Abstract
Background:
The manual segmentation of intact blood-brain barrier (BBB) regions in the stroke brain is cumbersome, due to the coexistence of infarction, large blood vessels, ventricles, and intact BBB regions, specifically in areas with weak signal enhancement following contrast agent injection.
Hypothesis:
That from dynamic susceptibility contrast (DSC)-MRI alone, without user intervention, regions of weak BBB damage can be segmented based on the leakage-related parameter K2 and the extent of intact BBB regions, needed to estimate K2 values, determined.
Study Type:
Feasibility.
Animal Model:
Ten female Sprague-Dawley rats (SD, 200–250g) underwent 1-hour middle carotid artery occlusion (MCAO) and 1-day reperfusion. Two SD rats underwent 1-hour MCAO with 3-day and 5-day reperfusion.
Field Strength/Sequence:
7T; ADC and T1 maps using diffusion-weighted echo planar imaging (EPI) and relaxation enhancement (RARE) with variable repetition time (TR), respectively. dynamic contrast-enhanced (DCE)-MRI using FLASH. DSC-MRI using gradient-echo EPI.
Assessment:
Constrained nonnegative matrix factorization (cNMF) was applied to the dynamic -curves of DSC-MRI (<4 min) in a BBB-disrupted rat model. Areas of voxels with intact BBB, classified by automated cNMF analyses, were then used in estimating K1 and K2 values, and compared with corresponding values from manually-derived areas.
Statistical Tests:
Mean ± standard deviation of ΔT1-differences between ischemic and healthy areas were displayed with unpaired Student’s t-tests. Scatterplots were displayed with slopes and intercepts and Pearson’s r values were evaluated between K2 maps obtained with automatic (cNMF)- and manually-derived regions of interest (ROIs) of the intact BBB region.
Results:
Mildly BBB-damaged areas (indistinguishable from DCE-MRI (10 min) parameters) were automatically segmented. Areas of voxels with intact BBB, classified by automated cNMF, matched closely the corresponding, manually-derived areas when respective areas were used in estimating K2 maps (Pearson’s r = 0.97, 12 slices).
Data Conclusion:
Automatic segmentation of short DSC-MRI data alone successfully identified areas with intact and compromised BBB in the stroke brain and compared favorably with manual segmentation.
Level of Evidence:
3
Technical Efficacy:
Stage 1
DYNAMIC SUSCEPTIBILITY CONTRAST magnetic resonance imaging (DSC-MRI) is a quantitative MRI technique to estimate brain perfusion parameters, such as cerebral blood volume (CBV), cerebral blood flow (CBF), and mean transit time (MTT).1–3 When gadolinium chelates are used as a contrast agent (CA) for DSC-MRI, CA may extravasate through a leaky blood–brain barrier (BBB) in subjects with cerebrovascular diseases.4–6 The extravasated CA then alters tissue relaxation times, causing bias in DSC-MRI signal signal–time-curves. Thus, investigations have been successfully undertaken regarding systematic measures to compensate for the leakage effects, particularly for tumor DSC-MRI with conspicuous BBB leakage.6–8
While BBB leakage has also been assessed by dynamic contrast-enhanced (DCE)-MRI,9,10 for certain ischemic strokes with weak BBB leakage the standard CA dose (0.1 mmol/kg) may not be adequate to generate sufficiently early (<10 min) tissue T1 reduction for signal enhancement that can be used to determine small vessel permeability (Ktrans, the standard measure of BBB permeability).11 In such a case, alteration of T2*-based DSC-MRI signal–time-curves at an elevated CA dose (0.3 mmol/kg) due to shortening T1 from leaky BBB may provide a more sensitive metric when segmenting regions of weakly damaged BBB within a shorter acquisition time (<4 min). Specifically, when the capacity for an increased CA dose is limited for DCE-MRI, with its stronger transverse relaxations, particularly prominent at high field strengths and its longer acquisition times.12–14 Thus, we hypothesize that the estimation of the vessel wall leakage index K2 of CA extravasation5,6,8,15–18 from DSC-MRI (<4 min) at an elevated dose (0.3 mmol/kg) with an intermediate repetition time (TR) (300 msec) can successfully segment BBB-damaged regions in a stroke model with weak BBB damage.
Methodologically, the standard model of K1 (blood-volume index) and K2 (leakage index) determination from DSC-MRI requires an unaltered ΔR2* curve from a region with intact BBB as a reference input function.5,6,8,15–17 Therefore, the extent of intact BBB must be known. For tumors, the model typically defines normal regions as nonenhancing voxels after CA injection. However, this assumption may not be readily applicable in cases of weak BBB damage due to ischemic stroke, in which segmentation of normal regions is cumbersome due to the coexistence of infarction, large vessels, ventricles, and normal regions in the same MR slice without apparent signal enhancement via CA injection. Thus, the automatic segmentation of each region from DSC-MRI signal–time-curves may achieve user-independent and objective determination of K2 values, avoiding the need for auxiliary MR images (diffusion, T2, and angiography) for region segmentation in addition to DSC-MRI signal–time-curves.
Nonnegative matrix factorization (NMF), which is an unsupervised pattern recognition (PR) algorithm, is fast and can efficiently classify dynamic signals without user intervention and training data.19–23 Constrained NMF (cNMF) analysis can iteratively search nonnegative matrices of the basic temporal patterns and corresponding weights, which satisfies the data matrix based on multiplicative update algorithms with certain constraints.24 Previous constrained NMF-based classification of heterogeneous tumor perfusion from DCE-MRI signals was reportedly effective.19,20 Thus, PR analysis may enable segmentation of BBB-damaged areas, large blood vessels, ventricles, and normal regions in the stroke brain from DSC-MRI signal–time-curves alone, without additional information.
In this study we focused on the automation of K2 mapping (leakage index) using the cNMF pattern recognition approach to avoid user-dependent manual region of interest (ROI) selection, which appears to be robust in the detection of BBB leakage for models of stroke with weak BBB damage.
Materials and Methods
Animal Model
All experiments (n = 12) were approved by the Institutional Animal Care and Use Committee. The transient middle carotid artery occlusion (tMCAO) group of Sprague–Dawley (SD) rats (200–250 g of SD female rats from Orient Bio [Gyeonggi, Republic of Korea]) underwent 1-hour MCAO surgery with intraluminal monofilament (0.35-mm diameter filament, Doccol, Sharon, MA) with 1-day reperfusion. The BBB disruption level can be controlled based on occlusion/reperfusion times25,26 for the tMCAO model. For a 1-hour occlusion time, corresponding BBB permeability is reportedly increased until 7-day reperfusion.26 Rat models of 1-hour MCAO with 1-day reperfusion were then used as the stroke group with weak BBB-damage (n = 4) for DSC-MRI studies (0.3 mmol/kg, <4 min).11 Longitudinal T1 difference (ΔT1) maps and DCE-MRI (0.1 mmol/kg, <10 min) experiments were performed with 1-day (n = 3), 3-day (n = 1), and 5-day (confirmed ex vivo Evans blue staining, n = 1) reperfusion animal models to evaluate the level of BBB disruption for direct comparison with DSC-MRI. Additional DCE-MRI experiments at the elevated dose of 0.3 mmol/kg were performed for a 1-day reperfusion model (n = 2) to directly compare DSC-MRI with DCE-MRI studies at the same dose. Rats were anesthetized with isoflurane during MRI scans with a 7T MR scanner (Bruker Biospin, Germany). For anesthesia, 1–2% isoflurane in a mixture of O2/N2O (3:7) was maintained during MRI scans. The body temperature of rats was maintained near 37°C by circulating warm water. The breathing rates were maintained at about 40 breath/min. To facilitate the CA injection, a 26G catheter was inserted into the tail vein
MRI
After placement of the rat and shimming across the rat brain, multislice T2-weighted and diffusion-weighted MR images, as well as T1 maps, were acquired before DSC- or DCE-MRI with CA injection. After DSC- or DCE-MRI, additional T1 maps were acquired. All experiments were performed with a 7T MR scanner (Bruker Biospin).
T2-weighted MR images were obtained by using rapid acquisition with relaxation enhancement (RARE)27 with the following parameters: TR = 5000 msec, RARE factor = 4, effective echo time (TE) = 30 msec, number of averages (NA) = 2, field of view (FOV) = 30 × 30 mm2, matrix size = 256 × 256, number of slices (NS) = 20, slice gap (SG) = 0 mm, and slice thickness (ST) = 0.5 mm.
