Abstract
E2F transcription activity has been shown to play a critical role in cell growth control, regulating the expression of a variety of genes that encode proteins important for the initiation of DNA replication and cell cycle regulation. We have shown that the E2F3 locus encodes two protein products: the E2F3a product, which is tightly regulated by cell growth, and the E2F3b product, which is constitutively expressed throughout the cell cycle. To further explore the mechanism controlling the expression of the two E2F3 gene products, we analyzed the genomic sequences flanking the 5′ region of E2F3a and E2F3b. We find that a series of E2F binding sites confer negative control on the E2F3a promoter in quiescent cells, similar to the control of the E2F1 and E2F2 promoters. In addition, a group of E-box elements, which are Myc binding sites, confer responsiveness to Myc and are necessary for full activation of the E2F3a promoter in response to growth stimulation. Based on these results and past experiments, it appears that the E2F1, E2F2, and E2F3a genes are similarly regulated by growth stimulation, involving a combination of E2F-dependent negative control and Myc-mediated positive control. In contrast, the constitutive expression of the E2F3b gene more closely reflects the control of expression of the E2F4 and E2F5 genes.
Considerable effort has been devoted to the elucidation of the regulatory pathways that govern the control of gene activity in relation to cell growth. Such work has led to the delineation of a pathway involving the retinoblastoma tumor suppressor protein that appears to be critically important for the transition from quiescence and into S phase. The activity of the Rb protein is regulated by the action of G1 cyclin-dependent kinases (cdk), primarily the D-type cyclins and associated cdk4 kinase. The importance of the Rb pathway is emphasized by the fact that disruption of various components that regulate the cell cycle, which regulate these kinases, can lead to the development of cancer (11).
The regulatory function of the Rb tumor suppressor protein appears to largely depend on the ability of Rb to bind to and inhibit the family of E2F transcription factors (8, 23, 24). In addition to activating the transcription of a group of genes that encode proteins necessary for DNA synthesis, it has been shown that E2F transcription factors activate genes that encode proteins that are involved in initiation of replication and maintenance of cell cycle regulation, including Orc1, Cdc6, and Mcm and cyclin E, cyclin A, and cdc2, respectively (5, 22).
The E2F family of proteins includes six distinct E2F members and at least two heterodimer partners, DP1 and DP2 (5, 22). Whereas the levels of DP protein are generally in abundance and do not appear to be limiting for E2F activity, dramatic changes of E2F proteins have been observed. Although the expression of E2F4 and E2F5 genes appear to relatively constant throughout the cell cycle (6, 26), E2F1 and E2F2 gene expression has been found to dramatically increase in late G1 (10, 12, 21, 27). The complexity of E2F activity resulting from the formation of a variety of heterodimeric protein complexes has led to the speculation that individual E2F family members might have a distinct role in cellular growth. For example, individual E2F family members might integrate distinct signaling pathways within the cell to facilitate the orderly progression throughout the cell cycle. Therefore, individual E2F genes or proteins might respond to distinct extracellular growth factors or distinct signal transduction pathways. Indeed, recent evidence indicates that distinct roles can be ascribed to individual E2F family members. For example, E2F1 has been shown to play a critical and unique role in the induction of apoptosis (3, 13, 14, 25, 28, 30). E2F3 has also been suggested to play a role in transcription activation but, unlike E2F1, it appears to be important for the efficient induction of the S phase in cycling cells (18). In contrast, E2F4 and E2F5 proteins associate with the Rb-related p130 protein in quiescent cells, where the complexes appear to function as transcription repressors, preventing the expression of various genes encoding proteins important for cell growth.
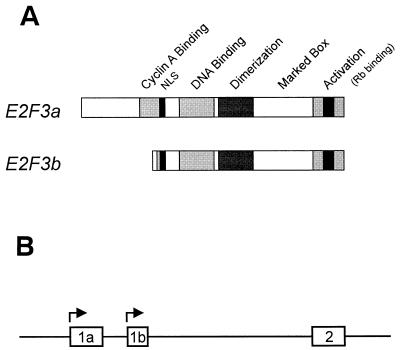
The E2F2 and E2F3 genes were originally identified by low-stringency hybridization as E2F1-related genes, constituting an E2F subfamily that shared sequence and function (16). We have now identified a novel E2F3 product that specifically interacts with Rb in quiescent cells (19). This novel product has been termed E2F3b and is encoded by a unique mRNA transcribed from an intronic promoter within the E2F3 locus. E2F3b RNA differs from the previously characterized E2F3 RNA, which is now termed E2F3a, by the nature of the initial coding exon (Fig. 1A). In contrast to the E2F3a product, which is tightly regulated by cell growth, the expression of E2F3b remains constant throughout the cell cycle. In addition, the E2F3b protein uniquely associates with Rb in quiescent cells.
