Abstract
The prefoldin complex (PFDc) was identified in humans as a co-chaperone of the cytosolic chaperonin T-COMPLEX PROTEIN RING COMPLEX (TRiC)/CHAPERONIN CONTAINING TCP-1 (CCT). PFDc is conserved in eukaryotes and is composed of subunits PFD1–6, and PFDc-TRiC/CCT folds actin and tubulins. PFDs also participate in a wide range of cellular processes, both in the cytoplasm and in the nucleus, and their malfunction causes developmental alterations and disease in animals and altered growth and environmental responses in yeast and plants. Genetic analyses in yeast indicate that not all of their functions require the canonical complex. The lack of systematic genetic analyses in plants and animals, however, makes it difficult to discern whether PFDs participate in a process as the canonical complex or in alternative configurations, which is necessary to understand their mode of action. To tackle this question, and on the premise that the canonical complex cannot be formed if one subunit is missing, we generated an Arabidopsis (Arabidopsis thaliana) mutant deficient in the six PFDs and compared various growth and environmental responses with those of the individual mutants. In this way, we demonstrate that the PFDc is required for seed germination, to delay flowering, or to respond to high salt stress or low temperature, whereas at least two PFDs redundantly attenuate the response to osmotic stress. A coexpression analysis of differentially expressed genes in the sextuple mutant identified several transcription factors, including ABA INSENSITIVE 5 (ABI5) and PHYTOCHROME-INTERACTING FACTOR 4, acting downstream of PFDs. Furthermore, the transcriptomic analysis allowed assigning additional roles for PFDs, for instance, in response to higher temperature.
Prefoldins contribute to Arabidopsis growth and response to the environment through mechanisms that are dependent on or independent of the canonical prefoldin complex.
Introduction
Prefoldins (PFDs) are conserved proteins present in archaea and in eukaryotes that were identified in humans and in yeast as part of a hexameric complex, called PFD complex (PFDc; Geissler et al., 1998; Vainberg et al., 1998). PFDs can be classified into α- or β-type depending on their structure (Supplemental Figure S1A; Arranz et al., 2018). In eukaryotes, there are two α-type (PFD3 and PFD5) and four β-type (PFD1, PFD2, PFD4, and PFD6) PFDs, whereas in archaea only one PFD per type is found. The PFDc adopts a jellyfish-like structure in which two α-subunits occupy a central position allowing the binding of four β-subunits (Siegert et al., 2000; Martin-Benito et al., 2002). In eukaryotes, the arrangement of the different subunits within the complex appears to be conserved (Gestaut et al., 2019).
Currently, the best characterized function of the PFDc is in proteostasis, as cochaperone of the chaperonin T-COMPLEX PROTEIN RING COMPLEX (TRiC)/CHAPERONIN CONTAINING TCP-1 (CCT) in the folding of tubulins and actin (Gestaut et al., 2019). Yeast genes involved in microtubule biogenesis (gim)/pfd mutants affecting different subunits of the complex show similar cytoskeleton-related defects, such as reduced α-tubulin levels (Geissler et al., 1998; Vainberg et al., 1998), which are not aggravated when several gim/pfd mutations are combined (Siegers et al., 1999). Tubulin- and actin-related defects are also observed in pfd mutants in other organisms. Knockdown of PFD genes in Caenorhabditis elegans, except PFD4 that is divergent in this species, causes impaired cell division and embryo lethality due to defects in the rate of microtubule (MT) polymerization (Lundin et al., 2008). Hypomorphic alleles of the Drosophila MERRY-GO-ROUND locus, which encodes PFD3, cause defects in the formation of the meiotic spindle due to reduced tubulin levels, with this reduction also being observed in fly DMEL-2 cells after knocking down PFD4 (Delgehyr et al., 2012). A missense mutation in the PFDN5 gene causes developmental alterations in the central nervous system in mice, which are associated with reduced accumulation of α-tubulin and β-actin (Lee et al., 2011). In Arabidopsis (Arabidopsis thaliana), pfd mutations provoke defects in the arrangement of cortical MTs and in the formation of the phragmoplast, leading to impaired cell elongation and division, respectively (Gu et al., 2008; Rodriguez-Milla and Salinas, 2009; Perea-Resa et al., 2017). In summary, the similar phenotypes caused by mutations in individual PFD genes are consistent with the idea that the function of the PFDc is impaired when a subunit is missing. This view is further supported by the unique arrangement of subunits within the complex, which is achieved by specific protein–protein interactions (Gestaut et al., 2019).
Genetic analyses in yeast have shown that other functions of PFDs are not performed by the canonical PFDc. All PFDs, except GIM2/PFD3 and GIM4/PFD2, are required for transcription elongation of long genes and bind chromatin in a transcription-dependent manner (Millan-Zambrano et al., 2013). Furthermore, no additivity was found when combining affected gim/pfd mutants, which suggested that these PFDs may exert this role by being part of an alternative complex. In the same line, only GIM2/PFD3, GIM3/PFD4, and GIM1/PFD6 proteins are required for the transcription of genes in response to osmotic or oxidative stress (Amorim et al., 2017).
The implication of PFDs in diverse cellular processes is well documented (Liang et al., 2020). Nonetheless, the lack of systematic genetic analyses makes it difficult to discern whether a particular role is exerted by the canonical PFDc or by individual subunits. For example, genetic and molecular analyses in human and mice cell lines indicate that PFDN5 acts as bridge protein that recruits a corepressor complex to the c-Myc transcription factor (TF) (Fujioka et al., 2001; Satou et al., 2001). Furthermore, PFDN5 is also required to recruit components of the RNAPII phosphorylation and splicing complexes to transcribed genes in humans (Payán-Bravo et al., 2021). Nonetheless, it has not been demonstrated whether it is the only PFD involved or whether these roles are performed by the PFDc. In Arabidopsis, PFD4 promotes the proteasomal degradation of ELONGATED HYPOCOTYL 5 (HY5), and accordingly, HY5 protein levels are higher in pfd4 mutants compared to the wild-type (Perea-Resa et al., 2017). HY5 levels are also increased in pfd3 and in pfd5 mutants, indicating that these subunits are also involved.
To understand the roles of PFDs in cellular processes, one of the issues that we need to clarify is whether they act as a canonical PFDc or as individual subunits. To address this question in Arabidopsis, and since the function of the complex is impaired if one subunit is missing, we have generated a mutant defective in the six PFDs and compared its growth habit and its behavior under abiotic stress conditions with those of the individual mutants. Furthermore, we have identified additional functions for PFDs based on a transcriptomic analysis of the sextuple mutant.
Results
The PFDc is formed in vivo in Arabidopsis
We first investigated whether the Arabidopsis PFDs can adopt the structure of their orthologs in yeast and humans. The structure of the Arabidopsis PFDs could be modeled in silico based on the structure of their human orthologs (Supplemental Figure S1A) and assembled to form the jellyfish-like complex (Gestaut et al., 2019; see the top view in Figure 1A and the comparison of the two complexes in Supplemental Figure S1B). The accuracy of the modeling was assessed using the Predicted Native Overlap, which refers to the fraction of the Cα atoms in the model that are predicted to be within 3.5 Å of their corresponding position in the known structure (Eramian et al., 2008). The fraction was higher than 0.3 for all PFDs (Supplemental Figure S1C), which is the indicative cutoff of good models (Eramian et al., 2008; Melo et al., 2009). The high similarity between complexes suggests that the PFDc would adopt the same 3D arrangement in Arabidopsis and in humans in vivo.
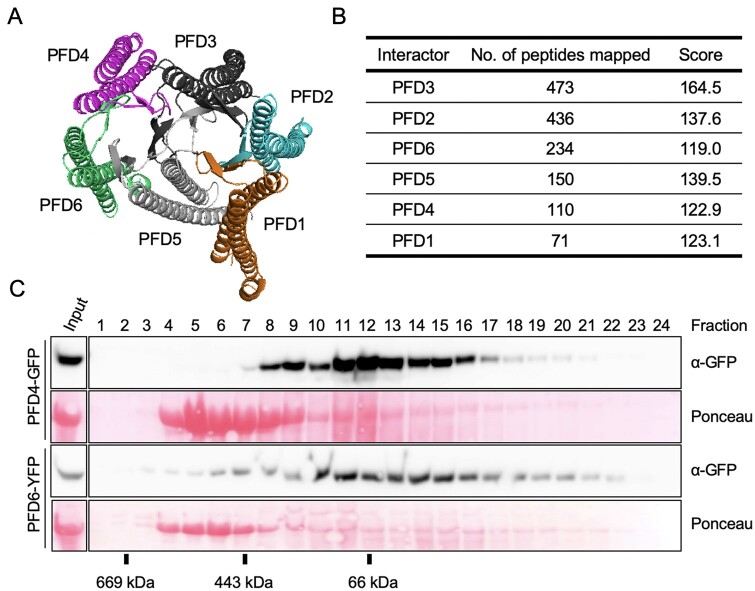
Figure 1.
The Arabidopsis PFDc. A, Predicted structure of the Arabidopsis PFDc using the human PFDc as template for modeling. B, Identification of the Arabidopsis PFDc in vivo. The table summarizes the average number of peptides and the Mascot score corresponding to each PFD subunit after TAP of GS-PFD3 (two replicates). C, Gel filtration fractions were analyzed by western blot and the fusion proteins revealed with anti-GFP antibodies. The Ponceau staining shows the large subunit of rubisco.
