Abstract
Iron (Fe) is an essential mineral element that governs the composition of natural plant communities and limits crop yield in agricultural ecosystems due to its extremely low availability in most soils, particularly at alkaline pH. To extract sufficient Fe from the soil under such conditions, some plants, including Arabidopsis (Arabidopsis thaliana), secrete Fe-mobilizing phenylpropanoids, which mobilize sparingly soluble Fe hydroxides by reduction and chelation. We show here that ectopic expression of the peptides IRONMAN (IMA1) and IMA2 improves growth on calcareous soil by inducing biosynthesis and secretion of the catecholic coumarin 7,8-dihydroxy-6-methoxycoumarin (fraxetin) via increased expression of MYB72 and SCOPOLETIN 8-HYDROXYLASE, a response that is strictly dependent on elevated environmental pH (pHe). By contrast, transcription of the cytochrome P450 family protein CYP82C4, catalyzing the subsequent hydroxylation of fraxetin to sideretin, which forms less stable complexes with iron, was strongly repressed under such conditions. We concluded that IMA peptides regulate processes supporting Fe uptake at both acidic and elevated pH by controlling gene expression upstream of or in concert with a putative pHe signal, adapting the plant to prevailing edaphic conditions. This regulatory pattern confers tolerance to calcareous soils by extending the pH range in which Fe can be efficiently absorbed from the soil. Our results further suggest that pHe calibrates the activities of components of the Fe deficiency response, accentuating processes that are most efficient under the prevailing conditions. Altering the expression of IMA peptides provides a route for generating plants adapted to calcareous soils.
Ectopic expression of IRONMAN peptides improves growth under iron-limiting conditions by inducing responses to limited iron availability in accordance with the environmental pH.
Introduction
In natural ecosystems, growth on calcareous, that is, carbonate-rich, alkaline soils, is restricted to so-called calcicole (chalk-loving) plants, while calcifuge (chalk-fleeing) species are excluded from such habitats (Grime and Hodgson, 1969; Lee, 1998). Among other factors, the ability to thrive on calcareous substrates has been attributed to a particularly efficient strategy to extract iron (Fe), an essential mineral nutrient with extremely limited solubility in most conditions, from the soil. In aerated soils, alkalinity decreases the availability of Fe to levels that are several orders of magnitude below the requirement of the plants (Schwab and Lindsay, 1983; Tyler, 1996). Species that are not well adapted to calcareous soils, for example, crops such as fruit trees and soybean or taxa which originate from acidic habitats, develop lime-induced chlorosis caused by insufficient uptake, maldistribution, or immobilization of Fe (Zohlen and Tyler, 2004).
The underlying causes for the superior Fe acquisition efficiency of calcicole species still remain largely enigmatic. Generally, gramineous species (Poaceae) have been regarded as being more Fe-efficient under alkaline condition than nongrasses, a distinction that has been associated with the mechanism by which Fe is mobilized by grass roots. In contrast to all other land plants, grasses have adopted a chelation-based Fe uptake system that relies on the synthesis and secretion of Fe-chelating phytosiderophores, a mechanism that has been designated as strategy II (Römheld and Marschner, 1986). Phytosiderophores are mugineic acid derivates that form soluble complexes with ferric Fe, which are fairly stable over a wide pH range (Shi et al., 1988). Conversely, nongrasses have adopted a Fe acquisition strategy which depends on the orchestrated action of enzymatic reduction of ferric chelates, H+-ATPase-mediated acidification of the rhizosphere, and secretion of low-molecular-weight Fe-mobilizing compounds such as flavins or phenylpropanoids, a system that is referred to as strategy I (Römheld and Marschner, 1986). In Arabidopsis, chelated ferric Fe is reduced by the plasma membrane-bound FERRIC REDUCTION OXIDASE2 (FRO2; Robinson et al., 1999) and the liberated Fe2+ is subsequently taken up from the soil solution by IRON-REGULATED TRANSPORTER1 (IRT1; Eide et al., 1996; Vert et al., 2002). The FRO2-mediated reduction of ferric Fe is supported by proton extrusion via the P-type H+-ATPase AHA2 (Santi and Schmidt, 2009), a process that creates a slightly acidic pH milieu in the apoplast, which is favorable for the uptake of Fe. FRO2-mediated ferric reduction is compromised at circumneutral or alkaline pH ranges (Susín et al., 1996; Lucena et al., 2007; Waters et al., 2018), rendering the strategy I system ineffective under such conditions.
In Arabidopsis, the genes mediating the Fe deficiency responses are under the control of a heterodimer consisting of the master transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT; bHLH29) and one out of four clade Ib bHLH proteins (bHLH38, bHLH39, bHLH100, and bHLH101) (Colangelo and Guerinot, 2004; Yuan et al., 2008; Sivitz et al., 2012; Wang et al., 2013). The expression of FIT, in turn, and that of all Ib bHLH proteins, is controlled by UPSTREAM REGULATOR OF IRT1 (URI; bHLH121) in concert with IAA-LEUCINE-RESISTANT3 (ILR3; bHLH105) (Kim et al., 2019; Gao et al., 2020; Lei et al., 2020). FIT is negatively regulated by the clade IVa bHLH transcription factors bHLH18, bHLH19, bHLH20, and bHLH25 (Cui et al., 2018), and by the E3 ligases BTSL1 and BTSL2 (Rodríguez-Celma et al., 2019), which target FIT for degradation via the 26S proteasome. A recently discovered family of peptides designated IRONMAN/FE-UPTAKE-INDUCING PEPTIDE (IMA/FEP) control Fe uptake, presumably via regulation of clade Ib bHLH proteins (Grillet et al., 2018; Hirayama et al., 2018). IMA/FEP peptides are ubiquitous across land plants and share a highly conserved consensus motif at the C-terminus, which is necessary and sufficient for IMA function. The expression of IMA1 and IMA2 is directly controlled by bHLH121 (Gao et al., 2020). Ectopic expression of IMA1 overrides the repression of Fe uptake by sufficient internal Fe levels, induces ferric reduction activity, and improves growth on media with low Fe solubility (Grillet et al., 2018). High ferric reduction activity is, however, insufficient to confer calcicole behavior (Schmidt and Fühner, 1998; Terés et al., 2019), indicating that other mechanisms are employed to enable plants to thrive under such conditions. Recent studies have shown that growth of nongrasses on calcareous soils requires the synthesis and secretion of species-specific, low-molecular-weight compounds, which increase the solubility of ferric hydroxides by reduction and chelation and greatly extend the pH range in which Fe can be efficiently mobilized (reviewed by Chen et al., 2017; Tsai and Schmidt, 2017; Robe et al., 2020b). In Arabidopsis, Fe deficiency triggers a massive increase in the abundance of enzymes of the phenylpropanoid pathway and an accumulation of the hydroxycoumarin scopoletin (Lan et al., 2011). Scopoletin, however, does not possess Fe-mobilizing properties, which suggests that other compounds derived from this pathway are critical for the acquisition of Fe.
The Fe(II)- and 2-oxoglutarate-dependent dioxygenase FERULOYL-COA 6′-HYDROXYLASE1 (F6′H1) catalyzes the ortho‐hydroxylation of feruloyl‐CoA into 6‐hydroxyferuloyl‐CoA, the first committed step in the biosynthesis of catecholic coumarins derived from scopoletin, and was identified in a genetic screen as being essential for survival on alkaline soil (Schmid et al., 2014). In the light, 6-hydroxyferuloyl-CoA is converted into scopoletin, a reaction that occurs partially spontaneously. In light-protected organs such as roots, scopoletin biosynthesis is dependent on enzymatic catalysis via COUMARIN SYNTHASE (COSY; Vanholme et al., 2019). Similar to F6′H1, COSY activity is indispensable for growth on alkaline substrates (Vanholme et al., 2019), suggesting that compounds which are produced downstream of scopoletin are required for the mobilization of Fe. Hydroxylation of scopoletin mediated by another Fe(II)- and 2-oxoglutarate-dependent dioxygenase, SCOPOLETIN 8-HYDROXYLASE (S8H), yields 7,8-dihydroxy-6-methoxycoumarin (fraxetin), a potent Fe chelator (Siwinska et al., 2018; Tsai et al., 2018, Rajniak et al., 2018). A further hydroxylation step, mediated by CYTOCHROME P450 FAMILY 82 SUBFAMILY C POLYPEPTIDE4 (CYP82C4), converts fraxetin into a novel catecholic coumarin referred to as 5,7,8‐trihydroxy‐6‐methoxycoumarin (sideretin; Rajniak et al., 2018). Secretion of scopoletin, fraxetin, and sideretin occurs via PLEIOTROPIC DRUG RESISTANCE9 (PDR9; Fourcroy et al., 2014; Ziegler et al., 2017; Rajniak et al., 2018).
Fraxetin and sideretin constitute the major Fe-mobilizing coumarins derived from this pathway and feature adjacent hydroxyl groups which bind Fe efficiently (Schmid et al., 2014; Rajniak et al., 2018; Tsai et al., 2018; Robe et al., 2020b). In particular, fraxetin secretion appears to be important for plant edaphic adaptation. Growth and chlorophyll content corresponded with fraxetin secretion across a suite of Arabidopsis accessions grown on high pH media with restricted Fe availability, indicating that this process is critical for the ability to thrive on alkaline soils (Tsai et al., 2018). Correspondingly, the better Fe uptake of Arabidopsis demes with high carbonate content in their native habitat relative to demes native to silicious soils was found to be due to higher proton extrusion and higher fraxetin secretion (Terés et al., 2019). Supplementing the growth media with fraxetin or growing sensitive plants from silicious soils alongside carbonate-adapted plants enhanced Fe and chlorophyll levels of the sensitive plants, underscoring a key role for Fe-mobilizing coumarins in the mobilization of Fe from recalcitrant pools (Terés et al., 2019).
Here, we set out to explore the basis on which IMA is conferring calcicole behavior to Arabidopsis thaliana, a calcifuge species. We demonstrate that overexpression of either IMA1 or IMA2 is sufficient to activate all Fe deficiency responses except for fraxetin secretion, which is modulated by the environmental hydrogen activity (pHe). Elevated pHe promotes the expression of S8H and represses the transcription of CYP82C4, thereby activating a separate, calcicole branch of the strategy I-type Fe deficiency response.
