Abstract
Introduction
More than 180 million people have been infected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and more than 4 million coronavirus disease-2019 (COVID-19) patients have died in 1.5 years of the pandemic. A novel therapeutic vaccine (NASVAC) has shown to be safe and to have immunomodulating and antiviral properties against chronic hepatitis B (CHB).
Materials and methods
A phase I/II, open-label controlled and randomized clinical trial of NASVAC as a postexposure prophylaxis treatment was designed with the primary aim of assessing the local and systemic immunomodulatory effect of NASVAC in a cohort of suspected and SARS-CoV-2 risk-contact patients. A total of 46 patients, of both sexes, 60 years or older, presenting with symptoms of COVID-19 were enrolled in the study. Patients received NASVAC (100 μg per Ag per dose) via intranasal at days 1, 7, and 14 and sublingual, daily for 14 days.
Results and discussion
The present study detected an increased expression of toll-like receptors (TLR)-related genes in nasopharyngeal tonsils, a relevant property considering these are surrogate markers of SARS protection in the mice model of lethal infection. The HLA-class II increased their expression in peripheral blood mononuclear cell's (PBMC's) monocytes and lymphocytes, which is an attractive property taking into account the functional impairment of innate immune cells from the periphery of COVID-19-infected subjects. NASVAC was safe and well tolerated by the patients with acute respiratory infections and evidenced a preliminary reduction in the number of days with symptoms that needs to be confirmed in larger studies.
Conclusions
Our data justify the use of NASVAC as preemptive therapy or pre-/postexposure prophylaxis of SARS-CoV-2 and acute respiratory infections in general. The use of NASVAC or their active principles has potential as immunomodulatory prophylactic therapies in other antiviral settings like dengue as well as in malignancies like hepatocellular carcinoma where these markers have shown relation to disease progression.
How to cite this article
Fleites YA, Aguiar J, Cinza Z, et al. HeberNasvac, a Therapeutic Vaccine for Chronic Hepatitis B, Stimulates Local and Systemic Markers of Innate Immunity: Potential Use in SARS-CoV-2 Postexposure Prophylaxis. Euroasian J Hepato-Gastroenterol 2021;11(2):59–70.
Keywords: Immunity, Innate, Postexposure, Prophylaxis, SARS-CoV-2, Therapy
Introduction
Severe acute respiratory syndrome coronavirus 2 (At present (June 26, 2021), more than 180 million people have been infected by SARS-CoV-2 and more than 4 million COVID-19 patients have died. Due to the complex pathological lesions in the lungs and multiple organs,1,2 the survivors of severe disease often results in disabling sequels.
A great effort is ongoing to mitigate SARS-CoV-2 pandemic at national and international levels by the diagnosis and isolation of SARS-CoV-2-infected patients3 and the introduction of new vaccines. However, the access to vaccines is still limited and the currently approved ones are now facing the challenge of resisting the new variants. On the contrary, treatments for SARS-CoV-2 have shown limited efficacy to prevent death. Several drugs have been repurposed for treatment of COVID-19;4 however, the benefits remain unclear in most cases. Considering these realities, it seems that SARS-CoV-2 may remain for a long time with the potential to evolve into much more transmissible variants that may eventually escape from vaccines or natural immunity, resulting in new infection outbreaks.
The reasons behind SARS-CoV-2 symptomatic infection, disease progression, and mortality are still unclear.5,6 Available information indicates that host immunity may be an important determinant of the outcome. Host immunity may determine if one will be asymptomatic, develop mild-to-moderate disease, or progress to severe COVID-19. Supportive evidence regarding the critical role of host immunity become clear because elderly people with compromised immunity or those with comorbidities are prone to develop the severe disease.5,6 Accordingly, immunomodulators have also been used for treatment.7,8
The innate immunity acts as the first-line of defense against viral infection and starts with the activation of pattern-recognition receptors (PRRs). The PRRs are located on endosomal membranes and the cytosol. These receptors are involved in the detection of viral components or replication intermediates known as pathogen-associated molecular patterns (PAMPs). Complex interactions among the viruses, viral receptors, PRRs, and PAMPs determine the initial step of viral infection. Cells of the innate immune system downregulate viral replication. To overcome this initial defensive system, SARS-CoV-2 has developed multiple evasive strategies,9 it is recognized that the evolution of new variants is linked to the increased capacity of the virus to evade the cell surveillance system.10
A novel therapeutic vaccine for chronic hepatitis B (CHB) (HeberNasvac, also known as NASVAC) has shown to be safe and to have immunomodulating and antiviral properties in phases I, II, and III clinical trials.11–13 Recently, safety and efficacy of NASVAC have been confirmed in normal individuals and patients with CHB in Japan.14–16 This product has already received a sanitary license in Cuba, and it has been recently used for SARS-CoV-2 postexposure prophylaxis.17 It is recognized that the induction of mucosal immunity may “shut the door” on SARS-CoV-2 by preventing the initial steps of infection.18 A nonspecific vaccine (HeberNasvac) has inspired other vaccine candidates aimed at inducing innate and adaptive immune responses by nasal administration with the combination of hepatitis B core antigens (HBcAg) and receptor-binding domain (RBD).19
The present study was designed to assess the local and systemic innate immune stimulation by NASVAC in hospitalized patients with respiratory symptoms or close contacts of COVID-19 during the first semester of 2020 in Cuba. NASVAC was given by intranasal (IN) and sublingual (SL) routes, to target the innate immunity at the SARS-CoV-2 portal of entry.
Materials and Methods
A phase I/II, open-label, controlled and randomized clinical trial of NASVAC as a postexposure prophylaxis treatment (RPCE00000326-En), was designed with the primary aim of assessing the local and systemic immunomodulatory effect of NASVAC in a cohort of suspected and SARS-CoV-2 risk-contact patients hospitalized at the “Luis Diaz Soto” Hospital (Havana, Cuba). A total of 46 patients, of both sexes, 60 years or older, presenting with symptoms of COVID-19 were enrolled in the study. Patients received NASVAC (100 μg per Ag per dose) via IN (at days 1, 7, and 14) and SL, daily (from days 1–14) for 14 days. Due to the PCR-delayed assessment program—on day 5 after the arrival to the hospital, according to the Ministry of Health protocol, the initial virological/serological status was unknown until day 5 after the inoculations. Thus, the study was conducted in patients with symptoms (78%), and half of them with serology positive to SARS-CoV-2 antigens; only in three cases, the polymerase chain reaction (PCR)-based test was detected as positive 5 days after hospitalization: two in patients from the control group and one in the treated group.
The study excluded the patients with an established lung disease, pneumonia, severe respiratory disease, patients with alanine aminotransferase (ALT) levels above twice the maximum level of normality, abnormal levels of serum bilirubin and serum creatinine, patients taking immunosuppressive drugs or chemotherapy 1 month before enrollment, hemoglobin <10 gm/dL, platelets <100.000/μL, and pregnancy or planning pregnancy. Patients with mental and/or psychiatric disorders unable to provide informed consent were also excluded. The patient profile is given in Table 1.
Table 1.
