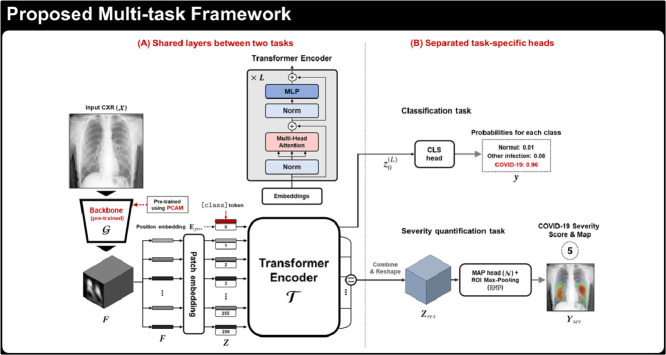
Graphical abstract
Keywords: Coronavirus disease-19, Chest X-ray, Vision transformer, Multi-task learning
MSC: 41A05, 41A10, 65D05, 65D17
Abstract
Developing a robust algorithm to diagnose and quantify the severity of the novel coronavirus disease 2019 (COVID-19) using Chest X-ray (CXR) requires a large number of well-curated COVID-19 datasets, which is difficult to collect under the global COVID-19 pandemic. On the other hand, CXR data with other findings are abundant. This situation is ideally suited for the Vision Transformer (ViT) architecture, where a lot of unlabeled data can be used through structural modeling by the self-attention mechanism. However, the use of existing ViT may not be optimal, as the feature embedding by direct patch flattening or ResNet backbone in the standard ViT is not intended for CXR. To address this problem, here we propose a novel Multi-task ViT that leverages low-level CXR feature corpus obtained from a backbone network that extracts common CXR findings. Specifically, the backbone network is first trained with large public datasets to detect common abnormal findings such as consolidation, opacity, edema, etc. Then, the embedded features from the backbone network are used as corpora for a versatile Transformer model for both the diagnosis and the severity quantification of COVID-19. We evaluate our model on various external test datasets from totally different institutions to evaluate the generalization capability. The experimental results confirm that our model can achieve state-of-the-art performance in both diagnosis and severity quantification tasks with outstanding generalization capability, which are sine qua non of widespread deployment.
1. Introduction
The novel coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has emerged as one of the deadliest viruses of the century, resulting in about 137 million people infected with over 2.9 million death worldwide as of April 2021. In the light of the unprecedented pandemic of COVID-19, public health systems have faced many challenges, including scarce medical resources, which are pushing healthcare providers to face the threat of infection (Ng et al., 2020). Considering its ominously contagious nature, the early screening of COVID-19 infection becoming increasingly important to avert the further spread of disease and thereby reduce the burden on the saturated health care system.
Currently, the real-time polymerase chain reaction (RT-PCR) is considered as the gold standard in the diagnosis of COVID-19 for its high sensitivity and specificity (Tahamtan and Ardebili, 2020), but it takes several hours and even days depending on regions to get the exam results due to overstressed laboratories. Since the majority of patients with confirmed COVID-19 present positive radiological findings, the radiologic examinations can be useful for rapid screening of disease (Shi et al., 2020). Although computed tomography (CT) scan has excellent sensitivity and specificity for COVID-19 diagnosis (Bernheim et al., 2020), the use of CT is a major burden because of its high cost and potential for cross-contamination in the radiology suite. Therefore, Chest X-ray (CXR) holds many practical advantages as a primary screening tool in the pandemic situation. In addition, CXR is useful for follow-up, which should be inexpensive and low in radiation exposure, to assess response to treatment.
Consequently, many studies have reported early application of CXR deep learning for diagnosis (Wang, Lin, Wong, 2020, Hemdan, Shouman, Karar, Narin, Kaya, Pamuk, Oh, Park, Ye, 2020) or severity quantification of COVID-19 (Cohen, Dao, Roth, Morrison, Bengio, Abbasi, Shen, Mahsa, Ghassemi, Li, et al., 2020, Signoroni, Savardi, Benini, Adami, Leonardi, Gibellini, Vaccher, Ravanelli, Borghesi, Maroldi, Farina, Zhu, Shen, Abbasi, Hoshmand-Kochi, Li, Duong, 2020, Wong, Qiu Lin, Wang, Chung, Shen, Abbasi, Hoshmand-Kochi, Duong, 2020), but they suffered from ineradicable drawbacks of poor generalization capability stemming from the scanty labelled COVID-19 data (Hu, Jacob, Parker, Hawkes, Hurst, Stoyanov, 2020, Zech, Badgeley, Liu, Costa, Titano, Oermann, 2018, Roberts, Driggs, Thorpe, Gilbey, Yeung, Ursprung, Aviles-Rivero, Etmann, McCague, Beer, et al., 2021). The stable generalization performance on unseen data is indispensable for widespread adoption of the system (Roberts et al., 2021).
One of the most commonly used measures to solve this problem is to build a robust model with innumerable training data (Chen et al., 2020a). However, although plenty of CXRs of COVID-19 is taken all around the world every day, available datasets are still limited due to lack of the expert labels and the difficulties in sharing patient data outside the hospital for privacy issues. The situation becomes even worse in the current pandemic situation, hindering the collaboration between different hospitals in different countries. As a result, several methods have been proposed to mitigate the problem by transfer learning (Apostolopoulos and Mpesiana, 2020), weakly supervised learning (Zheng, Deng, Fu, Zhou, Feng, Ma, Liu, Wang, 2020, Wang, Deng, Fu, Zhou, Feng, Ma, Liu, Zheng, 2020), and anomaly detection (Zhang et al., 2020), but their performances are still suboptimal.
The previous studies mostly utilize convolutional neural network (CNN) models, which were not specially designed for manifestations of COVID-19 which can be characterized by bilateral involvement, peripheral and lower zone dominance of ground-glass opacities, and patchy consolidations (Cozzi et al., 2020). Although CNN architecture has shown to be superb in many vision tasks, it may not be optimal for problems requiring high-level CXR disease classification, where global characteristics like multiplicity, distribution, and patterns have to be considered. This is due to the intrinsic locality of pixel dependencies in the convolution operation.
To overcome the similar limitation of CNN in computer vision problems that require the integration of global relationship between pixels, Vision Transformer (ViT) equipped with the Transformer architecture (Vaswani et al., 2017) was proposed to model long-range dependency among pixels through the self-attention mechanism, showing the state-of-the-art (SOTA) performance in the image classification task (Dosovitskiy et al., 2020). Since the Transformer was originally invented for natural language processing (NLP) in order to attend different positions of the input sequence within a corpus and compute a representation of that sequence, the choice of an appropriate corpus is the prerequisite for the Transformer design.
In the original paper (Dosovitskiy et al., 2020), two ViT models were suggested utilizing either direct pixel-patch embedding or feature embedding by ResNet backbone as corpora for Transformer. A problem occurs here, however, that neither the direct pixel-patch embedding nor feature embedding from ResNet may not be the optimal input embedding for the CXR diagnosis of COVID-19. Fortunately, several large-scale CXR data sets are constructed before the COVID-19 pandemic and are publicly available. For example, CheXpert (Irvin et al., 2019), a large dataset that contains over 220,000 CXR images, provides labeled common low-level CXR findings (e.g. consolidation, opacity, edema, etc.), which is also useful for the diagnosis of infectious disease. Moreover, an advanced CNN architecture has been suggested using the same dataset (Ye et al., 2020), which uses probabilistic class activation map (PCAM) pooling to leverage the class activation map to enhance the localization ability as well as classification performance. To take the maximum advantage of both the dataset and the network architecture for COVID-19, here we propose a novel ViT architecture which utilizes this advanced CNN architecture as a feature extractor for low-level CXR feature corpus, upon which Transformer is trained for downstream tasks of diagnosis by utilizing the self-attention mechanism in Transformer.
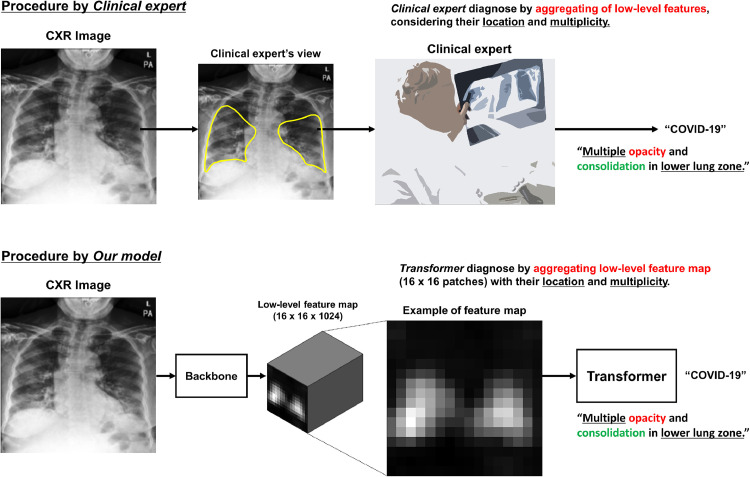
It is worth mentioning that our network is basically identical to the text classification task with Transformer architecture, where the Transformer not only adds meaning but also takes into account the location and relationship of words to classify at the sentence-level. Moreover, our method emulates the clinical experts who determine the final diagnosis of CXR (e.g. normal, bacterial pneumonia, COVID-19 infection, etc.) by comprehensively considering the low-level features with their pattern, multiplicity, location, and distribution (e.g. Multiple opacities and patch consolidations exist with lower lung zone dominance: high probability for COVID-19) as illustrated in Fig. 1 .
Fig. 1.
The analogy between the diagnosis by a clinical expert and by our method.
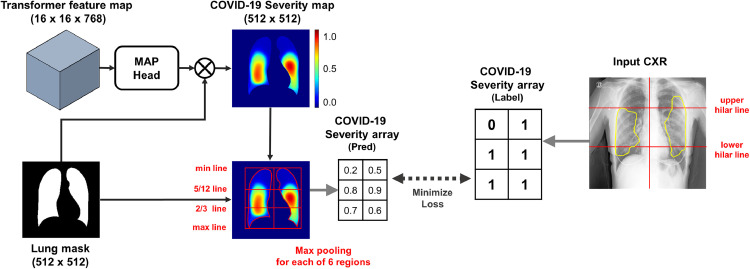
Another important contribution of this paper is to show that our ViT framework can be also used for COVID-19 severity quantification and localization, enabling the serial follow-up of severity and thereby assisting the treatment decision of clinicians (Cohen et al., 2020b). The severity of COVID-19 can be determined by quantifying the extent of COVID-19 involvement. Recently, array-based simple severity annotations where 1 or 0 is assigned to every 6 subdivisions of lungs are proposed by Toussie et al. (2020), and we are interested in utilizing this weak labeling approach for severity quantification. As the Transformer output already incorporates the long-range relationship between regions through self-attention, we use this Transformer output to design a light-weighted network that can accurately quantify and localize the COVID-19 extents from weak labels. Specifically, we adopt the region of interest (ROI) max-pooling of the output Transformer feature to bridge the severity map and simple array. Consequently, in addition to the global severity score from 0 to 6, our model can create an intuitive severity level map where each pixel value explicitly means the likelihood of the presence of a COVID-19 lesion using the weak array-based labels.
Finally, we have integrated the developed classification and severity quantification models into multi-task learning (MTL) framework to enable a single versatile model to perform the classification and severity quantification simultaneously, to better offer a more straightforward application of the developed system as well as improving the performances of individual tasks by sharing robust representation between related tasks.
In summary, our main contributions are as follows.
-
•
A novel ViT model for COVID-19 is proposed by leveraging the low-level CXR feature corpus that contains the representations for common CXR findings with the pre-built large-scale dataset.
-
•
We have not limited our model to classification but expanded our model to quantify severity to provide clinicians with clinical guidelines for making treatment decisions.
-
•
The classification and severity quantification models were integrated into a single multi-task model for straightforward applicability, which also improved the performances of both tasks.
-
•
We experimentally demonstrated that our method outperforms the previous models for COVID-19 as well as other CNN and Transformer-based architectures especially in terms of the generalization on unseen data.
The remainder of this paper is organized as follows. Section 2 summarizes the related works. Section 3 and Section 4 describes the proposed framework and datasets, respectively. Experimental results are presented in Section 5. Finally, we conclude this work in Section 6.
