DRAM-mediated mechanism participates in lipid-induced exosome secretion from hepatic cells in NAFLD.
Abstract
The biogenesis and diagnostic value of exosomes in nonalcoholic fatty liver disease (NAFLD) are unclear. In this study, we revealed that the plasma exosome level was higher in patients with NAFLD than that in healthy controls. Damage-regulated autophagy modulator (DRAM) was identified as one of the genes related to exosome secretion in patients with NAFLD. Then, loss or knockdown of DRAM down-regulated exosome secretion from hepatic cells using a knockout mouse model and a knockdown cell model. DRAM knockout reversed high-fat diet–induced increase of secreted exosomes. Furthermore, DRAM knockdown inhibited fatty acid (FA)–induced lysosomal membrane permeabilization and lysosome inhibitor reversed the down-regulation of exosome release in DRAM knockout mice. Last, FA-induced DRAM interacted with stomatin and promoted its lysosomal localization to enhance exosome secretion from hepatic cells. We revealed a DRAM-mediated mechanism for exosome secretion and provided the foundation for plasma exosomes as a potential biomarker for NAFLD.
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) characterized by excessive fat deposition in liver cells refers to a clinicopathological syndrome that is closely related to insulin resistance and genetic susceptibility (1). The natural history of NAFLD ranges from NAFL to nonalcoholic steatohepatitis (NASH), and the latter might process to cirrhosis and hepatocellular carcinoma (2). NAFLD is becoming one of the most common causes of chronic liver diseases and one of the main reasons for liver transplantation (3). Therefore, the early diagnosis of NAFLD is particularly important. The current gold standard for diagnosing NAFLD is liver biopsy, but it is an invasive examination that has the risk of bleeding, pain, and even death (4). Plasma biomarkers have advantages in speed and convenience for clinical screening, and it is urgent to find a plasma biomarker with high sensitivity and specificity for the diagnosis of NAFLD.
At present, a number of studies have found that extracellular vesicles (EVs) and their contents might be potential biomarkers for diseases (5–7). EVs are membranous vesicles that are secreted from cells and can be mainly classified into microvesicles and exosomes according to the production pathways and sizes. Exosomes are vesicles with a size of 30 to 150 nm that are derived from multivesicular bodies (MVBs) and released by the fusion of MVB with the plasma membrane (8). Exosomes contain a lot of biological molecules such as protein, DNA, and RNA, which are key mediums for cellular communications (9). Previous studies have shown that the number and the content of exosomes could reflect the physiological or pathological state of the cells that release them, and exosomes could affect the behavior of recipient cells through their contents (10, 11). Sun et al. (12) revealed that the number of exosomes could be used as a potential biomarker for cognitive impairment in patients with HIV infection. Liu et al. (13) found that the number of exosomes was positively correlated with the pathological stages of non–small cell lung cancer and had an indicative effect on the prognosis. These suggested that the number of exosomes could change during the development of diseases and might be a potential biomarker for diseases.
Previous studies have shown that exosomes are significantly increased in mice treated with high-fat diet (HFD) and hepatocytes treated with fatty acids (FAs) (14). We used the Gene Expression Omnibus (GEO) database and Gene Set Enrichment Analysis (GSEA) and screened out damage-regulated autophagy modulator (DRAM) as one of the genes that were positively correlated with exosome secretion in patients with NAFLD. DRAM is a lysosomal protein and is regulated by TP53 (15). Studies also showed that FA could induce an increase in the expression of DRAM in hepatic cells (16). These suggested that DRAM might be related with exosome secretion and disease progression of NAFLD.
In this study, we aimed to investigate the role and mechanism of DRAM in exosome release during the NAFLD occurrence. Using the GEO database and GSEA, DRAM was identified as one of the genes related to the exosome secretion in patients with NAFLD. CRISPR-Cas9 technology was used to construct the DRAM knockout mouse, and the results showed that loss of DRAM could inhibit HFD-induced release of exosomes. Further investigations showed that lipid-induced DRAM could recruit stomatin (STOM) to the lysosomes, lead to the lysosomal membrane permeabilization (LMP), and further promote the release of exosomes from hepatic cells. These data could provide the DRAM-mediated mechanism for lipid-induced release of exosomes from hepatic cells, and the number of plasma exosomes might be used as one of the diagnostic indicators for patients with NAFLD.
RESULTS
Plasma exosome level was increased in patients with NAFLD
To evaluate the level of plasma exosomes in patients with NAFLD, nanoparticle tracking analysis (NTA) after exosome isolation, bicinchoninic acid (BCA) assays for protein quantification of exosomes after exosome isolation, and phosphatidylserine enzyme-linked immunosorbent assay (ELISA) were used to detect the exosome level in plasma of patients with NAFLD and healthy controls. The process of phosphatidylserine ELISA for detecting the level of plasma exosomes is shown in Fig. 1A. The quality of plasma exosomes isolated by ultracentrifugation was assessed by transmission electron microscopy, NTA, and Western blotting assays for exosome markers including CD9 and CD63 (Fig. 1, B to D). Moreover, we did not detect protein expressions of the cytosolic marker tubulin and the microvesicle marker matrix metallopeptidase 2 (MMP2) in isolated exosomes from plasma samples (Fig. 1D). Then, we analyzed the correlations of the detection results using three methods, and we revealed that the detection results between ELISA and NTA, ELISA and BCA, and BCA and NTA were all positively correlated (Fig. 1, E to G). These suggested that the three methods for detecting plasma exosome level had similar detection capabilities. However, phosphatidylserine ELISA for directly detecting the level of plasma exosomes was faster and simpler. Subsequently, we further analyzed the data using phosphatidylserine ELISA and found that the plasma exosome level in patients with NAFLD was significantly higher than that in healthy controls (Fig. 1H). Meanwhile, the results of Western blotting showed that the protein levels of exosome markers including CD9 and CD63 were significantly increased in patients with NAFLD compared with healthy controls (Fig. 1I). By analyzing the markers derived from different tissues, we found that the expression of liver-derived exosome marker cytochrome P450 family 2 subfamily E member 1 (CYP2E1) was significantly higher in patients with NAFLD, while the small intestine–derived exosome marker glycoprotein A33 (GPA33) and the adipocyte-derived exosome marker FA binding protein 4 (FABP4) were not statistically different between the two groups (Fig. 1I). These data suggested that the level of liver-derived exosomes was increased in patients with NAFLD.
