast, the exon 1 probe only detected the larger, slower-migrating E2F3 transcript that was regulated by growth. It would thus appear that the smaller, constitutively expressed E2F3 RNA is lacking sequences derived from exon 1.
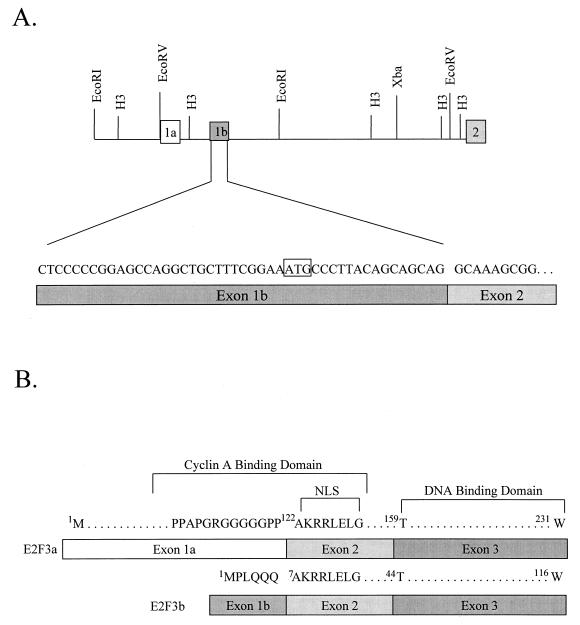
Since only the smaller transcript, lacking exon 1, is present in quiescent cells, we created a cDNA library using RNA from quiescent MEF cells in order to clone and characterize this smaller E2F3 gene product. Using primers from exons 4 and 5, we amplified 5′ RACE products from the library, subcloned these into pBS, and then sequenced the inserts. Two of nine clones contained sequence representing exons 2, 3, and 4, as well as an additional six codons upstream of exon 2 beginning with an ATG (Fig. 2A). The sequence unique to the shorter transcript was aligned to the E2F3 genomic sequence and found to lie within intron 1, 1.1 kb downstream of the known exon 1. We have now referred to the original first exon as exon 1a, and we have designated the unique sequence as exon 1b (Fig. 2A).
FIG. 2.
Alternate 5′ exons encode E2F3 RNAs. (A) Genomic structure and organization of the E2F3 locus. The deduced position of the exon 1b, as identified by RACE analysis and sequence comparison, is indicated in relation to the initial E2F3 exon, now termed exon 1a, and in relation to exon 2. The DNA sequence of the longest RACE product is presented. (B) Comparison of the amino acid sequence encoded by E2F3 exon 1a and exon 1b. The sequences homologous to the previously defined cyclin A binding domain of E2F1 are indicated as well as the sequence corresponding to the nuclear localization signal.
A comparison of the amino acid sequence encoded by exon 1a and exon 1b is shown in Fig. 2B. Most notably, the E2F3b protein lacks the N-terminal extension found in the E2F1, E2F2, and E2F3a proteins. Otherwise, the two E2F3 proteins are identical in sequence, which includes the DNA binding domain, a domain involved in dimerization with the DP1 partner protein, and the transcription activation-Rb binding domain. In addition, both proteins share the nuclear localization sequence that is encoded at the beginning of exon 2. Thus, the two proteins would be predicted to bind to identical DNA sequences and partner with DP1 equivalently, as well as to interact with Rb. Moreover, both proteins would have the intrinsic ability to localize to the nucleus. The proteins differ in the N terminus in that E2F3a includes the potential cyclin A binding domain characterized in E2F1 (3, 15, 16), whereas a portion of the putative domain would be lost in E2F3b. Moreover, the ubiquitin targeting domain described in E2F1 (19) would also be conserved in E2F3a but not in E2F3b.
Distinct promoters control the expression of E2F3a and E2F3b.
