Abstract
Warty fruit in cucumber (Cucumis sativus L.) is an important quality trait that greatly affects fruit appearance and market value. The cucumber wart consists of fruit trichomes (spines) and underlying tubercules, in which the existence of spines is prerequisite for tubercule formation. Although several regulators have been reported to mediate spine or tubercule formation, the direct link between spine and tubercule development remains unknown. Here, we found that the basic Helix-Loop-Helix (bHLH) gene HECATE2 (CsHEC2) was highly expressed in cucumber fruit peels including spines and tubercules. Knockout of CsHEC2 by the CRISPR/Cas9 system resulted in reduced wart density and decreased cytokinin (CTK) accumulation in the fruit peel, whereas overexpression of CsHEC2 led to elevated wart density and CTK level. CsHEC2 is directly bound to the promoter of the CTK hydroxylase-like1 gene (CsCHL1) that catalyzes CTK biosynthesis, and activated CsCHL1 expression. Moreover, CsHEC2 physically interacted with GLABROUS3 (CsGL3, a key spine regulator) and Tuberculate fruit (CsTu, a core tubercule formation factor), and such interactions further enhanced CsHEC2-mediated CsCHL1 expression. These data suggested that CsHEC2 promotes wart formation by acting as an important cofactor for CsGL3 and CsTu to directly stimulate CTK biosynthesis in cucumber. Thus, CsHEC2 can serve as a valuable target for molecular breeding of cucumber varieties with different wart density requirements.
Fruit wart is a key external quality trait, and CsHECATE2 promotes wart formation by interacting with a spine regulator and a tubercule factor to directly stimulate cytokinin biosynthesis in cucumber
Introduction
Cucumber (Cucumis sativus L.), a member of the Cucurbitaceae family, is one of the most important vegetable crops worldwide (Huang et al., 2009; Guo et al., 2020). Cucumber fruit is the edible organ with important economic value that can be consumed fresh or processed into pickles. The surface of cucumber fruit is often covered with spines, tubercules and bloom trichomes, which are highly specialized structures derived from epidermal cells. The fruit spine (type II trichome, the dominant type in cucumber) is composed of a spherical or conical base and a sharp spiny stalk (Chen et al., 2014). The tubercule, generally located beneath the spine base, is an arched structure derived from several layers of surface cells (Supplemental Figure S1A; Yang et al., 2014; Che and Zhang, 2019). The bloom trichome (type I trichome) has a glandular structure that produces fine white powdery secretions and is responsible for the rough outer appearance of the fruit (Supplemental Figure S1B; Samuels et al., 1993). When spines are combined with tubercules, the cucumber fruit has a warty trait. Warty fruit is an important quality trait that greatly affects fruit appearance and market value in cucumber. In 2019, China accounts for 80.1% of the total cucumber production worldwide (FAOSTAT), and the majority of cucumber fruits in China display warty trait. Thus, dissecting the regulatory mechanism of wart formation is of great significance for cucumber breeding with desired external quality.
Fruit spines are regulated by a complex gene network involving multiple transcription factors and endogenous phytohormones. Several spine mutants have been identified, including the completely glabrous mutants glabrous 3 (csgl3) and trichome-less (tril), and the reduced spine density mutant few spines 1 (fs1) in cucumber (Pan et al., 2015; Cui et al., 2016; Wang et al., 2016; Zhang et al., 2016; Du et al., 2020). Interestingly, the csgl3, tril and fs1 mutants are allelic and caused by different forms of mutations in the same gene Csa6G514870. The csgl3 mutants were due to insertion of a 5005-bp long terminal repeat retrotransposon in the fourth exon (Pan et al., 2015) or three single-nucleotide transitions in the fourth exon (Cui et al., 2016). The tril mutant was caused by a 5008-bp retrotransposon insertion following the first exon (Wang et al., 2016), and the fs1 mutant resulted from a 812-bp fragment substitution in the promoter region (Zhang et al., 2016). The gene Csa6G514870 encodes a member of the class IV homeodomain leucine zipper (HD-ZIP) transcription factor that plays an essential role in spine initiation and development (Du et al., 2020). Unlike the completely abolished spine in the fruit surface from csgl3 and tril mutants, fruit spines in csgl1, tiny branched hair (tbh) and micro-trichome tubercule (mict) mutants exhibited reduced size and aberrant morphology (Chen et al., 2014; Li et al., 2015; Zhao et al., 2015). However, the distribution and density of the deformed trichomes were unaffected, suggesting that CsGL1/TBH/Mict functions in trichome development rather than the initiation process (Li et al., 2015). Similarly, map-based cloning revealed that CsGL1/TBH/Mict encodes the same Class I HD-ZIP transcription factor, and loss-of-function mutants were due to a 2,649-bp genomic deletion spanning the first and second exons (Li et al., 2015; Zhao et al., 2015). In addition, several genes regulating spine initiation and development have been identified by means of reverse genetics. Overexpression of CsMYB6, a MIXTA-like MYB transcription factor, reduced the density of spines in cucumber. Further analyses indicated that CsMYB6 directly binds to the promoter region of CsTRY, a homolog of TRIPTYCHON in Arabidopsis (Arabidopsis thaliana), and inhibits its expression, and CsMYB6 also interacts with CsTRY protein, suggesting that CsMYB6 and CsTRY may function as a module to negatively regulate trichome initiation in cucumber (Yang et al., 2018). TRANSPARENT TESTA GLABRA1 (CsTTG1), a WD40 repeat-containing gene, acts as a positive regulator of fruit spine initiation and differentiation in cucumber. Biochemical and genetic analyses showed that CsTTG1 regulates fruit spine formation through direct protein-protein interaction with CsGL1 (Chen et al., 2016).
Fruit tubercule is a composite trait that includes tubercule initiation, tubercule shape, and tubercule size. The developmental process of fruit tubercule can be generally divided into three stages: initiation (earlier than 2 d before anthesis [DBA]), development (2 DBA to 13 d postanthesis [DPA]), and senescence (after 13 DPA; Yang et al., 2014). Genetic studies showed that the warty phenotype is dominant to the non-warty fruit trait, and that fruit tubercule trait is controlled by a single dominant gene (CsTu), which encodes a C2H2 zinc finger transcription factor (Wang et al., 2007; Zhang et al., 2010; Yang et al., 2014). Importantly, the spine gene CsGL1 is epistatic over CsTu, as CsTu is not expressed in the csgl1 mutant with no spine and no tubercule, while spine was unchanged upon mutation of CsTu (Cao et al., 2001; Yang et al., 2014). Further, among all the reported mutants with disrupted spine initiation, no tubercule was observed (Chen et al., 2014; Li et al., 2015; Cui et al., 2016; Liu et al., 2016; Du et al., 2020). All the reports suggested that the existence of spines is prerequisite for tubercule formation. Recently, cucumber Tubercule size 1 (CsTS1) gene was identified to encode an oleosin family protein that controls the size of tubercule. Sequence variation of CsTS1 promoter indicated that low expression levels of CsTS1 were associated with the 22 small-warty or non-warty cucumber lines. Biochemical analyses showed that CsTu directly binds to the CsTS1 promoter and enhances its transcriptional activity. These results suggested that CsTS1 acts in the tubercule size control and CsTu–CsTS1 regulatory module coordinately promotes fruit wart initiation and development in cucumber (Yang et al., 2014, 2019).
In addition, phytohormones such as gibberellin (GA), auxin (indole-3-acetic acid [IAA]), and cytokinin (CTK) have been shown to modulate fruit spine and tubercule development in cucumber. Transcriptome and hormone assay results showed that CsTu functions in CTK biosynthesis by indirectly promoting the expression of two CTK hydroxylase-like (CHL) genes, thereby stimulating cell division and ultimately causing the initiation of fruit tubercule (Yang et al., 2014). The auxin content was significantly increased in the 35S:CsTS1 tubercules but significantly decreased in the CsTS1-RNAi tubercules, and the expression of CsTS1 was induced by auxin treatments, suggesting that CsTS1 regulates tubercule development through increasing auxin signaling and auxin levels in cucumber (Yang et al., 2019). The GA biosynthesis gene CsGA20ox1 was found to be a negative regulator for fruit spine development and act as a potential downstream gene of CsGL1 (Li et al., 2015). Despite of its importance in cucumber breeding and production, the genetic and regulatory mechanisms of wart formation are very limited.
In this study, we identified a cucumber fruit wart regulator, a bHLH family gene HECATE2 (CsHEC2), which was highly expressed in fruit peels including spines and tubercules. Furthermore, we showed CsHEC2 directly interacts with CsGL3 and CsTu, and such interactions can further enhance CsHEC2-mediated CsCHL1 (a CTK biosynthesis enzyme) transcriptional activation, thereby resulting in CTK accumulation in fruit peels. Together, we uncover a transcriptional regulation module, CsGL3- and/or CsTu-CsHEC2, that promotes wart formation by directly activating the CTK biosynthesis gene CsCHL1 in cucumber.
