ABSTRACT
Gastric cancer is one of the top three leading causes of cancer-related death in the world. Evidence indicated that miR-4677-3p was dysregulated and involved in modulating invasion and migration in multiple types of cancer cells. The aim of this research is to explore the function and mechanism of miR-4677-3p in the development of gastric cancer. In this study, we discovered that miR-4677-3p was down-regulated in gastric cancer tissues and cells. Over-expression of miR-4677-3p suppressed the proliferation, migration and invasion of gastric cancer cells. Furthermore, miR-4677-3p directly bond to CEMIP 3ʹUTR region and inhibited CEMIP expression. CEMIP promoted cell proliferation, migration and invasion of gastric cancer cells via accelerating PI3K/AKT signaling pathway. siCEMIP or PI3K/AKT signaling inhibitor (Akti-1/2 and LY294002) partly reversed the effects of miR-4677-3p on the cellular growth and metastasis of gastric cancer. In general, miR-4677-3p regulated the development of gastric cancer through CEMIP-PI3K/AKT signaling pathway axis. This study verified the function and molecular mechanism of miR-4677-3p in gastric cancer cells, and may provide a potential diagnosis/prognosis target for patients with gastric cancer.
KEYWORDS: Gastric cancer, miR-4677-3p, metastases
Introduction
Gastric cancer is one of most common cancers with high mortality in the world [1–5]. The highest incidence rates of gastric cancer mostly occur in developing countries, especially East Asia, East Europe, and South America [6]. Although there are many studies focused on gastric cancer, the recurrence rate of patients was still high. Therefore, it is urgent to elucidate the underlying molecular mechanism of gastric cancer for its diagnosis and treatment.
MicroRNAs (miRNAs) is a class of non-coding single stranded RNA molecule with approximate 22 nucleotides [2,3,7–10], and regulates the expressions of target genes by binding to the 3ʹUTR region [7,9,11]. miRNAs plays a vital role in mammals’ cell processes, such as cell proliferation, differentiation, migration, invasion and apoptosis [7,12]. Reports defined the pivotal function of miRNAs in the underlying mechanism of regulating cancer progression [7,13,14]. It has been reported that many micro-RNAs, such as miR-744, miR-223, miR-125, miR-21, miR-101 and miR-130b, were related to the proliferation, migration and invasion of gastric cancer cells [11,15–19]. A recent study revealed that miR-4677-3p was down-regulated in human lung cancer, nonspecific peripheral T cell lymphoma and oral squamous cell carcinoma [20,21], and mediated the invasion and migration of lung adenocarcinoma cells [20]. But the specific function of miR-4677-3p in gastric cancer has not been reported till now.
Cell migration-including protein (CEMIP) is a poorly characterized protein, and it is associated with poor survival of cancer patients. It has been defined that CEMIP was highly expressed in cancer and promoted cell proliferation, migration, EMT progress and anti-apoptosis effects [22–30]. The elevated expression of CEMIP was closely associated with a poor prognosis of multiple human cancers, including prostate cancer, gastric cancer, and CRC [22,24,29,31,32]. Besides, there is an evidence showed that CEMIP was up-regulated about 29.69 times in gastric cancer [33].
Plentiful studies have identified the activation of PI3K/AKT signaling pathway as a contributor of cellular growth, metabolism and metastasis [34–36]. Besides, the PI3K/AKT pathway also affects multiple functions of gastric cancer cells, including proliferation, migration and invasion [37]. Dysregulation of PI3K/AKT pathway genes, such as AKT1, AKT2 and PIK3CA, is related to the recurrence of gastric cancer [38]. Therefore, PI3K/AKT signaling pathway plays a pivotal role in human gastric cancer development.
Based on these reports, our study verified the expression profile of miR-4677-3p during gastric cancer and aimed to explore the effect of miR-4677-3p and CEMIP, as well as the underlying molecular mechanism, in regulating the development of gastric cancer. Hoping to find a valuable biomarker for gastric cancer therapy and prognosis.
Materials and methods
Tissue specimens
Total 30 pairs of human gastric cancer tissues were recruited in this research and the adjacent non-tumor tissues sample were used as control. All tissues were acquired from the first affiliated hospital of Xi’an Jiaotong University, between January and July 2019. These tissues were promptly snap-frozen in liquid nitrogen after collecting, and then stored at −80°C. None of the patients had received radiotherapy or chemotherapy before. And all of them provided the written informed consents. Our study was approved by the ethics committee of the first affiliated hospital of Xi’an Jiaotong University.
Cell culture
We collected BGC-823, SGC-7901, HGC-27, MGC-803, AGS and MKN45 (human gastric cancer cell lines) and human gastric epithelial cell line, GES-1, from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China). Total cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) (both from Gibco; Thermo Fisher Scientific, Waltham, MA, USA). And the cells were then incubated in a humid atmosphere with 5% CO2 at 37°C.
