ABSTRACT
Being one of the most prevalent malignancies, hepatocellular carcinoma (HCC) threatens the health of population all over the world. Numerous researches have confirmed that long noncoding RNAs (lncRNAs) play an important role in tumor progression. Nonetheless, the mechanisms of unc-5 netrin receptor B antisense RNA 1 (UNC5B-AS1) in HCC remain obscure. Thus, this study aims to investigate the regulatory role and mechanism of UNC5B-AS1 in HCC cells. In our research, UNC5B-AS1 was subjected to gene expression analysis by RT-qPCR. Biological functions of UNC5B-AS1 in HCC cells were measured by MTT, colony formation, EdU and transwell assays. The combination between UNC5B-AS1, lysine demethylase 2A (KDM2A) and miR-4306 was validated by mechanism assays. Result showed UNC5B-AS1 was upregulated in HCC tissues and cells, contributing to the development of cancer staging and survival rate of HCC patients. Moreover, UNC5B-AS1 deficiency inhibited the proliferation, migration and epithelial–mesenchymal transition (EMT) of HCC cells. Furthermore, UNC5B-AS1 could interact with miR-4306 in HCC cells. Similarly, KDM2A was proved as the target gene of miR-4306. Finally, miR-4306 downregulation or KDM2A overexpression reversed the prohibitive role of UNC5B-AS1 knockdown in HCC progression. In short, UNC5B-AS1 accelerates the proliferation, migration and EMT of HCC cells via the regulation of miR-4306/KDM2A axis.
KEYWORDS: HCC, UNC5B-AS1, miR-4306, KDM2A
Introduction
HCC, as one of the most familiar cancers, is the main reason of cancer-caused death cases in China [1]. In the past few years, the amount of diagnosed HCC patients has been augmented. Although ameliorations were made on treatment strategies, outcomes of current HCC therapies are still in an unsatisfactory condition [2,3]. The five-year survival rate of HCC patients is no higher than 30% [4]. Accordingly, comprehending the molecular essence in HCC is urgently required.
LncRNAs, containing over 200 nucleotides, are a group of noncoding RNAs [5]. Previously, lncRNAs were considered as the transcription noise that did not exert any biological function in cells. However, a flow of recent researches have confirmed that lncRNAs do serve for crucial biological aims [6]. Importantly, accumulating evidences also indicate that lncRNAs participate in the progression of human cancers and exert the regulatory function through diverse mechanisms, such as regulating gene expression, sponging microRNAs (miRNAs) and altering DNA structure [7,8]. It has been reported that lncRNAs can serve as tumor promoters or suppressors to regulate the progression of cancers, including HCC [9,10]. The aberrant expression of lncRNAs is frequently observed in multifarious cancers [11]. For example, expression of lncRNA-n336928 has proven to be related to tumor staging and grading of bladder cancer and patients’ survival rate [12]. LncRNA-ATB participates in the development of colorectal cancer and is certificated as being critical for the prognosis [13]. Also, the overexpressed lncRNA CCHE1 predicts the poor prognosis of HCC and contributes to the carcinogenesis by activating ERK/MAPK pathway [14].
UNC5B-AS1 has been verified to exert the carcinogenic effect in multiple human cancers. For instance, UNC5B-AS1 was reported to accelerate the proliferative, migratory and invasive capacities of papillary thyroid cancer cells [15]. Besides, UNC5B-AS1 could facilitate the progression of prostate cancer via competitively binding to caspase-9 [16]. Furthermore, UNC5B-AS1 expedites ovarian cancer cell growth through the regulation of H3K27me on NDRG2 via EZH2 [17]. However, the role of UNC5B-AS1 in HCC is still unclear.
In our study, we explored the function of UNC5B-AS1 and its molecular mechanism in HCC cells, which might offer a new insight into therapy targets for HCC treatment.
Materials and methods
Tissue samples
Sixty-five pairs of HCC tissues and the matched normal tissues were acquired from HCC patients who had received surgeries at the Second Affiliated Hospital of Fujian Medical University between June 2013 and July 2018. None of the patients in this study were subjected to chemotherapy or radiotherapy in advance to surgery. Snap-frozen tissue samples from patients were maintained in liquid nitrogen immediately after being excised from patients and preserved at −80°C. All patients have signed written informed consent. Furthermore, this study was permitted by the ethics committee of the Second Affiliated Hospital of Fujian Medical University.
Cell culture
Normal human hepatocytes (LO-2) and human HCC cell lines (HepG2, Huh7, Hep3B and SK-HEP-1) were used in this research. HepG2, Hep3B and SK-HEP-1 cell lines from Cell Bank of the Chinese Academy of Sciences (Shanghai, China) were cultivated in DMEM (Thermo Fisher Scientific, Waltham, MA, USA). LO-2 and Huh7 cell lines from LianMai Bioengineering Co., Ltd (Shanghai, China) were kept in RPMI-1640 medium (Thermo Fisher Scientific). All cells were placed at 37°C in humidified incubators with 5% CO2 and supplied with 10% fetal bovine serum (FBS; Thermo Fisher Scientific), 100 IU/ml penicillin and 100 µg/ml streptomycin (Sigma-Aldrich, St. Louis, MO, USA) for culturing. The mediums were replaced every 3 days.
