Perivascular macrophages orchestrate the expansion of pericyte-like mesenchymal cells to create a proangiogenic niche in cancer.
Abstract
Tumor-associated macrophages (TAMs) are a highly plastic stromal cell type that support cancer progression. Using single-cell RNA sequencing of TAMs from a spontaneous murine model of mammary adenocarcinoma (MMTV-PyMT), we characterize a subset of these cells expressing lymphatic vessel endothelial hyaluronic acid receptor 1 (Lyve-1) that spatially reside proximal to blood vasculature. We demonstrate that Lyve-1+ TAMs support tumor growth and identify a pivotal role for these cells in maintaining a population of perivascular mesenchymal cells that express α-smooth muscle actin and phenotypically resemble pericytes. Using photolabeling techniques, we show that mesenchymal cells maintain their prevalence in the growing tumor through proliferation and uncover a role for Lyve-1+ TAMs in orchestrating a selective platelet-derived growth factor–CC–dependent expansion of the perivascular mesenchymal population, creating a proangiogenic niche. This study highlights the inter-reliance of the immune and nonimmune stromal network that supports cancer progression and provides therapeutic opportunities for tackling the disease.
INTRODUCTION
The stroma in cancer is composed of a variety of nonmalignant immune and mesenchymal cell populations that facilitate tumor progression (1). Although a variety of protumoral processes have been characterized for the individual cell populations that compose the stroma, it is apparent that these cells are not functioning autonomously but in concert with one another as part of a wider network of cross-communication to facilitate disease progression (2). Identifying nonredundant signaling pathways within the stromal network is desirable, as therapeutically targeting these signals could result in an unraveling of the stromal support network upon which the malignant tumor cells rely.
Macrophages form a major part of the stromal cell infiltrate in solid tumors (3) and are highly plastic to their environment, which creates phenotypic and functional diversity within the population (1, 4–10). Tumors exploit the plastic nature of macrophages, which we collectively refer to as tumor-associated macrophages (TAMs), to facilitate disease progression by promoting angiogenesis (11, 12), immune suppression (1, 13), chemotherapeutic resistance (14–16), and tumor cell migration and metastasis (4, 17–21). Although the protumoral function of TAMs is well described, less is known about their “cellular” networks and the niches they support. This study highlights the inter-reliance of the immune-mesenchymal stromal network in cancer. Unlike immune cells that are readily recruited into the tumor microenvironment from the systemic circulation (19, 22), we demonstrate that mesenchymal stromal populations rely on local proliferation to expand with tumor growth. Using the spontaneous MMTV-PyMT murine model of breast cancer, we characterize a subpopulation of TAMs that are defined by their selective expression of lymphatic vessel endothelial hyaluronic acid receptor 1 (Lyve-1) and spatial proximity to blood vasculature. We demonstrate that perivascular Lyve-1+ TAMs play a fundamental role in orchestrating the expansion of a population of pericyte-like mesenchymal cells that create a proangiogenic perivascular niche. This study highlights a role for perivascular TAMs (pvTAMs) in shaping the heterogeneity of the mesenchymal cell population in cancer and implicates their expression of platelet-derived growth factor–CC (PDGF-CC) as a therapeutic target in the process.
RESULTS AND DISCUSSION
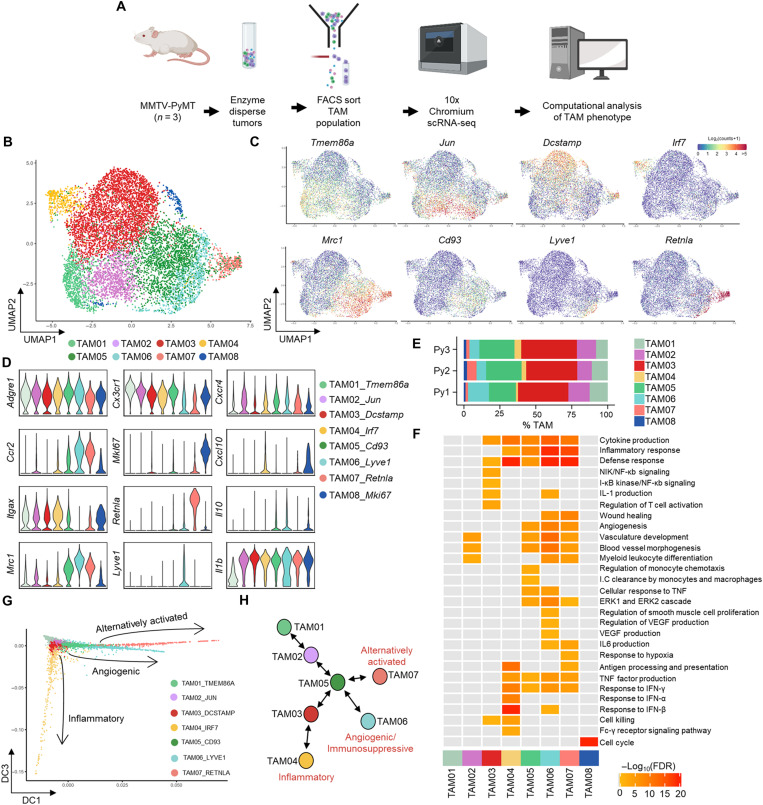
To resolve the phenotypic heterogeneity of TAMs within the tumor microenvironment, CD45+Ly6G−CD11b+F4/80hi cells, which represented a broad gating strategy to encompass all TAMs, were sorted using fluorescence-activated cell sorting (FACS) from enzyme-dispersed tumors from MMTV-PyMT mice (fig. S1A) (23). The TAMs were then subjected to the droplet-based 10x Genomics Platform for single-cell RNA sequencing (scRNA-seq; Fig. 1A). A total of 9039 TAMs were sequenced across three individual tumors. Unsupervised graph-based clustering of the transcriptomes, visualized using UMAP (24), revealed eight distinct transcriptomic TAM clusters (Fig. 1, B to D, and fig. S1, B and C). The presence of these transcriptomic clusters, despite the tumors being spontaneous, was conserved across the three tumors analyzed (Fig. 1E). Gene Ontology (GO) analysis of the transcriptional programs within these clusters revealed diversity in both the number and type of biological pathways that were active. One cluster (TAM08) represented a highly proliferative TAM state, indicating that TAMs are capable of proliferation in the tumor microenvironment; however, the transcriptome of this TAM subset was dominated singularly by cell cycle–associated genes and so was not carried forward for further functional analysis (Fig. 1F and fig. S1D). The TAM clusters with few enriched GO terms, which appeared to be the least polarized in their gene expression profile (TAM01 and 02), represented almost a quarter of TAMs within the tumor (23.3 ± 3.4% of all TAMs analyzed), suggesting that a notable proportion of TAMs remain relatively unspecialized in their role (Fig. 1, E and F, and fig. S1E). Trajectory inference analysis using Slingshot (25) and diffusion maps was used to align the seven identified clusters by transcriptomic similarity into a polarization model with three clusters—TAM04, TAM06, and TAM07—predicted as possible polarization extremes compared to the relatively unspecialized TAM01, between which all MMTV-PyMT TAM transcriptomes fall between (Fig. 1, G and H, and fig. S2). The three clusters representing possible polarization extremes were analyzed for their enrichment of M1/M2 (26) programs using the marker gene list of Orecchioni et al. (27). This analysis highlighted TAM04 to be skewed toward an inflammatory (M1-like) transcriptome (Fig. 2, A and B), which was more enriched for expression of inflammatory genes representative of a cellular response to type 1 interferons such as Irf7 and Isg15. TAM06 and TAM07 had a more protumoral (M2-like) transcriptome (Fig. 2, A and B). TAM06 was more enriched for anti-inflammatory genes such as Il10, whereas both TAM06 and TAM07 were enriched in Ccl2, Mmp19, Hb-egf, and Mrc1 (the gene for MRC1/CD206) (28). However, TAM06 and TAM07 were functionally distinct in many of their enriched GO biological pathways, with a preferential skewing of TAM06 toward angiogenic processes and TAM07 toward immune regulation, highlighting a specialized subdivision of roles within the tumor (Fig. 2, C and D). Flow cytometry analysis of gated F4/80hi TAMs stained for markers identified within the scRNA-seq analysis confirmed that similar TAM subpopulations could be distinguished using the predicted protein markers in MMTV-PyMT tumors. TAM06 and TAM07 subsets predicted by the scRNA-seq analysis could be differentiated on the basis of their expression level of CD206, major histocompatibility complex class II (MHCII), and the lymphatic vessel endothelial hyaluronic acid receptor 1 (Lyve-1) (Fig. 2, E and F) into the predicted polarization extremes of CD206hiMHCIIloLyve-1+ (Lyve-1+ TAMs; TAM06) and CD206intMHCIIhiLyve-1− (TAM07). The Lyve-1+ TAM subset accounted for 10.7 ± 3.5% of total TAMs and 1.4 ± 0.4% of live cells within the tumor (Fig. 2F). To validate that the populations identified in the scRNA-seq and flow cytometry data were equivalent, the FACS-gated populations were subjected to bulk population RNA-seq alongside CD206−MHCloF4/80hi TAMs as a comparator group. Principal components analysis (PCA) confirmed these populations to be transcriptionally distinct (Fig. 2G). Comparing the bulk population RNA-seq to that of the scRNA-seq populations validated close concordance between the identified populations across a range of predicted marker genes (Fig. 2H). Lyve-1+ TAMs also selectively expressed the transcription factor Maf (fig. S2D) and CD206intMHCIIhiLyve-1− TAMs expressed the transcription factor Retnla (Fig. 1D), which may indicate that these transcription factors play a role in polarization identity.
Fig. 1. scRNA-seq of TAMs in MMTV-PyMT tumors reveals three distinct polarization pathways.
(A) Schematic outlining the scRNA-seq experimental workflow that was conducted for n = 3 individual MMTV-PyMT tumors and mice, sequencing a total of 9039 cells using the 10x Genomics’ Chromium platform. (B) UMAP plot of sequenced TAMs colored by their associated cluster identity. (C) UMAP visualizations of predicted marker gene expression for distinct TAM clusters in (B). (D) Violin plots of selected genes associated with TAM cluster identity seen in (B). (E) Relative proportion of each TAM cluster across the individual MMTV-PyMT tumors analyzed. (F) Heatmap representing significantly up-regulated GO pathway terms in one or more TAM clusters. (G and H) Scatter plot of single cells projected into two dimensions using diffusion maps, where each cell (dot) is colored by cluster identity, labeled with diffusion component (DC) space annotation representing lineage trajectories predicted by the Slingshot package (G) and schematic map of each TAM cluster’s location along the respective trajectories (H).