Results
CsHEC2 is a nucleus-localized transcriptional activator
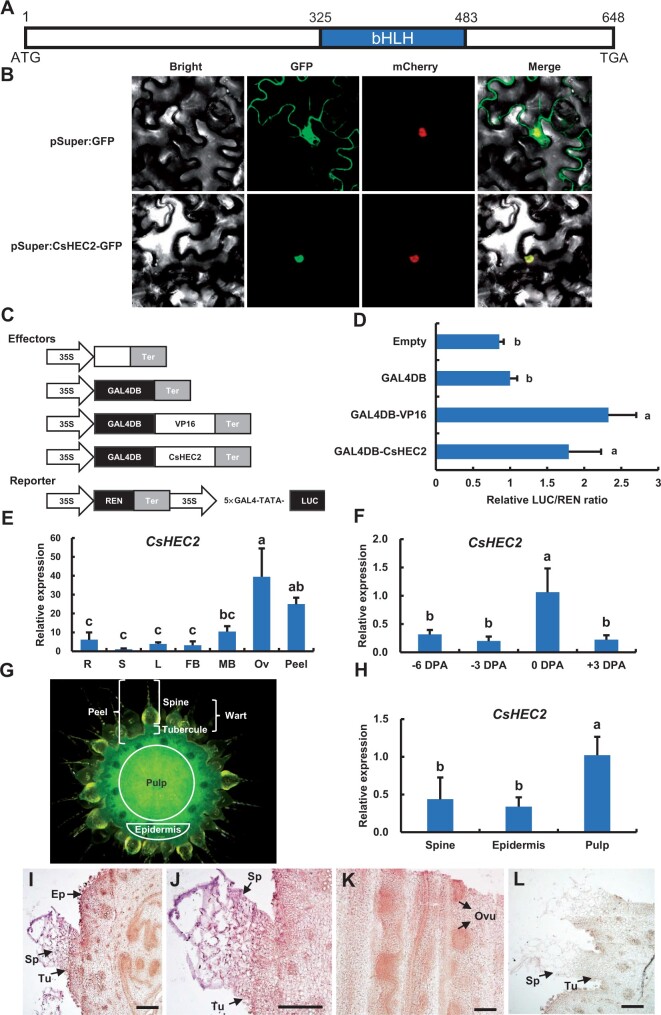
HEC transcription factors were shown to play important roles during the development of female reproductive organ and meristem regulation in Arabidopsis (Gremski et al., 2007; Schuster et al., 2014, 2015), while the functions of HEC genes in cucumber are largely unknown. Our recent study revealed that C. sativus Irregular Vasculature Patterning (CsIVP), the cucumber HEC3 gene, was highly expressed in vascular tissues and functions in organ morphogenesis and downy mildew resistance in cucumber (Yan et al., 2020). But, the functions of CsHEC1/2 in cucumber are unknown. In this study, we focus on the characterization and functional analyses of CsHEC2 (Csa2G285890). Sequence analysis showed that the full-length coding sequence (CDS) of CsHEC2 was 648 bp, with a single exon and a conserved bHLH domain (Figure 1A), similar to that of HEC family genes in Arabidopsis (Gremski et al., 2007). The CsHEC2 protein was localized in the nucleus using transient expression analyses in Nicotiana benthamiana leaves (Figure 1B). A dual-luciferase (LUC) reporter (DLR) system was performed to test the transcriptional activity of CsHEC2 (Ohta et al., 2001; Hellens et al., 2005). The full-length CDS of CsHEC2 was fused with the GAL4 DNA-binding domain (GAL4DB) as an effector, the transcriptional activation motif Herpes Simplex Virus 16 (VP16) was used as a positive control, while GAL4DB was used as a negative control (Figure 1C). Compared with the empty vector and GAL4DB controls, the relative intensity of LUC/REN activity significantly increased upon co-transformation of GAL4DB-CsHEC2 with the reporter (Figure 1, C and D), which was similar to that of VP16 positive control, indicating that CsHEC2 may act as a transcriptional activator in cucumber.
Figure 1.
Subcellular localization, transcriptional activity and expression pattern analysis of CsHEC2. A, Schematic diagram of the gene structure of CsHEC2. The blue box represents the bHLH domain of 53 amino acids and Arabic numerals represent the position of base pairs sequence. B, Subcellular localization indicating CsHEC2-GFP fusion protein located in the nucleus of N. benthamiana leaves. Empty GFP driven by the Super promoter was used as a control. The fluorescent signals of GFP channel indicate GFP position. The fluorescent signals of mCherry channel indicate mCherry-labeled nuclear marker (NF-YA4-mCherry) position. C, Schematic illustration for CsHEC2 transcriptional activity assay in (D). GAL4DB, GAL4 DNA binding domain; 5×GAL4-TATA, five GAL4-binding sites; VP16, Herpes Simplex Virus protein 16; REN, Renilla LUC; Ter, nopaline synthase terminator. VP16 was used as a positive control. The REN gene driven by the 35S promoter was used as an internal reference to normalize the LUC values. D, Relative LUC activities in N. benthamiana leaves indicating the transcriptional activation activity of CsHEC2. Values are means ± SD (n = 5). The different lowercase letters indicate significant differences (P < 0.01) by one-way ANOVA analysis with Tukey’s HSD test. E, F and H, Expression analyses of CsHEC2 in different cucumber organs (E), ovary peel at different developmental stages (F), and different parts of ovary at anthesis (H). R, young root; S, stem; L, leaf; FB, female bud; MB, male bud; Ov, ovary at anthesis. Error bars represent ±SD (n = 3). The different lowercase letters indicate significant differences (P < 0.05) by one-way ANOVA analysis with Tukey’s HSD test. G, Cross section view of a WT ovary at anthesis in (H). The epidermis consists of exocarp and tubercules. The fruit peel consists of epidermis and spines. I–L, In situ hybridization of CsHEC2 showing expression signals in the spine, tubercule, epidermis, and ovule of cucumber. I, Cross section of a representative ovary at anthesis, (J) Magnified view of the spine and tubercule in (I), (K) Longitudinal section of a cucumber ovary. L, The CsHEC2 sense probe was hybridized as a control. Ep, epidermis; Sp, spine; Tu, tubercule; Ovu, ovule. Scale bars represent 200 μm in (I) to (L).
CsHEC2 is highly expressed in spine and tubercule of cucumber fruit
To explore the expression pattern of CsHEC2, reverse transcription quantitative polymerase chain reaction (RT-qPCR) was performed in different cucumber organs (Figure 1E). Transcripts of CsHEC2 were greatly enriched in the ovary and fruit peel (Figure 1E). The fruit peel in cucumber consists of the wart and the epidermis (Figure 1G), in which the wart is composed of spines and tubercules (Che and Zhang, 2019). Although the transcript abundance of CsHEC2 was continuously detected from the initial stage of tubercule development to developmental stages of wart (Yang et al., 2014), the transcript level of CsHEC2 was the highest in fruit peels at anthesis (Figure 1F). Among the three parts of ovary at anthesis (Figure 1G), CsHEC2 was found to be expressed at a higher level in the pulp than in the spine or epidermis (Figure 1H). In situ hybridization further confirmed that CsHEC2 transcripts were enriched in spines, tubercules, and epidermis of ovary (Figure 1, I and J). In the longitudinal sections of cucumber ovary, strong expression of CsHEC2 was detected in developing ovules (Figure 1K). No signal was detected upon hybridization with the sense probe of CsHEC2 (Figure 1L). These results implied a potential function of CsHEC2 in regulating fruit wart formation in cucumber.
Knockout of CsHEC2 results in decreased wart density in cucumber fruit
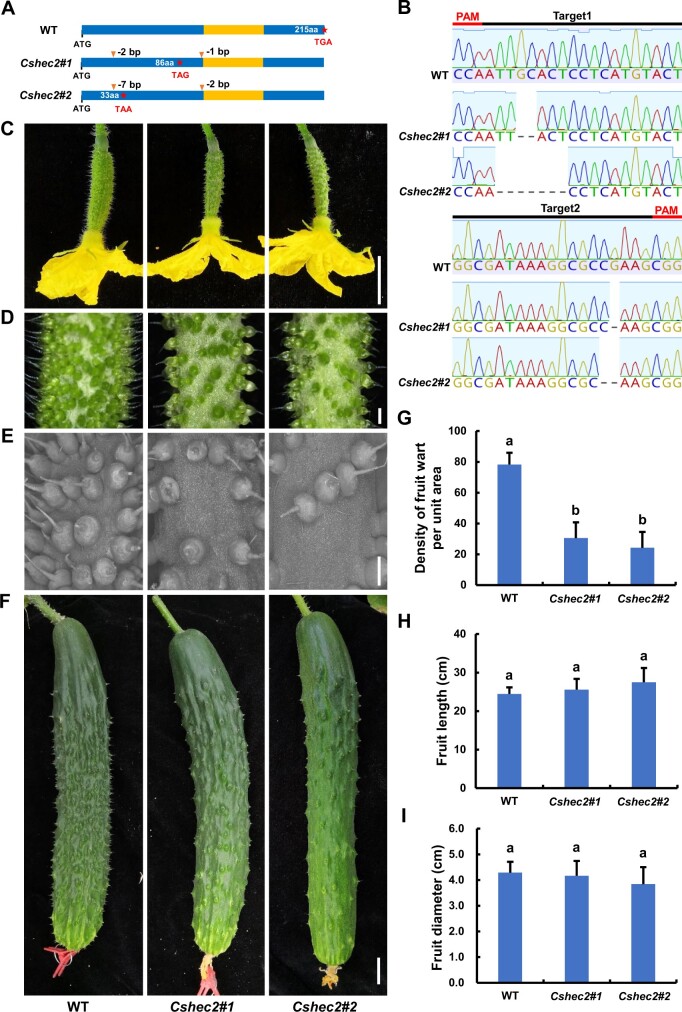
To investigate the function of CsHEC2 in cucumber, we used CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated system 9) system to knock out CsHEC2 as previously described (Hu et al., 2017). Two null mutants Cshec2#1 (a homozygous allele with a 2-bp and a 1-bp deletions) and Cshec2#2 (a homozygous allele with a 7-bp and a 2-bp deletions) were obtained (Figure 2, A and B), and both mutants generated a premature stop codon resulting in a truncated protein of 86 amino acids and 33 amino acids, respectively (Figure 2A). To explore any off-target effect, we searched the genomic sequences for potential off-target sites of CsHEC2 single-guide RNA (sgRNA), and cloned and sequenced these PCR products in the homozygous T2 mutants. No off-target mutations were found in the examined plants (Supplemental Table S1). Compared with the wild-type (WT) plants, the number of warts on the fruit surface was greatly reduced in both Cshec2#1 and Cshec2#2 mutants at anthesis (Figure 2, C–E) and 10 DPA (Figure 2F). No visible differences were detected in the appearance and morphological characteristics of spines and tubercules between WT and mutant plants (Figure 2, C–F). Quantification data showed that the fruit wart density at anthesis displayed a 61% reduction in Cshec2#1 plant and a 69% reduction in Cshec2#2 plant as compared to that in WT plant (Figure 2G). No significant differences were detected in fruit length (Figure 2H) or fruit diameter (Figure 2I) between WT and Cshec2 mutants. Interestingly, no changes were observed in the density of trichomes on the abaxial or adaxial surface of leaves produced from WT and mutant lines (Supplemental Figure S2). These results suggested that CsHEC2 specifically functions in wart density regulation in the cucumber fruit.