Fig. 1. Plasma exosome level was measured by three methods in patients with NAFLD.
(A) The diagram of phosphatidylserine ELISA for detection of plasma exosome level. TIM4 is a receptor of phosphatidylserine that is on the surface of exosomes. (B) The representative morphological image of plasma exosomes obtained by ultracentrifugation under the transmission electron microscope. (C) Results of NTA for size distribution of exosomes in patients with NAFLD. (D) Western blotting assays for exosome markers including CD63 and CD9, the cytosolic marker tubulin, and the microvesicle marker MMP2. (E to G) The correlations among the detection results of BCA assay, NTA, and phosphatidylserine ELISA (n = 31). (H) The levels of plasma exosomes in patients with NAFLD (n = 19) and healthy controls (HC; n = 12) using phosphatidylserine ELISA. (I) Western blotting analysis for CD9, CD63, cytochrome P450 family 2 subfamily E member 1 (CYP2E1), glycoprotein A33 (GPA33), and FA binding protein 4 (FABP4) of isolated plasma exosomes from patients with NAFLD and HC. Data were presented as means and SD. Statistical significance was calculated by t test. *P < 0.05; **P < 0.01.
DRAM was positively correlated with exosome secretion–related genes in liver tissues of patients with NAFLD
To further investigate the genes correlated with the increased exosomes in patients with NAFLD, we analyzed gene expressions in liver tissues of patients with NAFLD using a dataset (GSE89632) from the GEO database. The differentially expressed genes were selected and the cohort was divided into the high and low gene expression groups according to the cutoff value. Using GSEA, we found that exosome secretion–related genes were significantly enriched in the DRAM high-expression group of patients with NAFL and healthy controls (normalized enrichment score = 1.49, P < 0.05; Fig. 2A). In addition, the expression levels of RAB27B, VAMP3, and YKT6, which could promote exosome secretion, were significantly higher in liver tissues of patients with NAFL than that of healthy controls (Fig. 2, B to D). The expression levels of RAB27B, VAMP3, and YKT6 were also significantly higher in the DRAM high-expression group than that in the DRAM low-expression group (Fig. 2, E to G). Then, we analyzed the expression levels of DRAM in healthy controls, patients with NAFL, and patients with NASH. The results showed that DRAM expression in liver tissues of patients with NAFL was significantly higher than that of healthy controls (Fig. 2H), while there was no statistically significant difference of DRAM level in liver tissues between the patients with NASH and the healthy controls (fig. S1). Receiver operating characteristic (ROC) curve was plotted for DRAM expression level in liver tissues of patients with NAFL and healthy controls. The area under the ROC curve (AUC) was 0.731, while the sensitivity and specificity were respectively 60 and 83.3% at the optimal cutoff value (Fig. 2I). These data suggested that patients with NAFL and healthy controls could be distinguished well according to the expression level of DRAM in liver tissues. Furthermore, significant positive correlations between the DRAM expression level in liver tissues and plasma ALT, plasma AST, or liver steatosis were observed (Fig. 2, J, K, and L). In summary, DRAM might be closely related to the secretion of exosomes and the occurrence of NAFLD.
Fig. 2. DRAM expression in liver tissues was correlated with exosome secretion–related genes in patients with NAFLD.
(A) The samples were divided into the DRAM high-expression group (DRAM-high) and the DRAM low-expression group (DRAM-low) according to the cutoff value. GSEA showed that genes related to exosome secretion were enriched in DRAM-high. NES, normalized enrichment score. (B to D) Expression levels of the genes including RAB27B, VAMP3, and YKT6 in liver tissues of HC (n = 24) and patients with NAFL (NAFL; n = 20). (E to G) Expression levels of RAB27B, VAMP3, and YKT6 in liver tissues of the DRAM-high (n = 16) and the DRAM-low (n = 28). (H) The mRNA expressions of DRAM in the HC and NAFL group. (I) The receiver operating characteristic curve and area under the receiver operating characteristic curve (AUC) value of DRAM level in liver tissues to differentiate NAFL from HC. (J to L) The correlations between DRAM expression and NAFLD indicators such as plasma ALT, plasma AST, and the degree of liver steatosis. Data were presented as means and SD. Statistical significance was calculated by t test. *P < 0.05; **P < 0.01; ***P < 0.001.
Liver-derived exosome level was decreased in plasma of DRAM knockout mice
To investigate the effects of DRAM on the level of plasma exosomes, CRISPR-Cas9 technology was used to construct a DRAM knockout mouse model (Fig. 3A), and the heterozygous mice (DRAM+/−) were mated to obtain DRAM wild-type (DRAM+/+) mice and DRAM knockout (DRAM−/−) mice (Fig. 3B). The mRNA expression of DRAM in liver tissues was analyzed to verify the construction of DRAM knockout mice (Fig. 3C). Next, we detected the level of plasma exosomes in wild-type and DRAM knockout mice using phosphatidylserine ELISA, and the results showed that the level of plasma exosomes in DRAM knockout mice was lower than that in wild-type mice (Fig. 3D). After isolation of plasma exosomes from mice of the two groups, NTA and BCA assays also showed that the level of plasma exosomes in DRAM knockout mice was significantly decreased (Fig. 3, E and F). Results of Western blotting showed that exosome markers including CD9 and CD63 in plasma exosomes were significantly decreased in DRAM knockout mice compared with that in wild-type mice (Fig. 3G). Furthermore, exosome markers derived from different tissues were also measured by Western blotting; the results showed that the expression of liver-derived exosome marker CYP2E1 in the plasma exosomes of DRAM knockout mice was significantly lower than that of wild-type mice, while there were no significant differences of the small intestine–derived exosome marker GPA33 and adipocyte-derived exosome marker FABP4 between the two groups of mice (Fig. 3G). These data suggested that DRAM knockout in mice might inhibit the exosome secretion from liver into the plasma.
Fig. 3. DRAM knockout affected the plasma exosome level of mice.
(A) The diagram for the construction of DRAM knockout mice. (B) Illustration of the mating strategy for DRAM knockout mice. (C) The mRNA expressions of DRAM in liver tissues of wild-type (DRAM+/+) mice and DRAM knockout (DRAM−/−) mice (n = 5). (D) Phosphatidylserine ELISA for detecting the levels of plasma exosomes in wild-type mice and DRAM knockout mice (n = 5). (E and F) ExoQuick plasma prep and exosome precipitation kit was used to extract exosomes from same amount of plasma in wild-type mice and DRAM knockout mice (n = 5). The plasma exosome level was measured by BCA assay and NTA. (G) Western blotting analysis for CD9, CD63, CYP2E1, GPA33, and FABP4 of isolated plasma exosomes from the wild-type and DRAM knockout mice. Data were presented as means and SD. Statistical significance was calculated by t test. *P < 0.05; **P < 0.01; ND, not detected.