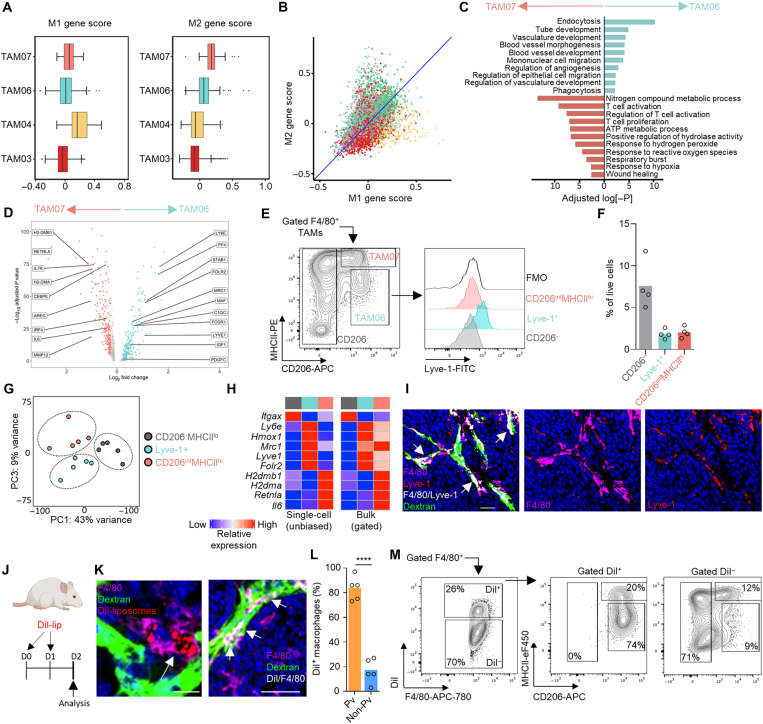
Fig. 2. Lyve-1 marks a subset of TAMs that reside proximal to blood vasculature.
(A and B) Box and whisker plots (A) and scatter plot (B) showing normalized mean M1- and M2-associated gene scores across the indicated TAM clusters identified using scRNA-seq. (C and D) Subset unique, significantly up-regulated GO terms (C) and individual genes (D) between the two subsets of protumoral TAM. (E) FACS-gated live [7-aminoactinomycin D–negative (7AAD−)] F4/80hi TAMs from enzyme-dispersed MMTV-PyMT tumors separated on the basis of CD206 and MHCII expression (left) and assessed for Lyve-1 expression (right; color-shaded histograms) against that of the fluorescence minus one staining (FMO) control (open black line). (F) Quantification of the gated populations in (E) (n = 4 tumors). (G) PCA plot of the 2000 most variable genes from the bulk-sequenced TAM populations (n = 5 tumors), using CD206− and MHCII− TAMs as a comparator. (H) Heatmaps comparing the relative expression of selected differentially expressed genes identified in the scRNA-seq (left) and bulk RNA-seq (right); population color is indicative of the populations identified in (G). (I) Representative image of a frozen section of MMTV-PyMT tumor showing DAPI (nuclei; blue); intravenous dextran marking vasculature (green), F4/80 (magenta), and Lyve-1 (red); and colocalizing pixels for Lyve-1 and F4/80 (white); scale bars, 25 μm. (J to M) Schematic for experimental approach to label pvTAMs using Dil-labeled liposomes (J). (K) Representative images of frozen sections of MMTV-PyMT tumors showing DAPI (nuclei; blue); intravenous dextran marking vasculature (green), Dil (red), and F4/80 (magenta); and Dil/F4/80 colocalizing pixels (white) (right panel alone); scale bars, 25 μm (left) and 50 μm (right). (L) Quantification of the spatial location of Dil+ F4/80+ TAMs (n = 5 mice). (M) Analysis of the surface phenotype of Dil+/− TAM from enzyme-dispersed tumors within the F4/80+ gate. Box and whisker plots; boxes show median and quartiles. Bar charts represent mean, and the dots show individual tumors and mice. ****P < 0.0001.
Lyve-1 has traditionally been considered a marker of lymphatic endothelium (29) but has also been used as a marker on tissue-resident macrophages (30–35) and TAMs (36). It has been demonstrated that macrophages expressing Lyve-1 can be found to spatially reside proximal to vasculature (34) and immunofluorescence staining of tissue sections from MMTV-PyMT tumors confirmed Lyve-1+ TAMs to be a population of perivascular cells (Fig. 2I). As Lyve-1 expression can be found on macrophages in homeostatic tissues (30–35), we investigated whether an equivalent macrophage population could be identified in the mammary gland before tumor onset. Using the flow cytometry gating strategy of Franklin et al. (22) for mammary tissue macrophages, an equivalent macrophage population (based on CD206 and MHCII markers) could not be found in the mammary gland before tumor onset (fig. S3A). Although a population of Lyve-1+ macrophages does reside in the mammary gland, its phenotype is distinct from that of the Lyve-1+ TAM population, most notably in a lower expression of CD206 (fig. S3, A and B). Although this study does not rule out a direct link between Lyve-1+ macrophages in the mammary gland and the TAM population that arises in the tumor, their exact origin remains to be determined. However, these data do at least suggest that the phenotype of Lyve-1+ TAMs has been polarized by the tumor microenvironment. GO pathway analysis also suggested that Lyve-1+ TAMs were highly endocytic (Fig. 2C). Liposomes containing the fluorescent lipophilic dye 1′-dioctadecyl-3,3,3′,3"tetramethylindocarbocyanine perchlorate (Dil) have previously been used to study pvTAM development (19), and we predicted that they could represent a tool to preferentially label the Lyve-1+ TAM subset. We developed a labeling protocol that could selectively mark pvTAMs. Confocal analysis of the labeled tumors demonstrated that Dil-liposomes specifically labeled a population of pvTAMs (Fig. 2, J to M, and fig. S4, A to C) and ex vivo characterization of the Dil-labeled TAMs in enzyme-dispersed MMTV-PyMT tumors confirmed the vast majority of labeled cells to be that of the Lyve-1+ TAM subset (Fig. 2M).
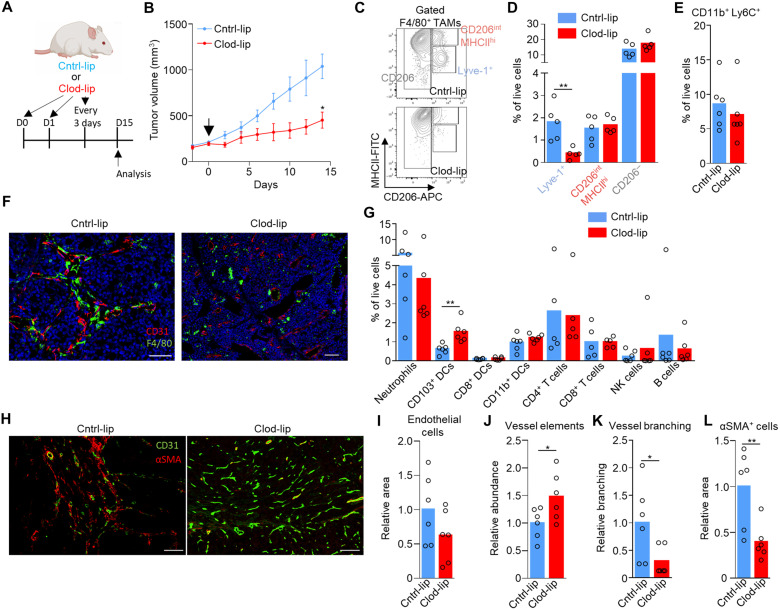
As the liposome labeling protocol preferentially labeled Lyve-1+ TAMs (Fig. 2M and fig. S4, B and C), we used clodronate-filled liposomes (37) under an equivalent administration protocol as a means to selectively deplete the population and investigate their possible role in tumor progression. Depletion of these cells in MMTV-PyMT tumors resulted in a significant slowing of tumor growth (Fig. 3, A and B), highlighting a fundamental role for these cells in tumor progression. Even over the long-term administration of clodronate-filled liposomes, which displayed little sign of toxicity in the animals (fig. S4D), provided a preferential depletion of Lyve-1+ TAMs, sparing CD206− and CD206−MHCIIhi TAM populations (Fig. 3, C and D) and CD11b+Ly6C+ monocytes (Fig. 3E). Furthermore, using immunofluorescence imaging, there was an observable selective spatial loss of pvTAMs within the clodronate-filled liposome treated mice (Fig. 3F), where most of TAMs surrounding blood vessels were no longer observable. To understand the mechanism through which Lyve-1+ pvTAMs promote tumor progression (Fig. 3B), we first phenotyped the immune-infiltrate of the tumors. Loss of Lyve-1+ pvTAMs did not change the abundance of any immune cell populations analyzed within the tumor microenvironment, other than a statistically significant increase in the abundance of the migratory CD11c+CD103+dendritic cells (Fig. 3G and fig. S4E), which contribute to cytotoxic T lymphocyte recruitment in the tumor (38) and priming of the antitumor immune response (39). However, there was no increase in CD8+ or CD4+ T cell recruitment after depletion of Lyve-1+ pvTAMs (Fig. 3G). Perivascular macrophages are known to play a role in angiogenesis (6), and the Lyve-1+ pvTAM population expressed pathways associated with proangiogenic functions (Fig. 2C), which could account for the control of tumor growth observed when the TAM subset was depleted (Fig. 3B). Immunofluorescence analysis of these tumors had shown no overall change in density of endothelial cells within the tumor (Fig. 3, H and I), but the tumors themselves were smaller (Fig. 3B). Further analysis of sections from MMTV-PyMT tumors stained for CD31+ endothelial cells and perivascular α-smooth muscle actin (αSMA)–expressing stromal cells revealed a change in vessel architecture, where depletion of Lyve-1+ pvTAMs resulted in an increase in the number of individual vessel elements in the tumor (Fig. 3J), with the vessel elements appearing smaller and less branched (Fig. 3K). However, most notably, there was a loss of αSMA+ stromal cells proximal to vasculature (Fig. 3, H and L). Although there is evidence that fibroblasts can be phagocytic (40), neither CD45−CD90+ cancer-associated fibroblasts (CAFs) (41) nor CD45− or CD31+ endothelial cells had up-taken the liposomes (fig. S4B), excluding any direct killing effect of the clodronate on these populations and highlighting a potential role of Lyve-1+ pvTAMs in maintaining this stromal population.
Fig. 3. Lyve-1+ pvTAM depletion slows tumor growth and is associated with a concurrent loss of perivascular αSMA+ stromal cells.
(A) Schematic for experimental approach and dosing strategy to deplete Lyve-1+ TAMs using clodronate-filled liposomes. Arrows represent days of treatment. (B) Growth curves of MMTV-PyMT tumors in mice treated with control PBS-filled liposomes (Cntrl-lip) or clodronate-filled liposomes (Clod-lip) as shown in (A); arrow marks the initiation of treatment (cohorts of n = 6 mice). (C to L) Tumors from (B) were excised at day 15 (after treatment initiation; n = 5 to 6 tumors) and analyzed. (C) Representative contour plot gating of live (7AAD−) CD45+Ly6C−F4/80+ TAMs from enzyme-dispersed MMTV-PyMT tumors measured by flow cytometry and the abundance of the gate subsets (D). (E) Abundance of live (7AAD−) CD45+CD11b+Ly6C+ monocytes. (F) Representative image of a frozen section of MMTV-PyMT tumor from mice treated with control- or clodronate-filled liposomes stained with DAPI (nuclei; blue) and antibodies against F4/80 (green) and CD31 (red). Scale bars, 50 μm (left) and 100 μm (right). (G) Abundance of major immune cell types in the tumor microenvironment measured by flow cytometry. (H to L) Representative image of a frozen section of MMTV-PyMT tumor stained with antibodies against CD31 (green) and αSMA (red) [scale bars, 100 μm (left and right)] (H), and the quantification of relative CD31+ pixel area (I), number of distinct CD31+ endothelial vessel elements as assessed using immunofluorescence analysis of stained frozen tissue sections (a total of n = 12 sections, across n = 6 mice per condition) (J), vessel branch points (K), and αSMA+ pixel area (L). A total of n = 12 sections were analyzed across the six tumors in each cohort. Growth curve is presented as mean ± SEM, bar charts represent mean, and the dots show individual data points from individual tumors and mice. *P < 0.05 and **P < 0.01.