Apparent diffusion coefficient (ADC) maps were acquired by using diffusion-weighted echo planar imaging (EPI)28 with the following parameters: TR = 5000 msec; number of segments = 4; effective TE = 20 msec; b-values = 200, 400, 600, and 1000 smm−2; NA = 1; FOV = 30 × 30 mm2; matrix size = 96 × 96; NS = 3; SG = 0.2 mm; and ST = 1 mm. Three ADC maps along the x, y, and z directions were averaged to obtain mean ADC values.
T1 maps were obtained by using RARE with variable TR (RAREVTR)29 with the following parameters: TR = 80, 150, 200, 400, 800, 1200, 1600, 2000, 2500, 3000, and 4500 msec; RARE factor = 4; effective TE = 4.58 msec; NA = 1; FOV = 30 × 30 mm2; matrix size = 96 × 96; NS = 3; SG = 0.2 mm; and ST = 1 mm.
DSC-MRI perfusion maps were acquired by using gradient-echo EPI sequence with the following pulse sequence parameters5: TR = 300 msec, effective TE = 17 msec, NA = 1, FOV = 30 × 30 mm2, matrix size = 96 × 96, NS = 3, SG = 0.2 mm, ST = 1 mm, bandwidth = 3.5 × 105 Hz, number of segments = 1, flip angle (FA) = 35°, number of repetitions (NR) = 800, and temporal resolution = 0.3 sec. The dose of the administered CA gadoterate meglumine (Gd-DOTA) was 0.3 mmol·kg−1 (130 μl). Pre- and post-CA injection acquisition times were 1 and 3 min, respectively.
DCE-MRI data were acquired by using fast low-angle shot (FLASH)30 with the following parameters: TR = 35 msec, TE = 1.9 msec, NA = 1, FOV = 30 × 30 mm2, matrix size = 96 × 96, NS = 3, SG = 0.2 mm, ST = 1 mm, FA = 30°, NR = 180, and temporal resolution = 3.36 sec. The Gd-DOTA injection dose for DCE-MRI was 0.1 mmol·kg−1, followed by a 0.2 mmol·kg−1 flush 15 min after DCE-MRI acquisition to sequentially obtain ΔT1 maps and improve contrast. The cumulative injected dose was 0.3 mmol·kg-1. Pre- and post-CA injection acquisition times were 1 and 9 min, respectively. Additional DCE-MRI experiments (n = 2) with the injection dose of 0.3 mmol/kg (same dose as DSC-MRI) were acquired for 20 min.
Ex Vivo Assessment of BBB Breakdown and Extent of Ischemia
Triphenyl tetrazolium chloride (TTC) and Evans blue stain were performed for estimating infarction31 and BBB disrupted region,32 respectively. Evans blue leakage in areas of damaged BBB was assessed ex vivo in a 5-day reperfusion model, to validate the association of postinjection T1 changes and BBB disruption. Post-MRI scans, a 3% Evans blue solution (3 ml/kg of body weight) was injected via the tail vein. After 30 min, the brain was perfused with 4°C saline and axially sectioned with 2-mm thickness. The slices were stained with a 1% TTC solution at 25°C for 10 min to detect the extent of the ischemic lesion by S.H.J. (4 years of experience). To validate the association between BBB-disrupted lesion from Evans blue leakage and ΔT1 map, respectively, thresholded masks were generated and the Dice coefficient was calculated between them.
Data Analysis
To obtain relative CBV (rCBV) maps, was converted from the DSC-MRI signal, where S(t) is the DSC-MRI signal and Spre is an averaged preinjection signal in DSC-MRI. By fitting ΔR2*(t) with the gamma variate function, rCBV values were calculated from the area of the fitted curve.1
For ADC calculations, each gradient direction of the ADC map was estimated with the following equation: S = S0 × e–ADC × b.28,33 Representative ADC maps were computed by averaging three gradient directions of ADC maps.
Vessel permeabilities were evaluated using an extended Tofts model to obtain Ktrans, Ve, and Vp maps from DCE-MRI signal–time-curves.30 The extended Tofts model was applied with the following equation: , where Ct and Cp represent CA concentration of tissue and blood, respectively. Cp was estimated from the internal carotid artery for each animal.
To confirm CA leakage, the ΔT1 map was computed by subtracting the postinjection T1 map from the preinjection T1 map, with ΔT1 longitudinally calculated at selected timepoints up to 85 min after CA injection.
To obtain K1 and K2 maps from DSC-MRI signal–time-curves, a ΔR2* vs. time curve of each voxel was fitted with arrival time and time scaling corrections16: , where represents an averaged ΔR2* curve for brain tissue regions with intact BBB, α is a time scaling factor, and τ is a time offset. K1 and K2 values then reflect cerebral blood volume and CA leakage, respectively. To calculate the curve, the ROIs were drawn manually on striatum of contralateral hemisphere by considering the T2-weighted image, ADC and rCBV maps, and then ΔR2* curves within ROIs were averaged.
To automatically segment the tissue region of intact BBB, cNMF was applied to ΔR2*(t) curves generated from DSC-MRI of rat stroke models. Detailed cNMF algorithms were described in a previous study,19,20,24 and were largely followed here, except for signal normalization. Briefly, to find nonnegative matrices S and W, representing the basic temporal curves and corresponding weights, respectively, that satisfy modeling of the data matrix V ≈ WS, multiplicative update algorithms were used. For initialization, W was constructed with random numbers (from 0 to 1) and the constrained (nonnegative) pseudoinversion matrix of W was allocated to initial S. Updates of W and S were performed by multiplicative update algorithms: . After each update process of W or S, negative elements in W or S were set to a small positive number ε (=2.2204×10−16, MatLab, R2016b, MathWorks, Natick, MA). This procedure was continued until the iteration error was lower than 10-8. Brain tissue selected within the skull boundary was used for cNMF analysis.
For large-vessel segmentation (high blood volume fraction), cNMF with k (number of the pattern) = 1 was applied to the unnormalized ΔR2*(t) curves. As the signal of the ΔR2*(t) curves reflects the blood volume for a healthy brain, the weight of such (k = 1) cNMF represents the blood volume fraction for each voxel. For the validation, the cNMF weights map were compared with the corresponding rCBV and K1 map. The scatterplot between cNMF weight and K1 values were plotted within the ROI of a healthy brain region.
Separately, for the segmentation of voxels with intact BBB, cNMF with k = 2 was applied to a normalized ΔR2*(t) curve. Normalization was performed to minimize blood volume heterogeneity (signal maximum in any specific pixel dependent on CA agent concentration and blood volume) by dividing the decaying stages of ΔR2*(t) with their maximum values, as shown in Supporting Information Fig. S1. Briefly, the signal of ΔR2*(t) curves in each voxel is scaled such that the maximum signal is equal to 1 for cNMF pattern recognition for voxels with differential K2 segmentation, as shown in Supplementary Fig. S1. Then, for k = 2, the cNMF weights difference map was defined by the difference between W matrix values of the first (fast decay) component and the second (slow decay) component from cNMF analyses. For validation, the cNMF weights difference map was compared with the ΔT1 and K2 map. The scatterplot between cNMF weights difference and K2 values were plotted within the ROI of the BBB-damaged region.
Automatic segmentation was then performed based on the cNMF weights (k = 1) and cNMF weights difference map (k = 2) to respectively delineate the region of large vessel and damaged BBB. When the cNMF weights map of k = 1 was larger than 1 (empirically set based on comparison with the corresponding rCBV map), the region was classified as a large vessel region. Regions in which cNMF weights difference maps of k = 2 were larger than 0.1, between 0 and 0.1, and smaller than 0 were defined as BBB-disrupted (or ventricles), mixture, and intact BBB regions, respectively (empirically set based on comparison with the corresponding ΔT1 map). The validity of these thresholds are shown in Supporting Information Fig. S2. The ventricles were further segmented based on the arrival time of contrast agent.34 Finally, the tissue region of intact BBB was then segmented by excluding large-vessel areas (from k = 1 cNMF) from intact BBB regions (from k = 2 cNMF).
To compare enhancement patterns of ΔR2* curves or decay patterns of normalized ΔR2* curves according to different tissue types, for short DSC-MRI acquisition (<4 min) with an elevated CA dose (0.3 mmol/kg), five different ROIs (sinus, ventricle, infarction, contralateral cortex, contralateral striatum) were drawn based on the T2-weighted image, and ADC and rCBV maps. Each ROI, depicting a representative region for each tissue type, was chosen to be 3 × 3 voxels, to keep the ROI the same size for the different tissue types, including the distinct anatomical structures of smaller size, such as ventricles and sinus.