2. Related works
2.1. Vision transformer
Transformer (Vaswani et al., 2017), which was originally invented for NLP, is a deep neural network based on a self-attention mechanism that facilitates appreciably large receptive fields. After demonstrating its astounding performance, not only has Transformer become a de facto standard practice in NLP, but it has also motivated the computer vision community to explore its applications in computer vision by taking advantage of the long-range dependency between pixels (Khan et al., 2021).
The ViT was the first major attempt to apply a pure Transformer directly to an image, suggesting that it can completely replace the standard convolution operations by achieving SOTA performance. However, the experimental results showed that training the vanilla ViT model requires a huge computational cost. Therefore, the authors also suggested hybrid architecture by conjugating CNN backbone (e.g. ResNet) to Transformer. With the feature extracted by ResNet, the Transformer can mainly focus on modeling the global attention. The experimental results suggest that it was able to achieve higher performance with the hybrid approach with a relatively small amount of computations.
After the introduction of ViT, the application of Transformer in computer vision has become an active area of investigation, resulting in many variant models of ViT showing SOTA performance in a variety of vision tasks including object detection (Zhu et al., 2020b), classification (Dosovitskiy, Beyer, Kolesnikov, Weissenborn, Zhai, Unterthiner, Dehghani, Minderer, Heigold, Gelly, et al., Chen, Radford, Child, Wu, Jun, Luan, Sutskever, 2020), segmentation (Zheng et al., 2020b), and so on.
2.2. Probabilistic class activation map pooling
Class activation map (CAM) is a sort of class-specific saliency map obtained by quantifying the contribution of a particular area of an image to the prediction of the network. The most useful aspect of CAM is that it enables the localization of the important area only with weak labels, namely image-level supervision. Despite its excellent localization ability, most previous works utilized CAM to generate heatmaps for lesion localization and visualization during inference. To leverage the localization ability of CAM to enhance the performance of the network itself, one recent study utilized the CAM during training in CXR classification and localization tasks (Ye et al., 2020). They devised a novel global pooling operation that explicitly leverages the CAM in a probabilistic manner and is known as PCAM pooling. Different from standard approaches that use CAM for direct localization, they bound it with an additional fully connected layer and sigmoid function to get probabilities for each CXR findings. Then, the normalized attention weights were obtained from these output probabilities to make weighted feature maps containing more useful representations for each class. They showed that PCAM pooling operation can enhance both localization and diagnostic performance of the model and achieved first place in the 2019 CheXpert Challenge. For a detailed process of the PCAM operation, please refer to Appendix A.
2.3. COVID-19 severity quantification
To build an automated algorithm for severity quantification, pixel-level annotation such as lesion segmentation labels can offer plentiful information. However, this type of labeling methods are labor-intensive and collecting large data with this pixel-level annotated label is not feasible under the global pandemic of COVID-19. To alleviate the problem, simplified severity annotation methods, such as score-based and array-based methods, have been proposed. For example, Cohen et al. (2020a) suggested a geographic extent score and a lung opacity score based on a rating system of lung edema proposed by Warren et al. (2018). A geographic extent score assigns scores that range from 0 to 4, while a lung opacity score assigns values of 0 to 3 based on the severity of involvement in each lung area. Borghesi and Maroldi (2020) designed Brixia score, another array-type severity labeling method, dividing lung with anatomic landmarks and assign a score of 0–3 to each subdivision. Similarly, Toussie et al. (2020) suggested an array-based severity score for COVID-19. After dividing both lungs into six divisions, each area is assigned a value of 0 or 1, depending on the presence of COVID-19 involvement, which adds up to overall severity of 0 to 6. We adopted the array-based annotation method suggested by Toussie et al. (2020) for severity quantification of COVID-19.
2.4. Deep learning models for COVID-19
Upon rapid spread of COVID-19, there have been numerous approaches to enable automated diagnosis and severity prediction of COVID-19. For diagnosis, Wang et al. (2020a) proposed COVIDNet that adopted a lightweight projection-expansion-projection-extension design and long-range connectivity to improve representational capacity and showed good performance compared with standard CNN models. Khan et al. (2020) proposed CoroNet which was based on Xception (Chollet, 2017) network pre-trained on ImageNet and subsequently fine-tuned with COVID-19 data. Similarly, Minaee et al. (2020) proposed Deep-COVID in which various data augmentations were used and the last layer of standard CNNs were fine-tuned for COVID-19 data. Using DarkNet-19 (Redmon and Farhadi, 2017) used for object detection framework, Ozturk et al. (2020) proposed DarkCOVIDNet.
To quantify the severity of COVID-19 infection on CXR, Cohen et al. (2020a) devised a network pre-trained to classify 7 pathologies and trained to perform linear regression to predict the severity scores. Kwon et al. (2020) proposed CheXNet that pre-trained on ImageNet and subsequently trained to predict COVID-19 severity with their custom dataset. Finally, Li et al. (2020) introduced PXS-score based on a convolutional Siamese network pre-trained on CheXpert dataset, where two separate images are taken as inputs and passed through twinned CNN and Euclidean distance between two outputs are used for calculating the severity scores.
As described above, however, the previous approaches are mainly based on the standard CNN model pre-training and transfer learning from the irrelevant dataset (e.g. ImageNet), and therefore do not guarantee an optimal generalization performance for COVID-19.
3. Proposed framework
One of the novel contributions of our approach is to show that we can maximize the performance of the Transformer model by using the low-level CXR corpus that comes from the backbone network trained with a large well-curated public record to produce common CXR findings. As the backbone network is trained with a large number of data, the subsequent models using this backbone for classification and severity quantification tasks are less prone to overfitting, even with a smaller number of labeled cases. This is shown to improve the generalization capability of the network.
After devising the model for classification and severity quantification of COVID-19, we further integrated these two models into a single multi-task model that can do two tasks simultaneously to offer better applicability as well as to improve the performances of individual tasks.
3.1. Pre-training backbone network for low-level feature corpus
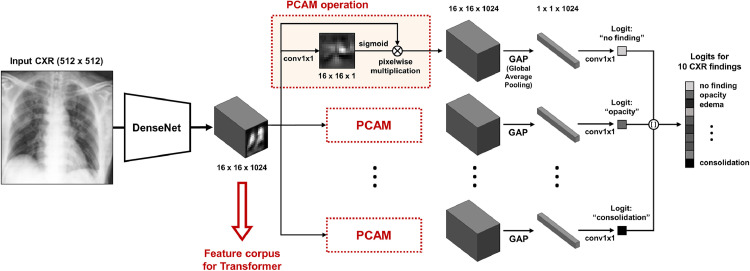
As a backbone network to extract low-level features, we used the modified version of the network proposed by Ye et al. (2020). Firstly, the backbone network was pre-trained to classify 10 common low-level findings with a large public dataset. As depicted in Fig. 2 , feature maps in each layer can be the candidates for utilizable feature embedding for the subsequent Transformer, and we experimentally found that the common embedding before the PCAM operation comprises of most useful information. Nevertheless, care should be exercised since the PCAM operation for specific low-level CXR findings (e.g. lung opacity, consolidation, etc.) turns out to be crucial to achieving the optimal embedding at the intermediate level, as PCAM aligns these features to obtain better performances. Through this operation, more prominent feature representations are embedded for each low-level entity, and combining these low-level feature representations to yield high-level results of classification and severity quantification with the Transformer is one of the key ideas of our method. More detailed experimental results about the role of PCAM operation will be provided within ablation studies of Section 5.6.2.
Fig. 2.
Backbone network to extract low-level CXR feature corpus.
3.2. Vision transformer for COVID-19: shared layer
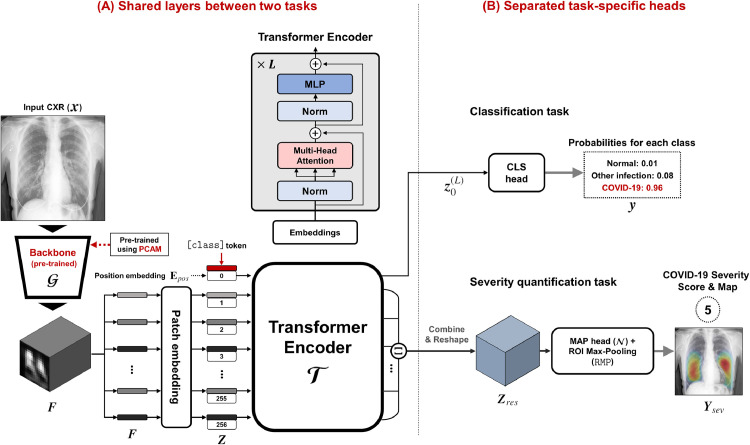
The overall framework and the architecture of our ViT model is provided in Fig. 3 . Since our model use the same pre-trained backbone and Transformer architecture for two tasks, shared backbone layer can be defined as in Fig. 3 (A). Specifically, for a given size input image , the backbone network generates size feature maps :
| (1) |
Here, the feature tensor is defined as
| (2) |
where denotes a -dimensional embedded representation of low-level features at the th encoded block. These feature vectors are used to construct the low-level CXR feature corpora for Transformer.
Fig. 3.
Proposed multi-task Vision Transformer model for diagnosis and severity quantification of COVID-19 on CXR, which consists of (A) shared backbone and Transformer and (B) task-specific heads for each task.
Then, similar to Bidirectional Encoder Representations from Transformers (BERT) (Devlin et al., 2018), our ViT uses Transformer encoder layers to the input embedding. Specifically, since the Transformer encoder utilizes constant latent vector of dimension , the extracted dimension feature is first projected to a dimension feature using 1 1 convolution kernel. We then prepended learnable [class] token embedding vector to projected feature tensor. This leads to the following composite projected feature tensor:
| (3) |
A positional embedding that has the same shape to the projected feature tensor is then added to encode a notion of the sequential order:
This is then used as an input to a Transformer composed of successive encoder layers:
| (4) |
where and denotes the th encoder layer. The encoder layers used in our model are the same as standard Transformer which consists of repeated layers of multi-head self-attention (MSA), multi-layer perceptron (MLP), layer normalization (LN), and residual connections in each block, as shown in Fig. 3 (A).
Then, the first column of represents the Transformer attended feature vector with respect to the [class] token, which is used for the classification task. The rest of the Transformer output also produces feature embedding at each block position by taking into account long-range relations between the blocks. Therefore, we conjecture that this information is useful for the severity quantification, as severity is determined by both local and global manifestations of the disease.
3.3. Vision transformer for COVID-19: classification
Simply adding linear classifiers to [class] token as the classification head, we can obtain the diagnosis result of the input CXR image (see Fig. 3 (A)).
For the interpretability of the classification model, we adopted a visualization method of saliency map tailored for ViT suggested by Chefer et al. (2020), which computes relevancy for the Transformer network. Specifically, unlike the traditional approaches of gradient propagation methods (Selvaraju, Cogswell, Das, Vedantam, Parikh, Batra, 2017, Smilkov, Thorat, Kim, Viégas, Wattenberg, Srinivas, Fleuret) or attribution propagation methods (Bach, Binder, Montavon, Klauschen, Müller, Samek, 2015, Gu, Yang, Tresp, 2018), which rely on the heuristic propagation along with attention graph or the obtained attention maps, the method in Chefer et al. (2020) calculate the local relevance with deep Taylor decomposition, which is then propagated throughout the layers. This relevance propagation method is especially useful for models based on Transformer architecture, as it overcomes the problem of self-attention operations and skips connections.
3.4. Vision transformer for COVID-19: Severity quantification
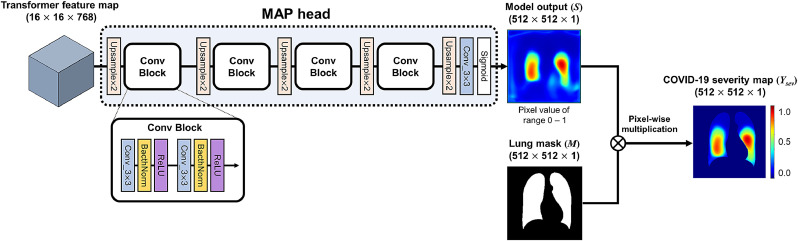
As shown in Fig. 3(B), reshaped output features except for [class] token are combined by an additional lightweight network to produce the COVID-19 severity map.
Specifically, as shown in Figs. 3(b) and 4 , we first extract the Transformer output except the [class] token position:
| (5) |
which is used as an input to the map head network
| (6) |
Fig. 4.
