Abstract
The basic helix-loop-helix (bHLH) transcription factors are a large group of proteins suggested to control key events in the development of B lymphocytes as well as of other cellular lineages. To examine how bHLH proteins activate a B-lineage-specific promoter, I investigated the ability of E47, E12, Heb, E2-2, and MyoD to activate the λ5 surrogate light chain promoter. Comparison of the functional capacity of the E2A-encoded E47 and E12 proteins indicated that even though both were able to activate the λ5 promoter and act in synergy with early B-cell factor (EBF), E47 displayed a higher functional activity than E12. An ability to act in synergy with EBF was also observed for Heb, E2-2, and MyoD, suggesting that these factors were functionally redundant in this regard. Mapping of functional domains in EBF and E47 revealed that the dimerization and DNA binding domains mediated the synergistic activity. Electrophoretic mobility shift assay analysis using the 5′ part of the λ5 promoter revealed formation of template-dependent heteromeric complexes between EBF and E47, suggesting that the synergistic mechanism involves cooperative binding to DNA. These findings propose a unique molecular function for E47 and provide overlapping expression with EBF as a molecular mechanism to direct B-cell-specific target gene activation by bHLH proteins.
The B-lymphoid differentiation pathway is a complex process critically dependent on the correct initiation of transcription from stage- and lineage-specific genes (18, 30, 49). The regulation of gene expression is governed by a number of transcription factors, of which some have been shown to be essential for normal B-cell development in mice (12, 59). Among these essential factors are the paired domain factor B-cell-specific activator protein (BSAP) (45, 65), the Zn finger protein IKAROS (67), the Ets protein Pu.1 (36, 55, 56), the helix-loop-helix protein early B-cell factor (EBF) (31) and the basic helix-loop-helix (bHLH) E2A proteins (3, 73). BSAP (1), Pu.1 (25), IKAROS (13), and EBF (15) display tissue-restricted expression patterns, revealing that they are directly involved in the regulation of B-cell-specific genes. In contrast, the E2A-encoded bHLH proteins E47 and E12 (19, 39) are broadly expressed (50), making it difficult to understand how they regulate B-lineage-specific genes. The bHLH proteins belong to an evolutionarily conserved family of transcription factors (5, 38). They participate in such diverse events as sex determination (47) and the formation of the peripheral nervous system in Drosophila (23), as well as in the development of B lymphocytes (5) and muscle in mice (70). They share the common feature of having an HLH dimerization domain adjacent to a basic domain (66). These form a structure with the ability to bind a DNA core site, the E-box (CANNTG) (10), after either homo- or heterodimerization with other bHLH proteins (41, 42). In myogenesis, activation of muscle-specific genes is achieved mainly by heterodimerization between ubiquitously expressed bHLH proteins like E12 or E47 and myocyte-specific bHLH proteins such as MyoD, myogenin, and herculin (29, 40). No lineage-specific bHLH protein has been found in B cells. Instead, cells of the B lineage contain high levels of a number of broadly expressed bHLH proteins (E47, E12, E2-2, and Heb) (2) and a B-cell-specific homodimer of E47 (B-cell factor 1 [BCF1]) (57, 60).
Site selection experiments have suggested that both MyoD and E47 homodimers bind to a similar DNA sequence, making it difficult to understand how these factors discriminate between muscle and B-lineage-restricted target genes (7, 62). One explanation has come from experiments comparing the ability of MyoD and E47 to activate the immunoglobulin heavy-chain (IgH) intron enhancer in nonlymphoid cells (52, 69). These experiments suggested that E47-specific activation was dependent on regions surrounding the core DNA binding site, since these prevented functional interaction of MyoD with the enhancer (69). This provides a model where tissue specificity is regulated in cis by the composition of the E-box and the surrounding sequences (9, 22, 69). E47 and E12 have been suggested to be involved in the regulation of a large number of B-cell-restricted genes (8, 24, 53, 58). However, the majority of these genes are expressed also in pre-T cells, limiting their use as model systems to study B-cell-restricted activation by E2A proteins. One exemption is the promoter of the pre-B-cell-specific λ5 gene (26, 27), where E47 acts in synergy with EBF to stimulate transcription (58). This control element mediates stage- and lineage-specific expression of reporter genes both in transient transfections (32, 33) and in transgenic mice (34). This makes the λ5 promoter a useful model system to investigate B-cell-specific gene activation by bHLH proteins. The promoter contains four E-boxes and at least three EBF binding sites (58). EBF is a transcription factor expressed in B cells, adipocytes, stromal cells, and the central nervous system (15, 68). It binds DNA as a homodimer with a large DNA binding domain including a Zn coordination motif (16). The dimerization is dependent on two α-helixes with homology to helix 2 in the bHLH transcription factor family (15, 16, 64). EBF has been suggested to be essential for the development of B cells since homologous disruption of the coding gene results in a differentiation block at the pro-B-cell stage (31). The importance of E47-EBF cooperation in the promotion of the B lineage is also shown from experiments in mice transheterozygous for mutations both in the E2A and EBF genes. These animals display an enhanced B-cell developmental block at the pre-B-cell stage compared to the single heterozygote mutant mice (46).
To examine how bHLH proteins interact with the promoter of a pre-B-cell-specific gene, I here investigate the ability of a set of bHLH proteins to activate the λ5 promoter alone or in combination with EBF. These experiments suggest that even though the studied class I bHLH proteins, E47, E12, Heb, and E2-2, differ in their quantitative ability to activate the promoter, they all act in synergy with EBF to stimulate transcription. This ability was also shared with MyoD, proposing that the mechanisms regulating tissue specificity of the λ5 promoter differ from those of the IgH intron enhancer. The synergy depended on functional DNA binding and dimerization domains, while the transactivation domains of EBF or E47 could be substituted for by that of the herpes simplex virus VP16 protein. EBF and E47 also appeared to bind DNA in a cooperative fashion, resulting in heteromeric complexes. These data suggest that tissue-specific activation of the λ5 promoter is achieved by overlapping expression of bHLH proteins and EBF, proposing a novel mechanism for the ability of ubiquitously expressed proteins to activate B-cell-specific target genes.
MATERIALS AND METHODS
Tissue culture conditions.
HeLa cells were grown in RPMI medium supplemented with 7.5% fetal calf serum, 10 mM HEPES, 2 mM pyruvate, and 50 μg of gentamicin per ml (complete RPMI media) (all purchased from Life Technologies AB, Täby, Sweden) at 37°C and 5% CO2.
Plasmids and constructs.
The expression plasmids were based on the eukaryotic expression vector cDNA3 (Invitrogen, BV, NV Leek, The Netherlands) which places the inserted cDNA under the control of a cytomegalovirus promoter. To obtain myc-epitope-tagged proteins, I cloned six copies of the myc epitope (51, 61), recognized by the 9E10 mouse monoclonal antibody, ClaI to blunt, EcoRI into BamHI to blunt, and EcoRI of cDNA3 to form the MD3 vector. cDNAs encoding Syrian hamster E47 or E12 (14) were cloned either into the cDNA3 vector or in frame with the myc tag BamHI site of MD3. The E47 forced homodimer (E47FD) construct was generated by joining two copies of a Syrian hamster E47 cDNA with a glycine linker as previously described (58). The E47-E12 forced dimer was constructed by subcloning of the carboxy-terminal E47 and the glycine linker (XhoI-XbaI) from the E47-E47 dimer into pGem3Z. This plasmid was digested with ApaI and XbaI and was ligated to an ApaI-XbaI fragment from Syrian hamster E12 (14). The resulting E12 cDNA was released by digestion with NdeI and XbaI and was ligated into the E47FD plasmid digested with XbaI and partial NdeI to create an E47-E12 forced dimer. The Heb expression plasmid encoded a myc-tagged human Heb protein (21). E2-2 (20) and MyoD (28) expression plasmids were constructed by insertion of full-length mouse cDNAs as EcoRI fragments into the EcoRI site of the cDNA3 expression vector. Δ1 to Δ3 deletions of E47 were introduced by 18 cycles of high-fidelity PCR (Boehringer) from the cDNA3 E47 construct by using the following sense primers: 47Δ1, 5′ TGCAGGATCCGCCGCCATGCGGCGGAGAGCTGCAGACAG; 47Δ2, 5′ TGCAGGATCCGCCGCCATGTTAGGTGACGGCTCGTCC; and 47Δ3, 5′ TGCAGGATCCGCCGCCATGGGCACCCGAGGGACTACATGGC. The primers introduced a BamHI site used to clone the truncations in frame with the myc tag of MD3. A standard SP6 primer directed against the cDNA3 was used as antisense primer, allowing for digestion with XbaI to clone the fragments BamHI to XbaI in the MD3 vector. 47Δ4 was obtained by cloning a NheI-XbaI fragment in frame with the myc tags in MD3. E47Δ5 was constructed by NheI-XbaI digestion of the myc-tagged full-length hamster E47, followed by blunting and religation. VP16EΔ4 was created by cloning a PCR product encoding the VP16 transactivation domain (herpes simplex virus type 1 amino acids 411 to 479) produced by 18 cycles of high-fidelity PCR (Boehringer) by using a Gal-4–VP16-encoding plasmid as template and by cloning the following primers into the BamHI-EcoRI sites of MD3: VP16 sense, 5′ ATGGGATCCCTCGAGATGGCACCCAAGAAGAAGCGG, and Vp16 Nhe, 5′ AGAGAATTCGCTAGCCCAATCGATCCCACCGTACTC. The NheI-XbaI (to blunt) fragment from E47 was then cloned in frame by ligation into an NheI-EcoRV-digested MD3VP16. The T7-tagged EBF protein has been described previously (58). myc-tagged EBF proteins were produced by cloning EBF-encoding cDNAs into the blunted BamHI site and the XbaI site of MD3 to form the fusion proteins. Full-length EBF was cloned by 18 cycles of PCR by using high-fidelity Taq (Boehringer), a full-length EBF cDNA plasmid (EBF17 [15]) as a template, the EBF sense primer 5′ AAAGAGATCTCATATGTTTGGGATCCAGGAAAGC, and an antisense SP6 primer. The PCR product was digested with NdeI, blunted, and redigested with XbaI to be cloned into the MD3 vector. EBFΔ1 (EΔ1) was obtained by PCR by using EBF sense primer and Δ1 antisense primer sequence 5′ AGCTCTAGAGACCGAACTGTTAGCAAGGGC. The resulting PCR product was cloned NdeI to blunt XbaI as described above. EΔ2 was obtained by cloning a KpnI (to blunt)-XbaI-digested full-length PCR product (as described above) into a BamHI (to blunt)-XbaI-digested MD3. EΔ3 protein was obtained by BamHI digestion of the full-length EBF-containing plasmid followed by blunting and religation. The EΔ4 protein was obtained by cloning of a KpnI (to blunt)-XbaI fragment of EBF into a BamHI (to blunt)-XbaI MD3. The EΔ1VP16 protein was obtained by introduction of the VP16 transactivation domain into the XbaI site in the carboxy terminus of EΔ1.