Figure 2.
Knockout of CsHEC2 by CRISPR/Cas9 resulted in reduced density of fruit wart in cucumber. A, Genotype identification of Cshec2 knockout mutants indicated the Cshec2#1 allele with 2-bp and 1-bp deletion and the Cshec2#2 allele with 7- and 2-bp deletion as shown in (B), both generating a premature stop codon and resulting in a truncated protein of 86 amino acids and 33 amino acids in length, respectively. The orange box represents the bHLH domain. B, Sanger sequencing chromatogram analysis of the transgene-free homozygous T2 mutants. Black lines represent targeted sequences and red lines indicate the protospacer-adjacent motif sequences. Dashed lines in chromatograms indicate nucleotide deletions. C–F, Phenotypic characterization of Cshec2 knockout plants in cucumber. C, Cucumber ovaries at anthesis. D, Magnified view of warts on the ovary at anthesis. E, Scanning electron micrographs of warts on the ovary at anthesis. F, Cucumber fruits at 10 DPA. The columns from left, middle, and right represent WT, Cshec2#1, and Cshec2#2 plants (in C–F), respectively. Scale bar: (C) and (F) 2 cm, (D) and (E) 1 mm. G–I, Quantification analysis of fruit wart density (G), fruit length (H) and fruit diameter (I) in WT and two Cshec2 mutants. Fruit warts of 40 mm2 surface area on the ovary at anthesis were counted in (G) and error bars represent ±SD (n ≥ 15); Fruit length (H) and fruit diameter (I) were measured in cucumber fruits at 10 DPA, and error bars represent ±SD (n ≥ 5). The different lowercase letters indicate significant differences (P < 0.01) by one-way ANOVA analysis with Tukey’s HSD test.
CsHEC2 regulates fruit wart formation through mediating CTK biosynthesis and metabolism in cucumber
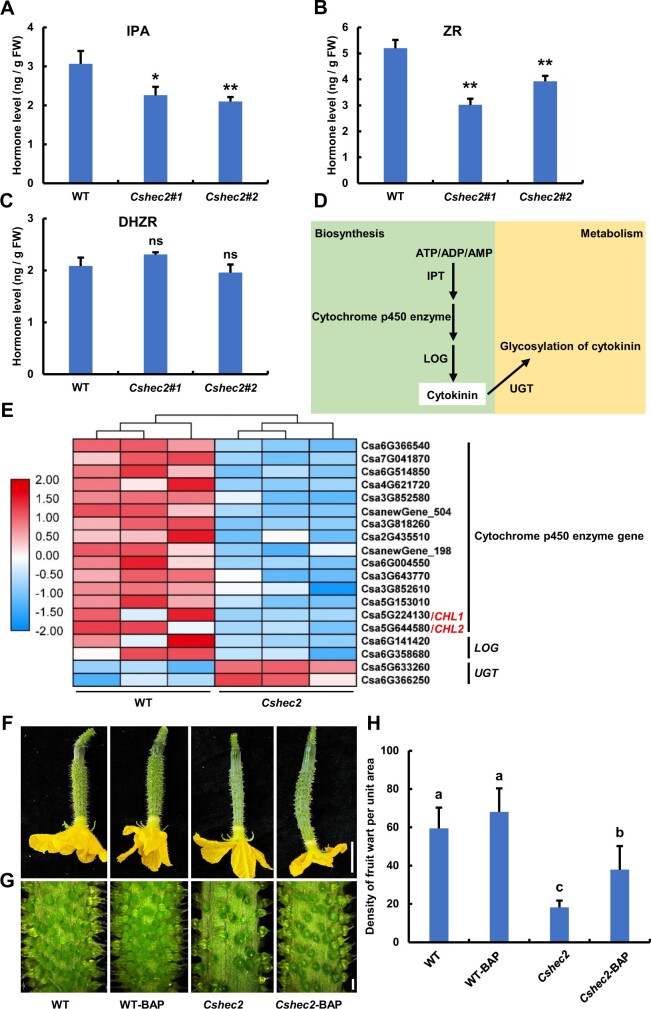
Previous studies indicated that auxin, CTK, and GA signaling pathways are involved in cucumber spine or tubercule formation (Yang et al., 2014; Li et al., 2015; Xie et al., 2018; Che and Zhang, 2019; Yang et al., 2019). Thus, we measured the levels of endogenous auxin (IAA), CTK (isopentenyl adenosine [IPA]; trans-zeatin riboside, ZR; dihydrogen ZR [DHZR]) and gibberellic acid (GA3) in the fruit peels of WT and Cshec2 mutants. Interestingly, only the levels of IPA and ZR were significantly decreased in the Cshec2 mutant compared with WT (Figure 3, A and B), while the levels of DHZR, IAA, and GA3 were unaltered (Figure 3C;Supplemental Figure S3). Therefore, we speculated that CsHEC2 may regulate fruit wart production through CTK pathway.
Figure 3.
Knockout of CsHEC2 led to reduced endogenous CTK content and disturbed expression of CTK pathway genes. A–C, The levels of endogenous CTK, (A) IPA, (B) ZR, and (C) DHZR were measured in WT and Cshec2 plants. Values are means ± SD of three independent biological replications. Significance analysis compared to WT was performed with the two-tailed Student’s t test (**P < 0.01, *P < 0.05, ns, no significant difference). D, Schematic diagram of a simplified biosynthesis and metabolic pathway for CTK. E, Heat map of the CTK pathway and related DEGs in WT and Cshec2 lines. Three biological replicates were performed and the colored bar on the left of the map represents fold change (log2 value). F–H, Representative morphology of cucumber ovaries at anthesis (F), magnified view of ovary at anthesis (G) and fruit wart density quantification (H) of WT and Cshec2 plants treated with 100 μM BAP. Values are means ± SD (n ≥ 15). The different lowercase letters indicate significant differences (P < 0.05) by one-way ANOVA analysis with Tukey’s HSD test. Scale bar: (F) 2 cm, (G) 1 mm.
To comprehensively analyze the regulatory network, RNA sequencing (RNA-seq) experiments were conducted on female buds at 7 DBA. Illumina high-throughput sequencing produced about 44.9–55.6 million paired-end reads for each sample (Supplemental Table S2). The principal component analysis (PCA) of the six RNA-seq datasets showed two distinct groups corresponding to WT and Cshec2 mutant samples (Supplemental Figure S4A), indicating good repeatability. Transcriptomic data analysis showed that 293 and 1295 genes were up- and down-regulated in Cshec2 mutants relative to WT, respectively (Supplemental Figure S4B; Supplemental Table S3), consistent with the role of CsHEC2 acting as a transcriptional activator (Figure 1, C and D). As shown in the heat map, fifteen cytochrome P450 enzyme genes and two LONELY GUY genes were significantly down-regulated in Cshec2 mutants (Figure 3, D and E), which can catalyze the biosynthesis of active form of CTK from ATP/ADP/AMP (Kurakawa et al., 2007; Schaller et al., 2015). Among them, two cytochrome P450 enzyme family genes, CsCHL1 (Csa5G224130) and CsCHL2 (Csa5G644580), were previously reported to be up-regulated by tubercule initiation gene CsTu and facilitated CTK biosynthesis in cucumber (Yang et al., 2014). In addition, two uridine diphosphate glycosyltransferase (UGT) genes were significantly up-regulated in Cshec2 mutants, which function in CTK conjugation metabolism to convert active CTKs into inactive glycosylated modified CTKs (Bajguz and Piotrowska, 2009). RT-qPCR was used to verify the transcriptomic data of several selected genes and displayed similar expression patterns (Figure 3E;Supplemental Figure S5, A–D).
To verify the positive role of CTK in wart formation in cucumber, exogenous CTK application was performed by treatment of WT and Cshec2 plants with 100 uM N6-Benzyladenine (BAP). Compared to the control group (0 uM BAP), BAP treatment was able to partially restore the Cshec2 phenotype (Figure 3, F and G), resulting an increase in wart density by 107.8% in Cshec2 fruits (Figure 3H). Taken together, our results suggested that CsHEC2 may regulate wart formation by promoting CTK biosynthesis and inhibiting CTK conjugation in cucumber fruit.
