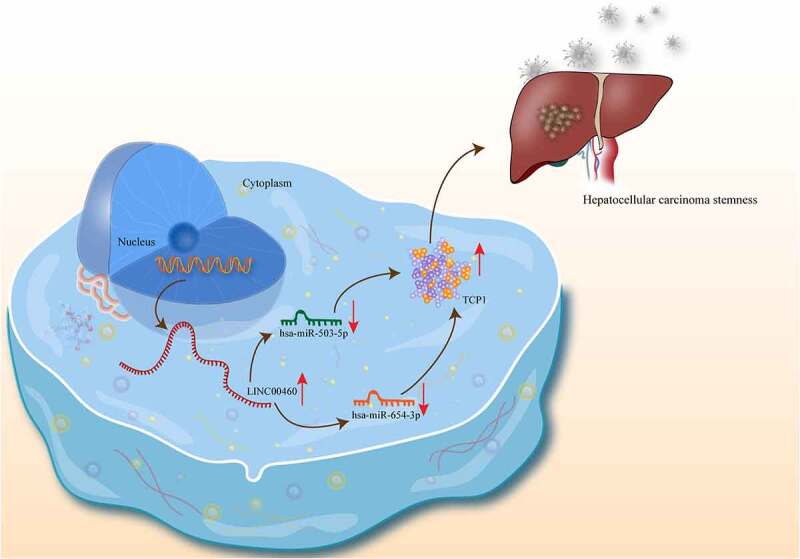
ABSTRACT
Abundant researches have stated that long noncoding RNAs (lncRNAs) are crucial molecules in intricate progression of various cancers in terms of their influence on cell stemness. However, no research has discussed the role of LINC00460 in the stemness of hepatocellular carcinoma (HCC). RT-qPCR and western blot were utilized to respectively examine the RNA and protein levels. Aldehyde dehydrogenase 1 (ALDH1) assays and sphere formation assay were performed to detect cell stemness property in vitro and in vivo subcutaneous xenograft tumor assay was performed to detect tumor growth. Interaction between RNAs was explored by luciferase reporter assays and RNA pull-down assays. Our results showed that LINC00460 was markedly over-expressed in HCC and silencing LINC00460 impaired cell stemness. Additionally, LINC00460 knockdown curbed proliferation, migration, invasion and epithelial-to-mesenchymal transition (EMT) and drove apoptosis of HCC cells. Further, LINC00460 bound to miR-503-5p and miR-654-3p to protect t-complex 1 (TCP1) from being inhibited by miR-503-5p/miR-654-3p. Rescue experiments confirmed the effect of LINC00460/miR-503-5p/miR-654-3p/TCP1 on HCC cell stemness. In conclusion, LINC00460 aggravated cell stemness in HCC via targeting miR-503-5p/miR-654-3p and TCP1, suggesting that LINC00460 may work as a potential signature for cell stemness in HCC.
KEYWORDS: LINC00460, miR-503-5p, miR-654-3p, TCP1, stemness, hepatocellular carcinoma
Introduction
As a primary liver cancer with a high mortality rate, HCC has the second highest cancer-induced mortality and represents a crucial health problem around the world [1]. With a poor prognosis caused by high recurrence rate and metastasis rate, HCC was mainly found in men under the age of 60. While transplantation and surgical resection are the most effective treatments for HCC patients in the early stage, development of a more appropriate therapy requires a multidisciplinary approach [1,2]. Although the pathogenesis of HCC and efficiency of novel anticancer drugs have been studied with some achievements, the recurrence and metastasis in HCC still cannot be perfectly solved. Accumulating data suggest that cancer stem cells (CSCs) which possess the same self-renewal, differentiation and proliferating property to take responsibilities for refactoring and propagating ability of the disease [3–5]. As a result, the targeted inhibition markers on CSCs potential stemness need to be explored.
Long noncoding RNA refers to lncRNA having nucleotides longer than 200 nt and possessing an open reading coding frame of 50–100 nt [6]. With a structure similar to mRNA, lncRNA is expressed specifically in terms of time and space, while some lncRNAs can generate ployA tails after shear processing [7]. As recorded, lncRNAs have crucial regulatory mechanisms in different cancers, including the regulation of cancer cell stemness. For instance, MSC-regulated lncRNA MACC1 antisense RNA 1 (MACC1-AS1) facilitates chemoresistance as well as stemness in gastric cancer via fatty acid oxidation [8]. LncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) enhances gastric cancer cell stemness via boosting SRY-box transcription factor 2 (SOX2) mRNA stability [9]. LncRNA papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) restrains the stemness of cancer cell and tumor growth in gastric cancer through exerting reciprocity with long intergenic non-protein coding RNA (Linc-pint) [10]. LINC00511/miR-185-3p/E2F1/Nanog homeobox (Nanog) axis contributes to the stemness and tumorigenesis in breast cancer [11]. Long intergenic non-coding RNA 460 (LINC00460) has been discovered to function in HCC with several different mechanisms. For example, LINC00460/miR-485-5p/p21 activated kinase 1 (PAK1) axis has promotion effect on the development of HCC [12]. LINC00460/miR-342-3p/anterior gradient 2 (AGR2) axis facilitates the progression of HCC [13]. However, whether LINC00460 influenced stemness of HCC cells is unknown.
Thus, in this study, we focused on the role of LINC00460 on HCC cell stemness with its interaction with some other miRNAs and target genes in HCC. This study might provide a new LINC00460-regulated mechanism in HCC cells, opening new perspectives for HCC treatment.
Materials and methods:
Cell lines
HCC cell lines (Huh-7, SK-HEP-1, PLC/PRF15, and Hep3b) and the human normal hepatic cell line (HL-7702) were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and incubated in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), 5% CO2 and penicillin-streptomycin at 37°C.