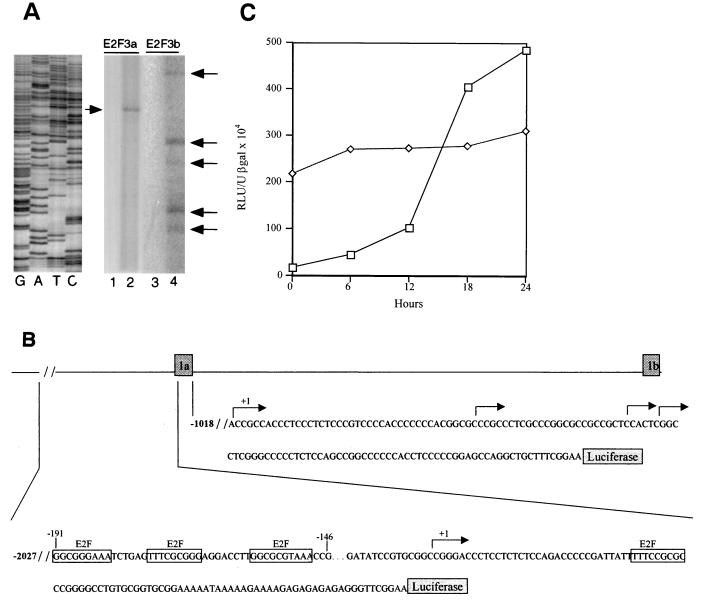
Using RACE as well as primer extension analysis, the sequence corresponding to the 5′ end of E2F3a and E2F3b mRNAs was identified (Fig. 3A and B). An examination of the DNA sequence flanking the E2F3a transcription start site as well as the E2F3b transcription start site revealed the presence of a number of potential binding sites for a variety of transcription factors. Most notably, the putative E2F3a promoter sequence contained multiple potential E2F binding elements in the 5′-flanking region of the E2F3a gene. There are three E2F recognition sites found in the upstream promoter and one site found in the 5′ UTR. Like many other E2F-regulated genes, there is an Sp1 recognition sequence very close to the E2F binding sites. In addition, we noted the presence of E-box elements that represent potential Myc binding sites. In contrast, although an analysis of the E2F3b promoter sequence revealed the presence of potential binding sites for several transcription factors, there were no sites for E2F nor Myc.
FIG. 3.
Cell cycle regulation of the E2F3a and E2F3b promoters. (A) Mapping of E2F3a and E2F3b transcription initiation sites by primer extension analysis. Primers specific for the E2F3a and the E2F3b transcripts (see Materials and Methods) were used with RNA from MEF arrested at G1/S. The major E2F3a extension product (lane 2) and the E2F3b extension products (lane 4) are indicated by arrows. Control reactions lacking MEF RNA are shown in lanes 1 and 3. The size of the primer extension products was compared to a set of DNA sequencing reactions as shown on the left. (B) Genomic organization of the E2F3a and E2F3b promoters. The genomic region including exons 1a and 1b is shown together with DNA sequence immediately upstream of each exon. The positions of transcription start sites, as inferred from the primer extension mapping, are indicated by arrows. In each case, the +1 site represents the longest primer extension product. The site of fusion of each promoter fragment to the luciferase reporter is shown as well as the E2F recognition sites in the E2F3a upstream sequence. (C) Analysis of E2F3a (□) and E2F3b (◊) promoter activity following the stimulation of cell growth. REF52 cells were transfected with 4 μg of E2F3a-luciferase or E2F3b-luciferase, together with 2 μg of the CMV–β-galactosidase vector as an internal control. Transfected cells were brought to quiescence by serum starvation for 48 h and then stimulated by the addition of fresh medium with serum. Samples were taken at the indicated times and assayed for luciferase and β-galactosidase activity. Luciferase activity was normalized to β-galactosidase activity.
To analyze the functional capacity of the sequences flanking exons 1a and 1b, the DNA fragments were placed in an expression vector utilizing the luciferase gene as a reporter. Each was then transfected into growing REF52 cells; the cells were then growth arrested by serum starvation and stimulated to reenter the cell cycle by the addition of serum, and then extracts were prepared every 4 h after serum addition and assayed for luciferase activity, which was normalized to a β-galactosidase internal control. As shown by the data in Fig. 3C, the two promoter constructs displayed very different properties. E2F3a promoter activity was low in quiescent and early-G1 cells and then increased in activity following serum stimulation; the activity in late G1 cells was approximately 15-fold higher than that in quiescent cells. This pattern of promoter activity closely parallels the accumulation of the E2F3a RNA as seen in the Northern blot analysis of RNA from serum stimulated cells (Fig. 1).
In contrast to the pattern of activity of the E2F3a promoter, the E2F3b promoter was active in quiescent cells and the activity of the promoter remained constant throughout the cell cycle (Fig. 3C), again directly reflecting the pattern of accumulation of the E2F3b RNA as seen in the Northern analysis (Fig. 1).