DRAM up-regulation promoted the secretion of exosomes in vitro
Next, we further confirmed the effects of DRAM on exosome secretion in vitro. The exosomes from cell culture supernatant of hepatic cell line HepG2 transfected with DRAM expression plasmid or empty vector were extracted. The results revealed that the levels of CD9 and CD63 in exosomes of cell culture supernatant were significantly increased in HepG2 cells with DRAM overexpression (Fig. 4A). The protein level of exosomes in DRAM-overexpressed HepG2 cells was significantly higher than that in the control group measured by BCA (Fig. 4B). It was also found that the level of exosomes secreted from HepG2 cells was significantly increased after overexpression of DRAM using phosphatidylserine ELISA (Fig. 4C). At the same time, after silencing DRAM, the protein levels of CD9 and CD63 were significantly reduced (Fig. 4D). Results of phosphatidylserine ELISA also showed that the level of exosomes secreted from HepG2 cells was significantly decreased after knocking down DRAM (Fig. 4E). In summary, we verified that DRAM could promote the exosome secretion of hepatic cells in vitro.
Fig. 4. DRAM affected exosome secretion from hepatic cells.
(A) Western blotting assays for protein levels of CD9, CD63, and Flag-DRAM in cell lysates and exosomal lysates from cell culture supernatant of HepG2 cells transfected with empty vector or Flag-DRAM expression plasmid. (B and C) The exosome levels from cell culture supernatant of HepG2 cells transfected with empty vector or Flag-DRAM expression plasmid were measured by BCA assay or phosphatidylserine ELISA. (D) Western blotting assays for protein levels of CD9, CD63, and DRAM in cell lysates and exosomal lysates from cell culture supernatant of HepG2 cells transfected with control small interfering RNAs (siRNAs) (si-con) or siRNAs targeting DRAM (si-DRAM). (E) Phosphatidylserine ELISA for the level of exosomes in cell culture supernatant of HepG2 cells transfected with si-con or si-DRAM. Data were presented as means and SD of three independent experiments. Statistical significance was calculated by t test. *P < 0.05.
Down-regulation of DRAM inhibited lipid-induced increase of exosome secretion
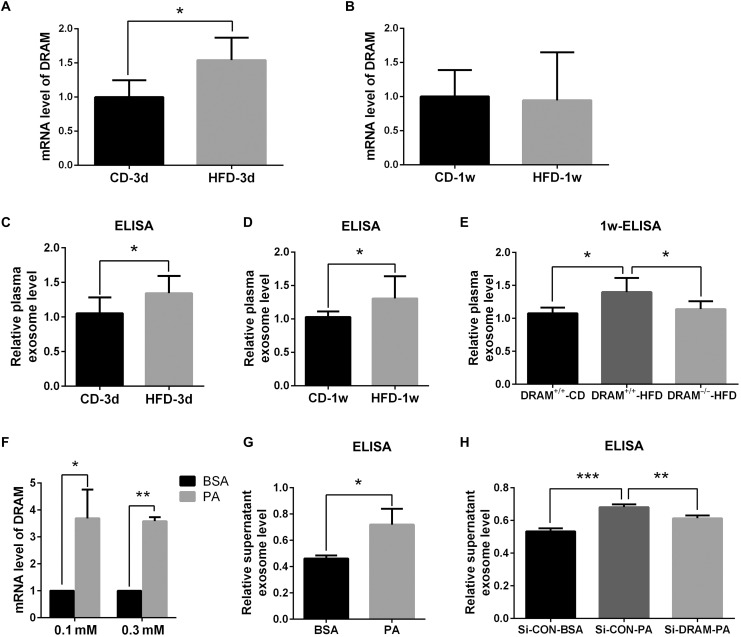
Previous studies have revealed that HFD and FA could increase the level of exosomes (14). To further explore the role of DRAM in HFD- or FA-induced exosome secretion of hepatic cells, first, we fed the wild-type mice with HFD and measured the DRAM expression in liver tissues at different time points. We found that DRAM expression of liver tissues was significantly increased at 3 days after HFD feeding (Fig. 5A). However, there was no statistical difference in DRAM expression of liver tissues between the HFD and control diet (CD) group at 1 week (Fig. 5B). Phosphatidylserine ELISA was used to detect the level of plasma exosomes, and it was found that the level of plasma exosomes in the HFD group was significantly higher than that in the CD group at 3 days and 1 week after HFD or CD feeding (Fig. 5, C and D). These results indicated that DRAM expression was increased in the liver of mice fed with HFD within a short period of time, accompanied by an increase in plasma exosome level. Then, we fed the wild-type and DRAM knockout mice with HFD or CD for 1 week, and found that knockout of DRAM could inhibit the HFD-induced increase of the plasma exosome level (Fig. 5E).
Fig. 5. DRAM affected the HFD- or FA-induced secretion of exosomes from hepatic cells in vivo and in vitro.
(A and B) The mRNA expression of DRAM in liver tissues of wild-type mice fed with CD or HFD at different time points (n = 3). Statistical significance was calculated by t test. (C and D) Phosphatidylserine ELISA for measuring the level of plasma exosomes in wild-type mice fed with CD or HFD at different time points (n = 4). Statistical significance was calculated by t test. (E) Phosphatidylserine ELISA for the levels of plasma exosomes in wild-type mice and DRAM knockout mice fed with HFD or CD for 1 week (n = 5). Statistical significance was calculated by one-way analysis of variance (ANOVA) followed by Tukey’s comparisons. (F) The mRNA levels of DRAM in HepG2 cells stimulated with 0.1 or 0.3 mM palmitic acid (PA). Statistical significance was calculated by t test. (G) Phosphatidylserine ELISA for the levels of exosomes in cell culture supernatant of HepG2 cells treated with 0.3 mM PA. Statistical significance was calculated by t test. (H) Phosphatidylserine ELISA for the exosome levels in cell culture supernatant of PA-treated HepG2 cells transfected with si-con or si-DRAM. Statistical significance was calculated by one-way ANOVA followed by Tukey’s comparisons. Data were presented as means and SD. *P < 0.05; **P < 0.01; ***P < 0.001.