We next determined if the PFD proteins associate in vivo. For that purpose, we performed tandem affinity purification (TAP) using an Arabidopsis PSB-D cell suspension line expressing the PFD3 protein fused to the C-t end of the GSrhino-TAP tag (Van Leene et al., 2015). After the two sequential immunopurification steps, the top PFD3 interactors identified in two replicates were the other five PFDs (Figure 1B), suggesting that the PFDc is formed in vivo. To further test this possibility, we reasoned that PFDs should elute in fractions compatible with the molecular weight of the PFDc in a gel-filtration chromatography. We subjected extracts of seedlings expressing either pPFD4::PFD4-GFP (Perea-Resa et al., 2017) or p35S::PFD6-YFP (Esteve-Bruna et al., 2020) to gel filtration. As shown in Figure 1C, the elution profiles of both proteins indicated that they are part of protein complexes of a range of molecular weights that includes that of PFDc (∼130 kDa including the fusion protein). Protein staining with Ponceau S showing the elution profile of the rubisco large subunit, which is consistent with the molecular weight of the rubisco complex (∼ 540 kDa), served as control of the analysis.
The Arabidopsis 6x pfd mutant
In order to investigate the PFDs’ contribution to Arabidopsis development and response to the environment, and to differentiate between processes regulated by the PFDc or by individual subunits, we decided to generate a sextuple mutant defective in the activity of the six PFDs. Mutants for PFD genes have been described in Arabidopsis, except for PFD1 (Gu et al., 2008; Rodriguez-Milla and Salinas, 2009; Perea-Resa et al., 2017; Esteve-Bruna et al., 2020). We identified a T-DNA mutant for the PFD1 gene in the GABI-Kat collection (Kleinboelting et al., 2012). The pfd1 mutant carries the T-DNA inserted in the third exon of the gene (Supplemental Figure S2A) and is null, or highly hypomorphic, as evidenced by the inability to amplify the full-length transcript by reverse transcription polymerase chain reaction (RT-PCR; Supplemental Figure S2B). With mutants available for all PFD genes, we generated the pfd1 pfd2 pfd3 pfd4 pfd5 pfd6 sextuple mutant (hereafter referred to as 6x pfd) by genetic crosses (see “Materials and methods” for details). An RNA-seq analysis of the sextuple mutant (see below) confirmed that pfd1, pfd2, pfd3, and pfd4 alleles are null or highly hypomorphic and that pfd6 carries the reported missense mutation (Supplemental Figure S2C). Nonetheless, it also showed that the PFD5 gene is transcribed in the mutant, albeit at a reduced level (∼40% of wild-type; Supplemental Figure S2, C and D). This result contrasts with the absence of full-length PFD5 transcript previously reported in the pfd5 mutant (Rodriguez-Milla and Salinas, 2009). The insertion site is in the third intron (Supplemental Figure S2E), suggesting that the T-DNA may be processed in a fraction of PFD5 pre-mRNAs and that this occurs more often in the sextuple mutant than in the pfd5.
PFDs participate in the organization of cortical MTs exclusively as canonical complex
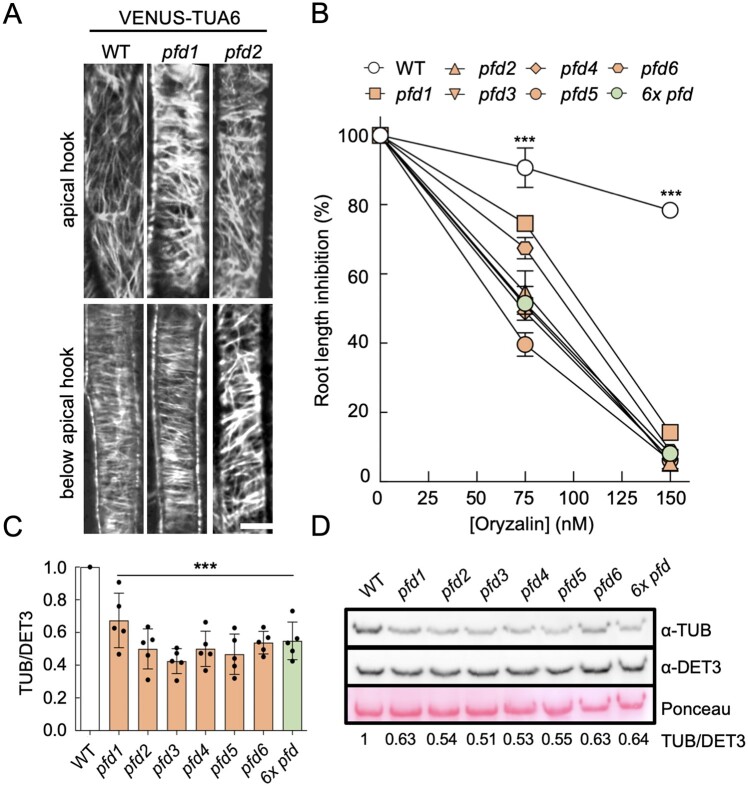
Defects in the organization of cortical MTs have been described in Arabidopsis for the pfd3, pfd4, pfd5, and pfd6 mutants (Gu et al., 2008; Rodriguez-Milla and Salinas, 2009; Perea-Resa et al., 2017). To determine if pfd1 and pfd2 mutations also cause defects in the organization of cortical MT, we introduced the MT marker pUBQ10::Venus-TUA6 (Salanenka et al., 2018) into both mutant backgrounds by genetic crosses and imaged MTs by confocal microscopy in 3-d-old etiolated seedlings. In the wild-type, cortical MTs were disorganized in apical hook cells and organized in elongating cells below the hook (Figure 2A). Similar organization was observed in the pfd1 mutant, while they were randomly arranged in both types of cells in pfd2 seedlings, similarly to pfd6 (Gu et al., 2008). We analyzed other traits dependent on tubulin folding: the sensitivity to the MT-depolymerizing drug oryzalin and tubulin levels. pfd1 seedlings showed a response to oryzalin similar to the other pfd mutants, including pfd2, although reduced sensitivity was observed at the lowest concentration (Figure 2B). α-tubulin levels were reduced in all mutants (Figure 2, C and D). These results show a similar behavior for all pfd mutants regarding MT-related phenotypes. Nonetheless, our results also indicate that PFD1 seems dispensable for the PFDc function in elongating cells below the apical hook.
Figure 2.
MT alterations in pfd mutants. A, Representative confocal images of VENUS-TUA6 in wild-type (WT), pfd1, and pfd2 hypocotyl cells (four hypocotyls were imaged per genotype). Scale bar = 10 μm. B, Effect of oryzalin on root elongation. The graph shows the average of two biological replicates (n = 8–16 roots per genotype, treatment and biological replicate). Error bars indicate standard error of mean. ***P < 0.001 in a two-way ANOVA test. C, Levels of α-tubulin relative to DET3. Data are the average of five independent experiments. ***P < 0.001 compared to the WT in Dunnet’s multiple comparison test after ANOVA test. D, Representative western blot showing α-tubulin levels in the WT and in pfd mutants. DET3 was used as loading control. The ratio of tubulin/DET3 of this representative blot is shown.
Although the function of MT folding is attributed to the PFDc (Gestaut et al., 2019), the lack of systematic analysis makes it difficult to rule out a complex-independent role for the individual subunits. The 6x pfd mutant showed the same sensitivity to oryzalin and the same reduction of α-tubulin levels as the individual pfd mutants (Figure 2, B–D), indicating that PFDs participate in MT-related processes exclusively as part of the PFDc.
PFDc-dependent and -independent contributions of PFDs to organ growth
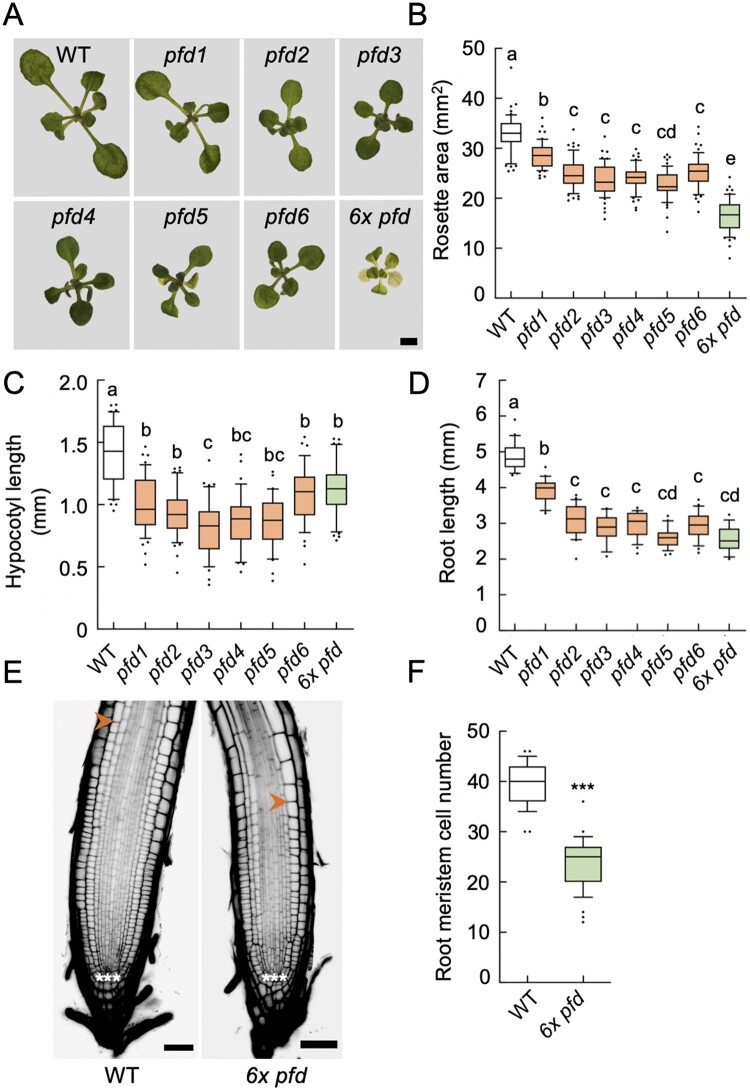
Reduced growth is a common trait of pfd mutants (Gu et al., 2008; Rodriguez-Milla and Salinas, 2009; Perea-Resa et al., 2017; Esteve-Bruna et al., 2020). To determine if PFDs’ contribution to organ growth is mediated by the PFDc, we analyzed the size of the rosette and the hypocotyl and root length in individual pfd mutants and in the 6x pfd. The rosette size was reduced to a similar extent in individual pfd mutants and further reduced in the 6x pfd (Figure 3, A and B; Supplemental Figure S3). MT-related defects leading to altered cell division and/or expansion may contribute to rosette growth alterations in pfd mutants. Nonetheless, the fact that the sextuple mutant exhibits a further reduction in size indicates that other processes, probably unrelated to MTs and redundantly controlled by two or more PFDs, are also altered in this mutant. Therefore, it seems that PFDs contribute to the rosette growth in two ways, dependent on and independent of the PFDc.
