Abstract
Objective
The aim of this study was to use whole-genome sequencing to characterize Klebsiella pneumoniae SKp2F and Klebsiella variicola SKv2E, both carrying blaKPC, co-isolated from the same sputum specimen.
Methods
Antimicrobial susceptibility testing was performed using microbroth dilution. Biofilm formation was determined by crystal violet staining and virulence was measured by a serum killing assay. Whole-genome sequencing of SKp2F and SKv2E was performed using an Illumina sequencer and the genetic characteristics were analyzed by computer.
Results
SKp2F and SKv2E were sensitive only to tigecycline and polymyxin among the tested antibiotics. The biofilm-forming ability of SKv2E is stronger than that of SKp2F. The grades of serum resistance of SKp2F and SKv2E are 4 and 3. MLST analysis of the 6,115,610 bp and 5,403,687 bp of SKv2E and SKp2F showed associations with ST1615 and ST631, respectively. SKv2E carried 13 resistance genes (blaKPC-2, blaTEM-1A, blaLEN17, aadA16, arr-3, qnrB4, oqxA/B, dfrA27, sul1, tetD, fosA, qacEΔ1) and SKp2F carried 23 (blaKPC-2, blaCTX-M-3, blaTEM-1B, blaCTX-M-65, blaSHV-27, aac(6ʹ)-IIa, rmtB, arr-3, aph(3ʹ)-Ia, aadA16, qnrS1, aac(6ʹ)-Ib-cr, qnrB91, oqxA/B, mph(A), tet(A), fosA, dfrA27, and two copies of qacEΔ1-sul1). Most of them were carried by various mobile genetic elements, such as IncFIB(K)/IncFII(K)/IncFII(Yp), IncFII(K) plasmid, Tn6338, and In469. Both SKv2E and SKp2F carried a large number of virulence factors, including type 1 and 3 fimbriae, capsule, aerobactin (iutA), ent siderophore (entABCDEFS, fepABCDGfes), and salmochelin (iroE/iroEN). SKv2E also carried type IV pili (pilW), fimbrial adherence (steB, stfD), and capsule biosynthesis gene (glf).
Conclusion
blaKPC-2-carrying K. variicola and K. pneumoniae, which carried multiple resistance genes, virulence factors, and highly similar mobile genetic elements, were identified from the same specimen, indicating that clinical samples may carry multiple bacteria. We should avoid misidentification, and bear in mind that resistance genes carrying mobile genetic elements can be transmitted or integrated between bacteria in the same host.
Keywords: Klebsiella variicola, Klebsiella pneumoniae, carbapenem-resistant Enterobacteriaceae, CRE, bla KPC
Introduction
Carbapenemase-producing Enterobacteriaceae (CPE) have become a global concern owing to their ability to hydrolyze carbapenems and most β-lactam antibiotics, posing a serious threat to human health and a significant challenge to clinical treatment.1,2 The Klebsiella pneumoniae carbapenemase (KPC) and metallo-β-lactamases are the two major groups of carbapenemases produced by the most of the carbapenemase-resistant Enterobacteriaceae (CRE) strains, because they carry the carbapenemase code genes such as blaKPC and blaNDM.3–6
The blaKPC and blaNDM gene-carrying strains always co-harbor many other types of resistance genes, such as extended-spectrum β-lactamase (ESBL) genes (blaCTX-M, blaSHV, and blaTEM), fluoroquinolone resistance genes (qnrA, qnrB, qnrS, andoqxA/B), and aminoglycoside resistance genes (rmtA, rmtB, and rmtC), resulting in high resistance to almost all kinds of commonly used antibiotics.7–10 These notorious resistance genes are usually carried by various mobile genetic elements, such as plasmids, integrons, and transposons, which can be transmitted between intraspecific or interspecific microorganisms.11–13 In recent years, there has been a high incidence of co-infection with more than two different multi-drug-resistant bacteria in the same patient, which brings a serious threat to patients14–17 because the variety of bacteria in the co-infection can be misdiagnosed or misidentified.18,19 For example, many types of Klebsiella species or subspecies (eg, Klebsiella variicola, Klebsiella quasipneumoniae subsp. quasipneumoniae, Klebsiella quasipneumoniae subsp. similipneumoniae, Klebsiella quasivariicola, Klebsiella africanensis, and Klebsiella variicola subsp. tropicalensis) have been identified and reported, which make up the Klebsiella pneumoniae complex. However, in more and more reports of K. pneumoniae infection, in recent years, cases of Klebsiella variicola infection are increasingly being found.18 Because of the morphological similarity between species in the K. pneumoniae complex, some Klebsiella species are always misidentified as K. pneumoniae.20,21 Klebsiellapneumoniae is an opportunistic pathogen that can lead to serious hospital infection and community-acquired infections. Klebsiella variicola is also an opportunistic pathogen, responsible for infections such as blood infections, respiratory tract infections, and urinary tract infections (UTIs), and blood infection caused by K. variicola has a higher mortality rate than that caused by K. pneumoniae.22 This tells us that a precise diagnosis is important for infection control.