Demographic and initial characteristics of patients enrolled in the phase I/II study
| Variables | CIGB2020 N = 24 | Control N = 22 | Total N = 46 | p value (test) |
|---|---|---|---|---|
| Age (years) | 0.094 (ANOVA) | |||
| Mean (SD) | 67.3 (6.6) | 70.1 (6.8) | 68.6 (6.7) | |
| Min–Max | 60.0–86.0 | 60.0–86.0 | 60.0–86.0 | |
| Sex (male/female) | 0.238 (Chi-sq) | |||
| Male (%) | 13 (54.2) | 9 (40.9) | 22 (47.8) | |
| Female (%) | 11 (45.8) | 13 (59.1) | 24 (52.2) | |
| Racial background | 0.433 (Fisher) | |||
| Caucasian (%) | 18 (75.0) | 13 (59.1) | 31 (67.4) | |
| African (%) | 3 (12.5) | 1 (4.5) | 3 (6.5) | |
| Chinese (%) | 0 (0) | 1 (4.5) | 1 (2.17) | |
| Mixed (%) | 3 (12.5) | 7 (31.8) | 11 (23.9) | |
| Weight (kg) | 0.997 (ANOVA) | |||
| Mean (SD) | 73.2 (13.9) | 73.2 (13.6) | 73.2 (13.6) | |
| Min–Max | 45.9–100.0 | 52.0–100.0 | 45.9–100.0 | |
| Height (m) | 0.295 (ANOVA) | |||
| Mean (SD) | 1.66 (1.11) | 1.63 (0.09) | ||
| Min; Max | 1.50–1.98 | |||
| BMI (m/cm 2 ) | 0.356 (ANOVA) | |||
| Mean (SD) | 26.4 (3.37) | 27.5 (3.55) | 26.9 (3.46) | |
| Min–Max | 19.6–33.4 | 21.7–33.1 | 19.6–33.4 | |
| Symptoms at day 0 (%) | 18 (75.0) | 16 (72.7) | 36 (78.3) | 0.861 (Chi-sq) |
| SARS-CoV-2 (PCR+) * | 1 (4.1) | 2 (9.1) | 3 (6.5) | 0.499 (Chi-sq) |
| SARS-CoV-2 (ELISA) | 12 (50.0%) | 9 (40.9) | 21 (45.6) | 0.534 (Chi-sq) |
*Conducted 5 days after the hospitalization
Formulation and Antigens
HeberNasvac® (NASVAC) is a liquid formulation comprising the hepatitis B surface antigens (HBsAg) and the nucleocapsid (core, HBcAg) of the hepatitis B virus (HBV), produced by recombinant DNA technology as virus-like particles (Center for Genetic Engineering and Biotechnology, CIGB, Havana, Cuba). NASVAC contains 100 μg of each antigen in a final volume of 1.0 mL in saline–phosphate buffer (PBS), pH 7.0. No other additives, preservatives, or stabilizers are included. The antigens and the formulation were produced and released under good manufacturing practice (GMP) conditions at the production facilities of the CIGB.
Oropharyngeal Scraping
Samples of oropharyngeal scrapings were taken from the tonsil's region, using Ayre's spatulas under the in-house established biosafety-approved protocol. Samples were collected on days preimmune, after the third (day 4) and seven immunizations (day 8). These samples were conserved in sample preservation solution (Miltenyi Biotec, Cologne, Germany) and processed the same day for RNA extraction and purification.
Collection of Peripheral Blood Mononuclear Cells
Blood samples were collected on days 0, 4, and 8 by venipuncture of the median cubital vein and distributed in cell preparation tubes (BD Vacutainer CPTTM)-heparin tubes for peripheral blood mononuclear cell (PBMC) isolation (BD 362753), ethylenediaminetetraacetic acid (EDTA) tubes for fluorescence activated single cell sorting assay (FACS) and hematology evaluations (BD 367841), and clot activator– containing tubes for biochemical analyses (BD 367812). In general, mucosal and blood samples were collected according to dedicated procedures approved by the Biosafety Department at CIGB and after training of healthcare workers at the clinical site to handle the extraction of the samples from potentially infected patients.
Flow Cytometry Evaluation
Samples of 50 μL of whole blood were stained using an optimized cocktail including anti-CD14 PE and anti-human leucocyte antigen DR (HLADR) FITH (PARTEC, Germany), anti-CD33 PECy7.7 and anti-CD15 VB515 (BD, United States), and anti-CD11b-APC (Miltenyi, Germany). A Sysmex-Partec flow cytometry was used, cells were gated to single events, and CD14+ cells were selected from CD33-positive events (monocytes). After exclusion of CD33+ and CD15+ cells, lymphocytes were gated in a forward scatter–side scatter (FSC–SSC) dot plot. Both monocytes and lymphocytes were later analyzed for HLA-DR expression and mean fluorescence intensity (MFI) was recorded.
Genes and Primers
Ol igonucleot ides were used to ampl i f y f ragment s of the human toll-like receptors (TLR3) genes (Direction: 5′ CGAGT TAAGAGGCTGGAATGGT 3′, Ant i sense: 5′ GCCAGGAATGGATGAGGTCAGA 3′), which generates an amplicon of 176 base pairs (bp); TLR7 (Direction: 5′ TGTCGACGCATCAAAAGCAT 3′, Antisense: 5′ GTGGAAATTGCCCTCGTTGT 3′), which generates an amplicon of 101 bp; and TLR8 (Direction: 5′ TCTGCATGAGGTTGTCGTGA 3′, Antisense: 5′GTCGTCGTCGTCGTGCGTGCGTGA 3C) from 103 bp.20 As a constitutive gene for normalization, the glucuronidase beta (GusB) gene (Direction: 5′ CGTGGTTGGAGAGCT CATTTGGAA 3′, Antisense: 5′ ATTCCCCAGCACTCTCGTCGGT 3′) was used, which generates a 73 bp amplicon. All PCR results were verified for height using DNA molecular weight markers.
Virology, Hematology, and Blood Chemistry
SARS-CoV-2 RNA presence was assessed using automatic system and PCR kits (Roche Diagnostics, Switzerland) in both studies. Clinical laboratory hematology and hemochemistry parameters (reactive C-protein, transaminases, creatinine, glycemia, and hemochemical profile in general based) were evaluated following each hospital-validated procedures.
SARS-CoV-2 ELISA for Serological Studies
SARS-CoV-2-specific serology was studied in the Cuban trial using a standardized in-house enzyme linked immunosorbent assay (ELISA) protocol (Analytical Lab, CIGB). Briefly, Costar 3590 plates were coated with 100 μL of 1 μg per well of recombinant SARS-CoV-2 nucleocapsid protein (rNP) or the RBD protein in coating buffer (0.1 M carbonate/bicarbonate buffer pH 9.6). The plates were incubated for 1 hour at 37°C. After washing three times with distilled water and 0.05% Tween 20, the coated plates were blocked with 250 μL per well of blocking solution (PBS, 2% skim milk powder, 0.05% tween 20, 2.5% goat sera) for 1 hour at 37°C. Subsequently, 100 μL of test sample and controls at 1/20 dilution in sample buffer (PBS, 0.2% skim milk powder, 0.05% tween 20, 0.25% goat sera), were added per well and incubated for 30 minutes at 37°C. After the washing step, specific immunoglobulins (IgA, IgM, or IgG) antibodies were detected using 100 μL per well of a secondary antibody peroxidase-conjugated (CIGB-SS, Cuba) diluted in PBS-T solution (2.68 mM KCl, 1.47 mM KH2PO4, 136.89 mM NaCl, 8.1 mM Na2HPO4, and 0.05% Tween 20). Substrate reaction was carried out using 100 μL per well of substrate solution (TMB-3,3′,5,5′-tetramethylbenzidine, Cat. # T0440, Sigma Life Sciences, United States) incubating plates at room temperature for 10 minutes. The reaction was stopped by adding stop solution (2 M H2SO4). Microtiter plates were read at a wavelength of 450 nm using the UMELISA reader (PR-521, Tecnosuma Internacional, Havana, Cuba).
