ABSTRACT
Coccidioides immitis and Coccidioides posadasii are causative agents of Valley fever, a serious fungal disease endemic to regions with hot, arid climate in the United States, Mexico, and Central and South America. The environmental niche of Coccidioides spp. is not well defined, and it remains unknown whether these fungi are primarily associated with rodents or grow as saprotrophs in soil. To better understand the environmental reservoir of these pathogens, we used a systematic soil sampling approach, quantitative PCR (qPCR), culture, whole-genome sequencing, and soil chemical analysis to identify factors associated with the presence of C. immitis at a known colonization site in Washington State linked to a human case in 2010. We found that the same strain colonized an area of over 46,000 m2 and persisted in soil for over 6 years. No association with rodent burrows was observed, as C. immitis DNA was as likely to be detected inside rodent holes as it was in the surrounding soil. In addition, the presence of C. immitis DNA in soil was correlated with elevated levels of boron, calcium, magnesium, sodium, and silicon in soil leachates. We also observed differences in the microbial communities between C. immitis-positive and -negative soils. Our artificial soil inoculation experiments demonstrated that C. immitis can use soil as a sole source of nutrients. Taken together, these results suggest that soil parameters need to be considered when modeling the distribution of this fungus in the environment.
IMPORTANCE Coccidioidomycosis is considered a highly endemic disease for which geographic range is likely to expand from climate change. A better understanding of the ecological niche of Coccidioides spp. is essential for generating accurate distribution maps and predicting future changes in response to the changing environment. Our study used a systematic sampling strategy, advanced molecular detection methods, and soil chemical analysis to identify environmental factors associated with the presence of C. immitis in soil. Our results demonstrate the fungus can colonize the same areas for years and is associated with chemical and microbiological soil characteristics. Our results suggest that in addition to climate parameters, soil characteristics need to be considered when building habitat distribution models for this pathogen.
KEYWORDS: Valley fever, Coccidioides, coccidioidomycosis
INTRODUCTION
Coccidioidomycosis, or Valley fever, is a serious fungal disease that affects tens of thousands of people in the United States each year and is also prevalent in Mexico and Central and South America (1, 2). Most cases are caused by the inhalation of airborne spores (arthroconidia) of the two closely related species of soil-dwelling fungi, Coccidioides immitis and Coccidioides posadasii. Although in approximately 60% of cases, patients are asymptomatic, the remaining 40% of cases experience symptoms that range from flu-like illness to life-threatening disseminated infection and meningitis (2). Unlike most other fungal pathogens that primarily affect immunocompromised patients, Coccidioides spp. can infect people with a healthy immune system (1). In addition, this fungus can infect dogs, cats, horses, camelids, rodents, and many other mammals (3).
In the United States, the vast majority of coccidioidomycosis occurs in Arizona and California’s Central Valley (1). Apart from these two states, the disease is endemic in parts of Texas, New Mexico, Nevada, and Utah (2). Over the past decade, locally acquired cases of coccidioidomycosis have been identified in south central Washington State, where the fungus was also isolated from soil, and the disease is now recognized as endemic (4, 5). Maps that accurately depict the geographic distribution of Coccidioides spp. are essential to promote awareness about this disease among health care providers and the public. Accurate maps provide a basis for assessing potential risk for exposures to Coccidioides spp. via natural processes and human activities that generate dusts containing the fungus. Additionally, these maps help promote timely diagnosis and treatment of coccidioidomycosis.
The first maps describing the distribution of coccidioidomycosis were constructed using results from skin testing performed in the1940 to 1950s (6). The U.S. Centers for Disease Control and Prevention (CDC) has since augmented these maps with data from the state public health surveillance programs so that newly identified areas of endemicity in Washington and Utah are included (https://www.cdc.gov/fungal/diseases/coccidioidomycosis/maps.html#approximate). However, accurately identifying and mapping areas with low prevalence of coccidioidomycosis remains challenging for the following reasons: (i) health care providers in low-prevalence areas often do not suspect, nor accurately diagnose, the disease; (ii) it is difficult to distinguish between locally acquired and travel-associated coccidioidomycosis cases, as the disease may develop months and even years after the exposure; and (iii) most people diagnosed with coccidioidomycosis have travel history to areas of endemicity (2, 7).
Environmental habitat modeling provides a useful tool for mapping the current distribution of Coccidioides spp. and predicting its future change in response to the changing environment. Several models have been developed using annual temperature, rainfall, vegetation cover, soil texture, soil moisture, salinity, pH, geochemical composition, and other parameters as the main variables governing the distribution of this fungus (8–13). Mean annual temperatures, precipitation patterns, and vegetation cover were shown to be most frequently associated with the Coccidioides species habitat (8–11). In addition, Dobos et al. incorporated soil salinity into a predictive habitat model that was able to predict areas in Washington State where C. immitis was found in soil (12). Ocampo-Chavira et al. found an overlap between Coccidioides spp. and the distribution of the desert woodrat, Neotoma lepida (13). However, the ecological niche of Coccidioides spp. is not yet fully established, and there is an ongoing debate in the medical mycology community whether these fungi can grow as saprotrophs in soil or must depend on animals and their habitats for survival (14–16). The answer to this question is important for creating accurate habitat distributions models: if Coccidioides spp. persist in soil, then geospatial, physical, and chemical characteristics of soil likely govern its distribution in the environment; however, if these fungi depend on animal hosts for survival and propagation, then their distribution is likely driven by the distribution of the host species.
The role of small animals in the life cycle of Coccidioides spp. was proposed in 1942 by Emmons (17). Although this hypothesis fell out of favor by the end of the 1960s after several authors observed correlations between the prevalence of Coccidioides species and soil characteristics (18), interest in this hypothesis was revived in 2009 after genomic analysis identified several unique features of the Coccidioides species genome (19). This analysis demonstrated that these fungi, in contrast to other soil-dwelling fungi, have expanded gene families of proteinase and lipase enzymes needed for digestion of animal tissues. Additionally, the analysis observed only a few cellulase and pectinase genes, which are needed for the degradation of plant materials (19); similar genomic features were recently identified in other dimorphic pathogenic fungi (20). The hypothesis involving rodents also provides a plausible explanation for the uneven distribution of Coccidioides spp. in the areas of endemicity observed by earlier investigators using culture (21), and it explains the affinity of this fungus for animal burrows (14).
In this study, we used a systematic sampling approach, molecular detection, and soil chemical analysis to identify factors associated with the presence of C. immitis. The study was conducted at a known positive site in Washington State, where a patient was infected by subcutaneous inoculation when falling from an all-terrain vehicle (ATV) (4, 22). Live C. immitis was isolated from this site and confirmed to be identical to the patient’s isolate by whole-genome sequencing. This sampling approach was designed to test specific hypotheses about factors that control distributions of C. immitis.
RESULTS
Spatial distribution of the Coccidioides sp. DNA detected by nested qPCR.