To study the effects of ROI segmentation of the tissue region of intact BBB regions on the determination of K2 or K1 maps, five different ROIs (infarctions from diffusion map, ventricles from T2-weighted MR image, large vessels from blood volume map, manual ROI of intact BBB region, and automatic ROI of intact tissue region from cNMF analyses) were used as . To show how important the choice of the tissue region of intact BBB, respective K1 and K2 maps were compared among the ROIs. K1 and K2 maps derived from manual ROIs of the tissue region of intact BBB in the contralateral striatum area, which were collectively selected from diffusion maps, T2-weighted MR images and angiograms were considered as reference values for the direct comparisons with the rest.
To study the effects of the acquisition time of DSC-MRI on cNMF analysis, perfusion parameters (K1, K2, cNMF weight with k = 1 and cNMF weight difference with k = 2) were computed with the intentionally limited number of data (the number of used data corresponded to the following acquisition times: 0.5, 1, 1.5, 2, 2.5, 2.85 min). The K1 and K2 map with a manually drawn ROI of a healthy brain region are compared with cNMF weights and cNMF weights difference maps.
Statistical Analysis
To verify Ktrans and Ve differences between ischemic and healthy areas, and also to validate the association between ΔT1 differences and K2 values from cNMF segmented lesions of BBB damage and healthy areas, the mean and standard deviation (SD) of corresponding values were displayed and accompanying significance analyses were run with unpaired Student’s t-tests with a significance level of 0.05 using MatLab. To validate the linearity of K1 and K2 maps obtained with the automatic ROI of the intact BBB region from cNMF analyses with respect to the reference maps, scatterplots were displayed with slopes and intercepts and Pearson’s r values were evaluated.
MCAO occlusion/reperfusion times, MRI acquisitions, and the number of animals used are summarized in Table 1.
TABLE 1.
Summarized experimental parameters for each tMCAO group
| Occlusion/reperfusion times of MCAO model | MRI acquisitions | Number of animals (comments) |
|---|---|---|
| 1h-MCAO/1-day reperfusion | DCE-MRI (0 mmol/kg), ΔT1 map | n = 1 (control study without contrast agent injection) |
| 1h-MCAO/1-day reperfusion | DCE- MRI (0.1 mmol/kg), ΔT1 map | n = 3 |
| 1h-MCAO/1-day reperfusion | DCE- MRI (0.3 mmol/kg), ΔT1 map | n = 2 |
| 1h-MCAO/1-day reperfusion | DSC- MRI (0.3 mmol/kg), ΔT1 map | n = 4 |
| 1h-MCAO/3-day reperfusion | DCE- MRI (0.1 mmol/kg), ΔT1 map | n = 1 (control case for conspicuous BBB-leakage) |
| 1h-MCAO/5-day reperfusion | ΔT1 map | n = 1 (Evans blue matching) |
Results
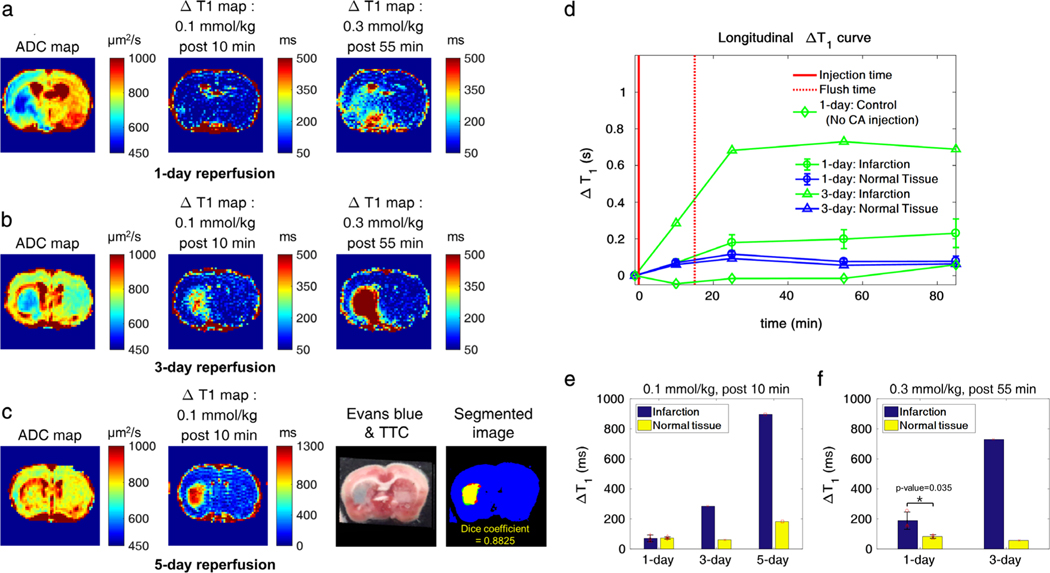
As shown in Fig. 1a, lesions with reduced ADC values were observed, but there were no noticeable changes in the 0.1 mmol/kg-injected ΔT1 maps (Fig. 1a, post-10 min CA injection). However, there were increases of ΔT1 on the ipsilateral hemisphere in the 0.3 mmol/kg-injected (cumulative) ΔT1 map (Fig. 1a, post-55 min CA injection). For direct comparisons, corresponding maps were generated for the 1-hour MCAO with 3-day and 5-day reperfusion after surgery, respectively, where increases of ΔT1 were observed in the ipsilateral brain at 10 min post-0.1 mmol/kg CA injection (Fig. 1b,c). The hyperintensity area on the ΔT1 map (>850 msec) in the ipsilateral lesion colocalized (Dice coefficient = 0.8825) with the corresponding Evans blue staining (Fig. 1c, 4th column), validating the use of ΔT1 values as standard BBB leakage index for the 1-hour MCAO reperfusion model. Longitudinal ΔT1 values were compared between the infarct and the normal area for the 1-day and 3-day reperfusion model in Fig. 1d. As shown for control experiments, no increase of ΔT1 in the ipsilateral lesions was observed for 1-hour MCAO with 1-day reperfusion without CA injection. The differences in ΔT1 values between the infarct and the healthy areas are plotted in Fig. 1e,f for 0.1 mmol/kg (10 min post-CA injection) and 0.3 mmol/kg injection (55 min post-CA injection), respectively. The 1-hour MCAO with 1-day reperfusion models exhibit negligible ΔT1 changes (P = 0.887) within 10 min post-0.1 mmol/kg CA injection. An elevated CA dose of 0.3 mmol/kg was required to increase ΔT1 changes (P = 0.035), highlighting the weak BBB damage of the 1-day reperfusion model compared with the 3-day and 5-day reperfusion models.
FIGURE 1:
ΔT1 maps for the MCAO models. a,b: ADC and ΔT1 maps at injection doses of 0.1 mmol/kg (post-10 min) and 0.3 mmol/kg (cumulative, post-55 min) for a 1-day and 3-day reperfusion model, respectively. c: The ADC and the ΔT1 map for a 0.1 mmol/kg CA injection dose post-10 min for a 5-day reperfusion model. Corresponding Evans blue staining results are shown to illustrate the association of elevated region of ΔT1 maps with respect to the area with BBB damage. Red regions indicate TTC-stained brain tissue. BBB damaged regions from Evans blue (red mask) and ΔT1 maps (green mask) are respectively shown in the segmented image. The yellow mask represents the overlapping (Dice coefficient = 0.8825) regions between Evans blue and ΔT1 maps. d: Longitudinal ΔT1 curves up to 85 min postinjection for infarcted and healthy brain regions in a 1-day and 3-day reperfusion model, where the solid red line marks the 0.1 mmol/kg CA injection point and the dotted red line the injection of the additional 0.2 mmol/kg CA flush. The longitudinal ΔT1 curve of infarction for a non-CA-injected 1-day reperfusion model (negative control) demonstrates the lack of significant ΔT1 increases, due to ongoing ischemia (d, green diamond line). e,f: Corresponding ΔT1 values for different models (n = 3 for the 1-day mean ± SD, and n = 1 each for the 3-day and 5-day reperfusion model, respectively) for 0.1 mmol/kg (post-10 min) and 0.3 mmol/kg (post-55 min), respectively.
