Abstract
Given the worldwide burden of neglected tropical diseases, there is ongoing need to develop novel anthelmintic agents to strengthen the pipeline of drugs to combat these burdensome infections. Many diseases caused by parasitic flatworms are treated using the anthelmintic drug praziquantel (PZQ), employed for decades as the key clinical agent to treat schistosomiasis. PZQ activates a flatworm transient receptor potential (TRP) channel within the melastatin family (TRPMPZQ) to mediate sustained Ca2+ influx and worm paralysis. As a druggable target present in many parasitic flatworms, TRPMPZQ is a promising target for a target-based screening campaign with the goal of discovering novel regulators of this channel complex. Here, we have optimized methods to miniaturize a Ca2+-based reporter assay for Schistosoma mansoni TRPMPZQ (Sm.TRPMPZQ) activity enabling a high throughput screening (HTS) approach. This methodology will enable further HTS efforts against Sm.TRPMPZQ as well as other flatworm ion channels. A pilot screen of ~16,000 compounds yielded a novel activator of Sm.TRPMPZQ, and numerous potential blockers. The new activator of Sm.TRPMPZQ represented a distinct chemotype to PZQ, but is a known chemical entity previously identified by phenotypic screening. The fact that a compound prioritized from a phenotypic screening campaign is revealed to act, like PZQ, as an Sm.TRPMPZQ agonist underscores the validity of TRPMPZQ as a druggable target for antischistosomal ligands.
Author summary
The drug praziquantel is used to treat diseases caused by parasitic flatworms. Praziquantel is an old drug, and there is a need to identify novel treatments that retain desirable features and improve weaknesses in the mode of PZQ action. One way to do this is to identify new drugs that exploit vulnerabilities in the same drug target but work in slightly differently ways. Here, we have optimized high throughput screening methods to pharmacologically profile a parasitic flatworm ion channel targeted by PZQ. We have identified several new chemical structures that interact with this channel complex. These ligands provide new opportunity for developing tools to manipulate flatworm biology and potentially new trajectories for anthelmintic drug development.
Introduction
Over a billion people worldwide require chemotherapy for neglected tropical diseases (NTDs, [1]). Schistosomiasis, a disease caused by infection by parasitic flatworms known as schistosomes, is one of several NTDs targeted for elimination as a public health problem in the World Health Organization 2021–2030 NTD road map [1]. Schistosomiasis, as well as several other parasitic flatworm infections [2], are treated using the anthelmintic drug praziquantel (PZQ). PZQ has remained an effective treatment for schistosomiasis over four decades of clinical use [3], underpinning recent mass drug administration (MDA) campaigns aimed at decreasing infections and morbidity in vulnerable populations. Alternatives to PZQ are however needed. PZQ has several features that could be improved and the threat of drug resistance, potentially accelerated by the rollout of MDA initiatives, persists [4–6].
We recently discovered that PZQ activates a Ca2+-permeable ion channel in Schistosoma mansoni that belongs to the melastatin family of transient receptor potential (TRP) channels (christened Sm.TRPMPZQ [7,8]). PZQ also acts a potent activator of TRPMPZQ in other PZQ-sensitive parasites [9]. TRP channels, which act as non-selective cation channels, are appealing targets for anthelmintic drug discovery owing both to their important physiological roles in sensory physiology as well as their druggability [10–15]. However, little is currently known about the pharmacology of flatworm TRP channels. Efforts to profile these channels will be important for validating tools to selectively manipulate worm physiology, as well as for anthelmintic development [13–15]. Insight to date suggests schistosome TRP channels, like other flatworm targets [16], exhibit a different pharmacological profile compared with their closest mammalian counterparts. For example, Schistosoma mansoni TRPA1 responds to capsaicin, a human TRPV ligand [17]. Sm.TRPMPZQ, which harbors a TRPM8-like binding pocket in the voltage-sensor like domain (transmembrane helices S1-S4, [9]), is not activated by the human TRPM8 agonists menthol and icilin [7]. Customization of these TRP channels to parasite-specific functions may underpin this divergence and specialization.
To characterize flatworm TRP channel pharmacology, it would be helpful to establish methods for screening individual channels against diverse drug libraries. Sm.TRPMPZQ is a good candidate for optimizing such an approach given it is a targeted by PZQ, and mediates a large, sustained Ca2+ signal in heterologously expressing cells. In collaboration with the Molecular Screening Center at Scripps Research in Florida, we optimized target-based screening approaches for Sm.TRPMPZQ with the goal of discovering other ligands that engage this ion channel complex [18]. The hope is that by optimizing high-throughput screening (HTS) methods for TRPMPZQ, new chemotypes distinct from PZQ can be identified including ligands that act at different sites on the channel relative to the transmembrane PZQ-binding pocket [9]. These could include allosteric modulators, or ligands that interact with the pore-forming domain (S5-S6). Such ‘hits’ could then be further iterated and evaluated as leads for anthelmintic development.
Here, we executed a pilot screen of ~16,000 compounds against Sm.TRPMPZQ which identified numerous antagonists as well as a single novel activator of Sm.TRPMPZQ. Interestingly this Sm.TRPMPZQ activator is a known chemical entity previously prioritized from a phenotypic screen of schistosome worms. These data underscore first, the feasibility of a high throughput screening (HTS) approach for flatworm TRP channels, and second convergence of target- and phenotype-based screening approaches on the same ligand here revealed to act as a Sm.TRPMPZQ agonist.
Results
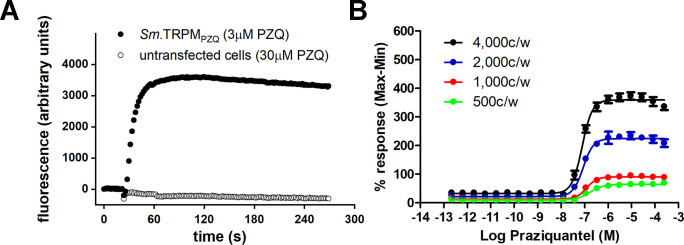
Activation of Sm.TRPMPZQ heterologously expressed in HEK293 cells was resolved by following changes in fluorescence emission of a synthetic Ca2+ indicator over time [7]. In cells, transiently expressing Sm.TRPMPZQ and seeded into a 96-well plate, addition of ±PZQ (3 μM) caused a rapid, dose-dependent increase in fluorescence (Fig 1A, [7]). Addition of higher concentrations of PZQ (≤30 μM) to untransfected HEK293 cells failed to elicit any change in basal fluorescence (Fig 1A, [7]).
Fig 1. Optimization of assay conditions for monitoring Sm.TRPMPZQ activity.