Cell transfection and treatments
miR-4677-3p mimic and miR-4677-3p inhibitor were synthesized, and pcDNA3.1-CEMIP (CEMIP-p) and CEMIP small interfering RNA (siRNAs), siCEMIP, were constructed by RiboBio (Guangzhou, China). Mimic negative control (NC), inhibitor NC, pcDNA3.1 and NC siRNA were used as controls in our research. All cell lines were cultured in 6-well plates at a density of 5 × 105 cells/well and incubated overnight. The transfection of mimics and vectors was performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA), and incubated for another 48 h in a moist condition with 5% CO2 at 37°C.
PI3K/AKT signaling inhibitors, Akti-1/2 and LY294002, were purchased from MedChemExpress (Shanghai, China). After transfection assay, both Akti-1/2 and LY294002 were added to incubate cells for 24 h (10 µM). Then, cells were collected for the subsequent determinations.
RNA isolation and quantitative RT-PCR
TRIzol® Reagent (Invitrogen, USA) was used to extract the total RNA of samples (including tissues and cell lines). The procedure was performed following the instruction of the RNA extracted kit. Then, cDNAs were generated from the total RNAs by using a PrimeScript RT Master Mix kit (Takara, Dalian, China). Subsequently, a StepOnePlus™ RealTime PCR system (Applied Biosystems, Foster City, CA, USA) was applied to carry out the real-time PCR process using the SYBR Premix Ex Taq™ (Takara Bio, Otsu, Japan). All the primers used for real-time PCR were listed in Table 1. The data of real-time PCR were calculated by the method of 2−ΔΔCt.
Table 1.
The primer sequences for qRT-PCR
| Name | Sequence (5ʹ-3ʹ) |
|---|---|
| miR-4677-3p Forward | CTGTGAGACCAAAGAACTACTCGC |
| miR-4677-3p Reverse | CTCTACAGCTATATTGCCAGCCAC |
| CEMIP Forward | GCTCTGGGATTTAAGGCAGC |
| CEMIP Reverse | ATTGGAGCCATGGACTGTGA |
| U6 Forward | CTCGCTTCGGCAGCA CA |
| U6 Reverse | AACGCTTCACGAA TTTGCGT |
| GADPH Forward | TGCACCACCAACTGCTTAGC |
| GADPH Reverse | GGCATGGACTGTGGTCATGAG |
Western blotting
The tissue samples and cells were lysed using RIPA lysis buffer (Genstar, China). The total proteins were extracted from cell lysates after centrifuging at 4°C 15,000 rpm for 15 min. The concentration of protein was measured using BCA Protein Assay kit (Genstar, China) and the proteins were separated by SDS-PAGE (10% polyacrylamide gels), then transferred onto PVDF membranes (Millipore, Boston, MA, USA). The PVDF membrane was blocked in 5% evaporated skim milk for 1 h, and then incubated in TBST solution which contained the primary antibodies at 4°C overnight. After washing five times in TBST solution, we incubated the membrane again in TBST which containing secondary antibody at room temperature for 1 h and imagined using StarSignal Plus Chemiluminescent Assay Kit (Genstar, China). β-actin and GADPH were used as the internal controls. The primary antibodies used in this study including anti-CEMIP, anti-PI3K, anti-pPI3K, anti-AKT, anti-pAKT, anti-AKT2 and anti-pAKT2. Horseradish peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG (1:1000) were used as secondary antibody in our research.
MTT assay
Cell proliferation was examined by the MTT Cell Proliferation and Cytotoxicity Assay Kit (Beyotime, China). The specific procedure was according to the protocol of manufacturer. In brief, the cultured cells (2 × 104 cells/mL) were incubated in 96-well plates containing DMEM. The incubation was lasted for 0 h, 24 h, 48 h, 72 h, 96 h. Then, MTT solution was supplemented for another incubation (5 mg/mL; 4 h) at 37°C. After discarding the solutions, 150 µL DMSO was used to dissolve formazan crystal. The final OD value was detected at 490 nm on a microplate reader (Applied Biosystems, Shanghai, China). Every test was conducted in triplicate and repeated three times.
Transwell assay
Cell migration was measured by Transwell assay using Transwell chambers (8 mm pore; BD Biosciences, San Jose, CA USA). 1 × 105 cells in 100 mL serum-free medium were put in the upper chambers without Matrigel, and 500 mL medium contained 10% FBS was placed in the lower chambers. After a 24 h-incubation at 37°C, cells on the top chambers were removed. The cells migrated to the lower chamber were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet for 15 min at room temperature, and then counted in at least 3 randomly selected fields under an inverted phase-contrast microscope (Olympus, Tokyo, Japan).
Cell invasion was evaluated by coating the chambers with 50 mL Matrigel (BD Biosciences). 1 × 105 cells in 100 mL serum-free medium were put in the upper chambers coated with Matrigel. 500 mL complete medium contained 10% FBS was placed in the lower chambers. After incubation, cells invading to the lower chamber were counted following the same procedure of cell migration assay.