The procedure of severity prediction and labeling. (A) Map head and ROI max-pooling of the proposed framework. (B) Our severity annotation method for severity quantification on CXRs.
Then, the network output is multiplied pixel-wise with the segmentation mask , generating the severity map . Finally, ROI max-pooling (RMP) is applied to provide the severity mask :
| (7) |
where denotes the Hadamard product. In detail, the lung was divided into a total of six subdivisions, by dividing the right and left lungs into three subdivisions (upper, middle, lower zone) with 5/12 and 2/3 lines. Next, the largest values within each six subdivision were assigned as predicted values of the severity array. Then, the map head network is trained by minimizing the error of the estimated severity array with respect to the weakly annotated severity label as in Fig. 4.
For details of the model output and post-processing for the severity array, refer to Appendix B.
To generate the lung segmentation mask, we used the method introduced by Oh and Ye (2021). In contrast to the existing approaches that are prone to under-segmentation for the severely infected lung with large consolidations, this novel approach enables the accurate segmentation of abnormal lung as well as normal lung area by learning common features using a single generator with AdaIN layers. Since a single generator is used for all these tasks by simply changing the AdaIN codes, the generator can synergistically learn the common features to improve segmentation performance for abnormal CXR data.
3.5. Multi-task learning
Since the classification and severity quantification model shares the same layers other than task-specific heads, we trained and evaluated the model with MTL as well as single-task learning (STL) for both tasks. By the MTL framework, we aimed not only to offer a simpler configuration for better applicability but also to improve the performances of two relevant tasks, COVID-19 classification and severity quantification, by learning more robust feature representation shared between the two related tasks as suggested in the previous studies (Zhang and Yang, 2017).
4. Datasets
Datasets used for this study can be divided into three: dataset for pre-training backbone, the datasets for classification, datasets for severity quantification.
4.1. Dataset for pre-training
For the pre-training of the backbone network to extract the low-level CXR features, we used CheXpert dataset containing 10 labeled CXR findings: no finding, cardiomegaly, opacity, edema, consolidation, pneumonia, atelectasis, pneumothorax, pleural effusion, and support device. With a total of 224,316 CXR images from 65,240 subjects, the 32,387 lateral view images were excluded, leaving 29,420 posterior-anterior (PA) and 161,427 anterior-posterior (AP) view data available. With this large number of CXRs, it was able to train the backbone network robust to the variation in subjects, which is one of the key strengths of our model.
4.2. Datasets for classification
Table 1 summarizes dataset resources and partitioning used for classification. To train and evaluate the Transformer model, we utilized both public datasets containing labeled cases of normal and infectious disease (Valencian Region Medical Image Bank [BIMCV] (De La Iglesia Vayá et al., 2020), Brixia (Signoroni et al., 2020b), National Institutes of Health [NIH] (Wang et al., 2017), CheXpert) and deliberately collected CXR data from four hospitals (Asan Medical Center [AMC], Seoul, Korea; Chonnam National Univerity Hospital [CNUH], Daejeon, Korea; Yeungnam University Hospital [YNU], Daegu, Korea; Kyungpook National University Hospital [KNUH], Daegu, Korea) labeled by board-certified radiologists for this study. Finally, the integrated dataset was divided into three label classes including normal, other infections (e.g. bacterial infection, tuberculosis), and COVID-19 infection, considering the application in the real clinical setting. Both PA and AP view CXRs were utilized to build and evaluate our model in a view-agnostic setting. We used three institutional data (CNUH, YNU, KNUH) as external test datasets to evaluate the generalization capability by using data collected from independent hospitals with different devices and settings, and other data for training and internal validation of the models.
Table 1.
Datasets and label distribution for classification.
| View | Total | External test |
Training and validation |
||||||
|---|---|---|---|---|---|---|---|---|---|
| CNUH | YNU | KNUH | AMC | NIH | Brixia | BIMCV | CheXpert | ||
| All views | |||||||||
| Normal | 26,846 | 417 | 300 | 400 | 8978 | 7158 | - | 93 | 9500 |
| Other infection | 1672 | 58 | 220 | 400 | 994 | - | - | - | - |
| COVID-19 | 5755 | 81 | 286 | 293 | - | - | 4313 | 782 | - |
| Total images | 34,273 | 556 | 806 | 1093 | 9972 | 7158 | 4313 | 875 | 9500 |
| PA view | |||||||||
| Normal | 13,649 | 320 | 300 | 400 | 8861 | 3768 | - | - | - |
| Other infection | 1468 | 39 | 144 | 308 | 977 | - | - | - | - |
| COVID-19 | 2431 | 6 | 8 | 80 | - | - | 1929 | 408 | - |
| Total images | 17,548 | 365 | 452 | 788 | 9838 | 3768 | 1929 | 408 | - |
| AP view | |||||||||
| Normal | 13,197 | 97 | - | - | 117 | 3390 | - | 93 | 9500 |
| Other infection | 204 | 19 | 76 | 92 | 17 | - | - | - | - |
| COVID-19 | 3324 | 75 | 278 | 213 | - | - | 2384 | 374 | - |
| Total images | 16,725 | 191 | 354 | 305 | 134 | 3390 | 2384 | 467 | 9500 |
4.3. Datasets for severity quantification
Table 2 summarizes dataset resources and global severity levels. Similar to diagnosis, the PA and AP view data were integrated and utilized without division for severity quantification task since there is the possibility that follow-up images may be obtained with both PA and AP view even in a single patient. Two board-certified radiologists labeled the severity for three institutional datasets (CNUH, YNU, KNUH) using the array-based severity labeling method of Toussie et al. (2020) as in Fig. 6. We also utilized publicly available data, Brixia dataset, after translating its severity score the same as that of the institutional datasets. We alternately used one institutional dataset as an external testset and trained the models with two remaining datasets together with Brixia dataset to evaluate the generalization capability in various external settings. Besides, 12 COVID-19 cases from BIMCV dataset were used to compare the severity map generated by our model to those annotated by clinical experts.
Table 2.
Datasets and label distribution for severity quantification.
| Severity | Total | CNUH | YNU | KNUH | Brixia |
|---|---|---|---|---|---|
| 1 | 361 | 26 | 63 | 25 | 247 |
| 2 | 521 | 11 | 59 | 22 | 429 |
| 3 | 448 | 8 | 25 | 18 | 397 |
| 4 | 920 | 7 | 35 | 31 | 847 |
| 5 | 774 | 12 | 18 | 29 | 715 |
| 6 | 1758 | 17 | 86 | 171 | 1484 |
| Total | 4782 | 81 | 286 | 296 | 4119 |
Fig. 6.
Examples of severity quantification results of our models on the external dataset.
Details of the patient and CXR image characteristics of four hospitals (CNUH, YNU, KNUH, AMC) datasets are provided in Appendix C.
4.4. Details of implementation and evaluation
The CXR images were preprocessed via histogram equalization, Gaussian blurring with kernel, normalization, and finally resized to . As our backbone network, the modified version of the network proposed by Ye et al. (2020), comprises the DenseNet-121 baseline followed by PCAM operations. Among several layers of intermediate feature maps, we used the feature map of size just before the PCAM operation. For subsequent Transformer architecture, we used a standard Transformer model with 12 layers and 12 heads per layer.
For pre-training of the backbone network, Adam optimizer with a learning rate of 0.0001 was used. We trained the backbone network for 160,000 optimization steps with a step decay scheduler with a batch size of 8. Data augmentations including random flipping, rotation, translation were performed to increase the variability of training data during pre-training. For the classification task, stochastic gradient descent (SGD) optimizer with momentum 0.9 was used with a learning rate of 0.001. A max gradient norm of 1 was applied to stabilize training. We trained the model for 10,000 optimization steps with a cosine warm-up scheduler (warm-up steps = 500) with a batch size of 16. For the severity quantification task, a map head with five upsizing convolution layers is used, with the last block followed by sigmoid non-linearity which squashes output into [0–1] range. Training of severity quantification model was done with SGD optimizer with a learning rate of 0.003 for 12,000 optimization steps with constant learning rate, and batch size of 4 was used. These optimal hyperparameters were determined experimentally. Similar to pre-training, various data augmentation (horizontal flipping, rotation, translation, and scaling) was performed to increase the training data for both tasks. As the loss functions, binary cross-entropy (BCE) losses were used for each class label for pre-training and classification task, while BCE losses for each location array within a CXR were used for severity quantification task.
In the MTL setting, the shared layers were trained with the optimizer, scheduler, and hyperparameter to those of the classification task. Considering the scales of loss from each task, the losses from task-specific heads were scaled to 1:5 for classification and severity quantification to balance their influence to the shared network layers.
Since our model was trained using both PA and AP CXRs, the classification, and severity quantification performances were evaluated in a view-agnostic manner with both PA and AP images. However, we also evaluated and provided the model performances for PA and AP images separately for the classification task, in which the diagnostic performance could differ significantly according to CXR views. We used the area under the receiver operating characteristic curve (AUC) as the evaluation metrics for diagnostic performance of the classification model, but also calculated sensitivity, specificity, and accuracy after adjusting the thresholds to meet the sensitivity value of , if possible. As evaluation metrics for severity quantification, we used the Mean Squared Error (MSE) as the main metric, but the Mean Absolute Error (MAE), Correlation Coefficient (CC), and score were also measured and compared. The performance metrics were reported with estimated 95% confidence intervals (CIs). Model performances were compared statistically using AUC with DeLong test (DeLong et al., 1988) for classification task and using MSE with paired t-test for severity prediction task, respectively. Statistically significant differences were defined as .
All experiments including preprocessing, development, and evaluation of the model, were performed using Python version 3.7 and PyTorch library version 1.7 on NVIDIA Tesla V100, Quadro RTX 6000, RTX 3090, and RTX 2080 Ti.
5. Experimental results
5.1. Benefit of the multi-task learning approach
We first evaluated whether the model trained with the MTL approach provides better performance than two task-specific models trained with the standard STL approach. As shown in Tables 3 and 4 , the multi-task model for two tasks outperformed the expert model trained exclusively for each task with statistical significance, for both classifications and severity prediction tasks. Hence, the following experiments were mainly conducted under the MTL setting, and other models used for comparison were also implemented with the MTL approach for a fair comparison.
Table 3.
Comparison of the classification performances of single task model for classification and multi-task model for two tasks.
| Models | External dataset 1 (CNUH) |
External dataset 2 (YNU) |
External dataset 3 (KNUH) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| AUC (95% CI) |
AUC (95% CI) |
AUC (95% CI) |
|||||||
| Normal | Others | COVID-19 | Normal | Others | COVID-19 | Normal | Others | COVID-19 | |
| Multi-task model | 0.968 (0.954–0.981) | 0.926 (0.893–0.959) | 0.953 (0.935–0.971) | 0.973 (0.964–0.983) | 0.935 (0.914–0.955) | 0.884 (0.861–0.906) | 0.961 (0.950-0.972) | 0.861 (0.837–0.885) | 0.898(0.878–0.918) |
| Single-task model | (0.893–0.943) | 0.901 (0.850-0.951) | (0.838–0.914) | 0.969 (0.959–0.979) | 0.925 (0.903–0.947) | (0.882–0.922) | (0.876–0.914) | 0.861 (0.837–0.885) | (0.777–0.838) |
Note:, denote the better performance of our model, while , denote worse performance of our model with statistical significance (, ). CI: confidence interval
Table 4.
Comparison of the severity quantification performances of single-task model for classification and multi-task model for two tasks.
| Models | External dataset 1(CNUH) |
External dataset 2(YNU) |
External dataset 3(KNUH) |
|---|---|---|---|
| MSE (95% CI) | MSE (95% CI) | MSE (95% CI) | |
| Multi-task model | 1.441(0.760-2.122) | 1.435(1.195–1.676) | 1.458(1.147–1.768) |
| Single-task model | 1.645(0.969–2.320) | 1.417(1.138–1.695) | (1.372–2.090) |
Note: denotes the better performance of our model with statistical significance (). CI, confidence interval.
5.2. Diagnostic performance on external test datasets
The detailed diagnostic performances of the proposed model are provided in Table 5 . On average of 3 label classes (normal, other infection, COVID-19), our model showed stable performances regardless of external data with the mean AUCs of 0.949, 0.931, 0.907, sensitivities of 90.2%, 87.0%, 85.1%, specificities of 84.9%. 86.2%, 83.7%, and accuracy of 86.8%, 86.5%, 84.1% for three labels in three external institutions, which confirmed the stability in performance even with a view-agnostic setting and outstanding generalization capability in clinical situations with different devices and settings. The diagnostic performances our model evaluated only on PA and AP view images are also provided in Appendix D.