Here, we report and characterize K. variicola and K. pneumoniae strains that were co-isolated from a sputum sample of a female inpatient, which both carried the carbapenemase-producing gene blaKPC.
Materials and Methods
Bacteria Isolation, Identification, and Antimicrobial Susceptibility Testing
Klebsiella variicola strain SKv2E and Klebsiella pneumoniae SKp2F were isolated from the same sputum specimen of a 69-year-old female patient, who was admitted with chronic obstructive pulmonary disease and pulmonary infection to the Department of Respiratory Medicine at The Second Affiliated Hospital of Xiamen Medical College, in November 2020. The species were identified using the VITEK 2 compact system and 16S rRNA and rpoB sequencing.20 The results of the 16S rRNA and rpoB sequencing displayed overlapping peaks,17,20 indicating the co-existence of two or more types of bacteria. Thereafter, we purely cultured the colony and chose five colonies randomly to sequence again, which finally confirmed the presence of K. variicola strain SKv2E and K. pneumoniae SKp2F.
In vitro, antimicrobial susceptibility testing of SKv2E and SKp2F against antimicrobial agents (OXOID), including ampicillin, aztreonam, ceftazidime, ciprofloxacin, ceftriaxone, cefuroxime, cefepime, gentamicin, imipenem, meropenem, polymyxin B, sulfamethoxazole-trimethoprim, and tigecycline, was performed by a broth microdilution method, according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI, M100-S27) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/).
String, Biofilm Formation Assay, and Serum Killing Activity Testing
To test the mucoviscosity phenotype, the colony of strains SKv2E and SKp2F was cultured on a blood agar plate overnight at 37°C for 24 hours, stretched by an inoculating loop. The strain formed a viscous string of >5 mm which was designated as mucoviscous. The biofilm formation assay was conducted according to our previous method.23 To address the virulence of the two strains, the human serum killing activity was defined using a previously described method.6
Whole Genome Sequencing and Analysis
Genomic DNA of K. variicola strain SKv2E and K. pneumoniae strain SKp2F was extracted using a DNA extraction kit (Sangong, China). The 300-bp paired-end library was constructed using the standard Illumina DNA sample preparation instructions. Then, it was sequenced on an Illumina MiSeq systems sequencer (Majorbio, China). The readings were assembled de novo and gene prediction was performed with a Glimmer 3.02 (http://www.cbcb.umd.edu/software/glimmer/). Annotation of the K. variicola SKv2E and K. pneumoniae SKp2F genomes was achieved using the NCBI Prokaryotic Genome Annotation Pipeline. The pairwise alignment was performed by a blast search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The resistome was identified using ResFinder 2.1 (https://cge.cbs.dtu.dk/services/ResFinder/) (minimum threshold for identity, 85%; minimum coverage, 60%).24 The virulence factors were predicted using the VFanalyzer of VFDB (http://www.mgc.ac.cn/VFs/).25
Conjugation Assay
To determine whether the blaKPC was carried by a conjugative plasmid, K. variicola SKv2E and K. pneumoniae SKp2F were cultured in Luria–Bertani (LB) broth as the donor, and azide-resistant E. coli strain J53 was used as the recipient. The transconjugants were selected on LB agar plates containing sodium azide (100 μg mL−1) and meropenem (1 μg mL−1). The presence of the blaKPC resistance gene in transconjugants was confirmed by PCR.6 The antimicrobial susceptibility of transconjugants was determined by the microbroth dilution method.14 The replicon F of the transconjugants was determined according to the previous method, based on the whole genome sequencing (WGS) analysis.26
Results
In Vitro Assay of Antimicrobial Susceptibility, Hypermucoviscosity, Biofilm, and Serum Resistance Assay
As shown in Table 1, SKv2E and SKp2F were resistant to all of the test antibiotics except for polymyxin B and tigecycline. String testing showed that SKv2E and SKp2F were non-hypermucoviscous strains. The two strains were biofilm-forming isolates, with SKv2E and SKp2F having optical density values (OD595) of 1.93 and 1.65, respectively. In the serum killing assay, the grades of SKv2E and SKp2F were 4 and 3, respectively (Table 2).