We collected 278 soil samples along 9 transects radiating from the center, T1 to T9, which was selected at the site of a 2010 ATV accident that led to coccidioidomycosis (22). Because at the time of sampling, the breadth of C. immitis colonization at the site was unknown, the first 74 samples were collected at 1-m intervals within 4 m from the center, and the remaining samples were collected at 10-m intervals within 100 m from the center. In addition to regular soils, rodent burrows were sampled each time they were encountered along the transects. Coccidioides species DNA was detected by single-tube (ST) nested quantitative PCR (qPCR) in 70 (25%) of the tested 278 samples (Fig. 1). Of those, 39 (56%) were located within 4 m of the center (Fig. S2 in the supplemental material), 63 (90%) were within 50 m of the center, and 7 (10%) were >50 m from the center (Fig. 1). Detection of Coccidioides species DNA was associated with shorter distance from the center (median, 3.5 m to the center for Coccidioides-positive samples versus 50 m in Coccidioides-negative samples, P < 0.001) and ATV tracks (66% for Coccidioides-positive versus 52% for Coccidioides-negative samples, P = 0.045) (Table 1). Detection was negatively associated with vegetation (34% for Coccidioides-positive versus 52% for Coccidioides-negative samples, P = 0.011). Of 278 samples, 27 (10%) were collected from rodent burrows, and the proportion of positive samples from rodent burrows was not statistically different between the Coccidioides-positive and -negative samples (both 10%, P = 0.952). Of seven positive samples collected >50 m from the center, three were from animal burrows, and four were from regular soils. No other associations were detected between Coccidioides species DNA and an observed characteristic (specific transect, pH, elevation, and the presence of an animal burrow).
FIG 1.
Distribution of C. immitis along the nine transects. Study site where a combination of a radial and an interrupted belt transect method was used. Nine 100-m transects were sampled in 10-m intervals; squares represent 1-m2 plots (left and right), and circles represent rodent burrows within or near 1-m2 plots. Transects are labeled T1 to T9. Plots and rodent burrows where samples were qPCR positive for Coccidioides species DNA are shown in red. Plots that were qPCR positive and from which C. immitis isolates were obtained are marked with blue marks. Examples of vegetation encountered in different areas of the transect are shown in Fig. S1 in the supplemental material.
TABLE 1.
Characteristics of soil samples by C. immitis colonization (n = 278)
| Characteristic | Data for: |
P valuec | ||
|---|---|---|---|---|
| All samples | Coccidioides-positive samples | Coccidioides-negative samples | ||
| Total (no. ([%]) | 278 | 70 (25) | 208 (75) | |
| Transect (median [IQR])b | 5 (3–7) | 6 (3–7) | 5 (3–7) | 0.380 |
| Distance (median [IQR] [m]) | 40 (4–70) | 3.5 (2–20) | 50 (20–80) | <0.001 |
| Altitude (median [IQR] [ft]) | 360 (353–364) | 362 (356–365) | 360 (351–364) | 0.095 |
| Rodent burrows (no. [%])a | 27 (10) | 7 (10) | 20 (10) | 0.952 |
| Leachate pH (median [IQR]) | 7.4 (7.0–7.9) | 7.4 (6.9–7.9) | 7.4 (7.0–7.8) | 0.762 |
| Vegetation (no. [%])a | 132 (47) | 24 (34) | 108 (52) | 0.011 |
| ATV track (no. [%])a | 154 (55) | 46 (66) | 108 (52) | 0.045 |
Data indicate no. of samples taken from type of site and proportion of samples out of total.
IQR, interquartile range.
Bold indicates a P value of <0.05.
Distribution of the Coccidioides sp. within 10 m from the center of the transects. Center is shown with a large red circle; squares represent 1-m2 plots (left and right), and small circles represent rodent burrows within or near 1-m2 plots. Distances (in meters) are labeled in yellow. Plots and rodent burrows where samples were qPCR positive for Coccidioides species DNA are shown in red. This figure is a graphical representation; transects and sampled areas are not to scale. Download FIG S2, PDF file, 0.09 MB (90.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Types of vegetation encountered at the study site. (A and B) ATV tracks located at the center and east of the center. (C and D) Low grass area in the north. (E) Dry ravine. (F) Marsh area in the south. Download FIG S1, PDF file, 0.4 MB (400.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Isolation and whole-genome sequencing of C. immitis from soil.
Four isolates were recovered from soil samples A391 (90 m, T3), A432 (10 m, T5), and A502 (30 m, T7) (Fig. 1). Two isolates were recovered from A502. Single-nucleotide polymorphism (SNP) analysis demonstrated that genomes of these isolates were nearly identical to those of isolates recovered in 2014 (B11001), those from soil collected in 2010 (B10992 and B10996), and the original clinical isolate from the patient, B10637 (Fig. 2). The range of pairwise SNP differences between any environmental or clinical isolate recovered from this site was 0 to 6 SNPs. The largest difference, 6 SNPs, was observed between genomes of A391 and A502 isolates from this study. B10637, the clinical isolate from 2010, differed from all environmental isolates from this site by ≤4 SNPs. Other clinical isolates from this clade (referred to as the Washington clade) that were obtained from patients who did not have exposure to this site differed by >200 SNPs, while genomes of C. immitis from California differed by >10,000 SNPs (49).
FIG 2.
Genetic relationships among C. immitis isolates recovered from 2010 to 2016 from the study site. Isolates in red are from samples collected during the survey. B10637 is from the clinical specimen of the patient infected in the ATV accident in 2010. B10992 and B10996 are environmental isolates recovered in 2013 from soils collected in 2010 at the site of the accident. B11001 is an environmental isolate recovered in 2014 from the same site. B11034 and B13956 are from other locally acquired cases in Washington (49). Isolates from this study are in red. Remaining isolates are from California coccidioidomycosis cases. The tree scale is in SNPs.
Analysis of soil leachate characteristics by analytical chemistry.
To identify soil leachate characteristics associated with C. immitis colonization, we selected 207 soil samples. Except for a single sample selected from the center of the study site, all samples were collected at 10- to 100-m distances from the center. The filtered deionized water leachates obtained from these samples were tested by ion chromatography for detection of common anions, dissolved organic carbon (DOC), and inductively coupled plasma optical emission spectrometry (ICP-OES) and inductively coupled plasma mass spectrometry (ICP-MS) for detection of trace elements. The full leachate data are tabulated in the USGS ScienceBase data catalog (23). The leachates had near-neutral pH (7.0 to 7.9), with nitrate the dominant anion in most samples (1.3 to 4.8 mg/liter) and calcium (2,160 to 5,750 μg/liter) and potassium (2,150 to 5,750 μg/liter) the dominant cations. DOC was measured at levels from 3.5 to 8 mg/liter. A relative standard deviation (RSD) of up to 50% between duplicate samples was found for anions and DOC. The levels of cations and anions measured in this study are substantially lower than those measured for water leachates of soils collected from transects across the United States and Canada (24) due to the substantially longer extraction times (up to 20 h) used by the authors.
The presence of the Coccidioides sp. DNA was statistically associated with elevated concentrations of boron (B), calcium (Ca), magnesium (Mg), sodium (Na), and silicon (Si) in soil leachates but was not associated with other anions and DOC (Table 2).
TABLE 2.