(A) Representative fluorescence trace showing the kinetics of the Ca2+ signal resulting from addition of PZQ (3 μM) to HEK293 cells transiently expressing Sm.TRPMPZQ (closed symbols), or addition of a higher concentration of PZQ (30 μM) to untransfected HEK293 cells (open symbols). (B) Concentration response curves of PZQ-stimulated Ca2+ signals in Sm.TRPMPZQ expressing HEK293 cells resolved at various densities of cells per well (c/w) measured in suspension in 1,536 well plates at room temperature. Each point on each curve represents the average of responses from 16 replicates which were also run in 2 separate experiments to achieve average and standard deviation.
In order to support a large scale HTS, we trialed various conditions to support miniaturization of this basic reporter assay into smaller volumes necessary for screening in 1536-well format. Miniaturization of the screening assay was aided by the large amplitude of the Sm.TRPMPZQ–dependent Ca2+ transient (change of fluorescence/basal fluorescence, ΔF/F = 12.3±2.1 for PZQ signals at Sm.TRPMPZQ versus ΔF/F = 4.5±1.5 for ATP signals through endogenous receptors as measured by confocal Ca2+ imaging [7]) and the non-desensitizing nature of the PZQ-evoked Ca2+ signal, which was sustained over several minutes (Fig 1A). Experimental performance was compared (i) using various densities of Sm.TRPMPZQ-transfected cells assayed in suspension, (ii) at different temperatures (room temperature (RT) versus 37°C), and (iii) using either freshly transfected cells, or thawed stocks of previously frozen transfected cells. Each condition was trialed in a 1,536 well format using the fluorescent dye calcium-5 (Kd for Ca2+ = 390 nM). Assay performance under these screening conditions was compared by calculating the Z’ factor (Z’), a widely used indicator of HTS assay robustness [19], as well as by monitoring the dynamic range of the assay (signal[Fmax]:basal[Fbasal], S:B). Z’ values over 0.5 are considered to be a prerequisite for an excellent HTS.
Assays in the 1536 well format demonstrated that increasing the cell count per well resulted in increased signal and dynamic range (Fig 1B), as well as a decrease in the measured EC50 (from 135±15 nM at 1000 cells/well, to 79±2 nM at 4000 cells/well, Table 1). The temperature at which responses were recorded (RT versus 37°C) did not change the sensitivity of Sm.TRPMPZQ under these assay conditions (Table 1). Optimal conditions for executing the assay were selected as 4000 cells/well in suspension at room temperature, where assay performance exceeded needed parameters (Table 1). Similar assay performance was achieved using either fresh cells (EC50 = 79±2 nM, Z’ = 0.82±0.03, S:B = 10.05±0.08, Table 1) or thawed cells from previously frozen stocks (EC50 = 90±10 nM, Z’ = 0.86±0.02, S:B = 10.55±0.08) under the same assay conditions. Therefore, to further minimize assay variation, pilot screens were all performed using a single batch of transfected cells prepared in bulk and then frozen. This facilitated execution of screens and removed transfection efficiency as an experimental variable. Data from all of these trials are summarized in Table 1.
Table 1. Assay metrics under specified conditions.
EC50, Z’ and S:B were determined for Sm.TRPMPZQ activation by PZQ at indicated cell densities and temperatures. Conditions in bold text indicate the final conditions selected for the screen. For each condition, responses were found to be positively cooperative (Hill coefficients range of 1.5–2).
| Cell density | Temperature | EC50 (nM) | Z’ | S:B |
|---|---|---|---|---|
| 500 c/w | RT | 195±25 | 0.31±0.37 | 5.55±0.73 |
| 500 c/w | 37°C | 202±4 | 0.44±0.14 | 7.28±0.28 |
| 1000 c/w | RT | 135±15 | 0.48±0.07 | 5.73±0.06 |
| 1000 c/w | 37°C | 144±2 | 0.53±0.08 | 7.41±0.01 |
| 2000 c/w | RT | 95±7 | 0.69±0.04 | 8.23±0.17 |
| 2000 c/w | 37°C | 108±2 | 0.79±0.03 | 8.04±0.39 |
| 4000 c/w | RT | 79±2 | 0.82±0.03 | 10.05±0.08 |
| 4000 c/w | 37°C | 80±1 | 0.83±0.01 | 9.24±1.23 |
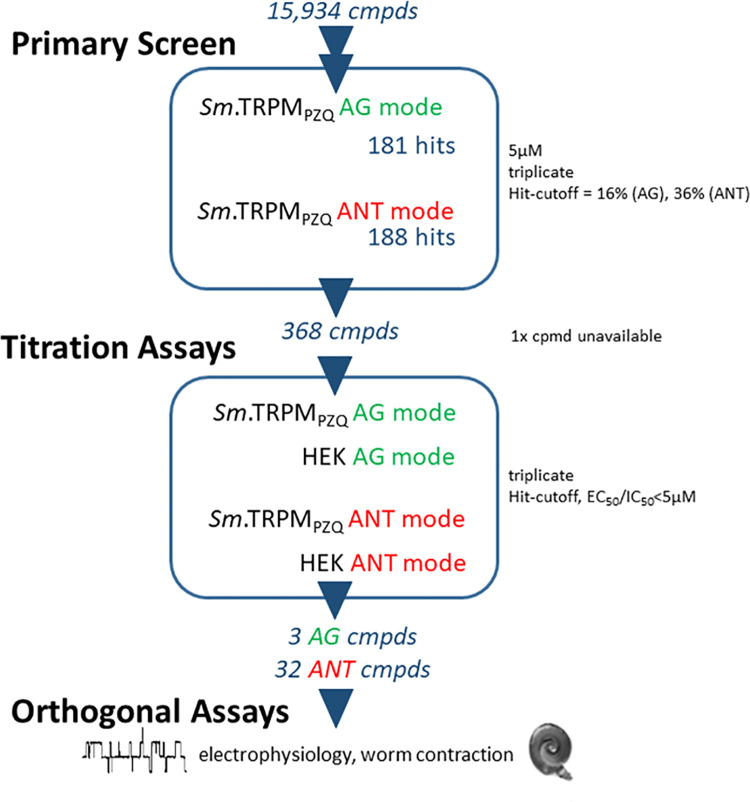
After optimizing conditions for miniaturization into a 1,536 well format, we proceeded to execute a screening campaign against a total of 15,984 compounds. The screened libraries comprised LOPAC1280 (1280 compounds), the Pathogen Box (400 compounds) and a subset of the in-house Scripps Drug Discovery Library, which included the Maybridge Hitfinder library. The screening pipeline consisted of (i) a primary, fixed concentration screen (5 μM) performed in triplicate in both ‘agonist’ and ‘antagonist’ mode, followed by (ii) titration and finally (iii) counter-screening assays of all putative ‘hits’. For the primary screen, following compound addition and completion of the ‘agonist’ (AG) mode read of 3 minutes, the ‘antagonist’ (ANT) mode commenced by PZQ addition (at an EC80 concentration) to wells that contain either compounds or DMSO (‘low control’). The output was compared to the response of wells with DMSO without PZQ stimulation (‘high control’). A summary of the workflow and observed compound attrition through each of these steps is shown schematically in Fig 2. Single point scatterplots from all compounds tested in the primary screen are shown in Fig 3.