Reporter plasmids were based on the pGL3 luciferase vector (Promega). The λ5 (pGL3λ5) and the basal fos reporter (pGL3fos) have been described previously (58). The μE2-E5 reporter was generated by blunt cloning of an oligonucleotide with two copies of the μE2-μE5-containing region of the IgH intron enhancer. The annealed oligonucleotides μE2-5 sense, (5′ AGAACACCTGCAGCAGCTGGCAGGAGAACACCTGCAGCAGCTGGCAGG) and μE5-2 antisense (5′ CCTGCCAGCTGCTGCAGGTGTTCTCCTGCCAGCTGCTGCAGGTGTTCT) were inserted into the SmaI site of the pGL3fos plasmid. The λ5 promoter E-box reporter was obtained by ligation of two copies of an oligonucleotide spanning the region −331 to −290 of the λ5 promoter (λ5E sense, 5′ TCTTGTTCCATGGGGCAGGTGTTCAGTTGCTCTCTACGGC, and λ5E antisense, 5′ GCCGTAGAGAGCAACTGAACACCTGCCCCATGGAACAAGA, harboring two E-boxes) into the SmaI site of the pGL3fos reporter plasmid. The λ5 fos reporter was obtained by cloning a KpnI-BstEII to blunt fragment from pGL3λ5 into a SmaI-KpnI-digested pGL3fos. This construct was then digested with ApaI and KpnI and was religated to yield the Apa 3′ fos and was digested with ApaI and XhoI followed by religation to obtain the Apa 5′ fos construct. The λ5 shuffled fos plasmid was obtained by ligation of a KpnI-EcoRV fragment from the pGL3λ5 reporter into the Apa 5′ fos plasmid partially NcoI (to blunt)-KpnI-digested to result in sticky blunt cloning. The point-mutated λ5 promoters were generated by PCR with mutated oligonucleotides. EBF site mutations 1, 1 plus 2, and 1 plus 2 plus 3, as well as E-box mutant 1, have been described earlier (58). The E-box mutants 2, 3, and 2 plus 3 were generated by PCR by using mutated sense oligonucleotides together with the GL2 antisense oligonucleotide directed against the luciferase gene and the pGL3λ5 as the template (λ5 E2M, 5′ AGCGGTACCC TGCAGAGAC TC T TGT TCCATGGGGCAGGTGT TAGGT TGC TCTCTACGGC; λ5 E3M, 5′ AGCGGTACCCTGCAGAGACTCTTGTTCCATGGGGTCGGTGTTCAGTTGC; and λ5 E2 plus 3M, 5′ AGCGGTACCCTGCAGAGAC TC T TGT TCCATGGGGTCGGTGT TAGGT TGC TC TCTACGGC). The structures of the reporter plasmids were verified by sequencing.
Transient transfections and luciferase assays.
A total of 250,000 HeLa cells were grown overnight in 1 ml of complete RPMI medium in a 24-well plate. The cells were washed once with serum-free medium (OPTIMEM; Life Technologies), and 800 μl of the serum-free medium was added for transfection. Five microliters of Lipofectin (Life Technologies) was diluted in 100 μl of serum-free medium, was incubated for 45 min at room temperature, and was mixed with the DNA diluted in 100 μl of serum-free medium. The mixture was incubated for 25 min, and the combined volume of 200 μl was added to the HeLa cells. The cells were then incubated in a CO2 incubator at 37°C for 6 h, after which the transfection medium was removed and replaced by complete RPMI medium. The cells were harvested after 40 h, and protein extracts were prepared directly in the 24-well plates by using 80 μl of cell lysis buffer (SDS AB; Promega, Falkenberg, Sweden). Luciferase assays were then conducted with 20 μl of the obtained extracts and 200 μl of luciferase assay reagent (Promega). This procedure resulted in protein extracts of even quality, as judged by Western blotting and repeated transfections of cytomegalovirus-controlled reporter constructs.
Protein extracts, recombinant proteins, and EMSA.
Nuclear extracts were prepared according to Schreiber et al. (54). Recombinant T7-tagged EBF or E2A proteins were generated by coupled in vitro transcription and translation by using a reticulocyte lysate kit (Promega) in the presence or absence of [35S]methionine. Two microliters of a 15-μl reaction mixture was loaded for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and 1.5 μl of the mixture was used for electrophoretic mobility shift assay (EMSA).
DNA probes were labeled with [γ-32P]ATP by incubation with T4 polynucleotide kinase (Life Technologies) and were annealed and purified on a 5% polyacrylamide Tris-borate-EDTA (TBE) gel. The λ5 promoter fragments were obtained by digestion of the pGL3λ5 plasmids with NcoI and either EcoRV or ApaI and by fill-in labeling of the NcoI site with [α-32P]dCTP by incubation with Klenow enzyme. The probes were purified on 5% polyacrylamide–TBE gels, were eluted in Tris-EDTA buffer (pH 8.0), and were precipitated. Nuclear extract or in vitro-transcribed and -translated protein was incubated with labeled probe (20,000 cpm, 3 fmol) for 30 min at room temperature in binding buffer (10 mM HEPES [pH 7.9], 70 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, 2.5 mM MgCl2) with 0.75 μg of poly(dI/dC) (Pharmacia). DNA competitors or antibodies were added 10 min before the addition of the DNA probe. The samples were separated on a 5% polyacrylamide–TBE gel, which was then dried and subjected to autoradiography. A Fuji BAS-III BioImager analyzer was used for quantification of the obtained complexes. The percentage value was calculated by analysis of the relative radioactive content in each of the obtained protein-DNA complexes as well as in the free probe. After subtraction of background activity, the obtained values were added to give the total radioactive content in each lane. The percent value was then calculated by dividing the amount in each complex by that in the whole lane. Oligonucleotides used for electrophoretic mobility shift assays were as follows: mb-1 EBF sense, 5′ GAGAGAGACTCAAGGGAATTGTGG; mb-1 EBF antisense, 5′ CCACAATTCCCTTGAGTCTCTCTC; μE5 sense, 5′ GGCCAGAACACCTGCAGACG; and μE5 antisense, 5′ CGTCTGCAGGTGTTCTGGCC. The 9E10 anti-myc antibody was purchased from Santa Cruz Biotech.
Western blotting.
Protein extracts were separated by SDS–10% PAGE gels and were blotted onto nylon membranes by semidry electroblotting. The membranes were then incubated for 1 h in phosphate-buffered saline supplemented with 0.5% Tween 20 (PBST) and 5% nonfat dry milk and then washed twice for 10 min with PBST. The membranes were then incubated with the primary mouse monoclonal anti-myc antibody 9E10 (Santa Cruz Biotech) or the polyclonal rabbit anti-E2A antibody (Santa Cruz Biotech) in PBST for 1 h. The filters were then washed twice for 15 min (each wash) in PBST before the addition of a 1:2,000 dilution of the secondary horseradish peroxidase-conjugated antibody (anti-mouse antibody for detection of 9E10 anti-myc antibody and anti-rabbit antibody for detection of the anti-E47 antibody, both from Santa Cruz Biotech). After 1 h of incubation with the secondary antibody, the membranes were washed twice for 20 min (each wash) in PBST. Detection of the secondary antibody was obtained by using an enhanced chemiluminescence system (Amersham). All the steps were performed at room temperature.
RESULTS
E2A proteins have partially redundant functions in the regulation of the λ5 promoter.
