Abstract
The mitochondrial matrix of the yeast Saccharomyces cerevisiae contains two molecular chaperones of the Hsp70 class, Ssc1 and Ssq1. We report that Ssc1 and Ssq1 play sequential roles in the import and maturation of the yeast frataxin homologue (Yfh1). In vitro, radiolabeled Yfh1 was not imported into ssc1-3 mutant mitochondria, remaining in a protease-sensitive precursor form. As reported earlier, the Yfh1 intermediate form was only slowly processed to the mature form in Δssq1 mitochondria (S. A. B. Knight, N. B. V. Sepuri, D. Pain, and A. Dancis, J. Biol. Chem. 273:18389–18393, 1998). However, the intermediate form in both wild-type and Δssq1 mitochondria was entirely within the inner membrane, as it was resistant to digestion with protease after disruption of the outer membrane. Therefore, we conclude that Ssc1, which is present in mitochondria in approximately a 1,000-fold excess over Ssq1, is required for Yfh1 import into the matrix, while Ssq1 is necessary for the efficient processing of the intermediate to the mature form in isolated mitochondria. However, the steady-state level of mature Yfh1 in Δssq1 mitochondria is approximately 75% of that found in wild-type mitochondria, indicating that this retardation in processing does not dramatically affect cellular concentrations. Therefore, Ssq1 likely has roles in addition to facilitating the processing of Yfh1. Twofold overexpression of Ssc1 partially suppresses the cold-sensitive growth phenotype of Δssq1 cells, as well as the accumulation of mitochondrial iron and the defects in Fe/S enzyme activities normally found in Δssq1 mitochondria. Δssq1 mitochondria containing twofold-more Ssc1 efficiently converted the intermediate form of Yfh1 to the mature form. This correlation between the observed processing defect and suppression of in vivo phenotypes suggests that Ssc1 is able to carry out the functions of Ssq1, but only when present in approximately a 2,000-fold excess over normal levels of Ssq1.
Hsp70s are ubiquitous molecular chaperones found in all major organelles of eukaryotes that function by binding transiently to exposed hydrophobic sequences in unfolded or partially folded proteins (3, 11, 36). Hsp70s' chaperone activity is driven by their inherent ATPase activity, which regulates binding and release of substrate polypeptides. Through this mechanism, Hsp70s function in diverse cellular processes such as protein folding, protein translocation across membranes, and assembly and disassembly of multimeric structures (6, 19, 21).
Ssc1, an essential heat shock protein of the 70-kDa class (Hsp70), is a key component of the mitochondrial protein import machinery of Saccharomyces cerevisiae (15, 23, 45). Mitochondrial import is a multistep process necessary for the biogenesis of most mitochondrial proteins (30, 31, 37, 39). The vast majority of mitochondrial proteins are translated in the cytosol with an N-terminal presequence that targets the preprotein to receptors on the outer mitochondrial membrane. After passage through a proteinaceous channel of the outer mitochondrial membrane, the presequence is then driven across the inner mitochondrial membrane through a protein channel and into the matrix by the membrane potential. Cooperating with cochaperones and components of the inner mitochondrial membrane, matrix-localized Ssc1 binds the emerging preprotein while processing proteases cleave the presequence. Ssc1, acting in an ATP-dependent manner, is required for the further translocation of the remaining preprotein across the inner mitochondrial membrane. Subsequent folding of the imported mitochondrial protein within the matrix is, at least in some cases, dependent upon Ssc1 and other molecular chaperones.
Although for many years Ssc1 was the only known Hsp70 of mitochondria, a second matrix-localized Hsp70, Ssq1, has recently been identified (40). Ssq1 is not essential; however, cells lacking Ssq1 grow extremely slowly at low temperatures such as 23°C. While Ssq1 function is not required for the proper import or processing of several mitochondrial proteins including cytochrome b2 (Cytb2) and the Fe/S protein of the cytochrome bc1 (Cytbc1) complex (40), recent studies demonstrate a role for Ssq1 in the maturation of Yfh1 (26), a nucleus-encoded mitochondrial protein involved in iron homeostasis (1, 5, 32). In in vitro import assays, the Yfh1 precursor was processed in two steps by the mitochondrial processing peptidase (MPP), generating first an intermediate and then the functional mature form (4). Unlike wild-type mitochondria, mitochondria lacking Ssq1 accumulated the intermediate form of Yfh1, which was only slowly processed to the mature form (26). Thus far, Yfh1 is the only substrate identified whose maturation is affected by the absence of Ssq1.
Yfh1 is the orthologue of the human protein frataxin. Mutations in the frataxin gene are associated with the neurodegenerative disease Friedreich's ataxia (7). Iron accumulates within the affected cells, and the activity of Fe/S-containing proteins, such as aconitase, is reduced (27, 35). Accumulation of iron, along with a reduction in the activity of Fe/S-containing proteins, has also been observed for mitochondria isolated from yeast strains with mutations in YFH1 (1, 12). It is thought that the increased levels of iron result in increased production of oxygen radicals within mitochondria (16). Since Fe/S centers are particularly sensitive to oxidative damage (13), the decrease in the activity of Fe/S-containing enzymes has been attributed indirectly to iron accumulation. Consistent with this idea, yfh1 cells are more sensitive to oxidative agents than are wild-type cells (1).
The phenotypes of Δyfh1 and Δssq1 cells are very similar. Both accumulate iron, have low activity of Fe/S-containing enzymes, and are hypersensitive to oxidative agents. To better understand the function of Ssq1 and its relationship to Ssc1 and Yfh1, we analyzed the translocation and maturation of Yfh1 in more detail. Efficient maturation of Yfh1 requires the sequential action of Ssc1 and Ssq1. Ssc1 is required for translocation across the inner membrane, and subsequently Ssq1 is needed for efficient processing by MPP. Overexpression of the abundant Ssc1 partially suppresses the defects caused by the absence of the minor Hsp70 Ssq1, indicating that these two Hsp70s partially overlap in function.
MATERIALS AND METHODS
Yeast strains, plasmids, media, and chemicals.