Cell transfection
HepG2 or Huh7 cells at 50%~80% of confluence were collected and transfected with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) for 48 h. The short-hairpin RNAs (shRNAs) specifically aiming at UNC5B-AS1 (sh-UNC5B-AS1#1, sh-UNC5B-AS1#2) or sh-NC, along with the KDM2A overexpression plasmid (pcDNA3.1/KDM2A) or the empty pcDNA3.1 vector, were synthesized by GenePharma (Shanghai, China). The miR-4306 mimics, miR-4306 inhibitor, NC mimics and NC inhibitor were produced by RiboBio (Guangzhou, Guangdong, China). Bio-repeats were run in triplicate.
Real-time quantitative PCR (RT-qPCR)
Using TRIzol (Invitrogen) in light of the directions from the manufacturer, total RNAs were isolated from tissues and cells to be reversely transcribed into complementary DNAs (cDNAs). Then, qPCR was conducted with the application of the SYBR Green Master Mix Kit (TaKaRa, Tokyo, Japan), with GAPDH and U6 as endogenous controls for cytoplasmic and nuclear genes, respectively. Results were analyzed by means of the 2−ΔΔCt method. Bio-repeats were conducted in triplicate.
MTT assay
Cell viability of transfected HepG2 or Huh7 cells was measured via MTT assays. Cells were transplanted into 96-well plates, each well containing 5 × 103 cells. Then, 5 mg/mL MTT (Sigma-Aldrich) was supplemented into each well, and cells were cultured for another 4 h. 150 μl dimethyl sulfoxide (DMSO, Sigma-Aldrich) was then added. Plates were oscillated slowly so as to make all the crystal substance dissolved. Optical density (OD) was detected at 490 nm via a SpectraMax i3x microplate reader (Molecular Devices, Silicon Valley, CA, USA). Bio-repeats were performed in triplicate.
Colony formation assay
Transfected HCC cells were placed into 6-well plates and cultivated for 14 days. Following that, cells were fixed by 4% paraformaldehyde (PFA; Sangon Biotech, Shanghai, China) and stained by 1 mL 0.1% crystal violet solution (Sigma-Aldrich). Thereafter, culture plates were photographed. Colonies in each well were observed and counted manually. Bio-repeats were performed in triplicate.
EdU assay
EdU assay was performed on the basis of the user guide of EdU kit (RiboBio, Guangzhou, China). After transfection, HepG2 or Huh7 cells were inoculated into 96-well plates and subjected to EdU staining. After that, cell nuclei were dyed with DAPI solution. In the end, the EdU positive cells were visualized with the help of a fluorescence microscope (Olympus, Tokyo, Japan). Bio-repeats were conducted in triplicate.
Transwell assay
Transwell assays were conducted with the application of Transwell chambers (Corning, Corning, NY) to assess the migration of cells. Culture medium containing 10% FBS was supplemented into the lower chambers, while the serum-free medium into the upper chambers in which cells were plated for incubation. After 24 h of cultivation, the cells migrating to the lower chambers were subjected to fixation, followed by the treatment with 0.1% crystal violet for staining. At last, the migrated cells were observed using a microscope. Bio-repeats were run in triplicate.
RNA pull down assay
In accordance with the protocols of the manufacturer, Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher) was utilized to perform this assay. After the extraction of the proteins, magnetic beads were added for precipitating the RNA–RNA complex. Subsequently, RNAs in the precipitates were extracted and analyzed via RT-qPCR. Bio-repeats were conducted in triplicate.
Luciferase reporter assay
The UNC5B-AS1 sequence or KDM2A 3ʹUTR with conjectured binding sites of miR-4306 was sub-cloned into the pmirGLO dual-luciferase vector (Promega, Madison, WI, USA). The wild-type luciferase vectors UNC5B-AS1-WT and KDM2A 3ʹUTR-WT were constructed. By use of the GeneArt™ Site-Directed Mutagenesis System (Thermo Fisher Scientific), the mutant vectors UNC5B-AS1-Mut and KDM2A 3ʹUTR-Mut were generated. HepG2 or Huh7 cells were severally co-transfected with above luciferase reporter vectors and miR-4306 mimics or NC mimics, respectively. Luciferase signals were monitored by the Dual Luciferase Reporter Assay Kit (Promega) after two whole days of co-transfection. Bio-repeats were run in triplicate.