RNA Purification, cDNA, and PCR for TLR Gene Expression Studies
RNA purification was performed in the phase I/II clinical trial using the RNeasy Mini Kit (250) purification kit (Qiagen, Gilden, Germany), following kit instructions. RNA was quantified using a NanoDrop spectrophotometer (Thermo-Scientific, Waltham, Massachusetts, United States). The RT reaction to obtain the cDNA was carried out according to kit instructions (Quantitect Reverse Transcription Kit) (Qiagen, Gilden, Germany). Patient samples with paired RNA available from days 0 and 4 (13 per group) as well as from days 0 and 8 (18 from treated and 12 from control groups) were selected for PCR assessment. Some samples were unable to render the required amount of RNA, reducing the sample size; however, the minimum planned size of 10 per group was achieved in both cases for local and systemic immunity assessments.
To study the relative expression of genes of interest, the “Quantitect SYBR Green PCR kit” from Qiagen, Germany, containing “Hot Start Taq Polymerase” was used. The specific oligonucleotides were synthesized in the CIGB and diluted to 10 pmol/mL for use in the PCR. The following amounts of reagents were combined for a PCR: 7.5 mL of the SYBR Green mixture from the kit used, 1 μL of each of the two oligonucleotides (10 pmol/mL), and 0.5 mL of water. A reaction mixture was manufactured with these reagents in the amount required according to the number of PCRs. Finally, 10 mL of each PCR tube were taken from this mixture, to which 5 mL of the 1/10 diluted cDNA was added, and thus the final volume of the PCR was completed. For negative controls, instead of DNA, PCRs were completed with 5 mL of water. The PCRs were accomplished in a PCR equipment End-time Eppendorf Master Cycler Gradient, Eppendorf, Germany. The reaction conditions were 95°C for 15 minutes, followed by 40 cycles of 94°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute. Finally, 5 minutes were added to the program, at 72°C. The PCR product was visualized by DNA electrophoresis on 2.5% agarose (Caledon, Georgetown, Canada) gels staining with ethidium bromide; the molecular weight marker “Bench Top 1Kb DNA ladder” (Promega, Madison, Wisconsin, United States) was used.
Semiquantitative Analysis of the Gene Expression
The analysis of the results was carried out by visual inspection of the bands in the agarose gels using a semiquantitative method based on the analysis of endpoint PCR optimized for high sensitivity detection. Briefly, the same amount of cDNA obtained from purified RNA samples was used in the endpoint PCR to compare the gene expression between days 0 and 4, and days 0 and 8. The paired samples were analyzed in the same PCR and were run on the same gel. The reporter gene for normalization (GusB) was also run with the paired samples in the same PCR and gel. In case of different intensities in the GusB comparison, a correction in the semiquantitative scale was applied in correspondence with the difference.
The result considered as an “increase in expression” received a value of “1”, in the case of “strong increase” (values of 2 or 3) if the increase in intensity between bands on days 4 and 8 was appreciable or very notable with respect to day 0, respectively, running in parallel, side by side. It was considered as a “decrease in stimulation” (value of −1) or “strong decrease” (value of −2 or −3) if the reduction in intensity between the bands on days 4 and 8 was appreciable or very notable compared to the bands on day 0, respectively, run in parallel, side by side.
The GusB gene has been previously validated by our group as a constitutive gene whose expression remains stable with the vaccine treatment during pharmacological studies on in vitro quantitative or semiquantitative studies. The photographic analysis was conducted in paired samples receiving the same optical treatment. The visual analysis was performed by two blinded experts to assess the difference in terms of intensity of the paired bands for numeric assignment.
Clinical Progression
Patients’ clinical progression (symptoms, X-ray) was assessed according to the hospital-established protocols. In addition, the protocol included an in deep analysis of adverse events, in terms of type, duration, intensity, and imputability, after each immunization.
Endpoints
The primary endpoint of the phase I/II clinical trial at Cuba was the stimulation of local and systemic markers of innate immune response before and after treatment with NASVAC, also between NASVAC-treated and untreated subjects. Specifically, the frequency of patients with increased gene expression of RNA sensing TLRs3, 7, and 8 in scraping of the oropharyngeal mucosa (tonsils) before (day 0) and after (days 4th and 8th), and between NASVAC-treated and control volunteers. The study was conducted in RNA samples, pairing days 0 vs 4, and days 0 vs 8. The impact of the treatment on cell activation and antigen presentation function of immune cells was studied by flow cytometry in PBMC samples, detecting the expression of HLA class II in the membranes of lymphocytes and monocytes.
Statistics
Frequency analysis was conducted using the Chi-square test; the proportion between the results of the vaccinated and control groups (frequencies of TLR increasing or reducing gene expression) was analyzed. To assess the change in FACs studies, between days 0 and day 4, and between days 0 and day 8, the Wilcoxon paired test was used. To compare between groups, a Kruskall–Wallis test followed by “a posteriori” Dunn test was implemented. For the analysis of quantitative variables in general, the groups were compared using analysis of variance (ANOVA) or the nonparametric Kruskall–Wallis test. Differences were considered statistically significant (p <0.05), very significant (p <0.01), or highly significant (p <0.001).
Results
Demographic Variables
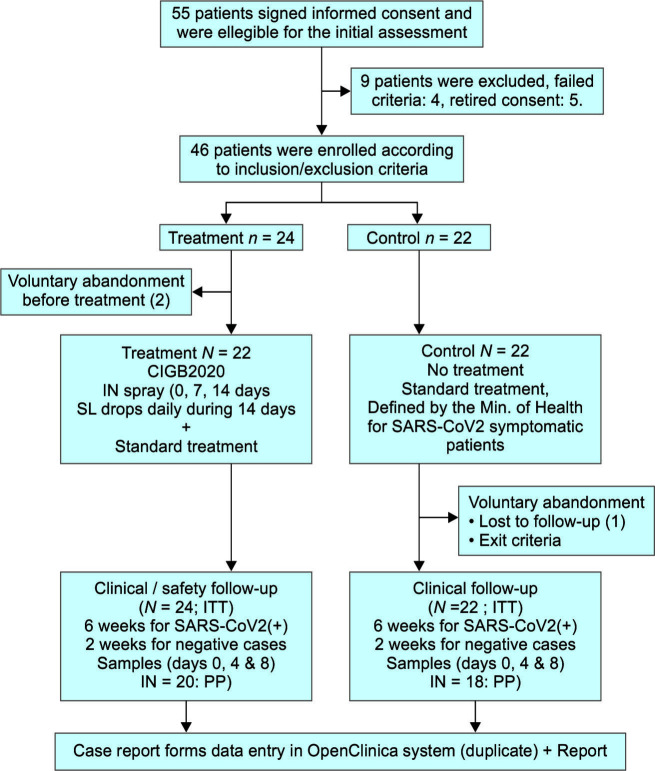
A total of 46 patients were recruited, 24 of them were assigned to the treatment group and 22 to the control group. Two patients in the control group abandoned the study before treatment start and 20 patients completed the treatment (Flowchart 1).
Flowchart 1.
Phase I/II clinical trial: patient selection
No significant differences were detected in terms of age, sex, racial composition, weight, height, body mass index, or pathological antecedents. Most patients were contacts of SARS-CoV-2-positive patients (75% in the treated group and 81.8% in the control group), and also most patients were symptomatic at the study start (83.3% in the treated patients and 72.7% in the control group) (Table 1). Regarding toxic habits, smoking and coffee intake were the most frequent and were also balanced between groups; alcohol intake was infrequent and occasional.