Overexpression of CsHEC2 led to increased wart density and CTK level in cucumber fruit peel
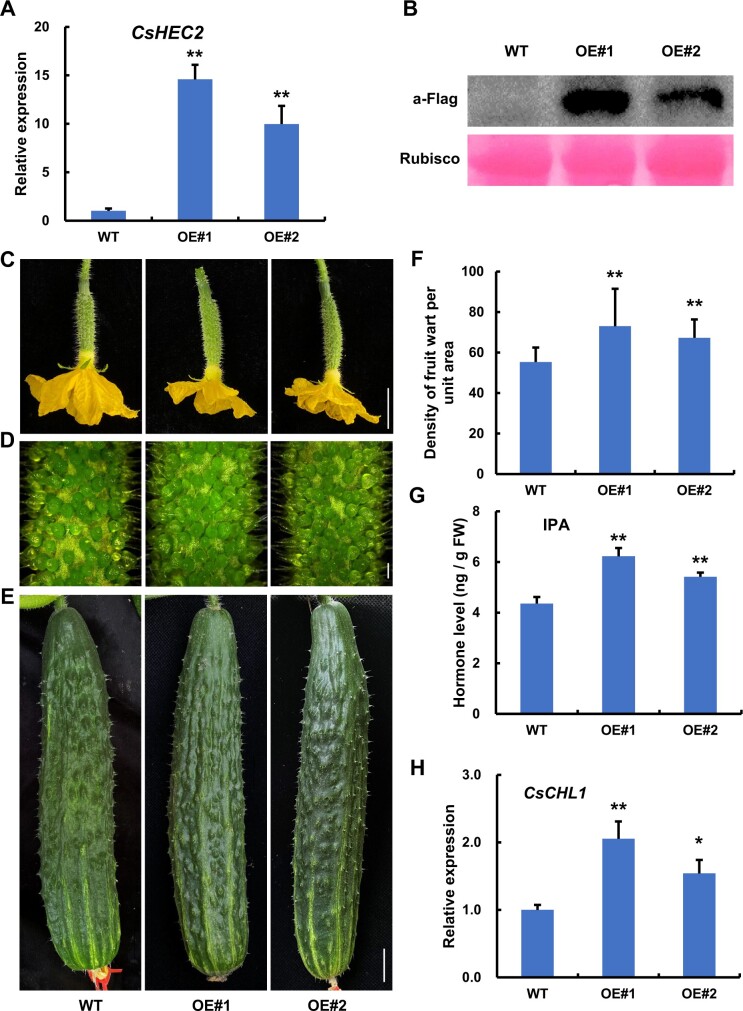
To further characterize the function of CsHEC2 in cucumber, the CsHEC2 overexpression vector (35S:CsHEC2-Flag) was constructed for genetic transformation. Four overexpression lines were obtained, and two of them (OE#1, OE#2) were selected for phenotypic observation. RT-qPCR showed that the expression of CsHEC2 was significantly increased in the transgenic lines (Figure 4A). Immunoblot analyses demonstrated that CsHEC2 protein accumulated at high levels in both OE transgenic lines (Figure 4B). Compared with the WT plants, the wart number on the fruit surface was greatly increased in CsHEC2-OE lines (Figure 4, C–E). Quantification data showed that the fruit wart density displayed a 31.9% increase in OE#1 line and a 21.6% increase in OE#2 line as compared to that in WT plant at anthesis (Figure 4F). Moreover, the level of CTK IPA was increased 42.8% and 24.2% in the fruit peels of OE#1 and OE#2 lines (Figure 4G), respectively, while no consistent changes were observed in the levels of ZR, DHZR, IAA, and GA3 (Supplemental Figure S6). These data supported that CsHEC2 promotes wart formation through activates CTK accumulation in cucumber fruit.
Figure 4.
Overexpression of CsHEC2 resulted in increased wart density and CTK levels in fruit peels of cucumber. A, RT-qPCR analysis of CsHEC2 expression in CsHEC2-OE lines. Values are means ± SD (n = 3). B, Immunoblot analysis of CsHEC2 protein levels in CsHEC2-OE plants. CsHEC2-Flag was detected using anti-Flag antibody. Rubisco large subunit stained by Ponceau S was used as a loading control. C–E, Phenotypic characterization of CsHEC2-OE plants. C, Cucumber ovaries at anthesis. D, Magnified view of warts on the ovary at anthesis. E, Cucumber fruits at 10 DPA. Scale bar: (C) and (E) 2 cm, (D) 1 mm. F, Quantification analysis of fruit wart density in WT and CsHEC2-OE plants. Fruit warts of 40 mm2 surface area on the ovary at anthesis were counted and error bars represent ±SD (n ≥ 14). G, The levels of endogenous CTK (IPA) were measured in WT and CsHEC2-OE plants. H, RT-qPCR analysis of CsCHL1 expression in CsHEC2-OE lines. Values are means ± SD (n = 3). Significance analysis compared to WT was performed with the two-tailed Student’s t test (**P < 0.01, *P < 0.05).
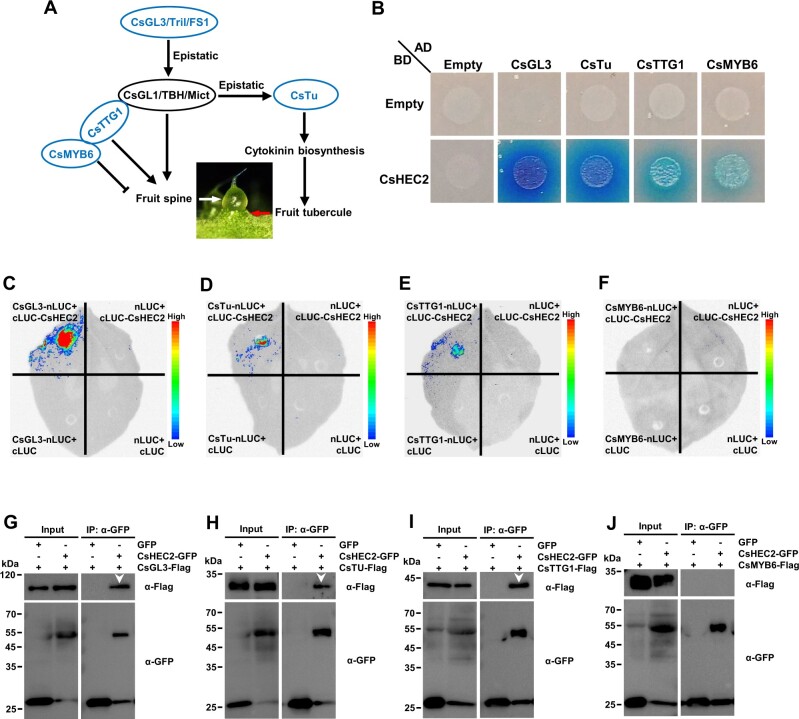
CsHEC2 interacts with wart initiation regulator CsGL3, CsTu, and CsTTG1
Considering that knockout and overexpression of CsHEC2 resulted in changes in fruit wart density, but not wart morphology (Figures 2 and 4), CsHEC2 may participate in wart initiation in cucumber. Several transcription factors have been reported to be involved in regulation of fruit spine or tubercule development in cucumber (Che and Zhang, 2019), in which CsGL3 (Pan et al., 2015; Cui et al., 2016; Wang et al., 2016), CsMYB6 (Yang et al., 2018), and CsTTG1 (Chen et al., 2016) regulate the initial development of fruit spines, and CsTu (Yang et al., 2014) regulates tubercule initiation by promoting CTK biosynthesis (Figure 5A). To investigate whether CsHEC2 interacts with these known regulators, yeast two-hybrid assays were performed. The results showed that CsHEC2 interacted strongly with CsGL3 and CsTu, while weakly with CsTTG1 and CsMYB6 (Figure 5B). To further confirm these protein interactions in vivo, firefly LUC complementation imaging (LCI) assays (Figure 5, C–F) and co-immunoprecipitation (Co-IP) analyses (Figure 5, G–J) were performed in N. benthamiana leaves. As shown in (Figure 5, C–J), CsHEC2 did interact with CsGL3, CsTu, and CsTTG1, but not with CsMYB6. These results indicated that CsHEC2 may function in fruit wart initiation through directly interacting with known regulators including CsGL3, CsTu, and CsTTG1 in cucumber.
Figure 5.
CsHEC2 interacted with CsGL3, CsTu, and CsTTG1 at protein level. A, Simplified regulatory network of fruit spine and tubercule development in cucumber. Blue regulators represent genes involved in fruit spine and tubercule initiation. B, Yeast two-hybrid assay showing that CsHEC2 interacts strongly with CsGL3 and CsTu, while CsHEC2 interacts weakly with CsTTG1 and CsMYB6 on SD-Leu/-Trp/-His/-Ade/X-α-gal selective medium by observing the growth of transformants. The combinations containing pGADT7 or pGBKT7 empty were used as negative controls. C–F, LCI assays showing that CsHEC2 interacts with CsGL3 (C), CsTu (D) and CsTTG1 (E) in N. benthamiana leaves, but CsHEC2 does not interact with CsMYB6 (F). cLUC-CsHEC2 was co-transformed with CsGL3-nLUC or CsTu-nLUC or CsTTG1-nLUC, respectively. The remaining combinations were used as controls. G to J, Co-IP analyses showing that CsHEC2 interacts with CsGL3 (G), CsTu (H), and CsTTG1 (I), but not with CsMYB6 (J). The indicated constructs were expressed in N. benthamiana leaves, and Co-IP was performed using anti-GFP antibody. The white arrowheads indicate specific interaction bands.