Figure 3.
Effect of pfd mutations in organ growth. A, Representative images of 14-d-old rosettes of the indicated genotypes grown in 1/2 MS plates under continuous light. Scale bar = 2 mm. B–D, Box plots showing the rosette area (n = 21–26) in 14-d-old plants (B), the hypocotyl length in 7-d-old etiolated seedlings (n = 34–47) (C), and the root length (n = 16–24) of 7-d-old seedlings grown in LD photoperiod (D). E, Images of representative root tips. The meristem extends between cells marked with three asterisks (the quiescent center) and the arrowhead. Scale bar = 50 µm. F, Number of meristematic cells in roots (n = 29 roots for the wild-type (WT) and n = 39 roots for 6x pfd) of 8-d-old seedlings grown under continuous light. For (B–D) and (F), horizontal lines inside boxes indicate the median and the box limits indicate the upper and lower quartiles. Whiskers extend 1.5× the interquartile range, leaving outliers outside whiskers. Genotypes with different letters show significant differences at P < 0.05 according to ANOVA with Tukey’s Honestly Significant Difference (HSD) test. ***P < 0.001 in a Student’s t test when comparing mutant with the WT.
The analysis of the hypocotyl and root length revealed a similar reduction in the size of both organs in individual pfd and sextuple mutants (Figure 3, C and D). Despite the slight but significant differences among genotypes, the lack of an additive effect in the 6x pfd mutant suggests that PFDs act as canonical complex to promote the growth of both organs. At least part of the contribution of the PFDc to hypocotyl elongation may be mediated by its role in the organization of cortical MTs, since the growth of this organ is almost entirely mediated by cell expansion (Gendreau et al., 1997). In addition to cell expansion, cell divisions occurring in the root meristem also contribute to the growth of this organ. Indeed, the number of meristematic cells was strongly reduced in 6x pfd roots (Figure 3, E and F). Hence, PFDs would contribute to root growth as PFDc, which mediates at least MT-dependent cell division.
The PFDc contributes to the regulation of flowering time
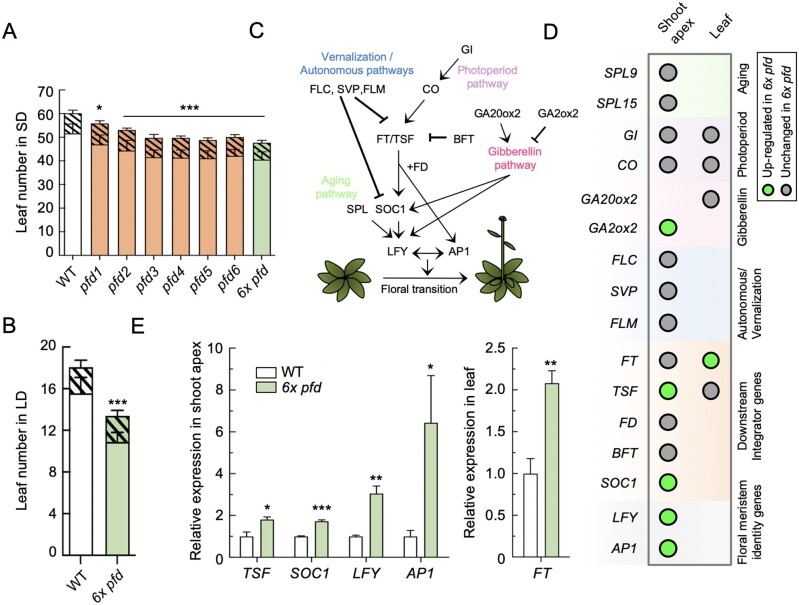
We reasoned that the increased expression of PFD genes in the vegetative rosette and in the shoot apex, before and after the transition to flowering, would be compatible with a role for PFDs in flowering time regulation (Supplemental Figure S4; Winter et al., 2007). To test this hypothesis and to determine eventually whether this role is performed by the PFDc, we measured the flowering time of individual pfd and sextuple mutants grown in short days (SDs). All mutants flowered earlier than the wild-type (Figure 4A). The phenotype was similar for all mutant lines, albeit the effect of the pfd1 mutation was milder. Importantly, the absence of additive effects in the 6x pfd suggests that the activity of PFDs on flowering time is exerted by the PFDc. This effect is independent of the photoperiod, since early flowering was also observed when the 6x pfd mutant was grown under long days (LDs; Figure 4B;Supplemental Figure S5A).
Figure 4.
Flowering phenotype of pfd mutants. A, B, Leaf number at bolting of plants grown in sd (n = 9–12) (A) or in LD (n = 12) (B). Open and filled bars represent rosette and cauline leaves, respectively. Error bars indicate standard deviation of each kind of leaves. * and *** indicate P < 0.05 and 0.001, respectively, compared to the wild-type (WT) in Dunnet’s multiple comparison tests after ANOVA tests when the total number of leaves is considered. C, Major pathways controlling flowering time in Arabidopsis. Arrows and blunted lines denote positive and negative interactions, respectively. D, Summary of RT-qPCR results. Circles indicate the genes analyzed in the shoot apex and/or the second oldest rosette leaf of 14-d-old wild-type and 6x pfd plants grown under LD. E, Expression of misregulated genes in 6x pfd plants compared to the WT. Data are mean from three biological replicates. Error bars represent standard error from means. *, **, and *** represent P < 0.05, 0.01, and 0.001 in t tests, respectively.
Next, to try understanding how the PFDc contributes to the flowering time regulation, we investigated whether the expression of key regulatory genes is altered in the 6x pfd mutant. The analysis included SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 (SPL9) and SPL15 (aging pathway); FLOWERING LOCUS M (FLM), FLOWERING LOCUS C, and SHORT VEGETATIVE PHASE (vernalization and autonomous pathways); GIGANTEA (GI) and CONSTANS (CO; photoperiod pathway); GIBBERELLIN 20-OXIDASE 2 (GA20ox2) and GIBBERELLIN 2-OXIDASE 2 (GA2ox2; gibberellin pathway); FLOWERING LOCUS T (FT), FD, TWIN SISTER OF FT (TSF), BROTHER OF FT AND TFL1, and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1; integrator genes); and LEAFY (LFY) and APETALA 1 (AP1; meristem identity genes; Fornara et al., 2010; Figure 4C). We analyzed their expression by reverse transcription quantitative polymerase chain reaction (RT-qPCR) in shoot apices and/or in the second oldest rosette leaf from 14-d-old plants grown in LD. Among representative genes of different pathways acting upstream of the integrator and floral meristem identity genes, only the expression of GA2ox2 was significantly altered in the mutant (Figure 4D;Supplemental Figure S5, B and C). We identified, however, the integrator FT-TSF module as the main target of the PFDc. The expression of FT in leaves and of TSF in the shoot apex was higher in the sextuple mutant than in the wild-type, which would explain the higher transcript levels of the downstream genes SOC1, LFY, and AP1 that lead to the floral transition (Figure 4, D and E; Supplemental Figure S5, B and C). These results suggest that the early flowering of the 6x pfd mutant is associated to increased FT-TSF activity. The PFDc, therefore, is required to delay flowering by attenuating the expression of integrator genes.
Different contributions of PFDs to the response to abiotic stress
The activity of PFD3, PFD4, and PFD5 is required for root growth under salt stress (Rodriguez-Milla and Salinas, 2009; Esteve-Bruna et al., 2020). The root growth of pfd2 and pfd6 mutants was affected by the 100 mM NaCl treatment in a similar way to that of the pfd3, pfd4, or pfd5 mutants, while it was less affected in the pfd1 (Figure 5A). Interestingly, 6x pfd seedlings behaved as the individual pfd, suggesting that PFDs contribute to the response to high salt as PFDc.
Figure 5.
Response to stress in pfd mutants. A–C, Percentage of root length inhibition in the presence of 100-mM NaCl (A), 12-mM LiCl (B), or 300-mM mannitol (C). Bars represent mean from four (A) or three (B, C) biological replicates. The number of roots analyzed per genotype and biological replicate were 9–13 (A), 5–7 (B), and 10–13 (C). Error bars indicate standard error of mean. * and *** indicate P < 0.01 and 0.001, respectively, compared to the wild-type (WT) in Dunnet’s multiple comparison tests after ANOVA. D, Freezing tolerance assay of cold-acclimated plants. Two-week-old wild-type, pfd1, pfd2, pfd3, pfd4, pfd5, pfd6, and 6x pfd plants grown at 20°C were transferred to 4°C for 5 d and subsequently exposed to −12°C for 6 h. Survival rates were determined after 1 week of recovering at 20°C. Error bars indicate standard error of mean from five biological replicates (n = 7–26 per genotype and biological replicate). *, **, and *** asterisks indicate P < 0.05, 0.01, and 0.001, respectively, compared to the WT in Dunnet’s multiple comparison tests after ANOVA tests. E, Representative plates after recovery.