Fig 2. Sm.TRPMPZQ screening pipeline.
All HTS assays were performed in 1,536 well format. A fixed concentration (nominally 5 μM) primary screen was followed by titration assays and counter-screening in untransfected HEK cells. All assays were performed in triplicate in both agonist (AG) and antagonist (ANT) mode. From the original 15,934 compounds (cmpds) screened, the pipeline yielded 3 putative agonist ‘hits’ (two hits were identified as PZQ present as a ligand in the screened libraries) and 32 putative antagonist ‘hits’. Selected compounds were validated using electrophysiology and by monitoring contraction of schistosome worms ex vivo.
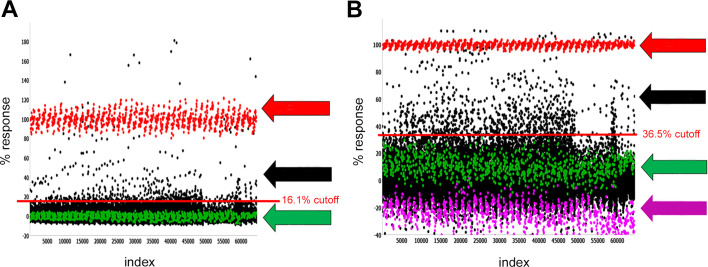
Fig 3. Primary HTS assay data.
Scatterplot of all 15,934 compounds tested in (A) agonist mode and (B) antagonist mode. Each dot graphed represents the activity of a well containing test compound (black, sample field) or controls (red high control; green, low control). Arrows (right) indicated the high control (red), low control (green) and the sample field (black, datapoints between the calculated cutoffs (red lines) and high control). For the agonist screen, high control reflected responses to PZQ (10 μM, red) and the low control represented responses to DMSO (green). For the antagonist screen, the high control reflected responses to DMSO (red) and the low control represented responses to submaximal PZQ (500 nM, green). The response to a maximal PZQ concentration (ECMAX, 10 μM) is also indicated (magenta). The EC80 stimulation is qualified as a percentage of the response to the high control PZQ EC100 (‘ECMAX’). All plates are assessed for robustness with Z’ >0.5 and ECStim between 70–95%.
Assay performance in the primary agonist screen met quality thresholds (Z’ = 0.71±0.04, S:B = 11.40±0.04) and the observed sensitivity to PZQ was consistent with the prior assay optimization trials (EC50 for PZQ = 101±3 nM). Assay performance in the primary antagonist screen also met quality thresholds (Z’ = 0.72±0.04, S:B = 9.4±0.4). Hit cut-offs for both screening modes was determined using an interval-based algorithm [20,21]. The sample field in the agonist screen was taken as a percentage response (‘hit-cutoff’ >16%) which identified 181 putative ‘hits’ for further progression. The initial sample field in the antagonist screen was calculated as a ‘hit-cutoff’ >36%, which identified 188 putative ‘hits’ for further evaluation.
A total of 368 compounds were then advanced to titration screening from the primary screen ‘agonist’ and ‘antagonist’ assays (1 compound was not commercially available). Each of these 368 compounds was profiled as a 10-point dose-response analysis run in triplicate against untransfected HEK cells (‘counterscreen’) as well as HEK cells expressing Sm.TRPMPZQ. Assay performance in these titration assays met required specifications (S1 Table). After curve-fitting, compounds displaying an EC50>5 μM (agonist) or IC50 >5 μM (antagonist) were considered ‘inactive’ and not studied further. The surviving ‘hits’ that progressed through this pipeline comprised 3 potential agonists and 32 potential antagonists. Full details of these hits and associated assay data are provided in S2 and S3 Tables.
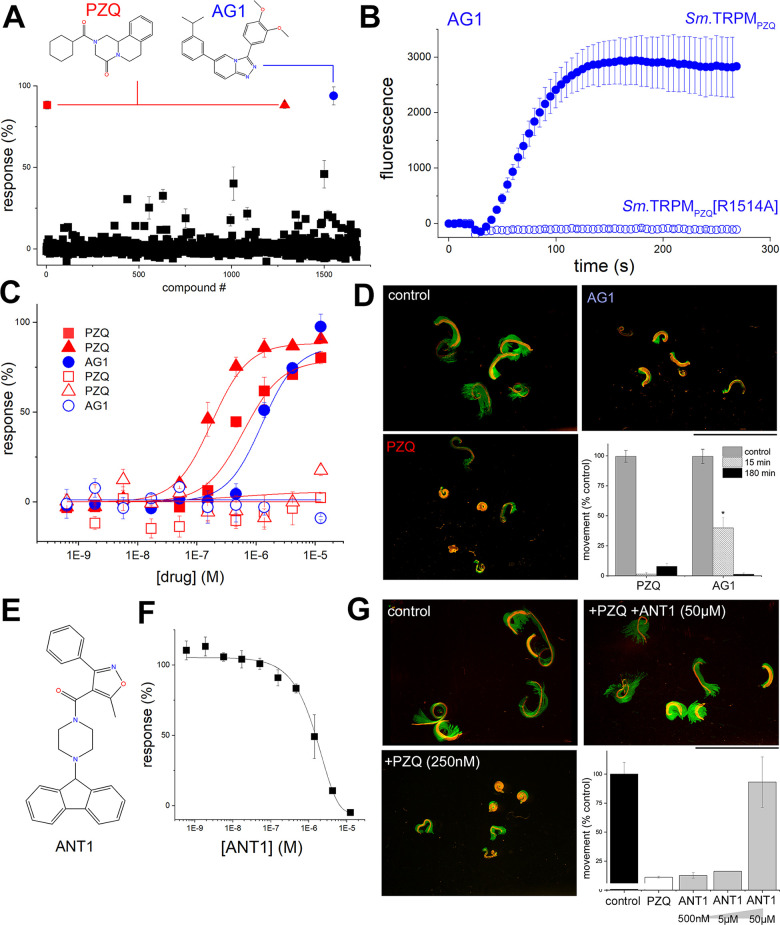
All three agonist hits were evaluated in further detail. Two of the agonist hits were identified from the Pathogen Box, and one from LOPAC1280. The primary screening data and titration analysis from the plates containing these compounds were extracted from the screening dataset. Two of these hits were identified as PZQ (red symbols, Fig 4A) as both the Pathogen Box and LOPAC1280 libraries contained PZQ as a test ligand. A third agonist ‘hit’ (christened ‘AG1’, blue circle), from the Pathogen Box, elicited strong activation of Sm.TRPMPZQ (Bmax = 93.9±5.6%) in the primary screen (Fig 4A). AG1 is a known chemical entity (3-(3,4-diimethoxyphenyl)-6-(3-(propan-2-yl)phenyl)-[1,2,4]triazolo[4,3-a]pyridine, MMV688313; Fig 