Luciferase reporter assay
The mutation of CEMIP was introduced into the miR-4677-3p binding site. The wild type (WT) and mutant type (MUT) 3ʹ UTR segments of CEMIP gene were cloned onto the firefly luciferase reporter vector. The constructed vectors and miR-4677-3p or control mimics were co-transfected into the HEK-293 cells using Lipofectamine 2000 (Invitrogen). And the transfected HEK-293 cells were incubated for 24 h, and then the activity of luciferase was measured using the Dual-Luciferase Reporter Assay System (Promega). The experiment was repeated at least three times.
Statistical analysis
All assays were duplicated at least thrice. GraphPad Prism software (version 5; GraphPad software, Inc., La Jolla, CA, USA) and SPSS version 22.0 (SPSS, USA) were used to analyze our results. The data were exhibited as mean ± standard deviations (SD). The statistical analysis of the significance of differences between groups was carried out with the analysis of variance (ANOVA) along with paired two-tailed student t-test. Spearman’s correlation analysis was used to evaluate the correlations between target gene expressions. P values < 0.05 were identified as the statistically significant difference in this study.
Results
miR-4677-3p was downregulated in gastric cancer tissues and cells
Firstly, we examined the expression of miR-4677-3p in gastric cancer tissues and cells by qRT-PCR. The results showed that the expression of miR-4677-3p was significantly downregulated in gastric cancer tissues compared with adjacent normal tissues (Figure 1a). Decreased miR-4677-3p expression was also observed in gastric cancer cell lines, especially BGC-823 and SGC-7901 cells (Figure 1b).
Figure 1.
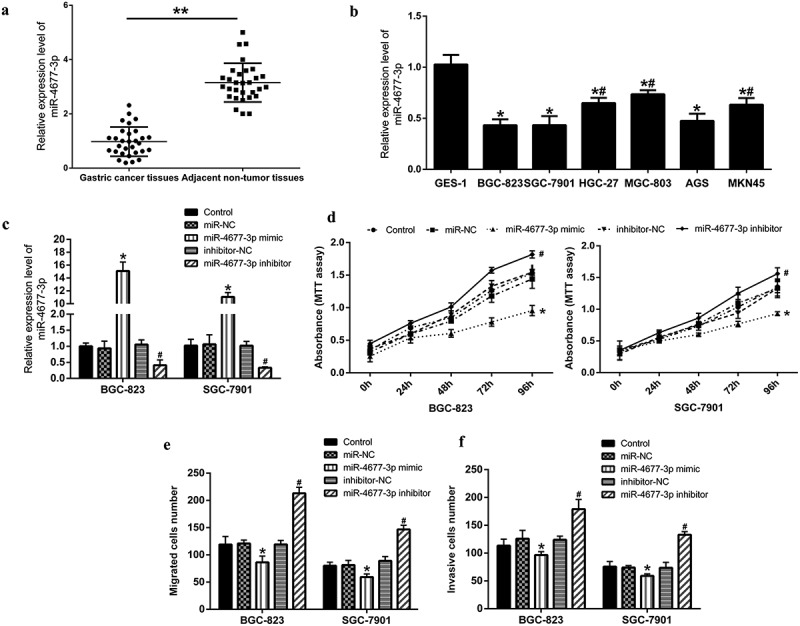
Over-expression miR-4677-3p suppressed the proliferation, migration and invasion of gastric cancer cells. a and b. miR-4677-3p was down-regulated in gastric cancer tissues and cells; in a, ** P < 0.01 compared with gastric cancer tissues; in b, * P < 0.05 compared with GES-1 cells and # P < 0.05 compared with BGC-823 cells. c. Expression of miR-4677-3p was measured after transfected with miR-4677-3p mimic and inhibitor. d. Cell proliferation was detected by MTT assay. e and f. Cell migration and invasion were examined using Transwell assay. * P < 0.05 compared with miR-NC; # P < 0.05 compared with inhibitor-NC
miR-4677-3p regulated the proliferation and metastases of gastric cancer cells
To identify the association between miR-4677-3p and gastric cancer development, miR-4677-3p mimic and inhibitor was transfected into two gastric cancer cell lines (BGC-823 and SGC-7901). We verified that miR-4677-3p was up-regulated by miR-4677-3p mimic and down-regulated by miR-4677-3p inhibitor both in BGC-823 and SGC-7901 (Figure 1c). Furthermore, the results of MTT assay showed that cell proliferation in miR-4677-3p-mimic group was depressed, but promoted in miR-4677-3p inhibitor group (Figure 1d). Through Transwell assay, we found that cell migration and invasion were promoted by miR-4677-3p inhibitor, and inhibited by miR-4677-3p mimic (Figure 1e, f). These evidences turned out that miR-4677-3p regulates proliferation, migration and invasion of gastric cancer cells.