The hypersensitive response of pfd mutants to NaCl could be caused by either the ionic or the osmotic component of the treatment. Previous findings show that the sensitivity of pfd3 and pfd5 mutants to NaCl is likely Na+-specific, because they are not hypersensitive to LiCl or mannitol (Rodriguez-Milla and Salinas, 2009). Nevertheless, a general effect on ionic or osmotic stress by redundantly acting PFDs cannot be ruled out. We measured the root length of individual pfd and 6x pfd mutants in the presence of 12-mM LiCl or 300-mM mannitol. LiCl stress inhibited root growth in a similar way in all genotypes, ruling out any involvement of PFDs in the response to nonspecific ionic stress (Figure 5B). Interestingly, the 6x pfd seedlings were more tolerant to the mannitol treatment, whereas a wild-type response was observed for individual pfd mutants (Figure 5C). These results indicate that the response of the plant to osmotic stress does not require the participation of the PFDc, but rather two or more PFD subunits that redundantly attenuate the response. Moreover, it indicates that it is unlikely that there is contribution of osmotic stress to the effect of high NaCl in pfd mutants. The contribution of PFDs to salt and osmotic stress responses is not mediated by transcriptional or post-transcriptional regulation of PFD genes (Supplemental Figure S6, A–C).
PFD3, PFD4, and PFD5 attenuate the acclimation to low temperatures (Perea-Resa et al., 2017). The individual pfd1 and pfd2 mutants showed the same freezing tolerance as pfd3, pfd4, and pfd5, whereas a wild-type response was observed for pfd6 (Figure 5, D and E). The 6x pfd plants showed a behavior similar to that of pfd1 to pfd5. The lack of additive effect in the 6x pfd mutant leads us to propose that PFDs contribute to freezing tolerance as PFDc, despite the pfd6 mutant shows a wild-type response. One explanation for this discrepancy would be that the missense pfd6 mutation causes a temperature-sensitive allele whose effect is compensated at low temperature (Figures 2–4).
PFDs mostly contribute to gene expression independently of the PFDc
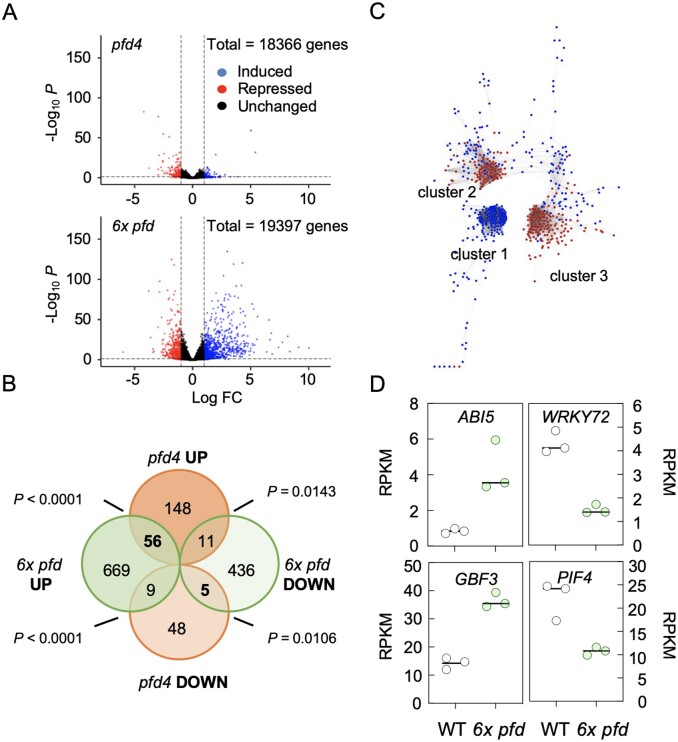
Impaired activity of PFDs results in altered gene expression (Esteve-Bruna et al., 2020). To determine whether PFDs participate in gene expression as canonical complex, we compared the differential expressed genes (DEGs) identified by RNA-seq in the pfd4 and 6x pfd mutants grown in vitro compared to the wild-type. A total of 1,186 DEGs, 734 up- and 452 downregulated, were identified in the 6x pfd mutant ([log2 FC] ≥ 2, P < 0.05; Figure 6, A and B; Supplemental Data Set S1). The relatively high number of DEGs in the mutant highlights the relevance of PFDs’ activity for gene expression.
Figure 6.
Transcriptomic analysis of the 6x pfd mutant. A, Volcano plots highlighting the DEGs in 6x pfd and pfd4 mutants (only genes with ≥1 RPKM in three replicates of mutants and/or the wild-type are shown). B, Venn diagram comparing DEGs in pfd4 and 6x pfd mutants. C, Cytoscape image of the coexpression network of DEGs in the 6x pfd mutant. Red and blue dots indicate downregulated and upregulated genes, respectively. D, Plots showing the expression level of the indicated genes extracted from the RNA-seq analysis. Each dot represents a replicate.
The number of DEGs was significantly lower in the pfd4 mutant (198 DEGs, [log2 FC] ≥ 2, P < 0.05; Figure 6, A and B; Supplemental Data Set S2; Esteve-Bruna et al., 2020). The fact that the transcriptional alterations are more severe in the 6x pfd mutant than in the pfd4 suggests that the global contribution of individual PFDs to gene expression is greater than that of the PFDc. Among the common genes, the upregulated ones followed the same trend (Figure 6B;Supplemental Figure S7A). It is important to note, however, that the majority of DEGs in pfd4 seedlings were not misexpressed in the 6x pfd mutant (Figure 6B). This could be explained if there is an antagonistic relationship between the different PFDs in certain conditions in their role as independent proteins: either the defects caused by loss of single PFDs are mutually cancelled in higher order mutants, or loss of one PFD causes an increase in the accumulation/activity of other PFDs.
PFDs regulate gene expression through different TFs
How the impairment in PFD activity is translated into transcriptional defects is not understood, and may depend on the context. To identify putative TFs that could inform on the transcriptional role of PFDs, we first examined the architecture of the coexpression network of the DEGs found in the 6x pfd mutant. We used the tool CORNET 3.0 (De Bodt et al., 2012) to determine the pairwise coexpression values among the 1,186 DEGs in the 6x pfd mutant in 454 microarray experiments. The analysis resulted in a network with 636 nodes and 9,646 edges (Supplemental Data Set S3), mostly organized into three compact gene clusters (Figure 6C). Interestingly, cluster 1 was exclusively formed by upregulated genes, while the two others mostly included downregulated genes (Supplemental Data Set S4). The highest coexpression values were observed between genes of the same cluster (see Supplemental Figure S7B for a larger image of clusters). This organization would be consistent with genes in each cluster being coregulated by a few TFs. Then, we searched for the putative upstream TF regulators using the TF2Network tool (Kulkarni et al., 2018). We found that 80% of genes in cluster 1 are coexpressed with ABA INSENSITIVE 5 (ABI5) and 69% of them are direct targets of this TF. Importantly, ABI5 itself was among the upregulated genes in the 6x pfd mutant (Figure 6D;Supplemental Data Set S1). ABI5 is a bZIP TF that plays a positive role in abscisic acid (ABA) signaling (Skubacz et al., 2016) and, in agreement with this, a gene ontology (GO) analysis showed that the category “response to ABA stimulus” was overrepresented in cluster 1 (P = 1.5 × 10−11; Supplemental Figure S7C). Between 39% and 63% of downregulated genes of cluster 2 were coexpressed with several AtWRKY TFs (AtWRKY65, 36, 72, 35, 29, 9, and 59; cited from the most to the least coexpressed). AtWRKY72 was also misexpressed in the 6x pfd mutant (Figure 6D;Supplemental Data Set S1). The latter and AtWRKY29 have been characterized, being related to defense against pathogens (Zhou et al., 2004; Bhattarai et al., 2010). The analysis of cluster 3 showed poor coexpression values with TFs (less than 17%). Nonetheless, it revealed that 66% and 33% of genes were direct targets of G-BOX BINDING FACTOR 3 (GBF3) and PHYTOCHROME-INTERACTING FACTOR 4 (PIF4), respectively. These TFs were also misexpressed in the 6x pfd mutant (Figure 6D;Supplemental Data Set S1). GBF3 is a bZIP TF that contributes to the plant’s response to abiotic stress (Ramegowda et al., 2017), and PIF4 is a well characterized bHLH TF that transmits information about ambient light and temperature to hormone and growth pathways (Choi and Oh, 2016). In summary, the in silico analyses predicted that part of DEGs in the 6x pfd mutant may be targets of ABI5, WRKY72, GBF3, and PIF4.