Safety
In general, NASVAC was safe and well tolerated. The adverse events were studied in vaccinated volunteers. A total of eight different adverse events were detected in five patients (20.8% of treated patients). Local adverse events consisted in nasal drops (0.58%), sneezing (0.3%), and otalgia (0.3%). Fever (0.3%) and asthenia (0.3%) were reported as systemic adverse events. None of them had serious adverse events. Half of the adverse events (50%) were unrelated, 25% possibly related, and 25% related to the product. Most adverse events disappear without treatment (62.5%) or with a pharmacological treatment (32.5%). In one case, the patient required hospitalization due to pneumonia, which was related to the basal condition. In summary, the administration of CIGB2020 simultaneously by IN and SL routes was safe and well tolerated under the study conditions and no significant changes appeared in the hematological and blood chemistry analysis between groups.
Clinical Progression of the Patients
A similar proportion of symptoms were reported at the study start (75.0 vs 72.7%) (Table 1). The mean duration of symptoms was longer in the control group (13.7 days vs 10.1 days, p = 0.508, ANOVA). The PCR assessment of SARS-CoV-2 infection was assayed between the 4th and the 5th day of the study start and detected one positive patient in the treatment group and two in the control group. The serological study of the patients using ELISA detected 50% positivity to any SARS-CoV-2 Ab in the treated group and 41% in the control group, suggesting a crossed reaction with previous CoV infection exists or the patients already passed the peak of viremia and detection. There was no difference in the evolution of respiratory symptoms. Among the three positive patients, one from the control group developed pneumonia, diarrhea, edema, and fatigue and after discharge, fatigue and shortage of breath remained for 42 days. The other two patients remained with mild symptoms.
Local Immune Stimulation
The impact of NASVAC on the gene expression of RNA-sensing TLRs was studied comparing the frequency of patients with increased gene expression on days 4 and 8 compared to day 0. A significantly higher proportion of patients with increased gene expression of TLR3, TLR7, and TLR8 was detected in NASVAC-treated patients compared to the control nontreated group between days 0 and 4 (p = 0.0003) as well as between days 0 and 8 (p <0.0001) when the three RNA receptors (TLR3, TLR7, and TLR8) were taken together (Table 2). A significantly higher proportion of patients with increased gene expression was detected in the case of TLR3 and TLR7 (p = 0.001 and 0.047, respectively) between days 0 and 4. In the case of TLR8, a trend was observed (p = 0.210). In the same individualized analysis between days 0 and 8, from significant to highly significant differences were detected for the frequency of patients increasing each TLR (TLR3, p = 0.007; TLR7, p = 0.0003; and TLR8, p = 0.017) (Table 2).
Table 2.
Frequency of patients with “increased” or “strongly increased” gene expression (GE) of TLR genes
| TLR with “increased” GE |
Day 0 vs day 4 Treated vs not-treated
|
Day 0 vs day 8 Treated vs not-treated
|
||
|---|---|---|---|---|
| Total (increased) | Total (increased) | Total (increased) | Total (increased) | |
| TLR3 | 13 (9) | 13 (1) | 18 (8) | 12 (0) |
| 0.001 (**) | 0.007 (**) | |||
| TLR7 | 13 (8) | 13 (3) | 18 (12) | 12 (0) |
| 0.047 (*) | 0.0003 (***) | |||
| TLR8 | 13 (10) | 13 (7) | 18 (9) | 12 (1) |
| 0.210 (n.s.) | 0.017 (*) | |||
| All RNA receptors | 39 (27) | 39 (11) | 54 (29) | 36 (1) |
| 0.0003 (***) | <0.0001 (***) | |||
| TLR with “strongly increased” GE | Total (strongly increased) | Total (strongly increased) | Total (strongly increased) | Total (strongly increased) |
| TLR3 | 13 (4) | 13 (0) | 18 (5) | 12 (0) |
| 0.029 (*) | 0.045 (*) | |||
| TLR7 | 13 (2) | 13 (0) | 18 (3) | 12 (0) |
| 0.141 (n.s.) | 0.136 (n.s.) | |||
| TLR8 | 13 (3) | 13 (1) | 18 (3) | 12 (1) |
| 0.277 (n.s.) | 0.511 (n.s.) | |||
| All RNA receptors | 39 (9) | 39 (1) | 54 (11) | 36 (1) |
| 0.006 (**) | <0.016 (*) | |||
p <0.05 (*), p <0.01 (**) and p <0.001 (***) refer to statistically significant, very significant or highly significant differences, respectively
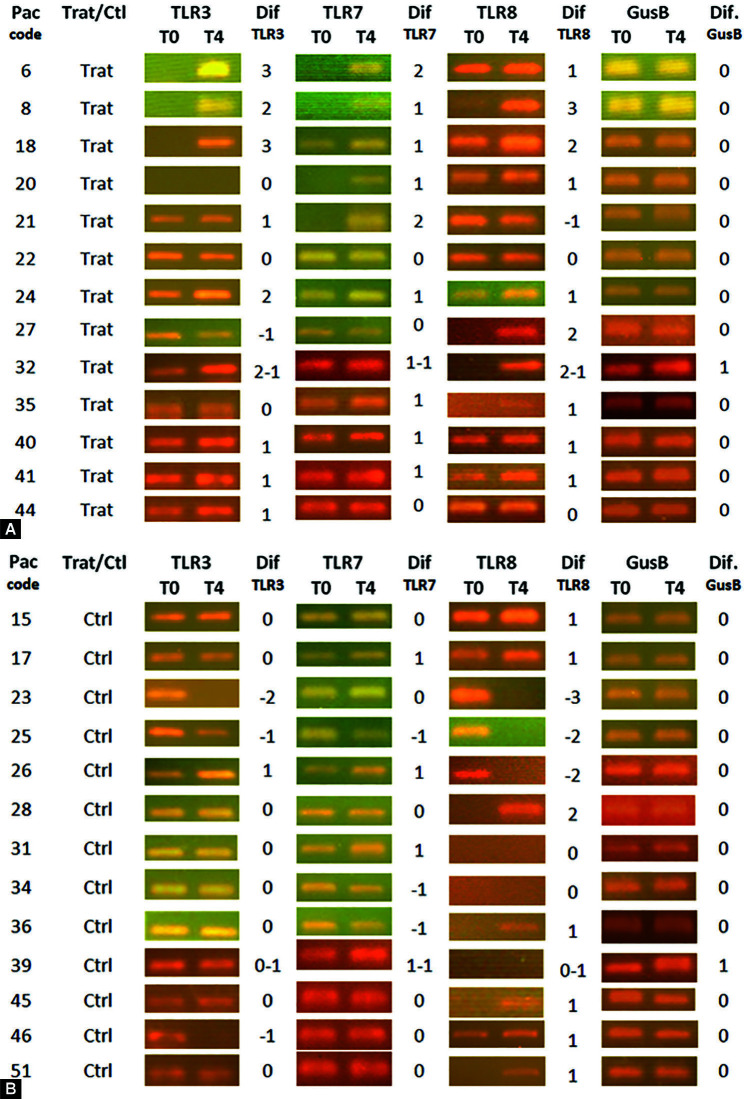
In general, the housekeeping gene GusB remained stable during the study. The similar intensity of GusB amplicons between patients and dates confirmed the quantity and quality of cDNA used in PCR. Between days 0 and 4, only two patients showed increases in GusB gene expression (one per group) as shown in Figure 1A. A correction was made in the analysis of TLRs to suppress this effect (−1). Between days 0 and 8, no difference in GusB gene expression was detected, confirming the stability of GusB gene expression on time as shown in Figure 2A.
