Abstract
Hypoplastic left heart syndrome (HLHS) is one of the most lethal congenital heart defects, and remains clinically challenging. While surgical palliation allows most HLHS patients to survive their critical heart disease with a single-ventricle physiology, many will suffer heart failure, requiring heart transplantation as the only therapeutic course. Current paradigm suggests HLHS is largely of hemodynamic origin, but recent findings from analysis of the first mouse model of HLHS showed intrinsic cardiomyocyte proliferation and differentiation defects underlying the left ventricular (LV) hypoplasia. The findings of similar defects of lesser severity in the right ventricle suggest this could contribute to the heart failure risks in surgically palliated HLHS patients. Analysis of 8 independent HLHS mouse lines showed HLHS is genetically heterogeneous and multigenic in etiology. Detailed analysis of the Ohia mouse line accompanied by validation studies in CRISPR gene-targeted mice revealed a digenic etiology for HLHS. Mutation in Sap130, a component of the HDAC repressor complex, was demonstrated to drive the LV hypoplasia, while mutation in Pcdha9, a protocadherin cell adhesion molecule played a pivotal role in the valvular defects associated with HLHS. Based on these findings, we propose a new paradigm in which complex CHD such as HLHS may arise in a modular fashion, mediated by multiple mutations. The finding of intrinsic cardiomyocyte defects would suggest hemodynamic intervention may not rescue LV growth. The profound genetic heterogeneity and oligogenic etiology indicated for HLHS would suggest that the genetic landscape of HLHS may be complex and more accessible in clinical studies built on a familial study design.
Keywords: Congenital heart disease, Hypoplastic left heart syndrome, Genetics, Mouse model, Oligogenic etiology
Congenital heart disease (CHD) is one of the common birth defects, affecting up to 1% of live births and is a leading cause of pre- and postnatal death. With recent advances in medical and surgical management, the population of CHD survivors is growing and there are now more adults with CHD in the US than infants born each year with CHD [1]. Hypoplastic left heart syndrome (HLHS), is a severe CHD involving hypoplasia of the left ventricle (LV), aorta, and mitral valve. Without treatment, it is uniformly fatal, but it is now survivable with a three-stage surgical palliation that recruits the right ventricle (RV) to be the systemic pumping chamber [2, 3]. This single-ventricle circulation now allows more HLHS patients to survive their critical heart disease, but the 5-year survival rate is still only 50–70% [4, 5].
The current clinical paradigm assumes HLHS is a valve disease with LV hypoplasia arising from the hemodynamic effects of restricted aortic blood flow. As most HLHS diagnosis is now made prenatally, pre-conception risk assessments may provide opportunities for in utero intervention [6-8]. Indeed, a clinical trial has been pursued with in utero aortic valvuloplasty performed to restore aortic blood flow in fetuses with evolving HLHS. However, this 10-year clinical trial has yielded little evidence of fetuses with evolving HLHS undergoing conversion from single to biventricular physiology [9]. Substantive progress in the development of therapeutic intervention will require deeper understanding of the underlying causes for the cardiac maldevelopment and the pathogenic mechanism causing the LV hypoplasia.
One of the continuing challenges underlying the poor prognosis for HLHS patients is clinical management of the single RV [10]. This RV serving as the systemic pumping chamber performs well for some patients, others will develop heart failure, resulting in the need for cardiac transplantation. The reason for the differing outcomes remains unknown and as more HLHS patients survive their critical heart disease, there is increasing urgency to understand the factors driving heart failure risk in HLHS [11-13]. Such insights are needed to guide the development of therapeutic interventions to improve the long-term prognosis for patients with HLHS.
Genetic Etiology of HLHS
The current poor understanding of the disease mechanism in HLHS in part reflects the paucity of knowledge regarding the genetic causes of HLHS and the lack of a genetic animal model of HLHS. HLHS is a CHD that is particularly well documented to have a genetic etiology. Family studies have shown a high recurrence risk for CHD among siblings [14-20], a high relative risk of 36.9 reported for first-degree relatives, and an overall heritability of 0.71–0.90 [21]. HLHS is also highly associated with various syndromes such as Turner and Jacobsen [22, 23], as well as trisomy 13, 18, and 9 [24]. Analysis in small kindreds or limited HLHS cases has identified several candidate genes including heterozygous mutations in NKX2.5 [25], GJA1 [26], NOTCH1 [27], HAND1 [28], and RBFOX2 [29], while two recent reports support a recessive inheritance of HLHS [30, 31]. Findings from two family-based genetic studies suggest HLHS and associated CHD lesions are genetically heterogeneous with an oligogenic etiology. In one study, statistical modeling suggested HLHS most likely has a digenic etiology [19, 32]. Although human epidemiologic studies support a genetic origin of HLHS, until recently, no HLHS mouse model had ever been generated. This deficiency has recently been addressed with the recovery of HLHS mutant mice from a large-scale mouse mutagenesis screen. This has led to new insights into the genetic and developmental etiology of HLHS [33].
Recovery of HLHS Mutant Mice
A genetic model of HLHS was generated for the first time with recovery of HLHS mutant mice from a large-scale ENU mutagenesis screen [29, 33, 34]. The previous failure to generate HLHS mutant mice was thought to reflect the inadequacy of the short gestation period of mice (18.5 days) to orchestrate the hemodynamic remodeling required for the evolution of HLHS. However, the difficulty in generating HLHS mutant mice was actually due to the obligate multigenic etiology of HLHS and its profound genetic heterogeneity [19, 33-35]. Thus in a large-scale mouse forward genetic screen, we screened 100,000 fetal mice from 3000 G1 pedigrees using fetal echocardiography. We recovered nine HLHS mutants from among 100,000 fetuses scanned. They are derived from eight independent mutant pedigrees, with HLHS being the rarest CHD phenotype recovered in the screen.
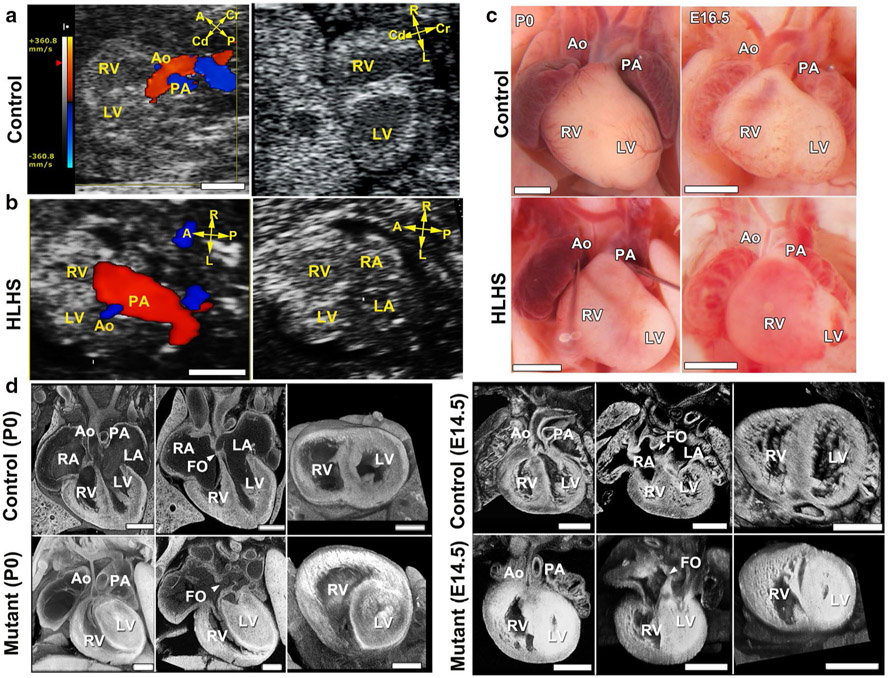
HLHS mutants were diagnosed by fetal hemodynamic assessments, which typically showed a narrowed aorta (blue, Fig. 1b, left panel), and with 2D imaging showing a small LV with poor contractility (Fig. 1b) (see Supplementary Movies 1, 2 in Liu et al. [34]). Confirmation by necropsy and histopathology showed a small LV with a hypoplastic aorta and mitral valve (MV)—the three essential features of HLHS (Fig. 1c, d; Supplementary Movie 3, 4 in Liu et al. [34]). The LV is often muscle-bound with obliteration of the chamber lumen (Fig. 1d). Analysis of the HLHS mutant mice from the eight lines showed the extent of LV and aortic/MV hypoplasia spanned a spectrum (Fig. 2), similar to the findings clinically. Unlike other CHD phenotypes recovered in the screen, HLHS was observed with a low transmission ratio that indicated non-Mendelian or complex genetic inheritance.