CEMIP was the direct target of miR-4677-3p
In order to further explore the regulatory mechanism of miR-4677-3p in gastric cancer development. Starbase and Targetscan were used to predict the potential target gene of miR-4677-3p. We found that there was a specific binding motif of miR-4677-3p in CEMIP 3ʹUTR region (Figure 2a). Next, CEMIP mRNA wild-type (wt CEMIP 3ʹUTR) or mutant form (mut CEMIP 3ʹUTR) vectors were constructed. The results of luciferase reports showed that miR-4677-3p mimic and wt CEMIP 3ʹUTR co-transfected cell showed significant lower luciferase activity than co-transfected cells in other group (Figure 2b). What’s more, the protein expression of CEMIP in miR-4677-3p inhibitor cells was higher than inhibitor-NC cells, and decreased expression of CEMIP was showed in miR-4677-3p mimic cells (Figure 2c). The CEMIP expression was also detected in human gastric cancer tissues and cell lines by qRT-PCR and western blot. The consequences show that the expression of CEMIP was dramatically elevated in gastric cancer tissues and cells than normal tissues and cells (Figure 2d and e). Spearman’s correlation analysis further proved that the expression of CEMIP was negatively correlated with miR-4677-3p in gastric cancer tissues (Pearson r = −0.5071, P < 0.001) (Figure 2f).
Figure 2.
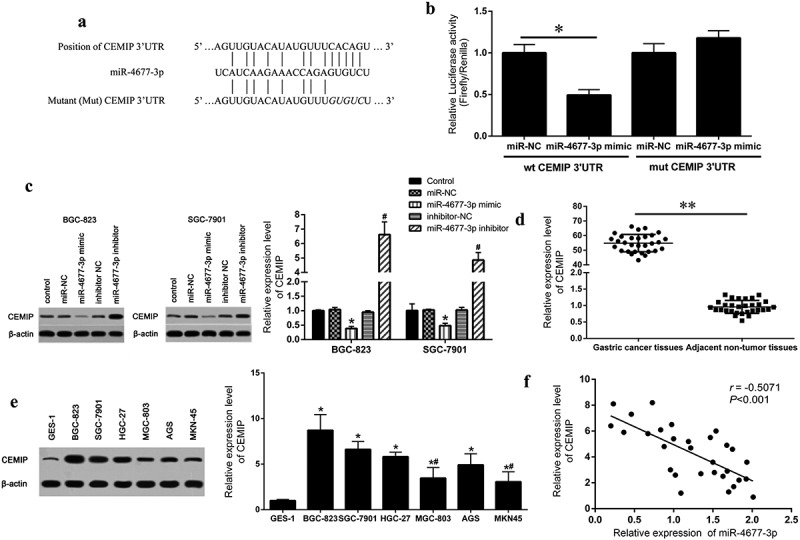
CEMIP was the direct target of miR-4677-3p. a. The binding site between miR‐4677-3p‐3p and CEMIP was predicted on bioinformatic software. b. Luciferase reporter assay was performed by transfecting with WT CEMIP 3ʹUTR or MUT CEMIP 3ʹUTR along with miR-4677-3p mimic or scramble mimic; * P < 0.05. c. CEMIP protein expression in miR-4677-3p mimic- and inhibitor-transfected cells. * P < 0.05 compared with miR-NC and # P < 0.05 compared with inhibitor-NC. d and e. CEMIP expression in gastric cancer tissues and cells was detected using qRT-PCR and western bloting; in d, ** P < 0.01; in e, * P < 0.05 compared with GES-1 and # P < 0.05 compared with BGC-823. f. miR-4677-3p expression was negatively correlated with CEMIP in gastric cancer tissues, based on Pearson’s correlation curve
CEMIP regulated cell proliferation and metastases through PI3K/AKT signaling pathway
Next, we over-expressed/silenced CEMIP in BGC-823 and SGC7901 cell lines, and identified that the expression of CEMIP was increased/decreased (Figure 3a). The results of MTT and Transwell assays implied that silencing CEMIP suppressed the proliferation and metastases of gastric cancer cells, whereas over-expression of CEMIP showed an opposite function (Figure 3b-d). A study verified that PI3K/AKT signaling pathways is regulated by CEMIP [39]. Then, we detected the impact of CEMIP on PI3K/AKT signaling pathways in gastric cancer and found that overexpressing CEMIP increased the ratio of p-PI3K/PI3K, p-AKT/AKT and p-AKT2/AKT2 (Figure 3e). Furthermore, Akti-1/2 and LY294002 (inhibitor in PI3K/AKT signaling pathway) partly reversed the effect of CEMIP overexpression on the activation of PI3K/AKT signaling in BGC-823 cells (Figure 3b-d).
Figure 3.