FIG. 1.
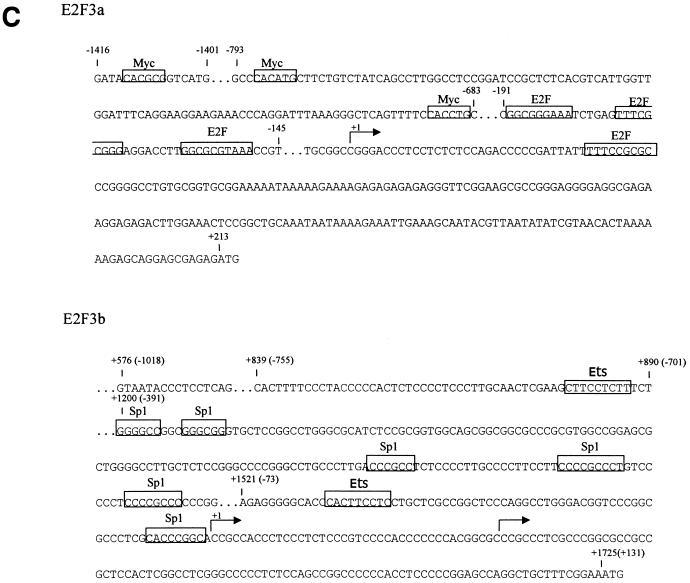
E2F3 genomic organization. (A) Schematic depiction of the domain organization of the E2F3a and E2F3b products. (B) Schematic depiction of the E2F3 genomic organization at the sites specifying initiation of transcription. The exon structures defining the E2F3a and E2F3b transcripts, which involve utilization of alternate transcription start sites and thus distinct initial exon sequences, have been defined (19). (C) DNA sequence in the 5′-flanking region of the mouse E2F3 gene. The +1 transcription site for both E2F3a and E2F3b is based on the longest clone sequenced from the RACE analysis as well as on primer extension analysis. Boxed sequences represent putative E2F and Myc binding sites as well as other consensus sites.
We have now explored the mechanisms controlling the expression of both E2F3a and E2F3b during a cell proliferative event through the isolation and analysis of the promoter elements for these two loci. Sequence analysis of the E2F3a flanking sequence reveals a variety of known transcription factor binding sites, including both E-box elements and E2F binding sites. In contrast, the E2F3b flanking sequence contains no such elements. We also find that the E2F3a promoter is subject to E2F-dependent negative regulation in quiescent cells, as was previously demonstrated for the E2F1 and E2F2 promoters, and that Myc also contributes to the positive activation of the E2F3a promoter. In contrast, the E2F3b promoter is constitutively active in quiescent and growing cells, reflecting the fact that the E2F3b product is not regulated as a function of cell growth.
MATERIALS AND METHODS
Cell culture.
REF52 cells were grown in Dulbecco modified Eagle medium (DMEM) containing 10% serum (5% fetal bovine serum and 5% calf serum). To bring cells to quiescence, cultures at 30% confluence (24 h after plating) were incubated in DMEM containing 0.25% serum for 48 h. To stimulate cell growth, the cells were fed with fresh medium containing 20% serum.
Construction of plasmids.