The transcriptome of the 6x pfd mutant reveals additional functions for PFDs
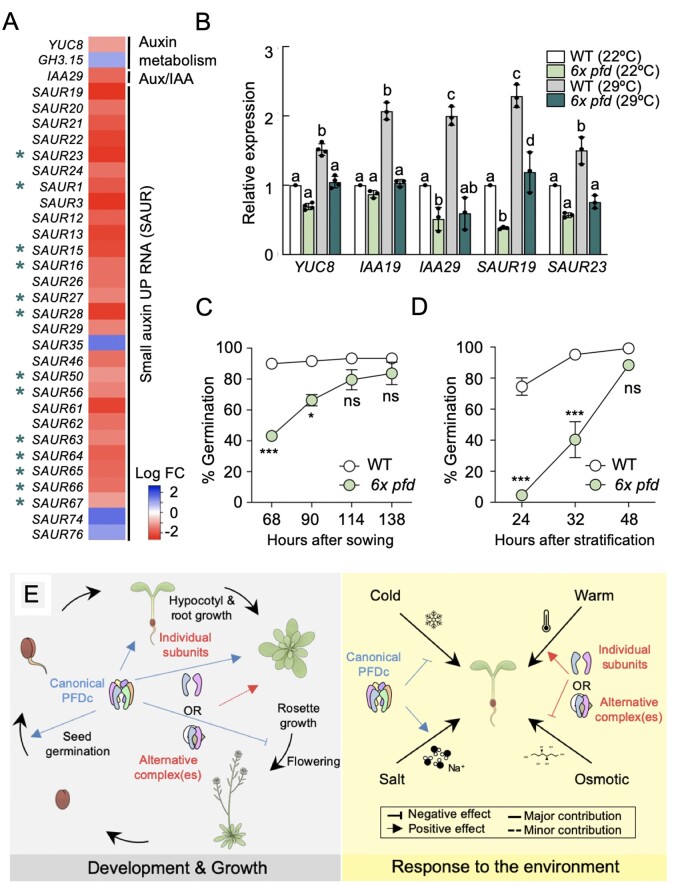
To identify additional biological functions affected by PFDs at the transcriptional level, we searched for enriched GO categories in the DEGs of 6x pfd seedlings (Supplemental Figure S7C and Supplemental Data Set S1). The GO category “response to auxin stimulus” was enriched in cluster 3 (P = 3.0 × 10−9). Particularly striking is the presence of 13 SMALL AUXIN UP-REGULATED RNA (SAUR) genes (Stortenbeker and Bemer, 2019) among those downregulated in this cluster (marked with an asterisk in Figure 7A). Indeed, another 13 SAUR together with the auxin biosynthesis gene YUCCA 8 (YUC8) and the auxin signaling gene AUXIN/INDOLE-3-ACETIC ACID 29 (IAA29), not represented in the microarray compendium used for coexpression analyses, also appeared downregulated in the 6x pfd mutant (Figure 7A;Supplemental Data Set S1). These results suggest that impairment of PFDs’ activity affects the auxin biosynthesis and signaling pathway.
Figure 7.
Additional functions for PFDs. A, Heatmap showing the behavior of auxin-related genes misregulated in 6x pfd seedlings. Asterisks mark SAUR genes included in cluster 3. B, Expression of auxin-related genes in response to 2 h at 29°C. Data are mean of three biological replicates ± standard error of mean. Different letters show significant differences at P < 0.05 according to ANOVA with Newman–Keuls test. C, D, Germination rates of nonstratified (C) and stratified (D) wild-type and 6x pfd seeds. Error bars represent standard error of mean from two (C) or seven (D) biological replicates. In (C), 53–59 seeds were analyzed per genotype and biological replicate. Data in (D) are also shown in Supplemental Figure S8 (n = 28–39 per genotype and biological replicate). * and *** represent P < 0.05 and 0.001 in Bonferroni tests after ANOVA tests, respectively. ns = no significant differences. E, Model depicting the different modes of action of PFDs in Arabidopsis.
The TF2Network analysis revealed that PIF4 probably controls the expression of genes in cluster 3. PIF4 is important for the expression of auxin biosynthesis and signaling genes, especially in response to warm temperature (Quint et al., 2016). In agreement with this, the enrichment of the GO category “response to auxin stimulus” was most significant among the direct targets of PIF4 in this cluster (P = 1.2 × 10−15). The reduced expression of PIF4 in the 6x pfd mutant (Figure 6D) may contribute to the low expression of the auxin genes. Nonetheless, we reasoned that the consequences of low PIF4 expression would be most obvious if we expose the 6x pfd mutant to warm temperature. To test this, we selected five genes whose induction by warm temperature is PIF4-dependent: YUC8 (Sun et al., 2012), IAA19 (Huai et al., 2018), IAA29 (Koini et al., 2009), and SAUR19 and 23 (Franklin et al., 2011). Notably, the induction at 29°C was mostly impaired for all genes in the mutant, except for SAUR19 (Figure 7B). These results indicate that PFDs are required for the proper molecular response of seedlings to warm temperature. The fact that SAUR19 still responds to the temperature shift, despite it is dependent on PIF4 (Franklin et al., 2011), suggests that PFDs may affect the temperature response through other pathways in addition to those involving this TF. The contribution of PFDs to the temperature response is not mediated by transcriptional regulation of PFD genes (Supplemental Figure S6D).
Since GO terms related to germination were enriched in the DEGs in the 6x pfd mutant (Supplemental Figure S7C and Supplemental Data Set S1) and all PFD genes are transcriptionally active in imbibed seeds (Supplemental Figure S4), we wondered whether PFDs have a role in this process. To determine the germination capacity of 6x pfd seeds, we compared their germination rate with that of the wild-type with or without stratification at 4°C for 72 h. The 6x pfd seeds exhibited a delay in germination regardless of stratification (Figure 7, C and D). The role of PFDs in germination might depend on the PFDc because seeds of all individual pfd mutants showed a delay in germination similar to that of 6x pfd seeds (Supplemental Figure S8).
The enrichment of GO categories related to abiotic stress among DEGs in 6x pfd seedlings (Supplemental Figure S7C; Supplemental Data Set S1) suggests that this mutant may constitutively manifest a stress response. To investigate this possibility, we compared the transcriptome of 6x pfd seedlings with that of wild-type seedlings exposed to either 150-mM NaCl or 4°C for 24 h (Esteve-Bruna et al., 2020). The meta-analysis revealed that 56.7% of the DEGs in the mutant were altered in the wild-type exposed to either of the two stresses (Supplemental Figure S9A and Supplemental Data Set S5), and that many of them behaved similarly (Supplemental Figure S9B). GO categories over-represented in the set of 163 DEGs common to the three conditions included response to abiotic stimuli and response to hormones ABA and JA (Supplemental Figure S9C and Supplemental Data Set S5). In particular, several genes encoding ABA receptors and those coding for the protein phosphatases that act as negative regulators of the ABA-signaling pathway were downregulated and upregulated, respectively, in the 6x pfd mutant (Supplemental Figure 10, A and B). These results, together with the upregulation of ABA-responsive genes in the mutant (Supplemental Figure 10C), suggest that PFDs are required to repress the ABA-related transcriptional response. In agreement with this, the transcriptional behavior is not a response to elevated ABA levels in the mutant, which are actually reduced compared to the wild-type (Supplemental Figure 10D), likely due to enhanced ABA catabolism (Supplemental Figure 10E). In summary, PFDs seem to be required to maintain adequate expression levels of many stress-related genes under nonstressful conditions, which is consistent with the role observed for PFDs under these stress conditions.
Discussion
Determining whether PFDs participate in a particular process as members of the canonical PFDc or, conversely, in alternative configurations, i.e. other complexes or as individual subunits, provides clues to their mode of action. The analysis of the 6x pfd mutant has allowed us to identify processes that are dependent and independent of the activity of the canonical PFDc (Figure 7E). Joint action with the TRiC/CCT would be expected in those processes that require the participation of the canonical complex (Gestaut et al., 2019). We have found that several growth-involving processes depend on PFDc, in which it probably participates through the folding of at least tubulins. For other processes also depending on the PFDc, such as the plant’s response to salt stress or the regulation of flowering time, the mechanistic connection with tubulins is not so obvious. Interestingly, altering MT polymerization affects gene expression in plants (Sangwan et al., 2001), and actin, another bona fide substrate of PFDc-TRiC/CCT, performs roles in the nucleus related to transcription (Blettinger et al., 2004). This opens up the possibility that the effect of the PFDc on the expression of the flowering integrator genes or of genes involved in salt stress is mediated through these two protein substrates. Alternatively, the PFDc may act through actin-related proteins, some of which are subunits of chromatin remodeling complexes that regulate the expression of FT (March-Diaz and Reyes, 2009; Kumar et al., 2012) and also of stress genes (Wang et al., 2019). In fact, CENTRACTIN, an actin-related protein belonging to a cytosolic complex, is substrate for TRiC/CCT chaperonin in vertebrates (Melki et al., 1993).
PFDc-TRiC/CCT may fold substrates other than tubulins or actin that could also mediate the function of the PFDc. Although not supported by functional or genetic analyses, interactomic approaches suggest that PFDc-TRiC/CCT assist the folding of the histone deacetylase HDAC1 in the nucleus of human cells (Banks et al., 2018). Similar approaches have identified the PFDc and TRiC/CCT complexes associated to the TARGET OF RAPAMYCIN (TOR) kinase complex in Arabidopsis (Van Leene et al., 2019). In Drosophila, TRiC/CCT interacts physically with members of the TOR complex and is required for TOR function, probably by participating in its assembly (Kim and Choi, 2019). That the TOR complex is substrate of the PFDc-TRiC/CCT in plants is an interesting possibility that awaits investigation.
We have identified other processes in which the action of PFDs does not involve the canonical complex (Figure 7E). Genetic analyses indicate that two or more PFDs act redundantly to promote rosette growth or to attenuate the response to osmotic stress. In these cases, it is more difficult to anticipate their mode of action. They may participate as individual subunits or as part of alternative complexes. For example, our previous results show that PFDs can participate in proteostasis independently of the PFDc. PFD4 acts as adaptor to mediate the stabilization of the spliceosome complex LIKE-SM 2–8 by the chaperone Hsp90, a process in which PFD2 does not appear to be involved (Esteve-Bruna et al., 2020). Their participation in alternative complexes is a very exciting possibility that has been proposed in yeast to explain the role of PFDs in transcriptional elongation, although the identity of the complex is currently unknown (Millan-Zambrano et al., 2013). A PFD-like complex formed by PFD2 and PFD6 together with the PFD-like proteins UNCONVENTIONAL PREFOLDIN RPB5 INTERACTOR 1, UBIQUITOUSLY EXPRESSED TRANSCRIPT, p53 AND DNA DAMAGE-REGULATED PROTEIN 1, and ASDURF has been identified in animals (Chaves-Perez et al., 2018). It is therefore reasonable to think that the putative Arabidopsis PFD-like complex may participate in PFDc-independent processes requiring PFD2 and/or PFD6. Defining the in vivo interactome of PFDs in different pfd mutant backgrounds would allow identifying putative alternative complexes and their partners, helping therefore to delineate PFDc-independent mechanisms for PFDs’ action.