Table 1.
Determination of Minimum Inhibitory Concentration (MIC) for K. variicola SKv2E and K. pneumoniae SKp2F and Their blaKPC Transconjugants
| MIC (μg/mL) | AMP | ATM | CAZ | CIP | CRO | CXM | SAM | FEP | GEN | IPM | MEM | SXT | TGC | PB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SKv2E | ≥32 | ≥32 | ≥64 | ≥4 | ≥16 | ≥32 | ≥32 | ≥64 | ≥16 | ≥8 | ≥8 | ≥160 | 0.5 | 1 |
| SKp2F | ≥32 | ≥32 | ≥64 | ≥4 | ≥16 | ≥32 | ≥32 | ≥64 | ≥16 | ≥8 | ≥8 | ≥320 | 0.5 | 0.5 |
| J53-pSKv2E-blaKPC | ≥32 | ≥32 | ≥32 | ≥4 | ≥16 | ≥32 | ≥32 | ≥32 | ≥16 | ≥8 | ≥8 | 160 | 0.25 | 1 |
| J53-pSKp2F-blaKPC | ≥32 | ≥32 | ≥32 | ≥4 | ≥16 | ≥32 | ≥32 | ≥32 | ≥16 | ≥8 | ≥8 | 160 | 0.25 | 0.5 |
| J53 | 1 | 0.125 | 0.25 | 0.125 | 1 | 0.25 | 0.5 | 0.25 | 0.5 | 0.125 | 0.25 | 4 | 0.125 | 0.5 |
Abbreviations: AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CRO, ceftriaxone; CXM, cefuroxime; SAM, ampicillin–sulbactam; FEP, cefepime; GEN, gentamicin; IPM, imipenem; MEM, meropenem; SXT, sulfamethoxazole–trimethoprim; TGC, tigecycline; PB, polymyxin B.
Table 2.
Genome Characteristics of K. variicola SKv2E and K. pneumoniae SKp2F
| Isolate | SKv2E | SKp2F |
|---|---|---|
| Genome length (bp) | 6,115,610 | 5,403,687 |
| No of scaffolds | 126 | 45 |
| No of tRNA | 79 | 84 |
| No of rRNA | 15 | 14 |
| No of ncRNA | 10 | 14 |
| No of CDs | 5089 | 5231 |
| MLST | ST1615 | ST631 |
| Resistance genes | blaKPC-2, blaTEM-1A, blaLEN17, aadA16, arr-3, qnrB4, oqxA/B, dfrA27, sul1, tetD, fosA, qacEΔ1 | blaKPC-2, blaCTX-M-3, blaTEM-1B, blaCTX-M-65, blaSHV-27, aac(6ʹ)-IIa, rmtB, arr-3, aph(3ʹ)-Ia, aadA16, qnrS1, aac(6ʹ)-Ib-cr, qnrB91, oqxA/B, mph(A), tet(A), fosA, dfrA27, two copies of qacEΔ1-sul1 |
| Grade of human serum resistance | 4 | 3 |
| String testing | Non-hypermucoviscous | Non-hypermucoviscous |
| Mean biofilm formation (OD595) | 1.93 | 1.65 |
| Plasmid replicons | Col(pHAD28), Col440I, IncFIB(K), IncFII(K), IncFII(Yp), IncHI1B | IncFII(K) |
Genome Characteristics of Strains SKv2E and SKp2F
The assembled WGS of K. variicola SKv2E and K. pneumoniae SKp2F produced 126 and 45 scaffolds, respectively, which resulted in estimated draft genomes 6,115,610 bp and 5,403,687 bp in length, with a total of 5130 and 4740 coding sequences (Table 2). Multi-locus sequence typing (MLST) analysis of the WGS data indicated that SKv2E belongs to ST1615, while SKp2F was found to be associated with ST631.