Figs 1A and B.
Amplicon bands resulting from endpoint PCR analyzed in gel electrophoresis to assess the gene expression of RNA sensing receptors (TLR3, TLR7, and TLR8) in scraping of the oropharyngeal mucosa (tonsils) before (day 0) and after (day 4). The value of “1” was assigned to a detectable increase in the expression; “2 and 3” in case of strong increase; “−1” to a detectable decrease; and “−2 and −3” in case of strong decrease. All samples of the same patient were run in parallel, side by side in the same gel. (A) Comparison between days 0 and 4 in CIGB2020- treated patients; (B) Comparison between days 0 and 4 in control patients
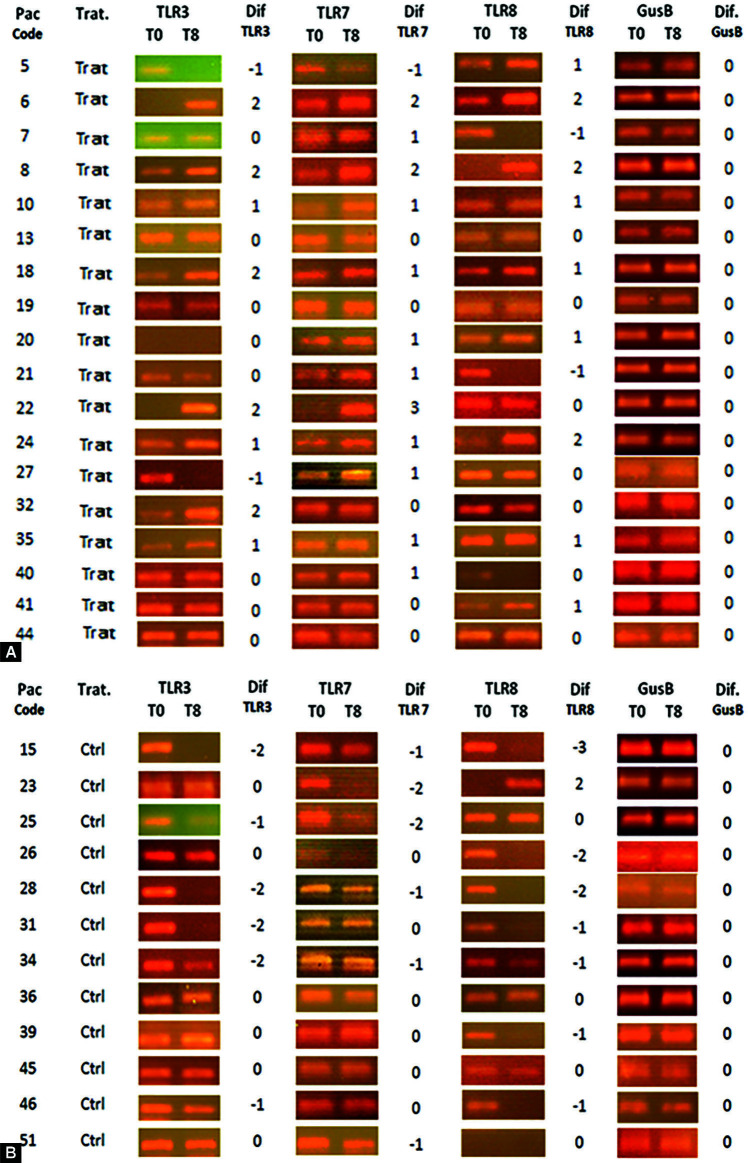
Figs 2A and B.
Amplicon bands resulting from endpoint PCR analyzed in gel electrophoresis to assess the gene expression of RNA sensing receptors (TLR3, TLR7, and TLR8) in scraping of the oropharyngeal mucosa (tonsils) before (day 0) and after (day 8). The value of “1” was assigned to a detectable increase in the expression; “2 and 3” in case of strong increase; “−1” to a detectable decrease; and “−2 and −3” in case of strong decrease. All samples of the same patient were run in parallel, side by side in the same gel. (A) Comparison between days 0 and 8 in CIGB2020- treated patients; (B) Comparison between days 0 and 8 in control patients
A significantly higher number of patients showing “strong increase” was detected on day 4 (p = 0.006) and 8 (p = 0.016), compared to day 0, when the three TLRs were pooled (Table 3). A significantly greater proportion of patients with strong increase of TLR3 in the group treated with CIGB2020, both at days 4 (p = 0.029) and 8 (p = 0.045) compared to day 0 (Table 2).
Table 3.
Frequency of patients with “decreased” or “undetectable” gene expression of TLR genes
| TLR with GE decreased |
Day 0 vs day 4 Treated vs not-treated
|
Day 0 vs day 8 Treated vs not-treated
|
||
|---|---|---|---|---|
| Total (decr.) | Total (decr.) | Total (decr.) | Total (decr.) | |
| TLR3 | 13 (1) | 13 (4) | 18 (2) | 12 (6) |
| 0.135 (n.s.) | 0.018 (*) | |||
| TLR7 | 13 (0) | 13 (3) | 18 (1) | 12 (6) |
| 0.065 (n.s.) | 0.005 (**) | |||
| TLR8 | 13 (1) | 13 (4) | 18 (1) | 12 (7) |
| 0.135 (n.s.) | 0.001 (**) | |||
| All RNA receptors | 39 (2) | 39 (11) | 54 (4) | 36 (19) |
| 0.006 (**) | <0.0001 (***) | |||
| TLR with GE undetectable | Total (undet.) | Total (undet.) | Total (undet.) | Total (undet.) |
| TLR3 | 13 (1) | 13 (2) | 18 (3) | 12 (3) |
| 0.539 (n.s.) | 0.576 (n.s.) | |||
| TLR7 | 13 (0) | 13 (0) | 18 (0) | 12 (2) |
| — (n.s.) | 0.073 (n.s.) | |||
| TLR8 | 13 (0) | 13 (6) | 18 (3) | 12 (7) |
| 0.005 (**) | 0.017 (*) | |||
| All RNA receptors | 39 (1) | 39 (8) | 54 (6) | 36 (12) |
| 0.013 (*) | 0.009 (**) | |||
Note: p <0.05 (*), p <0.01 (**) and p <0.001 (***) refer to statistically significant, very significant or highly significant differences, respectively
The individual data regarding expression of TLR gene of day 1 vs day 4 have been shown in Figures 1A and B; and between days 1 and 8 have been shown in Figures 2A and B. In control group patients, only few patients increased their gene expression for any TLR at day 4, and a generalized absence of increase was detected at day 8. On the contrary, a generalized reduction of TLR gene expression was observed in the control group, both at day 4 (p = 0.006) and 8 (p <0.0001) compared to day 0, when the three TLRs were pooled (Table 3).
A higher proportion of patients with reduction of TLR3 gene expression (p = 0.018), TLR7 (p = 0.005), and TLR8 (p = 0.0014) was detected at day 8 compared to day 0, in the control group. The comparison between days 0 and 4 only showed a nonsignificant trend in the same analysis in favor of the control group (TLR3, p = 0.13; TLR7, p = 0.06; and TLR8, p = 0.13). These results denote a trend effect by day 4 in the way to reduction to normal low values after 8 days in the nontreated patients (Table 3).
