Abstract
Environmental stresses cause an increased number of unfolded or misfolded proteins to accumulate in the endoplasmic reticulum (ER), resulting in ER stress. To restore ER homeostasis and survive, plants initiate an orchestrated signaling pathway known as the unfolded protein response (UPR). Asparagine-rich protein (NRP) 1 and NRP2, two homologous proteins harboring a Development and Cell Death domain, are associated with various stress responses in Arabidopsis (Arabidopsis thaliana), but the relevant molecular mechanism remains obscure. Here, we show that NRP1 and NRP2 act as key pro-survival factors during the ER stress response and that they inhibit cell death. Loss-of-function of NRP1 and NRP2 results in decreased tolerance to the ER stress inducer tunicamycin (TM), accelerating cell death. NRP2 is constitutively expressed while NRP1 is induced in plants under ER stress. In Arabidopsis, basic leucine zipper protein (bZIP) 28 and bZIP60 are important transcription factors in the UPR that activates the expression of many ER stress-related genes. Notably, under ER stress, bZIP60 activates NRP1 by directly binding to the UPRE-I element in the NRP1 promoter. These findings reveal a pro-survival strategy in plants wherein the bZIP60–NRPs cascade suppresses cell death signal transmission, improving survival under adverse conditions.
One-sentence summary: Asparagine-rich protein 1 and 2 containing a Development and Cell Death domain inhibit cell death to promote plant survival under environmental stress conditions.
Introduction
Because of their sessile nature, plants are constantly subjected to a variety of environmental stresses caused by abiotic and biotic factors. These adverse environmental cues (heat, drought, salt, cold, etc.) disturb cell homeostasis and cause the accumulation of misfolded or unfolded proteins in the endoplasmic reticulum (ER), causing ER stress (Liu et al., 2007b; Deng et al., 2011; Gidalevitz et al., 2011; Moreno et al., 2012). In order to adjust the protein folding capacity of ER and promote cell survival, organisms have evolved a conserved mechanism: the unfolded protein response (UPR), which restores ER homeostasis or ultimately induces cell death under irremediable ER stress conditions (Hetz et al., 2020). This evolutionarily conserved mechanism has been revealed in eukaryotes from yeast to angiosperms and mammals (Mori et al., 1992; Tirasophon et al., 1998; Iwata and Koizumi, 2005).
In mammals, the ER-localized stress sensors consist of the protein kinase RNA-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1; Harding et al., 1999; Shen et al., 2002; Tirasophon et al., 1998). In plants, the functional orthologs of IRE1(IRE1a/b) and ATF6 (basic leucine zipper protein 28 [bZIP28]) have been identified, and their regulatory mechanisms involved in the UPR are similar to those in mammals, in principle. Under normal conditions, the membrane-anchored transcription factors bZIP28 and IRE1a/b, the so-called “first responders,” are retained in the ER by interacting with chaperone binding immunoglobulin protein (BiP). Upon mild ER stress, BiP dissociates from the lumen-facing domain, thus liberating bZIP28 from the endoplasmic membranes to the Golgi where bZIP28 undergoes proteolytic processing mediated by sites 1 and site-2 proteases (S1P and S2P; Liu et al., 2007a; Srivastava et al., 2013; Sun et al., 2013a, 2015). Recently, it has been reported that the proteolytic activation of bZIP28 in Arabidopsis (Arabidopsis thaliana) does not require S1P (Iwata et al., 2017). On the other hand, the activation of IRE1a/b triggers an unconventional cytosolic splicing of the bZIP60 mRNA, resulting in the removal of the C-terminal transmembrane domain of bZIP60 due to a frameshift (Deng et al., 2011; Nagashima et al., 2011). The spliced bZIP60 and bZIP28 proteins are subsequently re-localized to the nucleus for the transcriptional modulation of downstream UPR target genes, such as the chaperone protein genes BiP/ERdj/CNX/CRT, or NAC transcription factor genes NAC062/103/089 (Kamauchi et al., 2005; Sun et al., 2013b; Yang et al., 2014a, 2014b; Song et al., 2015; Kim et al., 2019). However, reducing the overall frequency of translation via the PERK pathway in response to ER stress remains to be confirmed in plants.
Mild ER stress can be relieved by the “proximal UPR,” as mentioned above, while prolonged or severe stress triggers the “terminal UPR” signaling to ultimately commit cells to programmed death (Hetz and Papa, 2018; Hetz et al., 2020). Interestingly, the ER stress sensors IRE1, ATF6, and PERK that can restore ER homeostasis are also implicated in the activation of caspases in ER stress-induced cell death (Nakagawa et al., 2000). As the “initiator” and “executor,” caspases are essential for triggering cell death (Piszczek and Gutman, 2007; Kurokawa and Kornbluth, 2009). There are three groups of caspase-like proteases also involved in cell death in plants: metacaspases (MCs), vacuolar processing enzymes (VPEs), and saspases (Piszczek and Gutman, 2007; Hao et al., 2008). In Arabidopsis, NAC089, which is induced by bZIP28/60, can promote caspase-3/7-like activity and regulate cell death-related downstream genes including MC5 (Yang et al., 2014b). Two soybean (Glycine max) transcription factors, GmNAC81/30, activate cell death by enhancing the expression of VPE, a protease with caspase-1-like activity, in response to ER stress and osmotic stress (Mendes et al., 2013).
It has been reported that the NRP-mediated cell death pathway is a plant-specific branch of ER stress response. NRPs are asparagine-rich proteins that contain a Development and Cell Death (DCD) domain at the C-terminus (Tenhaken et al., 2005). DCD domain-containing proteins have been identified in algae and plants but not in bacteria, fungi, and animals (Tenhaken et al., 2005). Based on previous reports, NRPs may be closely related to various stress responses since NRPs are induced in salt, osmotic, cold, and ER stress, and so on (Reis and Fontes, 2012). In soybean, GmERD15 activates the expression of GmNRP-B in response to ER stress (Alves et al., 2011). GmNRP-A/B can trigger the expression of GmNAC81/30, thereby activating the caspase-like activity and promoting cell death (Mendes et al., 2013). Overexpression of GmBiP in Nicotiana benthamiana can prevent the NRP-mediated cell death signals from spreading out under ER stress (Reis et al., 2011; Carvalho et al., 2014).
Although DCD domain-containing proteins in Arabidopsis have been reported years ago, their detailed function is still obscure. Recently, two Arabidopsis DCD domain-containing protein genes, NRP1 and NRP2, were demonstrated to promote cell death and senescence (Reis et al., 2016), which is consistent with the hypothetical role of GmNRPs in soybean (Mendes et al., 2013). However, these observations were mostly established on transient expression evidence in N. benthamiana leaves, and the real biological role of NRP genes in plants remains to be verified. Contrary to the previous reports, in this study, we reveal that NRP1 and NRP2 function as pro-survival factors in ER-stress response and negatively regulate cell death and senescence in Arabidopsis. Although the overexpression and loss-of-function of either NRP1 or NRP2 have no effect on growth and ER stress response, loss-of-function of both NRP1 and NRP2 can remarkably induce cell death and promote senescence. Furthermore, we demonstrate that NRP1 is activated by bZIP60, a UPR key regulator, via directly binding to the NRP1 promoter during ER stress response, while NRP2 is constitutively expressed in plants. Our findings support that NRP1 and NRP2, with distinct expression patterns, play important roles in modulating ER stress response by the inhibition of cell death.
Results
NRP1 and NRP2 are essential for ER stress tolerance in Arabidopsis
In order to study the biological role of NRP genes in stress response, we obtained the Arabidopsis T-DNA insertion mutants of NRP1 (At5g42050) and NRP2 (At3g27090) from the Arabidopsis Biological Resource Center (ABRC) (Supplemental Figure S1A). Reverse transcription-PCR(RT-PCR) analysis confirmed that nrp1 and nrp2 are null alleles (Supplemental Figure S1B). These two mutants were visually indistinguishable from Col wild-type plants (Supplemental Figure S2). However, the nrp1 nrp2 double mutant (nrpD) exhibited a markedly precocious senescence phenotype compared with Col and the single mutants at 21 d after germination (Supplemental Figure S2). Consistently, quantitative RT-PCR (RT-qPCR) showed that the expression of SAG12 and SEN4, the known senescence genes, was also dramatically induced in nrpD (Supplemental Figure S3). As senescence process is generally accompanied by ER stress and autophagy (Xiong et al., 2005; Reis et al.,2011; Liu et al., 2012; Carvalho et al.,2014; Pluquet et al., 2015), we further examined the transcription of several marker genes involved in ER stress response and autophagy process. Notably, the expression of BIP3 and CNX1, the indicator genes of UPR (Chen and Brandizzi, 2013), and that of ATG8a and ATG18a, the indicator genes of autophagy (Liu et al., 2012), were also more induced in nrpD compared with that in Col wild-type after 3 weeks’ growth (Supplemental Figure S3). This demonstrated that a chronic ER stress occurs in adult nrpD plants. However, we observed no ER stress-triggered mRNA splicing of the UPR key regulator bZIP60, which generally happens in ER stress response, during plant senescence (Supplemental Figure S4, A and B), indicating that there is a difference between these two processes.