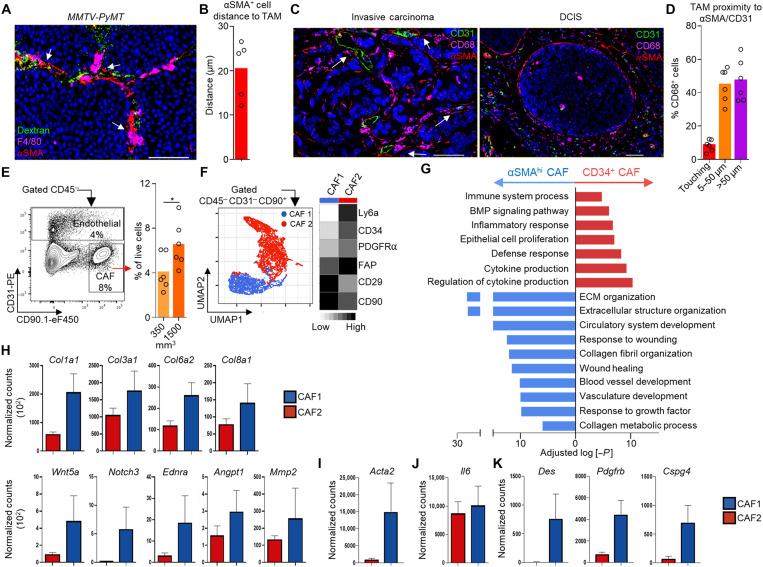
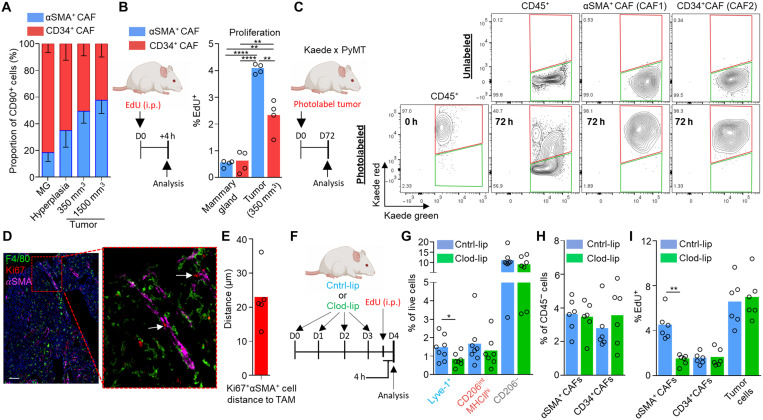
Staining tissue sections from MMTV-PyMT tumors for the αSMA+ cells and F4/80+ TAMs placed these populations in a close spatial arrangement with CD31+ vasculature providing opportunity for interactions and suggested a “niche” formation (Fig. 4A). The median distance between αSMA+ cells and F4/80+ TAMs was only 20 ± 7 μm (less than a cell thickness), highlighting the close association between these cells (Fig. 4B). A similar niche was also found in human invasive breast cancer (Fig. 4C), where 44.7 ± 10.2% of TAMs could be found in a perivascular niche with αSMA+ cells and 8.4 ± 3% TAMs could be found in direct contact with αSMA+ cells (Fig. 4D). This immune-mesenchymal niche was not present within ductal carcinoma in situ (DCIS) tissue (Fig. 4C) but could be found around the vessels in the adjacent tissue (albeit at a lower abundance than found within the tumor; fig. S5, A and B). Lyve-1+ macrophages have been identified in healthy tissue in close proximity to αSMA-expressing smooth muscle cells in the mouse aorta, where they play a homeostatic functional role in modulating collagen production that influences arterial tone (35). To further investigate these perivascular Lyve-1+ TAM-dependent αSMA+ cells, we characterized the heterogeneity of a broad pool of tumor-associated mesenchymal stromal cells (collectively termed CAFs) using flow cytometry within enzyme-dispersed MMTV-PyMT tumors. The CD45−CD31−CD90+ population accounted for 4.0 ± 1.6% of total live cells within 350-mm3 tumors, and their abundance increased as tumors progressed (Fig. 4E). We screened the CD45−CD90+ population for cell surface markers associated with mesenchymal subsets, including Ly6a, CD34, PDGFRα (PDGF receptor α), fibroblast activation protein-α (FAP), and CD29 (41–45). Clustering of the multiparametric flow cytometry data using UMAP (24) and FlowSOM (46) distinguished two distinct subsets (Fig. 4F). The first subset “CAF1” was CD29hiCD34−Ly6a−FAPloPDGFRαlo, and the second “CAF2” was CD29loCD34+Ly6a+FAPhiPDGFRαhi (Fig. 4F). Although the two subsets of CAFs could have been separated on the basis of any of the markers used, the two populations were FACS-sorted on the basis of their differing expression of CD34 for bulk RNA-seq to confirm the αSMA-expressing population (fig. S5C). CD34 was selected to separate these CAF subsets and has been previously demonstrated to negatively correlate with αSMA expression in CAFs in models of pancreatic adenocarcinoma (47). This analysis demonstrated clear transcriptional differences in these subsets (Fig. 4G and fig. S5, D and E). The CD34+ CAF population was functionally more skewed toward inflammation-related processes, while the CD34− CAF displayed an extracellular matrix and proangiogenesis-related program (Fig. 4, G and H, and fig. S5E). The CD34− CAF population also selectively expressed high levels of αSMA (Acta2) (Fig. 4I). These CAF subsets were largely similar to those identified in pancreatic ductal adenocarcinoma (42); however, there were also key differences such as Il6 was not a discriminatory marker for the CAF populations in MMTV-PyMT tumors (Fig. 4J). The CD34− CAF population also expressed Des, Pdgfrb, and Cspg4 (Fig. 4K), which are genes that are often associated with pericytes, a population of specialized vessel-associated cells (48, 49). To confirm the presence of pericyte markers desmin (Des), PDGFRβ (Pdgfrb), and NG2 (Cspg4) at the protein level in these cells, immunofluorescence staining of tissue sections from MMTV-PyMT mice confirmed that the perivascular αSMA+ cells were also desmin+ (fig. S5F), and ex vivo flow cytometry confirmed the presence of surface PDGFRβ (fig. S5G) and NG2 (fig. S5H). CD34− CAFs expressed PDGFRα, albeit low relative to the CD34+ population (Fig. 4F and fig. S5G), which is regarded as a broad marker of fibroblasts. The pericyte marker NG2 and fibroblast marker PDGFRα colocalized at the protein level on the CD34− CAFs, where higher NG2 expression was associated with relatively higher PDGFRα expression within the population (fig. S5H), suggesting the population may represent either a “pathological” pericyte phenotype or a pericyte-like CAF population. Pericytes are important to angiogenesis, supporting vessel stabilization and endothelial cell survival (50). The CD34− CAF population also displays similarities in gene expression to vasculature-associated “vCAFs” recently characterized in MMTV-PyMT tumors, although vCAFs did not have detectable surface protein expression of NG2 (45). A phenotypically similar pericyte-like CAF population expressing CD29, PDGFRβ, and high levels of αSMA has also been identified in human breast cancer (43). Heterogeneous expression of CD34 differentiated CAF populations across different ectopic tumor models including B16, LL2, and orthotopic 4T1 (fig. S5, I and J). Because of αSMA representing a defining feature of these cells, we elected to refer to these cells here as “αSMA+ CAFs.” Analyzing the abundance of the CAF populations over the different stages of tumor progression from the healthy mammary gland, hyperplasia, and the growing tumor revealed a relative increase in the abundance of the αSMA+ CAFs within the broader CAF population over tumor progression, suggesting a preferential selection of this subset within the tumor microenvironment (Fig. 5A). To elucidate the route through which these cells were accumulating in the tumor, we first explored local proliferation and pulsed mice bearing MMTV-PyMT tumors with 5-ethynyl-2′-deoxyuridine (EdU) to label actively proliferating cells. Although both CD34+ CAF and αSMA+ CAF populations displayed evidence of proliferation by comparison with healthy mammary gland, the αSMA+ CAFs were proliferating at a significantly faster rate (Fig. 5B). To address whether the proliferation was sufficient to account for their preferential expansion with tumor growth, we used the Kaede mouse (51) crossed to the MMTV-PyMT model. Using this approach, we were able to photoconvert all tumor and stromal cells within a 100-mm3 tumor from Kaede-green to Kaede-red (Fig. 5C). Analyzing tumors 72 hours after photoconversion demonstrated that CD45+ stromal cells predominantly displayed Kaede-green, highlighting the continual recruitment of hematopoietic stromal cells to the tumor from the periphery (19, 22). In contrast, both CD34+ CAFs and αSMA+ CAF populations remained Kaede-red, which indicated that both CAF populations derived from a tumor-resident source of cells and was not dependent on recruitment (Fig. 5C). Therefore, the rapid proliferation of the αSMA+ CAFs relative to CD34+ CAFs may also contribute to the dynamics of CAF heterogeneity over tumor growth (Fig. 5A).
Fig. 4. Lyve-1+ TAMs form a perivascular niche with proangiogenic pericyte-like αSMA+ CAFs.
(A) Representative image of a frozen section of MMTV-PyMT tumor stained with DAPI (nuclei; blue) and antibodies against F4/80 (magenta) and αSMA (red); functional vasculature was labeled in vivo using intravenous dextran-FITC (green). (B) Quantification of αSMA+ cell median distance from F4/80+ TAM from immunofluorescence images (n = 5). (C) Representative image of an FFPE section from human invasive ductal mammary carcinoma (left) and DCIS (right) stained with DAPI (nuclei; blue) and antibodies against CD31 (green), CD68 (magenta), and αSMA (red); images representative of four to six patients. (D) Quantification of the spatial position of CD68+ TAM in proximity to SMA+ stroma touching CD31+ vessels (<50 μm is regarded perivascular) across multiple regions of interest (n = 5 tumors). (E) Representative flow cytometry gating strategy for live (7AAD−) CD45− cells and CD31+ endothelial cells and CD90+ CAFs (left) and the abundance of CAFs at different tumor volumes (right); n = 6 mice per condition. (F) Identification of CAF subsets by unsupervised clustering from multiparametric flow cytometry data using the markers shown in the heatmap (right). UMAP plot shows individual cells colored by their unsupervised clustering assignment (left); n = 4 mice. (G to K) Bulk RNA-sequenced CAF subsets from MMTV-PyMT tumor (n = 5 mice) transcriptomes were investigated. GO pathway analysis and plot show the selected GO terms based on differentially expressed genes of the two CAF subsets (G), and bar plots depict normalized gene expression values for the indicated genes associated with angiogenesis (H), Acta2 (I), and Il6 (J) and pericyte-associated markers (K). Differences in gene expression in (H), (I), and (K) are all P < 0.0001. Bar charts represent mean, error bars represent SD, and the dots show individual data points from individual tumors and mice. Scale bars, 50 μm. *P < 0.05.
