ABSTRACT
Hepatocellular carcinoma (HCC) is a common type of primary liver cancer. Circular RNAs (circRNAs) have been demonstrated to be a crucial player in multiple cancers. However, a large number of circRNAs remain to be explored. Our study focused on investigating hsa_circ_0004018 in HCC. Firstly, we conducted quantitative reverse transcription PCR (RT-qPCR) to find that circ_0004018 was down-regulated in HCC cells. Western blot analysis was performed to detect the protein levels of phosphatase and tensin homologue (PTEN) and related factors of PI3K/AKT signaling pathway. From the results of functional assays, we found that overexpression of circ_0004018 significantly inhibited the proliferative and migratory capacities of HCC cells. The regulatory mechanism of circ_0004018 in HCC was determined by RNA immunoprecipitation (RIP), RNA pull-down, and luciferase reporter assays, thereby we knew that circ_0004018 regulated PTEN by sequestering microRNA-1197 (miR-1197) to modulate PI3K/AKT signaling pathway. Finally, rescue assays verified that circ_0004018 participated in modulation of cell proliferation and migration in HCC via sponging miR-1197 and regulating PTEN. In conclusion, circ_0004018 suppresses the proliferation and migration of HCC cells via sponging miR-1197 to inactivate the PTEN/PI3K/AKT signaling pathway.
Abbreviations: HCC: Hepatocellular carcinoma; circRNAs: Circular RNAs; PTEN: Phosphatase and tensin homologue; miR-1197: microRNA-1197; ceRNA: competitive endogenous RNA; ATCC: American Type Culture Collection; EMEM: Eagle’s Minimum Essential Medium; RT-qPCR: Quantitative real-time PCR; EdU: 5-ethynyl-20-deoxyuridine; FISH: Fluorescent in situ hybridization; RIP: RNA immunoprecipitation; 3ʹ-UTR: 3ʹ-untranslated region; Wt: wild-type; Mut; mutant type; gDNA: genomic DNA; Act D: Actinomycin D; PI3K: phosphatidylinositol-3-kinase; AKT: protein kinase; lncRNAs: long non-coding RNAs
KEYWORDS: Hepatocellular carcinoma, hsa_circ_0004018, miR-1197, PTEN, PI3K/AKT signaling pathway
Introduction
As a common malignancy, hepatocellular carcinoma (HCC) is featured with a high incidence and mortality [1]. At the same time, many factors may contribute to its occurrence. Despite many improvements in HCC treatment, HCC remains difficult to be cured and has poor prognosis [2]. Thus, developing effective biomarkers for HCC diagnosis and treatment is urgent.
Circular RNAs (circRNAs) are recognized to be a set of noncoding RNAs, which originate from back-splicing of linear primary transcripts [3]. Increasing studies have validated that circRNAs have been identified as important molecular biomarkers for multiple cancers, such as pancreatic ductal adenocarcinoma, colorectal cancer, and breast cancer [4,5]. CircRNAs have also been reported to be associated with HCC in different ways [6]. For example, Huang et al. have demonstrated that circRNA-100338 participates in HCC via mTOR signaling pathway [7]. Zhu et al. have verified the role of Qki5-circZKSCAN1-FMRP-CCAR1-Wnt signaling axis in HCC stemness [8]. Moreover, based on data of the previous study, circ_0004018 has been validated to sponge miR-660-3p to modulate TEP1 expression, and thereby impedes the progression of liver fibrosis [9]. Additionally, circ_0004018 expression level in HCC was testified to be significantly lower compared with para-tumorous tissues, and its potential correlation with HCC was suggested [10]. However, the function of circ_0004018 involved in HCC progression remains largely unknown.
Notably, competitive endogenous RNA (ceRNA) network in which circRNAs are significant components has been identified in the pathogenesis of HCC [11]. For instance, He et al. have demonstrated that hsa_circ_0000517 acts as a ceRNA in the advancement of HCC [12]. Liu et al. have proved that circ-DENND4C sequesters miR-195-5p and enhances TCF4 expression to influence malignant phenotypes in HCC [13]. Bai et al. have confirmed that circFBLIM1 serves as an oncogene in HCC via modulating miR-346/FBLIM1 axis [14].
In the present study, we aimed to explore the biological function of circ_0004018 in HCC and unveil the related mechanism in order to find out potential therapeutic targets against HCC.
Materials and methods
Cell culture
HCC cell lines (Huh7, Hep3B, SNU-182, and SK-HEP-1) and normal epithelial cell line (THLE-3) were chosen for this study. Among them, Huh7 and Hep3B were purchased from Yaji Biotechnology Co., Ltd. (Shanghai, China) and maintained in RPMI-1640 and MEM respectively. SNU-182, SK-HEP-1 cell lines, as well as THLE-3 cell line, were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA) and separately cultured in RPMI-1640, Eagle’s Minimum Essential Medium (EMEM) and BEGM. All cells were cultured in mediums containing 10% FBS with 5% CO2 at 37°C.