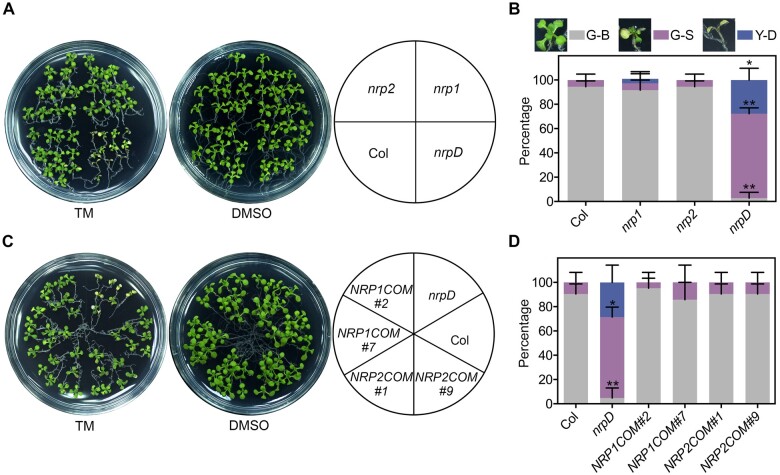
To further explore the possible role of NRPs in ER stress adaptation, 7-d-old Arabidopsis seedlings were treated with 1 μg/mL ER stress inducer tunicamycin (TM), which blocks N-glycosylation and causes protein misfolding in the ER. Unlike the previous report that both nrp1 and nrp2 exhibited more tolerance to ER stress than the wild-type (Reis et al., 2016), we observed no visible difference between nrp1/2 and Col under TM-triggered ER stress by multiple examinations. On the contrary, compared with Col, nrp1, and nrp2, the double mutant nrpD exhibited a noticeable growth retardation, chlorosis, and partial death (Figure 1, A and B). The ratios of green-small (G-S) and yellow-dead (Y-D) seedlings in nrpD were ∼70% and ∼30%, respectively, which were far higher than other genotypes (Figure 1, A and B). Furthermore, either the genomic complementation of NRP1 or NRP2 gene driven by the native promoter (termed as NRP1COM and NRP2COM) can completely rescue the nrpD phenotype to the Col extent (Figure 1, C and D; Supplemental Figure S2), confirming that loss-of-function of NRP1/2 is indeed responsible for the hypersensitivity of nrpD to TM. These observations suggested that NRP1 and NRP2 may play a redundant and essential role in ER stress response in Arabidopsis. However, there was no significant difference in the expression of BIP3 and ATG18a between Col and nrpD with a short term of TM treatment (4–8 h; Supplemental Figure S5). Therefore, we speculated that NRP1/2 may participate mainly in the regulation of cell death during late stage of ER stress response.
Figure 1.
Double mutant of NRP1 and NRP2 exhibits reduced tolerance to the ER stress inducer TM. A and B, TM sensitivity of Col, nrp1, nrp2, and nrpD. C and D, TM sensitivity of the complementation transgenic lines nrpD pNRP1:NRP1-mVenus (NRP1COM #2 and #7) and nrpD pNRP2:NRP2-mVenus (NRP2COM #1 and #9). Seven-day-old seedlings were transferred to 1/2 MS medium containing 1 μg/mL TM (DMSO as mock) for 10 d, and the picture was taken (A and C). The percentages of green-big (G-B), G-S, and Y-D seedlings were calculated (B and D). The images next to the boxes display the phenotype of plants in the three groups. Data represent means ± standard deviation (sd) of biological triplicates. Asterisks indicate significant differences compared with Col (Student’s t test, *P < 0.01; **P < 0.001).
Loss-of-function of NRP1 and NRP2 enhances cell death
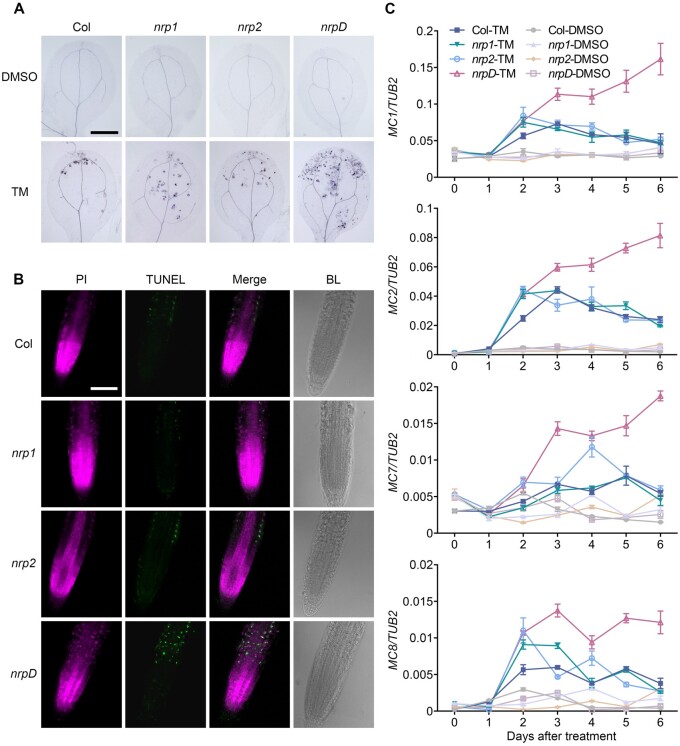
Although a previous study has reported that the loss-of-function of NRP1 or NRP2 enhanced ER stress tolerance and attenuated ER stress-induced cell death (Reis et al., 2016), we clearly observed the opposite phenotype in the double mutant nrpD, as described above. To further verify the cell death phenotype mediated by NRPs, the seedlings of Col, nrp1, nrp2, and nrpD were stained with trypan blue, which only stains dead cells (van Wees, 2008). The TM-treated cotyledons of nrpD showed obviously larger patches of darkly stained cells compared with those of Col, nrp1, and nrp2 (Figure 2A). The terminal deoxynucleotidyl transferase-mediated dUTP nick and labeling (TUNEL) assay (Yang et al., 2014b) further confirmed the cell death result with remarkably stronger TUNEL-positive signals in the root cell of nrpD treated with TM (Figure 2B). bzip60 and ire1a ire1b, two typical ER stress-related mutants, were used as additional controls to evaluate the validity of cell death assays under ER stress (Supplemental Figure S6), respectively. These observations indicated that a higher degree of cell death occurs in nrpD under ER stress.
Figure 2.
TM accelerates cell death in double mutant nrpD. A, The cotyledons of TM-treated Col, nrp1, nrp2, and nrpD seedlings were stained with trypan blue. B, DNA breakage in the Col, nrp1, nrp2, and nrpD roots was detected by TUNEL assay. Seven-day-old seedlings were treated with 1 μg/mL TM (DMSO as mock) for 2 d, and collected for trypan blue staining (cotyledons) or TUNEL assay (roots). Bar = 2 mm in (A), and 100 μm in (B). C, The expression pattern of cell death-related genes. Seven-day-old Col, nrp1, nrp2, and nrpD seedlings were treated with 1 μg/mL TM (DMSO as mock) for 6 d, and collected every day after treatment for RNA extraction. The Tubulin 2 gene (TUB2) was used as an internal control for calculation of relative gene expression level. Data represent means ± sd of biological triplicates.