Subsequently, we investigated the role of DRAM in exosome secretion from the FA-treated hepatic cells. First, HepG2 cells were treated with palmitic acid (PA) and the lipid accumulation in HepG2 cells with PA treatment was observed in fig. S2. The expression of DRAM was significantly up-regulated after PA treatment (Fig. 5F). We also found that the level of exosomes secreted from HepG2 cells was significantly increased after PA treatment (Fig. 5G). Further analysis indicated that silencing DRAM could inhibit the effects of PA on exosome secretion from HepG2 cells (Fig. 5H). These results indicated that DRAM knockdown could also inhibit FA-induced increase of exosome secretion from hepatic cells.
Silencing DRAM inhibited FA-induced LMP in hepatic cells to decrease the release of exosomes
When LMP was induced and the degradation of MVB containing exosomal precursors through lysosomes was inhibited, the release of exosomes could be promoted through the interaction between the MVB and the plasma membrane (17, 18). DRAM is a lysosomal membrane protein, and we further explored whether DRAM could affect the secretion of exosomes by regulating LMP. First, we used lysosomal galectin 3 (LGALS3) puncta assays to evaluate the effect of DRAM on LMP. LGALS3 could enter the lysosomes from cytoplasm after LMP. Our results showed that the location of LGALS3 in HepG2 cells with DRAM overexpression changed from diffuse cytoplasmic staining to the puncta in lysosomes (Fig. 6A). Meanwhile, compared with the control group, the level of cathepsin B (CTSB) was increased in the cytoplasm while its level in lysosomes was decreased in HepG2 cells after DRAM overexpression, which suggested that DRAM might induce LMP to increase the release of CTSB from lysosomes to cytoplasm (Fig. 6B). Further analysis showed that LGALS3 aggregation in lysosomes was observed in HepG2 cells with FA treatment and silencing DRAM could inhibit the FA-induced LGALS3 aggregation in lysosomes (Fig. 6C). Simultaneously, CTSB level in the cytoplasm of HepG2 cells was increased and its lysosomal level was reduced after FA stimulation, while CTSB diffusion from lysosomes to cytoplasm induced by FA was significantly inhibited by knockdown of DRAM (Fig. 6D). In view of the results above, we revealed that DRAM could induce LMP and DRAM knockdown could inhibit FA-induced LMP. Furthermore, the wild-type and DRAM knockout mice were injected with Bafilomycin A1 (Baf), which was a lysosomal inhibitor that could inhibit lysosomal function. Our results showed that Baf injection compensated for the decrease in exosome secretion caused by DRAM knockout (Fig. 6E). These suggested that the inhibition of lysosomal function could reverse DRAM knockout–induced down-regulation of exosome release. Immunofluorescence assays showed that the colocalization of MVB marker CD63 and lysosome marker lysosomal-associated membrane protein 1 (LAMP1) was increased in liver tissues of DRAM knockout mice compared to that of wild-type mice (fig. S3). This indicated that knockout of DRAM might promote the interaction between the MVB and lysosomes, increase the degradation of MVB through lysosomes, and inhibit the release of exosomes.
Fig. 6. DRAM affected FA-induced LMP and exosome secretion of hepatic cells.
(A) LGALS3–enhanced green fluorescent protein (EGFP) expression plasmid was cotransfected with empty vector or DRAM plasmid into the HepG2 cells, and the aggregation of LGALS3 was observed under a fluorescence microscopy. Scale bars, 10 μm. Cells with the aggregative puncta staining were counted and calculated in the right diagram. Statistical significance was calculated by t test. (B) Western blotting assays for the CTSB levels of lysosomes and cytoplasm in HepG2 cells transfected with empty vector or DRAM expression plasmid. (C) LGALS3-EGFP expression plasmid was cotransfected with si-con or si-DRAM into bovine serum albumin (BSA)– or PA-treated HepG2 cells. The aggregation of LGALS3 was observed under a fluorescence microscope. Scale bars, 10 μm. Cells with the aggregative puncta staining were counted and calculated in the right diagram. Statistical significance was calculated by one-way ANOVA followed by Tukey’s comparisons. (D) Western blotting for the CTSB levels of lysosomes and cytoplasm in BSA- or PA-treated HepG2 cells transfected with si-con or si-DRAM. (E) Phosphatidylserine ELISA for the levels of plasma exosomes in wild-type mice and DRAM knockout mice intraperitoneally injected with Bafilomycin A1 or dimethyl sulfoxide (DMSO) (n = 3). Statistical significance was calculated by t test. Data were presented as means and SD. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant.
DRAM interacted with STOM to promote exosome secretion of hepatic cells
Previous studies have reported that STOM could interrupt membrane integrity and increase biofilm permeability (19). It has also been pointed out that STOM is mainly distributed in plasma membrane and LAMP2-positive perinuclear vesicles such as late endosomes and lysosomes (20). Our analysis revealed the interaction between DRAM and STOM through coimmunoprecipitation (co-IP) assays (Fig. 7, A and B). Immunofluorescence assay showed that DRAM and STOM might be colocalized in hepatic cells (Fig. 7C). Meanwhile, the colocalizations of DRAM and STOM were also observed in liver tissues of mice fed with both CD and HFD (fig. S4A). Then, we found that STOM protein level in lysosomes was increased and its level in cytoplasm was decreased in HepG2 cells with DRAM overexpression (Fig. 7D). At the same time, we identified that the protein level of STOM in lysosomes was increased in hepatic cells after the stimulation of FA, while silencing DRAM inhibited the increase of lysosomal STOM level induced by FA (Fig. 7E). Aggregated LGALS3 puncta and increased protein level of CTSB in cytoplasm were observed in HepG2 cells with STOM overexpression, which indicated that STOM could induce LMP (fig. S4, B and C). Furthermore, we silenced STOM in HepG2 cells with DRAM overexpression (Fig. 7F) and found that silencing STOM could reverse the increased exosome release in hepatic cells caused by DRAM overexpression (Fig. 7G). Therefore, we revealed that FA-induced DRAM might affect LMP and exosome secretion by recruiting STOM to the lysosomes.
Fig. 7. DRAM interacted with STOM and affected its lysosomal localization.