Fig. 1.
HLHS phenotype in Ohia mutants. a, b Ultrasound color-flow imaging of normal fetus (a), showing robust flow from the aorta (Ao) and pulmonary artery (PA). In the HLHS mutant (b), the aorta showed only a narrow flow stream, whereas the pulmonary artery showed robust flow. 2D imaging revealed hypoplastic LV (b), as compared with the normal-sized LV in the control (a). c Newborn (P0) or E16.5 hearts from wild-type and HLHS mutants. Hypoplastic aorta and LV are visible in the HLHS mutant. d Histopathology showing the cardiac anatomy of HLHS mutant and littermate control at birth (P0) and E14.5. Compared with controls, the HLHS mutant exhibited hypoplastic aorta and aortic valve atresia, hypertrophied LV with no lumen, and MV stenosis and patent foramen ovale (FO), arrowhead. LA left atrium, RA right atrium, A anterior, P posterior, L left, R right, Cr cranial, Cd caudal. Scale bars: a, b 0.5 mm; c 1 mm; d 0.5 mm. Adapted with permission from Liu et al. [34]
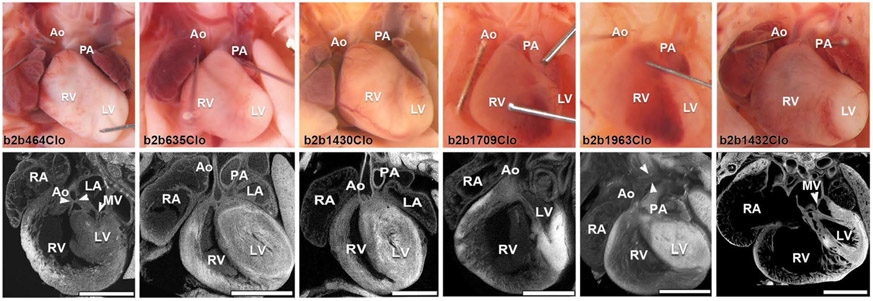
Fig. 2.
HLHS phenotypes observed in six of the eight HLHS mutant lines recovered from the mutagenesis screen. Top row shows necropsy photo and bottom, confocal histopathology of the same heart, showing a wide spectrum of disease associated with HLHS in these different mutant lines. Ao aorta, MV mitral valve, LV left ventricle, RV right ventricle, PA pulmonary artery, RA right atrium, LA left atrium. Adapted with permission from Liu et al. [34]
Hemodynamic Uncoupling of Aortic Flow and the Small LV Phenotype
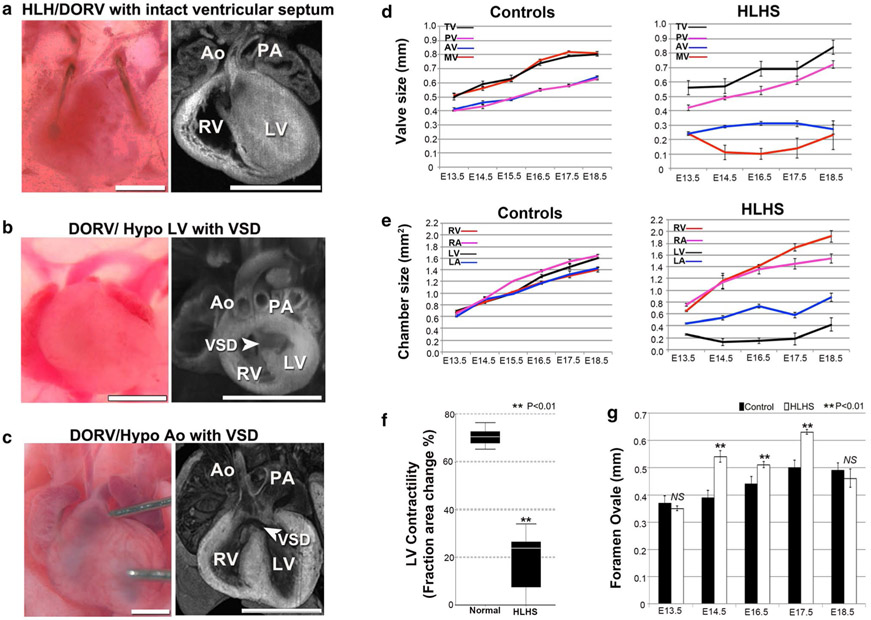
The mutant line Ohia, which yielded two HLHS mutants from the initial screen, was further analyzed by extensive breeding and phenotype/genotype analyses. This confirmed the heritability of the HLHS phenotype and in addition, yielded mutants with related CHD phenotypes, such as isolated hypoplastic LV, aortic arch anomalies and hypoplastic aorta, bicuspid aortic valve (BAV), or outflow tract malalignment defects. It is of significance to note that some Ohia mutants had double outlet RV with hypoplastic LV with normal-sized aorta, or hypoplastic aorta with normal-sized LV (Fig. 3a-c). These phenotypes demonstrated the hemodynamic uncoupling of the aortic flow from the small LV phenotype, indicating HLHS in the Ohia mutant line is not driven by hemodynamic factors.
Fig. 3.
HLHS and other CHD phenotypes in the Ohia mutant line. a–c Ohia mutants with isolated hypoplasia of the aorta or LV. Some Ohia mutants exhibit DORV with hypoplasia of both the aorta and LV (a), while others can exhibit DORV with hypoplastic LV, but normal-sized Ao (b), or DORV with hypoplastic aorta, but normal LV (c). These independent occurrences of hypoplasia of the LV vs. Ao would suggest these anatomical defects are not hemodynamically driven in Ohia mutants. Scale bar, 1 mm. d–g Delineating cardiac development in Ohia HLHS mutants using fetal echocardiography. d, e Developmental profile of the growth of cardiac valves and chambers in normal vs. HLHS embryos (n = 15 embryos per group). Data shown are mean ± s.e.m. LA left atrium, LV left ventricle, RA right atrium, RV right ventricle, MV mitral valve, TV tricuspid valve, AV aortic valve, PV pulmonary valve. f Fetal ultrasound measurements show LV contractility is decreased in HLHS mutant fetuses (n = 15 embryos per group). Data shown are median with interquartile range. Wilcoxon rank-sum test, P = 0.000003. g Measurements of the foramen ovale (FO) showed FO size was not restrictive in HLHS mutants. Data shown are mean ± s.e.m. Unpaired Student’s t test: E13.5, n = 12 (control = 6, HLHS = 6) P = 0.385, t = −0.908, df = 10; E14.5, n = 18 (control = 12, HLHS = 6), P = 0.000084, t = 5.222, df = 16; E16.5, n = 27 (control = 15, HLHS = 12), P = 0.001, t = 3.7, df = 25; E17.5, n = 21 (control = 12, HLHS = 9), P = 3.4633E – 9, t = 10.262, df = 19; E18.5, n = 12 (control = 6, HLHS = 6), P = 0.453, t = −0.782, df = 10. NS not significant. Adapted with permission from Liu et al. [34]
This is supported by further longitudinal interrogation conducted with fetal echocardiography at E13.5–18, which showed only right-sided heart structures grew with development of the HLHS mutant fetuses (Fig. 3d), while normal littermates showed steady growth of all four cardiac valves/chambers (Fig. 3e) (see Supplementary Movie 1, 2 in Liu et al. [34]). The HLHS mutants also showed poor LV contractility (Fig. 3f), but with sustained left-to-right flow through the foramen ovale (FO) (see Supplementary Movie 1). The previous hypothesized premature closure or restriction of the FO driving the LV hypoplasia in HLHS was not observed in Ohia HLHS mutants (Fig. 3g) [36]. Indeed quantification using the ultrasound imaging data showed enlargement rather than restriction in the opening of the FO. Together these findings indicate that LV hypoplasia in Ohia HLHS mutants is not due to hemodynamic effects from restricted blood flow from the aorta or the FO.