Plasmid transfection
PcDNA3.1 (+) circRNA Mini Vectors were subcloned with circ_0004018 to construct pcDNA3.1-circ_0004018 for overexpression and empty pcDNA3.1 (+) circRNA Mini Vector (hereafter termed as pcDNA3.1) was regarded as the negative control. MiR-1197 mimics and control mimics were purchased from GenePharma (Shanghai, China). In addition, the short hairpin RNAs (shRNAs) targeting PTEN (sh-PTEN#1, sh-PTEN#2), as well as control shRNA (sh-NC) were generated by Genechem (Shanghai, China). Lipofectamine 3000 (2.0 μl/50 μl; Invitrogen) was used for cell transfection.
Quantitative reverse transcription PCR (RT-qPCR) analysis
In accordance with the protocols of suppliers, total RNA was isolated with 1 ml TRIzol Reagent (Introgen, Carlsbad, CA, USA) and reverse transcribed into cDNA by means of RevertAid First Strand cDNA Synthesis Kit (Thermo fisher, IL, USA). SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) was applied to conduct PCR analysis. The relative expression of genes was determined by 2−ΔΔCt method. GAPDH and U6 served as internal controls of cytoplasm and nucleus, respectively. Primer sequences were clearly listed in Table 1.
Table 1.
Primer sequences used in RT-qPCR were listed
| Sequences for RT-qPCR primers | |
|---|---|
| Gene | Primer sequence |
| linear-SMYD4 | F GGGGTACCAGAGACCTGTCG R AGCAACCAGACGCACATTCT |
| circ_0004018 | F AGCAGCAGTGACACCTGAAT R GACACGTCTGTGTGTTGTATGG |
| PTEN | F CAGCAGCTTCTGCCATCTCT R TGCTTTGAATCCAAAAACCTTACT |
| miR-1197 | Sequence: taggacacatggtctacttct Reverse transcription stem loop primer CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGC agaagt Stem loop universal reverse primer CTCAACTGGTGTCGTGGA F CCGAGtaggacacatggtct |
| miR-378 g | Sequence: actgggcttggagtcagaag Reverse transcription stem loop primer CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGC cttctg Stem loop universal reverse primer CTCAACTGGTGTCGTGGA F TCGCAactgggcttggagt |
| GAPDH | F GACAGTCAGCCGCATCTTCT R GCGCCCAATACGACCAAATC |
| U6 | F TCCCTTCGGGGACATCCG R AATTTTGGACCATTTCTCGATTTGT |
5-ethynyl-20-deoxyuridine (EdU) staining assay
EdU assay was performed as previously described [15]. Cells were put on sterile coverslips in 96-well plates with 5 × 104 cells in each well. EdU kit from RiboBio (Guangzhou, China) was used following guidebook. Cell nucleus was double-stained with 50 μM EdU solution and 500 μl DAPI dye (Beyotime, Shanghai, China). Images were acquired by fluorescence microscope (Olympus, Tokyo, Japan).
Colony formation assay
HCC cells were incubated in 6-well plates at a density of 200 cells at 37°C for 14 days to detect colony formation ability. Subsequent to fixation with 4% paraformaldehyde, colonies were stained with 0.1% crystal violet. The stained cell colonies were counted manually.
Transwell migration assay
Cell samples were planted to the upper chamber for migration assay. The lower chamber was supplemented with 1% FBS. The cultivation process lasted 24 hours. Migrated cells into bottom were then fixed with 4% paraformaldehyde and dyed with 0.1% crystal violet. Finally, the migrated cells were observed and photographed under a microscope.
Subcellular fractionation
Subcellular fractionation was conducted with the employment of Cytoplasmic and Nuclear RNA Purification Kit (NORGEN, Thorold, ON, Canada). After cell cytoplasm and nucleus were separated by centrifugation, the expression of circ_0004018 in different fragments was detected by RT-qPCR. GAPDH or U6 was separately treated as the cytoplasmic control or the nuclear control.
Fluorescent in situ hybridization (FISH)
FISH assay was performed as previously described [16]. After fixation in 4% PFA for 15 min at 37°C and then permeabilized with 0.5% Triton X-100, HCC cells were hybridized with circ_0004018-specific FISH probe in buffer and counterstained with 500 μl Hoechst solution. Images were obtained with a confocal laser microscope (Olympus).
RNA pull down assay
With the aim to investigate the interaction between circ_0004018 and miR-1197, RNA pull down assay was adopted. Biotin-labeled circ_0004018 (Bio-circ_0004018), with a biotinylated nonsense sequence as the negative control (Bio-NC), was mixed with the cell lysates overnight, and then magnetic beads were added. The bound RNAs were purified for RT-qPCR analysis.
RNA immunoprecipitation (RIP)
With the aim to explore the relationship among circ_0004018, miR-1197 and PTEN in SNU-182 and SK-HEP-1 cells, RNA-binding protein immunoprecipitation kit (Millipore, Burlington, MA, USA) was used for RIP assay. HCC cell lysates were immunoprecipitated with anti-Ago2 (Abcam) or with the negative control anti-IgG (Abcam). After protein digestion, the precipitation of RNA was isolated. Finally, RT-qPCR was performed to analyze the extracted RNA precipitates.