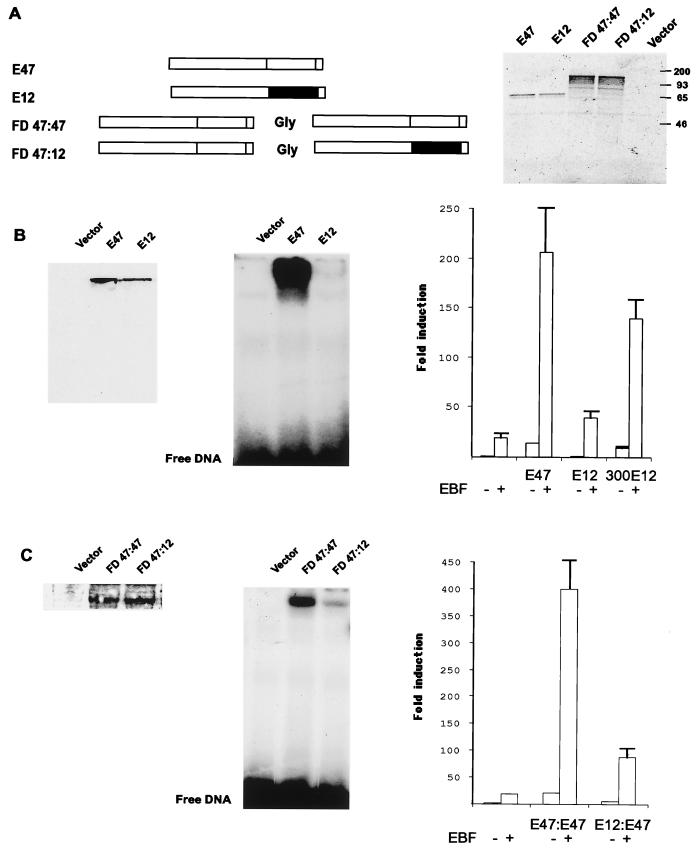
The E2A gene encodes two transcription factors (E12 and E47) that are generated by alternative splicing (39, 41). The findings that B cells contain a lineage-specific homodimer of E47 (57, 60) and that the absence of E47, but not E12, results in a B-cell developmental block (4) have led to the suggestion that E47 might have a unique function in the regulation of B-cell-specific genes. To examine if E47 plays a unique role in the regulation of the λ5 promoter, I made transient transfections of E2A-encoding expression plasmids and a λ5 promoter-controlled luciferase reporter gene. The functional ability of the B-cell-specific E47 homodimer (BCF1) (57, 60) and the ubiquitously formed E47-E12 heterodimer (41) was investigated by the production of forced dimers by the fusion of two Syrian hamster E2A proteins (14) by a glycine linker (Fig. 1A) (44, 58). The integrity of the dimers was investigated by in vitro translation of E47, E12, FD47-47, and FD47-12 followed by separation of the radioactive products by SDS-PAGE (Fig. 1A). Translation of either E47 or E12 resulted in products migrating with apparent molecular masses of about 70 kDa, while both the E47-E47 and the E47-E12 forced dimers migrated with apparent molecular masses of about 130 kDa. This suggests that the forced dimers were translated into intact fusion proteins. To allow for an estimation of protein expression levels in the transfected cells, E12 and E47 were fused to a myc (9e10) tag (51). Figure 1B shows a Western blot of HeLa cell nuclear extracts with the anti-myc antibody. Transfection of the tag results in a band of about 5 kDa (data not shown), while both E47 and E12 are detected as bands migrating with apparent molecular masses of about 70 kDa. This indicated that E47 and E12 were expressed at comparable levels. To compare the ability of these two proteins to interact with DNA, I carried out EMSAs with an IgH E-box element, μE5, and nuclear extracts from the transfected HeLa cells (Fig. 1B). No binding activity could be observed in the nuclear extract from HeLa cells transfected with the myc tag alone, while a strong bandshift was observed in extracts from the E47-transfected cells. The use of nuclear extracts from E12-transfected cells resulted in a faint bandshift, supporting the previous findings that E47 and E12 interact differentially with DNA (62). To investigate if E47 and E12 differ in their abilities to functionally interact with the λ5 promoter, transient transfections were performed by using 50 ng of E47- or E12-encoding expression plasmid in combination with 150 ng of EBF expression plasmid. Inclusion of EBF-encoding plasmid resulted in a 19-fold activation of the reporter gene, while E47 and E12 induced the reporter gene 14- and 2-fold, respectively. The combination of EBF and E47 induced the reporter gene 206-fold, and the combination of EBF and E12 induced the reporter gene 39-fold. Addition of a higher amount (300 ng) of E12 expression plasmid induced the reporter 10-fold in the absence and 139-fold in the presence of EBF. A fos basal promoter was induced less than twofold by the expression of the E2A proteins and EBF, suggesting that the activation was specific (data not shown). Expression of the forced dimers in the HeLa cells was detected by Western blotting using an antibody directed against E2A proteins (Fig. 1C). This resulted in a band of approximately 130 kDa in nuclear extracts from the transfected cells. The same nuclear extracts were then used in an EMSA with the μE5 E-box. The E47-E47-transfected cells contained more μE5 binding activity than the E47-E12-transfected cells, suggesting that the E47 homodimer interacts more efficiently with the μE5 binding site than the E47-E12 heterodimer. The ability of the forced dimers to activate the λ5 promoter was examined as described above. While 100 ng of expression plasmid encoding the E47-E47 homodimer induced the reporter gene 21-fold, the combination with 150 ng of EBF-encoding plasmid resulted in a 413-fold increase of reporter activity. The same experiment using the E47-E12 heterodimer resulted in 6.2- and 89.6-fold inductions, respectively. This suggests that both of the E2A-encoded proteins have the ability to activate the λ5 promoter alone or in cooperation with EBF but that E47 is more potent than E12 in this respect.
FIG. 1.
E2A proteins are partially redundant in their ability to activate the λ5 promoter in synergy with EBF. The left part of panel A shows schematic drawings indicating the structure of the E2A proteins and the E2A forced dimers. The right part shows an autoradiogram from an SDS-PAGE gel with the products obtained after in vitro translation of the E2A proteins and the forced dimers in the presence of [35S]methionine. The left part of panel B shows a Western blot obtained with a 9E10 anti-myc tag antibody and 10 μg of nuclear extracts from HeLa cells transfected with tagged E47 or E12 proteins as indicated. The second gel shows an EMSA analysis with the μE5 E-box from the IgH intron enhancer and 5 μg of the same nuclear extracts as in the Western blot. The right part shows a diagram indicating the luciferase activity relative to that of 200 ng of λ5 promoter-controlled reporter plasmid in the presence of 200 ng of empty expression plasmid, after Lipofectin-mediated transfection of 50 ng of tagged E47 or E12 with 150 ng of empty (−) or EBF-encoding (+) cDNA3 into HeLa cells. The induction obtained with 300 ng of E12 was related to the activity of the reporter transfected with 450 ng of empty cDNA3. The data shown are based on three representative transfection experiments. Error bars indicate standard deviations. The left part of panel C shows the resulting Western blot when nuclear extracts from transiently transfected HeLa cells were probed for the presence of E2A forced dimers by an anti-E2A antibody. The second radiogram shows an EMSA using a labeled μE5 E-box and the same nuclear extracts as in the Western blot. The right part of the panel shows diagrams indicating the luciferase activity obtained after transient transfections of 200 ng of the λ5 reporter plasmid with 150 ng of either empty or E47-E47- or E47-E12-encoding plasmids and 150 ng of empty (−) or EBF-encoding (+) expression plasmid in HeLa cells. The reporter activity obtained with 300 ng of empty expression plasmid was set as 1, and inductions were calculated from three representative transfection experiments. Error bars indicate standard deviations.
Heb, E2-2, and MyoD have the ability to functionally interact with the λ5 promoter and act in synergy with EBF.
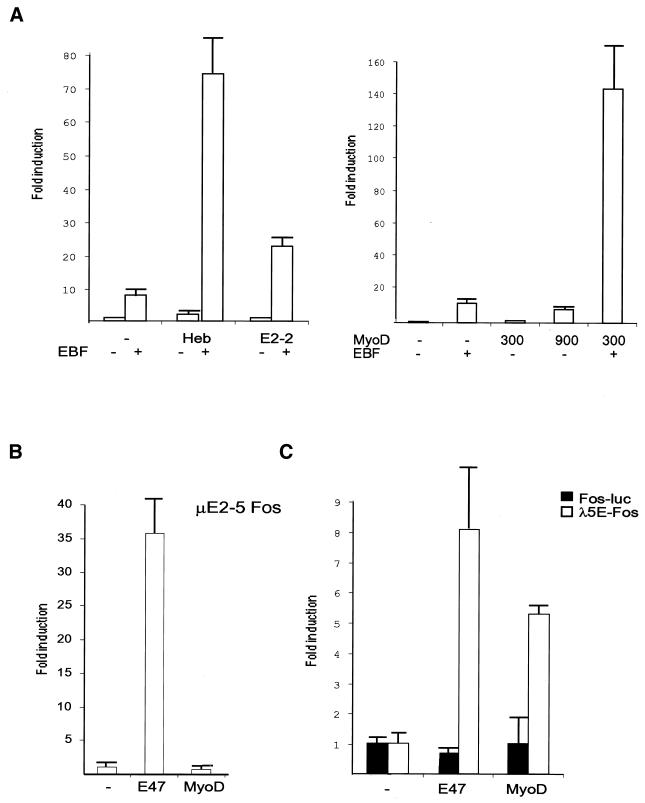
The finding that homologous disruption of the E2A gene by the insertion of a Heb-encoding cDNA rescues B-cell differentiation (71) suggests a molecular redundancy among E2A and Heb proteins. To examine this redundancy at a molecular level, I made transient transfections with the λ5 promoter reporter construct together with bHLH proteins and EBF (Fig. 2A). Inclusion of 150 ng of EBF together with 300 ng of empty expression plasmid induced the reporter gene 10-fold. The lower induction by EBF, as compared to Fig. 1, is probably explained by a larger total amount of DNA being included in this experiment. This was done to partially compensate for the lower expression levels of Heb, E2-2, and MyoD as compared to those obtained with the myc-tagged E2A proteins (data not shown). The inclusion of 300 ng of Heb-encoding expression plasmid activated the reporter twofold in the absence and 73-fold in the presence of EBF. Transfection of E2-2-encoding expression plasmid alone did not significantly affect the expression of the reporter gene, while the combination of EBF and E2-2 resulted in a 22-fold up-regulation. This indicates that both Heb and E2-2 are capable of functionally interacting with EBF and activating the λ5 promoter. To investigate the ability of a myogenic bHLH protein to activate the λ5 promoter, transfections of the λ5 reporter construct in combination with a MyoD-encoding expression vector were performed (Fig. 2A). Addition of 300 ng of MyoD expression plasmid resulted in a twofold induction of reporter activity, while the inclusion of 900 ng of MyoD expression plasmid induced the reporter eightfold. The combination of 300 ng of MyoD and 150 ng of EBF expression plasmids enhanced the reporter activity 141-fold. This indicates that the ability of bHLH proteins to activate the λ5 promoter alone or together with EBF is conserved and not restricted to E2A proteins.
FIG. 2.
MyoD and class I bHLH proteins have the ability to activate the λ5 promoter together with EBF. Panel A shows a diagram of the relative luciferase activity after transfection of 200 ng of λ5 promoter-controlled luciferase reporter plasmid with 300 ng of expression plasmid encoding either Heb, E2-2, or MyoD and 150 ng of either empty (−) or EBF-encoding (+) expression plasmids into HeLa cells. The fold induction was based on the activity obtained when the reporter plasmid was transfected together with 450 ng of empty expression plasmid or 900 ng of empty plasmid when 900 ng of MyoD plasmid was used. The data shown are based on three transfection experiments. Error bars indicate standard deviations. Panel B shows the relative induction of 200 ng of luciferase reporter gene under the control of a basal promoter and two copies of a μE5-μE2 combination from the IgH intron enhancer after transfection of 600 ng of empty, E47-expressing (58), or MyoD-encoding expression plasmid into HeLa cells. The induction was based on data from three representative transfections, and the error bars indicate standard deviations. (C) Relative inductions of 200 ng of reporter constructs controlled either by a basal fos promoter or two copies of the E3-E2 combination (58) from the 5′ part of the λ5 promoter cloned 5′ of the basal fos promoter after transfection with E47 or MyoD expression plasmids in HeLa cells as indicated. The data are collected from three representative transfections, and the error bars indicate the standard deviations.