Our results indicate that PFDs are required to maintain adequate levels of gene expression and that at least part their action is mediated by a few TFs (Figure 6). The mechanisms through which PFDs affect the expression of these genes are, however, a matter for future research. We anticipate that an important contribution will be indirect, through their participation in cellular proteostasis, as mentioned above. For instance, they can regulate the stability of TFs or complexes involved in different stages of gene expression, as already demonstrated in Arabidopsis for the TF HY5 (Perea-Resa et al., 2017) or the spliceosome complex LSM2–8 (Esteve-Bruna et al., 2020). Following this line, the stability or the recruitment to larger complexes of the TFs identified by the TF2Network analysis (Figure 6B) might depend on PFDs. Nonetheless, other possibilities cannot be excluded. In humans, PFD5 acts as a bridge protein that helps recruit a co-repressor complex to c-Myc (Fujioka et al., 2001; Satou et al., 2001) and PFD1 binds to the transcription start site of the Cyclin A gene to repress its expression (Wang et al., 2017), while, in the worm C. elegans, PFD6 activates the TF DAF-16/FOXO by physical interaction (Son et al., 2018). Therefore, it is possible that plant PFDs also contribute directly to the gene expression process, performing functions similar to those described in animals. Indeed, a fraction of the Arabidopsis PFD5 is located in the chromatin (Locascio et al., 2013), as occurs in yeast and animals where PFDs regulate various aspects of transcription (Millan-Zambrano et al., 2013; Payán-Bravo et al., 2021). Although this location would be compatible with a role for the Arabidopsis PFD5 in proteostasis, exerted locally at this location, it is likely that it also reflects the direct involvement of PFDs in transcriptional regulation.
PFDs are required for normal development of animals and their malfunction, due to mutation or misexpression, is normally associated to cancer or disease (Liang et al., 2020) or causes embryo lethality (Lundin et al., 2008; Delgehyr et al., 2012). Arabidopsis pfd mutants, including the 6x pfd, are viable and do not show apparent developmental defects beyond reduced size and early flowering, indicating that PFDs’ activity is mostly dispensable for normal development in this species (this work; Gu et al., 2008; Rodriguez-Milla and Salinas, 2009; Perea-Resa et al., 2017; Esteve-Bruna et al., 2020). Rather, their role in plants appears to be more relevant for properly interpreting environmental challenges (Rodriguez-Milla and Salinas, 2009; Perea-Resa et al., 2017; Esteve-Bruna et al., 2020). In fact, we show here that plants defective in the six PFDs perform better than the wild-type when exposed to osmotic stress, which adds to known defects in the response to high salt or low temperature. It is important to note that our results also expand this view. We show that they are not only required to respond to stress, but also to respond to environmental changes that, in principle, do not represent a stressful scenario for the plant, such as a moderate increase in the ambient temperature. This is further supported by the finding of enriched GOs related to environmental and stress responses in the gene set misregulated in the 6x pfd mutant, and with the fact that the TFs identified by coexpression analyses acting downstream of PFDs mainly participate in environmental responses. The different contribution of PFDs to the plant’s response to environmental changes, i.e. positive for salt stress or warm temperature and negative for cold or osmotic stress, is probably an indication of the varied modes of action through which these proteins act, which are just beginning to glimpse.
In summary, our results place PFDs as relevant players in the plant’s response to environmental changes. Furthermore, the genetic analyses provide evidence that PFDs’ action is not always mediated by the canonical PFDc and clues about their possible mode of action. The genetic resources generated in this work will help deciphering the mechanisms through which these versatile proteins act.
Materials and methods
Plant material and growth conditions
Arabidopsis (A. thaliana) accession Columbia-0 (Col-0) was used as the wild-type. Most mutants and transgenic lines have been described: pfd2 (Esteve-Bruna et al., 2020), pfd3 and pfd5 (Rodriguez-Milla and Salinas, 2009), pfd4 (Perea-Resa et al., 2017), pfd6-1 (Gu et al., 2008), pRGA::GFP-RGA (Silverstone et al., 2001), pPFD4::PFD4-GFP (Perea-Resa et al., 2017), p35S::PFD6-YFP (Esteve-Bruna et al., 2020), and pUBQ10::VENUS-TUA6 (Salanenka et al., 2018). The pfd1 mutant (GK-689A09) was obtained from the Nottingham Arabidopsis Stock Centre.
To grow seedlings in vitro, seeds were surface-sterilized, sown on plates with half-strength Murashige and Skoog (MS; Duchefa) media, pH 5.7, that includes 1% (w/v) sucrose and 8 g L−1 agar (control media), and stratified at 4°C for 3–4 d. Plates were exposed to continuous light (50–60 μmol m−2 s−1) or LD photoperiod (16 h of 90 μmol m−2 s−1) at 22°C. For hypocotyl length measurements and TUA6-VENUS visualization, seedlings were grown without sucrose.
To obtain the 6x pfd mutant, all double mutant combinations were first prepared by crossing individual pfd. Then, double mutants were used to obtain five triple mutants (pfd1,2,3; pfd1,3,5; pfd2,3,5; pfd2,4,6, and pfd3,5,6). Triple mutants were used to obtain three quadruple mutants (pfd1,2,3,5; pfd1,3,5,6, and pfd2,3,5,6). Crosses between triple and quadruple mutants, and between two quadruples, were performed to obtain three quintuples (pfd1,2,3,5,6; pfd1,3,4,5,6, and pfd2,3,4,5,6) and the sextuple mutant. All mutant combinations were genotyped in F2 generations with primers listed in Supplemental Table S1.
The pUBQ10pro::VENUS-TUA6 pfd1 and pUBQ10::VENUS-TUA6 pfd2 lines were obtained by crossing and confirmed by genotyping in F2 or F3 generations with the primers listed in Supplemental Table S1.
Protein structure prediction
The 3D structure of each Arabidopsis PFD was obtained by homology with the human PFDs (PDB code 6NR8; Gestaut et al., 2019) using Modeller (release 9.23; Webb and Sali, 2016). The 3D structure of the Arabidopsis PFDc was assembled, using the human PFDc (PDB code 6NR8) as template, and visualized with PyMOL 2.4 software. The evaluation of the structural models was carried out at the SaliLab Model Evaluation Server (https://modbase.compbio.ucsf.edu/evaluation//).
Tandem affinity purification
The GSrhino-PFD3 fusion under the control of the 35S promoter was generated and used to transform Arabidopsis PSB-D cell suspension cultures. The sequential affinity purification was performed as described (Van Leene et al., 2015). Proteolysis and peptide isolation, acquisition of mass spectra, and protein identification was carried out at the Unidad de Proteómica (CNB, Madrid, Spain).
Gel-filtration chromatography and western blot analysis
For gel-filtration chromatography, protein extracts of 7-d-old pPFD4::PFD4-GFP and p35S::PFD6-YFP seedlings grown under continuous light were prepared in extraction buffer (50-mM Tris–HCl, pH 7.5, 150-mM NaCl, 10-mM MgCl2, 10% [v/v] glycerol, 0.5% [v/v] Nonidet P-40, 2-mM PMSF, and 1x protease-inhibitor cocktail) and loaded onto a Superose™ 6 Increase column (GE Healthcare). Twenty-four fractions of 0.5 mL were collected and precipitated as described (Esteve-Bruna et al., 2020). Proteins were separated in 12% (w/v) sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane by western blotting. Membranes were stained with Ponceau S solution and then incubated with anti-GFP antibody (JL-8, Takara Bio Clontech, 1:5000).
To determine tubulin levels in pfd mutants, 7-d-old seedlings were grown under continuous light at 22°C. To determine GFP-RGA, PFD4-GFP, and PFD6-YFP levels in the presence of salt or mannitol, 7-d-old seedlings grown in continuous light were exposed to 100-mM NaCl or 300-mM mannitol in liquid media for 0, 8, or 24 h. Ground frozen tissue from whole seedlings was homogenized in extraction buffer. Total proteins were separated in 12% (w/v) SDS–PAGE, transferred to PVDF membranes by Western blotting and visualized with anti-GFP (JL-8, Takara Bio Clontech, 1:5,000), anti-α-tubulin (Invitrogen, 1:1,000), or anti-DET3 (1:10,000, provided by Prof. Dr Karin Schumacher). Quantification of protein bands in Western blots was performed using FIJI (https://fiji.sc/).
Confocal microscopy
To determine MT organization, seeds of pUBQ10pro::VENUS-TUA6 in wild-type, pfd1, and pfd2 backgrounds were germinated in MS media without sucrose and grown vertically in darkness for 3 d at 22°C. MT were visualized in cells at the apical hook and immediately below by using a Nikon Eclipse Ti2 inverted microscope, equipped with a Yokogawa Spinning Disk Field Scanning Confocal System (https://www.microscope.healthcare.nikon.com/en_EU/products/confocal-microscopes/csu-series/specifications). The objective used was the oil immersion CFI 60 x H Plan Apocr λ oil W.D. 0.13 mm N.A. 1.40. VENUS was excited by a 488-nm single-mode optical fiber laser (120 mW, power 60.0%) and the collection bandwith at 525–550 nm. Images were collected with a Photometrics Prime BSI CMOS camera (https://www.photometrics.com/products/prime-family/primebsi) with an exposure time of 100–200 ms with a 1×1 binning (2048 × 2048 pixels). The NIS-Element AR (Nikon, Japan, http://www.nis-elements.com/) was used as platform to control microscope, laser, camera, and post-acquisition analyses. Images were denoised by using the Denoise.ai algorithm (https://denoise.laboratory-imaging.com/process) and then analyzed using FIJI software.