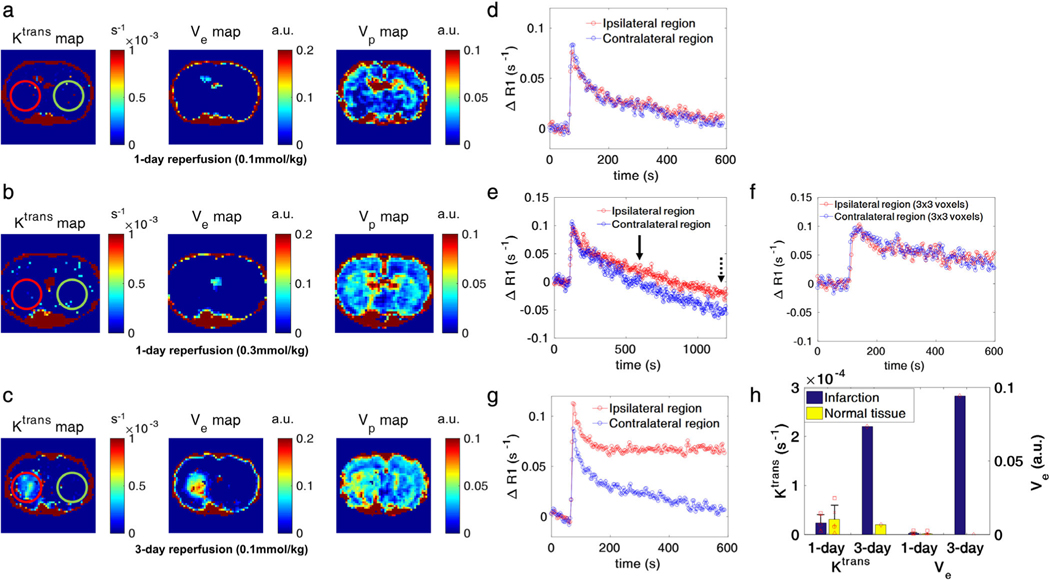
Injecting a higher 0.3 mmol/kg CA dose (equivalent to the dose of the DSC-MRI experiments) in one injection did not improve the detectability of subtle BBB damage in the first 10 min post-CA injection of the DCE-MRI in the 1-hour MCAO with 1-day reperfusion model (Fig. 2a,b). In both CA injection dose regimens (0.1 mmol/kg followed by 0.2 mmol/kg flush or 1 × 0.3 mmol/kg), ΔT1 (or ΔR1) values differed between infarcted and healthy brain regions only later (post-55 min of CA injection) in the DCE-MRI for the 1-day reperfusion model (Figs. 1, 2), with a clear deviation of ΔR1 curves between the ipsilateral infarcted and contralateral healthy brain being observed only after ∼10 min or more of DCE-MRI acquisition (Figs. 1d, 2e). These late curve detectable ΔT1 (or ΔR1) differences did not translate into noticeable differences (P = 0.631, 0.583, respectively) in the Ktrans and Ve values (Fig. 2h). Using a 1-hour MCAO rat with 3-day reperfusion model, the modest or not observable CA leakage and ΔT1 (or ΔR1) differences become detectable at early times post-CA injection by DCE-MRI, as represented by the higher Ktrans, Ve, and Vp in the infarction than the contralateral healthy brain (Fig. 2c,g,h). These data confirm the previously observed11,26 difficulties in segmenting subtle BBB-damage with a short DCE-MRI (<10 min) acquisition at both CA doses of 0.1 mmol/kg and 0.3 mmol/kg.
FIGURE 2:
DCE-MRI experimental results for the MCAO models. a,b: Permeability (Ktrans), extracellular extravascular space volume (Ve), and blood volume (Vp) maps from 1-hour MCAO with 24-hour reperfusion models at injection doses of 0.1 mmol/kg and 0.3 mmol/kg, respectively. c: Corresponding plots for a 1-hour MCAO with 3-day reperfusion model. d,e,g: ROI-based ΔR1 curves (red line: ipsilateral region, blue line: contralateral region) of the ROIs marked on the Ktrans maps (red circle: ipsilateral region, green circle: contralateral region) in a–c. The number of voxels in the ROI was ∼60 voxels for each case. f: Averaged ΔR1 curve from 3 × 3 voxels in ischemic lesion for comparison with corresponding DSC-MRI time–intensity curve. For the 0.3 mmol/kg dose of ΔR1 curves (e), 10 min and 20 min after CA injection are marked as solid and dotted line, respectively. g: The Ktrans and Ve values of infarction and contralateral healthy brain for the 1-day (n = 5, mean ± SD) and 3-day (n = 1) reperfusion models. There was no significant Ktrans and Ve value differences between normal tissue and infarction with the 1-day reperfusion model (n = 5, P = 0.631/0.583, unpaired t-test).
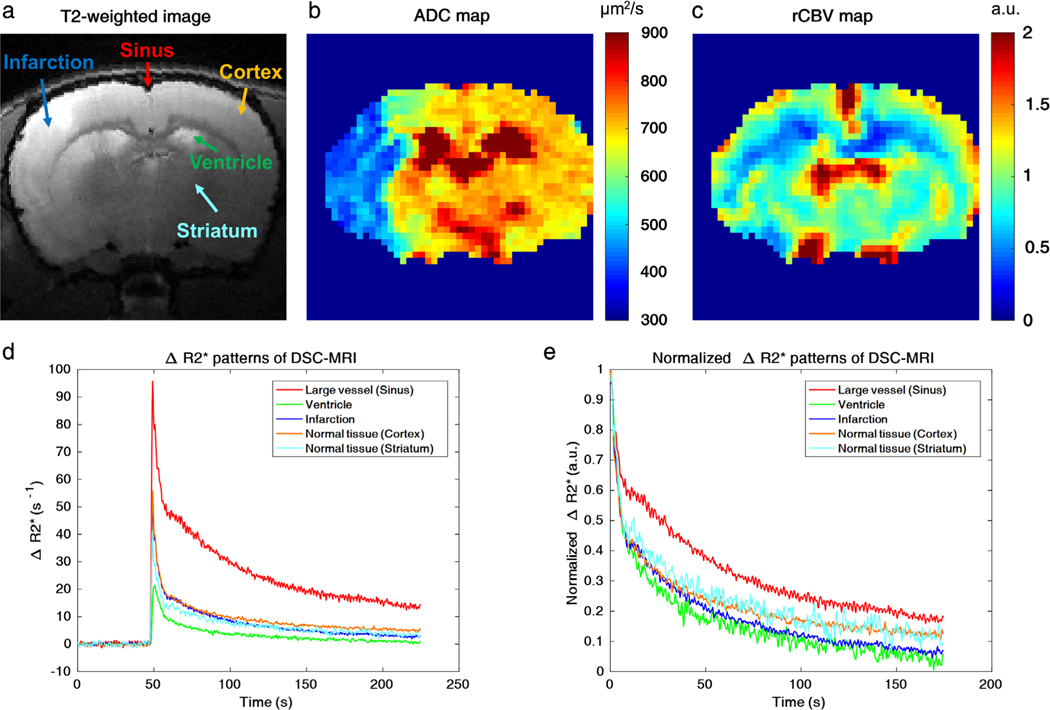
Five different ROIs (sinus, ventricle, infarction, contralateral cortex, contralateral striatum, colored arrows in Fig. 3a) were marked on the T2-weighted image (Fig. 3a), based on ADC (Fig. 3b) and rCBV (Fig. 3c) maps. Raw ΔR2* graphs in Fig. 3d showed that sinus and ventricle regions exhibited the largest and smallest peak enhancements among the five ROIs, respectively. For normalized ΔR2* curves, ventricle and infarction regions exhibited the fastest signal decays, while the sinus showed the slowest signal decay when compared with signals of the contralateral normal cortex and striatum regions (Fig. 3e).
FIGURE 3:
Dynamic ΔR2* patterns of five different brain regions. a: Five different ROIs on a T2-weighted MR image (red arrow: sinus, green arrow: ventricle, blue arrow: infarction, orange arrow: cortex, cyan blue arrow: striatum). b,c: ADC and rCBV maps, respectively. d,e: Raw and normalized ΔR2* time-intensity curves based on the five ROIs from Fig. 3a, respectively (red line: sinus, green line: ventricle, blue line: infarction, orange line: cortex, cyan blue line: striatum). The ROIs have the same voxel size (3 × 3 voxels) and the arrows indicate the center of the respective ROIs.