Data for select water-leachable chemical constituents by the Coccidioides sp. DNA
| Chemical | No. of samples | Median (interquartile range [IQR]) of: |
P valuea | |
|---|---|---|---|---|
| Coccidioides-positive samples | Coccidioides-negative samples | |||
| Nitrate (mg/liter) | 207 | 2.12 (1.28–4.04) | 2.79 (1.52–4.84) | 0.143 |
| Dissolved organic C (mg/liter) | 207 | 4.35 (3.55–8.03) | 4.12 (3.64–7.08) | 0.616 |
| B (μg/liter) | 97 | 8 (6–12) | 6 (6–8) | 0.027 |
| Ca (μg/liter) | 205 | 4,460 (2,950–5,040) | 3,130 (1,620–4,380) | 0.003 |
| Fe (μg/liter) | 147 | 54 (29–99) | 45 (31–101) | 0.670 |
| K (μg/liter) | 205 | 3,970 (2,150–5,610) | 4,220 (3,310–5,750) | 0.085 |
| Mg (μg/liter) | 205 | 655 (509–973) | 475 (316–630) | <0.001 |
| Na (μg/liter) | 205 | 496 (338–1,610) | 293 (186–528) | 0.002 |
| S (μg/liter) | 202 | 196 (138–351) | 167.5 (118–244.5) | 0.135 |
| Si (μg/liter) | 205 | 944 (865–1,130) | 830 (727–997) | <0.001 |
Bold indicates a P value of <0.05.
Targeted amplified sequencing of ITS.
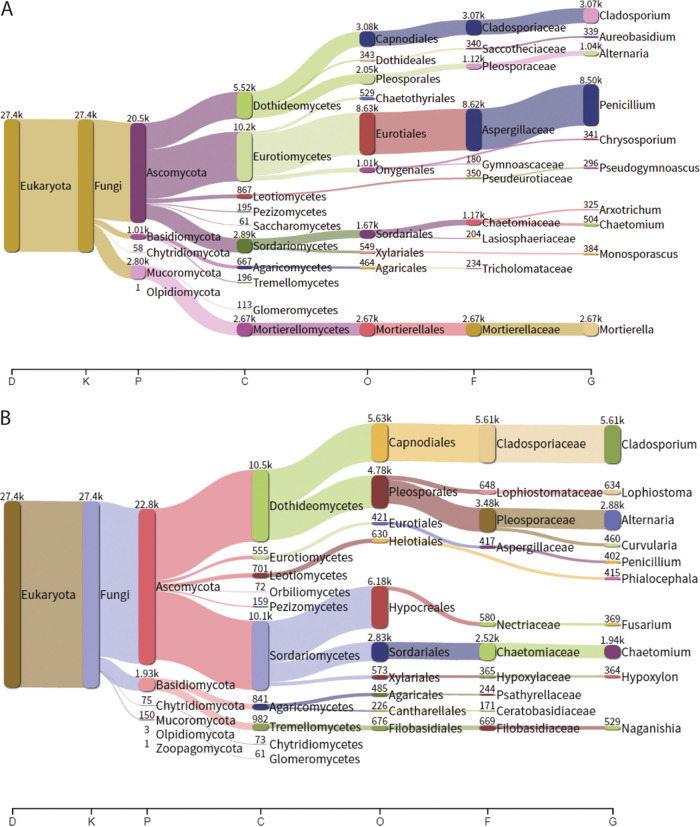
To provide an independent confirmation for the qPCR results and to identify fungal genera associated with the presence of the C. immitis in soil, we performed internal transcribed spacer 1 (ITS1) amplicon sequencing. Soils positive for the Coccidioides sp. by qPCR results were taxonomically classified in batch using Kraken 2 (Fig. 3A) as well as soils negative for the Coccidioides sp. by qPCR (Fig. 3B). For soils positive for the Coccidioides sp. by qPCR, 0.28% of reads were classified as the Coccidioides sp. by Kraken 2 in contrast to 0.08% of Coccidioides species reads in soils negative by qPCR. The predominant fungal phyla in soils positive for a Coccidioides sp. were Ascomycota (84.09%), Mucoromycota (11.51%), and Basidiomycota (4.16%). Meanwhile, the most abundant phyla in soils negative for a Coccidioides sp. were Ascomycota (91.34%) and Basidiomycota (7.74%). Characterizing the fungal community further, the most abundant families in soils positive for a Coccidioides sp. were Aspergillaceae (42.84%), Cladosporiaceae (15.23%), Mortierellaceae (13.26%), Chaetomiaceae (5.80%), and Pleosporaceae (5.56%). In contrast, the most abundant families in soils negative for the Coccidioides sp. were Cladosporiaceae (32.78%), Pleosporaceae (20.34%), and Chaetomiaceae (14.7%). Furthermore, the dominant fungal genera in Coccidioides species-positive soils were Penicillium (40.09%), Cladosporium (14.46%), Mortierella (12.58), Alternaria (4.89%), and Chaetomium (2.38%). Dominant fungal genera in soils negative for the Coccidioides sp. were Cladosporium (32.04%), Alternaria (16.44%), Chaetomium (11.09%), Lophiostoma (3.62%), Naganishia (3.02%), and Curvularia (2.63%). In addition, the bivariate analysis identified that a soil saprotroph, Aureobasidium, was positively associated with the presence Coccidioides species DNA in soil (P < 0.01), while a plant endophyte, Phialocephala, was negatively associated with Coccidioides sp. (P < 0.01) (Table S1).
FIG 3.
Fungal community for soils positive and negative for C. immitis. The fungal community of soils positive (A) and negative (B) for Coccidioides sp. DNA by qPCR was assessed by ITS1 amplicon sequencing.
Sample distribution (no. [%]) by fungi type and Coccidioides status in 280 soil samples. *, UNITE database-assigned ITS sequence to genus Paracoccidioides endemic to South America; this sequence likely belongs to another member of Ajellomycetaceae. **, Bold indicates a P value of <0.05. Download Table S1, DOCX file, 0.02 MB (17.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Propagation of C. immitis in soil under laboratory conditions.
To test whether C. immitis can propagate with soil as the sole source of nutrients, soil samples from a residential backyard in Atlanta, Georgia, were examined. The backyard had no signs of rodent activity. Samples were inoculated with ∼500 arthroconidia, and C. immitis DNA levels were monitored for 6 weeks. In sterile soil, the estimated number of C. immitis cells increased 200 times within 1 week after inoculation and remained at the level of 10,000 to 100,000 cells per gram of soil for the remainder of the 6-week duration of the experiment, indicating active proliferation (Fig. 4). In nonsterile (native) soil, the estimated concentration of arthroconidia decreased 25 times after 1 week and diminished below the qPCR detection level 4 weeks after the inoculation (Fig. 4).
FIG 4.
Propagation of C. immitis B10992 strain in soil. Sterile (gray) and native (black) soil samples were inoculated with C. immitis arthroconidia and monitored for 6 weeks during incubation. DNA levels were measured by qPCR analysis.