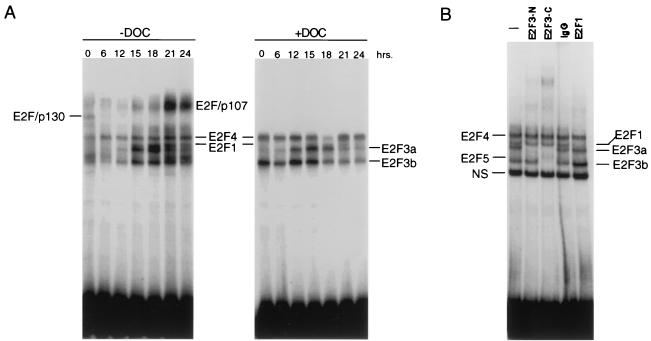
E2F3b DNA binding activity does not fluctuate following stimulation of cell growth.
In addition to the distinct transcription control for the two E2F3 products, the lack of N-terminal sequences in the E2F3b protein, which at least for E2F1 are important for controlling the accumulation of the protein and DNA binding activity during the cell cycle, would predict that the two E2F3 proteins might exhibit distinct patterns of accumulation. Indeed, direct assays for E2F activity in extracts of quiescent cells and cells stimulated to proliferate revealed a distinct pattern for the two activities. Similar to observations of previous experiments, E2F activity can be seen to fluctuate in response to stimulation of cell growth (Fig. 4A). This includes the loss of the p130 complex, the appearance of a p107 complex, and the accumulation of various E2F activities. Treatment of the extracts with deoxycholate eliminated the p130 complex, as well as the p107 complex, and allowed a better analysis of the total E2F activity. In addition to the E2F4 activity, it was also clear that DOC treatment led to the release of substantial E2F3b activity, particularly from the quiescent cells sample (Fig. 4A). The identity of the E2F bands was confirmed by antibody supershift assays using an extract from G1/S cells and antibodies specific for either the N terminus or the C terminus of E2F3 (Fig. 4B). Thus, the pattern of E2F activities that can be observed independent of association with Rb family proteins includes constant amounts of E2F4 and E2F3b as well as the regulated accumulation of E2F3a and E2F1.
FIG. 4.
Control of E2F3 DNA binding activity following stimulation of cell growth. (A) E2F DNA binding activity following stimulation of cell growth. REF52 cells were brought to quiescence and then stimulated by the addition of medium containing 10% serum. Cells were harvested at the indicated times following the stimulation of growth, nuclear extracts were prepared, and E2F DNA binding activity was measured using electrophoretic mobility shift assays as previously described. A sample of each nuclear extract was also treated with DOC and then analyzed for E2F DNA binding activity. (B) Identification of distinct E2F3 activities. A nuclear extract sample from the 18-h time point presented in the left panel (G1/S sample) was incubated without antibody (lane 1), an E2F3 antibody specific for the N terminus of E2F3a (lane 2), and E2F3 C-terminus-specific antibody that recognizes both E2F3a and E2F3b (lane 3), control IgG (lane 4), or an E2F1-specific antibody prior to being subjected to gel shift analysis. The E2F4- and E2F5-specific bands indicated on the DNA gel shift have been identified using specific antibodies against the respective proteins (data not shown).
The E2F3b protein is a specific partner for Rb in quiescent cells.
Given the distinct pattern of expression of the two forms of E2F3, together with the structural differences in the two proteins, we have looked for properties of the two proteins that might suggest functional differences. Clearly, one distinction is the pattern of expression; the E2F3b transcript is produced in quiescent cells, whereas the E2F3a transcript is only produced in proliferating cells. Previous work has detailed the cell-cycle-regulated accumulation of E2F3 DNA binding activity that reflects that of the E2F3a protein (18). We have now examined the state of the E2F3b protein in quiescent cells as an approach to understanding its function.