Results
IMA1 and IMA2 are closely co-expressed putative paralogs
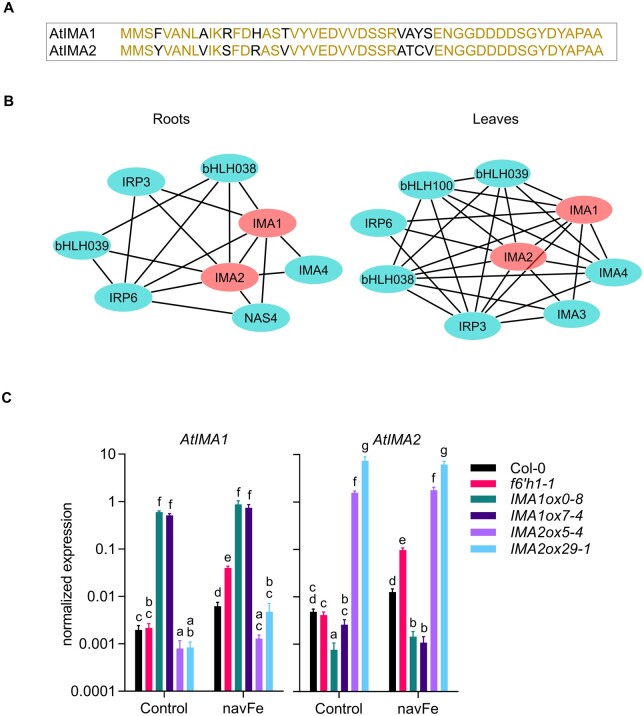
IMA1 and IMA2 are highly similar peptides harboring a conserved C terminus that has been associated with Fe uptake across land plants (Grillet et al., 2018). The two peptides share 82% sequence identity and are robustly induced by Fe deficiency in both roots and shoots (Figure 1A; Rodríguez-Celma et al., 2013a, 2013b). Publicly available RNA-seq data reveal similar organ- and tissue-level expression of both genes (Supplemental Figure S1). To infer organ-specific co-expression networks around IMA1 and IMA2, we downloaded and normalized a total of 5,556 RNA-seq data sets from the Sequence Read Archive hosted by the National Center for Biotechnology Information. Networks were constructed with the MACCU toolbox (Lin et al., 2011) based on pair-wise comparison of the co-expression relationships of Arabidopsis genes expressed in either roots or leaves with a Pearson coefficient >0.7 using tissue-specific data sets (1,194 for roots and 641 for leaves). In both roots and leaves, IMA1 and IMA2 are co-expressed with a suite of other putative or validated regulators of cellular Fe homeostasis, such as IMA4, bHLH38, bHLH39, IRON-RESPONSIVE PROTEIN3 (IRP3; At2g14247), and IRP6 (At5g05250; Figure 1B). Together with IMAs, IRPs were identified in transcriptional surveys as being highly Fe responsive in both roots and shoots (Figure 1A; Rodríguez-Celma et al., 2013a). IRP1 and IRP2 have been renamed to IMA1 and IMA2 after elucidation of their function (Grillet et al., 2018); the biological roles of IRP3–IRP6 remain to be elucidated. In roots, IMA1/IMA2 is further co-expressed with NICOTIANAMINE SYNTHASE4 (NAS4). In leaves, the expression of IMA1/IMA2 is associated with IMA3 and bHLH100 (Figure 1B).
Figure 1.
Sequences, co-expression networks, and expression levels of IMA1 and IMA2 in transgenic plants. A, Alignment of IMA1 and IMA2 peptides. Red fonts indicate identical amino acids, black fonts denote differences between the sequences. B, Co-expression networks around IMA1 and IMA2 in roots and leaves. Networks were constructed with the MACCU toolbox (Lin et al., 2011) against customized organ-specific databases. Genes with a Pearson coefficient of >0.7 for pairwise expression were included in the network. C, Expression of IMA1 and IMA2 in the wild-type, f6′h1 mutant plants, and transgenic lines overexpressing either IMA1 (IMA1ox) or IMA2 (IMA2ox). Expression was measured in roots of 12-d-old seedlings grown on ES (Control) or navFe media for 3 d after 9 d of precultivation on ES media. (Each bar represents the mean ± se of four independent experiments. Distinct letters above the bar graph indicate significant differences (P < 0.05) in two-way analysis of variance (ANOVA) followed by Tukey’s HSD test.
For further studies on the function of IMA1 and IMA2, we generated transgenic lines that expressed either gene under the control of the CaMV 35S promoter, leading to an increase in IMA1 transcript levels between ca. 260- and 300-fold in IMA1 overexpression lines (IMA1ox) and 320- to 1,500-fold of IMA2 in IMA2ox lines, compared to the wild-type (Figure 1C). In wild-type plants, growth on media with restricted Fe availability (nonavailable Fe [navFe] media; 10 μM FeCl3, pH 7.0) increased IMA1 and IMA2 expression by ca. three-fold; no further increase in IMA expression was observed in IMA1ox lines. Mutants harboring a defect in the expression of F6′H1 showed >20-fold increased transcript levels of both IMA genes relative to wild-type plants when grown under navFe conditions, possibly reflecting a severely low Fe status of the mutant plants (Figure 1C). Notably, overexpression of either of the two IMA genes under study appeared to antagonize the expression of the other. A plausible explanation is that the additional Fe acquired due to the increased abundance of IMA peptides represses the transcription of other IMA genes, while Fe-responsive genes downstream of and dependent on IMA are still induced.
Overexpression of IMA1/IMA2 supports Fe uptake at elevated pH
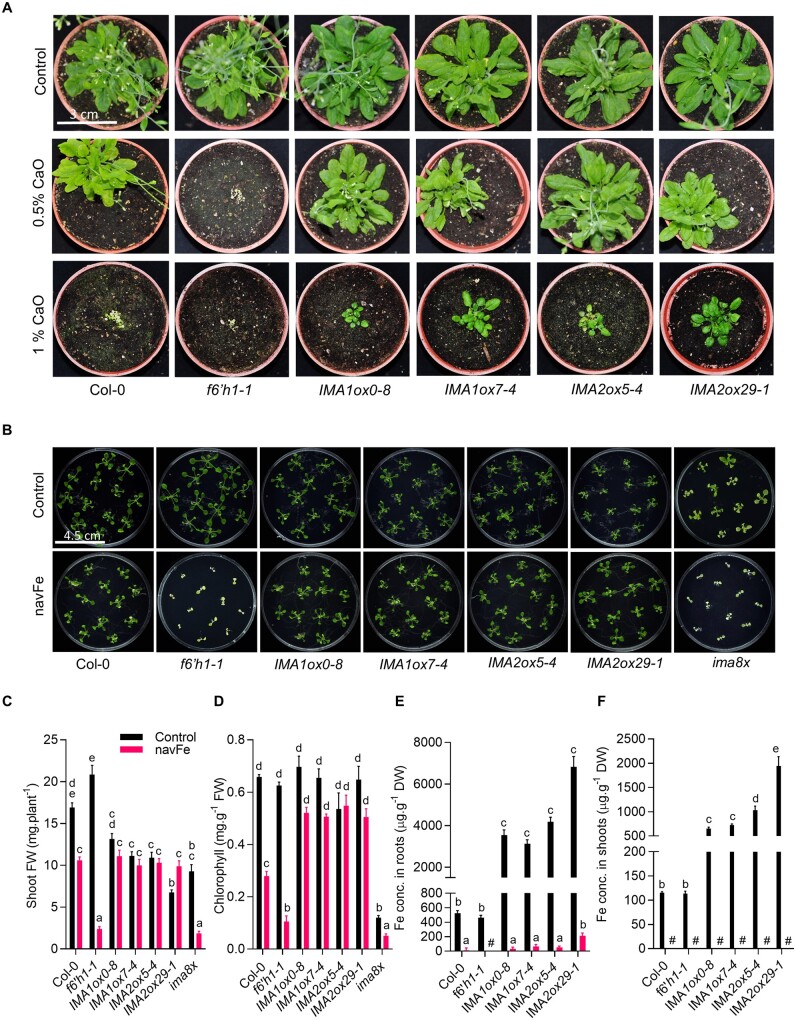
Growing plants on soil supplemented with calcium oxide (CaO) resulted in pronounced growth reduction in wild-type plants, which was more severe at higher (1%) CaO levels (Figure 2A). Plants grown on alkaline soil produced chlorotic leaves, indicative of restricted Fe availability. Mutants defective in the expression of F6′H1 showed a more severe phenotype than the wild-type, indicating that secretion of coumarins is critical for survival under such conditions. IMA1ox and IMA2ox lines showed markedly improved growth when compared to wild-type plants, a discrepancy which was more obvious in soil supplemented with 1% CaO (Figure 2A). IMAox plants grown under such conditions were bigger and greener than the heavily chlorotic wild-type plants, an observation which is likely attributable to improved Fe acquisition.
Figure 2.
Overexpression of IMA1 and IMA2 confers tolerance to alkaline conditions. A, Phenotypes of plants grown for 8 weeks on alkaline soil supplemented with CaO. B, Phenotypes of plants grown on ES (50 µM Fe-EDTA, pH 5.5; Control) or navFe (10 μM FeCl3, pH 7.0) media for 2 weeks. C, Shoot FW. D, Chlorophyll concentration. E and F, Fe concentration in roots (E) and shoots (F). DW, Dry weight; C–F, Each bar represents the mean ± se of four (seven for shoot FW) independent experiments. Distinct letters above the bar graphs indicate significant differences (P < 0.05) in two-way ANOVA followed by Tukey’s HSD test. Hash denotes not detected.
Growing plants on navFe media reduced growth of the wild-type by about 40% (Figure 2, B and C). When grown on media supplemented with 50 μM Fe-EDTA at pH 5.5, f6′h1 mutant plants were significantly bigger than the wild-type, matching previous findings (Rodríguez-Celma et al., 2013b). Growth of f6′h1 mutants on navFe media was, however, massively impaired, leading to ∼90% decrease in shoot fresh weight (FW) at the end of the experimental period (Figure 2, B and C). IMA1/IMA2ox lines grew slower than the wild–type under Fe-sufficient conditions (∼40% reduction in shoot FW on average), but did not show any significant growth reduction on navFe media. Octuple ima8x mutants, defective in the expression of all eight IMA genes, were highly chlorotic under all conditions, a phenotype that was more severe when grown on navFe media, where ima8x resembled f6′h1 mutant plants (Figure 2, B and C). Growth on navFe media further caused a pronounced decrease in chlorophyll concentration in wild-type and, more dramatically, in f6′h1 mutant plants. IMA1/IMA2ox lines exhibited a much less severe reduction in chlorophyll levels. On average, chlorophyll concentration of IMAox lines was decreased by 17% versus 58% in wild-type and 83% in f6′h1 mutant plants (Figure 2D). The lowest chlorophyll concentration was observed in ima8x mutants (Figure 2D).