4A) and represents a chemotype distinct from PZQ. Addition of AG1 (100 μM) resulted in a sustained cytoplasmic Ca2+ signal in cells expressing Sm.TRPMPZQ resembling the action of PZQ (compare Fig 4B with Fig 1A). AG1 action was also assessed against a Sm.TRPMPZQ channel mutant (Sm.TRPMPZQ[R1514A]) in the fourth transmembrane spanning helix (TM4) that blocks PZQ action by ablating interactions necessary to shape the PZQ binding pocket [9]. As expected, PZQ did not elevate Ca2+ in cells expressing Sm.TRPMPZQ[R1514A] (Fig 4B). However, this binding site mutant also ablated AG1 activity (Fig 4B), suggesting that AG1 also acts as an orthosteric ligand. Concentration-response curve analysis revealed AG1 acted as a full agonist of Sm.TRPMPZQ (EC50 = 1.6±0.3 μM) in Ca2+ flux assays, with no activity observed on counter-screening in naïve HEK cells (Fig 4C). The potency of AG1 was lower than observed with either sample of PZQ (EC50s of 177±21 nM, 619±225 nM) present in two libraries screened under identical conditions (Fig 4C). Both ligands were then re-sourced for validation assays and activities were re-assessed from independently procured samples (PZQ, EC50 = 406 nM; AG1, EC50 = 9.2 μM). Finally, the action of AG1 was studied against adult schistosome worms isolated from infected mice. Addition of AG1 to schistosome worms ex vivo evoked a sustained contraction (Fig 4D), although the kinetics of onset of the AG1-evoked contraction were slower than observed with PZQ. Collectively, these data identify AG1, a distinct chemical entity from PZQ, as a novel Sm.TRPMPZQ agonist.
Fig 4. Identification of new chemotypes active at Sm.TRPMPZQ.
(A) Primary screen of Sm.TRPMPZQ in agonist mode measuring peak Ca2+ signal amplitude in response to 1,678 compounds (LOPAC1280, Pathogen Box) tested at 5μM final concentration. Structures of ‘hits’ (PZQ, red; AG1, blue) are shown. (B) Kinetics of a response of wild-type Sm.TRPMPZQ to AG1 (30μM, closed circles), or the binding pocket mutant Sm.TRPMPZQ[R1514A] to AG1 (open circles). (C) Analysis of the three primary screen hits via full concentration response curves for PZQ (compounds #5, #1287) and AG1 (compound #1552) in HEK293 cells transiently transfected with Sm.TRPMPZQ (solid symbols) or untransfected controls (open). Data represent mean±sd of triplicate samples. (D) Images of adult schistosome worms with single frame image (red) overlayed with maximum intensity projection (green) of a time-lapse series to illustrate worm movement and effects of PZQ (500 nM) and AG1 (1 μM) on worm motion 15 mins after treatment. Graph shows effects of drugs on worm mobility after a 15 min and 180 min exposure (*, p<0.01). Data are analyzed from n≥3 independent infections. (E) Structure of a putative Sm.TRPMPZQ blocker (ANT1) from HTS screening activities. (F) Concentration-dependent blockade of Sm.TRPMPZQ dependent Ca2+ signals (evoked by 500 nM PZQ) by increasing concentrations of ANT1. (G) Images of adult schistosome worms with single frame image (red) overlayed with maximum intensity projection of a time-lapse series to illustrate worm movement (green) and the effect of PZQ (250 nM) as well as PZQ in the presence of ANT1 (50μM). Data are captured after 24 hours of incubation. Inset, quantitative analysis of data from n = 3 independent infections.
Screening in antagonist mode identified several 32 potential blockers of Sm.TRPMPZQ (S2 Table). To begin analysis of this larger set of ligands, we investigated that action of one of the more potent blockers, the compound ANT1 (1-(9H-fluoren-9-yl)-4-(5-methyl-3-phenyl-1,2-oxazole-4-carbonyl)piperazine, Fig 4E). ANT1 blocked PZQ-evoked Ca2+ signals (IC50 of 1.3±0.3 μM) mediated by Sm.TRPMPZQ (Fig 4F). When applied to intact worms ANT1 did not change worm motility, but at high concentrations (50μM) enhanced recovery of schistosome worms incubated in the continued presence of PZQ compared to worms treated with PZQ alone (Fig 4G).