PJ53 is isogenic to W303: trp1-1/trp1-1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 ade2-1/ade2-1 can1-100/can1-100 GAL2+/GAL2+ met2-Δ1/met2-Δ1 lys2-Δ2/lys2-Δ2 (20). PJ43B is a wild-type haploid derivative of PJ53. The haploid Δssq1 strain was made by first replacing one of the SSQ1 alleles of PJ53 with the LYS2 gene (Δssq1::LYS2) (40) and then sporulating the heterozygous diploid. Δssq1/pSSC1 is the haploid Δssq1 strain carrying the SSC1 gene on the centromeric plasmid, pRS316 (40, 42). PK83 carries the ssc1-3 temperature-sensitive allele, and PK83 is its isogenic wild-type strain (15). The strain BJ3497 (pep4::HIS3 ura3-52 hisΔ200 [22]), which is defective in proteinase A, was used for expression of the GST-Ssc1 (29) and GST-Ssq1HA (this study) fusion proteins.
To construct a yeast strain with a YFH1 deletion, we first obtained a copy of YFH1 by PCR amplifying genomic DNA from position −426 to +754 using Pfu1 polymerase (Stratagene, La Jolla, Calif.). YFH1 was then cloned into the pRS vectors, pRS306 and pRS316 (42), which carry the URA3 marker. The HIS3 gene was used to replace the entire protein coding sequence of YFH1, and the resulting pRS306YFH1::HIS3 plasmid was used to disrupt YFH1 in the diploid PJ53 by the standard two-step disruption procedure (34). Δyfh1::HIS3 derivatives were verified by PCR amplification.
To construct an expression vector for in vitro transcription-translation of YFH1, a 738-bp SnaBI-HindIII fragment, containing the coding region for YFH1 plus 63 bp of upstream DNA, was cloned into the pGEM-7zf+ vector (Promega, Madison, Wis.) digested with Ecl136II and HindIII.
To construct an expression vector for the GST-Ssq1HA fusion protein, we utilized the vector pRD56CS-SSC1, which expresses an in-frame fusion between glutathione S-transferase (GST) and the mature part of Ssc1 (29). Using pRD56CS-SSC1, we replaced the coding sequence of Ssc1, except for 16 amino acids at the N terminus of the mature protein which result in 4 amino acids that differ from the Ssq1 sequence in this region, with Ssq1HA (40).
Unless otherwise indicated, yeast were grown in YP medium (1% yeast extract, 2% peptone) with 2% glucose, 2% galactose, or 3% glycerol and 2% ethanol as the carbon source. Low-iron medium (0.67% yeast nitrogen base without iron, copper, and amino acids [Bio 101, Inc., Carlsbad, Calif.]) was supplemented with amino acids, 1 μm copper sulfate, and 2% galactose as the carbon source. For hydrogen peroxide sensitivity testing, 4 mM H2O2 was added to sterilized YP medium with 2% galactose prior to pouring plates.
All chemicals, unless stated otherwise, were purchased from Sigma (St. Louis, Mo.).
Translocation of proteins into and fractionation of mitochondria.
Mitochondria were isolated from PK82, PK83, PJ43B, Δssq1, and Δssq1/pSSC1 strains as previously described (15). [35S]methionine-labeled precursor proteins were synthesized in vitro using a coupled rabbit reticulocyte lysate system (Promega). Methods for import experiments have been described previously (15). Briefly, 50 μg of mitochondrial protein for the PJ43B, Δssq1, Δssq1/pSSC1, PK82, or PK83 strain was incubated for 5 min at 25°C, or 15 min at 37°C for PK82 and PK83. Mitochondrial samples were then incubated at 25°C with lysate containing 35S-labeled Yfh1 or Cytb2167DHFR (15) for the indicated times before addition of a mixture of 0.25 μM valinomycin, 4 μM antimycin A, and 10 μM oligomycin which results in dissipation of the membrane potential. Half of each sample was then treated with proteinase K at a final concentration of 100 μg/ml for 15 min on ice. Twenty-five micrograms of mitochondrial protein was then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
For fractionation of mitochondria, radiolabeled Yfh1 was imported into 200 μg of mitochondrial protein isolated from wild-type (PJ43B) or Δssq1 yeast strains. After import, mitochondrial samples were divided in half and reisolated by centrifugation for 5 min at 16,000 × g at 4°C. Each pellet was resuspended in SEM (250 mM sucrose, 1 mM EDTA, 10 mM MOPS [morpholinepropanesulfonic acid], pH 7.2) or EM (1 mM EDTA, 10 mM MOPS, pH 7.2) buffer at a concentration of 50 μg of mitochondrial protein/500 μl of buffer and incubated on ice for 15 min. Mitochondria and mitoplasts, obtained by incubating isolated mitochondria in hypotonic EM buffer, were treated with and without 50 μg of proteinase K for 15 min on ice. Samples were centrifuged for 15 min at 14,000 × g to isolate the mitochondria or mitoplasts. Samples were separated by SDS-PAGE, transferred to nitrocellulose, and subjected to PhosphorImager analysis (Molecular Dynamics). Samples were then analyzed by immunoblot analysis using polyclonal antibodies against Cytb2 (this study) or Mge1 (29) using the Renaissance detection kit from New England Nuclear Labs (Boston, Mass.).
Respiratory enzyme assays and measurement of mitochondrial iron.
Activities of the respiratory enzymes, succinate dehydrogenase-cytochrome c oxidoreductase (SDH-Cytbc1) and Cytbc1, were measured in mitochondria isolated from PJ43B, Δssq1, and Δssq1/pSSC1 yeast strains grown to an optical density at 600 nm of 0.5 to 1.0 at 34°C in YP medium containing 2% galactose (15). SDH-Cytbc1 activity was measured using succinate as the substrate as previously described (46), except that 2,6-dichloroindophenol was used in place of cytochrome c (41). Cytbc1 activity was measured using reduced ubiquinone as a substrate. The reduction of cytochrome c was determined as previously described (17, 41). Cellular aconitase activity was measured by monitoring the decrease in the absorbance of the substrate cis-aconitate at 240 nm as described previously (10, 41).
Mitochondrial iron levels were determined as previously described (41, 44). Briefly, iron levels in a suspension of purified mitochondria (25 μl) containing 200 to 400 μg of mitochondrial protein in buffer A (600 mM sorbitol, 10 mM Tris-HCl, pH 7.5) were determined colorimetrically with ferene. Unless otherwise indicated, samples were prepared by wet ashing with ultrapure H2SO4 and H2O2 (Merck, Whitehouse Station, N.J.) and neutralization following previously described procedures (2, 24). Suspensions of mitochondria isolated from yeast strains grown in low-iron-containing medium were prepared by solubilization using 1% SDS in acetate buffer, pH 4.3, as previously described (44). These two methods of sample preparation resulted in similar values when the same mitochondrial preparations were analyzed. Data were normalized to the protein content of the mitochondrial samples. Protein determinations were performed using the Bio-Rad protein assay from Bio-Rad Laboratories (Hercules, Calif.) with ovalbumin as a standard.