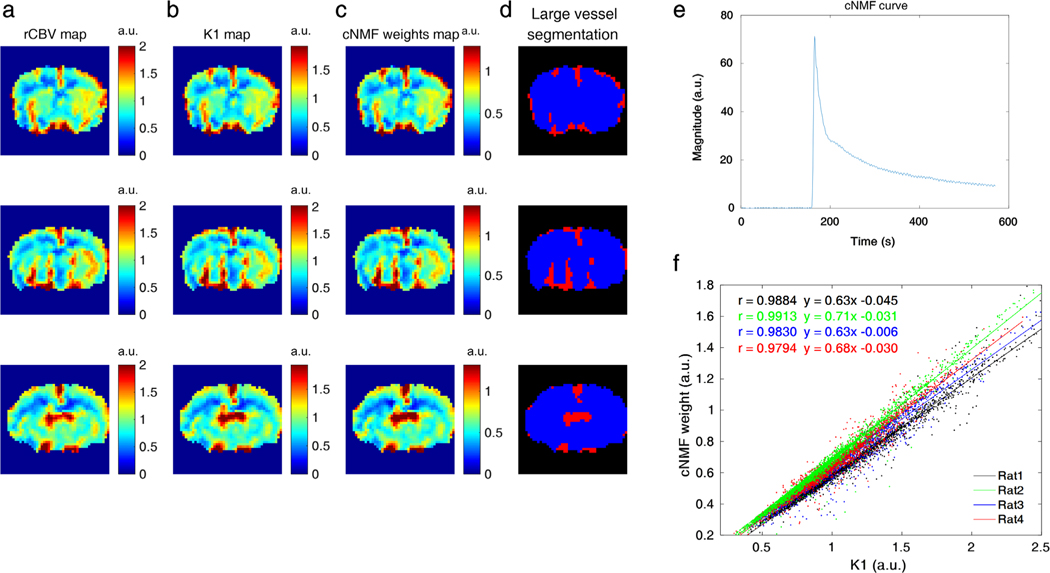
Figure 4 shows the cNMF with k = 1 results for large vessel segmentation. As shown in Fig. 4a–c, the cNMF weights map exhibited similar contrast on both the rCBV and K1 maps. When cNMF weights values were larger than 1, the voxels were then assigned to the large vessel region (marked as red color in Fig. 4d). A representative curve from cNMF (k = 1) is shown in Fig. 4e. For all cases, cNMF weight values related strongly to K1 values (Fig. 4f, Pearson’s r = 0.9884, 0.9913, 0.9830, and 0.9794). A linear but not identical correlation is assumed for the relationship between cNMF weight (k = 1) and K1, as both values were calculated based on different relative scales.
FIGURE 4:
a–c: rCBV, K1, and cNMF-weight maps with k (the number of patterns) = 1, respectively. d: The segmented large vessel regions (red = large vessels). Each row of a–d corresponds to different adjacent slices of a representative rat (rat2) brain scan. e: A representative cNMF curve with k = 1. f: Scatterplots between cNMF weights and K1 values for four different rats (dot: data, line: correlation curve, black: rat1, green: rat2, blue: rat3, red: rat4).
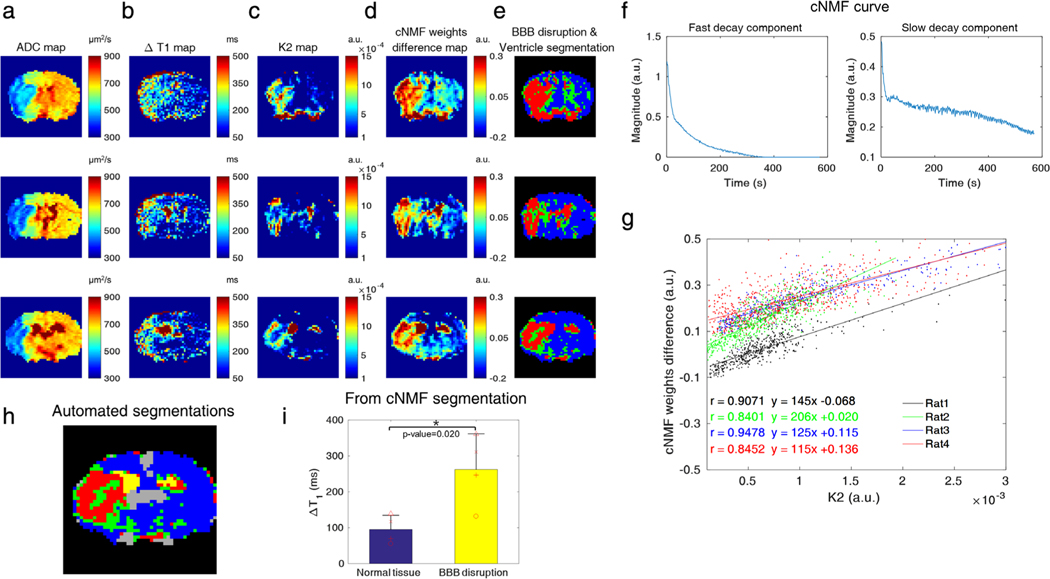
Figure 5 shows the cNMF with k = 2 results for ventricle and BBB-disrupted region segmentation. Infarction and BBB-disrupted regions were detected by ADC mapping (Fig. 5a) and ΔT1 mapping (55 min post-0.3 mmol/kg CA injection for DSC-MRI, Fig. 5b), respectively. The cNMF weights difference map and K2 map exhibited similar contrast, as shown in Fig. 5c,d. Figure 5e shows the segmentation results of BBB-disrupted (red), mixture (green), and intact BBB (blue) regions. Representative cNMF curves of fast-decay (first) and slow-decay (second) components are shown in Fig. 5f. The K2 and cNMF weights difference correlated strongly, as shown in Fig. 5g (Pearson’s r = 0.9071, 0.8401, 0.9478, and 0.8452). Again, a linear but not identical correlation was expected between cNMF weights difference (k = 2) and K2, as both values are calculated based on different relative scales. Red colored regions from the automatic cNMF segmentation primarily coincided with regions of BBB damage (ΔT1 > 250 msec) from the corresponding ΔT1 map in the same row. Conversely, regions from ΔT1 maps generally encompassed larger areas in the ipsilateral brain than areas from the cNMF segmentations, except for ventricle areas selected by cNMF segmentations. The automated segmentation (yellow: ventricle, red: BBB-damaged regions, green: a mixture of healthy and BBB-damaged brain, blue: healthy brain, gray: large vessels) of a representative slice are shown enlarged in Fig. 5h. The values of ΔT1 from cNMF classified BBB-damaged regions (red) were significantly different (P = 0.020) from those of corresponding normal tissue, as shown in Fig. 5i, indicating the association of BBB-damage with DSC-MRI derived leakage index (K2).
FIGURE 5:
a–d: The ADC map, ΔT1 map, K2 map, and cNMF-weights difference map with k (the number of patterns) = 2, respectively. e: The segmentation results to identify the BBB-damaged and ventricle regions (red: ventricle and BBB-damaged regions, green: a mixture of healthy and BBB-damaged brain, blue region: healthy brain). Each row of a–e corresponds to different adjacent slices of the same rat (rat2) and slices as displayed in Fig. 4a–d. f: Representative cNMF curves with k = 2 (left figure: fast decay component, right figure: slow decay component). g: Scatterplots between cNMF weight and K2 for four different rats (dot: data, line: correlation curve, black: rat1, green: rat2, blue: rat3, red: rat4). The ipsilateral regions that had larger K2 values (manual ROI-derived K2) than 10−4 were used for the scatterplots to relate K2 and cNMF weights. h: cNMF segmentations of five areas (yellow: ventricle, red: BBB-damaged regions, green: a mixture of healthy and BBB-damaged brain, blue: healthy brain, gray: large vessels). i: The values of ΔT1 from cNMF classified BBB-damaged regions (red) and corresponding normal tissue, indicating the association of BBB-damage with DSC-MRI-derived leakage index (K2). There was a significant ΔT1 difference between healthy brain tissue and BBB-damaged regions, as derived from the cNMF segmentation (n = 4, mean ± SD, P = 0.020, unpaired t-test).