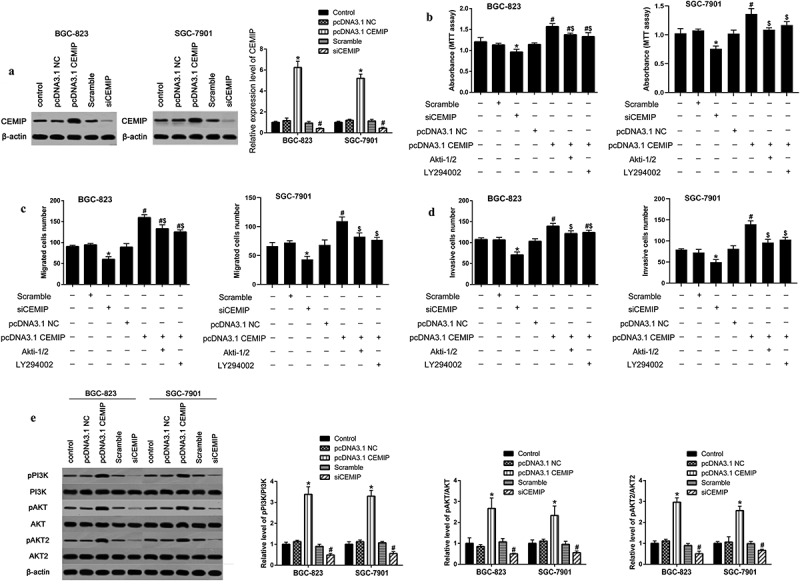
CEMIP regulated cell proliferation and metastasis through PI3K/AKT pathway. a. Protein expression of CEMIP in over-expressing and silencing cell lines; * P < 0.05 compared with pcDNA3.1 NC and # P < 0.05 compared with scramble siRNA. b. Cell proliferation was measured by MTT assay. c and d. Transwell assay was conducted to evaluate cell migration and invasion; * P < 0.05 compared with scramble siRNA, # P < 0.05 compared with pcDNA3.1 NC; $ P < 0.05 compared with pcDNA3.1 CEMIP. e. CEMIP increased the ratio of p-PI3K/PI3K, p-AKT/AKT and p-AKT2/AKT2. * P < 0.05 compared with pcDNA3.1 NC and # P < 0.05 compared with scramble siRNA
miR-4677-3p regulated the development of gastric cancer through CEMIP-PI3K/AKT signaling pathway
Finally, we investigated the involvement of CEMIP-PI3K/AKT signaling pathway in miR-4677-3p regulating the development of gastric cancer. The results implied that the ratio of p-PI3K/PI3K, p-AKT/AKT and p-AKT2/AKT2 were increased in miR-4677-3p inhibitor cell line, while silencing CEMIP counteracted this increasement (Figure 4a). Furthermore, siCEMIP transfection and PI3K/AKT signaling inactivation both partly reversed the contribution of miR-4677-3p inhibitor on cell proliferation, migration and invasion of gastric cancer cells (Figure 4b-d).
Figure 4.
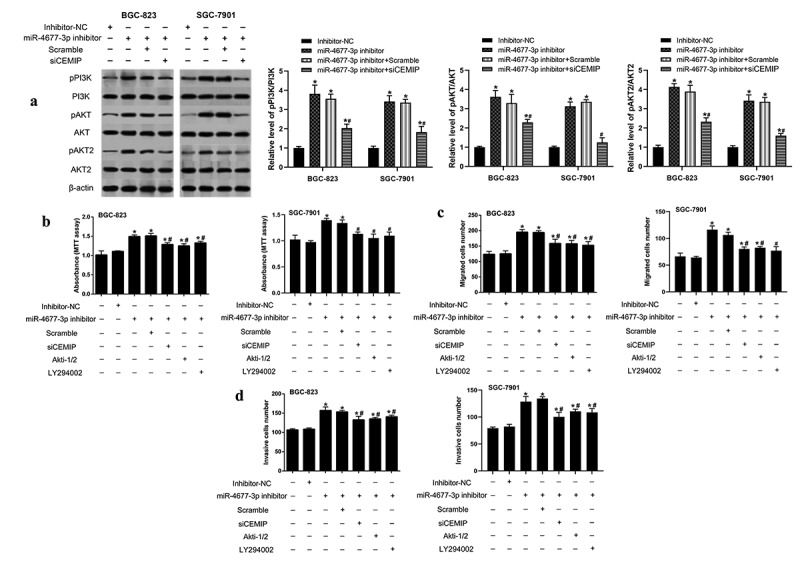
miR-4677-3p regulated the process of gastric cancer through CEMIP-PI3K/AKT signaling pathway. a. In miR-4677-3p inhibitor-transfected cell line, the ratio of p-PI3K/PI3K, p-AKT/AKT and p-AKT2/AKT2 were detected using western blotting. b. MTT assay was performed for the cell proliferation. c and d. Cell migration and invasion abilities of BGC-823 and SGC-7901 cell were measured by Transwell assay. * P < 0.05 compared with inhibitor NC, # P < 0.05 compared with miR-4677-3p inhibitor and scramble group
Discussion
Emerging studies showed that microRNAs were closely related to the development of gastric cancer [40]. Evidence indicated that miR-183-5p.1 expression was elevated in gastric cancer tissues and promoted the proliferation, migration and invasion of gastric cells [41]. The downregulation of miR-339-5p was found in the tissues and was correlated with the unfavorable prognosis of gastric cancer patients, and its overexpression significantly inhibited the malignancy of gastric cancer cells [42]. Recently, the alteration of miR-4677-3p was suggested to play a vital role in various tumors [13,43]. Similarly, the study of Lin and Berillo indicated that the expression of miR-4677-3p was downregulated in peripheral T-cell lymphoma, lung cancer and the early oral squamous cell carcinoma [21,43]. What’s more, miR-4677-3p was also indicated to inhibit cell proliferation, migration and invasion through regulating ZEB1 in lung adenocarcinoma [20]. In the present study, miR-4677-3p was found to be downregulated and decrease the ability of cell proliferation, migration and invasion of gastric cancer cells.