Table 5.
Diagnostic performance of the proposed model in various external test datasets from three different institutions.
| Metrics | External dataset 1 (CNUH) |
External dataset 2 (YNU) |
External dataset 3 (KNUH) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Normal | Others | COVID-19 | Normal | Others | COVID-19 | Normal | Others | COVID-19 | |
| AUC(95% CI) | 0.968(0.954–0.981) | 0.926(0.893–0.959) | 0.953(0.935–0.971) | 0.973(0.964–0.983) | 0.935(0.914–0.955) | 0.884(0.861–0.906) | 0.961(0.950-0.972) | 0.861(0.837–0.885) | 0.898(0.878–0.918) |
| Sensitivity(95% CI) | 93.5(90.7–95.7) | 84.5(72.6–92.7) | 92.6(84.6–97.2) | 95.3(92.3–97.4) | 85.5(80.1–89.8) | 80.1(75.0–84.5) | 91.5(88.3–94.0) | 79.7(75.5–83.6) | 84.0(79.3–88.0) |
| Specificity(95% CI) | 87.8(81.1–92.7) | 82.5(78.9–85.8) | 84.4(80.8–87.6) | 88.9(85.9–91.5) | 87.9(85.0–90.4) | 81.7(78.1–85.0) | 90.6(88.2–92.7) | 78.6(75.4–81.6) | 82.0(79.2–84.6) |
| Accuracy(95% CI) | 92.1(89.5–94.2) | 82.7(79.3–85.8) | 85.6(82.4–88.4) | 91.3(89.2–93.2) | 87.2(84.7–89.5) | 81.1(78.3–83.8) | 90.9(89.1–92.6) | 79.0(76.5–81.4) | 82.5(80.1–84.7) |
Note: CI: confidence interval.
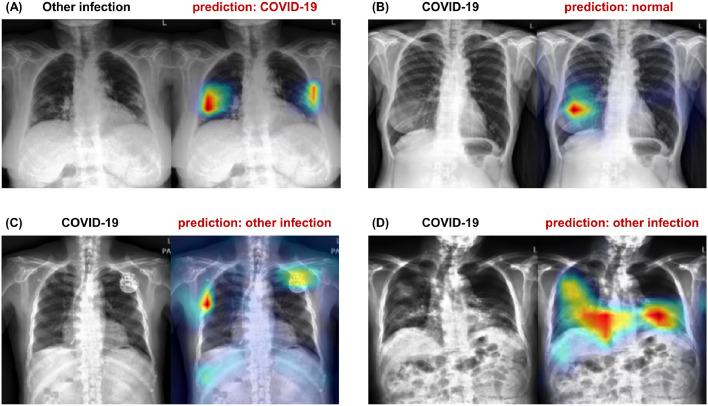
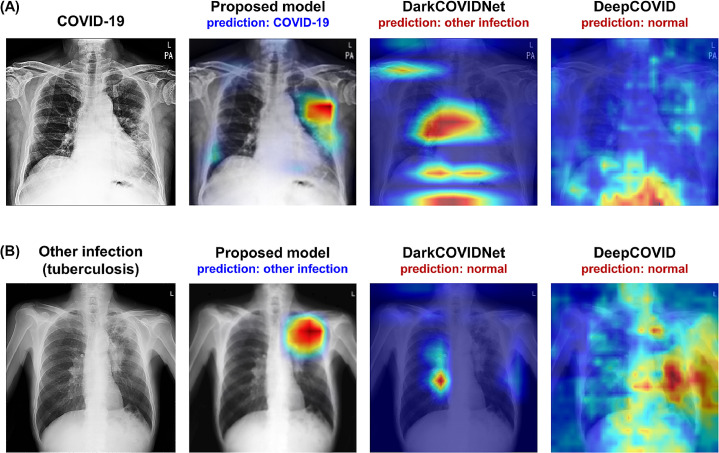
5.3. Model interpretability results
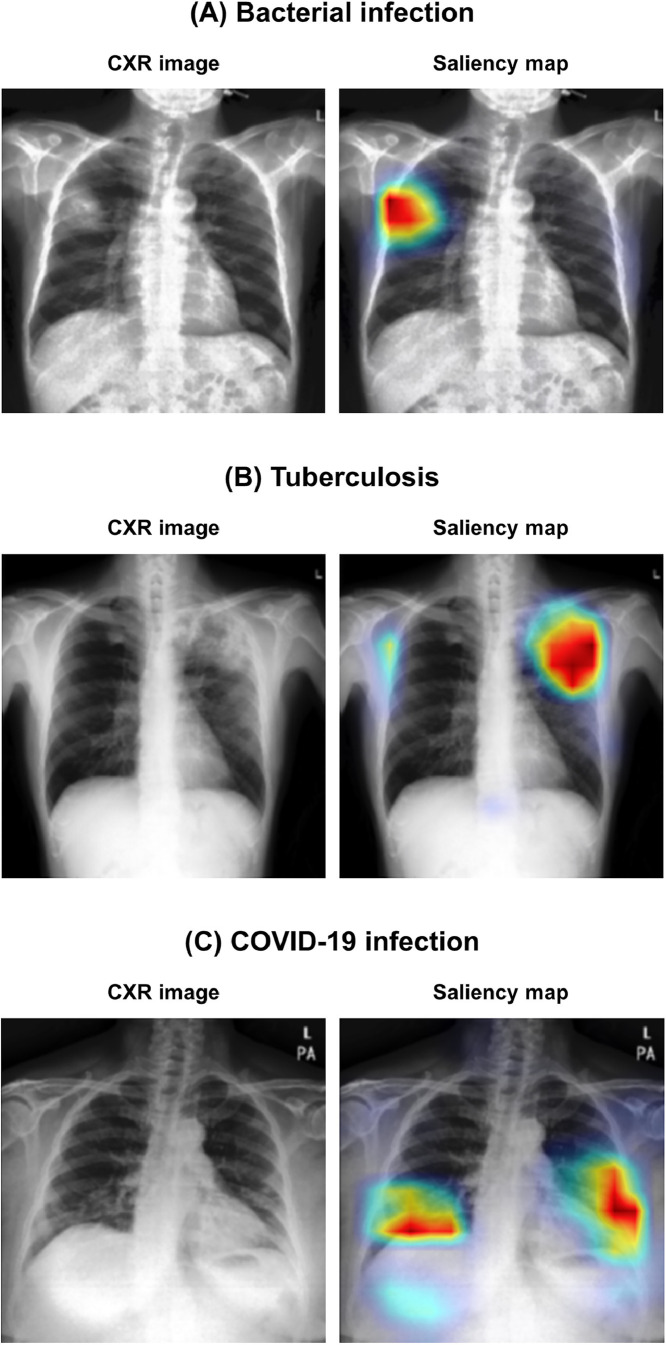
Figure 5 exemplifies the visualization of saliency maps for each disease class in the external test datasets. As shown in the examples, our model well-localized a focal infected area either by a bacterial infection (Fig. 5(a)) or tuberculosis (Fig. 5(b)), while it was also able to delineate the multi-focal lesions in the periphery of both lower lungs in Fig. 4(c), which is typical findings for COVID-19 pneumonia.
Fig. 5.
Examples of visualization results for each disease class. (A) Bacterial infection, (B) tuberculosis, and (C) COVID-19 infection.
5.4. Severity quantification results on external test datasets
The results of severity quantification of our model are shown in Table 6 . Our model showed the MSE of 1.441, 1.435, 1.458, the MAE of 0.843, 0.943, 0.890, correlation coefficient of 0.800, 0.830, 0.731, and score of 0.634, 0.633, 0.485 in three external institutions. Brixia dataset contains a consensus subset of 150 CXR images labeled by five independent radiologists. Within this subset, the average MSE between the consensus severity score calculated from majority voting and each radiologist’s rating is 1.683. As a result, the MSEs of 1.441, 1.435, and 1.458 in three external institutions show our model’s performance comparable to or better than those of experienced radiologists and generalization capability in the clinical environment.
Table 6.
Severity quantification performance of the proposed model in various external test datasets from three different institutions.
| Metrics | External dataset 1(CNUH) | External dataset 2(YNU) | External dataset 3(KNUH) |
|---|---|---|---|
| MSE(95% CI) | 1.441(0.760-2.122) | 1.435(1.195–1.676) | 1.458(1.147–1.768) |
| MAE (95% CI) | 0.843(0.653–1.033) | 0.943(0.857–1.029) | 0.890(0.796–0.984) |
| CC (95% CI) | 0.800 (0.705–0.867) | 0.830(0.790-0.863) | 0.731(0.673–0.780) |
| R (95% CI) | 0.634(0.512–0.756) | 0.633(0.566–0.700) | 0.485(0.404–0.566) |
Note: CI: confidence interval.
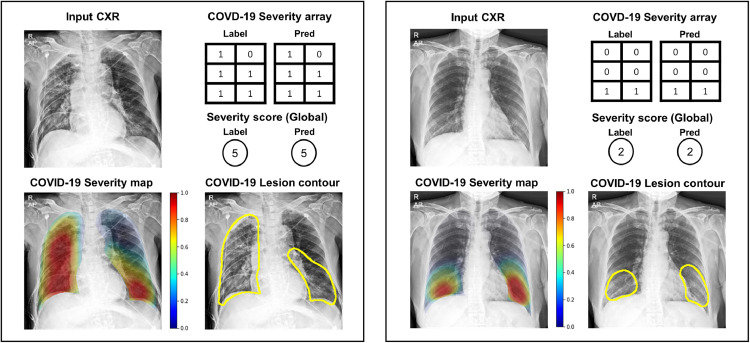
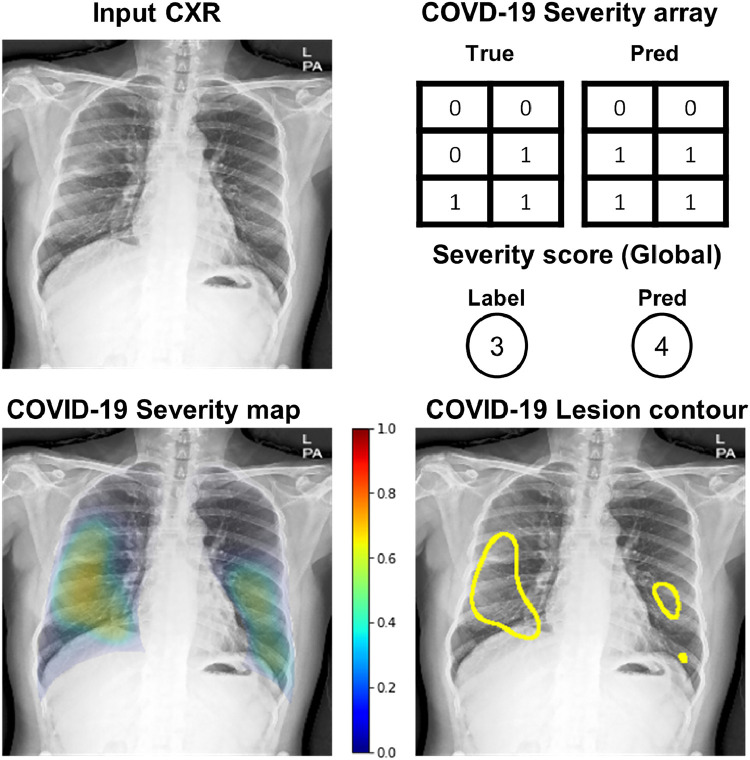
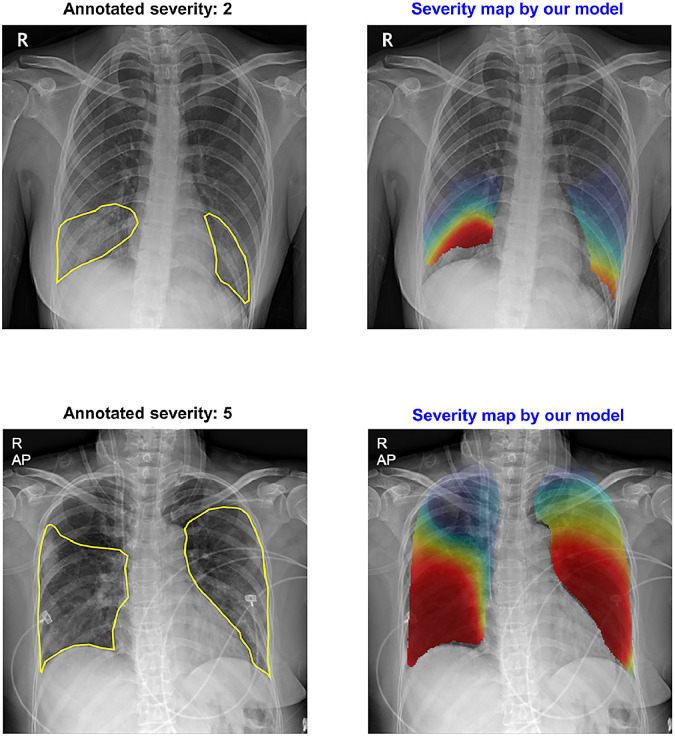
Figure 6 illustrates the examples of severity quantification, including the predicted scores, arrays, maps, and lesion contours in one of the external test datasets, which confirms that not only can our model correctly predict global severity, but it also generates an intuitive severity map that highlights the affected area, which can also be used to contour lesions.