Fig. 5. Lyve-1+ TAMs orchestrate αSMA+ CAF expansion within the perivascular niche of the tumor.
(A) Abundance of the respective CAF populations during distinct stages of tumor progression; n = 6 mice per stage. MG, mammary gland. (B) Schematic for experimental approach and dosing EdU into MMTV-PyMT mice to assess in vivo proliferation (left) and proportion EdU+ cells within each CAF subset (right). i.p., intraperitoneally. (C) Established tumors in Kaede MMTV-PyMT mice were photoconverted to Kaede-red, and then at 72 hours after photoconversion, tumors were analyzed (schematic left) for their respective Kaede-red/green proportion using flow cytometry for evidence of peripheral recruitment (Kaede-green cells). A representative unconverted tumor is shown for comparison (right top). (D and E) Representative image of a frozen section of MMTV-PyMT tumor stained with antibodies against F4/80 (green), αSMA (magenta), and the proliferation marker Ki67 (red). White arrows show αSMA+Ki67+ cells in contact with F4/80+ TAMs (D) and quantification of Ki67+αSMA+ cell median distance from F4/80+ TAMs quantified from immunofluorescence images across multiple tumors (n = 5) (E). (F to I) Schematic for experimental approach and dosing strategy to acutely deplete Lyve-1+ pvTAM with clodronate-filled liposome treatment for 4 days (F). (G) Abundance of TAM populations following control- or clodronate-filled liposome treatment (n = 6 mice Cntrl-lip and n = 5 mice Clod-lip). (H) Abundance of CD45− cell populations (cohorts of n = 6 mice) after 4 days of treatment with either control- or clodronate-filled liposomes. (I) Proportion of EdU+ cells within each CD45− cell subset (cohorts of n = 6 mice). Bar charts represent mean, error bars represent SD, and the dots show individual data points from individual tumors and mice. *P < 0.05, **P < 0.01, and ****P < 0.0001.
Immunofluorescence analysis for Ki67, a marker of proliferation (52), on αSMA+ cells, which we had identified as perivascular, confirmed a close spatial relationship between proliferating Ki67+αSMA+ CAFs and F4/80+ TAMs (Fig. 5D), which were a median distance of 22.6 ± 8.5 μm (within a cell thickness) from each other (Fig. 5E). To investigate whether Lyve-1+ pvTAMs might be implicated in the expansion of αSMA+ CAFs, we analyzed the incorporation of EdU after the depletion of Lyve-1+ pvTAMs using clodronate-filled liposomes (Fig. 5, F and G). Despite no observable drop in the proportion of αSMA+ CAFs within the tumor over the short-term acute treatment regimen (Fig. 5H), depleting Lyve-1+ pvTAMs significantly diminished the high rate of proliferation of the αSMA+ CAF population (Fig. 5I). Conversely, the proliferation rate of the CD34+ CAF and tumor cell compartments remained unaffected by the loss of Lyve-1+ pvTAMs (Fig. 5I).
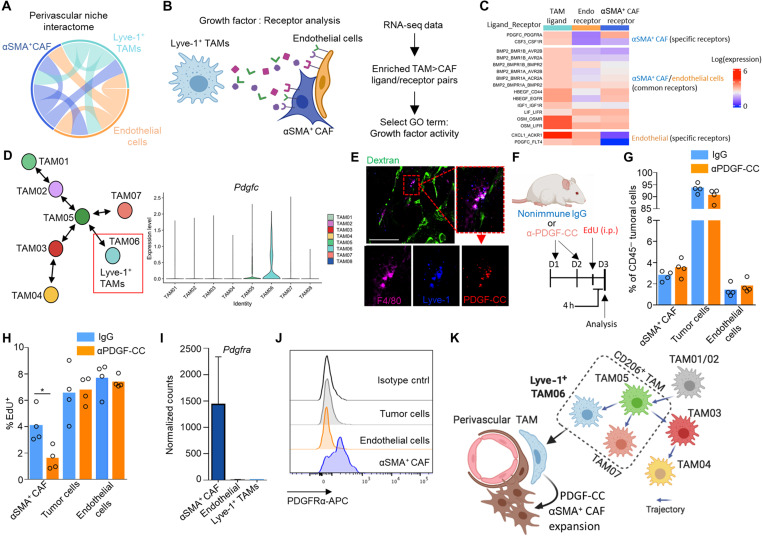
To resolve how Lyve-1+ pvTAMs could be orchestrating αSMA+ CAF expansion within the perivascular niche, we used CellPhoneDB, a manually curated repository and computational framework to map the possible biological ligand:receptor interactions within RNA-seq datasets (53) between the Lyve-1+ pvTAMs, αSMA+ CAFs, and CD31+ endothelial cells (which were all bulk-population RNA-sequenced) to construct an interactome of the major cell types in the perivascular niche (Fig. 6A). There were a total of 653 possible unique ligand:receptor interactions between these three cell types, highlighting the range of potential cross-talk between these populations in constructing the perivascular niche (fig. S6A). To refine this list, we selected for known mitogenic non–integrin-mediated ligands that were enriched in Lyve-1+ pvTAMs compared to other TAM populations and could interact with receptors specifically expressed on αSMA+ CAFs and not endothelial cells (Fig. 6, B and C). This highlighted the selective cross-talk between these two proximal cells involving Pdgfc (54) expressed by the Lyve-1+ pvTAM signaling to Pdgfra on the αSMA+ CAFs within the perivascular niche (Fig. 6C). More broadly, the Lyve-1+ TAM subset was a major source of Pdgfc in the tumor (Fig. 6D and fig. S6B) and Lyve-1+ TAMs could be found expressing PDGF-C in the perivascular niche (Fig. 6E).
Fig. 6. Lyve-1+ TAMs communicate to αSMA+ CAFs in the perivascular niche via a pro-proliferative PDGF-CC:PDGFR-α interaction.
(A) Circos plot showing predicted cross-talk of perivascular ligand-receptor interactions as identified by CellPhoneDB from the respective RNA-seq datasets. Outer sectors and links between sectors are weighted according to the total number of annotated ligand-receptor interactions between each respective cell type. (B) Schematic representing the method of cell type ligand-receptor interactome generation. (C) Heatmap showing the Lyve-1+ TAM and αSMA+ CAF population-specific secretome generated using data from (A) and the method outlined in (B) diagram displaying the ligand:receptor pairs between Lyve-1+ TAMs and αSMA+ CAFs and endothelial cells. The analysis highlighted a unique PDGF-CC:PDGFRα interaction specific to Lyve-1+ TAMs and αSMA+ CAFs. (D) Schematic map of each TAM cluster’s location along the respective trajectories marking the Lyve-1+ TAM population (left) and violin plots of Pdgfc expression associated with TAM clusters (right). (E) Representative image of a frozen section of MMTV-PyMT tumor stained with antibodies against F4/80 (magenta), Lyve-1 (blue), and PDGF-CC (red); the vessels are marked by dextran (green). Scale bar 50mm. (F to H) Schematic for experimental approach and dosing strategy to acutely inhibit PDGF-CC signaling using an anti–PDGF-CC neutralizing antibody (F). Abundance of indicated cell populations (G). Proportion of EdU+ cells within each CD45− cell subset (cohorts of n = 4 mice) (H). (I) Bar plot depicting normalized gene expression values for Pdgfra in the bulk RNA-sequenced populations (left) across n = 5 mice. (J) Representative histograms of surface PDGFRα staining on the indicated cells against isotype antibody staining of gated populations using flow cytometry analysis from enzyme-dispersed MMTV-PyMT tumors. (K) Schematic overview of the perivascular niche. Images in (B) and (K) were created using BioRender software. Bar charts represent mean, and the dots show individual data points from individual tumors and mice; error bars represent SD. *P < 0.05.
PDGFRs form either homo- or heterodimers between the α and β receptor subunit (αα, αβ, and ββ), and a homodimer of PDGF-C (PDGF-CC) selectively signals through PDGFRαα and PDGFRαβ dimers (55), which has been demonstrated to be a mitogenic and migratory factor for human dermal myofibroblasts (56, 57). In an elegant series of in vitro studies, macrophages have been demonstrated to form close stable homeostatic “circuits” with fibroblasts through their secretion of PDGF-BB as a means to maintain relative cell number (58). In cancer, these circuits, which rely on contact between the macrophage and fibroblast (58), could underpin the expansion of the perivascular niche due to the continual recruitment of monocytes to the tumor, allowing a reciprocal expansion of the fibroblast population in the niche environment. To assess whether PDGF-CC may play a role in orchestrating the expansion of the αSMA+ CAF population within the perivascular niche, we administered neutralizing antibodies to PDGF-CC (57), within an acute treatment regimen, in tumor-bearing MMTV-PyMT mice (Fig. 6F). Neutralization of PDGF-CC did not affect the abundance of the cell populations at the acute time point (Fig. 6G) but did diminish EdU incorporation of the αSMA+ CAFs, but not in the tumor cells or CD45−CD31+ endothelial cells within the vascular niche (Fig. 6H). This highlighted that the expansion of perivascular αSMA+ CAFs was PDGF-CC dependent and could account for the role of Lyve-1+ pvTAMs in orchestrating expansion of the population during tumor progression. As a population of perivascular fibroblasts has been implicated in recruiting macrophages to the perivascular niche (19), these observations in the current study highlight a potential reciprocal interactions between TAMs and mesenchymal populations in niche formation. PDGF-CC is a prognostic factor for poor survival in breast cancer (59) and has been demonstrated to be important to angiogenesis (60, 61). Within the perivascular niche, the αSMA+ CAFs selectively expressed PDGFRα (Fig. 6, I and J), alongside PDGFRβ (fig. S5G), and, hence, were the only cell to be capable of responding to PDGF-CC. Tumors grow slower in MMTV-PyMT Pdgfc−/− mice and display increased necrotic areas and evidence of hemorrhage (59). In accordance with our observations in murine models, using The Cancer Genome Atlas (TCGA), we observed an enrichment for a αSMA+ CAF signature (using genes identified in the murine population) above that of healthy tissue in human breast cancer (fig. S6C), and the αSMA+ CAF signature also positively correlated with PDGFC expression within the tumor (fig. S6D). This study raises an interesting parallel to the observations by Shook et al. (57), that macrophages expressing PDGF-CC support the expansion of αSMA+ myofibroblast populations in the wound healing response, a stromal response that shares many similarities to that of cancer (4, 62).