CsHEC2 directly binds to the promoter of CsCHL1 and activates its expression
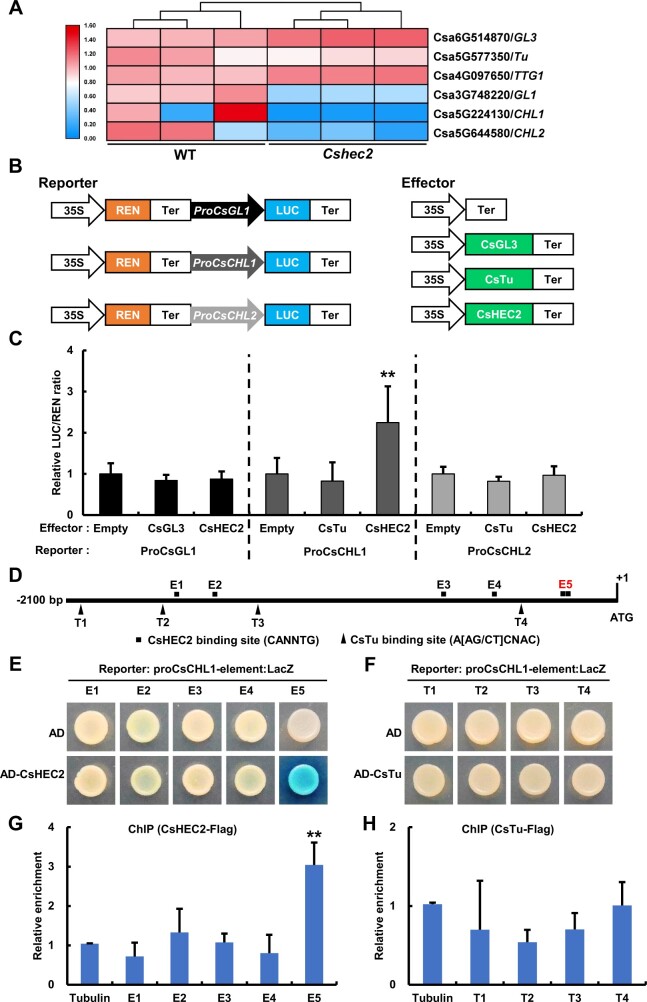
To further explore the relationship of CsHEC2 with these known regulators, RT-qPCR analyses were performed, and no significant changes were observed for the expression of CsGL3, CsTu, and CsTTG1 in WT and Cshec2 mutants. Similarly, these three genes displayed no differential changes in the RNA-seq transcriptome data (Figure 6A;Supplemental Fig. S5, E–G). However, the expression levels of CsGL3 downstream gene CsGL1 (Li et al., 2015; Pan et al., 2015), as well as the CsTu downstream genes CsCHL1 and CsCHL2 (Yang et al., 2014), were significantly reduced in Cshec2 mutants (Figure 6A;Supplemental Figure S5, A and H). To explore any direct regulation between CsGL3, CsTu, CsHEC2 and CsGL1, CsCHL1, and CsCHL2, DLR assay was performed in N. benthamiana leaves (Figure 6B). The results showed that co-expression of CsHEC2 with ProCsCHL1:LUC resulted in an increased LUC/REN ratio activity (Figure 6C). However, when co-expressed ProCsGL1:LUC with CsGL3 or CsHEC2, and ProCsCHL2:LUC with CsTu or CsHEC2, the LUC activity showed no significant changes (Figure 6C). Previous studies showed that bHLH transcription factors bind to the consensus sequence CANNTG (Massari and Murre, 2000; Toledo-Ortiz et al., 2003) and that C2H2 zinc finger proteins bind to the consensus sequence A[AG/CT]CNAC (Sun et al., 2015; Han et al., 2020). We scanned approximately 2 kb promoter regions of CsCHL1 to look for possible binding sites of CsHEC2 and CsTu. Five putative CsHEC2 binding sites (E1-E5) and four putative CsTu binding sites (T1-T4) were identified in the CsCHL1 promoter (Figure 6D). Y1H assay showed that CsHEC2 can directly bind to the CsCHL1 promoter via the E-box elements located in E5 (Figure 6E), while CsTu cannot (Figure 6F). Chromatin immunoprecipitation (ChIP)-PCR assays were performed to confirm the binding results in vivo, using cucumber protoplast system (Figure 6, G and H). The E5 fragment in CsCHL1 was significantly enriched after immunoprecipitation based anti-Flag antibodies recognizing CsHEC2-Flag protein-DNA complexes (Figure 6G). We also detected the expression of CsCHL1 by in situ hybridization and the results showed that the transcripts of CsCHL1 were highly accumulated in spine, tubercule, and epidermis of developing ovary (Supplemental Figure S7). Together with the significant down-regulation of CsCHL1 expression in Cshec2 mutants (Figure 6A;Supplemental Figure S5A) and up-regulation of CsCHL1 expression in CsHEC2-OE lines (Figure 4H), these data supported that CsCHL1 was a direct target gene of CsHEC2 rather than CsTu, and CsHEC2 activates CsCHL1 expression in cucumber.
Figure 6.
CsHEC2 directly binds to CsCHL1 and activates CsCHL1 expression. A, Heat map showing the expression of three CsHEC2 interactors and their putative downstream genes in WT and Cshec2 lines. The colored bar on the left of the map represents fold change (log2 value). B, Schematic illustration of LUC/REN assay for transcriptional activity assay as shown in (C). The REN gene driven by the 35S promoter was used as an internal reference to normalize the LUC values. The effector of empty vector was used as the control. C, Transient expression assays in N. benthamiana leaves showing that CsHEC2 significantly activated CsCHL1 expression. The LUC/REN ratio represents the LUC activity relative to the internal control REN. Values are means ± SD (n = 6). D, Schematic diagram of the distribution of putative CsHEC2 binding sites and CsTu binding sites in CsCHL1 promoter. E1-E5 are the putative binding sites of CsHEC2, T1-T4 are the putative binding sites of CsTu. Adenine residues of translation start codon (ATG) were located position +1. E and F, Yeast one-hybrid assay showing that CsHEC2 directly binds to E5, two adjacent E-box elements in CsCHL1 promoter (E), while CsTu does not bind to T1–T4 elements in CsCHL1 promoter (F). G and H, ChIP-PCR assay showing that CsHEC2 binds to the CsCHL1 promoter via E5 element in vivo (G), but CsTu does not bind to CsCHL1 promoter (H). Values are means ± SD (n = 3). Significance analysis (C, compared to empty vector; G and H, compared to Tubulin control) was performed with the two-tailed Student's t test (**P < 0.01).
CsGL3 and CsTu enhance the CsHEC2-mediated CsCHL1 transcriptional activation
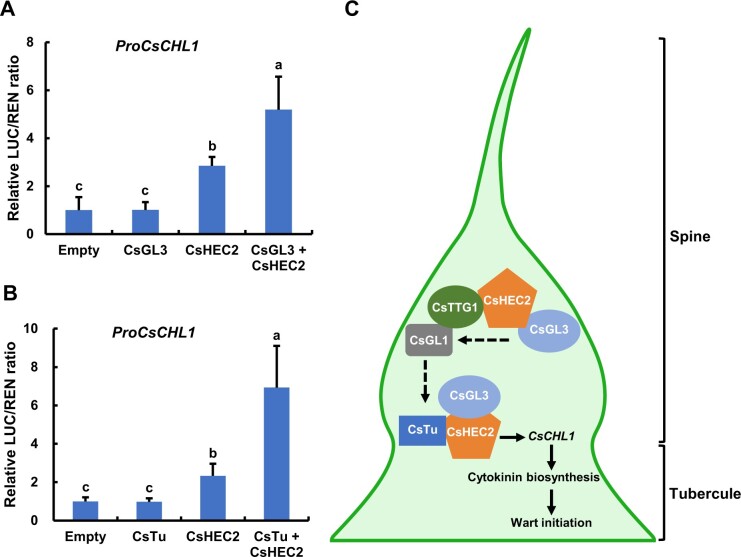
Given that CsHEC2 interacted with CsGL3 and CsTu, the core initiation regulators of spine and tubercule, respectively, we next investigate whether such interactions affect the downstream target CsCHL1 of CsHEC2 by DLR assays. The expression of CsHEC2 protein led to an increased LUC/REN ratio of ProCsCHL1, which was further enhanced by co-expression of CsHEC2 and CsGL3 proteins (Figure 7A). Similarly, when ProCsCHL1:LUC reporter was co-expressed with both CsHEC2 and CsTu, the activation of LUC activity was significantly increased compared with CsHEC2 alone (Figure 7B). These data suggested that CsHEC2 functions as a key cofactor for CsGL3 and CsTu to control wart formation via CsCHL1-mediating CTK biosynthesis in cucumber.
Figure 7.
CsGL3 and CsTu enhanced the CsHEC2-mediated CsCHL1 transcriptional activation. A and B, Transient transcriptional activity assays in N. benthamiana leaves showed that CsGL3 (A) and CsTu (B) further enhanced CsCHL1 transcriptional activation regulated by CsHEC2. Values are means ± SD (n = 5). The different lowercase letters indicate significant differences (P < 0.01) by one-way ANOVA analysis with Tukey’s HSD test. C, A working model of CsHEC2 regulation of fruit wart formation in cucumber. CsHEC2 promotes spine initiation and subsequent development through interaction with CsGL3 and CsTTG1 to indirectly stimulate expression of CsGL1. Meanwhile, CsHEC2 induces fruit tubercule formation via interacting with CsGL3 and CsTu and directly activates the expression of CTK biosynthesis gene CsCHL1 in cucumber.
Discussion
The bHLH transcription factor CsHEC2 is a wart formation regulator in cucumber
Trichomes are specialized structures in the aerial parts of plants derived from epidermal cells, and serve as the protective barrier from surrounding stresses, such as UV radiation, low temperature, insect herbivory, and pathogen invasion (Werker, 2000; Wagner et al., 2004). The unicellular and branched trichome of Arabidopsis is one of the best-studied models to explore the gene regulatory network underlying cell fate determination and cell morphogenesis. There are three groups of players in this regulatory complex, namely R2R3-MYB transcription factors (Glabra1, also named Glabrous1; Oppenheimer et al., 1991 or MYB23 [Kirik et al., 2005]), bHLH transcription factors (GL3; Payne et al., 2000 or Enhancer of Glabra3 [Zhang et al., 2003]), and the WD40 repeat protein TTG1 (Galway et al., 1994; Walker et al., 1999), that form an MYB-bHLH-WD40 complex to positively regulate trichome formation by inducing the class IV HD-ZIP gene GL2 expression (Larkin et al., 2003; Morohashi et al., 2007; Ishida et al., 2008; Wang and Chen, 2008). Unlike that in Arabidopsis, trichomes of cucumber are multicellular and unbranched, in which those covered the surface of fruit are called spines. Fruit spines and tubercules together constitute the warty trait, which determines the appearance quality and final market value in cucumber (Pan et al., 2015). Although several genes and hormones have been reported to regulate spine or tubercule formation, no bHLH gene was found to be involved in wart formation in cucumber yet.