(A and B) Co-IP analysis in HepG2 cells transfected with Flag-DRAM and STOM-Myc plasmids using anti-Flag or anti-Myc antibody. Western blotting assays were conducted using anti-Flag and anti-STOM antibodies. (C) HepG2 cells were cotransfected with Flag-DRAM and STOM-Myc plasmids, and Flag-DRAM was stained with green fluorescence and STOM-Myc was stained with red fluorescence. The yellow color of merged picture indicated colocalization. Scale bars, 10 μm. (D) Western blotting for STOM levels of lysosomes and cytoplasm in HepG2 cells transfected with empty vector or DRAM expression plasmid. (E) Western blotting for STOM levels of lysosomes in BSA- or PA-induced HepG2 cells transfected with si-con or si-DRAM. (F) Western blotting for the levels of STOM and Flag-DRAM in HepG2 cells transfected with empty vector, Flag-DRAM expression plasmid, si-con, or si-STOM. (G) Phosphatidylserine ELISA for the exosome levels in HepG2 cells transfected with the empty vector, Flag-DRAM expression plasmid, si-con, or si-STOM. Data were presented as means and SD of three independent experiments. Statistical significance was calculated by one-way ANOVA followed by Tukey’s comparisons. *P < 0.05; **P < 0.01.
DISCUSSION
Currently, the plasma indicator with good sensitivity and specificity in diagnosis of NAFLD is still needed. In the past few years, exosomes and their contents have been considered as potential biomarkers for diseases (21–23). Previous studies in vivo had shown that the number of EVs or exosomes secreted from hepatic cells with PA treatment was significantly increased (14, 24, 25). The level of plasma EVs or exosomes was also significantly increased in HFD-treated mice and patients with NAFLD (14, 26). In this study, we confirmed that the plasma exosome level was higher in patients with NAFLD than that in healthy controls, which indicated that plasma exosome level may be a potential biomarker for patients with NAFLD. Our experiments also revealed that phosphatidylserine ELISA for measuring the level of plasma exosomes had a similar detection capability compared with NTA and BCA assay after exosome isolation. Phosphatidylserine ELISA was a simpler and faster method, and it would be more suitable for large-scale diagnosis for patients with NAFLD.
Through the analysis of the GEO database, we found that the expression level of DRAM in liver tissues of patients with NAFL was significantly higher than that of healthy controls. Using the expression level of DRAM in liver tissues as an indicator to distinguish patients with NAFL from healthy controls, the value of AUC could reach 0.731 (Fig. 2I). These data indicated that the expression of DRAM in liver tissues might be able to diagnose patients with NAFLD in the population.
DRAM as a lysosomal membrane protein could participate in TP53-induced autophagy and could be induced by FA treatment (15, 16). It has been revealed that DRAM knockout could decrease HFD-induced glucose intolerance and the accumulation of adipose tissues in mice (27). The results of in vivo experiments showed that DRAM expression in liver tissues was significantly increased at 3 days and unchanged at 1 week after HFD feeding (Fig. 5, A and B). Another study also showed that the expression level of DRAM was higher in hepatic cells treated with 400 μM oleic acid (OA) than that in control cells, while DRAM level was significantly decreased in hepatic cells treated with higher concentrations (800 or 1200 μM) of OA compared with cells treated with 400 μM OA (28). Two points should be considered in this issue. On the one hand, DRAM could be induced by FA, starvation, adriamycin in high glucose, and virus infection, which indicated that DRAM might be one of the stress-induced genes (16, 29–31). Expressions of some stress-induced genes could be temporarily increased under the stresses, while the temporary increase of these genes could play important roles in stress-induced changes in cellular processes (32, 33). For example, NF-E2–related factor 2 (Nrf2) expression could be temporarily increased and then decreased, and knockdown of Nrf2 inhibited angiogenesis in cerebral microvascular endothelial cells under hypoxic conditions (34). On the other hand, DRAM is a downstream gene of TP53, and it has been reported that TP53 could play dual roles during the progression of NAFLD (35). The disease spectrum of NAFLD ranges from NAFL to NASH and might process to cirrhosis. The expression and role of DRAM might also be different in various stages of NAFLD. Our results found that the expression level of DRAM in liver tissues of patients with NAFL was significantly higher than that of healthy controls, while the difference of DRAM in liver tissues between the patients with NASH and healthy controls was not statistically significant (Fig. 2H and fig. S1). These data suggested that DRAM level might be increased in the early stage during the occurrence of NAFLD. In a future study, we would further investigate whether DRAM could play different roles in various stages of NAFLD.
Until now, some molecules have been reported to affect the process of exosome biogenesis. Loss of sirtuin 1 was revealed to inhibit the acidification of lysosomes, lead to impaired function of lysosomes, and promote the secretion of exosomes (18). Yan et al. (36) showed that AMPKα1 could regulate the secretion of exosomes by inhibiting tumor susceptibility 101 (TSG101) expression. In this study, our results showed that DRAM could also promote the secretion of exosomes from hepatic cells in vitro and in vivo. DRAM is a lysosomal protein and lysosomes could play an essential role in the secretion of exosomes. Studies have revealed that impaired lysosomal function reduced the degradation of MVB containing exosomal precursors through lysosomes, led to more MVB fusion with the plasma membrane, and increased the release of exosomes from cells (18). Our research also focused on the mechanism of DRAM involved in mediating the secretion of exosomes by promoting LMP. In addition, we also revealed the interaction between DRAM and STOM and the DRAM-mediated lysosomal localization of STOM. STOM is a protein that can change membrane integrity and is highly expressed in liver tissues. We would focus on the influence of STOM on lipid accumulation of hepatic cells in a future study.
In summary, our study showed that lipid-induced DRAM in hepatic cells could interact with STOM, promote its lysosomal localization, and induce LMP, which could reduce lysosomal degradation of MVB and promote the release of exosomes (Fig. 8). Our study not only revealed the DRAM-mediated mechanism for lipid-induced exosome secretion but also suggested that the plasma exosome level might act as a potential biomarker for patients with NAFLD. Phosphatidylserine ELISA method, which captures exosomes by phosphatidylserine to detect the level of exosomes, provides greater feasibility for the use of plasma exosomes as a clinical diagnostic indicator.
Fig. 8. Diagram for the role of DRAM in exosome secretion from hepatocytes during the occurrence of NAFLD.
HFD or FA could up-regulate the expression of DRAM in hepatocytes. DRAM further recruited STOM to the lysosomes and induced LMP, which impaired the lysosomal function and the degradation of MVB in lysosomes to promote the secretion of exosomes.