This study characterizes a biologically important subset of TAMs selectively expressing Lyve-1. We demonstrate that the Lyve-1+ pvTAM subset, which only accounts for 1.4 ± 0.4% of live tumoral cells, is pivotal to tumor growth. We define a previously unidentified role for pvTAMs in directing the expansion of a perivascular pericyte-like mesenchymal population to form a proangiogenic niche that is facilitated by a selective PDGFRα:PDGF-CC cross-talk (Fig. 6K). This study highlights the inter-reliance of stromal populations and the importance of the immune system in orchestrating nonimmune stromal cell reactions in cancer, which provides therapeutic opportunities for unraveling the complexity of the stromal support network and niches that underpin tumor progression.
MATERIALS AND METHODS
Mice
MMTV-PyMT mice used in this study were on an FVB/N background. Balb/c and C57Bl/6 wild-type mice were obtained from Charles River. Female C57Bl/6 homozygous Kaede mice (51) were crossed with male MMTV-PyMT (FVB background) mice, and the F1 offspring were used experimentally. Cohort sizes were informed by prior studies (4, 14). All mice used for experiments were female and randomly assigned to treatment groups. Mice were approximately 21 to 26 g when tumors became palpable. Experiments were performed in at least duplicate, and for spontaneous MMTV-PyMT tumor studies, individual mice were collected on separate days and all data points are presented.
Tumor studies
Murine 4T1 mammary adenocarcinoma, Lewis lung carcinoma (LL2), and B16-F10 melanoma cells were obtained from the American Type Culture Collection. Cells (2.5 × 105) in 100 μl of RPMI were injected by subcutaneous injection into the mammary fat pad of syngeneic Balb/c (4T1) or C57Bl/6 (B16-F10 and LL2) female mice that were 6 to 8 weeks of age. In studies using MMTV-PyMT mice, tumors arose spontaneously. When tumors became palpable, volumes were measured every 2 days using digital caliper measurements of the long (L) and short (S) dimensions of the tumor. Tumor volume was established using the following equation: Volume = (S2 × L)/2. MMTV-PyMT/Kaede mice were photo-labeled under anesthesia, and individual tumor mice were exposed to a violet light (405-nm wavelength) through the skin for nine 20-s exposure cycles with a short 5-s break interval between each cycle. Black cardboard was used to shield the rest of the mouse throughout the photoconversion procedure. Mice for 0-hour time points were culled immediately after photoconversion. This photoconversion approach was adapted from that used to label peripheral lymph nodes (63). Tumor tissues for flow cytometry analyses were enzyme-digested to release single cells as previously described (41). In brief, tissues were minced using scalpels, and then single cells were liberated by incubation for 60 min at 37°C with collagenase I (1 mg/ml) from Clostridium histolyticum (Sigma-Aldrich) and deoxyribonuclease (DNase) I (0.1 mg/ml; AppliChem) in RPMI (Gibco). Released cells were then passed through a 70-μm cell strainer before staining for flow cytometry analyses. Viable cells were numerated using a hemocytometer with trypan blue (Sigma-Aldrich) exclusion. For drug treatments, drugs were freshly prepared on the day of injection and administered by intraperitoneal injection using a 26-gauge needle. For EdU experiments, mice were intraperitoneally injected with EdU (50 mg/kg) dissolved in Dulbecco’s phosphate-buffered saline (PBS) and sacrificed 4 hours after injection. To liposome-deplete Pv macrophages, MMTV-PyMT mice were intraperitoneally injected with 150 μl of either clodronate- or PBS-filled liposomes (Anionic Clophosome, FormuMax) on the indicated days. To label PvTAM, MMTV-PyMT mice were intraperitoneally injected with 150 μl of Dil fluorescent tracing liposomes (Anionic Clophosome, FormuMax). To neutralize PDGF-CC, we used an antibody that has previously been used in vivo to block PDGF-CC signaling (57), and mice were intraperitoneally injected with 100 μg of a goat anti–PDGF-C neutralizing antibody (AF1447, Bio-Techne) solubilized in PBS on days −2 and −1 before analysis.
Murine tissue staining
Mouse mammary tumors were fixed overnight in 4% paraformaldehyde, followed by overnight dehydration in 30% sucrose before embedding in optimal cutting temperature (OCT) compound and snap-freezing in liquid nitrogen. Frozen sections from these tumors were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature and were washed in tris-buffered saline (100 mM tris and 140 mM NaCl), 0.05% Tween 20 (pH 7.4) (TBST) and blocked with TBST, 10% donkey serum (Sigma-Aldrich), and 0.2% Triton X-100. Immunofluorescence was performed as previously described (4). Antibodies and dilutions against the following targets were used: F4/80 1:100 (C1:A3-1, Bio-Rad), αSMA 1:100 (AS-29553, AnaSpec), CD31 1:100 (MEC13.1, BioLegend), CD31 1:100 (ab28364 Abcam), mKi67 1:100 (AF649, R&D Systems), CD34 1:100 (RAM34, Invitrogen), desmin 1:100 (PA5-19063, Invitrogen), and PDGF-C 1:100 (AF1447, R&D Systems). Primary antibodies were detected using antigen-specific donkey immunoglobulin G (IgG), used at 1:200: Alexa Fluor 405 anti-rabbit IgG, Alexa Fluor 488 anti-rabbit IgG, Alexa Fluor 488 anti-rat IgG, Alexa Fluor 568 anti-rabbit IgG, Alexa Fluor 568 anti-goat IgG, Alexa Fluor 647 anti-rabbit IgG (Thermo Fisher Scientific), NL657 anti-rat goat IgG (R&D Systems), and Cy3 anti-sheep donkey IgG (Jackson ImmunoResearch) were also used. Viable blood vessels were visualized in mice through intravenous injection of fluorescein isothiocyanate (FITC)–conjugated dextran (MW 20,000, Thermo Fisher Scientific) 20 min before sacrifice. Nuclei were stained using 4′,6-diamidino-2-phenylindole,dihydrochloride (DAPI; 1.25 μg/ml; Thermo Fisher Scientific). Images were acquired using a Nikon Eclipse Ti-E Inverted spinning disk confocal with associated NIS Elements software. Quantitative data were acquired from the images applied to Fig. 2 and were generated using NIS Elements.
For quantitative data generated for murine tissue staining in Fig. 4, a CellProfiler v3.0 (64) pipeline was used to identify cells as DAPI+ “primary objects” (i.e., all cells) via thresholding analysis. TAM and CAF DAPI+ “secondary objects” were then generated by cell surface and cytoplasmic staining of the protein markers F4/80 and αSMA, respectively. XY location and nuclear Ki67 staining status of the identified CAF and TAM object data were exported from the CellProfiler pipeline, and nearest neighbor identification was carried out using the FNN R package (https://CRAN.R-project.org/package=FNN).
Human tissue staining
Formalin-fixed paraffin-embedded (FFPE) human breast adenocarcinoma tissue sections of 4 μm were incubated at 60°C for 1 hour, before being deparaffinized with Tissue-Tek DRS2000, Sakura. Heat-induced antigen retrieval was performed using a pressure cooker (MenaPath Access Retrieval Unit, PASCAL). The slides were immersed in modified citrate buffer (pH 6) and gradually heated to 125°C. Excess of antigen retrieval buffer was washed first with distilled water followed by PBS, before incubation of the slides in blocking buffer containing 0.5% Triton and 5% donkey serum (Sigma-Aldrich) for 30 min at room temperature. The sections were then probed with anti-CD68 1:100 (KP1, Invitrogen), anti-αSMA 1:200 (1A4, Sigma-Aldrich), and anti-CD31 1:100 (EP3095, Abcam) diluted in blocking buffer overnight at 4°C. After further washing, sections were stained for 2 hours with donkey IgG antibodies purchased from Jackson ImmunoResearch and used at 1:600: Alexa Fluor 647 anti-mouse IgG and Alexa Fluor 488 anti-rabbit IgG. After washing in PBS, the sections were incubated with anti-αSMA conjugated with Cy3 probe 1:200 (1A4, Sigma-Aldrich). Counterstaining was performed with 1:2000 DAPI (Cell Signaling Technology) for 5 min, followed by a wash step using PBS. Mounting medium (FluorSave, Millipore) was applied to the slides. Images were acquired using a Nikon Eclipse Ti-E Inverted spinning disk confocal with associated NIS Elements software or Olympus slide scanner VS120-S6-W. VSI (Olympus) images were analyzed using VisioPharm analysis software. Briefly, regions of interest (ROIs) were drawn manually to select all areas containing tumor (DCIS and/or invasive breast cancer) in each image. Subsequently, a threshold algorithm-based Application Protocol Package (APP) was developed. Threshold classification method was based on custom-defined input bands (FITC, Cy3, and Cy5). This APP allowed to segment the original image, based on the expression of the predefined markers. Postprocessing steps were added to the APP to remove noise, to better define the vasculature coexpressing CD31 and αSMA markers, and to distinguish between PvTAMs, located within 50-μm radius from the vascular structures and the rest of the macrophages.
Flow cytometry
Flow cytometry was performed as previously described (1). The following antibodies against the indicated antigen were purchased from Thermo Fisher Scientific and were used at 1 μg/ml unless stated otherwise: CD3ε allophycocyanin (APC) and phycoerythrin (PE) (145-2C11), CD4 FITC (RM4-5), CD8β eFluor 450 (H35-17.2), CD11b APC–eFluor 780 (M1/70), CD11b BV510 (M1/70), CD11c APC (N418), CD16/32 (2.4G2; Tonbo Biosciences), CD19 APC (6D5; BioLegend), CD29 APC (eBioHMb1-1), CD31 eFluor 450 and PE (390), CD34 FITC and APC (RAM34), CD45 APC–eFluor 780, FITC, and peridinin chlorophyll protein (PerCP)–Cy5.5 (30-F11), CD90.2 eFluor 450 (53-2.1), CD90.1 eFluor 450 (HIS51), CD90.1 BV510 (OX-7), CD103 PE (2E7), CD206 APC (FAB2535A; Bio-Techne), F4/80 PE (BM8; BioLegend), F4/80 BV421 (BM8; BioLegend), FAP (10 μg/ml; AF3715, Bio-Techne), Ly6C PE and eFluor 450 (HK1.4), Ly6G FITC (1A8; BioLegend), Lyve-1 Alexa Fluor 488 (ALY7), MHCII PE, FITC, and eFluor 450 (M5/114.15.2), NG2 Alexa Fluor 488 (AB5320A4; Millipore), NK1.1 APC (PK136), PDGFRα APC and PerCP-Cy5.5 (APA5), PDGFRβ PE (APB5), and Ly6A/E Alexa Fluor 700 (D7). Where stated, the following corresponding isotype control antibodies at equivalent concentrations to that of the test stain were used: Armenian hamster IgG APC (eBio299Arm), goat IgG APC and PE (Bio-Techne), rat IgG2a APC, PE, and FITC (eBR2a), and rat IgG2b APC and eFluor 450 (eB149/10H5). Intracellular stains were performed as previously described (1). Dead cells and red blood cells were excluded using 7-amino actinomycin D (1 μg/ml; Sigma-Aldrich), Fixable Viability Dye eFluor 780 (Invitrogen), or DAPI alongside anti–Ter-119 PerCP-Cy5.5 or APC–eFluor 780 (Ter-119; Invitrogen). The FAP primary antibody was detected with a secondary biotin-conjugated anti-goat/sheep mouse IgG and 1:1000 streptavidin PE-Cy7 (Thermo Fisher Scientific). EdU was detected using the Click-IT Plus Flow Cytometry Assay with Alexa Fluor 488 (Thermo Fisher Scientific) in accordance with the manufacturers’ specifications. Briefly, cells were stained with cell surface antibodies and then fixed and permeabilized, and the click chemistry reaction was performed as specified with Alexa Fluor 488–conjugated picolyl azide to identify EdU incorporated into the genomic DNA. Cells were sorted to acquire pure populations using FACSAria (BD Biosciences). Data were collected on BD FACSCanto II (BD Biosciences) or BD LSR Fortessa (BD Biosciences). Data were analyzed using FlowJo software (BD biosciences). Unsupervised clustering of flow cytometry data was performed using the ImmunoCluster package (65). Briefly, the single-cell data were asinh-transformed with cofactor of 150 and clustering was performed with an ensemble method using FlowSOM (46) and ConsensusClusterPlus (66) to k = 8 clusters, based on the elbow criterion, which were manually merged on the basis of expression profiles into biologically meaningful populations as previously outlined (67). Dimensionality reduction for visualization purposes was performed with UMAP (24).