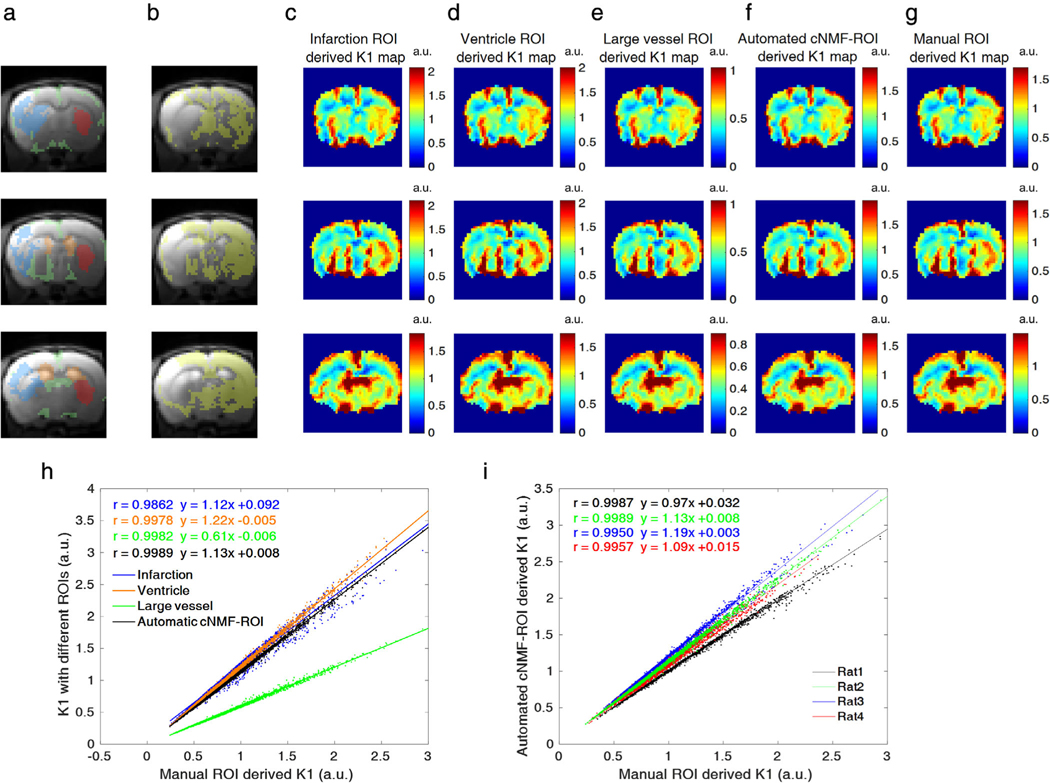
As shown in Fig. 6a–g, five different ROIs (infarction (blue), ventricle (orange), large vessel (green), manual-ROI (red), automated cNMF-ROI (yellow)) were overlaid on the anatomical MR image. For all cases, K1 maps exhibited similar contrast to each other, regardless of variations in ROI. As shown in Fig. 6h, all K1 maps related strongly to manually drawn ROIs (Pearson’s r = 0.9862, 0.9978, 0.9982, and 0.9989). Additionally, correlation coefficients between K1 from manual ROIs and automatic cNMF ROIs were > 0.99 for all rats (Fig. 6i, Pearson’s r = 0.9987, 0.9989, 0.9950, and 0.9957).
FIGURE 6:
a,b: Five different ROIs (blue: BBB-damaged regions, orange: ventricles, green: large vessels, red: manual ROI, yellow: automated cNMF-ROI). c–g: Corresponding K1 maps from five different ROIs, when a respective ROI was chosen as an intact BBB region. Each row of a–g corresponds to different slices of a representative rat (rat2) brain scan. h: Scatterplots between K1 values derived from four different ROIs and reference (manual) ROIs (dot: data, line: correlation curve, blue: BBB-damaged, orange: ventricle, green: vessel, black: automated cNMF-ROI) for a representative rat (rat2) averaged for all slices. i: Scatterplots between K1 values derived from four automated cNMF-ROIs and reference (manual) ROIs for four different rats (dot: data, line: correlation curve, black: rat1, green: rat2, blue: rat3, red: rat4).
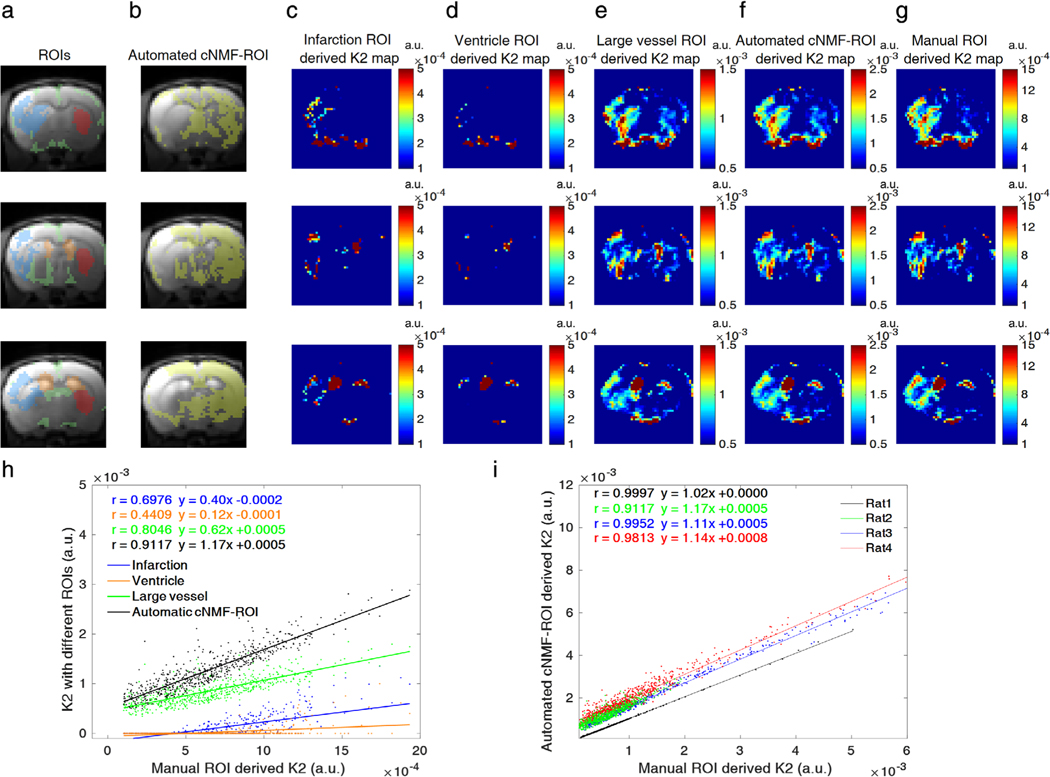
However, when infarction and ventricle regions were considered as intact BBB regions, the corresponding K2 values in infarction lesion were underestimated in comparison to K2 values derived from manual ROI, as shown in Fig. 7c,d. Scatterplots between K2 values derived from various ROIs and manual ROIs showed that the values derived from infarction and ventricle regions exhibited lower correlation (blue and orange color in Fig. 7h, Pearson’s r = 0.6976 and 0.4409, respectively). When large vessel regions were selected as healthy brain regions in the fitting, as shown in Fig. 7e, the Pearson’s r was 0.8046, slightly lower than that of the automatic ROI (black color in Fig. 7h, Pearson’s r = 0.9117). Automated cNMF ROI-derived K2 values (Fig. 7f) were highly correlated with respect to manual ROI (Fig. 7g) for all rats (12 slices from four rats), as plotted in Fig. 7i (Pearson’s r = 0.9997, 0.9117, 0.9761, and 0.9813).
FIGURE 7:
a,b: Five different ROIs (blue: BBB-damaged region, orange: ventricles, green: large vessels, red: manual ROI, yellow: automated cNMF-ROI). c–g: Corresponding K2 maps from five different ROIs, when denoted ROI is chosen as an intact BBB region. Each row of a–g corresponds to different slices of a representative rat (rat2) brain scan. h: Scatterplots between K2 values derived from four different ROIs and reference (manual) ROIs (dot: data, line: correlation curve, blue: infarction, orange: ventricle, green: vessel, black: automated cNMF-ROI) for a representative rat (rat2) averaged for all slices. i: Scatterplots between K2 values derived from four automated cNMF-ROIs and reference (manual) ROIs for four different rats (dot: data, line: correlation curve, black: rat1, green: rat2, blue: rat3, red: rat4). The ipsilateral regions that had larger K2 values (manual ROI-derived K2) than 10−4 were used for the scatterplots to relate manual ROI-derived K2 and cNMF ROI-derived K2.
The scan-time durations for fitting and PR analysis influenced the estimation of perfusion parameters (K1, K2) and cNMF weights, as shown in Supporting Information Figs. S3 and S4, respectively. K1 and cNMF weight maps were relatively robust to changes of acquisition times post-CA injection, as shown in Supporting Information Fig. S3a,b. K1 values (K1, 0.5 min), which were fitted from data within 0.5-min postinjection, correlated highly to the full data (K1, 2.85 min, Pearson’s r = 0.9765). For cNMF weights (cNMF0.5 min, k = 1), the Pearson’s r value with cNMF2.85 min was 0.9733. This trend was consistent for other animals, as shown in Supporting Information Fig. S3c,d. However, K2 values were highly sensitive to the length of the acquisition. The K2, 0.5 min map showed little permeability-oriented contrast. Permeability contrast became consistent for acquisition times over 2 min, as shown in Supporting Information Fig. S4a. Similar behavior was observed for the maps of the cNMF weights, as shown in Supporting Information Fig. S4b. The Pearson’s r values for K2, 0.5 min with K2, 2.85 min and cNMF0.5 min, k = 2 with cNMF2.85 min, k = 2 were 0.3147 and 0.1259, respectively (Supporting Information Fig. S4c,d). The Pearson’s r values for K2, 2 min with K2, 2.85 min and cNMF2 min, k = 2 with cNMF2.85 min, k = 2 were 0.9488 and 0.9636, respectively. This trend was consistent for other rats, as shown in Supporting Information Fig. S4c,d. Subsequently, cNMF analysis of DSC-MRI did not require a longer scan time than necessary for regular model-based fitting. At least 2 min acquisition of DSC-MRI is required both for K2 fitting and cNMF analysis to obtain BBB-leakage related contrast in this reperfusion model.