Complex Genetic Etiology for HLHS
Unlike other CHD phenotypes recovered in the screen, HLHS was observed with low transmission ratio indicating non-Mendelian inheritance. Whole exome sequencing analysis for mutation recovery showed that none of the eight HLHS mutant lines have mutations in the same genes. Together these findings indicate HLHS is multigenic and genetically heterogeneous, consistent with findings from human studies. This likely explains why HLHS had not been seen in mice previously, as any single gene knockout (KO) would not suffice, nor multiple KO gene combinations unless they are gene combinations known a priori to cause HLHS. In comparison, every pedigree in the mouse mutagenesis screen by design harbored 50–100 deleterious mutations, and with the screening of 3000 pedigrees, this greatly increased the odds of having mutation combinations that can cause HLHS, accounting for our success in recovering HLHS mutant mice. Nevertheless, we note HLHS is the rarest CHD phenotype recovered in our screen, with only nine HLHS mice recovered from over 100,000 fetal mice screened.
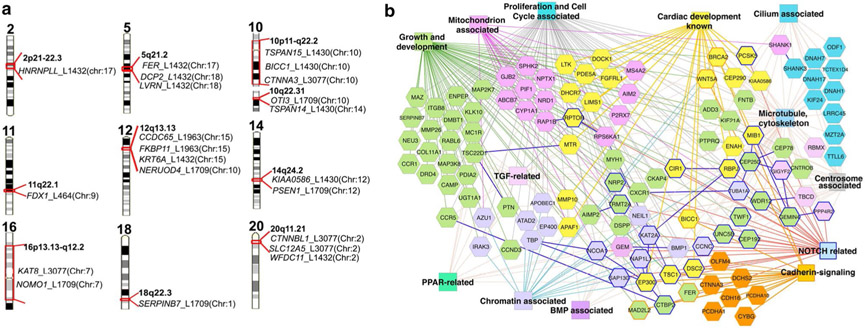
Exome sequencing of the eight HLHS mutant lines yielded 355 coding/splicing mutations, with 61 being homozygous, and none shared between the eight lines. Interestingly, five genes related to Notch signaling were recovered—Rbpj, Ep300, Nap1l1, Ncoa1, and Tspan12, consistent with previous studies indicating an important role for the Notch pathway in the genetic etiology of HLHS. We observed that mutations in five of the HLHS mouse lines are in genes situated in 10 of the 13 human chromosome intervals linked to HLHS (Fig. 4) [35, 37, 38]. Significant enrichment is observed when two or more of the mouse HLHS genes are interrogated across these human HLHS chromosome linkage intervals (P = 5.6 × 10−10) [34]. Together these findings demonstrate the clinical relevance of the mutations recovered from the HLHS mutant mice. They point to a multigenic etiology for HLHS that involves a profound degree of genetic heterogeneity. This would suggest that the clinical studies into the genetic landscape of HLHS may be difficult with the use of a standard case–control study design.
Fig. 4.
Mutations recovered from HLHS mutant mice and HLHS patients. a Mouse HLHS mutations in human chromosome regions linked to HLHS and left ventricular outflow tract obstruction (LVOTO). Genes recovered from mouse HLHS mutant lines found in human chromosome regions linked to HLHS/LVOTO are shown [8, 9]. The mouse chromosome location of each gene is indicated in parenthesis. The mouse mutant line is identified as L with three or four digit line identifier. b ToppCluster-generated interactome network of multi-hit genes recovered from subjects with HLHS and HLHS mutant mice. Genes (hexagons) associated with heart development and various gene ontology biological processes (color rectangles) are shown with corresponding color. Dark blue-outlined hexagons are NOTCH-related genes. Protein–protein interactions are indicated as solid dark blue lines between genes. Adapted with permission from Liu et al. [34]
Digenic Etiology of HLHS in the Ohia Mutant Line
Extensive breeding and phenotype/genotype analysis of the Ohia mutant mice identified HLHS arising from mutations in two genes, Sap130 and Pcdha9. Sap130, sin3A-associated protein 130, is a component of the Sin3A histone deacetylase (HDAC) repressor complex, while Pcdha9 is a protocadherin mediating cell–cell adhesion well described in the development of the nervous system [39-41]. The Sap130 mutation is a point mutation that causes an anomalous splice causing an in-frame 35 amino acid deletion, while the Pcdha9 mutation is a missense (P793L) mutation. Antibody staining showed the truncated Sap130 protein is expressed and is translocated into the nucleus. Analysis of a Sap130 KO mouse revealed that Sap130 deficiency causes peri-implantation lethality, while the Ohia Sap130 mutant can survive to term. These findings suggest the Ohia Sap130 mutation is likely hypomorphic.
Analysis of the Ohia mutant mice showed that the small LV phenotype can be elicited by the Sap130 mutation alone, while Pcdha9 played an essential role in the aortic hypoplasia and valvular defects in HLHS. Interestingly, the Pcdha9 mutation alone can cause BAV (Fig. 5a-c), a phenotype well described as highly associated with HLHS and often seen among family members of patients with HLHS [19, 42]. Sufficiency of Sap130/Pcdha9 mutations in causing HLHS was validated with replication of the HLHS phenotype in transgenic mice harboring Sap130/Pcdha9 mutation generated by CRISPR gene editing [34]. The CRISPR-targeted Sap130 allele expressed the same anomalously spliced Sap130 transcripts as seen in the Ohia mutants. The Pcdha9 CRISPR-targeted allele harbored a missense mutation near the missense mutation found in the Ohia mutant, suggesting both likely exert loss-of-function (LOF) effects.
Fig. 5.
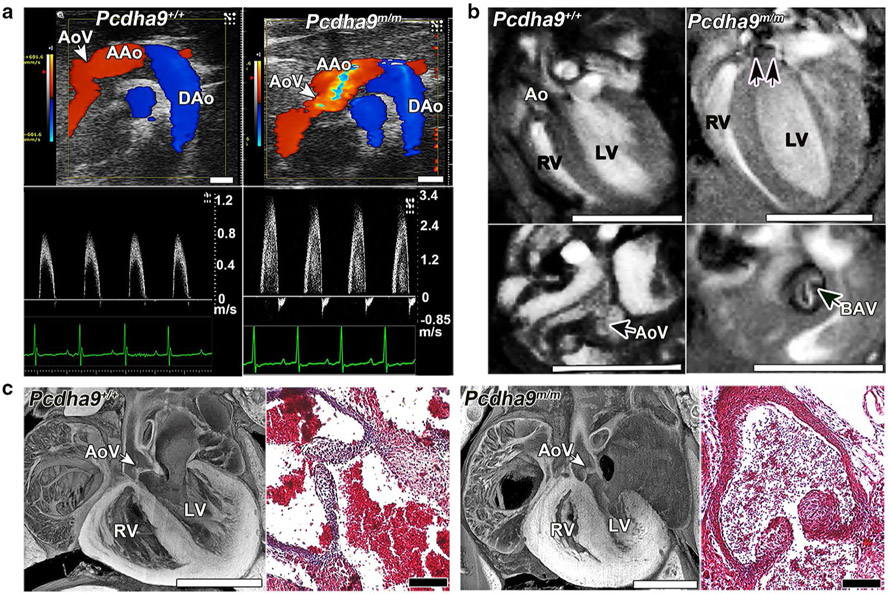
Bicuspid aortic valve phenotype in homozygous Pcdha9 mutant mice. a Echocardiography of adult Pcdha9 mutant vs. C57BL6 wild-type mice. Color-flow (top) and spectral Doppler (bottom) imaging, showing a high-velocity jet (> 3 m/s) across the aortic valve, thus indicating aortic stenosis (Supplementary Video 5). b Functional magnetic resonance imaging (MRI) showing the bicuspid aortic valve. MRI of homozygous Pcdha9 mutant mouse from (a), showing BAV with doming (double arrows) during systole (see Supplementary Videos 6, 7). LV hypertrophy, as determined by M-mode imaging (LV anterior wall/posterior wall: 0.45/0.64 mm in control vs. 0.91/0.91 mm in mutant). c Histopathology of newborn Pcdha9m/m vs. wild-type mice. Compared with wild type, the mutant mouse showed immature thickened aortic valves with increased trichrome staining, thus indicating abnormal matrix deposition. Adapted with permission from Liu et al. [34]
Regarding phenotype–genotype correlation, it is worth pointing out that in the Ohia HLHS mutant mice, we observed a spectrum of left-sided lesions as described above. This phenotype variability was replicated in the CRISPR gene-targeted mice, indicating it is inherent to the Sap130/Pcdha9 mutations. This is not unlike the often observed phenotype variability associated with most KO and mutant mouse models of CHD and likely reflects the stochastic processes regulating cardiovascular development. These findings would suggest that detailed subclassification of structural phenotypes that may have clinical utility for the management of patient care might not yield significant insights into phenotype–genotype correlations for advancing our understanding of the genetic etiology of HLHS.