Quantitative real-time polymerase chain reaction
mRNA was extracted from FACS-sorted cell populations using the TRIzol method and converted to complementary DNA (cDNA) then amplified using a CellAmp Whole Transcriptome Amplification Kit (Real Time) Ver.2 kit (Takara) according to the manufacturers’ protocol. mRNA of interest was measured using the SuperScript III Platinum One-Step qRT-PCR Kit (Thermo Fisher Scientific) according to the manufacturers’ protocol with the primers/probes Actb Mm02619580_g1 and Pdgfc Mm00480295_m1 (Thermo Fisher Scientific). Expression is represented relative to the housekeeping gene Actb. Gene expression was measured using an ABI 7900HT Fast Real-Time PCR instrument (Thermo Fisher Scientific).
Single-cell RNA-seq
TAMs (CD45+Ly6G−CD11b+F4/80hi) were sorted from enzyme-digested MMTV-PyMT tumors, and a total of 10,502 TAMs were sequenced from three MMTV-PyMT tumors and run through the 10x Genomics Chromium platform. An average of 43,000 reads per cell, a median of 2400 genes, and a median unique molecular identifier (UMI) count of 9491 per cells were obtained. Single-cell suspensions were prepared as outlined in the 10x Genomics Single Cell 3′ V3 Reagent kit user guide (10x Genomics). Briefly, samples were washed with PBS (Gibco) with 0.04% (w/v) bovine serum albumin (Sigma-Aldrich) and resuspended in 1 mL of PBS and 0.04% (w/v) bovine serum albumin. Sample viability was assessed using trypan blue (Thermo Fisher Scientific) exclusion and an EVE automated cell counter (AlphaMetrix) in duplicate to determine the appropriate volume for each sample to load into the Chromium instrument. The sorted TAMs were loaded onto the Chromium Instrument (10x Genomics) to generate single-cell barcoded droplets according to the manufacturers’ protocol using the 10x Genomics Single Cell 3′ V3 chemistry. cDNA libraries were prepared as outlined by the Single Cell 3′ Reagent Kit v3 user guide, and each of the three resulting libraries was sequenced on one lane each of HiSeq 2500 (Illumina) in rapid mode.
scRNA-seq data processing and analysis
The raw sequenced data were processed with Cell Ranger analysis pipeline version 3.0.2 by 10x Genomics (http://10xgenomics.com/). Briefly, sequencing reads were aligned to the mouse transcriptome mm10 using the STAR aligner (68). Subsequently, cell barcodes and unique molecular identifiers underwent Cell Ranger filtering and correction. Reads associated with the retained cell barcodes were quantified and used to build a transcript count tables for each sample. Downstream analysis was performed using the Seurat v3 R package (69). Before analysis, we first performed quality control filtering with the following parameters: Cells were discarded on the following criteria: where fewer than 800 unique genes detected, reads composed greater than 12% mitochondrial-associated gene transcripts, and cells whose number of transcripts detected per cell was greater than 65,000 for samples 1 and 2 and 60,000 for sample 3. All genes that were not detected in at least 10 single cells were excluded. On the basis of these criteria, the final dataset contained 9615 TAMs with 25,142 detected genes. The data were first normalized using the LogNormalize function and a scale factor of 10,000. The 2000 genes with highest variance were selected with the FindVariableGenes function. To minimize the effect of cell cycle–associated genes in the dimensionality reduction and clustering, cell cycle–associated genes defined by the GO term “Cell Cycle” were removed from the variable gene dataset, resulting in 1765 variable genes. PCA was used on the highly variable genes to reduce the dimensionality of the feature space, and 35 significant principal components were selected for downstream analysis. To reduce biases caused by technical variation, sequencing depth, and capture efficiency, the three sequencing samples were integrated using the Seurat integration method (69) as specified. Clusters were identified by a graph-based shared nearest neighbor (SNN) clustering approach within Seurat using the resolution parameters 0 to 1 in steps of 0.1, followed by analysis using the Clustree R package (70). Last, we used a resolution parameter of 0.4 to define 10 clusters. Differentially expressed genes were identified using the FindAllMarkers function, where the genes must be detected in a minimum of 25% of cells and have a logFC threshold of 0.25. After identifying marker genes, we excluded two clusters that contained suspected contaminating epithelial cells (enriched in Epcam, Krt18, and Krt8) and dying low-quality cells (enriched in mitochondrial genes and ribosomal subunit genes). Ultimately, we identified eight relevant clusters. All gene scores were calculated from the integrated Seurat data object, where the data were scaled using the scaleData () function across all TAM transcriptomes. The genes composing the M1 and M2 gene scores were derived from Orecchioni et al. (27), specifically from table S1. Genes present in the gene signatures that were not detected by any cells in the dataset were discarded. The gene score was calculated per cell as the mean of the scaled gene expression across the M1- or M2-associated genes, and the individual cell gene scores are plotted and grouped by TAM cluster. We used the Slingshot R package (25) to investigate inferred polarization trajectories in our TAM population. Briefly, dimensionality reduction was performed using diffusion maps with the Destiny R package (71) using the significant PCA principal components used for clustering. A lineage trajectory was mapped into the diffusion space using the first 15 diffusion components by Slingshot, and each cell was assigned a pseudotime value based on its predicted position along the predicted trajectories. We selected the cluster TAM01 as the base state for the trajectory because it had the lowest M1/M2 activation–associated gene score among the terminal trajectory branch clusters, no discriminating up-regulated GO pathways, and the fewest differentially expressed genes and represented the most naïve TAM transcriptomic base state. To detect nonlinear patterns in gene expression over pseudotime trajectory, we used the top variable gene set and regressed each gene on the pseudotime variable we generated, using a general additive model (GAM) with the GAM R package (CRAN - Package gam; r-project.org). Heatmaps were generated with the ComplexHeatmap package (72).
Bulk RNA-seq
Cells were sorted directly into RLT plus buffer (Qiagen) supplemented with 2-β-mercaptoethanol (BME) (Gibco), and lysates were immediately stored at −80°C until used. RNA was extracted with the RNeasy Plus Micro Kit (Qiagen) as per the manufacturers’ protocol, in addition to on-column DNase digestions specified by the manufacturer (Qiagen). cDNA was generated and amplified using the SMARTseq v4 Ultra Low Input RNA Kit (Clontech) on the contactless Labcyte liquid handling system (Beckman Coulter Life Sciences). Two hundred nanograms of amplified cDNA was used from each sample where possible to generate libraries using the Ovation Ultralow Library System V2 Kit (NuGEN). In brief, cDNA was fragmented through sonication on Covaris E220 (Covaris Inc.), repaired, and polished followed by ligation of indexed adapters. Adapter-ligated cDNA was pooled before final amplification to add flow cell primers. Libraries were sequenced on HiSeq 2500 (Illumina) for 100 paired-end cycles in rapid mode.
Bulk RNA-seq data processing and analysis
Prealignment quality control (QC) for each sample, independently for forward and reverse reads, was performed using the standalone tool FastQC. Reads were trimmed using Trimmomatic (73) and aligned to the reference genome (mm10) using HISAT2 (74). Polymerase chain reaction (PCR) duplicates were removed using SAMtools (75). Counts were generated using the GenomicAlignment (76) package using the mm10 reference genome. Before performing differential gene expression analysis, genes with very low expression were discarded. Differential expression analysis was performed with DESeq2 (77) package in R. The test statistics’ P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg. Genes with adjusted P values lower than 0.05 and absolute log2 fold change greater than 1 were considered significant. PCA plots were generated using regularized log-transformed (rlog) data.
GO pathway enrichment analysis
Enriched pathways were identified based on cluster (scRNA-seq) or population (bulk RNA-seq) differentially expressed gene lists as input using the web implementation of the gProfiler tool, using the g:GOst module (www.biit.cs.ut.ee/gprofiler/). We used pathway gene sets from the “biological processes” (GO:BP) database of GO (www.geneontology.org/) to find overrepresentation of information from the GO terms. All P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg.
Ligand:receptor mapping analysis
Ligand:receptor mapping was performed with the online implementation of the CellPhoneDB v1.0 tool (www.cellphonedb.org/) (78) run without the statistical method. Cell type ligand:receptor interactome was generated with each of the five replicates of the sorted cell population–derived bulk RNA-seq transcriptomes as input, selecting only genes with expression of 16 normalized counts or greater as input. The resulting interaction list was filtered by selecting non–integrin-mediated interactions and TAM ligands that were enriched in the TAM06 scRNA-seq population in the ligand:receptor pairs, finally selecting for ligands present in the GO term “growth factor activity” that were investigated further as potential candidates.
Computational analysis of cancer patient data
RSEM normalized expression datasets from TCGA were downloaded from the Broad Institute Firehose resource (https://gdac.broadinstitute.org/) and analyzed using custom R scripts. The CAF1 gene expression signature was generated by taking the mean normalized log2-transformed expression value of the component signature genes. The CAF1 gene signature genes were selected from the top 25 differentially expressed CAF1 genes by log fold change as the maximum set for which a significant positive correlation was observed between all genes and ACTA2 (αSMA). The final gene set was as follows: ACTA2, MMP13, LRRC15, COL10A1, SPON1, and COL1A1.
Statistics
Normality and homogeneity of variance were determined using a Shapiro-Wilk normality test and an F test, respectively. Statistical significance was then determined using a two-sided unpaired Student’s t test for parametric or Mann-Whitney test for nonparametric data using GraphPad Prism 8 software. A Welch’s correction was applied when comparing groups with unequal variances. Statistical analysis of tumor growth curves was performed using the “compareGrowthCurves” function of the statmod software package (79). No outliers were excluded from any data presented.