Western blot
The complete proteome was subjected to extraction in the lysis buffer. Concentration of proteins was tested by employing a BCA protein assay kit (Solarbio, Billerica, MA, USA). After the electrophoresis on 15% SDS-PAGE (Bio-Rad Laboratories, Hercules, CA, USA) for separation, proteins were subjected to transference onto polyvinylidene fluoride membrane (PVDF; Amersham, Munich, Germany) blocked with 5% nonfat milk. The membrane was first cultured with the following listed primary antibodies: anti-KDM2A antibodies (1/1000, ab191387; Abcam), anti-E-cadherin antibodies (1/1000, ab76055; Abcam), anti-N-cadherin antibodies (1/1000, ab76057; Abcam), anti-Vimentin antibodies (1/1000, ab8978; Abcam) and anti-GAPDH antibodies (1/1000, ab8245; Abcam). After the washes, membranes were incubated with secondary antibodies. Blots were visualized by enhanced chemiluminescence (ECL; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Bio-repeats were conducted in triplicate.
Statistical analysis
The experimental data were shown as mean ± SD of three independent experiments involving triplicate technical repetitions. Through the GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA) and SPSS 23.0 (IBM Corp., Armonk, NY, USA), statistical analysis was then conducted. Student’s t-test or one-way/two-way analysis of variance (ANOVA) was adopted to analyze the significance of the differences between two groups or among more than two groups. P < 0.05 was the criterion of statistical significance. Kaplan–Meier curve and the log-rank test were used for analyzing survival rate.
Results
UNC5B-AS1 is upregulated in HCC tissues and cells, and closely correlated with the survival rate
To confirm that UNC5B-AS1 acted as an oncogene in HCC, first of all, UNC5B-AS1 expression was evaluated in HCC tissues and cells. Through the StarBase (http://starbase.sysu.edu.cn/index.php) database, we found that UNC5B-AS1 expression in liver hepatocellular carcinoma (LIHC) tissues was much higher than that in normal tissues (Figure 1a). Then, we further detected UNC5B-AS1 expression in HCC tissues and cell lines. As shown in Figure 1(b,c), UNC5B-AS1 was remarkably upregulated in tumor tissues and HCC cell lines compared to that in corresponding control groups. Moreover, UNC5B-AS1 was obviously expressed in HepG2 and Huh7 cells most. Moreover, UNC5B-AS1 expression was positively associated with terminal stage of HCC (Figure 1d). Besides, the HCC patients with higher expression of UNC5B-AS1 possessed shorter survival time than those with lower expression (Figure 1e). To sum up, UNC5B-AS1 was high-expressed in HCC tissues and cells, and closely linked to the survival rate of HCC patients.
Figure 1.
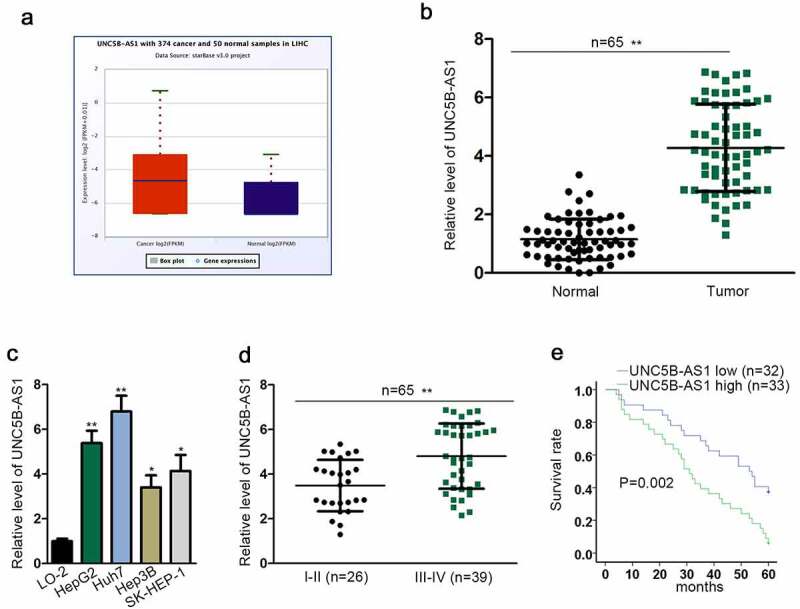
UNC5B-AS1 is upregulated in HCC tissues and cells, and closely associated with the survival rate
(A) UNC5B-AS1 expression in LIHC tissues and normal tissues obtained from starBase (http://starbase.sysu.edu.cn/index.php). (B) RT-qPCR detected UNC5B-AS1 expression in HCC tumor tissues and the matched normal ones. (C) UNC5B-AS1 expression in HepG2, Huh7, Hep3B, SK-HEP-1 and LO-2 was measured by RT-qPCR. (D) RT-qPCR tested UNC5B-AS1 expression in HCC tissues at different clinical stages (I–II and III–IV). (E) The overall survival rate of 5 years of HCC patients with high or low UNC5B-AS1 expression was displayed by Kaplan–Meier curve and the log-rank test. *P < 0.05, **P < 0.01.