Histochemical detection of Fe by Perls staining revealed massive accumulation of Fe in roots and shoots of both IMA1ox and IMA2ox lines under Fe-sufficient conditions and when grown on media supplemented with excess Fe (400 µM Fe-EDTA), suggesting that both genes play at least partly redundant roles in the regulation of Fe uptake (Supplemental Figure S2). Growing plants on navFe media abolished Fe staining completely in both wild-type and f6′h1 mutant plants. In IMA1/IMA2ox lines, staining was weak but still above the detection limit (Supplemental Figure S2), indicating that under such conditions, Fe uptake is less restricted in lines overexpressing either IMA1 or IMA2. Quantifying Fe levels in the genotypes under investigation supported the semi-quantitative analysis and revealed a massive increase in root and shoot Fe concentration in IMA1/IMA2ox lines under Fe-sufficient conditions, relative to wild-type plants (Figure 2, E and F). When grown on navFe media, Fe was barely (in roots) or not at all (in shoots) detectable (Figure 2, E and F). These very low Fe levels were likely caused by prolonged growth on navFe media, which leads to exhaustion of the internal Fe pools. To validate the supposition that the ameliorated chlorosis of IMA1/IMA2ox lines grown on navFe media was due to improved Fe acquisition, we analyzed plants that were pregrown on Fe-sufficient media for 7 d and subsequently transferred to navFe media for an additional 5 d. Fe levels remained low in all genotypes under these conditions, but IMA1/IMA2ox lines exhibited an on average >30-fold increase in Fe levels in roots and in shoots, suggesting that IMA peptides particularly support Fe uptake under conditions of restricted Fe availability (Supplemental Figure S3).
IMA regulates Fe acquisition at different levels
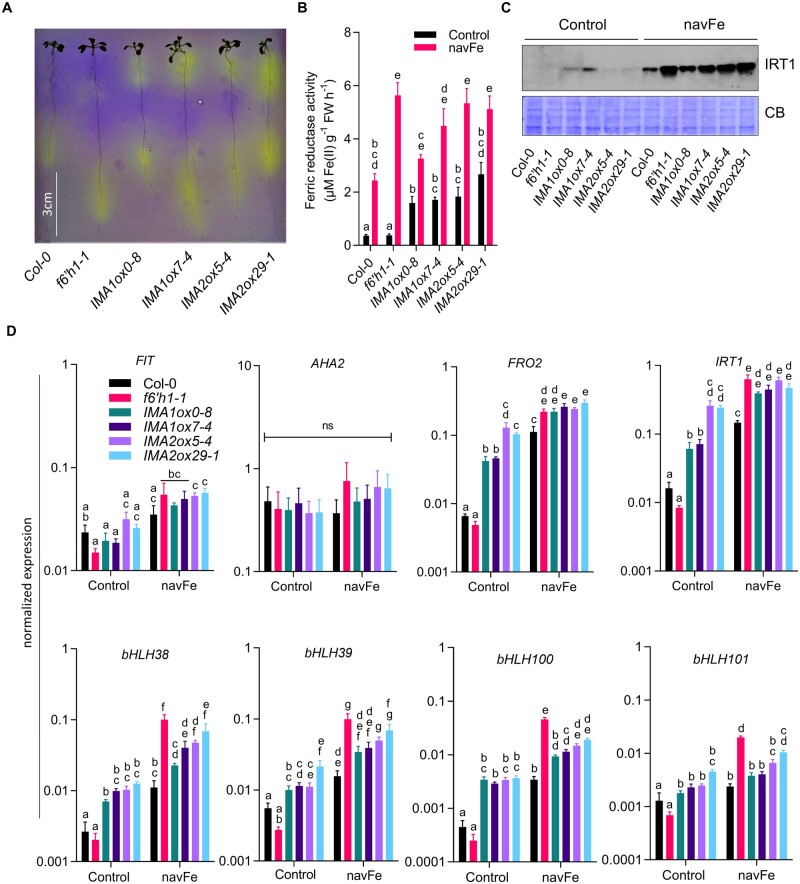
To investigate the effect of IMA1 and IMA2 on the different Fe acquisition processes, we first analyzed the capacity of the lines under study to acidify the rhizosphere. Under Fe-sufficient conditions, a low constitutive acidification confined to the root tip was recorded for wild-type and f6′h1 mutant plants; roots of f6′h1 plants were slightly more active (Figure 3A). IMA1/IMA2ox lines showed a more pronounced response, covering a larger root area (Figure 3A). The enzymatic reduction of ferric chelates is a key step in the strategy I-type of Fe acquisition. In wild-type plants, in vivo Fe chelate reduction (FCR) activity was increased by seven-fold when grown on navFe media (Figure 3B). While FCR activity of f6′h1 mutant plants was similar to wild-type plants under Fe-sufficient conditions, the mutant showed a much higher (ca. 15-fold) increase in activity when subjected to navFe conditions, possibly caused by the lack of Fe acquired via the secretion of Fe-mobilizing coumarins (Figure 3B). In both IMA1ox and IMA2ox lines, FCR activity was significantly increased under Fe-sufficient (ca. five-fold) and navFe conditions (ca. two-fold) relative to wild-type plants. Western blot analysis revealed that the Fe2+ transporter IRT1 is expressed at low levels under Fe-sufficient conditions, with a slight increase in IMA1ox and, to a lesser extent, in IMA2ox lines, when compared to wild-type plants (Figure 3C; Supplemental Figure S4). Under navFe conditions, the expression pattern of IRT1 protein resembled that of the in vivo FCR activity; the highest IRT1 accumulation was observed in f6′h1 plants and IMAox lines (Figure 3C).
Figure 3.
Fe acquisition processes are enhanced in IMA1 and IMA2 overexpression lines. A, Rhizosphere acidification by 8-day-old plants grown on ES media after transferring to plates containing the pH indicator bromocresol purple for 24 h. A representative image of three independent experiments is shown. Acidifcation is indicated by yellow color around the roots. B, Ferric reductase activity of 12-day-old plants grown on either ES (Control) or navFe media for 3 days, after 9 days of precultivation on ES media. C, Level of IRT1 protein (∼34 kDa) in root samples from 12-day-old plants grown on either ES or navFe media for 3 days, after 9 days of precultivation on ES media. Coomassie blue (CB) was used as loading control. Per lane, 10 µg of total protein was loaded. An image representative of three independent experiments is shown. D, Expression of major Fe uptake genes in roots of 12-day-old seedling grown on ES or navFe media for 3 days, after 9 days of precultivation on ES media. Gene expression was determined by RT-qPCR using ΔCT method and expression of elongation factor 1 alpha as an internal control. Each bar in (B) and (D) represents the mean ± se of four independent experiments. Distinct letters above the bar graph indicate significant differences (P < 0.05) in two-way ANOVA followed by Tukey’s HSD test.
Notably, transcript levels of the master regulator FIT and the P-type H+-ATPase AHA2 did not significantly differ among the treatments or genotypes under investigation, indicative of a chiefly posttranslational regulation of gene activity (Jahn et al., 2002; Meiser et al., 2011; Figure 3D). Instead, expression of FRO2 and IRT1 massively increased in IMA1/IMA2ox lines under all Fe regimes, with higher transcript levels under navFe conditions. When grown on navFe media, FRO2 and IRT1 mRNA levels in f6′h1 plants were higher than in the wild-type and were at the level of IMAox lines (Figure 3D). Genes encoding clade Ib bHLH proteins showed a pattern similar to FRO2 and IRT1, with a significant increase in IMAox lines under Fe-sufficient conditions. When grown on navFe media, all genes of the Ib bHLH clade, except bHLH101, were expressed at significantly higher levels in IMAox lines when compared with the wild-type.
Together, these data show that higher IMA levels activate proton extrusion, ferric reduction, and Fe uptake at various regulatory levels and in conjunction with the Fe status of the plants. The data further indicate that the higher FRO2 activity and IRT1 protein levels of f6′h1 mutants are insufficient for survival at elevated pH, suggesting that the secretion of Fe-mobilizing coumarins is indispensable for plants to thrive under such conditions. It can further be concluded that the improved growth of IMAox lines on media with low Fe availability is not solely caused by induction of the AHA2-FRO2-IRT1 module.
The composition of secreted coumarins is dependent on the pH/Fe regime
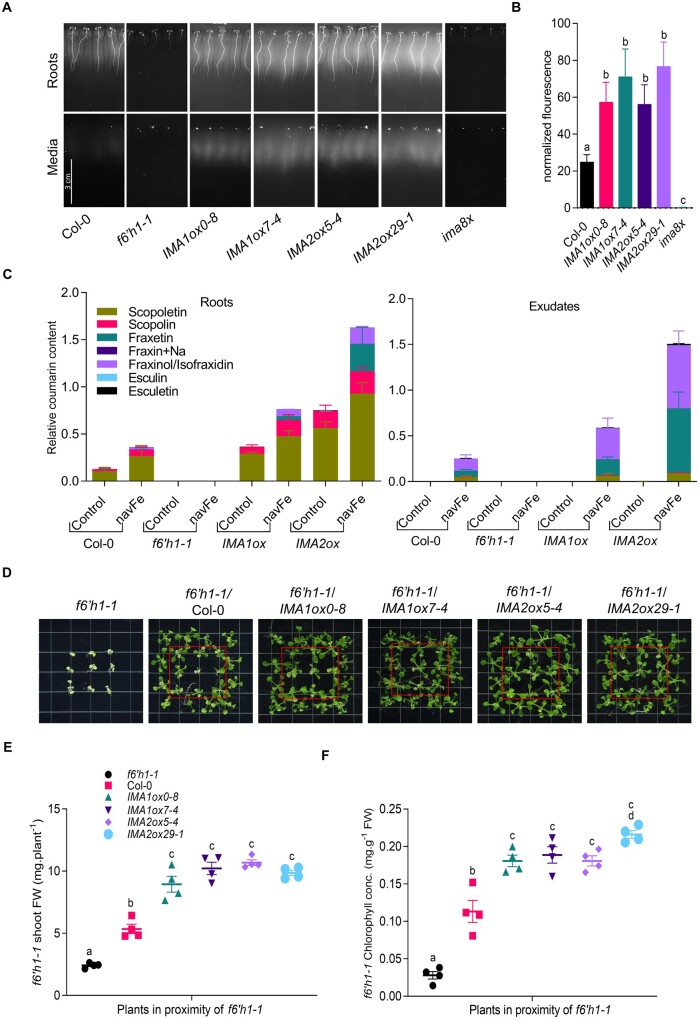
The quantities of coumarins in roots and in the media can be noninvasively approximated by detecting autofluorescence as an inherent property of some compounds such as scopoletin, scopolin, and esculin. Under Fe-sufficient conditions, only a faint signal was observed in roots of all genotypes; IMAox lines emitted a slightly more intense signal (Supplemental Figure S5A). When grown on navFe media, a significant increase in autofluorescence was noted for IMA1/IMA2ox lines relative to wild-type plants (Figure 4, A and B). In both roots and media of f6′h1 and ima8x mutants, fluorescent coumarins remained below the detection limit under all conditions. Media of IMA1/IMA2ox lines showed not only pronounced fluorescence, but also a yellowish color, possibly attributable to the formation of fraxetin–Fe complexes (Rajniak et al., 2018; Supplemental Figure S5B).
Figure 4.