The finding that MyoD was able to functionally interact with the λ5 promoter is interesting, since studies of the IgH intron enhancer have shown that E-boxes in this control element are nonresponsive to functional activation by myogenic bHLH proteins (69). Based on this, it has been suggested that a major component in tissue-specific gene regulation is a cis repression mechanism. To investigate the function of λ5 promoter E-boxes, I made a set of reporter constructs containing two repeats of either the μE5-μE2 E-boxes from the IgH intron enhancer or the distal E-boxes, E2 and E3 (Fig. 5), from the λ5 promoter cloned 5′ of a basal fos promoter. Transient transfections of these reporter constructs together with expression plasmids coding for either E47 (58) (600 ng) or MyoD (600 ng) showed that while the heavy-chain E-box reporter was induced 35-fold by E47, no induction could be observed after expression of MyoD (Fig. 2B). In contrast, the λ5 promoter E-box reporter was induced eightfold by E47 and 5.3-fold by MyoD (Fig. 2C). The activity of the basal fos promoter was not significantly affected by the expression of the bHLH proteins. This suggests that, in contrast to the IgH intron enhancer, the λ5 promoter is not isolated against activation by myogenic bHLH proteins. Instead, tissue-specific gene activation of λ5 may be a result of overlapping expression of bHLH proteins and EBF.
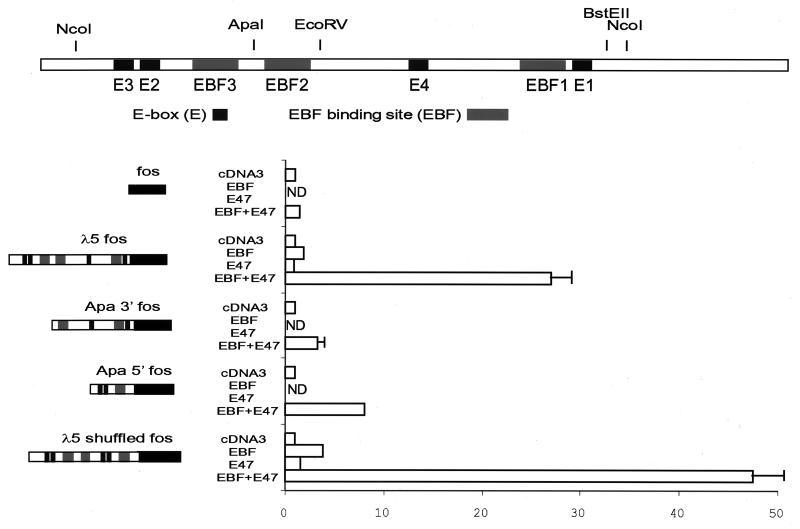
FIG. 5.
Maximal synergy between EBF and E47 is dependent on multiple binding sites within the λ5 promoter. The top part of panel A shows a schematic drawing of the λ5 promoter. Relevant restriction sites as well as binding sites for EBF (gray boxes) and E47 (black boxes) are indicated. The lower part shows the resulting luciferase activity when 200 ng of the indicated λ5 reporter constructs were transiently cotransfected with the indicated expression plasmids (150 ng of EBF and/or 300 ng of E47) into HeLa cells. ND, not done. The reporter activity obtained with 450 ng of empty expression plasmid was set as 1, and data are collected from three representative transfections. Error bars indicate standard deviations.
The functional cooperation between E47 and EBF is dependent upon transactivation domains but is mediated by DNA binding and dimerization domains.
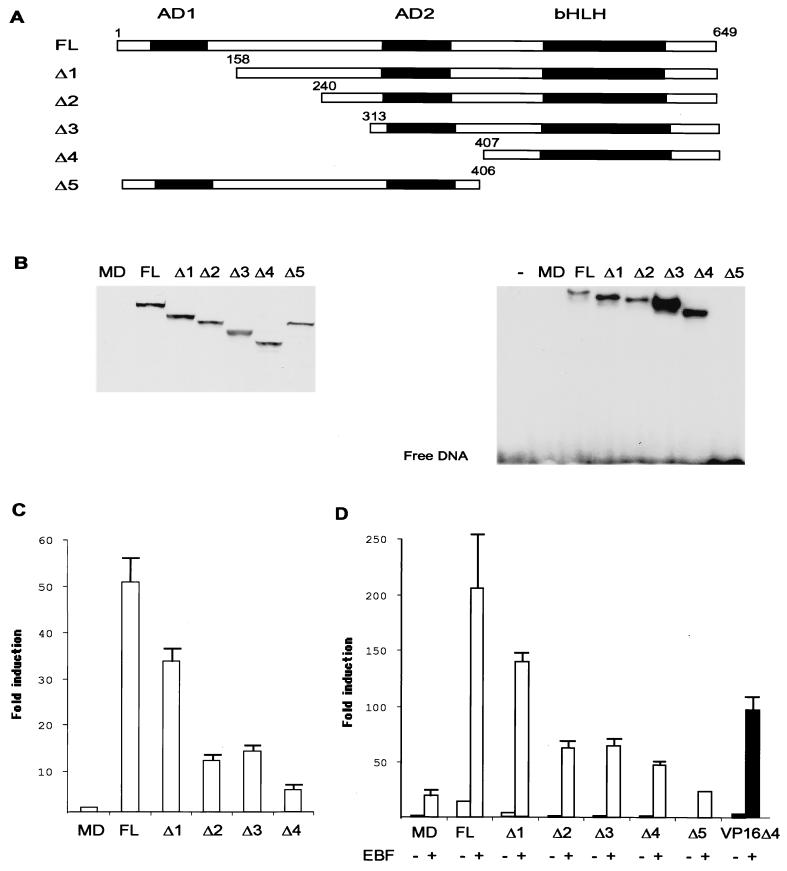
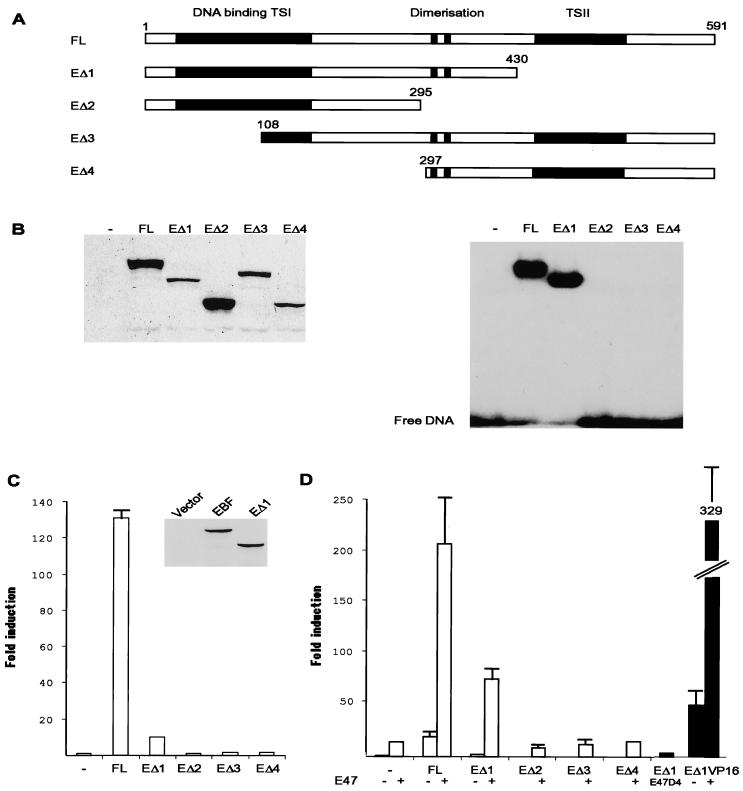
Having established that EBF has the ability to act in synergy with several bHLH family proteins, I wanted to investigate the underlying molecular mechanism. To this end, sequential deletions in the E47 protein were made (Fig. 3A) in frame with an amino-terminal myc (9E10) tag. Δ1 deletes the amino-terminal transactivation domain AD1 (35), and Δ2 and Δ3 delete increasing parts of the amino terminus of E47. The Δ4 deletion also lacks the AD2 activation domain (48), while Δ5 contains a carboxy-terminal deletion, resulting in a protein lacking the DNA binding bHLH domain (39). The expression levels and cellular location of these proteins were investigated by Western blotting nuclear extracts from transfected HeLa cells with a 9E10 anti-myc monoclonal antibody (Fig. 3B). This indicated that all the truncated proteins were expressed at comparable levels. The same nuclear extracts were then used in an EMSA with the μE5 E-box-spanning duplex oligonucleotide. Bands could be detected in nuclear extracts from cells transfected with the full-length as well as the Δ1 to Δ4 proteins. Δ1, Δ3, and Δ4 appeared to interact with DNA more efficiently than the wild-type E47, while Δ5 was unable to interact with DNA. The ability of these truncated proteins to activate the λ5 promoter was examined by transfection of 250 ng of λ5 promoter reporter plasmid with 300 ng of protein-encoding expression plasmids (Fig. 3C). Full-length E47 activated the promoter 50-fold, while Δ1 was able to induce the reporter 33-fold. Δ2, Δ3, and Δ4 protein activated the reporter 11-, 14-, and 5-fold, respectively. The ability of the truncated proteins to cooperate with EBF was examined by transfection of 150 ng of EBF-encoding plasmid together with 50 ng of E47 expression plasmid and 250 ng of λ5 promoter reporter plasmid (Fig. 3D). EBF alone was able to induce the reporter 19-fold, 50 ng of E47 induced the reporter 14-fold, and the combination of EBF and E47 resulted in a 206-fold activation. Transfection of the Δ1 protein resulted in a fourfold increase of reporter activity, while the combination with EBF induced the reporter 140-fold. Transfections of 50 ng of Δ2 and Δ3 induced the reporter 1.2- and 1.3-fold while the Δ4 protein did not induce the reporter (0.94-fold). The combination with EBF resulted in 62-, 64-, and 48-fold inductions, respectively. Cotransfection of Δ5 and EBF resulted in a 24-fold induction of reporter activity. This indicated that even though the full functional synergy between EBF and E47 requires the full transactivation potential of E47, a cooperation also occurs in the absence of the strong transactivation domains. To confirm this, I made a VP16 transactivation domain, E47Δ4 fusion protein (VP16Δ4). The plasmid encoding this protein was transfected alone or together with EBF in an independent experiment. Fifty nanograms of VP16Δ4 induced the λ5 reporter fourfold, while cotransfection with 150 ng of EBF resulted in a 97-fold induction, supporting the idea that the synergistic mechanism is independent of the AD1 and AD2 transactivation domains.