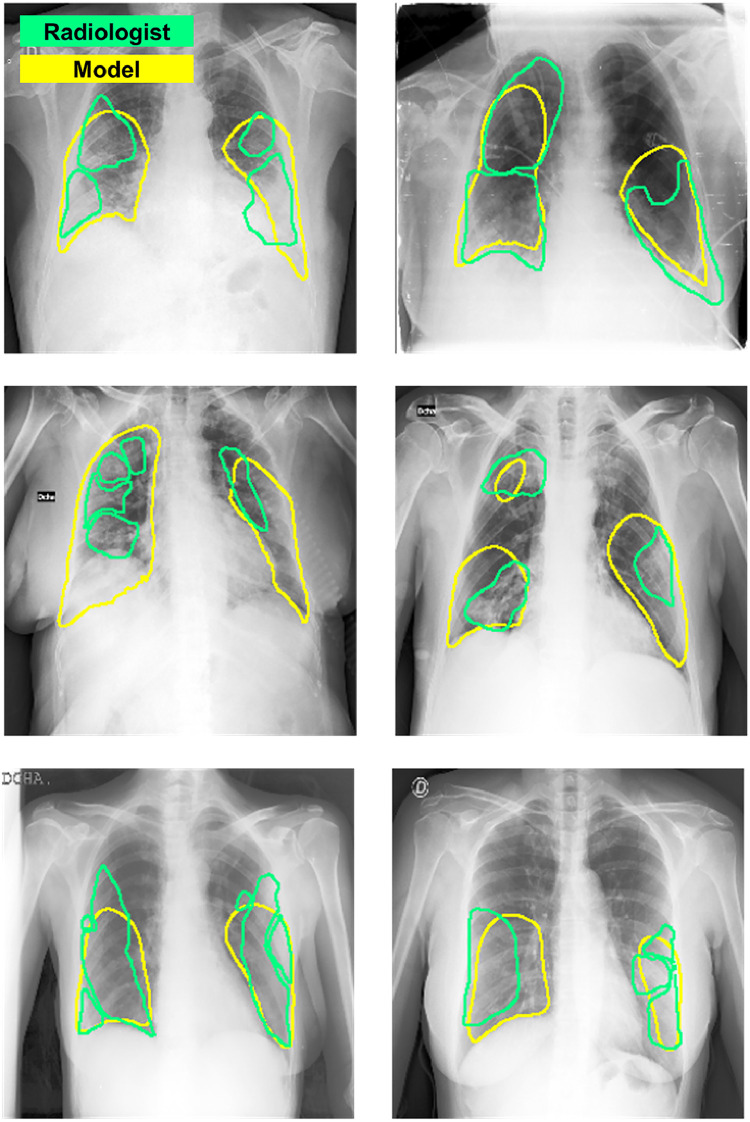
Finally, Fig. 7 exemplifies the comparison between the ground truth segmentation label of the involved area and the model’s prediction of involvement in BIMCV dataset. As shown in the figure, the model generally well-localized the areas of involvement.
Fig. 7.
Comparison of localization results in BIMCV dataset. Green: radiologist’s annotation. Yellow: model’s prediction after thresholding. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
5.5. Comparison with CNN and transformer-based models
To compare the performance with the other baseline and SOTA CNN-based models, we adopted the following models: ResNet-50, ResNet-512, DenseNet-121 as the baseline CNN-based models, and EfficientNet-B7, NASNet-A-Large, SE-Net-154 as the SOTA CNN-based models. For comparison with other Transformer-based models, we used ViT (ViT-B-16) and hybrid ViT (R50-ViT-B-16) models. All models underwent the same pre-training process on CheXpert dataset and were subsequently trained, evaluated with datasets and settings the same as the proposed model for a fair comparison. As suggested in Tables 7 and 8 , our model outperformed or at least comparable to both the SOTA CNN-based models as well as the baseline CNN-based models with statistical significance. When compared to Transformer-based models, our model showed statistically better performance than other Transformer-based models. Note that our model showed superior performance not only to the models with less complexity (e.g. ResNet-50, DenseNet-121) but also to those with more complex architectures (e.g. NASNet-A-Large, SE-Net-154, ViT models). These results suggest that our model offers better generalization performances in both classification and severity quantification tasks compared with the existing model architectures, which did not result from increased complexity.
Table 7.
Comparison of the classification performance with various baseline and SOTA CNN-based models, and Transformer-based models.
| Models (Params) | External dataset 1 (CNUH) |
External dataset 2 (YNU) |
External dataset 3 (KNUH) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| AUC (95% CI) |
AUC (95% CI) |
AUC (95% CI) |
|||||||
| Normal | Others | COVID-19 | Normal | Others | COVID-19 | Normal | Others | COVID-19 | |
| Proposed model (79.402M) | 0.968 (0.954–0.981) | 0.926 (0.893–0.959) | 0.953 (0.935–0.971) | 0.973 (0.964–0.983) | 0.935 (0.914–0.955) | 0.884 (0.861–0.906) | 0.961 (0.950-0.972) | 0.861 (0.837–0.885) | 0.898 (0.878–0.918) |
| Baseline CNN-based | |||||||||
| ResNet-50 (30.378M) | 0.946(0.926–0.966) | 0.919 (0.887–0.951) | 0.916 (0.888–0.944) | 0.973 (0.963–0.982) | 0.879 (0.851–0.907) | 0.847 (0.821–0.873) | 0.954 (0.942–0.966) | 0.719 (0.685–0.752) | 0.876 (0.855–0.896) |
| ResNet-152 (65.014M) | 0.938 (0.916–0.960) | 0.930 (0.890-0.971) | 0.903 (0.874–0.932) | 0.975 (0.965–0.985) | 0.876 (0.849–0.904) | 0.833 (0.806–0.861) | 0.970 (0.961–0.979) | 0.724 (0.691–0.756) | 0.852 (0.829–0.874) |
| DenseNet-121 (13.034M) | 0.931 (0.906–0.955) | 0.898 (0.849–0.947) | 0.900 (0.868–0.933) | 0.969 (0.959–0.980) | 0.898 (0.873–0.923) | 0.846 (0.820-0.872) | 0.956 (0.945–0.967) | 0.852 (0.828–0.877) | 0.804 (0.777–0.831) |
| SOTA CNN-based | |||||||||
| EfficientNet-B7 (71.052M) | 0.931 (0.909–0.954) | 0.920 (0.878–0.963) | 0.879 (0.842–0.915) | 0.981 (0.973–0.989) | 0.871 (0.844–0.898) | 0.863 (0.839–0.887) | 0.951 (0.939–0.964) | 0.850 (0.826–0.874) | 0.825 (0.798–0.852) |
| NASNet-A-Large (98.262M) | 0.943 (0.923–0.964) | 0.907 (0.869–0.945) | 0.898 (0.866–0.931) | 0.986(0.980-0.992) | 0.912 (0.887–0.937) | 0.846 (0.821–0.872) | 0.943 (0.929–0.956) | 0.893 (0.874–0.911) | 0.861 (0.838–0.884) |
| SENet-154 (121.434M) | 0.959 (0.942–0.975) | 0.929 (0.893–0.964) | 0.896 (0.864–0.928) | 0.977 (0.968–0.985) | 0.917 (0.895–0.938) | 0.838 (0.812–0.864) | 0.974 (0.965–0.983) | 0.911(0.894–0.929) | 0.847 (0.824–0.870) |
| Transformer-based | |||||||||
| ViT-B/16 (91.727M) | 0.820 (0.775–0.866) | 0.865 (0.810-0.919) | 0.701 (0.635–0.767) | 0.940 (0.924–0.956) | 0.830 (0.797–0.864) | 0.789 (0.758–0.821) | 0.945 (0.932–0.958) | 0.815 (0.787–0.843) | 0.825 (0.797–0.852) |
| R50-ViT-B/16 (105.58M) | 0.948 (0.931–0.966) | 0.915 (0.877–0.954) | 0.886 (0.854–0.917) | 0.975 (0.966–0.984) | 0.889 (0.863–0.915) | 0.851 (0.826–0.876) | 0.980(0.973–0.987) | 0.901 (0.882–0.919) | 0.845 (0.822–0.868) |
Note:, denote the better performance of our model, while , denote worse performance of our model with statistical significance (, ). CI: confidence interval.
Table 8.
Comparison of the severity quantification performance with various baseline and SOTA CNN-based models, and Transformer-based models.
| Models (Params) | External dataset 1 (CNUH) | External dataset 2 (YNU) | External dataset 3 (KNUH) |
|---|---|---|---|
| MSE (95% CI) | MSE (95% CI) | MSE (95% CI) | |
| Proposed model (79.402M) | 1.441 (0.760-2.122) | 1.435 (1.195–1.676) | 1.458 (1.147–1.768) |
| Baseline CNN-based | |||
| ResNet-50 (30.378M) | 1.489 (1.016–1.963) | 2.133 (1.847–2.419) | 2.128 (1.837–2.418) |
| ResNet-152 (65.014M) | 1.330 (0.930-1.729) | 2.034 (1.738–2.331) | 1.977 (1.672–2.282) |
| DenseNet-121 (13.034M) | 1.580 (1.027–2.133) | 1.650 (1.412–1.888) | 1.546 (1.257–1.836) |
| SOTA CNN-based | |||
| EfficientNet-B7 (71.052M) | 1.457 (0.780-2.135) | 2.673 (2.258–3.089) | 1.270 (1.000–1.540) |
| NASNet-A-Large (98.262M) | 1.401 (0.927–1.875) | 2.882 (2.449–3.314) | 1.894 (1.593–2.196) |
| SENet-154 (121.434M) | 1.163 (0.698–1.628) | 1.890 (1.597–2.184) | 1.899 (1.566–2.231) |
| Transformer-based | |||
| ViT-B/16 (91.727M) | 2.529 (1.536–3.522) | 3.112 (2.643–3.580) | 2.067 (1.691–2.442) |
| R50-ViT-B/16 (105.58M) | 1.257 (0.734–1.780) | 1.874 (1.618–2.130) | 1.538 (1.203–1.873) |
Note:, denote the better performance of ours with statistical significance (, ). CI: confidence interval.
5.6. Comparison with previous models in related works
We also compared our model with the tailored models in the related works of Section 2.4. The tailored models for comparison were implemented and trained using the settings proposed in the original papers (e.g. pre-training, hyperparameters, etc.) on our dataset the same as the proposed model for a fair comparison. As shown in Tables 9 and 10 , our model considerably outperformed previous models proposed in the related works for both COVID-19 classification and severity quantification. Although a few models showed reasonable performances in some test datasets (e.g. DarkCOVIDNet in YNU dataset and CheXNet in CNUH dataset), they failed to show stable performances over various external test datasets. The unstable performances of previous models for COVID-19 on various external test setting account for why the deep learning models readily developed for automated diagnosis and severity prediction of COVID-19 not lead to the widespread application.
Table 9.