Overexpression of IMA1 and IMA2 supports the secretion of Fe-mobilizing coumarins under navFe conditions. A, Accumulation of fluorescent coumarins in roots and media of 7-d-old plants grown on navFe media. Photos were captured using 365 nm as the excitation wavelength, before and after removing plants from the media. Representative images of three independent experiments are shown. B, Quantification of root exudate fluorescence. Each bar represents the mean ± se of four independent experiments. Distinct letters above the bar graph indicate significant differences (P < 0.05) in one-way ANOVA followed by Tukey’s HSD test. C, Relative abundance of selected coumarins in roots and exudates of plants grown on ES (Control) or navFe media. Bars represent the relative content of different coumarin compounds presented as peak area normalized to the number of roots and internal standard peak area. Values are mean relative coumarin content ± se obtained from three independent experiments. For coumarin extraction from roots, circa 20 plants were used for each replicate. D, Rescue of the phenotype of f6′h1-1 mutant plants (in red boxes) by wild-type, IMA1ox, or IMA2ox plants grown alongside mutant plants on navFe media. Representative images of four independent experiments are shown. E, Shoot FW of rescued f6′h1-1 plants. F, Total chlorophyll concentration of leaves from rescued f6′h1-1 plants. Grouped interleaved scatter plot show sem of the replicates.
Under Fe-sufficient conditions, targeted coumarin analysis identified scopoletin and its glycoside scopolin as the main phenolic compounds in roots. In contrast to the lack of fluorescence under Fe-sufficient conditions, IMAox lines showed a marked increase in the level of scopolin and scopoletin, compounds that constitute the major contributors to root fluorescence (Döll et al., 2018; Figure 4C). The lack of fluorescence in the roots under Fe-sufficient, slightly acidic conditions can be attributed to a pH-dependent shift in the spectra of fluorescent coumarins such as scopoletin, an assumption that was confirmed by UV–Vis spectroscopy (Supplemental Figure S6, A and B). When grown on navFe media, small amounts of fraxinol/isofraxidin were detected in the wild-type in addition to scopolin and scopoletin, which account for the largest part of the detected coumarins. In roots of IMA1/IMA2ox lines, all identified coumarins were increased relative to the wild-type, including the Fe-mobilizing compound fraxetin, which was detectable only in very low quantities in the wild-type. IMA1ox and IMA2ox lines accumulated 28- and 200-fold more fraxetin in the roots than the wild-type, respectively (Figure 4C).
As for exudates, fraxinol/isofraxidin and fraxetin were the most prominent compounds for wild-type plants, and scopoletin was detected in minor amounts. Similar to what was observed in roots, media of IMA1/IMA2ox lines contained much higher levels of coumarins than that of wild-type plants, and this effect was particularly pronounced when fraxetin levels are considered. IMA1ox and IMA2ox lines secreted 3- and 10-fold more fraxetin into the media than wild-type plants, an observation which matches the better performance of these lines on alkaline soil. In contrast, scopoletin levels did not differ much among the lines, except for f6′h1 mutants, which neither produced nor secreted any detectable amount of the coumarins covered in the analysis (Figure 4C). The difference in fraxetin secretion between wild-type plants and IMA1/IMA2ox lines was also observed in experiments aimed at rescuing the mutant phenotype of f6′h1 plants. Growing f6′h1 mutant plants alongside Columbia-0 (Col-0) doubled the shoot FW and increased chlorophyll concentration of the mutant by approximately four-fold (Figure 4, D–F). Replacing wild-type plants with IMA1/IMA2ox lines further improved growth and chlorophyll concentration of f6′h1 plants, supporting the notion that the coumarins secreted by IMA1/IMA2ox lines are biologically active and qualitatively or quantitatively superior to those of the wild-type (Figure 4, D–F).
Together, these data indicate that secretion of Fe-mobilizing compound requires an unidentified Fe deficiency/high pH signal; in contrast to the pronounced induction of proton extrusion, FCR activity, and IRT1 accumulation in the presence of Fe, no coumarins are secreted by IMA1/IMA2ox lines under Fe-sufficient conditions.
The expression of genes involved in coumarin biosynthesis is controlled by pHe and Fe
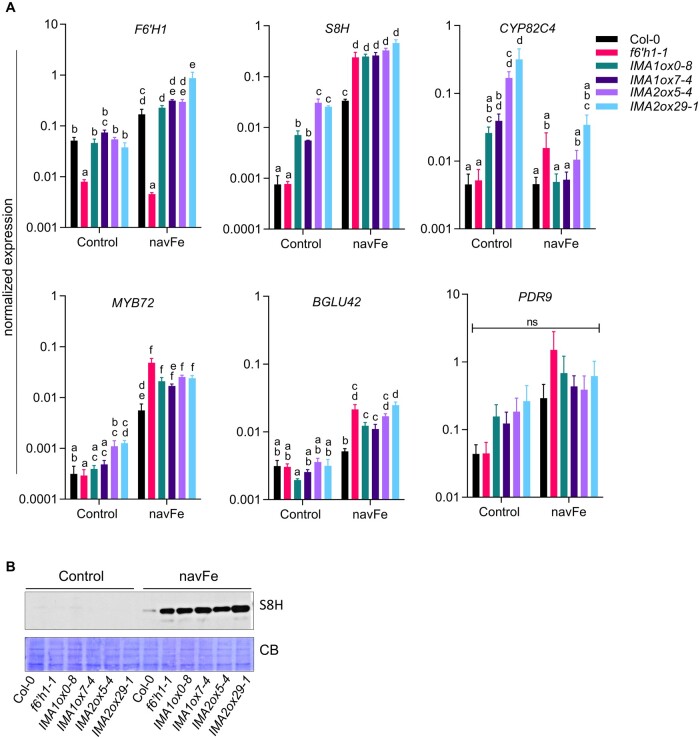
To further investigate the importance of the Fe/pH regime for the secretion of coumarins, we determined the transcript levels of genes involved in coumarin secretion under Fe-sufficient (pH 5.5) and navFe conditions (pH 7.0). Under Fe-sufficient conditions, the expression of F6′H1 did not change in lines overexpressing IMA1/IMA2, relative to wild-type plants (Figure 5A). Growing plants on navFe media increased F6′H1 mRNA levels significantly, with no apparent differences among the genotypes, except for f6′h1 mutant plants, which showed low transcript levels under all conditions. Expression of S8H, which mediates the conversion of scopoletin to fraxetin, was increased by overexpression of IMA under Fe-sufficient conditions, and strongly induced in all lines under navFe conditions, with higher expression in all IMAox lines relative to wild-type plants. Conspicuously, an increase in S8H transcript levels relative to wild-type plants was also observed in f6′h1 mutants under navFe, but not under Fe-sufficient conditions, possibly due to more depleted Fe levels of the plants under these conditions (Figure 5A). The strong induction of S8H was consistent with higher accumulation of S8H protein in the f6′h1 mutant and IMAox lines (Figure 5B; Supplemental Figure S4). While overexpression of IMA1 and IMA2 highly increased CYP82C4 transcript levels under (acidic) Fe-sufficient conditions, such an increase was observed neither in wild-type nor in IMAox plants when plants were grown under navFe conditions. Instead, in all lines transcription remained at the low levels observed under control conditions (Figure 5A).
Figure 5.
Expression of coumarin biosynthesis-related genes in IMA1/IMA2ox lines. RT-qPCR analysis of gene expression in roots of 12-d-old seedlings grown on ES (Control) or navFe media for 3 d, after 9 d preculture on ES media. Gene expression was determined using the ΔCT method with elongation factor 1 alpha as an internal control. Each bar represents the mean ± se of four independent experiments. Distinct letters above the bar graph indicate significant differences (P < 0.05) in two-way ANOVA followed by Tukey’s HSD test. B, Level of S8H protein (∼40 kDa) in root samples from 12-d-old plants grown on either ES or navFe media for 3 d, after 9 d of precultivation on ES media. CB was used as loading control. Per lane, 10 µg of total protein was loaded. An image representative of three independent experiments is shown.
The expression pattern of MYB72, a transcription factor that is critical for growth under Fe-limiting, alkaline conditions (Palmer et al., 2013), resembled that of S8H, except for a lack of pronounced induction of MYB72 by overexpression of IMA1/IMA2 under Fe-sufficient conditions (Figure 5A). Induction of the MYB72-dependent BETA GLUCOSIDASE42 (BGLU42), which mediates the deglycosylation of scopoletin, and possibly other coumarin glycosides, as a necessary step before secretion into the rhizosphere (Zamioudis et al., 2014), was observed under navFe conditions, but BGLU42 transcript levels did not differ among the genotypes when grown on Estelle and Somerville (ES) media. No statistically significant changes were observed for the expression of the coumarin exporter PDR9, although a trend toward higher transcript levels was observed under navFe conditions (Figure 5A).