The E2F3a luciferase expression vector was generated by subcloning the 2-kb promoter fragment of E2F3a from SacI/HaeII-digested 3kbH3pBS into the SacI/HindIII-digested pGL2Basic (Promega). The primers GAGAGAGATCTTTCCGAAAGCAGCCTGG and GAGAGAGATCTAATACCCTCCTCAGCG containing nested BglII sites were used to generate a 1.0-kb E2F3b promoter fragment via PCR. The PCR product was digested with BglII and subcloned into BglII-digested pGL2Basic. The E2F3a (E2F−) plasmid was created using the GeneEditor (Promega) in vitro site-directed mutagenesis kit as specified by the manufacturer. The following point mutations were incorporated (the wild-type sequence is followed by mutant sequence): E2F site 1, GCGGGAAA to GCTTGAAA; E2F site 2, TTTCGCGGG to TTTCAAGGG; and E2F site 3, GCGCGTAA to GCTTGTAA. The E2F3a (Myc−) plasmid was created using the Transformer (Clontech) site-directed mutagenesis kit as specified by the manufacturer. Briefly, the unique SalI site in the pGL2Basic vector backbone was altered using the selection oligonucleotide 5′-CAAGGGCATCGGTCCACGGATCCAGACAT-3′. The following mutations were made: Myc site 1, CACGCG to ATCGAT; Myc site 2, CACATG to ATCAAT; and Myc site 3, CACCTG to ATCCAT. The primers GAGAGAGGTACCCCCCTCCCTTGCAAC containing a KpnI site at the 5′ end and GGCCCGGAGAGCAAGGCCCC were used to generate the −733 E2F3b fragment via PCR. The PCR product was digested with KpnI/SacII and subcloned into KpnI/SacII-digested E2F3b luciferase expression vector. The −350 E2F3b fragment was generated via XhoI/SacII digestion of E2F3b followed by a religation of the 6.0-kb fragment. Cytomegalovirus (CMV)-Myc was a gift from S. Hann (Vanderbilt University) (7). The control vector pRc-CMV was purchased from Invitrogen. All constructs were confirmed by sequencing.
Northern analysis.
Poly(A)+ RNA was isolated from an equal amount of total RNA and processed for Northern analysis as described previously (2).
Promoter transfection assays.
REF52 cells were transfected with the various luciferase expression vectors, together with CMV–β-galactosidase as an internal control, as previously described (27). After transfection, cells were brought to quiescence by serum starvation. For serum stimulation studies, cells were stimulated to grow following the addition of 20% serum. Cells were harvested at various times after stimulation, and luciferase and β-galactosidase assays were performed as described previously (27).
Gel mobility shift assays.
Gel mobility shift assays to detect E2F binding activity or Myc binding were performed as previously described (18, 27). For E2F binding, an end-labeled plasmid DNA fragment from the dihydrofolate reductase (DHFR) promoter containing two E2F recognition sites was used as a probe. Oligonucleotides 5′-GGCGGAGATATGCAAATATGG-3′ and 5′-AGGAAGCTGCTGCTGACAATG-3′ were used as primers to generate two 110-bp PCR fragments spanning positions −113 to −123 of the E2F3a wild-type and E2F3a E2F mutant promoters. The E2F wild-type and E2F mutant PCR fragments were used as unlabeled competitors. Myc binding assays used E-box containing annealed oligonucleotides 5′-GATCCTGACGACCACGTGGTCTTACG-3′ and 5′-GATCCGTAAGACCACGTGGTCGTCAG-3′ as a probe and E-box mutant annealed oligonucleotides 5′-GATCCTGACGACATCGATGTCTTACG-3′ and 5′-GATCCGTAAGACATCGATGTCGTCAG-3′ as a control. Unlabeled competitors were generated by PCR from the wild-type and Myc mutant E2F3a promoters. Primers 5′-GCGTCAGCTGAACCTTCTACC-3′ and 5′-GTGTCAAAGGCAAGTTGGACC-3′ generated a 174-bp PCR fragment corresponding to positions −829 to −655 which contains Myc sites 1 and 2. Primers 5′-GTCTCATTCTCCCAGTTCTGGG-3′ and 5′-ATTGTGGGGCTGGAGAAATGG-3′ generated a 130-bp PCR fragment corresponding to positions −1444 to −1314 containing Myc site 3.
RESULTS
Analysis of promoter sequences controlling expression of the E2F3 locus.
Our recent experiments have demonstrated a complexity in the E2F3 locus whereby the use of alternate initial coding exons (termed exon 1a and exon 1b) gives rise to two distinct transcripts and protein products (19) (Fig. 1). Given the organization of the E2F3 locus, together with the distinct regulation of the two E2F3 RNAs, we have now sought to identify the mechanisms responsible for the control of the expression of these two RNAs through analysis of their respective promoter sequences.