Vector construction and cell transfection
Three interference sequences were designed in accordance with the sequence of LINC00460 and these sequences were inserted into the pLKO.1 vector to construct three interference vectors named, respectively, as sh-LINC00460-1, −2, −3 were constructed. Besides, the sequence of LINC00460 was inserted into pcDNA3.1 vector to form pcDNA3.1-LINC00460 with the empty pcDNA3.1 vector as the negative control. The miR-503-5p/miR-654-3p mimics and NC mimics were obtained from ABM biology (Canada). LINC00460-WT/Mut Luciferase vector was formed by means of inserting the sequence of LINC00460 with wild type and mutant type of miR-503-5p/miR-654-3p sites into Luciferase reporter vector.
For transfection, the Huh-7 and Hep3b cells were paved in the 24-well plates. Next day, cells were transfected at about 30% confluence with the application of Lipofectamine 2000 on the basis of manufacturer’s instructions. The cell status was surveyed in 6 h after transfection.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted with Trizol reagent. Then, complementary DNA (cDNA) was acquired from reverse transcription of RNA using PrimeScript RT master mix (Perfect Real Time) (Takara, Cat#: RR0036A) or miScript II RT Kit (QIAGEN, Cat#: 218,160). The data on gene expression was detected by SYBR Green PCR Master Mix (Applied Biosystems, Cat#: 4,309,155) at 95°C for 10 minutes; 95°C for 30 seconds; and 60°C for 30 seconds. 2−ΔΔCt method was applied to calculate the relative expression. GAPDH and U6 were regarded as internal references.
Western blot
First, the cells whose confluence had reached more than 80% were collected, and then, the proteins which were extracted in Radioimmunoprecipitation assay (RIPA) lysis buffer and isolated with SDS-polyacrylamide gel electrophoresis (SDS-PAGE) were transferred on Polyvinylidene Fluoride (PVDF) membranes. The membranes were blocked with 5% skim milk at normal temperature, then the proteins could be incubated at 4°C with primary antibody, and secondary antibody was incubated after TBST washing. Protein bands were detected by electrochemiluminescence (ECL) luminous liquid. Antibodies used in western blot were anti-CD44 (ab189524, Abcam, UK), anti-SOX2 (ab92494, Abcam, UK), anti-Oct4 (ab181557, Abcam, UK), anti-Nanog (ab109250, Abcam, UK), anti-ZO1 (ab276131, Abcam, UK), anti-E-cadherin (ab231303, Abcam, UK), anti-N-cadherin (ab76011, Abcam, UK), anti-TCP1 (#3561S, Cell Signaling Technology, USA), anti- IRE1 (ab37073, Abcam, UK), anti-CHOP (#2895, Cell Signaling Technology, USA), anti-GRP78 (ab21685), anti-p-eIF2α (ab32157, Abcam, UK), anti-eIF2α (ab169528, Abcam, UK) and anti-GAPDH (ab8245, Abcam, UK).
Fluorescence in situ hybridization (FISH)
The cultured cells were fixed in paraformaldehyde (PFA) solution for 15 minutes and then treated with 0.5% TritonX-100 for 15 minutes. After treatment with prehybridization solution, the cells were hybridized with Digoxigenin (DIG) labeled probe overnight at 37°C. After washing and blocking, the nuclei were stained with DAPI (4ʹ, 6-diamidino-2-phenylindole). The probe images were observed with confocal microscopy.
RNA pull-down assay
Secondary structures were constructed by adding Structure Buffer into 1 µg bio-miR-503-5p/miR-654-3p, and then these RNAs were heated at 95°C for 2 minutes along with 3-minute ice-bath. After 30-minute stationary treating at room temperature, 15 µL streptavidin beads were added into RNAs to incubate the mixture 2 h at 4°C. The complexes pulled by magnetic beads were treated with cell lysis buffer under 4°C followed by a brief centrifugation. Then, RT-qPCR was performed to detect the enrichment of RNA which was extracted by Trizol.
Luciferase reporter assay
The full-length of wild-type (WT) or mutated (Mut) LINC00460 or the 3ʹUTR sequence of TCP1 containing the binding sites of miR-503-5p/miR-654-3p was sub-cloned into the pmirGLO vector to construct the LINC00460-WT/Mut or TCP1-WT/Mut vector. After that, these vectors were respectively co-transfected with miR-503-5p/miR-654-3p mimics into the Huh-7 and Hep3b cells. Luciferase activity was determined by Dual-Luciferase Reporter Assay System, with Renilla luciferase gene as a control.
Sphere formation assay
The transfected Huh-7 and Hep3b cells were cultured in the ultra-low attachment 6-well plates and cultured in medium. About 12 d later, sphere formation was observed and counted under a microscope.
In vivo subcutaneous
Nude mice (5-wk-old, female) were acquired from Hospital. Twelve mice were divided into two groups, six in control group and six in the experimental group. Huh-7 and Hep3b cells transfected with sh-NC or sh-LINC00460 were subcutaneously injected into each mouse and the growth of tumor was examined every 4 d. Twenty-eight days later, mice were sacrificed and tumorigenesis was detected. Animal study was approved by the Ethics Committee of Jingmen First People’s Hospital.
ALDH1 assay
The ALDH1 activity was examined by ALDH1 Kit (GOY-E7193, eBioscience, USA) in the transfected Huh-7 and Hep3b cells and the ALDH1 Kit was utilized with a series of procedures under guidance. Bio-repeats were implemented in triplicate.
Colony formation assay
Cells (1 × 103) were seeded in each one of the 6 wells of a plate and underwent incubation for 14 d at 37°C. After two rounds of PBS wash, cells were fixed by 4% formaldehyde and sent to crystal violet staining. The colonies formed by over 50 cells were manually counted in triplicate assays.