Finally, on day 4, it was observed that a higher proportion of patients from the nontreated group with undetectable signals for any of the TLRs (p = 0.013) were analyzed together. At day 8, a very significant increase in the proportion of TLRs with undetectable signals was found in the control group (p = 0.009), another variable suitable to evidence the reduction in the level of stimulation with the time. Among all RNA receptors, the TLR8 was the one with the higher number of undetectable results, with significant differences at both time points (Table 3).
Systemic Immune Stimulation
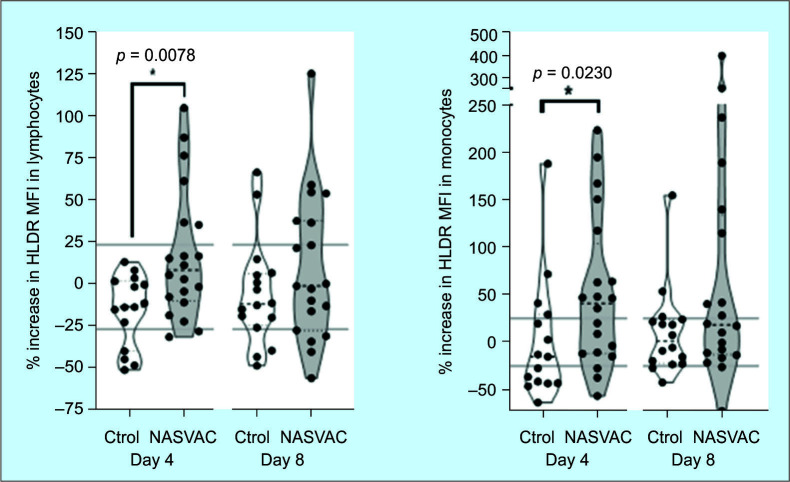
The expression of HLA-DR in monocytes significantly increased (p <0.05, Dunn test) in the group treated with CIGB2020 at day 4 after treatment started. The difference was also significant in the fluorescence mean change, further confirming the stimulation effect of CIGB2020 at day 4. In the analysis at day 8th, an important trend was detected both in proportions and fluorescence mean; however, the differences were not statistically significant (p >0.05) as shown in Figure 3. The analysis of lymphocytes also evidenced a superior frequency of patients with HLA-DR expression as well as a marked increase in the fluorescence mean in lymphocytes from patients treated with CIGB2020 comparing day 4 vs day 0 (p <0.05) as shown in Figure 3. There was an increasing trend in frequency and fluorescence mean at day 8, but no significant differences as in the case of monocytes. No increase was detected neither in the concentration nor in the percentages of total monocytes/lymphocytes in PBMC between groups.
Fig. 3.
Increase in HLA-DR expression in lymphocytes and monocytes of patients treated with HeberNasvac or in the control group between (day 0 vs 4 and day 0 vs 8). In both cases, the HLA-DR antigen presentation marker increased their frequency and intensity
Discussion
The increase in the gene expression of innate immune receptors (RNA-sensing TLR3, TLR7, and TLR8) in tonsils of patients with 60 years and older after NASVAC treatment is consistent with the multi-TLR agonist effect found by in vitro HBcAg stimulation of HepaRG cell line and also from the literature.21–23 The rationale of stimulating the TLRs at the nasopharyngeal tonsils is supported by the dramatic evidence of survival when TLR3, 7/8, or 9 agonists were nasally administered to mice in the model of lethal infection with SARS,24,25 characterizing novel surrogate markers of protection.
Recent transcriptomic profiling studies of SARS-CoV-2 in vitro infected cells recommend the use of TLR3 and TLR7/8 agonists for early immune stimulation.26 Studies in bovine coronaviruses have shown the downregulation of TLR3 gene expression compared to bovine rotavirus, further evidencing that TLR3 is a target during CoV mechanisms of infection.27 Evasion or impairment of innate sensing mechanisms or their activation pathways are common mechanisms of respiratory RNA viruses and may also justify their recurrence.9,28
Simultaneous stimulation of MyD88-dependent and independent signaling pathways (associated to TLR7/TLR8 and TLR3, respectively) results in synergistic antiviral responses.29–31 Previous studies have described HBcAg agonist effect on TLR2 and TLR7;22,23 however, there is no previous report of TLR3 gene expression stimulation or activation related to HBcAg. NASVAC comprises HBcAg with the unique characteristic of having a higher proportion of RNA to protein in respect to others in the literature. Such high levels have been detected using deep sequencing. The presence of dsRNA is justified by the composition of the RNA within the particles.21
The impact of NASVAC on the gene expression of innate immunity receptors was detected as early as the first sampling on day 4th. The effect was still detectable at the same levels at least up to day 8th. These early stimulations are characteristic of innate immunity responses and constitute an attractive property for using NASVAC in pre-/postexposure prophylaxis or preventive therapy. In these settings, a rapid expression of innate immunity sensors and the increase of antigen-presenting capabilities during initial infection are required to trigger a fast and effective adaptive response, compensating the evasive mechanisms of the coronavirus and other acute respiratory pathogens.9,28
At present, most of the clinical trials indexed in the field of pre-/postexposure prophylaxis or as preemptive therapy are based on chloroquine or hydroxychloroquine. However, recently published results of large trials in post-exposure prophylaxis (PEP) setting have shown limited efficacy.32,33 Other studies exploring preventive capacity of bacillus Calmette-Guérin (BCG) innate immune stimulation have started in several countries. Considering that BCG is a vaccine with production shortage, limited availability,34 and adverse event may be expected as a living vaccine,35 it has a limited scope as a generalized prophylaxis.
Local stimulation by SL vaccination has been used before to prevent recurrent respiratory infections.36–39 The Food and Drug Administration (FDA) approved the start of a phase II trial to the nasal influenza vaccine candidate T-COVID (Altimmune, USA), a nasally administered adenovirus vaccine previously developed for preventing influenza was repurposed as a SARS-CoV-2 preemptive therapy. The study was based on the capacity of the adenovirus-based influenza vaccine to prevent death in mice lethally infected with a nonrelated respiratory pathogen after IN and not after parenteral administration,40,41 further supporting the relevance of the local- and not parenteral stimulation.
We highlight that the stimulatory effect observed in the aged patients of our study may be modeled by the study exploring the effect of multiple TLRs on 12 and 22 months old mice lethally infected with SARS.25 COVID-19 mortality is higher in the elderly; also vaccines are less responsive in this setting. Age-related alterations in innate immune responses after stimulation of pathogen recognition receptors42 may explain these findings.
A recent study confirmed the reduced expression of HLA-DR on monocytes and myeloid dendritic cells (mDCs) in patients with severe COVID-19 disease.43 The increase of HLA class II expression in monocytes and lymphocytes of NASVAC-treated aged volunteers suggests that these patients may overcome this pathogenic mechanism of SARS-CoV-2. In addition, these results are consistent with previous in vitro and ex vivo experiences, demonstrating the stimulatory effect of NASVAC on the innate and adaptive immune system.44,45 The increase in the gene expression of TLRs, HLA class I/II, cytokines, and costimulatory molecules has been detected in vitro in the HepaRG model.21 All these effects are consistent with the antiviral and immunomodulatory effect of this product in the setting of CHB and CoV.11–13,17 A significant outcome of TLR3, TLR7, and TLR8 signaling pathways is the production of interferons. Encouraging results have been obtained with the use of type I interferon (IFN) in SARS-CoV-2 treatment.46 Moreover, a recent article evidenced a potential preexposure prophylactic effect on CHB patients previously treated with PegIFN.47 Taken together, HeberNasvac may act at two levels: (a) overcoming the innate immunity evasive activity of the virus9,10 improving viral RNA detection and (b) by direct antiviral effect as an efficient IFN inducer.17,21
The pDCs, mDCs, and CD14+ monocytes of COVID-19-infected individuals are less responsive to stimulation with bacterial/viral ligand cocktail in comparison to healthy controls (comprising TLR2, 4, and 5 ligands and TLR3 and 7/8, respectively). It is considered that the innate immune cells in the periphery of COVID-19-infected subjects have functional impairment.48
The effects described in the present article are attractive properties for the intended use in pre- and postexposure prophylaxis of SARS-CoV-2. The synergistic stimulation of MyD88- dependent and independent pathways (via TLR3, TLR7, and TLR8) has shown the impact on decreasing viremia and clinical improvement in the setting of infectious diseases like dengue49 and may find applicability in the field of hepatocellular carcinoma50,51 where TLR3 expression correlates with apoptosis, proliferation, angiogenesis, and predicts prognosis.