DISCUSSION
We used a systematic environmental sampling approach, advanced molecular detection methods, culture, analytical chemistry, and whole-genome sequencing to describe the distribution of C. immitis at the known colonization site in Washington State. Our results demonstrated that a single strain of C. immitis colonized an area of over 46,000 m2 and persisted in soil for over 6 years. We tested different environmental factors and identified chemical and microbiological signatures associated with the presence of C. immitis DNA in soil. Our results were consistent with those reported in earlier studies, which described uneven distribution of Coccidioides species in the environment (21) and persistence of these fungi at certain sites for years (25). Additionally, these results suggested that soil chemistry and microbial compositions affect the distribution of C. immitis in soils (26, 27).
To better understand the distribution of C. immitis at the study site, we used an unbiased sampling method which has been developed by ecologists for maximizing the utility of collecting samples for the subsequent statistical analysis (28). To maximize our chances of capturing both positive and negative samples, the study was conducted at the site in Washington State, where C. immitis was previously detected in soil and linked to human infection (22). To ensure accuracy of our results, we used three different methods for detection of the Coccidioides sp. in soil. First, soil samples were screened with an ST-nested qPCR assay, which is a modification of the Cocci DX qPCR assay developed for clinical testing (29). ST-nested qPCR targets the same unique transposable element in the Coccidioides species genome as Cocci DX but uses a different set of nested qPCR primers to increase sensitivity. The performance of ST-nested qPCR is similar to that of CocciENV DX, which is another modification of the original Cocci DX assay (30, 31). Second, for the independent confirmation of ST-nested qPCR results, ITS1 regions of rRNA genes from all soil samples were amplified and sequenced. Similar to other investigators (15), we observed that ITS1 sequencing was not as sensitive as qPCR; however, qPCR-positive samples were five times more likely to be positive by ITS than the qPCR-negative samples. Because qPCR and ITS1 sequencing target different regions of the genome, finding samples positive by both assays bolstered confidence in these results. Furthermore, the highest number of Coccidioides species ITS1 reads was detected in a soil sample collected from a rodent burrow that had the lowest threshold cycle (CT) value in ST-nested qPCR (data not shown). Finally, we used a subset of positive qPCR soils for culture and were able to recover isolates of C. immitis from three samples. Since culturing of Coccidioides spp. from soil is technically challenging and the recovery rate is extremely low (32), only a subset of positive soil samples was used for culture. Despite a small sample, the whole-genome sequencing data of isolates recovered from soil demonstrated near identity among isolates from sampling locations more than 100 m apart. Isolates from soils collected from 2010 to 2016 were no more than 6 SNPs different from each other and a clinical isolate recovered from a patient in 2010. These results suggest that C. immitis in the soil at this site has low genetic variability.
We observed a correlation between the presence of Coccidioides species DNA in a soil sample and distance from the center. However, both qPCR- and culture-positive samples were detected as far as 100 and 90 m from the center, respectively. The farthest qPCR-positive sample was identified 100 m from the center and was collected from a rodent burrow on T1; however, another sample, which was positive by qPCR and culture, was collected from a site without any rodent activity 90 m from the center. A significant association was observed between the presence of Coccidioides species DNA and ATV tracks and between Coccidioides species DNA and the lack of vegetation. However, both factors (ATV tracks and lack of vegetation) were also associated with distance, as the center of the sampling site had more ATV tracks and less vegetation. The vegetation at the site consisted of low grass and small deciduous trees and did not fit a profile of vegetation types described by others for Coccidioides species habitats (Fig. S1 in the supplemental material) (33). Positive samples were collected from the bare ground of ATV tracks and grass and from under trees. These results are consistent with the data presented by others who showed no association between Coccidioides spp. and a specific vegetation type (33).
Although Coccidioides species DNA was identified in several soil samples collected from rodent burrows, no statistically significant association between rodent habitats and C. immitis was detected at this site. Equal proportions of burrows and regular soil samples were positive for the Coccidioides sp.; however, the overall area occupied by the burrows likely constituted a smaller portion of the total ground on the grid; therefore, the rate/cm2 may be higher in burrows. Even on the periphery of the sampling area, Coccidioides species DNA was as likely to be detected inside the burrows as it was in the regular soil, suggesting that this fungus can colonize soil. Since all soil samples were collected at a depth of 15 to 20 cm, it is unlikely that the detection of the Coccidioides sp. in regular soil was the result of arthroconidia present on the surface.
Our soil inoculation experiments confirmed earlier observations that Coccidioides spp. can use soil as a sole source of nutrients (34). In the first week after inoculation of sterile soil, the amount of C. immitis DNA increased 200 times and remained at a level of 10,000 for100,000 arthroconidia per gram of soil for the duration of the experiment. However, 1 week after inoculation of nonsterile soil, the amount of C. immitis DNA decreased 10 times and dropped below the qPCR detection level 4 weeks after inoculation. The most likely reason for the decline of C. immitis in nonsterile soils was the competition with soil microbiota from Georgia soil, to which C. immitis is not adapted. Competition with soil microbes has been proposed as the main factor controlling the distribution of Coccidioides spp. in soil by many investigators (18, 26, 27, 35, 36). Since the study site had numerous rodent burrows, suggestive of widespread rodent activity, we could not exclude the possibility that soil outside burrows might have been contaminated with urine, feces, or other rodent substances. Therefore, to test whether C. immitis can use soil as a sole source of nutrients, we used soil from a site in Georgia without any evidence of rodent colonization. In the future, it would be informative to test the ability of C. immitis to grow in soil from the study site.
Our analysis identified significant associations between the presence of Coccidioides species DNA and increased concentrations of B, Ca, Mg, Na, and Si in qPCR-positive compared to qPCR-negative soil samples. These findings are consistent with those reported in earlier studies (27). Elconin et al. showed that C. immitis was more likely to be isolated from soils with higher concentrations of Ca, Na, Mg, and Cl (27). It has also been shown that Coccidioides spp. were frequently isolated from soils rich in boron (33). To our knowledge, increased Si concentration has not been previously shown to be associated with Coccidioides spp. Although most Si in soil is bound in a form of insoluble minerals, such as crystalline silica (quartz) and aluminosilicates (e.g., clays, feldspars), soluble Si is available in a form of silicate salts that are often bound to Na, Ca, Mg, and other cations, which may explain why Si was associated with the increased concentrations of cations in our study (38). It also has been proposed that the presence of silicate salts alters availability of the essential minerals such as K, Mb, and Mg to plants (38). The effect of silicates on soil microbial communities has not been investigated to our knowledge. The short extraction time used in this study, necessitated by the need to carry out the leaching procedure in a biosafety level 3 facility, led to several constituents in most samples being below the analytical detection limit. It may also have contributed to the excessive RSD for anions and DOC. In addition, the levels of cations and anions measured in this study were substantially lower than those measured for water leachates of soils collected from transects across the United States and Canada (24), most likely due to the substantially longer extraction times (up to 20 h) used by the authors. It would be useful in future studies to carry out longer extraction times.