Transwell assays
HCC cells were in each insert of a transwell chamber (Corning, Cambridge, USA) without (migration assay) or with (invasion assay) matrigel (356,234; Millipore, Bedford, MA). The lower chambers contained 1% FBS. After 1-d incubation at 37°C, cells at upper membrane surface were removed by a cotton tip, whereas cells at lower surface were stained by 0.1% crystal violet. The number of migratory or invaded cells each field under a microscope (Olympus, Tokyo, Japan) was calculated.
Terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling (TUNEL)
Transfected HCC cells were cultured overnight and rinsed twice in PBS. Then, cells fixed by 4% paraformaldehyde were permeabilized in 0.25% Triton‐X 100. Thereafter, TUNEL assays were performed under manufacturer’s instructions (Roche). In brief, HCC cells underwent 45 min-incubation in terminal deoxynucleotidyl transferase (TdT) reaction cocktail, followed by the addition of Click‐iT reaction cocktail. Then, nucleus was stained by DAPI. Rate of TUNEL-stained cells under each field of fluorescence microscopy (DMI4000B; Leica Microsystems GmbH) was calculated.
Statistical analyses
Data of each assay are presented as mean ± SD. Bio-repeats were implemented in triplicate. A SPSS 22.0 package system was used to perform all statistical analyses. The mean values were compared by Student’s t-test, one-way ANOVA and two-way ANOVA followed by Dunnett or Tukey. Data were statistically significant with P less than 0.05.
Results
LINC00460 is over-expressed in HCC cell lines
To explore the role of LINC00460 in HCC cells, the expression of LINC00460 was first detected by RT-qPCR analysis in HCC cell lines (Huh-7, SK-HEP-1, PLC/PRF15, and Hep3b) and the human normal hepatic cell line (HL-7702). The results showed that LINC00460 was highly expressed in HCC cell lines, especially in Huh-7 and Hep3b cell lines (Figure 1a). Therefore, LINC00460 participated in HCC.
Figure 1.
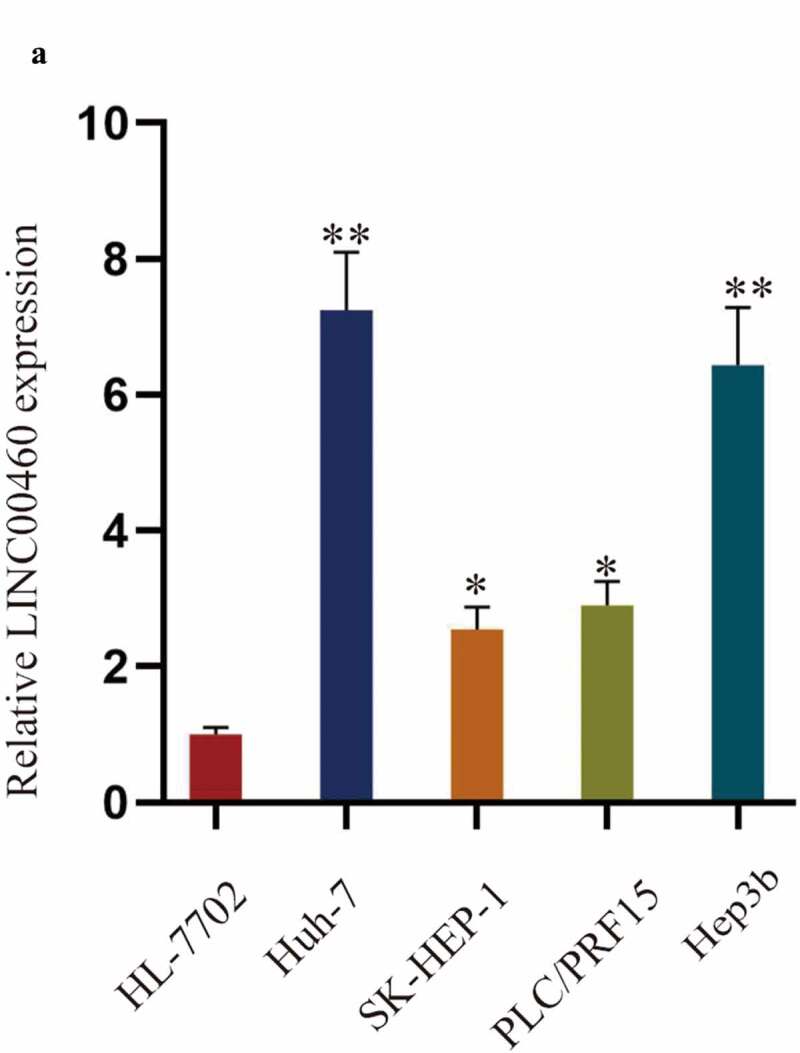
LINC00460 is over-expressed in HCC cell lines. A. The expression of LINC00460 was examined by RT-qPCR analysis in HCC cell lines (Huh-7, SK-HEP-1, PLC/PRF15, and Hep3b) and the human normal hepatic cell line (HL-7702). **P < 0.01, *P < 0.05; one-way ANOVA and Tukey
LINC00460 positively regulates the malignant behaviors of HCC cells
Subsequently, we further studied the role of LINC00460 in HCC cells. First, we silenced LINC00460 by sh-LINC00460-1/2/3 and overexpressed it by pcDNA3.1-LINC00460 in Huh-7 and Hep3b cells. The decline of LINC00460 by sh-LINC00460-1/2/3 and increase of LINC00460 by pcDNA3.1-LINC00460 in Huh-7 and Hep3b cells were confirmed by RT-qPCR, and sh-LINC00460-2/3 knocked down LINC00460 more significantly (Figure 2a). Thus, sh-LINC00460-2/3 were utilized in the subsequent experiments.
Figure 2.