UNC5B-AS1 knockdown inhibits HCC cell proliferation, migration and EMT
After confirming the upregulation of UNC5B-AS1 in HCC cells and tissues, we continued to evaluate its biological function. In advance to loss-of-function experiments, the knockdown effects of sh-UNC5B-AS1#1/2 were assessed in HepG2 and Huh7 cells (Figure 2a). Subsequently, MTT assay was conducted. We discovered that the absorbance value was gradually declined in sh-UNC5B-AS1#1/2-transfected cells, indicating cell proliferation could be inhibited by UNC5B-AS1 depletion (Figure 2b). Then, it was displayed from colony formation assay that colony number was notably decreased after UNC5B-AS1 was knocked down in cells (Figure 2c). Furthermore, EdU-positive cells were also reduced when UNC5B-AS1 was silenced in cells, which further verified the restrained cell proliferation by the knockdown of UNC5B-AS1 (Figure 2d). Besides, transwell assay was utilized to detect cell migration. The results indicated that UNC5B-AS1 depletion hampered HCC cell migratory capability since the number of migrated cells was declined in transfected cell groups (Figure 2e). Meanwhile, shortage of UNC5B-AS1 suppressed EMT of HCC, according to the decline in the protein level of N-cadherin as well as Vimentin, and the rise in E-cadherin (figure 2f). Taken together, the mentioned results validated that UNC5B-AS1 was an oncogene in HCC cells, and UNC5B-AS1 downregulation restrained the proliferation, migration and EMT of HCC cells.
Figure 2.
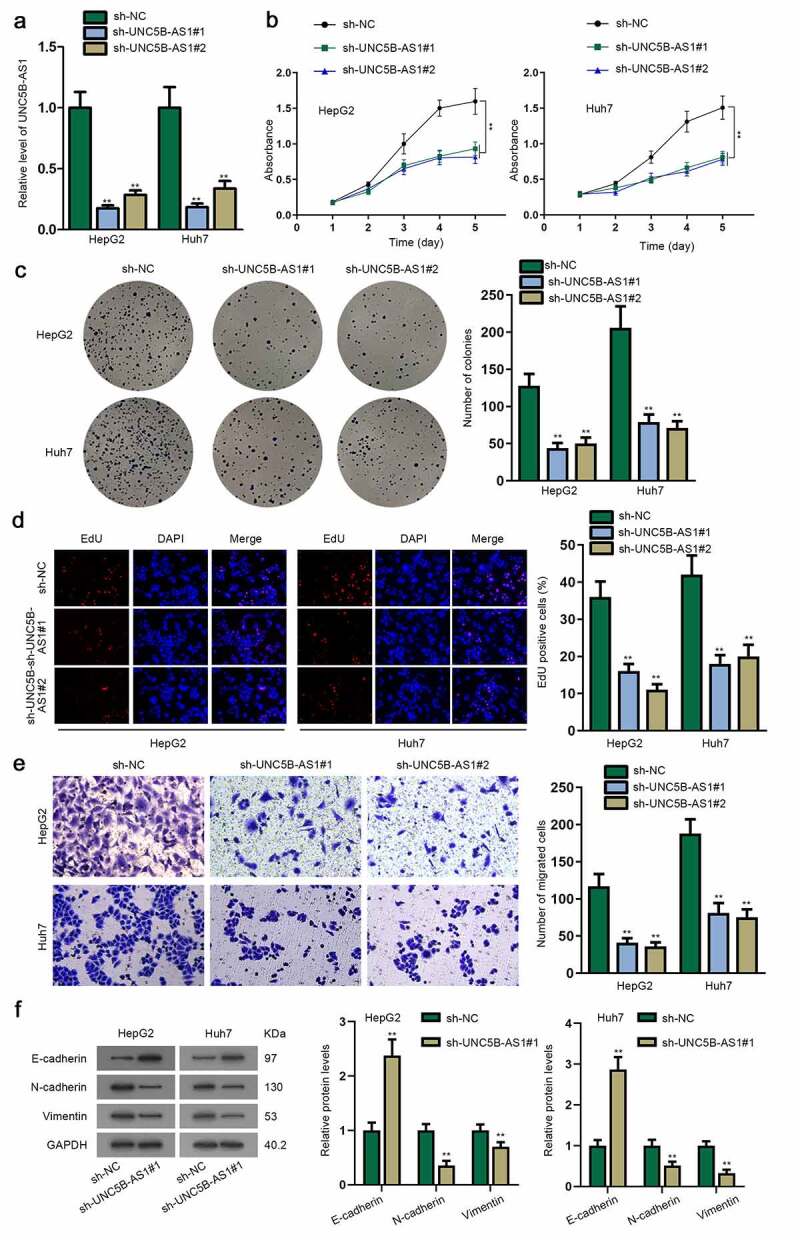
UNC5B-AS1 knockdown inhibits HCC cell proliferation, migration and EMT
(A) RT-qPCR detected the efficacy of sh-UNC5B-AS1#1/2. (B-D) MTT, colony formation and EdU assays were applied for detecting the ability of HCC cells transfected with sh-UNC5B-AS1#1/2 to proliferate. (E) Transwell assay was utilized to measure cell migration in the sh-UNC5B-AS1#1/2-transfected groups and negative control groups. (F) E-cadherin, N-cadherin and Vimentin were measured in transfected HCC cells through western blot assay, with GAPDH as an internal control. **P < 0.01.