Egeberg et al. proposed that higher salinity affects the soil microbial community, making it more amenable for Coccidioides spp., and they isolated several strains of bacteria and fungi that were antagonistic to C. immitis in culture (27). A recent study by Lauer et al. identified soil bacteria antagonistic to C. immitis (36). Our results identified differences in the microbial communities between C. immitis-positive and -negative soils. C. immitis was positively associated with fungi from genera Penicillium, Aureobasidium, and Mortierella, which are saprophytes frequently isolated from soil. At the same time, C. immitis was negatively correlated with fungi from genera Chaetomium, Curvularia, Alternaria, and Phialocephala, which are commonly associated with plants. These results were not surprising, as the presence of the Coccidioides sp. in these soils correlated with ATV tracks and lack of vegetation, and many fungal genera that were overrepresented in Coccidioides-negative soils are known to colonize plant rhizospheres, which suggests that the difference in mycobiomes between C. immitis-positive and -negative soils may be due to the difference in vegetation cover rather than presence or absence of the fungus. Overall, all factors associated with the presence of C. immitis in our study were also associated with distance from the sampling center, making it difficult to establish the causative relationships between these parameters and the Coccidioides sp. Our study was limited to a single C. immitis colonization site in Washington State. A recent study in Mojave Desert, California, reported that Coccidioides species DNA is more likely to be found in native undisturbed soils than cultivated soil but did not identify physical and chemical soil parameters associated with the presence of C. immitis, which may suggest that the associations observed in our study may be strain or site specific (37). Similar studies need to be conducted in other areas to determine whether observed relationships will hold for other locations colonized by other strains of C. immitis and C. posadasii. It is possible that the observed associations are site, strain, or species specific.
The debate about the role of animals and their habitats in the life cycle of Coccidioides has been ongoing for over 70 years and has been often framed as a competition of the two opposing hypotheses, one positing that Coccidioides spp. are soil saprotrophs and the other stating that they persist almost exclusively an animal pathogen (14, 35). Our results show that C. immitis can use soil as the sole source of nutrients. We also observed no association between the presence of C. immitis DNA and animal burrows and identified a potential chemical and microbiological soil signature associated with C. immitis colonization, which provide evidence for the saprotrophic lifestyle hypothesis. Contrary to our results, recent findings by Kolleth et al. demonstrated that C. posadasii, a sibling species found in Arizona, is more likely to be detected inside animal burrows than the topsoil and can use animal hair as the sole source of nutrients (15). The apparent dissimilarities between the findings of the two studies may be due to the differences between the two species or testing sites: while we investigated a C. immitis colonization site in Washington State, Kollath et al. studied C. posadasii colonization sites in Arizona. Different results might also be attributable to different sampling strategies: while Kollath et al. specifically targeted animal burrows, we used a less biased transect sampling design. In addition, these seemingly contradictory results may also indicate that both soil propagation and animal infection are essential for Coccidioides species life cycle.
Early investigators suggested that desert rodents and other animals contribute to the spread of Coccidioides spp. to the new sites: the fungus first grows on the corpse of an animal that dies of infection; however, if soil chemical and microbiological composition are conducive, it spreads to nearby soils, establishing a permanent colonization site (18, 25). Our results, as well as those of Kollath et al., are consistent with this model. Taken together, these observations suggest that although rodents may be important components of the Coccidioides species life cycle, soil parameters need to be taken into consideration when modeling Coccidioides species habitats. Notably, two recent distribution models provided evidence supporting both of these hypotheses. Dobos et al. used soil salinity as one of the variables and was able to predict areas in Washington State where C. immitis is present (12), while Ocampo-Chavira et al. discovered an overlap between Coccidioides spp. and the distribution of desert woodrat, Neotoma lepida (13). Both parameters need to be considered when modeling distribution and spread of this pathogen in the future.
MATERIALS AND METHODS
Study site and survey methods.
A combination of a radial and an interrupted belt transect method was used to survey the approximately 46,500-m2 area at an all-terrain vehicle (ATV) park in Washington State in September 2016. The site of the ATV accident that led to coccidioidomycosis infection in 2010 (4, 22) was designated the center of the study site. Nine 100-m radii with a central angle of 40° were measured from the center (Fig. 1). Along each 100-m radius, an interrupted belt transect was employed where soil within two 1-m2 plots (left and right) was sampled in increments of 10 m, The first 10 m from the center was an exception, where left and right 1-m2 plots were sampled at 1, 2, 3, 4, and 10 m. If a sampling plot was in an area unsafe to sample, a situation that arose occasionally in difficult terrain, soil was sampled as close to the intended plot as possible. If a rodent hole was identified in a 1-m2 plot or within 0.5 m of the 1-m2 plot, the rodent hole was sampled. For each 1-m2 plot, the following data were collected: altitude, presence of vegetation (yes/no), and along the ATV track (yes/no).
The site included a large area of exposed soil from ATV tracks in the center and east of the center (Fig. 1; Fig. S1A and B in the supplemental material), low grass area north of the center (Fig. S1C and D), a dry ravine with deciduous trees (Fig. S1E) transitioning to grassy marsh (Fig. S1F) south of the center, and a bordering orchard east of the center. No formal vegetation survey was performed. Numerous rodent burros were observed.
Soil collection.
Composite soil samples were collected from each 1-m2 plot; specifically, soil was collected from three different points at a depth of 10 to 20 cm. Soil samples from rodent burrows were collected separately at a similar depth (one per burrow) and not mixed with regular soils. After each sample collection, the sampling tool was disinfected with 10% bleach and rinsed with deionized water. Samples were transported at room temperature and stored at 4°C before DNA extraction.
DNA extraction.
For C. immitis cultures, DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Gaithersburg, MD) according to the manufacturer’s instructions. For soil samples, DNA was extracted using the PowerLyzer power soil DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) and following the manufacturer’s instructions with the following modification: instead of the recommended 0.25 g, approximately 0.75 g of soil was loaded into the bead tube for extraction. The FastPrep-24 5G (MP Biomedicals, LLC., Santa Ana, CA, USA) high-speed benchtop homogenizer was used for optimal homogenization and cell lysis. Samples were homogenized for seven 1-min cycles at 6 m/s with 5-min rest breaks between each cycle. Upon addition of elution buffer to the spin filter, samples sat at room temperature for 5 min before centrifugation. Different soil aliquots were used for DNA extraction, culture, and chemical analysis.
qPCR.
C. immitis was detected by amplifying a 249-bp region of a transposable element using a TaqMan-based single-tube (ST) nested qPCR assay previously described (29). Briefly, reactions were performed using a Rotor-Gene 6000 thermocycler (Qiagen, Valencia, CA, USA) with a thermal program consisting of two amplification phases distinguished by their annealing temperature. CT values were calculated using a manual threshold setting at a fluorescence value of 10−1.0. Positive and nontemplate controls were included in each qPCR run. qPCRs were performed in triplicates from the same DNA extract. The qPCR assay does not differentiate between C. immitis and C. posadasii species; therefore, soils positive by this assay were referred as positive for Coccidioides species DNA, although no evidence of C. posadasii was detected in these samples.
Culture.