Environmental pH modulates gene transcription
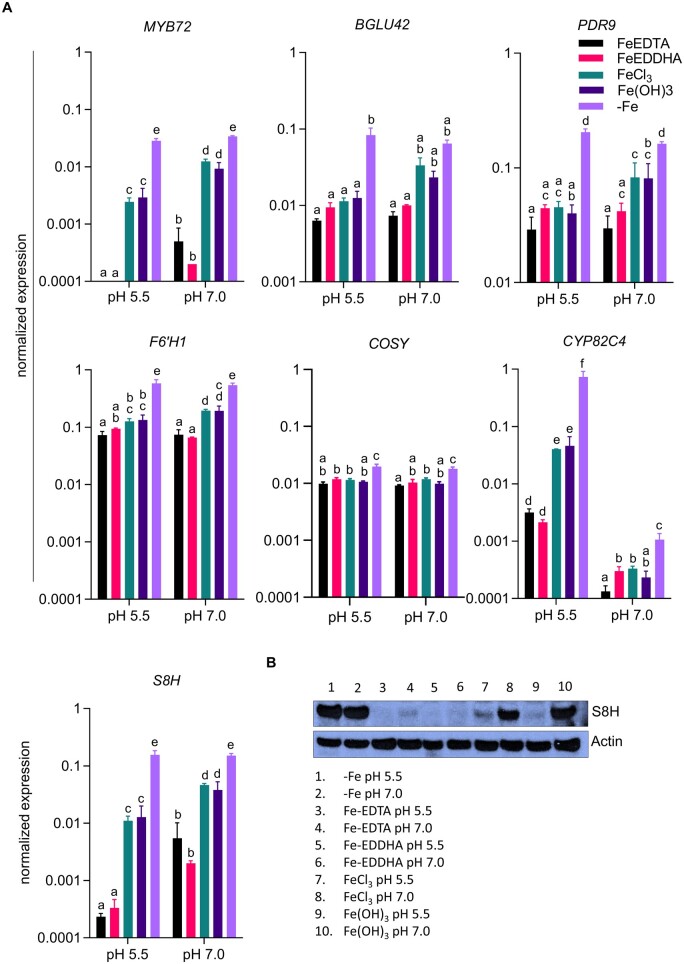
To distinguish the effect of pHe from that of Fe availability, we determined the transcript levels of genes involved in the secretion of Fe-mobilizing coumarins at various Fe/pH regimes. The activity of Fe in the media is dependent on the stability of the Fe compound, which in turn is strongly affected by pH. To account for this effect, we grew plants on Fe-deficient media (+ferrozine), and media supplemented with various Fe sources, including Fe-EDTA (unstable at elevated pH), Fe-ethylenediamine-N,N′-bis (FeEDDHA; stable over the pH range under investigation), FeCl3 (low solubility particularly at elevated pH), and precipitated Fe(OH)3 (very low solubility over the whole pH range; Boxma, 1981; Norvell, 1991; Lucena and Chaney, 2006). Seedlings grown on media containing Fe sources that are stable at elevated pH (FeEDDHA or Fe-EDTA) showed no or very mild chlorosis and no changes in Fe concentration when compared to plants grown at pH 5.5. However, supplementing the media with FeCl3 caused dramatic decreases in chlorophyll levels and Fe concentration (Supplemental Figure S7). At pH 5.5, expression of MYB72 was strongly induced by Fe deficiency and, to a lesser extent, by low Fe availability (Figure 6A). At pH 7.0, MYB72 transcript levels were much higher under all conditions, except for Fe-deficient media where expression appears to reach maximum transcription rates which cannot be further enhanced. All coumarin-related genes showed the highest transcript levels on Fe-deficient media. The transcription of BGLU42, F6′H1, and COSY was not much affected by the various growth conditions (Figure 6A). Instead, S8H expression was strongly induced by Fe deficiency at pH 5.5 and by supplementing the media with FeCl3 or Fe(OH)3 (Figure 6A). Growth on media with elevated pH increased the message levels of S8H under all conditions (except for Fe-deficient media), particularly when plants were grown on media supplemented with less soluble Fe compounds. Generally, the S8H expression pattern resembles that of MYB72 (Figure 6A). Western blot analysis mirrored the high induction of S8H by Fe deficiency at both acidic and elevated pH and the pH-dependent expression in all treatments, with protein abundance increasing with decreasing stability of the added Fe compounds (Figure 6B). While expression of CYP82C4 showed a pattern similar to that of S8H at pH 5.5, growth at pH 7.0 strongly downregulated transcript levels in all treatments (Figure 6A). Although inducibility by Fe deficiency was still observable at neutral pH, expression levels of CYP82C4 were >600-fold lower at pH 7.0 when compared to pH 5.5, suggesting that at elevated pH, fraxetin is favored over sideretin production. The expression of FIT, IMA1, and IMA2 was not much affected by the Fe/pH regimes under study, except for a strong induction of all three genes by the absence of Fe, which was not influenced by pH. Interestingly, expression of IMA3 showed a different pattern, being strongly repressed by stable Fe compounds (Fe-EDTA and Fe-EDDHA) at elevated pH (Supplemental Figure S8). This decrease at elevated pH, albeit less pronounced, was also observed for IMA1 and IMA2 when grown on media supplemented with Fe-EDDHA.
Figure 6.
Media pH and Fe availability control the expression of coumarin-related genes. Plants were directly germinated for 14 d on either Fe-deficient media (with 120 µM ferrozine) or media supplemented with either Fe-EDTA (40 µM), FeEDDHA (40 µM), FeCl3 (40 µM), or precipitated Fe(OH)3 (40 µM) at pH 5.5 or pH 7.0. A, Expression of MYB72, BGLU42, PDR9, F6′H1, COSY, CYP82C4, and S8H in roots. Gene expression was determined using the ΔCT method with elongation factor 1 alpha as an internal control. Each bar represents the mean ± se of three independent experiments. Distinct letters above the bar graph indicate significant differences (P < 0.05) in two-way ANOVA followed by Tukey’s HSD test. B, Expression of S8H protein (∼40 kDa) in root samples. Actin (∼45 kDa) was used as loading control. Per lane, 20 µg of total protein were loaded. An image representative of three independent experiments is shown.
In conclusion, the results from these experiments suggest that depending on the external pH, IMA1/IMA2 support the biosynthesis and secretion of different coumarins. Under acidic conditions, sideretin production is favored through upregulation of both S8H and CYP82C4. At elevated pH and in the absence of available Fe, S8H is highly induced while CYP82C4 expression is strongly repressed, supporting the production of fraxetin, which forms more stable complexes with Fe under such conditions.
Discussion
IMA overexpression confers calcicole behavior
The Arabidopsis genome harbors eight IMA genes, of which only two (IMA1/FEP3 and IMA3/FEP1) have been functionally explored (Grillet et al., 2018; Hirayama et al., 2018). IMA1 and IMA2 share an identical C-terminal consensus motif characteristic of all IMA peptides across angiosperms, exhibit similar expression patterns, and induce Fe acquisition responses in a similar manner and to a comparable extent, suggesting that the two genes are to a large degree genetically redundant. In leaves, IMA1 and IMA2 are tightly co-expressed with IMA4 and genes that have been associated with cellular Fe homeostasis. Notably, in roots, expression of IMA1/2 is closely associated with NAS4, a putative target of MYB72 (Palmer et al., 2013). Homozygous myb72 and nas4 mutants display similar phenotypes, featuring lower chlorophyll and decreased Fe levels when compared with the wild-type. Overexpression of NAS4 rescued myb10myb72 mutants (Palmer et al., 2013), indicating that NAS4 is of particular importance for the adaptation to alkaline conditions with low Fe availability.
The enzymatic reduction of ferric chelates is the rate-limiting step of strategy I-type Fe uptake, and determines the efficiency by which Fe is taken up (Grusak et al., 1990). Under Fe-sufficient (and slightly acidic) conditions, elevated IMA levels increase the expression of FRO2 and in vivo FCR activity, which is causative for increased Fe levels in all plant parts, including seeds (Grillet et al., 2018). However, activated FCR activity and increased proton extrusion of f6′h1 plants are insufficient to alleviate the pronounced growth inhibition of the mutant on media with elevated pH, indicating that the secretion of Fe-mobilizing coumarins is indispensable under such conditions. In IMAox lines, fraxetin secretion was massively increased under permissive conditions (low Fe status of the plants and elevated pH), an increase that was much more pronounced than that observed for other Fe acquisition responses. Conspicuously, under Fe-sufficient conditions, an IMA-dependent induction was observed for the synthesis of scopolin and scopoletin but not for fraxetin, indicating that IMA-mediated fraxetin production requires both a low Fe status and high pHe, thereby prioritizing the secretion of fraxetin over that of the upstream (scopoletin) and downstream products (sideretin). At elevated pH, a concurrent, unweighted induction of all steps of the coumarin pathway may be counterproductive for Fe acquisition, since sideretin secretion will reduce fraxetin levels and thus compromise the mobilization of Fe from recalcitrant pools in soils.
Similar to IMAox lines, in wild-type plants, the most pronounced accumulation and secretion of fraxetin was observed under the combined influence of elevated pHe and Fe deficiency; no such pattern was observed for scopoletin (Sisó-Terraza et al., 2016). A pH-dependent increase in the secretion of phenolic compounds was also reported for soybean and mung bean plants (Waters et al., 2018; Nair et al., 2020), suggesting that the pH response is conserved in some, if not most, nongrass species. We show here that in the presence of the pH-stable Fe source Fe-EDDHA, an increase in media pH from 5.5 to 7.0 led to a markedly higher (ca. six-fold on average) expression of S8H. Under similar conditions, CYP82C4 transcript levels were decreased by the same factor. At elevated pH, the Fe deficiency-induced expression of CYP82C4 was compromised, amplifying the difference in transcript levels between pH 5.5 and pH 7.0 by >600-fold. Similar to elevated pH ranges, under acidic conditions, an indiscriminate induction of all steps of the coumarin biosynthetic pathway would be disadvantageous for plant fitness; fraxetin synthesis would cause an unfavorable decrease of the phytoalexin scopoletin and decrease the production of sideretin, which would deplete the amount of chelated Fe as a substrate for FRO2-mediated ferric reduction. It should be noted that the expression of both S8H and CYP82C4 is controlled by FIT (Colangelo and Guerinot, 2004) and inducible by Fe deficiency at the transcript and protein level under acidic conditions (Lan et al., 2011; Grillet et al., 2018). This finding implies that trans-acting factors can modulate or abolish FIT-dependent induction in response to pHe. In contrast to yeast, where ambient pH is perceived by a sensor complex at the cell surface and communicated via a well-described signaling cascade to drive the expression of genes adapting the cell to an alkaline environment (Obara and Kihara, 2014; Peñalva et al., 2014), such pHe signaling has not yet been described in plants (Raven, 1990; Tsai and Schmidt, 2021). However, changes in media pH were shown to rapidly and thoroughly alter gene expression profiles in Arabidopsis roots, strongly suggesting that plants are capable of monitoring and responding to changes in pHe (Lager et al., 2010; Tsai and Schmidt, 2020). It is noteworthy that some of the genes under study did not show sensitivity toward changes in environmental pH, excluding nonspecific secondary effects of pHe on gene expression.
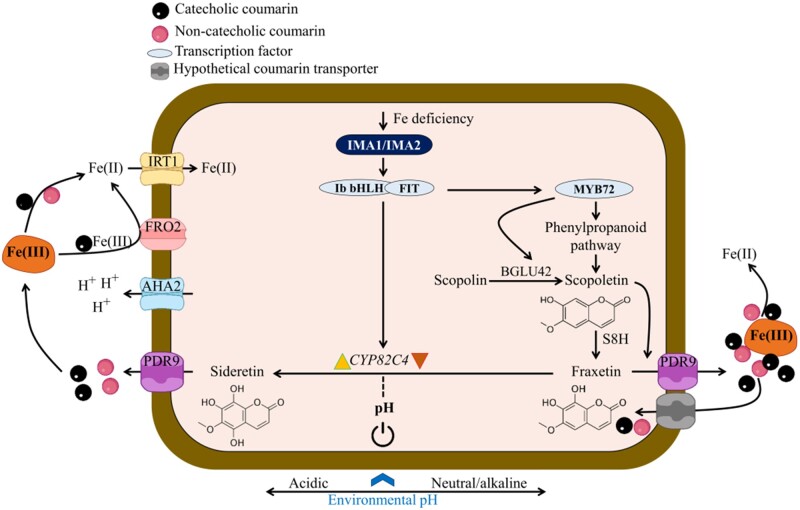
Fe deficiency responses fractionate into a calcifuge and a calcicole branch
The strategy I-type Fe deficiency response constitutes a co-operative entity, which orchestrates Fe acquisition processes at low Fe availability. Our data imply that different branches of the Fe deficiency syndrome are calibrated in a pHe-dependent manner, accentuating processes that are most efficient under a given set of edaphic conditions (Figure 7). The balance between the two branches of the Fe deficiency response is tipped by the pHe, inducing a “calcifuge” suite of processes at acidic pH, and a “calcicole” response at elevated pH. Under acidic conditions, Fe is mainly acquired via enzymatic reduction and taken up through FRO2/IRT1. It can be assumed that CYP82C4 activity and sideretin production are closely associated with the AHA2-FRO2-IRT1 module, a supposition that is corroborated by the extremely close correlation of IRT1 and CYP82C4 expression (http//:atted.jp), and the identification of CYP82C4 as a potential interactor of IRT1 (Martín-Barranco et al., 2020). Sideretin exhibits maximal Fe-mobilizing pH capacity at lower pH ranges when compared to fraxetin (Rajniak et al., 2018; Tsai et al., 2018), implying that sideretin secretion is part of the calcifuge branch. A pH-dependent prioritization of the Fe acquisition process is of ecological importance, since it profoundly increases the range of hydrogen activities in which Fe can be assimilated from the soil.