FIG. 3.
The transactivation domains of E47 are essential for full function but not for cooperation with EBF. Panel A shows a schematic drawing of the full-length and truncated E47 proteins that were fused to an amino-terminal 9E10 myc tag. Functional domains in E47 are indicated by black boxes and represent the transactivation domains AD1 and AD2 and the bHLH domain as indicated. The numbers indicate the amino acid positions for the truncations. The figure is not drawn to scale. (B) The left panel shows Western blot analysis with 9E10 anti-myc antibody and 10-μg nuclear extracts from HeLa cells transiently transfected with 800 ng of the indicated E47 protein. The right panel shows an autoradiogram from EMSA using 5-μg nuclear extracts from transiently transfected HeLa cells and an end-labeled oligonucleotide encompassing the μE5 E-box as indicated. (C) Diagram indicating the relative luciferase activity obtained after transient transfections of 200 ng of the λ5 reporter plasmid with 300 ng of E47-encoding plasmids as indicated. The reporter activity obtained with 300 ng of empty expression plasmid was set as one, and the data were calculated from three representative transfection experiments. Error bars indicate standard deviations. (D) Diagram representing the relative luciferase activity obtained when 200 ng of the λ5 reporter plasmid was transfected with 50 ng of E47-encoding plasmids in combination with 150 ng of empty (−) or 9E10-tagged EBF-encoding (+) expression plasmid in HeLa cells. The black bars indicate that these data were collected from an independent transfection experiment. The reporter activity obtained with 200 ng of empty expression plasmid was set as 1, and data are collected from three representative transfections. Error bars indicate standard deviations.
To further examine the mechanisms underlying the functional synergy between EBF and E47, sequential deletions of the EBF protein were fused to an amino-terminal 9E10 myc tag (Fig. 4A). EΔ1 is devoid of the carboxy-terminal transactivation domain but retains its dimerization and DNA binding domains. The EΔ2 protein lacks the dimerization domain, while the EΔ3 and EΔ4 proteins are devoid of the DNA binding domain (15, 16). The nuclear levels of the proteins were examined by Western blot analysis of nuclear extracts from transiently transfected HeLa cells using the anti-myc 9E10 monoclonal antibody (Fig. 4B). All of the proteins could be detected in this experiment even though the protein levels were varying. DNA binding by full-length and EΔ1 proteins, as well as the inability of EΔ2 to -4 to bind DNA, was confirmed by EMSA with the mb-1 promoter EBF binding site (Fig. 4B) and nuclear extracts from transfected HeLa cells. Transfection of 800 ng of full-length EBF together with 250 ng of λ5 promoter reporter plasmid into HeLa cells resulted in a 130-fold induction of luciferase activity (Fig. 4C). Transfection with 800 ng of EΔ1, lacking the carboxy-terminal transactivation domain, resulted in a 10-fold activation of the reporter plasmid, even though the Western blot analysis with the anti-myc antibody suggested that EBF and EΔ1 were present in comparable amounts in the cellular extracts from the transfected cells (Fig. 4C). No activation could be detected after expression of EBF deletants with impaired ability to bind DNA, either due to disruption of the dimerization (Δ2) or the DNA binding domains (Δ3 and Δ4). The ability of the proteins to functionally interact with E47 was examined by transfection of 50 ng of 9E10-tagged E47 in combination with 150 ng of the indicated EBF protein. The addition of 150 ng of full-length EBF resulted in a 19-fold induction of reporter activity, while the combination with E47 induced the reporter 206-fold (Fig. 4D). Performing the same experiment with the EΔ1 protein resulted in 2- and 75-fold inductions, respectively. The combination of E47 and EΔ2 to -4 did not induce reporter activity above the 14-fold induction observed with 50 ng of E47 alone. In contrast to what was observed for E47, where the same relative loss of functional activity was noted both in the presence and absence of EBF, the 10-fold loss of functional activity observed for the EΔ1 protein alone was not completely reflected in the threefold loss of function when combined with E47. One possible explanation for this could be that the interaction of EBF with E47 enhances the functional ability of the context-dependent transactivation domain TSI (16), which is retained in the EΔ1 protein. To investigate this possibility, I cotransfected 150 ng of EΔ1 with E47Δ4 in an independent experiment. This combination resulted in a fourfold induction of the λ5 promoter, suggesting that EBF TSI is unable to support high levels of transcription even in the presence of a bHLH domain. To further investigate this, the TSII domain of EBF was substituted by the transactivation domain from VP16, creating a fusion protein between EΔ1 and VP16. This protein was tested for its ability to activate the λ5 promoter in an independent transfection experiment. One hundred fifty nanograms of the fusion protein resulted in a 52-fold up-regulation of promoter activity, and the inclusion of 50 ng of 9E10-tagged E47 induced the promoter 329-fold. Thus, it is possible to substitute the carboxy-terminal transactivation domain of EBF with a heterologous transactivation domain and still retain functional synergy with E47. These data suggest that both DNA binding and transactivation domains in EBF participate to obtain full function but that the DNA binding and dimerization domains are sufficient to allow functional cooperation with E47.
FIG. 4.
The carboxy-terminal transactivation domain of EBF is important for full functional activity, but DNA binding and dimerization domains are sufficient to mediate synergy with E47. Panel A shows a schematic drawing of the full-length and truncated EBF proteins that were fused to an amino-terminal 9E10 myc tag. Functional domains in EBF are indicated by black boxes and represent the DNA binding, dimerization, and transactivation domains TSI and TSII as indicated. The numbers indicate the amino acid positions for the introduced truncations. The figure is not drawn to scale. (B) The left panel shows Western blot analysis with 9E10 anti-myc antibody and 10-μg nuclear extracts from HeLa cells transiently transfected with 800 ng of the indicated EBF protein. The right panel shows an autoradiogram from EMSA using 5-μg nuclear extracts from transiently transfected HeLa cells and an end-labeled oligonucleotide encompassing the mb-1 promoter EBF binding site as indicated. (C) Diagram indicating the relative luciferase activity obtained after transient transfections of 200 ng of the λ5 reporter plasmid with 800 ng of EBF encoding expression plasmids as indicated. The reporter activity obtained with 800 ng of empty expression plasmid was set as 1, and data were calculated from three representative transfections. Error bars indicate standard deviations. The radiogram presents a Western blot using 9E10 antibody and the same protein extract as used for the luciferase assays. (D) Diagram representing the relative luciferase activity obtained when 200 ng of the λ5 reporter plasmid was transfected with 150 ng of EBF-encoding plasmids in combination with 50 ng of empty (−) or 9E10-tagged E47-encoding (+) expression plasmid in HeLa cells. The black bars indicate that these data were collected from an independent transfection experiment. The reporter activity obtained with 200 ng of empty expression plasmid was set as 1, and data are collected from three representative transfections. Error bars indicate standard deviations.
Functional synergy between EBF and E47 demands multiple binding sites but not the specific configuration of the λ5 promoter.
The λ5 promoter contains at least three EBF binding sites and four E-boxes (Fig. 5), making it difficult to predict the essential features of this control element to allow for functional cooperation between EBF and bHLH proteins. To examine this, I made luciferase reporter constructs where the natural λ5 initiator was substituted with a basal fos promoter (Fig. 5). The activity of this construct was induced twofold by the inclusion of 150 ng of EBF expression plasmid, while no significant induction was observed after the addition of 300 ng of E47 expression plasmid (untagged E47 [58]). The combination of the two factors induced the reporter 28-fold, suggesting that this element is also responsive to synergistic activation in the context of a TATA box-containing promoter. To estimate the relative contribution to the synergy of the different EBF and E47 binding sites in the λ5 promoter, I cloned the 3′ and the 5′ ApaI fragments upstream of basal fos promoters. The 3′ Apa fragment, containing two E-boxes and two EBF sites, was induced threefold, and the 5′ fragment responded to the addition of 150 ng of EBF and 300 ng of E47 by an eightfold increase of functional activity. These data suggest that even though the 5′ part of the promoter supported synergistic activity, the full functional synergy between EBF and E47 does not appear to be a result of any single binding site, but rather of several binding sites within the promoter. To examine whether the full synergistic activity was strictly dependent on promoter structure or, rather, on the number of binding sites, I made a construct with a composite promoter by adding the 5′ EcoRV-KpnI fragment to the KpnI-ApaI construct. This resulted in the same number of binding sites as the wild-type promoter but in an altered configuration. This promoter was induced fourfold by EBF and twofold by E47, while the combination of the factors resulted in a 47-fold increase in functional activity. This suggests that functional synergy between EBF and E47 can be mediated by the 5′ region of the λ5 promoter and is dependent on the number of binding sites rather than on any specific configuration of the control element.
EBF and E47 bind to the λ5 promoter in a cooperative manner.