Proteolytic enzymes, such as MCs, are linked to developmental and stress-induced cell death (Coll et al., 2010). We thus examined the cell death status by analyzing the expression of several MC genes in response to long-term ER stress treatment (Piszczek and Gutman, 2007). In the Col, nrp1, and nrp2 seedlings, the expression level of MC1, MC2, MC7, and MC8 began to increase after the first day of TM treatment and reached the maximum level at the second day, and then declined gradually to the basal level in the following days (Figure 2C). In contrast, the expression of these genes increased continuously and presented a higher level in nrpD spanning the treatment time (Figure 2C). Consistent with the phenotype, the NRP1COM and NRP2COM lines presented an identical expression pattern of MC1 and MC2 to that in Col (Supplemental Figure S7). Other MC genes (MC3, MC5, and MC9) were not significantly induced by TM, while the expression of MC4 and MC6 only showed an increasing pattern in nrpD compared with other genotypes at the fifth day of treatment (Supplemental Figure S8). Therefore, our findings, different from the previous report (Reis et al., 2016), suggested that NRP1 and NRP2 act redundantly as pro-survival factors to enhance the ER stress tolerance and repress cell death in plants.
bZIP60 is required for ER stress-induced NRP1 expression
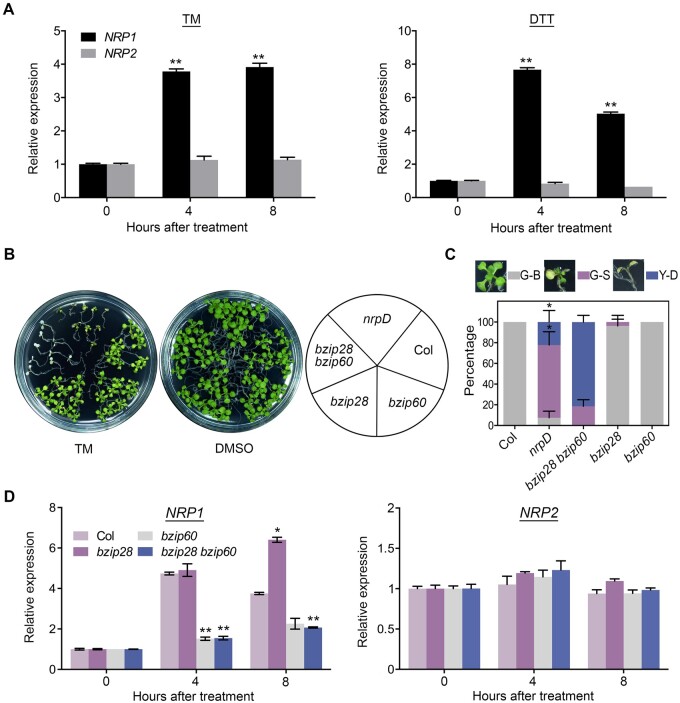
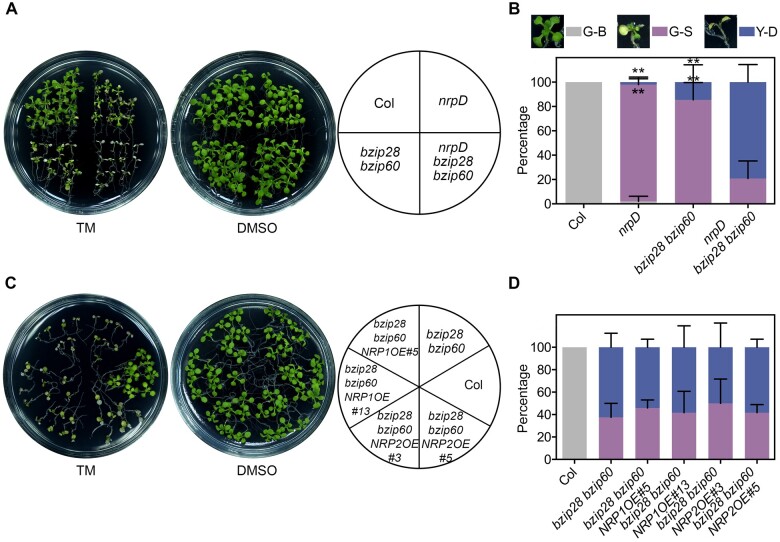
To further investigate the role of NRP1 and NRP2 in ER stress response, we performed qRT-PCR to detect their expression. NRP1, but not NRP2, was significantly upregulated by either TM or dithiothreitol (DTT), another ER stress inducer (Figure 3A). As DTT triggers ER stress by preventing disulfide bond formation, a different mechanism from that of TM (Braakman et al., 1992), it was further confirmed that the induction of NRP1 by ER stress could result from the accumulation of universal misfolded proteins in the ER, rather than the inactivation of specific proteins. These results indicated that NRP1 and NRP2 diverge in expression patterns during ER stress response.bZIP60 and bZIP28 are important regulators in ER stress response (Iwata et al., 2010; Liu and Howell, 2010). Interestingly, like nrpD, the bzip28 bzip60 double mutant also showed hypersensitivity to TM (with a more severe cell death phenotype than nrpD), while there was no obvious phenotype in bzip28 or bzip60 single mutants (Sun et al., 2013a; Figure 3, B and C). Considering the regulatory role of bZIP60/28 in ER stress, we wondered whether these two genes function in the NRP1/2 pathway in ER stress response. To this end, we examined the expression of NRP1/2 and bZIP60/28 in different genotypes. No significant difference in the bZIP60 and bZIP28 transcription or bZIP60 mRNA splicing was observed between the wild-type and nrpD subjected to mock or TM treatment (Supplemental Figures S4C and S9), indicating that bZIP60/bZIP28 are not regulated by NRP1/2. Notably, the TM-triggered induction of NRP1 observed in Col and bzip28 was remarkably impaired in bzip60 single and bzip28 bzip60 double mutant (Figure 3D). In contrast, NRP2 transcripts presented no significant change among Col, bzip28, bzip60, and bzip28 bzip60 before or after TM treatment (Figure 3D). Although a higher expression level of NRP1 was observed in bzip28 compared with that in Col after 8 h of TM treatment, these analyses may indicate that the expression of NRP1 is activated by bZIP60, but not bZIP28, and that NRP2 is not involved in such regulation.
Figure 3.
Gene expression of NRP1 in ER stress is regulated by bZIP60. A, The relative expression level (treatment/mock) of NRP1and NRP2 in Col. Seven-day-old seedlings were treated with 1 μg/mL TM (DMSO as mock), or 2 mM DTT (H2O as mock) for indicated time. Asterisks indicate significant differences compared with 0 h (Student’s t test, **P < 0.001). B and C, TM sensitivity of Col, nrpD, bzip28, bzip60, and bzip28 bzip60. Seven-day-old seedlings were treated with TM for 10 d, and the picture was taken (B). The percentages of G-B, G-S, and Y-D seedlings were calculated (C). The images next to the boxes display the phenotype of plants in the three groups. Asterisks indicate significant difference between nrpD and bzip28 bzip60 (Student’s t test, *P < 0.01). D, The expression level (treatment/mock) of NRP1and NRP2 in Col, bzip28, bzip60, and bzip28 bzip60. Seven-day-old seedlings were treated with 1 μg/mL TM (DMSO as mock) for indicated time. Asterisks indicate significant differences compared with Col (Student’s t test, *P < 0.01; **P < 0.001). TUB2 was used as an internal control of the expression analyses. A and C–D, Data represent means ± sd of biological triplicates.
It has been reported that bZIP60 is also involved in salt stress response (Henriquez-Valencia et al., 2015; Li et al., 2017). Different from that in ER stress, bZIP60 can regulate salt-responsive gene expression under salt stress without IRE1-mediated mRNA splicing (Henriquez-Valencia et al., 2015). Here, we found that NRP1 and NRP2 were also involved in the regulation of salt stress response. Compared with Col and the single mutants, the nrpD double mutant exhibited markedly less tolerance to salt stress (Supplemental Figure S10). Expression analysis showed that both NRP1 and NRP2 were upregulated after 4 h under salt stress, and the induction of NRP1 presented a greater level than that of NRP2 (Supplemental Figure S11). Interestingly, there was no significant difference in the transcription level of NRP1 and NRP2 between Col, bzip28, bzip60, and bzip28 bzip60 (Supplemental Figure S11), suggesting that the salt stress-triggered induction of NRP1 and NRP2 is independent of bZIP60 and bZIP28.