Luciferase reporter assay
Circ_0004018 or PTEN mRNA 3ʹ-untranslated region (3ʹ-UTR) sequence of wild-type (Wt) and mutant type (Mut) of miR-1197 with luciferase reporter vector was generated as pmirGLO-circ_0004018-Wt/Mut and pmirGLO-PTEN 3ʹ-UTR-Wt/Mut respectively. Later, the reporter plasmids were co-transfected with control mimics and miR-1197 mimics into HCC cells. After 48 h, the Dual-Luciferase Reporter Gene Assay Kit (Beyotime, Shanghai, China) was applied to measure luciferase activity. The luciferase activity of the experimental reporter was normalized to Renilla luciferase (internal control).
Western blot analysis
The cell lysis buffer (100 μl) was utilized firstly to abstract total protein from cells. After that, protein samples were separated with SDS-PAGE and later transferred to PVDF membranes (Millipore, Billerica, MA, USA). Afterward, 5% nonfat milk was exploited for membrane sealing-off and primary antibodies were used to treat membranes at 4°C overnight. After washed with PBS (1 ml) ×3, the blots were then incubated with secondary antibody for 1 h at dark room. The antibodies against PTEN, PI3K, p-PI3K, AKT, p-AKT and GAPDH were all obtained from Abcam. GAPDH served as the internal control. The protein concentrations were quantified by Chemiluminescence system (GE Healthcare, Chicago, USA).
Statistical analysis
SPSS 22.0 statistical software package was use to analyze experimental data. All data were expressed as mean ± standard error of the mean (SEM). Differences between two groups or more groups were compared by Student’s t-test or analysis of variance (ANOVA). Each independent experiment was conducted at least three times. Differences were indicated to be statistically significant when P < 0.05.
Results
Circ_0004018 is obviously down-regulated in HCC cells
In accordance with the results on circRNADisease (http://cgga.org.cn:9091/circRNADisease/), circ_0004018 expression was down-regulated in HCC (Figure 1(a)). Besides, there are several reports demonstrating the anti-tumor effect of circ_0004018 in HCC [17,18]. In addition, from the results of RT-qPCR analysis, we found that compared with normal cell line (THLE-3), circ_0004018 was obviously down-regulated in HCC cell lines (Huh7, Hep3B, SNU-182, and SK-HEP-1) (Figure 1(b)). Furthermore, RT-qPCR confirmed that linear-SMYD4 level was observably reduced after adding RNase R while circ_0004018 was resistant to RNase R digestion, testifying the circular characteristics of circ_0004018 (Figure 1(c)). In addition, agarose gel electrophoresis (AGE) was conducted to further corroborate the circular structure of circ_0004018. It turned out cDNA of circ_0004018 could be detected both in divergent and convergent primers, suggesting its ring form (Figure 1(d)). The expression level of circ_0004018 and linear-SMYD4 was detected in SNU-182 and SK-HEP-1 cells treated with the transcription inhibitor, Actinomycin D (Act D) to examine the stability of circ_0004018. It was displayed in the results that after Act D treatment, the half-life of circ_0004018 was much longer than that of linear SMYD4, suggesting that circ_0004018 was much more stable than linear-SMYD4 (Figure 1(e)). All data testified the presence of circ_0004018 in HCC cells.
Figure 1.
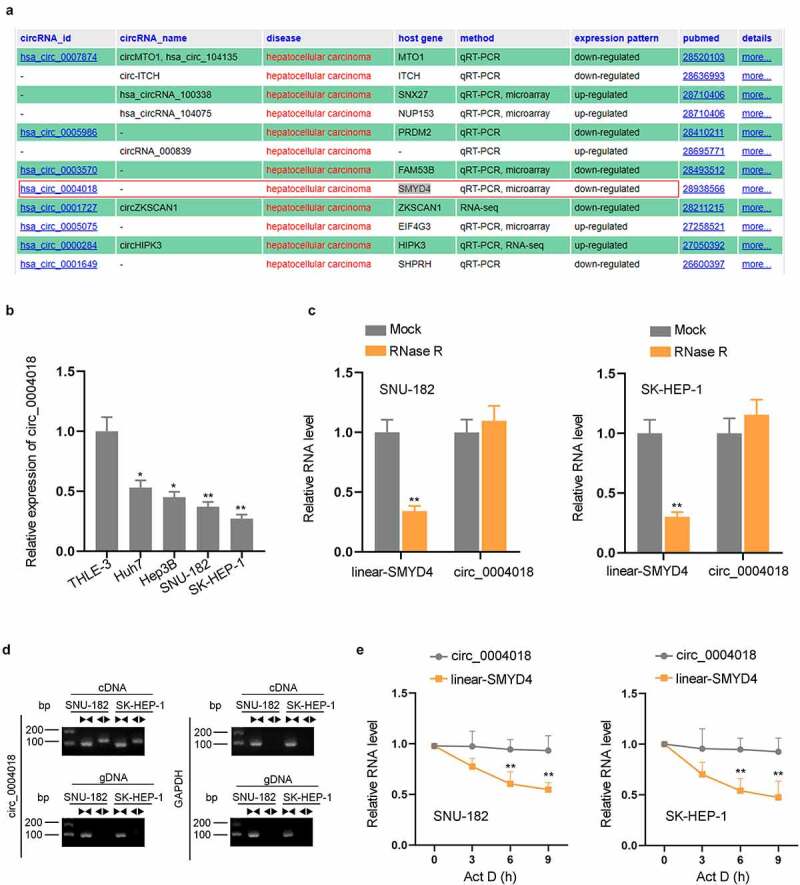