Comparison of the classification performance of the proposed model with COVID-19 classification models in related works.
| Models (Params) | External dataset 1 (CNUH) |
External dataset 2 (YNU) |
External dataset 3 (KNUH) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| AUC (95% CI) |
AUC (95% CI) |
AUC (95% CI) |
|||||||
| Normal | Others | COVID-19 | Normal | Others | COVID-19 | Normal | Others | COVID-19 | |
| Proposed model (79.402M) | 0.968 (0.954–0.981) | 0.926 (0.893–0.959) | 0.953 (0.935–0.971) | 0.973 (0.964–0.983) | 0.935 (0.914–0.955) | 0.884 (0.861–0.906) | 0.961 (0.950-0.972) | 0.861 (0.837–0.885) | 0.898 (0.878–0.918) |
| CoroNet (33.969M) | 0.772 (0.731–0.813) | 0.760 (0.689–0.830) | 0.834 (0.796–0.872) | 0.803 (0.774–0.833) | 0.708 (0.667–0.749) | 0.812 (0.780-0.844) | 0.701 (0.669–0.732) | 0.753 (0.723–0.783) | 0.847 (0.823–0.871) |
| COVIDNet (11.750M) | 0.787 (0.740-0.834) | 0.744 (0.667–0.822) | 0.636 (0.575–0.697) | 0.715 (0.681–0.750) | 0.586 (0.542–0.630) | 0.844 (0.816–0.872) | 0.665 (0.633–0.696) | 0.491 (0.456–0.526) | 0.651 (0.611–0.690) |
| DarkCOVIDNet (1.164M) | 0.749 (0.697–0.802) | 0.708 (0.633–0.784) | 0.843 (0.784–0.902) | 0.952 (0.938–0.966) | 0.898 (0.873–0.923) | 0.901 (0.879–0.922) | 0.466 (0.432–0.499) | 0.562 (0.522–0.602) | 0.479 (0.444–0.514) |
| DeepCOVID (11.178M) | 0.711 (0.660-0.762) | 0.701 (0.625–0.777) | 0.791 (0.742–0.841) | 0.893 (0.871–0.916) | 0.751 (0.714–0.789) | 0.844 (0.817–0.870) | 0.690 (0.659–0.721) | 0.625 (0.589–0.660) | 0.770 (0.737–0.803) |
Note:, denote the better performance of our model w with statistical significance (, ). CI: confidence interval.
Table 10.
Comparison of the severity quantification performance of the proposed model with COVID-19 classification models in related works.
| Models (Params) | External dataset 1 (CNUH) | External dataset 2 (YNU) | External dataset 3 (KNUH) |
|---|---|---|---|
| MSE (95% CI) | MSE (95% CI) | MSE (95% CI) | |
| Proposed model (79.402M) | 1.441 (0.760-2.122) | 1.435 (1.195–1.676) | 1.458 (1.147–1.768) |
| CheXNet (6.961M) | 1.457 (0.854–2.059) | 5.182 (4.472–5.892) | 1.891 (1.398–2.384) |
| Cohen (6.966M) | 3.268 (2.548–3.987) | 3.668 (3.249–4.086) | 2.043 (1.724–2.361) |
| PXS (7.979M) | 4.227 (3.196–5.259) | 4.014 (3.533–4.495) | 4.965 (4.558–5.372) |
Note:, denote the better performance of ours with statistical significance (, ). CI: confidence interval.
5.7. Simulation of application under real-world prevalence
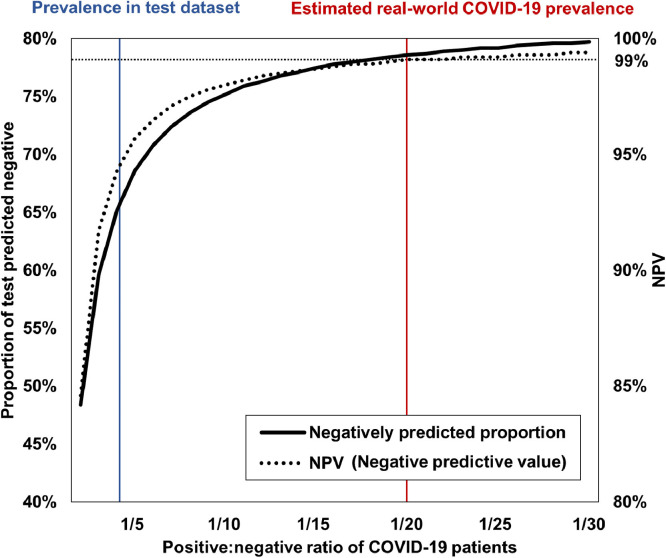
In experiments of the diagnostic model, the results should be interpreted with caution, since the actual prevalence of the disease is not the same as in the experimental dataset collected for the study. That is to say, in our case, the prevalence of 26.9% for COVID-19 in the external test set is quite higher than the real-world prevalence of COVID-19 in any country. Therefore, we evaluated the performance metrics under a range of disease prevalences of COVID-19 in the external test datasets using bootstrapping with replacement. As shown in Fig. 8 , the proportion of predicted negative and negative predicted value (NPV) for COVID-19 drastically increase with decreasing COVID-19 prevalence to real-world reported ranges (Yiannoutsos et al., 2021), from NPV of 93.7% to 99.1% and negatively predicted proportion of 63.9% to 78.6%. Thus, this simulation suggests that under the real-world prevalence of COVID-19, about 80% of the RT-PCR test can be spared with the application of the proposed model as a screening tool with an NPV over 99%.
Fig. 8.
Simulation under the different prevalence of COVID-19.
5.8. Analysis of failure cases of the proposed model
To have a better understanding of the model’s misprediction, we exemplified the failure cases by the proposed model for both classification and severity quantification tasks. As shown in Fig. 9 , though our model failed to offer the correct predictions for the failure cases, its confusion could be explained with cogent interpretations, and it attends on the lesion of interest in many cases. Similarly, for severity quantification, it provided the severity array come close to the label annotation, even in case of the wrong prediction as in Fig. 10 .
Fig. 9.
Examples of the failure cases of the proposed model for the classification task. (A) The model misclassified a case of tuberculosis as COVID-19, as the location and distribution of the consolidative lesions resemble those of COVID-19 (lower and peripheral distribution of patch consolidations). (B) The model failed to diagnose a faint COVID-19 lesion in the right lower lobe of the patients, possibly due to the fact that the COVID-19 lesion was concealed by the opacity of breast tissue. (C) The model failed to diagnose in a mild COVID-19 case, showing the confusion by the support device. (D) A severe COVID-19 case was confused as other infection, in which an opacity was located at an unusual location for COVID-19 involvement (right middle lobe), but the model retained proper attention to the abnormal lesions.
Fig. 10.
Example of the failure case of the proposed model for severity quantification task. The model confused a faint opacity in the right middle lobe as COVID-19 involvement, yielding an overall score higher than the label. Nevertheless, its prediction came close to the label annotation.
In addition, we further exemplified the cases in which the previous classification models failed while the proposed model offered the correct predictions for comparison in Appendix E.
5.9. Ablation studies
To get better understanding about the contribution of individual components within our model, we conducted a series of ablation studies as provided in Tables 11 and 12 . More details are as follows.
Table 11.
Ablation study results for classification performance.
| Methods | External dataset 1 (CNUH) |
External dataset 2 (YNU) |
External dataset 3 (KNUH) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| AUC (95% CI) |
AUC (95% CI) |
AUC (95% CI) |
|||||||
| Normal | Others | COVID-19 | Normal | Others | COVID-19 | Normal | Others | COVID-19 | |
| Proposed model | 0.968 (0.954–0.981) | 0.926 (0.893–0.959) | 0.953 (0.935–0.971) | 0.973 (0.964–0.983) | 0.935 (0.914–0.955) | 0.884 (0.861–0.906) | 0.961 (0.950-0.972) | 0.861 (0.837–0.885) | 0.898 (0.878–0.918) |
| w/o pre-train | 0.898 (0.868–0.927) | 0.869 (0.815–0.922) | 0.848 (0.806–0.891) | 0.962 (0.950-0.975) | 0.919 (0.896–0.942) | 0.895 (0.873–0.917) | 0.887 (0.868–0.907) | 0.811 (0.784–0.837) | 0.812 (0.783–0.841) |
| w/o Transformer | 0.935 (0.914–0.957) | 0.893 (0.846–0.939) | 0.914 (0.887–0.942) | 0.964 (0.953–0.975) | 0.899 (0.875–0.924) | 0.846 (0.820-0.872) | 0.925 (0.910-0.940) | 0.791 (0.762–0.820) | 0.794 (0.765–0.822) |
| w/o PCAM | 0.943 (0.921–0.965) | 0.874 (0.817–0.931) | 0.911 (0.882–0.941) | 0.968 (0.957–0.979) | 0.911 (0.886–0.937) | 0.892 (0.870-0.915) | 0.938 (0.924–0.952) | 0.868 (0.845–0.890) | 0.817 (0.788–0.846) |
| w/o position embedding | 0.965 (0.950-0.980) | 0.939 (0.909–0.969) | 0.940 (0.918–0.963) | 0.976 (0.967–0.985) | 0.925 (0.902–0.947) | 0.893(0.871–0.914) | 0.963 (0.952–0.973) | 0.861 (0.838–0.885) | 0.878 (0.856–0.901) |
Note:, denote the better performance of our model, while , denotes worse performance of our model with statistical significance (, ). CI: confidence interval.
Table 12.
Ablation study results for severity quantification performance.
| Methods | External 1 (CNUH) | External 2 (YNU) | External 3 (KNUH) |
|---|---|---|---|
| MSE (95% CI) | MSE (95% CI) | MSE (95% CI) | |
| Proposed model | 1.441 (0.760-2.122) | 1.435 (1.195–1.676) | 1.458 (1.147–1.768) |
| w/o pre-train | 1.737 (1.037–2.437) | 2.319 (1.907–2.732) | 1.835 (1.402–2.268) |
| w/o Transformer | 1.544 (0.882–2.206) | 1.977 (1.682–2.272) | 1.374 (1.091–1.657) |
| w/o PCAM | 1.436 (0.777–2.095) | 2.205 (1.881–2.529) | 1.353 (1.058–1.648) |
| w/o position embedding | 1.447 (0.838–2.056) | 1.504 (1.257–1.750) | 1.522 (1.237–1.807) |
Note:, denote the better performance of ours with statistical significance (, ). CI: confidence interval.
5.9.1. Pre-training backbone on large CXR datasets
Pre-training the backbone on a pre-built large CXR dataset (CheXpert dataset) to extract low-level features is one of the key ideas of our method. Therefore, we conducted experiments to compare the performances of the proposed model with and without CheXpert pre-trained weights both in the internal validation dataset and the external test datasets. As shown in Tables 13 and 14 , the experimental results suggest the performance increases with pre-training were prominent in the external test datasets, while the improvement was not prominent, and even better performance without pre-trained backbone was observed in the internal validation dataset. Combined together, these results demonstrate that the model without CheXpert pre-trained weights is more prone to overfitting, supporting our arguments that pre-training the backbone on large-scale CXR is a crucial component of the model in terms of better generalization capability.
Table 13.
Diagnostic performance of the proposed model with and without pre-trained backbone weights on CheXpert dataset.
| Metrics | Internal Validation |
External dataset 1 (CNUH) |
External dataset 2 (YNU) |
External dataset 3 (KNUH) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC |
AUC |
AUC |
AUC |
|||||||||
| Normal | Others | COVID-19 | Normal | Others | COVID-19 | Normal | Others | COVID-19 | Normal | Others | COVID-19 | |
| w pre-train | 0.977 | 0.975 | 0.986 | 0.968 | 0.926 | 0.953 | 0.973 | 0.935 | 0.885 | 0.961 | 0.861 | 0.898 |
| w/o pre-train | 0.992 | 0.977 | 0.998 | 0.898 | 0.869 | 0.848 | 0.962 | 0.919 | 0.895 | 0.887 | 0.811 | 0.812 |
Note:, denote the better performance of our model, while , denote worse performance of our model with statistical significance (, ).
Table 14.
Severity quantification performance of the proposed model with and without pre-trained backbone weights on CheXpert dataset.
| Methods | Internal validation | External 1 (CNUH) | External 2 (YNU) | External 3 (KNUH) |
|---|---|---|---|---|
| MSE | MSE | MSE | MSE | |
| w pre-train | 0.528 | 1.441 | 1.435 | 1.458 |
| w/o pre-train | 0.607 | 1.737 | 2.319 | 1.835 |
Note:, denote the better performance of ours with statistical significance (, ).
5.9.2. PCAM operation
To support our claim that PCAM operation enables the backbone network to embed better feature representations for subsequent tasks, we conducted an ablation study with and without PCAM operation. The experimental results in Tables 11 and 12 show that the model with PCAM operation shows better performances both for classification and severity prediction tasks, but the benefit was more prominent for classification task. These findings are consistent with the intuition that PCAM operation would be more useful in classification tasks where the robust representations for various low-level features can be more directly related to the final diagnosis of a given CXR.