To further investigate the role of NRPs in other stress responses, we treated the mutant seedlings under a mild heat stress (37°C). Like that under salt and ER stress, nrpD exhibited less tolerance to heat stress (Supplemental Figure S12, A and C), and transcript levels of both NRP1 and NRP2 were increased, although NRP2 was more remarkably induced by heat stress (Supplemental Figure S12B). In contrast, NRP1 was regulated by bZIP60, like it was under ER stress (Supplemental Figure S13). These observations suggest that NRPs likely participate in the regulation of ER and heat stress response in a same manner. Since the mRNA of active bZIP60 requires an IRE1a/b-mediated unconventional splicing in ER stress response (Nagashima et al., 2011), we next evaluated the expression of NRP1 in ire1a ire1b. As expected, the bZIP60 mRNA splicing was abolished in this double mutant (Deng et al., 2011; Nagashima et al., 2011). Strikingly, the expression of NRP1 was less changed in ire1a ire1b compared with that in Col wild-type under either ER stress, high salinity, or heat stress (Supplemental Figure S13). Thus, it was indicated that IRE1a/b is not required for the regulation of bZIP60–NRP1 cascade, and that the NRP genes might be regulated by multiple regulators under diverse stresses.
bZIP60 activates NRP1 expression by binding to UPRE-I element
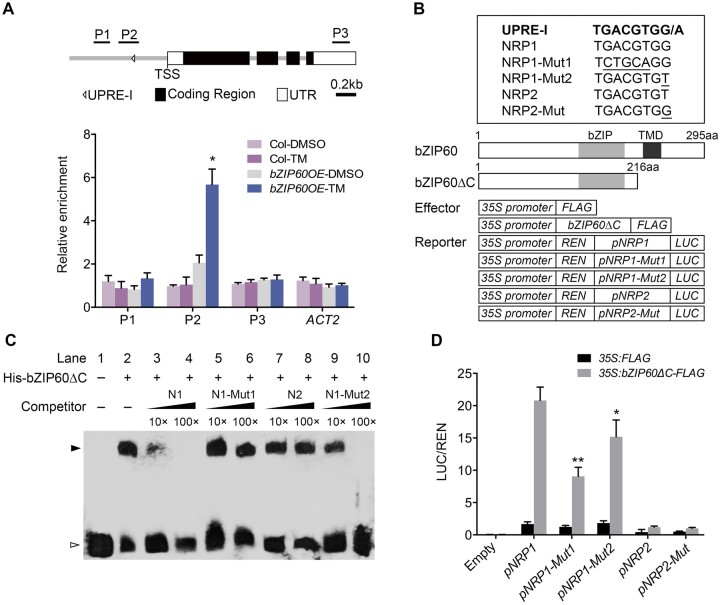
To investigate how bZIP60 regulates the transcription of NRP1 under ER stress, we performed ChIP-qPCR assay with transgenic plants harboring a FLAG-bZIP60 fusion gene driven by the cauliflower mosaic virus (CaMV) 35S promoter (bZIP60OE). The results showed that the P2 region of the NRP1 promoter was enriched significantly by bZIP60 in the TM-treated seedlings (Figure 4A). Likewise, bZIP60 also bound to the same promoter region in the seedlings under heat stress (Supplemental Figure S14B). In contrast, bZIP60 was not associated with the NRP1 promoter when the seedlings were subjected to salt stress (Supplemental Figure S14A), which further supported that the induction of NRP1 by salt stress is independent of bZIP60. Consistent with the result that NRP2 was not regulated by bZIP60 (Figure 3D), bZIP60 did not bind to the NRP2 promoter region (Supplemental Figure S15A).
Figure 4.
bZIP60 activates the expression of NRP1 by directly binding to its promoter. A, ChIP analysis of bZIP60 binding to the NRP1 promoter. The upper part shows schematic diagram of the NRP1 genomic regions. P1–P3 indicate fragments for ChIP-qPCR amplification. The lower part shows ChIP analysis of bZIP60 binding to the NRP1 genomic regions upon the precipitation with anti-FLAG antibody. Seven-day-old seedlings of 35S:FLAG-bZIP60 and Col were treated with 1 μg/mL TM (DMSO as mock) for 4 h and harvested for ChIP assay. The enrichment of Actin 2 (ACT2) genomic fragment was used as the negative control. PP2A was used as an internal control for ChIP analysis. The values are means ± sd of biological triplicates. Asterisks indicate significant differences compared with mock (Student’s t test, *P < 0.01). B, The upper panel shows the UPRE-I element sequence and its mutated sequences used for EMSA (C) and transient expression assay (D). The lower panel shows the schematic diagrams of the effector and reporter constructs used in transient expression assay in (D). aa, amino acid. C, EMSA experiment detecting the protein–DNA binding. The purified His-bZIP60ΔC was incubated with the biotin-labeled NRP1 DNA fragments (40 bp) (Lane 2–10). The un-labeled NRP1/2 DNA and various mutated NRP1 DNA were use as cold competitors. Lane 3, 10× un-labeled NRP1 (N1); lane 4, 100× unlabeled NRP1; lane 5, 10× unlabeled mutated form NRP1-Mut1 (N1-Mut1); lane 6, 100× unlabeled mutated form NRP1-Mut1; lane 7, 10× unlabeled form NRP2 (N2); lane 8, 100× unlabeled form NRP2; lane 9, 10× unlabeled mutated form NRP1-Mut2 (N1-Mut2); lane 10, 100× unlabeled mutated form NRP1-Mut2. Black and white arrow heads point to the positions of shifted bands and free probes, respectively. D, Transient expression assay of the expression of NRP1 regulated by bZIP60. pNRP1:LUC (pNRP1), pNRP1-Mut1:LUC (pNRP1-Mut1), pNRP1-Mut2:LUC (pNRP1-Mut2), pNRP2:LUC (pNRP2), pNRP2-Mut:LUC (pNRP2-Mut), and empty vector (Empty) were co-transformed with 35S:bZIP60ΔC-FLAG effectors (35S:FLAG as a control) into Arabidopsis Col mesophyll protoplasts and cultured overnight. The LUC activity was calculated by relative LUC activity (LUC/REN). The values are means ± sd of biological triplicates. Asterisks indicate significant differences between pNRP1:LUC and pNRP1-Mut1:LUC or pNRP1-Mut2:LUC (Student’s t test, *P < 0.05; **P < 0.01).
We next searched the NRP1 promoter region for known ER stress-responsive cis-elements (Sun et al., 2013b), and found that one copy of UPRE-I (TGACGTGG/A) element was present on the minus strand (−176 to −183, relative to the transcription start site [TSS]) within the P2 region of the NRP1 promoter (Figure 4, A and B). Previous studies have reported that the UPRE elements can be recognized by bZIP60 (Iwata and Koizumi, 2005; Sun et al., 2013b); thus, it is likely that bZIP60 activates NRP1 by binding to this cis-element. To test this hypothesis, electrophoretic mobility shift assay (EMSA) was performed with a biotinylated NRP1 DNA probe containing UPRE-I and recombinant bZIP60 N-terminal protein (His-bZIP60ΔC) that is sufficient to activate the expression of ER stress-responsive genes (Iwata and Koizumi, 2005; Figure 4B). His-bZIP60ΔC bound to the labeled NRP1 probe to form a shift band, which was notably counteracted by excess cold competitors, but not by the un-labeled NRP1 probe containing a mutated UPRE-I (NRP1-Mut1) (Figure 4, B and C), suggesting that the UPRE-I element is necessary for the binding of bZIP60 to the NRP1 promoter.
Our foregoing results demonstrated that NRP2 is constitutively expressed and not regulated by bZIP60 under ER stress (Figure 3, A and D; Supplemental Figure S15A). Interestingly, the NRP2 DNA region also contains a UPRE-I-like element (+62 to +69, relative to the TSS). This element (TGACGTGT) differs only in one base from that of NRP1 (TGACGTGG; Figure 4B). EMSA showed that bZIP60ΔC cannot bind to the UPRE-I-like element in NRP2 promoter (Supplemental Figure S15B). We thus speculated that the mismatched base (G–T) in UPRE-I-like element may cause the nonrecognition. As expected, the unlabeled NRP2 was unable to compete with the NRP1 probe (Figure 4, B and C). Nevertheless, a high concentration (100×) of mutated NRP1 cold probe that contains a UPRE-I-like element (NRP1-Mut2; Figure 4B) could attenuate the shifted band (Figure 4C), indicating that the key mismatched base (G–T) in the UPRE-I-like element, probably together with the flanking sequence, leads to the nonrecognition of bZIP60 from the NRP2 promoter.