Resistome and Virulence Factors of SKv2E and SKp2F
The WGS data confirmed the presence of blaKPC-2 carried by SKv2E; in addition, other resistance genes related to resistance to β-lactams (blaTEM-1A, blaLEN17), aminoglycosides (aadA16, arr-3), fluoroquinolones (qnrB4, oqxA/B), trimethoprim (dfrA27, sul1), tetracycline (tetD), fosfomycin (fosA), and benzylkonium (qacEΔ1) were identified. SKp2F also carried blaKPC-2, along with other resistance genes related to resistance to β-lactams (blaCTX-M-3, blaKPC-2, blaTEM-1B, blaCTX-M-65, blaSHV-27), aminoglycosides (aac(6ʹ)-IIa, rmtB, aph(3ʹ)-Ia, aadA16), fluoroquinolones (qnrS1, aac(6ʹ)-Ib-cr, qnrB91, and oqxA/B), phenicol (floR), rifamycin (arr-3), macrolide (mph(A)), tetracycline (tet(A)), fosfomycin (fosA), and trimethoprim (dfrA27, sul1). Furthermore, both SKv2E and SKp2F carried a large number of virulence factors, including type 3 fimbriae (mrkABCDFHIJ), type 1 fimbriae (fimABCDEFGHIK), capsule coding genes, rscAB (virulence regulation genes), aerobactin (iutA), ent siderophore (entABCDEFS and fepABCDGfes), and salmochelin (iroE/iroEN). Type IV pili (pilW), fimbrial adherence determinants (steB, stfD), and capsule biosynthesis and transport genes (glf) were also identified from SKv2E (Supplementary Table S1).
Plasmid Transferability of blaKPC-2
Conjugation assays showed that both of the blaKPC genes were successfully transferred to azide-resistant E. coli J53. It was found that the blaKPC genes were carried by plasmid, designated pSKv2E-KPC and pSKp2F-KPC, respectively. The transconjugants of SKv2E and SKp2F were named J53-pSKv2E-blaKPC and J53-pSKp2F-blaKPC. Plasmid replicon typing showed that the replicons of the blaKPC-carrying plasmid of SKv2E are IncFIB(K), IncFII(K), and IncFII(Yp), and the replicon of SKp2F is IncFII(K).
Genetic Context of the Resistance Gene-Carrying Regions
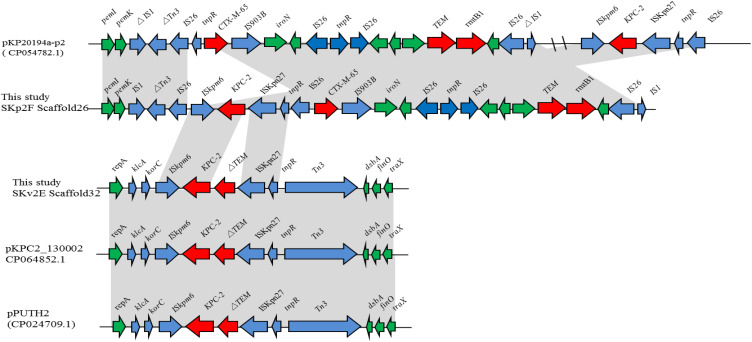
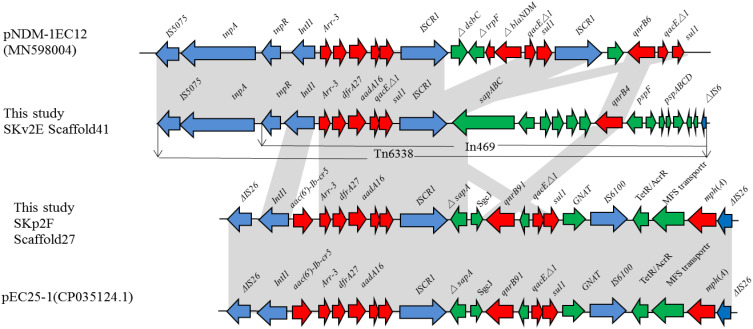
For K. variicola strain SKv2E, 10 of the 13 resistance genes were carried by scaffold27, scaffold32, and scaffold41. Sequence analysis showed that blaKPC was found in the 65,049-bp-long scaffold32, with a G+C content of 54.16%. The blaKPC gene was carried by klcA-korC-ISKpn6-blaKPC-2-blaTEM-ISKpn27-Tn3. This region is the same blaKPC-carrying region in plasmids pPUTH2 (CP024709.1) and pKPC2_130002 (CP064852.1) (Figure 1). Furthermore, the 44,418-bp-long scaffold41 of SKv2E carried six resistance genes (qnrB4, arr-3, dfrA27, qacEΔ1, sul1, and aadA16), which were harbored by the partial integron In469 (Figure 2).