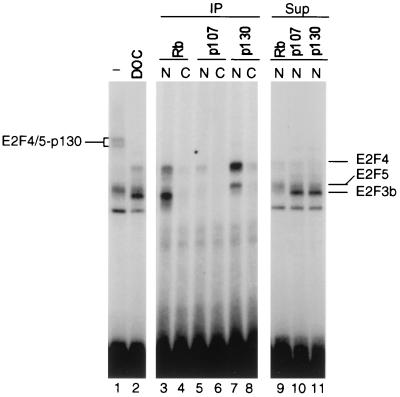
To determine the nature of the interactions of the E2F3b protein with respect to Rb family members, we used immunoprecipitation assays as shown in Fig. 5. Samples of a quiescent cell extract were immunoprecipitated under nondenaturing conditions with antibodies specific to Rb, p107, or p130, and then E2F activity was released by treatment with DOC. DNA binding assays of the DOC-released material revealed an association of Rb with the E2F3b protein as well as small amounts of E2F4 (lane 3). The fact that there was little or no E2F-Rb complex evident in the direct gel shift assay, in spite of the evidence for an association of E2F3b with Rb by immunoprecipitation and DOC release, likely reflects a reduced DNA binding affinity of the E2F-Rb relative to the free E2F protein. In contrast to the association of Rb with E2F3b, p130 was exclusively associated with E2F4 and E2F5 (lane 7) with no evidence for an interaction with E2F3b. The small amount of p107 found in quiescent cells was also associated with E2F4 and E2F5 but not with E2F3b (lane 5). The evident specificity of these interactions was further strengthened by the observations that the E2F3b protein remained in the supernatants of the p107 or p130 immunoprecipitates (lanes 10 and 11) and that essentially no E2F3b protein remained in the supernatant of the Rb immunoprecipitate (lanes 9 to 11). Based on these results, we conclude that the E2F3b protein is the predominant partner for Rb in quiescent cells, whereas E2F4 and E2F5 are the specific partners for p130 as well as the small amounts of p107 found in quiescent cells.
FIG. 5.
E2F3b is specifically associated with Rb in quiescent cells. Nuclear (N) and cytoplasmic (C) extracts prepared from quiescent cells were immunoprecipitated with antibodies specific for the indicated Rb family member protein. The immunoprecipitates (IP) were treated with DOC to release the associated E2F activities as described in Materials and Methods. The released material was assayed for E2F DNA binding activity by gel mobility shift (lanes 3 to 8). In addition, the supernatants (Sup) from the immunoprecipitations were also assayed for E2F activity (lanes 9 to 11). Finally, a sample of the nuclear extract was directly treated with DOC and then assayed for E2F binding activity (lanes 1 and 2).
DISCUSSION
The study of cell proliferation control has demonstrated the critical role for E2F transcriptional activity in the regulation of various genes that encode proteins essential for DNA replication and cell cycle regulation. These studies have also detailed the complexity of E2F and Rb activity, defining six distinct members of the E2F family of proteins and three members of the Rb family; further work has begun to define functional specificity of this complexity, with unique roles played by different E2F family members (4, 22). The results we describe here now provide evidence for further complexity whereby the E2F3 locus is seen to generate two distinct products through the utilization of distinct promoters. This transcriptional control of E2F3 expression results in the production of related proteins that are expressed with very different kinetics during the course of cell proliferation. Most importantly, the association of the E2F3b protein with Rb in quiescent cells suggests a basis for specificity in the repression of transcription that is evident from the analysis of cells deficient in each of the Rb family members (11).
The E2F3 locus provides further complexity to the E2F family.
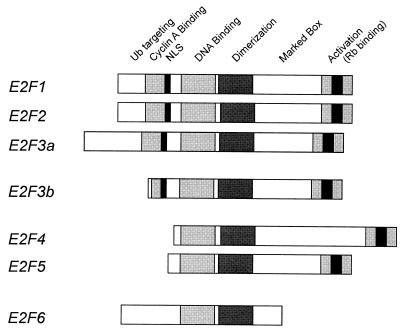
The identification of the E2F3b protein as an alternative product of the E2F3 locus provides further insight into the evolution of the structural and functional complexity of the E2F family (Fig. 6). Like E2F1, E2F2, and E2F3a, E2F3b does not interact with either p130 or p107; rather, E2F3b interacts specifically with Rb. Unlike E2F1, E2F2, and E2F3a, the E2F3b product is constitutively expressed, accumulating in both quiescent and proliferating cells. Thus, in this regard, E2F3b shares properties in common with the E2F4 and E2F5 products. In addition, the E2F3b protein lacks the N-terminal extension that is found in the E2F1, E2F2, and E2F3a proteins. Although an understanding of the precise role of these N-terminal amino acids in the function of these proteins is still developing, several observations are pertinent. First, the ability of cyclin A-cdk2 to control the DNA binding activity of the E2F1 protein is dependent on the interaction of the kinase with a motif found in these N-terminal sequences (3, 15, 16). Second, the ubiquitin-mediated degradation of E2F1 is dependent on sequences found in the N terminus of E2F1 (19). Taken together, these observations suggest that because of the absence of these regulatory elements found in E2F1, E2F2, and E2F3a, the accumulation of E2F3b is regulated quite differently from these three E2F proteins, whose accumulation is tightly governed by cell proliferation control.
FIG. 6.
Relationship of E2F3b to the E2F family. Schematic representation of the entire E2F family with conserved domains indicated.