Circ_0004018 is obviously down-regulated in HCC cells. (a) Down-regulation of circ_0004018 in HCC was demonstrated in circRNADisease database. (b). Measurement of circ_0004018 expression in different cell lines was performed via RT-qPCR. (c). RNA levels of circ_0004018 and linear-SMYD4 were detected after RNase R treatment in SNU-182 and SK-HEP-1. (d). AGE was conducted to test the expression of circ_0004018. (e). ActD was added to examine the stability of circ_0004018 and linear-SMYD4 in SNU-182 and SK-HEP-1 cells. *P < 0.05, **P < 0.01
Up-regulation of circ_0004018 suppresses cell proliferation and migration in HCC
Functional assays were implemented with the aim to ravel out circ_0004018 impact on HCC cell proliferation and migration. Firstly, we examined the overexpression efficiency of pcDNA3.1-circ_0004018 (Figure 2(a)). Then gain-of-function experiments were performed in cells with or without pcDNA3.1-circ_0004018 transfection. EdU and colony formation assays were carried out for observation on HCC cell proliferation. An overt reduction in number of EdU positive cells and cell colonies could be discovered in response to the transfection of pcDNA3.1-circ_0004018, which suggested that circ_0004018 up-regulation significantly hindered cell proliferation in HCC (Figure 2(b–c)). In addition, cell migratory capacity was assessed by transwell assay. Similarly, we found that relative to the control group, the number of migrated cells was much smaller after circ_0004018 was up-regulated, indicating that overexpression of circ_0004018 inhibited HCC cell migration (Figure 2(d)). In a word, circ_0004018 negatively modulated HCC cell proliferation and migration.
Figure 2.
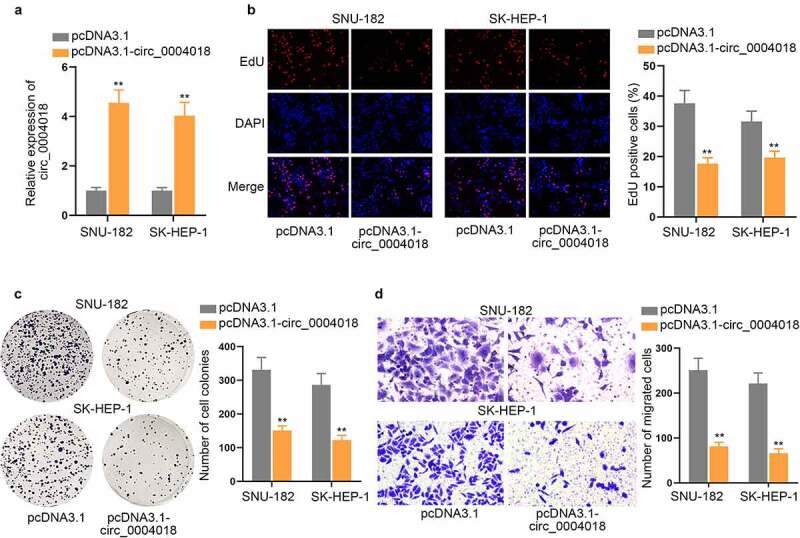
Up-regulation of circ_0004018 suppresses cell proliferation and migration in HCC. (a) The expression of circ_0004018 was analyzed after pcDNA3.1-circ_0004018 transfection into cells via RT-qPCR. (b-c). Cell proliferation was observed by means of EdU and colony formation assays in response to an augment in circ_0004018 expression. (d). Transwell migration assay was implemented with the aim to assess HCC cell migration under the influence of circ_0004018 overexpression. **P < 0.01
Circ_0004018 serves as miR-1197 sponge in HCC cells
Subcellular fractionation and FISH assays were conducted to observe the distribution of circ_0004018 in HCC cells and we discovered that circ_0004018 was preferentially enriched in the cytoplasm (Figure 3(a–b)). Previous reports have confirmed that circRNAs can modulate gene expression dependent on sponging miRNAs in the cytoplasm [19]. In this case, we conjectured that circ_0004018 could also serve as miRNA sponge. Based on the condition of CLIP Data ≥ 3, starBase (http://starbase.sysu.edu.cn/) predicted 2 potential miRNAs (miR-1197 and miR-378 g). RNA pull down assay indicated that miR-1197, instead of miR-378 g, was pulled down by Bio-circ_0004018 probe (Figure 3(c)). Hence, we selected miR-1197 for further study. It can be understood from a recent literature that miR-1197 is sponged by circ_0000429 in Non-small-cell lung carcinoma [20]. As shown in Figure 3(d), the binding site between circ_0004018 and miR-1197 was predicted by starBase. SNU-182 and SK-HEP-1 cells were transfected with miR-1197 mimics to enhance the expression of miR-1197 (Figure 3(e)). Luciferase reporter assay displayed that miR-1197 mimics dramatically decreased the luciferase activity of circ_0004018-Wt rather than that of circ_0004018-Mut in HCC cells (Figure 3(f)). The finding certified the targeted binding between circ_0004018 and miR-1197. To be concluded, miR-1197 was sequestered by circ_0004018 in HCC cells.