PCDHA Mutation and Bicuspid Aortic Valve
BAV can be observed in Ohia HLHS mutants harboring the Sap130/Pcdha9 mutations, and also in isolation in mice harboring only the Pcdha9 mutations. BAV is the most common congenital anomaly observed in humans, affecting 1–2% of the adult human population [43]. Interestingly, a 16.7-kb deletion spanning the PCDHA9 and PCDHA10 genes is found in multiple human populations and reported to have a 1–8% allele frequency [44]. While this deletion is said to be present in unaffected individuals, we note BAV is typically not diagnosed until late adult life and would not affect reproductive fitness. Notably, African Americans have a lower frequency of the PCDHA deletion allele compared to Europeans, and they also have a lower BAV incidence (P = 0.001) [45]. Mice with deletion of Pcdha2 through Pcdha11 are reported to be viable, but have not been examined for cardiac phenotype [46, 47]. Our studies showed some Pcdha9 mutant mice with BAV are adult viable, but can succumb to sudden death from aortic insufficiency from BAV pathology. However, this is observed with low penetrance, suggesting other epigenetic or environmental factors may contribute to the emergence of pathology. Further studies are warranted to determine the risk of BAV in human subjects with the PCDHA deletion allele.
Intrinsic Cardiomyocyte Proliferation and Differentiation Defects
We investigated the causes for the HLHS-LV hypoplasia using the Ohia HLHS mutant embryos. For these studies, hearts at different development stages (E14.5 to term) were analyzed for cardiomyocyte cell proliferation using phosphohistone H3 (PH3) and Ki67. Interestingly, while cardiomyocytes labeled with PH3 increased, Ki67 positive cardiomyocytes decreased in the HLHS-LV tissue, findings reminiscent of a previous study of human HLHS fetal heart tissue [48]. In contrast, in the HLHS-RV, Ki67 was also decreased, but no change was observed for PH3. This was associated with a reduction in cardiomyocytes in anaphase–telophase in the HLHS-LV tissue, indicating a block in mitotic cell cycle progression, but this was not observed in the HLHS-RV tissue. TUNEL labeling also showed an increase in apoptosis in the HLHS-LV but not RV tissue. Electron microscopy revealed a cardiomyocyte differentiation defect, with the HLHS-LV exhibiting short myofilament bundles with more branching and poorly defined Z-bands. This was associated with a mitochondrial maturation defect indicated by a low-density matrix, sparse cristae, and a reduction in mitochondrial size and shape. While similar changes were observed in the RV, the changes were of lesser magnitude. Overall, these findings suggest the LV hypoplasia in HLHS is associated with intrinsic cardiomyocyte cell proliferation and differentiation defects linked to a mitochondrial maturation defect. These changes are not likely secondary to hemodynamic effects caused by the structural heart defects, since they can be observed at early stages of development (E14.5, equivalent to 7–8 week gestation human fetus). In addition, we recently observed these same cell proliferation and cardiomyocyte differentiation defects in cardiomyocytes generated in vitro from the differentiation of induced pluripotent stem cells (iPSC) derived from the Ohia HLHS mutant fibroblasts. Together these findings suggest that the small LV phenotype in Ohia HLHS mutants is elicited by intrinsic cardiomyocyte defects.
Metabolic Pathways Recovered by RNA-seq and ChIP-seq Analyses
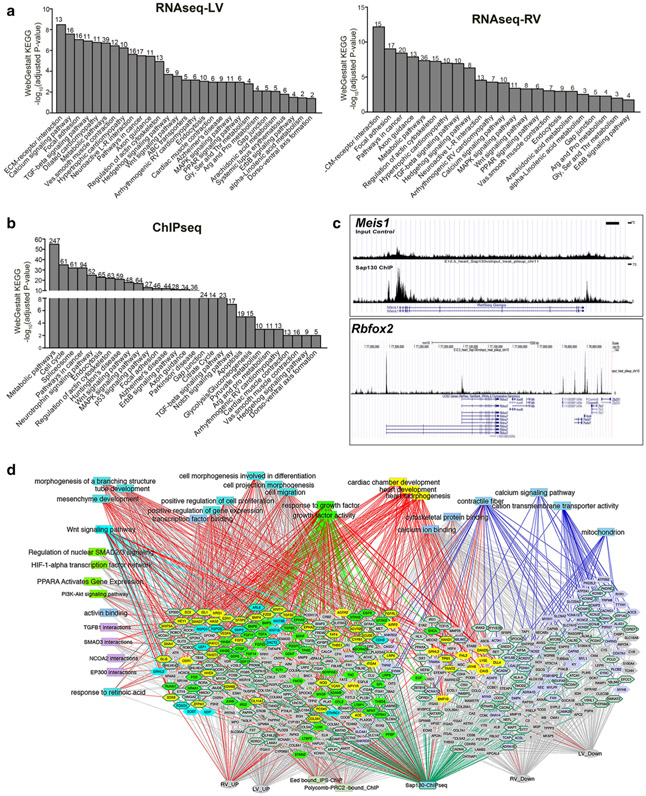
Gene expression changes in the Ohia HLHS heart tissue was profiled using RNA-seq analysis of RV and LV heart tissue obtained from embryos at E13.5, 14.5, and 17.5. Gene enrichment analysis identified metabolic and mitochondria-related pathways, growth factor signaling, cancer pathway, calcium signaling, cardiac muscle contraction, and dilated/hypertrophic cardiomyopathy (Fig. 6a). Similar pathways were recovered in the HLHS-LV and RV tissues, but the HLHS-RV showed lower fold changes. Using antibody to Sap130, we also conducted chromatin immunoprecipitation sequencing analysis to identify Sap130 target genes in embryonic heart tissue. This analysis yielded many of the same pathways identified by RNA-seq, with the top pathway being metabolic pathways, followed by cell cycle, spliceosome, and pathways in cancer (Fig. 6c). We note that metabolic pathways and pathways in cancer were shared in common between the RNA-seq and ChIP-seq analyses. Also observed among Sap130 target genes are Notch pathway components such as Rbpj, Mib1, Ep300, and Notch1. Particularly notable is recovery of Meis1, a gene that regulates mitochondrial metabolism [49] and postnatal cardiomyocyte cell cycle arrest [50], and Rbfox2, a RNA-binding protein recently identified as a HLHS candidate gene [29] (Fig. 6c). Rbfox2 is known to regulate alternative splicing and RNA processing, consistent with the studies by Ricci et al. showing the dysregulation of RNA splicing and mRNA processing in human HLHS-RV tissue [51]. In the study by Ricci et al., spliceosome and various metabolic pathways were also identified as significantly affected pathways in the newborn HLHS-RV tissue, findings that overlap the pathway enrichment analysis of the Sap130 ChIP-seq data [52].
Fig. 6.