Purification of GST-Ssq1HA and GST-Ssc1 proteins.
BJ3497 cells harboring the GST-SSC1 (29) or GST-SSQ1-HA (this study) overexpression plasmid were grown to an optical density at 600 nm of 3.0 in 6 liters of synthetic complete medium without uracil containing 2% galactose at 30°C. Cells were harvested by centrifugation for 5 min at 2,700 × g. The cells were washed with cold water and resuspended in 90 ml of buffer P (20 mM NaPi [pH 7.3], 150 mM NaCl, 1% [vol/vol] Triton X-100, 0.1% β-mercaptoethanol, 1 mM phenylmethanesulfonyl fluoride, 1 mM TPCK [l-chloro-3{4-tosylamido}-4-phenyl-2-butanone], and 2 μg each of pepstatin A and leupeptin per ml). The cell suspension was frozen in liquid nitrogen, thawed on ice, and lysed using a French press at 20,000 lb/in2. The mixture was centrifuged at 100,000 × g for 60 min. The cleared extract was loaded directly onto a 5-ml glutathione agarose column equilibrated with buffer P containing 0.1% Triton X-100 and 10% (vol/vol) glycerol. The column was washed with 10 volumes of buffer P containing 1 M NaCl, 0.1% Triton X-100, and 10% glycerol and subsequently with 10 volumes of the same buffer containing 150 mM NaCl. Protein was eluted with buffer P containing 20 mM glutathionine adjusted to pH 7.3 and 10% glycerol.
Protein quantitation using immunoblot analysis.
To determine Yfh1 levels, mitochondrial proteins purified from wild-type (PJ43-2B) and Δssq1 strains were subjected to SDS-PAGE, transferred to nitrocellulose (Protran; Schleicher & Schuell Co., Keene, N.H.), and immunoblotted with antibodies to Yfh1 and Mge1 (29). Relative concentrations of Yfh1 were determined by densitometrically comparing Yfh1 and Mge1 signals on exposed film (X-O; Eastman Kodak Co., Rochester, N.Y.) using Ofoto (Light Source Computer Images, Inc.) and ScanAnalysis (Biosoft) software programs. Ssc1 and F1β levels were determined as described above, except that mitochondrial protein purified from the Δssq1/pSSC1 yeast strain was also examined and polyclonal antibodies against Ssc1 (38) and F1β (this study) were utilized. For quantitation of Ssq1 and Ssc1, fixed amounts of mitochondrial protein purified from the Δssq1/pRS316SSQ1-HA strain (40) mixed with varying amounts of purified GST-Ssc1 or GST-Ssq1HA were analyzed as described above, except that monoclonal antibodies against hemagglutinin (HA) or polyclonal antibodies against Ssc1 (38) were utilized. Densitometric signals from serial dilutions of GST-purified proteins were compared to the signals from a constant dilution of mitochondria, and the relative amounts of Ssc1 and Ssq1 were determined by comparing the level per microgram of mitochondrial protein. Similar results were obtained using a different polyclonal antiserum generated from the last 14 amino acids of Ssc1 (29).
To generate rabbit antisera, we utilized the pGEX-KT vector system (18), which expresses an in-frame fusion between GST and amino acids 165 to 335 for Cytb2 and 319 to the last amino acid of the coding sequence for F1β. Each fusion protein was expressed in Escherichia coli and purified by adsorption to glutathione-agarose beads. Eluate from the beads was used to inoculate rabbits. Grazia Isaya (Mayo Clinic) kindly provided Yfh1 antiserum.
RESULTS
Ssc1 is required for import of radiolabeled Yfh1 into isolated mitochondria.
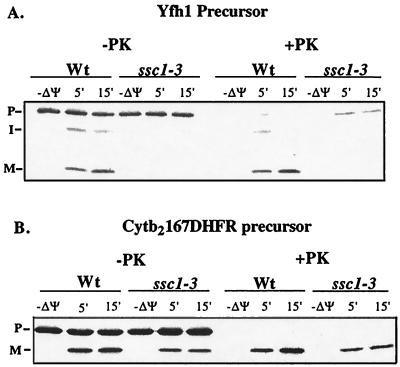
To evaluate the contributions of Ssc1 to the maturation of Yfh1, we tested whether functional Ssc1 was necessary for the import of Yfh1 into mitochondria. Mitochondria were isolated from yeast strains carrying a temperature-sensitive allele of SSC1, ssc1-3, grown at the permissive temperature of 25°C. Isolated mitochondria were preincubated at 37°C for 15 min to induce the mutant phenotype (15) and then incubated at 25°C with 35S-labeled Yfh1. In wild-type mitochondria, Yfh1 was processed to the mature form, which was resistant to exogenously added proteinase K (Fig. 1A) (26). In ssc1-3 mitochondria, Yfh1 was not processed to either the intermediate or the mature form. Furthermore, the precursor form was accessible to exogenously added proteinase K, indicating that Yfh1 preprotein was not imported into mitochondria in the absence of Ssc1 function (Fig. 1A). Despite the inability to import Yfh1, the ssc1-3 mitochondria used in this experiment were translocation competent, as demonstrated by the import and processing of Cytb2167DHFR (Fig. 1B), a fusion protein construct lacking the intact Cytb2 heme binding domain, thus making its maturation independent of Ssc1 function (45). We conclude that Ssc1 is required for the translocation of the precursor form of radiolabeled Yfh1 into isolated mitochondria, as it is for many matrix-localized mitochondrial proteins (30).
FIG. 1.
Import of Yfh1 requires Ssc1 function. The import of radiolabeled Yfh1 preprotein (A) and that of Cytb2167DHFR preprotein (B) into isolated mitochondria from wild-type (Wt) and ssc1-3 cells in the absence and the presence of a membrane potential (ΔΨ) were compared. To induce the mutant phenotype of Ssc1-3, mitochondria were preincubated for 15 min at 37°C. After 5 or 15 min, further import was stopped by dissipating the membrane potential. Samples were then divided; half were treated with proteinase K (+PK), and half were not (−PK). P, precursor; I, intermediate; M, mature.
Relative levels of Ssc1 and Ssq1 within mitochondria.