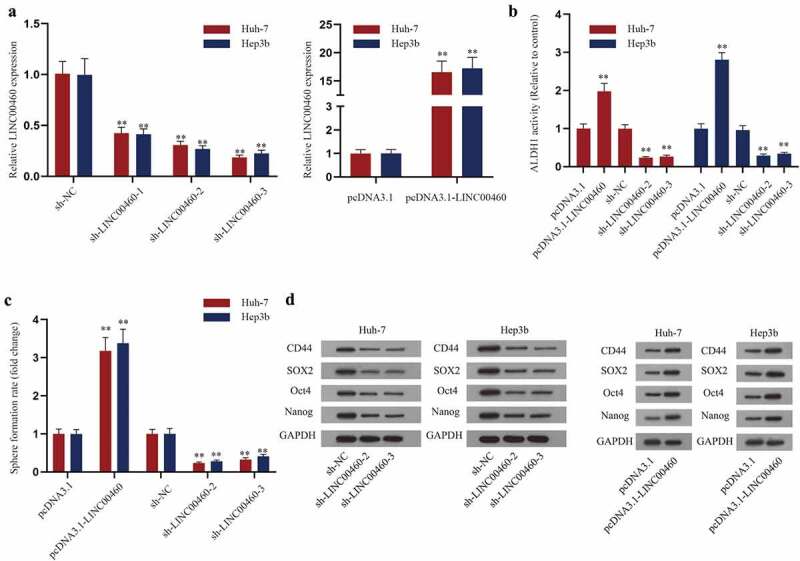
LINC00460 regulates the stemness formation of HCC. A. The interference efficiency of sh-LINC00460-1/2/3 and overexpression efficiency of pcDNA3.1-LINC00460 were verified by RT-qPCR analysis. B. The activity of ALDH1 was detected in Huh-7 and Hep3b cells with downregulated or upregulated LINC00460. C. The sphere formation efficiency of each transfection group of HCC cells normalized to control group was detected. D. Western blot analysis detected the levels of CD44, SOX2, Oct4 and Nanog with LINC00460 overexpression or knockdown. **P < 0.01; Student’s t-test, one-way ANOVA and Dunnett/Tukey
Then, we analyzed the effect of LINC00460 on HCC cell stemness. As shown in Figure 2b, the ALDH1 activities were increased by LINC00460 overexpression and decreased by sh-LINC00460-2/3 in HCC cells. Then, we found that LINC00460 overexpression facilitated sphere formation of HCC cells, while LINC00460 knockdown impeded facilitated sphere formation of HCC cells (Figure 2c). Additionally, western blot results showed that upregulating LINC00460 enhanced the levels of stemness marker proteins (CD44, SOX2, Oct4 and Nanog) while downregulating LINC00460 suppressed the levels of these proteins (Figure 2d). To sum up, LINC00460 overexpression strengthens the stemness of HCC cells and LINC00460 downregulation inhibits HCC cell stemness.
Furthermore, we explored the influence of LINC00460 on HCC cell proliferation, invasion, migration, apoptosis and EMT. Colony formation assays verified that LINC00460 positively regulated cell proliferation as LINC00460 upregulation increased colony numbers and its downregulation decreased colony numbers in HCC cells (Fig. S1A). Besides, transwell assays proved that the invasive and migratory abilities of HCC cells were strengthened by LINC00460 overexpression and were weakened by LINC00460 silence (Fig. S1B-C). As for cell apoptosis, TUNEL staining demonstrated that LINC00460 knockdown increased total apoptotic rate, while LINC00460 overexpression decreased apoptotic rate of HCC cells, indicating that LINC00460 negatively regulated cell apoptosis of HCC (Fig. S1D). Meanwhile, western blot analyses were conducted to detect the level of EMT-related proteins, ZO-1 and E-cadherin (EMT negatively related proteins), N-cadherin and Vimentin (EMT positively related proteins) (Fig. S1E). The results showed that LINC00460 upregulation increased the level of N-cadherin and Vimentin, decreased the level of ZO-1 and E-cadherin while downregulated LINC00460 caused the completely opposite results. In conclusion, LINC00460 facilitates HCC cell proliferation, invasion, migration, EMT, stemness and suppresses the apoptosis of HCC cells.
The effect of LINC00460 on stem phenotype of HCC in vivo
By the construction of xenograft model, effect of LINC00460 on HCC tumor growth was assessed. Data showed that tumor weight and volume in mice were smaller in mice injected with HCC cells with LINC00460 knockdown at day 28 after injection (Figure 3a-b). Growth curve depicted that HCC tumor grew slower in mice of sh-LINC00460-2/3 group compared with sh-NC group (Figure 3c). The results indicated that knockdown of LINC00460 curbed tumor growth of HCC in vivo. Meanwhile, western blot assays performed with the xenograft model showed that the level of proteins associated with cell stemness was reduced with LINC00460 knockdown (Figure 3d). Taken together, down-regulated LINC00460 inhibited stemness of HCC in vivo.
Figure 3.