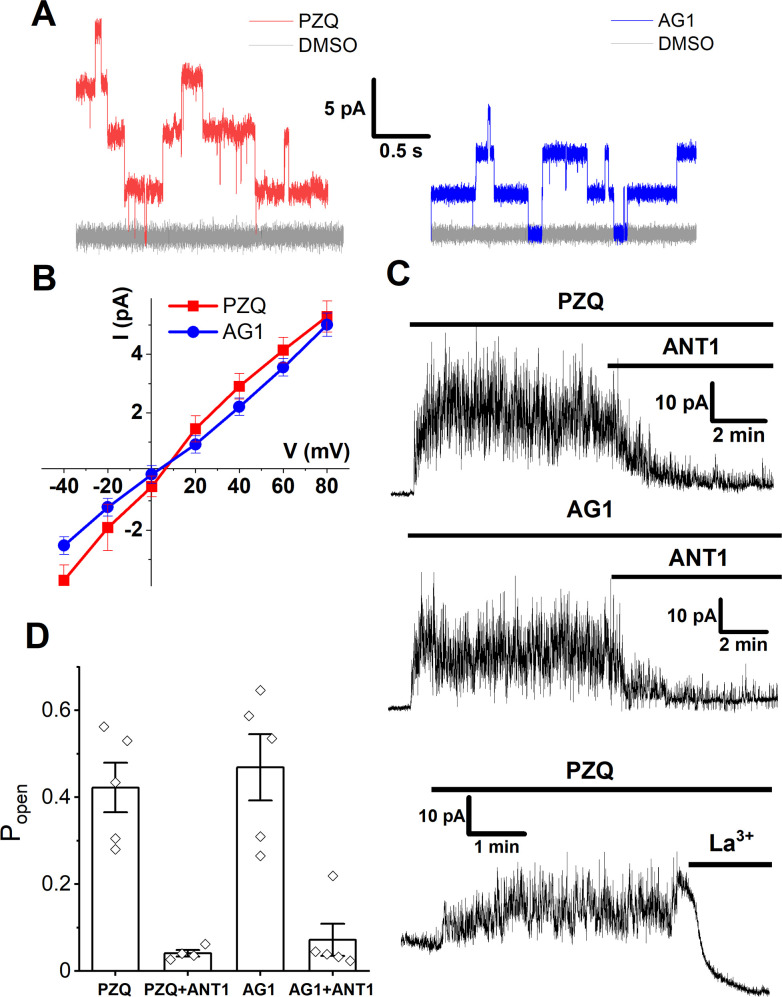
To validate ANT1 using an orthogonal assay, ANT1 action versus PZQ and AG1 was assessed using an electrophysiological approach. Single channel recordings of Sm.TRPMPZQ activity were made in Ca2+-free buffer following addition of either PZQ or AG1 (10 μM) to a cell-free patch in ‘inside-out’ recording mode. Both PZQ and AG1 evoked step-wise openings of Sm.TRPMPZQ (Fig 5A) with channel activation defined by linear I-V relationship in response to both agonists (Fig 5B). The mean slope single-channel conductance for PZQ- and AG1- activated Sm.TRPMPZQ was 76 ± 8 and 61 ± 4 pS respectively (mean ± SD). Addition of PZQ, or AG1, evoked sustained, non-desensitizing Sm.TRPMPZQ currents, which were blocked by subsequent addition of ANT1 in a time-dependent manner (Fig 5C). Analysis of single channel open probability (Popen) from these records demonstrated ANT1 decreased Popen at Sm.TRPMPZQ channels activated by either agonist (Fig 5D). After ANT1 application, brief channel openings persisted in the presence of PZQ or AG1 that were not seen in the presence of PZQ after addition of the pore blocker La3+ [7]. This observation is again suggestive of a competitive interplay between ANT1 and the channel activators, PZQ or AG1, within the orthosteric binding pocket of Sm.TRPMPZQ.
Fig 5. Electrophysiological analysis of new Sm.TRPMPZQ chemotypes.
(A) Sm.TRPMPZQ channel fluctuations evoked by PZQ (red) or AG1 (blue) both applied at 10 μM compared to vehicle responses (grey, 0.1% DMSO). Recordings were made in Ca2+-free solution at a clamping potential of +60mV in an inside-out configuration. DMSO and the test drug (PZQ or AG1) were added sequentially to the same patch. (B) Current (I)-voltage (V) relationship of Sm.TRPMPZQ activated with PZQ (10 μM, red) or AG1 (10 μM, blue). (C) Effect of ANT1 (top and middle, 50 μM) or La3+ (bottom, 10 mM) addition on Sm.TRPMPZQ activity evoked by PZQ (10 μM) or AG1 (10 μM) measured in cell-free mode at +40mV using an inside-out configuration. (D) Measurements of single channel open probability (Popen) under the indicated conditions. Welch’s t-test: PZQ vs ‘PZQ+ANT1’, p = 0.002; AG1 vs ‘AG1+ANT1’, p = 0.004.
In summary, these data reveal that AG1 behaves a novel activator of Sm.TRPMPZQ and ANT1 as a blocker of PZQ (or AG1) action at Sm.TRPMPZQ. Both these ligands have structures different from PZQ, evidencing the druggability of Sm.TRPMPZQ by novel chemotypes.
Discussion
Here, we report successful miniaturization of a reporter assay (6μl assay volume) to monitor the activity of a parasitic flatworm ion channel, Sm.TRPMPZQ. This approach will enable further HTS campaigns to provide insight about the properties and regulation of this recently discovered flatworm ion channel that is targeted by PZQ [8,9]. Critically, the ability to screen diverse libraries provides opportunity to discover new chemotypes active at this channel complex. This is important as the structure-activity relationship of PZQ has long been known be ‘tight’ with even minor modifications to the ligand causing a marked decrease in activity [9]. This has frustrated efforts to rationally engineer PZQ derivatives with enhanced properties, or improved metabolic stability. Alternative chemotypes to PZQ are required, both to mitigate some of the deficiencies of this stalwart therapeutic, as well as to bolster the global drug development pipeline [4]. Our knowledge of the properties of the PZQ binding site in TRPMPZQ derive from recent mutagenesis and modeling studies guided by analyses of responses to PZQ and related derivatives [9]. However, an expansive body of work on the human TRPM8 channel evidences accommodation of various chemotypes within a malleable transmembrane binding pocket [22,23], that overlaps with the binding site for PZQ characterized in Sm.TRPMPZQ [9]. Indeed (S)-PZQ engages this binding pocket in hTRPM8 [24,25]. Such data justify screening for new chemotypes active at flatworm TRPMPZQ channels.
For these reasons, we optimized methods to allow a pilot screen of ~16,000 compounds at Sm.TRPMPZQ. After titration and counter-screening, a single novel activator of Sm.TRPMPZQ−for simplicity referred to as AG1 –was identified and validated in an orthogonal assay via single channel recording. Application of AG1 to schistosome worms ex vivo also caused a rapid contraction (Fig 4D). Interestingly, AG1 is not a novel chemical entity, it was previously prioritized in an earlier phenotypic screen. Mansour et al. executed a screen of ~300,000 compounds against larval, juvenile, and adult schistosomes [26]. AG1 (LSHTM-1507) was one of seven hits prioritized from the phenotypic screening pipeline [26] all of which displayed activity against each of the three life cycle stages, a good cytotoxicity window and structural attractiveness for further derivatization [27]. A second ‘hit’ from the same screen (LSHTM-1945) is a closely related chemical structure, and a third compound (LSHTM-1956) was a derivative of a mammalian TRPC6 inhibitor [28]. AG1 was also identified as an anti-schistosomal compound in previous screens of the Pathogen Box executed by independent laboratories [29]. That a phenotypic screen of ~300,000 compounds [26], inherently blind to the mode of action of many of the screened ligands, should identify one (possibly more) Sm.TRPMPZQ activators highlights an impressive convergence of phenotypic and target-based screening methods and underscores the relevance of TRPMPZQ as a druggable target. These data support the execution of a larger target-based screen to identify Sm.TRPMPZQ activators, especially given the low hit rate for agonist discovery observed from this foundational screening effort (1 from ~16,000 compounds).