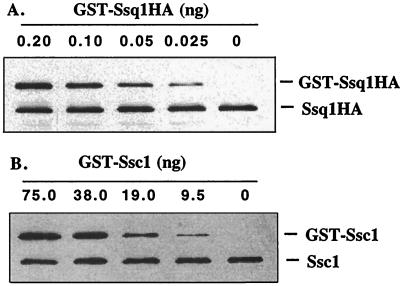
The above results, coupled with previously reported data, indicate that both Ssc1 and Ssq1 function in the maturation of Yfh1 (26). To better understand the functional relationship between Ssc1 and Ssq1, we determined the relative amounts of the two proteins in wild-type mitochondria. Mitochondrial lysates were first separated on an SDS-polyacrylamide gel and stained using Coomassie blue dye. Densitometric analysis of the stained gel indicated that Ssc1 protein made up approximately 2% of the mitochondrial protein profile (data not shown), consistent with previously published values (33). To estimate the amount of Ssc1 relative to Ssq1 in isolated mitochondria, we performed immunoblot analysis on samples containing fixed amounts of mitochondrial protein mixed with known amounts of purified GST-Ssc1 or GST-Ssq1HA as described in Materials and Methods (Fig. 2). We currently do not have antibodies specific to Ssq1, and therefore, we utilized an HA-tagged version of Ssq1 for our analysis which is functional in vivo (40). Ssc1 and Ssq1 were detected by immunoblot analysis using polyclonal antibodies specific to Ssc1 or monoclonal antibodies specific for the HA epitope, respectively. By comparison of the signal from isolated mitochondria with that of purified GST fusion proteins, we calculated that Ssc1 is 1,000 to 2,000 times more abundant than Ssq1 in wild-type mitochondria. Taken together, these data suggest that Ssc1 and Ssq1 make up approximately 1.0 to 2.0% and 0.001 to 0.002% of mitochondrial protein, respectively.
FIG. 2.
Determination of the relative levels of Ssc1 and Ssq1 in mitochondria. Immunoblot band intensities of predetermined concentrations of purified GST fusion proteins were compared with immunoblot band intensities from mitochondrial proteins isolated from Δssq1/pRD-SSQ1HA yeast cells. Mitochondrial proteins (10.0 [A] or 0.5 [B] μg) and varying amounts of purified GST-Ssq1HA (A) or GST-Ssc1 (B) were separated by electrophoresis and subjected to immunoblot analysis using monoclonal antibodies specific for HA (A) or polyclonal antibodies specific to Ssc1 (B) to determine the relative levels of Ssc1 and Ssq1 in mitochondria. Similar results were obtained using two independent mitochondrial preparations and five separate quantitation analyses.
The intermediate and mature forms of radiolabeled Yfh1 are located within the matrix of wild-type and Δssq1 mitochondria.
Although Ssq1 is of low abundance, it plays an important role in the maturation of Yfh1 (26). According to previous studies confirmed in this report, Yfh1 can be imported into mitochondria isolated from strains carrying mutations in SSQ1. However, it accumulates as a protease-resistant intermediate form (Fig. 3A) (26). To better define the contribution of Ssq1 to Yfh1 maturation, we thoroughly analyzed the state of the intermediate form of Yfh1. We determined the intramitochondrial location of the intermediate form to ascertain whether it is completely translocated into the matrix or whether it exists as an intermembrane space intermediate, exposing the amino-terminal presequence to the matrix but retaining the remainder of the protein in the intermembrane space.
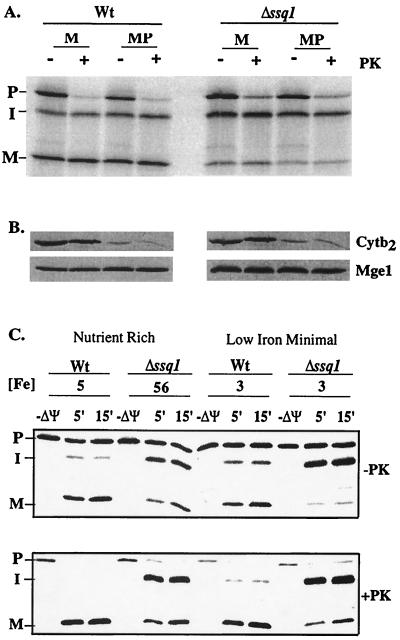
FIG. 3.
The intermediate and mature forms of radiolabeled Yfh1 are located within the matrix of wild-type (Wt) and Δssq1 mitochondria. (A) Yfh1 preprotein synthesized in a reticulocyte lysate was added to mitochondria isolated from wild-type and Δssq1 yeast cells at 25°C. After 7.5 min, further import was stopped by dissipating the membrane potential. Mitochondria (M) and mitoplasts (MP) were then prepared. Equivalent amounts of mitochondria and mitoplasts were either treated (+PK) or not treated (−PK) with proteinase K. Samples were separated by electrophoresis and transferred to nitrocellulose, and the amounts of radiolabeled Yfh1 were compared. P, precursor; I, intermediate; M, mature. (B) Immunoblot analysis was performed using antibodies specific for Cytb2 or Mge1 after separation by electrophoresis. (C) Mitochondria were isolated from wild-type (Wt) or Δssq1 cells grown in either nutrient-rich or low-iron minimal medium. The concentration of iron (picomoles of Fe per microgram of mitochondrial protein) in each preparation of mitochondria is indicated ([Fe]). Imports of radiolabeled Yfh1 preprotein into these mitochondria in the absence and in the presence of a membrane potential (ΔΨ) were compared. After 5 or 15 min, further import was stopped by dissipating the membrane potential. Samples were then divided; half were treated with proteinase K (+PK), and half were not (−PK). P, precursor; I, intermediate; M, mature.
Mitochondria isolated from wild-type and Δssq1 strains were fractionated following the import of radiolabeled Yfh1. In both wild-type and Δssq1 mitochondria, the intermediate and mature forms of Yfh1 remained protease resistant following a hypotonic treatment that ruptures the outer, but not the inner, mitochondrial membrane, forming mitoplasts. Under these conditions, proteins of the intermembrane space, such as Cytb2, are released from mitochondria and any residual protein is degraded by added protease, while matrix proteins, such as Mge1, remain resistant to protease (Fig. 3B). The intermediate and mature forms of Yfh1 were resistant to digestion with proteinase K in both wild-type and Δssq1 mitoplasts, as well as intact mitochondria, indicating that the intermediate and mature forms of Yfh1 are located within the matrix (Fig. 3A). In several experiments, a decrease in the intensity of the radiolabeled intermediate and mature form of Yfh1 in Δssq1 mitochondria was observed. To determine if this effect was specific for Yfh1, we cotranslocated a commonly used fusion protein which localizes to the matrix, Su9DHFR. A similar decrease in the signals of the two proteins was observed in the mitoplast preparations (data not shown), suggesting that Δssq1 mitochondria are slightly more susceptible to breakage during the procedure used to generate mitoplasts than are wild-type mitochondria.