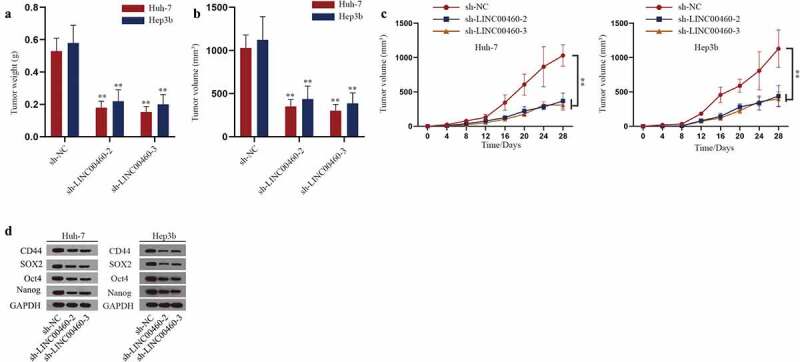
The effect of LINC00460 on stemness and growth of HCC in vivo. A-B. The tumor weight and volume were measured in sh-LINC00460-1/2 groups versus sh-NC group. C. The tumor volume in mice was analyzed in sh-LINC00460-2/3 groups versus sh-NC group every 4 d. D. Western blot analysis detected the levels of CD44, SOX2, Oct4 and Nanog in tumors from mice of each group. **P < 0.01; one-way ANOVA and Dunnett
MiR-503-5p and miR-654-3p interact with LINC00460 and play an inhibitory role on HCC cell stemness
The subcellular localization of lncRNA contributes to the understanding of the regulatory pattern of lncRNA to a certain extent. Thus, in order to explore the mechanism of LINC00460 in liver cancer, we first performed FISH assay to analyze the subcellular localization of LINC00460 in Huh-7 and Hep3b cells (Figure 4a). The results proved that LINC00460 was mainly located in the cytoplasm of HCC cells. In recent years, a large number of studies have reported that cytoplasmic lncRNA can participate in the regulation of tumor biological behaviors in the mode of ceRNA [14]. Therefore, we screened the target miRNA of LINC00460 in HCC. According to the ENCORI website (http://starbase.sysu.edu.cn/ CLIP-Date≥1; Degradome-Data≥0; pan-Cancer≥0), miR-320b, miR-3064-5p, miR-6504-5p, miR-654-3p, miR-503-5p were found as the candidate miRNAs of LINC00460 (Figure 4b). Then, we determined the target miRNA related to LINC00460 in HCC cells. RT-qPCR analysis showed that the expression of miR-503-5p and miR-654-3p were notably increased by LINC00460 knockdown (Figure 4c). Next, we confirmed miR-503-5p and miR-654-3p overexpression by miR-503-5p mimics and miR-654-3p mimics (Figure 4d). Interaction between LINC00460 and miR-503-5p/miR-654-3p was then detected. Consequently, miR-503-5p mimics and miR-654-3p mimics distinctly lowered the luciferase activity of LINC00460-WT while the luciferase activity of mutant LINC00460 hardly changed (Figure 4e-f). Furthermore, RNA pull-down assays proved that both miR-503-5p and miR-654-3p expressed a high enrichment with bio-LINC00460 (Figure 4g). In a word, both miR-503-5p and miR-654-3p were the targets of LINC00460 in HCC cells.
Figure 4.
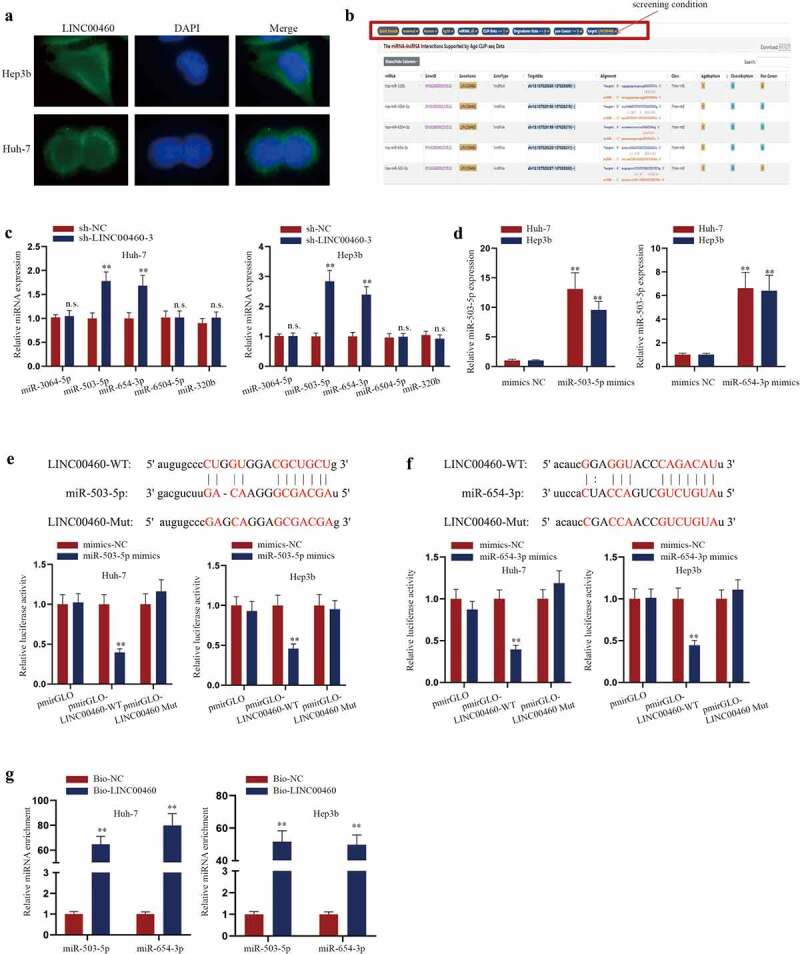
LINC00460 targets miR-503-5p and miR-654-3p. A. FISH image of the distribution of LINC00460 (green) in the cytoplasm and nucleus of HCC cells. B. ENCORI website (http://starbase.sysu.edu.cn/) was applied to predict the candidate miRNAs for LINC00460. C. RT-qPCR analysis was used to detect the expressions of miRNAs with LINC00460 knockdown. D. The overexpression efficiency of miR-503-5p/miR-654-3p mimics was detected by RT-qPCR analysis. E-F. The combination of LINC00460 and miR-503-5p/miR-654-3p was analyzed by luciferase reporter assays. The binding sites between LINC00460 and miR-503-5p/miR-654-3p were obtained from ENCORI. G. The combination between LINC00460 and miR-503-5p/miR-654-3p was by RNA pull-down assays analyzed the enrichment of miR-503-5p/miR-654-3p. **P < 0.01; Student’s t-test, two-way ANOVA and Tukey/Dunnett
Then, we assessed the role of miR-503-5p and miR-654-3p on HCC cell stemness. First, miR-503-5p and miR-654-3p were found to express lower in HCC cell lines (Huh-7 and Hep3b) than in the human normal hepatic cell line (HL-7702) (Fig. S2A). The knockdown efficiency of miR-503-5p/miR-654-3p inhibitor was verified by RT-qPCR analyses (Fig. S2B). As shown in Fig. S2C and S2D, downregulated miR-503-5p and miR-654-3p increased ALDH1 activity and sphere formation in HCC cells, while upregulated miR-503-5p and miR-654-3p exerted reversed results. Then, western blot assays showed that the protein levels of CD44, SOX2, Oct4 and Nanog were increased in miR-503-5p/miR-654-3p inhibitor groups and were curbed in miR-503-5p/miR-654-3p mimics groups (Fig. S2E). Collectively, miR-503-5p and miR-654-3p were low expressed in HCC cells and suppressed HCC cell stemness.