Five grams of soil sample were transferred to a sterile 15-ml sterile tube, mixed with 5 ml of sterile phosphate-buffered saline (PBS), and vortexed. The tubes were then agitated horizontally at medium speed on a rocker table for 2 h at 25°C and allowed to settle for 1 min. Fifty microliters of the supernatant were spread on a modified yeast extract medium (15 g/liter yeast extract, 2 ml/liter Hutner’s trace elements, 0.5 g/liter cycloheximide, 0.05 g/liter chloramphenicol, 0.05 g/liter gentamicin, and 20 g/liter agar). The plates were incubated at 37°C and observed for growth at 3, 5, and 7 days. Small (2-mm) to medium (10-mm) discrete white fuzzy colonies were selected before the plates became confluent and transferred on Sabouraud dextrose agar slants supplemented with 0.05 g/liter of cycloheximide and 0.05 g/liter of chloramphenicol. The slants were incubated at 37°C and observed for growth every 2 days. Isolates with colony morphology consistent with Coccidioides spp. were confirmed by ITS sequencing.
Whole-genome sequencing and SNP analysis.
Genomic libraries were constructed and barcoded using the NEBNext Ultra DNA library prep kit for Illumina (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s instructions. Genomic libraries were sequenced on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) using the HiSeq rapid SBS kit v2, 500 cycles (Illumina). Isolate B10637 served as the reference for SNP analysis (2). Read data were aligned to the reference using Burrow-Wheeler Aligner version 0.7.7 (http://bio-bwa.sourceforge.net), and variants were identified using SAMtools version 0.1.18 (http://samtools.sourceforge.net). We filtered variants using the publicly available SNP analysis pipeline NASP (39) (http://tgennorth.github.io/NASP) to remove positions that had less than 10× coverage, less than 90% variant allele calls, or those identified by Nucmer as being within duplicated regions in the reference. A maximum-parsimony phylogeny was constructed using MEGA7 (http://www.megasoftware.net), and bootstrapping was performed with 1,000 reiterations.
Analytical chemistry.
To identify soil leachate characteristics associated with Coccidioides species colonization, deionized water leachates were prepared from 207 samples for chemical analysis, including anions, dissolved organic carbon (DOC), and major cations and trace metals following the protocol outlined by Hageman et al. (40). The first 74 samples collected at 1-m intervals within 4 m from the center were excluded because they were collected in close proximity to each other. Briefly, 6 g of soil and 120 ml of high-pressure liquid chromatography (HPLC)-grade water were added to a 500-ml high-density polyethylene (HDPE) bottle, and the solution was shaken by hand for 5 min. The bottle was placed upright to allow the leachate to settle for 10 min. Pipetting from the top of the leachate, 50 ml were removed and filtered through a 0.22-μm nitrocellulose filter. The filtered leachate was added to a 125-ml (HDPE) bottle and sent to the U.S. Geological Survey (USGS) lab in Denver, Colorado, for chemical analyses. Three types of blank samples were produced and analyzed: blank 1 contained HPLC-grade water only, blank 2 contained HPLC-grade water shaken in the HDPE bottle, and blank 3 contained HPLC-grade water shaken and filtered through the 0.22-μm nitrocellulose filter.
Ion chromatography for major anions was conducted by high-performance ion chromatography (HPIC) using the Thermo Scientific Dionex ICS-5000+ capillary HPIC system to measure the anions fluoride (detection limit, 0.04 mg/liter), chloride (0.60 mg/liter), nitrate (0.06 mg/liter), and sulfate (0.80 mg/liter). The Shimadzu total organic carbon analyzer with a Shimadzu autosampler was used to measure DOC concentrations. Detection limits for the lower and upper reporting limit were 0.2 mg/liter and 25 mg/liter, respectively. Samples with DOC concentrations above the upper reporting limit were diluted and reanalyzed. Inductively coupled plasma optical emission spectrometry (ICP-OES) and inductively coupled plasma mass spectrometry (ICP-MS) were used to measure cations and trace metals on sample splits acidified by trace metal-grade concentrated nitric acid. Data passed quality metrics if the concentration of an element was above the lower limit of determination (LOD) and the calculated relative standard deviation (RSD) of duplicate samples was ≤15%. Elements measured and reporting limits included rhenium (0.01 ppb), sulfur (20 ppb), antimony (5 ppb), scandium (0.2 ppb), selenium (0.5 ppb), tin (0.02 ppb), strontium (0.01 ppb), tantalum (0.05 ppb), tellurium (0.01 ppb), thorium (0.1 ppb), titanium (0.1 ppb), thallium (0.005 ppb), uranium (0.001 ppb), vanadium (0.05 ppb), tungsten (0.02 ppb), yttrium (0.01 ppb), zinc (1 ppb), and zirconium (0.05 ppb).
ITS1 amplicon sequencing and analysis.
Using genomic DNA extracted from soil samples, the nuclear ribosomal RNA gene (rRNA gene) internal transcribed spacer 1 (ITS1) region was amplified using the primer pair ITS1-F (CTTGGTCATTTAGAGGAAGTAA) and ITS2 (GCTGCGTTCTTCATCGATGC) (41, 42). PCR mixtures for amplification contained 10 μl of Invitrogen Platinum Hot Start PCR master mix (Thermo Fisher Scientific, Waltham, MA, USA), 0.5 μl of 10 μM forward primer, 0.5 μl of 10 μM reverse primer, 13 μl of PCR-grade water, and 1 μl of template DNA. PCR conditions were as follows: denaturation at 94°C for 1 min, 35 amplification cycles of 30 s at 94°C, 30 s at 52°C, and 30 s at 68°C, followed by a 10-min final extension at 68°C. Duplicate amplifications were quantified fluorescently with the Qubit double-stranded DNA (dsDNA) high-sensitivity (HS) assay kit (Thermo Fisher Scientific) and diluted to equimolar concentration. Libraries were generated using the Nextera XT index kits (Illumina, San Diego, CA, USA) and sequenced on the Illumina MiSeq platform in 2 × 250 bp paired-end reads at the CDC Genome Sequencing Laboratory. Sequencing runs for 278 samples yielded amplicon data with an average mean Phred-scaled quality score per sample of 32.3. Soil microbiome analysis was performed using Kraken 2 (43). A custom fungal k-mer-based database was generated using the default Kraken 2 fungi database and the UNITE general FASTA release for Fungi (44–46) classifications. Pavian was used for classifications (47).
Soil inoculation with C. immitis.
Soil collected from a residential backyard in Georgia was divided in half (samples A and B). Sample A was autoclaved at 120°C to inhibit all microorganisms; sample B was left intact. Both soils were inoculated with ∼500 arthroconidia of B10992 strain that was isolated from soil at the study site in 2013 and incubated at 25°C for 6 weeks inside a biosafety level 3 laboratory. Each week, aliquots of 0.5 g soil were removed and tested by qPCR as described above. The number of C. immitis cells in soil was calculated using a published calibration curve with the assumption that one arthroconidium is equivalent to one mycelial cell (30). The experiment was performed in triplicates.
Statistical analyses.
All analyses were done in SAS v9.4. For the characteristics in quantitative scale, the median value with interquartile range (IQR) was presented. The bivariate association between Coccidioides species colonization and a characteristic was assessed using a chi-square test for dichotomized variables and a Wilcoxon rank-sum test for quantitative variables. A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
We gratefully acknowledge Suzette Morman for her substantial contributions to the planning of the water leaching tests in the CDC biosafety level 3 (BSL3) facility, as well as the interpretation of the analytical results for the water leaching tests. We also wish to acknowledge Heather Hill and all other local health jurisdiction and health care facility staff who supported this project.