To analyze the functional capacity of the sequences flanking exons 1a and 1b, the DNA fragments were placed in an expression vector utilizing the luciferase gene as a reporter. REF52 cells were transfected with the reporter along with a CMV-driven β-galactosidase plasmid as an internal control; the cells were brought to quiescence by serum starvation and then stimulated to re-enter the cell cycle by the addition of fresh medium with serum. Cell samples were then harvested at various times after serum addition, extracts were prepared, and assays for luciferase and β-galactosidase were performed.
As seen in our initial experiments, the sequences flanking the 5′ region of the E2F3b transcription start site exhibited promoter activity which did not vary through the cell cycle (Fig. 2). Although we have not precisely delineated the sequence elements that are essential for E2F3b promoter activity, it is also clear from the analysis of several truncations of the promoter sequence that although there is some reduction in promoter activity upon deletion of 5′ flanking sequence, substantial activity can be found within the 5′ 350 nucleotides (Fig. 2).
FIG. 2.
Sequences required for E2F3b promoter activity. (A) Schematic depiction of E2F3b promoter constructs. Luciferase reporter constructs containing the indicated 5′-flanking sequence were used for assays of promoter activity. (B) REF52 cells were transfected with 4 μg of the indicated E2F3b-luciferase plasmids together with 2 μg of CMV–β-galactosidase. Transfected cells were treated as described in Materials and Methods and harvested at the indicated time points. Luciferase activity was normalized to the β-galactosidase activity. Symbols: □, E2F3b (−1018); ■, E2F3b (−733); ○, E2F3b (−350).
E2F-dependent control of E2F3a promoter activity.
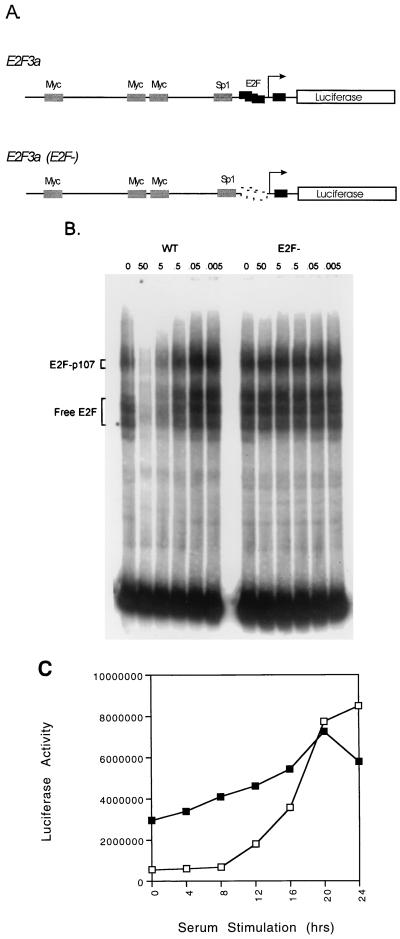
Based on previous studies that have demonstrated a role for E2F elements in the growth-regulated activation of a variety of promoters, we investigated the role of these sites in E2F3a promoter activity using an expression construct in which the E2F sites were eliminated by mutation. REF52 cells were transfected with either the wild-type E2F3a-luciferase construct or the E2F3a (E2F−)-luciferase construct, each together with a CMV–β-galactosidase internal control. Transfected cells were growth arrested by the removal of serum and then stimulated to enter the cell cycle by the addition of fresh medium with 20% serum. In contrast to the constant activity of the E2F3b promoter, the sequences flanking the E2F3a start site conferred tight growth regulation (Fig. 3), reflecting the pattern of accumulation of the E2F3a RNA. The absence of promoter activity in the quiescent cells appears to reflect E2F-dependent negative control since mutation of the E2F binding sites in the E2F3a promoter resulted in a marked elevation of the activity of the promoter in quiescent cells. The mutant promoter exhibited a fivefold increase in activity in quiescent cells compared to the activity of the wild-type promoter. After serum stimulation, the mutant promoter only increased a further 1.9-fold compared to the 15-fold increase in activity of the wild-type promoter. We thus conclude that the activity of the E2F3a promoter is subject to E2F-mediated negative regulation in quiescent cells, similar to the previous results for the E2F1 and E2F2 promoters, likely due to the repression mediated by an E2F-Rb family complex.