Figure 7.
Model for the pH-dependent induction of the calcifuge (left) and calcicole (right) branches of the Fe-deficiency response. The expression of IMA genes is induced by a low Fe status of the plant, triggering the induction of two distinct suites of responses in correspondence with pHe. Environmental pH dictates the biosynthesis of either sideretin or fraxetin by supporting the expression of CYP82C4 at low pHe and repressing CYP82C4 transcription at elevated pHe (indicated by yellow and brown arrowheads). At acidic pH, IMAs support the acquisition of Fe from Fe hydroxides by inducing proton extrusion, sideretin secretion, FCR, and uptake of Fe2+ via IRT1. The calcicole branch is positively regulated by MYB72. At neutral or alkaline pH ranges, IMAs induce the biosynthesis and secretion of fraxetin via PDR9, and Fe is mobilized via reduction and chelation. The β-glucosidase BGLU42 is prioritized over other BGLUs and required to facilitate the secretion of scopoletin, and presumably other coumarins, by catalyzing their deglycosylation. Re-uptake of coumarins into the roots as such or as a coumarin–Fe complex is mediated by an as yet unidentified transporter (Robe et al., 2020a). The two branches are not mutually exclusive, but prioritized by the prevailing pHe. The blue arrow indicates a putative pH signal, tipping the balance (denoted by black circle) of induction (yellow arrowhead) and repression (brown arrowhead) of CYP82C4.
The transcription factor MYB72 was found to be critical for growth on alkaline soil (Palmer et al. 2013). In this study, we show that MYB72 expression is highly pH-dependent, exhibiting a pattern similar to S8H but contrary to CYP82C4, which places MYB72 in the calcicole branch of the response. The β-glucosidase BGLU42 is a putative target of MYB72 (Palmer et al., 2013), is highly responsive to Fe deficiency (Grillet et al., 2018), and was shown to deglycosylate scopolin (Ahn et al., 2010; Stringlis et al., 2018). Interestingly, under navFe conditions, overexpression of IMA1/2 significantly increases BGLU42 expression relative to wild-type plants (Figure 5), while the expression of three other scopoletin-hydolyzing β-glucosidases from the same clade, BGLU21, BGLU22, and BGLU23, was decreased under such conditions (Ahn et al., 2010; Tsai and Schmidt, 2020). Moreover, BGLU21 and BGLU22 are downregulated in IMA1ox lines under Fe-deficient conditions (Grillet et al., 2018), suggesting that BGLU42 could be involved in the deglycosylation of coumarins, including fraxetin, at elevated pHe. It may thus be assumed that high pH triggers a module of the Fe-deficiency response that comprises MYB72, S8H, and BGLU42, which supports the secretion of fraxetin and confers calcicole behavior (Figure 7). An open question is whether secreted coumarins can be transported back into the cell together, either as such or as a complex with Fe. Uptake of the secreted coumarins, analogously to phytosiderophores released by grasses, would be ecologically advantageous in view of energy conservation and the low efficiency of enzymatic Fe3+ reduction at neutral or alkaline pH. Glycosylated coumarins were detected in f6′h1 plants when grown alongside wild-type plants, or when scopoletin, esculetin, or fraxetin was provided exogenously, suggesting that coumarins can be taken up and stored as glycosides in the vacuole (Robe et al., 2020a). Whether or not the coumarins taken up from the soil solution can be transported into the cell as coumarin–Fe complex by an auxiliary Fe uptake system remains to be elucidated.
Conclusions
In summary, we show here that high IMA1/IMA2 levels partly overrule the downregulation of Fe uptake at sufficient internal Fe levels and induce rhizosphere acidification, Fe3+ reduction, Fe2+ uptake, and the biosynthesis and secretion of scopoletin by acting as transcriptional activators. A complex, pH-dependent regulation was observed for the secretion of Fe-mobilizing catecholic coumarins, prioritizing the production of fraxetin over sideretin at elevated pH. Notably, at elevated pH, f6′h1 (defective in secreting catecholic coumarins) and ima8x mutants (defective in all Fe deficiency responses) showed similar phenotypes, indicating that a functional AHA2-FRO2-IRT1 module is insufficient to enable growth under such conditions.
Root-secreted coumarins not only act directly on Fe uptake via mobilization of recalcitrant Fe pools, but also indirectly by retracting pathogens and attracting beneficial bacteria, which in turn improve the access to mobile Fe in soils (Stringlis et al., 2019; Voges et al., 2019; Harbort et al., 2020). While such beneficial effects on plant growth were shown for scopoletin and fraxetin, sideretin does not appear to play such a role (Harbort et al., 2020). It may be concluded that the dual function of fraxetin in Fe acquisition is critical for conferring fitness at elevated pH. Manipulating the expression of IMA/FEP peptides constitutes a promising approach to generate germplasm adapted to low Fe availability. Ectopic expression of IMAs results in an increase in Fe in both strategies I and II crops such as rice (Oryza sativa) and tomato (Solanum lycopersicum), with increased Fe content in grains and fruits, respectively (Grillet et al., 2018; Kobayashi et al., 2020), making IMAs a promising candidate for improving Fe acquisition of virtually all crops. In particular, crops grown on calcareous soils may profit from improved growth and nutritional quality of edible plant parts.
Materials and methods
Plant materials and growth conditions
Seeds of Arabidopsis (A. thaliana (L.) Heynh) accession Col-0 and f6′h1-1 (Salk_132418C) T-DNA insertion mutants were obtained from the Arabidopsis Biological Resource Center (Ohio State University). The lines carrying CaMV pro35S::IMA1, and ima8x octuple mutants have been described previously (Grillet et al., 2018). Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998) was used to transform wild-type plants with pH2GW7 (obtained from the VIB-UGent Center for Plant Systems Biology) containing CaMV 35S promoter-driven IMA2 CDS (using the primers listed in Supplemental Table S1), yielding IMA2 overexpression lines. Overexpression in transgenic plants was confirmed by RT-qPCR. Plants were grown in a growth chamber on solid nutrient media as described by Estelle and Somerville (1987) (ES media), composed of 5 mM KNO3, 2 mM MgSO4, 2 mM Ca(NO3)2, 2.5 mM KH2PO4, 70 µmM H3BO3, 14 µM MnCl2, 1 µM ZnSO4, 0.5 µM CuSO4, 0.01 µM CoCl2, 0.2 µM Na2MoO4, 1% (w/v) MES, and 1.5% (w/v) sucrose, solidified with 0.4% Gelrite pure (Kelco). Unless stated otherwise, media were supplemented with 50 µM Fe-EDTA or 400 µM Fe-EDTA (Fe excess; Fe2+). The pH was adjusted to 5.5. Media with different Fe sources were prepared by replacing Fe-EDTA with 40 μM of either Fe-EDDHA, FeCl3, or Fe(OH)3. For Fe-deficient media, 120 μM ferrozine was added to the media. For preparing navFe media, 10 μM FeCl3 was added as an Fe source and the pH was adjusted to 7.0 with KOH. For all media adjusted to pH 7.0, MOPS (1 g/L) was used as buffering agent instead of MES. Seeds were surface-sterilized with 30% (v/v) commercial bleach containing 6% NaClO and 70% (v/v) ethanol for 5 min, followed by five rinses with sterile MQ water, and stratified for 2 d in the dark at 4°C before transferring to a growth chamber. Plants were grown at 21°C under continuous illumination (50 μmol m2s−1). Soil of variable alkalinity was prepared by mixing 0.5% or 1% CaO (w/w) with peat-based soil.
RT-qPCR
Unless stated otherwise, 9-d-old plants were transferred from ES media to either fresh ES or navFe media for 3 d. At the end of the experimental period, samples were immediately frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and treated with DNase using TURBO DNA-free kit (Ambion, Austin, TX, USA). cDNA was synthesized using DNA-free RNA with oligo-dT (20) primers and SuperScript III First-Strand Synthesis System for RT-qPCR (Invitrogen, Waltham, MA, USA). After incubation at 50°C for 1 h and 70°C for 15 min, 1 μL of RNase H was added and samples were incubated for 20 min at 37°C. The first-strand cDNA was used as a PCR template in a 10 μL reaction system using the SYBR Green PCR Master Mix (Applied Biosystems, Waltham, MA, USA) with programs recommended by the manufacturer in an AB QuantStudio Real-Time PCR System (Applied Biosystems). Three independent replicates were performed for each sample. The ΔCT method was used to determine the relative amount of gene expression, with the expression of elongation factor 1 alpha (EF1α; At5g60390) as an internal control. Primers used for RT-qPCR are listed in Supplemental Table 1.
Biomass and chlorophyll concentration
For biomass determination, whole shoots of 12- or 14-d-old seedlings grown on ES or navFe media were harvested and chlorophyll was extracted with 80% acetone. Total chlorophyll was calculated from absorbance measured at 645 and 663 nm using a protocol from Wellburn (1994).
Fe concentration
Total Fe concentration in shoots and roots was determined using a protocol described by Pan et al. (2015). Shoot and root samples (20–30 seedlings per sample) were oven-dried at 55°C for 2 d and digested with 65% (v/v) HNO3 at 100°C for 6 h, followed by 30% (v/v) H2O2 at 56°C for 2 h. Digested samples were mixed in assay solution containing 1 mM bathophenanthroline disulfonate (BPDS), 0.6 M sodium acetate, and 0.48 M hydroxylamine hydrochloride. The concentration of the resulting Fe2+–BPDS3 complex was measured at A535 using a PowerWave XS2 microplate spectrophotometer (BioTek, Winooski, VT, USA). The Fe concentration in different samples was calculated by plotting OD values against a standard curve made with FeCl3.