As shown in Supporting Information Fig. S5 for certain cases, the hyperintensity area on ΔT1 maps is markedly larger and encompasses the region with elevated K2 values of DSC-MRI. Furthermore, the regions with higher ΔT1 (red area in the second column) are generally observed to coincide with regions exhibiting elevated K2 (red area in the third column).
Discussion
Although the K1, K2 model of DSC-MRI is widely used to measure true CBV (K1 index) without CA extravasation effects,6,8,17,35 numerous studies also attempted to evaluate vascular permeability (K2 index) of neurovascular disease,5,6,8,15–18 which requires the robust delineation of tissue region of intact BBB. Often, nonenhancing voxels with CA injection were selected to obtain curves of intact BBB from DSC-MRI,6 because vessel permeability information cannot be obtained from anatomical/structural images. However, for brain tissue with mild BBB-damaged lesions, such as the 1-hour MCAO 24-h reperfusion model, it turns out to be difficult to objectively segment tissue regions of intact BBB even with short DCE-MRI acquisition. Consequently, incorrectly chosen curves are observed to deteriorate the following K2 estimation. Namely, K2 values were observed to be sensitive to both ROI selection for curve and scan-time durations. Thus, an automatic pattern recognition method would facilitate the robust segmentation of the tissue region of intact BBB, and cNMF-based segmentation is observed to be effective for distinguishing the region of mild BBB-leakage from DSC-MRI.
On the other hand, our results also manifest the underlying difficulties of absolute intersubject comparisons of conventional K2 values of DSC-MRI. The issues mainly come from different patterns of curve between subjects. Several factors, such as CA injection speed, the physiological condition of a subject, and resulting heterogeneity of ΔR2* patterns even in healthy tissue may affect the shape of curve from intact BBB. Therefore, as shown from varying intercepts of scatterplots between automatic- and manual (striatum)-ROI derived K2 values among different rats, the user-independent voxel selections of intact BBB regions may be a more objective procedure when intersubject comparisons are necessary, especially when the location of the lesion is unknown a priori.
It was also worthwhile to compare the sensitivity of various MR protocols in detecting mild BBB damage. A dose of 0.3 mmol/kg Gd-DOTA was chosen for the DSC-MRI in this study to obtain a good contrast-to-noise ratio, as commonly used in preclinical applications.36–39 While 0.1 mmol/kg CA is a typical dose for most preclinical DCE-MRI studies, in our study K2 from DSC-MRI (<4 min) appeared to be more sensitive to the BBB-damaged region than Ktrans from DCE-MRI (<10 min), even with the same CA dose. High temporal resolution (0.3 sec) with good signal contrast of ΔR2* curves from the elevated injection dose appears to be advantageous in detecting BBB-leakage related blood volume alterations of DSC-MRI in a relatively short acquisition time (<4 min). On the other hand, due to the leakage of CA, there is the possibility of complementary T2*-lengthening, as the susceptibility difference between blood vessel and tissue areas may be reduced. The extent of T2*-lengthening and the effect on the DSC-MRI time curve due to CA leakage should be explored, as it would reduce ΔR2*-contrast in the same manner as the shortening of T1.
Within the scope of this study, the overlap of elevated K2 and large ΔT1 regions warrants the validity of K2 as a vessel wall leakage index, as the difference in T1 values before and after (55 min) CA injection at the dose of 0.3 mmol/kg of reperfused brain should clearly indicate the region with leaking BBB as validated by representative Evans Blue staining results. As a control, there was no increase of ΔT1 in ipsilateral lesions without any CA injection for the corresponding model as well during the time-courses of MRI experiments. However, caution should be exercised when making direct one-to-one comparisons and interpretations between with ΔT1 and K2 values from DSC-MRI. Variations of CA concentration in imaging voxels due to differing perfusion and diffusion times of CA after injection (55 min for ΔT1, 2–3 min for DSC-MRI) may confound the direct comparisons.
Limitations of this study are as follows. First, because it has been reported that preinjection of CA before DSC-MRI acquisition significantly reduces leakage-oriented DSC-MRI signal alterations,11 longitudinal studies between DCE-MRI and DSC-MRI were not pursued in the same animal model. The availability of simultaneous DCE- and DSC-MRI acquisitions at an elevated dose (∼0.3 mmol/kg) may provide more direct comparisons between DCE- and DSC-MRI signal–time-curves. Second, we measured absolute T1 values before and after CA injection, which would shorten if CA leaked into the interstitial space through BBB openings of similar size. Notably, ΔT1 changes involve confounding factors, such as blood volume, interstitial space, and CA diffusion characteristics other than vessel wall permeability. Thus, direct comparisons between histology and in vivo MRI require caution, beyond issues related to in vivo – ex vivo alignment and vastly different imaging volumes. Third, when comparing the sensitivity of Ktrans and K2, the larger number of animals for various occlusion/reperfusion times of the MCAO model should be further investigated. In this study, we focused on the automation of K2 mapping for the 1-hour MCAO 24-hour reperfusion model, which has mild BBB-damage.
In conclusion, cNMF-based pattern recognition analysis was applied to dynamic ΔR2* curves of DSC-MRI (<4 min) for a rat model of 1-hour MCAO with 1-day reperfusion demonstrating weak BBB disruption. Pixels were automatically segmented as BBB-damaged, ventricles, large blood vessels, and normal tissue regions from signal–time-curves of DSC-MRI. By comparing K1 and K2 parameters derived from automatic (classified from cNMF-based segmentation of DSC-MRI alone) and manual (selected from T2-weighted images, ADC maps, and CBV maps) ROIs of intact BBB region, the usefulness of cNMF-based segmentation for the automation of voxel selections with differential vessel wall leakage index (K2) was demonstrated, particularly in evaluating DSC-MRI parameters for a stroke model with weak BBB damage. Clinically, this type of analysis might improve the observer-independent assessment of the stroke area, especially at early timepoints of stroke occurrence, where DCE-MRI has limited detectability.40
Supplementary Material
Acknowledgments
Contract grant sponsor: National Research Foundation of Korea (NRF); Contract grant sponsor: Korean government; Contract grant numbers: 2018M3C7A1056887 and 2018R1A6A1A03025810; Contract grant sponsor: Memorial Sloan-Kettering Cancer Support; Contract grant number: P30 CA008748.