This work was made possible through support from the Advanced Molecular Detection (AMD) initiative. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention. This article has been peer reviewed and approved for publication consistent with USGS Fundamental Science Practices (48). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Contributor Information
Anastasia P. Litvintseva, Email: frq8@cdc.gov.
Aaron P. Mitchell, University of Georgia
REFERENCES
- 1.Brown J, Benedict K, Park BJ, Thompson GR, III.. 2013. Coccidioidomycosis: epidemiology. Clin Epidemiol 5:185–197. doi: 10.2147/CLEP.S34434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCotter OZ, Benedict K, Engelthaler DM, Komatsu K, Lucas KD, Mohle-Boetani JC, Oltean H, Vugia D, Chiller TM, Sondermeyer Cooksey GL, Nguyen A, Roe CC, Wheeler C, Sunenshine R. 2019. Update on the epidemiology of coccidioidomycosis in the United States. Med Mycol 57:S30–S40. doi: 10.1093/mmy/myy095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker BM. 2018. Coccidioidomycosis in animals, p 81–114. In Sybren de Hoog G, Guillot J, Verweij PE, Seyedmousavi S (ed), Emerging and epizootic fungal infections in animals. Springer, Cham, Switzerland. [Google Scholar]
- 4.Marsden-Haug N, Goldoft M, Ralston C, Limaye AP, Chua J, Hill H, Jecha L, Thompson IIG, Chiller T. 2013. Coccidioidomycosis acquired in Washington State. Clin Infect Dis 56:847–850. doi: 10.1093/cid/cis1028. [DOI] [PubMed] [Google Scholar]
- 5.Marsden-Haug N, Hill H, Litvintseva AP, Engelthaler DM, Driebe EM, Roe CC, Ralston C, Hurst S, Goldoft M, Gade L, Wohrle R, Thompson GR, Brandt ME, Chiller T, Centers for Disease Control and Prevention (CDC) . 2014. Coccidioides immitis identified in soil outside of its known range–Washington, 2013. MMWR Morb Mortal Wkly Rep 63:450. [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards PQ, Palmer CE. 1957. Prevalence of sensitivity to coccidioidin, with special reference to specific and nonspecific reactions to coccidioidin and to histoplasmin. Dis Chest 31:35–60. doi: 10.1378/chest.31.1.35. [DOI] [PubMed] [Google Scholar]
- 7.Hubber J, Person A, Jecha L, Flodin-Hursh D, Stiffler J, Hill H, Bassham S, McCotter O, Oltean H. 2020. Knowledge, attitudes, and practices regarding coccidioidomycosis among healthcare providers in four counties in Washington State, 2017. Med Mycol 58:730–736. doi: 10.1093/mmy/myz111. [DOI] [PubMed] [Google Scholar]
- 8.Baptista‐Rosas RC, Hinojosa A, Riquelme M. 2007. Ecological niche modeling of Coccidioides spp. in western North American deserts. Ann N Y Acad Sci 1111:35–46. doi: 10.1196/annals.1406.003. [DOI] [PubMed] [Google Scholar]
- 9.Gorris M, Cat L, Zender C, Treseder K, Randerson J. 2018. Coccidioidomycosis dynamics in relation to climate in the southwestern United States. GeoHealth 2:6–24. doi: 10.1002/2017GH000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorris ME, Treseder KK, Zender CS, Randerson JT. 2019. Expansion of coccidioidomycosis endemic regions in the United States in response to climate change. GeoHealth 3:308–327. doi: 10.1029/2019GH000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weaver E, Kolivras KN, Thomas RQ, Thomas VA, Abbas KM. 2020. Environmental factors affecting ecological niche of Coccidioides species and spatial dynamics of valley fever in the United States. Spat Spatiotemporal Epidemiol 32:100317. doi: 10.1016/j.sste.2019.100317. [DOI] [PubMed] [Google Scholar]
- 12.Dobos RR, Benedict K, Jackson BR, McCotter OZ. 2021. Using soil survey data to model potential Coccidioides soil habitat and inform Valley fever epidemiology. PLoS One 16:e0247263. doi: 10.1371/journal.pone.0247263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ocampo-Chavira P, Eaton-Gonzalez R, Riquelme M. 2020. Of mice and fungi: Coccidioides spp. distribution models. J Fungi 6:320. doi: 10.3390/jof6040320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor JW, Barker BM. 2019. The endozoan, small-mammal reservoir hypothesis and the life cycle of Coccidioides species. Med Mycol 57:S16–S20. doi: 10.1093/mmy/myy039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kollath DR, Teixeira MM, Funke A, Miller KJ, Barker BM. 2020. Investigating the role of animal burrows on the ecology and distribution of Coccidioides spp. in Arizona soils. Mycopathol 185:145–159. doi: 10.1007/s11046-019-00391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Rocío Reyes-Montes M, Pérez-Huitrón MA, Ocaña-Monroy JL, Frías-De-León MG, Martínez-Herrera E, Arenas R, Duarte-Escalante E. 2016. The habitat of Coccidioides spp. and the role of animals as reservoirs and disseminators in nature. BMC Infect Dis 16:550. doi: 10.1186/s12879-016-1902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emmons CW. 1942. Isolation of Coccidioides from soil and rodents. Public Health Rep (1896–1970) 57:109–111. doi: 10.2307/4583988. [DOI] [Google Scholar]
- 18.Ajello L. 1967. Comparative ecology of respiratory mycotic disease agents. Bacteriol Rev 31:6–24. doi: 10.1128/br.31.1.6-24.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, Maiti R, Kodira CD, Neafsey DE, Zeng Q, Hung C-Y, McMahan C, Muszewska A, Grynberg M, Mandel MA, Kellner EM, Barker BM, Galgiani JN, Orbach MJ, Kirkland TN, Cole GT, Henn MR, Birren BW, Taylor JW. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res 19:1722–1731. doi: 10.1101/gr.087551.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muñoz JF, McEwen JG, Clay OK, Cuomo CA. 2018. Genome analysis reveals evolutionary mechanisms of adaptation in systemic dimorphic fungi. Sci Rep 8:4473. doi: 10.1038/s41598-018-22816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swatek F, Omieczynski D, Plunkett O. Coccidioides immitis in California, p 255–264. University of Arizona Press, Tucson, AZ. [Google Scholar]
- 22.Litvintseva AP, Marsden-Haug N, Hurst S, Hill H, Gade L, Driebe EM, Ralston C, Roe C, Barker BM, Goldoft M, Keim P, Wohrle R, Thompson GR, Engelthaler DM, Brandt ME, Chiller T. 2015. Valley fever: finding new places for an old disease: Coccidioides immitis found in Washington State soil associated with recent human infection. Clin Infect Dis 60:e1–e3. doi: 10.1093/cid/ciu681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow NADK, Gade L, McCotter O, Hurst S, Salamone A, Wohrle R, Clifford W, Kim S, Salah Z, Oltean HAL, Plumlee GS. 2021. Factors influencing distribution of Coccidioides immitis in soil, Washington State, 2016 — data release. https://www.sciencebase.gov/catalog/item/615a1567d34e0df5fba0ab44. doi: 10.5066/P9KEWAT0. [DOI] [PMC free article] [PubMed]
- 24.Garrett RG, Hall G, Vaive J, Pelchat P. 2009. A water-leach procedure for estimating bioaccessibility of elements in soils from transects across the United States and Canada. Applied Geochem 24:1438–1453. doi: 10.1016/j.apgeochem.2009.04.014. [DOI] [Google Scholar]