Figure 3.
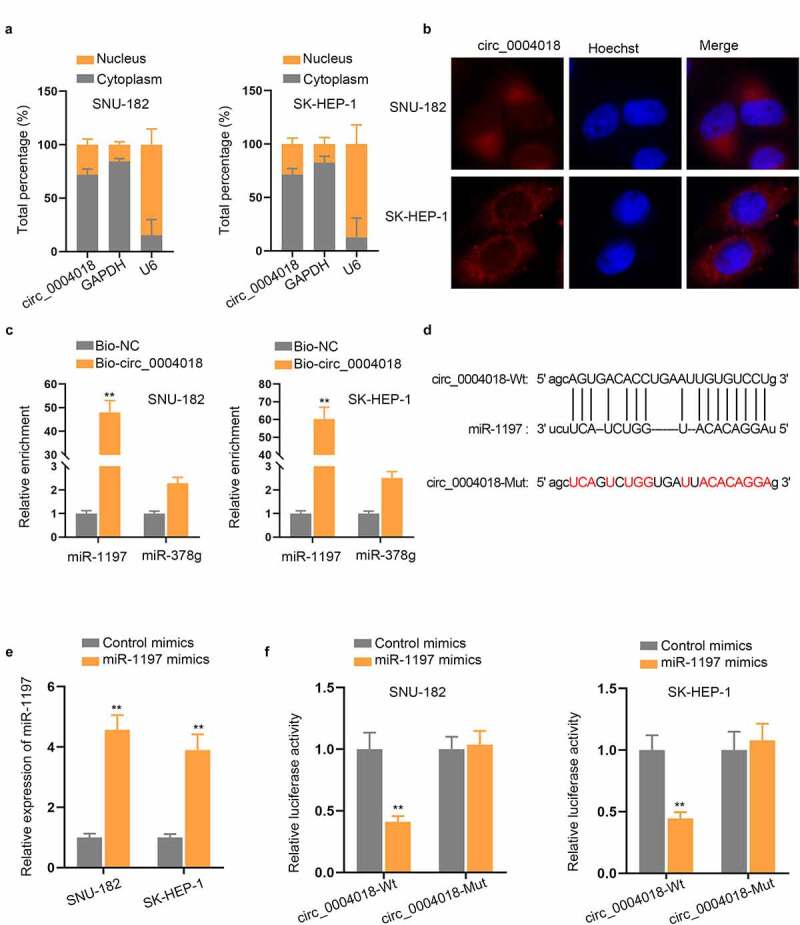
Circ_0004018 serves as miR-1197 sponge in HCC cells. (a-b). The distribution of circ_0004018 in the nucleus and cytoplasm of HCC cells was examined through subcellular fractionation and FISH assays. (c). RNA pull down verified the strong affinity of circ_0004018 and miR-1197. (d). The predicted binding site of miR-1197 within the sequence of circ_0004018 was presented. (e). Enhanced miR-1197 expression level was observed in miR-1197 mimics-transfected HCC cells based on RT-qPCR results. (f). The binding of circ_0004018 and miR-1197 was further verified with the help of luciferase reporter assay. **P < 0.01
Circ_0004018 positively modulates PTEN expression to inactivate PI3K/AKT signaling pathway by sequestering miR-1197
PTEN has been reported to be a tumor suppressor in HCC [21,22]. Additionally, PTEN has been defined as a negative regulator of the phosphatidylinositol-3-kinase (PI3K)/protein kinase (AKT) pathway [23]. Intriguingly, through starBase, we found PTEN was a target gene of miR-1197. Thus, we speculated that circ_0004018 might regulate PTEN expression to inactivate PI3K/AKT signaling pathway via sponging miR-1197. From western blot analysis, we discovered that overexpression of circ_0004018 exactly enhanced PTEN protein level while cutting down the protein levels of p-PI3K and p-AKT (Figure 4(a)), suggesting that circ_0004018 could inactivate PI3K and AKT signaling pathway via modulating PTEN expression. The binding sites for PTEN 3ʹ-UTR on miR-1197 were demonstrated in Figure 4(b). RIP assay manifested that circ_0004018, miR-1197 and PTEN were all highly precipitated in Ago2 antibody (Figure 4(c)). Luciferase reporter assays unveiled that miR-1197 mimics effectively lessened the luciferase activity of PTEN 3ʹ-UTR-Wt, while PTEN 3ʹ-UTR-Mut group had no significant change (Figure 4(d)). More interestingly, the protein level of PTEN was prominently enhanced by circ_0004018 overexpression and then restored by miR-1197 mimics transfection (Figure 4(e)). In summary, circ_0004018 inactivated PI3K/AKT signaling pathway via enhancing PTEN expression.