We further performed dual-luciferase (LUC) reporter assay using the promoter regions of NRP1 (∼1 kb) and NRP2 (∼2 kb) fused with firefly LUC gene as reporters. Various reporters were co-transformed with 35S:FLAG and 35S:bZIP60ΔC-FLAG into Arabidopsis mesophyll protoplasts, respectively (Figure 4B). The LUC activity driven by the NRP1 promoter was remarkably induced by bZIP60ΔC (Figure 4D). Mutation of the UPRE-I at the NRP1 promoter (pNRP1-Mut1:LUC and pNRP1-Mut2:LUC) significantly attenuated this induction (Figure 4, B and D). In contrast, the LUC activity in pNRP2:LUC was not induced even if the promoter contained an identical UPRE-I sequence to NRP1 (pNRP2-Mut; Figure 4, B and D). In addition, we examined the activity of the unspliced bZIP60, and found that the full-length bZIP60 can activate the expression of NRP1 in ire1a ire1b double mutant protoplasts where the bZIP60 mRNA is not spliced (Supplemental Figure S16). Therefore, it can be concluded that the UPRE-I in the NRP1 promoter is the key element for bZIP60 binding, and its flanking sequence may also play an important role in the bZIP60 recognition.
NRP1 and NRP2 are not sufficient to rescue the loss-of-function of bZIP28/60 in ER stress response
NRP1 and NRP2 were reported to promote senescence and cell death when ectopically expressed in N. benthamiana leaves or protoplasts (Reis et al., 2016), but our study revealed a presumably opposite role of NRPs in cell death regulation. To further verify the biological function of NRPs in plants, we generated stable NRP1 and NRP2 overexpression transgenic lines and confirmed their overexpression by qRT-PCR (Supplemental Figure S17A). The overexpression of both NRP1 and NRP2 can rescue the susceptibility of nrpD to TM (Supplemental Figure S17, B and C), suggesting that these genes are functional alleles. Inconsistent with the transient assay in previous reports, no visible morphological difference between Col and the overexpression lines was observed under either normal growth conditions or TM treatment with different concentrations (Supplemental Figures S2 and 18).
To investigate the genetic relationship between bZIP60 and NRP1, we generated various combined genetic backgrounds of NRP1, NRP2, bZIP28, and bZIP60 by crossing. Under TM treatment, the phenotype of nrp1 bzip60 was similar to that of Col; and nrp1 bzip28 bzip60 was similar to bzip28 bzip60; and nrpD bzip60 was similar to nrpD (Supplemental Figure S19, A and B). These observations supported that there is functional redundancy between NRP1 and NRP2, and between bZIP60 and bZIP28. Notably, the nrpD bzip28 bzip60 quadruple mutant exhibited a more severe growth retardation and cell death phenotype than nrpD and bzip28 bzip60 (Figure 5, A and B). This could be explained by bZIP28 and bZIP60 regulating multiple downstream genes besides NRP1. Unexpectedly, neither the overexpression of NRP1 nor that of NRP2 could rescue the hypersensitivity of bzip28 bzip60 to TM (Figure 5, C and D). Studies have demonstrated that bZIP28 and bZIP60 present overlapping but partially independent functions in chronic ER stress response (Ruberti et al., 2018). In addition, bZIP60 regulates diverse downstream genes, such as transcription factors NAC089/103/062 and chaperone proteins BiP/CNX (Iwata and Koizumi, 2005; Sun et al., 2013b; Yang et al., 2014a, 2014b), functioning through a complex regulatory network. Thus, it is suggested that the NRP genes may be necessary but not sufficient for the bZIP28/60-mediated tolerance to ER stress.
Figure 5.
TM tolerance analysis of plants with combined NRP1/2 and bZIP28/60 genetic background. The sensitivity of nrpD bzip28 bzip60 (A and B), bzip28 bzip60 NRP1OE, and bzip28 bzip60 NRP2OE (C and D) to the ER stress inducer TM was detected. Seven-day-old seedlings were transferred to 1/2 MS medium containing 0.5 μg/mL (A and B) or 1 μg/mL (C and D) TM (DMSO as mock) for 10 d, and the pictures were taken (A and C). The percentages of G-B, G-S, and Y-D seedlings were calculated (B and D). The images next to the boxes display the phenotype of plants in the three groups. Data represent means ± sd of biological triplicates. Asterisks indicate significant differences between nrpD, bzip28 bzip60, and nrpD bzip28 bzip60 (Student’s t test, **P < 0.001).
Discussion
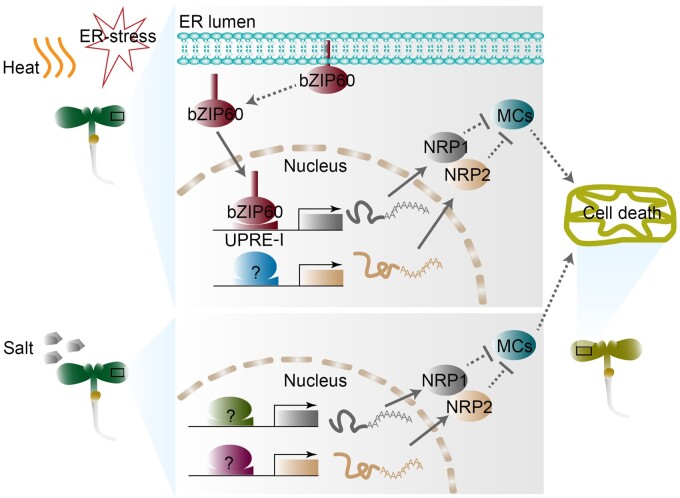
As the assembly factory for a third of the total proteome in cells (Wallin and von Heijne, 1998), the ER is the center of growth and various stress signal transmission (Nawkar et al., 2018; Hetz et al., 2020). Thus, ER stress caused by adverse conditions is related to cell fate (Hetz and Papa, 2018). However, the regulatory mechanisms in the determination of survival or death of cells under irreversible ER stress remain elusive in yeast and mammals; meanwhile in plants, even the understanding of UPR is largely limited. Here, we uncovered that NRP1/2, specific in algae and plants, function as pro-survival factors involved in the inhibition of cell death under chronic ER stress. Furthermore, bZIP60 serves as the activator of NRP1, but not NRP2, by directly binding to the UPRE-I element in the NRP1 promoter in ER stress response. The regulation of bZIP60 to NRP1 is independent of IRE1a/b-mediated mRNA splicing, indicating an unidentified mechanism that triggers the release of bZIP60 protein from ER. Under salt stress, the expression of NRP1/2 is regulated by unknown factors other than bZIP60 (Figure 6). These findings revealed the positive role of NRP1/2 in ER stress tolerance. Notably, previous studies have reported a contradictory observation that NRPs may promote senescence and cell death in plants (Costa et al., 2008; Reis et al., 2016). However, the conclusions in these studies were mainly established on ectopic transient expression systems using N. benthamiana leaves and protoplasts. More genetic evidence would be required to clarify the biological role of NRPs in plants. Meanwhile, the seedling age (2 weeks) for TM treatment in previous studies also differed from our study. By providing biological and genetic evidence in plants, our findings reveal that NRP1 and NRP2 play important roles in modulating ER stress response by inhibiting but not promoting cell death.
Figure 6.
Hypothetical model of the mechanism by which NRP1/2 regulate cell death in various stress responses. Under ER stress and heat stress, the expression of NRP1 activated by bZIP60 is independent of the IRE1a/b splicing pathway, while during salt stress, the expression of NRP1 is regulated by unknown factors (green item with question mark) other than bZIP60. NRP2 is regulated by unknown factors (blue and pink items with question marks) under these stresses. In turn, NRP1/2 represses the transcription of MC genes, thereby inhibiting cell death. Dotted lines indicate unknown regulations.
When plants suffer from ER stress, the S1P/S2P-bZIP28 and IRE1-bZIP60 signaling pathways are activated in a similar pattern to that described in yeast and mammalian cells (Iwata and Koizumi, 2005; Liu and Howell, 2010). As an important upstream transcription factor, bZIP60 cooperates with bZIP28 to activate the downstream genes in UPR (Song et al., 2015). In this study, we found that NRP1 is a direct target of bZIP60 in ER stress response. Interestingly, this regulatory model is not applicable to salt stress, although both genes are involved in the regulation of salt stress response (Henriquez-Valencia et al., 2015; Supplemental Figures S10 and S11). NRPs are reported to be involved in numerous stress responses (Costa et al., 2008; Hoepflinger et al., 2011), implying a general role of NRPs in modulating cell death during these responses. Like the regulation cascade of bZIP60-NRP1 in UPR, it is quite likely that the expression of NRPs is also regulated by the key transcription factors under different stresses. In contrast to NRP1, NRP2 is constitutively expressed under ER stress and is not regulated by bZIP60/28, but it presents a redundant function with NRP1 in the inhibition of cell death.