In this study, we used CRISPR/Cas9 technology to knock out the bHLH transcription factor CsHEC2 and found that the density of fruit wart in cucumber was significantly reduced (Figures 1 and 2). Overexpression of CsHEC2 resulted in significant increase in wart density (Figure 4). Importantly, Cshec2 mutants showed no significant changes in fruit length (Figure 2H) or fruit diameter (Figure 2I) compared to WT. Therefore, CsHEC2 is a bHLH gene functions specifically for fruit wart formation in cucumber. Our recent studies showed that the HEC3 subfamily gene CsIVP functions in organ morphogenesis by directly promoting vascular-related regulators expression, and in downy mildew resistance by salicylic acid-mediated pathway in cucumber (Yan et al., 2020). In the evolution of HEC family genes, the HEC3 group and the HEC1/2 clades were produced by a duplication event prior to the origin of angiosperms (Ortiz-Ramírez et al., 2018; Yan et al., 2020). In Arabidopsis, mutations of HEC genes led to developmental defects in the stigma, septum, and transmitting tract (Gremski et al., 2007), and HEC genes buffered auxin and CTK signals during gynoecium development and shoot stem cell control (Schuster et al., 2014, 2015). No roles of HECs have been reported in trichome development yet. The functional divergence of HEC genes between cucumber and Arabidopsis may be due to neofunctionalization after replication. Besides, unlike the pepo fruit of cucumber, the Arabidopsis fruit belongs to the silique that fails to produce any spines or tubercules on its fruit surface. Therefore, studying the function of HEC2 homologs in different fruit types with distinct trichome characteristics will help to understand the cause of gene functional differentiation.
CsHEC2 regulates spine formation through interaction with CsGL3 and CsTTG1
During spine development, the CsGL3/Tril/FS1 gene has been shown to play a core role in spine initiation through gene dosage effects (Pan et al., 2015; Cui et al., 2016; Wang et al., 2016; Zhang et al., 2016; Du et al., 2020). The Class I HD-ZIP transcription factor CsGL1/TBH/Mict functions in trichome differentiation rather than the initiation process (Li et al., 2015; Zhao et al., 2015). Genetic analyses showed that CsGL3 has an epistatic effect on CsGL1 and the expression of CsGL1 depends on CsGL3 in cucumber (Pan et al., 2015). Here, we found that CsHEC2 directly interacted with CsGL3 at the protein level (Figure 5, B, C, and G). We also found that the expression of CsGL3 was unaffected in Cshec2 mutant, while the expression of CsGL1 had a significant decrease (Figure 6A;Supplemental Figure S5, E and H). Our DLR assay showed that CsGL3 and CsHEC2 were unable to directly activate the expression of CsGL1 gene (Figure 6C). In addition, we also detected the direct protein interaction between CsHEC2 and CsTTG1 (Figure 5, B, E, and I), which regulates the initial development of fruit spines through interaction with CsGL1 (Chen et al., 2016). Therefore, CsHEC2 may form a protein complex with CsGL3 and CsTTG1 to control spine initiation and subsequent development via indirectly promoting the expression of CsGL1 (Figure 7C).
CsHEC2 promotes tubercule formation through interaction with CsTu to directly stimulate CTK biosynthesis in the fruit peel of cucumber
Previous studies showed that CsTu is required for warty fruit phenotype and probably functions through activating the CTK hydroxylase-like genes, CsCHL1 and CsCHL2 (Zhang et al., 2010; Yang et al., 2014). However, the specifical mechanism of how CsTu regulates CsCHL1/2 expression remains unknown. CTK has been shown to play an important role in nodule development, in which a local increase in CTK activates cortical cell division, and exogenous CTK treatment is sufficient to induce the dedifferentiation and division of legume cortical cells, resulting in the formation of nodule primordia (Cooper and Long, 1994; Mathesius et al., 2000; Oldroyd, 2007; Dong et al., 2020). Similar to that in nodule, occurrence of fruit wart formation was reported to be associated with high CTK content (Yang et al., 2014). In this study, we found that CTK content was significantly lower in the Cshec2 mutant (Figure 3, A and B), and greatly increased in the CsHEC2-OE lines (Figure 4G). Meanwhile, the expression level of CsCHL1 was substantially decreased in Cshec2 mutants (Figure 3E;Supplemental Figure S5A), and elevated in CsHEC2-OE lines (Figure 4H). Exogenous CTK treatment partially restored the wart density phenotype in the Cshec2 mutant (Figure 3, F–H). Importantly, we found that CsHEC2 directly binds to the CsCHL1 promoter and facilitates its transcriptional activation, while CsTu could not (Figure 6, B–H). We further showed that the protein interactions of CsHEC2 with CsGL3 and CsTu reinforced the CsHEC2-mediated CsCHL1 transcriptional activation (Figure 7, A and B). Together, our data suggested that CsHEC2 promotes fruit tubercule formation via cooperating with CsGL3 and CsTu to directly activate the expression of CsCHL1 and thereby increases CTK accumulation in cucumber (Figure 7C). Our work provides a valuable gene target for molecular breeding of cucumber varieties with different wart density requirements. Future studies with applications of genomics and CRISPR/Cas9 technology to explore the function of putative additional players will help enrich the regulatory network underlying fruit wart formation, which will serve as the gene resources for accelerating improvement of fruit appearance quality in cucumber.
Materials
Inbred line XTMC (Northern China type) was used for expression analysis and genetic transformation. After seeds germinated in an incubator, the cucumber seedlings were transferred to the greenhouse of China Agricultural University in Beijing, where they were grown under standard water management and pest control. Cotyledons of cucumber seedlings growing for about 10 d in growth chamber were used for protoplast isolation and transformation. Nicotiana benthamiana plants were grown in a growth chamber set at 24°C under a long-day condition (16-h light/8-h dark). The fully expanded young leaves of N. benthamiana about 6-week-old plants were used for biochemical experiments.
RNA extraction and RT-qPCR
Total RNA was extracted from different cucumber tissues at different developmental stages using an Eastep Super isolation Kit (Promega, Madison, WI, USA) and cDNA was synthesized using a FastKing gDNA Dispelling RT SuperMix Kit (Tiangen Biotech, Beijing, China) according to the manufacturer's protocol. RT-qPCR was performed to determine the expression levels of genes by using the TB Green®Premix Ex Taq™ II (Takara, Shiga, Japan) on the CFX384 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Three biological replicates and three technical replicates were performed for each gene. The cucumber ubiquitin extension protein gene (CsaV3_5G031430) was used as an internal reference to normalize the expression results (Wan et al., 2010). The primer information is listed in Supplemental Table S3.
Subcellular localization
The full-length CDS without the stop codon of CsHEC2 was cloned into the pSUPER1300 vector and fused with the green fluorescent protein (GFP) to produce a CsHEC2–GFP fusion protein. The empty pSUPER1300 vector was served as a control. The resultant constructs were introduced into the Agrobacterium tumefaciens strain GV3101. Nicotiana benthamiana leaves were co-transformed with the GFP-fusion construct and the nuclear location marker (NF-YA4-mCherry; Zhang et al., 2019). After 48h infiltration, fluorescence signals were visualized at excitation/emission wavelength of 488/510 nm (GFP), 552/610 nm (mCherry), using a confocal laser scanning microscope (Leica SP8, Germany). The primer information is listed in Supplemental Table S3.
Cucumber transformation
To generate the construct used for CRISPR/Cas9-edited plants of CsHEC2, the specific sgRNA target sites were selected by the sgRNA design web (http://crispr.hzau.edu.cn/CRISPR2/). The PCR fragment harboring two target sites was amplified using four partially overlapping primers and then inserted in the binary CRIPSR/Cas9 vector pKSE402G using BsaI site and T4 Ligase (New England Biolabs, Ipswich, MA, USA; Xing et al., 2014; Hu et al., 2017). To obtain CsHEC2 overexpression lines, full-length CDS of CsHEC2 was amplified and cloned into pCAMBIA1300-Flag vector to generate the Pro35S:CsHEC2-Flag construct. The D35S-GFP expression cassette was inserted into the Pro35S:CsHEC2-Flag construct using the EcoRI site to generate the overexpression vector (pCAMBIA1300-Flag+GFP). The resultant constructs were transferred into the A. tumefaciens strain EHA105 and then transformed into cucumber by Agrobacterium-mediated cotyledon method as previously described (Hu et al., 2017).
The GFP of pKSE402G or modified overexpression vector was used as a reporter to select putative positive buds in T0 transgenic plants. Genomic DNA was extracted from plants using the CTAB method. The target gene and potential off-target sites were respectively amplified with specific primers, and the resultant PCR products were sequenced and aligned by Geneious software. The primer information is listed in Supplemental Table S3.