Just like PZQ, AG1 was more active against adult worms than juveniles [26], and the activity reported in phenotypic assays (IC50 of ~1.6 μM, [26]) mirrors the activity range for Sm.TRPMPZQ activation in vitro (1.3 μM, Fig 4B). However, AG1 represents a discrete chemotype from PZQ. It is a triazolopyridine derivative and a considerably more hydrophobic ligand (Mr = 373.4, XLogP3-AA = 5.5, TPSA = 48.6 Å2) compared with PZQ (Mr = 312.4, XLogP3-AA = 2.7, TPSA = 40.6 Å2). The higher hydrophobicity of AG1 may contribute to the slower kinetics of action on intact worms ex vivo (Fig 4D). PZQ derivatives previously shown to activate Sm.TRPMPZQ displayed attributes tightly clustered around those of the parent ligand (average properties of agonists with EC50<10 μM, Mr = 323, average LogP = 2.3 [9]). However, the physiochemical properties of AG1 are not dissimilar from other classes of ligands that activate human TRPM8 (hTRPM8, [23]). It is therefore exciting to have discovered a novel agonist chemotype which activates Sm.TRPMPZQ (Fig 4). Preliminary mutagenesis data with AG1 (Fig 4B), and the similar effects of ANT1 on AG1- and PZQ-evoked Sm.TRPMPZQ activity (Fig 5C), suggest that AG1, like PZQ, functions as an orthosteric ligand occupying the transmembrane binding pocket. Further mutagenesis work will be needed to expand these observations and resolve the relative binding poises of the two drugs. This comparison may hold significance as binding site mutants that impair PZQ binding may not necessarily impair the interaction of AG1, or other channel activators. This is known for hTRPM8 ligands [9]—for example, the conservative replacement hTRPM8[R842K] inhibits PZQ activation of hTRPM8 but has no effect on activation by cooling agent WS-12. Similarly, hTRPM8[H845W] abolishes responses to icilin but not PZQ [9]. This could prove an important observation in the context of engaging TRPMPZQ channels refractory to PZQ, for example in Fasciola species [9].
The screen also identified a larger number of potential Sm.TRPMPZQ blockers (32 from 16,000 compounds), which require further investigation to define their mode of action. For example, do these ligands act as pore blockers or as binding site antagonists? The utility of Sm.TRPMPZQ blockers (as opposed to Sm.TRPMPZQ activators) as anthelmintics is unproven, however their study and optimization has merit in providing selective tools for understanding worm physiology and the role of Sm.TRPMPZQ in vivo. Here, we validated the blocking activity of a single of these candidates (ANT1), which antagonized PZQ action in the Ca2+ reporter (Fig 4F), electrophysiological (Fig 5C and 5D) and a worm contraction assay (Fig 4G). ANT1, like PZQ, contains a piperazin-1-yl methanone substructure. This structural resemblance, and the dissimilar action of ANT1 and La3+ on PZQ-evoked responses (Fig 5C and 5D), suggests ANT1 also acts as an orthosteric ligand.
In conclusion, this work has optimized methods for executing a highly miniaturized, large scale small molecule screen against a parasitic flatworm ion channel targeted by PZQ. Discovery of new chemotypes using these methods will help decipher features of Sm.TRPMPZQ ligands critical for anthelmintic efficacy, and provide a workflow for screening campaigns at other parasitic flatworm ion channels.
Materials and methods
Ethics statement
All animal experiments followed ethical regulations approved by the MCW IACUC committee (AUA00006079/6476).
Reagents
Cell culture reagents were from Invitrogen. Lipofectamine 2000 was from ThermoFisher. Libraries were sourced as follows: LOPAC1280 (Sigma), the Pathogen Box (Medicines for Malaria), Scripps Drug Discovery Library (Scripps Institute). FLIPR Calcium 5 assay kits were from Molecular Devices. Additional supply of AG1 (3-(3,4-Dimethoxyphenyl)-6-(3-propan-2-ylphenyl)-[1,2,4]triazolo[4,3-a]pyridine; PubChem CID:122196572, C23H23N3O2) was sourced from Evotec and ANT1 (1-(9H-fluoren-9-yl)-4-(5-methyl-3-phenyl-1,2-oxazole-4-carbonyl)piperazine; PubChem CID:2813918, C28H25N3O2) was sourced from Maybridge. All other routinely used chemical reagents were purchased from Sigma.
HTS screening workflow
HEK cells were transfected with cDNA (400μg/ml DNA) encoding Sm.TRPMPZQ using a Maxcyte STX electroporation system and incubated overnight at 37°C. This system is uniquely capabable of large scale transfections providing one batch of cells ready for large scale HTS [30]. Briefly, the HEK cells were resuspended at 108/ml in Maxcyte electroporation buffer. Plasmid DNA was added to the cells in buffer to achieve a concentration of 400 μg/ml from a stock of 5mg/ml (in TE buffer). This mixture was loaded into the appropriate cassette, inserted into the machine, and cells electroporated. Cells were then frozen to provide a single supply of transfected cells for all subsequent assays. On the day of the assays, cells were thawed in a 37°C waterbath, and resuspended in DMEM + 10% FBS and counted. The cells were pelleted by centrifugation at 170xg for 5 minutes. The media was aspirated, and the remaining pellet was resuspended in 1x HBSS with 20 mM HEPES and 1% DMSO to achieve 1.33x106cells/ml. This cell suspension was then dispensed at 3 μl/well to the 1536 well plates (Greiner Bio One part 789072) at a density of 4,000 cells/well. Measurements of fluorescence intensity (λex = 470 nm, λem = 535 nm) were made using a FLIPR (Molecular Devices FLIPR Tetra). A read of basal fluorescence values was made (5s) prior to addition of compounds or controls (30 nl). In both ‘agonist’ and ‘antagonist mode’, the total assay volume was 6μl (1% DMSO). For the primary screen, each compound was tested at a single concentration in triplicate, and responses compared on a per plate basis by evaluating the percentage response of each compound versus the ‘high control’ (response to EC100 PZQ (10 μM) for the agonist screen, DMSO for the antagonist screen)) corrected for ‘low control’ (response to DMSO for the agonist screen, response to EC80 PZQ (500 nM) in the presence of DMSO or compounds (nominally 5 μM) for the antagonist screen). Ligand performance was categorized using an algorithm based on the activity of the entire dataset to define a ‘hit-cutoff’ parameter for both agonist-mode and antagonist-mode screening. Any compound with greater percent activation than this cutoff was assigned as ‘active’ prior to titration and counter-screening. Titration assays were executed using the same reagents, protocols and detection systems, done as 10-point concentration-response curves, performed in triplicate. Counter-screening was performed using untransfected HEK293 cells for comparison with results testing the same compounds effect on cells expressing Sm.TRPMPZQ, with ATP used as the positive control. For each compound, activity was plotted against compound concentration and data were fitted with a sigmoidal function. In order to be considered a hit at this stage a compound must achieve a EC50<5 μM for agonists and IC50 <5 μM for antagonists. All compounds selected for titration were subjected to LC-MS analysis to confirm mass and sample purity. ChemAxon (https://www.chemaxon.com) academic licensing was provided for the use of Instant JChem (ver. 15.10.12.0) to perform compound mining of the libraries utilized in this effort.