Previous reports indicated that the proteins containing a DCD domain are expressed throughout the life cycle and under stressful conditions in Arabidopsis (Tenhaken et al., 2005; Hoepflinger et al., 2011; Zhou et al., 2017). It could be speculated that the constitutive expression pattern of NRP2 is mainly responsible for a basal function in cells to maintain normal homeostasis in mildly fluctuating environments. In contrast, the expression of NRP1 induced by ER stress might ensure cell homeostasis in response to the “emergencies” or “major safety events” under adverse conditions. Such a “belt and braces” mechanism involving different regulatory patterns may be an important strategy for the survival and stress adaptation in plants.
Despite the fact that the expression of NRP1 is regulated directly by bZIP60 during ER stress, unexpectedly, NRP1/2 overexpression cannot rescue the hypersensitivity of bzip28 bzip60 to TM (Figure 5, C and D). Recent work has elucidated the mechanisms by which bZIP60 and bZIP28 activate the expression of downstream NAC transcription factor genes ANAC062/103/089 to regulate ER stress response (Sun et al., 2013b; Yang et al., 2014a, 2014b). NRPs were also reported to mediate cell death by regulating NAC genes in soybean (Faria et al., 2011; Mendes et al., 2013). Considering the promoting effect of NAC089 on cell death (Yang et al., 2014b), we examined the expression of NAC089 in nrpD, and intriguingly found that NAC089 was not regulated by NRP1/2 (Supplemental Figure S20). Our observations showed that MC1/2/7/8—unlike MC5, which is activated by NAC089 (Yang et al., 2014b)—are involved in NRP1/2-mediated cell death (Figure 2C; Supplemental Figure S8). Thus, it seems that NRP1/2 are not linearly coupled to the bZIP60/28-NAC pathway, but an independent branch of ER stress signaling regulated by bZIP60. It is noteworthy that the mRNA of active bZIP60 requires an IRE1a/b-mediated unconventional splicing in UPR (Nagashima et al., 2011). Notably, the expression of NRP1, same as that of Bax Inhibitor-1 (BI-1), an anti-apoptotic molecule conserved in mammals and plants (Watanabe and Lam, 2008; Lebeaupin et al., 2020), was unaltered in ire1a ire1b double mutant compared with the wild-type (Supplemental Figures S13 and S21), whereas the activation of NA089/103 and BIP3 can be inhibited by loss-of-function of IRE1a and IRE1b (Nagashima et al., 2011). In contrast, both NRP1 and BI-1 were still activated by bZIP60 (Figure 3D; Supplemental Figure S21). These observations suggest that the anti-death molecules, BI-1 and NRP1, share a similar transcriptional regulation pattern mediated by bZIP60 and which is independent of IRE1.
In mammalian cells, the pro-apoptotic genes such as death receptor 5 (DR5) are activated by PERK, which induces the caspase activity to trigger apoptosis under overwhelming ER stress (Hetz et al., 2020). Upon mild ER stress, IRE1 can degrade the DR5 mRNA to inhibit apoptosis via the IRE1-dependent decay to maintain cellular health (Hetz et al., 2020). Thus, the fate of cells under prolonged ER stress depends on the coordination between the PERK and the IRE1 pathway (Chang et al., 2018). The mechanism by which cell fate is determined under chronic mild ER stress remains poorly understood in plants. The expression pattern of the cell death-related genes MC1/2/7/8 presented the initial rapid increase and the subsequent gradual decline, implying that the cell survival in plants with chronic ER stress may also be modulated by the coordination among the various components (Figure 2C). In nrpD, however, such coordination appeared to be out of control, resulting in a continuous increase of the expression of MC1/2/7/8 which eventually led to cell death. Loss-of-function of NRPs not only reduces the tolerance of plants to ER stress, but also causes spontaneous ER stress during normal growth process (Supplemental Figure S3). It is not clear whether the precocious senescence in nrpD is triggered by the spontaneous ER stress, or vice versa, which is so far a controversial issue (Pluquet et al., 2015). Besides, prolonged or unresolved ER stress can induce an autophagy event (Srivastava et al., 2018). As a degradation process facilitating the recycling of cellular components, autophagy is required for maintaining cell homeostasis for plant growth. It has been reported that autophagy contributes to mitigating ER stress (Liu et al., 2012), whereas it also promotes cell death (Minina et al.,2014). Collectively, NRP1/2 is likely to act as the modulators that inhibit the spread of cell death signals. Whether and how NRPs mediate the processes of ER stress, autophagy, and senescence in plants requires a further investigation.
Materials and methods
Plant material and growth conditions
The Arabidopsis (A. thaliana) T-DNA mutants nrp1 (SALK_041306), nrp2 (GK_520C04), bzip60 (SALK_050203, Sun et al, 2013a), bzip28 (SALK_132285, Sun et al, 2013a), ire1 (SALK_018112; Deng et al., 2011), and ire1b (SAIL_238_F07; Deng et al., 2011) were obtained from the ABRC. The double mutants, nrpD, bzip28 bzip60, and ire1a ire1b were generated by crossing nrp1 with nrp2, bzip28 with bzip60, and ire1a with ire1b, respectively. The quadruple mutant nrpD bzip28 bzip60 were generated by crossing nrpD with bzip28 bzip60. Seeds were incubated at 4°C for 2 d and then grown at 22°C under 16-h light/8-h dark photoperiod conditions.
Plasmid construction and plant transformation
To generate the 35S:NRP1-6HA and 35S:NRP2-6HA overexpression transgenic lines, the coding regions of NRP1 and NRP2 were cloned into pGreen-35S:6HA (Liu et al., 2016). To generate the pNRP1:NRP1-mVenus and pNRP2:NRP2-mVenus transgenic lines, the genomic fragment containing ∼1 kb (NRP1) or ∼2-kb (NRP2) upstream and coding region was cloned into pGreen-mVenus plasmid. To generate the 35S:3FLAG-bZIP60 transgenic lines, the bZIP60 coding sequence was cloned into the pGreen-35S:3FLAG vector. By using Agrobacterium-mediated transformation, the constructs 35S:NRP1-6HA, 35S:NRP2-6HA and 35S:3FLAG-bZIP60 were introduced into Arabidopsis Col background, and the pNRP1:NRP1-Venus and pNRP2:NRP2-Venus constructs were introduced into nrpD background. These transgenic plants were selected with Basta. Primers used for the cloning are listed in Supplemental Table S1.
Stress treatment and cell death assay
Surface-sterilized seeds were plated on one-half-strength Murashige and Skoog (1/2 MS) medium containing 1% (w/v) sucrose at 4°C for 2 d in the dark and then grown at 22°C under a 16-h light/8-h dark cycle before treatment. For chronic ER stress phenotype analyses, the 7-d-old seedlings were transferred to the 1/2 MS mediums containing 0.5 or 1 μg/mL TM (dissolved in DMSO) for another 10 days. For salt stress phenotype analyses, the 7-d-old seedlings were transferred to the 1/2 MS mediums containing 150 mM NaCl for another 7 d. For heat stress phenotype analyses, the 10-d-old seedlings were incubated at 37°C for 1 d then grown at 22°C for 4 d. According to the plant size, leaf color, and survival, the seedlings were divided into three groups: big-green, small-green, and Y-D. The percentage of plants in each group was calculated using at least 20 samples per genotype and three biological replicates. For cell death assay, seedlings were collected after 2 d of treatment with 1 μg/mL TM for trypan blue staining or TUNEL labeling as described below. More than 20 samples were used for each cell analysis experiment, and repeated experiments were carried out for 3 times.
For trypan blue staining, cotyledons were stained with trypan blue as described previously (van Wees, 2008).
For TUNEL staining, roots were detached and fixed overnight in 4% (v/v) paraformaldehyde at room temperature. TUNEL reaction was performed in a microcentrifuge tube using TUNEL BrightGreen Apoptosis Detection Kit (Vazyme, Nanjing, China ) according to the manufacturer’s instructions. To visualize the nuclei in root cells, samples were stained using 10 µg/mL propidium iodide (PI, Sigma-Aldrich, St Louis, MO, USA) in PBS buffer solution for 10 min. The TUNEL-positive and PI-stained nucleus were observed with laser confocal fluorescence microscopy (Leica SP8) fitted with the configuration: excitation at 488 nm (TUNEL) and 587 nm (PI).