Study approval
All experiments involving animals were approved by the Animal and Welfare and Ethical Review Boards of King’s College London and the University of Birmingham and the Home Office UK. Human breast adenocarcinoma tissue was obtained with informed consent under ethical approval from the King’s Health Partners Cancer Biobank (REC reference 12/EE/0493).
Acknowledgments
We thank Y. Haque (KCL) and the NIHR BRC flow cytometry platform at Guy’s and St Thomas’ Biomedical Research Centre for cell sorting and flow cytometry assistance, the Nikon Imaging Centre (KCL) for use of their facilities and assistance with confocal microscopy analyses, A. Saxena (KCL) for support and useful expert discussion regarding the RNA sequencing analyses, U. Niazi (KCL) for bioinformatic support, and A. Ramsay and A. Ivetic (KCL) for helpful discussions. We thank Y. Miwa (Tsukuba University), O. Kanagawa (RCAI, RIKEN), and M. Tomura (Osaka Ohtani University) for the Kaede mice. An earlier version of this manuscript was posted on bioRxiv: https://www.biorxiv.org/content/10.1101/2020.10.30.361907v2.
Funding: This work was funded by a grant from the European Research Council (335326) and Cancer Research UK (DCRPGF\100009). P.G. was supported by a grant from the Wellcome Trust (101529/Z/13/2). J.W.O. and J.E.A. are supported by the UK Medical Research Council (MR/N013700/1) and are KCL members of the MRC Doctoral Training Partnership in Biomedical Sciences. PyMT × Kaede studies were supported by a Cancer Immunology Project Award (C54019/A27535) from Cancer Research UK awarded to D.R.W. The research was supported by the Cancer Research UK King’s Health Partners Centre and Experimental Cancer Medicine Centre at King’s College London, and the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
Author contributions: J.W.O. and J.N.A. conceived the project, designed the approach, performed experiments, interpreted the data, and wrote the manuscript. J.E.A., I.D., E.J.H., I.B., J.C., T.M., P.G., D.S., and R.N. designed the approach, performed experiments, and interpreted the data. S.E.P., T.N., F.D., S.K., D.R.W., and T.L. designed experiments, interpreted the data, and provided key expertise.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The transcriptomic datasets that support the findings of this study are available through the Gene Expression Omnibus: GSE160561 (bulk RNA-seq) and GSE160641 (scRNA-seq). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Legends for tables S1 to S5
Figs. S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
REFERENCES AND NOTES
- 1.Arnold J. N., Magiera L., Kraman M., Fearon D. T., Tumoral immune suppression by macrophages expressing fibroblast activation protein-α and heme oxygenase-1. Cancer Immunol. Res. 2, 121–126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garner H., de Visser K. E., Immune crosstalk in cancer progression and metastatic spread: A complex conversation. Nat. Rev. Immunol. 20, 483–497 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Noy R., Pollard J. W., Tumor-associated macrophages: From mechanisms to therapy. Immunity 41, 49–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muliaditan T., Caron J., Okesola M., Opzoomer J. W., Kosti P., Georgouli M., Gordon P., Lall S., Kuzeva D. M., Pedro L., Shields J. D., Gillett C. E., Diebold S. S., Sanz-Moreno V., Ng T., Hoste E., Arnold J. N., Macrophages are exploited from an innate wound healing response to facilitate cancer metastasis. Nat. Commun. 9, 2951 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassetta L., Fragkogianni S., Sims A. H., Swierczak A., Forrester L. M., Zhang H., Soong D. Y. H., Cotechini T., Anur P., Lin E. Y., Fidanza A., Lopez-Yrigoyen M., Millar M. R., Urman A., Ai Z., Spellman P. T., Hwang E. S., Dixon J. M., Wiechmann L., Coussens L. M., Smith H. O., Pollard J. W., Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 35, 588–602.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis C. E., Harney A. S., Pollard J. W., The multifaceted role of perivascular macrophages in tumors. Cancer Cell 30, 365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian B., Deng Y., Im J. H., Muschel R. J., Zou Y., Li J., Lang R. A., Pollard J. W., A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLOS ONE 4, e6562 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friebel E., Kapolou K., Unger S., Núñez N. G., Utz S., Rushing E. J., Regli L., Weller M., Greter M., Tugues S., Neidert M. C., Becher B., Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell 181, 1626–1642.e20 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Zhang L., Li Z., Skrzypczynska K. M., Fang Q., Zhang W., O'Brien S. A., He Y., Wang L., Zhang Q., Kim A., Gao R., Orf J., Wang T., Sawant D., Kang J., Bhatt D., Lu D., Li C.-M., Rapaport A. S., Perez K., Ye Y., Wang S., Hu X., Ren X., Ouyang W., Shen Z., Egen J. G., Zhang Z., Yu X., Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181, 442–459.e29 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Murray P. J., Allen J. E., Biswas S. K., Fisher E. A., Gilroy D. W., Goerdt S., Gordon S., Hamilton J. A., Ivashkiv L. B., Lawrence T., Locati M., Mantovani A., Martinez F. O., Mege J. L., Mosser D. M., Natoli G., Saeij J. P., Schultze J. L., Shirey K. A., Sica A., Suttles J., Udalova I., van Ginderachter J. A., Vogel S. N., Wynn T. A., Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Palma M., Venneri M. A., Galli R., Sergi L. S., Politi L. S., Sampaolesi M., Naldini L., Tie2 identifies a hematopoietic lineage of proangiogenic monocytes required for tumor vessel formation and a mesenchymal population of pericyte progenitors. Cancer Cell 8, 211–226 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Lin E. Y., Li J. F., Bricard G., Wang W., Deng Y., Sellers R., Porcelli S. A., Pollard J. W., Vascular endothelial growth factor restores delayed tumor progression in tumors depleted of macrophages. Mol. Oncol. 1, 288–302 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Etzerodt A., Tsalkitzi K., Maniecki M., Damsky W., Delfini M., Baudoin E., Moulin M., Bosenberg M., Graversen J. H., Auphan-Anezin N., Moestrup S. K., Lawrence T., Specific targeting of CD163+ TAMs mobilizes inflammatory monocytes and promotes T cell-mediated tumor regression. J. Exp. Med. 216, 2394–2411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muliaditan T., Opzoomer J. W., Caron J., Okesola M., Kosti P., Lall S., van Hemelrijck M., Dazzi F., Tutt A., Grigoriadis A., Gillett C. E., Madden S. F., Burchell J. M., Kordasti S., Diebold S. S., Spicer J. F., Arnold J. N., Repurposing tin mesoporphyrin as an immune checkpoint inhibitor shows therapeutic efficacy in preclinical models of cancer. Clin. Cancer Res. 24, 1617–1628 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeNardo D. G., Brennan D. J., Rexhepaj E., Ruffell B., Shiao S. L., Madden S. F., Gallagher W. M., Wadhwani N., Keil S. D., Junaid S. A., Rugo H. S., Hwang E. S., Jirström K., West B. L., Coussens L. M., Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 1, 54–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchem J. B., Brennan D. J., Knolhoff B. L., Belt B. A., Zhu Y., Sanford D. E., Belaygorod L., Carpenter D., Collins L., Piwnica-Worms D., Hewitt S., Udupi G. M., Gallagher W. M., Wegner C., West B. L., Wang-Gillam A., Goedegebuure P., Linehan D. C., DeNardo D. G., Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 73, 1128–1141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeNardo D. G., Barreto J. B., Andreu P., Vasquez L., Tawfik D., Kolhatkar N., Coussens L. M., CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16, 91–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian B. Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L. R., Kaiser E. A., Snyder L. A., Pollard J. W., CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arwert E. N., Harney A. S., Entenberg D., Wang Y., Sahai E., Pollard J. W., Condeelis J. S., A unidirectional transition from migratory to perivascular macrophage is required for tumor cell intravasation. Cell Rep. 23, 1239–1248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyckoff J. B., Wang Y., Lin E. Y., Li J. F., Goswami S., Stanley E. R., Segall J. E., Pollard J. W., Condeelis J., Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 67, 2649–2656 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Harney A. S., Arwert E. N., Entenberg D., Wang Y., Guo P., Qian B. Z., Oktay M. H., Pollard J. W., Jones J. G., Condeelis J. S., Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage-derived VEGFA. Cancer Discov. 5, 932–943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franklin R. A., Liao W., Sarkar A., Kim M. V., Bivona M. R., Liu K., Pamer E. G., Li M. O., The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guy C. T., Cardiff R. D., Muller W. J., Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12, 954–961 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becht E., Innes L. M., Healy J., Dutertre C.-A., Kwok I. W. H., Ng L. G., Ginhoux F., Newell E. W., Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Street K., Risso D., Fletcher R. B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S., Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills C. D., Kincaid K., Alt J. M., Heilman M. J., Hill A. M., M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 164, 6166–6173 (2000).10843666 [Google Scholar]
- 27.Orecchioni M., Ghosheh Y., Pramod A. B., Ley K., Macrophage polarization: Different gene signatures in M1(LPS+) vs. classically and M2(LPS-) vs. alternatively activated macrophages. Front. Immunol. 10, 1084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boskovic J., Arnold J. N., Stilion R., Gordon S., Sim R. B., Rivera-Calzada A., Wienke D., Isacke C. M., Martinez-Pomares L., Llorca O., Structural model for the mannose receptor family uncovered by electron microscopy of Endo180 and the mannose receptor. J. Biol. Chem. 281, 8780–8787 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Jackson D. G., Prevo R., Clasper S., Banerji S., LYVE-1, the lymphatic system and tumor lymphangiogenesis. Trends Immunol. 22, 317–321 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Ensan S., Li A., Besla R., Degousee N., Cosme J., Roufaiel M., Shikatani E. A., el-Maklizi M., Williams J. W., Robins L., Li C., Lewis B., Yun T. J., Lee J. S., Wieghofer P., Khattar R., Farrokhi K., Byrne J., Ouzounian M., Zavitz C. C. J., Levy G. A., Bauer C. M. T., Libby P., Husain M., Swirski F. K., Cheong C., Prinz M., Hilgendorf I., Randolph G. J., Epelman S., Gramolini A. O., Cybulsky M. I., Rubin B. B., Robbins C. S., Self-renewing resident arterial macrophages arise from embryonic CX3CR1(+) precursors and circulating monocytes immediately after birth. Nat. Immunol. 