Besides, our results also found that CEMIP was a direct target gene of miR-4677-3p. The expression of CEMIP was negatively correlated with miR-4677-3p in gastric cancer tissues. The regulatory mechanism of miRNAs was illustrated to promote the degradation of target mRNAs or inhibiting gene translation by binding to their 3`UTR [42]. For example, miR-765 was found to target basic leucine zipper ATF-like transcription factor 2, a tumor suppressor, and then regulated the cell apoptosis induced by sensitivity alteration of anticancer drug [44]. Another evidence indicated that sirtuin 1 was the direct downstream target of miR-12129 confirmed by luciferase and rescue assay, and miR-12129/sirtuin 1 axis blocked cell cycle and inhibited cell proliferation during gastric cancer progression [45]. Similarly, the research of Wang and Zhang suggested that miR-29 c-3p and miR-216a directly bond to and inhibited the expression of CEMIP [26,46]. Moreover, CEMIP was reported to accelerate cell survival in breast cancer [23]. Overexpressing CEMIP facilitated the migration and invasion of prostate cancer cells [24]. Another study indicated that CEMIP downregulation suppressed tumor invasion of colorectal cancer [26].
It was well-known that PI3K/AKT signaling pathway played an important role in multiple cancers [47–49]. PI3K, AKT1 and AKT2, the key components of PI3K/AKT signaling pathway, exerted regulatory effects on cellular invasion, apoptosis, survive, metabolism and proliferation [50]. AKT2 was upregulated and promoted the invasion of ovarian cancer cells [51,52]. Numerous reports defined that PI3K, AKT1 and AKT2 were dramatically hyper-phosphorylated in cancer cells compared with normal cells [35,51,52]. Activating the PI3K/AKT pathway accelerated the motility and invasion of gastric cancer cells [53]. Besides, CEMIP could promote the activation of PI3K/AKT pathway and then exacerbated tumor process of ovarian cancer, as well as intrahepatic cholangiocarcinoma [35,39]. Our study also revealed that via activating PI3K/AKT pathway, CEMIP overexpression promoted the pathological progress of gastric cancer.
Taken together, our research suggested that miR-4677-3p was down-regulated in gastric cancer tissues and cells. Inhibiting miR-4677-3p elevated the expression of CEMIP by directly targeting to its 3`UTR, and promoted the proliferation, migration and invasion of gastric cancer cells via activating PI3K/AKT pathway. Generally, miR-4677-3p regulated the processes of gastric cancer cells through CEMIP-PI3K/AKT axis. This may provide a novel evidence for the biomarker target of gastric cancer therapy and prognosis.
Funding Statement
The study was funded by National Natural Science Foundation of China (81602611).
Disclosure statement
The authors declare that there are no competing interests associated with the manuscript.
Author contribution
SXH designed the study. CM, DZ, YRL, MDR, WGM and GFL performed the experiments. DZ and WHM analyzed the data. CM wrote the manuscript.