LINC00460 regulates HCC cell stemness through miR-503-5p/miR-654-3p/TCP1 axis
Afterward, we predicted the target mRNA of miR-503-5p and miR-654-3p on ENCORI (http://starbase.sysu.edu.cn/) (Figure 5a), and found that TCP1 is the common target of both miR-503-5p and miR-654-3p. For further confirmation, RNA pull-down assays and the luciferase reporter assays were carried out to analyze the binding between miR-503-5p/miR-654-3p and TCP1. As shown in Figure 5b, TCP1 was notably enriched in bio-miR-503-5p/miR-654-3p groups. As shown in Figure 5c, the luciferase activity of TCP1 was decreased after the up-regulation of miR-503-5p/miR-654-3p. Taken together, miR-503-5p and miR-654-3p could target TCP1 in HCC cells. RIP assay showed that LINC00460, miR-503-5p, miR-654-3p and TCP1 co-existed in the Ago2-RISC complex, which suggested that LINC00460 has the potential to regulate TCP1 via targeting miR-503-5p and miR-654-3p as a ceRNA (Figure 5d). Next, we further determined the effect of LINC00460/miR-503-5p/miR-654-3p axis on TCP1. The results indicated that both sh-LINC00460-3 and miR-503-5p/miR-654-3p mimics could lessen the level of TCP1 (Figure 5e). In addition, the rescue experiments certified that LINC00460 overexpression increased TCP1 expression, whereas miR-503-5p or miR-654-3p overexpression decreased TCP1 expression, and the effect LINC00460 overexpression on TCP1 expression was partially reversed by single transfection of miR-503-5p or miR-654-3p mimics, and was fully reversed by co-overexpression of miR-503-5p and miR-654-3p (figure 5f), indicating that LINC00460 regulated TCP1 dependent on miR-503-5p and miR-654-3p. To sum up, LINC00460 regulates TCP1 via targeting miR-503-5p and miR-654-3p in a ceRNA mode.
Figure 5.
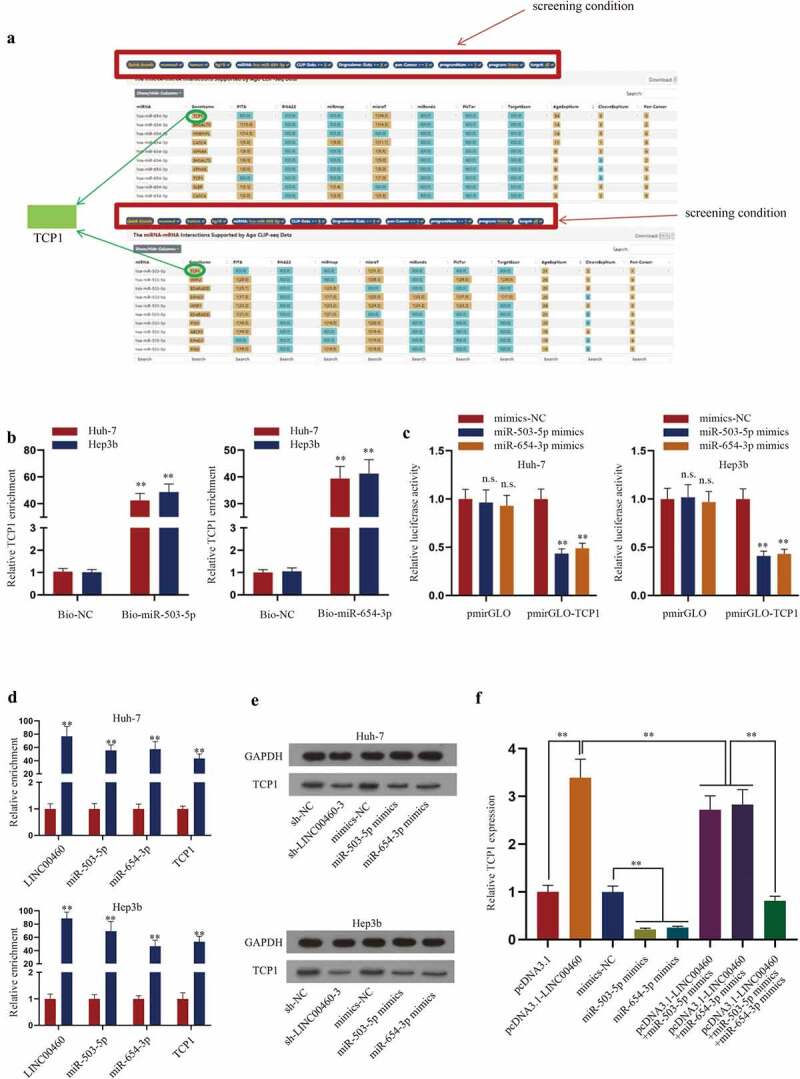
TCP1 is the target gene of LINC00460 and miR-503-5p/miR-654-3p. A. ENCORI website (http://starbase.sysu.edu.cn/) was applied to predict mRNAs targeted by miR-503-5p/miR-654-3p. B. The combination between TCP1 and miR-503-5p/miR-654-3p was analyzed by RNA pull-down assays. C. The effect of miR-503-3p/miR-654-3p on TCP1 was analyzed by luciferase reporter assays. D. Enrichment of LINC00460, miR-503-5p, miR-654-3p and LINC00460 in Ago2 RIP precipitates versus IgG RIP precipitates was detected by RT-qPCR. E. The protein level of TCP1 was examined by western blot after LINC00460 knockdown or miR-503-5p/miR-654-3p overexpression. F. Expression of TCP1 was detected by RT-qPCR in Hep3b cells transfected with pcDNA3.1, pcDNA3.1-LINC00460, mimics-NC, miR-503-5p mimics, miR-654-3p mimics, pcDNA3.1-LINC00460+ miR-503-5p mimics, pcDNA3.1-LINC00460+ miR-654-3p mimics, or pcDNA3.1-LINC00460+ miR-503-3p mimics+miR-654-3p mimics. **P < 0.01, ***P < 0.001; Student’s t-test, two-way ANOVA/one-way ANOVA and Dunnett/Tukey
Furthermore, the effect of LINC00460/miR-503-5p/miR-654-3p/TCP1 axis on HCC cell stemness was analyzed by rescue experiments. Consequently, sh-LINC00460-3 reduced ALDH1 activity, sphere formation and levels of stemness markers (CD44, SOX2, Oct4, and Nanog); such effect was partly reversed by single transfection of miR-503-5p or miR-654-3p inhibitor and was fully reversed by co-transfection of miR-503-5p inhibitor plus miR-654-3p inhibitor (Fig. S3A-C), indicating that LINC00460 regulated HCC cell stemness via miR-503-5p and miR-654-3p.