MiR-4306 is targeted by UNC5B-AS1 in HCC cells
For further exploration of the regulatory mechanism of UNC5B-AS1 in HCC, we aimed to find the miRNA targeted by UNC5B-AS1. Herein, five miRNAs with the highest possibility to combine with UNC5B-AS1 are listed in Figure 3a through the prediction of miRDB and NCBI. Then, via RNA pull down assay, miR-4306 was significantly pulled down by the biotinylated UNC5B-AS1, while other miRNAs almost remained unchanged (Figure 3b). Thus, miR-4306 was selected for the following analysis. Utilizing RT-qPCR analysis, we found out miR-4306 was underexpressed in LIHC tissues compared with normal tissues (Figure S1A). As shown in Figure S1B, miR-4306 was negatively correlated with the terminal stage of HCC. Moreover, the overall survival rate was lower in HCC patients with lowly expressed miR-4306 (Figure S1C). We subsequently conducted correlation analysis and found out that miR-4306 had a negative correlation with miR-4306 in both HCC tissues and normal tissues (Figure S1D). Then, binding sites between UNC5B-AS1 and miR-4306 were predicted by bioinformatics (Figure 3c). The luciferase reporter assay indicated that the luciferase activity of UNC5B-AS1-WT was reduced by miR-4306 mimics while that of UNC5B-AS1-Mut remained almost unaffected, which confirmed their binding (Figure 3d). In addition, RT-qPCR depicted that miR-4306 expression was evidently downregulated in HCC cell lines (Figure 3e). Besides, it was validated that knockdown of UNC5B-AS1 increased miR-4306 expression in HCC cells (figure 3f). Overall, miR-4306 is targeted by UNC5B-AS1 in HCC cells.
Figure 3.
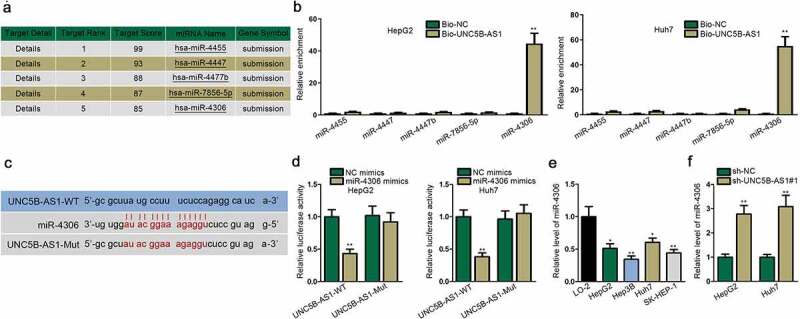
MiR-4306 is targeted by UNC5B-AS1 in HCC cells
(A) Five downstream miRNA candidates (hsa-miR-4455, hsa-miR-4447, hsa-miR-4477b, hsa-miR-7856-5p and hsa-miR-4306) of UNC5B-AS1 were screened out by miRDB (http://mirdb.org/) and NCBI (https://www.ncbi.nlm.nih.gov/). (B) The relative enrichment of five miRNA candidates was detected in RNA pull down assay. (C) The binding sites between UNC5B-AS1 and miR-4306 were predicted by bioinformatics. (D) Luciferase reporter assay confirmed the combination and binding site between UNC5B-AS1 and miR-4306. (E) miR-4306 expression was tested by RT-qPCR in HCC cells lines and normal human hepatocytes. (F) The effects of silenced UNC5B-AS1 on miR-4306 expression were assessed by RT-qPCR. *P < 0.05, **P < 0.01.
KDM2A is targeted by miR-4306 in HCC cells
To explore the target gene of miR-4306, we initially selected two messenger RNAs (mRNAs), E2F6 and KDM2A, based on TargetScan, miRDB and miRTarBase databases (Figure 4a). KDM2A was significantly upregulated by miR-4306 inhibitor than E2F6 in HCC cells (Figure 4b). Furthermore, the upregulation of KDM2A in LIHC tissues was verified by the data from StarBase database (Figure S1E). We then used RT-qPCR and found out KDM2A was overexpressed in LIHC tissues compared with normal tissues (Figure S1F). As shown in Figure S1G, KDM2A was positively associated with the terminal stage of HCC. The overall survival rate was lower in HCC patients with higher expression of KDM2A (Figure S1H). We subsequently conducted correlation analysis and discovered that KDM2A was negatively associated with miR-4306, and positively correlated with UNC5B-AS1 in both HCC tissues and normal tissues (Figure S1I-J). As displayed by Figure 4c, there were three binding sites between KDM2A and miR-4306, which were confirmed by luciferase reporter assay (Figure 4d). Furthermore, UNC5B-AS1 depletion decreased the level of KDM2A mRNA and protein in HCC cells (Figure 4e & Figure S2A). Also, miR-4306 overexpression silenced the protein level of KDM2A, while miR-4306 ablation exhibited the opposite effect on KDM2A expression (Figure S2B). Furthermore, miR-4306 inhibitor offset the prohibitive function of sh-UNC5B-AS1 on the expression of KDM2A mRNA and protein, which indicated that UNC5B-AS1 could regulate KDM2A expression via targeting miR-4306 in HCC cells (figure 4f & Figure S2C). To sum up, KDM2A is targeted by miR-4306 in HCC cells, and its expression is positively regulated by UNC5B-AS1.