RNA-seq and Sapq130 ChIP-seq Pathway Enrichment Analysis. a–c Differentially expressed genes identified by RNA-seq analysis of Ohia mutant HLHS-LV (a) vs. RV (b) heart tissue, and ChIP-seq (c) analysis of Sap130 target genes in embryonic heart tissue were analyzed by WebGestalt KEGG pathway enrichment analysis. Interestingly, all three datasets yielded similar gene-enriched pathways. d ToppCluster network analysis of genes (hexagons) that are differentially expressed in the hearts of HLHS mice based on RNA-seq analysis, or identified as Sap130 target genes by ChIP-seq analysis of E12.5 mouse heart. Genes highlighted in yellow are associated with cardiac development terms. Associations of these genes are indicated by red edges into other functional categories that include GO biological process and cellular component, pathways, or mouse knockout allele phenotype associations as determined by ToppGene. Genes that are Sap130 targets are shown surrounded with green edges. Genes associated with other enriched functional categories such as growth factor signaling, Wnt signaling, or mitochondrial function are colored individually with matching edges, except when they are linked to cardiac morphogenesis, in which case the edges are in red. Adapted with permission from Liu et al. [34]
ToppGene Network Analysis
ToppGene network analysis [53] showed that many down-regulated genes are related to cardiac differentiation, including contractile fiber, mitochondria, cation transport and ion channels, and calcium binding/signaling. In contrast, genes up-regulated were involved in mesenchyme determination, heart development, and tube/branching morphogenesis, many overlapping with Wnt and Tgfβ signaling (Fig. 6d). The combined pathway enrichment analysis using data generated from exome sequencing, RNA-seq, and ChIP-seq analyses showed convergence of pathways indicating HLHS may arise from the disruption of early events in mesenchyme/heart development involving perturbation of Notch, Tgfβ, and Wnt signaling. The mitochondrial and cardiomyocyte differentiation defects, and premature cell cycle arrest with abnormal activation of the hypertrophy program, could underlie the muscle-bound phenotype seen in Ohia HLHS mutants. Mitochondrial fusion and calcium signaling, processes that regulate cardiomyocyte differentiation, are also known to regulate Notch signaling [54]. Regarding the restriction of defects to left-sided heart structures in HLHS, it is interesting to note Notch signaling is known to play a role in left–right patterning [55] and Brg1, a component of the SWI/SNF chromatin-modulating complex implicated in left–right patterning [56], is also known to interact with the Sin3A complex, suggesting a potential mechanistic basis for the restriction of lesions to left-sided heart structures [51-53, 55-58].
Sap130 and Sin3A Regulation of Pcdha9 and EMT
To interrogate the Sap130–Pcdha9 interactions driving the pathogenesis of HLHS, we generated a cardiomic expression atlas using the combined microarray data obtained from 118 heart-related samples at different stages of development available from public databases [57-64]. This analysis showed Pcdha9 expression in E9.5 embryos was restricted to the cardiac cushion, which we confirmed by real-time PCR analysis. In contrast, Sap130 was more broadly expressed. Using the microarray data, Pcdha9-co-regulated genes were identified. This yielded many genes associated with cardiac valve development, including Notch pathway components, genes mediating epithelial–mesenchymal transformation (EMT), and mesenchymal cell differentiation. Genes identified as Sap130 co-regulated included many associated with the mitotic cell cycle, mitotic checkpoint control, and chromatin-regulatory components (Eed, Ezh2, and Suz12) of the Polycomb repressive complex 2 (PRC2). Previous studies showed SIN3A together with HDAC, PRC2, and lysine-specific demethylase 1 (LSD1) plays important roles in the repression of cadherin to promote EMT [65-67]. Together, these findings suggest Sap130 epigenetically regulates EMT mediating cardiac cushion development. Consistent with this, we observed the PCDHA gene cluster has multiple enhancers with SIN3A-binding sites. SIN3A-binding sites also have been mapped within the PCDHA cluster by ChIP-seq analysis.
Most unexpected was the finding in the original Ohia HLHS mutant mouse, not only Sap130 and Pcdha9 mutations, but also mutations in Sin3A and Snai1 among other mutations identified by exome sequencing analysis. While Sap130 and Pcdha9 mutations were homozygous, the Sin3A and Snai1 mutations were heterozygous. Snai1 is a transcription factor known to promote EMT by forming a complex with Sin3A and HDAC to repress E-cadherin expression. The presence of mutations in Snai1, Sin3A, Sap130, and Pcdha9 all in the same HLHS mutant is unlikely to have occurred by chance. While only the Sap130 and Pcdh9 mutations are required to generate HLHS, the original Ohia mutant harboring all four mutations survived to term. In contrast, the Ohia mutant with only the Sap130/Pcdha9 mutations dies at mid-gestation (E13.5–14.5). Such mid-gestation lethality is also observed in the CRISPR Sap130/Pcdha9 gene-targeted mice. Thus the Sin3A and Snai1 mutations may act as protective modifiers suppressing the more severe deleterious effects of the Sap130/Pcdha9 mutations, thereby allowing HLHS mutants to survive to term. These findings suggest that the complex genetics of HLHS may involve the interplay of pathogenic mutations and protective modifiers that together determine the final phenotypic disease outcome.
Role of Sin3A/Sap130 and Rbfox2 in Stem Cell Survival, Muscle Differentiation, and EMT Regulation
It is notable that Sin3A/Sap130 and Rbfox2 share many biological functions in common, suggesting insights into the mechanism of HLHS pathogenesis may emerge from a better understanding of the functions of these two genes. Thus Sin3A/Sap130 and Rbfox2 are both known to regulate muscle differentiation [68, 69] and metabolism [51, 70] and both are required for stem cell survival. Hence, deficiency in either of these genes may contribute to LV hypoplasia via disturbance of stem cell survival and muscle differentiation. Additionally, as both genes play important roles in EMT [71, 72], this could drive the valve defect phenotypes associated with HLHS, with presence of mutations in Pcdha9 in combination with Sap130 increasing the penetrance of valve defects in the Ohia mutant mice.
As Rbfox2 and Sap130/Sin3A are both known to regulate splicing, this also would point to the important role of altered splicing in HLHS disease pathogenesis. Indeed alternative splicing has been shown to regulate cardiac precursor proliferation and differentiation [73], with mice deficient in Rbm24 (RNA-binding motif 24) or Srsf10 (serine–arginine-rich splicing factor 10) exhibiting early embryonic lethality with impaired cardiogenesis [74, 75]. The observation of genome wide alternative splicing in the human fetal heart further suggests a role for splicing in the regulation of heart development and the pathogenesis of CHD [76]. In addition, various cardiac diseases are observed with mutations in splicing factors [77], such as dilated cardiomyopathy in patients and mice with RBM20 (RNA-binding motif protein 20) mutations [78, 79]. This may reflect the essential role of alternative splicing in the physiological regulation of cardiac muscle differentiation and function, such as in the critical regulation of calcium handling [75, 76].
Clinical Study of the Genetic Etiology of HLHS
To gain further insights into the genetic etiology of HLHS, we conducted exome sequencing of 68 HLHS patients. Rare LOF variants were recovered and compared to LOF variants in 95 ethnically matched CEU 1000 genomes control subjects (MAF < 0.01). Genes harboring these mutations were compared to those recovered from the HLHS mutant mice. This analysis showed more genes were shared between the 68 HLHS patients and eight HLHS mutant mice than expected at random (P = 0.013; Table 1). More genes were found to be shared among the 68 HLHS patients (P = 9.1 × 10−24), but not between the control CEU subjects [80] (Table 1). ToppGene network analysis of genes shared between the HLHS patients and mice showed enrichment of pathways that regulate heart development, chromatin, cell proliferation/cell cycle, mitochondrial associated, TGFβ/BMP and Notch signaling, and centrosome/cilia (Fig. 4b). The cilia finding is of particular interest given we previously observed a significant enrichment for cilia-related genes in our large-scale mouse forward genetic screen for CHD-causing mutations [29].
Table 1.