Perls staining for Fe(III)
Two-week-old Arabidopsis seedlings were vacuum infiltrated with Perls solution (2% HCl and 2% K4Fe(CN)6) for 30 min as described by Roschzttardtz et al. (2009). Samples were then rinsed 3 times with distilled water. Following staining, chlorophyll was removed from leaf samples with 80% acetone. Samples were rinsed thrice with distilled water and micrographs were taken with a Zeiss Lumar V12 Stereo microscope.
Rhizosphere acidification
For visualizing rhizosphere acidification, seedlings were germinated vertically on ES media with 0.5 g/L MES (instead of 1.0 g/L used in ES media) for 7 d and then carefully transferred to 1% BACTO™ agar media containing 0.006% Bromocresol Purple and 0.2 mM CaSO4 for 24 h. The pH was adjusted to 6.1 with NaOH. Pictures were taken under white light at the end of the experimental period.
Ferric chelate reduction activity
In vivo root FCR activity was measured on 12-d-old seedlings after 3 d of growth on either Fe-replete or Fe-deplete media as described previously (Grillet et al., 2014). FCR activity was determined individually on roots of 5–10 intact seedlings. Plants were incubated in the dark for 30 min with mild shaking in a 2 mL assay solution consisting of 100 µM Fe-EDTA and 300 µM BPDS in 10 mM MES at pH 5.5. Spectrophotometric absorbance at 535 nm was recorded with a PowerWave XS2 plate reader (BioTek Instruments) to determine Fe2+–BPDS3 concentrations using a standard curve. Experiments were conducted at least 3 times independently.
Protein extraction and western blot analysis
Total protein from liquid-nitrogen-ground root tissues was extracted as described previously (Tsai et al., 2018). Equal amounts of total protein were loaded and separated on Nu-PAGE 4%–12% Bis–Tris gels (Invitrogen). Proteins were immune-detected using anti-IRT1 (Agrisera, Vännäs, Sweden; AS11 1780) or anti-S8H (Tsai et al., 2018) as primary antibodies followed by detection with secondary antibodies (Agrisera; AS09 602 and GE Healthcare, Chicago, IL, USA; A934-100UL, respectively). The chemiluminescence signal detection was performed using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA, USA) on X-ray film. Monoclonal anti-actin (plant) antibody detected by the secondary antibody (GE Healthcare; NA931-1ML) was used as a loading control (Sigma-Aldrich, St Louis, MO, USA; A0480).
Detection of fluorescent compounds in roots and media
The accumulation of fluorescent compounds in the roots and secreted into the media was visualized with a Bio Spectrum 600 imaging system (UVP) at 365 nm excitation wavelength, SYBR Gold 485 to 655 nm as emission filter, and exposure time set to 9 s as described previously (Tsai et al., 2018). The fluorescent intensity was quantified using the ImageJ tool. To record the UV–Vis spectra, a Varian Carry 5000 spectrophotometer equipped with a 1-cm quartz cuvette was used.
Coumarin extraction from roots and media
Coumarins were extracted following a protocol adapted from Tsai et al. (2018). For each sample, 18–20 roots frozen in liquid nitrogen were finely ground using glass beads in a Tissue Lyzer II (Qiagen). Subsequently, extraction was performed by adding 500 µL of 80% (v/v) methanol containing 0.5 ppm 4-methylumbelliferone as an internal standard into the finely ground samples. Samples were vigorously vortexed for 15 min. Following centrifugation at 13,200 rpm for 5 min, supernatants were collected in light-protected Eppendorf tubes. The pellets were subjected to re-extraction to obtain a final pooled volume of 1 mL. For coumarin extraction from growth media, roots were removed from the plates and the media were oven-dried at 50°C for 2 d, and subsequently incubated in 10 mL of 80% (v/v) methanol containing 0.1 ppm 4-methylumbelliferone for 30 min on a shaker. The extract was collected without disturbing the media debris. The extraction process was repeated twice. The samples were centrifuged at 13,200 rpm for 10 min and supernatants were used for targeted UPLC analysis.
Targeted coumarin analysis
Targeted coumarin profiling and data analysis was done as previously described (Tsai et al., 2018). UPLC–QTOF–MS analysis was performed on an Acquity UPLC system (Waters, Milford, MA, USA) and a SYNAPT G2 high-definition mass spectrometry system (Waters) with an electrospray ionization interface, ion mobility, and time-of-flight system. Spectra were collected in the positive ionization (ES+) mode. To obtain the relative coumarin levels, the metabolite peak area was normalized to the internal standard peak area and the number of roots.
Statistical analyses
All statistical analyses were conducted in R version 4.0.3 (R Core Team, 2020; https://www.eea.europa.eu/data-and-maps/indicators/oxygen-consuming-substances-in-rivers/r-development-core-team-2006). Data transformation and one/two-way analysis of variance were carried out with the reshape2 version 1.4.4 (Wickham, 2007), stats (R Core Team, 2020), and multcomp version 1.4-16 (Hothorn et al., 2008) packages, respectively. Post-hoc analysis was performed with function TukeyHSD; compact letter display of all pair-wise comparisons was obtained with glht function. Plots were generated with GraphPad Prism version 9.
Accession numbers
Arabidopsis genome initiative locus identifiers for the genes mentioned in this article are as follows: IMA1, At1g47400; IMA2, At1g47395; IMA3, At2g30766; IMA4, AT1G07367; IRP3, At2g14247; IRP4, At1g13609; IRP5, At3g56360; IRP6, At5g05250; AHA2, At4g30190; S8H, At3g12900; F6′H1, At3g13610; FIT, At2g28160; FRO2, At1g01580; IRT1, At4g19690; MYB72, At1g56160; CYP82C4, At4g31940; bHLH38, At3g56970, bHLH39, At3g56980; bHLH100, At2g41240; bHLH101, At5g04150; EF1α, At5g60390; BGLU42, At5g36890; PDR9, At3g53480; COSY, At1g28680; NAS4, At1g56430; bHLH18, At2g22750; bHLH19, At2g22760; bHLH20, At2g22770; bHLH25, At4g37850; URI, At3g19860; ILR3, At5g54680, BTSL1, At1g74770; and BTSL2, At1g18910, BGLU21, At1g66270; BGLU22, At1g66280; BGLU23, At3g09260.
Supplemental data
The following supplemental materials are available in the online version of this article.
Supplemental Table S1. Primers used in this study.
Supplemental Figure S1. Expression patterns of IMA1 and IMA2.
Supplemental Figure S2. Perls staining of Col-0, f6′h1-1, IMA1ox, and IMA2ox seedlings.
Supplemental Figure S3. Phenotypes and Fe concentrations of Col-0, f6′h1-1, IMA1ox, and IMA2ox seedlings.
Supplemental Figure S4. Full scans of western blots.
Supplemental Figure S5. Autofluorescence of seedlings and media.
Supplemental Figure S6. pH dependence of scopoletin fluorescence.
Supplemental Figure S7. Phenotype and Fe concentration of wild-type seedlings grown on media supplemented with different Fe sources at pH 5.5 and pH 7.0.
Supplemental Figure S8. RT-qPCR analysis of key genes involved in Fe homeostasis.
Supplementary Material
Acknowledgments
The authors would like to thank Tushar S. Jadhav (Institute of Chemistry, Academia Sinica, Taipei) for insightful discussion on UV–Vis spectra analysis and Dr Louis Grillet (Department of Agricultural Chemistry, National Taiwan University, Taipei) for generating the IMA2 overexpression lines without tag constructs. We thank the Plant Tech Core Facility at ABRC for protoplast isolation and transformation, the Small Molecule Metabolomics core facility, sponsored by the Institute of Plant and Microbial Biology, and the Scientific Instrument Center, Academia Sinica, for providing technical assistance, useful discussion, and support with data acquisition and analysis of Synapt HDMS experiments. We also acknowledge the use of the QuantStudio 12K Flex Real-Time PCR System at the Genomic Technology Core Facility at IPMB. The authors are most grateful for the continued support provided by Dr Wendar Lin from the Bioinformatics Core Facility at IPMB, Academia Sinica.
Funding
This work was supported by a grant from the Ministry of Science and Technology to W.S. (grant No.: 108-2311-B-001 -033 -MY3).
Conflict of interest statement. None declared.
C.K.G., H.H.T., and W.S. designed the research; C.K.G. and H.H.T. performed and analyzed experiments; W.S. and C.K.G. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Wolfgang Schmidt (wosh@gate.sinica.edu.tw).