Phenotypic analyses
The quantification of the sensitivity to oryzalin was carried out by measuring the length of the primary root of 7-d-old seedlings grown in LD on vertical plates supplemented with increasing concentrations of oryzalin (0, 75, and 150 nM). Root length was measured using FIJI.
To determine rosette area, seeds were sown on 140-mm diameter Petri dishes at low density (40 seed per plate) and grown under continuous light. Plates were photographed with a digital camera 14 d after germination. The measurement was obtained using the FIJI plug-in Rosette tracker from at least three biological replicates (20 seedlings each). For hypocotyl length measurements, seeds were germinated in white light for 8 h and then grown in vertical plates in darkness for 7 d. Hypocotyl length was measured with FIJI.
For flowering-time measurements, seeds were sown on pots, stratified for 7 d at 4°C and grown under SD (8-h light/16-h dark) or LD (16-h light/8-h dark) photoperiods at 22°C. Flowering time was recorded as the number of rosette and cauline leaves or days at bolting.
For dormancy assay, seed lots to be compared were freshly harvested on the same day from individual plants grown in identical conditions. Seeds were sown immediately without stratification and incubated under continuous light. For the germination assay, freshly harvested seeds were sown and stratified for 3 d at 4°C. The percentage of seeds with an emerged radicle was determined at different time points.
Stress tolerance assays
NaCl, LiCl, and mannitol tolerance was analyzed by transferring 4-d-old seedlings grown on vertical MS plates under LD conditions to new MS plates supplemented with or without 100-mM NaCl, 12-mM LiCl, or 300-mM mannitol and incubated vertically for 4 d. Root length was measured using FIJI. Tolerance to freezing temperatures was determined as follows: 2-week-old plants grown on plates under LD photoperiod at 20°C were transferred to 4°C for 5 d and subsequently exposed to −12°C for 6 h. Survival rates were determined after 1 week of recovering at 20°C.
RNA extraction and RT-qPCR
For expression analysis of flowering-related genes, seeds were sown on soil and grown in LD. Shoot apex and the second oldest leaf from 14-d-old plants were collected. For expression analysis of auxin-related genes, seedlings were grown for 5 d under continuous light at 22°C and then transferred to 29°C for 2 h. For PFD expression in the presence of salt or mannitol, 7-d-old seedlings grown in continuous light were exposed to 100-mM NaCl or 300-mM mannitol in liquid media for 0, 8, or 24 h. Total RNA was extracted using a RNA-extraction kit (Machery-Nagel) and treated with DNase I on column (Machery-Nagel). cDNA was synthesized with the PrimeScript 1st strand cDNA Synthesis Kit (Takara), and used as a template for RT-qPCR assays employing the SYBR Premix Ex Taq II (Takara) with primers listed in Supplemental Table S1. The relative expression values were calculated using the At1g13320 (PDF2-1) gene as a reference, using the ΔΔCT method. All assays were performed with at least two biological replicates, each including three technical replicates.
For RNA-seq experiments, two type of samples were collected: (1) 7-d-old wild-type and 6x pfd seedlings grown under continuous light at 22°C and (2) wild-type and pfd4 seedlings grown under LD conditions for 2 weeks at 22°C. Total RNA was extracted with the RNeasy Plant Mini Kit (Qiagen), and the RNA concentration and integrity (RIN) were measured in a RNA nanochip (Bioanalyzer, Agilent Technologies 2100). The preparation of the libraries and the subsequent sequencing in an Illumina NextSeq 500 platform was carried out by the Genomic Service of the University of Valencia.
RNA-seq analysis
Read trimming was performed with cutadapt. Approximately, 20 million 75-bp paired-end reads per sample were generated and >90% reads were aligned to the TAIR10 Col-0 reference genome using HISAT2 with default parameters. htseq-count was used for read counting and DESeq2 for identifying DEGs as those that display absolute value of log2 fold chance (logFC) > 1 and P adjusted value <0.05.
Heatmaps from RNA-seq data were performed using the http://www1.heatmapper.ca/expression/ website. Volcano plots were constructed with the EnhancedVolcano R package. GO terms were obtained from AgriGO v2 and then were filtered with REVIGO for doing scatter plots. The Integrative Genomics Viewer was used to visualize reads from alignment files.
Microarray-based expression analysis
Data for PFD gene expression were gathered from the Arabidopsis eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Heatmaps were generated using the Matrix2png tool (https://matrix2png.msl.ubc.ca/).
The coexpression analysis was done with CORNET (https://bioinformatics.psb.ugent.be/cornet/versions/cornet3.0/). Options used were: Pearson correlation method, correlation coefficient >0.7, retrieve top 170 genes, show pairwise correlations only. Microarray Compendium 1 TAIR10 (454 experiments with bias toward cell cycle, growth, and development) was used as source data.
ABA quantification
Seedlings were grown for 7 d in continuous light. The ground tissue (about 150 mg of frozen seedlings) was resuspended in 80% (v/v) methanol-1% (v/v) acetic acid containing the internal standard deuterium-labeled hormone (2H6-ABA) and mixed by shaking during 1 h at 4°C. The extract was kept at −20°C overnight. After centrifugation, the supernatant was dried in a vacuum evaporator. The dry residue was dissolved in 1% (v/v) acetic acid and passed through a reverse phase Oasis HLB column. The final residues were dissolved in 5% (v/v) acetonitrile-1% (v/v) acetic acid. ABA hormone was then separated using an autosampler and reverse-phase Ultra High Performance Liquid Chromatography (2.6 µm Accucore RP-MS column, 50 mm length × 2.1 mm i.d.; ThermoFisher Scientific) with a 5%–50% (v/v) acetonitrile gradient containing 0.05% (v/v) acetic acid, at 400 µL·min−1 over 14 min. The ABA was analyzed with a Q-Exactive mass spectrometer (Orbitrap detector; ThermoFisher Scientific) by targeted selected ion monitoring. The concentrations of ABA in the extracts were determined using embedded calibration curves and the Xcalibur 4.0 and TraceFinder 4.1 SP1 programs.
Statistical analysis
P-values in overlapping DEGs from two genotypes or conditions were calculated using hypergeometric tests. The rest of P-values were obtained from one-way analysis of variance (ANOVA) tests followed by multiple comparison tests when more than two genotypes were compared together. t Tests were performed instead when comparing only two genotypes.
Accession numbers
Raw sequences (fastq files) and differential expression gene tables used in this article have been deposited in the Gene Expression Omnibus database (accession no. GSE138432). PFD1 (At2g07340), PFD2 (At3g22480), PFD3 (At5g49510), PFD4 (At1g08780), PFD5 (At5g23290), and PFD6 (At1g29990).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Table S1 . Oligonucleotides used in this work.
Supplemental Figure S1 . Structure of the human and Arabidopsis PFDc.
Supplemental Figure S2 . The pfd mutant alleles.
Supplemental Figure S3 . pfd mutant plants grown on soil.
Supplemental Figure S4 . Heatmap showing the absolute expression levels of the six PFD genes at different stages of Arabidopsis development.
Supplemental Figure S5 . The 6x pfd mutant flowers early.
Supplemental Figure S6 . PFDs’ transcripts and proteins levels in response to environmental changes.
Supplemental Figure S7 . RNA-seq analysis of 6x pfd seedlings.
Supplemental Figure S8 . The PFDc contributes to seed germination.
Supplemental Figure S9 . Constitutive stress signature in 6x pfd seedlings.
Supplemental Figure S10 . Expression of ABA-related genes in 6x pfd seedlings.
Supplemental Data Set S1 . DEGs and enriched GO categories in 6x pfd seedlings.
Supplemental Data Set S2 . DEGs in pfd4 seedlings.
Supplemental Data Set S3 . Gene pairs and correlation values used to construct the coexpression network.
Supplemental Data Set S4 . List of genes in clusters 1, 2, and 3 in the coexpression network.
Supplemental Data Set S5 . Comparison of DEGs in the 6x pfd mutant and in wild-type seedlings treated with salt or low temperature.
Supplementary Material
Acknowledgments
We thank Dr Cristina Úrbez (IBMCP, Spain) for her help with the Arabidopsis PSB-D cell suspensions, Dr Esther Carrera for the ABA quantification carried out at the Plant Hormone Quantification Service (IBMCP, Spain), and Drs Jiri Friml (IST, Austria) and Karin Schumacher (University of Heidelberg, Germany) for seeds of pUBQ10::Venus-TUA6 and for the anti-DET3 antibody, respectively. Part of imaging analyses was carried out at NOLIMITS, an advanced imaging facility established by the Università degli Studi di Milano (Italy).
Funding
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness and “Agencia Estatal de Investigación”/FEDER/European Union (BIO2013-43184-P to D.A. and M.A.B., and BIO2016-79133-P and PID2019-109925GB-I00 to D.A.). N.B.-T., A.S.-M., and A.P.-A. were recipient of Ministerio de Economía y Competitividad (BES-2014-068868), EU MSCA-IF (H2020-MSCA-IF-2016-746396) and Ministerio de Educación (FPU17/05186) fellowships, respectively.
Conflict of interest statement. None declared.
Senior author.
N.B.-T., D.E.-B., A.S.-M., M.A.B., and D.A. designed the experiments; N.B.-T., D.E.-B., A.S.-M., R.M.E.-A., F.R., C.C.-L., C.N.-R., A.P.-A., and D.A. performed the experiments; N.B.-T., D.E.-B., A.S.-M., R.M.E-A., F.R., J.F., C.C.-L., C.N.-R., A.P.-A., P.C., J.S., M.A.B. and D.A. analyzed the data; N.B.-T., D.E.-B., M.A.B., and D.A. prepared the figures; D.A. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is David Alabadí (dalabadi@ibmcp.upv.es).