Figure 4.
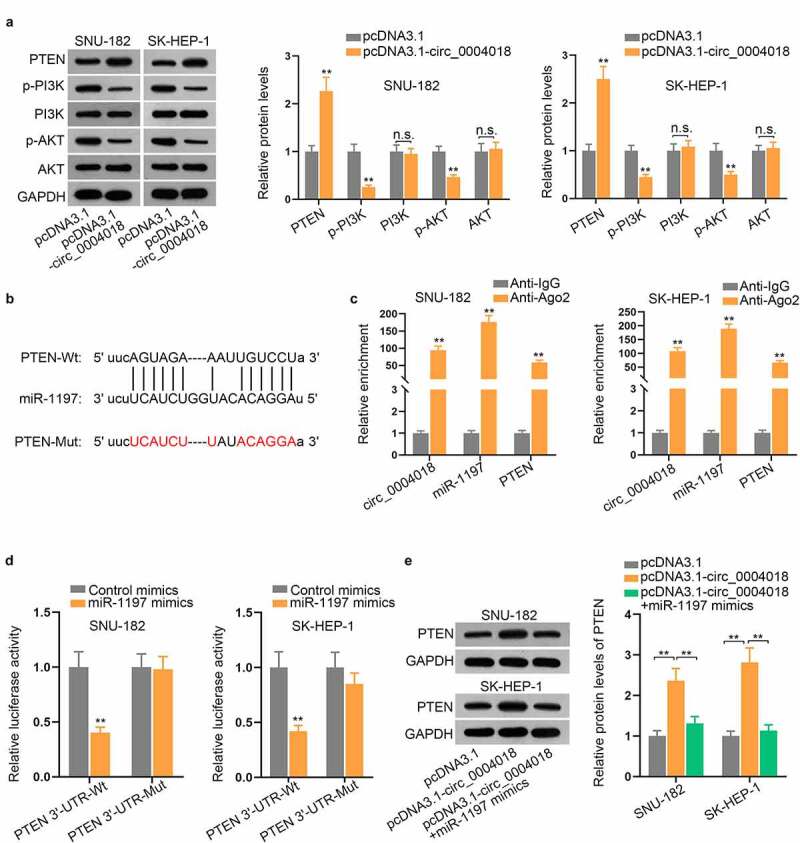
Circ_0004018 sequesters miR-1197 and modulates PTEN expression to inactivate PI3K/AKT signaling pathway. (a) The protein levels of PTEN and related factors of PI3K/AKT pathway were analyzed by western blot after circ_0004018 was overexpressed. (b). The binding sequence of miR-1197 and PTEN was shown. (c). RIP assay exhibited the enrichment of circ_0004018, miR-1197, as well as PTEN in Ago2 group. (d). The binding between PTEN and miR-1197 was raveled out with the help of luciferase reporter assay. €. PTEN protein level was tested in cells with the transfection of different plasmids, including pcDNA3.1, pcDNA3.1-circ_0004018 and pcDNA3.1-circ_0004018+ miR-1197 mimics. **P < 0.01. n.s.: no significance
Up-regulation of miR-1197 or down-regulation of PTEN rescues the suppressive impacts of circ_0004018 overexpression on HCC cell proliferation and migration
To elucidate the validity of ceRNA mechanism among circ_0004018, miR-1197 and PTEN, rescue assays were implemented in SK-HEP-1 cells. We firstly knocked down the expression of PTEN (Figure 5(a)). The results of cell proliferation assays disclosed that circ_0004018 overexpression restrained cell proliferation in HCC and this effect was rescued by miR-1197 up-regulation or PTEN down-regulation (Figure 5(b–c)). Transwell migration assay also revealed that miR-1197 augment or PTEN depletion rescued the repressed cell migratory capacity caused by upregulation of circ_0004018 (Figure 5(d)). Overall, upregulation of miR-1197 or downregulation of PTEN recovered the suppressed cell proliferation and migration caused by circ_0004018 overexpression in HCC.
Figure 5.