17, 159–168 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Cho C. H., Koh Y. J., Han J., Sung H. K., Jong Lee H., Morisada T., Schwendener R. A., Brekken R. A., Kang G., Oike Y., Choi T. S., Suda T., Yoo O. J., Koh G. Y., Angiogenic role of LYVE-1-positive macrophages in adipose tissue. Circ. Res. 100, e47–e57 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Pinto A. R., Paolicelli R., Salimova E., Gospocic J., Slonimsky E., Bilbao-Cortes D., Godwin J. W., Rosenthal N. A., An abundant tissue macrophage population in the adult murine heart with a distinct alternatively-activated macrophage profile. PLOS ONE 7, e36814 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu H., Chen M., Reid D. M., Forrester J. V., LYVE-1-positive macrophages are present in normal murine eyes. Invest. Ophthalmol. Vis. Sci. 48, 2162–2171 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Chakarov S., Lim H. Y., Tan L., Lim S. Y., See P., Lum J., Zhang X. M., Foo S., Nakamizo S., Duan K., Kong W. T., Gentek R., Balachander A., Carbajo D., Bleriot C., Malleret B., Tam J. K. C., Baig S., Shabeer M., Toh S. A. E. S., Schlitzer A., Larbi A., Marichal T., Malissen B., Chen J., Poidinger M., Kabashima K., Bajenoff M., Ng L. G., Angeli V., Ginhoux F., Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363, eaau0964 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Lim H. Y., Lim S. Y., Tan C. K., Thiam C. H., Goh C. C., Carbajo D., Chew S. H. S., See P., Chakarov S., Wang X. N., Lim L. H., Johnson L. A., Lum J., Fong C. Y., Bongso A., Biswas A., Goh C., Evrard M., Yeo K. P., Basu R., Wang J. K., Tan Y., Jain R., Tikoo S., Choong C., Weninger W., Poidinger M., Stanley E. R., Collin M., Tan N. S., Ng L. G., Jackson D. G., Ginhoux F., Angeli V., Hyaluronan Receptor LYVE-1-expressing macrophages maintain arterial tone through hyaluronan-mediated regulation of smooth muscle cell collagen. Immunity 49, 1191 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Dollt C., Becker K., Michel J., Melchers S., Weis C. A., Schledzewski K., Krewer A., Kloss L., Gebhardt C., Utikal J., Schmieder A., The shedded ectodomain of Lyve-1 expressed on M2-like tumor-associated macrophages inhibits melanoma cell proliferation. Oncotarget 8, 103682–103692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buiting A. M., Zhou F., Bakker J. A., van Rooijen N., Huang L., Biodistribution of clodronate and liposomes used in the liposome mediated macrophage 'suicide' approach. J. Immunol. Methods 192, 55–62 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Spranger S., Dai D., Horton B., Gajewski T. F., Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31, 711–723.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts E. W., Broz M. L., Binnewies M., Headley M. B., Nelson A. E., Wolf D. M., Kaisho T., Bogunovic D., Bhardwaj N., Krummel M. F., Critical role for CD103(+)/CD141(+) dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in melanoma. Cancer Cell 30, 324–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall S. E., Savill J. S., Henson P. M., Haslett C., Apoptotic neutrophils are phagocytosed by fibroblasts with participation of the fibroblast vitronectin receptor and involvement of a mannose/fucose-specific lectin. J. Immunol. 153, 3218–3227 (1994). [PubMed] [Google Scholar]
- 41.Kraman M., Bambrough P. J., Arnold J. N., Roberts E. W., Magiera L., Jones J. O., Gopinathan A., Tuveson D. A., Fearon D. T., Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 330, 827–830 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Öhlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A. S., Ponz-Sarvise M., Corbo V., Oni T. E., Hearn S. A., Lee E. J., Chio I. I. C., Hwang C.-I., Tiriac H., Baker L. A., Engle D. D., Feig C., Kultti A., Egeblad M., Fearon D. T., Crawford J. M., Clevers H., Park Y., Tuveson D. A., Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa A., Kieffer Y., Scholer-Dahirel A., Pelon F., Bourachot B., Cardon M., Sirven P., Magagna I., Fuhrmann L., Bernard C., Bonneau C., Kondratova M., Kuperstein I., Zinovyev A., Givel A.-M., Parrini M.-C., Soumelis V., Vincent-Salomon A., Mechta-Grigoriou F., Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 33, 463–479.e10 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Davidson S., Efremova M., Riedel A., Mahata B., Pramanik J., Huuhtanen J., Kar G., Vento-Tormo R., Hagai T., Chen X., Haniffa M. A., Shields J. D., Teichmann S. A., Single-cell RNA sequencing reveals a dynamic stromal niche that supports tumor growth. Cell Rep. 31, 107628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartoschek M., Oskolkov N., Bocci M., Lövrot J., Larsson C., Sommarin M., Madsen C. D., Lindgren D., Pekar G., Karlsson G., Ringnér M., Bergh J., Björklund Å., Pietras K., Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat. Commun. 9, 5150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Gassen S., Callebaut B., Van Helden M. J., Lambrecht B. N., Demeester P., Dhaene T., Saeys Y., FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Feig C., Jones J. O., Kraman M., Wells R. J. B., Deonarine A., Chan D. S., Connell C. M., Roberts E. W., Zhao Q., Caballero O. L., Teichmann S. A., Janowitz T., Jodrell D. I., Tuveson D. A., Fearon D. T., Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 20212–20217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Armulik A., Genove G., Betsholtz C., Pericytes: Developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 21, 193–215 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Crisan M., Yap S., Casteilla L., Chen C. W., Corselli M., Park T. S., Andriolo G., Sun B., Zheng B., Zhang L., Norotte C., Teng P. N., Traas J., Schugar R., Deasy B. M., Badylak S., Bűhring H. J., Giacobino J. P., Lazzari L., Huard J., Péault B., A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3, 301–313 (2008). [DOI] [PubMed] [Google Scholar]
- 50.De Palma M., Biziato D., Petrova T. V., Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 17, 457–474 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Tomura M., Yoshida N., Tanaka J., Karasawa S., Miwa Y., Miyawaki A., Kanagawa O., Monitoring cellular movement in vivo with photoconvertible fluorescence protein "Kaede" transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 105, 10871–10876 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller I., Min M., Yang C., Tian C., Gookin S., Carter D., Spencer S. L., Ki67 is a graded rather than a binary marker of proliferation versus quiescence. Cell Rep. 24, 1105–1112.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vento-Tormo R., Efremova M., Botting R. A., Turco M. Y., Vento-Tormo M., Meyer K. B., Park J. E., Stephenson E., Polański K., Goncalves A., Gardner L., Holmqvist S., Henriksson J., Zou A., Sharkey A. M., Millar B., Innes B., Wood L., Wilbrey-Clark A., Payne R. P., Ivarsson M. A., Lisgo S., Filby A., Rowitch D. H., Bulmer J. N., Wright G. J., Stubbington M. J. T., Haniffa M., Moffett A., Teichmann S. A., Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X., Pontén A., Aase K., Karlsson L., Abramsson A., Uutela M., Bäckström G., Hellström M., Boström H., Li H., Soriano P., Betsholtz C., Heldin C. H., Alitalo K., Östman A., Eriksson U., PDGF-C is a new protease-activated ligand for the PDGF α-receptor. Nat. Cell Biol. 2, 302–309 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Gilbertson D. G., Duff M. E., West J. W., Kelly J. D., Sheppard P. O., Hofstrand P. D., Gao Z., Shoemaker K., Bukowski T. R., Moore M., Feldhaus A. L., Humes J. M., Palmer T. E., Hart C. E., Platelet-derived growth factor C (PDGF-C), a novel growth factor that binds to PDGF α and β receptor. J. Biol. Chem. 276, 27406–27414 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Jinnin M., Ihn H., Mimura Y., Asano Y., Yamane K., Tamaki K., Regulation of fibrogenic/fibrolytic genes by platelet-derived growth factor C, a novel growth factor, in human dermal fibroblasts. J. Cell. Physiol. 202, 510–517 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Shook B. A., Wasko R. R., Rivera-Gonzalez G. C., Salazar-Gatzimas E., López-Giráldez F., Dash B. C., Muñoz-Rojas A. R., Aultman K. D., Zwick R. K., Lei V., Arbiser J. L., Miller-Jensen K., Clark D. A., Hsia H. C., Horsley V., Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 362, eaar2971 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou X., Franklin R. A., Adler M., Jacox J. B., Bailis W., Shyer J. A., Flavell R. A., Mayo A., Alon U., Medzhitov R., Circuit design features of a stable two-cell system. Cell 172, 744–757.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roswall P., Bocci M., Bartoschek M., Li H., Kristiansen G., Jansson S., Lehn S., Sjölund J., Reid S., Larsson C., Eriksson P., Anderberg C., Cortez E., Saal L. H., Orsmark-Pietras C., Cordero E., Haller B. K., Häkkinen J., Burvenich I. J. G., Lim E., Orimo A., Höglund M., Rydén L., Moch H., Scott A. M., Eriksson U., Pietras K., Microenvironmental control of breast cancer subtype elicited through paracrine platelet-derived growth factor-CC signaling. Nat. Med. 24, 463–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X., Kumar A., Zhang F., Lee C., Li Y., Tang Z., Arjunan P., VEGF-independent angiogenic pathways induced by PDGF-C. Oncotarget 1, 309–314 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crawford Y., Kasman I., Yu L., Zhong C., Wu X., Modrusan Z., Kaminker J., Ferrara N., PDGF-C mediates the angiogenic and tumorigenic properties of fibroblasts associated with tumors refractory to anti-VEGF treatment. Cancer Cell 15, 21–34 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Dvorak H. F., Tumors: Wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 315, 1650–1659 (1986). [DOI] [PubMed] [Google Scholar]
- 63.Dutton E. E., Gajdasik D. W., Willis C., Fiancette R., Bishop E. L., Camelo A., Sleeman M. A., Coccia M., Didierlaurent A. M., Tomura M., Pilataxi F., Morehouse C. A., Carlesso G., Withers D. R., Peripheral lymph nodes contain migratory and resident innate lymphoid cell populations. Sci. Immunol. 4, eaau8082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quin C. M., Goodman A., Chernyshev V., Kamentsky L., Cimini B. A., Karhohs K. W., Doan M., Ding L., Rafelski S. M., Thirstrup D., Wiegraebe W., Singh S., Becker T., Caicedo J. C., Carpenter A. E., CellProfiler 3.0: Next-generation image processing for biology. PLOS Biol. 16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Opzoomer J. W., Timms J. A., Blighe K., Mourikis T. P., Chapuis N., Bekoe R., Kareemaghay S., Nocerino P., Apollonio B., Ramsay A. G., Tavassoli M., Harrison C., Ciccarelli F., Parker P., Fontenay M., Barber P. R., Arnold J. N., Kordasti S., ImmunoCluster provides a computational framework for the nonspecialist to profile high-dimensional cytometry data. eLife 10, e62915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilkerson M. D., Hayes D. N., ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nowicka M., Krieg C., Crowell H. L., Weber L. M., Hartmann F. J., Guglietta S., Becher B., Levesque M. P., Robinson M. D., CyTOF workflow: Differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res. 6, 748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zappia L., Oshlack A., Clustering trees: A visualization for evaluating clusterings at multiple resolutions. Gigascience 7, giy083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Angerer P., Haghverdi L., Büttner M., Theis F. J., Marr C., Buettner F., Destiny: Diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M. T., Carey V. J., Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Efremova M., Vento-Tormo M., Teichmann S. A., Vento-Tormo R., CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Elso C. M., Roberts L. J., Smyth G. K., Thomson R. J., Baldwin T. M., Foote S. J., Handman E., Leishmaniasis host response loci (lmr1-3) modify disease severity through a Th1/Th2-independent pathway. Genes Immun. 5, 93–100 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends for tables S1 to S5
Figs. S1 to S6
Tables S1 to S5