FIG. 3.
E2F-dependent regulation of the E2F3a promoter. (A) Schematic depiction of the E2F3a wild-type promoter and the promoter containing alterations in the E2F binding elements. (B) E2F binds to the E2F3a promoter. Gel mobility shift assays were performed with a nuclear extract from G1/S-arrested mouse embryo fibroblasts and an end-labeled DNA fragment derived from the DHFR promoter that contains two overlapping E2F binding sites as the probe. Competition assays were performed as described in Materials and Methods using the indicated amount (in nanograms) of PCR products derived from either the wild-type E2F3a promoter sequence spanning the E2F sites or the mutant promoter in which the E2F sites were altered by point mutations. The positions of either the E2F-p107 complex or free E2F complexes are indicated. (C) REF52 cells were transfected with 4 μg of the wild-type E2F3a-luciferase plasmid (□) or 4 μg of the E2F3a (E2F−)-luciferase plasmid (■), together with 2 μg of CMV–β-galactosidase. Transfected cells were processed as described in Fig. 2. Luciferase activity was normalized to β-galactosidase activity.
A role for Myc in the control of E2F3a promoter activity.
Based on the observation that the E2F3a is cell cycle regulated, together with the fact that sequence analysis revealed the presence of several E-box elements that can serve as binding sites for the Myc protein (Fig. 4A and B), we have investigated the role of Myc in the regulation of E2F3a promoter activity. As an initial approach, we determined the effect of overexpression of Myc protein on the activity of the E2F3a promoter. REF52 cells were transfected with the E2F3a expression vector together with increasing amounts of a Myc expression vector. Cells were harvested and then extracts were assayed for luciferase activity. As shown in Fig. 4C, coexpression of Myc protein led to a stimulation of E2F3a promoter activity. In contrast, Myc had no effect on the activity of the E2F3b promoter, a finding consistent with the absence of E-box elements within the E2F3b promoter sequence.
FIG. 4.
Myc activates the E2F3a promoter but not the E2F3b promoter. (A) Schematic depiction of the E2F3a wild-type promoter and the promoter containing alterations in the Myc binding elements. (B) Myc binds to the E2F3a promoter. Gel mobility shift assays for Myc binding were performed as described in Materials and Methods using baculovirus-produced Myc and Max proteins and an end-labeled double-stranded oligonucleotide that contains an E-box sequence (27). Specificity of binding was demonstrated by competition with cold wild-type DNA probe (WT) or a mutant version of the probe in which the E-box element was disrupted by mutation (Mut). Competition assays to demonstrate binding to the sites in the E2F3a promoter were performed using PCR products containing the wild-type sequence of the proximal two sites (Site1,2) or the distal site (Site3) or mutant versions of each (Mut1,2 and Mut3). (C) The E2F3a promoter, but not the E2F3b promoter, is activated by Myc. REF52 cells were transfected with 8 μg of either the E2F3a-luciferase plasmid or the E2F3b-luciferase plasmid, together with the indicated amount (in micrograms) of a CMV-driven Myc expression vector along with 2 μg of CMV–β-galactosidase. When less than 3 μg of CMV-Myc was used, the control vector pRc-CMV was included to bring the total amount of CMV vector added to 3 μg. Transfected cells were incubated in low-serum medium for 48 h, at which time cells were harvested, extracts were prepared, and luciferase and β-galactosidase activities were measured. (D) Activation of wild-type E2F3a by Myc is dependent on intact E-box elements. REF52 cells were transfected with 8 μg of either wild-type E2F3a-luciferase or the E-box-site mutant E2F3a (E-box−)-luciferase plasmid in which the E-box sequences were altered by mutation as described in Materials and Methods. Each plasmid was cotransfected with either 10 μg of CMV-Myc or 10 μg of the control pRc-CMV, together with 2 μg of β-galactosidase.