5.9.3. Role of transformer
Another key idea of our approach is that the Transformer is capable of properly combining the extracted low-level features to yield high-level outputs. To validate this argument, the ablation study without the Transformer was conducted, which is identical to train and evaluate the performance of the CNN backbone (DenseNet-121 equipped with PCAM) without a Transformer body, which was trained in a multi-task manner for the classification and severity quantification tasks. As provided in Tables 11 and 12, the performances were significantly deteriorated without the Transformer architecture, both for the classification and the severity prediction tasks, proving that the Transformer architecture plays a key role within our method.
5.9.4. Positional embedding
Since a recent study has suggested that ViT model works decently without the positional embeddings (Chen et al., 2021), we performed an ablation study with and without the positional embeddings. As shown in Tables 11 and 12, the model without the positional embedding showed no statistical difference in severity prediction task, but provided slightly lower performances for classification task in some datasets. This is consistent with the intuition that the positional information has meaning for diagnosis of disease (e.g. tuberculosis often involves the apex of lungs, but COVID-19 more often presents in the lower periphery), but may not be important to yield a summed severity score overall lung areas which can be considered to be permutation-invariant.
6. Discussion and conclusion
Increasing concerns on the overestimation of the deep learning model for COVID-19 now bring the real-world applicability of the models into question. As pointed out in recent literature (Wynants et al., 2020), although hundreds of deep learning models for automated diagnosis of COVID-19 have been suggested so far, most of them did not work well in a real-world application. Most of them were sensitive to specific settings of image acquisition, overfit to unimportant findings of image (Roberts et al., 2021) and therefore showed unpardonable performance deterioration in a different setting. Similarly, in this study, we have observed that previously suggested models for both COVID-19 classification and severity quantification showed unsatisfactory generalization performances in various external data. Our model, on the other hand, showed stable performances in various external test datasets with different settings and even regardless of PA and AP view (see Appendix D and Appendix F). This finding is important since it will broaden the actual applicability of the developed model in the clinical setting.
In the current pandemic situation, our method holds great promise as a screening tool. As shown in the simulation of real-world COVID-19 prevalence (see Fig. 8), it could reliably deprioritize the population with a low risk of infection using readily obtainable CXRs. With NPV over 99%, the model could spare up to 80% of the tested population from the molecular test, thereby prioritize the limited medical resources to subjects more likely to have COVID-19. In this respect, the application of our model would be of great value in the resource-constrained area. Supposing it is used along with the molecular test, it could be utilized to isolate the suspected subjects waiting for RT-PCR results, as it was reported that positive radiological findings precede positive RT-PCR results in a substantial portion (308 out of 1,014) of patients (Ai et al., 2020). In addition, since our model also provides the estimated severity of COVID-19 infection, it is possible to give guidance in treatment decisions or to evaluate the response using our model for severity prediction of consecutive CXRs.
In summary, we developed a novel ViT model that leverages low-level CXR feature corpus for diagnosis and severity quantification of COVID-19. The novelty of this work is to decouple the overall framework into two steps, the first is to pre-train the backbone network to classify low-level CXR findings with the prebuilt large-scale dataset to embed optimal feature corpus, which was then leveraged in the second step by Transformer for high-level diagnosis of disease including COVID-19. By maximally utilizing the benefit of the large-scale dataset containing more than 220,000 CXR images, the overfitting problem of neural networks with limited numbers of COVID-19 cases can be substantially alleviated. In addition, we also adapted the proposed method to severity quantification problem, demonstrating a performance similar to that of clinical experts, thereby expanding its application in the clinical setting. Not confined to devising the model for each task, we enabled a novel ViT model to be a multi-task model that can be used for both classification and severity prediction, offering a simpler configuration and better performances for individual tasks. We performed extensive experiments on various external institutions to demonstrate the superior generalization performance of the proposed model over the existing models for COVID-19 as well as other CNN and Transformer-based architectures, which is the sine qua non of widespread adoption of the system.
Finally, we believe that the novel concept of making higher-level diagnoses by aggregating low-level feature corpus, which is readily available with pre-built datasets, can be applied to quickly develop a robust algorithm against the newly emerging pathogen, since it is expected to share the common low-level CXR features with existing diseases.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported in part by the National Research Foundation (NRF) of Korea under Grant NRF-2020R1A2B5B03001980, and in part by KAIST Mobile Clinic Module Project under Grant MCM-2021-N11210024.
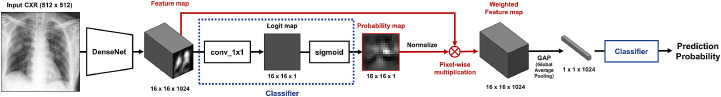
Appendix A. Details of PCAM Operation
Figure A1 depicts the detailed process of the PCAM operation. First, the feature map from backbone network is transformed to a probability map using 1 1 convolution and sigmoid layer. This probability map is then normalized and pixel-wise multiplied with feature map to generate weighted feature map. Finally, the weighted feature map is reduced with global average pooling and passed to final classifier to provide prediction probability.
Fig. A1.
Detailed process of Probabilistic Class Activation Map (PCAM) pooling.
Appendix B. Details of Model Output for Severity Array
Using the feature map from the Transformer, a map head leveraging the five upsizing convolution layers followed by a sigmoid layer generates an output with a range of [0–1]. This is subsequently multiplied by lung mask to provide severity map suitable for the shape of lung as shown in Fig. B1
Fig. B1.
Details of the model output and post-processing for severity array in the severity quantification task.
Appendix C. Details of four hospital datasets
The details of patient and CXR characteristics of four hospital data deliberately collected for this study are provided in Table C1.
Table C1.
Details on patient characteristics and CXR images.
| Data | CNUH | YNU | KNUH | AMC |
|---|---|---|---|---|
| Details on patient characteristics | ||||
| Age | 47.9 17.2 | 57.4 18.5 | 53.8 18.9 | 46.2 14.5 |
| Sex | Male (45.7%), Female (45.1%), N/A (9.2%) | Male (52.6%), Female (47.3%), N/A (0.2%) | Male (29.8%), Female (33.6%), N/A (36.6%) | Male (48.9%), Female (47.1%),N/A (3.9%) |
| COVID-19 cases | 81 | 286 | 293 | - |
| COVID-19 severity | 3 (1–6) | 3 (1–6) | 6 (1–6) | - |
| CT positive cases | N/A (100%) | N/A (100%) | Positive (2.0%), N/A (98.0%) | - |
| Country | South Korea | South Korea | South Korea | South Korea |
| Details on CXR images | ||||
| Number of images | 365 | 806 | 1093 | 9972 |
| View | PA (65.6%), AP (34.4%) | PA (56.1%), AP (43.9%) | PA (72.1%), AP (27.9%) | PA (98.7%), AP (1.3%) |
| Modality | CR (95.0%), N/A (5.0%) | CR (99.9%), N/A (0.1%) | CR (100%) | CR (3.7%), DX (96.3%) |
| Exposure time (msec) | 6.7 3.4 | 16.5 7.7 | 12.1 8.3 | 8.9 3.9 |
| Tube current (mA) | 473.3 198.1 | 307.8 36.4 | 311.8 39.6 | 298.9 43.7 |
| Bits | 12 (12–14) | 12 (12-12) | 12 (12–14) | 14 (10–15) |
Note: Values are presented as mean standard deviation or median (range).
Appendix D. Classification Results According to Views
Tables D.2 and D.3 shows the classification results evaluated exclusively on PA and AP view CXRs, respectively. For both PA and AP view images, our model provided stable performances, although the diagnostic performance with AP view images was slightly lower than PA view images. Nonetheless, it still showed good performance (AUC ) in the external test dataset, considering the fact that the diagnosis of infectious disease using only AP view image is not standard and usually deteriorates the diagnostic performance.
Table D.2.
Diagnostic performance of the proposed model in various external test datasets from three different institutions (PA).
| Metrics | External dataset 1 (CNUH) |
External dataset 2 (YNU) |
External dataset 3 (KNUH) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Normal | Others | COVID-19 | Normal | Others | COVID-19 | Normal | Others | COVID-19 | |
| AUC (95% CI) | 0.977 (0.959–0.995) | 0.969 (0.948–0.990) | 0.936 (0.870-1.000) | 0.970 (0.951–0.990) | 0.968 (0.950-0.985) | 0.936 (0.841–1.000) | 0.961 (0.947–0.975) | 0.891 (0.865–0.916) | 0.903 (0.864–0.942) |
| Sensitivity (95% CI) | 91.6 (88.0–94.4) | 89.7 (75.8–97.1) | 83.3 (35.9–99.6) | 94.3 (91.1–96.7) | 91.0 (85.1–95.1) | 87.5 (47.4–99.7) | 91.5 (88.3–94.0) | 83.8 (79.2–87.7) | 87.5 (78.2–93.8) |
| Specificity (95% CI) | 91.1 (78.8–97.5) | 89.6 (85.7–92.7) | 83.6 (79.3–87.3) | 92.8 (87.4–96.3) | 91.9 (88.3–94.7) | 86.7 (83.2–89.7) | 90.7 (87.4–93.4) | 84.0 (80.4–87.1) | 84.2 (81.3–86.8) |
| Accuracy (95% CI) | 91.5 (88.2–94.2) | 89.6 (86.0–92.5) | 83.6 (79.4–87.2) | 93.8 (91.2–95.8) | 91.6 (88.6–94.0) | 86.7 (83.3–89.7) | 91.1 (88.9–93.0) | 83.9 (81.1–86.4) | 84.5 (81.8–87.0) |
Note: CI: confidence interval.
Table D.3.
Diagnostic performance of the proposed model in various external test datasets from three different institutions (AP).
| Metrics | External dataset (CNUH) |
||
|---|---|---|---|
| Normal | Others | COVID-19 | |
| AUC (95% CI) | 0.918 (0.875–0.960) | 0.809 (0.714–0.904) | 0.879 (0.829–0.929) |
| Sensitivity (95% CI) | 88.7 (80.6–94.2) | 73.7 (48.8–90.9) | 85.3 (75.3–92.4) |
| Specificity (95% CI) | 88.3 (80.0–94.0) | 64.0 (56.3–71.1) | 84.5 (76.6–90.5) |
| Accuracy (95% CI) | 88.5 (83.1–92.6) | 64.9 (57.7–71.7) | 84.8 (78.9–89.6) |
Note: CI: confidence interval.
Appendix E. Analysis of Failure Cases in Previous Models
For analysis of the failure cases, we additionally analyzed the failure cases of the previous models using our model for comparison. The previous models were visualized with the methods proposed in the original papers. Note that COVIDNet and CoroNet could not be implemented since they did not provide the details of the model visualizations. As shown in Fig. E1 , our model successfully predicted the correct label and localized the lesion in the failure cases of the previous models for both COVID-19 and other infections. Similarly, for severity quantification, our model more correctly predicted ground truth severity annotation than the previous models as in Fig. E2 . In addition, the severity map generated by our model predicted the locations of the COVID-19 involvement with the high agreement.
Fig. E1.
Examples of success with our method when the previous classification models fail. (A) Ground truth is COVID-19, but the previous COVID-19 classification models failed to make correct diagnoses. On the contrary, our model makes a correct diagnosis of COVID-19. (B) Similarly, when the previous COVID-19 classification models make wrong diagnoses, our model is able to make a correct diagnosis of other infections.
Fig. E2.
Examples of success with our method when the previous severity quantification models fail. (A) Annotated severity score is 2, but other models fail to make the correct prediction (CheXNet, Cohen, PXS scores are 4, 5, 3). On the contrary, our model predicts a correct severity score while providing a severity map with high agreement. (B) Also in the severe case with a score of 5, our model makes a correct prediction of severity while other models fail (CheXNet, Cohen, PXS scores are 6, 4, 2).
Appendix F. Further Evaluation on Other Datasets
We have further evaluated the generalization performance of our model in other publicly available datasets. For classification, we used Actualmed COVID-19 CXR Dataset (DarwinAI et al.) containing 155 PA and 30 AP CXRs. This dataset contains 58 COVID-19 cases and 127 non-COVID-19 cases. Note that classification metrics could only be calculated in COVID-19, since the dataset contain only COVID-19 and non-COVID-19 labels. For severity quantification, COVID-19 Image Data Collection (Cohen et al., 2020c) was used which contains 163 images annotated with Brixia severity score. These scores are converted in accordance with our severity scoring method. After conversion, it contains 14 (8.6%), 16 (9.8%), 16 (9.8%), 19 (11.7%), 19 (11.7%), 79 (48.5%) cases of severity score 1, 2, 3, 4, 5, 6, respectively.