- 25.Maddy K. 1965. Observations on Coccidioides immitis found growing naturally in soil. Ariz Med 22:281–288. [PubMed] [Google Scholar]
- 26.Maddy KT. Ecological factors possibly relating to the geographic distribution of Coccidioides immitis, p 144–157. [Google Scholar]
- 27.Egeberg RO, Elconin AE, Egeberg MC. 1964. Effect of salinity and temperature on Coccidioides immitis and three antagonistic soil saprophytes. J Bacteriol 88:473–476. doi: 10.1128/jb.88.2.473-476.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burnham KP, Anderson DR, Laake JL. 1980. Estimation of density from line transect sampling of biological populations. Wildlife Monographs 72:3–202. doi: 10.1002/bimj.4710240306. [DOI] [Google Scholar]
- 29.Chow NA, Griffin DW, Barker BM, Loparev VN, Litvintseva AP. 2016. Molecular detection of airborne Coccidioides in Tucson, Arizona. Med Mycol 54:584–592. doi: 10.1093/mmy/myw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gade L, McCotter OZ, Bowers JR, Waddell V, Brady S, Carvajal JA, Sunenshine R, Komatsu KK, Engelthaler DM, Chiller T, Litvintseva AP. 2020. The detection of Coccidioides from ambient air in Phoenix, Arizona: evidence of uneven distribution and seasonality. Med Mycol 58:552–559. doi: 10.1093/mmy/myz093. [DOI] [PubMed] [Google Scholar]
- 31.Bowers J, Parise K, Kelley E, Lemmer D, Schupp J, Driebe E, Engelthaler D, Keim P, Barker B. 2019. Direct detection of Coccidioides from Arizona soils using CocciENV, a highly sensitive and specific real-time PCR assay. Med Mycol 57:246–255. doi: 10.1093/mmy/myy007. [DOI] [PubMed] [Google Scholar]
- 32.Barker BM, Litvintseva AP, Riquelme M, Vargas-Gastélum L. 2019. Coccidioides ecology and genomics. Med Mycol 57:S21–s29. doi: 10.1093/mmy/myy051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisher FS, Bultman MW, Johnson SM, Pappagianis D, Zaborsky E. 2007. Coccidioides niches and habitat parameters in the southwestern United States: a matter of scale. Ann N Y Acad Sci 1111:47–72. doi: 10.1196/annals.1406.031. [DOI] [PubMed] [Google Scholar]
- 34.Lubarsky R, Plunkett O. 1955. Some ecologic studies of Coccidioides immitis in soil, p 308–310. In Therapy of fungus diseases. An international symposium. Little, Brown, & Co., Boston, MA. [Google Scholar]
- 35.Lacy GH, Swatek FE. 1974. Soil ecology of Coccidioides immitis at Amerindian middens in California. Appl Microbiol 27:379–388. doi: 10.1128/am.27.2.379-388.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lauer A, Baal JD, Mendes SD, Casimiro KN, Passaglia AK, Valenzuela AH, Guibert G. 2019. Valley fever on the rise—searching for microbial antagonists to the fungal pathogen Coccidioides immitis. Microorganisms 7:31. doi: 10.3390/microorganisms7020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lauer A, Etyemezian V, Nikolich G, Kloock C, Arzate AF, Sadiq Batcha F, Kaur M, Garcia E, Mander J, Kayes Passaglia A. 2020. Valley fever: environmental risk factors and exposure pathways deduced from field measurements in California. Int J Environ Res Public Health 17:5285. doi: 10.3390/ijerph17155285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greger M, Landberg T, Vaculík M. 2018. Silicon influences soil availability and accumulation of mineral nutrients in various plant species. Plants 7:41. doi: 10.3390/plants7020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, Driebe EM, Drees KP, Hicks ND, Williamson CHD. 2016. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genom 2:e000074. doi: 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hageman PL. 2007. US Geological Survey field leach test for assessing water reactivity and leaching potential of mine wastes, soils, and other geologic and environmental materials. Series 5-D3. U.S. Geological Survey, Reston, VA. [Google Scholar]
- 41.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 42.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications, vol 18. Academic Press, London, UK. [Google Scholar]
- 43.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:1–13. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kõljalg U, Larsson K-H, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Vrålstad T, Ursing BM. 2005. UNITE: a database providing web‐based methods for the molecular identification of ectomycorrhizal fungi. New Phytol 166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- 45.Abarenkov K, Henrik Nilsson R, Larsson K-H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U. 2010. The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol 186:281–285. doi: 10.1111/j.1469-8137.2009.03160.x. [DOI] [PubMed] [Google Scholar]
- 46.Nilsson RH, Abarenkov K, Larsson K-H, Kõljalg U. 2011. Molecular identification of fungi: rationale, philosophical concerns, and the UNITE database. Open Applied Informat J 5:81–86. doi: 10.2174/1874136301005010081. [DOI] [Google Scholar]
- 47.Breitwieser FP, Salzberg SL. 2020. Pavian: interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics 36:1303–1304. doi: 10.1093/bioinformatics/btz715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.U.S. Geological Survey. 2011. U.S. Geological Survey fundamental science practices. https://pubs.er.usgs.gov/publication/cir1367.
- 49.Oltean HN, Etienne KA, Roe CC, Gade L, McCotter OZ, Engelthaler DM, Litvintseva AP. 2019. Utility of whole-genome sequencing to ascertain locally acquired cases of coccidioidomycosis, Washington, USA. Emerg Infect Dis 25:501–506. doi: 10.3201/eid2503.181155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of the Coccidioides sp. within 10 m from the center of the transects. Center is shown with a large red circle; squares represent 1-m2 plots (left and right), and small circles represent rodent burrows within or near 1-m2 plots. Distances (in meters) are labeled in yellow. Plots and rodent burrows where samples were qPCR positive for Coccidioides species DNA are shown in red. This figure is a graphical representation; transects and sampled areas are not to scale. Download FIG S2, PDF file, 0.09 MB (90.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Types of vegetation encountered at the study site. (A and B) ATV tracks located at the center and east of the center. (C and D) Low grass area in the north. (E) Dry ravine. (F) Marsh area in the south. Download FIG S1, PDF file, 0.4 MB (400.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Sample distribution (no. [%]) by fungi type and Coccidioides status in 280 soil samples. *, UNITE database-assigned ITS sequence to genus Paracoccidioides endemic to South America; this sequence likely belongs to another member of Ajellomycetaceae. **, Bold indicates a P value of <0.05. Download Table S1, DOCX file, 0.02 MB (17.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.