Figure 1.
Schematic mapping of the genetic characteristics of the resistance gene (blaKPC-2)-carrying region in strain K. variicola SKv2E and K. pneumoniae SKp2F. The construction of the sequence comparison was performed using blast (http://blast.ncbi.nlm.nih.gov). Genes are shown as arrows, and their orientations of transcription are indicated by the arrowheads.
Figure 2.
Schematic mapping of the genetic characteristics of resistance gene (arr-3, dfrA27, aadA16, qnrB)-carrying region in strain K. variicola SKv2E and K. pneumoniae SKp2F. The construction of the sequence comparison was performed using blast (http://blast.ncbi.nlm.nih.gov). Genes are shown as arrows, and their orientations of transcription are indicated by the arrowheads.
For K. pneumoniae strain SKp2F, the WGS data confirmed that blaKPC was found in the 21,330-bp-long scaffold26, along with several other resistance genes (blaCTX-M-65, blaTEM-1, rmtB), with a G+C content of 51.77%. The blaKPC gene-carrying context (ISKpn6-blaKPC-2–ISKpn27) and (blaCTX-M-65, blaTEM-1, rmtB) gene-harboring regions were both the same as the corresponding region of plasmid pKP20194a-p2 (CP054782.1). The 16,914-bp-long scaffold27 of SKp2F carried 10 resistance genes (arr-3, dfrA27, aadA16, aac(6ʹ)-Ib-cr, qnrB91, mph(A), and two copies of qacEΔ1-sul1). The linear structure of this resistance gene-carrying region is similar to several plasmids, such as pKSH203-CTX-M-3 (CP034325.1), pEC25-1 (CP035124.1), pM297-1.2 (CP051492.1), and pHC139-5copy (CP061843.1) (Figure 2). Furthermore, the resistance region (IntI1-aac(6ʹ)-Ib-cr5-arr-3-dfrA27-aadA16-qacEΔ1-sul1-ISCR1) in this scaffold was similar to the arr-3, dfrA27, aadA16, qacEΔ1-sul1-carrying scaffold41 of SKv2E, with the difference of a resistance gene aac(6ʹ)-Ib-cr5 inserted between IntI1 and arr-3 (Figure 2). Four other resistance genes (tet(A), floR, blaTEM-1B, and blaCTX-M-3) were carried by the 14,502-bp-long scaffold28, which is the same as in many plasmids, such as pHKU49_CIP (MN543570.1) and pRGF99-1-75k (CP075554.1).
Discussion
Misidentification of bacterial infections from the same sample is a serious problem, which often affects the infection control and the therapeutic outcome.17,20 In recent years, several Klebsiella species or subspecies (eg, K. variicola, K. quasipneumoniae subsp., K. quasivariicola, and K. africanensis) have been increasingly identified from clinical samples.18 Because of the morphological similarity between these Klebsiella species, some other non-K. pneumoniae species are being misidentified as K. pneumoniae.20,21 It is well known that these Klebsiella species, as well as K. pneumoniae, are opportunistic pathogens responsible for infections, and blood infection has also been shown to be caused by other Klebsiella species; for example, K. variicola has a higher pathogenicity than K. pneumoniae.22 This tells us that precise diagnosis is important in infection control. In this study, we isolated K. variicola and K. pneumoniae, which both carry blaKPC and other resistance genes, from the same patient using the VITEK 2 compact system and 16S rRNA and rpoB sequencing.