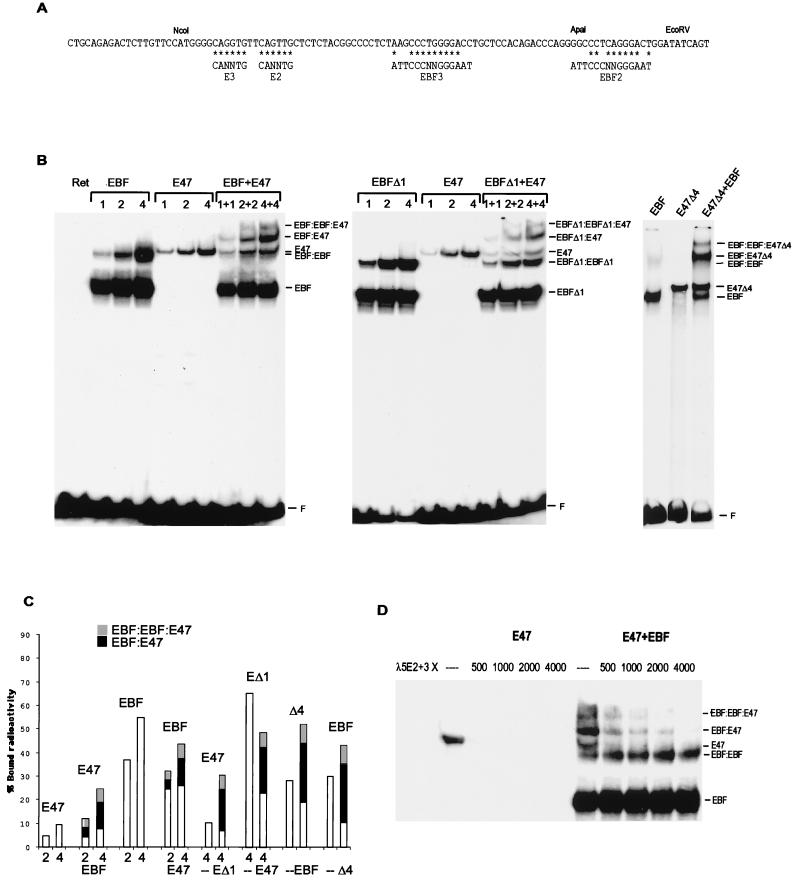
The deletion analysis indicated that the functional cooperation between EBF and E47 was based on an interaction between the DNA binding and dimerization domains rather than on synergy between transactivation domains. One possible explanation would be that the factors bind to DNA cooperatively. To investigate how EBF and E47 interact with the 5′ part of the λ5 promoter, I used an 85-bp fragment containing two defined EBF binding sites and two E-boxes obtained by digestion of the promoter with NcoI and EcoRV (Fig. 6A). This fragment was end labeled with Klenow enzyme and used in an EMSA with increasing amounts of in vitro-translated EBF and/or E47 (Fig. 6B). Inclusion of EBF resulted in two protein DNA complexes composed of EBF dimers (EBF) or tetramers (EBF-EBF) (Fig. 6B), while the addition of E47 resulted in one prominent complex (E47). The combination of EBF and E47 resulted in two additional high-molecular-weight complexes (EBF-E47 and EBF-EBF-E47). This indicated that EBF and E47 form heteromeric complexes on the λ5 promoter. The suggested molecular composition of the complexes was supported by antibody supershift experiments (data not shown), DNA competitions (data not shown), and studies of mutant λ5 promoters (Fig. 7A). To estimate the relative amounts of each of the formed complexes, I measured the radioactivity present in the obtained bands in a Fuji BAS-III BioImager (Fig. 6C). The use of 2 or 4 μl of in vitro-translated EBF resulted in 37 or 55% of the labeled DNA binding to EBF, respectively. The same amounts of E47 resulted in 4.8 or 9.6% of the probe being found in complex with E47. The mixture of 2 μl of EBF and 2 μl of E47 resulted in the EBF-EBF-E47 complex containing 3.4%, the EBF-E47 complex containing 4.7%, the E47 complex containing 4.1%, the EBF-EBF complex containing 3%, and the EBF complex containing 21% of the probe. The corresponding results of using 4 μl of each protein preparation were 6.5% EBF-EBF-E47, 11.0% EBF-E47, 7.1% E47, 3.9% EBF-EBF, and 24% EBF. This suggests that the total amount of radioactivity in complex with E47 increased from 4.8 or 9.6% in the absence to 12.2 or 24.6% in the presence of EBF. The amount of EBF bound to DNA when combined with E47 was slightly reduced from 37 or 58% in the absence to 32.1 or 43.5% in the presence of E47. This loss is probably due to the fact that the addition of E47 has a negative effect on the formation of the tetrameric EBF complex (EBF-EBF), possibly by competing for DNA-bound EBF to create EBF-E47 tetramers. Thus, the combination of EBF and E47 results in a 2.5-fold increase in E47 DNA binding (Fig. 6C) activity and a preferential formation of ternary complexes, even in the presence of free DNA.
FIG. 6.
EBF and E47 form a stabilized ternary complex on the λ5 promoter. (A) DNA sequence of the 5′ region of the λ5 promoter with indicated EBF binding sites and E-boxes. Asterisks indicate base pair matches to the consensus binding sites. (B) Autoradiograms from EMSA with an end-labeled λ5 5′ NcoI-EcoRV fragment and increasing amounts of in vitro-translated EBF or/and E47 proteins as indicated. The middle panel shows an EMSA with the same probe and increasing amounts of in-vitro-translated EBFΔ1 and/or E47. The amount of protein in each reaction was normalized by the addition of nonprogrammed reticulocyte lysate. The right panel displays an EMSA with the λ5 promoter probe and 7.5 μg of nuclear extracts from transfected HeLa cells in combination with 1.5 μl of in vitro-translated EBF. The gels displayed are representative of two experiments. F, free DNA. (C) A diagram compiling the data obtained by densitometric analysis of the obtained EMSA complexes. The bars represent the total relative amount of radioactivity in complex with protein and are divided into sections according to the presence of the protein in homo- (white) or heteromeric (black and grey) complexes as indicated. (D) Autoradiogram of an EMSA where the preformed complexes between E47 and/or EBF on the λ5 promoter have been distorted by the addition of increasing amounts of unlabeled oligonucleotides spanning the E2 and E3 boxes in the λ5 promoter as indicated. The gel displayed is representative of three experiments. The probe is not shown, since the gel has been cut to save space.
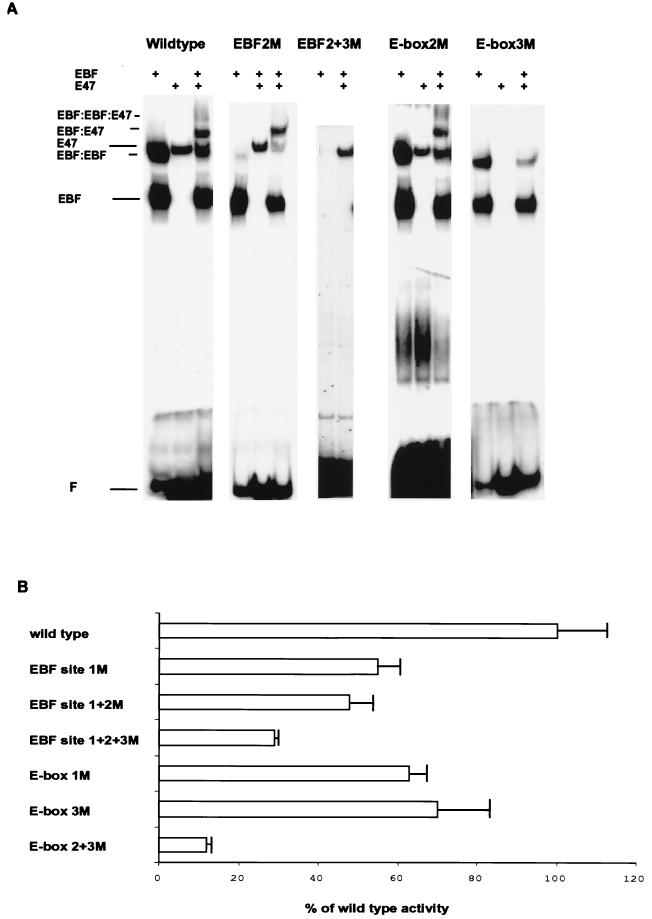
FIG. 7.
The formation of ternary complexes and functional synergy between EBF and E47 demands functional binding sites for both factors. (A) Autoradiograms from EMSAs with end-labeled wild-type or mutated 5′ NcoI-EcoRV fragments and 4 μl of in vitro-translated recombinant EBF and/or E47 as indicated. The gels displayed each represent one out of three experiments. F, free DNA. (B) The resulting luciferase activity when 200 ng of the indicated λ5 promoter reporter constructs was transiently cotransfected with 150 ng of EBF- and 50 ng of E47-encoding expression plasmids into HeLa cells. The reporter activity obtained with the wild-type promoter was set as 1, and data were collected from three representative transfections. Error bars indicate standard deviations.
To further investigate the formation of the ternary complex, EMSAs were conducted with the λ5 promoter fragment and in vitro-translated truncated EBF proteins. The complex formation pattern generated by EBFΔ1 (Fig. 4A, EΔ1) protein in combination with E47 largely resembled that of full-length EBF (Fig. 6B). Quantitation of two independent experiments in the BioImager (Fig. 6C) suggested that the combination of 4 μl of EBFΔ1 and 4 μl of E47 resulted in an EBFΔ1-EBFΔ1-E47 complex containing 6.2% and a EBFΔ1-E47 complex containing 16.3%. The E47 complex contained 7.7%, the EBFΔ1-EBFΔ1 complex contained 4.3%, and the EBFΔ1 complex contained 24% of the radioactivity. The total amount of bound E47 increased from 10.3% in the absence to 30.2% in the presence of EBFΔ1 (Fig. 6C). No complex formation in addition to the E47 complex was observed by using either in vitro-translated EΔ2 or EΔ3 proteins or nuclear extracts from cells transfected with these truncated proteins (Fig. 4B) (data not shown). A slightly different approach was used to investigate the ability of the bHLH domain of E47 (Fig. 3A, E47Δ4) to form ternary complexes with EBF, because the EBF-E47Δ4 complex migrated with the same mobility as the EBF-EBF complex, forcing me to use lower amounts of EBF to reduce the formation of the latter complex. Mixing the nuclear extract from cells transfected with the E47-Δ4 protein with 1.5 μl of in vitro-translated EBF resulted in two large complexes (EBF-EBF-E47Δ4 and EBF-E47Δ4) that could not be detected in the nuclear extract alone (Fig. 6B). The presence of E47Δ4 protein in these complexes was confirmed by the addition of anti-myc antibody, suggesting that this is a ternary complex formed by double occupancy of EBF and E47Δ4 (data not shown). Quantitative analysis of two independent experiments as described above revealed that the EBF complex contained 11% and the E47Δ4 complex contained 20% of the total radioactivity (Fig. 6B). The ternary complexes contained 32% of the total amount of radioactivity. In two parallel reactions containing unprogrammed reticulocyte lysate (E47Δ4) or nuclear extract from HeLa cells transfected with empty vector (EBF), the relative amount of radioactivity detected in complex with E47Δ4 was 28% and the relative amount of radioactivity detected in complex with EBF was 30% (Fig. 6C). Thus, the total amount of bound E47 increased from 28 to 52% in the presence of EBF. In this experiment, there was also an increase of EBF binding from 30 to 43% in the presence of E47 (Fig. 6C). EMSA with nuclear extracts from E47Δ5-transfected cells and recombinant EBF did not result in any ternary complex formation (data not shown). This suggests that the formation of ternary complexes on this region of the λ5 promoter is dependent on the DNA binding and dimerization domains of both EBF and E47.