Δssq1 mitochondria accumulate 10-fold more iron than normal wild-type levels of 3 to 7 pmol of Fe/μg of mitochondrial protein (26, 41). To determine whether the high amounts of iron cause the observed processing defect of Yfh1 in Δssq1 mitochondria, mitochondria were isolated from wild-type and Δssq1 yeast strains grown in low-iron-containing media. Under these conditions, both Δssq1 and wild-type mitochondria have approximately 3 pmol of Fe/μg of mitochondrial protein (Fig. 3C). The maturation of radiolabeled Yfh1 was monitored using mitochondria isolated from wild-type and Δssq1 yeast strains grown in nutrient-rich and low-iron minimal media (Fig. 3C). Radiolabeled Yfh1 was processed efficiently in wild-type mitochondria, while the processing defect of Yfh1 persisted in Δssq1 mitochondria, indicating that lack of Ssq1 function, not iron, is responsible for the Yfh1 processing defect associated with Δssq1 mitochondria.
Taken together, these results suggest that the abundant Ssc1 is required for translocation of the Yfh1 preprotein across the inner membrane, while the rare Ssq1 is needed for the subsequent posttranslocational processing of the intermediate to the functional mature form.
Levels of Yfh1 in Δssq1 mitochondria.
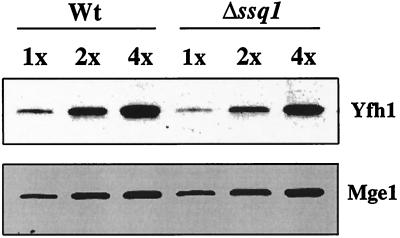
In in vitro translocation experiments, Δssq1 mitochondria inefficiently process the matrix-localized intermediate form of Yfh1 to the mature form (Fig. 3). To determine whether this in vitro observation translates into lowered amounts of Yfh1 in Δssq1 mitochondria in vivo, we directly determined the relative levels of Yfh1 in mitochondria isolated from wild-type and Δssq1 mitochondria using Yfh1-specific antibodies. Relative to the control, Mge1, Yfh1 levels in Δssq1 mitochondria were about 75% of those in wild-type mitochondria (Fig. 4). Thus, there is only a minor reduction in the steady-state level of mature Yfh1 in Δssq1 mitochondria. Therefore, the inefficient processing of Yfh1 observed in the in vitro import assay is likely revealing a kinetic effect that is not detected in an analysis of steady-state levels.
FIG. 4.
Comparison of Yfh1 levels. Mitochondrial proteins (5.0, 10.0, and 20.0 μg) isolated from either wild-type (Wt) or Δssq1 yeast cells were separated by electrophoresis, transferred to nitrocellulose, and subjected to immunoblot analysis using polyclonal antibodies specific to Yfh1 and Mge1.
We asked if increasing the level of mature Yfh1 in Δssq1 cells suppressed the growth defects caused by the lack of Ssq1 function. Δssq1 cells carrying YFH1 on either a low-copy centromeric or a high-copy-number 2μm plasmid, which increases the amount of mature Yfh1, grew no better than Δssq1 cells carrying only control vectors (data not shown). Together, these results suggest that the inefficiency of Yfh1 processing is not responsible for Δssq1 phenotypes and is, therefore, not the sole defect of Δssq1 cells.
Increased levels of Ssc1 overcome the Yfh1 processing defect in Δssq1 mitochondria.
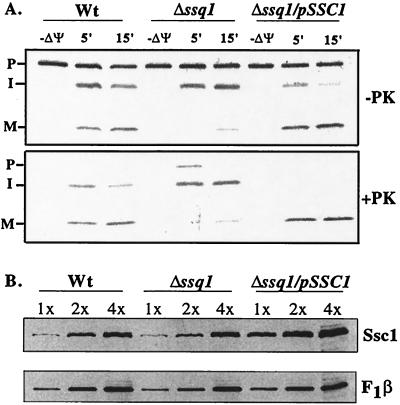
Previously, we reported that an additional copy of SSC1 partially suppressed the cold-temperature growth phenotype of Δssq1 yeast strains (40). Therefore, we tested whether overexpression of Ssc1 could also overcome the processing defect of Yfh1 in Δssq1 mitochondria. Radiolabeled Yfh1 was imported into mitochondria isolated from wild-type and Δssq1 yeast strains, as well as Δssq1/pSSC1, a yeast strain with SSQ1 deleted carrying an additional copy of SSC1 on a centromeric plasmid (40). Unlike Δssq1 mitochondria, Δssq1/pSSC1 mitochondria were able to process the intermediate form of Yfh1 to the mature form with nearly the same efficiency as that of wild-type mitochondria (Fig. 5A). Therefore, increased levels of Ssc1 restored the processing of Yfh1 to near-wild-type efficiencies even in the absence of Ssq1, suggesting that, when in excess, Ssc1 can compensate for Ssq1 function.
FIG. 5.
A twofold increase in the level of Ssc1 overcomes the processing defect of Yfh1 in Δssq1 mitochondria. (A) Imports of radiolabeled Yfh1 preprotein into mitochondria isolated from wild-type (Wt), Δssq1, and Δssq1/pSSC1 cells in the absence and in the presence of a membrane potential (ΔΨ) were compared. After 5 or 15 min, further import was stopped by dissipating the membrane potential. Samples were then divided; half were treated with proteinase K (+PK), and half were not (−PK). P, precursor; I, intermediate; M, mature. (B) The levels of Ssc1 in mitochondria isolated from wild-type (Wt), Δssq1, and Δssq1/pSSC1 yeast strains were compared. Mitochondrial proteins (0.5, 1.0, and 2.0 μg) were separated by electrophoresis, transferred to nitrocellulose, and subjected to immunoblot analysis using polyclonal antibodies specific to Ssc1 and F1β.
We compared the level of Ssc1 in mitochondria isolated from wild-type, Δssq1, and Δssq1/pSSC1 yeast strains. A series of twofold dilutions of mitochondrial proteins isolated from the three yeast strains were tested by immunoblot analysis using polyclonal antibodies specific to Ssc1. Densitometric analysis of the immunoblots using F1β as a control revealed that the amount of Ssc1 was approximately twofold higher in Δssq1/pSSC1 mitochondria than in wild-type and Δssq1 mitochondria (Fig. 5B). This twofold increase in Ssc1 levels indicates that an approximately 2,000-fold excess of Ssc1 over normal levels of Ssq1 suppresses the processing defect of the Yfh1 precursor in Δssq1 mitochondria.