References
- Ahn YO, Shimizu BI, Sakata K, Gantulga D, Zhou Z, Bevan DR, Esen A (2010) Scopolin-hydrolyzing-glucosidases in roots of Arabidopsis. Plant Cell Physiol 51: 132–143 [DOI] [PubMed] [Google Scholar]
- Boxma R (1981) Effect of pH on the behaviour of various iron chelates in sphagnum (moss) peat. Commun Soil Sci Plant Anal 12: 755–763 [Google Scholar]
- Chen YT, Wang Y, Yeh KC (2017) Role of root exudates in metal acquisition and tolerance. Curr Opin Plant Biol 39: 66–72 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Chen CL, Cui M, Zhou WJ, Wu HL, Ling HQ (2018) Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Mol Plant 11: 1166–1183 [DOI] [PubMed] [Google Scholar]
- Döll S, Kuhlmann M, Rutten T, Mette MF, Scharfenberg S, Petridis A, Berreth DC, Mock HP (2018) Accumulation of the coumarin scopolin under abiotic stress conditions is mediated by the Arabidopsis thaliana THO/TREX complex. Plant J 93: 431–444 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fettt J, Guerinott ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle MA, Somerville C (1987) Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. MGG Mol Gen Genet 206: 200–206 [Google Scholar]
- Fourcroy P, Sisó-Terraza P, Sudre D, Savirón M, Reyt G, Gaymard F, Abadía A, Abadia J, Álvarez-Fernández A, Briat JF (2014) Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol 201: 155–167 [DOI] [PubMed] [Google Scholar]
- Gao F, Robe K, Bettembourg M, Navarro N, Rofidal V, Santoni V, Gaymard F, Vignols F, Roschzttardtz H, Izquierdo E, et al. (2020) The transcription factor bHLH121 interacts with bHLH105 (ILR3) and its closest homologs to regulate iron homeostasis in Arabidopsis. Plant Cell 32: 508–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet L, Lan P, Li W, Mokkapati G, Schmidt W (2018) IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat Plants 4: 953–963 [DOI] [PubMed] [Google Scholar]
- Grillet L, Ouerdane L, Flis P, Hoang MTT, Isaure MP, Lobinski R, Curie C, Mari S (2014) Ascorbate efflux as a new strategy for iron reduction and transport in plants. J Biol Chem 289: 2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JP, Hodgson JG (1969) An investigation of the ecological significance of lime-chlorosis by means of large-scale comparative experiments. InRorison IH, ed, Ecological Aspects of the Mineral Nutrition of Plants, Blackwell, Oxford, pp 67–99 [Google Scholar]
- Grusak MA, Welch RM, Kochian LV (1990) Does iron deficiency in Pisum sativum enhance the activity of the root plasmalemma iron transport protein? Plant Physiol 94: 1353–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbort CJ, Hashimoto M, Inoue H, Niu Y, Guan R, Rombolà AD, Kopriva S, Voges MJEEE, Sattely ES, Garrido-Oter R. et al. (2020) Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host Microbe 28: 825–837.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Lei GJ, Yamaji N, Nakagawa N, Ma JF (2018) The putative peptide gene FEP1 regulates iron deficiency response in Arabidopsis. Plant Cell Physiol 59: 1739–1752 [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biometrical J 50: 346–363 [DOI] [PubMed] [Google Scholar]
- Jahn TP, Schulz A, Taipalensuu J, Palmgren MG (2002) Post-translational modification of plant plasma membrane H+-ATPase as a requirement for functional complementation of a yeast transport mutant. J Biol Chem. 277: 6353–6358 [DOI] [PubMed] [Google Scholar]
- Kim SA, LaCroix IS, Gerber SA, Guerinot ML (2019) The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc Natl Acad Sci USA 116: 24933–24942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nagano AJ, Nishizawa NK (2020) Iron deficiency-inducible peptide-coding genes OsIMA1 and OsIMA2 positively regulate a major pathway of iron uptake and translocation in rice. J Exp Bot 72: 2196–2211 [DOI] [PubMed] [Google Scholar]
- Lager I, Andréasson O, Dunbar TL, Andreasson E, Escobar MA, Rasmusson AG (2010) Changes in external pH rapidly alter plant gene expression and modulate auxin and elicitor responses. Plant Cell Environ 33: 1513–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Shiau JY, Wu YC, Lin W, Schmidt W (2011) ITRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol 155: 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA (1998) The Calcicole-calcifuge problem revisited. Adv Bot Res 29: 1–30 [Google Scholar]
- Lei R, Li Y, Cai Y, Li C, Pu M, Lu C, Yang Y, Liang G (2020) bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining Fe homeostasis in Arabidopsis. Mol Plant 13: 634–649 [DOI] [PubMed] [Google Scholar]
- Lin WD, Liao YY, Yang TJW, Pan CY, Buckhout TJ, Schmidt W (2011) Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiol 155: 1383–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena C, Romera FJ, Rojas CL, García MJ, Alcántara E, Pérez-Vicente R (2007) Bicarbonate blocks the expression of several genes involved in the physiological responses to Fe deficiency of Strategy I plants. Funct Plant Biol 34: 1002. [DOI] [PubMed] [Google Scholar]
- Lucena JJ, Chaney RL (2006) Synthetic iron chelates as substrates of root ferric chelate reductase in green stressed cucumber plants. J Plant Nutr 29: 423–439 [Google Scholar]
- Martín-Barranco A, Spielmann J, Dubeaux G, Vert G, Zelazny E (2020) Dynamic control of the high-affinity iron uptake complex in root epidermal cells. Plant Physiol 184: 1236–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser J, Lingam S, Bauer P (2011) Posttranslational regulation of the iron deficiency basic helix-loop-helix transcription factor FIT is affected by iron and nitric oxide. Plant Physiol 157: 2154–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RM, HanumanthaRao B, Vemula A, Rathore A, Brumbarova T, Ivanov R (2020) Availability of soil iron determines the distribution strategy and seed iron content in mungbean (Vigna radiata) plants. Plant Soil 446: 413–423 [Google Scholar]
- Norvell WA (1991) Reactions of metal chelates in soils and nutrient solutions. InMortvedt JJ, ed, Micronutrients in Agriculture, Soil Science Society of America, Madison, WI, pp 187–227 [Google Scholar]
- Obara K, Kihara A (2014) Signaling events of the Rim101 pathway occur at the plasma membrane in a ubiquitination-dependent manner. Mol Cell Biol 34: 3525–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML (2013) MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet 9: e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan IC, Tsai HH, Cheng YT, Wen TN, Buckhout TJ, Schmidt W (2015) Post-transcriptional coordination of the Arabidopsis iron deficiency response is partially dependent on the E3 ligases RING DOMAIN LIGASE1 (RGLG1) and RING DOMAIN LIGASE2 (RGLG2). Mol Cell Proteomics 14: 2733–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva MA, Lucena-Agell D, Arst HN (2014) Liaison alcaline: pals entice non-endosomal ESCRTs to the plasma membrane for pH signaling. Curr Opin Microbiol 22: 49–59 [DOI] [PubMed] [Google Scholar]
- Rajniak J, Giehl RFH, Chang E, Murgia I, Von Wirén N, Sattely ES (2018) Biosynthesis of redox-active metabolites in response to iron deficiency in plants. Nat Chem Biol 14: 442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA (1990) Sensing pH? Plant, Cell Environ 13: 721–729 [Google Scholar]
- Robe K, Conejero G, Gao F, Lefebvre-Legendre L, Sylvestre-Gonon E, Rofidal V, Hem S, Rouhier N, Barberon M, Hecker A. et al. (2020a) Coumarin accumulation and trafficking in Arabidopsis thaliana: a complex and dynamic process. New Phytol 229: 2062–2079 [DOI] [PubMed] [Google Scholar]
- Robe K, Izquierdo E, Vignols F, Rouached H, Dubos C (2020b) The coumarins: secondary metabolites playing a primary role in plant nutrition and health. Trends Plant Sci 26: 248–259 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Chun Pan I, Li W, Lan P, Buckhout TJ, Schmidt W (2013a) The transcriptional response of Arabidopsis leaves to Fe deficiency. Front Plant Sci 4: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Connorton JM, Kruse I, Green RT, Franceschetti M, Chen YT, Cui Y, Ling HQ, Yeh KC, Balk J (2019) Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling. Proc Natl Acad Sci USA 116: 17584–17591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Lin WD, Fu GM, Abadía J, López-Millán AF, Schmidt W (2013b) Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol 162: 1473–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V, Marschner H (1986) Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol 80: 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Conéjéro G, Curie C, Mari S (2009) Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol 151: 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S, Schmidt W (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Schmid NB, Giehl RFH, Döll S, Mock HP, Strehmel N, Scheel D, Kong X, Hider RC, von Wirén N (2014) Feruloyl-CoA 6′-Hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol 164: 160–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W, Fühner C (1998) Sensitivity to and requirement for iron in Plantago species. New Phytol 138: 639–651 [Google Scholar]
- Schwab AP, Lindsay WL (1983) Effect of redox on the solubility and availability of iron. Soil Sci Soc Am J 47: 201–205 [Google Scholar]
- Shi WM, Chino M, Youssef RA, Mori S, Takagi S (1988) The occurrence of mugineic acid in the rhizosphere soil of barley plant. Soil Sci Plant Nutr 34: 585–592 [Google Scholar]
- Sisó-Terraza P, Luis-Villarroya A, Fourcroy P, Briat JF, Abadía A, Gaymard F, Abadía J, Álvarez-Fernández A (2016) Accumulation and secretion of coumarinolignans and other coumarins in Arabidopsis thaliana roots in response to iron deficiency at high pH. Front Plant Sci 7: 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz AB, Hermand V, Curie C, Vert G (2012) Arabidopsis bHLH100 and bHLH101 Control iron homeostasis via a FIT-independent pathway. PLoS One 7: e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwinska J, Siatkowska K, Olry A, Grosjean J, Hehn A, Bourgaud F, Meharg AA, Carey M, Lojkowska E, Ihnatowicz A (2018) Scopoletin 8-hydroxylase: a novel enzyme involved in coumarin biosynthesis and iron-deficiency responses in Arabidopsis. J Exp Bot 69: 1735–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis IA, De Jonge R, Pieterse CMJ (2019) The age of coumarins in plant-microbe interactions. Plant Cell Physiol 60: 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis IA, Yu K, Feussner K, De Jonge R, Van Bentum S, Van Verk MC, Berendsen RL, Bakker PAHM, Feussner I, Pieterse CMJ (2018) MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci USA 115: E5213–E5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susín S, Abadía A, González-Reyes JA, Lucena JJ, Abadía J (1996) The pH requirement for in vivo activity of the iron-deficiency-induced “turbo” ferric chelate reductase: a comparison of the iron-deficiency-induced iron reductase activities of intact plants and isolated plasma membrane fractions in sugar beet. Plant Physiol 110: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terés J, Busoms S, Perez Martín L, Luís-Villarroya A, Flis P, Álvarez-Fernández A, Tolrà R, Salt DE, Poschenrieder C (2019) Soil carbonate drives local adaptation in Arabidopsis thaliana. Plant Cell Environ 42: 2384–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Schmidt W (2017) One way. Or another? Iron uptake in plants. New Phytol 214: 500–505 [DOI] [PubMed] [Google Scholar]
- Tsai HH, Schmidt W (2021) The enigma of environmental pH sensing in plants. Nat Plants 7: 106–115 [DOI] [PubMed] [Google Scholar]
- Tsai HH, Rodríguez-Celma J, Lan P, Wu YC, Vélez-Bermúdez IC, Schmidt W (2018) Scopoletin 8-hydroxylase-mediated fraxetin production is crucial for iron mobilization. Plant Physiol 177: 194–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Schmidt W (2020) pH-dependent transcriptional profile changes in iron-deficient Arabidopsis roots. BMC Genomics 21: 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler G (1996) Soil chemical limitations to growth and development of Veronica officinalis L. and Carex pilulifera L. Plant Soil 184: 281–289 [Google Scholar]
- Vanholme R, Sundin L, Seetso KC, Kim H, Liu X, Li J, De Meester B, Hoengenaert L, Goeminne G, Morreel K, et al. (2019) COSY catalyses trans–cis isomerization and lactonization in the biosynthesis of coumarins. Nat Plants 5: 1066–1075 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot M, Lou Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges MJEEE, Bai Y, Schulze-Lefert P, Sattely ES (2019) Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc Natl Acad Sci USA 116: 12558–12565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ (2013) Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant 6: 503–513 [DOI] [PubMed] [Google Scholar]
- Waters BM, Amundsen K, Graef G (2018) Gene expression profiling of iron deficiency chlorosis sensitive and tolerant soybean indicates key roles for phenylpropanoids under alkalinity stress. Front Plant Sci 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn AR (1994) The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144: 307–313 [Google Scholar]
- Wickham H (2007) Reshaping data with the reshape package. J Stat Softw 21: 1–20 [Database] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Hanson J, Pieterse CMJ (2014) β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol 204: 368–379 [DOI] [PubMed] [Google Scholar]
- Ziegler J, Schmidt S, Strehmel N, Scheel D, Abel S (2017) Arabidopsis transporter ABCG37/PDR9 contributes primarily highly oxygenated coumarins to root exudation. Sci Rep 7: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohlen A, Tyler G (2004) Soluble inorganic tissue phosphorus and calcicole-calcifuge behaviour of plants. Ann Bot 94: 427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.