Total RNA extraction and RT-qPCR
Total RNAs were extracted from 7-d-old seedlings treated with 1 μg/mL TM or 2 mM DTT (Braakman et al., 1992) by using Eastep Super Total RNA Extraction Kit (Promega, Madison, WI, USA) according to the manufacturer’s instruction. The cDNA was synthesized by using MMLV-RTase (Promega). RT-qPCR was performed on LightCycler 480 thermal cycler system (Roche, Basel, Switzerland) with KAPA SYBR Fast qPCR Kit Master Mix (KAPA BIO, Wilmington, MA, USA). The relative gene expression was quantified using the comparative Ct method (2−△△Ct), and Tubulin 2 (TUB2) was used as an internal control. The primers used for gene expression analysis are listed in Supplemental Table S1.
Transient expression assay
For the dual-LUC assay, the genomic fragment containing ∼1-kb (NRP1) or ∼2-kb (NRP2) upstream relative to the coding region was cloned into the pGreenII 0800-LUC reporter plasmid (Song et al., 2014), respectively. To generate the 35S:bZIP60ΔC-FLAG effector, the truncated coding sequence of bZIP60 (Iwata and Koizumi, 2005) was cloned into the pPZPY122-3FLAG (from ABRC) vector. Primers used for the constructs are listed in Supplemental Table S1. Protoplasts were isolated from 4-week-old Arabidopsis mesophyll cells. Various constructs of effector and reporter were transfected into Arabidopsis protoplasts according to the protocol, as described previously (Wu et al., 2009). The activity of firefly LUC and renilla LUC (REN) were quantified by using Dual-LUC Reporter Assay Kit (Promega) according to the manufacturer’s instructions. Relative LUC activity was calculated by normalizing against the REN activity.
ChIP analysis
The ChIP assay was performed as described previously (Liu et al., 2016). Seven-day-old 35S:FLAG-bZIP60 and Col seedlings were treated with 1 μg/mL TM (DMSO as mock) or 150 mM NaCl for 4 h, and fixed in 1%(v/v) formaldehyde under vacuum on ice for 10 min. The nuclear proteins and chromatins were isolated from fixed samples. The chromatins were sonicated to generate DNA fragments with an average size of 500 bp and immunoprecipitated by anti-FLAG (F3165; Sigma, St Louis, MO, USA) at 4°C for 2 h. The co-immunoprecipitated DNA was recovered and analyzed by qPCR with KAPA SYBR Fast qPCR Kit Master Mix (KAPA BIO). Relative enrichment folds were calculated by normalizing the amount of a target DNA fragment to that of a genomic fragment of PP2A (AT1G13320) and then against the respective input DNA amount. The enrichment of Actin 2 (ACT2) genomic fragment was used as a negative control. All primers used in ChIP assays are listed in Supplemental Table S1.
EMSA
The EMSA was performed using the LightShift Chemiluminescent EMSA kit (Pierce, Waltham, MA, USA) as described previously (Yang et al., 2014b). The 40-bp 5′-end biotinylated oligonucleotide containing UPRE-I in the P2 region of the NRP1/2 promoter was used as a probe, while unlabeled probes containing the native NRP1 UPRE-I (TGACGTGG), mutated NRP1 UPRE-I (TCTGCAGG/TGACGTGT), or NRP2 UPRE-I-like (TGACGTGT), were used as cold competitors (Figure 4C). For His-bZIP60ΔC construct, the truncated bZIP60ΔC was cloned into pQE30 and expressed in the Escherichia coli strain rosetta. Recombinant His-bZIP60ΔC protein was purified by Ni-NTA agarose beads (Qiagen, Hilden, Germany) and used for protein–DNA binding. Probes used for EMSA are listed in Supplemental Table S2.
Accession numbers
NRP1 (AT5G42050), NRP2 (AT3G27090), bZIP28 (AT3G10800), bZIP60 (AT1G42990), SAG12 (AT5G45890), SEN4 (AT4G30270), MC1 (AT1G02170), MC2 (AT4G25110), MC3 (AT5G64240), MC4 (AT1G79340), MC5 (AT1G79330), MC6 (AT1G79320), MC7 (AT1G79310), MC8 (AT1G16420), MC9 (AT5G04200), NAC089 (AT5G22290), IRE1a (AT2G17520), IRE1b (AT5G24360), TUB2 (AT5G62690), PP2A (AT1G13320), and ACT2 (AT3G18780).
Supplemental data
The following supplemental materials are available in the online version of this article.
Supplemental Figure S1. Schematic diagram and identification of T-DNA insertion mutants nrp1 and nrp2.
Supplemental Figure S2. Loss-of-function of NRP1 and NRP2 leads to precocious senescence.
Supplemental Figure S3. The expression analysis of senescence, ER stress, and autophagy-related marker genes in Col, nrp1, nrp2, and nrpD at different growth stages.
Supplemental Figure S4. bZIP60 mRNA splicing analysis.
Supplemental Figure S5. The expression analysis of BIP3 and ATG18a in Col and nrpD.
Supplemental Figure S6 The trypan blue staining and TUNEL assay of bzip60 and ire1a ire1b.
Supplemental Figure S7. The expression analysis of MC1 and MC2 in Col and the complementation transgenic lines.
Supplemental Figure S8. The expression analysis of MC genes in Col and nrpD.
Supplemental Figure S9. The expression analysis of bZIP60 and bZIP28 in Col and nrpD.
Supplemental Figure S10. The double mutant nrpD exhibits less tolerance to salt stress.
Supplemental Figure S11. The expression analysis of NRP1and NRP2 in Col, bzip28, bzip60, and bzip28 bzip60 under salt stress.
Supplemental Figure S12. NRP1 and NRP2 are involved in heat stress response.
Supplemental Figure S13. The expression analysis of NRP1in ire1a ire1b mutant under ER stress, high salinity stress, and heat stress.
Supplemental Figure S14. ChIP analysis of bZIP60 binding to the NRP1 promoter under salt and heat stress.
Supplemental Figure S15. ChIP and EMSA analysis of bZIP60 binding to the NRP2 genomic regions.
Supplemental Figure S16. Transient expression assay of the expression of NRP1 regulated by bZIP60.
Supplemental Figure S17. The NRP1/2 overexpression plants can rescue the nrpD susceptibility to TM.
Supplemental Figure S18. The TM sensitivity of NRP1/2 overexpression plants is similar to that of Col wild-type.
Supplemental Figure S19. The TM sensitivity of nrp1 bzip60, nrpD bzip60, and nrp1 bzip28 bzip60.
Supplemental Figure S20. The expression analysis of NAC089 in Col, nrp1, nrp2, and nrpD.
Supplemental Figure S21. The expression analysis of BI-1 in Col, bzip60, ire1a ire1b, and nrpD.
Supplemental Table S1. Primers used in this study.
Supplemental Table S2. Probes used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Chengwei Yang for providing bzip60 seeds.
Funding
This work was supported by China Postdoctoral Science Foundation (2019M663146) and the Guangzhou Municipal Science and Technology Project (202002030057).
Conflict of interest statement. The authors declare no conflict of interest.
Y.Y. and X.H. designed the research. Y.Y., X.L., W.Z., Q.Q., L.Z., S.L., and Y.L. performed research. Y.Y. X.L., and X.H. analyzed data. Y.Y. and X.H. wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Xingliang Hou (houxl@scib.ac.cn).