Increased levels of Ssc1 suppress the iron accumulation in Δssq1 mitochondria.
Previous observations demonstrated that Δssq1 mitochondria accumulate iron (26, 41). Wild-type mitochondria contain approximately 5.0 pmol of Fe/μg of mitochondrial protein, while Δssq1 mitochondria accumulated approximately 10-fold more iron than did wild-type mitochondria (Table 1). Therefore, we tested whether increasing the level of Ssc1 also suppressed the accumulation of iron in Δssq1 mitochondria. Increasing the level of Ssc1 significantly reduced the amount of iron that accumulated in the absence of Ssq1 function. However, iron levels remained 2.5-fold higher than wild-type levels (Table 1).
TABLE 1.
Mitochondrial iron content and respiratory enzyme activitiesa
| Strain | Iron contentb | Activity
|
||
|---|---|---|---|---|
| Aconitasec | Ubi-Cytbc1d | SDH-Cytbc1e | ||
| Wild type | 5.4 ± 1.0 | 546 ± 77 | 918 ± 133 | 91 ± 12 |
| Δssq1 | 56.2 ± 12.9 | 37 ± 15 | 304 ± 38 | 7 ± 1 |
| Δssq1/pSSC1 | 13.6 ± 1.0 | 323 ± 33 | 827 ± 156 | 49 ± 8 |
Values obtained are averages of at least three independent preparations ± standard deviations.
Iron content is picomoles of iron per microgram of mitochondrial protein.
Values are shown as nanomoles of aconitate converted per minute per milligram of cell lysate.
Values are shown as nanomoles of cytochrome c reduced per minute per milligram of mitochondrial protein. Ubi, ubiquinone.
Values are shown as nanomoles of 2,6-dichloroindophenol reduced per minute per milligram of mitochondrial protein.
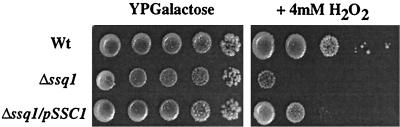
When present in excess, iron interacts with O2, generating reactive oxygen species via iron-catalyzed Fenton chemistry. These toxic hydroxyl radicals can cause oxidative damage to cellular macromolecules such as DNA, proteins, or lipids (13, 16). One indication that yeast strains are oxidatively stressed is an enhanced growth inhibition in the presence of oxidative reagents, such as H2O2. We tested the sensitivity of wild-type, Δssq1, and Δssq1/pSSC1 strains to H2O2 and observed that Δssq1 strains were extremely sensitive to H2O2 compared to wild-type cells (Fig. 6), consistent with previously reported data (41). The additional copy of SSC1 partially overcame the hypersensitivity of Δssq1 cells to H2O2 but did not restore growth to wild-type levels (Fig. 6). This increased sensitivity of Δssq1/pSSC1 strains to H2O2 compared to that of the wild type correlates with the higher-than-normal levels of iron observed in this strain, suggesting that, even at increased levels, Ssc1 does not completely replace Ssq1 function.
FIG. 6.
Sensitivity to H2O2. Wild-type (Wt), Δssq1, and Δssq1/pSSC1 cells were serially diluted 10-fold and spotted onto YP-galactose plates with or without 4 mM H2O2 and incubated at 34°C for 4 days.
Increased levels of Ssc1 partially restore activity of Fe/S-containing mitochondrial respiratory enzymes in Δssq1 mitochondria.
The highly reactive hydroxyl radicals generated from excess free iron can damage the Fe/S centers of Fe-containing mitochondrial proteins, resulting in a decrease in their activity (13). Δssq1 mitochondria show a decrease in the enzymatic activities of several respiratory components containing Fe/S clusters: aconitase, Cytbc1, and SDH (41, 43). Therefore, we tested whether the overproduction of Ssc1 restored the activity of these proteins. The activity of aconitase is reduced to 7% in Δssq1 cells, but increasing the level of Ssc1 restored the activity of aconitase in Δssq1/pSSC1 cells to 58% of wild-type levels (Table 1). Using isolated mitochondria, we measured the activity of certain complexes of the electron transport chain that contain proteins with Fe/S centers, the Cytbc1 complex and the SDH complex. Assaying Cytbc1 activity directly by supplying the reaction with reduced ubiquinone, we determined that Δssq1 and Δssq1/pSSC1 mitochondria maintained 33 and 90% of wild-type Cytbc1 activity, respectively (Table 1). To measure the activity of the SDH complex, we assayed SDH-Cytbc1 activity. The rate-limiting step of ubiquinone-mediated mitochondrial electron transport is diffusion of the ubiquinones within the inner membranes (8, 28). Therefore, the measured activity of the SDH-Cytbc1 assay is indicative of the activity of the SDH complex itself. We found that the Δssq1 mitochondria possess only 8% of wild-type SDH-Cytbc1 activity, suggesting a strong defect in SDH activity itself, consistent with previously reported data (41, 43). The Δssq1/pSSC1 mitochondria had 54% of the SDH-Cytbc1 activity of wild-type mitochondria. We conclude that increasing Ssc1 levels approximately twofold can partially restore the activity of Fe/S-containing proteins. This suppression may well be the indirect result of lowering the free iron level within Δssq1 mitochondria.
DISCUSSION
The in vitro data reported here support a sequential mode of action of the two mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1; first, import across the mitochondrial inner membrane requires Ssc1 function, and second, processing of the intermediate to the mature form of the protein within the matrix is facilitated by Ssq1. It is likely that Ssc1 drives import of Yfh1 in a manner similar to the way it functions in the import of many polypeptides (30). The effect of Ssq1 on Yfh1 maturation is likely direct, as reducing the amount of iron accumulating in Δssq1 mitochondria did not alleviate the processing defect. However, the mechanism of a specific role of Ssq1 in facilitating the second cleavage of Yfh1 by MPP is unresolved (4). Perhaps it binds Yfh1 at a specific site(s), maintaining the intermediate form in a conformation that allows the second cleavage site to become accessible to MPP. In the absence of Ssq1, the Yfh1 intermediate may fold into a conformation that masks this cleavage site.