References
- [1].Diao N, Li Y, Yang J, et al. High expression of HMBOX1 contributes to poor prognosis of gastric cancer by promoting cell proliferation and migration. Biomed Pharmacother. 2019;115:108867. [DOI] [PubMed] [Google Scholar]
- [2].Liao Z, Li Y, Zhou Y, et al. MicroRNA-197 inhibits gastric cancer progression by directly targeting metadherin. Mol Med Rep. 2018;17:602–611. [DOI] [PubMed] [Google Scholar]
- [3].Li C, Li X, Gao S, et al. MicroRNA-133a inhibits proliferation of gastric cancer cells by downregulating ERBB2 expression. Oncol Res Featuring Preclinical Clin Cancer Ther. 2017;25(7):1169–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. [DOI] [PubMed] [Google Scholar]
- [5].Correa P. Gastric cancer: overview. Gastroenterol Clin North Am. 2013;42(2):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jemal A, Bray F, Mm C, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [7].Yan J, Guo X, Xia J, et al. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med Oncol. 2014;31(3):879. [DOI] [PubMed] [Google Scholar]
- [8].Slezak-Prochazka I, Durmus S, Kroesen B-J, et al. MicroRNAs, macrocontrol: regulation of miRNA processing. Rna. 2010;16(6):1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350. [DOI] [PubMed] [Google Scholar]
- [10].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- [11].Xu AJ, Fu LN, Wu HX, et al. MicroRNA‑744 inhibits tumor cell proliferation and invasion of gastric cancer via targeting brain‑derived neurotrophic factor. Mol Med Rep. 2017;16(4):5055–5061. [DOI] [PubMed] [Google Scholar]
- [12].Bartel DP. MicroRNAs: target recognition and regulatory functions. cell. 2009;136(2):215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang Q, Jie Z, Ye S, et al. Genetic variations in miR-27a gene decrease mature miR-27a level and reduce gastric cancer susceptibility. Oncogene. 2014;33(2):193. [DOI] [PubMed] [Google Scholar]
- [14].Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang BG, Li JF, Yu BQ, et al. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol Rep. 2012;27(4):1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nishida N, Mimori K, Fabbri M, et al. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725–2733. [DOI] [PubMed] [Google Scholar]
- [17].Li X, Zhang Y, Zhang H, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9(7):824–833. [DOI] [PubMed] [Google Scholar]
- [18].Lai KW, Koh KX, Loh M, et al. MicroRNA-130b regulates the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer. 2010;46(8):1456–1463. [DOI] [PubMed] [Google Scholar]
- [19].Wang H-J, Ruan H-J, He X-J, et al. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer. 2010;46(12):2295–2303. [DOI] [PubMed] [Google Scholar]
- [20].Zhong Y, Wang J, Lv W, et al. LncRNA TTN‐AS1 drives invasion and migration of lung adenocarcinoma cells via modulation of miR‐4677‐3p/ZEB1 axis. J Cell Biochem. 2019;120(10):17131–17141. [DOI] [PubMed] [Google Scholar]
- [21].Lin Y, Chen W-M, Wang C, et al. MicroRNA profiling in peripheral T-cell lymphoma, not otherwise specified. Cancer Biomarkers. 2017;18(4):339–347. [DOI] [PubMed] [Google Scholar]
- [22].Wang X-D, Lu J, Lin Y-S, et al. Functional role of long non-coding RNA CASC19/miR-140-5p/CEMIP axis in colorectal cancer progression in vitro. World J Gastroenterol. 2019;25(14):1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Banach A, Jiang Y-P, Roth E, et al. CEMIP upregulates BiP to promote breast cancer cell survival in hypoxia. Oncotarget. 2019;10(42):4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang P, Song Y, Sun Y, et al. AMPK/GSK3β/β-catenin cascade–triggered overexpression of CEMIP promotes migration and invasion in anoikis-resistant prostate cancer cells by enhancing metabolic reprogramming. FASEB J. 2018;32(7):3924–3935. [DOI] [PubMed] [Google Scholar]
- [25].Liang G, Fang X, Yang Y, et al. Silencing of CEMIP suppresses Wnt/β-catenin/Snail signaling transduction and inhibits EMT program of colorectal cancer cells. Acta Histochem. 2018;120(1):56–63. [DOI] [PubMed] [Google Scholar]
- [26].Zhang D, Zhao L, Shen Q, et al. Down‐regulation of KIAA1199/CEMIP by miR‐216a suppresses tumor invasion and metastasis in colorectal cancer. Int J Cancer. 2017;140(10):2298–2309. [DOI] [PubMed] [Google Scholar]
- [27].Jia S, Qu T, Wang X, et al. KIAA1199 promotes migration and invasion by Wnt/β-catenin pathway and MMPs mediated EMT progression and serves as a poor prognosis marker in gastric cancer. PLoS One. 2017;12(4):e0175058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Evensen NA, Li Y, Kuscu C, et al. Hypoxia promotes colon cancer dissemination through up-regulation of cell migration-inducing protein (CEMIP). Oncotarget. 2015;6(24):20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fink SP, Myeroff LL, Kariv R, et al. Induction of KIAA1199/CEMIP is associated with colon cancer phenotype and poor patient survival. Oncotarget. 2015;6(31):30500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Evensen NA, Kuscu C, Nguyen H-L, et al. Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J Natl Cancer Inst. 2013;105(18):1402–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Koga A, Sato N, Kohi S, et al. KIAA1199/CEMIP/HYBID overexpression predicts poor prognosis in pancreatic ductal adenocarcinoma. Pancreatology. 2017;17(1):115–122. [DOI] [PubMed] [Google Scholar]