Sharing genes with loss-of-function mutations in HLHS patients and mice (adapted with permission from Liu et al. [34])
| Comparisona |
No. shared genes with unique LOF |
P valueb | Inference | |||
|---|---|---|---|---|---|---|
| Group 1 | Groups 2 | Observed | Expected | Observed/ expected |
||
| hHLHS | mHLHS | 33 | 22 | 1.50 | 0.013 | More sharing than by chance |
| CEU | mHLHS | 28 | 22 | 1.27 | 0.21 | Chance sharing |
| hHLHS | 247 | 69 | 3.57 | 9.1 × 10−24 | More sharing than by chance | |
| CEU | 116 | 265 | 0.44 | 4.5 × 10−16 | Less sharing in CEU than expected | |
| hHLHS | CEU | 306 | 453 | 0.67 | 4.2 × 10−7 | Less sharing in CEU than HLHS |
Comparison includes 68 HLHS (hHLHS) patients, and 95 1000 genomes ethic-matched CEU subjects, and eight independently derived HLHS mutant mice (mHLHS). The HLHS patients had 1278 genes with 1736 unique LOF, while the 1000 genomes CEU subjects had 1372 genes with 1550 unique LOF. Unique LOF are seen in only a single subject in the HLHS or CEU cohort. The eight HLHS mutant mice in total had 329 mutations in 329 genes
Comparison between groups was performed with χ2 goodness of fit tests
Summary and Future Prospects
We showed HLHS has a multigenic etiology, which likely accounts for the previous failure to obtain HLHS mutant mouse models. Our analysis revealed a digenic etiology for HLHS in the Ohia HLHS mutant mouse line, consistent with the 2-loci model previously proposed for HLHS based on human clinical studies [32]. In Ohia mutants, Sap130 was shown to drive the LV hypoplasia, likely by perturbing cardiomyocyte proliferation and differentiation, while protocadherin mutation may drive the aortic valve defects. A paradigm shift is indicated in which the complex phenotype of HLHS may arise in a combinatorial fashion, with intrinsic cardiomyocyte defects underlying the LV hypoplasia. These findings suggest a familial study design might be better suited for interrogating the genetic etiology of HLHS. As our findings also indicated HLHS is not solely a valve disease as previously proposed [19], this has important implications for therapeutic interventions, suggesting in utero aortic angioplasty will not be effective [81]. Also of interest is the indication of the disruption of RNA processing, findings that align with the notion that cell intrinsic defects underlie the pathogenesis of HLHS. Overall, these findings suggest that novel therapeutic intervention will emerge as new insights are obtained into the molecular mechanism of disease pathogenesis in HLHS, interventions that may help prevent heart failure or reduce heart failure risk among HLHS patients.
Acknowledgements
This work was supported by funding from NIH U01-HL098180, R01-HL132024, S10-OD010340 (CWL), and Children’s Heart Foundation (LJM and DWB) and Junior Cooperative Society (DWB).
Footnotes
Conflict of interest All authors declare no conflicts of interest.
Ethical Approval All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All studies were pursued under animal protocols approved by the University of Pittsburgh and the Jackson Laboratory Institutional Animal Care and Use Committees. All procedures performed in studies involving human participants were carried out with informed consent under a human study protocol approved by the University of Pittsburgh Institutional Review Board and in accordance with the ethical standards of the 1964 Helsinki Declaration and its later amendments.
Informed Consent Informed consent was obtained from all individual participants included in the study.
References
- 1.Moodie D (2011) Adult congenital heart disease: past, present, and future. Tex Heart Inst J 38:705. [PMC free article] [PubMed] [Google Scholar]
- 2.Delmo Walter EM, Hubler M, Alexi-Meskishvili V, Miera O, Weng Y, Loforte A, Berger F, Hetzer R (2009) Staged surgical palliation in hypoplastic left heart syndrome and its variants. J Card Surg 24:383–391 [DOI] [PubMed] [Google Scholar]
- 3.Gordon BM, Rodriguez S, Lee M, Chang RK (2008) Decreasing number of deaths of infants with hypoplastic left heart syndrome. J Pediatr 153:354–358 [DOI] [PubMed] [Google Scholar]
- 4.Feinstein JA, Benson DW, Dubin AM, Cohen MS, Maxey DM, Mahle WT, Pahl E, Villafane J, Bhatt AB, Peng LF, Johnson BA, Marsden AL, Daniels CJ, Rudd NA, Caldarone CA, Mussatto KA, Morales DL, Ivy DD, Gaynor JW, Tweddell JS, Deal BJ, Furck AK, Rosenthal GL, Ohye RG, Ghanayem NS, Cheatham JP, Tworetzky W, Martin GR (2012) Hypoplastic left heart syndrome: current considerations and expectations. J Am Coll Cardiol 59:S1–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kern JH, Hayes CJ, Michler RE, Gersony WM, Quaegebeur JM (1997) Survival and risk factor analysis for the Norwood procedure for hypoplastic left heart syndrome. Am J Cardiol 80:170–174 [DOI] [PubMed] [Google Scholar]
- 6.Donofrio MT, Moon-Grady AJ, Hornberger LK, Copel JA, Sklansky MS, Abuhamad A, Cuneo BF, Huhta JC, Jonas RA, Krishnan A, Lacey S, Lee W, Michelfelder EC Sr, Rempel GR, Silverman NH, Spray TL, Strasburger JF, Tworetzky W, Rychik J, American Heart Association Adults with Congenital Heart Disease Joint Committee of the Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology CoCS, Anesthesia, Council on Cardiovascular, Stroke Nursing (2014) Diagnosis and treatment of fetal cardiac disease: a scientific statement from the American Heart Association. Circulation 129:2183–2242 [DOI] [PubMed] [Google Scholar]
- 7.Verheijen PM, Lisowski LA, Plantinga RF, Hitchcock JF, Bennink GB, Stoutenbeek P, Meijboom EJ (2003) Prenatal diagnosis of the fetus with hypoplastic left heart syndrome management and outcome. Herz 28:250–256 [DOI] [PubMed] [Google Scholar]
- 8.Rychik J (2014) Hypoplastic left heart syndrome: can we change the rules of the game? Circulation 130:629–631 [DOI] [PubMed] [Google Scholar]
- 9.Freud LR, McElhinney DB, Marshall AC, Marx GR, Friedman KG, Pedro J, Emani SM, Lafranchi T, Silva V, Wilkins-Haug LE (2014) Fetal aortic valvuloplasty for evolving hypoplastic left heart syndrome: postnatal outcomes of the first 100 patients. Circulation 130:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nawaytou HM, Peyvandi S, Brook MM, Silverman N, Moon-Grady AJ (2016) Right ventricular systolic-to-diastolic time index: hypoplastic left heart fetuses differ significantly from normal fetuses. J Am Soc Echocardiogr 29:143–149 [DOI] [PubMed] [Google Scholar]
- 11.Arnold RR, Loukanov T, Gorenflo M (2014) Hypoplastic left heart syndrome—unresolved issues. Front Pediatr 2:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oechslin EN, Harrison DA, Connelly MS, Webb GD, Siu SC (2000) Mode of death in adults with congenital heart disease. Am J Cardiol 86:1111–1116 [DOI] [PubMed] [Google Scholar]
- 13.Khairy P, Fernandes SM, Mayer JE Jr, Triedman JK, Walsh EP, Lock JE, Landzberg MJ (2008) Long-term survival, modes of death, and predictors of mortality in patients with Fontan surgery. Circulation 117:85–92 [DOI] [PubMed] [Google Scholar]
- 14.Nora JJ, Nora AH (1988) Update on counseling the family with a first-degree relative with a congenital heart defect. Am J Med Genet 29:137–142 [DOI] [PubMed] [Google Scholar]
- 15.Maestri NE, Beaty TH, Liang KY, Boughman JA, Ferencz C (1988) Assessing familial aggregation of congenital cardiovascular malformations in case-control studies. Genet Epidemiol 5:343–354 [DOI] [PubMed] [Google Scholar]
- 16.Briard ML, Chauvet ML, Le Merrer M, Frezal J (1984) Epidemiological and genetic study of 3 congenital cardiopathies with neonatal disclosure. Archives francaises de pediatrie 41:313–321 [PubMed] [Google Scholar]