In conclusion, the present phase I/II study detected an increased expression of TLR3, TLR7, and TLR8 genes in nasopharyngeal tonsils, a significant property considering these are surrogate markers of SARS protection in the mice model of lethal infection. The stimulation of HLA-class II in PBMC's monocytes and lymphocytes further increases the importance of these findings, taking into account the functional impairment of innate immune cells from the periphery of COVID-19-infected subjects.
The present study supports the use of NASVAC as preemptive therapy or pre-/postexposure prophylaxis of SARS-CoV-2, or both, to prevent infection or to arrest the progression to severe COVID-19. The safety of NASVAC in patients suffering from acute respiratory infections also warrants the start of larger studies. The use of this product or their active principles has potential as immunomodulatory prophylactic therapies in other antiviral settings like dengue as well as in malignancies like hepatocellular carcinoma, where these markers have shown relation to disease progression.
Acknowledgments
Our gratitude to Ms Olivia Rubido, Cytologist, for his valuable contribution with the scraping technique and Mr Fabio Aguilar, Media Techniques Intern, for his technical support in image processing.
Orcid
Mamun A Mahtab https://orcid.org/0000-0003-3728-3879
Julio C Aguilar https://orcid.org/0000-0003-0166-4784
Footnotes
Source of support: Nil
Conflict of interest: None
References
- 1.Bugra A, Das T, Arslan MN, et al. Postmortem pathological changes in extrapulmonary organs in SARS-CoV-2 rt-PCR–positive cases: a single-center experience. Irish J Med Sci. (1971) 2021:1–11. doi: 10.1007/s11845-021-02638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Damiani S, Fiorentino M, De Palma A, et al. Pathological post‐mortem findings in lungs infected with SARS‐CoV‐2. J Pathol. 2021;253(1):31–40. doi: 10.1002/path.5549. [DOI] [PubMed] [Google Scholar]
- 3.Walensky RP, Del Rio C. From mitigation to containment of the COVID-19 pandemic: putting the SARS-CoV-2 genie back in the bottle. JAMA. 2020;323(19):1889–1890. doi: 10.1001/jama.2020.6572. [DOI] [PubMed] [Google Scholar]
- 4.Sultana J, Crisafulli S, Gabbay F, et al. Challenges for drug repurposing in the COVID-19 pandemic era. Front Pharmacol. 2020;11:588654. doi: 10.3389/fphar.2020.588654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhry R, Dranitsaris G, Mubashir T, et al. A country level analysis measuring the impact of government actions, country preparedness and socioeconomic factors on COVID-19 mortality and related health outcomes. EClinicalMedicine. 2020:100464. doi: 10.1016/j.eclinm.2020.100464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmad T, Chaudhuri R, Joshi MC, et al. COVID-19: The emerging immunopathological determinants for recovery or death. Front Microbiol. 2020;11:588409. doi: 10.3389/fmicb.2020.588409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islam MA, Mazumder MA, Akhter N, et al. Extraordinary survival benefits of severe and critical patients with COVID-19 by immune modulators: the outcome of a clinical trial in Bangladesh. Euroasian J Hepatogastroenterol. 2020;10(2):68–75. doi: 10.5005/jp-journals-10018-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortegiani A, Ippolito M, Greco M, et al. Rationale and evidence on the use of tocilizumab in COVID-19: a systematic review. Pulmonology. 2021;27(1):52–66. doi: 10.1016/j.pulmoe.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikkert M. Innate immune evasion by human respiratory RNA viruses. J Innate Immun. 2020;12(1):4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorne LG, Bouhaddou M, Reuschl AK, et al. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B. 1.1. 7 UK variant. bioRxiv. 2021 doi: 10.1101/2021.06.06.446826. [DOI] [Google Scholar]
- 11.Fernández G, Sanchez AL, Jerez E, et al. Five-year follow-up of chronic hepatitis B patients immunized by nasal route with the therapeutic vaccine HeberNasvac. Euroasian J Hepatogastroenterol. 2018;8(2):133. doi: 10.5005/jp-journals-10018-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Mahtab M, Akbar SM, Aguilar JC, et al. Therapeutic potential of a combined hepatitis B surface antigen and core antigen vaccine in patients with chronic hepatitis B. Hepatol Int. 2013;7(4):981–989. doi: 10.1007/s12072-013-9486-4. [DOI] [PubMed] [Google Scholar]
- 13.Al Mahtab M, Akbar SMF, Aguilar JC, et al. Treatment of chronic hepatitis B naïve patients with a therapeutic vaccine containing HBs and HBc antigens (a randomized, open and treatment controlled phase III clinical trial). PLoS One. 2018;13(8):e0201236. doi: 10.1371/journal.pone.0201236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida O, Akbar SM, Kohara M, et al. Induction of anti-HBs and reduction of HBs antigen by nasal administration of a therapeutic vaccine containing HBs and HBc antigen (NASVAC) in patients with chronic hepatitis B. Boston, MA, USA: American Association for Study of the Liver; 2019. [Google Scholar]
- 15.Hiasa Y, Yoshida O, Guillen G, et al. The HB vaccine containing HBs and HBc antigen (NASVAC) can effectively induce anti-HBs antibody in non-responders to the prophylactic vaccine. Boston, MA, USA: American Association for Study of the Liver; 2019. [Google Scholar]
- 16.Yoshida O, Imai Y, Akbar SMF, et al. HBsAg reduction by nasal administration of A therapeutic vaccine containing HBsAg and HBcAg (NASVAC) in a patients with chronic HBV infection: the results of 18 months follow-up.. The Liver Meeting, AASLD; November 13–16, 2020. [Google Scholar]
- 17.Akbar SMF, Mahtab MA, Aguilar JC, et al. Repurposing NASVAC, a Hepatitis B therapeutic vaccine, for pre- and post-exposure prophylaxis of SARS-CoV-2 infection. J Antivir Antiretrovir. 2021;13(20):1000004. (S20:004). Available from: https://doi.org/10.21203/rs.3.rs-438628/v1 . [Google Scholar]
- 18.Mudgal R, Nehul S, Tomar S. Prospects for mucosal vaccine: shutting the door on SARS-CoV-2. Hum Vaccin Immunother. 2020;16(12):2921–2931. doi: 10.1080/21645515.2020.1805992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burki T. Behind Cuba's successful pandemic response. Lancet Infect Dis. 2021;21(4):465–466. doi: 10.1016/S1473-3099(21)00159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Yang Y, Sun J, et al. Expression of Toll-like receptors and their association with cytokine responses in peripheral blood mononuclear cells of children with acute rotavirus diarrhoea. Clin Exp Immunol. 2006;144(3):376–381. doi: 10.1111/j.1365-2249.2006.03079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patent Application: CU 2020-0028: Priority date: 20.04.2020. International presentation date: 20.04.2021.Viral nucleoproteins and formulations thereof.