Yeast two-hybrid assay
The full-length CDSs of CsHEC2, CsTu, CsGL3, CsTTG1, and CsMYB6 were cloned into pGADT7 or pGBKT7 vector. All reconstructed plasmids were verified by sequencing and then transformed into yeast strain AH109. Yeast two-hybrid assays were performed using the Matchmaker™ GAL4 Two-Hybrid System 3 & Libraries (Clontech, Mountain View, CA, USA) method, following the description of manufacturer’s instructions. The yeast transformants were grown on SD/-Leu/-Trp medium and protein interactions were assayed on selective medium SD/–Trp/–Leu/-His/-Ade with X‐α‐Gal. The primer information is listed in Supplemental Table S3.
LCI assay
The full-length CDS of CsHEC2 was cloned into pCAMBIA1300-cLUC, and the full-length CDSs without the stop codon of CsTu, CsGL3, CsTTG1, and CsMYB6 were cloned into pCAMBIA1300-nLUC. The prepared vectors were transferred into the Agrobacterium strain GV3101 and then co-infiltrated into N. benthamiana leaves together with the p19 Agrobacterium as described previously (Zhou et al., 2018). After 48-h infiltration, the abaxial sides of leaves were sprayed with 1 mM D-Luciferin, Potassium Salt (Biovision, San Francisco, CA, USA), and then the LUC activities were analyzed and captured using a CCD imaging system of MiniChemi 610 (Sagecreation). The primer information is listed in Supplemental Table S3.
Co-IP assay
The CDS of CsHEC2 was cloned into pCAMBIA1300-GFP vector, and the CDSs of CsTu, CsGL3, CsTTG1, and CsMYB6 were cloned into pCAMBIA1300-FLAG vector. The prepared vectors were transferred into the Agrobacterium strain GV3101 and then co-infiltrated into N. benthamiana leaves together with the p19 Agrobacterium as described previously (Zhou et al., 2018). After 48-h infiltration, samples were collected, ground into fine powder in liquid nitrogen and homogenized in extraction buffer (50 mM HEPES [pH 7.5], 150 mM KCl, 1 mM EDTA [pH 8.0], 2.5 mM MgCl2, 1 mM DTT, 0.5% (v/v) Triton X-100, 1× protease inhibitor cocktail [Roche, Basel, Switzerland]). Immunoprecipitation was performed using anti-GFP nanobody-coated agarose beads (KT-HEALTH, China, Catalog No. KTSM1301) at 4°C for 2 h. The beads were washed 6 times with washing buffer (50 mM HEPES [pH 7.5], 150 mM KCl, 1 mM EDTA, 0.2% (v/v) Trition-X 100, 1 mM DTT). The immunoprecipitates were separated by SDS-PAGE and detected by immunoblot with anti-GFP (TransGen Biotech, Beijing, China, Catalog No. HT801) or anti-FLAG (Sigma-Aldrich, Burlington, MA, USA; Catalog No. F3165) antibodies. The primer information is listed in Supplemental Table S3.
Yeast one-hybrid assay
The full-length CDSs of CsHEC2 and CsTu were cloned into pB42AD vector (effector). The 30-bp sequences around the E-box cis-element (CANNTG) of possible CsHEC2 binding site or sequences of possible CsTu binding site (A[AG/CT]CNAC) in the promoter regions of CsCHL1∼2 kb upstream of the start codon were repeated three times and cloned into the pLacZi2u vector (reporter). The combinations of indicated effectors and reporters were co-transferred into the yeast strain EGY48. The yeast assays were performed according to described previously (Li et al., 2010). Yeast positive transformants were selected on SD/-Trp-Ura agar plates and interactions of protein and DNA were assayed on SD/Gal/Raf/-Trp-Ura agar plates containing 40 ug/mL 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside by observing blue color development. The primer information is listed in Supplemental Table S3.
DLR assay
The transient transcriptional activity assay was analyzed in N. benthamiana leaves as described previously (Ohta et al., 2001; Xu et al., 2020). The full-length CDS of CsHEC2 was cloned into the effector GAL4DB vector to generate the GAL4DB-CsHEC2. The fusion effector plasmids (GAL4DB-CsHEC2 or positive control GAL4DB-VP16 or negative control GAL4DB or empty control) and reporter plasmid were introduced into Agrobacterium strain GV3101. Different combinations of Agrobacterium culture were co-injected into N. benthamiana leaves for transcriptional activity analysis.
To detect transient transcriptional activity of specific promoters, the full-length CDSs of CsHEC2, CsTu and CsGL3 were fused to the pGreen II 62-SK vector to generate effector constructs; the promoter of CsCHL1 (−1,890 bp), CsCHL2 (−1,792 bp), and CsGL1 (−2,082 bp) were cloned into the vector pGreen II 0800-LUC to generate the reporter constructs. The REN gene under the control of cauliflower mosaic virus 35S promoter in the pGreenII 0800-LUC vector was used as the internal control. The prepared vectors were introduced into Agrobacterium strain GV3101 (pSoup19) and then co-infiltrated in N. benthamiana leaves. After incubation for 48-h, the Firefly LUC and REN activities were assayed using the dual-LUC® reporter assay reagents (Promega, Madison, WI, USA; Hellens et al., 2005). The ratio of LUC to REN was calculated as the final transcriptional activity of the corresponding combination. The primer information is listed in Supplemental Table S3.
ChIP assay
The CDS of CsHEC2 or CsTu was cloned into PUC19-35S-FLAG-RBS vector. The cucumber protoplasts were transfected with the prepared plasmids and incubated overnight at 23°C. The samples were treated with the final concentration of 1% (v/v) formaldehyde to crosslink protein-DNA complexes, and the final concentration of 0.125 M glycine was added to stop the cross-linking. The cross-linked tissues were then used for ChIP experiments as described previously (Ding et al., 2015). ChIP reaction was performed using anti-FLAG M2 affinity agarose beads (Sigma-Aldrich, Burlington, MA, USA, Catalog No. A2220). The co-precipitated DNA was purified by QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany) and analyzed using RT-qPCR. Cucumber TUBULIN gene was used as an internal control. Three biological and three technical replicates were performed. The primers for ChIP-PCR are listed in Supplemental Table S3.
RNA-sequencing and data analysis
RNA-seq experiments were performed using the female buds at 7 DBA. Samples were collected from WT and Cshec2 mutant plants, and three biological replicates were prepared for each sample. RNA library construction and sequencing were performed by the Biomarker Technologies Corporation (Beijing, China) on an Illumina NovaSeq 6000 platform. After screening and trimming, clean reads were mapped to the cucumber genome (Chinese Long version 2.0) using the HISAT2 software (Huang et al., 2009; Li et al., 2011; Kim et al., 2015). Transcriptomic data were analyzed on the BMKCloud platform (www.biocloud.net). The sequencing data information was displayed in Supplemental Table S2. Differentially expressed genes (DEGs) were calculated using DESeq2 (Love et al., 2014) and the false discovery rate ˂0.05 and the fold change >1.5 as were set as the threshold.
Measurement of endogenous hormones
To measure the levels of endogenous auxin (IAA), CTK (IPA, ZR, and DHZR), and GA3, about 0.2 g of fruit peel samples at two DBA from WT, Cshec2 mutants or CsHEC2-OE lines were harvested and homogenized in 3 mL of 80% (v/v) methanol (containing antioxidant). Then, the extraction and quantification of phytohormones were performed using enzyme-linked immunosorbent assay as previously described (Wang et al., 2012). Three biological replicates were performed for each plant type.
Exogenous CTK treatments
CTK treatment was carried out as described previously (Xue et al., 2019). Plants of WT and Cshec2 were treated with 100 or 0 uM BAP (control) (MedChemExpress, Catalog No HY-B0941), a synthetic CTK, when the first true leaf has just emerged. The seedlings were sprayed every other day until they reached the five-true-leaf stage, then twice a week until fruit set. Each treatment was performed with six plants and the experiment was repeated twice independently.
Accession numbers
The accession number of all genes used in this paper is listed in Supplemental Table S5. Sequencing data were deposited with the Gene Expression Omnibus database at the National Center for Biotechnology Information under accession number GSE166286.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Morphological characteristics of cucumber fruit surface.
Supplemental Figure S2. Morphological characteristics of leaf surface in WT and CsHEC2 knockout lines.
Supplemental Figure S3. Contents of IAA and GA3 in the fruit peels of WT and CsHEC2 knockout lines.
Supplemental Figure S4. Global analysis of RNA-Seq data from WT and Cshec2 samples.
Supplemental Figure S5. RT-qPCR analysis of DEGs selected from the transcriptomic data.
Supplemental Figure S6. Contents of ZR, DHZR, IAA, and GA3 in the fruit peels of WT and CsHEC2-OE lines.
Supplemental Figure S7. In situ hybridization of CsCHL1 in cucumber.
Supplemental Table S1. Analysis of the potential off-target sites for CsHEC2 sgRNA.
Supplemental Table S2. Summary of transcriptome sequencing data.
Supplemental Table S3. List of DEGs between WT and Cshec2 female buds.
Supplemental Table S4. Primer information used in this study.
Supplemental Table S5. Accession number of genes used in this paper.
Supplementary Material
Acknowledgments
The authors are grateful to members of Zhang’s lab for technical assistance and discussions.
Funding
This work is supported by grants from the National Natural Science Foundation of China [32025033] and [31930097], the National Key Research and Development Program [2018YFD1000800], and the Chinese Universities Scientific Fund.
Conflict of interest statement. The authors declare that they have no conflicts of interest.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
X.Z., Z.W., and Z.Z. designed the research; Z.W., L.W., L.H., and Z.Z. performed the experiments; X.Z., Z.W., and Z.Z. wrote the paper; Z.C., X.L., S.W., L.L., J.C., W.S., and J.Z. provided experimental assistance; all the authors revised the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Xiaolan Zhang (zhxiaolan@cau.edu.cn).