MATERIALS AND METHODS
Human samples
Nineteen patients with NAFLD and 12 healthy subjects were recruited in this study. Patients with NAFLD were diagnosed by liver biopsy, computed tomography (CT), or color Doppler ultrasound after excluding alcohol and other clear liver damage factors (37). Healthy subjects have normal liver enzymes and abdominal CT results. The venous blood was taken from the participants after 8 hours of fasting. Every participant signed an informed consent form, and this study was approved by the Ethics Committee of Qingdao Municipal Hospital. Demographic and laboratory characteristics of the participants are listed in table S1.
Data collection and processing
The mRNA expressions and clinical data of 20 patients with NAFL, 19 patients with NASH, and 24 healthy controls were obtained from a dataset with the accession number GSE89632 of the GEO database. GSEA software (https://software.broadinstitute.org/gsea/index.jsp) was performed to calculate the enrichment of genes (38, 39). According to the optimal cutoff value of gene expression in liver tissues, the subjects were divided into the high and low gene expression groups.
Animal experiments
C57BL/6 mice with DRAM heterozygous knockout (DRAM+/−) were obtained from Shanghai Model Organisms (Shanghai, China). DRAM heterozygous knockout (DRAM+/−) mice were mated to generate homozygous (DRAM−/−) and wild-type (DRAM+/+) littermates. Eight-week-old male mice in the DRAM knockout group and wild-type group were fed with CD or HFD (60% fat, Research Diets, New Brunswick, NJ, USA). All mice were randomly selected and maintained under a 12-hour light/dark cycle. At the end of experiments, mice were euthanized after 6 hours of fasting. All animal experiments were carried out in accordance with the guideline approved by the Medical Experimental Animal Care Commission of Qingdao University.
Cell culture
Human hepatic cell line HepG2 was obtained from GeneChem (Shanghai, China). STR profiling and mycoplasma contamination testing were provided by GeneChem. HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Biological Industries, Kibbutz Beit Haemek, Israel). Cells were cultured in a humidified 5% CO2 incubator at 37°C. Exo-clear complete cell growth medium (System Biosciences, Palo Alto, CA, USA) was used for extraction of exosomes from cell culture supernatant.
Oil red O staining and intracellular triglyceride measurement
For FA treatment, 0.3 mM PA or bovine serum albumin solution (Kunchuang Biotechnology, Xian, China) was incubated with the cultured HepG2 cells. After 24 hours, the intracellular lipids were stained using oil red O staining kit (Solarbio, Beijing, China) and the content of intracellular triglycerides was detected by a triglyceride assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions.
Plasmids, siRNAs, and transfection agents
Small interfering RNAs (siRNAs) were synthetized from GenePharma (Shanghai, China). siRNAs were transfected using HiPerFect transfection reagent (Qiagen, Dusseldorf, Germany) according to the protocol from the manufacturer.
Expression plasmids for Flag-DRAM, STOM-Myc, and LGALS3–enhanced green fluorescent protein (EGFP) were obtained from Youbio Biological Technology (Hunan, China). Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) was used to transfect plasmids according to the instruction from the manufacturer.
RNA isolation and qRT-PCR
Total RNA was extracted from cells and tissues using RNAiso (Takara, Shiga, Japan). Then, RNA was converted to cDNA using PrimeScript RT reagent kit with genomic DNA Eraser (Takara). The quantification of gene expression was determined with the SYBR Green polymerase chain reaction (PCR) kit (Qiagen). The following primers were used for quantitative reverse transcription PCR (qRT-PCR) analysis in this study: DRAM (mouse): 5′-CAGCCTTCATCATCTCCTACG-3′ and 5′-ATGCAGAGAAGTTTATCATG-3′ (27), DRAM (human): 5′-TCAAATATCACCATTGATTTCTGT-3′ and 5′-GCCACATACGGATGGTCATCTCTG-3′ (16), β-actin (mouse): 5′-CAGCTTCTTTGCAGCTCCTT-3′ and 5′-CACGATGGAGGGAATACAG-3′, and β-actin (human): 5′-CATCCTCACCCTGAAGTACCCC-3′ and 5′-AGCCTGGATGCAA CGTACATG-3′.
Exosome isolation
For the extraction of exosomes from human plasma and cell culture media, first, cells were removed (300g for 10 min) and the supernatant was centrifuged at 2000g for 10 min to discard dead cells. Then, the supernatant or plasma was collected and centrifuged at 12,000g for 30 min to remove cell debris. Afterward, the supernatant or plasma was filtered through a centrifugal filter unit (Millipore, Billerica, MA, USA) to remove the apoptotic bodies and microvesicles. Next the supernatant or plasma was collected and ultracentrifuged at 110,000g for 2 hours to precipitate the exosomes. Subsequently, the exosomes were washed with phosphate-buffered saline (PBS) to remove contaminated proteins and the ultracentrifugation at 110,000g for 70 min was used to collect the exosomes. For the extraction of exosomes from plasma of mice, plasma was filtered through a centrifugal filter unit (Millipore), and ExoQuick plasma prep and exosome precipitation kit (System Biosciences) was used according to the instruction of the manufacturer.
Nanoparticle tracking analysis
The concentration and size of exosomes were measured using NanoSight NS300 (Malvern, Worcs, UK). The sample was diluted with PBS and injected into the beam path to capture the fast-moving diffraction light spot under the Brownian motion of each particle. Five digital videos of each sample were analyzed to determine the concentration and size of exosomes.
Electron microscopy
The exosomes resuspended in PBS were added dropwise to formvar/carbon-coated copper grid and stained with 3% aqueous phosphotungstic acid for 5 min at room temperature. Then, the grids were observed under a transmission electron microscope (JEOL, Tokyo, Japan).
Lysosomal LGALS3 puncta assay
To perform the LGALS3 puncta assay, LGALS3-EGFP expression plasmid was cotransfected with empty vector or DRAM plasmid into the HepG2 cells. After 48 hours, three areas were randomly selected to observe the fluorescence. There are at least 100 cells in each area and more than three puncta in one cell are defined as positive.
Lysosome isolation
The lysosome isolation kit (Genmed Scientifics Inc., TX, USA) was used to isolate lysosomes. In short, cells were mixed with lysis buffer and homogenized with a dounce homogenizer. The separation solution was added into the supernatant and centrifuged at 25,000g for 20 min. Then, the supernatant was collected as cytoplasm and the precipitate was collected as lysosome fraction.