To investigate if this could be an effect of altered kinetics in protein-DNA complex formation, I conducted EMSA analysis with different preincubation times of the labeled DNA and in vitro-translated proteins. The kinetics for the formation of the ternary complex followed that of EBF, suggesting that EBF and E47 bind the DNA independently (data not shown). To investigate if the ternary complex formation resulted in a stabilization of the protein-DNA complex, I carried out an EMSA competition assay of E47 bound to DNA alone or in complex with EBF (Fig. 6D). The recombinant proteins were incubated with the NcoI-EcoRV λ5 promoter fragment for 15 min, after which increasing amounts of unlabeled duplex oligonucleotides containing the λ5 E2 and E3 E-boxes were added. The binding of E47 alone to the probe was already severely reduced at a 500-fold excess of unlabeled oligonucleotide, while the EBF-E47 complex could be detected at a 2,000-fold excess of competitor. This indicates that the ternary complex between EBF and E47 has a higher stability than that obtained by E47 bound to DNA by itself.
Ternary complexes between EBF and E47 form predominantly by binding to EBF site 3 and E-box 3 in the λ5 promoter.
To investigate the template requirements for the formation of ternary complexes, I introduced point mutations in the EBF binding sites and the E-boxes in the λ5 promoter. The mutated fragments were end labeled and tested in EMSAs with in vitro-translated EBF and E47 (Fig. 7A). Mutation in EBF site 2 did not have any large effect on EBF, E47, or EBF-E47 complex formation, while the formation of EBF-EBF and EBF-EBF-E47 complexes was severely impaired. Mutation in both EBF sites 2 and 3 dramatically reduced EBF binding as well as ternary complex formation. Alterations in E-box 2 did not significantly affect either the binding of E47 to the promoter or the formation of any ternary complex. This was contrasted by a mutation in E-box 3, because this resulted in severely impaired ability to interact with E47 and to form ternary complexes between EBF and E47. This suggests that the ternary complex EBF-E47 is template dependent and formed primarily by interaction of E47 with E-box 3 and EBF with EBF site 3, while the formation of the EBF-EBF and EBF-EBF-E47 complexes requires the presence of two functional EBF binding sites.
To investigate the functional roles of the different EBF and E47 binding sites in the λ5 promoter (Fig. 7B), I transfected a set of point-mutated promoters coupled to a luciferase reporter gene into HeLa cells together with expression plasmids encoding EBF and E47 as above. Mutation in EBF site 1 (Fig. 5 and 7B) resulted in 55% of the activity of the wild-type promoter, while the combined mutation in EBF sites 1 and 2 reduced the activity to 48% of that of the wild type. The combined mutation of EBF sites 1, 2, and 3 resulted in a promoter induced to 30% of the wild-type activity (Fig. 7B). Introduction of point mutations in E-box 1 or 3 reduced the activity to 63 and 70%, respectively, while the combined mutation of E-box 2 and 3 reduced the functional activity to 12% of that of the wild-type promoter (Fig. 7B). Hence, even though some binding site redundancy is apparent, full functional synergy requires functional binding sites for both EBF and E47.
DISCUSSION
Here, I present data suggesting that the ability to activate a B-lineage-specific promoter is shared by class I bHLH proteins as well as by MyoD. The redundancy is, however, not complete, since the E2A proteins E47 and E12 differed in their quantitative ability to activate the λ5 promoter. The ability of MyoD to functionally interact with the E-boxes from the λ5 promoter, but not the μE2-μE5 E boxes from the IgH intron enhancer, suggests that combinatorial effects of EBF expression and bHLH proteins rather than the E-box composition can dictate B-cell specificity. Finally, I present data suggesting that even though the full function was dependent on the individual transactivation domains of both proteins, DNA binding and dimerization appear to be the most critical feature of E47 and EBF to obtain functional synergy, probably by forming a ternary complex with increased stability.
The E2A gene generates the two transcription factors E12 and E47 by alternative usage of the bHLH domain (39, 41). The gene has been shown to be essential for B-cell development by targeted disruption in mice. These animals display a pre-pro-B-cell differentiation block with surface expression of B220 and CD43 and mRNA production from the B29 gene and sterile μ0 transcripts but without expression of several other early B-cell markers (3). Another E2A-deficient mouse strain shows an even earlier developmental block, further supporting the relevance of E2A proteins in the earliest stages of B-cell development (73). The individual roles for E47 and E12 have been examined in transgenic mice, and they suggest that while E47 alone allowed for the formation of B cells, E12 only supported B-cell development to the pre-B-cell stage (4). Disruption of the E2-2 or Heb gene resulted in a reduction of number of B cells in fetal liver but with development of mature B lymphocytes (72). These findings would suggest that E47 and possibly the B-cell-specific E47 homodimer (BCF1) (2, 57) have a unique role in B-cell development. This is contradicted by the finding that functional replacement of the E2A gene by a Heb-encoding cDNA restores B-cell development in the absence of E2A-encoded gene products (71). This suggests that the unique role of E47 may result from the level of expression rather than from the biochemical and functional features of the protein. Such a dose dependency model is also supported by the findings that mice heterozygous for the E2A mutation develop fewer B cells than wild-type mice (73) and that this phenotype is enhanced when introduced on E2-2-deficient genetic background (72). Dosage effects have also been suggested from observations in Drosophila sex determination (47) and in myogenesis in mice (6). The findings I present in this report provide a possible molecular explanation for the apparently contradictory data obtained from the studies in transgenic mice, since E47 appears more able to induce a B-cell-specific target gene than E12. In a dose dependency model, E47 would provide a higher functional dose per molecule than E12. Therefore, compensation of E47 with the same level of E12 would result in a net loss of functional E protein activity. The apparently higher functional activity of an E47 homodimer, as compared to an E12-E47 heterodimer, also provides a possibly unique role for the B-cell-specific E47 homodimer BCF1 (2, 57) since this provides the highest functional dose. This may be a feature highly relevant for a lineage critically dependent on a high functional activity of bHLH proteins.
B-cell-restricted activation by broadly expressed bHLH proteins has been a rather puzzling phenomenon since no or small differences have been detected in the DNA binding specificity of B-cell-specific E47 and myocyte-specific MyoD complexes (7, 62). Instead, tissue-specific activation has been attributed to the fact that even though the protein binds to DNA, there are additional requirements for the target site to obtain functional activation (22, 69). This is probably the case for the IgH intron enhancer, where a VP16 MyoD fusion but not MyoD can activate transcription (69). This is also supported by the finding that E47 but not MyoD has the ability to activate germ line transcription from the IgH intron enhancer, Iμ, in fibroblasts (8). This is in contrast to findings obtained in this study, where both E47 and MyoD are capable of activating a B-lineage-restricted control element. However, the activity obtained with either of the bHLH proteins was rather low compared to that obtained when combined with EBF. This leads me to suggest that the tissue specificity of the λ5 promoter is a result of the combined expression of EBF and bHLH proteins, a combination likely to be rather unique for the B lineage. This does not explain the stage specificity of the λ5 promoter (32–34) even though a possible explanation rests in the finding that EBF levels are down-regulated in the B cell compared to the pre-B cell (11, 15, 17). Synergistic effects by overlapping expression patterns of transcription factors have also been reported from a large number of differentiation systems like Mef-MyoD in myogenesis (37) and C/EBFα–peroxisome proliferator-activated receptor γ2 (63) in adipogenesis. E47 has also been shown to act in synergy with the tissue-restricted Pu-interacting protein Pip to activate the Igλ enhancer (43). These collected findings suggest that B-lineage-specific activation by bHLH proteins may be accomplished either by cis repression mechanisms or by overlapping expression with tissue-restricted factors.
Functional synergy between transcription factors has been shown to be mediated by a number of distinct mechanisms. For instance, cooperation between Pip and E47 appears to be a result of enhanced DNA binding to adjacent sites, while the functional interaction between Mef2 and MyoD is dependent on interactions between DNA binding domains, but only on one functional binding site for either of the factors (37). Full functional synergy between EBF and E47 appeared to involve the transactivation as well as the DNA binding domains of the factors in combination with multiple binding sites for both proteins in the target promoter. However, a functional cooperation could also be observed in the absence of EBF or E47 transactivation domains, suggesting that the molecular mechanisms involve interactions between DNA binding and dimerization domains. One possible explanation for this is that the functional synergy is dependent on cooperative DNA binding. This was also supported by the disproportionally large amount and stability of heteromeric protein complexes formed on the λ5 promoter in EMSA. The formation of the ternary complexes in vitro appeared to be dependent on EBF site 3 and E-box 3. This is also in line with the notion that those binding sites carry the highest match to the defined consensus sites for the proteins (7, 64). The importance of these distinct sites for the functional activity was not as striking, indicating that other binding sites may compensate for these in the HeLa cells.
The findings that EBF and E47 were able also to act in synergy on templates structurally different from the λ5 enhancer and that mice transheterozygous for mutations in the E2A and EBF genes display a pre-B-cell differentiation block that cannot be completely explained by the absence of λ5 (46) suggest that these proteins may act in concert to activate other genes important for B-cell development. Identification of these genes will most probably increase our understanding of the molecular events involved in the progression of B-cell differentiation.
ACKNOWLEDGMENTS
I thank Y. Zhuang and R. Grosschedl for their kind gifts of plasmids, L. Erlandsson for critically reading the manuscript, and P. Åkerblad and D. Liberg for helpful discussions.
This work was funded by the Swedish Medical Research Council, the Swedish Cancer Foundation, the Åke Wibergs foundation, the Magnus Berwalls Foundation, the Kocks Foundation, the Österlunds Foundation, and the Crafoord Foundation.