To determine if the Myc-mediated activation of E2F3a promoter activity does indeed depend on the ability of Myc to bind to the promoter, we generated point mutations in the Myc recognition sequences located at −688 to −693, −779 to −784, and −1412 to −1417. As shown in Fig. 4D, the E2F3a promoter lacking the Myc sites exhibits a marked reduction of in promoter activity compared to the wild-type promoter, displaying only a twofold induction compared to the CMV control. Baseline activities of the wild-type E2F3a promoter and the E-box mutant E2F3a promoter were similar.
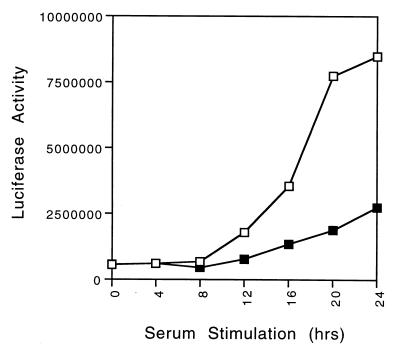
Given the ability of Myc to activate the E2F3a gene, as well as the evidence for a role of the Myc binding sites in mediating activation of the E2F3a promoter, we have investigated the potential role of these elements in the normal growth-activated induction of E2F3a promoter activity. REF52 cells were transfected with either the wild-type E2F3a-luciferase construct or the E2F3a (E-box−)-luciferase construct and then brought to quiescence by serum starvation. The quiescent cells were then stimulated to grow by the addition of fresh medium with 20% serum, and samples were then taken at various times and assayed for luciferase activity. As shown in Fig. 5, the wild-type promoter again reflected the pattern of accumulation of the endogenous E2F3a transcript. In contrast, the mutation of the Myc binding sites resulted in a three- to fourfold reduction of promoter activity during the time of peak activation. In light of the ability of Myc to stimulate E2F3a promoter activity, as well as the ability of Myc to induce the endogenous E2F3a transcript, together with the observation that mutation of the Myc binding sites impairs the growth-regulated activity of the E2F3a promoter, we conclude that the full activation of the E2F3a promoter following growth stimulation reflects a contribution from Myc.
FIG. 5.
E-box elements are required for full activity of the E2F3a promoter in response to stimulation of cell proliferation. Effects of mutation of Myc binding sites on growth stimulated the expression of the E2F3a promoter. REF52 cells were transfected with 4 μg of the wild-type E2F3a-luciferase plasmid (□) or the mutant E2F3a (E-box−)-luciferase plasmid (■), together with 2 μg of CMV–β-galactosidase. Transfected cells were treated as described in Fig. 2, and the luciferase and β-galactosidase activities were measured. The luciferase activity was normalized to the β-galactosidase activity.
Myc activates the endogenous E2F3a gene.
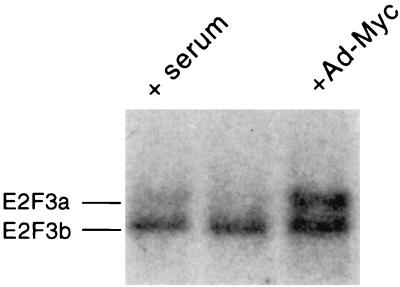
Given the ability of Myc to activate the E2F3a promoter, which is dependent on the Myc binding sites, and given the indication of specificity in this induction as seen by the lack of response of the E2F3b promoter, we investigated the ability of Myc to specifically activate the endogenous E2F3a gene. REF52 cells were starved for 48 h and then were either serum stimulated or infected with either Ad-Myc or the control virus Ad-CMV that lacks an insert. The cells were harvested either 20 h after serum stimulation or 26 h after virus infection, poly(A)+ RNA was isolated, and the RNA was analyzed by Northern blotting. As shown in Fig. 6, infection of REF52 cells with Ad-Myc resulted in the induction of the slower-migrating E2F3a RNA, which is equivalent to the induction that resulted from serum stimulation of the quiescent cells. In contrast, the level of the faster-migrating E2F3b RNA remained constant and was not affected by the expression of Myc protein. We thus conclude that Myc does have the capacity to induce the E2F3 gene and does so specifically through the activation of the E2F3a promoter.
FIG. 6.