However, Ssq1 is not uniquely required for the processing of the intermediate form of Yfh1, as increasing the levels of Ssc1 twofold suppresses this maturation defect. Since Ssc1 is normally 1,000 to 2,000 times more abundant than Ssq1, we were surprised that increasing the level of the very abundant Ssc1 twofold was able to significantly suppress the processing defect of Yfh1 in vitro. Normally, Ssc1 is likely occupied in carrying out chaperone functions in the mitochondrial matrix. Therefore, little Ssc1 might be free and available for interaction with Yfh1. Even though the additional copy of SSC1 results in only a twofold increase in the total amount of Ssc1 protein, doubling the protein concentration might dramatically increase the level of “free” Ssc1. For example, if 5% of Ssc1 is normally unbound to substrates and free for interaction, a twofold increase in total Ssc1 concentration would be equivalent to a 20-fold increase in available Ssc1.
Based on the assumption that the free pool of Ssc1 is normally relatively small, we propose two possible explanations for the suppression. (i) Ssq1 and Ssc1 could differ in the specificity of their interactions with peptides, with Ssq1 having a very high affinity for a critical sequence in Yfh1. According to this scenario, Ssc1 would be capable of interacting with this Yfh1 sequence, but with a much lower affinity than Ssq1. Thus, a higher concentration of Ssc1 would be required to attain effective interactions with Yfh1. (ii) On the other hand, sequences within Ssq1, outside the peptide binding cleft, may serve to target Ssq1 to a particular site within the mitochondrial matrix, which facilitates interaction with the incoming precursor Yfh1 polypeptides. For example, factors may exist which interact with both Ssq1 and the intermediate form of Yfh1. By this model, Ssq1 might not have an intrinsic higher affinity for Yfh1 but rather have a very high effective concentration near Yfh1 precursors. Such factors may associate with the mitochondrial inner membrane, since matrix fractions alone were unable to process Yfh1 efficiently in vitro (4). If this model is correct, very high levels of free Ssc1 in the matrix would be required to obtain the same effective concentration as Ssq1.
Overexpression of Ssc1 not only suppressed the in vitro Yfh1 processing defect but also partially suppressed the in vivo phenotypes of iron accumulation, cold sensitivity, and reduced activity of Fe/S-containing proteins. This cosuppression lends credence to the idea that the similarity of ssq1 and yfh1 mutant phenotypes is at least partially due to the role of Ssq1 in Yfh1 maturation. However, the steady-state level of Yfh1 in mitochondria lacking Ssq1 is 75% of that found in wild-type mitochondria, raising the question of whether this small reduction in Yfh1 levels is sufficient to cause the observed phenotypes of ssq1 mutants. Since increasing the level of mature Yfh1 did not suppress the growth phenotypes of ssq1 mutant cells, the cause of the multiple ssq1 mutant phenotypes must be more complicated than simply the reduction in the efficiency of processing of the intermediate form of Yfh1. One can envision two possible explanations for this result. (i) Ssq1 facilitates the proper folding of mature Yfh1. Therefore, the mature form of Yfh1 is not folded properly in Δssq1 mitochondria. To test this possibility, we assessed aggregation and altered protease sensitivity of Yfh1 in Δssq1 mitochondria. However, no differences were observed in these experiments between wild-type and Δssq1 mitochondria (data not shown). Therefore, at this time, there are no experimental data to support this model. (ii) Ssq1 is involved in functions in addition to the maturation of Yfh1. These functions could involve the maturation of other proteins, which have yet to be identified, or other roles of Ssq1 in the matrix as discussed below.
The phenotypes of ssq1 mutant strains are partially suppressed by a 2,000-fold excess of Ssc1 over normal levels of Ssq1, suggesting that, even at increased levels, Ssc1 is not completely replacing Ssq1 function. Perhaps Ssc1 levels must be increased more than twofold to achieve full suppression. However, we found that Δssq1 cells harboring a high-copy 2μm plasmid containing SSC1 did not enhance suppression over that seen with a centromeric plasmid. In fact, expression of Ssc1 from this SSC1::2μm plasmid inhibited growth of wild-type cells (data not shown). Possibly, excess Ssc1 inappropriately binds to folded proteins, causing deleterious effects on the cell.
On the other hand, the partial suppression of ssq1 defects by Ssc1 suggests that Ssq1 has a specific function in mitochondria that cannot be replaced by Ssc1. For example, Ssq1 could play a unique role in the maintenance and/or assembly of Fe/S proteins (41, 43), which would explain not only the partial suppression of Δssq1 defects but also the inability of near-wild-type levels of mature Yfh1 to suppress Δssq1 phenotypes as well. Genetic data support a connection between Ssq1 and other proteins, Nfu1, Isu1 and Isu2, and Nfs1, believed to be involved in the assembly and/or maintenance of Fe/S centers (25, 41, 43). In nitrogen-fixing bacteria, orthologues of Isu1, Isu2, Nfu1, and Nfs1 are required for the synthesis of the Fe/S cluster of nitrogenase (9, 14, 48). Interestingly, orthologues of these genes along with genes encoding an hsp70 and a DnaJ-like protein reside within the same operon in Azotobacter vinelandii and non-nitrogen-fixing bacteria, suggesting the existence of a general macromolecular system for the biogenesis of Fe/S proteins (47). Therefore, it is possible that Ssq1 plays a chaperone-type role in maturation of proteins containing Fe/S clusters. For example, Ssq1 might assist in the assembly of a multimeric complex, composed of the proteins mentioned above, which functions in the assembly or repair of Fe/S proteins. However, at this time, a direct role for Ssq1 or any of these gene products in the assembly or repair of Fe/S proteins remains to be clearly separated from the indirect effects that may be caused by an alteration in iron homeostasis. Clearly, more work needs to be done to resolve the connections between the complex phenotypes described for Δssq1 strains.
ACKNOWLEDGMENTS
We thank Grazia Isaya for generously providing the Yfh1 antibodies and A. Dancis for helpful suggestions.
This work was supported by the National Institutes of Health (GM27870) to E.A.C. and the Biotechnology Training Program (5T32GM08349) and the Cremer Basic Sciences Fellowship Fund (C.V.). The work of J.M. was partially supported by the Polish State Committee for Scientific Research Project 6P04A06017.