References
- Alves MS, Reis PA, Dadalto SP, Faria JA, Fontes EP, Fietto LG (2011) A novel transcription factor, ERD15 (Early Responsive to Dehydration 15), connects endoplasmic reticulum stress with an osmotic stress-induced cell death signal. J Biol Chem 286: 20020–20030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Helenius J, Helenius A (1992) Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J 11: 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho HH, Silva PA, Mendes GC, Brustolini OJ, Pimenta MR, Gouveia BC, Fontes EP (2014) The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events. Plant Physiol 164: 654–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TK, Lawrence DA, Lu M, Tan J, Harnoss JM, Marsters SA, Liu P, Sandoval W, Martin SE, Ashkenazi A (2018) Coordination between two branches of the unfolded protein response determines apoptotic cell fate. Mol Cell 71: 629–636 [DOI] [PubMed] [Google Scholar]
- Chen Y, Brandizzi F (2013) Analysis of unfolded protein response in Arabidopsis. Methods Mol Biol 1043: 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P (2010) Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397 [DOI] [PubMed] [Google Scholar]
- Costa MD, Reis PA, Valente MA, Irsigler AS, Carvalho CM, Loureiro ME, Aragão FJ, Boston RS, Fietto LG, Fontes EP (2008) A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant-specific asparagine-rich proteins to promote cell death. J Biol Chem 283: 20209–20219 [DOI] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA 108: 7247–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria JA, Reis PA, Reis MT, Rosado GL, Pinheiro GL, Mendes GC, Fontes EP (2011) The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol 11: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Prahlad V, Morimoto RI (2011) The stress of protein misfolding: from single cells to multicellular organisms. CSH Perspect Biol 3: a009704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X, Qian J, Xu S, Son X, Zhu J (2008) Location of caspase 3-like protease in the development of sieve element and tracheary element of stem in Cucurbita moschata. J Integr Plant Biol 50: 1499–1507 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274 [DOI] [PubMed] [Google Scholar]
- Henriquez-Valencia C, Moreno AA, Sandoval-Ibañez O, Mitina I, Blanco-Herrera F, Cifuentes-Esquivel N, Orellana A (2015) bZIP17 and bZIP60 regulate the expression of BiP3 and other salt stress responsive genes in an UPR-independent manner in Arabidopsis thaliana. J Cell Biochem 116: 1638–1645 [DOI] [PubMed] [Google Scholar]
- Hetz C, Papa FR (2018) The unfolded protein response and cell fate control. Mol Cell 69: 169–181 [DOI] [PubMed] [Google Scholar]
- Hetz C, Zhang K, Kaufman RJ (2020) Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol 21: 421–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepflinger MC, Pieslinger AM, Tenhaken R (2011) Investigations on N-rich protein (NRP) of Arabidopsis thaliana under different stress conditions. Plant Physiol Biochem 49: 293–302 [DOI] [PubMed] [Google Scholar]
- Iwata Y, Ashida M, Hasegawa C, Tabara K, Mishiba KI, Koizumi N (2017) Activation of the Arabidopsis membrane-bound transcription factor bZIP28 is mediated by site-2 protease, but not site-1 protease. Plant J 91: 408–415 [DOI] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N (2005) An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA 102: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Sakiyama M, Lee M-H, Koizumi N (2010) Transcriptomic response of Arabidopsis thaliana to tunicamycin-induced endoplasmic reticulum stress. Plant Biotechnol 27: 161–171 [Google Scholar]
- Kamauchi S, Nakatani H, Nakano C, Urade R (2005) Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J 272: 3461–3476 [DOI] [PubMed] [Google Scholar]
- Kim S, Choi Y, Kwon C, Yun HS (2019) Endoplasmic reticulum stress-induced accumulation of VAMP721/722 requires CALRETICULIN 1 and CALRETICULIN 2 in Arabidopsis. J Integr Plant Biol 61: 974–980 [DOI] [PubMed] [Google Scholar]
- Kurokawa M, Kornbluth S (2009) Caspases and kinases in a death grip. Cell 138: 838–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeaupin C, Blanc M, Vallee D, Keller H, Bailly-Maitre B (2020) Bax Inhibitor-1: between stress and survival. FEBS J 287: 1722–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wei H, Liu L, Yang X, Zhang X, Xie Q (2017) Unfolded protein response activation compensates endoplasmic reticulum-associated degradation deficiency in Arabidopsis. J Integr Plant Biol 59: 506–521 [DOI] [PubMed] [Google Scholar]
- Liu JX, Howell SH (2010) bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH (2007a) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH (2007b) Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J 51: 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hu PW, Huang MK, Tang Y, Li YG, Li L, Hou XL (2016) The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat Commun 7: 12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC (2012) Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes GC, Reis PA, Calil IP, Carvalho HH, Aragão FJ, Fontes EP (2013) GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc Natl Acad Sci USA 110: 19627–19632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina EA, Bozhkov PV, Hofius D (2014) Autophagy as initiator or executioner of cell death. Trends Plant Sci 19: 692–697 [DOI] [PubMed] [Google Scholar]
- Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, Chen Y, Brandizzi F, Dong X, Orellana A, et al. (2012) IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One 7: e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Sant A, Kohno K, Normington K, Gething MJ, Sambrook JF (1992) A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J 11: 2583–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y, Mishiba K, Suzuki E, Shimada Y, Iwata Y, Koizumi N (2011) Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner B A, Yuan J (2000) Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 403: 98–103 [DOI] [PubMed] [Google Scholar]
- Nawkar GM, Lee ES, Shelake RM, Park JH, Ryu SW, Kang CH, Lee SY (2018) Activation of the transducers of unfolded protein response in plants. Front Plant Sci 9: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piszczek E, Gutman W (2007) Caspase-like proteases and their role in programmed cell death in plants. Acta Physiol Plant 29: 391–398 [Google Scholar]
- Reis PA, Carpinetti PA, Freitas PP, Santos EG, Camargos LF, Oliveira IH, Silva JC, Carvalho HH, Dal-Bianco M, Soares-Ramos JR, et al. (2016) Functional and regulatory conservation of the soybean ER stress-induced DCD/NRP-mediated cell death signaling in plants. BMC Plant Biol 16: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis PA, Fontes EP (2012) N-rich protein (NRP)-mediated cell death signaling: a new branch of the ER stress response with implications for plant biotechnology. Plant Signal Behav 7: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis PA, Rosado GL, Silva LA, Oliveira LC, Oliveira LB, Costa MD, Alvim FC, Fontes EP (2011) The binding protein BiP attenuates stress-induced cell death in soybean via modulation of the N-rich protein-mediated signaling pathway. Plant Physiol 157: 1853–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluquet O, Pourtier A, Abbadie C (2015) The unfolded protein response and cellular senescence. A review in the theme: cellular mechanisms of endoplasmic reticulum stress signaling in health and disease. Am J Physiol Cell Physiol 308: C415– C–425. [DOI] [PubMed] [Google Scholar]
- Ruberti C, Lai YS, Brandizzi F (2018) Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP28 and bZIP60, as well as an antagonizing function of BAX-Inhibitor 1 upon the pro-adaptive signaling mediated by bZIP28. Plant J 93: 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R (2002) ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 3: 99–111 [DOI] [PubMed] [Google Scholar]
- Song S, Huang H, Gao H, Wang J, Wu D, Liu X, Yang S, Zhai Q, Li C, Qi T, Xie D (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song ZT, Sun L, Lu SJ, Tian Y, Ding Y, Liu JX (2015) Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc Natl Acad Sci USA 112: 2900–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Deng Y, Shah S, Rao AG, Howell SH (2013) BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 25: 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Li Z, Russo G, Tang J, Bi R, Muppirala U, Chudalayandi S, Severin A, He M, Vaitkevicius SI, et al. (2018) Response to persistent ER stress in plants: a multiphasic process that transitions cells from prosurvival activities to cell death. Plant Cell 30: 1220–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Lu SJ, Zhang SS, Zhou SF, Sun L, Liu JX (2013a) The lumen-facing domain is important for the biological function and organelle-to-organelle movement of bZIP28 during ER stress in Arabidopsis. Mol Plant 6: 1605–1615 [DOI] [PubMed] [Google Scholar]
- Sun L, Yang ZT, Song ZT, Wang MJ, Sun L, Lu SJ, Liu JX (2013b) The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J 76: 274–286 [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang SS, Lu SJ, Liu JX (2015) Site-1 protease cleavage site is important for the ER stress-induced activation of membrane-associated transcription factor bZIP28 in Arabidopsis. Sci China Life Sci 58: 270–275 [DOI] [PubMed] [Google Scholar]
- Tenhaken R, Doerks T, Bork P (2005) DCD - a novel plant specific domain in proteins involved in development and programmed cell death. BMC Bioinformatics 6: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ (1998) A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev 12: 1812–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees S (2008) Phenotypic analysis of Arabidopsis mutants: trypan blue stain for fungi, oomycetes, and dead plant cells. CSH Protoc 3: pdb.prot4982 [DOI] [PubMed] [Google Scholar]
- Wallin E, von Heijne G (1998) Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci 7: 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Lam E (2008) Arabidopsis Bax inhibitor-1: a rheostat for ER stress-induced programmed cell death. Plant Signal Behav 3: 564–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis Sandwich-a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Yang ZT, Lu SJ, Wang MJ, Bi DL, Sun L, Zhou SF, Song ZT, Liu JX (2014a) A plasma membrane-tethered transcription factor, NAC062/ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis. Plant J 79: 1033–1043 [DOI] [PubMed] [Google Scholar]
- Yang ZT, Wang MJ, Sun L, Lu SJ, Bi DL, Sun L, Song ZT, Zhang SS, Zhou SF, Liu JX (2014b) The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet 10: e1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Zhu T, Han L, Liu M, Xu M, Liu Y, Han D, Qiu D, Gong Q, Liu X (2017) The asparagine-rich protein NRP interacts with the Verticillium effector PevD1 and regulates the subcellular localization of cryptochrome 2. J Exp Bot 68: 3427–3440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.