Figure 4.
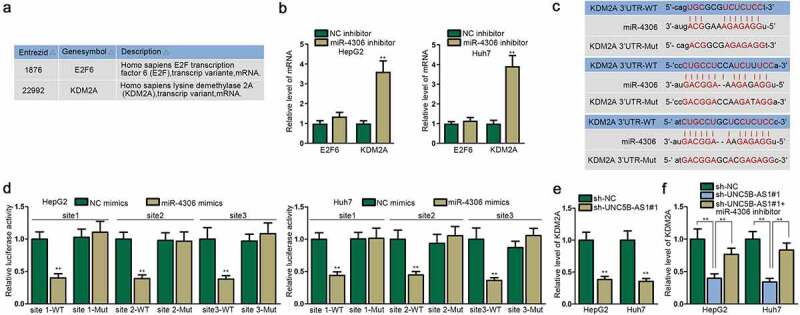
KDM2A is targeted by miR-4306 in HCC cells
(A) E2F6 and KDM2A were filtered out as the downstream gene of miR-4306 according to TargetScan (http://www.targetscan.org/mamm_31/), miRDB (http://mirdb.org/) and miRTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php). (B) E2F6 and KDM2A expressions were assessed by RT-qPCR in cells after the transfection with miR-4306 inhibitor. (C) Three binding sites between miR-4306 and KDM2A were predicted through bioinformatics. (D) The binding sites between miR-4306 and KDM2A were verified by luciferase reporter assay. (E-F) KDM2A mRNA expression was measured by RT-qPCR when UNC5B-AS1 and miR-4306 were silenced in HCC cells. **P < 0.01.
UNC5B-AS1 promotes the proliferation and EMT of HCC cells by regulating miR-4306/KDM2A axis
Rescue experiments were implemented to confirm whether UNC5B-AS1 propels the proliferation and EMT of HCC via regulating miR-4306/KDM2A axis. Firstly, the overexpression efficiency of KDM2A was measured in HepG2 cells transfected with pcDNA3.1/KDM2A (Figure 5a). Then, MTT, colony formation assay and EdU assay illustrated that miR-4306 inhibitor or pcDNA3.1/KDM2A reversed the suppressive effects of sh-UNC5B-AS1 on cell proliferation in HCC (Figure 5(b,c,d)). Besides, transwell assay also proved that cell migration repressed by UNC5B-AS1 depletion could be recovered by miR-4306 inhibitor or KDM2A overexpression (Figure 5e). Moreover, western blot assay elucidated that miR-4306 downregulation or KDM2A overexpression countervailed the inhibitive influence of UNC5B-AS1 insufficiency on EMT of HCC cells (figure 5f). Broadly speaking, the above results validated UNC5B-AS1 promotes the proliferative and migratory abilities, and EMT of HCC cells by modulating miR-4306/KDM2A axis.
Figure 5.
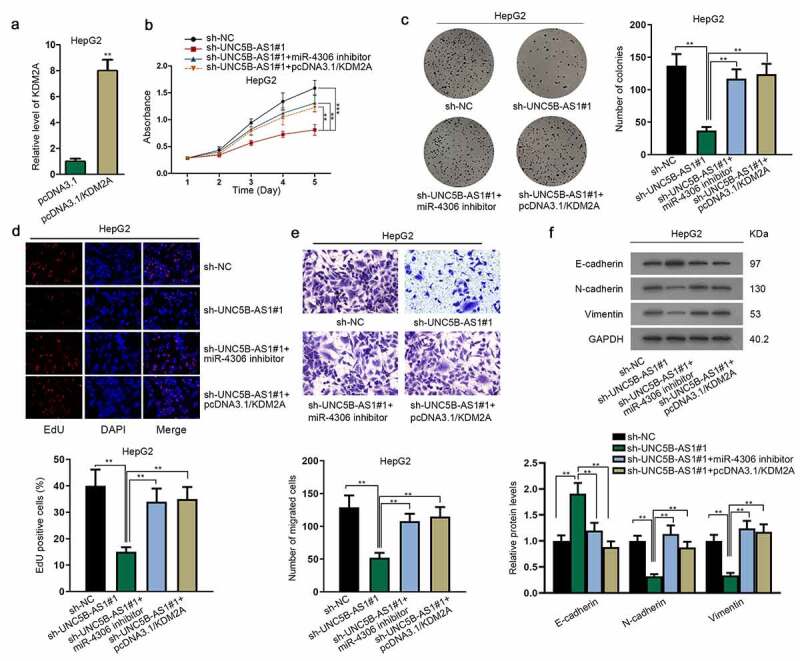
UNC5B-AS1 promotes HCC cell proliferation, migration and EMT by regulating miR-4306/KDM2A axis
(A) The overexpression efficacy of pcDNA3.1/KDM2A was detected by RT-qPCR. (B-D) The proliferation of different transfected cells was subjected to the detection of MTT, colony formation and EdU assays. (E) Transwell assay was applied for detecting cell migration in different groups. (F) Western blot assay assessed the EMT of HCC cells in different groups, with GAPDH as an internal control. **P < 0.01.