- 17.Dennis NR, Warren J (1981) Risks to the offspring of patients with some common congenital heart defects. Am J Med Genet 18:8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loffredo CA, Chokkalingam A, Sill AM, Boughman JA, Clark EB, Scheel J, Brenner JI (2004) Prevalence of congenital cardio-vascular malformations among relatives of infants with hypoplastic left heart, coarctation of the aorta, and d-transposition of the great arteries. Am J Med Genet Part A 124A:225–230 [DOI] [PubMed] [Google Scholar]
- 19.Hinton RB Jr, Martin LJ, Tabangin ME, Mazwi ML, Cripe LH, Benson DW (2007) Hypoplastic left heart syndrome is heritable. J Am Coll Cardiol 50:1590–1595 [DOI] [PubMed] [Google Scholar]
- 20.Laursen HB (1980) Some epidemiological aspects of congenital heart disease in Denmark. Acta Paediatr Scand 69:619–624 [DOI] [PubMed] [Google Scholar]
- 21.McBride KL, Marengo L, Canfield M, Langlois P, Fixler D, Belmont JW (2005) Epidemiology of noncomplex left ventricular outflow tract obstruction malformations (aortic valve stenosis, coarctation of the aorta, hypoplastic left heart syndrome) in Texas, 1999–2001. Birth Defects Res A 73:555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natowicz M, Kelley RI (1987) Association of Turner syndrome with hypoplastic left-heart syndrome. Am J Dis Child 141:218–220 [DOI] [PubMed] [Google Scholar]
- 23.Grossfeld PD, Mattina T, Lai Z, Favier R, Jones KL, Cotter F, Jones C (2004) The 11q terminal deletion disorder: a prospective study of 110 cases. Am J Med Genet Part A 129A:51–61 [DOI] [PubMed] [Google Scholar]
- 24.Feinstein JA, Benson DW, Dubin AM, Cohen MS, Maxey DM, Mahle WT, Pahl E, Villafañe J, Bhatt AB, Peng LF (2012) Hypoplastic left heart syndrome: current considerations and expectations. J Am Coll Cardiol 59:S1–S42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, Grossfeld P, Fatkin D, Jones O, Hayes P, Feneley M, Harvey RP (2003) Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol 41:2072–2076 [DOI] [PubMed] [Google Scholar]
- 26.Dasgupta C, Martinez AM, Zuppan CW, Shah MM, Bailey LL, Fletcher WH (2001) Identification of connexin43 (alpha1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE). Mutat Res 479:173–186 [DOI] [PubMed] [Google Scholar]
- 27.Iascone M, Ciccone R, Galletti L, Marchetti D, Seddio F, Lincesso AR, Pezzoli L, Vetro A, Barachetti D, Boni L, Federici D, Soto AM, Comas JV, Ferrazzi P, Zuffardi O (2012) Identification of de novo mutations and rare variants in hypoplastic left heart syndrome. Clin Genet 81:542–554 [DOI] [PubMed] [Google Scholar]
- 28.Reamon-Buettner SM, Ciribilli Y, Inga A, Borlak J (2008) A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Hum Mol Genet 17:1397–1405 [DOI] [PubMed] [Google Scholar]
- 29.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, Jin SC, Deanfield J, Giardini A, Porter GA Jr, Kim R, Bilguvar K, Lopez-Giraldez F, Tikhonova I, Mane S, Romano-Adesman A, Qi H, Vardarajan B, Ma L, Daly M, Roberts AE, Russell MW, Mital S, Newburger JW, Gaynor JW, Breitbart RE, Iossifov I, Ronemus M, Sanders SJ, Kaltman JR, Seidman JG, Brueckner M, Gelb BD, Goldmuntz E, Lifton RP, Seidman CE, Chung WK (2015) De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350:1262–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theis JL, Hrstka SC, Evans JM, O’Byrne MM, de Andrade M, O’Leary PW, Nelson TJ, Olson TM (2015) Compound heterozygous NOTCH1 mutations underlie impaired cardiogenesis in a patient with hypoplastic left heart syndrome. Hum Genet 134:1003–1011 [DOI] [PubMed] [Google Scholar]
- 31.Theis JL, Zimmermann MT, Evans JM, Eckloff BW, Wieben ED, Qureshi MY, O’Leary PW, Olson TM (2015) Recessive MYH6 mutations in hypoplastic left heart with reduced ejection fraction. Circ Cardiovasc Genet 8:564–571 [DOI] [PubMed] [Google Scholar]
- 32.McBride KL, Pignatelli R, Lewin M, Ho T, Fernbach S, Menesses A, Lam W, Leal SM, Kaplan N, Schliekelman P, Towbin JA, Belmont JW (2005) Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: segregation, multiplex relative risk, and heritability. Am J Med Genet Part A 134A:180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Francis R, Kim AJ, Ramirez R, Chen G, Subramanian R, Anderton S, Kim Y, Wong L, Morgan J (2014) Interrogating congenital heart defects with noninvasive fetal echocardiography in a mouse forward genetic screen. Circ Cardiovasc Imaging 7:31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Yagi H, Saeed S, Bais AS, Gabriel GC, Chen Z, Peterson KA, Li Y, Schwartz MC, Reynolds WT, Saydmohammed M, Gibbs B, Wu Y, Devine W, Chatterjee B, Klena NT, Kostka D, de Mesy Bentley KL, Ganapathiraju MK, Dexheimer P, Leatherbury L, Khalifa O, Bhagat A, Zahid M, Pu W, Watkins S, Grossfeld P, Murray SA, Porter GA Jr, Tsang M, Martin LJ, Benson DW, Aronow BJ, Lo CW (2017) The complex genetics of hypoplastic left heart syndrome. Nat Genet 49:1152–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinton RB, Martin LJ, Rame-Gowda S, Tabangin ME, Cripe LH, Benson DW (2009) Hypoplastic left heart syndrome links to chromosomes 10q and 6q and is genetically related to bicuspid aortic valve. J Am Coll Cardiol 53:1065–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taketazu M, Barrea C, Smallhorn JF, Wilson GJ, Hornberger LK (2004) Intrauterine pulmonary venous flow and restrictive foramen ovale in fetal hypoplastic left heart syndrome. J Am Coll Cardiol 43:1902–1907 [DOI] [PubMed] [Google Scholar]
- 37.Martin LJ, Pilipenko V, Kaufman KM, Cripe L, Kottyan LC, Keddache M, Dexheimer P, Weirauch MT, Benson DW (2014) Whole exome sequencing for familial bicuspid aortic valve identifies putative variants. Circ Cardiovasc Gene 7:677–683 [DOI] [PubMed] [Google Scholar]
- 38.Martin LJ, Ramachandran V, Cripe LH, Hinton RB, Andelfinger G, Tabangin M, Shooner K, Keddache M, Benson DW (2007) Evidence in favor of linkage to human chromosomal regions 18q, 5q and 13q for bicuspid aortic valve and associated cardiovascular malformations. Hum Genet 121:275–284 [DOI] [PubMed] [Google Scholar]
- 39.Chen WV, Maniatis T (2013) Clustered protocadherins. Development 140:3297–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T (2005) Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet 37:171–176 [DOI] [PubMed] [Google Scholar]
- 41.Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, Tapia JC, Shapiro L, Honig B, Maniatis T (2014) Single-cell identity generated by combinatorial homophilic interactions between alpha, beta, and gamma protocadherins. Cell 158:1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewin MB, McBride KL, Pignatelli R, Fernbach S, Combes A, Menesses A, Lam W, Bezold LI, Kaplan N, Towbin JA, Belmont JW (2004) Echocardiographic evaluation of asymptomatic parental and sibling cardiovascular anomalies associated with congenital left ventricular outflow tract lesions. Pediatrics 114:691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin PS, Kloesel B, Norris RA, Lindsay M, Milan D, Body SC (2015) Embryonic development of the bicuspid aortic valve. J Cardiovasc Dev Dis 2:248–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noonan JP, Li J, Nguyen L, Caoile C, Dickson M, Grimwood J, Schmutz J, Feldman MW, Myers RM (2003) Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster. Am J Hum Genet 72:621–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandra S, Lang RM, Nicolarsen J, Gayat E, Spencer KT, Mor-Avi V, Hofmann Bowman MA (2012) Bicuspid aortic valve: inter-racial difference in frequency and aortic dimensions. JACC Cardiovasc Imaging 5:981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasegawa S, Hamada S, Kumode Y, Esumi S, Katori S, Fukuda E, Uchiyama Y, Hirabayashi T, Mombaerts P, Yagi T (2008) The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol Cell Neurosci 38:66–79 [DOI] [PubMed] [Google Scholar]
- 47.Suo L, Lu H, Ying G, Capecchi MR, Wu Q (2012) Protocadherin clusters and cell adhesion kinase regulate dendrite complexity through Rho GTPase. J Mol Cell Biol 4:362–376 [DOI] [PubMed] [Google Scholar]