References
- Amorim AF, Pinto D, Kuras L, Fernandes L (2017) Absence of Gim proteins, but not GimC complex, alters stress-induced transcription. Biochim Biophys Acta - Gene Regul Mech 1860:773–781 [DOI] [PubMed] [Google Scholar]
- Arranz R, Martin-Benito J, Valpuesta JM (2018) Structure and function of the cochaperone prefoldin. Adv Exp Med Biol 1106:119–131 [DOI] [PubMed] [Google Scholar]
- Banks CAS, Miah S, Adams MK, Eubanks CG, Thornton JL, Florens L, Washburn MP (2018) Differential HDAC1/2 network analysis reveals a role for prefoldin/CCT in HDAC1/2 complex assembly. Sci Rep 8:13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai KK, Atamian HS, Kaloshian I, Eulgem T (2010) WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J 63:229–240 [DOI] [PubMed] [Google Scholar]
- Blettinger BT, Gilbert DM, Amberg DC (2004) Actin up in the nucleus. Nat Rev Mol Cell Biol 5:410–415 [DOI] [PubMed] [Google Scholar]
- De Bodt S, Hollunder J, Nelissen H, Meulemeester N, Inzé D (2012) CORNET 2.0: integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol 195:707–720 [DOI] [PubMed] [Google Scholar]
- Chaves-Perez A, Thompson S, Djouder N (2018) Roles and functions of the unconventional prefoldin URI. Adv Exp Med Biol 1106:95–108 [DOI] [PubMed] [Google Scholar]
- Choi H, Oh E (2016) PIF4 integrates multiple environmental and hormonal signals for plant growth regulation in Arabidopsis. Mol Cells 39:587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Wieland U, Rangone H, Pinson X, Mao G, Dzhindzhev NS, McLean D, Riparbelli MG, Llamazares S, Callaini G, et al. (2012) Drosophila Mgr, a prefoldin subunit cooperating with von Hippel Lindau to regulate tubulin stability. Proc Natl Acad Sci USA 109:5729–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramian D, Eswar N, Shen M-Y, Sali A (2008) How well can the accuracy of comparative protein structure models be predicted? Protein Sci 17:1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Bruna D, Carrasco-López C, Blanco-Touriñán N, Iserte J, Calleja-Cabrera J, Perea-Resa C, Úrbez C, Carrasco P, Yanovsky MJ, Blázquez MA, et al. (2020) Prefoldins contribute to maintaining the levels of the spliceosome LSM2-8 complex through Hsp90 in Arabidopsis. Nucleic Acids Res 48: 6280–6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141:550. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A 108:20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Taira T, Maeda Y, Tanaka S, Nishihara H, Iguchi-Ariga SMM, Nagashima K, Ariga H (2001) NM-1, a c-Myc-binding protein, is a candidate for a tumor suppressor in leukemia/lymphoma and tongue cancer. J Biol Chem 276:45137–45144 [DOI] [PubMed] [Google Scholar]
- Geissler S, Siegers K, Schiebel E (1998) A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J 17:952–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Hofte H (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114:295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D, Roh SH, Ma B, Pintilie G, Joachimiak LA, Leitner A, Walzthoeni T, Aebersold R, Chiu W, Frydman J (2019) The chaperonin TRiC/CCT associates with prefoldin through a conserved electrostatic interface essential for cellular proteostasis. Cell 177:751–765 e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Deng Z, Paredez AR, DeBolt S, Wang ZY, Somerville C (2008) Prefoldin 6 is required for normal microtubule dynamics and organization in Arabidopsis. Proc Natl Acad Sci U S A 105:18064–18069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai J, Zhang X, Li J, Ma T, Zha P, Jing Y, Lin R (2018) SEUSS and PIF4 coordinately regulate light and temperature signaling pathways to control plant growth. Mol Plant 11:928–942 [DOI] [PubMed] [Google Scholar]
- Kim AR, Choi KW (2019) TRiC/CCT chaperonins are essential for organ growth by interacting with insulin/TOR signaling in Drosophila. Oncogene 38:4739–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40:D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19:408–413 [DOI] [PubMed] [Google Scholar]
- Kulkarni SR, Vaneechoutte D, Van de Velde J, Vandepoele K (2018) TF2Network: predicting transcription factor regulators and gene regulatory networks in Arabidopsis using publicly available binding site information. Nucleic Acids Res 46:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484:242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Smith RS, Jordan W, King BL, Won J, Valpuesta JM, Naggert JK, Nishina PM (2011) Prefoldin 5 is required for normal sensory and neuronal development in a murine model. J Biol Chem 286:726–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Xia L, Oyang L, Lin J, Tan S, Yi P, Han Y, Luo X, Wang H, Tang L, et al. (2020) The functions and mechanisms of prefoldin complex and prefoldin-subunits. Cell Biosci 10:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Blázquez M, Alabadí D (2013) Dynamic regulation of cortical microtubule organization through prefoldin-DELLA interaction. Curr Biol 23:804–809 [DOI] [PubMed] [Google Scholar]
- Lundin VF, Srayko M, Hyman AA, Leroux MR (2008) Efficient chaperone-mediated tubulin biogenesis is essential for cell division and cell migration in C. elegans. Dev Biol 313:320–334 [DOI] [PubMed] [Google Scholar]
- March-Diaz R, Reyes JC (2009) The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol Plant 2:565–577 [DOI] [PubMed] [Google Scholar]
- Martin-Benito J, Boskovic J, Gomez-Puertas P, Carrascosa JL, Simons CT, Lewis SA, Bartolini F, Cowan NJ, Valpuesta JM (2002) Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J 21:6377–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R, Vainberg IE, Chow RL, Cowan NJ (1993) Chaperonin-mediated folding of vertebrate actin-related protein and γ-tubulin. J Cell Biol 122:1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo F, Sánchez R, Sali A (2009) Statistical potentials for fold assessment. Protein Sci 11: 430–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan-Zambrano G, Rodriguez-Gil A, Penate X, de Miguel-Jimenez L, Morillo-Huesca M, Krogan N, Chavez S (2013) The prefoldin complex regulates chromatin dynamics during transcription elongation. PLoS Genet 9:e1003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payán-Bravo L, Fontalva S, Peñate X, Cases I, Guerrero-Martínez JA, Pareja-Sánchez Y, Odriozola-Gil Y, Lara E, Jimeno-González S, Suñé C, et al. (2021) Human prefoldin modulates co-transcriptional pre-mRNA splicing. Nucleic Acids Res 49: 6267–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C, Rodriguez-Milla MA, Iniesto E, Rubio V, Salinas J (2017) Prefoldins negatively regulate cold acclimation in Arabidopsis thaliana by promoting nuclear proteasome-mediated HY5 degradation. Mol Plant 10:791–804 [DOI] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2:15190. [DOI] [PubMed] [Google Scholar]
- Ramegowda V, Gill US, Sivalingam PN, Gupta A, Gupta C, Govind G, Nataraja KN, Pereira A, Udayakumar M, Mysore KS, et al. (2017) GBF3 transcription factor imparts drought tolerance in Arabidopsis thaliana. Sci Rep 7:9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Milla MA, Salinas J (2009) Prefoldins 3 and 5 play an essential role in Arabidopsis tolerance to salt stress. Mol Plant 2:526–534 [DOI] [PubMed] [Google Scholar]
- Salanenka Y, Verstraeten I, Löfke C, Tabata K, Naramoto S, Glanc M, Friml J (2018) Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane. Proc Natl Acad Sci U S A 115:3716–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan V, Foulds I, Singh J, Dhindsa RS (2001) Cold-activation of Brassica napus BN115 promoter is mediated by structural changes in membranes and cytoskeleton, and requires Ca 2+ influx. Plant J 27:1–12 [DOI] [PubMed] [Google Scholar]
- Satou A, Taira T, Iguchi-Ariga SM, Ariga H (2001) A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J Biol Chem 276:46562–46567 [DOI] [PubMed] [Google Scholar]
- Siegers K, Waldmann T, Leroux MR, Grein K, Shevchenko A, Schiebel E, Hartl FU (1999) Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J 18:75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I (2000) Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell 103:621–632 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13:1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubacz A, Daszkowska-Golec A, Szarejko I (2016) The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front Plant Sci 7:1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son HG, Seo K, Seo M, Park S, Ham S, An SWA, Choi ES, Lee Y, Baek H, Kim E, et al. (2018) Prefoldin 6 mediates longevity response from heat shock factor 1 to FOXO in C. elegans. Genes Dev 32:1562–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stortenbeker N, Bemer M (2019) The SAUR gene family: the plant’s toolbox for adaptation of growth and development. J Exp Bot 70:17–27 [DOI] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) Pif4-mediated activation of yucca8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet 8:e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ (1998) Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93:863–873 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Eeckhout D, Cannoot B, De Winne N, Persiau G, Van De Slijke E, Vercruysse L, Dedecker M, Verkest A, Vandepoele K, et al. (2015) An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat Protoc 10: 169–187 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Han C, Gadeyne A, Eeckhout D, Matthijs C, Cannoot B, De Winne N, Persiau G, Van De Slijke E, Van de Cotte B, et al. (2019) Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat Plants 5: 316–327 [DOI] [PubMed] [Google Scholar]
- Wang D, Shi W, Tang Y, Liu Y, He K, Hu Y, Li J, Yang Y, Song J (2017) Prefoldin 1 promotes EMT and lung cancer progression by suppressing cyclin A expression. Oncogene 36:885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gao S, Peng X, Wu K, Yang S (2019) Roles of the INO80 and SWR1 chromatin remodeling complexes in plants. Int J Mol Sci 20:4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A (2016) Comparative protein structure modeling using MODELLER. InCurrent Protocols in Bioinformatics. John Wiley, New York, pp 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV (2007) Electronic fluorescent pictograph browser for exploring and analyzing large-scale biological data sets. PLoS One 2:718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Menke FLH, Yoshioka K, Moder W, Shirano Y, Klessig DF (2004) High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and defense gene expression. Plant J 39:920–932 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.