Coimmunoprecipitation
HepG2 cells were cotransfected with Flag-DRAM expression plasmid and STOM-Myc expression plasmid. Then, cells were collected and lysed by NP-40 lysis buffer (50 mM tris, 100 mM NaCl, 1% NP-40, 10% glycerol, 1 mM Na3VO4, 10 mM sodium fluoride (NaF), and 1.5 mM EDTA). The primary antibody or normal immunoglobulin G (Santa Cruz Biotechnology, Dallas, TX, USA) was respectively incubated with the cell lysate overnight. Then, Protein A/G Plus-Agarose (Santa Cruz Biotechnology) was added to the lysate and incubated. The precipitate was collected and Western blotting was performed.
Phosphatidylserine ELISA
The collected supernatant or plasma was passed through a centrifugal filter unit (Millipore) to remove the apoptotic bodies and microvesicles. Then, the exosome level in plasma or cell supernatant was quantified by a phosphatidylserine ELISA kit according to the manufacturer’s instruction (Wako, Osaka, Japan).
Immunofluorescence
HepG2 cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Liver tissues were fixed with 4% paraformaldehyde and embedded in paraffin. Then, the primary and secondary antibodies were subsequently incubated with the cells and tissues. 4′,6-Diamidino-2-phenylindole (Servicebio, Wuhan, China) was used to stain the nuclei. The images were observed and captured under a florescence microscope (Nikon, Tokyo, Japan), and they were quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Western blotting
Cells and exosomes were lysed in radioimmunoprecipitation assay (RIPA) buffer (SolarBio) containing the protease inhibitor cocktail (Merck, Darmstadt, Germany). Proteins were separated by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membrane (Millipore). The membrane was blocked with 5% milk and incubated with primary antibody overnight at 4°C. Then, the membrane was incubated with the secondary antibody. The antibodies used in this study are listed in table S2.
BCA assay
The BCA kit (Cwbio, Beijing, China) was used to measure the protein level of exosomes and cells. In short, RIPA buffer was used to extract proteins from cells or exosomes. Then, the lysates were mixed with the working solution and incubated at 37°C for 30 min. The absorbance was detected at 562 nm using a microplate analyzer (Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analysis
Quantitative data were presented as means and SD. Student’s t test was used to evaluate the difference between two groups. The significant differences among three groups were analyzed by one-way analysis of variance. Pearson correlation analysis was used to assess the correlation between two parameters. Spearman correlation analysis was used to evaluate the correlation of the ranked data. ROC curve was used to determine the cutoff value, sensitivity, and specificity. AUC was used to evaluate the diagnostic accuracy of DRAM expression level. The statistical analysis was performed using GraphPad Prism 8.0 software (GraphPad Software, San Diego, CA, USA). All P values were considered statistically significant when <0.05.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (grant numbers 81873976 and 31770837) and the Key Research and Development Program of Shandong Province (grant number 2019GSF108148).
Author contributions: Conceptualization and design: L.Z. and Y.X. Methodology development and data acquisition: J.Z., J.T., M.W., Y.W., M.D., and X.M. Data analysis: J.Z., B.S., and S.L. Writing—original draft: L.Z. and J.Z. Writing—review and editing: Z.Z., L.C., Y.X., and K.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Tables S1 and S2
REFERENCES AND NOTES
- 1.Brunt E. M., Wong V. W.-S., Nobili V., Day C. P., Sookoian S., Maher J. J., Bugianesi E., Sirlin C. B., Neuschwander-Tetri B. A., Rinella M. E., Nonalcoholic fatty liver disease. Nat. Rev. Dis. Primers. 1, 15080 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wang X. J., Malhi H., Nonalcoholic fatty liver disease. Ann. Intern. Med. 169, ITC65–ITC80 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Pais R., Barritt A. S. IV, Calmus Y., Scatton O., Runge T., Lebray P., Poynard T., Ratziu V., Conti F., NAFLD and liver transplantation: Current burden and expected challenges. J. Hepatol. 65, 1245–1257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J. H., Cai J. J., She Z. G., Li H. L., Noninvasive evaluation of nonalcoholic fatty liver disease: Current evidence and practice. World J. Gastroenterol. 25, 1307–1326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong M. E., Chen Y., Xiao Y., Xu L., Zhang G., Lu J., Qiu H., Ge W., Wu B., Serum extracellular vesicles contain SPARC and LRG1 as biomarkers of colon cancer and differ by tumour primary location. EBioMedicine 50, 211–223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng S., Wang Y., Liu S., Chen T., Hu Y., Zhang G., Zhang X., Yu B., Extracellular vesicles: A potential biomarker for quick identification of infectious osteomyelitis. Front. Cell. Infect. Microbiol. 10, 323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K., Ota Y., Kogure T., Suzuki Y., Iwamoto H., Yamakita K., Kitano Y., Fujii S., Haneda M., Patel T., Ota T., Circulating extracellular vesicle-encapsulated HULC is a potential biomarker for human pancreatic cancer. Cancer Sci. 111, 98–111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abels E. R., Breakefield X. O., Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 36, 301–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalluri R., LeBleu V. S., The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Souza-Schorey C., Clancy J. W., Tumor-derived microvesicles: Shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 26, 1287–1299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kok V. C., Yu C. C., Cancer-derived exosomes: Their role in cancer biology and biomarker development. Int. J. Nanomedicine 15, 8019–8036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun B., Dalvi P., Abadjian L., Tang N., Pulliam L., Blood neuron-derived exosomes as biomarkers of cognitive impairment in HIV. AIDS 31, F9–F17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q., Xiang Y., Yuan S., Xie W., Li C., Hu Z., Wu N., Wu L., Yu Z., Bai L., Li Y., Plasma exosome levels in non-small-cell lung cancer: Correlation with clinicopathological features and prognostic implications. Cancer Biomark. 22, 267–274 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Liu X.-L., Pan Q., Cao H.-X., Xin F.-Z., Zhao Z.-H., Yang R.-X., Zeng J., Zhou H., Fan J.