- [32].Shostak K, Zhang X, Hubert P, et al. NF-κB-induced KIAA1199 promotes survival through EGFR signalling. Nat Commun. 2014;5(1):5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mao Y, Zhao Q, Yin S, et al. Genome‐wide expression profiling and bioinformatics analysis of deregulated genes in human gastric cancer tissue after gastroscopy. Asia‐Pacific J Clin Oncol. 2018;14(2):e29–e36. [DOI] [PubMed] [Google Scholar]
- [34].Mayer IA, Arteaga CL. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med. 2016;67(1):11–28. [DOI] [PubMed] [Google Scholar]
- [35].Sang H, Li T, Li H, et al. Gab1 regulates proliferation and migration through the PI3K/Akt signaling pathway in intrahepatic cholangiocarcinoma. Tumor Biol. 2015;36(11):8367–8377. [DOI] [PubMed] [Google Scholar]
- [36].Tapia O, Riquelme I, Leal P, et al. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 2014;465(1):25–33. [DOI] [PubMed] [Google Scholar]
- [37].Riquelme I, Tapia O, Leal P, et al. miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in gastric cancer through regulation of the PI3K/AKT/mTOR pathway. Cell Oncol. 2016;39(1):23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fang W-L, Huang K-H, Lan Y-T, et al. Mutations in PI3K/AKT pathway genes and amplifications of PIK3CA are associated with patterns of recurrence in gastric cancers. Oncotarget. 2016;7(5):6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shen F, Z-h Z, Liu Y, et al. CEMIP promotes ovarian cancer development and progression via the PI3K/AKT signaling pathway. Biomed Pharmacother. 2019;114:108787. [DOI] [PubMed] [Google Scholar]
- [40].Jiang F, Shen XB. miRNA and mRNA expression profiles in gastric cancer patients and the relationship with circRNA. Neoplasma. 2019;66(6):879–886. [DOI] [PubMed] [Google Scholar]
- [41].Jun LJ, Shen J, Yue H, et al. miRNA1835p.1 promotes the migration and invasion of gastric cancer AGS cells by targeting TPM1. Oncol Rep. 2019;42:2371–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang C, Huang Y, Zhang J, et al. MiRNA-339-5p suppresses the malignant development of gastric cancer via targeting ALKBH1. Exp Mol Pathol. 2020;115:104449. [DOI] [PubMed] [Google Scholar]
- [43].Berillo O, Regnier M, Ivashchenko A. Binding of intronic miRNAs to the mRNAs of host genes encoding intronic miRNAs and proteins that participate in tumourigenesis. Comput Biol Med. 2013;43(10):1374–1381. [DOI] [PubMed] [Google Scholar]
- [44].Lin W, Miao Y, Meng X, et al. miRNA-765 mediates multidrug resistance via targeting BATF2 in gastric cancer cells. FEBS Open Bio. 2020;10(6):1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang W, Liao K, Liu D. MiRNA-12129 suppresses cell proliferation and block cell cycle progression by targeting SIRT1 in GASTRIC cancer. Technol Cancer Res Treat. 2020;19:153303382092814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang L, Yu T, Li W, et al. The miR-29c-KIAA1199 axis regulates gastric cancer migration by binding with WBP11 and PTP4A3. Oncogene. 2019;38(17):3134. [DOI] [PubMed] [Google Scholar]
- [47].Gao Y, Xiao X, Zhang C, et al. Melatonin synergizes the chemotherapeutic effect of 5‐fluorouracil in colon cancer by suppressing PI 3K/AKT and NF‐κB/iNOS signaling pathways. J Pineal Res. 2017;62(2):e12380. [DOI] [PubMed] [Google Scholar]
- [48].Li W, Ma J, Ma Q, et al. Resveratrol inhibits the epithelial-mesenchymal transition of pancreatic cancer cells via suppression of the PI-3K/Akt/NF-κB pathway. Curr Med Chem. 2013;20(33):4185–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xing H, Weng D, Chen G, et al. Activation of fibronectin/PI-3K/Akt2 leads to chemoresistance to docetaxel by regulating survivin protein expression in ovarian and breast cancer cells. Cancer Lett. 2008;261(1):108–119. [DOI] [PubMed] [Google Scholar]
- [50].Wang Q, Chen X, Hay N. Akt as a target for cancer therapy: more is not always better (lessons from studies in mice). Br J Cancer. 2017;117(2):159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hers I, Vincent EE, Tavaré JM. Akt signalling in health and disease. Cell Signal. 2011;23(10):1515–1527. [DOI] [PubMed] [Google Scholar]
- [52].Noske A, Kaszubiak A, Weichert W, et al. Specific inhibition of AKT2 by RNA interference results in reduction of ovarian cancer cell proliferation: increased expression of AKT in advanced ovarian cancer. Cancer Lett. 2007;246(1–2):190–200. [DOI] [PubMed] [Google Scholar]
- [53].Kang MH, Kim JS, Seo JE, et al. BMP2 accelerates the motility and invasiveness of gastric cancer cells via activation of the phosphatidylinositol 3-kinase (PI3K)/Akt pathway. Exp Cell Res. 2010;316(1):24–37. [DOI] [PubMed] [Google Scholar]