Schistosome mobility assays
Adult schistosomes were harvested from the mesenteric vasculature of female Swiss Webster mice previously infected (~49 days earlier) with S. mansoni cercariae (NMRI strain) by the Schistosomiasis Resource Center at the Biomedical Research Institute (BRI, Rockville, MD). Harvested schistosomes were washed in RPMI 1640 supplemented with 5% heat inactivated FBS (Gibco), HEPES (25 mM) and penicillin-streptomycin (100 units/mL). After isolation, worms were incubated overnight (37°C/5% CO2) in vented petri dishes (100x25 mm). Movement assays were performed using male and female worms in six well dishes (~5 individual worms/3ml media per well). Video recordings of worm motility (4 frames/sec) were captured using a Zeiss Discovery v20 stereomicroscope coupled to a QiCAM 12-bit cooled color CCD camera controlled by Metamorph imaging software [31].
Electrophysiology
For electrophysiological analysis, a HEK293 cell line stably expressing Sm.TRPMPZQ [7] was generated using the Flp-In T-REx system (K650001, Thermofisher Scientific, Carlsbad, CA). Flp-In T-REx HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), streptomycin (100 μg/ml), L-glutamine (290 μg/ml), blasticidin (10 μg/ml), and hydromycin B (100 μg/ml). HEK293 cells were plated onto glass coverslips and Sm.TRPMPZQ expression induced by addition of tetracycline (2 μg/ml) to the media 24–48 hours before recording. Prior to recording, coverslips were secured within a recording chamber of an Olympus BX51WI upright microscope. Cells were bathed in a solution containing 145 mM KCl, 10 mM HEPES, 1 mM EGTA (pH 7.2, 310–315 mOsm/kg with sucrose). The pipette solution contained 140 mM LiCl, 10 mM HEPES, 1 mM EGTA (pH 7.4, 280–285 mOsm/kg with sucrose). Patch pipettes were made of borosilicate glass (BF150-110-10, Sutter Instrument, Novato, CA) pulled on a vertical puller (Narishige, Amityville, NY, Model PC-10) with resistances 8–10 MΩ. After gigaseal formation, membrane patches were excised from cells and current recorded in voltage-clamp mode in ‘inside-out’ configuration using a MultiClamp 700B amplifier and Digidata 1440A digitizer (Molecular Devices, Sunnyvale, CA). Signals were filtered with an 8-pole Bessel low pass filter at 1 kHz, and sampled at 20 kHz. Open probability (Popen) was assessed in single channel search mode using Clampfit 10 software. All recordings were done at room temperature. Data represent mean±standard error for ≥3 independent experiments.
Supporting information
Assay performance measured across 12 plates for 368 compounds in titration assays in untransfected HEK cells (counterscreen) and HEK cells expressing Sm.TRPMPZQ.
(DOCX)
Data from 3 compounds prioritized after titration assays. Compound descriptors and screening data in agonist and antagonist modes in HEK cells, and HEK cells expressing Sm.TRPMPZQ are shown.
(XLSX)
Data from 32 compounds prioritized after titration assays. Compound descriptors and screening data in agonist and antagonist modes in HEK cells, and HEK cells expressing Sm.TRPMPZQ are shown.
(XLSX)
Acknowledgments
Schistosome-infected mice were provided by the NIAID Schistosomiasis Resource Center at the Biomedical Research Institute (Rockville, MD) through NIH-NIAID Contract HHSN272201000005I for distribution via BEI Resources. We thank Medicines for Malaria Venture (MMV, Switzerland) for supply of the Pathogen Box.
Data Availability
All data are in the manuscript and its supporting information files.
Funding Statement
This work was supported by NIH R01-AI145871 and R01-AI155405 (to JSM), NIH F31-AI145091 (to NAY) and the Marcus Family (to JSM). This work was also supported in part by a NIH S10 instrument award (1S10OD025282-01) that provided Scripps Research with the FLIPR Tetra system integrated into HTS operations (to TPS,LS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Organization WH. Ending the neglect to attain the Sustainable Development Goals–A road map for neglected tropical diseases 2021–2030. World Health Organ Monogr Ser. 2020. [Google Scholar]
- 2.Chai JY. Praziquantel treatment in trematode and cestode infections: an update. Infect Chemother. 2013;45(1):32–43. doi: 10.3947/ic.2013.45.1.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukushige M, Chase-Topping M, Woolhouse MEJ, Mutapi F. Efficacy of praziquantel has been maintained over four decades (from 1977 to 2018): A systematic review and meta-analysis of factors influence its efficacy. PLoS Negl Trop Dis. 2021;15(3):e0009189. doi: 10.1371/journal.pntd.0009189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spangenberg T. Alternatives to Praziquantel for the Prevention and Control of Schistosomiasis. ACS Infect Dis. 2020. doi: 10.1021/acsinfecdis.0c00542 [DOI] [PubMed] [Google Scholar]
- 5.Cupit PM, Cunningham C. What is the mechanism of action of praziquantel and how might resistance strike? Future medicinal chemistry. 2015;7(6):701–5. doi: 10.4155/fmc.15.11 [DOI] [PubMed] [Google Scholar]
- 6.Chan JD, Zarowiecki M, Marchant JS. Ca2+ channels and praziquantel: A view from the free world. Parasitol Int. 2013;62(6):619–28. doi: 10.1016/j.parint.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park SK, Gunaratne GS, Chulkov EG, Moehring F, McCusker P, Dosa PI, et al. The anthelmintic drug praziquantel activates a schistosome transient receptor potential channel. J Biol Chem. 2019;294(49):18873–80. doi: 10.1074/jbc.AC119.011093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park SK, Marchant JS. The Journey to Discovering a Flatworm Target of Praziquantel: A Long TRP. Trends Parasitol. 2020;36(2):182–94. doi: 10.1016/j.pt.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S-K, Friedrich L, Yahya NA, Rohr C, Chulkov EG, Maillard D, et al. Mechanism of praziquantel action at a parasitic flatworm ion channel. biorxiv. 2021. doi: 10.1101/2021.03.09.434291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue T, Yamashita T, Agata K. Thermosensory signaling by TRPM is processed by brain serotonergic neurons to produce planarian thermotaxis. J Neurosci. 2014;34(47):15701–14. doi: 10.1523/JNEUROSCI.5379-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arenas OM, Zaharieva EE, Para A, Vasquez-Doorman C, Petersen CP, Gallio M. Activation of planarian TRPA1 by reactive oxygen species reveals a conserved mechanism for animal nociception. Nat Neurosci. 2017;20(12):1686–93. doi: 10.1038/s41593-017-0005-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma S, Kashyap SS, Robertson AP, Martin RJ. Diethylcarbamazine activates TRP channels including TRP-2 in filaria, Brugia malayi. Commun Biol. 2020;3(1):398. doi: 10.1038/s42003-020-01128-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolstenholme AJ, Williamson SM, Reaves BJ. TRP channels in parasites. Adv Exp Med Biol. 2011;704:359–71. doi: 10.1007/978-94-007-0265-3_20 [DOI] [PubMed] [Google Scholar]