REFERENCES
- 1.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 2.Beinert H. Micro methods for the quantitative determination of iron and copper in biological material. Methods Enzymol. 1978;54:435–445. doi: 10.1016/s0076-6879(78)54027-5. [DOI] [PubMed] [Google Scholar]
- 3.Blond-Elguindi S, Cwirla S, Dower W, Lipshutz R, Sprang S, Sambrook J, Gething M J. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 4.Branda S, Cavadini P, Adamec J, Kalousek F, Taroni F, Isaya G. Yeast and human frataxin are processed to mature form in two sequential steps by the mitochondrial processing peptidase. J Biol Chem. 1999;274:22763–22769. doi: 10.1074/jbc.274.32.22763. [DOI] [PubMed] [Google Scholar]
- 5.Branda S, Yang Z, Chew A, Isaya G. Mitochondrial intermediate peptidase and the yeast frataxin homolog together maintain mitochondrial homeostasis in Saccharomyces cerevisiae. Hum Mol Genet. 1999;8:1099–1110. doi: 10.1093/hmg/8.6.1099. [DOI] [PubMed] [Google Scholar]
- 6.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 7.Campuzano V, Montermini L, Molto M D, Pianese L, Cossee M, Cavalcanti F, Monros E. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 8.Chazotte B, Hackenbrock C R. Lateral diffusion as a rate-limiting step in ubiquinone-mediated mitochondrial electron transport. J Biol Chem. 1989;264:4978–4985. [PubMed] [Google Scholar]
- 9.Dean D R, Bolin J T, Zheng L. Nitrogenase metalloclusters: structures, organization, and synthesis. J Bacteriol. 1993;175:6737–6744. doi: 10.1128/jb.175.21.6737-6744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fansler B, Lowenstein J M. Aconitase from pig heart. Methods Enzymol. 1969;13:26–30. [Google Scholar]
- 11.Flynn G, Pohl J, Flocco M, Rothman J. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 12.Foury F, Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- 13.Fridovitch I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 14.Fu W, Jack R F, Morgan T V, Dean D R, Johnson M K. nifU gene product from Azotobacter vinelandii is a homodimer that contains two identical [2Fe-2S] clusters. Biochemistry. 1994;33:13455–13463. doi: 10.1021/bi00249a034. [DOI] [PubMed] [Google Scholar]
- 15.Gambill B D, Voos W, Kang P J, Miao B, Langer T, Craig E A, Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein S, Meyerstein D, Czapski G. The Fenton reagents. Free Radic Biol Med. 1993;15:435–445. doi: 10.1016/0891-5849(93)90043-t. [DOI] [PubMed] [Google Scholar]
- 17.Graham L A, Trumpower B L. Mutational analysis of the mitochondrial Rieske iron-sulfur protein of Saccharomyces cerevisiae. J Biol Chem. 1991;266:22485–22492. [PubMed] [Google Scholar]
- 18.Hakes D J, Dixon J E. New vectors for high level expression of recombinant proteins in bacteria. Anal Biochem. 1992;202:293–298. doi: 10.1016/0003-2697(92)90108-j. [DOI] [PubMed] [Google Scholar]
- 19.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 20.James P, Pfund C, Craig E. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 21.Johnson J L, Craig E A. Protein folding in vivo: unraveling complex pathways. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- 22.Jones E W. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- 23.Kang P J, Ostermann J, Shilling J, Neupert W, Craig E A, Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy M C, Kent T A, Emptage M, Merkle H, Beinert H, Münck E. Evidence for the formation of a linear [3Fe-4S] cluster in partially unfolded aconitase. J Biol Chem. 1984;259:14463–14471. [PubMed] [Google Scholar]
- 25.Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight S A B, Sepuri N B V, Pain D, Dancis A. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- 27.Lamarche J B, Cote M, Lemieux B. The cardiomyopathy of Friedreich's ataxia morphological observations in 3 cases. Can J Neuro Sci. 1980;7:389–396. doi: 10.1017/s0317167100022927. [DOI] [PubMed] [Google Scholar]
- 28.Mathai J C, Sauna Z E, John O, Sitaramam V. Rate-limiting step in electron transport. J Biol Chem. 1993;268:15442–15454. [PubMed] [Google Scholar]
- 29.Miao B, Davis J E, Craig E A. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J Mol Biol. 1997;265:541–552. doi: 10.1006/jmbi.1996.0762. [DOI] [PubMed] [Google Scholar]
- 30.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 31.Pfanner N, Craig E A, Honlinger A. Mitochondrial preprotein translocase. Annu Rev Cell Dev Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- 32.Radisky D C, Babcock M C, Kaplan J. The yeast frataxin homologue mediates mitochondrial iron efflux. J Biol Chem. 1999;274:4497–4499. doi: 10.1074/jbc.274.8.4497. [DOI] [PubMed] [Google Scholar]
- 33.Rassow J, Maarse A, Krainer E, Kubrich M, Muller H, Meijer M, Craig E, Pfanner N. Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix Hsp70 and the inner membrane protein Mim44. J Cell Biol. 1994;127:1547–1556. doi: 10.1083/jcb.127.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothstein R. Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 35.Rötig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 36.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan K R, Jensen R E. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 38.Sakuragi S, Liu Q, Craig E. Interaction between the nucleotide exchange factor Mge1 and the mitochondrial Hsp70 Ssc1. J Biol Chem. 1999;274:11275–11282. doi: 10.1074/jbc.274.16.11275. [DOI] [PubMed] [Google Scholar]
- 39.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 40.Schilke B, Forster J, Davis J, James P, Walter W, Laloraya S, Johnson J, Miao B, Craig E. Cold-sensitivity of an S. cerevisiae mutant lacking a newly identified mitochondrial Hsp70 is suppressed by loss of mitochondrial DNA. J Cell Biol. 1996;134:603–614. doi: 10.1083/jcb.134.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schilke B, Voisine C, Beinert H, Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strain J, Lorenz C R, Bode J, Garland S, Smolen G A, Ta D T, Vickery L E, Culotta V C. Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae. J Biol Chem. 1998;273:31138–31144. doi: 10.1074/jbc.273.47.31138. [DOI] [PubMed] [Google Scholar]
- 44.Tangerås A, Flatmark T, Bäckström D, Ehrenberg A. Mitochondrial iron not bound in heme and iron-sulfur centers. Biochim Biophys Acta. 1980;589:162–175. doi: 10.1016/0005-2728(80)90035-3. [DOI] [PubMed] [Google Scholar]
- 45.Voos W, Gambill B D, Guiard B, Pfanner N, Craig E A. Presequence and mature parts of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J Cell Biol. 1993;123:119–123. doi: 10.1083/jcb.123.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson K, Bertoli E, Griffiths D E. Phase transitions in yeast mitochondrial membranes. Biochem J. 1975;146:401–407. doi: 10.1042/bj1460401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng L, Cash V L, Flint D H, Dean D R. Assembly of iron-sulfur clusters. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 48.Zheng L, White R H, Cash V L, Jack R F, Dean D R. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc Natl Acad Sci USA. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]