These Klebsiella species carry many types of carbapenemase-coding genes, such as blaKPC, blaNDM, and blaOXA48, leading to resistance to most commonly used antimicrobial agents and which causing serious threats to public health.5,27–31 With no exception for K. variicola SKv2E and K. pneumoniae SKp2F, antimicrobial susceptibility testing showed that these two strains were resistant to most commonly used antibiotics, such as the β-lactam antibiotics, fluoroquinolones, aminoglycosides, and others. For the virulence assay, we proved that K. variicola SKv2E has a higher pathogenicity than K. pneumoniae SKp2F via human serum killing testing, which was similar to previous research.20 This is because the type IV pili coding gene (pilW), colonization and immune evasion gene (glf), and fimbrial adherence determinant genes (steB, stfD) were determined from SKv2E, which may increase the grade of serum resistance or virulence.32,33 In addition, we identified the gene pilW, which encodes type IV pili, from K. variicola SKv2E, which may be beneficial to the formation of biofilm,34,35 and this may be a reason for K. variicola SKv2E having stronger biofilm-forming capability than K. pneumoniae SKp2F.
The transmission of antibiotic resistance genes and/or virulence factors by various mobile genetic elements (plasmids, integrons, and transposons)36,37 among the bacterial community is one of the major threats to human health. In this study, we found that the carbapenemase-coding blaKPC genes of SKv2E and SKp2F were carried by similar linear structures, ISKpn6-blaKPC-2-blaTEM-ISKpn27 and ISKpn6-blaKPC-2-ISKpn27, which had a high incidence in the blaKPC-carrying Klebsiella isolates.38–40 Moreover, other resistance genes (arr-3, dfrA27, aadA16, and qacEΔ1-sul1) were carried by the transposon Tn6338 and were confirmed in the genomes of both SKv2E and SKp2F. These results indicate that resistance genes carrying mobile genetic elements can be transmitted or integrated between bacteria in the same host.
Conclusions
We identified blaKPC-harboring K. variicola and K. pneumoniae from the same sample, and both carried multiple resistance genes, virulence factors, and various mobile genetic elements. Our results demonstrate that we should pay more attention to the bacteria identified. We also found that some mobile genetic elements from K. variicola and K. pneumoniae were highly similar. This indicates that these resistance genes carrying mobile genetic elements can be transmitted or integrated between bacteria in the same host.
Funding Statement
This research was funded by the grant from the Sichuan Province Science and Technology Project (2020YJ0338), the Science and Technology Strategic Cooperation Programs of Luzhou Municipal People’s Government, and Southwest Medical University (2020LZXNYDJ47).
Nucleotide Sequence Accession Numbers
These Whole Genome Shotgun projects have been deposited in DDBJ/EMBL/GenBank under the sequence accession numbers JAHRXK000000000 and JAHRXL000000000 for Klebsiella pneumoniae strain SKp2F and Klebsiella variicola strain SKv2E, respectively.
Ethical Approval
This study was conducted after agreement from the local ethics committee (no. 20180309059) and with the patient’s informed consent.
Disclosure
The authors have no conflicts of interest to declare.
References
- 1.Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, Pascual A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing enterobacteriaceae. Clin Microbiol Rev. 2018;31(2):e00079–e00117. doi: 10.1128/CMR.00079-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang F, Ding M, Yan X, et al. Carbapenem-resistant K. pneumoniae exhibiting clinically undetected amikacin and meropenem heteroresistance leads to treatment failure in a murine model of infection. Microb Pathog. 2021;160:105162. doi: 10.1016/j.micpath.2021.105162 [DOI] [PubMed] [Google Scholar]