As shown in Tables F1 and Table F2 , our model provided good performances in both COVID-19 classification and severity quantification tasks in these datasets from other sources.
Table F1.
Classification performance of the proposed model in other external datasets.
| Metrics | Actualmed COVID-19 CXR dataset |
|
|---|---|---|
| PA view | AP view | |
| AUC (95% CI) | 0.838 (0.757 - 0.919) | 0.875 (0.724 - 1.000) |
| Sensitivity (95% CI) | 81.3 (63.6 - 92.8) | 76.9 (56.4 - 91.0) |
| Specificity (95% CI) | 78.1 (69.7 - 85.0) | 100.0 (39.8–100.0) |
| Accuracy (95% CI) | 78.7 (71.4 - 84.9) | 80.0 (61.4 - 92.3) |
Note: CI: confidence interval.
Table F2.
Severity quantification performance of the proposed model in other external datasets.
| Metrics | COVID-19 Image Data Collection |
|---|---|
| MSE (95% CI) | 1.468 (1.089 - 1.847) |
| MAE (95% CI) | 0.890 (0.762 - 1.017) |
| CC (95% CI) | 0.746 (0.669 - 0.807) |
| R (95% CI) | 0.409 (0.295–0.523) |
Note: CI: confidence interval.
References
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos I.D., Mpesiana T.A. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach S., Binder A., Montavon G., Klauschen F., Müller K.-R., Samek W. On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE. 2015;10(7):e0130140. doi: 10.1371/journal.pone.0130140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., Diao K., Lin B., Zhu X., Li K., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020:200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesi A., Maroldi R. COVID-19 outbreak in italy: experimental chest X-ray scoring system for quantifying and monitoring disease progression. Radiol. Med. 2020;125(5):509–513. doi: 10.1007/s11547-020-01200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chefer, H., Gur, S., Wolf, L., 2020. Transformer interpretability beyond attention visualization. arXiv preprint arXiv:2012.09838.
- Chen L., Min Y., Zhang M., Karbasi A. International Conference on Machine Learning. PMLR; 2020. More data can expand the generalization gap between adversarially robust and standard models; pp. 1670–1680. [Google Scholar]
- Chen M., Radford A., Child R., Wu J., Jun H., Luan D., Sutskever I. International Conference on Machine Learning. PMLR; 2020. Generative pretraining from pixels; pp. 1691–1703. [Google Scholar]
- Chen, X., Xie, S., He, K., 2021. An empirical study of training self-supervised vision transformers. arXiv preprint arXiv:2104.02057.
- Chollet F. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Xception: deep learning with depthwise separable convolutions; pp. 1251–1258. [Google Scholar]
- Cohen J.P., Dao L., Roth K., Morrison P., Bengio Y., Abbasi A.F., Shen B., Mahsa H.K., Ghassemi M., Li H., et al. Predicting COVID-19 pneumonia severity on chest X-ray with deep learning. Cureus. 2020;12(7) doi: 10.7759/cureus.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J. P., Morrison, P., Dao, L., Roth, K., Duong, T. Q., Ghassemi, M., 2020b. COVID-19 image data collection: prospective predictions are the future. arXiv preprint arXiv:2006.11988 (3). https://github.com/ieee8023/covid-chestxray-dataset.
- Cohen, J. P., Morrison, P., Dao, L., Roth, K., Duong, T. Q., Ghassemi, M., 2020c. COVID-19 image data collection: prospective predictions are the future. arXiv preprint arXiv:2006.11988https://github.com/ieee8023/covid-chestxray-dataset.
- Cozzi D., Albanesi M., Cavigli E., Moroni C., Bindi A., Luvarà S., Lucarini S., Busoni S., Mazzoni L.N., Miele V. Chest X-ray in new coronavirus disease 2019 (COVID-19) infection: findings and correlation with clinical outcome. Radiol. Med. 2020;125:730–737. doi: 10.1007/s11547-020-01232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DarwinAI, Vision, Group, I. P. R., Ross, M., VanBerlo, B., Ebadi, A., Git, K.-A., Al-Haimi, A.,. Actualmed-covid-chestxray-dataset. https://github.com/agchung/Actualmed-COVID-chestxray-dataset. (Accessed on 08/13/2021).
- De La Iglesia Vayá, M., Saborit, J. M., Montell, J. A., Pertusa, A., Bustos, A., Cazorla, M., Galant, J., Barber, X., Orozco-Beltrán, D., García-García, F., et al., 2020. BIMCV COVID-19+: a large annotated dataset of RX and CT images from COVID-19 patients. arXiv preprint arXiv:2006.01174.
- DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988:837–845. [PubMed] [Google Scholar]
- Devlin, J., Chang, M.-W., Lee, K., Toutanova, K., 2018. BERT: pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805.
- Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., et al., 2020. An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929.
- Gu J., Yang Y., Tresp V. Asian Conference on Computer Vision. Springer; 2018. Understanding individual decisions of CNNs via contrastive backpropagation; pp. 119–134. [Google Scholar]
- Hemdan, E. E.-D., Shouman, M. A., Karar, M. E., 2020. COVIDX-Net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv preprint arXiv:2003.11055.
- Hu Y., Jacob J., Parker G.J., Hawkes D.J., Hurst J.R., Stoyanov D. The challenges of deploying artificial intelligence models in a rapidly evolving pandemic. Nat. Mach. Intell. 2020;2(6):298–300. [Google Scholar]
- Irvin J., Rajpurkar P., Ko M., Yu Y., Ciurea-Ilcus S., Chute C., Marklund H., Haghgoo B., Ball R., Shpanskaya K., et al. Proceedings of the AAAI Conference on Artificial Intelligence. vol. 33. 2019. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison; pp. 590–597. [Google Scholar]
- Khan A.I., Shah J.L., Bhat M.M. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput. Methods Programs Biomed. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, S., Naseer, M., Hayat, M., Zamir, S. W., Khan, F. S., Shah, M., 2021. Transformers in vision: a survey. arXiv preprint arXiv:2101.01169.
- Kwon Y.J., Toussie D., Finkelstein M., Cedillo M.A., Maron S.Z., Manna S., Voutsinas N., Eber C., Jacobi A., Bernheim A., et al. Combining initial radiographs and clinical variables improves deep learning prognostication in patients with COVID-19 from the emergency department. Radiology. 2020;3(2):e200098. doi: 10.1148/ryai.2020200098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.D., Arun N.T., Gidwani M., Chang K., Deng F., Little B.P., Mendoza D.P., Lang M., Lee S.I., OShea A., et al. Automated assessment and tracking of COVID-19 pulmonary disease severity on chest radiographs using convolutional siamese neural networks. Radiology. 2020;2(4):e200079. doi: 10.1148/ryai.2020200079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaee S., Kafieh R., Sonka M., Yazdani S., Soufi G.J. Deep-covid: predicting covid-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narin, A., Kaya, C., Pamuk, Z., 2020. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed]
- Ng K., Poon B.H., Kiat Puar T.H., Shan Quah J.L., Loh W.J., Wong Y.J., Tan T.Y., Raghuram J. COVID-19 and the risk to health care workers: a case report. Ann. Intern. Med. 2020;172(11):766–767. doi: 10.7326/L20-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- Oh, Y., Ye, J. C., 2021. Unifying domain adaptation and self-supervised learning for CXR segmentation via AdaIN-based knowledge distillation. arXiv preprint arXiv:2104.05892.
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmon J., Farhadi A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Yolo9000: better, faster, stronger; pp. 7263–7271. [Google Scholar]
- Roberts M., Driggs D., Thorpe M., Gilbey J., Yeung M., Ursprung S., Aviles-Rivero A.I., Etmann C., McCague C., Beer L., et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021;3(3):199–217. [Google Scholar]
- Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-CAM: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, china: a descriptive study. Lancet Infect. Dis. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoroni, A., Savardi, M., Benini, S., Adami, N., Leonardi, R., Gibellini, P., Vaccher, F., Ravanelli, M., Borghesi, A., Maroldi, R., Farina, D., 2020a. End-to-end learning for semiquantitative rating of COVID-19 severity on chest X-rays. arXiv preprint arXiv:2006.04603https://brixia.github.io/. [DOI] [PMC free article] [PubMed]
- Signoroni, A., Savardi, M., Benini, S., Adami, N., Leonardi, R., Gibellini, P., Vaccher, F., Ravanelli, M., Borghesi, A., Maroldi, R., et al., 2020b. End-to-end learning for semiquantitative rating of COVID-19 severity on chest X-rays. arXiv preprint arXiv:2006.04603. [DOI] [PMC free article] [PubMed]
- Smilkov, D., Thorat, N., Kim, B., Viégas, F., Wattenberg, M., 2017. SmoothGrad: removing noise by adding noise. arXiv preprint arXiv:1706.03825.
- Srinivas, S., Fleuret, F., 2019. Full-gradient representation for neural network visualization. arXiv preprint arXiv:1905.00780.
- Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussie D., Voutsinas N., Finkelstein M., Cedillo M.A., Manna S., Maron S.Z., Jacobi A., Chung M., Bernheim A., Eber C., et al. Clinical and chest radiography features determine patient outcomes in young and middle-aged adults with COVID-19. Radiology. 2020;297(1):E197–E206. doi: 10.1148/radiol.2020201754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., Kaiser, L., Polosukhin, I., 2017. Attention is all you need. arXiv preprint arXiv:1706.03762.
- Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. ChestX-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 2097–2106. [Google Scholar]
- Warren M.A., Zhao Z., Koyama T., Bastarache J.A., Shaver C.M., Semler M.W., Rice T.W., Matthay M.A., Calfee C.S., Ware L.B. Severity scoring of lung oedema on the chest radiograph is associated with clinical outcomes in ARDS. Thorax. 2018;73(9):840–846. doi: 10.1136/thoraxjnl-2017-211280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A., Qiu Lin Z., Wang L., Chung A.G., Shen B., Abbasi A., Hoshmand-Kochi M., Duong T.Q. COVIDNet-S: towards computer-aided severity assessment via training and validation of deep neural networks for geographic extent and opacity extent scoring of chest X-rays for SARS-CoV-2 lung disease severity. arXiv e-prints. 2020:arXiv–2005. [Google Scholar]
- Wynants L., Van Calster B., Collins G.S., Riley R.D., Heinze G., Schuit E., Bonten M.M., Dahly D.L., Damen J.A., Debray T.P., et al. Prediction models for diagnosis and prognosis of COVID-19: systematic review and critical appraisal. BMJ. 2020;369 doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, W., Yao, J., Xue, H., Li, Y., 2020. Weakly supervised lesion localization with probabilistic-cam pooling. arXiv preprint arXiv:2005.14480.
- Yiannoutsos C.T., Halverson P.K., Menachemi N. Bayesian estimation of SARS-CoV-2 prevalence in Indiana by random testing. Proc. Natl. Acad. Sci. 2021;118(5) doi: 10.1073/pnas.2013906118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech J.R., Badgeley M.A., Liu M., Costa A.B., Titano J.J., Oermann E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15(11):e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Xie, Y., Li, Y., Shen, C., Xia, Y., 2020. COVID-19 screening on chest X-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338.
- Zhang, Y., Yang, Q., 2017. A survey on multi-task learning. arXiv preprint arXiv:1707.08114.
- Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv. 2020 [Google Scholar]
- Zheng, S., Lu, J., Zhao, H., Zhu, X., Luo, Z., Wang, Y., Fu, Y., Feng, J., Xiang, T., Torr, P. H., et al., 2020b. Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. arXiv preprint arXiv:2012.15840.
- Zhu J., Shen B., Abbasi A., Hoshmand-Kochi M., Li H., Duong T.Q. Deep transfer learning artificial intelligence accurately stages COVID-19 lung disease severity on portable chest radiographs. PLoS ONE. 2020;15(7):e0236621. doi: 10.1371/journal.pone.0236621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Su, W., Lu, L., Li, B., Wang, X., Dai, J., 2020b. Deformable DETR: deformable transformers for end-to-end object detection. arXiv preprint arXiv:2010.04159.