- 48.Gaber N, Gagliardi M, Patel P, Kinnear C, Zhang C, Chitayat D, Shannon P, Jaeggi E, Tabori U, Keller G, Mital S (2013) Fetal reprogramming and senescence in hypoplastic left heart syndrome and in human pluripotent stem cells during cardiac differentiation. Am J Pathol 183:720–734 [DOI] [PubMed] [Google Scholar]
- 49.Tomoeda M, Yuki M, Kubo C, Yoshizawa H, Kitamura M, Nagata S, Nishizawa Y, Tomita Y (2011) Role of Meis1 in mitochondrial gene transcription of pancreatic cancer cells. Biochem Biophys Res Commun 410:798–802 [DOI] [PubMed] [Google Scholar]
- 50.Mahmoud AI, Kocabas F, Muralidhar SA, Kimura W, Koura AS, Thet S, Porrello ER, Sadek HA (2013) Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature 497:249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verma SK, Deshmukh V, Nutter CA, Jaworski E, Jin W, Wadhwa L, Abata J, Ricci M, Lincoln J, Martin JF, Yeo GW, Kuyumcu-Martinez MN (2016) Rbfox2 function in RNA metabolism is impaired in hypoplastic left heart syndrome patient hearts. Sci Rep 6:30896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ricci M, Xu Y, Hammond HL, Willoughby DA, Nathanson L, Rodriguez MM, Vatta M, Lipshultz SE, Lincoln J (2012) Myocardial alternative RNA splicing and gene expression profiling in early stage hypoplastic left heart syndrome. PLoS ONE 7:e29784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J, Bardes EE, Aronow BJ, Jegga AG (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37:W305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kasahara A, Cipolat S, Chen Y, Dorn GW 2nd, Scorrano L (2013) Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science 342:734–737 [DOI] [PubMed] [Google Scholar]
- 55.Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, Brueckner M, Khokha MK (2013) The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature 504:456–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, Chawengsaksophak K, Hamada H, Yost HJ, Rossant J, Bruneau BG (2007) Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc Natl Acad Sci USA 104:846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee MP, Yutzey KE (2011) Twist1 directly regulates genes that promote cell proliferation and migration in developing heart valves. PLoS ONE 6:e29758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rivera-Feliciano J, Lee KH, Kong SW, Rajagopal S, Ma Q, Springer Z, Izumo S, Tabin CJ, Pu WT (2006) Development of heart valves requires Gata4 expression in endothelial-derived cells. Development 133:3607–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakano H, Liu X, Arshi A, Nakashima Y, van Handel B, Sasidharan R, Harmon AW, Shin JH, Schwartz RJ, Conway SJ, Harvey RP, Pashmforoush M, Mikkola HK, Nakano A (2013) Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat Commun 4:1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Loo PF, Mahtab EA, Wisse LJ, Hou J, Grosveld F, Suske G, Philipsen S, Gittenberger-de Groot AC (2007) Transcription factor Sp3 knockout mice display serious cardiac malformations. Mol Cell Biol 27:8571–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manuylov NL, Tevosian SG (2009) Cardiac expression of Tnnt1 requires the GATA4-FOG2 transcription complex. Sci World J 9:575–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vijaya M, Manikandan J, Parakalan R, Dheen ST, Kumar SD, Tay SS (2013) Differential gene expression profiles during embryonic heart development in diabetic mice pregnancy. Gene 516:218–227 [DOI] [PubMed] [Google Scholar]
- 63.Martinez-Fernandez A, Li X, Hartjes KA, Terzic A, Nelson TJ (2013) Natural cardiogenesis-based template predicts cardiogenic potential of induced pluripotent stem cell lines. Circ Cardiovasc Genet 6:462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Magarin M, Schulz H, Thierfelder L, Drenckhahn JD (2016) Transcriptional profiling of regenerating embryonic mouse hearts. Genom Data 9:145–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peinado H, Portillo F, Cano A (2004) Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol 48:365–375 [DOI] [PubMed] [Google Scholar]
- 66.Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, Garcia de Herreros A, Peiro S (2008) Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 28:4772–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP (2010) The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 29:1803–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gallagher TL, Arribere JA, Geurts PA, Exner CR, McDonald KL, Dill KK, Marr HL, Adkar SS, Garnett AT, Amacher SL, Conboy JG (2011) Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev Biol 359:251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Oevelen C, Bowman C, Pellegrino J, Asp P, Cheng J, Parisi F, Micsinai M, Kluger Y, Chu A, Blais A, David G, Dynlacht BD (2010) The mammalian Sin3 proteins are required for muscle development and sarcomere specification. Mol Cell Biol 30:5686–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu M, Pile LA (2017) The transcriptional corepressor SIN3 directly regulates genes involved in methionine catabolism and affects histone methylation, linking epigenetics and metabolism. J Biol Chem 292:1970–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Braeutigam C, Rago L, Rolke A, Waldmeier L, Christofori G, Winter J (2014) The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene 33:1082–1092 [DOI] [PubMed] [Google Scholar]
- 72.Strobl-Mazzulla PH, Bronner ME (2012) A PHD12-Snail2 repressive complex epigenetically mediates neural crest epithelial-to-mesenchymal transition. J Cell Biol 198:999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salomonis N, Nelson B, Vranizan K, Pico AR, Hanspers K, Kuchinsky A, Ta L, Mercola M, Conklin BR (2009) Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput Biol 5:e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng Y, Valley MT, Lazar J, Yang AL, Bronson RT, Firestein S, Coetzee WA, Manley JL (2009) SRp38 regulates alternative splicing and is required for Ca(2+) handling in the embryonic heart. Dev Cell 16:528–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J, Hung LH, Licht T, Kostin S, Looso M, Khrameeva E, Bindereif A, Schneider A, Braun T (2014) RBM24 is a major regulator of muscle-specific alternative splicing. Dev Cell 31:87–99 [DOI] [PubMed] [Google Scholar]
- 76.Wang H, Chen Y, Li X, Chen G, Zhong L, Chen G, Liao Y, Liao W, Bin J (2016) Genome-wide analysis of alternative splicing during human heart development. Sci Rep 6:35520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van den Hoogenhof MM, Pinto YM, Creemers EE (2016) RNA splicing: regulation and dysregulation in the heart. Circ Res 118:454–468 [DOI] [PubMed] [Google Scholar]
- 78.Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, Michels VV, Olson TM (2009) Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol 54:930–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beraldi R, Li X, Martinez Fernandez A, Reyes S, Secreto F, Terzic A, Olson TM, Nelson TJ (2014) Rbm20-deficient cardiogenesis reveals early disruption of RNA processing and sarcomere remodeling establishing a developmental etiology for dilated cardiomyopathy. Hum Mol Genet 23:3779–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA (2012) An integrated map of genetic variation from 1092 human genomes. Nature 491:56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Freud LR, McElhinney DB, Marshall AC, Marx GR, Friedman KG, Del Nido PJ, Emani SM, Lafranchi T, Silva V, Wilkins-Haug LE, Benson CB, Lock JE, Tworetzky W (2014) Fetal aortic valvuloplasty for evolving hypoplastic left heart syndrome: postnatal outcomes of the first 100 patients. Circulation 130:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]