- 14.Bais S, Greenberg RM. TRP channels as potential targets for antischistosomals. International Journal for Parasitology Drugs and Drug Resistance. 2018;8(3):511–7. doi: 10.1016/j.ijpddr.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bais S, Greenberg RM. Schistosome TRP channels: An appraisal. International Journal for Parasitology Drugs and Drug Resistance. 2020;13:1–7. doi: 10.1016/j.ijpddr.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan JD, Grab T, Marchant JS. Kinetic profiling an abundantly expressed planarian serotonergic GPCR identifies bromocriptine as a perdurant antagonist. International Journal for Parasitology Drugs and Drug Resistance. 2016;6(3):356–63. doi: 10.1016/j.ijpddr.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bais S, Berry CT, Liu X, Ruthel G, Freedman BD, Greenberg RM. Atypical pharmacology of schistosome TRPA1-like ion channels. PLoS Negl Trop Dis. 2018;12(5):e0006495. doi: 10.1371/journal.pntd.0006495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baillargeon P, Fernandez-Vega V, Sridharan BP, Brown S, Griffin PR, Rosen H, et al. The Scripps Molecular Screening Center and Translational Research Institute. SLAS Discov. 2019;24(3):386–97. doi: 10.1177/2472555218820809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]
- 20.Otsuka Y, Airola MV, Choi YM, Coant N, Snider J, Cariello C, et al. Identification of Small-Molecule Inhibitors of Neutral Ceramidase (nCDase) via Target-Based High-Throughput Screening. SLAS Discov. 2021;26(1):113–21. doi: 10.1177/2472555220945283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith E, Janovick JA, Bannister TD, Shumate J, Ganapathy V, Scampavia L, et al. Rescue of mutant gonadotropin-releasing hormone receptor function independent of cognate receptor activity. Scientific Reports. 2020;10(1):10579. doi: 10.1038/s41598-020-67473-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diver MM, Cheng Y, Julius D. Structural insights into TRPM8 inhibition and desensitization. Science. 2019;365(6460):1434–40. doi: 10.1126/science.aax6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Muniz R, Bonache MA, Martin-Escura C, Gomez-Monterrey I. Recent Progress in TRPM8 Modulation: An Update. International journal of molecular sciences. 2019;20(11): doi: 10.3390/ijms20112618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunaratne GS, Yahya NA, Dosa PI, Marchant JS. Activation of host transient receptor potential (TRP) channels by praziquantel stereoisomers. PLoS Negl Trop Dis. 2018;12(4):e0006420. doi: 10.1371/journal.pntd.0006420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babes RM, Selescu T, Domocos D, Babes A. The anthelminthic drug praziquantel is a selective agonist of the sensory transient receptor potential melastatin type 8 channel. Toxicol Appl Pharmacol. 2017;336:55–65. doi: 10.1016/j.taap.2017.10.012 [DOI] [PubMed] [Google Scholar]
- 26.Mansour NR, Paveley R, Gardner JM, Bell AS, Parkinson T, Bickle Q. High Throughput Screening Identifies Novel Lead Compounds with Activity against Larval, Juvenile and Adult Schistosoma mansoni. PLoS Negl Trop Dis. 2016;10(4):e0004659. doi: 10.1371/journal.pntd.0004659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner JMF, Mansour NR, Bell AS, Helmby H, Bickle Q. The discovery of a novel series of compounds with single-dose efficacy against juvenile and adult Schistosoma species. PLoS Negl Trop Dis. 2021;15(7):e0009490. doi: 10.1371/journal.pntd.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. https://pubchem.ncbi.nlm.nih.gov/bioassay/2553 [Google Scholar]
- 29.Maccesi M, Aguiar PHN, Pasche V, Padilla M, Suzuki BM, Montefusco S, et al. Multi-center screening of the Pathogen Box collection for schistosomiasis drug discovery. Parasites & vectors. 2019;12(1):493. doi: 10.1186/s13071-019-3747-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith E, Davis-Gardner ME, Garcia-Ordonez RD, Nguyen TT, Hull M, Chen E, et al. High-Throughput Screening for Drugs That Inhibit Papain-Like Protease in SARS-CoV-2. SLAS Discov. 2020;25(10):1152–61. doi: 10.1177/2472555220963667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan JD, McCorvy JD, Acharya S, Johns ME, Day TA, Roth BL, et al. A Miniaturized Screen of a Schistosoma mansoni Serotonergic G Protein-Coupled Receptor Identifies Novel Classes of Parasite-Selective Inhibitors. PLoS Pathog. 2016;12(5):e1005651. doi: 10.1371/journal.ppat.1005651 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Assay performance measured across 12 plates for 368 compounds in titration assays in untransfected HEK cells (counterscreen) and HEK cells expressing Sm.TRPMPZQ.
(DOCX)
Data from 3 compounds prioritized after titration assays. Compound descriptors and screening data in agonist and antagonist modes in HEK cells, and HEK cells expressing Sm.TRPMPZQ are shown.
(XLSX)
Data from 32 compounds prioritized after titration assays. Compound descriptors and screening data in agonist and antagonist modes in HEK cells, and HEK cells expressing Sm.TRPMPZQ are shown.
(XLSX)
Data Availability Statement
All data are in the manuscript and its supporting information files.