-G., Lipotoxic hepatocyte-derived exosomal MicroRNA 192-5p activates macrophages through Rictor/Akt/Forkhead box transcription factor O1 signaling in nonalcoholic fatty liver disease. Hepatology 72, 454–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crighton D., Wilkinson S., O'Prey J., Syed N., Smith P., Harrison P. R., Gasco M., Garrone O., Crook T., Ryan K. M., DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126, 121–134 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Pang L., Liu K., Liu D., Lv F., Zang Y., Xie F., Yin J., Shi Y., Wang Y., Chen D., Differential effects of reticulophagy and mitophagy on nonalcoholic fatty liver disease. Cell Death Dis. 9, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tripathi A., Thangaraj A., Chivero E. T., Periyasamy P., Callen S., Burkovetskaya M. E., Guo M. L., Buch S., Antiretroviral-mediated microglial activation involves dysregulated autophagy and lysosomal dysfunction. Cells 8, 1168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latifkar A., Ling L., Hingorani A., Johansen E., Clement A., Zhang X., Hartman J., Fischbach C., Lin H., Cerione R. A., Antonyak M. A., Loss of Sirtuin 1 alters the secretome of breast cancer cells by impairing lysosomal integrity. Dev. Cell 49, 393–408.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J. H., Hsieh C. F., Liu H. W., Chen C. Y., Wu S. C., Chen T. W., Hsu C. S., Liao Y. H., Yang C. Y., Shyu J. F., Fischer W. B., Lin C. H., Lipid raft-associated stomatin enhances cell fusion. FASEB J. 31, 47–59 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Snyers L., Umlauf E., Prohaska R., Association of stomatin with lipid-protein complexes in the plasma membrane and the endocytic compartment. Eur. J. Cell Biol. 78, 802–812 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Peng X.-X., Yu R., Wu X., Wu S.-Y., Pi C., Chen Z.-H., Zhang X.-C., Gao C.-Y., Shao Y. W., Liu L., Wu Y.-L., Zhou Q., Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J. Immunother. Cancer 8, e000376 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang C., Hopfner F., Katsikoudi A., Hein R., Catli C., Evetts S., Huang Y., Wang H., Ryder J. W., Kuhlenbaeumer G., Deuschl G., Padovani A., Berg D., Borroni B., Hu M. T., Davis J. J., Tofaris G. K., Serum neuronal exosomes predict and differentiate Parkinson's disease from atypical parkinsonism. J. Neurol. Neurosurg. Psychiatry 91, 720–729 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan D. B. A., Armitage J., Teo T. H., Ong N. E., Shin H., Moodley Y. P., Elevated levels of circulating exosome in COPD patients are associated with systemic inflammation. Respir. Med. 132, 261–264 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Hirsova P., Ibrahim S. H., Krishnan A., Verma V. K., Bronk S. F., Werneburg N. W., Charlton M. R., Shah V. H., Malhi H., Gores G. J., Lipid-induced signaling causes release of inflammatory extracellular vesicles from hepatocytes. Gastroenterology 150, 956–967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang F., Chen Q., Wang W., Ling Y., Yan Y., Xia P., Hepatocyte-derived extracellular vesicles promote endothelial inflammation and atherogenesis via microRNA-1. J. Hepatol. 72, 156–166 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Kakazu E., Mauer A. S., Yin M., Malhi H., Hepatocytes release ceramide-enriched pro-inflammatory extracellular vesicles in an IRE1α-dependent manner. J. Lipid Res. 57, 233–245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beaumatin F., O'Prey J., Barthet V. J. A., Zunino B., Parvy J.-P., Bachmann A. M., O'Prey M., Kania E., Gonzalez P. S., Macintosh R., Lao L. Y., Nixon C., Lopez J., Long J. S., Tait S. W. G., Ryan K. M., mTORC1 activation requires DRAM-1 by facilitating lysosomal amino acid efflux. Mol. Cell 76, 163–176.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu K., Lou J., Wen T., Yin J., Xu B., Ding W., Wang A., Liu D., Zhang C., Chen D., Li N., Depending on the stage of hepatosteatosis, p53 causes apoptosis primarily through either DRAM-induced autophagy or BAX. Liver Int. 33, 1566–1574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garufi A., Pistritto G., Baldari S., Toietta G., Cirone M., D'Orazi G., p53-Dependent PUMA to DRAM antagonistic interplay as a key molecular switch in cell-fate decision in normal/high glucose conditions. J. Exp. Clin. Cancer Res. 36, 126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laforge M., Limou S., Harper F., Casartelli N., Rodrigues V., Silvestre R., Haloui H., Zagury J. F., Senik A., Estaquier J., DRAM triggers lysosomal membrane permeabilization and cell death in CD4+ T cells infected with HIV. PLOS Pathog. 9, e1003328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu K., Shi Y., Guo X. H., Ouyang Y. B., Wang S. S., Liu D. J., Wang A. N., Li N., Chen D. X., Phosphorylated AKT inhibits the apoptosis induced by DRAM-mediated mitophagy in hepatocellular carcinoma by preventing the translocation of DRAM to mitochondria. Cell Death Dis. 5, e1078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park S. W., Do H. J., Choi W., Kim J. H., Song H., Seo H. G., Kim J. H., GCNF regulates OCT4 expression through its interactions with nuclear receptor binding elements in NCCIT cells. J. Cell. Biochem. 119, 2719–2730 (2018). [DOI] [PubMed] [Google Scholar]
- 33.He M., Wang C., Long X. H., Peng J. J., Liu D. F., Yang G. Y., Jensen M. D., Zhang L. L., Mesencephalic astrocyte-derived neurotrophic factor ameliorates steatosis in HepG2 cells by regulating hepatic lipid metabolism. World J. Gastroenterol. 26, 1029–1041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y., Mao Y., Li H., Shen G., Nan G., Knockdown of Nrf2 inhibits angiogenesis by downregulating VEGF expression through PI3K/Akt signaling pathway in cerebral microvascular endothelial cells under hypoxic conditions. Biochem. Cell Biol. 96, 475–482 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Yan Z., Miao X., Zhang B., Xie J., p53 as a double-edged sword in the progression of non-alcoholic fatty liver disease. Life Sci. 215, 64–72 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Yan C., Tian X., Li J., Liu D., Ye D., Xie Z., Han Y., Zou M.-H., A high-fat diet attenuates AMPK α1 in adipocytes to induce exosome shedding and nonalcoholic fatty liver development in vivo. Diabetes 70, 577–588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chalasani N., Younossi Z., Lavine J. E., Charlton M., Cusi K., Rinella M., Harrison S. A., Brunt E. M., Sanyal A. J., The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 67, 328–357 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C., PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1 and S2