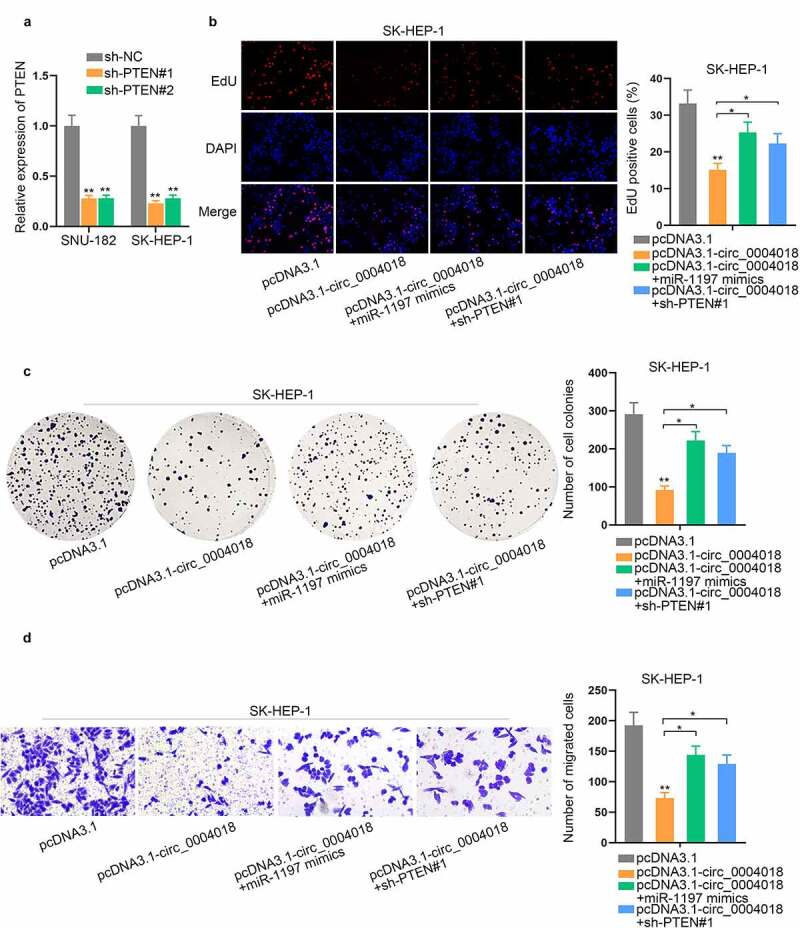
Up-regulation of miR-1197 or down-regulation of PTEN rescues the suppressive impacts of circ_0004018 up-regulation on HCC cell proliferation and migration. (a) Quantification and analysis of PTEN expression in SNU-182 and SK-HEP-1 cells transfected with sh-PTEN#1/2 were achieved via RT-qPCR. (b-c). Cell proliferative capacity was ascertained in different groups by conducting EdU and colony formation assays. (d). With the help of transwell assay, assessment in HCC cell migration was conducted under different conditions. *P < 0.05, **P < 0.01
Discussion
HCC is an appalling disease all over the world due to high incidence, frequent recurrence and poor survival rate. CircRNAs have been recognized as important participants in the occurrence and progression of HCC [24]. Circ_0004018 has been found to be significantly downregulated in liver fibrosis and suppresses the progression of liver fibrosis via modulating miR-660-3p/TEP118, [25]. In accordance with past research, circ_0004018 presents overtly low expression in HCC tissues than in their paired non-tumor tissues from HCC patients, and its link with miR-626 and the Wnt/β-catenin signaling pathway is also revealed [26]. Fu et al. have also discovered that circ_0004018 is down-regulated in HCC tissues and cell lines [10]. In our study, the primary object was the deep investigation into circ_0004018 role in HCC cell proliferation and migration. Consistent with the above reports, our experimental findings manifested that circ_0004018 expression in HCC cells was obviously lower than in THLE-3, and overexpression of circ_0004018 remarkably weakened the proliferative and migratory capacities of HCC cells.
Recent evidence has demonstrated the importance of ceRNA mechanism in diverse cancers. In brief, circRNAs or long non-coding RNAs (lncRNAs) have the capacity to regulate mRNA expression by competing for common miRNAs [27]. The interaction between circ_0004018 and several miRNAs in HCC has been demonstrated by Fu et al. [10]. Similarly, in this paper, we found that circ_0004018 was mainly accumulated in the cytoplasm of HCC cells, suggesting that circ_0004018 might exert its roles at post-transcriptional level. With the help of bioinformatics analysis and RNA pull down assay, miR-1197 was screened out as the candidate miRNA of circ_0004018. Based on previous literature, miR-1197 could be sponged by circ_0000429 and thereby involved in the specific regulatory mechanism of circ_0000429 in Non-small-cell lung carcinoma [20]. Consistently, the interaction between circ_0004018 and miR-1197 was verified based on luciferase reporter assays, s. Furthermore, PTEN was discovered as a target gene of miR-1197. More importantly, the protein level of PTEN was enhanced by circ_0004018 upregulation while counteracted by up-regulation of miR-1197.
As one of the most crucial oncogenic pathways, PI3K/AKT signaling pathway influences cancer development and progression via regulation of cell survival, cell death, and cell growth [28,29]. PTEN is a crucial component involved in PI3K/AKT signaling pathway. To be more specific, as a negative regulator of PI3K/AKT pathway, PTEN may lead to inactivation of PI3K/AKT signaling network [30]. In addition, PTEN is determined to be a valuable predictor for HCC recurrence and overall survival [31]. In this research, we found that circ_0004018 inactivated PI3K/AKT pathway to inhibit cell proliferation and migration in HCC via sponging miR-1197 and modulating PTEN expression.