Myc activates the endogenous E2F3a gene but not the E2F3b gene. REF52 cells were brought to quiescence and then infected with an adenovirus recombinant containing the c-myc gene (Ad-Myc) (right lane) or a control virus lacking an insert (Ad-CMV) (middle lane). An additional aliquot of cells was stimulated by serum addition (left lane). Poly(A)+ RNA was isolated and subjected to Northern analysis. E2F3a and E2F3b RNAs were detected using an E2F3 cDNA probe.
DISCUSSION
Based on a large body of previous work, the E2F family could be seen as two distinct subfamilies: the E2F1, E2F2, and E2F3 group versus the E2F4 and E2F5 group (5, 22). The E2F1 to -3 subfamily shares sequence and structural similarities as well as the ability to preferentially interact with Rb but not with p130 or p107 (16). The E2F1 to -3 genes also share a regulatory pattern that restricts their expression to proliferating cells (10, 12, 18, 21, 27). In contrast, the E2F4 and E2F5 proteins are more closely related with respect to sequence and structure, they uniquely bind to p130 and p107, and they are constitutively expressed without regard to the state of cell proliferation (1, 6, 9, 24). Our recent analysis of genetic complexity in the E2F3 locus (19), detailing the utilization of distinct exons to generate two related but distinct protein products, now adds to the overall complexity of the E2F family.
The characteristics of the novel E2F3b product places it at the juncture of the two E2F subfamilies. E2F3b shares sequence with the E2F1 to -3a group and, like E2F1 to -3a, the E2F3b protein uniquely associates with Rb. However, unlike E2F1 to -3 but similar to E2F4 and E2F5, the E2F3b protein lacks the N-terminal sequences of this group that confers posttranscriptional control of accumulation, and the E2F3b gene is not regulated with respect to the cell proliferation cycle but rather is constitutively expressed. The data we describe here now add to this comparison since it is clear that the expression pattern is dictated by the promoter elements that regulate the expression of the two E2F3 transcripts, and these promoters reflect the organization of the E2F subfamilies. Like E2F1 and -2, the E2F3a promoter is regulated by E2F-mediated negative control in quiescent cells. Moreover, like E2F1 and E2F2, E2F3a is subject to regulation by the Myc protein, dependent on E-box elements within the E2F3a promoter. In contrast, the E2F3b promoter is not controlled by cell growth and does not respond to Myc, and these properties coincide with an absence of E2F and Myc binding sites in the promoter.
Potential roles for the E2F3 activities in cell cycle control.
Although the two E2F3 products are largely identical, they do differ by the presence of an N-terminal domain that is found in E2F3a but is absent from E2F3b. A number of recent experiments, directed at an analysis of the function of N-terminal sequences of E2F1, suggest a role for these sequences in the regulated accumulation of E2F1 activity. This N-terminal region contains sequence that targets cyclin A-cdk2 to the protein, resulting in the phosphorylation of the DP1 heterodimeric partner which then leads to an inhibition of DNA binding activity (4, 15, 31). A distinct N-terminal domain of E2F1 is responsible for targeting the Skp2-containing SCF complex to the protein, resulting in ubiquitin-mediated degradation of E2F1 (20). Both events thus contribute to the regulated accumulation of E2F1 protein and activity during the cell cycle. Although equivalent experiments have not been performed for E2F2 or E2F3a, the similar patterns of accumulation of the proteins and the sequence similarity with E2F1 suggests that both E2F2 and E2F3a will be similarly regulated. In contrast, E2F3b has no such sequence, which is consistent with the constant presence of the E2F3b protein in quiescent and proliferating cells.
Despite these differences, E2F3b does share a nuclear localization sequence with E2F3a that allows the protein to accumulate in the nucleus. This property, together with the additional property of binding to Rb, creates a unique E2F-Rb complex in quiescent and growing cells with the potential to repress transcription of target genes. Given the fact that the remainder of the two E2F3 proteins are identical, one might speculate that whatever determines promoter specificity in the E2F family might be shared between these two E2F3 proteins. As such, it seems possible that the genes that are specifically regulated by E2F3a during the cell cycle might be specifically negatively regulated by E2F3b in quiescent cells.
E2F genes as Myc targets.