References
- 1.Calamante F, Thomas DL, Pell GS, Wiersma J, Turner R. Measuring cerebral blood flow using magnetic resonance imaging techniques. J Cereb Blood Flow Metab 1999;19:701–735. [DOI] [PubMed] [Google Scholar]
- 2.Cha S, Knopp EA, Johnson G, Wetzel SG, Litt AW, Zagzag D. Intracranial mass lesions: Dynamic contrast-enhanced susceptibility-weighted echo-planar perfusion MR imaging. Radiology 2002;223:11–29. [DOI] [PubMed] [Google Scholar]
- 3.Zierler KL. Theoretical basis of indicator-dilution methods for measuring flow and volume. Circ Res 1962;10:393–407. [Google Scholar]
- 4.Bang OY, Saver JL, Alger JR, et al. Patterns and predictors of blood-brain barrier permeability derangements in acute ischemic stroke. Stroke 2009;40:454–461 [DOI] [PubMed] [Google Scholar]
- 5.Haselhorst R, Kappos L, Bilecen D, et al. Dynamic susceptibility contrast MR imaging of plaque development in multiple sclerosis: Application of an extended blood-brain barrier leakage correction. J Magn Reson Imaging 2000;11:495–505 [DOI] [PubMed] [Google Scholar]
- 6.Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood-volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. Am J Neuroradiol 2006;27:859–867. [PMC free article] [PubMed] [Google Scholar]
- 7.Dósa E, Guillaume DJ, Haluska M, et al. Magnetic resonance imaging of intracranial tumors: Intra-patient comparison of gadoteridol and ferumoxytol. Neurooncology 2010;13:251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quarles CC, Ward BD, Schmainda KM. Improving the reliability of obtaining tumor hemodynamic parameters in the presence of contrast agent extravasation. Magn Reson Med 2005;53:1307–1316. [DOI] [PubMed] [Google Scholar]
- 9.Heye AK, Thrippleton MJ, Armitage PA, et al. Tracer kinetic modeling for DCE-MRI quantification of subtle blood-brain barrier permeability. Neuroimage 2016;125:446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villringer K, Sanz Cuesta BE, Ostwaldt A-C, et al. DCE-MRI blood-brain barrier assessment in acute ischemic stroke. Neurology 2017;88: 433–440. [DOI] [PubMed] [Google Scholar]
- 11.Jin SH, Kang MS, Cho H. Cerebral blood perfusion deficits by dynamic susceptibility contrast (DSC)-MRI with gadolinium chelates in rats with postischemic reperfusion without significant dynamic contrast enhanced (DCE)-MRI derived vessel permeabilities: A cautionary note. Plos One 2018;13:e0201076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heilmann M, Walczak C, Vautier J, et al. Simultaneous dynamic T1 and T2* measurement for AIF assessment combined with DCE MRI in a mouse tumor model. Magn Reson Mater Phys Biol Med 2007;20: 193–203. [DOI] [PubMed] [Google Scholar]
- 13.Fruytier AC, Magat J, Colliez F, et al. Dynamic contrast-enhanced MRI in mice at high field: Estimation of the arterial input function can be achieved by phase imaging. Magn Reson Med 2014;71:544–550. [DOI] [PubMed] [Google Scholar]
- 14.Han S, Cho H. Temporal resolution improvement of calibration-free dynamic contrast-enhanced MRI with compressed sensing optimized turbo spin echo: The effects of replacing turbo factor with compressed sensing accelerations. J Magn Reson Imaging 2016;44:138–147. [DOI] [PubMed] [Google Scholar]
- 15.Taoka T, Kawai H, Nakane T, et al. Application of histogram analysis for the evaluation of vascular permeability in glioma by the K2 parameter obtained with the dynamic susceptibility contrast method: Comparisons with Ktrans obtained with the dynamic contrast enhance method and cerebral blood volume. Magn Reson Imaging 2016;34.7:896–901. [DOI] [PubMed] [Google Scholar]
- 16.Leigh R, Jen SS, Varma DD, Hillis AE, Barker PB. Arrival time correction for dynamic susceptibility contrast MR permeability imaging in stroke patients. PLoS One 2012;7:e52656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donahue KM, Krouwer HG, Rand SD, et al. Utility of simultaneously acquired gradient-echo and spin-echo cerebral blood volume and morphology maps in brain tumor patients. Magn Reson Med 2000;43: 845–853. [DOI] [PubMed] [Google Scholar]
- 18.Bonekamp D, Deike K, Wiestler B, et al. Association of overall survival in patients with newly diagnosed glioblastoma with contrast-enhanced perfusion MRI: Comparison of intraindividually matched T1-and T2*-based bolus techniques. J Magn Reson Imaging 2015;42:87–96. [DOI] [PubMed] [Google Scholar]
- 19.Stoyanova R, Huang K, Sandler K, et al. Mapping tumor hypoxia in vivo using pattern recognition of dynamic contrast-enhanced MRI data. Transl Oncol 2012;5:437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han S, Stoyanova R, Lee H, et al. Automation of pattern recognition analysis of dynamic contrast-enhanced MRI data to characterize intratumoral vascular heterogeneity. Magn Reson Med 2018;79: 1736–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YCC, Ackerstaff E, Tschudi Y, et al. Delineation of tumor habitats based on dynamic contrast enhanced MRI. Sci Rep 2017;7:9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JS, Lee DD, Choi S, Park KS, Lee DS. Non-negative matrix factorization of dynamic images in nuclear medicine. Nuclear Science Symposium Conference Record, 2001 IEEE. 2001;4:2027–2030. [Google Scholar]
- 23.Wang D, Xiao M, Zhang Y, Wan M. Abdominal parametric perfusion imaging with respiratory motion-compensation based on contrast-enhanced ultrasound: In-vivo validation. Comput Med Imaging Graph 2018;65:11–21. [DOI] [PubMed] [Google Scholar]
- 24.Sajda P, Du S, Brown TR, et al. Nonnegative matrix factorization for rapid recovery of constituent spectra in magnetic resonance chemical shift imaging of the brain. IEEE Trans Med Imaging 2004;23: 1453–1465. [DOI] [PubMed] [Google Scholar]
- 25.Neumann-Haefelin T, Kastrup A, de Crespigny A, et al. Serial MRI after transient focal cerebral ischemia in rats: Dynamics of tissue injury, blood-brain barrier damage, and edema formation. Stroke 2000;31: 1965–1972. [DOI] [PubMed] [Google Scholar]
- 26.Lin CY, Chang C, Cheung WM, et al. Dynamic changes in vascular permeability, cerebral blood volume, vascular density, and size after transient focal cerebral ischemia in rats: Evaluation with contrast-enhanced magnetic resonance imaging. J Cereb Blood Flow Metab 2008;28: 1491–1501. [DOI] [PubMed] [Google Scholar]
- 27.Hennig J, Nauerth A, Friedburg H. RARE imaging: A fast imaging method for clinical MR. Magn Reson Med 1986;3:823–833. [DOI] [PubMed] [Google Scholar]
- 28.Meng X, Fisher M, Shen Q, et al. Characterizing the diffusion/perfusion mismatch in experimental focal cerebral ischemia. Ann Neurol 2004;55: 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee DK, Han SH, Cho H. Optimization of sparse phase encodings for variable-repetition-delay turbo-spin echo (TSE) T1 measurements for preclinical applications. J Magn Reson 2017;274:57–64. [DOI] [PubMed] [Google Scholar]
- 30.Roberts C, Issa B, Stone A, et al. Comparative study into the robustness of compartmental modeling and model-free analysis in DCE-MRI studies. J Magn Reson Imaging 2006;23:554–563. [DOI] [PubMed] [Google Scholar]
- 31.Busch E, Krüger K, Hossmann K-A. Improved model of thromboembolic stroke and rt-PA induced reperfusion in the rat. Brain Res 1997; 778:16–24. [DOI] [PubMed] [Google Scholar]
- 32.Uyama O, Okamura N, Yanase M, et al. Quantitative evaluation of vascular permeability in the gerbil brain after transient ischemia using Evans blue fluorescence. J Cereb Blood Flow Metab 1988;8:282–284. [DOI] [PubMed] [Google Scholar]
- 33.Stejskal EO, Tanner JE. Spin diffusion measurements: Spin echoes in the presence of a time-dependent field gradient. J Chem Phys 1965; 42:288–292. [Google Scholar]
- 34.Kao YH, Guo WY, Wu YT, et al. Hemodynamic segmentation of MR brain perfusion images using independent component analysis, thresholding, and Bayesian estimation. Magn Reson Med 2003;49: 885–894. [DOI] [PubMed] [Google Scholar]
- 35.Boxerman JL, Prah DE, Paulson ES, et al. The role of preload and leakage correction in gadolinium-based cerebral blood volume estimation determined by comparison with MION as a criterion standard. Am J Neuroradiol 2012;33:1081–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirestam R, Ryding E, Lindgren A, et al. Absolute cerebral blood flow measured by dynamic susceptibility contrast MRI: A direct comparison with Xe-133 SPECT. Magma 2000;11:96–103. [DOI] [PubMed] [Google Scholar]
- 37.Chen F, Feng Y, Zheng K, et al. Enhanced antitumor efficacy of a vascular disrupting agent combined with an antiangiogenic in a rat liver tumor model evaluated by multiparametric MRI. PLoS One 2012;7: e41140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mystkowska D, Tutas A, Jezierska-Wozniak K, et al. Usefulness of clinical magnetic resonance scanners for imaging experimental changes in laboratory rodents’ central nervous system. Polish Ann Med 2012;19: 43–49. [Google Scholar]
- 39.Chyi T, Chang C. Temporal evolution of 3-nitropropionic acid-induced neurodegeneration in the rat brain by T2-weighted, diffusion-weighted, and perfusion magnetic resonance imaging. Neuroscience 1999;92: 1035–1041. [DOI] [PubMed] [Google Scholar]
- 40.Welker K, Boxerman J, Kalnin XA, et al. ASFNR recommendations for clinical performance of MR dynamic susceptibility contrast perfusion imaging of the brain. Ame J Neuroradiol 2015;36:E41–E51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.