Additionally, effect of TCP1 in HCC was detected. According to RT-qPCR analysis, we found that TCP1 was highly expressed in HCC cells (Fig. S4A). After the knockdown efficiency of sh-TCP1-1/2/3 and the overexpression efficiency of pcDNA3.1-TCP1 were verified by RT-qPCR analysis (Fig. S4B), we evaluated the effects of TCP1 on HCC cell stemness. The results showed that TCP1 upregulation increased ALDH1 activity, sphere formation and the level of stemness marker proteins, while TCP1 knockdown caused the opposite results (Fig. S4C-E). In other words, TCP1 could facilitate the stemness of HCC cells.
In the meantime, rescue experiments of Figure S5A-C displayed that TCP1 overexpression could counteract the inhibition of miR-503-5p mimics on cell stemness, resulting in a notably increase on cell stmeness, while the co-transfection of miR-654-3p could re-curb this promotion. Therefore, we concluded that miR-503-5p and miR-654-3p co-targets TCP1 in the regulation of HCC cell stemness.
At last, the rescue experiments were implemented to verify LINC00460/TCP1 axis in HCC cells. We validated that TCP1 overexpression fully counteracted the inhibitory effect of sh-LINC00460-3 on ALDH1 activity, sphere formation and stemness markers (Fig. S6A-C), indicating that LINC00460 regulates HCC cell stemness by TCP1. In addition, a former study indicated that targeting TCP1 could induce endoplasmic reticulum (ER) stress in cancer cells [15], so we preliminarily tested whether LINC00460/TCP1 could affect markers related to ER stress in HCC cells. Expectedly, sh-LINC00460-3 induced levels of IRE1, CHOP, GRP78, and p-eIF2α, and such effect was counteracted by TCP1 overexpression, with total eIF2α level unchanged in each group (Fig. S6D), indicating that LINC00460/TCP1 potentially regulate ER stress in HCC cells. In conclusion, LINC00460 promoted HCC cell stemness via miR-503-5p/miR-654-3p/TCP1 axis.
Discussion
As shown in numerous outstanding cancer-related studies, the biological progression of HCC can be facilitated or restrained by abnormal lncRNAs expression. For instance, lncRNA MCM3AP antisense RNA 1 (MCM3AP-AS1) overexpression drove the growth of HCC and lncRNA DRHC curbed HCC invasion and proliferation [16,17]. Multiple reports have suggested LINC00460 as an onco-lncRNA in HCC and uncovered its effects on cell growth and metastasis in HCC cells [12,13,18]. Herein, we obtained data concordant with former findings, showing high expression level of LINC00460 in HCC cells. Also, we provided data to validating that LINC00460 positively influenced proliferation, migration, invasion and EMT, whereas negatively affected apoptosis in HCC cells.
Besides, cell stemness of HCC is also substantially discussed by former literatures. For instance, HCC chemo-resistance and stemness could be mediated by cytoplasmic polyadenylation element-binding protein 1 (CPEB1) [19]. In the meantime, miR-5188 [20], Cripto-1 [21] and interferon induced protein 44 like (IFI44L) [22] also have a regulatory influence on HCC stemness. Although LINC00460 has been linked with HCC by former works, our study was the first to link LINC00460 with stemness in HCC. We discovered that silenced LINC00460 reduced ALDH1 activity, tumor formation as well as stemness marker protein levels, while overexpression of LINC00460 resulted in a totally opposite condition. Taking these consequences together, we suggested the promoting effect of LINC00460 in HCC cell stemness. Additionally, animal studies indicated that LINC00460 depletion plays an inhibitory role in the aspect of tumor growth as well as the stemness of tumor cells.