Our previous studies have demonstrated an ability of Myc to activate the E2F1 and E2F2 genes (17, 27). The findings presented here now show that the E2F3a transcript is also induced by Myc, thus demonstrating that the transcription of each of the growth-regulated E2F genes is stimulated by Myc. The fact that Myc-mediated activation of E2F3a is dependent on the presence of the E-box elements within the E2F3a promoter suggests a direct role for Myc in the regulation of the E2F3a gene. Moreover, our recent experiments have demonstrated a role for both E2F1 and E2F2 in enabling Myc function since mouse embryo fibroblasts that carry either an E2F1 or an E2F2 null allele are impaired in their ability to respond to Myc-induced cell proliferation or Myc-induced apoptosis (G. Leone, R. Sears, S. J. Field, M. A. Thompson, H. Yang, Y. Fujiwara, M. E. Greenberg, S. Orkin, J. DeGregori, and J. R. Nevins, submitted for publication). Based on the observations presented here, as well as past experiments that have documented a role for E2F3 activity in cell cycle progression (18), we suspect that the same will be true for E2F3: that the ability of Myc to function in cell proliferation will be dependent on its ability to activate the E2F3a gene.
REFERENCES
- 1.Beijersbergen R L, Kerkhoven R M, Zhu L, Carlee L, Voorhoeve P M, Bernards R. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 1994;8:2680–2690. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- 2.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis and G1/S regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dynlacht B D, Moberg K, Lees J A, Harlow E, Zhu L. Specific regulation of E2F family members by cyclin-dependent kinases. Mol Cell Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson N. The regulation of E2f by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 6.Ginsberg D, Vairo G, Chittenden T, Xiao Z-X, Xu G, Wydner K L, DeCaprio J A, Lawrence J B, Livingston D M. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994;8:2665–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 7.Hann S, Dixit M, Sears R, Sealy L. The alternatively initiated c-Myc proteins differentially regulated transcription through a noncanonical DNA binding site. Genes Dev. 1994;8:2441–2452. doi: 10.1101/gad.8.20.2441. [DOI] [PubMed] [Google Scholar]
- 8.Hiebert S W. Regions of the retinoblastoma gene product required for its interaction with the E2F transcription factor are necessary for E2 promoter repression and pRb-mediated growth suppression. Mol Cell Biol. 1993;13:3384–3391. doi: 10.1128/mcb.13.6.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hijmans E M, Voorhoeve P M, Beijersbergen R L, van'T Veer L J, Bernards R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol Cell Biol. 1995;15:3082–3089. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsiao K-M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 11.Hunter T, Pines J. Cyclins and cancer II: cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 12.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 13.Kowalik T F, DeGregori J, Leone G, Nevins J R. E2F1-specific induction of apoptosis and p53 accumulation is modulated by mdm2. Cell Growth Differ. 1998;9:113–118. [PubMed] [Google Scholar]
- 14.Kowalik T F, DeGregori J, Schwarz J K, Nevins J R. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W G, Livingston D M. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 16.Lees J A, Saito M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leone G, DeGregori J, Sears R, Jakoi L, Nevins J R. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387:422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- 18.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, Miron A, Nevins J R. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marti, A., C. Wirbelauer, M. Scheffner, and W. Krek. Interaction between SCFSKP2 ubiquitin protein ligase and E2F-1 underlies regulation of E2F-1 degradation. Nat. Cell Biol., in press. [DOI] [PubMed]
- 21.Neuman E, Flemington E K, Sellers W R, Kaelin W G., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 23.Qian Y, Luckey C, Horton L, Esser M, Templeton D J. Biological function of the retinoblastoma protein requires distinct domains for hyperphosphorylation and transcription factor binding. Mol Cell Biol. 1992;12:5363–5372. doi: 10.1128/mcb.12.12.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin X-Q, Livingston D M, Ewen M, Sellers W R, Arany Z, Kaelin W G., Jr The transcription factor E2F-1 is a downstream target of RB action. Mol Cell Biol. 1995;15:742–755. doi: 10.1128/mcb.15.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin X-Q, Livingston D M, Kaelin W G, Adams P D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg R A. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sears R, Ohtani K, Nevins J R. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan B, Lee W-H. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vairo G, Livingston D M, Ginsberg D. Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 30.Wu X, Levine A J. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu M, Sheppard K-A, Peng C-Y, Yee A S, Piwnica-Worms H. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol Cell Biol. 1994;14:8420–8431. doi: 10.1128/mcb.14.12.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]