Discussion
HCC is listed in the top five most common cancers [18]. Most of HCC patients are diagnosed at the terminal stage, which increases the difficulty in treatment. Moreover, there is a high recurrent rate of HCC even after surgical resection, which results in the unsatisfactory prognosis of HCC [19]. Thus, it is in sore need of effective strategies to diagnose and treat HCC.
LncRNAs have been reported to modulate biological processes, like proliferation and invasion, of cancer cells [20]. Recent researches have attested that lncRNAs can be potential therapeutic targets for human cancers. Besides, it was found that several lncRNAs are closely related to cell proliferation, migration, as well as invasion in HCC [20–22]. In this study, UNC5B-AS1 was discovered to be upregulated in HCC tissues and cells, which was compactly associated with cancer staging and survival rate of patients. Moreover, the shortage of UNC5B-AS1 suppressed cell proliferation, migration and EMT in HCC, indicating that UNC5B-AS1 served as a tumor promoter in HCC.
MiRNAs with 20 ~ 25 nucleotides are small non-coding RNAs with high conservation [23]. It has been proved that miRNAs accompanied by lncRNAs are linked to cancer progression. For example, lncRNA Taurine-Upregulated Gene 1 (TUG1) accelerates malignant development of gastric cancer cells by negatively regulating miR-145-5p [24]. LncRNA MALAT1 depletion suppresses cell growth and migration in esophageal squamous cell carcinoma through modulating miR-101 and miR-217 [25]. In addition, the downregulation of lncRNA MEG3 impedes proliferation and apoptosis of cervical cancer cells via the modulation of miR-21 [26]. In the present research, UNC5B-AS1 was verified to bind with miR-4306, thus negatively regulating miR-4306 expression which was downregulated in HCC cells.
Competing endogenous RNA (ceRNA) is the mechanism involving lncRNA, miRNA and mRNA, having the potential to influence the initiation and progression of cancers [27]. For instance, lncRNA H19 could modulate FOXM1 expression through emulously binding to miR-342-3p in gallbladder cancer [28]. MALAT1 facilitates cell progression in breast cancer via competitively interacting with miR-1 against cdc42 [29]. Additionally, lncRNA BC032469 positively regulates the expression of hTERT through inhibiting the expression of miR-1207-5p and facilitates cell proliferation in gastric cancer [30]. KDM2A was confirmed to be an oncogene in several cancers, such as gastric cancer [31], cervical cancer [32] and glioblastoma [33]. In our research, it was found that KDM2A was the target gene of miR-4306 and confirmed that miR-4306 combines with KDM2A. Furthermore, the expression of KDM2A was positively or negatively regulated by UNC5B-AS1 or miR-4306 in HCC. The positive correlation between UNC5B-AS1 and KDM2A and the negative correlation between miR-4306 and UNC5B-AS1/KDM2A in HCC and adjacent benign tissues have been verified. Through the rescue experiments, it was validated that miR-4306 deficiency or KDM2A overexpression could countervail the repressive effect of UNC5B-AS1 depletion on cell progression in HCC.
Overall, this study verified that UNC5B-AS1 promotes the proliferation, migration and EMT of HCC cells via regulating miR-4306/KDM2A signaling pathway, which may cast new light on the treatment of HCC.
Supplementary Material
Acknowledgments
We appreciate the technical supports of lab members.