References
- Bajguz A, Piotrowska A (2009) Conjugates of auxin and cytokinin. Phytochemistry 70: 957–969 [DOI] [PubMed] [Google Scholar]
- Cao C, Zhang S, Guo H (2001) The genetic relationship between glabrous foliage character and warty fruit character of cucumber. Acta Hortic Sin 28: 565–566 [Google Scholar]
- Che G, Zhang X (2019) Molecular basis of cucumber fruit domestication. Curr Opin Plant Biol 47: 38-46 [DOI] [PubMed] [Google Scholar]
- Chen C, Liu M, Jiang L, Liu X, Zhao J, Yan S, Yang S, Ren H, Liu R, Zhang X (2014) Transcriptome profiling reveals roles of meristem regulators and polarity genes during fruit trichome development in cucumber (Cucumis sativus L.). J Exp Bot 65: 4943–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Yin S, Liu X, Liu B, Yang S, Xue S, Cai Y, Black K, Liu H, Dong M. et al. (2016) The WD-repeat protein CsTTG1 regulates fruit wart formation through interaction with the homeodomain-leucine zipper I protein mict. Plant Physiol 171: 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JB, Long SR (1994) Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. Plant Cell 6: 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Miao H, Ding LH, Wehner TC, Liu PN, Wang Y, Zhang SP, Gu XF (2016) A new glabrous gene (csgl3) identified in trichome development in cucumber (Cucumis sativus L.). PLoS One 11: e0148422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Yan S, Jiang L, Zhao W, Ning K, Zhao J, Liu X, Zhang J, Wang Q, Zhang X (2015) HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in Arabidopsis. PLoS Genet 11: e1005479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Zhu Y, Chang H, Wang C, Yang J, Shi J, Gao J, Yang W, Lan L, Wang Y. et al. (2020) An SHR-SCR module specifies legume cortical cell fate to enable nodulation. Nature 589: 586–590 [DOI] [PubMed] [Google Scholar]
- Du H, Wang G, Pan J, Chen Y, Xiao T, Zhang L, Zhang K, Wen H, Xiong L, Yu Y. et al. (2020) The HD-ZIP IV transcription factor Tril regulates fruit spine density through gene dosage effects in cucumber. J Exp Bot 71: 6297–6310 [DOI] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW (1994) The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF (2007) The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601 [DOI] [PubMed] [Google Scholar]
- Guo J, Xu W, Hu Y, Huang J, Zhao Y, Zhang L, Huang CH, Ma H (2020) Phylotranscriptomics in cucurbitaceae reveal multiple whole-genome duplications and key morphological and molecular innovations. Mol Plant 13: 1117–1133 [DOI] [PubMed] [Google Scholar]
- Han G, Wei X, Dong X, Wang C, Sui N, Guo J, Yuan F, Gong Z, Li X, Zhang Y. et al. (2020) Arabidopsis ZINC FINGER PROTEIN1 acts downstream of GL2 to repress root hair initiation and elongation by directly suppressing bHLH genes. Plant Cell 32: 206–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Li D, Liu X, Qi J, Gao D, Zhao S, Huang S, Sun J, Yang L (2017) Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol Plant 10: 1575–1578 [DOI] [PubMed] [Google Scholar]
- Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P. et al. (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Lee MM, Wester K, Herrmann U, Zheng Z, Oppenheimer D, Schiefelbein J, Hulskamp M (2005) Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132: 1477–1485 [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Larkin JC, Brown ML, Schiefelbein J (2003) How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu Rev Plant Biol 54: 403–430 [DOI] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y. et al. (2010) Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Cao C, Zhang C, Zheng S, Wang Z, Wang L, Ren Z (2015) The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J Exp Bot 66: 2515–2526 [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Z, Yan P, Huang S, Fei Z, Lin K (2011) RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genomics 12: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bartholomew E, Cai Y, Ren H (2016) Trichome-related mutants provide a new perspective on multicellular trichome initiation and development in cucumber (Cucumis sativus L). Front Plant Sci 7: 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Charon C, Rolfe BG, Kondorosi A, Crespi M (2000) Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol Plant Microbe Interact 13: 617–628 [DOI] [PubMed] [Google Scholar]
- Morohashi K, Zhao M, Yang M, Read B, Lloyd A, Lamb R, Grotewold E (2007) Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol 145: 736–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE (2007) Plant science. Nodules and hormones. Science 315: 52–53 [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493 [DOI] [PubMed] [Google Scholar]
- Ortiz-Ramírez CI, Plata-Arboleda S, Pabón-Mora N (2018) Evolution of genes associated with gynoecium patterning and fruit development in Solanaceae. Ann Bot 121: 1211–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Bo K, Cheng Z, Weng Y (2015) The loss-of-function GLABROUS 3 mutation in cucumber is due to LTR-retrotransposon insertion in a class IV HD-ZIP transcription factor gene CsGL3 that is epistatic over CsGL1. BMC Plant Biol 15: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels AL, Glass ADM, Ehret DL, Menzies JG (1993) The effects of silicon supplementation on cucumber fruit: changes in surface characteristics. Ann Bot 72:433–440 [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ (2015) The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27: 44–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C, Gaillochet C, Lohmann JU (2015) Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 142: 3343–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C, Gaillochet C, Medzihradszky A, Busch W, Daum G, Krebs M, Kehle A, Lohmann JU (2014) A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev Cell 28: 438–449 [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang A, Zhou Z, Zhao Y, Yan A, Bao S, Yu H, Gan Y (2015) GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol 206: 220–230 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GJ, Wang E, Shepherd RW (2004) New approaches for studying and exploiting an old protuberance, the plant trichome. Ann Bot 93: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399: 257–261 [DOI] [PubMed] [Google Scholar]
- Wang G, Qin Z, Zhou X, Zhao ZY (2007) Genetic analysis and SSR markers of tuberculate trait in Cucumis sativus. Chin Bull Bot 24: 168–172 [Google Scholar]
- Wang S, Chen JG (2008) Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol 49: 1792–1804 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li B, Du M, Eneji AE, Wang B, Duan L, Li Z, Tian X (2012) Mechanism of phytohormone involvement in feedback regulation of cotton leaf senescence induced by potassium deficiency. J Exp Bot 63: 5887–5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Nie JT, Chen HM, Guo CL, Pan J, He HL, Pan JS, Cai R (2016) Identification and mapping of Tril, a homeodomain-leucine zipper gene involved in multicellular trichome initiation in Cucumis sativus. Theor Appl Genet 129: 305–316 [DOI] [PubMed] [Google Scholar]
- Werker E (2000) Trichome diversity and development. Adv Bot Res 31: 1–35 [Google Scholar]
- Xie Q, Liu P, Shi L, Miao H, Bo K, Wang Y, Gu X, Zhang S (2018) Combined fine mapping, genetic diversity, and transcriptome profiling reveals that the auxin transporter gene ns plays an important role in cucumber fruit spine development. Theor Appl Genet 131: 1239–1252 [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhao X, Aiwaili P, Mu X, Zhao M, Zhao J, Cheng L, Ma C, Gao J, Hong B (2020) A zinc finger protein BBX19 interacts with ABF3 to affect drought tolerance negatively in chrysanthemum. Plant J 103: 1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Dong M, Liu X, Xu S, Pang J, Zhang W, Weng Y, Ren H (2019) Classification of fruit trichomes in cucumber and effects of plant hormones on type II fruit trichome development. Planta 249: 407–416 [DOI] [PubMed] [Google Scholar]
- Yan S, Ning K, Wang Z, Liu X, Zhong Y, Ding L, Zi H, Cheng Z, Li X, Shan H. et al. (2020) CsIVP functions in vasculature development and downy mildew resistance in cucumber. PLoS Biol 18: e3000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Cai Y, Liu X, Dong M, Zhang Y, Chen S, Zhang W, Li Y, Tang M, Zhai X. et al. (2018) A CsMYB6-CsTRY module regulates fruit trichome initiation in cucumber. J Exp Bot 69: 1887–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wen C, Liu B, Cai Y, Xue S, Bartholomew ES, Dong M, Jian C, Xu S, Wang T. et al. (2019) A CsTu-TS1 regulatory module promotes fruit tubercule formation in cucumber. Plant Biotechnol J 17: 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhang W, He H, Nie J, Bie B, Zhao J, Ren G, Li Y, Zhang D, Pan J. et al. (2014) Tuberculate fruit gene Tu encodes a C2 H2 zinc finger protein that is required for the warty fruit phenotype in cucumber (Cucumis sativus L.). Plant J 78: 1034–1046 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang L, Zheng S, Liu Z, Wu X, Gao Z, Cao C, Li Q, Ren Z (2016) A fragment substitution in the promoter of CsHDZIV11/CsGL3 is responsible for fruit spine density in cucumber (Cucumis sativus L.). Theor Appl Genet 129: 1289–1301 [DOI] [PubMed] [Google Scholar]
- Zhang S, Feng M, Chen W, Zhou X, Lu J, Wang Y, Li Y, Jiang CZ, Gan SS, Ma N, Gao J (2019) In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nat Plants 5: 290–299 [DOI] [PubMed] [Google Scholar]
- Zhang W, He H, Guan Y, Du H, Yuan L, Li Z, Yao D, Pan J, Cai R (2010) Identification and mapping of molecular markers linked to the tuberculate fruit gene in the cucumber (Cucumis sativus L.). Theor Appl Genet 120: 645–654 [DOI] [PubMed] [Google Scholar]
- Zhao JL, Pan JS, Guan Y, Zhang WW, Bie BB, Wang YL, He HL, Lian HL, Cai R (2015) Micro-trichome as a class I homeodomain-leucine zipper gene regulates multicellular trichome development in Cucumis sativus. J Integr Plant Biol 57: 925–935 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Bi G, Zhou JM (2018) Luciferase complementation assay for protein-protein interactions in plants. Curr Protoc Plant Biol 3: 42–50 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.