Being widely uncovered, the interaction between lncRNA and miRNA on the cell biological behavior of HCC is common, such as the inhibition of lncRNA HOX transcript antisense RNA (HOTAIR) on miR-122 expression in HCC [23]. Also, considering the widely reported mechanism of LINC00460 in the mode of ceRNA (competing endogenous RNA) in HCC [13,18], we further screened new target miRNAs and the downstream gene for LINC00460. By bioinformatics prediction, we identified and then confirmed that miR-503-5p and miR-654-3p could combine with LINC00460 in HCC cells. Later, we identified TCP1 as the target for miR-503-5p and miR-654-3p and demonstrated that LINC00460 competitively interacts with miR-503-5p or miR-654-3p to enhance TCP1 expression. Rescue assays showed that co-inhibition of miR-503-5p and miR-654-3p inhibitors fully reversed the effect of LINC00460 knockdown on inhibiting TCP1 and HCC cell stemness, suggesting that LINC00460 modulate TCP1 expression and stemness depending on these miRNAs. Additionally, we found through literature research that it was documented that targeting TCP1 could induce ER stress in cancer cells [15], so we preliminarily tested whether LINC00460/TCP1 axis influenced ER stress in HCC cells. Expectedly, our data first showed that inhibiting LINC00460/TCP1 axis induced ER stress-related markers in HCC cells, suggesting that LINC00460/TCP1 potentially regulated ER stress in HCC.
In conclusion, we first revealed that LINC00460/miR-503-5p/miR-654-3p/TCP1 axis aggravated cell stemness in HCC. With this finding, the potential role of LINC00460 as a biomarker in HCC stemness provides a possibility for optimizing the clinic treatment of HCC in the future. However, our research is still short of data of the association between LINC00460 expression and clinicopathological characteristics of HCC patients. Also, investigation on LINC00460/TCP1 axis regulating ER stress in HCC cells will be furthered in our future study.
Supplementary Material
Acknowledgments
We appreciated all supports.
Funding Statement
The work was supported by the Natural Science Foundation of Hubei Province (2016CFC732).
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the author(s).
Data availability statement
.The data will be available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, et al. Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol. 2016;25(2):74–85. . [DOI] [PubMed] [Google Scholar]
- [2].Miller ZA, Lee KS.. Screening for hepatocellular carcinoma in high-risk populations. Clin Imaging. 2016;40(2):311–314. [DOI] [PubMed] [Google Scholar]
- [3].Prasad S, Ramachandran S, Gupta N, et al. Cancer cells stemness: a doorstep to targeted therapy. Biochim Biophys Acta Mol Basis Dis. 2020;1866(4):165424. . [DOI] [PubMed] [Google Scholar]
- [4].Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6(9):674–687. [DOI] [PubMed] [Google Scholar]
- [5].Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47(Supplement 1):S2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Deniz E, Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct Integr Genomics. 2017;17(2–3):135–143. [DOI] [PubMed] [Google Scholar]
- [7].Dahariya S, Paddibhatla I, Kumar S, et al. Long non-coding RNA: classification, biogenesis and functions in blood cells. Mol Immunol. 2019;112:82–92. [DOI] [PubMed] [Google Scholar]
- [8].He W, Liang B, Wang C, et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38(23):4637–4654. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xiao Y, Pan J, Geng Q, et al. Lnc RNA MALAT 1 increases the stemness of gastric cancer cells via enhancing SOX 2 mRNA stability. FEBS Open Bio. 2019;9(7):1212–1222. . [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [10].Hong L, Wang H, Wang J, et al. <p>LncRNA PTCSC3 Inhibits Tumor Growth and Cancer Cell Stemness in Gastric Cancer by Interacting with lncRNA Linc-pint. Cancer Manag Res. 2019;11:10393–10399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lu G, Li Y, Ma Y, et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37(1):289. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tu J, Zhao Z, Xu M, et al. LINC00460 promotes hepatocellular carcinoma development through sponging miR-485-5p to up-regulate PAK1. Biomed Pharmacother. 2019;118:109213. [DOI] [PubMed] [Google Scholar]
- [13].Yang J, Li K, Chen J, et al. <p>Long Noncoding RNA LINC00460 Promotes Hepatocellular Carcinoma Progression via Regulation of miR-342-3p/AGR2 Axis. Onco Targets Ther. 2020;13:1979–1991. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [14].Qi X, Zhang D-H, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. [DOI] [PubMed] [Google Scholar]
- [15].Lin YF, Lee YF, Liang PH. Targeting β-tubulin: CCT-βcomplexes incurs Hsp90- and VCP-related protein degradation and induces ER stress-associated apoptosis by triggering capacitative Ca2+ entry, mitochondrial perturbation and caspase overactivation. Cell Death Dis. 2012;3(11):e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhuang R, Zhang X, Lu D, et al. lncRNA DRHC inhibits proliferation and invasion in hepatocellular carcinoma via c-Myb-regulated MEK/ERK signaling. Mol Carcinog. 2019;58(3):366–375. [DOI] [PubMed] [Google Scholar]
- [18].Hong H, Sui C, Qian T, et al. Long noncoding RNA LINC00460 conduces to tumor growth and metastasis of hepatocellular carcinoma through miR-342-3p-dependent AGR2 up-regulation. Aging (Albany NY). 2020;12(11):10544–10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xu M, Fang S, Song J, et al. CPEB1 mediates hepatocellular carcinoma cancer stemness and chemoresistance. Cell Death Dis. 2018;9(10):957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wei C, Yang C, Wang S, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lo RC, Leung CO-N, Chan KK-S, et al. Cripto-1 contributes to stemness in hepatocellular carcinoma by stabilizing Dishevelled-3 and activating Wnt/β-catenin pathway. Cell Death Differ. 2018;25(8):1426–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huang WC, Tung SL, Chen YL, et al. IFI44L is a novel tumor suppressor in human hepatocellular carcinoma affecting cancer stemness, metastasis, and drug resistance via regulating met/Src signaling pathway. BMC Cancer. 2018;18(1):609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cheng D, Deng J, Zhang B, et al. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine. 2018;36:159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
.The data will be available from the corresponding author on reasonable request.