Funding Statement
This study was supported by Medical innovation project of Fujian Health Committee (NO.2018-CX-35), Project of Quanzhou Science and technology bureau under grant (NO. 2018Z133).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Maluccio M, Covey A.. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–399. [DOI] [PubMed] [Google Scholar]
- [3].Colecchia A, Schiumerini R, Cucchetti A, et al. Prognostic factors for hepatocellular carcinoma recurrence. World J Gastroenterol. 2014;20:5935–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rahimi RS, Trotter JF. Liver transplantation for hepatocellular carcinoma: outcomes and treatment options for recurrence. Ann Gastroenterol. 2015;28:323–330. [PMC free article] [PubMed] [Google Scholar]
- [5].Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. [DOI] [PubMed] [Google Scholar]
- [6].Jathar S, Kumar V, Srivastava J, et al. Technological developments in lncRNA Biology. Adv Exp Med Biol. 2017;1008:283–323. [DOI] [PubMed] [Google Scholar]
- [7].Tu J, Zhao Z, Xu M, et al. LINC00707 contributes to hepatocellular carcinoma progression via sponging miR-206 to increase CDK14. J Cell Physiol. 2019;234:10615–10624. [DOI] [PubMed] [Google Scholar]
- [8].Liu B, Ye B, Yang L, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18:499–508. [DOI] [PubMed] [Google Scholar]
- [9].Hu Z, Huang P, Yan Y, et al. Hepatitis B virus X protein related lncRNA WEE2-AS1 promotes hepatocellular carcinoma proliferation and invasion. Biochem Biophys Res Commun. 2019;508:79–86. [DOI] [PubMed] [Google Scholar]
- [10].Xu Y, Zhang G, Zou C, et al. Long noncoding RNA DGCR5 suppresses gastric cancer progression by acting as a competing endogenous RNA of PTEN and BTG1. J Cell Physiol. 2019;234:11999–12010. [DOI] [PubMed] [Google Scholar]
- [11].Wu J, Du M, Zhang Q, et al. Long noncoding RNA UCA1 promotes the proliferation, invasion, and migration of nasopharyngeal carcinoma cells via modulation of miR-145. Onco Targets Ther. 2018;11:7483–7492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [12].Chen T, Xie W, Xie L, et al. Expression of long noncoding RNA lncRNA-n336928 is correlated with tumor stage and grade and overall survival in bladder cancer. Biochem Biophys Res Commun. 2015;468(4):666–670. [DOI] [PubMed] [Google Scholar]
- [13].Iguchi T, Uchi R, Nambara S, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35:1385–1388. [PubMed] [Google Scholar]
- [14].Peng W, Fan H. Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway. Biomed Pharmacothe. 2016;83:450–455. [DOI] [PubMed] [Google Scholar]
- [15].Wang Y, Bhandari A, Niu J, et al. The lncRNA UNC5B-AS1 promotes proliferation, migration, and invasion in papillary thyroid cancer cell lines. Hum Cell. 2019;32:334–342. [DOI] [PubMed] [Google Scholar]
- [16].Tan SF, Ni JX, Xiong H. LncRNA UNC5B-AS1 promotes malignant progression of prostate cancer by competitive binding to caspase-9. Eur Rev Med Pharmacol Sci. 2020;24:2271–2280. [DOI] [PubMed] [Google Scholar]
- [17].Wang H, Su H, Tan Y. UNC5B-AS1 promoted ovarian cancer progression by regulating the H3K27me on NDRG2 via EZH2. Cell Biol Int. 2020;44:1028–1036. [DOI] [PubMed] [Google Scholar]
- [18].Marquardt JU, Thorgeirsson SS. SnapShot: hepatocellular carcinoma. Cancer Cell. 2014;25:550.e1. [DOI] [PubMed] [Google Scholar]
- [19].Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. [DOI] [PubMed] [Google Scholar]
- [20].Chen DL, Ju HQ, Lu YX, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xiao JN, Yan TH, Yu RM, et al. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143:981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang JY, Weng MZ, Song FB, et al. Long noncoding RNA AFAP1-AS1 indicates a poor prognosis of hepatocellular carcinoma and promotes cell proliferation and invasion via upregulation of the RhoA/Rac2 signaling. Int J Oncol. 2016;48:1590–1598. [DOI] [PubMed] [Google Scholar]
- [23].Takahashi RU, Prieto-Vila M, Hironaka A, et al. The role of extracellular vesicle microRNAs in cancer biology. Clin Chem Lab Med. 2017;55:648–656. [DOI] [PubMed] [Google Scholar]
- [24].Ren K, Li Z, Li Y, et al. Long noncoding RNA Taurine-Upregulated Gene 1 promotes cell proliferation and invasion in gastric cancer via negatively modulating miRNA-145-5p. Oncol Res. 2017;25:789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang X, Li M, Wang Z, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang J, Yao T, Wang Y, et al. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol Ther. 2016;17:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang SH, Ma F, Tang ZH, et al. Long non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancer. J Exp Clin Cancer Res. 2016;35:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chou J, Wang B, Zheng T, et al. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. 2016;472:262–269. [DOI] [PubMed] [Google Scholar]
- [30].Lu MH, Tang B, Zeng S, et al. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–3534. [DOI] [PubMed] [Google Scholar]
- [31].Huang Y, Liu Y, Yu L, et al. Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumour Biol. 2015;36:271–278. [DOI] [PubMed] [Google Scholar]
- [32].Ou R, Zhu L, Zhao L, et al. HPV16 E7-induced upregulation of KDM2A promotes cervical cancer progression by regulating miR-132-radixin pathway. J Cell Physiol. 2019;234:2659–2671. [DOI] [PubMed] [Google Scholar]
- [33].Shou T, Yang H, Lv J, et al. MicroRNA‑3666 suppresses the growth and migration of glioblastoma cells by targeting KDM2A. Mol Med Rep. 2019;19:1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