- 3.Bonomo RA, Burd EM, Conly J, et al. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis. 2018;66(8):1290–1297. doi: 10.1093/cid/cix893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791–1798. doi: 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu L, Wang S, Zhang Z, et al. Whole genome sequence of bla(NDM) and bla(KPC) co-producing Klebsiella pneumoniae isolate KSH203 with capsular serotype K25 belonging to ST11 from China. J Glob Antimicrob Resist. 2020;20:272–274. doi: 10.1016/j.jgar.2020.01.006 [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Zhang J, Li Y, et al. Diversity and frequency of resistance and virulence genes in blaKPC and blaNDM co-producing Klebsiella pneumoniae strains from China. Infect Drug Resist. 2019;12:2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Zhang H, Zhang X, et al. Characterization of an NDM-19-producing Klebsiella pneumoniae strain harboring 2 resistance plasmids from China. Diagn Microbiol Infect Dis. 2019;93(4):355–361. doi: 10.1016/j.diagmicrobio.2018.11.007 [DOI] [PubMed] [Google Scholar]
- 8.Di Tella D, Tamburro M, Guerrizio G, Fanelli I, Sammarco ML, Ripabelli G. Molecular epidemiological insights into colistin-resistant and carbapenemases-producing clinical Klebsiella pneumoniae isolates. Infect Drug Resist. 2019;12:3783–3795. doi: 10.2147/IDR.S226416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu R, Li Q, Zhang F, Ding M, Liu J, Zhou Y. Characterisation of bla(NDM-5) and bla(KPC-2) co-occurrence in K64-ST11 carbapenem-resistant Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;27:63–66. doi: 10.1016/j.jgar.2021.08.009 [DOI] [PubMed] [Google Scholar]
- 10.Tang L, Huang J, She J, Zhao K, Zhou Y. Co-Occurrence of the bla (KPC-2) and Mcr-3.3 gene in aeromonas caviae SCAc2001 isolated from patients with diarrheal disease. Infect Drug Resist. 2020;13:1527–1536. doi: 10.2147/IDR.S245553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopotsa K, Osei Sekyere J, Mbelle NM. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann NY Acad Sci. 2019;1457(1):61–91. doi: 10.1111/nyas.14223 [DOI] [PubMed] [Google Scholar]
- 12.Kizny Gordon A, Phan HTT, Lipworth SI, et al. Genomic dynamics of species and mobile genetic elements in a prolonged blaIMP-4-associated carbapenemase outbreak in an Australian hospital. J Antimicrob Chemother. 2020;75(4):873–882. doi: 10.1093/jac/dkz526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Q, Yin Z, Zhao Y, et al. Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int J Antimicrob Agents. 2017;49(6):709–718. doi: 10.1016/j.ijantimicag.2017.01.021 [DOI] [PubMed] [Google Scholar]
- 14.Ding M, Shi J, Ud Din A, et al. Co-infections of two carbapenemase-producing Enterobacter hormaechei clinical strains isolated from the same diabetes individual in China. J Med Microbiol. 2021;70(3):001316. doi: 10.1099/jmm.0.001316. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, He F. Genomic analysis of two bacterial strains co-isolated from a urinary tract infection: NDM-1-producing Enterobacter cloacae accompanied by extended-spectrum β-lactamase-producing Escherichia coli. J Glob Antimicrob Resist. 2019;17:198–200. doi: 10.1016/j.jgar.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 16.Karad DD, Somani Y, Khande H, Yadav B, Kharat AS. Molecular characterization of a multidrug-resistant/pandrug-resistant nosocomial polymicrobial infection with Klebsiella pneumoniae, Providencia rettgeri, and Acinetobacter baumannii from Rural Maharashtra, India. Acta Biochim Pol. 2020;67(3):387–392. [DOI] [PubMed] [Google Scholar]
- 17.Garza-Ramos U, Moreno-Dominguez S, Hernández-Castro R, et al. Identification and characterization of imipenem-resistant Klebsiella pneumoniae and Susceptible Klebsiella variicola isolates obtained from the same patient. Microb Drug Resist. 2016;22(3):179–184. doi: 10.1089/mdr.2015.0181 [DOI] [PubMed] [Google Scholar]
- 18.Potter RF, Lainhart W, Twentyman J, et al. Population structure, antibiotic resistance, and uropathogenicity of Klebsiella variicola. mBio. 2018;9(6);e02481–e02518. doi: 10.1128/mBio.02481-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fontana L, Bonura E, Lyski Z, Messer W. The brief case: Klebsiella variicola-identifying the misidentified. J Clin Microbiol. 2019;57(1);e00826–e00918. doi: 10.1128/JCM.00826-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez J, Martínez L, Rosenblueth M, Silva J, Martínez-Romero E. How are gene sequence analyses modifying bacterial taxonomy? The case of Klebsiella. Int Microbiol. 2004;7(4):261–268. [PubMed] [Google Scholar]