- 22.Cooper A, Tal G, Lider O, et al. Cytokine induction by the hepatitis B virus capsid in macrophages is facilitated by membrane heparan sulfate and involves TLR2. J Immunol. 2005;175(5):3165–3176. doi: 10.4049/jimmunol.175.5.3165. [DOI] [PubMed] [Google Scholar]
- 23.Lee BO, Tucker A, Frelin L, et al. Interaction of the hepatitis B core antigen and the innate immune system. J Immunol. 2009;182(11):6670–6681. doi: 10.4049/jimmunol.0803683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumaki Y, Salazar AM, Wandersee MK, et al. Prophylactic and therapeutic intranasal administration with an immunomodulator, Hiltonol®(Poly IC: LC), in a lethal SARS-CoV-infected BALB/c mouse model. Antiviral Res. 2017;139:1–12. doi: 10.1016/j.antiviral.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J, Wohlford-Lenane C, Zhao J, et al. Intranasal treatment with poly (I•C) protects aged mice from lethal respiratory virus infections. J Virol. 2012;86(21):11416–11424. doi: 10.1128/JVI.01410-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasad K, Khatoon F, Rashid S, et al. Targeting hub genes and pathways of innate immune response in COVID-19: a network biology perspective. Int J Biol Macromol. 2020;163:1–8. doi: 10.1016/j.ijbiomac.2020.06.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aich P, Wilson HL, Kaushik RS, et al. Comparative analysis of innate immune responses following infection of newborn calves with bovine rotavirus and bovine coronavirus. J Gen Virol. 2007;88(10):2749–2761. doi: 10.1099/vir.0.82861-0. [DOI] [PubMed] [Google Scholar]
- 28.Nelemans T, Kikker t M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. 2019;11(10):961. doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagchi A, Herrup EA, Warren HS, et al. MyD88-dependent and MyD88- independent pathways in synergy, priming, and tolerance between TLR agonists. J Immunol. 2007;178(2):1164–1171. doi: 10.4049/jimmunol.178.2.1164. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Q, Egelston C, Gagnon S, et al. Using 3 TLR ligands as a combination adjuvant induces qualitative changes in T cell responses needed for antiviral protection in mice. J Clin Invest. 2010;120(2):607–616. doi: 10.1172/JCI39293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Q, Egelston C, Vivekanandhan A, et al. Toll-like receptor ligands synergize through distinct dendritic cell pathways to induce T cell responses: implications for vaccines. Proc Natl Acad Sci. 2008;105(42):16260–16265. doi: 10.1073/pnas.0805325105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skipper CP, Pastick KA, Engen NW, et al. Hydroxychloroquine in nonhospitalized adults with early COVID-19: a randomized trial. Ann Intern Med. 2020;173(8):623–631. doi: 10.7326/M20-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boulware DR, Pullen MF, Bangdiwala AS, et al. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N Engl J Med. 2020;383(6):517–525. doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curtis N, Sparrow A, Ghebreyesus TA, et al. Considering BCG vaccination to reduce the impact of COVID-19. Lancet. 2020;395(10236):1545–1546. doi: 10.1016/S0140-6736(20)31025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grange JM. Complications of bacille Calmette-Guérin (BCG) vaccination and immunotherapy and their management. Commun Dis Public Health. 1998;1(2):84–88. [PubMed] [Google Scholar]
- 36.Sánchez Ramón S, Manzanares M, Candelas G. MUCOSAL anti-infections vaccines: beyond conventional vaccines. Reumatol Clin. 2020;16(1):49–55. doi: 10.1016/j.reuma.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Sánchez-Ramón S, Conejero L, Netea MG, et al. Trained immunity-based vaccines: a new paradigm for the development of broad-spectrum anti-infectious formulations. Front Immunol. 2018;9:2936. doi: 10.3389/fimmu.2018.02936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sánchez-Ramón S, Pérez de Diego R, Dieli-Crimi R, et al. Extending the clinical horizons of mucosal bacterial vaccines: current evidence and future prospects. Curr Drug Targets. 2014;15(12):1132–1143. doi: 10.2174/1389450115666141020160705. [DOI] [PubMed] [Google Scholar]
- 39.Alecsandru D, Valor L, Sánchez-Ramón S, et al. Sublingual therapeutic immunization with a polyvalent bacterial preparation in patients with recurrent respiratory infections: immunomodulatory effect on antigen-specific memory CD4+ T cells and impact on clinical outcome. Clin Exp Immunol. 2011;164(1):100–107. doi: 10.1111/j.1365-2249.2011.04320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altimmune: waiting for their next move against COVID-19. News article. Jun 16, 2020. Available from: https://ir.altimmune.com/investors/news-articles .
- 41.Altimmune Receives Award from U.S. Department of Defense to Fund Phase 1/2 Clinical Trial of T-COVID™ in Outpatients with Early COVID-19. Press release. 29.06.2020. Available from: www.altimmune.com .
- 42.Metcalf TU, Cubas RA, Ghneim K, et al. Global analyses revealed age‐related alterations in innate immune responses after stimulation of pathogen recognition receptors. Aging Cell. 2015;14(3):421–432. doi: 10.1111/acel.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arunachalam PS, Wimmers F, Mok CK, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lobaina Y, Hardtke S, Wedemeyer H, et al. In vitro stimulation with HBV therapeutic vaccine candidate Nasvac activates B and T cells from chronic hepatitis B patients and healthy donors. Mol Immunol. 2015;63(2):320–327. doi: 10.1016/j.molimm.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Akbar SM, Yoshida O, Chen S, et al. Immune modulator and antiviral potential of DCs pulsed with both hepatitis B surface antigen and core antigen for treating chronic HBV infection. Antivir Ther. 2010;15(6):887–895. doi: 10.3851/IMP1637. [DOI] [PubMed] [Google Scholar]
- 46.Monk PD, Marsden RJ, Tear VJ, et al. Inhaled Interferon Beta COVID-19 Study Group. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med. 2021;9(2):196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akbar SMF, Al Mahtab M, Aguilar JC, et al. Role of pegylated interferon in patients with chronic liver diseases in the context of SARS-CoV-2 Infection. Euroasian J Hepatogastroenetrol. 2021;11(1):27–31. doi: 10.5005/jp-journals-10018-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varchetta S, Mele D, Oliviero B, et al. Unique immunological profile in patients with COVID-19. Cell Mol Immunol. 2021;18(3):604–612. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sariol CA, Martínez MI, Rivera F, et al. Decreased dengue replication and an increased anti-viral humoral response with the use of combined Toll-like receptor 3 and 7/8 agonists in macaques. PLoS One. 2011;6(4):e19323. doi: 10.1371/journal.pone.0019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan MM, Xu YY, Chen L, et al. TLR3 expression correlates with apoptosis, proliferation and angiogenesis in hepatocellular carcinoma and predicts prognosis. BMC Cancer. 2015;15(1):1–6. doi: 10.1186/s12885-015-1262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonnin M, Fares N, Testoni B, et al. Toll-like receptor 3 downregulation is an escape mechanism from apoptosis during hepatocarcinogenesis. J Hepatol. 2019;71(4):763–772. doi: 10.1016/j.jhep.2019.05.031. [DOI] [PubMed] [Google Scholar]