REFERENCES
- 1.Adams B, Dorfler P, Aguzzi A, Kozmik Z, Urbanek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 2.Bain G, Gruenwald S, Murre C. E2A and E2-2 are subunits of B-cell-specific E2-box DNA-binding proteins. Mol Cell Biol. 1993;13:3522–3529. doi: 10.1128/mcb.13.6.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain G, Maandag E C R, Izon D J, Amsen D, Kruisbeek A M, Weintraub B C, Kroop I, Schlissel M S, Feeney A J, van Roon M, van der Valk M, te Riel H P J, Berns A, Murre C. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 4.Bain G, Maandag E C R, te Riele H P J, Feeney A J, Sheehy A, Schlissel M, Shinton S A, Hardy R R, Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 5.Bain G, Murre C. The role of E-proteins in B- and T-lymphocyte development. Semin Immunol. 1998;10:143–153. doi: 10.1006/smim.1998.0116. [DOI] [PubMed] [Google Scholar]
- 6.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell T K, Weintraub H. Differences and similarities in the DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 8.Choi J K, Shen C-P, Radomska H S, Eckhardt L A, Kadesch T. E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO J. 1996;15:5014–5021. [PMC free article] [PubMed] [Google Scholar]
- 9.Davis R L, Cheng P F, Lassar A B, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 10.Ephrussi A, Church G M, Tonegawa S, Gilbert W. B lineage-specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985;227:134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- 11.Feldhaus A L, Mbangkollo D, Arvin K L, Klug C A, Singh H. BLyF, a novel cell-type- and stage-specific regulator of the B-lymphocyte gene mb-1. Mol Cell Biol. 1992;12:1126–1133. doi: 10.1128/mcb.12.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgopoulos K. Transcription factors required for lymphoid lineage commitment. Curr Opin Immunol. 1997;9:222–227. doi: 10.1016/s0952-7915(97)80139-2. [DOI] [PubMed] [Google Scholar]
- 13.Georgopoulos K, Moore D D, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258:808–812. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- 14.German M S, Blanar M A, Nelson C, Moss L G, Rutter W J. Two related helix-loop-helix proteins participate in separate cell-specific complexes that bind the insulin enhancer. Mol Endocrinol. 1991;5:292–299. doi: 10.1210/mend-5-2-292. [DOI] [PubMed] [Google Scholar]
- 15.Hagman J, Belanger C, Travis A, Turck C W, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 16.Hagman J, Gutch M J, Lin H, Grosschedl R. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagman J, Travis A, Grosschedl R. A novel lineage-specific nuclear factor regulates mb-1 gene transcription at the early stages of B cell differentiation. EMBO J. 1991;10:3409–3417. doi: 10.1002/j.1460-2075.1991.tb04905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson A, Calame K. Transcriptional regulation during B cell development. Annu Rev Immunol. 1998;16:163–200. doi: 10.1146/annurev.immunol.16.1.163. [DOI] [PubMed] [Google Scholar]
- 19.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer μE5/κE2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 20.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 21.Hu S-J, Olson E N, Kingston R E. HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA-binding activity of myogenic regulatory factors. Mol Cell Biol. 1992;12:1031–1042. doi: 10.1128/mcb.12.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, Blackwell T K, Kedes L, Weintraub H. Differences between MyoD DNA binding and activation site requirements revealed by functional random sequence selection. Mol Cell Biol. 1996;16:3893–3900. doi: 10.1128/mcb.16.7.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kageyama R, Nakanishi S. Helix-loop-helix factors in growth and differentiation of the vertebrate nervous system. Curr Opin Genet Dev. 1997;7:659–665. doi: 10.1016/s0959-437x(97)80014-7. [DOI] [PubMed] [Google Scholar]
- 24.Kee B L, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klemsz M J, McKercher S R, Celada A, van Beveren C, Maki R A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 26.Kudo A, Sakaguchi N, Melchers F. Organization of the murine Ig-related lambda 5 gene transcribed selectively in pre-B lymphocytes. EMBO J. 1987;6:103–107. doi: 10.1002/j.1460-2075.1987.tb04725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kudo A, Thalmann P, Sakaguchi N, Davidson W F, Pierce J H, Kearney J F, Reth M, Rolink A, Melchers F. The expression of the mouse VpreB/lambda 5 locus in transformed cell lines and tumors of the B lineage differentiation pathway. Int Immunol. 1992;4:831–840. doi: 10.1093/intimm/4.8.831. [DOI] [PubMed] [Google Scholar]
- 28.Lassar A B, Buskin J N, Lockshon D, Davis R L, Apone S, Hauschka S D, Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell. 1989;58:823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- 29.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronora A, Baltimore D, Weintraub H. Functional activity of myogenic HLH protein requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 30.Liberg D, Sigvardsson M. Transcriptional regulation in B cell differentiation. Crit Rev Immunol. 1999;19:127–153. [PubMed] [Google Scholar]
- 31.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 32.Martensson A, Martensson I L. Early B cell factor binds to a site critical for lambda5 core enhancer activity. Eur J Immunol. 1997;27:315–320. doi: 10.1002/eji.1830270145. [DOI] [PubMed] [Google Scholar]
- 33.Martensson I L, Melchers F. Pre-B cell-specific lambda 5 gene expression due to suppression in non pre-B cells. Int Immunol. 1994;6:863–872. doi: 10.1093/intimm/6.6.863. [DOI] [PubMed] [Google Scholar]
- 34.Martensson I L, Melchers F, Winkler T H. A transgenic marker for mouse B lymphoid precursors. J Exp Med. 1997;185:653–661. doi: 10.1084/jem.185.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massari M E, Jennings P A, Murre C. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol Cell Biol. 1996;16:121–129. doi: 10.1128/mcb.16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKercher S R, Torbett B E, Anderson K L, Henkel G W, Vestal D J, Baribault H, Klemsz M, Feeney A J, Wu G E, Paige C J, Maki R A. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 37.Molkentin J D, Olson E N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murre C, Bain G, van Dijk M A, Engel I, Furnari B A, Massari M E, Matthews J R, Quong M W, Rivera R R, Stuiver M H. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 39.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 40.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 41.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 42.Murre C, Voronova A, Baltimore D. B-cell- and myocyte-specific E2-box-binding factors contain E12/E47-like subunits. Mol Cell Biol. 1991;11:1156–1160. doi: 10.1128/mcb.11.2.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagulapalli S, Atchison M L. Transcription factor Pip can enhance DNA binding by E47, leading to transcriptional synergy involving multiple protein domains. Mol Cell Biol. 1998;18:4639–4650. doi: 10.1128/mcb.18.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neuhold L A, Wold B. HLH forced dimers: tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell. 1993;74:1033–1042. doi: 10.1016/0092-8674(93)90725-6. [DOI] [PubMed] [Google Scholar]
- 45.Nutt S L, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- 46.O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 47.Parkhurst S M, Bopp D, Ish-Horowicz D. X:A ratio, the primary sex-determining signal in Drosophila, is transduced by helix-loop-helix proteins. Cell. 1990;63:1179–1191. doi: 10.1016/0092-8674(90)90414-a. [DOI] [PubMed] [Google Scholar]
- 48.Quong M W, Massari M E, Zwart R, Murre C. A new transcriptional-activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol Cell Biol. 1993;13:792–800. doi: 10.1128/mcb.13.2.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reya T, Grosschedl R. Transcriptional regulation of B-cell differentiation. Curr Opin Immunol. 1998;10:158–165. doi: 10.1016/s0952-7915(98)80244-6. [DOI] [PubMed] [Google Scholar]
- 50.Roberts V J, Steenbergen R, Murre C. Localization of E2A mRNA expression in developing and adult rat tissues. Proc Natl Acad Sci USA. 1993;90:7583–7587. doi: 10.1073/pnas.90.16.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roth M B, Zahler A M, Stolk J A. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruezinsky D, Beckmann H, Kadesch T. Modulation of the IgH enhancer's cell type specificity through a genetic switch. Genes Dev. 1991;5:29–37. doi: 10.1101/gad.5.1.29. [DOI] [PubMed] [Google Scholar]
- 53.Schlissel M, Voronova A, Baltimore D. Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 1991;5:1367–1376. doi: 10.1101/gad.5.8.1367. [DOI] [PubMed] [Google Scholar]
- 54.Schreiber E, Matthias P, Müller M, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott E W, Fisher R C, Olson M C, Kehrli E W, Simon M C, Singh H. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 56.Scott E W, Simon M C, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 57.Shen C P, Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 59.Singh H. Gene targeting reveals a hierarchy of transcription factors regulating specification of lymphoid cell fates. Curr Opin Immunol. 1996;8:160–165. doi: 10.1016/s0952-7915(96)80053-7. [DOI] [PubMed] [Google Scholar]
- 60.Sloan S R, Shen C-P, McCarrick-Walmsley R, Kadesh T. Phosphorylation of E47 as a potential determinant of B-cell-specific activity. Mol Cell Biol. 1996;16:6900–6908. doi: 10.1128/mcb.16.12.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sogaard M, Tani K, Ye R R, Geromanos S, Tempst P, Kirchhausen T, Rothman J E, Sollner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 62.Sun X H, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- 63.Tontonoz P, Hu E, Spiegelman B M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. . (Erratum, 80:957, 1995.) [DOI] [PubMed] [Google Scholar]
- 64.Travis A, Hageman J, Hwang L, Grosschedl R. Purification of early-B-cell factor and characterization of its DNA-binding specificity. Mol Cell Biol. 1993;13:3392–3400. doi: 10.1128/mcb.13.6.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urbanek P, Wang Z Q, Fetka I, Wagner E F, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 66.Voronova A, Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J H, Nichogiannopoulou A, Wu L, Sun L, Sharpe A H, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 68.Wang M M, Reed R R. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 69.Weintraub H, Genetta T, Kadesch T. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev. 1994;8:2203–2211. doi: 10.1101/gad.8.18.2203. [DOI] [PubMed] [Google Scholar]
- 70.Yun K, Wold B. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr Opin Cell Biol. 1996;8:877–889. doi: 10.1016/s0955-0674(96)80091-3. [DOI] [PubMed] [Google Scholar]
- 71.Zhuang Y, Barndt R J, Pan L, Kelley R, Dai M. Functional replacement of the mouse E2A gene with a human HEB cDNA. Mol Cell Biol. 1998;18:3340–3349. doi: 10.1128/mcb.18.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2 and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]