- 21.Brisse S, Passet V, Grimont PAD. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol. 2014;64(Pt 9):3146–3152. [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez-Medina N, Barrios-Camacho H, Duran-Bedolla J, Garza-Ramos U. Klebsiella variicola: an emerging pathogen in humans. Emerg Microbes Infect. 2019;8(1):973–988. doi: 10.1080/22221751.2019.1634981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu L, Huang M, Zhang XZ, et al. Frequency of virulence factors in high biofilm formation blaKPC-2 producing Klebsiella pneumoniae strains from hospitals. Microb Pathog. 2018;116:168–172. doi: 10.1016/j.micpath.2018.01.030 [DOI] [PubMed] [Google Scholar]
- 24.Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. doi: 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47(D1):D687–D692. doi: 10.1093/nar/gky1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elisa E, Carloni F, Francesca E, et al. Comparative analysis of the standard PCR-Based Replicon Typing (PBRT) with the commercial PBRT-KIT. Plasmid. 2017;90:10–14. doi: 10.1016/j.plasmid.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 27.Han R, Shi Q, Wu S, et al. Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314. doi: 10.3389/fcimb.2020.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25(4):682–707. doi: 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi: 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 30.Nasri E, Subirats J, Sànchez-Melsió A, Mansour HB, Borrego CM, Balcázar JL. Abundance of carbapenemase genes (bla(KPC), bla(NDM) and bla(OXA-48)) in wastewater effluents from Tunisian hospitals. Environ Pollut. 2017;229:371–374. doi: 10.1016/j.envpol.2017.05.095 [DOI] [PubMed] [Google Scholar]
- 31.Ripabelli G, Sammarco ML, Scutellà M, Felice V, Tamburro M. Carbapenem-Resistant KPC- and TEM-producing Escherichia coli ST131 isolated from a hospitalized patient with urinary tract infection: first isolation in Molise Region, Central Italy, July 2018. Microb Drug Resist. 2020;26(1):38–45. doi: 10.1089/mdr.2019.0085 [DOI] [PubMed] [Google Scholar]
- 32.Godlee C, Cerny O, Durkin CH, Holden DW. SrcA is a chaperone for the Salmonella SPI-2 type three secretion system effector SteD. Microbiology. 2019;165(1):15–25. doi: 10.1099/mic.0.000732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esmailnia E, Amani J, Gargari SLM. Identification of novel vaccine candidate against Salmonella enterica serovar Typhi by reverse vaccinology method and evaluation of its immunization. Genomics. 2020;112(5):3374–3381. doi: 10.1016/j.ygeno.2020.06.022 [DOI] [PubMed] [Google Scholar]
- 34.Musafer HK, Kuchma SL, Naimie AA, Schwartzman JD, Al-Mathkhury HJ, O’Toole GA. Investigating the link between imipenem resistance and biofilm formation by Pseudomonas aeruginosa. Microb Ecol. 2014;68(1):111–120. doi: 10.1007/s00248-013-0361-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuchma SL, Griffin EF, O’Toole GA. Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J Bacteriol. 2012;194(19):5388–5403. doi: 10.1128/JB.00899-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3(9):722–732. doi: 10.1038/nrmicro1235 [DOI] [PubMed] [Google Scholar]
- 37.Reyes JA, Melano R, Cárdenas PA, Trueba G. Mobile genetic elements associated with carbapenemase genes in South American Enterobacterales. Braz J Infect Dis. 2020;24(3):231–238. doi: 10.1016/j.bjid.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Li F, Cui S, et al. Prevalence and distribution characteristics of bla(KPC-2) and bla(NDM-1) genes in Klebsiella pneumoniae. Infect Drug Resist. 2020;13:2901–2910. doi: 10.2147/IDR.S253631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekizuka T, Yatsu K, Inamine Y, et al. Complete genome sequence of a bla(KPC-2)-positive Klebsiella pneumoniae strain isolated from the effluent of an urban Sewage treatment plant in Japan. mSphere. 2018;3(5). doi: 10.1128/mSphere.00314-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao Y, Lazaro-Perona F, Falgenhauer L, et al. Insights into a novel bla(KPC-2)-encoding IncP-6 plasmid reveal carbapenem-resistance circulation in several Enterobacteriaceae species from wastewater and a hospital source in Spain. Front Microbiol. 2017;8:1143. doi: 10.3389/fmicb.2017.01143 [DOI] [PMC free article] [PubMed] [Google Scholar]