A unique role for E2F3 activity in cell growth control.
Several observations seem particularly relevant when considering the functional role of the E2F3b product. Of perhaps most significance are the observations of Dyson and colleagues that define a specificity in transcriptional repression in quiescent cells mediated by Rb family proteins (11). Whereas cyclin E and p107 transcription was derepressed in cells lacking Rb, there was no effect on the expression of various other E2F targets. In contrast, the combined loss of p107 and p130 led to derepression of transcription of several other E2F target genes but not of cyclin E or p107. As such, these results provide evidence for specificity in the action of Rb and p130 in transcription repression. Since the specificity of promoter selection in Rb-mediated repression most likely must reside in the DNA binding component of the Rb family member complex, namely, the associated E2F protein, our observation that Rb and p130 exhibit distinct E2F binding specificity in quiescent cells now provides a mechanism by which to consider the specificity of repression.
Previous work has suggested that individual E2F proteins may exhibit DNA binding specificity (29); thus, the specificity of Rb-mediated versus p130-mediated repression could indeed reflect the ability of distinct E2F proteins, in this case E2F3b versus E2F4, to select promoter sequences. In this regard, it is interesting to note that the cyclin E gene has been suggested to be a target for positive control by E2F3a in cycling cells (18), an observation of interest considering the fact that E2F3a and E2F3b share the identical DNA binding domain. Given the fact that E2F3b is the predominant partner for Rb in quiescent cells, together with the fact that it is the E2F protein that imparts DNA sequence specificity to the Rb repression, we suggest that E2F3b is likely involved in the process of controlling the expression of cyclin E, and other targets specifically repressed by Rb, in the quiescent cell.
ACKNOWLEDGMENTS
We thank Kaye Culler for assistance in the preparation of the manuscript.
G.L. was supported by a fellowship from the Medical Research Council of Canada, S.I. and R.S. were supported by the Howard Hughes Medical Institute, and M.A. was supported by a Merck/UNCF fellowship. This work was supported by the Howard Hughes Medical Institute and by a V Foundation grant to G.L.
REFERENCES
- 1.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis and G1/S regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dynlacht B D, Moberg K, Lees J A, Harlow E, Zhu L. Specific regulation of E2F family members by cyclin-dependent kinases. Mol Cell Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dyson N. The regulation of E2f by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 5.George C P, Kadonaga J T. Primer extension analysis of RNA. In: Krieg P A, editor. A laboratory guide to RNA isolation, analysis, and synthesis. New York, N.Y: Wiley-Liss, Inc.; 1996. pp. 133–139. [Google Scholar]
- 6.Helin K, Harlow E. The retinoblastoma protein as a transcriptional repressor. Trends Cell Biol. 1993;3:43–46. doi: 10.1016/0962-8924(93)90150-y. [DOI] [PubMed] [Google Scholar]
- 7.Herrera R E, Sah V P, Williams B O, Makela T P, Weinberg R A, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsiao K-M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 9.Hunter T. Braking the cycle. Cell. 1993;75:839–841. doi: 10.1016/0092-8674(93)90528-x. [DOI] [PubMed] [Google Scholar]
- 10.Hunter T, Pines J. Cyclins and cancer II: cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 11.Hurford R K, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda M-A, Jakoi L, Nevins J R. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 14.Kowalik T F, DeGregori J, Schwarz J K, Nevins J R. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W G, Livingston D M. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 16.Krek W, Xu G, Livingston D M. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 17.La Thangue N B. DRTF1/E2F: an expanding family of heterodimeric transcription factors implicated in cell-cycle control. Trends Biochem Sci. 1994;19:108–114. doi: 10.1016/0968-0004(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 18.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between SCFSKP2 ubiquitin protein ligase and E2F-1 underlies regulation of E2F-1 degradation. Nat Cell Biol. 1999;1:14–19. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- 20.Neuman E, Flemington E K, Sellers W R, Kaelin W G., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 22.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 23.Qin X-Q, Livingston D M, Kaelin W G, Adams P D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sears R, Ohtani K, Nevins J R. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shan B, Lee W-H. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherr C J. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 27.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 28.Smith E J, Leone G, DeGregori J, Jakoi L, Nevins J R. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 from a G1 cell state. Mol Cell Biol. 1996;16:6965–6976. doi: 10.1128/mcb.16.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao Y, Kassatly R F, Cress W D, Horowitz J M. Subunit composition determines E2F DNA-binding site specificity. Mol Cell Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 31.Wu X, Levine A J. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]