Overall, circ_0004018 was demonstrated to be significantly downregulated in HCC cells. Functionally, circ_0004018 restrained cell proliferation and migration in HCC. Moreover, circ_0004018 sponged miR-1197 and regulated the expression of PTEN, thereby inactivating the PI3K/AKT signaling pathway in HCC cells. Finally, rescue experiments substantiated that up-regulation of miR-1197 or down-regulation of PTEN rescued the suppressive impact of circ_0004018 on cell proliferation and migration in HCC, which further verified the circ_0004018/miR-1197/PTEN/PI3K/AKT pathway and might provide a potential biomarker for HCC diagnosis and treatment in the future.
Supplementary Material
Acknowledgments
We sincerely thank for the help provided by all lab personnel in this research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Khattab M, Fouad M, Ahmed E.. Role of biomarkers in the prediction and diagnosis of hepatocellular carcinoma. World J Hepatol. 2015;7:2474–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34:153–159. [DOI] [PubMed] [Google Scholar]
- [3].Santer L, Bär C, Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol Ther. 2019;27:1350–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gao JL, Chen G, He HQ, et al. [CircRNA as a new field in human disease research]. Zhongguo Zhong Yao Za Zhi. 2018;43:457–462. [DOI] [PubMed] [Google Scholar]
- [5].Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. [DOI] [PubMed] [Google Scholar]
- [6].Qiu L, Xu H, Ji M, et al. Circular RNAs in hepatocellular carcinoma: biomarkers, functions and mechanisms. Life Sci. 2019;231:116660. [DOI] [PubMed] [Google Scholar]
- [7].Huang XY, Huang ZL, Zhang PB, et al. CircRNA-100338 is associated with mTOR signaling pathway and poor prognosis in hepatocellular carcinoma. Front Oncol. 2019;9:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhu YJ, Zheng B, Luo GJ, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li S, Song F, Lei X, et al. hsa_circ_0004018 suppresses the progression of liver fibrosis through regulating the hsa-miR-660-3p/TEP1 axis. Aging (Albany NY). 2020;12:11517–11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fu L, Yao T, Chen Q, et al. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xiong DD, Dang YW, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].He S, Guo Z, Kang Q, et al. Circular RNA hsa_circ_0000517 modulates hepatocellular carcinoma advancement via the miR-326/SMAD6 axis. Cancer Cell Int. 2020;20:360. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [13].Liu X, Yang L, Jiang D, et al. Circ-DENND4C up-regulates TCF4 expression to modulate hepatocellular carcinoma cell proliferation and apoptosis via activating Wnt/β-catenin signal pathway. Cancer Cell Int. 2020;20:295. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [14].Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. 2018;37:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhuang X, Tong H, Ding Y, et al. Long noncoding RNA ABHD11-AS1 functions as a competing endogenous RNA to regulate papillary thyroid cancer progression by miR-199a-5p/SLC1A5 axis. Cell Death Dis. 2019;10:620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pan G, Mao A, Liu J, et al. Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 2020;53:e12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fu L, Jiang Z, Li T, et al. Circular RNAs in hepatocellular carcinoma: functions and implications. Cancer Med. 2018;7:3101–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hu J, Li P, Song Y, et al. Progress and prospects of circular RNAs in hepatocellular carcinoma: novel insights into their function. J Cell Physiol. 2018;233:4408–4422. [DOI] [PubMed] [Google Scholar]
- [19].Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. [DOI] [PubMed] [Google Scholar]
- [20].Wang JY, Zhang F, Hong L, et al. CircRNA_0000429 regulates development of NSCLC by acting as a sponge of miR-1197 to control MADD. Cancer Manag Res. 2021;13:861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang Z, Zhao Y, Wang Y, et al. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. Biomed Pharmacothe. 2019;116:108932. [DOI] [PubMed] [Google Scholar]
- [22].Fu X, Wen H, Jing L, et al. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci. 2017;108:620–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Milella M, Falcone I, Conciatori F, et al. PTEN: multiple functions in human malignant tumors. Front Oncol. 2015;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). 2018;10(8):258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhou XY, Liu H, Ding ZB, et al. lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics. 2020;112:1021–1029. [DOI] [PubMed] [Google Scholar]
- [26].Zhu P, Liang H, Huang X, et al. Circular RNA Hsa_circ_0004018 Inhibits Wnt/β-Catenin signaling pathway by targeting microRNA-626/DKK3 in hepatocellular carcinoma. Onco Targets Ther. 2020;13:9351–9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang J, Ren Q, Hua L, et al. Comprehensive analysis of differentially expressed mRNA, lncRNA and circRNA and their ceRNA networks in the longissimus dorsi muscle of two different pig breeds. Int J Mol Sci. 2019;20(5):1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 2014;4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].van de Stolpe A. Quantitative measurement of functional activity of the PI3K signaling pathway in cancer. Cancers (Basel). 2019;11(3):293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Haddadi N, Lin Y, Travis G, et al. PTEN/PTENP1: ‘Regulating the regulator of RTK-dependent PI3K/Akt signalling’, new targets for cancer therapy. Mol Cancer. 2018;17:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen D, Li Z, Cheng Q, et al. Genetic alterations and expression of PTEN and its relationship with cancer stem cell markers to investigate pathogenesis and to evaluate prognosis in hepatocellular carcinoma. J Clin Pathol. 2019;72:588–596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


